Abstract
Current practice in IBD is to classify patients based on clinical signs and symptoms and provide treatments accordingly. However, the response of IBD patients to available treatments is highly variable, highlighting clinically significant heterogeneity among patients. Thus, more accurate patient stratification is urgently needed to more effectively target therapeutic interventions to specific patients. Here we review the degree of heterogeneity in IBD, discussing how the microbiota, genetics, and immune system may contribute to the variation among patients. We highlight how molecular heterogeneity may relate to clinical phenotype, but in other situations may be independent of clinical phenotype, encouraging future studies to fill the gaps. Finally, we discuss novel stratification methodologies as a foundation for precision medicine, in particular a novel stratification strategy based on conserved genes across species. All of these dimensions of heterogeneity have potential to provide strategies for patient stratification and move IBD practice towards personalised medicine.
Keywords: IBD, heterogeneity in IBD, personalised medicine, molecular stratification of IBD patients, immunology
1. Clinical and symptomatic heterogeneity
Inflammatory bowel disease [IBD] is classically divided into ulcerative colitis [UC] and Crohn’s disease [CD]. The first descriptions of clinical cases likely to be UC were published as early as the 18th century, and a description of what was probably CD even more than a century earlier.1 At that time the pathophysiology and aetiology of these diseases were unknown, and it was believed that a pathogenic micro-organism was the causative agent,1 a view that changed around the 1950s, when IBD was proposed to have a complex immunological aetiology instead.1 Today, IBD is considered a multifactorial disease in which aberrant immune responses against commensal microbiota triggered by environmental factors occur in genetically susceptible hosts.
Increasingly, IBD is considered as a continuous disease spectrum rather than two distinct entities.2 Active disease is characterised by diarrhoea with blood and/or mucus, abdominal pain, signs of systemic toxicity during a severe flare, weight loss, and fatigue, symptoms which vary between patients. This variation exists even between individuals with the same clinical disease classification,3,4 revealing a high degree of heterogeneity already at the clinical level. The heterogeneity of these diseases was identified even in the earliest publications, one of which included some attempts to categorise affected patients.5 Variability of symptoms stems most notably from variable locations of inflammation in both diseases and variable disease behaviour in CD.3,4 However, some of the symptomatic variability cannot be explained by these factors.6 Also, ileal and ileocolonic CD seem separate from colonic CD which in turn is, according to some parameters, nearer to UC.2,7
Furthermore, IBD associated with primary sclerosing cholangitis [PSC-IBD] has a characteristic phenotype that is different from non-PSC-IBD8 and likely has a specific pathogenesis. PSC-IBD is also associated with specific clinical outcomes: higher risk of colectomy, colorectal cancer, and death compared with non-PSC-IBD8 [Table 1].
Table 1.
Possible and existing clustering strategies with clinical implication in IBD.
| Feature | Clustering strategies or suggested clustering strategies with clinical implication |
|---|---|
| Clinical | Montreal classification |
| Increased risk of colectomy, colorectal cancer, and death in PSC-IBD patients compared with non-PSC-IBD patients8 | |
| Genetics | At least four gene loci have been associated with prognosis in CD35 |
| NOD2 risk allele is associated with phenotype of ileitis and absence of colitis36 | |
| Variation in IL-6 is associated with phenotype of ileitis36 | |
| A variant of TL4 is associated with a risk of surgery at disease onset in CD36 | |
| Microbiota | Increase in Bifidobacterium, Collinsella, Lachnospira, Lachnospiraceae, Roseburia, and Eggerthella taxa and reduction in Phascolarctobacterium in CD anti-TNFα responders compared with non-responders61 |
| Increase in Faecalibacterium prausnitzii in UC and CD anti-TNFα responders compared with non-responders62,84 | |
| Increased Eggerthella, Clostridiales, and Oscillospira in CD patients with more quiescent disease and increase in Enterobacteriaceae, Enterobacteriaceae, and Klebsiella in those with more aggressive disease61 | |
| Altered microbial profile is linked to disease recurrence 1 year after ileocaecal resection in CD64 | |
| Lower abundance of Faecalibacterium prausnitzii in CD at the time of ileal resection associated with endoscopic recurrence at 6 months65 | |
| Lower abundance of Faecalibacterium prausnitzii in CD at the time of infliximab cessation associated with quicker relapse63 | |
| Immune system | Frequency of NKp44+ILC3 correlates with disease severity98 |
| Increase of TREG with increasing disease activity in UC and CD101,102 | |
| Greater accumulation of TREG in ileal mucosa in paediatric ileal CD patients compared with adult ileal CD patients, possibly contributing to relative rareness of isolated ileal enteritis in paediatric CD patients103 | |
| Variation in IL-22BP produced by CD4 T cells in association with response to anti-TNF response122 | |
