Abstract
Background and Aims
The potassium channel Kv1.3 is a potentially attractive therapeutic target in T cell-mediated inflammatory diseases, as the activity of antigen-activated T cells is selectively impeded by Kv1.3 inhibition. In this study, we examined Kv1.3 as a potential therapeutic intervention point for ulcerative colitis [UC], and studied the efficacy of DES1, a small-molecule inhibitor of Kv1.3, in vitro and in vivo.
Methods
Kv1.3 expression on T cells in peripheral blood mononuclear cells [PBMCs] isolated from donors with and without UC was examined by flow cytometry. In biopsies from UC patients, Kv1.3-expressing CD4+ T cells were detected by flow cytometry and immunohistochemistry. In vitro, we determined the ability of DES1 to inhibit anti-CD3-driven activation of T cells. In vivo, the efficacy of DES1 was determined in a humanised mouse model of UC and compared with infliximab and tofacitinib in head-to-head studies.
Results
Kv1.3 expression was elevated in PBMCs from UC patients and correlated with the prevalence of TH1 and TH2 T cells. Kv1.3 expression was also detected on T cells from biopsies of UC patients. In vitro, DES1 suppressed anti-CD3-driven activation of T cells in a concentration-dependent manner. In vivo, DES1 significantly ameliorated inflammation in the UC model and most effectively so when PBMCs from donors with higher levels of activated T cells were selected for reconstitution. The efficacy of DES1 was comparable to that of either infliximab or tofacitinib.
Conclusion
Inhibition of Kv1.3 [by DES1, for instance] appears to be a potential therapeutic intervention strategy for UC patients.
Keywords: Basic science, experimental models and pathophysiology, biomarkers
1. Introduction
Ulcerative colitis [UC] and Crohn’s disease [CD] belong to the family of chronic inflammatory bowel diseases [IBDs]. Both diseases cause abdominal pain and diarrhoea, but whereas UC is characterised by superficial mucosal inflammation and rectal bleeding,1 in CD inflammation penetrates transmurally and causes perforation, strictures, and fibrosis, and inflammation can affect the entire intestinal tract. For a long time, characterisation of inflammatory processes in UC and CD followed the paradigm that UC was a disease driven by TH2 cytokines2,3 whereas immunological processes in CD were defined by TH1 cytokines.4,5 This paradigm has now fallen, perhaps best demonstrated by the fact that the anti-interleukin [IL]-12/23 antibody ustekinumab has been approved for both CD and UC.6 In a recent study designed to characterise inflammation in IBD patients, we identified two parallel inflammatory processes: a pro-inflammatory process and a remodelling process, signified by activities of TH1 and TH2 cells and by activities of TH17 and monocytes [i.e., M1, M2], respectively. The coexistence of these two processes reflects that IBD is highly dynamic, with one or the other process typically predominating at a given point in time.7 Specific suppression of the [excess] activity of the immune cells involved in these processes offers an attractive therapeutic intervention point in IBD.
The voltage-gated potassium channel Kv1.3 has come into focus of interest as it provides a therapeutic point of intervention to suppress T cell activity and proliferation.8 Kv1.3 is known to be crucial for maintaining the membrane potential required for induction of inflammatory responses because Kv1.3 inhibition reduces calcineurin-mediated immune cell activation and proliferation. The channel is particularly upregulated in human activated effector memory T cells and central memory T cells, and also in class-switched B cells [CD27+ IgD–] and certain monocytes.9–12 Inhibition of Kv1.3 therefore offers a potential opportunity to specifically impair inflammatory immune cell activity and proliferation, including in antigen-activated T cells.
The peptide toxin ShK, isolated from the sea anemone Stichodactyla helianthus13 and derivatives thereof, and the small molecule PAP-114 were the first molecules identified as specific inhibitors of Kv1.3 reaching the clinic. They have shown efficacy in preclinical models of inflammatory diseases such as rheumatoid arthritis [RA], experimental autoimmune diabetes, and asthma15,16 and, ultimately, in early clinical trials with plaque psoriasis patients.17 Unfortunately, peptide toxins are not orally available, and the high lipophilicity of PAP-1 complicates its clinical development as an oral inhibitor. None of these molecules thus appears to have the potential to meet the needs of IBD patients who would benefit from a drug with oral availability, which is a major objective for successful clinical development of an inhibitor [e.g., of Kv1.3] for treating IBD.
A hurdle in the preclinical development of Kv1.3 inhibitors is the fact that Kv1.3 is not the only Kv1.x channel that regulates the membrane potential of mouse T cells.18,19 Therefore, most of the successful preclinical studies have so far been performed in well-established rat models that more closely resemble the clinically relevant scenario in humans, in that neither express redundant Kv1.x channels. Rather, the membrane potentials of human and rat T cells appear to be controlled by Kv1.3 along with KCa3.1.20 More recently, a humanised mouse model for UC [NSG-UC] has, however, been developed7,21 based on immuno-compromised NOD/ScidIL-2Rγ null [NSG] mice reconstituted with human peripheral blood mononuclear cells [PBMCs] from UC patients. In this model, symptoms of UC are induced by rectal application of ethanol, which likely disrupts the colonic mucosal and epithelial barriers, thus facilitating passage of luminal contents [e.g., antigens] into the colon. The effect of rectal co-administration of ethanol in our model is similar to that of rectally administered 2,4,6-trinitrobenzenesulphonic acid [TNBS], which is used to induce severe colonic inflammation in the TNBS model of colitis.22 Only cells obtained from donors with active inflammation, however, induce pronounced clinical symptoms in the mice.7,21,23 This chimeric model allows preclinical researchers to evaluate the efficacy of molecules with development potential to treat human IBD patient groups in a humanised model that more closely replicates the clinically relevant scenario in humans. Furthermore, the combination of patient profiling and studies in the NSG-UC mouse model might also offer a tool to predict responsiveness to therapeutics at the level of individual patients.7
In this study, we examined IBD as a potential disease to be targeted by Kv1.3 small-molecule inhibitors. We examined the expression of Kv1.3 on T cells in PBMCs from IBD patients as compared with non-IBD donors, and in biopsies from UC donors. We further demonstrated the ability of the small molecule DES1 to suppress anti CD3-mediated T-cell activation and secretion of cytokines and chemokines in vitro, and to ameliorate inflammation in the NSG-UC model in vivo. Finally, we examined responsiveness to DES1 in the NSG-UC mouse model reconstituted with two different subgroups of UC patients to assess which patient group might benefit most from treatment with DES1. The results indicate that DES1 is a potent suppressor of T cell activation and cytokine production in vitro, and appears to confer a broad immuno-modulatory effect on T cell activity in vivo, as reflected in its efficacy in the NSG-UC mouse model. The NSG-UC results also suggest that patients exhibiting high levels of activated CD4+ T cells may represent a particularly favourable target group for a DES1-based treatment of IBD.
2. Materials and Methods
2.1. Ethical considerations
Written consent was given by all donors. The study was approved by the Institutional Review Board [IRB] of the Medical Faculty at the University of Munich [2015–22 and 343-09]. Animal studies were approved by the committees of the government of Upper Bavaria, Germany [ROB-55.2–2532.Vet_02-15-74-2015] and performed in compliance with German Animal Welfare Laws.
2.2. Isolation of PBMCs and engraftment
As described previously, 60 mL of peripheral blood in trisodium citrate solution [S-Monovette, Sarstedt, Nürnberg, Germany] were collected from the arm vein of healthy donors or from IBD patients.24 The blood was diluted with Hank’s balanced salt solution [HBSS, Sigma Aldrich, Deisenhofen, Germany] in a 1:2 ratio. The suspension was loaded onto LeucoSep tubes [Greiner Bio One, Frickenhausen, Germany]. PBMCs were separated by centrifugation at 400 g for 30 min without acceleration. The interphase was extracted and diluted with phosphate-buffered saline [PBS] to a final volume of 40 mL. Cells were counted and centrifuged at 1400 g for 5 min. The cell pellet was resuspended in PBS at a concentration of 4 x 106 cells in 100 µL.
2.3. Cell culture
PBMCs of healthy donors were isolated from 10 mL of blood. The cell pellet was resuspended in Roswell Park Memorial Institute [RPMI] Medium [Thermo Fisher Scientific, Waltham, MA, USA] at a concentration of 1 × 106 cells/mL, as described previously.24 An additional 500 μL RPMI with 10% fetal calf serum [FCS] and 1% penicillin-streptomycin [Sigma-Aldrich, St Louis, MO, USA] were added to each well and sample. Wells containing PBMCs and RPMI, or PBMCs, RPMI, and anti-CD3 [0.31 ng/mL, BioLegend, San Diego, CA, USA], served as negative and positive controls, respectively. For analysis of the effect of DES1, compounds were dissolved in PBS and were added at increasing concentrations ranging from 300 to 1000 nM. ShK [Smartox Biotechnology, St Egreve, France] was dissolved in de-ionised water and added at a concentration of 100 nM with 0.1% BSA. Cells were incubated for 48 h. Following incubation, the content of each well was centrifuged at 1400 g for 5 min. The pellet was resuspended in 1 x PBS, 2 mM ethylendiaminetetraacetate [EDTA], and 2% FCS, [FACS buffer] and labelled for flow cytometry as listed in Tables S4 and S5, available as Supplementary data at ECCO-JCC online. For the cell proliferation assay, CellTrace [CSFE, Thermofisher, Darmstadt, Germany, Cat# C34571] was added according to the manufacturer’s instructions and cells were incubated for 1 week. Viability staining of the cell cultures was performed with fluorescent probes [Thermofisher, Darmstadt, Germany, CAT# 65-0865-14].
2.4. Flow cytometric analysis
Labelling of human leukocytes was performed according to Table S5. All antibodies were purchased from Biolegend [San Diego, USA] and used according to the manufacturer’s instructions. Flow cytometry was performed using Attune NXT [Thermofisher, Scientific, Darmstadt, Germany] and analysed with FlowJo 10.1-Software [FlowJo LLC, Oregon, USA].
2.5. Electrophysiology
Kv1.3 channels mediating outward delayed rectifying potassium current [encoded by the human KCNA3 gene] were cloned and expressed in CHO cells. Margatoxin [Sigma-Aldrich M8278] was used as positive control for the Kv1.3-mediated currents in this cell line [IC50 of ~300 pM]. The dose-response relationship of DES-006535 [DES1] was evaluated at concentrations of 0.003, 0.01, 0.03, 0.1, 0.3, 1, 3, and 10 μM in at least three distinct cells [n ≥ 3] and the duration of compound exposure at each concentration was at least 3 min. The compound concentrations were prepared by diluting stock solutions into an appropriate HEPES-buffered physiological saline extracellular solution with the following constituents [in mM]: NaCl, 140; KCl, 4; CaCl2, 2; MgCl2, 1; glucose, 5; HEPES, 10. A pH value of 7.4 was established with NaOH. The intracellular solution was similarly [in mM]: KCl, 50; KF, 60; NaCl, 10; EGTA, 20; HEPES, 10. A pH value of 7.2 was established with NaOH.
