Zhang, Kim, and colleagues reveal two macrophage subpopulations lining peritoneal surfaces that are distinct from peritoneal fluid macrophages. One population expresses LYVE1, resembles omental macrophages, and promotes expansion of ovarian cancer within the cavity in the absence of the omentum.
Abstract
Two resident macrophage subsets reside in peritoneal fluid. Macrophages also reside within mesothelial membranes lining the peritoneal cavity, but they remain poorly characterized. Here, we identified two macrophage populations (LYVE1hi MHC IIlo-hi CX3CR1gfplo/− and LYVE1lo/− MHC IIhi CX3CR1gfphi subsets) in the mesenteric and parietal mesothelial linings of the peritoneum. These macrophages resembled LYVE1+ macrophages within surface membranes of numerous organs. Fate-mapping approaches and analysis of newborn mice showed that LYVE1hi macrophages predominantly originated from embryonic-derived progenitors and were controlled by CSF1 made by Wt1+ stromal cells. Their gene expression profile closely overlapped with ovarian tumor-associated macrophages previously described in the omentum. Indeed, syngeneic epithelial ovarian tumor growth was strongly reduced following in vivo ablation of LYVE1hi macrophages, including in mice that received omentectomy to dissociate the role from omental macrophages. These data reveal that the peritoneal compartment contains at least four resident macrophage populations and that LYVE1hi mesothelial macrophages drive tumor growth independently of the omentum.
Introduction
Serous membranes line the peritoneal cavity as they generate a functional border for visceral organs. A major serosal surface includes the gut-associated mesentery that anatomically bridges the intestines and mesenteric lymph nodes (MLNs). The mesentery anchors the small intestine and colon and facilitates blood circulation and interstitial fluid flow through the mesenteric lymphatic vessels to maintain tissue homeostasis. The serous membranes of the mesentery are enriched in stromal cells such as fibroblasts and mesothelial cells that produce vitamin A metabolites that sustain peritoneal fluid macrophages (Buechler et al., 2019), as well as collagens, elastin, laminin, and glycoproteins that form a complex extracellular matrix (Jackson-Jones et al., 2020). Resident macrophages on the serosal surface of the liver (David et al., 2016) and within parietal peritoneal membranes (Uderhardt et al., 2019) have been described, and both populations play a role in governing recruitment of neutrophils following tissue injury. However, whether these membrane-associated macrophages are related to those in the peritoneal fluid in phenotype or origin is unknown. Indeed, the full phenotypic and gene expression profile of peritoneal membrane–associated macrophages has not been reported.
The peritoneal cavity contains serous fluid that hosts two types of resident macrophages that have been well characterized in recent years, the Gata6-dependent large peritoneal macrophages (LPMs; Gautier et al., 2014; Gautier et al., 2012; Ghosn et al., 2010; Okabe and Medzhitov, 2014; Rosas et al., 2014) and the IRF4-dependent small peritoneal macrophages (Ghosn et al., 2010; Kim et al., 2016). The LPMs float freely in peritoneal fluid and participate critically in the entrapment and clearance of microorganisms that might gain entry to the cavity after breach of the intestinal boundary (Zhang et al., 2019a). The biology of the peritoneal macrophage has long been linked to the immunologic properties of the omental adipose tissue located within the peritoneal cavity. The omentum is home to collections of organized immune cells called milky spots or fat-associated lymphoid clusters containing memory T cells (Han et al., 2017), B cells (Wu et al., 2019), natural killer T cells (Bénézech et al., 2015), dendritic cells, and innate lymphoid cells (Moro et al., 2010). Furthermore, the peritoneum can be affected by pathologies that include postsurgical adhesions, endometriosis, and metastases of tumors most commonly arising from the colon, appendix, ovaries, or stomach. Experimental mouse models have begun to explore the role of macrophages in these conditions (Hogg et al., 2021; Weiss et al., 2018; Xia et al., 2020; Zindel et al., 2021).
Epithelial ovarian cancer is the deadliest gynecological malignancy, with high-grade serous ovarian cancer (HGSOC) being the most common subtype. HGSOC can originate from both fallopian tube and ovarian surface epithelium (Zhang et al., 2019b) and is commonly associated with widespread peritoneal carcinomatosis (Lengyel, 2010). More than 60% of patients with ovarian cancers are diagnosed at an advanced stage with peritoneal metastases (Siegel et al., 2018). HGSOC metastasis often leads to malignant ascites that is predominantly composed of inflammatory cells that include macrophages, neutrophils, lymphocytes (Robinson-Smith et al., 2007; Singel et al., 2019), cancer-associated fibroblasts (Gao et al., 2019), and tumor cells. HGOSC metastasis typically involves the peritoneal cavity, including relevant adjacent tissues such as the omentum. In addition to the omentum, peritoneal serosa and mesentery are also common metastatic sites (Steinkamp et al., 2013); distant metastatic seeding to abdominal organs and to the lungs and pleura also occurs. Contributions of different immune cells to this metastatic evolution of cancer cells in the peritoneal environment remain understudied. Recently, Lawrence and colleagues (Etzerodt et al., 2020) characterized omental macrophages and concluded that a major subset of omental macrophages could account for the tumor-promoting role of the omentum.
Here, we profiled mesenteric membrane–associated macrophages and identified two distinct populations (LYVE1hi MHC IIlo-hi CX3CR1gfplo/− and LYVE1lo/− MHC IIhi CX3CR1gfphi subsets) that coexist in the mesothelial layer, generally resembling interstitial macrophages described in the lung previously (Chakarov et al., 2019; Gibbings et al., 2017). Mesenteric LYVE1hi macrophages were derived from embryonic precursors and were controlled by colony stimulating factor 1 (CSF1) derived from local stromal cells. These macrophages did not depend on GATA6 or IRF4 and maintained a life cycle distinct from that of peritoneal fluid macrophages. Bulk RNA sequencing (RNA-seq) analysis of LYVE1hi membrane-associated macrophages and its comparison to single-cell RNA-seq (scRNA-seq) of omental macrophages in ovarian tumors showed that LYVE1hi macrophages had a specialized gene expression pattern that correlated with the genes expressed by LYVE1hi omental macrophages during ovarian cancer progression (Etzerodt et al., 2020). Using in vivo ablation approaches in omentectomized mice, we unraveled the relationship between LYVE1hi macrophages, the omentum, and ovarian tumor progression. Our data indicate that LYVE1hi macrophages within peritoneal membranes like those associated with the mesentery promote tumor progression even in the absence of the omentum. Since omentectomy is standard in primary debulking surgery for ovarian cancer, our results raise the possibility that embryonic-derived mesenteric LYVE1hi macrophages may drive tumor progression and potentially disease recurrence after initial therapy for metastatic ovarian cancer.
Results
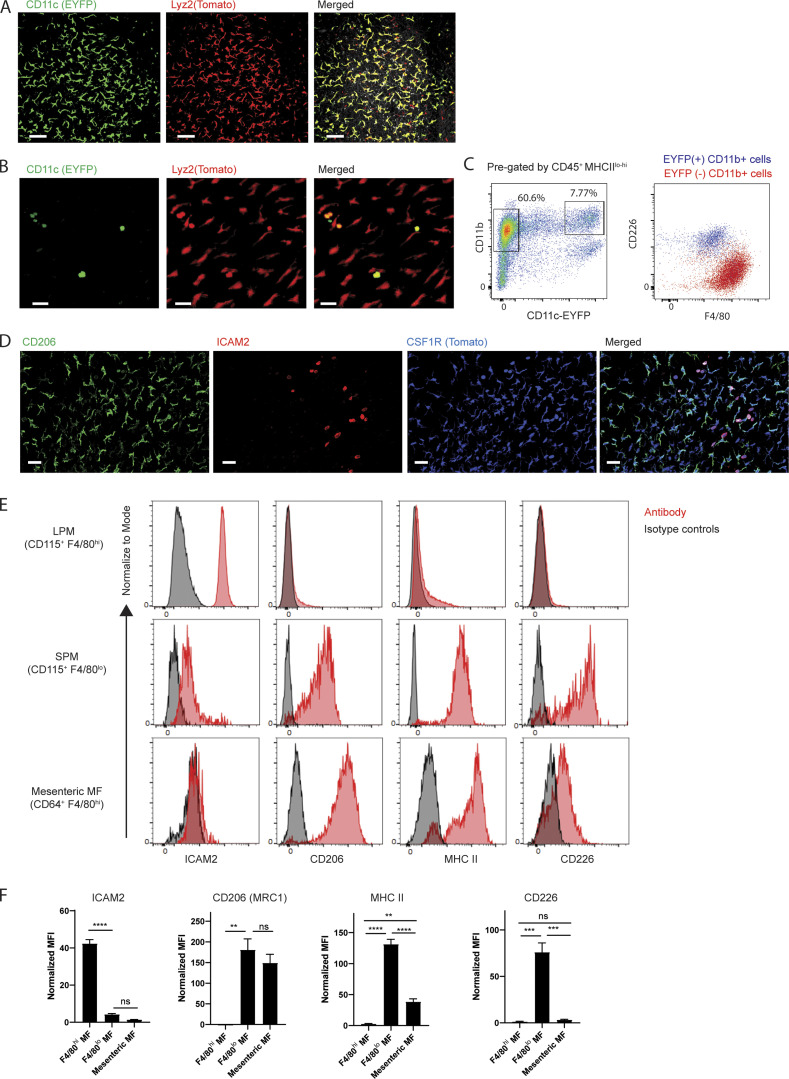
Mesenteric membrane–associated macrophages are phenotypically distinct from macrophages in the liver capsule and peritoneal cavity
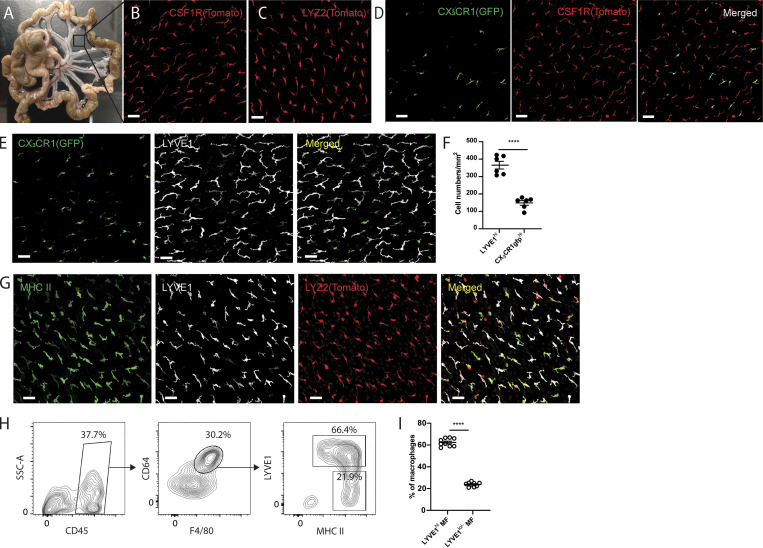
The vascular and lymphatic tracts that connect the intestine with the draining MLNs in mice are joined by avascular sheets of tissue known to be lined by mesothelium and accompanying fibroblasts (Fig. 1 A). Taking advantage of two myeloid-specific reporter strains, Csf1rCreERT2:Rosa26LSL-tdTomato and Lyz2Cre:Rosa26LSL-tdTomato mice, we observed elongated Tomato+ cells in these mesenteric membranes (Fig. 1, B and C). The cells were negative for enhanced YFP (EYFP) in CD11cEYFP mice (Lyz2Cre:Rosa26LSL-tdTomato:CD11cEYFP), distinguishing them from liver capsule macrophages (David et al., 2016) that were positive for both EYFP and Tomato reporters in the respective strains (Fig. S1 A). We did, however, observe a few round-shaped Tomato+ EYFP+ cells sparsely scattered on the mesenteric sheet (Fig. S1 B), and they were in close contact with Tomato+ cells (Video 1). In flow cytometric analysis of gut mesentery, CD11b+ EYFP+ cells corresponded to the previously described F4/80lo CD226+ MHC II+ macrophages (Kim et al., 2016), but most CD11b+ EYFP− cells were F4/80+ and did not express CD226 (Fig. S1 C). Instead, they expressed high levels of CD206, which is absent on peritoneal macrophages (Fig. S1 D). ICAM2, which marks peritoneal macrophages (Gautier et al., 2012), was observed only on a few round-shaped Tomato+ cells sitting atop the mesenteric sheet (Fig. S1 D). Flow cytometric analysis of F4/80hi and F4/80lo peritoneal macrophage subsets and mesenteric macrophages in the same mice confirmed that mesenteric macrophages are positive for CD206 and MHC II. They were negative for ICAM2 and CD226 (Fig. S1, E and F). The mean fluorescence intensity for MHC II expression in mesenteric macrophages was relatively lower than that of F4/80lo peritoneal macrophages (Fig. S1 F). Overall, it appears that mesenteric membrane–associated macrophages are phenotypically distinct from the two well-established serous fluid peritoneal macrophage populations, which only infrequently attach to the mesenteric membrane in unperturbed mice. An estimate of their number (see Materials and methods) at 106 within the peritoneal cavity suggests that their total numbers in the peritoneal cavity are approximately similar to the number of resident macrophages in the peritoneal fluid (1–2 × 106; Zhang et al., 2019a). By comparison, we estimated the number of macrophages in the omentum to be far lower, at 2 × 104 per omentum.
Figure 1.
