Abstract
Drug combination therapy is a promising strategy to treat complex diseases such as cancer and infectious diseases. However, current knowledge of drug combination therapies, especially in cancer patients, is limited because of adverse drug effects, toxicity and cell line heterogeneity. Screening new drug combinations requires substantial efforts since considering all possible combinations between drugs is infeasible and expensive. Therefore, building computational approaches, particularly machine learning methods, could provide an effective strategy to overcome drug resistance and improve therapeutic efficacy. In this review, we group the state-of-the-art machine learning approaches to analyze personalized drug combination therapies into three categories and discuss each method in each category. We also present a short description of relevant databases used as a benchmark in drug combination therapies and provide a list of well-known, publicly available interactive data analysis portals. We highlight the importance of data integration on the identification of drug combinations. Finally, we address the advantages of combining multiple data sources on drug combination analysis by showing an experimental comparison.
Keywords: machine learning, drug combination therapy, personalized medicine, bioinformatics, data integration
Introduction
Drug combination therapy has become a promising strategy for several complex diseases, such as cancer, diabetes and bacterial infections. This strategy can increase therapeutic efficacy, reduce toxicity and overcome drug resistance compared with single-drug administrations. Therefore, it is becoming an optimal option with increasing attention from researchers. However, there is limited information about effective drug combinations since screening all possible drug combinations is challenging and expensive. Thus, the computational prediction of combinatorial drug therapies is needed and essential to provide more sustainable treatment for the patients. To predict efficient drug combinations, computational approaches have been developed to date; however, they face several challenges that need to be solved, such as missing data, different data types and standardization. Consequently, machine learning (ML) models are increasingly being applied to efficiently explore the drug combinations from a large number of both approved and investigational chemical compounds.
In this review, we aim to systematically assess representative ML methods that have been proposed in recent years for understanding the drug combination therapies by grouping these methods into three categories. These methods have often been evaluated to overcome drug resistance in a variety of researches [1, 2]. To the best of our knowledge, there are already some reviews that have elucidated drug combination therapies, while these reviews emphasize different perspectives from this review or with a special focus on a particular biological problem [3–8]. On the other hand, this review mainly considers three perspectives as follows: (i) drug combination therapies from the viewpoint of developing ML methods, (ii) importance of data integration from different sources and (iii) publicly available interactive data analysis portals. The difference and detailed comparison between these reviews can be seen in Table 1.
Table 1.
Comparison with existing survey papers
| References | ML approaches | Toxicity | Systems biology approaches | Data integration | Data sources | Interactive data analysis portals | Experimental evaluation |
|---|---|---|---|---|---|---|---|
| Systems biology approaches for advancing the discovery of effective drug combinations [3] | ✓ | ✓ | ✓ | ||||
| Modeling of compound combination effects and applications to efficacy and toxicity: state-of-the-art, challenges and perspectives[4] | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Advances in computational approaches in identifying synergistic drug combinations [5] | ✓ | ✓ | |||||
| Predictive approaches for drug combination discovery in cancer [6] | ✓ | ✓ | ✓ | ||||
| Systems pharmacology: defining the interactions of drug combinations [7] | ✓ | ✓ | ✓ | ||||
| Artificial intelligence in drug combination therapy [8] | ✓ | ✓ | |||||
| This review | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
The outline of this review starts with a brief discussion of the significance of the synergy, efficacy and toxic effects of drug combinations. We provide information on relevant data sources commonly used for drug combination prediction methods and side information to improve the prediction accuracy. Then, we divide ML methods into three categories: drug combination sensitivity prediction, drug synergy prediction and drug synergy classification, and evaluate the methods in each category. To address the need for comparative studies, we show an experimental comparison of drug combination prediction methods. Finally, we discuss a list of well-known, publicly available software for analyzing combination data.
Drug combination therapy
Motivation
Drug combination therapies allow us to elucidate disease characteristics between patients caused by variation in therapeutic responses, define synergistic drug effects and minimize adverse drug reactions. The usage of drug combinations has multiple advantages over monotherapy, such as higher efficacy and lower toxicity [9, 10]. With monotherapy, it is really hard to treat complex diseases such as cancer [11, 12]. A single drug typically targets a single protein or pathway. Two clinically identical tumors rarely have commonly mutated genes, and so traditional therapies need to go beyond the ‘one disease, one drug, one target’ paradigm. Thus, combination therapy is rapidly becoming regular where single-drug treatments are ineffective. However, the identification of combinatorial drugs is expensive and time consuming since testing every possible combination of these drugs would be infeasible. The main questions that arise here are (1) identifying predictive biomarkers that may reveal the underlying mechanism of the drug combinations and (2) how to predict whether a known or new drug combination will be useful for a specific patient.
The existing computational methods can be divided into three categories: systems biology-based methods [3, 13], network-based methods [14, 15] and ML-based methods [8]. These methods mainly evaluate two important properties of drug combinations: sensitivity and synergy. Although these two can be roughly defined as predicting the correct treatment for the right patient, they have different measurement units. Sensitivity is the drug combination response in preclinical studies based on cell lines or patient-derived cells, which is usually measured in the unit of percentage inhibition of cell viability or growth. In contrast, synergy is defined as the degree of drug interactions where the effect of the combination is greater than that predicted by their individual potencies. Synergy is generally quantified through a selected reference model based on the properties of the dose–response curves, which describe the magnitude of the response of a drug, of specific drugs. Many approaches have been proposed using chemical, biological and molecular data to model sensitive and synergistic drug combinations, especially for cancer [16].
The effect of drug combination is measured by large-scale dose–response matrix experiments in various concentrations of single agents and combinations. The combination can be categorized as synergistic, additive or antagonistic based on the difference between the observed and the expected responses computed by a reference model [17]. Two compounds are considered synergistic when the combined effect is greater than that predicted by their single-agent potencies. If the effect of each drug neither decreases nor increases the sum of individual drug effects, it is called additive, known as noninteraction. In contrast to the synergistic effect, the combination is antagonistic if the sum is less than the response of individual agents. Synergistic combinations are preferable to delay the beginning of the resistance, whereas antagonistic combinations are useful for inhibiting the expansion of resistance [18]. Even though researchers focus on synergistic drug combinations, antagonism might be more beneficial for identifying the toxicity levels. There are four commonly used reference models for categorizing the level of drug combination synergism as follows: (i) Bliss independence: the most commonly used model, which provides a score under the assumption of each drug acts independently of the other. Each measurement above this score indicates synergy. The main drawback of the model is that it claims synergy when two identical drugs are combined; (ii) Loewe additivity: the idea behind the model is that the drugs can not interact with themselves and need to have equal individual drug maximum effects while computing the combination effect. However, this model is not applicable when a dose–effect curve is not available; (iii) Highest single agent: this model assumes that the combined effect should be superior to the effects achieved by the single drugs for synergism;and (iv) Chou–Talalay: this model relies on the linearity of the median-effect plot, which was proposed by Chou to linearize the dose and effect relations of all data points by plotting log(dose) versus log[fraction affected/fraction unaffected], which is generally not the case for other reference models. A significant limitation of the model is its dependence on accurate and well-defined dose–effect curves, which are not always available. A detailed explanation and comparison between these methods can be found in [1]. These models serve as a baseline to understand the interaction between drugs based on their single performance. However, each of these reference models has limitations and is not entirely suited for combinations of more than two drugs. In addition, just a reference model cannot consider how drugs may interact; thus, more complex mathematical models have emerged.
Drug synergy, efficacy and the toxic effects of drug combinations
Drug synergy can be defined as combining two or more chemical compounds to produce a more significant effect than an individual compound based on specific mathematical models. Clinical trials show higher synergy outcomes for proper combinations, such as more efficacy and less toxicity, and many approaches neglect the toxicity and efficacy of drug combinations [19]. There is a common misunderstanding that synergy and efficacy are treated as the same. Although synergy measures the degree of interaction, the efficacy of a drug combination depends on the extent of patient-to-patient variability and degree of correlation among monotherapy responses [20–22]. Therefore, it is possible that even though the combination is synergistic, the actual response might be inefficient to reach therapeutic efficacy, which may lead to prioritization of drug combinations that are unable to kill cancer cells despite strong synergy [23]. On the other hand, acquiring less toxicity by decreasing the drug doses allows for fewer adverse effects. These three major factors, i.e. synergy, efficacy and toxicity, should be considered together while categorizing drug combination therapies. Even though two drugs are not synergistic, they might still be beneficial by having higher efficacy and lower toxicity [24].