All currents were recorded with a whole-cell [i.e., CHO] configuration setup on the Patchliner platform with membrane resistance ≥1 MΩ, and steady state block was measured using a 20-pulse pattern with fixed amplitudes [depolarisation: +40 mV amplitude, 100-ms duration] repeated at 10-s intervals from a holding potential of –90 mV. Area under curve [AUC] was measured during the step to +40 mV. The steady state inhibition before and after compound application was used to calculate the percentage of current inhibited at each concentration.
2.6. Study protocol
NSG mice were obtained from Charles River Laboratories [Sulzfeld, Germany]. Mice were kept under specific pathogen-free conditions in individually ventilated cages in a facility controlled according to the Federation of Laboratory Animal Science Association [FELASA] guidelines. Following engraftment on Day 1, mice were presensitised by rectal application of 150 µL of 10% ethanol on Day 8 using a 1-mm cat catheter [Henry Schein, Hamburg, Deutschland]. The catheter was lubricated with Xylocain©Gel 2% [AstraZeneca, Wedel]. Rectal application was performed under general anaesthesia using 4% isoflurane. Post-application mice were kept at an angle of 30° to avoid ethanol dripping. On Day 15, the mice were challenged by rectal application of 50% ethanol following the protocol used on Day 8. For the drug treatments, DES1 [D. E. Shaw Research, New York, USA] and tofacitinib [Merck KGaA, Darmstadt, Germany] were administered daily over one period of 3 days and a second period of 4 days [Days 7–9 and 14–17, respectively], such that each period encompassed one of the two ethanol challenges [on Days 8 and 15]. The dosages were 5 mg/kg/day for DES1 and 5 mg/kg/day for tofacitinib. The dosage of infliximab [Janssen Biotech, Horsham, Pennsylvania, USA] was 6 mg/kg/day, and treatment was limited to Days 7 and 14 only. All therapeutics were dissolved in PBS and administered intraperitoneally.
2.7. Clinical activity score
The assessment of colitis severity was performed daily according to the following scoring system: loss of body weight: 0% [0], 0–5% [1], 5–10% [2], 10–15% [3], and 15–20% [4]; stool consistency: formed pellet [0], loose stool or unformed pellet [2], liquid stools [4]; behaviour: normal [0], reduced activity [1], apathy [4], and ruffled fur [1]; body posture: intermediately hunched posture [1], permanently hunched posture [2]. The scores were added daily into a total score with a maximum of 12 points per day. Animals who suffered from weight loss >20%, rectal bleeding, rectal prolapse, self-isolation, or a severity score >7 were euthanised immediately and not taken into account. All scores were added for statistical analysis.
2.8. Macroscopic colon score
After sacrificing the animals, the colon was removed, a photograph was taken and the colon was scored for macroscopic characteristics as follow: pellet: formed [0], soft [1], liquid [2]; length of colon: >10 cm [0], 8–10 cm [1], <8 cm [2]; dilation: no [0], minor [1], severe [2]; hyperaemia: no [0], yes [2]; necrosis: no [0], yes [2].
2.9. Histological analysis
At autopsy, samples from distal parts of the colon were fixed in 4% formaldehyde for 24 h before storage in 70% ethanol, and were routinely embedded in paraffin. Samples were cut into 3-µm sections and stained with haematoxylin and eosin [H&E].
Epithelial erosions were scored as follows7: no lesions [1], focal lesions [2], multifocal lesions [3], major damage with involvement of basal membrane [4]. Inflammation was scored as follows: infiltration of few inflammatory cells into the lamina propria [1], major infiltration of inflammatory cells into the lamina propria [2], confluent infiltration of inflammatory cells into the lamina propria [3], infiltration of inflammatory cells including tunica muscularis [4]. Fibrosis was scored as follows: focal fibrosis [1], multifocal fibrosis and crypt atrophy [2]. The presence of oedema, hyperaemia, and crypt abscess was scored with one additional point in each case. The scores for each criterion were added into a total score ranging from 0 to 12. Images were taken with an AxioVert 40 CFL camera [Zeiss, Oberkochen, Germany] using the Zeiss ZE n2 lite software. Figures show representative longitudinal sections in original magnification. In Adobe Photoshop CC, a tonal correction was used in order to enhance contrasts within the pictures.
2.10. Isolation of human leukocytes from mouse spleen and colon
To isolate human leukocytes, spleens were minced and cells filtrated through a 70-µL cell strainer followed by centrifugation at 1400 g for 5 min, and were re-suspended in FACS buffer. For further purification, cell suspensions were filtrated using a 35-µM cell strainer followed by labelling for flow cytometry analysis, as described previously.7
For isolation of lamina propria mononuclear cells [LPMC], a modified protocol of Weigmann25 was used. The washed and minced colon was pre-digested in an orbital shaker with slow rotation [40 g] at 37°C for 1 x 20 min in pre-digestion solution containing 1 x HBSS [Thermo Scientific, Darmstadt, Deutschland], 5 mM EDTA, 5% FCS, 100 U/mL pencillin-streptomycin [Sigma Aldrich, Deisenhofen, Germany]. Epithelial cells were removed by filtering through a nylon filter. After washing with RPMI, the remaining colon pieces were digested for 2 x 20 min in digestion solution containing 1 x RPMI [Thermo Scientific, Darmstadt, Deutschland], 10% FCS, 1 mg /mL collagenase A [Sigma Aldrich, Deisenhofen, Germany], 10 KU/mL Dnase I [Sigma Aldrich, Deisenhofen, Germany], 100 U/mL pencillin-streptomycin [Sigma Aldrich, Deisenhofen, Germany] in an orbital shaker with slow rotation [40 g] at 37°C.25
Isolated LPMC were centrifuged at 1400 g for 5 min and resuspended in FACS buffer. Cell suspensions were filtrated one more time using a 35-µM cell strainer for further purification before labelling the cells for flow cytometry analysis.
2.11. Isolation of human leukocytes from colon biopsies
To isolate human leukocytes from colonic biopsies of UC patients, biopsies were minced and cells filtered through a 70-μL cell strainer [Greiner Bio-One, Frickenhausen, Germany] followed by centrifugation at 1400 g for 5 min, and were resuspended in FACS buffer followed by labelling for flow cytometry analysis.
2.12. Detection of cytokines in cell culture supernatants, sera, and colon
The cell culture supernatant was collected after centrifuging the cell pellet of the cell culture after a 48-h incubation. The luminex assay was performed according to the manufacturer’s instructions using 50 µL of supernatant: hIFNγ Thermo Fisher Scientific Cat# EPXS010-10228–901, RRID:AB_2576179], hIL-4 [Thermo Fisher Scientific Cat# EPXS010-10225–901, RRID:AB_2576178], hIL-10 [Thermo Fisher Scientific Cat# EPXS010-10215–901, RRID:AB_2576174].
Whole blood was collected and allowed to clot at room temperature for 30 min. After a 10-min centrifugation at 2000 g and 4°C, the supernatant was transferred to a fresh polypropylene tube and used immediately or stored at –80°C.
To isolate human leukocytes from mouse colons, approximately 10-mm long sections of the terminal colon were dissected and cleaned of faeces with ice-cold PBS, and 500 µL of protease inhibitor cocktail [cOmplete, Roche, Penzberg, Germany] was added according to the manufacturer’s instructions. Samples were milled with a 5-mm stainless steel bead [Qiagen, Germany Hilden], centrifuged for 5 min at 3000 rpm, and supernatants were shock-frozen and stored at –80°C. MsCRP [Thermo Fisher Scientific, Cat# EPX01A-26045–901, RRID: AB_2575963]; msTGFß [Thermo Fisher Scientific, Cat# EPX01A-20608–901, RRID:AB_2575921]; msMCP-3 [Thermo Fisher Scientific, Cat# EPX01A-26006–901]; and msIL-6 [Thermo Fisher Scientific, Cat# EPXS010-20603–901 RRID:AB_2575933, Darmstadt, Germany] were detected by Luminex assay [MAGPIX; Luminex Corporation, Austin, TX, USA].
2.13. Detection of amino acids
Samples were prepared according to the manufacturer’s instructions. Following incubation of 100 µL of serum with internal standards for 5 min, 25 µL of 15% 5-sulphosalicylic acid was added and samples were centrifuged at 9000 g for 15 min at 4°C. Supernatants were filtered through a 0.2-µM membrane and 75 µL of lithium loading buffer was added. Samples were analysed by the amino acid analyser Biochrom 30+ [Biochrom, Cambridge, UK].
2.14. Immunohistochemistry
For immunochemistry [IHC] of the PBMCs, the cover-slides were coated with Poly-L-Lysine [PLL, 50 µL/mL, Sigma Aldrich, Deisenhofen, Germany] for 1 h at room temperature and washed with de-ionised water. Isolated PMBCs from healthy donors were plated onto the slides [106 cells in 100 µL] and incubated for 1 h at 37°C; 100 µL of 4% paraformaldehyde PFA [Sigma Aldrich, Deisenhofen, Germany] was added and incubated for 10 min at room temperature. Slides were washed twice with PBS, and blocking buffer [1% BSA in PBS] was added for 30 min at room temperature. Following the removal of blocking buffer, the first antibody was diluted in 100 µL blocking buffer [CD4 1:100, Kv1.3 1:200, for antibodies used see Table S5] and incubated overnight at 4°C, sealed with parafilm. Following two washing steps with PBS, the second antibody was added at a concentration of 1:400 in 100 µL blocking buffer for 1 h at room temperature. Slides were washed three times with PBS and sealed with cover slides with mounting medium [Anti-fade gold, Thermo Fisher, Darmstadt, Germany].
For the IHC of colon biopsies, the samples from different parts of the patients’ colons were fixed in 4% formaldehyde for 24 h before storage in 70% ethanol, and were embedded in paraffin. Samples were cut into 3-µm sections. After de-paraffinisation and rehydration with xylene and ethanol, an antigen retrieval in 1 mM EDTA was conducted. Incubation with antibodies followed the previous protocol.