Two distinct macrophage populations coexist in the avascular regions of mesenteric membranes. (A) Whole-mount images of gut mesentery in adult WT mice. Square box indicates a region of avascular mesenteric membrane. (B and C) Whole-mount images of the avascular mesenteric membrane from tamoxifen-induced Csf1rCreERT2:R26LSL-tdTomato mice (B) and Lyz2Cre:R26LSL-tdTomato mice (C). Scale bar, 50 µm. (D) Whole-mount images of mesenteric membrane from tamoxifen-induced Csf1rCreERT2:R26LSL-tdTomato:CX3CR1gfp mice. CX3CR1-GFP cells (left), Csf1r-expressing Tomato+ cells (middle), and merged pictures (right) for two distinct macrophage populations. Scale bar, 50 µm. (E) Immunohistochemistry analysis of a whole-mount mesenteric membrane from CX3CR1gfp/+ mouse stained for LYVE1. CX3CR1gfp expression (left), LYVE1 (middle), and merged pictures (right) for two distinct macrophage subsets. Scale bar, 50 µm. (F) Quantification of LYVE1hi CX3CR1gfplo/− macrophages and LYVE1lo/− CX3CR1gfphi macrophages in mesenteric membranes. Data are representative of three independent experiments (n = 3; mean ± SEM). Macrophages were quantified in two different regions of mesenteric membrane per mouse. Unpaired Student’s t test: ****, P < 0.0001. (G) Immunohistochemistry analysis of a whole-mount mesenteric membrane from Lyz2Cre:R26LSL-tdTomato mice stained with LYVE1 and MHCII. Scale bar, 50 µm. (H) Flow cytometric analysis of membrane-associated macrophages isolated from gut mesentery with CD45, F4/80, CD64, LYVE1, and MHC II staining. SSC, side scatter. (I) Frequency of LYVE1hi membrane-associated macrophages and LYVE1lo/− membrane-associated macrophages from flow cytometric analysis (H). Data are pooled from two independent experiments (n = 9; mean ± SEM). Unpaired Student’s t test: ****, P < 0.0001. All imaging data are representative of at least three independent experiments.
Figure S1.
Characterization of membrane-associated macrophages. (A) Two-photon images of liver capsule from Lyz2cre:R26LSL-tdTomato:CD11cEYFP mice. Scale bar, 70 µm. (B) Two-photon images of mesenteric membrane from Lyz2cre:R26Tomao:CD11cEYFP mice. Scale bar, 40 µm. (C) Representative flow cytometric analysis of gut mesentery in CD11cEFYP mice. CD11cEYFP and CD11b gating of CD45+ MHCIIlo-to-hi mesenteric cells (left). Overlay of CD11b+ EYFP− and CD11b+ EYFP+ cells (right; n = 6). (D) Whole-mount confocal images of Csf1rCreER:R26Tomato mice with CD206 and ICAM2 staining. Imaging data are representative of at least two independent experiments. Scale bar, 40 µm. (E) Flow cytometric analysis for ICAM2, CD206, MHC II, and CD226 expression in F4/80hi LPMs, F4/80lo small peritoneal macrophages (SPM) and F4/80hi mesenteric macrophages. Data are representative of at least two independent experiments. (F) Mean fluorescent intensity (MFI) values of ICAM2, CD206, MHC II, and CD226, which are normalized by isotype controls. Data are pooled from at least two independent experiments (n = 3–6 mice). Statistical analysis was performed by one-way ANOVA and Tukey’s multiple-comparison test: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Video 1.
Video reconstruction of interactions of Tomato-expressing macrophages under Lyz2 promoter with CD11cEYFP-expressing peritoneal macrophages. Two-photon intravital microscope visualized Tomato-expressing mesenteric membrane macrophage in contact with CD11c EYFP-expressing F4/80lo peritoneal macrophages.
Mesenteric membrane–associated macrophages consist of two distinct subsets
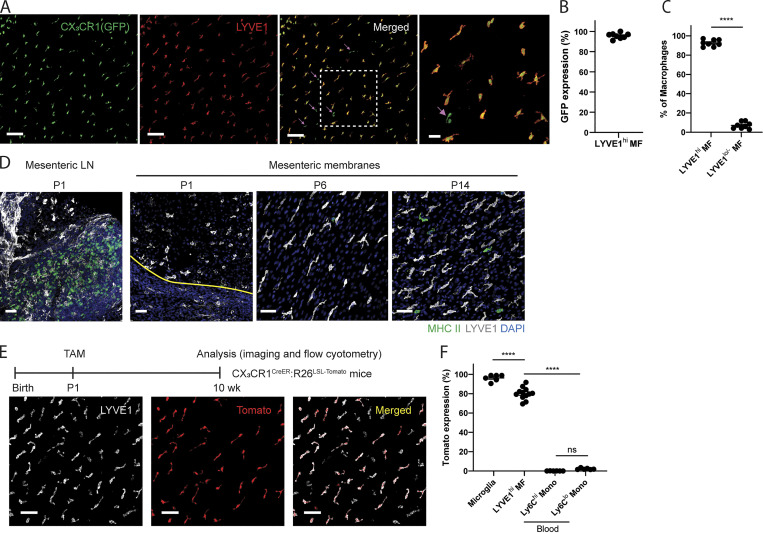
Some macrophage populations constitutively express GFP reporter in CX3CR1gfp/+ mice (Bain et al., 2014; Gibbings et al., 2017; Stamatiades et al., 2016; Williams et al., 2020; Zigmond et al., 2014; Zigmond et al., 2012), while other tissue-resident macrophages do not (Yona et al., 2013). To determine whether mesenteric membrane–associated macrophages express GFP reporter in CX3CR1gfp/+ mice, Csf1rCreERT2:Rosa26LSL-tdTomato mice were crossed with CX3CR1gfp mice to generate Csf1rCreERT2:Rosa26LSL-tdTomato:CX3CR1gfp/+ mice. In the mesenteric membranes of these dual reporter mice, we observed two distinct macrophage subsets: Tomato+ CX3CR1gfplo/− macrophages and Tomato+ CX3CR1gfphi macrophages (Fig. 1 D). To characterize these subsets further, mesenteric membranes were stained for LYVE1, as it was recently reported that two distinct LYVE1hi and LYVE1lo interstitial macrophage subsets are present together in some tissues (Chakarov et al., 2019; Etzerodt et al., 2020; Lacerda Mariano et al., 2020; Lim et al., 2018). In CX3CR1gfp/+ mice, we observed that most CX3CR1gfplo/− macrophages highly expressed LYVE1, while CX3CR1gfphi macrophages were negative or had low expression for LYVE1 (Fig. 1 E). LYVE1hi macrophages were the dominant population, compared with CX3CR1gfphi macrophages (Fig. 1 F).
To further characterize LYVE1hi and LYVE1lo/− macrophages, membrane-associated macrophages were stained for MHC II in Lyz2Cre:Rosa26LSL-tdTomato mice. Both LYVE1hi and LYVE1lo/− membrane-associated macrophages expressed MHC II, albeit more weakly in the LYVE1hi macrophages, in images from confocal microscopy (Fig. 1 G). Through flow cytometric analysis of single-cell suspensions generated from gut mesentery, we confirmed that F4/80+ CD64+ macrophages could be divided into two major populations: LYVE1hi MHC IIlo-hi and LYVE1lo/− MHC IIhi macrophages (Fig. 1 H). The frequency of the two macrophage subsets did not differ depending on whether they were examined by confocal microscopy or flow cytometry (Fig. 1, F and I).
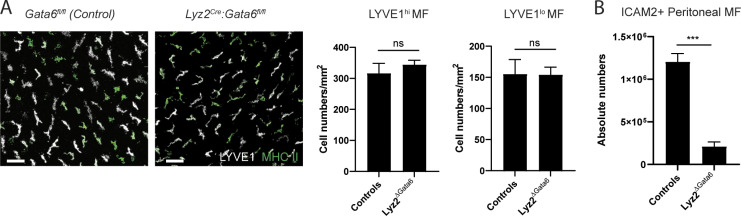
Both LYVE1hi and LYVE1lo/− membrane-associated macrophages were intact in the absence of Gata6 expression within macrophages (Lyz2cre:Gata6fl/fl mice; Fig. S2 A), whereas peritoneal macrophages were highly reduced (Fig. S2 B), indicating that the membranous macrophages were maintained independently from peritoneal fluid macrophages. Taken together, mesenteric membrane–associated macrophages consist of two distinct subsets: LYVE1hi CX3CR1lo/− MHC IIlo-hi macrophages (henceforth termed LYVE1hi macrophages) and LYVE1lo/− CX3CR1hi MHC IIhi macrophages (henceforth termed LYVE1lo/− macrophages).
Figure S2.
Mesenteric membrane–associated macrophages in Lyz2Cre:Gata6fl/fl mice. (A) Whole-mount images and quantification of membrane-associated macrophages of Lyz2cre:Gata6fl/fl mice and littermate controls. Scale bar, 50 µm. (B) Quantification of ICAM2+ peritoneal macrophages in Lyz2cre:Gata6fl/fl mice and littermate controls. Imaging data and flow cytometric analysis are representative of two independent experiments (n = 3 per genotype; mean ± SEM). Unpaired Student’s t test: ***, P < 0.001.
LYVE1hi membrane-associated macrophages are located on the barrier surfaces of many organs
In the mesentery, LYVE1hi macrophages, probed initially by immunostaining for LYVE1, were rather evenly distributed across the mesenteric surface in both the avascular and vascular areas of the mesentery (Fig. 2 A). The vascularized area was rich in adipose tissue housing the nerves, blood vessels, and lymphatic vessels, marked using tamoxifen-treated Prox1CreERT2:R26LSL-tdTomato mice (Fig. 2 A).
Figure 2.
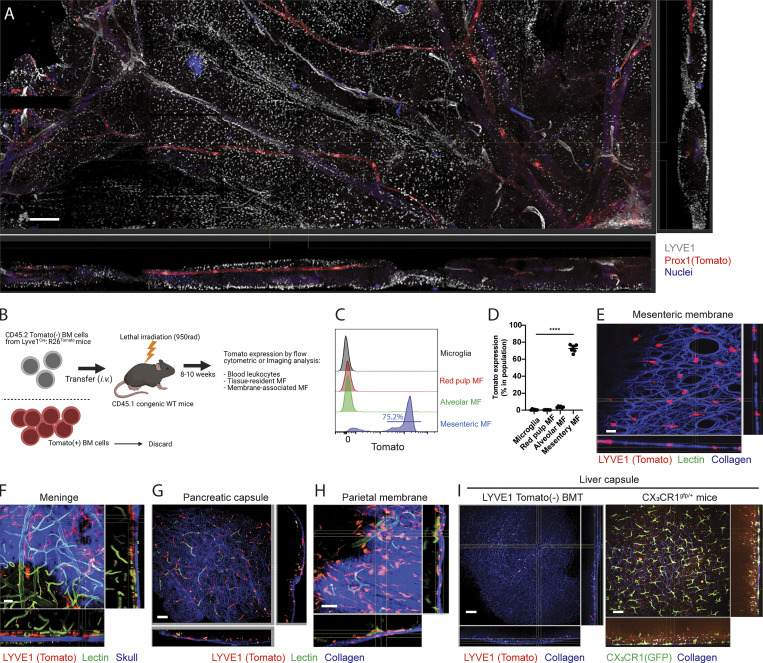
Lyve1hi membrane-associated macrophages are located in serous membrane of tissue parenchyma. (A) Whole-mount, confocal microscopy tile-scan reconstructions to examine the location and distribution of LYVE1hi mesenteric macrophages in tamoxifen-induced Prox1CreER: R26LSL-tdTomato mice (red, Prox1-expressing lymphatic collector; white, LYVE1-expressing macrophages and lymphatic capillaries; blue, nuclei). Scale bar, 400 µm. Images are representative of two independent experiments with scanning of a large region of tissue. (B) Schemes for Tomato− BM transplantation to whole-body-irradiated mice (created with BioRender.com). Sorted CD45.2 Tomato− BM cells from Lyve1cre:R26LSL-tdTomato mice were transplanted to irradiated congenic CD45.1 WT mice. Tomato reporter will label only adult macrophages with an active LYVE1 promoter in adulthood, bypassing embryo-restricted activity at this promoter. (C) Histogram showing Tomato reporter expression of tissue-resident macrophages in brain, spleen, lung, and gut mesentery of Tomato− BM transplanted CD45.1 recipient mice. (D) Quantification of Tomato expression in donor-derived tissue-resident macrophages of Tomato− BM transplanted CD45.1 mice. (C-D) Data are pooled from at least two independent experiments (n = 6, mean ± SEM). Statistical analysis was performed by one-way ANOVA and Tukey’s multiple-comparison test: ****, P < 0.0001. (E) Whole mount images of the mesenteric membrane in Tomato− BM transplanted chimeric mice (red, Tomato+ macrophages; blue, collagens imaged by second harmonic generation [SHG]). Scale bar, 20 µm. (F) Whole mount images of the meninge in Tomato− BM transplanted chimeric mice (red, Tomato+ macrophages; green, Alexa488-conjugated lectin injected blood vessels; blue, skull imaged by SHG). Note Tomato+ perivascular macrophages underneath skull. Scale bar, 20 µm. (G) Whole mount images of pancreas from Tomato− BM transplanted chimeric mice (red, Tomato+ macrophages; green, Alexa488-conjugated lectin injected blood vessels; blue, collagen bundles shown as SHG) Scale bar, 50 µm. (H) Whole-mount images of the parietal peritoneal membrane from Tomato− BM transplanted chimeric mice (red, Tomato+ macrophages; green, Alexa488-conjugated lectin injected blood vessels; blue, collagens shown by SHG). Scale bar, 50 µm, (E, G, and H) Note Tomato+ membrane-associated macrophages in collagen-enriched serosa membrane. (I) Comparison of liver capsular macrophages between Tomato− BM-transplanted chimeric mice (left) and CX3CR1gfp/+ mice (right); red, Tomato+ macrophages; green, CX3CR1gfp+ macrophages; blue, collagen. Scale bar, 50 µm. All two-photon microscopic images are representative of at least two independent experiments.