| A subset of ileal CD patients has distinct cellular profile in their inflamed ileum, with accumulation of IgG plasma cells, inflammatory mononuclear phagocytes, activated T cells, and stromal cells, which predicts response to anti-TNF treatment138 | |
| Increase in inflammatory fibroblasts and monocytes as well as DC2 subsets in UC patients predicts non-response to anti-TNF treatment139,140 | |
| Transcriptomics | Transcriptional risk score based on eQTLs data can predict development of stricturing and/or penetrating disease within 3 years from disease onset in paediatric CD34 |
| Upregulation of extracellular matrix accumulation-associated genes in ileum in paediatric CD patients who later developed stricturing disease behaviour131 | |
| Upregulation of genes associated with acute microbial immune responses in paediatric patients who later developed penetrating disease131 | |
| A transcriptional signature based on 17 genes in whole- blood samples from IBD patients can predict aggressive disease course132 | |
| High pre-treatment levels of OSM predict poor response to anti-TNF treatment137 | |
| Rectal transcriptomic profiles UC1 and UC2 predict response to anti-TNF therapy143 |
IBD, inflammatory bowel disease; PSC, primary sclerosing cholangitis; CD, Crohn’s disease; UC, ulcerative colitis IL, interleukin; ILC, innate lymphoid cell; NK, natural killer; TREG, regulatory T cell; BP, binding protein; TNF, tumour necrosis factor; Ig, Immunoglobulin; TLR, Toll-like receptor; OSM, oncostatin M; NOD2, nucleotide-binding oligomerisation domain-containing protein 2; eQTL, expression quantitative trait loci study.
Inflammation outside the gut, so called extraintestinal manifestations [EIMs], contribute to the clinical heterogeneity in IBD patients.9 Wide heterogeneity in EIMs exists when it comes to both clinical manifestations and underlying pathological mechanisms.10 It is still unknown why some patients get unifocal and the others multifocal inflammation, and the potential of these observations to assist patient stratification has not yet been exploited.9
Given the heterogeneity of the disease, classification and division of IBD into different entities is a complicated task and will require a better understanding at the cellular and molecular levels. In future, clinically observable phenotypes will likely be less relevant since molecular characterisation will likely provide deeper understanding of the diverse pathogenic mechanisms of IBD and may underpin therapeutic decision making. However, single-cell RNA sequencing [scRNA-seq] data on immune cell populations in both gut and blood confirm that the division into UC and CD is still biologically relevant, as distinguishing features can be observed between these groups.11
1.1. Age at disease onset
Based on age at IBD onset, patients can be defined into very early onset [VEO]-IBD that debuts before the age of 6 years, paediatric IBD that starts before age of 18, adult onset IBD that starts at ages 18 to 60, and elderly onset with debut at over 60 years age. In particular VEO-IBD, which is discussed in more detail below, is often a very different type of disease compared with later onset disease; however, there is also more general heterogeneity among patients based on the age of disease onset. In CD, ileocolonic location dominates in paediatric patients, whereas colonic location is the most common in elderly onset disease.12–15 However, in VEO-IBD, isolated colitis is the most common manifestation.16,17 Also, after 5–10 years diagnosis over half of the paediatric onset patients have stricturing or penetrating disease behaviour, whereas the number is only around 30% in elderly onset patients even after 15 years of follow-up.12,14,15 In UC, extensive colitis is more common in childhood compared with adult and elderly onset disease, where left-sided colitis dominates.13,15 Further, EIMs have been reported to be more common in paediatric onset and lower in elderly onset IBD.13–15 Together these differences in clinical manifestations indicate more severe disease with earlier onset.
There are also aetiological differences between different age groups. The genetic component has a greater influence in paediatric onset IBD.18 Positive first-degree relative family history has been reported to be slightly over 10% in the paediatric population in general and over 20% in VEO-IBD.16,17 In a French population-based registry, the rate of family history with IBD fell with increasing age of disease onset, with 16% and 13% positive family history for paediatric CD and UC patients, respectively, and 7% and 3% for CD and UC patients with elderly-onset disease.12 It is also clear that many environmental exposures are relevant for gut inflammation, for example smoking and polypharmacy, as well as hormonal factors differing between different age groups.18 The microbiota also undergoes changes throughout the lifetime.18 In paediatric patients, an increase in mucosa-associated aerobic and facultative-anaerobic bacteria has been reported in contrast to increase in anaerobic bacteria in adult onset disease, but otherwise microbial differences in relation to IBD disease onset have not been well studied.18–20 Similarly, the immune system is known to undergo many functionally relevant changes through the lifetime, but its significance in IBD is not well defined.18 Thus, IBD heterogeneity in relation to disease onset is an important future research area and may inform optimal management strategies for these different groups which may in future be classified as different diseases.