2.15. Statistical analysis
Statistical analysis was performed with R [A Language and Environment for Statistical Computing; R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/]. Data were analysed using Cumming plots26 using the dabestr package, and are used in this work for data presentation and comparison. Cumming plots are a new generation of data analysis with bootstrap-coupled estimation [DABEST] plots that move beyond p-values. These plots can be used to visualise large samples and multiple groups easily. For correlation analysis, Pearson’s product-moment correlation was performed and a 95% confidence interval was applied. Heatmap was performed using R. Variables were also represented with mean, standard deviation, and median values. A two-sided Student’s t test and a confidence level = 95 was used to compare binary groups and, for more than two groups, analysis of variance [ANOVA] followed by TukeyHSD was conducted. Principal component analysis [PCA] was performed using the plyr, ChemometricsWithR, maptools, car, and rgeos packages. Orthogonal partial least square discrimination analysis [oPLS-DA] was performed using the ropls package.
3. Results
3.1. Kv1.3 expression on T cells of IBD patients
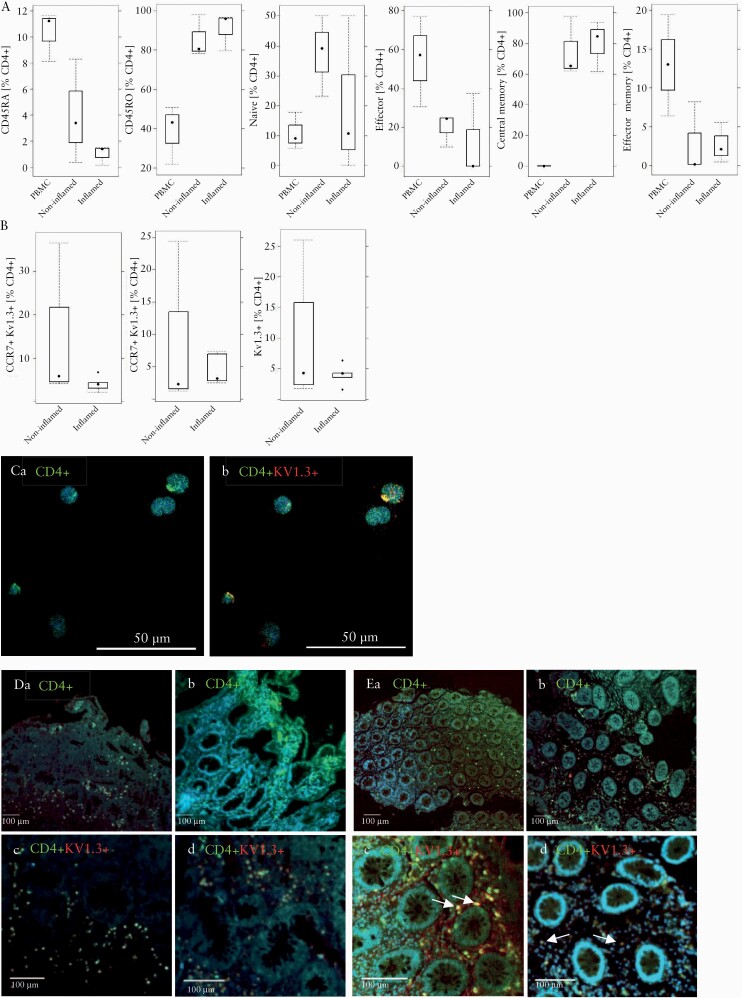
Kv1.3 expression has been shown to be greatly elevated in effector memory T cells in rheumatoid arthritis.16 Therefore, we first analysed in CD4+ T cells from IBD patients the relative frequencies of antigen-inexperienced [CD4+ CD45RA+], antigen-experienced [CD4+ CD45RO+], naïve [CD4+ CD45RA+, CD62L+, CCR7+], effector [CD4+ CD45RA+ CD62L- CCR7-], effector memory [CD4+ CD45RO+, CD62L-, CCR7-], and central memory cells [CD4+ CD45RO+, CD62L+, CCR7+]27 in the compartments of blood [i.e., PBMCs] and inflamed and non-inflamed colonic regions [for gating strategy see Supplementary Figure S1, available as Supplementary data at ECCO-JCC online]. As shown in Figure 1A, frequencies [in %] of antigen-experienced CD4+ cells were high in inflamed colon (% leukocytes 90 ± 9.41, mean ± standard deviation [SD]) and low in PBMCs [38.46 ± 15.0], as opposed to antigen-inexperienced CD4 T cells, which were high in PBMCs [10.31 ± 1.89] and low in inflamed colon [0.96 ± 0.76]. Central memory CD4 T cells represented the predominant population in the colon [80.00 ± 16.35], whereas the frequency of effector memory CD4+ T cells was low [<5%] in both the inflamed and non-inflamed colonic regions, which was also relatively lower than the 12.50 ± 21.56 found in the PBMC group. No significant difference between these T cell populations was observed between non-inflamed and inflamed regions of the colon. In summary, an inverse pattern of the relative prevalence of the T cell subtypes quantified here [aside from the naïve subtypes] appears to exist between the colon and PBMC compartments.
Figure 1.
Kv1.3 expression on T cells in PBMCs and colonic biopsies from IBD patients. [A] Distribution of subtypes of antigen experienced and inexperienced CD4+ T cells in PBMCs and colonic biopsies from IBD patients [n = 3]. Isolated leukocytes were subjected to flow cytometric analysis and frequencies of subtypes of CD4+ cells in isolated PBMCs, and inflamed and non-inflamed areas of the colon are depicted as boxplots. [B] Frequencies of Kv1.3-expressing effector and naïve/central memory cells in biopsies of IBD patients [n = 6] in inflamed and non-inflamed [n = 5] areas depicted as boxplots. Boxes represent upper and lower quartiles, whiskers represent variability, and outliers are plotted as individual points. [C] Qualitative IHC of CD4+ T cells expressing Kv1.3 in PBMCs. [a] CD4; [b] CD4 and Kv1.3. Qualitative IHC of colonic biopsies from UC [D, n = 1] and CD [E, n = 1] patients. [a] CD4, non-inflammatory region. [b] CD4, inflammatory region, [c] CD4 and Kv1.3, non-inflammatory region, [d] CD4 and Kv1.3, inflammatory region. CD4 was stained with Alexa Fluor 433 [green] and Kv1.3 with Alexa Fluor 647 [red], cell nuclei with Dapi [blue]. PBMC, peripheral blood mononuclear cells; IBD, inflammatory bowel disease; UC,ulcerative colitis; CD, Crohn’s disease.
To analyse Kv1.3 expression levels in the compartment of the colon, biopsies from non-inflammatory [n = 5] and inflammatory regions of UC patients [n = 6] were analysed by flow cytometric analysis. As shown in Figure 1B frequencies of Kv1.3-expressing central memory/naïve T cells, effector memory/effector cells were similar in inflamed and non-inflamed regions. This observation appears to corroborate a previous observation that non-inflammatory regions of the colon already exhibit a certain activation status.28 To further examine the expression of Kv1.3 in CD4+ T cells, PBMCs from a healthy donor were analysed by IHC as described in Materials and Methods. As shown in Figure 1Ca, PBMCs stained with anti CD4+ antibodies exhibited expression concentrated in focal areas of the outer membrane and Kv1.3 expression co-localised with CD4+ expression [Figure 1Cb]. To corroborate the expression of Kv1.3, biopsies from an inflamed region from a CD and from a UC patient were compared by IHC analysis. As shown in Figure 1D and E, co-expression of CD4+ and Kv1.3 was detected in non-inflamed regions and in inflamed regions in UC and CD biopsies. These results indicate that T cells are active in the immunological compartment of the colon and that this activity might be suppressed by a selective Kv1.3 inhibitor, thus suggesting Kv1.3 as a potentially attractive target for IBD treatment.
3.2. Development of a small-molecule inhibitor of Kv1.3
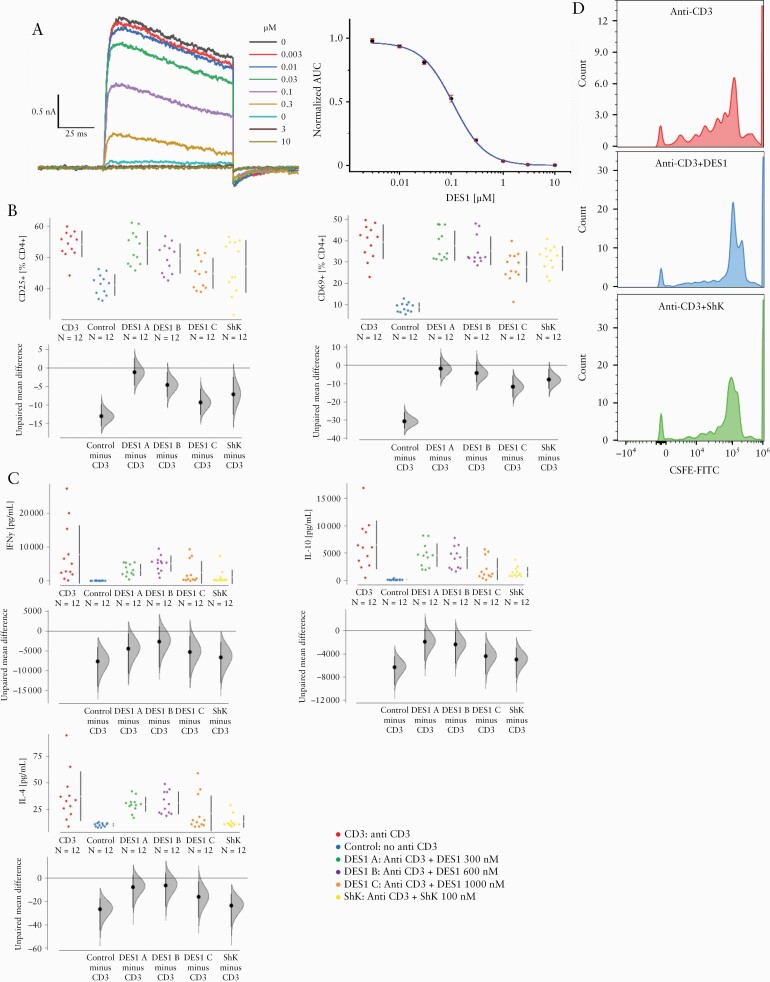
The small molecule inhibitor of Kv1.3, DES1, was designed to target and stabilise an inactive conformation of Kv1.3 that was discovered using long-timescale molecular dynamics [MD] simulations. DES1 was identified as part of a medicinal chemistry campaign that started from an MD-based virtual screening hit. After finding this hit, and guided in part by further MD simulations, we iteratively performed successive rounds of design aimed at maximising Kv1.3 potency and ion channel selectivity while maintaining physicochemical, drug metabolism, and pharmacokinetic properties that are suitable for in vivo administration. As shown in Table S1, available as Supplementary data at ECCO-JCC online, DES1 is a potent Kv1.3 inhibitor with exquisite selectivity against a variety of potassium, sodium, and calcium voltage-gated ion channels. We further tested the in vitro potency of DES1 by automated patch clamp experiments in mammalian CHO cells expressing the human or rat channel recombinantly. The resulting IC50 values were 111 ± 0.03 nM [human, n = 17; Figure 2A] and 155 ± 0.05 nM [rat, n = 2; the rat and mouse channels are nearly identical].