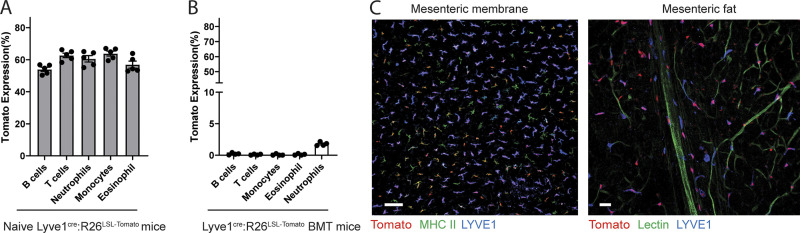
LYVE1 is transiently expressed in erythroid-myeloid progenitors (EMPs) during embryogenesis and in hematopoietic stem cells (HSCs) in adult hematopoiesis (Lee et al., 2016). As such, 50–70% of blood leukocytes expressed the Tomato reporter in Lyve1Cre:Rosa26LSL-tdTomato mice due to this embryonic history (Fig. S3 A). Thus, to visualize LYVE1-expressing macrophages selectively using reporter mice, we designed a bone marrow (BM) chimeric mouse model in which CD45.2 Tomato− BM cells were first isolated from Lyve1Cre:Rosa26LSL-tdTomato mice by FACS (Fig. 2 B). They were then transplanted into lethally irradiated CD45.1 congenic mice. 8–10 wk later, BM chimeric mice were analyzed by flow cytometry and microscopy. In contrast to unmanipulated Lyve1Cre:Rosa26LSL-tdTomato mice, Tomato reporter–expressing cells were not found among blood leukocytes in mice receiving the Tomato− BM transplant (Fig. S3 B). Other tissue-resident macrophages such as microglia, red pulp macrophages, and alveolar macrophages also were not labeled by the Tomato reporter due to marked radioresistance and lack of LYVE1 expression (microglia) or radiosensitivity and lack of LYVE1 expression (e.g., spleen or alveolar macrophages; Fig. 2, C and D). By contrast, mesenteric macrophages were mainly radiosensitive, and >70% were highly labeled by the Tomato reporter in flow cytometric analysis and imaging (Fig. 2, C–E; and Fig. S3 C).
Figure S3.
Comparison of blood leukocytes between naive Lyve1cre:R26LSL-tdTomato mice and Tomato− BM-transplanted mice. (A) Tomato expression of blood leukocytes in naive Lyve1cre:R26LSL-tdTomato mice (n = 5). (B) Tomato expression of blood leukocytes from Tomato− BM-transplanted mice (n = 4). Data are representative of at least two independent experiments. (C) Images of avascular region of a mesenteric membrane and the region containing adipose tissue in the BM-transplanted mice. Scale bars, 100 and 30 µm.
Across a range of organs, Lyve1Cre:Rosa26LSL-tdTomato BM-transplanted mice were visualized by two-photon microscopy immediately after intravenous injection of Alexa Fluor 488–conjugated lectins to label blood vasculature. Consistent with the recent report that LYVE1hi perivascular macrophages are resident in dura mater and pia mater as vascular-associated macrophages (Kim et al., 2021), we found Tomato-labeled meningeal perivascular macrophages near the blood vasculature underneath the skull (Fig. 2 F and Video 2). Tomato+ macrophages were also concentrated in the collagen-enriched barrier surface of the pancreas (Fig. 2 G) and the parietal peritoneal membrane (Fig. 2 H and Video 3), indicating that LYVE1-expressing macrophages were enriched at a number of barrier surfaces, including multiple mesothelial linings of the peritoneal cavity. However, Tomato+ macrophages were absent from the liver capsule (Fig. 2 I, left). At the liver surface, we detected only GFP+ cells in CX3CR1gfp/+ mice (Fig. 2 I, right).
Video 2.
Video reconstruction of Tomato-expressing macrophages driven by Lyve1 promoter in pia/dura mater of brain shown in Fig. 2 E. Tomato− BM cells of Lyve1cre:R26LSL-tdTomato mice were transplanted into irradiated WT mice. The mice were used to visualize meningeal perivascular macrophages. Tomato-expressing macrophages were associated with blood vasculature underneath the skull.
Video 3.
Video reconstruction of Tomato-expressing macrophages driven by LYVE1 promoter in the peritoneal parietal membrane shown in Fig. 2 G. Tomato− BM cells of Lyve1cre:R26LSL-tdTomato mice were transplanted into irradiated WT mice. 10 wk later, the mice were injected with Alexa Fluor 488–conjugated lectin, and the parietal peritoneal membrane was visualized. Tomato-expressing macrophages were mainly located in the collagen-enriched serosal membrane of the parietal peritoneum.
LYVE1hi membrane-associated macrophages constitutively display an alternatively activated macrophage (AAM) gene expression profile
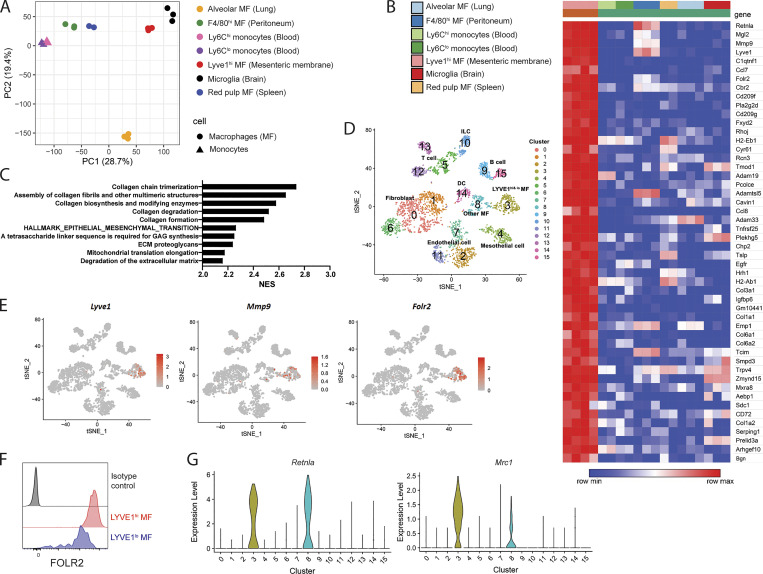
After bulk RNA-seq, principal component analysis (PCA) showed that LYVE1hi macrophages cluster together as replicates. They clustered distinctly from other tissue-resident macrophages including resident macrophages from peritoneal lavage, lung, spleen, and brain or two blood monocyte subsets (Fig. 3 A). Among the most highly up-regulated genes (≥16-fold compared with the other macrophage populations) in the mesenteric LYVE1hi macrophages were Mgl2 (CD301b), Mmp9, Lyve1, C1qtnf1, Folr2, Cbr2, and AAM-related genes such as Retnla (RELMα) and Mrc1 (CD206; Fig. 3 B). In addition to the up-regulation of canonical AAM-related genes, pathway analysis implemented by fast gene set enrichment analysis (fast GSEA) showed that pathways related to extracellular matrix organization such as collagen formation and degradation were enriched in mesenteric LYVE1hi macrophages (Fig. 3 C).
Figure 3.
LYVE1hi macrophages have their own gene expression patterns. (A) PCA of tissue-resident macrophages (alveolar macrophages, F4/80+ peritoneal macrophages, microglia, splenic red pulp macrophages, and LYVE1hi membrane-associated macrophages) and blood monocytes (Ly6Chi and Ly6Clo monocytes) obtained from RNA-seq dataset. PC, principal component. (B) Heatmap analysis of top 50 up-regulated genes of 12,000 genes expressed in LYVE1hi membrane-associated macrophages. Heatmap depicts mean expression intensity of mRNA transcripts for genes differentially expressed between LYVE1hi macrophages and other macrophages, including monocytes. (C) Pathway analysis of genes differentially expressed in LYVE1hi membrane-associated macrophages implemented by fast GSEA, showing top 10 enriched pathways from Reactome database and Molecular Signatures Database. NES, normalized enrichment score. (D) t-SNE plot displaying reanalyzed scRNA-seq of whole mesentery cells (accession no. GSE102665). (E) Expression of Lyve1, MMP9, and Folr2 on the t-SNE plot of scRNA-seq described in D. (F) Flow cytometric analysis showing FOLR2 expression of LYVE1hi and LYVE1lo mesenteric macrophages. Data are representative of three mice. (G) Violin plot of Retnla and Mrc1 expression obtained from scRNA-seq described in D.
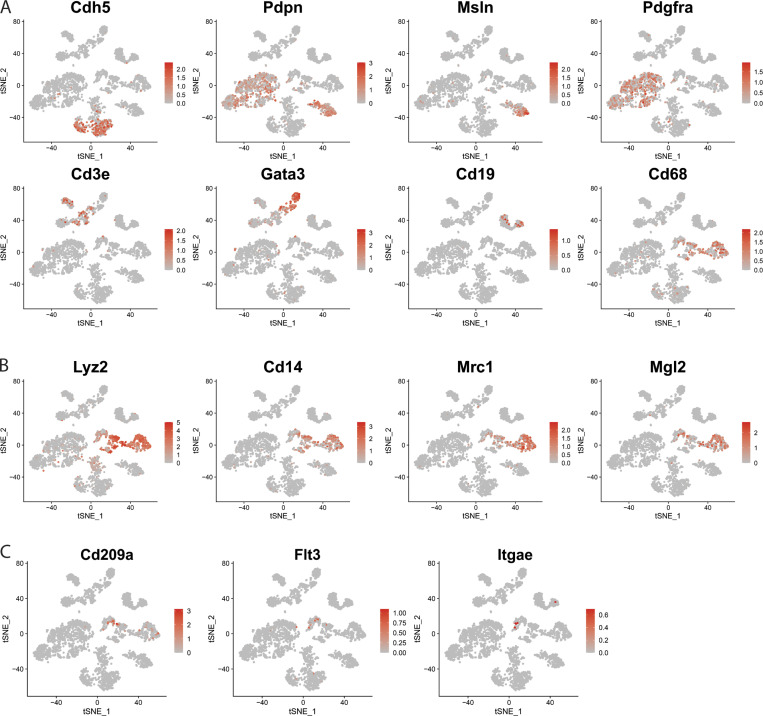
To further investigate and validate these findings, we reanalyzed an scRNA-seq dataset that examined whole mesenteric cells (GEO accession no. GSE102665; Koga et al., 2018), producing 16 clusters based on cell-specific gene expression representing a range of cell types (Figs. 3 D and S4 A). We defined clusters 3 and 8 of this t-distributed stochastic neighbor embedding (t-SNE) plot as macrophage populations due to expression of Cd68, Lyz2, Mrc1, Cd14, and Mgl2 (Fig. S4, A and B) and cluster 14 as a dendritic cell population due to coenrichment in genes such as Cd209, Flt3, and Itgae (CD103; Fig. S4 C). The Lyve1, Mmp9, and Folr2 mRNA transcripts that were up-regulated in bulk RNA-seq of LYVE1hi membrane-associated macrophages were selectively detected in cluster 3 (Fig. 3 E). Overall, cluster 3 corresponded to both LYVE1hi and LYVE1lo mesenteric macrophage populations, whereas other macrophages corresponded to cluster 8 (Fig. 3, D and E; and Fig. S4 B). Folate receptor 2 expression, encoded by Folr2 mRNA, was observed in LYVE1hi and LYVE1lo macrophages through flow cytometric analysis (Fig. 3 F), confirming that LYVE1+ macrophages, including both LYVE1hi and LYVE1lo subsets, belong to cluster 3 (Fig. 3, D–F). Other AAM-associated genes, Mrc1 and Retnla, characterized LYVE1-enriched macrophages in both datasets (Fig. 3 G), with many AAM genes including Retnla and Mrc1 also highly enriched in LYVE1lo/− mesenteric macrophages. Thus, mesenteric barrier membrane macrophages are oriented toward support of extracellular matrix remodeling, likely needed to build and maintain the barrier in which the cells reside.
Figure S4.
t-SNE plot identifying different cell populations in scRNA-seq of whole mesentery cells (accession no. GSE102665). (A) Signature genes that represent different cell populations of whole mesenteric cells. (B) t-SNE-plot for macrophage populations. (C) t-SNE-plot for dendritic cell populations.
LYVE1hi membrane-associated macrophages originate from embryonic precursors
Many peripheral tissue macrophages develop from embryonic precursors (EMPs or fetal liver monocytes; Gomez Perdiguero et al., 2015; Hoeffel et al., 2015; Yona et al., 2013). However, liver capsular macrophages are derived from circulating monocytes, not embryonic precursors (Sierro et al., 2017). Thus, we wondered whether LYVE1hi mesenteric membrane–associated macrophages were derived from embryonic precursors or were replenished from circulating monocytes. We first examined membrane-associated macrophages in newborn CX3CR1gfp/+ mice (postnatal day 0 [P0]), as most tissue-resident macrophages are established from CX3CR1-expressing precursor cells during embryogenesis (Yona et al., 2013). LYVE1hi macrophages were already established in the mesenteric membrane of these newborn mice (Fig. 4 A). Unlike LYVE1hi macrophages from adult CX3CR1gfp/+ mice, which harbored low expression of GFP reporter (Fig. 1, D and E), LYVE1hi macrophages in the mesentery of newborn mice highly expressed GFP in >90% of all mesenteric macrophages (Fig. 4 B). LYVE1lo/− CX3CR1gfp+ macrophages (Fig. 4 A, arrows) accounted for <10% of total macrophages in newborn mice (Fig. 4 C). In newborn mice, both LYVE1hi and LYVE1lo/− macrophages were more amoeboid in morphology and rarely expressed MHC II, although MHC II was highly expressed in cells in the MLN. By P6, these macrophages elongated, and MHC II expression in some LYVE1lo/− macrophages began to emerge within 2 wk after birth (Fig. 4 D). We conclude that mesenteric membrane–associated macrophages are established during embryogenesis but undergo postnatal adaptations in phenotype and morphology.
Figure 4.