Existing drug combination therapies
Drug combination therapies have become a powerful approach to fight against complex diseases in recent years. The primary motivation behind this approach is to find synergistic drug combinations while maximizing efficacy and reducing toxicity. With the recent advances, Food and Drug Administration (FDA)-approved new drug therapies include various novel drugs; thus, many successful treatments prevailed. Most of the approved FDA drug combinations target the treatment of infectious diseases [1]. For example, the FDA approved a pairwise drug combination of Dolutegravir and Lamivudine, which together blocks the HIV-1 multiplication for the treatment of HIV-1 infection in April 2019 [25]. In addition to this, much of the research and development is targeting different cancer types in recent years. One example is treatment of melanoma patients with the combination of Dabrafenib and Trametinib that can harbor BRAF V600E mutations was approved by the FDA in 2015 [26]. Moreover, the treatment of using both Vermurafenib, which targets BRAF, and Cobimetinib, which targets MAP2K1, has been shown to be synergistic for treating BRAF mutated melanoma and was also approved by FDA. Another combination was approved for diabetes treatment using Osiglitazone and Exenatide [27]. Osiglitazone is already an antidiabetic drug, but it increases the risk of myocardial infarction. Nevertheless, when two drugs are combined together, they decrease the risk of myocardial infarction [28]. Not only pairwise combinations but also triple and quadruple combinations are emerging recently. As an example, the combination of Oravirine, Lamivudine and Tenofovir was approved in 2018 to deal with HIV-1 infection for adults [29].
Data sources
A wide range of research programs has generated various databases on combinatorial drug therapy to accelerate the discovery of personalized multitargeted drug combinations in recent years. These advancements bring new opportunities for the application of large-scale ML methods in predicting drug combinations. Two of the most significant and oldest resources of the publicly available database for investigating drug combinations are US FDA and Drug Combination Database (DCDB) [30]. The FDA Orange Book [31] consists of drugs and combinations approved based on the FDA’s safety and effectiveness. The first combination was approved in the 1940s, and FDA updates the number of approved drug combinations every year. The dataset contains 419 drug combinations where 341 double, 67 triple, and 11 more than triple combinations, and 367 are structurally unique combinations from 328 unique small molecules by 2018 [29]. DCDB is known as the first database committed for the research of multicomponent drugs. This database includes 1363 drug combinations from 904 distinctive components from various clinical studies and FDA Orange Book. Around 20% of DCDB drug combinations are approved to be used in patients. Meanwhile, approximately 13% of the combinations are reported to be nonefficacious. Another essential resource is NCI-ALMANAC (A Large Matrix of Anti-Neoplastic Agent Combinations) [32], one of the pioneers in the characterization of drugs in vitro. NCI-ALMANAC is a library of cancer cell lines maintained by National Cancer Institute (NCI). The library provides FDA-approved drug combinations for targeting cancer and killing tumor cells from the NCI-60 cell lines obtained from nine cancer types. This data has >5000 pairs of drug combinations from 104 FDA-approved drugs and 60 cell lines. The ONEIL study [33] provided a comprehensive data source of drug combinations, which has been widely applied by many researchers recently. This source includes 38 drugs and their pairwise combinations, 92 208 drug combinations, and 583 distinct combinations against 39 cancer cell lines representing multiple cancer types. One of the latest drug combination datasets is AstraZeneca’s DREAM-AZ dataset [34], consisting of cell viability response measurements and synergy scores from 910 pairwise combinations of 118 drugs across 85 molecularly characterized cancer cell lines. The dataset is the result of a DREAM Challenge to evaluate computational strategies for predicting synergistic drug pairs and biomarkers. Although DCDB and FDA databases offer drug combinations for multiple diseases, NCI-ALMANAC, DREAM-AZ and the ONEIL study focus on oncological diseases. Another disease-specific repository is Antifungal Synergistic Drug Combination Database (ASDCD) [35], which is designed for synergistic antifungal drug combinations. ASDCD consists of published synergistic antifungal drug combinations, chemical structures, drug-targets, target-related signaling pathways, drug indications and other pertinent data. The database has 210 antifungal synergistic drug combinations and 105 individual drugs from >12 000 references.
Integrating data for predicting drug combinations
More detailed information on the dose–response effects of combinations is required in drug combination therapy applications since it is demonstrated that drug combination therapy is better than single-drug treatment. Therefore, it is necessary to consider the relationship among drugs, targets and diseases when using drug combination therapy. The integrative analysis of side information with rapid accumulation would enable us to analyze the relationships between the side information. The current computational methods rely on the selective incorporation of target features, pharmacogenomics and chemical property information.
Drug-related information plays a significant role in understanding the behavior of compound combinations, such as the similarity of structure and biochemical properties between drugs. Drug–target and drug–drug interactions can also be used to improve predictions of effective combination therapies. In particular, drug–drug interactions are essential since they might cause unexpected pharmacological effects, including adverse drug events. The existence of drug–target interactions in the same pathway is shown to be predictive of synergism [27]. It is also shown that they contribute to the identification of novel synergistic chemical pairs. In addition, adverse drug effects contribute to identifying novel synergistic chemical pairs, illustrated by a number of synergistic drug combinations reported for various diseases [36]. In recent years, gene expression profiles have also helped predict synergistic effects of drug combinations on cancer cell lines [37, 38]. Details of these data types and their related tools can be found in the review papers we provided in Table 1.
ML approaches
We categorize ML approaches for drug combination therapy into the following three problem settings: (1) drug combination sensitivity prediction, (2) drug synergy prediction and (3) drug synergy classification. First, ‘drug combination sensitivity prediction’ is to predict the sensitivity of two or more drugs under an experimental condition from the input of drug combination sensitivity values for multiple conditions. Sensitivity is a measure of treatment response that can be defined in the unit of percentage inhibition of cell viability or growth. Figure 1A illustrates this problem setting. The main task is to predict the unknown response of a drug pair and a cell line (missing entries in a matrix) for which other response values of drug pairs and cell lines are already given. This experimental setup is very similar to monotherapy, only includes more experiments such as considering four doses for each drug in a pairwise combination, 16 measurements are required, whereas four measurements are enough for monotherapy. Second, ‘drug synergy prediction’ is to measure the degree of the interaction between two or more drugs. Synergy is defined as a combination effect that is greater than the predicted effect of the individual drugs. This task has two different problem settings, and Figure 1B demonstrates two subtasks. The first subtask (Figure 1B1), ‘drug synergy score estimation’ is to compute synergy score of drug combinations. Synergy score is usually calculated as the deviation of the observed drug combination effect from the expected combination effect based on the properties of the dose–response curves of the single drugs. There are various reference models (baseline) for quantifying the level of drug combination synergism. The most common and prominent reference models based on performance of individual drugs are Loewe Additivity and Bliss Independence. The full dose–response matrices (full surface) predicted in the ‘drug combination sensitivity prediction’ task are necessary to estimate the reference model and compute a synergy score. In the second subtask (Figure 1B2), ‘drug synergy matrix completion’, the purpose is to predict unknown synergy scores of drug combinations and cell lines. The task can be considered as continuation of the first subtask since the synergy scores obtained in the first subtask can be used to fill the synergy matrix of combinations, and unknown synergy scores of drug combinations and cell lines are predicted. Third, ‘drug synergy classification’ identifies novel synergistic combinations when drugs interact with each other for several cell lines under multiple conditions (Figure 1C). This task basically can be considered as classifying the drug combinations whether they are synergistic, additive or antagonistic. There are different datasets consist of binary values, which are specifically designed for this task such as FDA Orange Book [29] and DCDB [30]. However, this task can be also considered as a continuation of ‘drug synergy prediction’. A continuous parameter of synergy score obtained from the reference model can be used to categorize the drug combinations as being synergistic, additive and antagonistic. In this setting, the synergy scores have usually been cut off according to a fixed threshold. We explain and summarize each of the three categories in detail in Table 2. In the rest of Section ML approaches, we briefly introduce the ML-based drug combination prediction methods, which are categorized into three categories: sensitivity prediction, synergy prediction and synergy classification methods. Table 3 summarizes the features of all methods shown in this survey.