Figure 2.
DES1 inhibits Kv1.3 activation in vitro. [A] Analysis of inhibition of Kv1.3 by automated patch clamp analysis in mammalian CHO cells recombinantly expressing the human channel. Whole-cell current recordings at increasing concentrations [left] and a single, representative dose-response curve for the human channel [right]; each data point is an average of triplicate recordings, and the area under curve [AUC] measured at the 20th pulse was used to determine the IC50. [B, C] DES1 inhibits T-cell activation and cytokine production in a concentration-dependent manner. PBMCs [1 E-06 cells] were incubated in 1.5 mL RPMI for 48 h in the absence [Control] or presence [CD3] of anti-CD3 antibodies [0.31 ng/mL] and increasing concentrations of DES1 [DES1 A: 300 nM, DES1 B:600 nM, DES1 C: 1000 nM]. ShK [100 nM] served as a control. [biological n = 2, technical n = 3] [B] Flow cytometric analysis of frequencies of activated CD4+ T cells [CD25+ and CD69+] depicted as Cumming plots. [C] Analysis of cytokine secretion by Luminex assay. The upper part of the plot presents each data point in a swarm plot. In the lower panel of the plots, the effect sizes are shown. The 0 point of the difference axis indicates the mean of the reference group [CD3]. The dots show the difference between groups [effect size]. The shaded curve shows the entire distribution of excepted sampling error for the difference between the means. The error bar in the filled circles indicates the 95% confidence interval [bootstrapped] for the difference between means. [D] CellTrace carboxyfluoresceinsuccinimidylester [CFSE] dilution of CD4+ T cells. PBMC, peripheral blood mononuclear cells.
3.3. Suppression of T cell activity and proliferation by DES1 ex vivo
To examine the effect of DES1 on the activity of T cells in vitro, PBMCs were isolated from blood samples from healthy donors and cultivated in the presence or absence of anti-CD3 antibody [0.31 ng/mL] for 48 h, as described in Materials and Methods [donors: n = 2, replicates: n = 2, measurement n = 3]. Concentrations of DES1 ranged from 300 to 1000 nM [DES1 A: 300 nM, DES1 B: 600 nM, DES1 C: 1000 nM]. Furthermore, 100 nM ShK was used as a positive control for inhibition of Kv1.3-mediated cytokine secretion [cytokine secretion was already significantly inhibited at only 10 nM ShK, and the expression of the surface activation markers was also affected by ShK at 10 nM, but insignificantly so; Figure S2, available as Supplementary data at ECCO-JCC online]. Frequencies of CD4+, CD25+, CD4+, and CD69+ T cells were analysed by flow cytometric analysis as markers of T cell activation.29 As expected, frequencies of both cell types increased significantly upon activation by anti-CD3 antibodies, as shown in Cumming plots26 [Figure 2B]. Additional analysis by ANOVA followed by TukeyHSD corroborated the significance [CD25+: p = 6E-06; CD69+: p = 0E-07]. DES1 inhibited the expression of these surface markers in a concentration-dependent manner, and the inhibition reached significance at 1000 nM [CD25+: p = 0.001; CD69+: p = 0.0005]. As T-cell activation by anti-CD3 antibodies results in cytokine release,30 supernatants were additionally analysed by Luminex assays for IFNy, IL-4, and IL-10 levels. As shown in Figure 2C, anti-CD3 significantly increased the secretion of cytokines [IFNy: p = 0.0006; IL-4: p = 0.002; IL-10 p = 2 E-05]. DES1 inhibited the secretion of these cytokines and the decline reached significance at a concentration of 1000 nM [IFNy: p = 0.02; IL-4: p = 0.03; IL-10: p = 0.0002].
In order to examine whether DES1 inhibited the proliferation of CD4+ T cells, proliferation was examined by CellTrace CFSE dilution as described in Materials and Methods. As shown in Figure 2D, DES1 and ShK inhibited, with significance, the proliferation of CD4+ T cells at concentrations of 600 nM and 100 nM, respectively.
3.4. Validation of DES1 in the NSG-UC mouse model
To validate DES1 as a potential therapeutic for treatment of UC, the efficacy of DES1 was tested in vivo and compared with the efficacy of the standard therapeutics infliximab and tofacitinib in head-to-head studies in the NSG-UC mouse [ms] model. Readouts were the clinical, colon, and histological scores, serum levels of tryptophan, and levels of msMCP3, msCRP, msTGFß, and msIL-6 chemokines and cytokines measured in the colon samples from the mice. NSG mice were reconstituted with PMBCs from UC patients [n = 6] exhibiting a Simple Clinical Colitis Activity Index [SCCAI31] between 3 and 12 and challenged as described in Materials and Methods [for patient characteristics and groups of animals, see Table S2 available as Supplementary data at ECCO-JCC online]. Seven days after reconstitution, the NSG-UC mice were divided into different groups: one was unchallenged control [Control]; the other groups were challenged with ethanol and treated by intraperitoneal injection with PBS [PBS], 5 mg/kg DES1 in PBS, 6 mg/kg infliximab in PBS, or with 5 mg/kg tofacitinib in PBS. Six to seven animals were assigned to each group. Mice were monitored for weight loss and diarrhoea throughout the experiment and scored according to a clinical score as described in the Materials and Methods [for complete dataset see Table S3 available as Supplementary data at ECCO-JCC online].
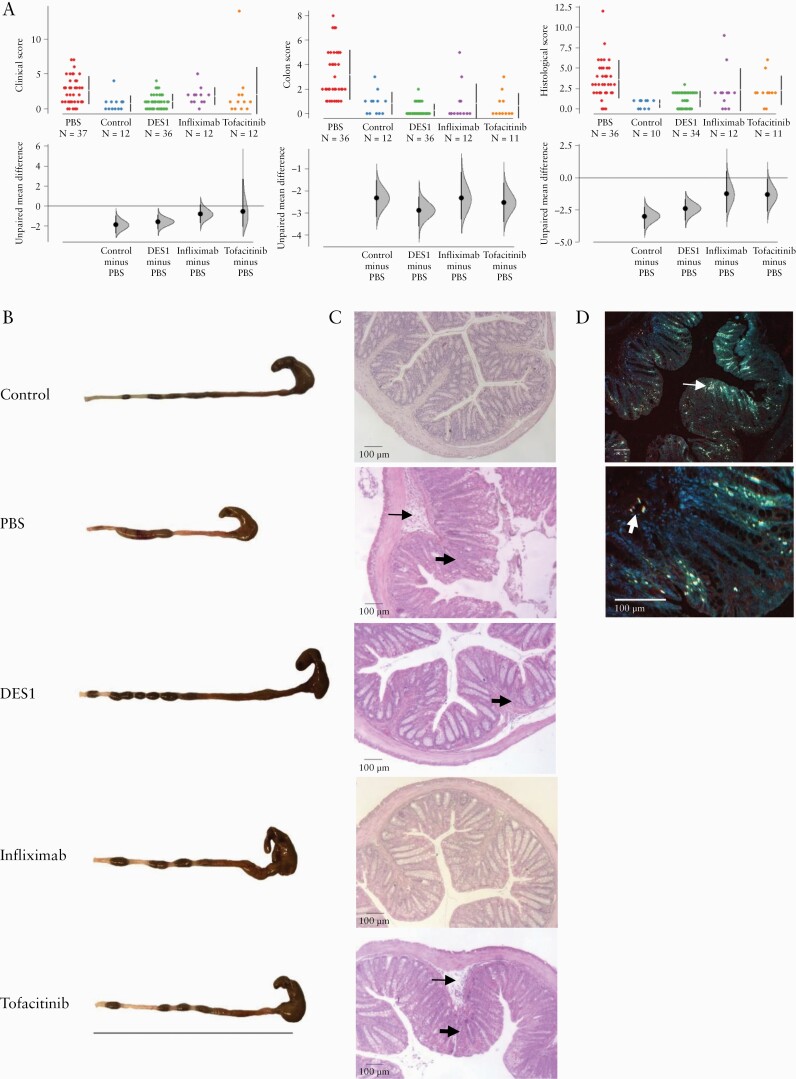
In response to rectal challenge with ethanol, the clinical score of mice in the PBS group increased significantly due to weight loss and occurrence of diarrhoea as compared with the unchallenged control group, and the differences in effect sizes were significant. All other groups benefited from the respective treatments and exhibited just a slight increase of the clinical score; however, only the effect sizes in the DES1 group were significantly different from the PBS group [Figure 3A].
Figure 3.
NSG-UC mice benefit from treatment with DES1, infiximab, and tofacitinib. NSG mice were reconstituted with PBMCs from UC donors [n = 6] and were left unchallenged [Control, n = 12], or challenged with 10% ethanol at Day 8, 50% ethanol at Day 15, and treated on Days 7–9 and 14–17 with PBS [PBS, n = 37], 5 mg/kg DES1 [DES1, n = 36], or 5 mg/kg tofacitinib [tofacitinib [n = 12]. Infliximab was administered on Days 7 and 14 at a concentration of 6 mg/kg in PBS. [A] Clinical, colon, and histological scores depicted as Cumming plots. The upper part of the plot presents each data point in a swarm plot. In the lower panel of the plots, the effect sizes are shown. The 0 point of the difference axis indicates the mean of the reference group [PBS]. The dots show the difference between groups [effect size]. The shaded curve shows the entire distribution of excepted sampling error for the difference between the means. The error bar in the filled circles indicates the 95% confidence interval [bootstrapped] for the difference between means. [B] Representative macrophotographs of NSG-UC mice. Bar corresponds to 10 cm. [C] Representative microphotographs of haematoxylin-stained colonic sections from NSG-UC mice. Arrows indicate oedema and influx of inflammatory cells, bold arrows indicate goblet cell loss and fibrosis. [D] IHC of CD4+ T cells expressing Kv1.3 in a histological section of a PBS-treated mouse. Arrow indicates CD4+ positive cells, bold arrows double-positive CD4+ Kv1.3+ cells. PBMC, peripheral blood mononuclear cells; UC,ulcerative colitis; IHC, immunohistochemistry.
At Day 21 mice were sacrificed and the colons were removed, inspected macroscopically, and scored according to a colon score described in Materials and Methods. As shown in Figure 3B the visual pathological appearance of the colons and the colon scores supported the observed clinical score. The colons of mice from the PBS group exhibited pathological alterations like unformed pellets, dilation and, in severe cases, shortening as compared with the control animals. All treated animals thus appeared to benefit from treatment compared with the untreated PBS group [Figure 3A].
