Graphical abstract
Keywords: Phlebotomus argentipes, Leishmania donovani, Salivary glands, Marker of exposure, IgG antibodies, Bangladesh
Highlights
-
•
Phlebotomus argentipes is a sole vector of Leishmania donovani in the Indian subcontinent.
-
•
40% of humans in the study area have IgG antibodies against P. argentipes saliva.
-
•
A correlation was found between IgG responses against P. argentipes saliva and rPagSP06.
-
•
rPagSP06 is a valid antigen to measure human exposure to P. argentipes.
Abstract
Phlebotomus argentipes is a predominant vector of Leishmania donovani, the protozoan parasite causing visceral leishmaniasis in the Indian subcontinent. In hosts bitten by P. argentipes, sand fly saliva elicits the production of specific anti-salivary protein antibodies. Here, we have utilised these antibodies as markers of human exposure to P. argentipes in a visceral leishmaniasis endemic area in Pabna district, Bangladesh. The use of whole salivary gland homogenate as an antigen to detect these antibodies has several limitations, therefore it is being superseded by the use of specific recombinant salivary proteins. We have identified three major P. argentipes salivary antigenic proteins recognised by sera of bitten humans, expressed them in a recombinant form (rPagSP04, rPagSP05 and rPagSP06) and tested their applicability in ELISA and immunoblot. One of them, PpSP32-like protein rPagSP06, was identified as the most promising antigen, showing highest resemblance and correlation with the IgG response to P. argentipes salivary gland homogenate. Furthermore, we have validated the applicability of rPagSP06 in a large cohort of 585 individuals and obtained a high correlation coefficient for anti-rPagSP06 and anti-P. argentipes saliva IgG responses. The anti-rPagSP06 and anti-P. argentipes salivary gland homogenate IgG responses followed a similar right-skewed distribution. This is the first report of screening human sera for anti-P. argentipes saliva antibodies using recombinant salivary protein. The rPagSP06 was proven to be a valid antigen for screening human sera for exposure to P. argentipes bites in a visceral leishmaniasis endemic area.
1. Introduction
Phlebotomus (Euphlebotomus) argentipes (Diptera: Psychodidae) is a sole vector of Leishmania donovani, the causative agent of visceral leishmaniasis (VL) in the Indian subcontinent (Maroli et al., 2013). Bangladesh belongs to the six countries which account for more than 90% of global VL cases (Alvar et al., 2012). Despite efforts to eliminate VL in the Indian subcontinent, more than 100,000 cases were reported in Bangladesh between 1994 and 2013 (Chowdhury et al., 2014). While in the Indian subcontinent VL is considered to be anthroponotic, the feeding preferences of P. argentipes are mainly zoophilic, with humans being their second choice after bovine blood (Chowdhury et al., 2016). In Bangladesh, P. argentipes is the predominant sand fly and the only species of the genus Phlebotomus occurring throughout the whole country, together with 10 Sergentomyia spp., while the presence of other two Phlebotomus spp. is markedly restricted: Phlebotomus papatasi has been reported only from Natore and Rajshahi districts and there is a single report of Phlebotomus sergenti from Tangail district (Hossain et al., 1993, Özbel et al., 2017).
The antibody response against P. argentipes saliva was first described in a hamster model (Ghosh and Mukhopadhyay, 1998) and high levels of specific anti-saliva IgG in bitten hamsters were recently demonstrated by Spitzova et al. (2020). Furthermore, the use of anti-salivary antibodies to estimate human exposure to P. argentipes bites, and consequently the risk of transmitting L. donovani, was demonstrated in two studies in India and Nepal (Clements et al., 2010, Gidwani et al., 2011). Interestingly, the average indoor density of the sand fly females correlated with the level of anti-saliva IgG antibodies of the inhabitants. This observation, together with the significant decrease in anti-P. argentipes IgG antibodies after 30 days of sand fly absence, led to the use of this anti-P. argentipes saliva response as a tool to evaluate the efficacy of insecticidal nets as a control strategy against VL (Gidwani et al., 2011).
Numerous studies have already proven the importance and benefits of using the human anti-sand fly saliva antibody response as a marker of vector exposure (Rohousova et al., 2005, Rohousova et al., 2018, Souza et al., 2010, Teixeira et al., 2010, Marzouki et al., 2015, Mondragon-Shem et al., 2015, Kammoun-Rebai et al., 2017). All of these studies used extract, lysate or homogenate of salivary glands as an antigen to detect the anti-saliva antibodies. More recently, laborious dissections of tiny salivary glands of sand flies were superseded by the expression of specific recombinant salivary proteins responsible for the antigenicity of sand fly saliva (Lestinova et al., 2017, Sumova et al., 2018).
Sand fly saliva contains a vast variety of biologically active compounds. The major salivary proteins of P. argentipes were identified by Edman degradation and included representatives of typical phlebotomine protein families such as yellow-related protein, antigen 5-related protein, PpSP32-like protein, apyrase, lufaxin-like protein, endonuclease-like protein and odorant-binding proteins (Anderson et al., 2006). The proteins responsible for the antigenic properties of P. argentipes saliva have to date been identified only by using experimentally bitten hamsters (Martín-Martín et al., 2013). Immunised hamsters reacted mainly with odorant-binding proteins, antigen 5-related proteins and apyrases. However, the antigenicity of salivary proteins was repeatedly shown to be species-specific, not only for the sand fly (Volf and Rohoušová, 2001, Rohousova et al., 2005, Thiakaki et al., 2005) but also for the host (Teixeira et al., 2010, Martín-Martín et al., 2012), therefore, proteins antigenic to the human host might differ from those antigenic to hamsters. Determining the P. argentipes human antigens in a VL endemic region in Bangladesh is the focus of the study presented here.
A number of control trials have been conducted in VL endemic parts of the Indian subcontinent including indoor residual spraying, long-lasting insecticide-impregnated bed nets, insecticide spraying of potential breeding sites and insecticide-treated wall lining or wall wash with lime (Mondal et al., 2016, Chowdhury et al., 2017, Chowdhury et al., 2019). Apart from the above-mentioned study by Gidwani et al. (2011), the outcome of the control trials was based either on a reduction in vector density or in VL incidence. The decrease in VL incidence may not be a relevant parameter to measure exposure to the vector since the prevalence of L. donovani infection in P. argentipes is usually low; Bhattarai et al. (2009) reported 0.5% Leishmania-positive sand flies in an endemic region in Nepal. A more sensitive method to evaluate the efficacy of anti-vector campaigns is to measure exposure to the bites of the vector in the affected population. It is therefore important to develop a suitable and reliable marker of exposure to P. argentipes as a tool to accurately estimate the exposure to vector bites in human populations living at risk of VL. In the present work, we identified three major antigens that were recognised by sera of humans bitten by P. argentipes, expressed them in a recombinant form and propose the PpSP32-like protein rPagSP06 as a relevant antigen to measure specific anti-P. argentipes IgG antibodies as a marker of exposure to this sand fly species.
2. Materials and methods
2.1. Ethics statement
Written informed consent was obtained from all adult volunteers who participated in the study. For inclusion of young children, written consent was obtained from their parents or guardians. Each volunteer was treated in accordance with international bio-ethical rules and laws for human sampling. Blood was sampled by medical doctors. This study was approved by the Ethical Review Committee of Mymensingh Medical College, Bangladesh, and Life Science Research Ethics and Safety, The University of Tokyo, Japan (Permission number:14-70). Ethical clearance was obtained from the Council for Scientific and Industrial Research (CSIR) Institutional Review Board (RPN008/CSIR-IRB/2019) for samples collected in Ghana. All data were anonymized.
2.2. Collection of field samples
A total of 532 serum samples were collected from 5th to 7th August 2014, in Pabna district, in Bangladesh. The endemic serum samples were obtained from people of both sexes (302 females and 230 males) and across all age groups. In total, 4% of the sampled individuals had suffered from VL in the past. Negative control serum samples were obtained from healthy donors from Japan (18 samples) and Accra, Ghana (35 samples). Serum samples were stored in aliquots at −80 °C. At the study site in Pabna, we also conducted sand fly collection (at the beginning of March and August 2014). Sand flies were collected using CDC light traps placed in and around randomly selected human dwellings overnight. The head and abdominal segments of the captured sand flies were cut off from the whole body and transferred on labelled slides in Swan solution. Captured specimens of genus Phlebotomus were morphologically identified using standard keys and descriptions based on the features of cibarium, pharynx and spermathecae in females, and genitalia, coxite and style in males (Lewis, 1982, LEWIS, 1987).