Figure 1 .

Schematic structures of combinatorial drug prediction methods. (A) Sensitivity prediction: predicting drug combination responses in preclinical studies based on cell lines or patient-derived cells (multiple concentrations of drugs can be considered for each cell line). (B) Synergy prediction: predicting the degree of drug interactions that contribute to the drug combination sensitivity. (C) Drug classification: identifying the degree of interaction between drugs (multiple concentrations of drugs can be considered for each cell line).
Table 2.
Summary of drug combination prediction tasks
| Drug combination prediction tasks | Subtasks | Input | Common metric | Metric type | Output | Task | Note | Ref.paper |
|---|---|---|---|---|---|---|---|---|
| Sensitivity prediction (Figure 1A) | Incomplete multi drug–dose response matrices | IC50, EC50, GI50 | Cont. | Complete multi drugdose response matrices | Predict missing entries in a matrix | Similar to monotherapy prediction, only more doses involved. Considering four doses for each drug in a pairwise combination, we require 16 measurements, four measurements are enough for monotherapy. | [39] | |
| Synergy prediction | Drug synergy score estimation (Figure 1B1) | Complete multi drug–dose response matrices (Full surface) | IC50, EC50, GI50 | Cont. | Synergy scores | Predict single score using reference models | The predicted complete dose–response matrices from sensitivity prediction task can be used to calculate combinatorial landscapes over the full concentration ranges using a reference model. | [40, 41] |
| Drug synergy matrix completion (Figure 1B2) | Incomplete single synergy matrix | CI | Cont. | Complete single synergy matrix | Predict missing entries in a matrix | The predicted synergy scores obtained from drug synergy score prediction task can be used to fill synergy matrix of combinations | [42] | |
| Synergy classification (Figure 1C) | Binary correspondence of drug combinations | CCI | Discrete | Single score (binary value) | Predict class of drug combinations, i.e synergistic or antagonistic | CI can be used to categorize the drug combinations as being synergistic, additive and antagonistic by defining threshold value. | [27] |
Note: IC50 = Measure of drug concentration where the response is reduced by half, EC50 = Measure of drug concentration that gives half-maximal response, GI50 = Measure of drug concentration that reduces total cell growth by %50, CI = Combination index, measure for deviation of the inhibitory effect of the combination from additivity, CCI = categorized combination index
Table 3.
Summary of drug combination prediction methods
| Prediction problem | Method | Disease | Drug combination source | Combination input type | Model | Outcome | Side information | Performance evaluation | Case study | Experimental validation |
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity prediction | Xia et al. [44] (BestComboScore) | Cancer | NCI-ALMANAC | % Growth | Deep learning | Single point ( % Growth) | GE, microRNA, Protein abundance (NCI-60 Database); Chemical structure (NCI-ALMANAC) | MSE, MAE, R2 | ✓ | |
| Zagidullin et al. [45] (DrugComb) | Cancer | NCI-ALMANAC, ONEIL, FORCINA, CLOUD | %Inhibition | Linear regression | Full matrix (% Inhibition) | Chemical structures (PubChem) | RMSE | ✓ | ||
| Julkunen et al. [39] (ComboFM) | Cancer | NCI-ALMANAC | % Growth | Factorization machines | Full matrix (% Growth) | Molecular fingerprints, GE, Drug concentrations (NCI-60) | RMSE, PCC, SC | ✓ | ✓ | |
| Synergy prediction | Huang et al. [46] (DrugComboRanker) | Cancer | GE (CMAP); Drug similarity (STITCH, OMIM, MEDLINE); PPI (BioGRID) | Genomic profile | BNMF | Single point (Rank) | – | Average enrichment score Geometric accuracy metric Relative number of well- known drug communities | ✓ | |
| Preuer et al. [37] (DeepSynergy) | Cancer | ONEIL | Synergy score quantified by Combenefit [40] | Deep learning | Single point (Synergy score) | GE (ArrayExpress); Chemical descriptors | MSE, RMSE, PCC, AUC, AUPR, ACC,BACC, PREC, TPR, TNR, Kappa | ✓ | ||
| Jeon et al. [47] | Cancer | ONEIL | Synergy score quantified by Combenefit [40] | Extremely randomized tree | Single point (Synergy score) | GM, GE,CNV (COSMIC); DTI (DrugBank, GDSC) | PCC, F1 scores | ✓ | ||
| Celebi et al. [42] | Cancer | DREAM-AZ | Summary measure | XGBoost | Single point (Synergy score) | DTI (SUPERFAMILY, Prosite, SMART, Pfam); Pathway (KEGG); GE (GDSC); Mutation, CNV (COSMIC); Chemical structures, Drug targets, Monotherapy (DREAM) | WAPCC, AUC | |||
| Ianevski et al. [48] (DECREASE) | Multiple | FIMM, DLBCL, ONEIL, NIH | Incomplete surface | cNMF & XGBoost | Single point (Synergy score) | – | RMSE | ✓ | ✓ | |
| Drug classification | Zhao et al. [49] | Multiple | FDA Orange book | Single point (Binary value) | Maximization of the F1 score | Single point (Confidence score) | DTI (STITCH, TTD DrugBank); Pathway (KEGG) | TPR | ✓ | |
| Li et al. [50] | Multiple | DCDB, TTD, Pubmed | Single point (Binary value) | Probability ensemble approach, Bayesian network | Single point (Binary value) | DDI (DrugBank); Drug side effect (SIDER) | AUC | ✓ | ✓ | |
| Iwata et al. [27] | Multiple | FDA, KEGG DRUG | Single point (Binary value) | Logistic regression | Single point (Binary value) | DTI (DrugBank, BindingDB, MATADOR, ChEMBL, PDSP-Ki, TTD, KEGG DRUG) | AUC, AUPR | ✓ | ||
| Chen et al. [51] (NLLSS) | Fungal | Literature search Web search | Single point (Binary value) | Laplacian Regularized Least Square | Single point (Rank) | Chemical structure (KEGG DRUG); DDI (DrugBank) | AUC | ✓ | ✓ | |
| Li et al.[52] (SyDRa) | Cancer | NCI-DREAM | Single point (Binary value) | Random forest | Single point (Binary value) | GE (CMAP, NCI-DREAM); Chemical structure (DrugBank); PPI (BioGRID); Pathway (MSigDB) | AUC score | |||
| Gayvert et al.[22] | Cancer | Held et al. [53] | Summary measure | Random forest | Single point (Four category) | Single drug screen ([53]) | AUC, TPR, TNR | ✓ | ✓ | |
| Liu et al. [54] | Multiple | DCDB | Single point (Binary value) | Gradient tree boosting | Single point (Binary value) | DTI (STITCH); Chemical structures (PubChem); PPI (STRING) | PREC, REC, MCC, F measure, AUC | ✓ | ||
| Karimi et al. [55] (Drug Combo Generator) | Cancer | FDA Orange book | Single point (Binary value) | Deep generative model | Single point (Network score) | Disease–gene associations (OMIM); Drug–disease association (CTD); PPI ([56]) | Network score[57] | ✓ | ||
| Sun et al. [58] (DTF) | Cancer | ONEIL | Summary measure | Deep tensor factorization | Single point (Probability score) | – | AUC, AUPR TPR, ACC, BACC, Kappa |
Notes: FDA = Food and Drug Administration [31], DTI = Drug–target interaction, STITCH [59], TTD = Therapeutic Target Database [60], KEGG = Kyoto Encyclopedia of Genes and Genomes [61],TPR = True postive rate, CMAP = Connectivity Map [62], BNMF = Bayesian non-negative matrix factorization, GE = Gene expression, OMIM = Online Mendelian Inheritance in Man [63], PPI = Protein–protein interaction, KEGG DRUG [64], BioGRID = Biological General Repository for Interaction Datasets [65], BindingDB = Binding Database [66], MATADOR = Manually Annotated Targets and Drugs Online Resource [67], ChEMBL [68], PDSP-Ki = Psychoactive Drug Screening Program [69], TTD = Therapeutic Target Database [70], AUC = Area under curve, AUPR = Area under precision–recall curve, DCDB = Drug Combination Database [30], DDI = Drug–drug interaction, SIDER = Side Effect Resource [71], NCI-DREAM [16], MSigDB = Molecular signatures database [72], ONEIL [33], ArrayExpress database [73], MSE = Mean square error, RMSE = Root mean square error, PCC = Pearson correlation score, ACC = Accuracy, BACC = Balanced accuracy, PREC = Precision, SyDRa = Synergistic Drug combination using Random forest algorithm, TNR = True negative rate, NCI ALMANAC [32], MAE = mean absolute error, R2 = Coefficient of determination, GM = Gene mutations, CNV = Copy number variationsCNV, COSMIC = Catalogue of Somatic Mutations in Cancer [73], GDSC = Genomics of Drug Sensitivity in Cancer [74], NIH LINCS = Library of Integrated Network-based Cellular Signatures, PCI = Probability concordance index, REC = Recall, MCC = Matthews correlation coefficient, FORCINA [75], CLOUD [76], XGBoost = gradient boosted decision tree, DREAM-AZ [34], WAPCC = weighted average Pearson correlation coefficient, cNMF = composite Nonnegative Matrix Factorization, FIMM = Finland Institute of Molecular Medicine, DLBCL [77], SC = Spearman correlation score, pAUC = partial AUC.