The histological analyses of the colons revealed severe disruption of the colon architecture in the PBS group, as indicated by a mixed infiltrate of inflammatory cells, development of oedema, loss of goblet cells, and of fibrosis in the PBS group. IHC of sections of the PBS-treated mice exhibited influx of CD4+ T cells into the mucosa, some of which expressed Kv1.3. Treatment with DES1, tofacitinib, or infliximab improved all these pathological manifestations with varying levels of efficacy and, further, demonstrated that DES1 had an efficacy comparable to infliximab and tofacitinib. Whereas the influx of inflammatory cells was much less pronounced in the DES1 group and the colon architecture appeared almost normal, mice treated with either infliximab or tofacitinib still exhibited clear signs of inflammation [Figure 3C]. All treated mouse groups, including the DES1-treated group, exhibited fibrosis, which indicated still ongoing inflammatory [e.g., wound-healing] processes. Colon sections were subjected to histological scoring as described in Materials and Methods. The histological score reflects and quantifies these previous observations of changes in the inflammatory processes upon therapeutic treatment [Figure 3A]. IHC analysis revealed the influx of CD4+ cells into the mucosa and the expression of Kv1.3 on cells.
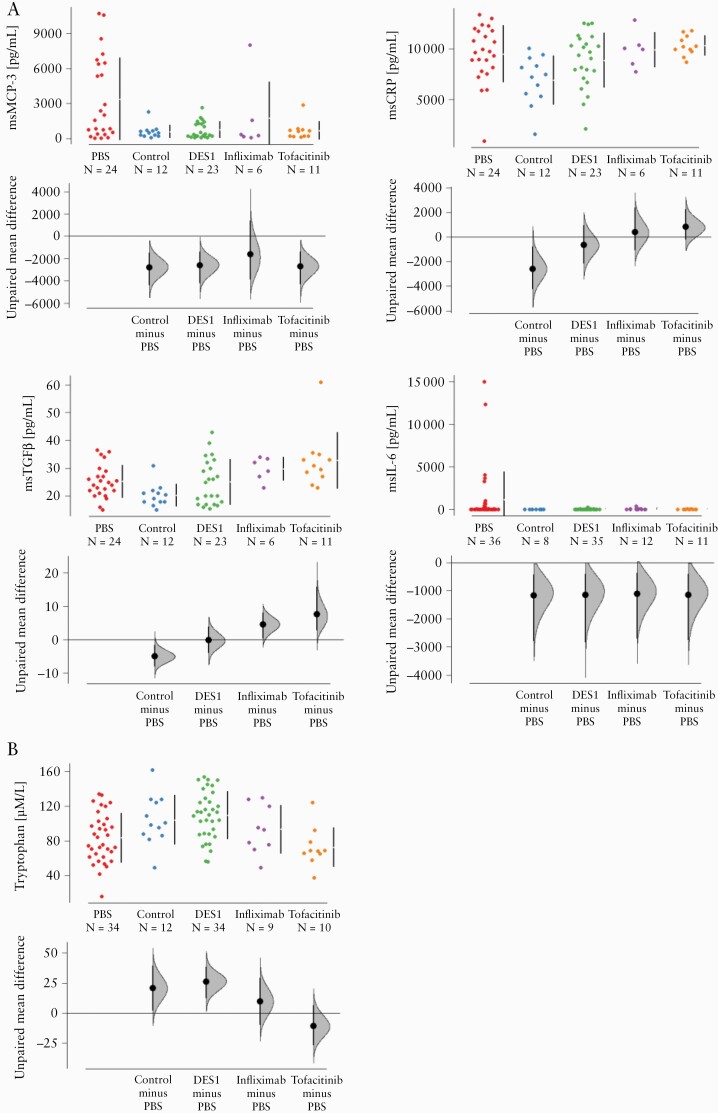
To further assess the anti-inflammatory activity of DES1, infliximab, and tofacitinib, colonic levels of the inflammatory markers msCRP, msMCP-3, msTGFbeta, and msIL-6 were analysed. These colonic markers had been previously identified as significant in analysis of sera of UC patients versus non-IBD patients32 and in the NSG-UC mouse model [work submitted for publication by L-MU]. Proteins of the colon were extracted and subjected to Luminex analysis. Upon challenge with ethanol, the levels of msMCP3, msCRP, and msTGFbeta all increased significantly [see Figure 4A] in the PBS group, whereas msIL-6 only increased in some of the challenged mice. Treatment with DES1, infliximab, or tofacitinib had a significant impact on colonic msMCP-3 levels; upon treatment, the MCP-3 levels in these groups almost reached the level of the control group. However, levels of msCRP and msTGFbeta did not decline, indicating ongoing inflammatory processes in all treated groups. As we have previously observed that inflammation had a significant impact on metabolism,24 serum tryptophan levels were measured as an additional marker for inflammation [tryptophan is an essential amino acid whose metabolites modulate inflammation].33 Challenge with ethanol led to a decrease of the tryptophan level, indicating increased tryptophan metabolism, and subsequent treatment with DES1 brought levels back to normal. This effect was not observed in the tofacitinib or infliximab groups, potentially supporting a broader efficacy of DES1 in reducing inflammation [Figure 4B].
Figure 4.
Treatment of NSG-UC mice with DES1, infliximab, and tofacitinib affect inflammatory markers. NSG-UC mice were treated as described in Figure 3. [A] msCRP-3, msTGFß, msCRP, msIL-6 levels in colon extracts of mice depicted as Cumming plots. [B] Serum tryptophan levels depicted as Cumming plots. The upper part of the plot presents each data point in a swarm plot. In the lower panel of the plots, the effect sizes are shown. The zero [0] point of the difference axis indicates the mean of the reference group [PBS]. The dots show the difference between groups [effect size]. The shaded curve shows the entire distribution of excepted sampling error for the difference between the means. The error bar indicates the 95% confidence interval [bootstrapped] for the difference between means [filled circles].
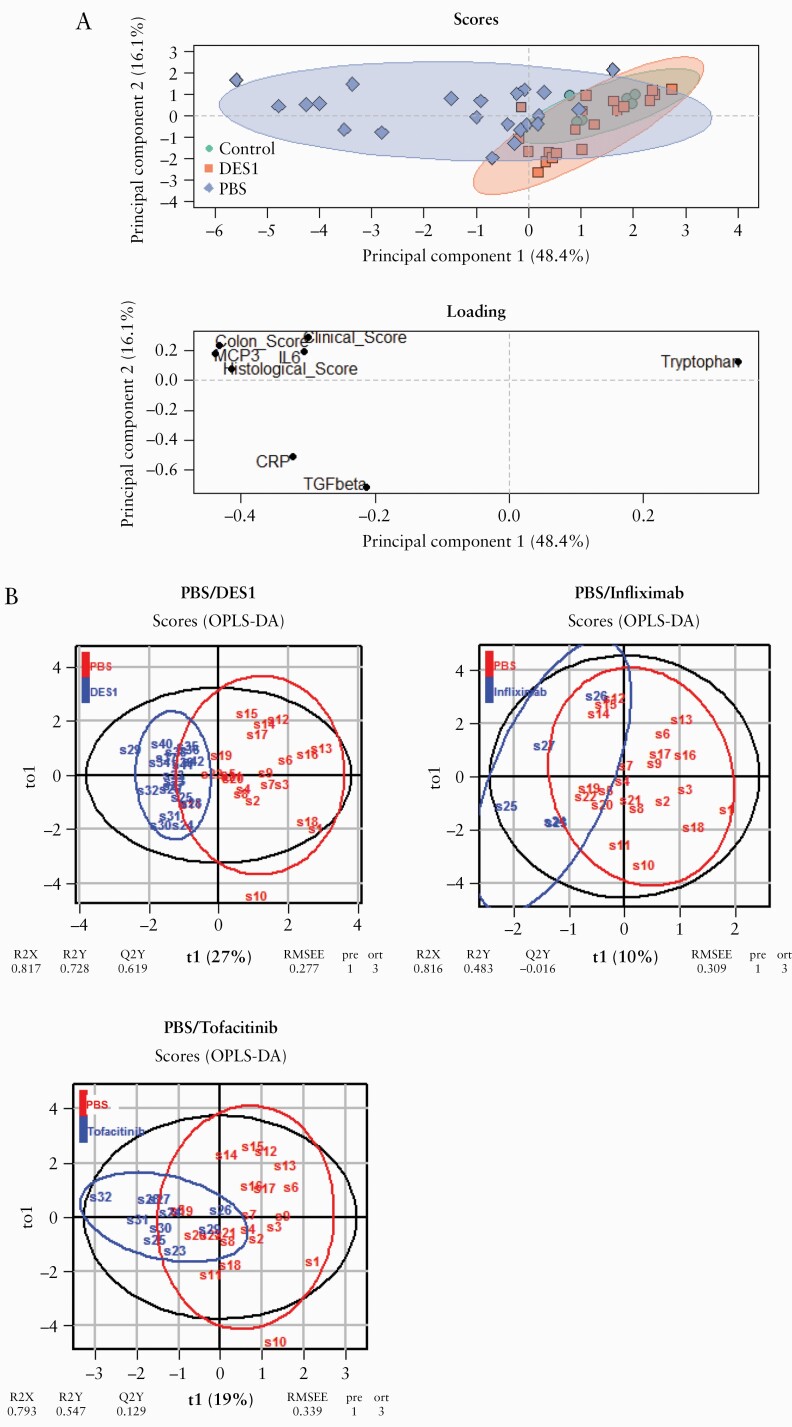
So far, all single parameters indicated efficacy of DES1. To combine all parameters in the evaluation of DES1, a principal component analysis [PCA] was performed. This technique is widely used to analyse inter-relation of variables, reduce the dimensionality in an interpretable way, emphasise variation, and capture patterns in a dataset.34 Here, the analysis included the clinical, histological, and colon scores and the levels of msMCP3, msCRP, msTGFbeta, msIL-6, and tryptophan. As shown in Figure 5A, mice of the control group clustered closely together, and upon challenge these dots were widely spread out, reflecting the variability in this group. Treatment with DES1 resulted in distinctly different clustering from the PBS group; however, these dots were not completely overlying the control group, suggesting that these mice did not revert to a completely non-inflammatory level upon treatment with DES1.
Figure 5.
DES1 treated NSG-UC mice are statistically distinct from the challenged control group. Mice were treated as described in Figure 3. The clinical, colon, and histological scores, levels of msIL-6, msTGFß, msCRP, msMCP-3, and tryptophan were used for modelling. [A] Principal component analysis [PCA]; [B] orthogonal partial least square discrimination analysis [oPLS-DA]. R2X: fraction of variation of X variables explained by the model; R2Y: fraction of variation of Y variables explained by the model, Q2Y: fraction of variation of the Y variables predicted by the model; RMSEE: root mean square error of estimation.