2.3. Sand fly salivary gland homogenate
A laboratory colony of P. argentipes originating from India (kindly donated by R. Molina in 2008) and P. papatasi from Turkey (established by our laboratory in 2005) were maintained in the insectary at the Department of Parasitology, Charles University, Czech Republic, under standard conditions (Lawyer et al., 2017). Antigen preparation was done as described previously (Sumova et al., 2018). Briefly, salivary glands of P. argentipes and P. papatasi were dissected from 4 to 6 days old female sand flies into 20 mM Tris buffer with 150 mM NaCl (20 glands per 20 μl of buffer) and stored at −80 °C. Before use, salivary glands were disrupted by three freeze and thaw cycles in liquid nitrogen to obtain the salivary gland homogenate (SGH).
2.4. Recombinant proteins
Three antigenic salivary proteins of P. argentipes were expressed in a mammalian expression system. The expressed proteins were the PpSP32-like protein rPagSP06 (GenBank accession number: ABA12138.1), yellow-related protein rPagSP04 (GenBank accession number: ABA12136.1) and antigen 5-related protein rPagSP05 (GenBank accession number: ABA12137.1) (Anderson et al., 2006). The gene constructs were prepared by isolating the total RNA from 1 day old P. argentipes females using a High Pure RNA Tissue Kit (Roche, Switzerland), and consequently synthetizing the cDNA with the anchored-oligo (dT)18 primers using the Transcriptor First Strand cDNA Synthesis Kit (Roche) following the manufacturer’s protocol. Transcripts were amplified by PCR and subcloned into the pTW5sec expression plasmid, a derivative of pTT5 (Durocher et al., 2002, Bláha et al., 2015). Proteins expressed using this plasmid contain additional ITG- and -GTHHHHHHHHG amino sequences at their N- and C-termini, respectively. Proteins were then transiently expressed in the human embryonic kidney 293S (HEK293S) GnTI- cell line (ATCC CRL-3022) as previously described (Bláha et al., 2015, Willen et al., 2019, Spitzova et al., 2020). Recombinant proteins were then purified by IMAC chromatography using HiTrap Talon Crude columns (GE Healthcare, USA) followed by size exclusion chromatography using a Superdex 200 Increase 10/300 GL column (GE Healthcare). Proteins were stored at −80 °C in PBS (pH 7.5) until use. Protein concentrations were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, USA) at 280 nm and calculated using the theoretical molar extinction coefficients and molecular weights of the proteins. The identity of the proteins was further verified by mass spectrometry.
2.5. ELISA
The assay was performed as described elsewhere (Sumova et al., 2018), with minor modifications. ELISA plates (ThermoFisher Scientific) were coated with P. argentipes or P. papatasi SGH (45 ng of proteins per well; corresponding to 0.2 and 0.14 gland per well for P. argentipes and P. papatasi, respectively), or recombinant proteins: rPagSP06 (0.1 µg/well), rPagSP04 (0.1 µg/well) or rPagSP05 (0.2 µg/well) diluted in 20 mM carbonate-bicarbonate buffer (pH 9.5) at 4 °C overnight. The concentration of recombinant proteins was determined based on preliminary experiments. After washing with PBS with 0.05% Tween 20 (PBS-Tw), plates were blocked with 6% non-fat dried milk (Bio-Rad, USA) diluted in PBS-Tw and incubated for 1 h at 37 °C. After another washing step, plates were incubated with human sera diluted 1:100 in 2% non-fat dried milk for 1.5 h at 37 °C. Plates were washed and incubated for 45 min at 37 °C with peroxidase-conjugated anti-human IgG antibody (Sigma-Aldrich, USA) diluted 1:1000 in PBS-Tw. The chromogenic reaction was developed for 6 min in McIlwain phosphate-citrate buffer (pH 5.5) with orthophenylendiamine (Sigma-Aldrich) and hydrogen peroxide in the dark. The reaction was stopped by adding 10% sulfuric acid and the O.D. values were measured at 492 nm using a Tecan Infinite M200 microplate reader. Each serum sample was run in duplicate. In each plate, the same positive and negative control sera were used to enable standardisation for inter-plate and inter-measurement variability.
2.6. Immunoblot
The immunogenicity of the P. argentipes SGH and recombinant salivary proteins, and potential cross-reaction with P. papatasi SGH was tested by an immunoblot assay. Salivary gland homogenate (5.6 μg of total salivary proteins per lane, equivalent to 25 and 17 glands of P. argentipes and P. papatasi respectively) and recombinant proteins rPagSP06, rPagSP04 and rPagSP05 (2 μg of each protein per lane) were separated on a 12% polyacrylamide gel under non-reducing conditions using a Mini-protean apparatus (Bio-Rad). Separated protein bands were either stained by Coomassie Blue (G-250) or transferred onto a nitrocellulose membrane using the iBLOT system (Invitrogen, USA) and then cut into strips. Strips were blocked in 5% non-fat dried milk diluted in Tris-buffered saline with 0.05% Tween 20 (TBS-Tw) overnight at 4 °C and subsequently incubated for 1.5 h with human sera diluted 1:100 in TBS-Tw. After the washing step with TBS-Tw, the strips were incubated for 1 h with peroxidase-conjugated anti-human IgG antibody diluted 1:1000 in TBS-Tw. The chromogenic reaction was developed using a substrate solution containing diaminobenzidine and hydrogen peroxide.
2.7. Endoglycosylation of rPagSP06
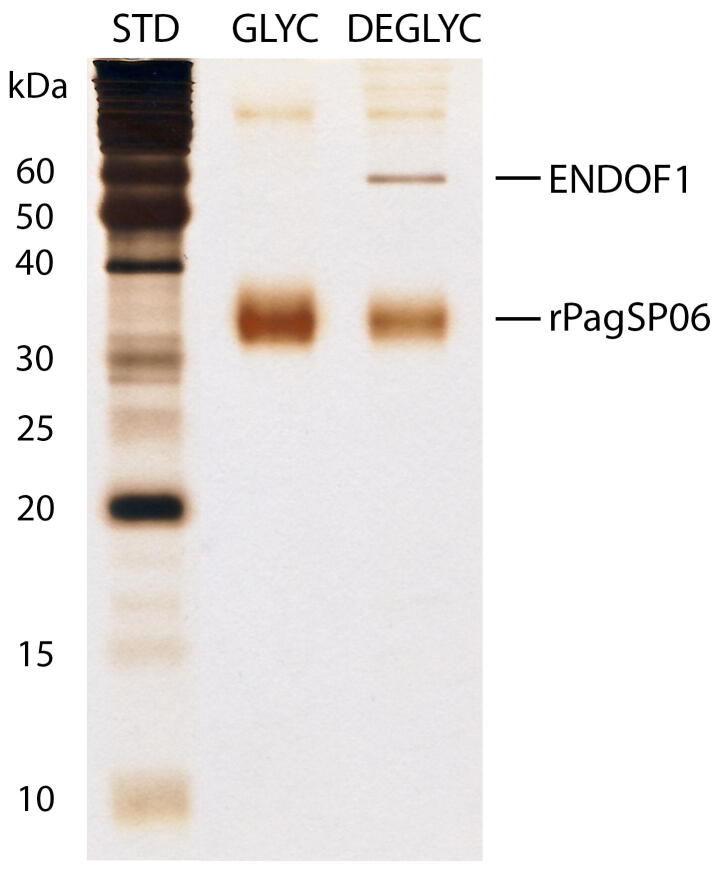
After the expression and purification process, the protein rPagSP06 formed a wide smear-like band on the gel. To clarify if this smear was caused by protein N-glycosylation, rPagSP06 was preincubated 10:1 with endoglycosidase F1 (Grueninger-Leitch et al., 1996; expressed at the Department of Biochemistry, Charles University, Czech Republic) at 37 °C for 20 h. The mixture containing 0.2 μg of protein was then electrophoretically separated on a 15 % polyacrylamide gel under non-reducing conditions using a Mini-protean apparatus. As a reference, the same amount of original rPagSP06 was used. The bands on the gel were visualised by silver staining using a standard protocol.
2.8. Statistical analysis and bioinformatics
Statistical analyses were performed using R software (http://cran.r-project.org/). The results were graphically presented using the ggplot2 package in R software (Wickham, 2009). Samples were tested in duplicate and the average of the duplicate O.D. values obtained by ELISA was used for downstream analyses if the coefficient of variation (CoV) between the duplicates did not exceed 20%. Two positive and two negative control samples were included in each ELISA plate to assess the inter-plate variability. The CoV between these inter-plate controls also did not exceed 20%. The cut-off values were calculated per assay and equaled the average O.D. value of the negative control samples plus 3 S.D. The non-parametric Kruskal–Wallis test with the Wilcoxon rank sum test with Holm correction as a post-hoc analysis was used to compare the median O.D. values between antigens. A P value of <0.05 was considered to be statistically significant. The percentages of positivity were compared between assays using the Chi-Square test. The non-parametric Spearman rank correlation test was used to assess correlations between total anti-P. argentipes SGH and anti-recombinant protein IgG antibody levels.