2. Aetiology
2.1. Genetics
IBD develops as a complex interaction between environment, genes, microbiota, and immune system.3,4 However, there are also more than 50 observed forms of IBD-like intestinal inflammation in which rare high-penetrance genetic variants cause a specific phenotype that is often associated with immune defects and VEO-IBD.21–24 For example, mutations in either IL-10 or the IL-10 receptor cause a phenotype of VEO-IBD with severe perianal disease and colitis.21,23 In contrast, the immunodysregulation polyendocrinopathy enteropathy X-linked [IPEX] syndrome is caused by a mutation on Foxp3 gene, leading to absence or reduction in regulatory T cells causing several autoimmune manifestations including intestinal inflammation.23 Also other immunodeficiencies or genetic disorders influencing, for example, intestinal epithelial barrier function (e.g., NF-kappa-B [NFκB] essential modulator deficiency), T and B cell maturation [e.g., common variable immune deficiency and severe combined immunodeficiency] or neutrophils [e.g., NADPH oxidase mutations causing chronic granulomatous disease] have been identified in VEO-IBD patients.21,23,25,26 Whole exome sequencing has shown to be useful in finding rare genetic variants contributing to IBD, which could be especially beneficial in studies on VEO-IBD.21,27 Importantly, such single gene defects provide insight into sporadic IBD and its heterogeneity, highlighting the impact of specific pathways on IBD pathogenesis which may be more difficult to detect in sporadic, multifactorial IBD.21–23,28 Also, these observations imply extensive inter-individual variation in the specific pathogenic pathways that lead to a single clinical phenotype. Indeed, in each individual, IBD likely develops from a unique combination of risk factors which interact and increase the cumulative risk of IBD.3,4,21,29
Genome-wide association studies [GWAS] have revealed over 260 loci that are associated with the risk of sporadic IBD, some of which are associated with the function of the immune system and have revealed relevant pathological pathways.30,31 There exists significant heterogeneity in GWAS hits between different demographic populations.32 For example, three coding variants of nucleotide-binding oligomerisation domain-containing protein [NOD]2, with odds ratio of 2.13–3.03 for IBD development in the European population, were not present in the East Asian population and there did not significantly contribute to the risk of IBD.32 One limitation in GWAS studies is that common variation does not account for all the heritability in IBD,27 as highlighted also in the previous section. So far, disease risk alleles discovered through GWAS have not been converted into new clinical strategies. This is likely due to the lack of assigned functions to IBD risk variants, many of which are located in non-coding regions.30 Integration of genetic and molecular data offers possibilities for clinically meaningful patient stratification; 30 for example, expression quantitative trait loci studies [eQTLs] can link hits on non-coding regions to functional genes.33 Indeed, Marigorta et al. found that transcriptional risk score based on eQTLs data could predict which paediatric CD patients will develop stricturing and/or penetrating disease within 3 years from disease onset, but a genetic risk score based on GWAS could not34 [Table 1]. Moreover, Lee et al. demonstrated that GWAS variants have no or minor impact on disease prognosis in CD but found instead four separate loci associated with prognosis in a within-cases GWAS.35 Thus, genetic analyses should not only be applied to prediction of susceptibility to developing IBD. However, some variants, such as NOD2, IL-6, and Toll-like receptor [TLR]4 have been associated with both susceptibility to CD and phenotypic features such as ileitis without colitis, ileitis, and surgery at disease onset, respectively.36
2.2. Heterogeneity in experimental models of IBD
Analogous to the heterogeneous group of human genetic defects that ultimately cause IBD-like syndromes, there is heterogeneity in the range of experimental mouse models which result in acute and chronic gut inflammation.37 The dextran sodium sulphate [DSS] murine model of colitis is one of the most widely used for its simplicity. DSS induces damage in epithelial cells with the consequent barrier disruption and bacterial translocation. Its initiation is dependent on signalling through NOD-like receptor protein 3 [NLRP3] inflammasome, which activates caspase-1 leading to interleukin [IL]-1β production.38 Further, DSS-induced IL-1β release was shown to require functional lysosomes and reactive oxygen species [ROS] production.38 Although it is considered a T cell-independent model, expression of both Th1 and Th2 as well as Th17-type cytokines has been reported to be dysregulated in the DSS model.39 By contrast, the oxazolone colitis, a model of UC at histological level, is mediated mainly by T helper [Th] 2-type response and is dependent on initiation by natural killer [NK] T cells, giving rise to high IL-13 production.40,41