In order to compare the efficacy of DES1 with infliximab or tofacitinib, an orthogonal partial least square discriminating analysis [oPLS-DA] was performed using the same parameters as in the PCA analysis. oPLS-DA is a latent variable regression method that evaluates the differences between groups. Here, oPLS-DA was used to evaluate the differences between the PBS group versus DES1, infliximab, or tofacitinib. The comparison of DES1 with the PBS group revealed a clear distinction, as indicated by a high R2X value [fraction of variation of X variables explained by the model] of 0.817, a high R2Y value [fraction of variation of Y variables explained by the model value] of 0.728, a high R2Q value [fraction of variation of the Y variables predicted by the model] of 0.619, and a low RMSEE [root mean square error of estimation] value of 0.277 [Figure 5B]. The pR2Y and the pR2Q values below 0.05 corroborated the validity of the oPLS model and indicated that the separation of the DES1 group from the PBS group was significant. The separation of infliximab and tofacitinib from the PBS group was less pronounced, as indicated by the lower overall R2X, R2Y, and R2Q values [infliximab: 0.816, 0.483, and –0.016, respectively; tofacitinib: 0.793, 0.547, and 0.129, respectively], and by the higher RMSEE values [infliximab: 0.309; tofacitinib: 0.339]. These results suggest that DES1 may be more efficacious than the other two compounds in the NSG-UC model.
3.5. Patient profiling
To assess whether the responsiveness to DES1 could be ascribed to a certain inflammatory profile of patients we first examined the expression levels of Kv1.3 on CD4+ T cells in PBMCs of IBD patients as compared with non-IBD donors [for basic patient demographics see Table 1].
Table 1.
Basic patient demographics,
| UC | CD | Non-IBD | |
|---|---|---|---|
| n = 20 | n = 7 | n = 8 | |
| Age [years] | |||
| Mean [SD] | 35.77 [13.57] | 49.28 [20.8] | 30.17 [15.33] |
| Range | 24–77 | 26–74 | 21–65 |
| Gender [% male] | 60 | 42 | 25 |
| Duration of UC/CD [years] | |||
| Mean [SD] | 10.56 [8.78] | 11.14 [7.05] | |
| Range | 1–23 | 0–20 | |
| SCCAI31/CDAI44 | |||
| Mean [SD] | 5.89 [3.26] | 144 [152.9] | |
| Range | 0–12 | 0–349 | |
| Treatment [current] | |||
| TNFα-blocker | 4 | 1 | |
| Vedolizumab | 8 | 3 | |
| Mesalazine | 6 | ||
| Glucocorticoids | 2 | 1 | |
| JAK Inhibitor | 2 | 1 |
IBD, inflammatory bowel disease; UC,ulcerative colitis; CD, Crohn’s disease; SD, standard deviation; SCCAI, Simple Clinical Colitis Activity Index; CDAI, Crohn’s Disease Activity Index.
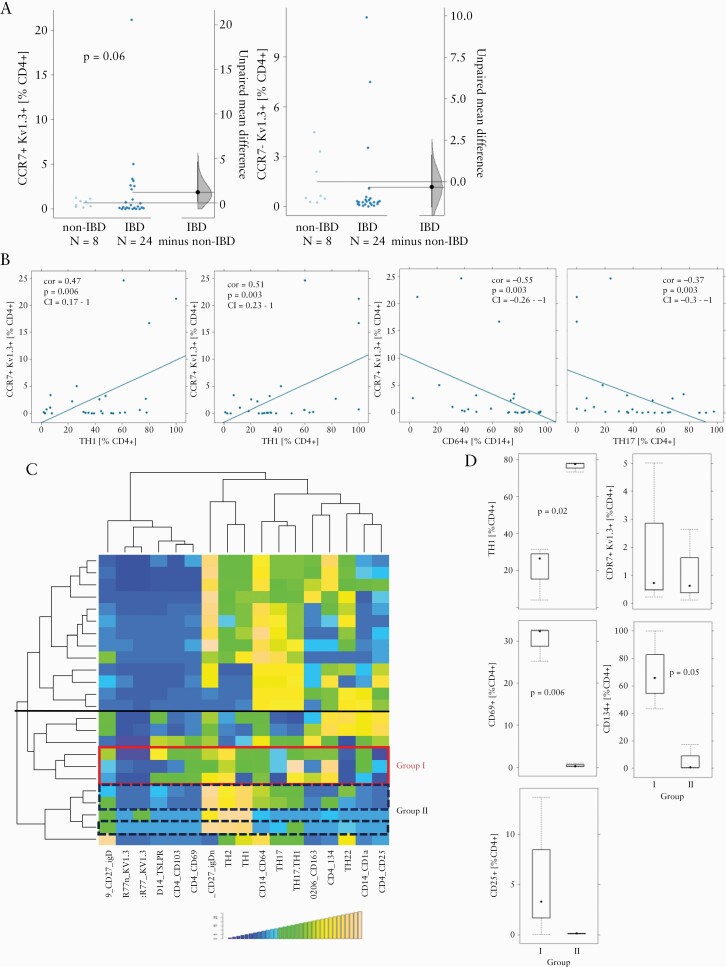
As shown in Figure 6A naïve/central memory CD4+ T cells of IBD patients displayed elevated levels of Kv1.3 expression and the difference almost reached significance. In contrast, levels of Kv1.3 expression were similar in effector/effector memory cells in both groups. We then analysed whether Kv1.3 expression correlated with a particular subgroup of inflammatory immune cells. As shown in Figure 6B, Kv1.3 expression levels correlated significantly with TH1 [cor = 0.47, p = 0.006, 95% CI = 0.17–1], TH2 [cor = 0.51, p = 0.003, 95% CI = 0.23–1], TH17 [cor = -0.37, p = 0.03, 95% CI = -0.3–1] and CD14+ CD64+ M1 monocytes [cor = -0.55, p = 0.003, 95% CI = -0.26–1]. The positive and negative correlations of Kv1.3 expression with the two pairs of immune cells, TH1, TH2 and TH17, CD14+, respectively, suggested that Kv1.3 expression plays a key role in the TH1/TH2-driven inflammation process, as described previously.7 The immunological profiling of the six UC donors selected for reconstitution in the NSG-UC mice indicated that these donors clustered in the TH1/TH2-driven process of inflammation [Figure 6C]. Closer inspection revealed, moreover, that the six donor profiles clustered in two different subgroups, referred to as group I and II, signified by their marked differences in relative frequencies of TH1 [p = 0.02] and activated CD4+ T cells (CD69+: p = 0.006; CD134+: p = 0.05; CD25+: p = n.s. [not sinificant]; Figure 6C. No difference was observed in the frequencies of Kv1.3-expressing naïve/central memory T cells. To assess whether patient responsiveness to DES1 differed between these two groups, mice were divided according to the donors clustering into groups I and II and analysed by oPLS-DA using, again, the clinical, colon, and histological scores, along with colonic and serum levels of IL-6 and tryptophan, respectively, as variables.
Figure 6.
Responsiveness to DES1 correlated with activated CD4+ T cells. [A] Frequencies of Kv1.3-expressing effector and memory/naive T cells in PBMCs from IBD [n = 27 patients and non-IBD [n = 8] donors depicted as Cumming plots.26 The left part of the plot presents each data point in a swarm plot. In the right part panel of the plots, the effect sizes are shown. The 0 point of the difference axis indicates the mean of the reference group [non-IBD]. The dots show the difference between groups [effect size]. The shaded curve shows the entire distribution of excepted sampling error for the difference between the means. The error bar in the filled circles indicates the 95% confidence interval [bootstrapped] for the difference between means. [B] Correlation analysis of frequencies of Kv1.3-expressing T cells with TH1, TH2, TH17, and M1 monocytes displayed as scatter plots [TH1 and TH2 frequencies originate from distinct gating procedures and their sum thus exceeds 100%]. Method = Pearson, numbers display Pearson’s product-moment correlation values [cor], p-values [p], and 95% confidence interval [CI]. [C] Hierarchically clustered heatmap of 24 patients. The horizontal black line separates the two main subgroups of inflammation. The coloured boxes indicate profiles of donors selected for reconstitution [red: group I; blue, dashed line: group II]. [D] Frequencies of subtypes of immune cells in group I and group II depicted as boxplots. Boxes represent upper and lower quartiles, whiskers represent variability, and outliers are plotted as individual points. PBMC, peripheral blood mononuclear cells; IBD, inflammatory bowel disease.
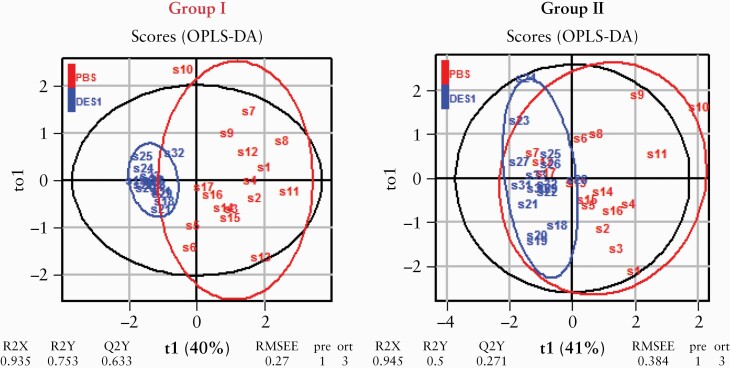
As shown in Figure 7 both DES1-treated groups clustered distinctly from the PBS-treated group, suggesting that patients with a an immunological profile clustering in the TH1/TH2-driven process of inflammation may be the preferential target group for a potential DES1-based treatment of UC. To visualise and quantify the separation of the groups, an OPLs-DA analysis was performed. In both models the separation of the groups was significant, as indicated by the pR2Y and pR2Q values below 0.05. Mice reconstituted with PBMCs from donors in group I benefited relatively more from the treatment as indicated by higher R2X [group I: 0.95 vs group II: 0.94], R2Y [group I: 0.75 vs group II: 0.5], higher R2Q [group I: 0.63; group II: 0.27], and higher RMSEE levels [group I: 0.27 vs group II: 0.38]. These data altogether indicate that elevated levels of activated CD4+ T cells may be taken as a potential indicator of UC patient responsiveness to DES1 treatment.
Figure 7.
Analysis of responsiveness to DES1 in mice reconstituted with PBMCs from subgroups of patients by orthogonal partial least square discrimination analysis [oPLS-DA]. R2X: fraction of variation of X variables explained by the model; R2Y: fraction of variation of Y variables explained by the model; Q2Y: fraction of variation of the Y variables predicted by the model; RMSEE: root mean square error of estimation. PBMC, peripheral blood mononuclear cells.