Putative protein N- and O-glycosylation sites were predicted using NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) and NetOGlyc 3.1 (http://www.cbs.dtu.dk/services/NetOGlyc-3.1/; (Julenius et al., 2004)) software.
3. Results
3.1. Antibody response in humans
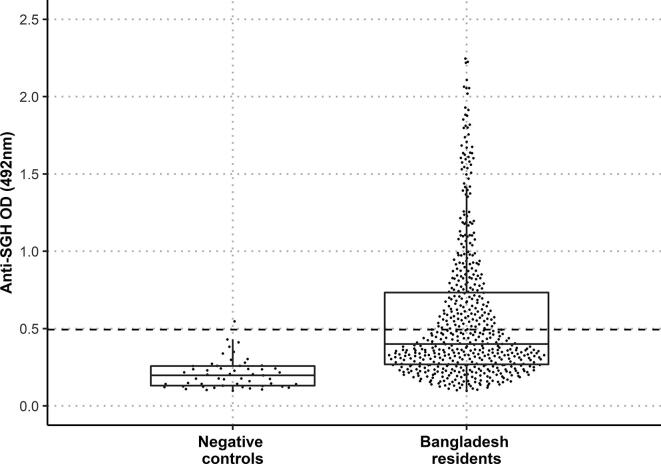
Specific IgG against saliva of P. argentipes in human serum samples from an endemic area in Bangladesh was measured by ELISA using P. argentipes SGH as an antigen. The cut-off value was calculated as the mean of the negative control samples plus 3 S.D. (O.D. = 0.493). Based on this cut-off, 40.2% of all tested volunteers had positive IgG responses against P. argentipes SGH (Fig. 1). The median O.D. value from all endemic human serum samples was 0.400 (interquartile range (IQR): (Q1: 0.269, Q3: 0.733)) and differed significantly from the negative control group (median = 0.198 (Q1: 0.131, Q3: 0.258)) (P < 0.0001).
Fig. 1.
Distribution of the anti-Phlebotomus argentipes salivary gland homogenate (SGH) IgG antibody responses of volunteers from Bangladesh (532 samples) and the negative control samples (53 samples). Dots represent O.D. values for individual serum samples measured by ELISA. The dashed line marks the cut-off value based on the negative control serum samples. The lower and upper borders of the boxplots are the 25th and 75th percentile of the O.D. values (Q1-Q3), the bold lines within the boxes represent the median values, the vertical bars indicate the minimum and maximum values calculated as Q1 ± 1.5 × IQR (interquartile range).
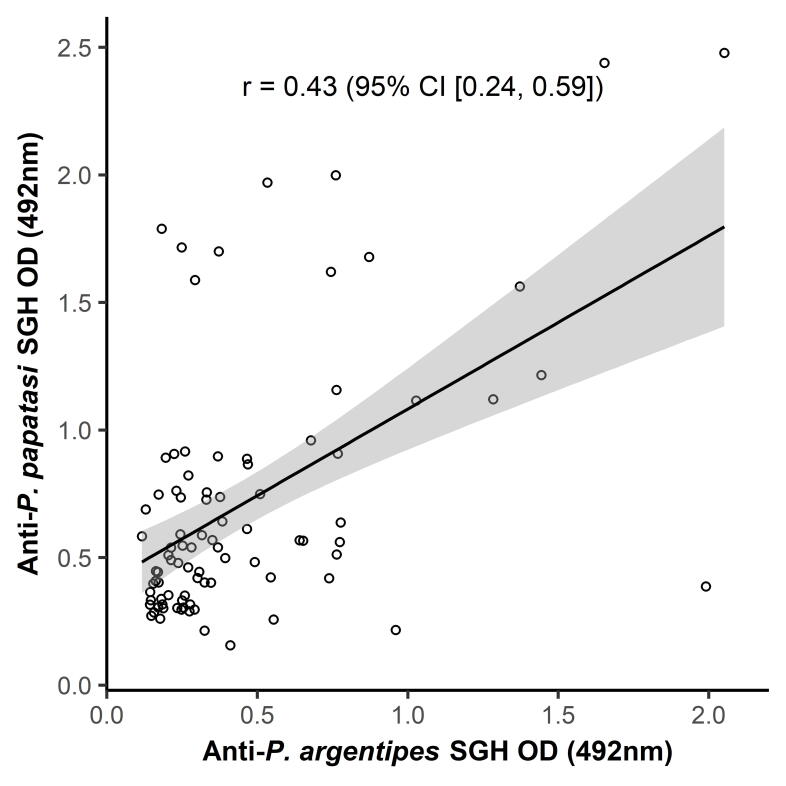
Sand fly collections confirmed that P. argentipes is a dominant species at the study site throughout the season; it formed 99.5% and 76.9% of Phlebotomus fauna in March and August, respectively, while P. papatasi was present in relatively low numbers (0.5% and 23.1% in March and August, respectively). Apart from the genus Phlebotomus, the sand fly fauna was also composed of Sergentomyia spp. (6.5% and 60% in March and August, respectively). Due to the presence of P. papatasi in our sand fly collection, an additional ELISA was performed using a randomly selected portion of the human serum samples and the P. papatasi SGH as an antigen (Supplementary Fig. S1). The anti-P. papatasi IgG response was correlated with the anti-P. argentipes SGH IgG response. The weak correlation (r = 0.43, P < 0.0001) measured between both responses suggests low cross-reactivity between the two species at the study site.
3.2. Identification of major P. argentipes salivary antigens
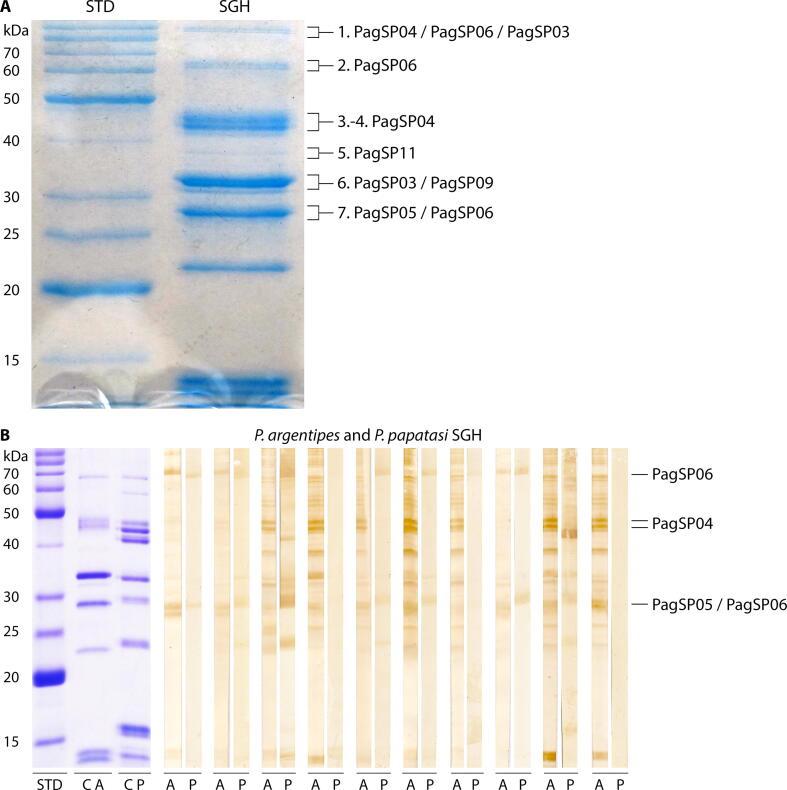
To identify major P. argentipes proteins, seven protein bands were excised from the Coomassie-stained gel and the corresponding proteins were identified by mass spectrometry (Fig. 2A). The identified proteins belong to the abundant sand fly protein families: apyrases (PagSP03), yellow-related proteins (PagSP04), antigen 5-related proteins (PagSP05), PpSP32-like proteins (PagSP06), lufaxin-like proteins (PagSP09) and endonucleases (PagSP11). The most antigenic proteins in P. argentipes SGH were distinguished by immunoblots using highly positive (non-pooled) human serum samples (ELISA O.D. values higher than 1.5). The IgG antibodies in these samples recognised with various intensities up to 15 different protein bands from less than 15 kDa to more than 80 kDa (Fig. 2B). From these proteins, we decided to select three major antigens prominently recognised by human sera, proteins PagSP06, PagSP04 and PagSP05 (marked in Fig. 2B). PpSP32-like protein PagSP06 was present in two protein bands of different sizes. The higher molecular weight of this protein is likely explained by a possible dimerization or by high glycosylation of this protein family (Marzouki et al., 2012, Vlkova et al., 2014); PagSP06 amino acid sequence has one putative N-glycosylation and five O-glycosylation sites.
Fig. 2.