Membrane-associated macrophages originate from embryonic precursors. (A) Whole-mount images of mesentery in newborn (P0) CX3CR1gfp/+ pups stained with LYVE1. Right-most image is the enlargement of the boxed area in the adjacent image. Arrows indicate LYVE1lo/− CX3CR1-GFP+ macrophages. Images are representative of two independent experiments. Scale bar, 100 µm; 30 µm (higher-magnification image). (B) CX3CR1-GFP expression within the Lyve1hi macrophage pool in the mesenteric membrane of newborn mice. GFP expression is quantified within total LYVE1hi macrophage population. (C) The frequency of LYVE1hi macrophages versus LYVE1lo/− macrophages in the mesenteric membrane of newborn mice. In B and C, data are representative of two independent experiments (n = 4; mean ± SEM). Membrane-associated macrophages were quantified in one to three different regions of mesenteric membrane per mouse. Unpaired Student’s t test: ****, P < 0.0001. (D) Whole-mount images of MLNs and mesenteric membranes of P1, P6, and P14 neonatal mice stained with LYVE1 and MHCII. White, LYVE1; green, MHCII; blue, DAPI. The yellow line in the P1 panel indicates the border of mesenteric vessels and mesenteric membrane. Images are representative of at least two independent experiments per time point. Scale bar, 50 µm. (E) Representative whole-mount images of adult CX3CR1CreERT2:R26LSL-Tomato mice in which tamoxifen was injected on P1 (white, LYVE1; red, Tomato reporter). Scale bar, 50 µm. (F) Tomato expression in microglia, LYVE1hi mesenteric membrane–associated macrophages and blood monocyte subsets. LYVE1hi membrane-associated macrophages were quantified in two different regions of membrane per mouse from confocal microscopy images. Microglia and blood monocytes were quantified via flow cytometric analysis. In E and F, data are pooled from two independent experiments (n = 6; mean ± SEM). Statistical analysis was performed by one-way ANOVA and Tukey’s multiple-comparison test: ****, P < 0.0001.
To determine if embryonic derived LYVE1hi macrophages are maintained through self-renewal, we performed tamoxifen-pulse labeling to track the LYVE1hi macrophages after birth. Accordingly, tamoxifen was injected i.p. into P1 pups of CX3CR1CreERT2:R26LSL-tdTomato mice. 8–10 wk later, Tomato reporter was visualized in LYVE1hi macrophages (Fig. 4 E). Tomato reporter remained high in LYVE1hi macrophages (80.08 ± 1.78%) and microglia (96.28 ± 1.39%), while it was negative at these time points in blood Ly6Chi monocytes (0.09 ± 0.02%) and Ly6Clo monocytes (2.19 ± 0.32%), suggesting that LYVE1hi macrophages are likely long-lived or self-renewed from P1-labeled LYVE1hi macrophages (Fig. 4 F). Collectively, these data suggest that most LYVE1hi macrophages originate from embryonic precursors and are then maintained locally for long periods.
Membrane-associated macrophages are controlled by CSF1 produced in Wt1-expressing stromal cells
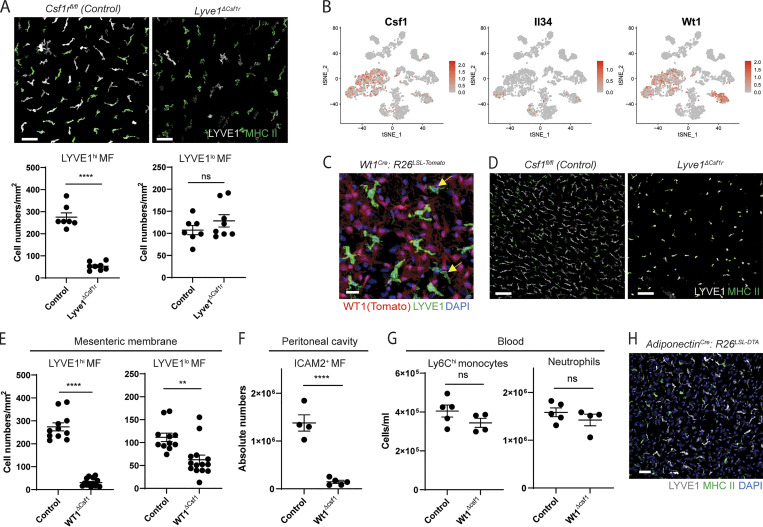
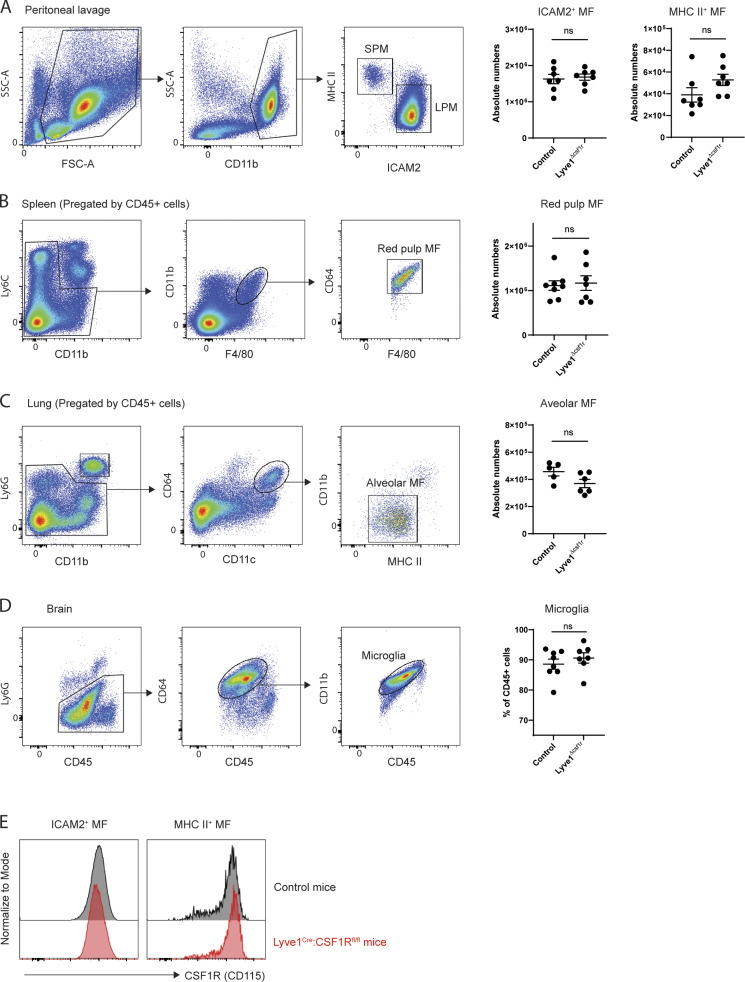
It is well established that CSF1 receptor signaling is crucial for the generation, differentiation, and survival of most tissue-resident macrophages (Cecchini et al., 1994; Dai et al., 2002; Ivanov et al., 2019; Williams et al., 2020). Thus, we investigated whether LYVE1hi macrophages required CSF1 receptor signaling. Indeed, in Lyve1Cre:Csf1rfl/fl mice (Lyve1ΔCsf1r mice), LYVE1hi macrophages were reduced by ∼80%. The number of LYVE1lo/− MHC IIhi macrophages was comparable to that of littermate controls (Fig. 5 A). Additionally, the number of other tissue-resident macrophages known to lack LYVE1 expression residing in peritoneum, spleen, lung, and brain of Lyve1ΔCsf1r mice was not significantly changed compared with standard WT mice (Fig. S5, A–D), and Csf1 receptor is normally expressed in peritoneal fluid macrophages of Lyve1ΔCsf1r mice (Fig. S5 E). These results imply that Csf1 receptor deficiency in the stage of EMP or HSC has no impact on the pool of tissue-resident macrophages, and Lyve1hi membrane-associated macrophages are selectively depleted by the lack of Csf1 receptor–mediated signaling in Lyve1ΔCsf1r mice. This outcome may arise because Csf1R would be necessary for persistence of macrophages that might arise from Cre-recombinase activated HSC/EMP progenitors, such that there would evolve a natural selection bias to greatly favor the seeding of tissue macrophages arising from Cre-recombinase nonactivated HSC/EMP progenitors (30–50% of populations). Then, expression of LYVE1 by tissue-resident macrophages would trigger selective loss of Csf1R, such that only LYVE1+ macrophages would be substantially impacted.
Figure 5.
Membrane-associated macrophages are controlled by CSF1 produced by stromal cells of serous membranes. (A) Whole-mount images of the two distinct membrane-associated macrophages and their quantification in Lyve1Cre:Csf1rfl/fl mice and littermate Csf1rfl/fl control mice. Scale bar, 50 µm. Data are representative of three independent experiments (n = 3 per genotype; mean ± SEM). Macrophages were quantified in multiple regions of mesenteric membrane per mouse. Unpaired Student’s t test: ****, P < 0.0001. (B) Expression pattern of Csf1, Il34, and Wt1 depicted on the t-SNE plot derived from scRNA-seq of whole mesenteric cells shown in Fig. 3 D. (C) Whole-mount images stained for LYVE1 in mesenteric membranes from WT1cre:R26LSL-tdTomato mice. Representative images of three independent experiments. Arrows indicate LYVE1lo/− macrophages (red, Wt1 Tomato reporter; green, LYVE1; blue, DAPI). Scale bar, 20 µm. (D) Whole-mount images of mesenteric membranes from Wt1Cre:Csf1fl/fl mice and littermate Csf1fl/fl control mice. Representative images of two independent experiments. Scale bar, 100 µm. (E) Quantification of LYVE1hi and LYVE1lo membrane-associated macrophages obtained from whole-mount images of Wt1Cre:Csf1fl/fl mice and littermate Csf1fl/fl control mice. Data are pooled from two independent experiments (Csf1fl/fl mice, n = 4; Wt1Cre:Csf1fl/fl mice, n = 6; mean ± SEM). Macrophages were quantified from multiple regions of mesenteric membrane per mouse. Unpaired Student’s t test: **, P < 0.01; ****, P < 0.0001. (F) Quantification of ICAM2+ macrophages in peritoneal cavity. Data are representative of at least three independent experiments (Csf1fl/fl mice, n = 4; Wt1Cre:Csf1fl/fl mice, n = 5; mean ± SEM). Unpaired Student’s t test: ****, P < 0.0001. (G) Quantification of Ly6Chi monocytes and neutrophils in blood. Data are representative of three independent experiments (Csf1fl/fl mice, n = 5; Wt1Cre:Csf1fl/fl mice, n = 4; mean ± SEM). Unpaired Student’s t test. (H) Confocal image from the mesenteric membrane of Adiponectincre: R26LSL-DTA mice and controls. Images are representative of two independent experiments. Scale bar, 50 µm.
Figure S5.
Quantification of tissue-resident macrophages of Lyve1Cre:Csf1rfl/fl and control mice. (A) Gating strategy of two peritoneal macrophage subsets and their quantification in Lyve1Cre:Csf1rfl/fl mice and control mice. FSC, forward scatter; SSC, side scatter. (B) Gating strategy of splenic red pulp macrophages and their quantification in Lyve1Cre:Csf1rfl/fl mice and control mice. (C) Gating strategy of alveolar macrophages and their quantification of in Lyve1Cre:Csf1rfl/fl mice and control mice. (D) Gating strategy and percentage of microglia in CD45+ brain leukocytes of Lyve1Cre:Csf1rfl/fl mice and control mice. (E) Representative histogram of CSF1R expression in ICAM2+ and MHC II+ peritoneal macrophage subsets analyzed in A. In A–E, data were pooled from two independent experiments (n = 5–8 mice). Statistical analysis was performed by unpaired Student’s t test.
CSF1 and IL34 are the ligands of CSF1 receptor (Wang et al., 2012), so we examined mRNA for CSF1 and IL34 in the scRNA-seq data from the whole mesentery. Csf1 in particular is highly up-regulated in Wilms tumor 1 homologue (Wt1)–expressing mesenteric fibroblasts and mesothelial cells (Fig. 5 B), and both LYVE1lo/− macrophages (yellow arrows) and LYVE1hi macrophages were located in proximity to Tomato-labeled stromal cells within the mesothelial membranes from Wt1cre:R26LSL-tdTomato mice (Fig. 5 C). To test whether membrane-associated macrophages were controlled by Csf1 produced by local stromal cells, we generated Wt1cre:Csf1fl/fl mice (Wt1ΔCsf1 mice) that would delete Csf1 in WT1-expressing fibroblasts and mesothelial cells. Both LYVE1hi macrophages in particular and, to a lesser extent, LYVE1lo/− macrophages were significantly reduced in the absence of Csf1 produced in stromal cells, with residual cells appearing rounded in morphology (Fig. 5, D and E). Consistent with a previous finding (Bellomo et al., 2020), F4/80+ ICAM2+ peritoneal macrophages were highly reduced in Wt1ΔCsf1 mice (Fig. 5 F). However, blood Ly6Chi monocytes and neutrophils of Wt1ΔCsf1 mice were comparable in number to their littermate controls (Fig. 5 G).
Some visceral white adipose tissues (WATs) are generated from Wt1-expressing cells (Chau et al., 2014). To test whether membrane-associated macrophages are controlled by WAT-derived CSF1, we studied Adiponectincre:R26LSL-DTA mice lacking adipocytes. LYVE1hi and LYVE1lo/− macrophages in mesenteric membranes were intact in these mice, implying that membrane-associated macrophages did not depend on adipocytes for maintenance (Fig. 5 H). Altogether, we conclude that CSF1 locally produced by fibroblasts and/or mesothelial cells within serous membranes controls the development and maintenance of macrophages within the membranes themselves and in the adjacent fluid cavities.