Drug combination sensitivity prediction
There has been a considerable improvement in ML models that can be used to predict drug responses for the last decades. Many of these previous studies have been successfully applied for single drug–response prediction [43]. However, starting with the release of the NCI-ALMANAC database [32], specifically for the cancer disease, the prediction of pairwise drug–response combination therapies has started to gain attention from researchers.
BestComboScore [44] is known as the first attempt that ML evaluation of paired drug response sensitivity prediction. The model applies deep neural network and intermediate integration, which means that multiple types of molecular and drug features are jointly trained in the first layer of the neural network. BestComboScore predicts the best growth inhibition seen in any experiment for a given drug pair and does not include drug concentrations as an input feature to reduce training data imbalance. The model was tested on different combinations of features to evaluate the relative importance of each feature. Empirical results reveal that all molecular features have potential over prediction; however, the most predictive capacity belongs to drug descriptors. Even though the model was designed for pairwise drug combinations, the model can be extended to more than two drugs in combination therapy.
DrugComb [45] is an open-access data portal that targets both sensitivity and synergy prediction of drug combinations. The portal stores comprehensive drug combination data sources from different databases and provides curation and standardization. DrugComb can also be considered a data analysis tool that allows users to visualize, analyze and annotate drug combination dose–response data. The model uses linear regression to provide sensitivity scores of drug combinations, considering drug chemical fingerprint information as predictors. The primary outcome is that chemical information can be an essential feature for explaining the sensitivity of drug combinations. This research can be improved by an integration pipeline for more heterogeneous sources. Furthermore, more advanced ML methods would be tested to enhance accuracy.
comboFM [39] is one of the recent studies that transfer drug combination dose–response data into a higher-order tensor. The outstanding discrimination with other studies is that comboFM includes various doses of drug combination responses into experiments. Besides, genomic descriptors of cell lines and chemical descriptors of each drug are integrated into the model. Then, higher-order factorization machines are applied to predict missing entries in the dose–response matrices, untested drug–drug–cell line triplets and new drug combinations. After dose–response matrices are completed, synergy scores are computed, resulting in a single score defined for each drug combination. All these data except concentration values and genomic descriptors are represented by binary values in a one-hot encoding form more than continuous values, which may cause loss of information sometimes. This design can be used as drug repurposing since new drug combinations can be predicted even without available combination measurements.
Drug synergy prediction
Many synergy prediction methods, which are reviewed under Section Drug synergy prediction, can also be regarded as a combinatorial drug response prediction method since they require fully measured dose–response matrices to calculate drug synergy scores. For example, one of the main objects of DECREASE [48], which is also reviewed in Section Drug synergy prediction, is to fill missing entries in the drug–dose response matrix first and then calculate synergy scores.
DrugComboRanker [46] predicts synergistic drugs that target different signaling modules of cancer-specific networks, by integrating genomic profiles of both drugs and cancers, aiming to combine drugs with existing therapy to reduce drug resistance. There are two main steps of the model: (1) drug functional network construction and (2) partitioning of the functional drug network into clusters using a Bayesian nonnegative matrix factorization. The functional drug network is constructed based on genomic profiling data of drugs. The second part discovers the clusters that share common responses to drug treatment and predicts the functional targets of drugs. Rich genomic profiles of drugs, diseases and network-based integration make this approach practical, whereas only the combinations in known disease–pathway interactions can be found.
DeepSynergy [37] is known as the first deep learning method to predict drug combination synergies. Compared with previous studies developed for small datasets, DeepSynergy uses more extensive synergy data [33] for prediction and incorporates chemical and genomic information as input information. The method has a normalization strategy because this model integrates heterogeneous data sources, which might cause information loss. Deep neural network algorithms are very well suited for large datasets, and the predictive performance of DeepSynergy can be improved if more data is available.
Incorporating prior information has already shown its predictive power in predicting drug synergy scores. However, the arising challenge here might be the curse of dimensionality, which means if we have more features than observations, this might cause that the model results in good performance only to its initial dataset, and not to any other data sets. Finding relevant features and understanding the importance of the side information make the prediction models much more manageable. There are two studies relevant to this point. These two examples of feature selection methods offer the opportunity for understanding the significance of feature selection in predicting drug synergies. A benchmark study [47] uses the genomic information of cancer cell lines, drug–targets and molecular information to predict the synergy between two drugs. The difficulty comes with high dimensionality, especially for genomic information. Jeon et al. [47] select only genes in cancer-related pathways by using a method called extremely randomized trees (ERT), which uses multiple decision trees without bagging to reduce the variance of a single decision tree. The other data-driven study for the prediction of drug synergies is made by [42]. This study uses an ensemble learning algorithm, XGBoost, to select biologically relevant and most predictive drug and cell line features to understand the biological factors underlying drug synergy. Experimental results reveal that monotherapy and genomic features are most informative. In contrast, target features have a minor effect on drug combination therapy among comprehensive pharmacological and molecular information.
DECREASE [48] is a comprehensive design that can identify synergistic and antagonistic combinations with a minimal set of measurements. It is a two-step, efficient ML model to predict drug synergies from drug–dose response values. Many studies require fully measured dose–response matrices for the computation of drug synergy scores. Some of them use single concentrations of an individual agent, which any outlier might have a drastic impact. DECREASE tries to solve these challenges by detecting outliers and handling missing values in drug–dose response data. As the first step, outliers are detected using the difference between the observed responses and the expected responses based on the Bliss independence model. Second, the composite Nonnegative Matrix Factorization algorithm is used to predict missing dose–response values. After predicting full dose–response matrices from a sparse input, the synergy scores are calculated by a reference model such as Bliss independence or Loewe additivity. The crucial difference with other models is that it does not incorporate any side information and does not attempt to do prediction to new experiments. DECREASE also implements an interactive web tool that enables testing different biological problems such as identifying bacterial, fungal or antiviral drug combination synergies. DECREASE might be improved with higher-order combination data rather than pairwise combinations in the future.
Drug synergy classification
One of the earlier ML approaches for the classification of drug combinations has started with [49], which presents a simple method to predict whether a drug pair is an effective combination by maximizing the F1 score. This method is one of the earlier approaches that integrate molecular and pharmacological data as side information to classify drug combinations. The model treats drug combinations as combinations of their corresponding features, including their target proteins, therapeutic effects and indication areas. Integration results imply that side-effects and pathways are not sufficient for drug combination prediction; however, drug target proteins, anatomical therapeutic chemical (ATC) codes and drug indications are informative.