Identification of Phlebotomus argentipes salivary antigens. (A) Identification of the antigenic P. argentipes salivary proteins. The 12% SDS–PAGE gel with a salivary gland homogenate (SGH) protein profile was stained by Coomassie Blue (G-250). The marked bands were excised from the gel and the major proteins were identified by mass spectrometry. The names of the proteins were adapted from Anderson et al. (2006). Proteins PagSP03 (GenBank accession number: ABA12135.1), PagSP09 (GenBank accession number: ABA12140.1) and PagSP11 (GenBank accession number: ABA12142.1) were not further studied. (B) Variability in the intensity of anti-P. argentipes and anti-Phlebotomus papatasi SGH IgG reactions of 10 P. argentipes-positive human serum samples on an immunoblot. Each serum sample was run with both antigens and the strips were placed next to each other. The identified P. argentipes salivary proteins are indicated. ‘A’ and ‘P’ mark the strips with separated protein profiles of P. argentipes and P. papatasi SGH, respectively. ‘C’ marks the strip stained by Coomassie Blue (G-250). Corresponding molecular weights (kDa) of the standard (BenchMark Protein Ladder, ThermoFisher Scientific) are indicated.
To further examine the potential cross-reactivity between P. argentipes and P. papatasi, the same human serum samples were also tested with the latter species. In comparison to P. argentipes, the IgG response against P. papatasi had much lower intensities in up to 10 different protein bands. A strong reaction was observed only for two serum samples. One of them strongly reacted with some of the P. papatasi yellow-related proteins (~42 kDa), and the other clearly recognised bands of ~67 kDa, ~42 kDa, ~30 kDa and ~24 kDa. The bands of ~67 kDa, ~30 kDa were also weakly recognised by some other serum samples. The region of ~67 kDa includes P. argentipes PagSP06, while the corresponding band in P. papatasi was not identifiable in the original study (Valenzuela et al., 2001). Antigenic P. argentipes proteins PagSP05/PagSP06 form a double band of ~28 kDa, while the reactive band in P. papatasi is shifted to ~30 kDa and it is not present in all tested sera (Fig. 2B).
The three aforementioned P. argentipes salivary proteins (PagSP06, PagSP04 and PagSP05) were expressed in a recombinant form in a mammalian expression system (HEK293S cells) and purified by two-phase chromatography. The identity of proteins was verified by mass spectrometry.
3.3. Antigenicity of selected recombinant proteins
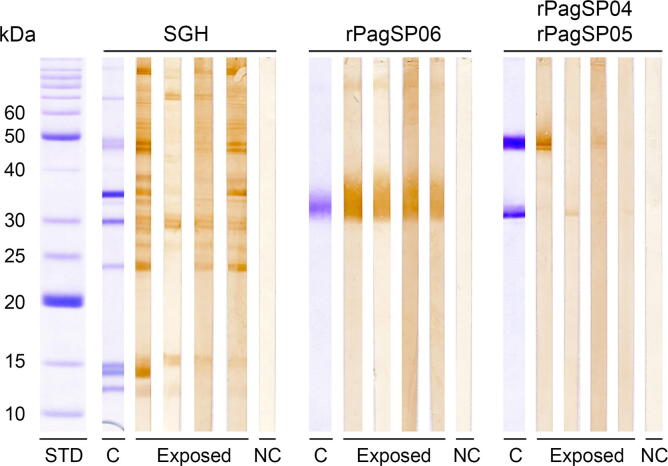
The antigenicity of the three P. argentipes recombinant proteins (rPagSP06, rPagSP04 and rPagSP05) was tested on an immunoblot with human serum samples positive to the SGH (Fig. 3). None of the proteins nor the SGH reacted with the negative control sample. The rPagSP06 protein was recognised with a similar intensity by all four anti-SGH positive serum samples. The rPagSP04 and rPagSP05 proteins strongly differed in molecular weight, allowing them to be tested simultaneously on the same strips. However, neither of them gave a satisfactory result: the rPagSP04 protein showed a strong reaction with one serum sample and faintly reacted with another one, and the rPagSP05 protein was very weakly recognised by two serum samples. The serum samples that positively reacted with rPagSP04 differed from the samples reacting with rPagSP05.
Fig. 3.
Immunoblot with Phlebotomus argentipes salivary gland homogenate (SGH), and recombinant P. argentipes salivary PpSP32-like protein rPagSP06, yellow-related protein rPagSP04 and antigen 5-related protein rPagSP05. The assay was performed with four anti-SGH positive sera (according to the ELISA; marked as exposed) and one negative control (NC) serum sample. rPagSP04 (~47 kDa) and rPagSP05 (~30 kDa) were loaded into the same well. ‘C’ marks the strips stained by Coomassie Blue (G-250). The corresponding molecular weights (kDa) of the standard (BenchMark Protein Ladder, ThermoFisher Scientific; stained by Coomassie Blue (G-250)) are indicated.
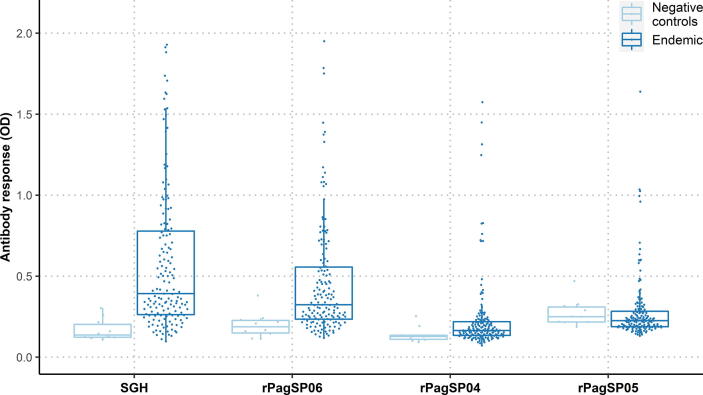
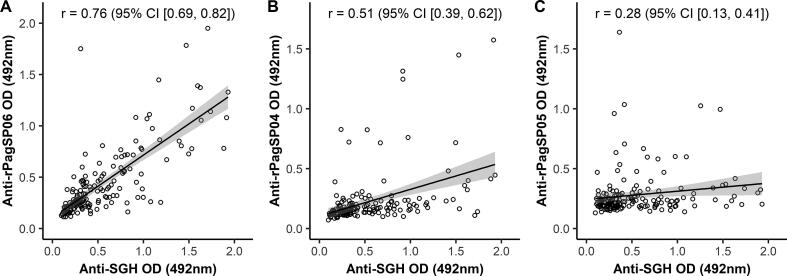
The antigenicity of the three selected recombinant proteins (rPagSP06, rPagSP04 and rPagSP05) was further evaluated by ELISA, for which a random selection of the human serum samples was tested (n = 155) (Fig. 4). Based on the cut-off values calculated from the mean of 10 negative control samples, 51.6%, 35.5%, 12.9%, and 8.4% of tested volunteers were considered IgG positive to P. argentipes SGH, rPagSP06, rPagSP04 and rPagSP05, respectively (Fig. 4). The means, S.D., medians, IQRs and cut-off values measured for individual antigens are summarised in Supplementary Table S1. The distribution of measured optical densities for both negative and endemic human serum samples is graphically visualised as a combined bee swarm-boxplot in Fig. 4. A significant difference was observed between the endemic and negative control samples for P. argentipes SGH (P < 0.0001), rPagSP06 (P < 0.001), and rPagSP04 (P < 0.05), while there was no difference for rPagSP05 (P = 0.197). The antibody responses of the endemic serum samples differed significantly between all antigens (P < 0.0001), though with lower significance between P. argentipes SGH and rPagSP06 (P < 0.05).
Fig. 4.
Distribution of measured ODs for residents of Bangladesh and negative control samples for all antigens tested. Distribution for the anti-Phlebotomus argentipes antibody response of 10 negative controls and 155 residents living in P. argentipes endemic areas in Bangladesh are shown for the P. argentipes salivary gland homogenate (SGH), and recombinant P. argentipes salivary PpSP32-like protein rPagSP06, yellow-related protein rPagSP04, and antigen 5-related protein rPagSP05. Dots represent O.D. values for individual serum samples measured by ELISA. The lower and upper borders of the boxplots are the 25th and 75th percentiles of the O.D. values (Q1-Q3), the bold lines within the boxes represent the median values, the vertical bars indicate the minimum and maximum values calculated as Q1 ± 1.5 × IQR (interquartile range).
The IgG responses against all three recombinant proteins were correlated with the anti-P. argentipes SGH IgG response. The strongest correlation (r = 0.76, P < 0.0001) was calculated for the PpSP32-like protein rPagSP06. Moderate (r = 0.51, P < 0.0001) and weak (r = 0.28, P < 0.0001) correlations were measured for the yellow-related protein rPagSP04 and the antigen 5-related protein rPagSP05, respectively (Fig. 5). Based on these results, proteins rPagSP04 and rPagSP05 were labelled poor markers of human exposure to P. argentipes and therefore excluded from further experiments.