Gene expression patterns in LYVE1hi mesenteric membrane–associated macrophages resemble those expressed by omental macrophages in ovarian tumors
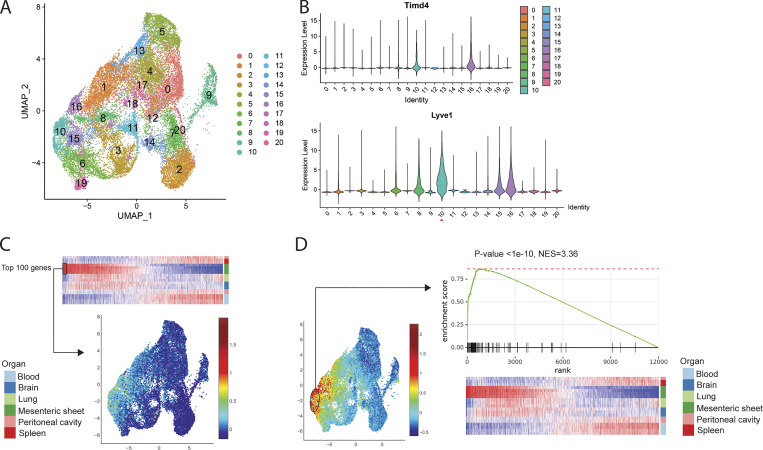
It was reported that tissue-resident macrophages play important roles in tumor progression by expressing profibrotic factors (Zhu et al., 2017). More recently, Etzerodt et al. (2020) showed that TIM4+ LYVE1hi omental macrophages promote ovarian tumor progression through scRNA-seq and a cell ablation model. We suspected that the LYVE1hi macrophages we identified lining the mesenteric membranes of the peritoneal cavity were related to the tumor-associated macrophages in the omentum. To test this possibility, we compared the RNA-seq data from LYVE1hi membrane-associated macrophages to the scRNA-seq data (ArrayExpress accession no. E-MTAB-8593) from omental macrophages performed in mice bearing experimental ovarian tumors. Tumor-associated omental macrophages were reanalyzed and divided into 21 different clusters based on their gene expression patterns (Fig. 6 A). Timd4 was up-regulated in clusters 10 and 16 and Lyve1 in clusters 6, 8, 10, 15, and 16, with highest expression in cluster 10 among these five clusters. This analysis placed the tumor-promoting TIM4+ LYVE1+ omental macrophages in cluster 10 of our reanalysis (Fig. 6 B). Next, the top 100 enriched genes expressed by LYVE1hi membrane-associated macrophages were compared with genes expressed in the scRNA-seq that we reanalyzed. As shown in our heatmap analysis, we confirmed that the gene expression pattern of LYVE1hi mesenteric membrane macrophages most closely matched genes expressed in cluster 10 (Fig. 6 C). Conversely, genes expressed in cluster 10 were highly matched to the LYVE1hi macrophages in GSEA (Fig. 6 D). We conclude that macrophages constitutively present in the mesenteric membrane express a pattern of genes previously associated with omental macrophages that drive tumor progression.
Figure 6.
Comparison of gene expressions between RNA-seq of Lyve1hi membrane-associated macrophages and scRNA-seq of Lyve1hi omental macrophages in ovarian tumor progression. (A) Reanalyzed uniform manifold approximation and projection (UMAP) plot of scRNA-seq data (ArrayExpress accession no. E-MTAB-8593) of F4/80+ CD64+ omental macrophages isolated 10 wk after ID8 ovarian tumor cell injection. (B) Violin plot of Timd4 and Lyve1 expression from the scRNA-seq dataset in A. (C) UMAP plot of scRNA-seq showing top 100 genes up-regulated in bulk RNA-seq datasets of LYVE1hi membrane-associated macrophages. (D) GSEA of RNA-seq data from Lyve1hi membrane-associated macrophages showing select genes enriched in Cluster 10 of scRNA-seq. NES, normalized enrichment score.
LYVE1hi mesothelial macrophages enhance omentum-independent ovarian tumor progression
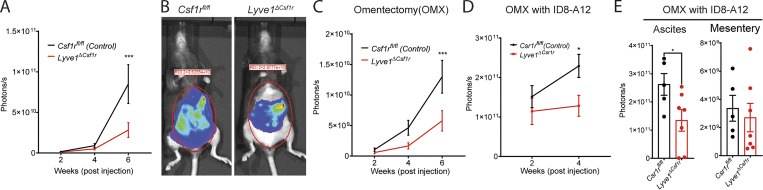
Given the overlap between omental macrophage gene expression and that of other macrophages in peritoneal membranes lining the cavity where the tumor was implanted, we wondered if the previous correlation in reduced tumor progression between removal of the omentum and the lack of LYVE1hi macrophages (Etzerodt et al., 2020) meant de facto that only omental macrophages inside the peritoneal cavity were implicated in tumor progression. In particular, we wondered if membrane-associated LYVE1hi macrophages beyond those in the omentum were relevant drivers of tumor progression. To examine these questions, luciferase/GFP-labeled murine epithelial ovarian tumor cells (ID8) were injected i.p. into LYVE1hi macrophage–ablated mice (Lyve1ΔCsf1r) or their littermate controls (Csf1rfl/fl) to model intraperitoneal metastatic HGSOC (Leinster et al., 2012; Lengyel et al., 2014). Tumor burden was monitored biweekly through noninvasive bioluminescence imaging. In the first 4 wk, peritoneal tumor burden was comparable between Lyve1ΔCsf1r mice and littermate controls (Fig. 7 A). During this period, the tumor was particularly localized to the omental fat region of the peritoneal cavity, spreading into the greater cavity space thereafter (Etzerodt et al., 2020). By 6 wk after implantation, tumor progression was significantly delayed if LYVE1hi macrophages were genetically ablated, compared with littermate controls (Fig. 7, A and B). These findings were similar to those previously reported (Etzerodt et al., 2020).
Figure 7.
Deficiency in LYVE1hi macrophages delays intraperitoneal expansion of ovarian cancer in an omentum-independent manner. (A) Quantification of bioluminescence signals at different time points after tumor implantation (Csf1rfl/fl mice, n = 12; Lyve1ΔCsf1r mice, n = 13; mean ± SEM). (B) Bioluminescence images of tumor-bearing mice at 6 wk after inoculation. (C) Quantification of bioluminescence signal at different time points after inoculation in omentectomized (OMX) mice (Csf1rfl/fl mice, n = 11; Lyve1ΔCsf1r mice, n = 11; mean ± SEM). (D) Quantification of bioluminescence signal at different time points after inoculation of the omentum-primed ID8-A12 cells in OMX mice (Csf1rfl/fl mice, n = 10; Lyve1ΔCsf1r mice, n = 12; mean ± SEM). (E) Quantification of bioluminescence signal of ascites and mesenteries 4 wk after inoculation of the omentum-primed ID8-A12 cells in OMX mice (mean ± SEM). Unpaired Student’s t test: *, P < 0.05; ***, P < 0.001. Statistical analysis was performed using one-way ANOVA (A, C, and D) and Student’s t test (E).
The key question was whether the role of macrophages in driving tumor progression was restricted and required the omentum. To investigate this issue, we performed surgical omentectomy and compared tumor progression between Lyve1ΔCsf1r mice and littermate controls, designing two sets of experiments to rigorously test whether LYVE1hi macrophages promoted ovarian tumors independently of the omentum. First, we used the ID8-Luc-GFP cell line to model intraperitoneal metastasis after omentectomy. Both Lyve1cre:Csf1rfl/fl mice and their littermates developed significantly less intraperitoneal metastasis of ovarian tumors than mice without omentectomy (Fig. 7, A and C), confirming the critical role of the omentum in ovarian tumor progression. However, in the absence of omentum, Lyve1ΔCsf1r mice still developed less intraperitoneal metastasis than littermate controls (Fig. 7 C), demonstrating an independent role for the LYVE1hi macrophages outside of the omentum. Second, as previously pioneered (Etzerodt et al., 2020), we developed omentum-primed ID8 cells by conditioning ID8 cells in WT mice for 12 wk. We termed recovered cells ID8-A12. ID8-A12 cells were inoculated into omentectomized Lyve1ΔCsf1r mice or littermate controls. Ovarian tumors were rapidly expanded in both genotypes within 4 wk after inoculation. However, ID8-A12 cells expanded significantly more slowly in Lyve1ΔCsf1r mice than in control mice (Fig. 7 D). The tumor within ascites rather than the mesentery accounted for the differences in tumor burden between the two genotypes (Fig. 7 E). Taken together, these data underscore that LYVE1hi macrophages promote intraperitoneal expansion of ovarian tumors independent of the omentum.
Discussion
The complexity of resting macrophage heterogeneity and specialization across and within given organs continues to evolve and grow. The present focus on the peritoneal cavity in this body of work highlights the intricate network of resident macrophages that occupy the body cavity. We and others have recognized the existence of two resident peritoneal macrophage subsets in the serosal fluid of the peritoneum, GATA6-dependent LPMs (Gautier et al., 2014; Okabe and Medzhitov, 2014; Rosas et al., 2014) and monocyte-derived, IRF4-dependent small peritoneal macrophages (Kim et al., 2016). Evidence of the existence of more than these two resident macrophages in the body cavity is found in the literature, particularly in the studies of Uderhardt et al. (2019) identifying fixed macrophages in the peritoneal lining membranes. However, the basic phenotype of these fixed macrophages has not been reported or compared with the fluid-borne macrophages, and thus even very recent reviews (Liu et al., 2021) and papers in the field focused on peritoneal macrophages (Louwe et al., 2021) have not placed these fixed macrophages into context. Instead, the omental fat-associated macrophages are typically the only macrophages routinely considered beyond the serous fluid macrophages within the peritoneal compartment (Liu et al., 2021). Here, we show that macrophages resembling omental macrophages are situated as fixed macrophages in the mesothelial linings of the peritoneum, including the vascularized and avascular parts of the mesentery, the parietal peritoneal membrane, and the surface of organs such as the pancreas.
Strikingly, phenotypes of the fixed macrophages observed were characterized by high expression of LYVE1 and low expression of MHC II in the first population and lower expression of LYVE1 but higher expression of MHC II in the second population. Indeed, it is striking that almost all organ surfaces including the meninges of the central nervous system are characterized by the presence of LYVE1+ macrophages, even when the major organ parenchymal macrophage is devoid of this marker. We note that the liver stands out as the exception to the pattern, having a phenotypically distinct macrophage type at the border surface (Sierro et al., 2017), which our findings confirmed. The LYVE1hi and LYVE1lo/− macrophages within the serous membranes are likely counterparts to the interstitial macrophages previously described in the lungs (Chakarov et al., 2019; Gibbings et al., 2017), artery wall (Lim et al., 2018), and mammary gland (Wang et al., 2020). Indeed, although it is clear that organs have unique resident macrophages, it is also emerging that some macrophage phenotypes are found more broadly across all organs. We argue that the cells we have characterized here should be called interstitial peritoneal macrophages to help distinguish them from serous fluid-borne macrophages of the peritoneal cavity and to simultaneously underscore their potential common features with the interstitial macrophages of other organs (Chakarov et al., 2019; Gibbings et al., 2017). As noted in these past studies and recapitulated in our present study in the peritoneal cavity, their phenotype is oriented toward an alternatively activated, or M2, state and to the maintenance and remodeling of extracellular matrix, perhaps especially relevant in light of the collagen- and matrix-rich environment these cells live within and their possible role in maintaining the exterior barrier of the associated membranes and organ surfaces. We show that they appear to be in a state of interdependence with neighboring stromal cells that serve as the crucial source of CSF1 that these macrophages require for persistence.
With the clear evidence that the phenotype of interstitial macrophage subsets lining the mesenteric and peritoneal membranes is distinct from the phenotype and life cycle of the fluid-borne macrophage subsets of the peritoneum, it is reasonable to regard the peritoneal cavity space as being influenced by four different types of resident macrophages. One begins to wonder, when this point is taken into account, how the invasion into injured organs by fluid-borne peritoneal macrophages (Wang and Kubes, 2016), and their influence on surgical adhesions (Zindel et al., 2021), is impacted by these interstitial macrophages. For example, cell deletion schemes that have targeted LPMs of the serous fluid are broad enough in their mechanism of action to have also deleted these interstitial macrophages in relevant anatomic spaces, but this point was not considered, as the presence of these macrophages was not clear at the time of the studies. It may also be the case that deletions of specific but still rather broadly expressed genes such as Rxra affected outcomes such as peritoneal cancer progression (Casanova-Acebes et al., 2020) due to deletions of the LYVE1+ serosal macrophages, as their status was not checked. On the other hand, it very well may be that serous fluid peritoneal macrophages and the LYVE1+ interstitial macrophages we describe here each have requisite roles in peritoneal tumor progression (Casanova-Acebes et al., 2020; Xia et al., 2020).
With respect to cancer progression, a recent study published compelling evidence that LYVE1hi macrophages were critical mediators of ovarian tumor cell expansion in the peritoneal cavity (Etzerodt et al., 2020). We show here that the macrophages we describe are a similar population to those studied by Etzerodt et al. (2020). Because the same authors reproduced that the presence of the omental fat tissue drove tumor progression and because they were able to identify interstitial peritoneal macrophages with the LYVE1hi phenotype in the omentum, they concluded that omental macrophages in particular were necessary for ovarian tumor progression in the peritoneal cavity. In our study, we posited that the role of these macrophages in affecting tumor expansion might not be restricted to the omentum. To address this issue, we surgically removed the omentum in two different experimental scenarios, with one scenario involving tumors that were allowed to be conditioned by omental factors that enhance the aggressiveness of the tumor but then reimplanted into mice wherein the omentum had been surgically removed. We could not remove the lesser omentum due to its key role in maintaining viability of portions of the stomach and spleen, so we cannot eliminate a role for lesser omental macrophages. However, we underscore that the number of omental macrophages is much smaller than those lining the peritoneal compartment overall. Furthermore, although it was not directly stated in the previous publication, it is highly unlikely that Etzerodt et al. (2020) removed the lesser omentum, due to its importance in physiology. Finally, patients with ovarian cancer metastasis to the peritoneum undergo resection of the omentum as routine debulking; thus, this study highlighting a role for LYVE1hi macrophages in tumor progression beyond the role of the greater omentum represents a clinically important finding (National Comprehensive Cancer Network, 2021). Multiple distinct tumor immune microenvironments can coexist within ovarian cancer from the same patient and likely contribute to heterogeneous responses of metastatic lesions to therapy (Jiménez-Sánchez et al., 2017). Seen in this light, our results raise the potential that mesothelial LYVE1hi macrophages could influence the growth of microscopic nonresected residual tumor and affect the response to adjuvant chemotherapy and cancer recurrence.