Iwata et al. [27] constructed a more complex predictive model than [49]. The framework uses logistic regression to predict beneficial drug combinations using target proteins and ATC drug codes and minimizes the loss function with L1 regularization to overcome overfitting. The model has three central prediction motivations: (i) the model should predict known drug–drug pairs, (ii) predict new combinations for drugs of known combinations and also (iii) predict new pairs without any known drug combination. Experimental assessment proves that predicting new combinations is challenging, and task (iii) is the hardest. Moreover, the ATC drug code is the most useful information, followed by target–protein and indication profiles. However, the shortcoming is that ATC information unobtainable for new drug candidate compounds, and complete information would improve the prediction of new combinations. The merit of the model is the L1 regularizer for interpretability, whereas the downside is lacking of disease context. The predictive performance would be enhanced by incorporating adverse drug–drug interactions that are valuable in drug development.
The integration of side data has proved to be promising on drug combination therapies, and so more side data were examined on probability ensemble approach (PEA) [50] to predict drug combination classes for analyzing both the efficacy and adverse effects of drug combinations. PEA is a systems pharmacology framework of using a Bayesian network to solve the missing data problem and incorporate different features even though some features are weakly informative. Given a pair of drugs, the model calculates drug similarity features and combines them using a Bayesian network into a likelihood ratio that represents its probabilistic similarity to the known interaction. PEA reveals the importance of each of the auxiliary sources by using not only drug properties but also the drug–targets in the protein–protein interaction network and the similarity of drug–targets. The main results of [50] are (1) the model with all features has higher performance than those with single features and (2) side-beneficial effects would enable the analysis of the relationships between drugs and combinations.
Network-based Laplacian regularized Least Square [51] distinguishes antifungal synergistic drug combinations from nonsynergistic ones by integrating drug–target interactions and drug chemical structures as side information. The model classifies drugs according to whether they have activity in the antifungal assay. If one drug shows activity, but the other does not, then the first drug is considered as the principal, and the latter is considered as the adjuvant. The idea behind the model is that principal drugs often have similar synergistic effects with adjuvant drugs. The framework has two classifiers for principal and adjuvant drugs separately and combines these two classifiers into a single classifier to obtain a final predictive result. Each classifier has its own drug similarity measures. Based on the classification score, drug combination pairs with high scores can be expected to have a high probability of being synergistic.
SyDRa [52] identifies synergistic anticancer drug combinations by using a random forest algorithm with three types of features: drug–chemical structure, drug–target network and pharmacogenomics. These feature combinations and labels of drug combinations are used as an input to random forest algorithm to distinguish synergistic and nonsynergistic drug combinations. The primary finding is that the pharmacogenomics features (especially similarity between gene expression profiles) contribute to drug synergism more than the other features. SyDRa uses a small training dataset compared with independent test sets, which might affect the robustness of the model. Kyoto Encyclopedia of Genes and Genomes pathways targeted by each drug in a drug combination play a vital role in SyDRa. Nevertheless, disease-specific pathways, such as breast cancer-related pathways, are not considered to improve performance.
A different approach was made by Gayvert et al. [22] to identify synergistic and effective drug combinations from drug combination efficacies and single-drug agent information. This study uses only single-agent knowledge as prior information. For each drug pair, input features are constructed by taking the mean and difference of the single-agent dose–response in each tested cell line. The reason behind the idea is that the synergy depends on the context and this makes it difficult to use the information of different cancers or genotypes. Importantly, this work directly concentrates on specific diseases such as mutant BRAF melanoma. Nevertheless, it is still difficult to generalize this work to all cancer types and this work has small training data that might cause overfitting and produce inaccurate results.
Gradient tree boosting (GTB) was utilized by a heterogeneous network-based inference to classify efficacious drug combinations using features derived from drug–protein heterogeneous network [54]. Protein networks play an essential role in treating complex diseases, and therefore they might be applied to decrease the activity level of carcinogenic genes while developing drug combinations. This method has the following three steps:
The model incorporates a drug similarity network, protein similarity network and known drug–protein associations into a drug–protein heterogeneous network.
Drug combination features are extracted by running a random walk with restart on the heterogeneous network.
Extracted features are trained on a GTB classifier to predict new drug combinations.
Like many other network-based methods, the assumption is that similar drugs are likely to interact with similar target proteins. However, there is always a possibility that new drugs or proteins might have low similarity and be located far in the feature space, which causes failure in predicting drug combinations. On the other hand, the model is pertinent to the increased size of drug combinations because of the nature of heterogeneous networks.
Deep Tensor Factorization (DTF) [58] combines two submodels: weighted tensor factorization and deep learning methods. DTF generates features using the output of the weighted tensor factorization method. The extracted latent features of drug synergy information are used as input features to train the deep neural network and finally predict the synergistic effect of drug pairs. DTF can also be used for predicting missing synergy scores in addition to the classification task. Experimental results demonstrate that DTF shows a rather similar performance to DeepSynergy [37], and there is no statistically significant difference, although DTF does not incorporate any side information. DTF uses only a single data source for a complex problem; however, incorporating more information might significantly improve performance.
Drug-Combo-Generator [55] is the first network-based deep generative model for overcoming drug resistance in drug combination design. Unlike discriminative models, the target is to define all drug combinations in the enormous chemical combinatorial space and evaluate combination effects instead of simply defining them as synergistic or antagonistic. The method has fundamentally two steps. First, prior knowledge of disease-related sources is jointly embedded into the system, such as gene–gene, disease–gene, disease–disease relationships, using hierarchical variational graph auto-encoders. The reason is relationships between disease–proteins and drug–targets in human protein–protein interactome might help to understand drug behavior such that targets of two drugs belong to the same disease module might cover different neighborhoods [57]. These embeddings create features for each disease in the second step, where a graph-set generator for reinforcement learning is trained to maximize the therapeutic efficacy for drug combinations in chemistry- and system-aware environments. The objective is to generate a set of drug combinations with similar distributions to the prior set of graphs. The advantage is that the model can generate higher-order combinations that might be favorable in the future, especially for infectious diseases.
Empirical comparison
We reviewed several methods for drug combination therapies in this survey, and it would also be interesting to compare them with one another. However, different studies use different data sources, scoring metrics and validation with little overlap. Tables 4, 5, 6 and 7 summarize the performance scores of sensitivity prediction, synergy prediction and classification of drug combinations, respectively, as reported by the original studies. The datasets used for each method can be found in Table 3. Unfortunately, we could not include all the methods we reviewed in the tables since results are shown only by pictures in several works.
Table 4.
Performance scores of drug combination sensitivity prediction methods referred to in this review
| Molecular and drug features | MSE | MAE |

|
|
|---|---|---|---|---|
| Xia et al. [44] (BestComboScore) | One-hot encoding | 0.5253 | 0.5709 | –1.001 |
| Gene expression, One-hot encoding | 0.2447 | 0.3999 | 0.1272 | |
| Gene expression, 500-dimensional noise | 0.2450 | 0.2450 | 0.1271 | |
| One-hot encoding, Dragon7 descriptors | 0.0292 | 0.1086 | 0.8892 | |
| Proteome, Dragon7 descriptors | 0.0303 | 0.1117 | 0.8844 | |
| microRNA, Dragon7 descriptors | 0.0275 | 0.1050 | 0.8952 | |
| Gene expression, Dragon7 descriptors | 0.0180 | 0.0906 | 0.9364 | |
| Gene expression, microRNA, Proteome, Dragon7 descriptors | 0.0158 | 0.0833 | 0.9440 |
The method which achieved the best score is in bold.
Table 5.
Performance scores of drug combination sensitivity prediction methods referred to in this review
| Methods | RMSE | Pearson | Spearman | ||
|---|---|---|---|---|---|
| Julkunen et al. [39] (comboFM) | New response matrix entries | comboFM-5 | 9.86 | 0.97 | 0.91 |
| comboFM-2 | 17.89 | 0.91 | 0.84 | ||
| comboFM-1 | 31.56 | 0.70 | 0.66 | ||
| Random forest | 10.91 | 0.97 | 0.91 | ||
| New response matrices | comboFM-5 | 10.39 | 0.97 | 0.91 | |
| comboFM-2 | 18.00 | 0.91 | 0.83 | ||
| comboFM-1 | 31.57 | 0.70 | 0.66 | ||
| Random forest | 12.23 | 0.96 | 0.90 | ||
| New drug combinations | comboFM-5 | 13.04 | 0.95 | 0.88 | |
| comboFM-2 | 19.37 | 0.89 | 0.81 | ||
| comboFM-1 | 31.79 | 0.69 | 0.66 | ||
| Random forest | 15.44 | 0.93 | 0.86 |
The method which achieved the best score is in bold.