Fig. 5.
Correlations of anti-Phlebotomus argentipes salivary gland homogenate (SGH) IgG response (O.D.) with the IgG responses (O.D.) against recombinant P. argentipes salivary PpSP32-like protein rPagSP06 (A), yellow-related protein rPagSP04 (B) and antigen 5-related protein rPagSP05 (C). The ELISA was performed with 155 randomly selected human serum samples from the endemic area and 10 negative control samples. The analyses were run by Spearman-Rank correlation. The correlation coefficients (r) and confidence intervals (CI) are indicated.
3.4. Evaluation of recombinant PagSP06 as a marker of human exposure to bites of P. argentipes
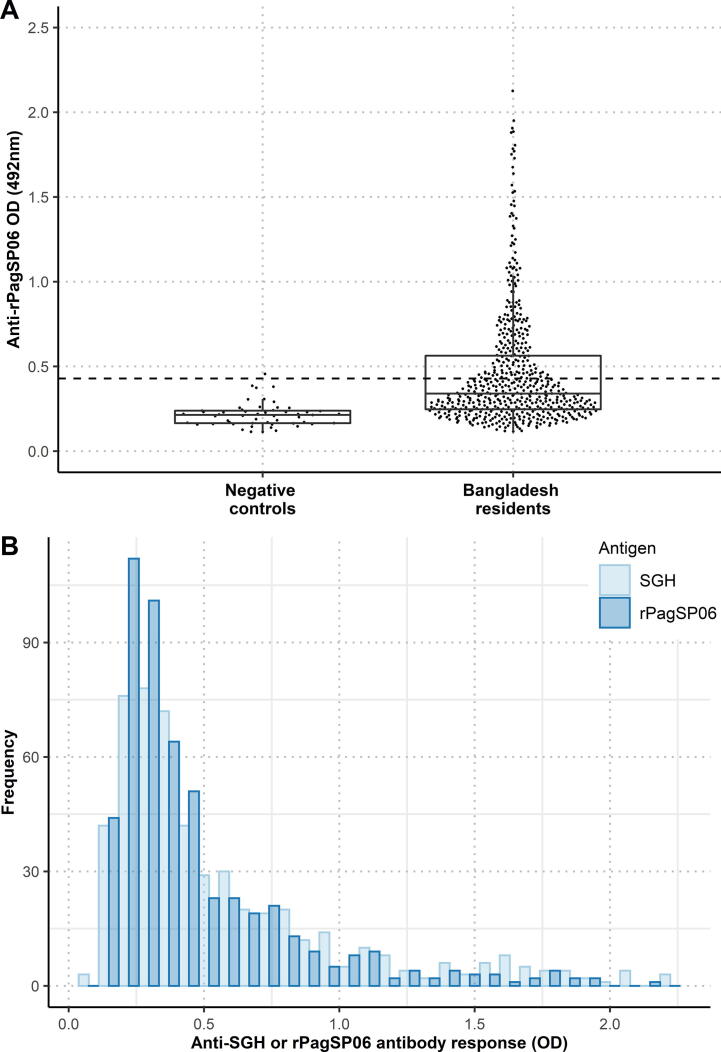
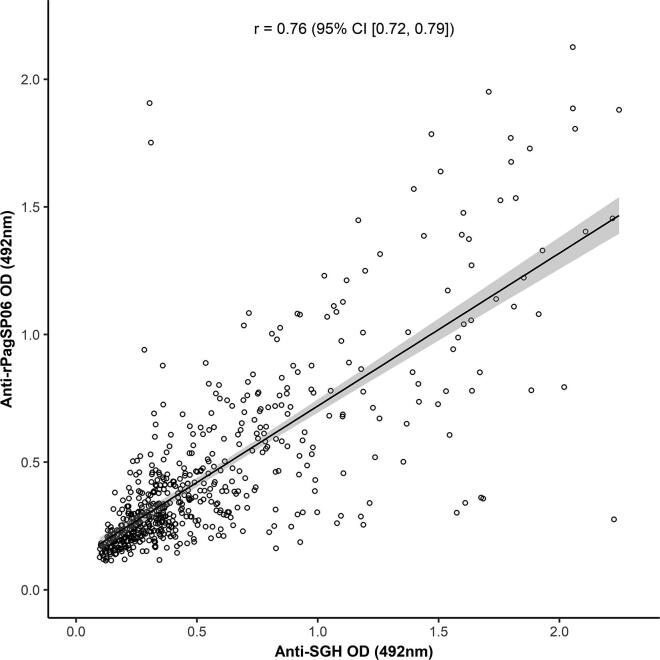
Antigenicity of rPagSP06 protein was further evaluated by ELISA with all available human serum samples (532 endemic and 53 negative controls) (Fig. 6A). The antibody responses of the endemic samples (median = 0.340 (Q1: 0.247, Q3: 0.563)) differed significantly from the negative controls (median = 0.214 (Q1: 0.165, Q3: 0.239)) (P < 0.0001). Based on the cut-off value calculated from the mean of negative control samples plus 3 S.D. (O.D. = 0.429), 36.1% of all tested volunteers were considered IgG positive to rPagSP06. Importantly, no significant difference was found between the percentage of individuals positive for SGH or rPagSP06 (P = 0.198). However, median O.D. values of the endemic samples did significantly differ between the rPagSP06 (median = 0.340 (Q1: 0.247, Q3: 0.563)) and P. argentipes SGH (median = 0.400 (Q1: 0.269, Q3: 0.733)) (P < 0.001). The antibody responses against both antigens did not differ for the negative control samples (medianSGH = 0.198 (Q1: 0.131, Q3: 0.258), medianrPagSP06 = 0.214 (Q1: 0.165, Q3: 0.239)) (P = 0.314). Importantly, the anti-SGH and anti-rPagSP06 IgG antibody response follow a similar right-skewed distribution (Fig. 6B), with most samples exhibiting an O.D. value lower than the cut-off value. Moreover, the anti-rPagSP06 IgG response to all collected serum samples strongly correlated with the anti-P. argentipes SGH IgG antibody response (Fig. 7). The correlation coefficient between these two assays was 0.76 (95% CI (0.72, 0.79), P < 0.0001).
Fig. 6.
Distributions of the antibody responses against Phlebotomus argentipes salivary gland homogenate (SGH) and a recombinant P. argentipes salivary PpSP32-like protein rPagSP06. (A) Boxplots of the anti-rPagSP06 IgG antibody responses of 532 volunteers from Bangladesh and 53 negative control samples. Dots represent O.D. values for individual serum samples measured by ELISA. The dashed line marks the cut-off value based on the negative control serum samples. The lower and upper borders of the boxplots are the 25th and 75th percentiles of the O.D. values (Q1-Q3), the bold lines within the boxes represent the median values, the vertical bars indicate the minimum and maximum values calculated as Q1 ± 1.5 × IQR (interquartile range). (B) Frequency distribution of the anti-P. argentipes SGH and anti-rPagSP06 OD values in the sampled population.
Fig. 7.
Correlations of anti-Phlebotomus argentipes salivary gland homogenate (SGH) IgG response (O.D.) with the IgG responses (O.D.) against recombinant P. argentipes salivary PpSP32-like protein rPagSP06. The ELISA was performed with 532 human serum samples from endemic area and 53 negative control samples. The analysis was run by Spearman-Rank correlation. The correlation coefficient (r) and confidence interval (CI) are indicated.
4. Discussion
Our study describes for the first known time the utilisation of a P. argentipes salivary recombinant protein to determine human exposure to this important VL vector. We focused our research on people living in Chatmohar Upazila, Pabna district, Bangladesh, which has the second highest number of VL patients after the most endemic Mymensingh district. An outbreak of VL occurred in the Pabna district in 1980 (Elias et al., 1989) and since then, new cases are reported every year. Between 2008 and 2014, the Pabna district was among the three most affected districts in Bangladesh with more than 7000 VL cases recorded during this period (Özbel et al., 2017). In our study performed in 2014, 4% of sampled individuals had suffered from VL in the past. These data illustrate the importance of establishing a method to determine the risk of contracting VL in this region. We focused our study on detection of anti-P. argentipes SGH antibodies as a marker of human exposure to the sole vector of L. donovani in the Indian subcontinent. In the villages where we performed our study, P. argentipes was a predominant captured Phlebotomus species with a percentage of 99.5% in March and 76.9% in August. In our study, 40.2% of all participants had IgG antibodies against P. argentipes SGH. The exposure and percentage of positive individuals were comparable with those previously published by other studies conducted in different endemic areas or even higher (Souza et al., 2010, Gidwani et al., 2011, Sumova et al., 2018). The high positivity rate obtained in the present study suggests a high P. argentipes biting rate and shows the necessity of using personal protective measures such as impregnated bed nets in this endemic area.