Our findings clearly point to a role for LYVE1hi macrophages in promoting tumor progression under conditions when omental macrophages are not relevant due to omental resection. This approach allows us to reveal the role of LYVE1hi macrophages beyond the omentum, but the design does not allow us to independently address whether the LYVE1hi macrophages of the omentum also contribute to the tumor expansion. We assume that, in presence of the omentum, they do. The question then turns to how these macrophages contribute to tumor progression. In general, M2-type macrophages are thought to drive tumor expansion, and one way they might do so is through extracellular matrix remodeling (DeNardo and Ruffell, 2019; Noy and Pollard, 2014), highly consistent with the phenotypic orientation of the LYVE1hi macrophages. However, one puzzle is that the ovarian tumor presence in peritoneal fluid rather than on the membrane is most impacted by the loss of LYVE1hi macrophages. A future direction will be to turn toward understanding how the LYVE1hi macrophages orchestrate an altered tumor response and whether they are directly involved in secreting relevant factors or act in other ways, such as conditioning the stromal cells nearby through cell–cell contact. A limitation of our study, and a common limitation affecting the previous study and many others in the field, is that while it is likely that the deletion of local peritoneal-lining LYVE1hi macrophages in the omentectomized mice accounts for the reduced tumor growth, we cannot be sure that the LYVE1hi body cavity macrophages per se are the ones at play in controlling tumor growth. It remains possible, albeit perhaps unlikely, that LYVE1hi macrophages resident in distal tissues play a key role.
LYVE1hi and LYVE1lo interstitial peritoneal macrophages express many common genes, including some signature genes, but others are not shared. In our depletion system, Lyve1ΔCsf1r mice lose LYVE1hi macrophages in the peritoneal lining mesentery and other membranes. However, LYVE1lo/− MHC II+ membrane-associated macrophages and other tissue-resident macrophages in peritoneum, spleen, lung, and brain are not deleted. In Lyve1ΔCsf1r mice, we nonetheless observed a reduction in tumor expansion, indicating that LYVE1lo/− macrophages are not functionally able to stand in for the LYVE1hi macrophages, at least when it comes to supporting tumor expansion, despite sharing location and some phenotypic similarity. It is interesting that, at birth, the LYVE1hi macrophages appear almost exclusively present, only to give way over time to sharing the space with the LYVE1lo population that also is prone to inducing MHC II. We suspect that the LYVE1lo/− macrophages are monocyte derived and can develop, if the right conditions exist, into the LYVE1hi macrophage population. We intend to investigate this question in the future.
In closing, we have defined the phenotype of fixed tissue macrophages in the membranes, especially the membranes of the mesentery, lining the peritoneal cavity. These findings in turn reveal the complexity of the resident macrophage pool that can influence the peritoneal space, with at least two macrophages in the fluid and two in the membranes with distinct phenotypes. Genetic loss of LYVE1hi macrophages through an approach that takes advantage of their dependence on CSF1 and CSF1R allows us to demonstrate that, beyond the omentum, LYVE1hi macrophages promote ovarian tumor growth in the peritoneal space. Future studies focused on the biology of resident peritoneal macrophages must take into account all of the relevant macrophages in the compartment.
Materials and methods
Mice
Mice were maintained in specific pathogen–free (SPF) barrier facilities with 12-h light–dark cycle by the Division of Comparative Medicine, Washington University School of Medicine (WUSM), or the Biological Resource Laboratory, University of Illinois at Chicago (UIC). All animal experiments and procedures were approved by the Institutional Animal Care and Use Committees at WUSM and UIC. Csf1rCreERT2 mice (FVB-Tg(Csf1r-cre/Esr1*)1Jwp/J; Qian et al., 2011), Lyz2cre mice (B6.129P2-Lyz2tm1(cre)Ifo/J; Clausen et al., 1999), R26LSL-tdTomato mice (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J; Madisen et al., 2010), CD11cYFP transgenic mice (B6.Cg-Tg(Itgax-Venus)1Mnz/J; Lindquist et al., 2004), CX3CR1gfp/+ mice (B6.129P2(Cg)-Cx3cr1tm1Litt/J; Jung et al., 2000), Prox1CreERT2 mice (Prox1tm3(cre/ERT2)Gco/J), Gata6fl/fl mice (Gata6tm2.1Sad/J; Sodhi et al., 2006), Lyve1cre mice (B6;129P2-Lyve1tm1.1(EGFP/cre)Cys/J; Pham et al., 2010), Csf1rfl/fl mice (B6.Cg-Csf1rtm1.2Jwp/J; Li et al., 2006), CD45.1 mice (B6.SJL-Ptprca Pepcb/BoyJ), and WT1cre mice (Wt1tm1(EGFP/cre)Wtp/J; Zhou et al., 2008) were purchased from The Jackson Laboratory. Csf1rCreERT2 mice and Gata6fl/fl mice were backcrossed to C57BL6 background using the Speed Congenics mouse genetics core at WUSM. CX3CR1CreERT2 mice were reconstituted from the cryopreserved sperm provided from S. Jung (Weizmann Institute of Science, Rehovot, Israel; Yona et al., 2013). Adiponectincre:R26LSL-DTA mice and Csf1fl/fl mice were kindly provided by Charles A. Harris (WUSM, St. Louis, MO) and Jean Jiang (University of Texas Health Science Center, San Antonio, TX; Harris et al., 2012), respectively.
Tamoxifen treatment
Tamoxifen diet (500 mg/kg; Envigo) was fed ad libitum to adult Csf1rCreERT2:Rosa26LSL-tdTomato, Csf1rCreERT2:Rosa26LSL-tdTomato:CX3CR1gfp/+, and Prox1CreERT2:Rosa26LSL-tdTomato mice for 3 wk. For fate mapping, 40 µg tamoxifen (Sigma-Aldrich) was injected i.p. into P1 pups of CX3CR1CreERT2:Rosa26LSL-tdTomato mice.
Cell isolation and staining for flow cytometry
Blood cells were collected by puncture of submandibular cheek vessels into 2 mM EDTA–containing tubes. RBCs were removed by lysis buffer (BD) in accordance with the manufacturer’s instructions. Blood leukocytes were then stained to detect surface Ly6G, CD11b, CD115, Siglec F, and Ly6C. Peritoneal cells were collected after injection of 5 ml HBSS containing 2 mM EDTA and 2% FBS into the peritoneal cavity. After blood and peritoneal cells were collected, mice were perfused with PBS. Mesenteric membranes were isolated from gut mesentery, including WAT. Brain, spleen, lung, and mesenteric membranes were enzymatically digested with collagenase types I and X (Sigma-Aldrich), hyaluronidase (Sigma-Aldrich), and DNase (Roche) in a gentle-shaking incubator (250 rpm, 37°C, 30 min), and then cells were filtered through 70-µm cell strainers. For analysis of microglia of the brain, cells were resuspended in 40% Percoll and subjected to density centrifugation (2,000 g, 20 min at 20°C with no break/acceleration). To process spleens, isolated cells were lysed with lysis buffer (BD). Total peritoneal cells, blood leukocytes, and tissue-resident macrophages were counted using an automated cell counter (Nexcelom). For quantification, these numbers were multiplied by the percentage of CD11b+ CD115+ Ly6C+ monocytes and CD11b+ Ly6G+ neutrophils in blood, CD45+ F4/80+ CD64+ CD11blo for red pulp macrophages in spleen, CD45+ CD11chi CD64+ CD11blo for alveolar macrophages in lung, CD64+ CD45lo CD11bhi for microglia in brain, and CD11b+ CD115+ macrophages stained with F4/80hi for LPM and F4/80lo for small peritoneal macrophages in peritoneum.
Single-cell suspensions collected from each tissue were maintained on ice for staining. Dead cells were identified by propidium iodide staining during flow cytometry. Antibodies purchased from BioLegend/Invitrogen or BD Biosciences were used as follows; CD45 (30F11), CD45.1 (A20), CD45.2 (104), CD11b (M1/70), CD115 (AFS98), CD102 (ICAM2; 3C4(MIC2/4)), MHC II (I-A/I-E; M5/114.15.2), Ly6C (HK1.4), Ly6G (1A8), CD11c (N418), CD170 (Siglec F; E50-2440), CD64 (FcRγI; X54-5/7.1), MerTK (DS5MMER), F4/80 (BM8), CD206 (Mrc1;C068C2), LYVE1 (ALY7), FOLR2 (10/FR2), and isotype controls (IgG2a κ chain, IgG1a, and IgG2b κ chain).
Cell sorting
To remove dead cells, cells stained with propidium iodide were gated out during cell sorting on a BD Aria II instrument. CD3ε, CD19, and Ly6G staining was used to exclude lymphocytes and neutrophils in some tissues. For selection of brain microglia by sorting, cells were stained with CD45, CD11b, CD64, F4/80, and CD206. For splenic red pulp macrophages, cells were stained with CD45, CD64, MerTK, F4/80, and CD11b. For lung alveolar macrophages, cells were stained with CD45, CD11c, Siglec F, CD64, and CD11b. For membrane-associated Lyve1hi macrophages of gut mesentery, cells were stained with CD45, F4/80, CD64, and LYVE1. For peritoneal macrophages, peritoneal cells were stained with CD11b, CD115, MHCII, and ICAM2. For Ly6Chi and Ly6Clo monocytes in blood, blood leukocytes were stained with CD11b, CD115, and Ly6C. For bulk RNA-seq, we double-sorted tissue-resident macrophages (1,000 cells/replicate), and the sorted cells were directly collected into LoBind tubes containing 5 µl of TCL lysis buffer (Qiagen) containing 1% β-mercaptoethanol, based on the Cell Preparation and Sorting Standard Operating Procedures of the Immunological Genome Project Consortium (ImmGen; https://www.immgen.org/ImmGenProtocols.html).
Ultra-low input (ULI) RNA-seq
Library preparation, quality controls and generation of data were performed by ImmGen according to the standard operating procedure for ULI RNA-seq. Data can be accessed through GEO accession no. GSE122108. Reads were aligned to the mouse genome GRCm38/mm10 primary assembly (GENCODE) and gene annotation Ver.M16 with STAR 2.5.4a. The raw read counts were generated by featureCounts (http://subread.sourceforge.net/) and normalized with DESeq2 package from Bioconductor. The top 12,000 genes ranked by average gene expression were selected for differential expression analysis using the DESeq2. Top 100 differentially expressed genes in Lyve1hi macrophage samples were used as gene signatures for the ensuing analysis. Heatmaps and PCA plots were generated using the Phantasus online service (Artyomov, 2021b).
For the analysis of the open source scRNA-seq datasets (accession nos. GSE102665 and E-MTAB-8593), the Seurat package (Butler et al., 2018) was used. Raw reads in each cell were first scaled by library size and then log-transformed. To improve downstream dimensionality reduction and clustering, any unwanted source of variation arising from the number of detected molecules was first regressed out. Highly variable genes were then identified and selected for PCA reduction of high-dimensional data. The top 10 principal components were selected for unsupervised clustering of cells. Clustering results are shown in a t-SNE plot from Single Cell Navigator online service (Artyomov, 2021a). GSEA was performed to test for the enrichment of cluster-specific gene sets at the top of the bulk RNA-seq genes ranked according to their differential expression significance (Lyve1hi macrophages versus all others). Violin and feature plots were generated using Seurat package.
Whole-mount imaging by confocal microscope
Mesenteries of adult mice were detached from the associated gut and fixed with 4% paraformaldehyde (Thermo Fisher Scientific) containing 30% sucrose (Fisher) overnight at 4°C. Gut mesenteries still attached to the intestine of neonatal mice (P0–14) were pinned on SYLGARD184 (Ellsworth; 4019862)-coated plates and fixed with 4% paraformaldehyde overnight at 4°C. After fixation, samples were stored at 4°C in PBS containing 0.01% sodium azide. For whole-mount imaging by confocal microscope, gut mesenteries were blocked in a solution containing 5% goat serum (Sigma-Aldrich; D9663) or 5% BSA (Sigma-Aldrich; A9576) overnight at 4°C. Samples were stained with rabbit-anti LYVE1 (Abcam; 14917), rat-anti MHC II (Invitrogen; M5/114.15.2), rat-Folr2 (BioLegend; 10/FR2), rat-anti CD206 (Bio-Rad; MR5D3), and rat-anti-ICAM2 (Invitrogen; 3C4(mIC2/4), Alexa Fluor 488) diluted in 1% BSA and incubated overnight 4°C with gentle agitation. Samples were washed with PBS then incubated with secondary antibodies conjugated with Alexa Fluor 488, Cy3, or Alexa Fluor 647/Cy5 (Jackson ImmunoResearch) overnight 4°C. After nuclei were further stained with bis Benzimide H 3342 (Sigma-Aldrich), macrophages in mesenteric membranes and mesenteric fat tissues were visualized on a confocal microscope (Leica SPE/inverted Leica SP8 or Zeiss 880). All images were collected using Leica LAS X software, and analysis was performed using Imaris software (Bitplane).
Estimation of serous membrane macrophage numbers in the peritoneal cavity and omentum
We estimated the approximate area of the parietal membrane (∼6 cm2), back of peritoneal wall (∼6 cm2), peritoneal membrane at the bottom of the peritoneal cavity (∼4 cm2), and surfaces of serous tissues (pancreas, ∼4 cm2; gut mesentery, ∼8-12 cm2; and diaphragm, ∼4 cm2). This area in total added up to 32–36 cm2. Our data show that Lyve1hi macrophages ranged from ∼245–380 cells/mm2 (Fig. 5 A). Therefore, we estimate that the total number of LYVE1hi macrophages may be ∼0.78–1.36 × 106 cells per mouse (∼106 cells). For estimating LYVE1hi macrophages in the omentum, we first made a single-cell suspension and counted the yield of LYVE1hi macrophages per mouse omentum, deriving 0.5–1 × 104. We then stained greater omentum tissue from three mice for LYVE1 macrophages. By immunostaining, we estimated there were 2 × 104 LYVE1hi macrophages, far less than in the peritoneal surfaces and mesenteries.
BM transplantation
BM cells were isolated from the tibia and femur of LYVE1Cre:R26LSL-tdTomato mice on a CD45.2 background. After lysis of RBC in BM cells, Tomato− cells were sorted using a FACS Aria II system (BD). Sorted BM cells (>95% purity) were injected i.v. into lethally irradiated (950 rad) CD45.1 congenic mice (1.5–3.0 × 106 cells/mouse). Recipient mice were euthanized and analyzed 8–10 wk after BM transplantation.