Alternatively, adaptive transfer of CD4+CD45RBhigh T cells [naive T cells] from a wild-type [WT] mouse into a recipient lacking T and B cells [RAG1- or RAG2-deficient mouse] may resemble CD.39,42,43 This model, commonly termed ‘T cell transfer model of colitis’, develops transmural pancolitis in combination with small bowel inflammation 5–8 weeks after cell transfer.42,43 In this model, inflammation is mainly of Th1 type and colitis development can be prevented by anti-interferon [IFN]γ or anti-tumour necrosis factor alpha [TNFα] antibodies, or by administration of recombinant interleukin [rIL]-10.43 However, the expression of some Th2- and Th17-type cytokines has also been reported to be dysregulated.39 Further, IL10-/- mice develop a spontaneous colitis after weaning that is mainly Th1 mediated, but also with contribution of a Th17 immune response.44–46 Also, cytokines IL-23 and IL-13 may be critical for the development of colitis in IL-10 knockout mice.44,47
Important differences in colonic transcriptomic profiles, possibly having functional relevance, have been described for different murine IBD models.39 For example, in IL10-/- mice where spontaneous colitis is accelerated with piroxicam [PAC IL10-/- model], the expression of genes associated with tissue remodelling were relatively high compared with the T cell transfer model of colitis, whereas those associated with bacterial sensing were lower.39 Generally, IBD-associated genes were upregulated in PAC IL10-/- to a higher extent compared with the T cell transfer model of colitis.39 Furthermore, the penetrance of the disease and mechanism of action might differ between animal facilities even if the same animal background and method are used, which is attributed to microbial variations.48,49 For example, it has been shown that different microbial communities in identical genetic backgrounds induce DSS-induced colitis by distinct mechanisms, namely either neutrophil- or T cell-dependent,49 thus indicating a high degree of experimental IBD heterogeneity dictated also by environmental factors, which is analogous to human IBD. Although this complicates the interpretation and generalisation of results from animal studies, it provides a great opportunity to study complex interactions between genetics, environmental, and immunological factors as well as heterogeneity in IBD. Moreover, the fact that environmental, particularly microbial, factors have turned out to be so significant in experimental IBD further emphasises the fundamental role of the genotype-environment interface in human IBD.
Animal models are a vital tool facilitating the disentanglement of the multiple pathogenetic elements of a complex diseases such as IBD. Each model facilitates focus on different aspects of IBD pathogenesis which may then be reconstructed into an enhanced understanding of the human disease. However, the potential of animal models to accurately recapitulate specific IBD phenotypes remains limited. The development of new models that reflect the pathogenesis of specific forms of IBD, such as PSC-IBD or fistulising CD, would greatly enhance the personalisation of IBD management.
2.3. Microbiota
Altered microbiota [dysbiosis] in gut is an extensively studied hallmark of IBD.50,51 The microbial signature in IBD is characterised by instability and more frequent fluctuations compared with healthy controls.50 Also changes in abundance of species, taxonomic changes, reduced diversity, and functional metabolic changes characterise microbiota in IBD.50,52–55
In colonic surgical samples, IBD patients had enrichment of Proteobacteria and the Bacillus subgroup of Firmicutes, whereas Bacteroidetes and the Lachnospiraceae subgroup of Firmicutes were depleted compared with non-IBD controls.56 Interestingly, approximately one-third of CD and one-fourth of UC patients had more pronounced alterations in microbiota, with overall lower proportion of Firmicutes and Bacterioidetes together with manifold increase in relative abundance of Proteobacteria and Actinobacteria compared with non-IBD controls.56
The heterogeneity in microbial signature observed in cross-sectional studies is complemented by longitudinal studies which also reveal instability of the microbiota within individual IBD patients, including states where the microbiota is indistinguishable of those observed in non-IBD controls.50,52 However, although the microbiota is highly heterogeneous within one individual at different time points, there are still possible clustering strategies based on microbiota composition.57
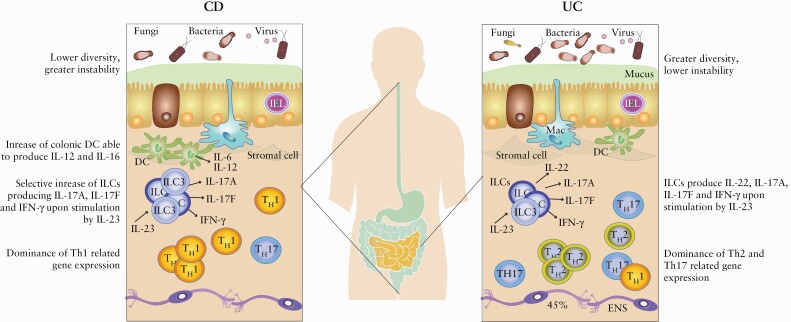
Generally, CD patients have more pronounced alterations in microbiota than UC patients, and this is most evident in ileal CD, especially after ileocaecal resection.50 For example, CD patients show both lower diversity48 and more pronounced fluctuations and inter-individual variation in microbial composition compared with UC50,58 [Figure 1]. Further, CD patients show greater abundance in Proteobacteria and Bacteroidetes and decrease in Clostridia compared with UC patients.59 In faecal samples from IBD and irritable bowel syndrome patients, as well as healthy controls, Vich Vila et al. could identify 477 different taxa of which 219 were associated with CD and 102 with UC, among which 15 were UC specific,60 further demonstrating differences between CD and UC patients in microbial composition.
Figure 1.