To identify the predominant protein eliciting the anti-P. argentipes antibodies, we selected three major salivary antigens, expressed them as recombinant proteins and tested the antibody responses against them as potential markers of anti-P. argentipes exposure in humans living in VL foci in Bangladesh. These three proteins were selected based on immunoblots performed with the P. argentipes SGH and highly anti-SGH positive serum samples. The proteins that showed the strongest reaction with these positive human serum samples were identified by mass spectrometry as yellow-related protein rPagSP04, antigen 5-related protein rPagSP05 and PpSP32-like protein rPagSP06. Both the immunoblot and the ELISA assays showed that the proteins rPagSP04 and rPagSP05 are not suitable to measure anti-P. argentipes exposure in humans, further supported by the low correlation of their measured antibody responses and the response against the whole SGH. Even more so, antibody responses against the rPagSP05 protein did not differ between endemic and negative control serum samples. These results are somewhat surprising as yellow-related, and antigen 5-related proteins are known as suitable exposure markers in other sand fly species. Yellow-related proteins were demonstrated as valid markers of human exposure to Lutzomyia longipalpis (Souza et al., 2010) and Phlebotomus orientalis (Sumova et al., 2018) and repeatedly of canine exposure to Phlebotomus perniciosus (Martín-Martín et al., 2014, Kostalova et al., 2015, Kostalova et al., 2017, Risueño et al., 2018, Willen et al., 2019, Maia et al., 2020). Antigen 5-related proteins were proposed as human exposure markers to Lutzomyia intermedia (Carvalho et al., 2017) and P. orientalis (Sumova et al., 2018). However, P. argentipes is a species of the subgenus Euphlebotomus and therefore phylogenetically distant from all above-mentioned sand flies, thus the antigenic properties of its salivary proteins differ.
In our study, the best results were obtained for PpSP32-like protein rPagSP06. This protein performed well in the preliminary assay when the antibody response against rPagSP06 strongly correlated with the anti-SGH IgG response. This led to its evaluation in the large-scale study with all available endemic human samples (532 samples) and negative control samples (53 samples). Importantly, ELISA results revealed significant differences in anti-rPagSP06 antibodies between the endemic and negative control serum samples. However, a difference between median antibody responses against rPagSP06 and SGH also was observed, whereas the positivity of the endemic serum samples did not statistically differ between the two assays. The latter observation is likely to be more important, as the difference in medians could be explained by a lower background produced in the assay with rPagSP06. Furthermore, a strong correlation coefficient of r = 0.76 was obtained between the IgG antibody response against rPagSP06 and the response against P. argentipes SGH. This correlation coefficient is comparable with a previous study that determined the best marker of human exposure to the African VL vector P. orientalis, particularly for the combination of antigen 5-related protein and yellow-related protein (r = 0.76–0.82) (Sumova et al., 2018). A much lower correlation was shown for the human IgG response against a combination of two yellow-related proteins and anti-Lu. longipalpis saliva (Souza et al., 2010).
The PpSP32-like protein family acquired its name from the protein PpSP32 which was firstly described in P. papatasi as a protein similar to the silk protein from a golden silk orb-weaver spider Trichonephila clavipes (Valenzuela et al., 2001). The PpSP32-like protein family (called ‘silk-related family’ by some authors) is present exclusively in sand flies and its function remains unknown. These proteins were found in sialotranscriptomes of all sand flies studied so far (Lestinova et al., 2017, Coutinho-Abreu and Valenzuela, 2018, Polanska et al., 2020). PpSP32-like proteins are usually highly glycosylated (Marzouki et al., 2012, Vlkova et al., 2014); this also applies to the P. argentipes PagSP06 which sequence contains one putative N-glycosylation and five O-glycosylation sites. The presence of N-glycosylation in rPagSP06 was verified by pre-incubation treatment with endoglycosidase-F1. In comparison to glycosylated recombinant protein, the de-glycosylated rPagSP06 creates on the gel a narrower protein band showing that this protein was expressed in several glycoforms (Supplementary Fig. S2).
A member of the PpSP32-like protein family was described as an immunodominant target of the antibody response to P. papatasi bites in humans (Marzouki et al., 2012). Its ability to serve as a marker of human exposure to P. papatasi was further validated in a large cohort study in Tunisia (Marzouki et al., 2015). The number of tested samples was similar to our study, but the authors obtained a lower correlation between anti-saliva and anti-PpSP32 protein (r = 0.54). PpSP32-like proteins are one of the most divergent sand fly protein families, they are conserved only in protein termini domains (Abdeladhim et al., 2016). Phlebotomus papatasi PpSP32 protein shares a 41.4 % amino acid identity with P. argentipes protein rPagSP06. In studies performed in India and Nepal where P. papatasi is common, authors encountered problems with cross-reaction between the IgG antibodies against P. argentipes and P. papatasi salivary lysates (Clements et al., 2010, Gidwani et al., 2011). In Bangladesh, however, P. papatasi is less common and we confirmed that in the Pabna district P. argentipes is a highly predominant Phlebotomus species. In our study, we tested the potential cross-reactivity between the P. argentipes and P. papatasi SGH. The weak correlation between the human IgG responses against the two sand fly species suggests a low cross-reactivity among them. This finding is in disagreement with the previous study of Clements et al. (2010), which found a strong correlation between IgG levels against P. argentipes and P. papatasi. This discrepancy could be explained by a higher abundance of P. papatasi in the study area of the former study. We also showed by immunoblot that the human sera highly exposed to P. argentipes recognised, to a lesser extent and with various intensities, certain P. papatasi salivary proteins. Importantly, the reason for this recognition might be either a partial cross-reactivity between the two species, or simultaneous exposure of the tested individuals to less abundant P. papatasi. The latter is more probable but both alternatives might be involved. Nevertheless, it would be very useful to examine the cross-reactions of rPagSP06 with human sera from areas where P. papatasi is the main human-biting sand fly. We assume that utilisation of the distinct recombinant protein would overcome the issue with whole salivary lysates, even in other regions of the Indian subcontinent.
In conclusion, PpSP32-like recombinant protein rPagSP06 was shown to be a suitable antigen to measure human exposure to P. argentipes in L. donovani foci in Bangladesh. The human IgG antibody response against this antigen could therefore serve as a marker to evaluate the anti-vector control campaigns and to assess other epidemiological measures to eradicate VL in the Indian subcontinent and other regions where P. argentipes serves as an important vector.
Acknowledgements
We gratefully thank Barbora Kalouskova and Ondrej Vanek from the Department of Biochemistry, Charles University, Czech Republic, for expression and purification of the recombinant proteins, and Karel Harant and Pavel Talacko from the Laboratory of Mass Spectrometry, Biocev, Charles University, for proteomic and mass spectrometric analyses. We appreciate the assistance of Vit Dvorak for language revision and Helena Kulikova and Lenka Krejcirikova from Charles University for excellent technical and administrative support. We also appreciate the support given by Sarkar Santana Rani and Fashiur Rahman from Mymensingh Medical College, and Shahidul Islam from Health Inspector of Chatmohor, Bangladesh, Bumpei Tojo, Yasutaka Osada, Sara Kato, Rina Hobo, and Keita Yoshii from The University of Tokyo, Japan, during the blood and sand fly collection period in Pabna, Bangladesh. This work was supported by the projects entitled Centre for Research of Pathogenicity and Virulence of Parasites [CZ.02.1.01/0.0/0.0/16_019/0000759] and Charles University Research Centre [UNCE/SCI/012‐204072/2018]. The research stay of Chizu Sanjoba at Charles University was funded by the European Union’s Horizon 2020 research and innovation program under grant agreement No 731060 (Infravec2). This study was also supported by a project entitled Research and Development of Prevention and Diagnosis for Neglected Tropical Diseases, supported by JST/JICA (Japan Science and Technology Agency / Japan International Cooperation Agency) and the Science and Technology Research Partnership for Sustainable Development, Japan. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpara.2021.05.006.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Correlation of anti-Phlebotomus argentipes salivary gland homogenate (SGH) IgG response (O.D.) with anti-Phlebotomus papatasi SGH IgG response (O.D.). The ELISA was performed with 108 randomly selected human serum samples from an endemic area and 11 negative control samples. For the correlation analysis, only the endemic sera which were over the cut-off level (calculated based on the negative serum samples) for at least one antigen were used (88 serum samples in total). The analysis was run by Spearman-Rank correlation. The correlation coefficient (r) and confidence interval (CI) is indicated.
Supplementary figure 2.
Endoglycosidase treatment of recombinant Phlebotomus argentipes salivary PpSP32-like protein rPagSP06. The effect of deglycosylation of the rPagSP06 protein was tested by SDSPAGE (15% gel, non-reducing conditions). Silverstained gel of the glycosylated (GLYC) and deglycosylated (DEGLYC) protein rPagSP06. ENDOF1 represents endoglycosidase F1. The corresponding molecular weights (kDa) of the standard (STD; BenchMark Protein Ladder, ThermoFisher Scientific) are indicated.