Table 6.
Performance scores of drug combination synergy prediction methods referred to in this review
| Methods | Pearson | |
|---|---|---|
| Preuer et al. [37] (DeepSynergy) | DNN |
0.73 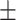 0.04 0.04
|
| Gradient boosting | 0.69 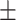 0.02 0.02 |
|
| Random forest |

|
|
| SVM |

|
|
| Elastic net |

|
|
| Baseline |
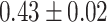
|
|
| Jeon et al. [47] | Elastic net | 0.65 |
| Ridge regression | 0.661 | |
| Kernel ridge regression (RBF) | 0.728 | |
| Random forest | 0.731 | |
| Extremely randomized tree | 0.738 |
The method which achieved the best score is in bold.
Table 7.
Performance scores for some of the drug combination classification models referred to in this review
| Methods or Datasets | AUC | |
|---|---|---|
| Iwata et al. [50] | Whole features | 0.90 |
| ATC codes of the drugs | 0.85 | |
| Drug side-effects | 0.72 | |
| kNN | 0.79 | |
| Chen et al. [51] (NLLSS) | Combined space classifier | 0.9054 |
| Principal space classifier | 0.8244 | |
| Adjuvant space classifier | 0.8328 | |
| Gayvert et al. [22] | BRAF-specific effectiveness | 0.8809 |
| General BRAF-effectiveness | 0.8630 | |
| BRAF synergy | 0.8683 | |
| Li et al.[52] (SyDRa) | SyDRa | 0.89 |
| Pharmacogenomics features | 0.83 | |
| CT (chemical similarity, drug target network) features | 0.73 | |
| Liu et al. [54] | GTB | 0.949 |
| kNN | 0.768 | |
| SVM | 0.859 | |
| Logistic | 0.520 | |
| Naive Bayes | 0.508 | |
| Random forest | 0.866 | |
| Adaboost | 0.866 | |
| LogistBoost | 0.808 | |
| Preuer et al. [37] (DeepSynergy) | DNN |
0.90 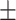 0.03 0.03
|
| Gradient Boosting |

|
|
| Random Forests |
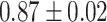
|
|
| SVM |

|
|
| Elastic Nets |
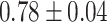
|
|
| Baseline |

|
|
| Sun et al. [58] (DTF) | DTF |

|
| DeepSynergy |
0.90 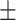 0.02 0.02
|
|
| Logistic regression |
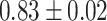
|
|
| CP-WOPT |
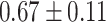
|
kNN = k-Nearest Neighbors, GTB = Gradient tree boosting, SVM = Support vector machines, DNN = Deep neural networks, CP-WOPT = CANDECOMP/PARAP-AC-Weighted OPTimazation, BRAF-specific effectiveness = combinations that achieve at least 50% growth inhibition within the genotypic group, General BRAF-effectiveness = combinations that achieve at least 70% growth inhibition.
The method which achieved the best score is in bold.
Drug combination sensitivity prediction
BestComboScore [44] is a study that evaluates the importance of each integrated dataset, and so in Table 4, the second column shows different combinations of datasets. Results show that all three molecular feature types provide marginal benefit, whereas most of the predictive feature is Dragon7 descriptors. On the other hand, using all data (gene expression, microRNA, proteome and chemical descriptors) is most advantageous in performance.
comboFM [39] is an extensive study on different designs such as predicting new dose–response matrix entries, dose–response matrices and drug combinations. Table 5 shows the performance results of these three setups. Comparisons are between comboFM-5, comboFM-2, comboFM-1 and random forest methods. Although comboFM-5 corresponds to the 5th-order factorization machine, comboFM-2 and comboFM-1 are equal to the 2nd-order and 1st-order factorization machines, respectively. comboFM-1 is also considered as ridge regression. Results demonstrate that comboFM-5 significantly outperforms comboFM-1 and comboFM-2 in both missing value and new drug combination prediction settings. Although predicting new combinations is a more challenging task, comboFM-5 has particularly high Pearson correlation scores in both settings.
Drug synergy prediction
Table 6 provides an experimental comparison of drug synergy prediction methods presented in this review. Pearson correlation score is the common evaluation score by three methods. DeepSynergy [37] outperforms the other ML methods significantly in terms of Pearson correlation. However, ERT in [47] also achieves almost the same score as Deepsynergy. The reason why ERT outperforms other methods might be the extreme randomness, which reduces the variance of the model and makes the model more robust particularly with noisy data. The performance of linear models is clearly lower than nonlinear models in both experiments. The detailed information about data sources can be found in Table 3. Table 6 demonstrates that ensemble learning algorithms such as gradient boosting and random forest have great potential besides deep learning algorithms.
Drug synergy classification
Table 7 shows the results of the comparison of area under the receiver operating characteristic curve (AUC) between the actual and predicted drug synergy scores of seven methods presented in this review. The second column shows comparison methods or comparison datasets. For example, in the first row, ‘whole feature’ means, those including all drug molecular and pharmacological features, are taken into account in [50]. DTF [78] and DeepSynergy [37] are both competitive based on the results and the predictions from the two approaches are complementary. The best performance was by GTB of [54], but all methods achieved around 90% or more of AUC, which implies the data was too easy to evaluate the method, and more tough data would be good to be used for evaluation. Besides, training the drug combination prediction problem as a classification problem might underestimate the actual situation.
Interactive data analysis portals
Table 8 is a list of publicly available software, interactive data portals for analysing combination data.
Table 8.
The list of interactive data portals and tools
| Link | |
|---|---|
| Combenefit [40] | https://sourceforge.net/projects/combenefit/ |
| SynergyFinder [41] | https://synergyfinder.fimm.fi |
| DeepSynergy [37] | http://www.bioinf.jku.at/software/DeepSynergy/ |
| DrugComb [45] | https://drugcomb.fimm.fi |
| DrugCombDB [79] | http://drugcombdb.denglab.org/ |
| SynToxProfiler [19] | https://syntoxprofiler.fimm.fi |
Combenefit [40] is the first free, open-source, advanced software package for the visualization and analysis of drug combinations. Combenefit incorporates three reference models; the Loewe, the Bliss and the highest single agent (HSA) models, which are mentioned in Section Drug combination therapy. The graphical interface of Combenefit provides model-based quantification of drug combinations in single and high-throughput settings. Combenefit was also used to generate one of the commonly used datasets for the AstraZeneca–Sanger Drug Combination Prediction DREAM challenge [34].
SynergyFinder [41] is known as the first publicly available open-source web application to assess the degree of synergy or antagonism. In addition to Combenefit, SynergyFinder includes one more reference model, zero interaction potency (ZIP), for synergy scoring. Using multiple reference models provides an unbiased evaluation of the significance of drug combinations. An interactive graphical interface offers a 2D or a 3D synergy map over the dose matrix.
DrugComb [45] is a free access computational tool that has the following four features:
(1) Free accessible computational tool that provides a web server for evaluation and visualization of accumulated, standardized and harmonized drug combination dose–response data.
(2) Users can analyze their own data.
(3) The sensitivity of drug combinations can be evaluated.
(4) Extensive database, including NCI ALMANAC [32], ONEIL [33], FORCINA [75] and CLOUD [76] datasets.
DeepSynergy [37] is a publicly available web application, which uses a deep learning model trained on the ONEIL data [33] to assess the synergy of drugs. The application provides predictions for untested drug combinations for a given cell line.
DrugCombDB [79] has a user-friendly website for evaluating the synergy of two drugs or antagonism by using the ZIP reference model and data visualization. DrugCombDB integrates drug combinations from high-throughput screening (HTS) assays of drug combinations, external databases and PubMed literature.
SynToxProfiler [19] is the first web tool available for analyzing drug combinations in terms of synergy, toxicity and efficacy. SynToxProfiler can rank synergistic drug pairs with higher efficacy and lower toxicity, using scores based on the integration of synergy, toxicity and efficacy scores. In addition, SynToxProfiler can deal with combinations of a multiple (>2) number of drugs.