Comparison of factors that differ between UC and CD. There are remarkable differences between patients with diagnoses UC and CD, but also great heterogeneity within these patient groups. Generally, CD patients have, for example, lower diversity and greater instability in their microbiota compared with UC patients.50,58,61 Also, in both CD and UC, ILCs produce IL-22, IL-17A, IL-17F, and IFN-γ upon stimulation by IL-23, but in CD there is a selective increase in IL-17A, IL-17F, and IFN-γ producing ILCs.92 Further, in CD, colonic DC show increased expression of IL-12 and IL-16 during inflammation compared with UC patients.86 Also, CD is characterised by intestinal dominance of Th1-related gene expression, whereas Th2- and Th17-related gene expression dominates in UC.108 CD, Crohn’s disease; UC, ulcerative colitis; IL, interleukin; Th, T helper cell; ILC, innate lymphoid cell; IEL, intestinal epithelial lymphocyte; Mac, macrophage; ENS, enteric nerve system; DC, dendritic cell; RA, retinoic acid; TNF, tumour necrosis factor.
CD patients who respond to anti-TNFα treatment have been shown to have increase in Bifidobacterium, Collinsella, Lachnospira, Lachnospiraceae, Roseburia, and Eggerthella taxa and reduction in Phascolarctobacterium compared with anti-TNFα non-responders61 [Figure 2, Table 1]. Also, higher pre-treatment abundance of Faecalibacterium prausnitzii predicted better response to anti-TNFα treatment in both UC52 and CD.62 Further, higher abundance of Faecalibacterium prausnitzii on infliximab treatment cessation predicted sustained remission in CD.63 Also, in CD, increased Eggerthella, Clostridiales, and Oscillospira have been associated with quiescent clinical disease whereas increase in Enterobacteriaceae, Enterobacteriaceae, and Klebsiella have been found to predict more aggressive disease course.61 Furthermore, post-surgical CD patients have significantly reduced microbiota diversity compared with patients who have not undergone surgery.61 Moreover, in CD, post-surgical disease recurrence 1 year after ileocaecal resection is associated with an altered microbial profile.64 Endoscopic recurrence of CD 6 months after ileal resection can also be predicted by lower abundance of Faecalibacterium prausnitzii at the time of the surgery.65
Figure 2.
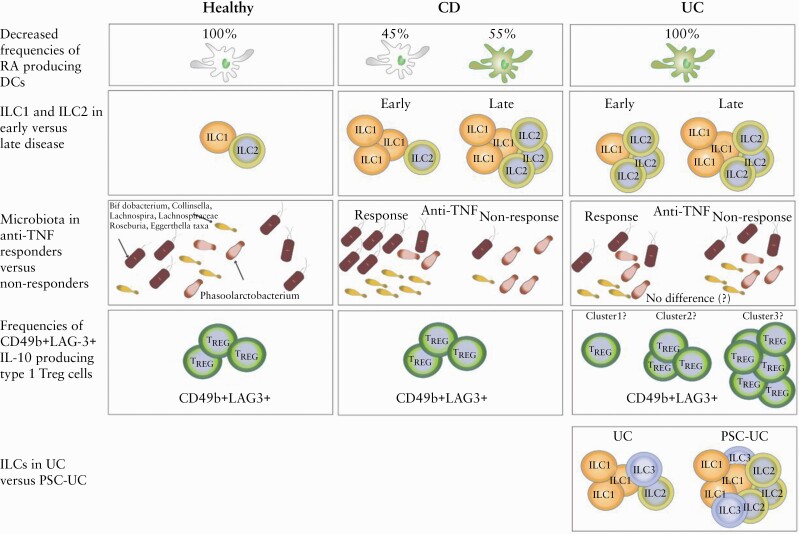
Examples of heterogeneity between UC, CD and healthy subjects. The first row illustrates how in UC all the DCs have decreased capacity to produce RA [green DCs] compared with healthy individuals [white DCs], whereas DCs from CD patients show heterogeneity in their RA production capacity.84 The second row illustrates that in early CD the abundance of colonic ILC1s is selectively increased, whereas in early UC the abundance of ILC2s is increased.98 Later in the disease course, ILC1s and ILC2s are increased compared with healthy individuals in both disease groups.98 The third row shows an example of microbiota-based clustering, where CD patients responding to anti-TNF treatment have increased abundance of Bifidobacterium, Collinsella, Lachnospira, Lachnospiraceae, Roseburia, and Eggerthella taxa and reduction in Phascolarctobacterium compared with anti-TNFα non-responders, whereas UC patients do not seem to show clustering based on these bacteria and anti-TNF response.61 The fourth row exemplifies a possible clustering strategy based on CD49b+LAG-3+ IL-10-producing type 1 TREG cells that show more variable frequencies in UC compared with CD.85 The fifth row illustrates that UC patients with PSC have higher abundance of colonic ILCs compared with UC patients without PSC.96 CD, Crohn’s disease; UC, ulcerative colitis; IL, interleukin; Th, T helper cell; ILC, innate lymphoid cell; DC, dendritic cell; RA, retinoic acid; TNF, tumour necrosis factor; PSC, primary sclerosing cholangitis.