To examine the effects of DES1 on the activation states of T cells in the spleen and colon, human leukocytes were isolated from these organs at autopsy and subjected to flow cytometric analysis [Supplementary Figure S3, available as Supplementary data at ECCO-JCC online]. We further parsed the data according to whether the donor was in group I or group II. In cells from the spleen, the only statistically significant effect in response to DES1 treatment was a reduction of effector memory CD4+ T cells in group I, whereas the levels of CD4+ CD69+ and CD4+ CD134+ T cells were not affected by DES1 treatment for either group. For colonic leukocytes, by way of contrast, levels of both CD4+ CD69+ and CD4+ CD134+ T cell subtypes decreased, albeit not significantly, with DES1 treatment and only for group I. This observation may nonetheless suggest that the apparent higher efficacy of DES1 observed for group I may be attributed to functional inhibition of apparently strongly Kv1.3-dependent CD4+ CD69+ and CD4+ CD134+ T-cell subpopulations. For the colon samples, the observed differences between the DES1-treated and PBS-treated groups were, however, not significant, due to the overall low number of leukocytes. Colonic segments from the individual mice within each treatment group of six mice, all reconstituted with PBMCs from the same donor, were thus pooled before isolation of leukocytes. With n being the total number of experiments, and with each experiment using PBMCs from a distinct donor, this analysis was based on the following number of [donor] replicates: group I: PBS n = 3, DES1 n = 3; group II: PBS n = 3; DES1 n = 3]. Interestingly, in group II colonic leukocytes, the number of effector memory T cells, whose function upon terminal differentiation may depend nearly exclusively on Kv1.3,11 declined upon DES1 treatment, but again without reaching significance, due to the limited number of samples.
4. Discussion
4.1. Expression of Kv1.3
Recently, the activity of T cells has become a compelling intervention point in IBD. The approved Janus kinase 1/3 inhibitor [JAK] tofacitinib, which relays the activation signals intracellularly, was at the forefront of development and is now used when treatment with biologics fails. Treatment with tofacitinib is, unfortunately, accompanied by potential side effects such as severe infections, risk of venous and arterial thromboembolic events, and elevated lipid levels.35 The central role of Kv1.3 channels in T cell activation, cytokine production, and proliferation suggest more subtle immune-modulatory routes of potential therapeutic intervention upstream of the JAK signalling pathways.36 Peptide toxins isolated from the sea anemone Stichodactyla helianthus were first identified as powerful inhibitors of Kv1.3, and subsequently a modified variant, ShK186 [i.e., dalazatide], was successfully tested in a Phase 1b clinical trial for plaque psoriasis.17 One drawback of a peptide-based IBD treatment is lack of oral availability, which could be challenging for patients who would need to integrate an injectable medication into their daily lives. Therefore, the successful clinical development of an orally available inhibitor of Kv1.3 has the potential to be an attractive treatment option for many IBD patients with active disease.
In this study, we validated Kv1.3 as a promising target of IBD treatment. Antigen-mediated T cell activation results in assembly of signalosomes, which cluster T cell receptors [TCRs] along with CD3, CD4/CD8, OX40, and interleukin receptors. A previous study demonstrated that Kv1.3 co-localises with CD4 at the cap of effector T cells,16 and the IHC stainings of PBMCs performed in this study are in line with these observations. Because interaction of TCRs with antigens induces Kv1.3 to localise at the outer T cell membrane,16 we assumed that IBD patients might also exhibit increased expression of Kv1.3 on their T cell membranes. Indeed, elevated Kv1.3 expression was found in naïve or central memory T cells [CD4+ CCR7+] isolated from PBMCs from IBD patients as compared with similarly isolated T cells originating from non-IBD patients. In the IBD patients, Kv1.3 expression levels correlated positively with the frequencies of TH1 and TH2 cells, but negatively with frequencies of TH17 and CD14+ CD64+ cells. This corresponds to our previous findings of the existence of a TH1/TH2-driven process of inflammation in UC,7 but it does not rule out that a TH17/CD14+-driven process of inflammation in UC patients could be positively affected by a Kv1.3-based therapeutic intervention.37
As the colon is the essential inflammatory compartment in IBD, we further analysed Kv1.3 expression levels in biopsies obtained from non-inflammatory and inflammatory colonic regions of UC patients. Here, we found no significant difference in Kv1.3 expression in effector memory or effector cells [CD4+ CCR7–] or central memory/naïve cells [CD4+ CCR7+], regardless of whether the biopsies were taken from non-inflammatory or inflammatory regions of the colon. This observation is somewhat in contrast to a previous finding of increased Kv1.3 channel activity [due to elevated Kv1.3 expression] in CCR7– effector memory cells in synovial fluid from rheumatoid arthritis [RA] patients.16 This difference of Kv1.3 expression levels in these T cell subsets, albeit between two distinct inflammatory diseases, could be explained by observations that non-inflamed regions of the colon are not necessarily healthy. Rather, the non-inflamed regions could represent an intermediate inflammatory state on the way to becoming highly inflamed,28 whereas such elevated inflammation might already be present in the entire joint of RA patients.16 IHC from the inflammatory region of the colon also confirmed the co-expression and co-localisation of Kv1.3 and CD4 on the caps of T cells, thus further corroborating that Kv1.3 might be an attractive therapeutic target in IBD.
4.2. Ex vivo efficacy of DES1
Exposure to anti-CD3 antibodies mimics the activation of the signalosome by antigens and induces expression of the activation markers IL-2R [CD25] and the galectin receptor [CD69].24,38 Although the function of the galectin receptor has yet to be elucidated, it is well established that IL-2/IL-2R mediate the proliferation of T cells.39 In addition, the activation of T cells by anti-CD3 antibodies results in increased expression of various cytokines, including IFNy.30 As shown in this study, DES1 and ShK are potent inhibitors of all three outcomes, as expected based on ~1000-fold differences in potencies against Kv1.3 obtained from patch clamp measurements. DES1 required ~10x–100x higher concentrations than ShK to elicit its ex vivo efficacy [IC50 values of ~100 nM vs ~50–500 pM for DES1 and ShK, respectively, against Kv1.3 obtained from electrophysiology with the channel expressed in mammalian cells].
4.3. In vivo efficacy of DES1
To prove that the in vitro activity can be translated to in vivo models, the efficacy of DES1 was tested in the NSG-UC mouse model and compared with the efficacy of infliximab and tofacitinib. Like infliximab and tofacitinib, treatment with DES1 reduced most of the pathological parameters such as the clinical, colon, and histological scores, and chemokine and cytokine levels of msMCP-3 and msIL-6, whereas the serum level of the inflammatory [metabolic] marker tryptophan returned to its normal level. msTGFß and msCRP levels remained unaffected, suggesting that some ongoing inflammation remained following DES1 treatment—as well as following treatment with infliximab or tofacitinib—in this model. TGFß is a promoter of fibrosis,40 and the residual inflammation may reflect ongoing remodelling and wound healing processes.15 PCA and oPLS-DA analyses indicate a distinct discrimination between the untreated challenged control group and the DES1-treated group. Moreover, they suggest that DES1 appears as efficacious as infliximab or tofacitinib while offering a new pathway of potential therapeutic intervention in IBD patients.
4.4. Patient profiling
The unsatisfactory translatability of data obtained in preclinical studies to later human trials presents a major challenge in drug development. This especially applies to IBD, as conventional animal models hardly reflect the variability of disease manifestations observed in patients.41 Studies in the NSG-UC mouse model in combination with immune profiling of donors offer the opportunity to narrow this gap.7 In this study, immune profiles of patients whose PBMCs were selected for reconstitution were correlated with responsiveness to DES1 in the NSG-UC mouse model. All these patients clustered in the TH1/TH2-driven group and all mice benefited from treatment with DES1. However, within this main group, patients clustered in two different subgroups signified by differences in levels of frequencies of TH1 and activated CD4+ T cells. When groups of mice reconstituted with these donors were divided according to the subgroups of patients, oPLS-DA analysis revealed that both groups responded significantly but that mice from the group displaying higher levels of activated T cells benefited more. This observation is in line with the mode of action of DES1 to suppress the activation of T cells. Analysis of colonic leukocytes supported this observation in that DES1 treatment decreased levels of activated T cells in the colon in group I. We did not observe this effect for splenic leukocytes. This discrepancy is most likely a result of the mice being reconstituted with donor PMBCs 7 days before the first treatment with DES1. Human T cells probably become activated immediately upon engraftment of the spleen [or are already active] and thus do not respond to DES1 treatment 7 days later, by which point their activity has decreased.
4.5. Limitations and conclusions
Building on our previous study,7 we have demonstrated here that the NSG-IBD mouse model is well suited to validate target molecules in the development of therapeutics for IBD [e.g., UC]. However, it is only an acute phase model and the human inflammatory cells are restricted to PMBCs; it therefore falls short of fully reflecting the lifelong human disease.7 A strength of this model, however, is that it preserves the immunological background of the donor [to a certain degree], and the combination of patient profiling and efficacy testing, for instance, in the NSG-UC mouse model, may be an important step for patient stratification in future clinical trials.
Within the limitations of the NSG-IBD model, we have here demonstrated that Kv1.3 inhibition appears to represent an attractive therapeutic intervention point in UC, and immuno-modulatory small molecule Kv1.3 inhibitors that suppress T cell activation, cytokine production, and proliferation could potentially prevent relapse and maintain remission of UC. However, we cannot conclude from these results that strongly Kv1.3-dependent T cells are the only immune cells targeted by DES1. For example, DES1 may also suppress the activity of B cells and monocytes, both known to express Kv1.3, and potentially upregulate Kv1.3 to become strongly [or even more] Kv1.3-dependent upon inflammation, thereby contributing to the observed in vivo efficacy reported here. The same caveat applies for polymorphonuclear cells, which are known to play an important role in UC, and may have also contributed to the efficacy reported here. These effects will have to be examined in future studies.
The ultimate therapeutic goal for all IBDs [of which UC and CD are the most prevalent in adults] is an individualised and disease phase-dependent therapy based on orally available drugs. Approximately 40% of all IBD patients fail to respond to currently available biologics,42,43 and when patients do respond, these treatments often lose efficacy over time due to immunogenicity [for example, this is often observed in anti-TNF-based treatment of IBD]. Small-molecule inhibitors like DES1, for which we demonstrated efficacy in a humanised mouse model of IBD, have the advantage that they do not risk triggering such immunogenicity and subsequent loss of response. Moreover, reversible small-molecule inhibitors have an ‘on/off’ mechanism on their target and can therefore swiftly be adapted to different disease phases without long wash-out times, which hamper switching to alternate treatment options
Tofacitinib and DES1 were applied intraperitoneally in this study to allow for direct comparison with infliximab, which cannot be administered orally. Follow-up studies that investigate the efficacy of orally administered DES1, or derivatives thereof, will be important to move towards addressing the unmet need of many IBD patients for safe, efficacious, and orally available drugs.