Whole-mount imaging using two-photon microscopy
Lyz2Cre:R26LSL-tdTomato:CD11cEYP mice and DyLight488-conjugated lectin (50 µg/mouse; Vector Laboratories; DL1174)–injected BM-transplanted mice were used for live imaging. Immediately after sacrificing BM-transplanted mice, Tomato reporter–labeled macrophages and DyLight488-labeled blood vasculatures of brain (including the skull), liver, pancreas, peritoneum, and gut mesenteries were visualized through the customized Leica SP8 two-photon microscope with a Mai Tai HP DeepSee laser (Spectra-Physics) and a 25×, 0.95-NA water-immersion objective. All images including the 3D video were collected by Leica LAS X software and generated by Imaris (Bitplane) software. Liver and gut mesentery of Lyz2Cre:R26LSL-tdTomato:CD11cEYP and CX3CR1gfp/+ mice were fixed with 4% paraformaldehyde (Affymetrix) overnight at 4°C, and tissues were stored in PBS containing 0.01% sodium azide at overnight at 4°C until ready to image.
Tumor implantation
The original ID8 cell line, derived from spontaneous in vitro malignant transformation of C57BL6 mouse ovarian surface epithelial cells (Roby et al., 2000), was modified to express GFP and firefly luciferase. These ID8 cells were cultured in RPMI 1640 with heat-inactivated FBS (10%), L-glutamine (2 mM), Hepes (25 mM), sodium pyruvate (1 mM), 2-mercaptoethanol (50 mM), penicillin-streptomycin (100 U/ml), and nonessential amino acids (Godoy et al., 2013). ID8-A12 ascites cells were generated by harvesting total ascites cells 12 wk after implanting 106 ID8 cells i.p. in C57BL6 mice. 106 ID8 or ID8-A12 cells were injected i.p. into mice of different genotypes with or without omentectomy. Biweekly bioluminescence imaging was performed to noninvasively quantify tumor burden in the peritoneal cavity.
IVIS imaging
In vivo bioluminescence imaging was performed on an IVIS 50 (PerkinElmer; Living Image 4.3.1), with exposures of 1 s to 1 min, binning 2–8, field of view 12.5 cm, f/stop 1, and open filter. D-Luciferin (150 mg/kg in PBS; Gold Biotechnology) was injected into the mice i.p. and imaged ventrally using isoflurane anesthesia (2% vaporized in O2). The total photon flux (photons/s) was measured from regions of interest using the Living Image 2.6 program.
Cells and mesenteries were imaged using the IVIS 50 with (PerkinElmer; Living Image 4.3.1) 1-s to 1-min exposure, bin 4–8, field of view 12 cm, f/stop 1, and open filter after addition of 150 µg/ml D-luciferin (Gold Biotechnology). For analysis, a grid was placed over the plate, and total photon flux (photons/s) was measured using Living Image 2.6.
Omentectomy
Operative omentectomy in mice was accomplished under general anesthesia by continuous inhalation of 2–3% isoflurane in 60% oxygen using a veterinary vaporizer, and then mice were placed on a heating pad in a supine position. Through a midline incision in the region of the stomach, the greater omentum was carefully exposed and removed via electrocautery. The midline incision was then closed with absorbable sutures in two layers. Mice were resuscitated with an i.p. injection of saline, given a local injection of analgesia, and then allowed to recover in a warmed incubator. Removal of both the entire greater and lesser omentum resulted in malperfusion of the stomach and spleen and thus was not feasible.
Online supplemental material
Figs. S1 and S2 accompany Fig. 1 and add additional information on characterization of mesenteric membrane macrophages, including comparison to the liver surface macrophages (Fig. S1) and demonstration of independence from LPM (Fig. S2). Fig. S3 accompanies Fig. 2 and illustrates the need to use a specialized BM transplant strategy to obtain faithful reporting of fluorescent tags to LYVE1hi macrophages in adult mice. Fig. S4 supplies additional information to Fig. 3 on the computational analysis of mesenteric membrane macrophages from a previously published dataset. Fig. S5 accompanies Fig. 5 and shows the lack of impact of the LYVE1cre:Csf1rfl/fl genotype on resident macrophage numbers in organs where macrophages do not express LYVE1. Video 1 illustrates contact between mesenteric membrane macrophages and peritoneal fluid macrophages. Video 2 and Video 3 show 3D rotational views of LYVE1hi macrophages in the meninges covering the brain and in the parietal peritoneal membrane to support the conclusion that they are preferentially positioned on the border of the tissue.
Acknowledgments
We thank Dr. Steffen Jung (Weizmann Institute of Science, Israel) and Charles Harris (WUSM) for providing mouse strains. We also thank all the members of the Randolph laboratory at WUSM and the Kim laboratory at UIC for helpful discussion or reading the manuscript. We thank Christophe Benoist and colleagues at ImmGen for collecting samples and generating data from ULI-RNAseq. We also thank WUSM and UIC Flow Cytometry Core. We are grateful for technical support from Julie Prior and Kathleen Duncan from the Molecular Imaging Center at WUSM.
This study was supported by National Institutes of Health grants R37 AI049653 (to G.J. Randolph); R01DK119147, DP1DK126190, and R01DK126753 (to K-W. Kim); R01AG045040 (to J.X. Jiang); R01CA188900 (to B.H. Segal); R00HL138163 (to J.W. Williams); T32DK077653 (to E.C. Erlich and E.J. Onufer); P50CA094056 (to WUSM Molecular Imaging Center); P30AR0737752 (to WUSM Rheumatic Disease Research Center); and P30CA091842 (to WUSM Siteman Cancer Center Small Animal Cancer Imaging shared resource); and Welch Foundation grant AQ-1507 (to J.X. Jiang). R.S. Czepielewski received support from the Lawrence C. Pakula, MD, IBD Research Fellowship (FA-2020-01-IBD-1).
Author contributions: Conceptualization: K-W. Kim, N. Zhang, G.J. Randolph; Investigation: K-W. Kim, N. Zhang, S.H. Kim, E.C. Erlich, E.J. Onufer, J. Kim, J. Ding, B.T. Saunders, J.R. Dominguez, R.S. Czepielewski, B.A. Helmink, J.W. Williams; Resources: G.J. Randolph, K-W. Kim, J.X. Jiang, B.H. Segal; Formal analysis and visualization: N. Zhang, K-W. Kim, S.H. Kim, J. Kim, J. Ding, A. Gainullina, B.T. Saunders, B.H. Zinselmeyer; Writing: K-W. Kim, N. Zhang, S.H. Kim, G.J. Randolph; Supervision: K-W. Kim, G.J. Randolph, N. Zhang. All authors edited the manuscript.
References
- Artyomov, M. 2021a.. Single Cell Navigator. https://artyomovlab.wustl.edu/scn/ (accessed October 26, 2021)
- Artyomov, M. 2021b.. Phantasus. https://artyomovlab.wustl.edu/phantasus/ (accessed October 26, 2021)
- Bain, C.C., Bravo-Blas A., Scott C.L., Perdiguero E.G., Geissmann F., Henri S., Malissen B., Osborne L.C., Artis D., and Mowat A.M.. 2014. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 15:929–937. 10.1038/ni.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellomo, A., Mondor I., Spinelli L., Lagueyrie M., Stewart B.J., Brouilly N., Malissen B., Clatworthy M.R., and Bajénoff M.. 2020. Reticular Fibroblasts Expressing the Transcription Factor WT1 Define a Stromal Niche that Maintains and Replenishes Splenic Red Pulp Macrophages. Immunity. 53:127–142.e7. 10.1016/j.immuni.2020.06.008 [DOI] [PubMed] [Google Scholar]
- Bénézech, C., Luu N.T., Walker J.A., Kruglov A.A., Loo Y., Nakamura K., Zhang Y., Nayar S., Jones L.H., Flores-Langarica A., et al. 2015. Inflammation-induced formation of fat-associated lymphoid clusters. Nat. Immunol. 16:819–828. 10.1038/ni.3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechler, M.B., Kim K.W., Onufer E.J., Williams J.W., Little C.C., Dominguez C.X., Li Q., Sandoval W., Cooper J.E., Harris C.A., et al. 2019. A Stromal Niche Defined by Expression of the Transcription Factor WT1 Mediates Programming and Homeostasis of Cavity-Resident Macrophages. Immunity. 51:119–130.e5. 10.1016/j.immuni.2019.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova-Acebes, M., Menéndez-Gutiérrez M.P., Porcuna J., Álvarez-Errico D., Lavin Y., García A., Kobayashi S., Le Berichel J., Núñez V., Were F., et al. 2020. RXRs control serous macrophage neonatal expansion and identity and contribute to ovarian cancer progression. Nat. Commun. 11:1655. 10.1038/s41467-020-15371-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini, M.G., Dominguez M.G., Mocci S., Wetterwald A., Felix R., Fleisch H., Chisholm O., Hofstetter W., Pollard J.W., and Stanley E.R.. 1994. Role of colony stimulating factor-1 in the establishment and regulation of tissue macrophages during postnatal development of the mouse. Development. 120:1357–1372. 10.1242/dev.120.6.1357 [DOI] [PubMed] [Google Scholar]
- Chakarov, S., Lim H.Y., Tan L., Lim S.Y., See P., Lum J., Zhang X.M., Foo S., Nakamizo S., Duan K., et al. 2019. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science. 363:eaau0964. 10.1126/science.aau0964 [DOI] [PubMed] [Google Scholar]
- Chau, Y.Y., Bandiera R., Serrels A., Martínez-Estrada O.M., Qing W., Lee M., Slight J., Thornburn A., Berry R., McHaffie S., et al. 2014. Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat. Cell Biol. 16:367–375. 10.1038/ncb2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, B.E., Burkhardt C., Reith W., Renkawitz R., and Förster I.. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8:265–277. 10.1023/A:1008942828960 [DOI] [PubMed] [Google Scholar]
- Dai, X.M., Ryan G.R., Hapel A.J., Dominguez M.G., Russell R.G., Kapp S., Sylvestre V., and Stanley E.R.. 2002. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 99:111–120. 10.1182/blood.V99.1.111 [DOI] [PubMed] [Google Scholar]
- David, B.A., Rezende R.M., Antunes M.M., Santos M.M., Freitas Lopes M.A., Diniz A.B., Sousa Pereira R.V., Marchesi S.C., Alvarenga D.M., Nakagaki B.N., et al. 2016. Combination of Mass Cytometry and Imaging Analysis Reveals Origin, Location, and Functional Repopulation of Liver Myeloid Cells in Mice. Gastroenterology. 151:1176–1191. 10.1053/j.gastro.2016.08.024 [DOI] [PubMed] [Google Scholar]
- DeNardo, D.G., and Ruffell B.. 2019. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 19:369–382. 10.1038/s41577-019-0127-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzerodt, A., Moulin M., Doktor T.K., Delfini M., Mossadegh-Keller N., Bajenoff M., Sieweke M.H., Moestrup S.K., Auphan-Anezin N., and Lawrence T.. 2020. Tissue-resident macrophages in omentum promote metastatic spread of ovarian cancer. J. Exp. Med. 217:e20191869. 10.1084/jem.20191869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q., Yang Z., Xu S., Li X., Yang X., Jin P., Liu Y., Zhou X., Zhang T., Gong C., et al. 2019. Heterotypic CAF-tumor spheroids promote early peritoneal metastatis of ovarian cancer. J. Exp. Med. 216:688–703. 10.1084/jem.20180765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., et al. Immunological Genome Consortium . 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13:1118–1128. 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, E.L., Ivanov S., Williams J.W., Huang S.C., Marcelin G., Fairfax K., Wang P.L., Francis J.S., Leone P., Wilson D.B., et al. 2014. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. J. Exp. Med. 211:1525–1531. 10.1084/jem.20140570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosn, E.E., Cassado A.A., Govoni G.R., Fukuhara T., Yang Y., Monack D.M., Bortoluci K.R., Almeida S.R., Herzenberg L.A., and Herzenberg L.A.. 2010. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc. Natl. Acad. Sci. USA. 107:2568–2573. 10.1073/pnas.0915000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings, S.L., Thomas S.M., Atif S.M., McCubbrey A.L., Desch A.N., Danhorn T., Leach S.M., Bratton D.L., Henson P.M., Janssen W.J., and Jakubzick C.V.. 2017. Three Unique Interstitial Macrophages in the Murine Lung at Steady State. Am. J. Respir. Cell Mol. Biol. 57:66–76. 10.1165/rcmb.2016-0361OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy, H.E., Khan A.N., Vethanayagam R.R., Grimm M.J., Singel K.L., Kolomeyevskaya N., Sexton K.J., Parameswaran A., Abrams S.I., Odunsi K., and Segal B.H.. 2013. Myeloid-derived suppressor cells modulate immune responses independently of NADPH oxidase in the ovarian tumor microenvironment in mice. PLoS One. 8:e69631. 10.1371/journal.pone.0069631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero, E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., Garner H., Trouillet C., de Bruijn M.F., Geissmann F., and Rodewald H.R.. 2015. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 518:547–551. 10.1038/nature13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, S.J., Glatman Zaretsky A., Andrade-Oliveira V., Collins N., Dzutsev A., Shaik J., Morais da Fonseca D., Harrison O.J., Tamoutounour S., Byrd A.L., et al. 2017. White Adipose Tissue Is a Reservoir for Memory T Cells and Promotes Protective Memory Responses to Infection. Immunity. 47:1154–1168.e6. 10.1016/j.immuni.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S.E., MacDougall M., Horn D., Woodruff K., Zimmer S.N., Rebel V.I., Fajardo R., Feng J.Q., Gluhak-Heinrich J., Harris M.A., and Abboud Werner S.. 2012. Meox2Cre-mediated disruption of CSF-1 leads to osteopetrosis and osteocyte defects. Bone. 50:42–53. 10.1016/j.bone.2011.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel, G., Chen J., Lavin Y., Low D., Almeida F.F., See P., Beaudin A.E., Lum J., Low I., Forsberg E.C., et al. 2015. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 42:665–678. 10.1016/j.immuni.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg, C., Panir K., Dhami P., Rosser M., Mack M., Soong D., Pollard J.W., Jenkins S.J., Horne A.W., and Greaves E.. 2021. Macrophages inhibit and enhance endometriosis depending on their origin. Proc. Natl. Acad. Sci. USA. 118:e2013776118. 10.1073/pnas.2013776118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, S., Gallerand A., Gros M., Stunault M.I., Merlin J., Vaillant N., Yvan-Charvet L., and Guinamard R.R.. 2019. Mesothelial cell CSF1 sustains peritoneal macrophage proliferation. Eur. J. Immunol. 49:2012–2018. 10.1002/eji.201948164 [DOI] [PubMed] [Google Scholar]