In conclusion, there are some overarching characteristics of the IBD microbiota but much inter- and intra-individual variation exists. Furthermore, the relationship between abundance of various species and the alteration in function is not well characterised.
2.4. Environment
Studies on monozygotic twins have shown significant discordance when it comes to both occurrence of IBD and phenotype of developed IBD, indicating the influence of environmental factors in addition to genetic factors.66,67 Early life events affecting the risk of IBD through changes in microbiota, especially antibiotic use and mode of feeding, are among the most frequently identified environmental risk factors.68 There is also evidence implicating urbanisation, air pollution, non-steroidal anti-inflammatory drug use, hypoxia, and diet in the risk of IBD.68
Since environmental factors affect individuals in different ways it is reasonable to propose that environmental pressures significantly account for the heterogeneity in IBD patients.69 As discussed above, the microbiota is associated with heterogeneity in IBD patients and, further, in healthy individuals there is robust evidence that diet is a significant environmental factor affecting gut microbial heterogeneity.70–72 Thus, it is likely that diet significantly contributes to heterogeneity also among IBD patients. Further, dietary compounds can have direct effects on the immune system and cause epigenetic modifications.70 Dietary studies have also particularly addressed gene-environment interactions as foundations for why environmental factors seem to affect individuals in different ways.69 For example, in the paediatric population, interactions between genotypes of polyunsaturated fatty acid [PUFA] metabolic genes CYP4F3 and FADS2 and dietary long chain n-6:n-3 PUFA intake ratio affect the risk of CD.73 Polymorphism in CYP4F3 has also been shown to modify the effect of n-6:n-3 PUFA intake ratio in the risk of adult UC.74 Further, the interaction between polymorphism of rs7657746 [IL-21] and potassium intake affects the risk of CD; there is increasing disease risk with increasing potassium intake in individuals with GG genotype, whereas people with GA and AA genotypes have decreasing risk of CD with increasing potassium intake.75
In the same way, in individuals with the GG genotype of UC susceptibility locus rs1801274, increasing intake of haem iron decreases the risk of UC, whereas the opposite is true for those with the AA phenotype.76 There are as yet no ways to cluster patients based on diet and other environmental exposures, but this may become an interesting field of research as incidence of IBD grows globally also in areas where IBD has previously been rare.
2.5. Immune system
Both innate and adaptive immune cells are implicated in the pathogenesis of IBD77 and balance between distinct immune responses likely lies at the root of much of the heterogeneity between patients [Figure 1]. In homeostasis, intestinal dendritic cells [DC] are specialised in producing retinoic acid [RA] during the process of imprinting naïve T lymphocytes in the intestinal mucosa,78–80 inducing the homing molecules C-C chemokine receptor type 9 [CCR9] and integrin α4β7, and serves as an adjuvant to generate Foxp3+ regulatory T cells [Foxp3+ TREG].81–83 It is believed that breakdown of this process might contribute to IBD pathogenesis, which is supported by the observation that IBD patients have lower numbers of RA-producing DCs.84 Interestingly, Magnusson et al. reported decreased frequencies of RA-producing cells in all 15 UC patients but in only four out of nine CD patients compared with control tissue,84 suggesting greater heterogeneity in RA-producing DC frequencies in CD [Figure 2]. In the same study, there was an overall decrease of CD141+CD103+DC and CD1c+CD103+DC in inflamed mucosa of IBD patients compared with non-inflamed mucosa from the same individual, but this was not observed in a subset of CD and in UC patients.84 Similar results have been reported for CD103+DC in total in CD.85 These differences are likely to have a functional role because CD103+DC decrease in CD patients after treatment with an anti-TNFα agent.84 Hart et al. showed that DC from both inflamed UC and CD intestinal tissue express significantly higher levels of Toll-like receptor [TLR]-2 and TLR-4 compared with intestinal DC from healthy controls.86 Further, they showed that intestinal DC from patients with active CD show significantly higher levels of the maturation/activation marker CD40 compared with both healthy controls and DC from non-inflamed tissue of CD patients, but levels decreased after anti-TNF treatment irrespective of the response.86 Also, DC from inflamed intestinal tissue of CD patients show significantly higher expression of both IL-6 and IL-12 compared with DC from both healthy controls and inflamed tissue of UC patient86 [Figure 1], suggesting heterogeneity in DC-produced cytokines.