Data Availability
The data underlying this article are available [and may be found in the article itself] except for the chemical structure of DES1 and the data underlying its development.
Supplementary Material
Acknowledgements
Our special thanks go to the donors. Without their commitment, this work could not have been possible. We thank the team in the animal facility for their excellent work and their enduring friendliness in stressful situations, and Igor Baburin and Steffen Hering at ChanPharm for helpful discussions and assistance in performing the electrophysiology experiments.
Funding
This work was funded by D. E. Shaw Research, LLC, New York, USA.
Conflict of Interest
This study was funded by D. E. Shaw Research. DES is the sole beneficial owner and Chief Scientist of D. E. Shaw Research, with which MØJ, FG, and VJ are affiliated; A-LU, AR, PW, and MS receive funding through an ongoing research agreement with D. E. Shaw Research; RG has a consultancy agreement with D. E. Shaw Research. D. E. Shaw Research has filed a patent application for DES1 and its use.
Author Contributions
A-LU: ex vivo experiments, animal studies, data analysis; MØJ, FG, VJ, DES: inhibitor design, study design, writing of the manuscript; MØJ: data analysis; AR IHC, animal study; MS, PW: animal study; LK: patient recruitment, patient history; MSie: study concept; FB: study concept, patient recruitment, patient history; RG: study concept, data analysis, writing of the manuscript; AA: study concept.
References
- 1. Conrad K, Roggenbuck D, Laass MW. Diagnosis and classification of ulcerative colitis. Autoimmun Rev 2014;13:463–6. [DOI] [PubMed] [Google Scholar]
- 2. Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, Strober W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity 2002;17:629–38. [DOI] [PubMed] [Google Scholar]
- 3. Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest 2007;117:514–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parronchi P, Romagnani P, Annunziato F, et al. Type 1 T-helper cell predominance and interleukin-12 expression in the gut of patients with Crohn’s disease. Am J Pathol 1997;150:823–32. [PMC free article] [PubMed] [Google Scholar]
- 5. Berrebi D, Besnard M, Fromont-Hankard G, et al. Interleukin-12 expression is focally enhanced in the gastric mucosa of pediatric patients with Crohn’s disease. Am J Pathol 1998;152:667–72. [PMC free article] [PubMed] [Google Scholar]
- 6. Aggeletopoulou I, Assimakopoulos SF, Konstantakis C, Triantos C. Interleukin 12/interleukin 23 pathway: biological basis and therapeutic effect in patients with Crohn’s disease. World J Gastroenterol 2018;24:4093–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jodeleit H, Caesar J, Villarroel Aguilera C, et al. The combination of patient profiling and preclinical studies in a mouse model based on NOD/Scid IL2Rγ null mice reconstituted with peripheral blood mononuclear cells from patients with ulcerative colitis may lead to stratification of patients for treatment with adalimumab. Inflamm Bowel Dis 2020;26:557–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin CS, Boltz RC, Blake JT, et al. Voltage-gated potassium channels regulate calcium-dependent pathways involved in human T lymphocyte activation. J Exp Med 1993;177:637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Winslow MM, Neilson JR, Crabtree GR. Calcium signalling in lymphocytes. Curr Opin Immunol 2003;15:299–307. [DOI] [PubMed] [Google Scholar]
- 10. Land J, Lintermans LL, Stegeman CA, et al. Kv1.3 channel blockade modulates the effector function of B cells in granulomatosis with polyangiitis. Front Immunol 2017;8:1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wulff H, Knaus HG, Pennington M, Chandy KG. K+ channel expression during B cell differentiation: implications for immunomodulation and autoimmunity. J Immunol 2004;173:776–86. [DOI] [PubMed] [Google Scholar]
- 12. Sarkar S, Nguyen HM, Malovic E, et al. Kv1.3 modulates neuroinflammation and neurodegeneration in Parkinson’s disease. J Clin Invest 2020;130:4195–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Castañeda O, Sotolongo V, Amor AM, et al. Characterization of a potassium channel toxin from the Caribbean Sea anemone Stichodactyla helianthus. Toxicon 1995;33:603–13. [DOI] [PubMed] [Google Scholar]
- 14. Schmitz A, Sankaranarayanan A, Azam P, et al. Design of PAP-1, a selective small molecule Kv1.3 blocker, for the suppression of effector memory T cells in autoimmune diseases. Mol Pharmacol 2005;68: 1254–70. [DOI] [PubMed] [Google Scholar]
- 15. Koshy S, Huq R, Tanner MR, et al. Blocking KV1.3 channels inhibits Th2 lymphocyte function and treats a rat model of asthma. J Biol Chem 2014;289:12623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beeton C, Wulff H, Standifer NE, et al. Kv1.3 channels are a therapeutic target for T cell-mediated autoimmune diseases. Proc Natl Acad Sci U S A 2006;103:17414–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tarcha EJ, Olsen CM, Probst P, et al. Safety and pharmacodynamics of dalazatide, a Kv1.3 channel inhibitor, in the treatment of plaque psoriasis: a randomized phase 1b trial. PLoS One 2017;12:e0180762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ishida Y, Chused TM. Lack of voltage sensitive potassium channels and generation of membrane potential by sodium potassium ATPase in murine T lymphocytes. J Immunol 1993;151:610–20. [PubMed] [Google Scholar]
- 19. Koo GC, Blake JT, Talento A, et al. Blockade of the voltage-gated potassium channel Kv1.3 inhibits immune responses in vivo. J Immunol 1997;158:5120–8. [PubMed] [Google Scholar]
- 20. Chiang EY, Li T, Jeet S, et al. Potassium channels Kv1.3 and KCa3.1 cooperatively and compensatorily regulate antigen-specific memory T cell functions. Nat Commun 2017;8:14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Palamides P, Jodeleit H, Föhlinger M, et al. A mouse model for ulcerative colitis based on NOD-scid IL2R γnull mice reconstituted with peripheral blood mononuclear cells from affected individuals. Dis Model Mech 2016;9:985–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Antoniou E, Margonis GA, Angelou A, et al. The TNBS-induced colitis animal model: an overview. Ann Med Surg [Lond] 2016;11:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al-Amodi O, Jodeleit H, Beigel F, Wolf E, Siebeck M, Gropp R. CD1a-expressing monocytes as mediators of inflammation in ulcerative colitis. Inflamm Bowel Dis 2018;24:1225–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jodeleit H, Al-Amodi O, Caesar J, et al. Targeting ulcerative colitis by suppressing glucose uptake with ritonavir. Dis Model Mech 2018;11:dmm036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weigmann B, Tubbe I, Seidel D, Nicolaev A, Becker C, Neurath MF. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat Protoc 2007;2:2307–11. [DOI] [PubMed] [Google Scholar]
- 26. Ho J, Tumkaya T, Aryal S, Choi H, Claridge-Chang A. Moving beyond P values: data analysis with estimation graphics. Nat Methods 2019;16:565–6. [DOI] [PubMed] [Google Scholar]
- 27. Benichou G, Gonzalez B, Marino J, Ayasoufi K, Valujskikh A. Role of memory T cells in allograft rejection and tolerance. Front Immunol 2017;8:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smillie CS, Biton M, Ordovas-Montanes J, et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 2019;178:714–30.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stingl LA, Sinska A, Landesmann U, Smolen JS. Induction of interleukin 2 receptiveness and proliferation in resting peripheral T cells by monoclonal anti-CD3 [T3] antibodies does not require the presence of macrophages. Clin Exp Immunol 1987;68:146–55. [PMC free article] [PubMed] [Google Scholar]
- 30. Sousa IG, Simi KCR, do Almo MM, et al. Gene expression profile of human T cells following a single stimulation of peripheral blood mononuclear cells with anti-CD3 antibodies. BMC Genomics 2019;20:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walmsley RS, Ayres RC, Pounder RE, Allan RN. A simple clinical colitis activity index. Gut 1998;43:29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jodeleit H, Milchram L, Soldo R, et al. Autoantibodies as diagnostic markers and potential drivers of inflammation in ulcerative colitis. PLoS One 2020;15:e0228615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amobi A, Qian F, Lugade AA, Odunsi K. Tryptophan catabolism and cancer immunotherapy targeting IDO mediated immune suppression. Adv Exp Med Biol 2017;1036:129–44. [DOI] [PubMed] [Google Scholar]
- 34. Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci 2016;374:20150202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sandborn WJ, Su C, Sands BE, et al. ; OCTAVE Induction 1, OCTAVE Induction 2, and OCTAVE Sustain Investigators. Tofacitinib as induction and maintenance therapy for ulcerative colitis. N Engl J Med 2017;376:1723–36. [DOI] [PubMed] [Google Scholar]
- 36. Nguyen A, Kath JC, Hanson DC, et al. Novel nonpeptide agents potently block the C-type inactivated conformation of Kv1.3 and suppress T cell activation. Mol Pharmacol 1996;50:1672–9. [PubMed] [Google Scholar]
- 37. Koch Hansen L, Sevelsted-Møller L, Rabjerg M, et al. Expression of T-cell KV1.3 potassium channel correlates with pro-inflammatory cytokines and disease activity in ulcerative colitis. J Crohns Colitis 2014;8:1378–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ziegler SF, Ramsdell F, Alderson MR. The activation antigen CD69. Stem Cells 1994;12:456–65. [DOI] [PubMed] [Google Scholar]
- 39. Crawley JB, Rawlinson L, Lali FV, Page TH, Saklatvala J, Foxwell BM. T cell proliferation in response to interleukins 2 and 7 requires p38MAP kinase activation. J Biol Chem 1997;272:15023–7. [DOI] [PubMed] [Google Scholar]
- 40. Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev 2003;83:835–70. [DOI] [PubMed] [Google Scholar]
- 41. Perše M, Cerar A. Dextran sodium sulphate colitis mouse model: traps and tricks. J Biomed Biotechnol 2012;2012:718617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peyrin-Biroulet L, Danese S, Argollo M, et al. Loss of response to vedolizumab and ability of dose intensification to restore response in patients with Crohn’s disease or ulcerative colitis: a systematic review and meta-analysis. Clin Gastroenterol Hepatol 2019;17:838–46.e2. [DOI] [PubMed] [Google Scholar]
- 43. West NR, Hegazy AN, Owens BMJ, et al. ; Oxford IBD Cohort Investigators. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med 2017;23:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Best WR, Becktel JM, Singleton JW, Kern F Jr. Development of a Crohn’s disease activity index. National Cooperative Crohn’s Disease Study. Gastroenterology 1976;70:439–44. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available [and may be found in the article itself] except for the chemical structure of DES1 and the data underlying its development.