References
- Abdeladhim M., V. Coutinho-Abreu I., Townsend S., Pasos-Pinto S., Sanchez L., Rasouli M., B. Guimaraes-Costa A., Aslan H., Francischetti I.M.B., Oliveira F., Becker I., Kamhawi S., Ribeiro J.M.C., Jochim R.C., Valenzuela J.G., Ghedin E. Molecular diversity between salivary proteins from New World and Old World sand flies with emphasis on Bichromomyia olmeca, the sand fly vector of Leishmania mexicana in Mesoamerica. PLoS Negl. Trop. Dis. 2016;10:e0004771. doi: 10.1371/journal.pntd.0004771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvar J., Vélez I.D., Bern C., Herrero M., Desjeux P., Cano J., Jannin J., Boer M.D., Kirk M. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.M., Oliveira F., Kamhawi S., Mans B.J., Reynoso D., Seitz A.E., Lawyer P., Garfield M., Pham M., Valenzuela J.G. Comparative salivary gland transcriptomics of sandfly vectors of visceral leishmaniasis. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai N.R., Das M.L., Rijal S., van der Auwera G., Picado A., Khanal B., Roy L., Speybroeck N., Berkvens D., Davies C.R., Coosemans M., Boelaert M., Dujardin J.-C. Natural infection of Phlebotomus argentipes with Leishmania and other trypanosomatids in a visceral leishmaniasis endemic region of Nepal. Trans. R. Soc. Trop. Med. Hyg. 2009;103:1087–1092. doi: 10.1016/j.trstmh.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Bláha J., Pachl P., Novák P., Vaněk O. Expression and purification of soluble and stable ectodomain of natural killer cell receptor LLT1 through high-density transfection of suspension adapted HEK293S GnTI− cells. Protein Expr. Purif. 2015;109:7–13. doi: 10.1016/j.pep.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Carvalho A.M., Fukutani K.F., Sharma R., Curvelo R.P., Miranda J.C., Barral A., Carvalho E.M., Valenzuela J.G., Oliveira F., De Oliveira C.I. Seroconversion to Lutzomyia intermedia LinB-13 as a biomarker for developing cutaneous leishmaniasis. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-03345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R., Chowdhury V., Faria S., Akter S., Dash A.P., Bhattacharya S.K., Maheswary N.P., Bern C., Akhter S., Alvar J., Kroeger A., Boelaert M., Banu Q., Oliveira F. Effect of insecticide-treated bed nets on visceral leishmaniasis incidence in Bangladesh. A retrospective cohort analysis. PLoS Negl. Trop. Dis. 2019;13:e0007724. doi: 10.1371/journal.pntd.0007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R., Faria S., Huda M.M., Chowdhury V., Maheswary N.P., Mondal D., Akhter S., Akter S., Khan R.K., Nabi S.G., Kroeger A., Argaw D., Alvar J., Dash A.P., Banu Q., Valenzuela J.G. Control of Phlebotomus argentipes (Diptera: Psychodidae) sand fly in Bangladesh: A cluster randomized controlled trial. PLoS Negl. Trop. Dis. 2017;11:e0005890. doi: 10.1371/journal.pntd.0005890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R., Kumar V., Mondal D., Das M.L., Das P., Dash A.P., Kroeger A. Implication of vector characteristics of Phlebotomus argentipes in the kala-azar elimination programme in the Indian sub-continent. Pathog. Glob. Health. 2016;110:87–96. doi: 10.1080/20477724.2016.1180775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R., Mondal D., Chowdhury V., Faria S., Alvar J., Nabi S.G., Boelaert M., Dash A.P., Ghedin E. How far are we from visceral leishmaniasis elimination in Bangladesh? An assessment of epidemiological surveillance data. PLoS Negl. Trop. Dis. 2014;8:e3020. doi: 10.1371/journal.pntd.0003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements M.F., Gidwani K., Kumar R., Hostomska J., Dinesh D.S., Kumar V., Das P., Müller I., Hamilton G., Volfova V., Boelaert M., Das M., Rijal S., Picado A., Volf P., Sundar S., Davies C.R., Rogers M.E. Measurement of recent exposure to Phlebotomus argentipes, the vector of Indian visceral leishmaniasis, by using human antibody responses to sand fly saliva. Am. J. Trop. Med. Hyg. 2010;82:801–807. doi: 10.4269/ajtmh.2010.09-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho-Abreu I.V., Valenzuela J.G. Comparative evolution of sand fly salivary protein families and implications for biomarkers of vector exposure and salivary vaccine candidates. Front. Cell. Infect. Microbiol. 2018;8 doi: 10.3389/fcimb.2018.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher Y., Perret S., Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30 doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M., Mizanur Rahman A.J.M., Khan N.I. Visceral leishmaniasis and its control in Bangladesh. Bull. World Health Organ. 1989;67:43–49. [PMC free article] [PubMed] [Google Scholar]
- Ghosh K.N., Mukhopadhyay J. The effect of anti-sandfly saliva antibodies on Phlebotomus argentipes and Leishmania donovani. Int. J. Parasitol. 1998;28:275–281. doi: 10.1016/S0020-7519(97)00152-5. [DOI] [PubMed] [Google Scholar]
- Gidwani K., Picado A., Rijal S., Singh S.P., Roy L., Volfova V., Andersen E.W., Uranw S., Ostyn B., Sudarshan M., Chakravarty J., Volf P., Sundar S., Boelaert M., Rogers M.E., Kamhawi S. Serological markers of sand fly exposure to evaluate insecticidal nets against visceral leishmaniasis in India and Nepal: a cluster-randomized trial. PLoS Negl. Trop. Dis. 2011;5:e1296. doi: 10.1371/journal.pntd.0001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueninger-Leitch F., D'Arcy A., D'Arcy B., Chène C. Deglycosylation of proteins for crystallization using recombinant fusion protein glycosidases. Protein Sci. 1996;5:2617–2622. doi: 10.1002/pro.5560051224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M.I., Khan S.A., Ameen M.-U. Phlebotomine sandflies of Bangladesh: recent surveys. Med. Vet. Entomol. 1993;7:99–101. doi: 10.1111/j.1365-2915.1993.tb00659.x. [DOI] [PubMed] [Google Scholar]
- Julenius K., Mølgaard A., Gupta R., Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2004;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Kammoun-Rebai W., Bahi-Jaber N., Naouar I., Toumi A., Ben Salah A., Louzir H., Meddeb-Garnaoui A., Olivier M. Human cellular and humoral immune responses to Phlebotomus papatasi salivary gland antigens in endemic areas differing in prevalence of Leishmania major infection. PLoS Negl. Trop. Dis. 2017;11:e0005905. doi: 10.1371/journal.pntd.0005905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostalova T., Lestinova T., Maia C., Sumova P., Vlkova M., Willen L., Polanska N., Fiorentino E., Scalone A., Oliva G., Veronesi F., Cristóvão J.M., Courtenay O., Campino L., Gradoni L., Gramiccia M., Volf P. The recombinant protein rSP03B is a valid antigen for screening dog exposure to Phlebotomus perniciosus across foci of canine leishmaniasis. Med. Vet. Entomol. 2017;31:88–93. doi: 10.1111/mve.12192. [DOI] [PubMed] [Google Scholar]
- Kostalova T., Lestinova T., Sumova P., Vlkova M., Rohousova I., Berriatua E., Oliva G., Fiorentino E., Scalone A., Gramiccia M., Gradoni L., Volf P., Debrabant A. Canine antibodies against salivary recombinant proteins of Phlebotomus perniciosus: A longitudinal study in an endemic focus of canine leishmaniasis. PLoS Negl. Trop. Dis. 2015;9:e0003855. doi: 10.1371/journal.pntd.0003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawyer P., Killick-Kendrick M., Rowland T., Rowton E., Volf P. Laboratory colonization and mass rearing of phlebotomine sand flies (Diptera, Psychodidae) Parasite. 2017;24:42. doi: 10.1051/parasite/2017041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestinova T., Rohousova I., Sima M., de Oliveira C.I., Volf P., Milon G. Insights into the sand fly saliva: Blood-feeding and immune interactions between sand flies, hosts, and Leishmania. PLoS Negl. Trop. Dis. 2017;11:e0005600. doi: 10.1371/journal.pntd.0005600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWIS D.J. Phlebotomine sandflies (Diptera: Psychodidae) from the Oriental Region. Syst. Entomol. 1987;12:163–180. doi: 10.1111/j.1365-3113.1987.tb00194.x. [DOI] [Google Scholar]