Platt et al. showed in murine colitis that the dominant population of inflammatory macrophages are distinct from gut-resident macrophages and are derived from blood monocytes in a CCR2-dependent manner.87 In the study by Magnusson et al., both CD and UC mucosa showed heterogeneity in the extent of accumulation of HLA-DRint macrophages in the inflamed in relation to the non-inflamed mucosa84 and similar results have been obtained by Dige et al. for CD,85 suggesting specific importance of these cells for inflammation in a subset of patients irrespective of the diagnosis. It is likely that these differences have a functional role in IBD, since the proportion of HLA-DRint macrophages of total lamina propria myeloid cells [LPMC] decreases in CD patients after anti-TNFα treatment.85 Moreover, the proportion of total colonic lamina propria myeloid cells which were HLA-DRhi macrophages was decreased in inflamed mucosa compared with non-inflamed mucosa in few patients despite the lack of significant difference in the total abundance of these cells.84
Innate lymphoid cells [ILCs] have lymphoid origin and mirror functions of T cells.77,88 Among the most well-known subtypes are ILC1s, ILC2s, and ILC3s. ILC1s function mainly in type 1 immunity reacting against tumours and intracellular microbes, and ILC2s function in type 2 immunity reacting against large extracellular pathogens and allergens.89 ILC3s in turn function mainly in type 3 immunity against extracellular microbes such as bacteria and fungi.89 Thus, ILC1s, ILC2s, and ILC3s mirror functions of Th1, Th2, and Th17 cells, respectively. There are data suggesting that ILCs are involved in intestinal inflammation in IBD by IL-23 driven induction of secretion of cytokines IL-17A, IL-17F, interferon-γ [IFN-γ], and granulocyte-macrophage colony-stimulating factor [GM-CSF].77,90–95 ILCs are also able to produce IL-22 and IL-26 upon stimulation by IL-23, but numbers of IL-17A-, IL-17F-, and IFN-γ-producing ILCs are selectively increased in the inflamed ileal and colonic tissue of CD patients compared with IL-22- and IL-26-producing cells92 [Figure 1].
However, one can observe marked heterogeneity between individual CD patients in the relative abundance of these cells,92 suggesting differential importance between CD patients. Moreover, many more alterations in ILC populations, including the dynamics of the changes along the disease course, have been described in IBD77 [Figure 2]. Also, ILCs are increased in the colon of patients with PSC-UC compared with both UC patients without PSC and controls96 [Figure 2]. Thus, it can be speculated that ILCs could contribute to higher risk of colon cancer in patients with PSC-UC since there are studies implicating ILCs in intestinal cancer development.96,97 NKp44+ILC3 are decreased in inflamed tissue and correlate with disease severity98 [Table 1]. In treatment-naïve, relatively newly diagnosed IBD, ILC1s are increased in inflamed gut tissue in CD patients whereas ILC2s are increased in inflamed gut tissue in UC patients,98 functionally mirroring their adaptive T cell counterparts [Figure 2]. In contrast, in patients with IBD established for at least 1 year before sampling, both ILC1s and ILC2s are increased in inflamed gut tissue in both UC and CD98 [Figure 2]. Also, there is accumulation of IL-17-producing ILC3s in inflamed tissue in CD but not in UC.92 Frequencies of ILCs do not differ between IBD patients and controls generally in blood and non-inflamed tissue.98 However, frequencies of ILC1s and ILC2s are increased among total ILCs in the blood of patients with PSC-UC compared with patients with UC and controls.96
In the adaptive immune system, regulatory T cells [TREG], including both Foxp3+ TREG and IL10+ Foxp3neg T regulatory type 1 [Tr1] cells, are key regulators of intestinal homeostasis, and dysfunction of these cells may be a key feature of IBD pathogenesis.77,99,100 Increased numbers of colonic and ileal Foxp3+ TREG have been shown in both active and inactive CD and in active, but not in inactive, UC but marked heterogeneity is observed between patients.101–103 Holmén et al. reported increase of TREG with increasing disease activity in UC,102 and same has also been reported for CD101 [Table 1]. Interestingly, Reikvam et al. showed that paediatric ileal CD patients have even greater accumulation of TREG in ileal mucosa compared with adult ileal CD patients, which the authors suggested may contribute to the relative rarity of isolated ileal enteritis in paediatric CD patients.103 TREG from IBD patients show approximately 60% decreased ability to suppress autologous T cell proliferation.104 One explanation could be increase in circulating RORγt+IL17+Foxp3+ TREG cells among IBD patients compared with heathy controls.104 Also, TREG in inflamed colonic lamina propria have increased apoptosis rate compared with TREG in non-inflamed tissue.105 This may have a functional role in the pathogenesis of IBD, since the apoptosis rate decreases after treatment with an anti-TNFα agents together with decreasing disease activity.105 Further, Foxp3 negative IL-10-producing Tr1 cells can be identified by the expression of the CD49b and lymphocyte-activation gene 3 [LAG-3] surface markers,106 which are decreased in CD and UC patients compared with controls.106,107