Conclusion
Drug combination therapy offers a promising approach to improve the effectiveness of treatments, which are often limited by drug resistance, toxicity and disease heterogeneity. However, identifying all possible combinations for synergistic interaction is infeasible due to the size of combinatorial space. In addition, a deeper understanding of the underlying biological impact is required to efficiently explore the sizeable synergistic space. Incorporation of biological and chemical knowledge would be the solution for this objective. Computational methods especially ML are becoming a crucial element of combination therapy research in this phase to integrate data from many different biomedical sources and understand what kind of effects they have on the prediction of compound combinations. In this light, we discussed the latest ML approaches for drug combination therapies. We also brought attention to the data that needs to be integrated for a comprehensive analysis to efficiently identify effective drug combinations and highlight the importance of side-effect management for avoiding unexpected toxicity. Finally, we provided an empirical comparison of methods presented in this review to understand the difference between methods and a list of publicly available software for analyzing combination data.
Using ML for combination therapy requires proper selection of inputs, so including chemical properties and bioactivity data is crucial to enhance the possibility of overcoming drug resistance. In addition to inputs, experimental results also demonstrate that model selection also has a big impact on the performance such that linear models result in lower performance compared with nonlinear models. Furthermore, ensemble learning algorithms such as gradient boosting and random forest have great potential besides deep learning algorithms. In all prediction settings, predicting new combinations, in which the predictions are made for drug combinations outside the training space with no available combination measurement, is a more challenging task than the other tasks but a more realistic case. The accurate drug combination predictions provide a promising approach for personalized treatments and accelerate the clinical use of drug combination therapies to increase therapeutic efficacies.
One possible way to improve the identification of more sustainable drug combinations might be to collaborate with cost-effective alternative strategies, particularly drug repurposing, which has also effectively found potential treatments. In this collaboration, drug repurposing can be used to identify the hit compounds and then drug combination test with the identified hit compounds to find effective drug combinations [78]. On the other hand, due to the immense data increase, big data problems are arising, and carefully chosen feature extraction methods are needed to improve the prediction accuracy [8]. There will be a need to develop more efficient computational methods for different diseases as expectations increase and opportunities emerge. Pairwise drug combinations are the most prominent; however, combinations of three or four drugs are emerging recently. These current and future situations demonstrate that ML methods are promising and have an exciting future for personalized drug combination therapies by utilizing different types of molecular and genomic data.
Key Points
Drug combination therapies allow us to elucidate disease characteristics between patients caused by variation in therapeutic responses, define synergistic drug effects and minimize adverse drug reactions.
Identification of combinatorial drugs is expensive and time consuming since testing every possible combination of these drugs would be infeasible.
We aim to systematically assess representative ML methods that have been proposed in recent years for understanding the drug combination therapies by grouping these methods into three categories.
Three major factors, i.e. synergy, efficacy and toxicity, should be considered together while categorizing drug combination therapies.
ML methods are becoming a crucial element of combination therapy research to integrate data from many different biomedical sources and understand what kind of effects they have on the prediction of compound combinations.
Funding
H.M. has been supported in part by JST ACCEL (JPMJAC1503); MEXT Kakenhi (16H02868, 19H04169). Academy of Finland (315896); and the Finnish Center for Artificial Intelligence FCAI (320181).
Contributor Information
Betül Güvenç Paltun, Department of Computer Science, Aalto University, Espoo, Finland; Helsinki Institute for Information Technology (HIIT), Finland.
Samuel Kaski, Department of Computer Science, Aalto University, Espoo, Finland; Helsinki Institute for Information Technology (HIIT), Finland; University of Manchester, UK.
Hiroshi Mamitsuka, Department of Computer Science, Aalto University, Espoo, Finland; Helsinki Institute for Information Technology (HIIT), Finland; Bioinformatics Center, Institute for Chemical Research, Kyoto University, Uji 6110011, Japan.
References
- 1. Pemovska T, Bigenzahn JW, Superti-Furga G. Recent advances in combinatorial drug screening and synergy scoring. Curr Opin Pharmacol 2018;42:102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lopez JS, Banerji U. Combine and conquer: challenges for targeted therapy combinations in early phase trials. Nat Rev Clin Oncol 2017;14(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ryall KA, Tan AC. Systems biology approaches for advancing the discovery of effective drug combinations. J Chem 2015;7(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bulusu KC, Guha R, Mason DJ, et al. Modelling of compound combination effects and applications to efficacy and toxicity: state-of-the-art, challenges and perspectives. Drug Discov Today 2016;21(2):225–38. [DOI] [PubMed] [Google Scholar]
- 5. Sheng Z, Sun Y, Yin Z, et al. Advances in computational approaches in identifying synergistic drug combinations. Brief Bioinform 2018;19(6):1172–82. [DOI] [PubMed] [Google Scholar]
- 6. Madani Tonekaboni SA, Soltan Ghoraie L, Manem VSK, et al. Predictive approaches for drug combination discovery in cancer. Brief Bioinform 2018;19(2):263–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hasselt JC, Iyengar R. Systems pharmacology: defining the interactions of drug combinations. Annu Rev Pharmacol Toxicol 2019;59:21–40. [DOI] [PubMed] [Google Scholar]
- 8. Tsigelny IF. Artificial intelligence in drug combination therapy. Brief Bioinform 2019;20(4):1434–48. [DOI] [PubMed] [Google Scholar]
- 9. Jia J, Zhu F, Ma X, et al. Mechanisms of drug combinations: interaction and network perspectives. Nat Rev Drug Discov 2009;8(2):111–28. [DOI] [PubMed] [Google Scholar]
- 10. Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol 2012;30(7):679–92. [DOI] [PubMed] [Google Scholar]
- 11. Liu H, Zhao Y, Zhang L, et al. Anti-cancer drug response prediction using neighbor-based collaborative filtering with global effect removal. Mol Ther Nucleic Acids 2018;13:303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Güvenç Paltun B, Kaski S, Mamitsuka H. Diverse: Bayesian data integrative learning for precise drug response prediction. IEEE/ACM Trans Comput Biol Bioinform 2021;1. [DOI] [PubMed] [Google Scholar]
- 13. Tang J, Karhinen L, Xu T, et al. Target inhibition networks: predicting selective combinations of druggable targets to block cancer survival pathways. PLoS Comput Biol 2013;9(9):e1003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang J, Aittokallio T. Network pharmacology strategies toward multi-target anticancer therapies: from computational models to experimental design principles. Curr Pharm Des 2014;20(1):23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jaeger S, Igea A, Arroyo R, et al. Quantification of pathway cross-talk reveals novel synergistic drug combinations for breast cancer. Cancer Res 2017;77(2):459–69. [DOI] [PubMed] [Google Scholar]
- 16. Bansal M, Yang J, Karan C, et al. A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol 2014;32(12):1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 2006;58(3):621–81. [DOI] [PubMed] [Google Scholar]
- 18. Saputra EC, Huang L, Chen Y, et al. Combination therapy and the evolution of resistance: the theoretical merits of synergism and antagonism in cancer. Cancer Res 2018;78(9):2419–31. [DOI] [PubMed] [Google Scholar]
- 19. Ianevski A, Timonen S, Kononov A, et al. Syntoxprofiler: an interactive analysis of drug combination synergy, toxicity and efficacy. PLoS Comput Biol 2020;16(2):e1007604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang J, Wennerberg K, Aittokallio T. What is synergy? The Saariselkä agreement revisited. Front Pharmacol 2015;6:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Palmer AC, Sorger PK. Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell 2017;171(7):1678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gayvert KM, Aly O, Platt J, et al. A computational approach for identifying synergistic drug combinations. PLoS Comput Biol 2017;13(1):e1005308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doroshow JH, Simon RM. On the design of combination cancer therapy. Cell 2017;171(7):1476–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fitzgerald JB, Schoeberl B, Nielsen UB, et al. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol 2006;2(9):458–66. [DOI] [PubMed] [Google Scholar]
- 25. Scott LJ. Dolutegravir/lamivudine single-tablet regimen: a review in HIV-1 infection. Drugs 2020;80(1):61–72. [DOI] [PubMed] [Google Scholar]
- 26. Spain L, Julve M, Larkin J. Combination dabrafenib and trametinib in the management of advanced melanoma with BRAFV600 mutations. Expert Opin Pharmacother 2016;17(7):1031–8. [DOI] [PubMed] [Google Scholar]