- Lewis D.J. A taxonomic review of the genus Phlebotomus (Diptera : Psychodidae) Bull. Br. Museum Nat. Hist. Entomol. 1982;45:121–209. [Google Scholar]
- Maia C., Cristóvão J., Pereira A., Kostalova T., Lestinova T., Sumova P., Volf P., Campino L. Monitoring Leishmania infection and exposure to Phlebotomus perniciosus using minimal and non-invasive canine samples. Parasites Vectors. 2020;13:1–12. doi: 10.1186/s13071-020-3993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroli M., Feliciangeli M.D., Bichaud L., Charrel R.N., Gradoni L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med. Vet. Entomol. 2013;27:123–147. doi: 10.1111/j.1365-2915.2012.01034.x. [DOI] [PubMed] [Google Scholar]
- Martín-Martín I., Molina R., Jiménez M. An insight into the Phlebotomus perniciosus saliva by a proteomic approach. Acta Trop. 2012;123:22–30. doi: 10.1016/j.actatropica.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Martín-Martín I., Molina R., Jiménez M. Identifying salivary antigens of Phlebotomus argentipes by a 2DE approach. Acta Trop. 2013;126:229–239. doi: 10.1016/j.actatropica.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Martín-Martín I., Molina R., Rohoušová I., Drahota J., Volf P., Jiménez M. High levels of anti-Phlebotomus perniciosus saliva antibodies in different vertebrate hosts from the re-emerging leishmaniosis focus in Madrid, Spain. Vet. Parasitol. 2014;202:207–216. doi: 10.1016/j.vetpar.2014.02.045. [DOI] [PubMed] [Google Scholar]
- Marzouki S., Abdeladhim M., Abdessalem C.B., Oliveira F., Ferjani B., Gilmore D., Louzir H., Valenzuela J.G., Ahmed M.B., Bates P.A. Salivary antigen SP32 is the immunodominant target of the antibody response to Phlebotomus papatasi bites in humans. PLoS Negl. Trop. Dis. 2012;6:e1911. doi: 10.1371/journal.pntd.0001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzouki S., Kammoun-Rebai W., Bettaieb J., Abdeladhim M., Hadj Kacem S., Abdelkader R., Gritli S., Chemkhi J., Aslan H., Kamhawi S., Ben Salah A., Louzir H., Valenzuela J.G., Ben Ahmed M., Bates P.A. Validation of recombinant salivary protein PpSP32 as a suitable marker of human exposure to Phlebotomus papatasi, the vector of Leishmania major in Tunisia. PLoS Negl. Trop. Dis. 2015;9:e0003991. doi: 10.1371/journal.pntd.0003991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal D., Das M.L., Kumar V., Huda M.M., Das P., Ghosh D., Priyanka J., Matlashewski G., Kroeger A., Upfill-Brown A., Chowdhury R., Acosta-Serrano A. Efficacy, safety and cost of insecticide treated wall lining, insecticide treated bed nets and indoor wall wash with lime for visceral leishmaniasis vector control in the Indian sub-continent: a multi-country cluster randomized controlled trial. PLoS Negl. Trop. Dis. 2016;10:e0004932. doi: 10.1371/journal.pntd.0004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon-Shem K., Al-Salem W.S., Kelly-Hope L., Abdeladhim M., Al-Zahrani M.H., Valenzuela J.G., Acosta-Serrano A. Severity of Old World cutaneous leishmaniasis is influenced by previous exposure to sandfly bites in Saudi Arabia. PLoS Negl. Trop. Dis. 2015;9:1–14. doi: 10.1371/journal.pntd.0003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özbel, Y., Sanjoba, C., Matsumoto, Y., 2017. Geographical distribution and ecological aspect of sand fly species in Bangladesh, in: Noiri, E., Jha, T. (Eds.), Kala Azar in South Asia. Springer International Publishing, Cham, pp. 199–209. 10.1007/978-3-319-47101-3_17.
- Polanska N., Ishemgulova A., Volfova V., Flegontov P., Votypka J., Yurchenko V., Volf P. Sergentomyia schwetzi: Salivary gland transcriptome, proteome and enzymatic activities in two lineages adapted to different blood sources. PLoS One. 2020;15:1–40. doi: 10.1371/journal.pone.0230537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risueño J., Spitzová T., Bernal L.J., Muñoz C., López M.C., Thomas M.C., Infante J.J., Volf P., Berriatua E. Longitudinal monitoring of anti-saliva antibodies as markers of repellent efficacy against Phlebotomus perniciosus and Phlebotomus papatasi in dogs. Med. Vet. Entomol. 2018;33:99–109. doi: 10.1111/mve.12343. [DOI] [PubMed] [Google Scholar]
- Rohousova I., Ozensoy S., Ozbel Y., Volf P. Detection of species-specific antibody response of humans and mice bitten by sand flies. Parasitology. 2005;130:493–499. doi: 10.1017/S003118200400681X. [DOI] [PubMed] [Google Scholar]
- Rohousova I., Talmi-Frank D., Vlkova M., Spitzova T., Rishpon K., Jaffe C.L., Volf P., Baneth G., Ephros M. Serological evaluation of cutaneous leishmania tropica infection in Northern Israel. Am. J. Trop. Med. Hyg. 2018;98:139–141. doi: 10.4269/ajtmh.17-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza A.P., Andrade B.B., Aquino D., Entringer P., Miranda J.C., Alcantara R., Ruiz D., Soto M., Teixeira C.R., Valenzuela J.G., de Oliveira C.I., Brodskyn C.I., Barral-Netto M., Barral A., Milon G. Using recombinant proteins from Lutzomyia longipalpis saliva to estimate human vector exposure in visceral leishmaniasis endemic areas. PLoS Negl. Trop. Dis. 2010;4:e649. doi: 10.1371/journal.pntd.0000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzova T., Sumova P., Volfova V., Polanska N., Poctova L., Volf P. Interactions between host biogenic amines and sand fly salivary yellow-related proteins. Parasit. Vectors. 2020;13:237. doi: 10.1186/s13071-020-04105-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumova P., Sima M., Spitzova T., Osman M.E., Guimaraes-Costa A.B., Oliveira F., Elnaiem D.-E., Hailu A., Warburg A., Valenzuela J.G., Volf P., Dutra W.O. Human antibody reaction against recombinant salivary proteins of Phlebotomus orientalis in Eastern Africa. PLoS Negl. Trop. Dis. 2018;12:e0006981. doi: 10.1371/journal.pntd.0006981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira C., Gomes R., Collin N., Reynoso D., Jochim R., Oliveira F., Seitz A., Elnaiem D.-E., Caldas A., de Souza A.P., Brodskyn C.I., de Oliveira C.I., Mendonca I., Costa C.H.N., Volf P., Barral A., Kamhawi S., Valenzuela J.G., Milon G. Discovery of markers of exposure specific to bites of Lutzomyia longipalpis, the vector of Leishmania infantum chagasi in Latin America. PLoS Negl. Trop. Dis. 2010;4:e638. doi: 10.1371/journal.pntd.0000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiakaki M., Rohousova I., Volfova V., Volf P., Chang K.-P., Soteriadou K. Sand fly specificity of saliva-mediated protective immunity in Leishmania amazonensis-BALB/c mouse model. Microbes Infect. 2005;7:760–766. doi: 10.1016/j.micinf.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Valenzuela J.G., Belkaid Y., Garfield M.K., Mendez S., Kamhawi S., Rowton E.D., Sacks D.L., Ribeiro J.M. Toward a defined anti-Leishmania vaccine targeting vector antigens: characterization of a protective salivary protein. J. Exp. Med. 2001;194:331–342. doi: 10.1084/jem.194.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlkova M., Sima M., Rohousova I., Kostalova T., Sumova P., Volfova V., Jaske E.L., Barbian K.D., Gebre-Michael T., Hailu A., Warburg A., Ribeiro J.M.C., Valenzuela J.G., Jochim R.C., Volf P., McDowell M.A. Comparative analysis of salivary gland transcriptomes of Phlebotomus orientalis sand flies from endemic and non-endemic foci of visceral leishmaniasis. PLoS Negl. Trop. Dis. 2014;8:e2709. doi: 10.1371/journal.pntd.0002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volf P., Rohoušová I. Species-specific antigens in salivary glands of phlebotomine sandflies. Parasitology. 2001;122:37–41. doi: 10.1017/S0031182000007046. [DOI] [PubMed] [Google Scholar]
- Wickham, H., 2009. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York.
- Willen L., Lestinova T., Kalousková B., Sumova P., Spitzova T., Velez R., Domenech E., Vaněk O., Gállego M., Mertens P., Volf P., Fujiwara R.T. Field study of the improved rapid sand fly exposure test in areas endemic for canine leishmaniasis. PLoS Negl. Trop. Dis. 2019;13:e0007832. doi: 10.1371/journal.pntd.0007832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.