- Jackson-Jones, L.H., Smith P., Portman J.R., Magalhaes M.S., Mylonas K.J., Vermeren M.M., Nixon M., Henderson B.E.P., Dobie R., Vermeren S., et al. 2020. Stromal Cells Covering Omental Fat-Associated Lymphoid Clusters Trigger Formation of Neutrophil Aggregates to Capture Peritoneal Contaminants. Immunity. 52:700–715.e6. 10.1016/j.immuni.2020.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Sánchez, A., Memon D., Pourpe S., Veeraraghavan H., Li Y., Vargas H.A., Gill M.B., Park K.J., Zivanovic O., Konner J., et al. 2017. Heterogeneous Tumor-Immune Microenvironments among Differentially Growing Metastases in an Ovarian Cancer Patient. Cell. 170:927–938.e20. 10.1016/j.cell.2017.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., and Littman D.R.. 2000. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114. 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.W., Williams J.W., Wang Y.T., Ivanov S., Gilfillan S., Colonna M., Virgin H.W., Gautier E.L., and Randolph G.J.. 2016. MHC II+ resident peritoneal and pleural macrophages rely on IRF4 for development from circulating monocytes. J. Exp. Med. 213:1951–1959. 10.1084/jem.20160486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.S., Kolesnikov M., Peled-Hajaj S., Scheyltjens I., Xia Y., Trzebanski S., Haimon Z., Shemer A., Lubart A., Van Hove H., et al. 2021. A Binary Cre Transgenic Approach Dissects Microglia and CNS Border-Associated Macrophages. Immunity. 54:176–190.e7. 10.1016/j.immuni.2020.11.007 [DOI] [PubMed] [Google Scholar]
- Koga, S., Hozumi K., Hirano K.I., Yazawa M., Terooatea T., Minoda A., Nagasawa T., Koyasu S., and Moro K.. 2018. Peripheral PDGFRα+gp38+ mesenchymal cells support the differentiation of fetal liver-derived ILC2. J. Exp. Med. 215:1609–1626. 10.1084/jem.20172310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda Mariano, L., Rousseau M., Varet H., Legendre R., Gentek R., Saenz Coronilla J., Bajenoff M., Gomez Perdiguero E., and Ingersoll M.A.. 2020. Functionally distinct resident macrophage subsets differentially shape responses to infection in the bladder. Sci. Adv. 6:eabc5739. 10.1126/sciadv.abc5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, L.K., Ghorbanian Y., Wang W., Wang Y., Kim Y.J., Weissman I.L., Inlay M.A., and Mikkola H.K.A.. 2016. LYVE1 Marks the Divergence of Yolk Sac Definitive Hemogenic Endothelium from the Primitive Erythroid Lineage. Cell Rep. 17:2286–2298. 10.1016/j.celrep.2016.10.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinster, D.A., Kulbe H., Everitt G., Thompson R., Perretti M., Gavins F.N., Cooper D., Gould D., Ennis D.P., Lockley M., et al. 2012. The peritoneal tumour microenvironment of high-grade serous ovarian cancer. J. Pathol. 227:136–145. 10.1002/path.4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel, E. 2010. Ovarian cancer development and metastasis. Am. J. Pathol. 177:1053–1064. 10.2353/ajpath.2010.100105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel, E., Burdette J.E., Kenny H.A., Matei D., Pilrose J., Haluska P., Nephew K.P., Hales D.B., and Stack M.S.. 2014. Epithelial ovarian cancer experimental models. Oncogene. 33:3619–3633. 10.1038/onc.2013.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Chen K., Zhu L., and Pollard J.W.. 2006. Conditional deletion of the colony stimulating factor-1 receptor (c-fms proto-oncogene) in mice. Genesis. 44:328–335. 10.1002/dvg.20219 [DOI] [PubMed] [Google Scholar]
- Lim, H.Y., Lim S.Y., Tan C.K., Thiam C.H., Goh C.C., Carbajo D., Chew S.H.S., See P., Chakarov S., Wang X.N., et al. 2018. Hyaluronan Receptor LYVE-1-Expressing Macrophages Maintain Arterial Tone through Hyaluronan-Mediated Regulation of Smooth Muscle Cell Collagen. Immunity. 49:1191. 10.1016/j.immuni.2018.12.009 [DOI] [PubMed] [Google Scholar]
- Lindquist, R.L., Shakhar G., Dudziak D., Wardemann H., Eisenreich T., Dustin M.L., and Nussenzweig M.C.. 2004. Visualizing dendritic cell networks in vivo. Nat. Immunol. 5:1243–1250. 10.1038/ni1139 [DOI] [PubMed] [Google Scholar]
- Liu, M., Silva-Sanchez A., Randall T.D., and Meza-Perez S.. 2021. Specialized immune responses in the peritoneal cavity and omentum. J. Leukoc. Biol. 109:717–729. 10.1002/JLB.5MIR0720-271RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louwe, P.A., Badiola Gomez L., Webster H., Perona-Wright G., Bain C.C., Forbes S.J., and Jenkins S.J.. 2021. Recruited macrophages that colonize the post-inflammatory peritoneal niche convert into functionally divergent resident cells. Nat. Commun. 12:1770. 10.1038/s41467-021-21778-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen, L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro, K., Yamada T., Tanabe M., Takeuchi T., Ikawa T., Kawamoto H., Furusawa J., Ohtani M., Fujii H., and Koyasu S.. 2010. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 463:540–544. 10.1038/nature08636 [DOI] [PubMed] [Google Scholar]
- National Comprehensive Cancer Network . 2021. National Comprehensive Cancer Network Guidelines 2021. https://www.nccn.org (accessed October 26, 2021)
- Noy, R., and Pollard J.W.. 2014. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 41:49–61. 10.1016/j.immuni.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe, Y., and Medzhitov R.. 2014. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 157:832–844. 10.1016/j.cell.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, T.H., Baluk P., Xu Y., Grigorova I., Bankovich A.J., Pappu R., Coughlin S.R., McDonald D.M., Schwab S.R., and Cyster J.G.. 2010. Lymphatic endothelial cell sphingosine kinase activity is required for lymphocyte egress and lymphatic patterning. J. Exp. Med. 207:17–27. 10.1084/jem.20091619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, B.Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L.R., Kaiser E.A., Snyder L.A., and Pollard J.W.. 2011. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 475:222–225. 10.1038/nature10138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Smith, T.M., Isaacsohn I., Mercer C.A., Zhou M., Van Rooijen N., Husseinzadeh N., McFarland-Mancini M.M., and Drew A.F.. 2007. Macrophages mediate inflammation-enhanced metastasis of ovarian tumors in mice. Cancer Res. 67:5708–5716. 10.1158/0008-5472.CAN-06-4375 [DOI] [PubMed] [Google Scholar]
- Roby, K.F., Taylor C.C., Sweetwood J.P., Cheng Y., Pace J.L., Tawfik O., Persons D.L., Smith P.G., and Terranova P.F.. 2000. Development of a syngeneic mouse model for events related to ovarian cancer. Carcinogenesis. 21:585–591. 10.1093/carcin/21.4.585 [DOI] [PubMed] [Google Scholar]
- Rosas, M., Davies L.C., Giles P.J., Liao C.T., Kharfan B., Stone T.C., O’Donnell V.B., Fraser D.J., Jones S.A., and Taylor P.R.. 2014. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 344:645–648. 10.1126/science.1251414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R.L., Miller K.D., and Jemal A.. 2018. Cancer statistics, 2018. CA Cancer J. Clin. 68:7–30. 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- Sierro, F., Evrard M., Rizzetto S., Melino M., Mitchell A.J., Florido M., Beattie L., Walters S.B., Tay S.S., Lu B., et al. 2017. A Liver Capsular Network of Monocyte-Derived Macrophages Restricts Hepatic Dissemination of Intraperitoneal Bacteria by Neutrophil Recruitment. Immunity. 47:374–388.e6. 10.1016/j.immuni.2017.07.018 [DOI] [PubMed] [Google Scholar]
- Singel, K.L., Emmons T.R., Khan A.N.H., Mayor P.C., Shen S., Wong J.T., Morrell K., Eng K.H., Mark J., Bankert R.B., et al. 2019. Mature neutrophils suppress T cell immunity in ovarian cancer microenvironment. JCI Insight. 4:e122311. 10.1172/jci.insight.122311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi, C.P., Li J., and Duncan S.A.. 2006. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC Dev. Biol. 6:19. 10.1186/1471-213X-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatiades, E.G., Tremblay M.E., Bohm M., Crozet L., Bisht K., Kao D., Coelho C., Fan X., Yewdell W.T., Davidson A., et al. 2016. Immune Monitoring of Trans-endothelial Transport by Kidney-Resident Macrophages. Cell. 166:991–1003. 10.1016/j.cell.2016.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkamp, M.P., Winner K.K., Davies S., Muller C., Zhang Y., Hoffman R.M., Shirinifard A., Moses M., Jiang Y., and Wilson B.S.. 2013. Ovarian tumor attachment, invasion, and vascularization reflect unique microenvironments in the peritoneum: insights from xenograft and mathematical models. Front. Oncol. 3:97. 10.3389/fonc.2013.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uderhardt, S., Martins A.J., Tsang J.S., Lämmermann T., and Germain R.N.. 2019. Resident Macrophages Cloak Tissue Microlesions to Prevent Neutrophil-Driven Inflammatory Damage. Cell. 177:541–555.e17. 10.1016/j.cell.2019.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., and Kubes P.. 2016. A Reservoir of Mature Cavity Macrophages that Can Rapidly Invade Visceral Organs to Affect Tissue Repair. Cell. 165:668–678. 10.1016/j.cell.2016.03.009 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Szretter K.J., Vermi W., Gilfillan S., Rossini C., Cella M., Barrow A.D., Diamond M.S., and Colonna M.. 2012. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 13:753–760. 10.1038/ni.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Chaffee T.S., LaRue R.S., Huggins D.N., Witschen P.M., Ibrahim A.M., Nelson A.C., Machado H.L., and Schwertfeger K.L.. 2020. Tissue-resident macrophages promote extracellular matrix homeostasis in the mammary gland stroma of nulliparous mice. eLife. 9:e57438. 10.7554/eLife.57438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, J.M., Davies L.C., Karwan M., Ileva L., Ozaki M.K., Cheng R.Y., Ridnour L.A., Annunziata C.M., Wink D.A., and McVicar D.W.. 2018. Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J. Clin. Invest. 128:3794–3805. 10.1172/JCI99169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J.W., Zaitsev K., Kim K.W., Ivanov S., Saunders B.T., Schrank P.R., Kim K., Elvington A., Kim S.H., Tucker C.G., et al. 2020. Limited proliferation capacity of aortic intima resident macrophages requires monocyte recruitment for atherosclerotic plaque progression. Nat. Immunol. 21:1194–1204. 10.1038/s41590-020-0768-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., Xu J., Tan J., Song Y., Liu L., Zhang F., Zhang Y., Li X., Chi Y., and Liu Y.. 2019. Mesenteric adipose tissue B lymphocytes promote local and hepatic inflammation in non-alcoholic fatty liver disease mice. J. Cell. Mol. Med. 23:3375–3385. 10.1111/jcmm.14232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, H., Li S., Li X., Wang W., Bian Y., Wei S., Grove S., Wang W., Vatan L., Liu J.R., et al. 2020. Autophagic adaptation to oxidative stress alters peritoneal residential macrophage survival and ovarian cancer metastasis. JCI Insight. 5:e141115. 10.1172/jci.insight.141115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona, S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., et al. 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 38:79–91. 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N., Czepielewski R.S., Jarjour N.N., Erlich E.C., Esaulova E., Saunders B.T., Grover S.P., Cleuren A.C., Broze G.J., Edelson B.T., et al. 2019a. Expression of factor V by resident macrophages boosts host defense in the peritoneal cavity. J. Exp. Med. 216:1291–1300. 10.1084/jem.20182024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Dolgalev I., Zhang T., Ran H., Levine D.A., and Neel B.G.. 2019b. Both fallopian tube and ovarian surface epithelium are cells-of-origin for high-grade serous ovarian carcinoma. Nat. Commun. 10:5367. 10.1038/s41467-019-13116-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B., Ma Q., Rajagopal S., Wu S.M., Domian I., Rivera-Feliciano J., Jiang D., von Gise A., Ikeda S., Chien K.R., and Pu W.T.. 2008. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 454:109–113. 10.1038/nature07060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Herndon J.M., Sojka D.K., Kim K.W., Knolhoff B.L., Zuo C., Cullinan D.R., Luo J., Bearden A.R., Lavine K.J., et al. 2017. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity. 47:323–338.e6. 10.1016/j.immuni.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond, E., Varol C., Farache J., Elmaliah E., Satpathy A.T., Friedlander G., Mack M., Shpigel N., Boneca I.G., Murphy K.M., et al. 2012. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 37:1076–1090. 10.1016/j.immuni.2012.08.026 [DOI] [PubMed] [Google Scholar]
- Zigmond, E., Bernshtein B., Friedlander G., Walker C.R., Yona S., Kim K.W., Brenner O., Krauthgamer R., Varol C., Müller W., and Jung S.. 2014. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 40:720–733. 10.1016/j.immuni.2014.03.012 [DOI] [PubMed] [Google Scholar]
- Zindel, J., Peiseler M., Hossain M., Deppermann C., Lee W.Y., Haenni B., Zuber B., Deniset J.F., Surewaard B.G.J., Candinas D., and Kubes P.. 2021. Primordial GATA6 macrophages function as extravascular platelets in sterile injury. Science. 371:eabe0595. 10.1126/science.abe0595 [DOI] [PubMed] [Google Scholar]