- 27. Iwata H, Sawada R, Mizutani S, et al. Large-scale prediction of beneficial drug combinations using drug efficacy and target profiles. J Chem Inf Model 2015;55(12):2705–16. [DOI] [PubMed] [Google Scholar]
- 28. Zhao S, Nishimura T, Chen Y, et al. Systems pharmacology of adverse event mitigation by drug combinations. Sci Transl Med 2013;5(206):206ra140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Das P, Delost MD, Qureshi MH, et al. A survey of the structures of US FDA approved combination drugs. J Med Chem 2018;62(9):4265–311. [DOI] [PubMed] [Google Scholar]
- 30. Liu Y, Hu B, Fu C, et al. DCDB: drug combination database. Bioinformatics 2010;26(4):587–8. [DOI] [PubMed] [Google Scholar]
- 31. Hare D, Foster T. The Orange Book: the Food and Drug Administration’s advice on therapeutic equivalence. Am Pharm 1990;(7):35–37. [DOI] [PubMed] [Google Scholar]
- 32. Holbeck SL, Camalier R, Crowell JA, et al. The National Cancer Institute ALMANAC: a comprehensive screening resource for the detection of anticancer drug pairs with enhanced therapeutic activity. Cancer Res 2017;77(13):3564–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O’Neil J, Benita Y, Feldman I, et al. An unbiased oncology compound screen to identify novel combination strategies. Mol Cancer Ther 2016;15(6):1155–62. [DOI] [PubMed] [Google Scholar]
- 34. Menden MP, Wang D, Mason MJ, et al. Community assessment to advance computational prediction of cancer drug combinations in a pharmacogenomic screen. Nat Commun 2019;10(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen X, Ren B, Chen M, et al. ASDCD: antifungal synergistic drug combination database. PloS one 2014;9(1):e86499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang H, Zhang P, Qu XA, et al. Systematic prediction of drug combinations based on clinical side-effects. Sci Rep 2014;4(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Preuer K, Lewis RP, Hochreiter S, et al. DeepSynergy: predicting anti-cancer drug synergy with deep learning. Bioinformatics 2018;34(9):1538–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo WF, Zhang SW, Feng YH, et al. Network controllability-based algorithm to target personalized driver genes for discovering combinatorial drugs of individual patients. Nucleic Acids Res 2021;49(7):e37–e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Julkunen HJ, Cichonska A, Gautam P, et al. comboFM: leveraging multi-way interactions for systematic prediction of drug combination effects. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Di Veroli GY, Fornari C, Wang D, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 2016;32(18):2866–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ianevski A, He L, Aittokallio T, et al. SynergyFinder: a web application for analyzing drug combination dose–response matrix data. Bioinformatics 2017;33(15):2413–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Celebi R, Don’t Walk OB, Movva R, et al. In-silico prediction of synergistic anti-cancer drug combinations using multi-omics data. Sci Rep 2019;9(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Güvenç Paltun B, Mamitsuka H, Kaski S. Improving drug response prediction by integrating multiple data sources: matrix factorization, kernel and network-based approaches. Brief Bioinform 2021;22(1):346–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xia F, Shukla M, Brettin T, et al. Predicting tumor cell line response to drug pairs with deep learning. BMC Bioinformatics 2018;19(18):71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zagidullin B, Aldahdooh J, Zheng S, et al. DrugComb: an integrative cancer drug combination data portal. Nucleic Acids Res 2019;47(W1):W43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang L, Li F, Sheng J, et al. DrugComboRanker: drug combination discovery based on target network analysis. Bioinformatics 2014;30(12):i228–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jeon M, Kim S, Park S, et al. In silico drug combination discovery for personalized cancer therapy. BMC Syst Biol 2018;12(2):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ianevski A, Giri AK, Gautam P, et al. Prediction of drug combination effects with a minimal set of experiments. Nat Mach Intell 2019;1(12):568–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao XM, Iskar M, Zeller G, et al. Prediction of drug combinations by integrating molecular and pharmacological data. PLoS Comput Biol 2011;7(12):e1002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li P, Huang C, Fu Y, et al. Large-scale exploration and analysis of drug combinations. Bioinformatics 2015;31(12):2007–16. [DOI] [PubMed] [Google Scholar]
- 51. Chen X, Ren B, Chen M, et al. NLLSS: predicting synergistic drug combinations based on semi-supervised learning. PLoS Comput Biol 2016;12(7):e1004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li X, Xu Y, Cui H, et al. Prediction of synergistic anti-cancer drug combinations based on drug target network and drug induced gene expression profiles. Artif Intell Med 2017;83:35–43. [DOI] [PubMed] [Google Scholar]
- 53. Held MA, Langdon CG, Platt JT, et al. Genotype-selective combination therapies for melanoma identified by high-throughput drug screening. Cancer Discov 2013;3(1):52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu H, Zhang W, Nie L, et al. Predicting effective drug combinations using gradient tree boosting based on features extracted from drug-protein heterogeneous network. BMC Bioinformatics 2019;20(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Karimi M, Hasanzadeh A, Shen Y. Network-principled deep generative models for designing drug combinations as graph sets. Bioinformatics 2020;36(Suppl 1):i445–i445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Menche J, Sharma A, Kitsak M, et al. Uncovering disease-disease relationships through the incomplete interactome. Science 2015;347(6224). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cheng F, Kovács IA, Barabási AL. Network-based prediction of drug combinations. Nat Commun 2019;10(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sun Z, Huang S, Jiang P, et al. DTF: Deep tensor factorization for predicting anticancer drug synergy. Bioinformatics 2020;36(16):4483–9. [DOI] [PubMed] [Google Scholar]
- 59. Kuhn M, Szklarczyk D, Franceschini A, et al. STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Res 2010;38(Suppl 1):D552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen X, Ji ZL, Chen YZT. Therapeutic target database. Nucleic Acids Res 2002;30(1):412–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kanehisa M, Goto S, Furumichi M, et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 2010;38(Suppl 1):D355–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lamb J, Crawford ED, Peck D, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313(5795):1929–35. [DOI] [PubMed] [Google Scholar]
- 63. Hamosh A, Scott AF, Amberger JS, et al. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005;33(Suppl 1):D514–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res 2007;36(Suppl 1):D480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stark C, Breitkreutz BJ, Reguly T, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 2006;34(Suppl 1):D535–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu T, Lin Y, Wen X, et al. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res 2007;35(Suppl 1):D198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Günther S, Kuhn M, Dunkel M, et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res 2007;36(Suppl 1):D919–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gaulton A, Bellis LJ, Bento AP, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 2012;40(D1):D1100–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Roth BL, Lopez E, Patel S, et al. The multiplicity of serotonin receptors: uselessly diverse molecules or an embarrassment of riches? Neuroscientist 2000;6(4):252–62. [Google Scholar]
- 70. Zhu F, Han B, Kumar P, et al. Update of TTD: therapeutic target database. Nucleic Acids Res 2010;38(Suppl 1):D787–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kuhn M, Campillos M, Letunic I, et al. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol 2010;6(1):343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liberzon A, Subramanian A, Pinchback R, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27(12):1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Iorio F, Knijnenburg TA, Vis DJ, et al. A landscape of pharmacogenomic interactions in cancer. Cell 2016;166(3):740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yang W, Soares J, Greninger P, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2012;41(D1):D955–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Forcina GC, Conlon M, Wells A, et al. Systematic quantification of population cell death kinetics in mammalian cells. Cell Systems 2017;4(6):600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Licciardello MP, Ringler A, Markt P, et al. A combinatorial screen of the cloud uncovers a synergy targeting the androgen receptor. Nat Chem Biol 2017;13(7):771. [DOI] [PubMed] [Google Scholar]
- 77. Griner LAM, Guha R, Shinn P, et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell–like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci 2014;111(6):2349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sun W, Sanderson PE, Zheng W. Drug combination therapy increases successful drug repositioning. Drug Discov Today 2016;21(7):1189–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu H, Zhang W, Zou B, et al. DrugCombDB: a comprehensive database of drug combinations toward the discovery of combinatorial therapy. Nucleic Acids Res 2020;48(D1):D871–81. [DOI] [PMC free article] [PubMed] [Google Scholar]