Interestingly, UC patients show more variable frequencies of these CD49b+LAG-3+ IL-10-producing Tr1 compared with CD patients107 [Figure 2], suggesting that frequencies of CD49b+LAG-3+ IL-10-producing Tr1 cells might allow stratification of UC but not CD patients. Furthermore, the same cell type can show great heterogeneity even within an individual. For example, IL-10-producing Foxp3neg CD4+ T cells are transcriptionally heterogeneous and can display both regulatory and pro-inflammatory activity.107
In contrast to the regulatory activity of TREGs, Th1, Th2, and Th17 lymphocytes have a pro-inflammatory phenotype.77 The expression of Th17- and Th2-related genes is shown to be increased in UC compared with CD, whereas the expression of Th1-related genes is increased in CD108 [Figures 1 and 3]. However, one can observe wide heterogeneity in the expression of specific cytokines also within patient groups. For example, expression of the Th2 cytokine IL-13 shows dichotomy among UC patients, and expression levels are associated with mucosal microbiota composition as well as clinical characteristics including sex, age at disease onset, steroid/immunosuppressive/anti-TNF-α drug use, and presence of extensive colitis [Butera, 2020].109 Also, there is heterogeneity between patients with both CD and UC in the intestinal expression of Th17 signature genes IL-22, IL-17A, IL-17F, and IL-26.92 The Th17-secreted cytokines are increased in the inflamed tissue of IBD patients, which correlates with infiltration of activated Th17 into the intestinal tissue.77 IL-17 acts as a paracrine signal to increase the production of additional pro-inflammatory cytokines, chemokines, and inflammatory mediators that eventually contribute to tissue damage.77 In intestinal lamina propria, CD14+CD163 low myeloid cells induce Th17 cell differentiation and IL-17 production in a way that is dependent on IL-1β, transforming growth factor [TGF]β, IL-6, and IL-23.77,110
Figure 3.
Summary of the most important known and speculated differences between UC1 and UC2 patient groups. Patients in the cluster UC1 show generally higher expression of genes related to neutrophil infiltration and degranulation as well as cytokine signalling, whereas gene expression signature in the cluster UC2 suggests high B cell activation.143 Further, patients in the cluster UC2 are more responsive to both infliximab and vedolizumab compared with patients in the UC2 cluster.143 UC, ulcerative colitis; IL, interleukin; TNF, tumour necrosis factors; TREM, triggering receptor expressed on myeloid cells; S100A8, S100 calcium binding protein A8; BCL2A1, B cell lymphoma 2 associated protein A1; CYP26B1, cytochrome P450 26B1; NOX, nicotinamide adenine dinucleotide phosphate oxidase; NR3C2, nuclear receptor subfamily 3 group C member 2; PARM, prostate androgen- regulated mucin-like protein.
Interestingly, the role of IL-23 in the pathogenesis of IBD seems to be heterogeneous and may depend on the genetic and/or environmental background, since the primary risk variant, as well as two secondary risk variants of IL-23 receptor [IL23R] gene which are significantly associated with the risk of IBD in European populations, are not significantly associated with the risk in East Asian populations.32 This is particularly remarkable since IL23R risk variants have the second largest effect size on the risk of IBD after NOD2 variants, in population with European ancestry.111 Further, risk variants in IL23R contribute to greater risk of developing CD versus UC (odds ratio [OR] = 1.4) though it is significantly associated with the risk of both diagnoses, suggesting variation in the importance of this risk variant based on disease phenotype.111 Blocking antibodies against IL-23 and IL-12 [ustekinumab] has been shown to be effective in treating CD and UC and is now licensed for the treatment of IBD, including in Europe, the USA, Canada, and Japan.112 Relatively good response to ustekinumab has been shown in CD patients resistant to anti-TNF therapy,113,114 highlighting CD heterogeneity in response to different biologic therapies and need for patient stratification. Initially, both preclinical and human studies indicated the importance of IL-12 and IL-23 in CD but not in UC.86,115–118 However, evidence of the clinical efficiency of ustekinumab in UC112 has recently led to the approval of this drug for UC, demonstrating that cytokine profiling of groups of patients does not directly translate into effective therapeutic strategies—although individual cytokine profiling may be more clinically informative.
IL-22 is part of the IL-10 superfamily and an important cytokine in the pathogenesis of IBD.77 It has heterogeneous functions and is involved in epithelial regeneration and healing as well as pro-inflammatory responses in the gut.77,119 IL-22 is elevated in IBD patients and levels correlate positively with disease activity in both CD and UC.120,121 However, IL-22 gene expression is highly heterogeneous in both inflamed and uninflamed intestinal tissue of IBD patients,122 suggesting a possibility for patient stratification. Not only IL22 itself but regulators of IL22 display heterogeneity in IBD patients. IL-22 activity can be controlled by IL-22-binding protein [IL-22BP] which blocks IL-22 receptor [IL-22R] activation.123,124 Interestingly, IL-22BP expression is increased in intestinal tissue from CD and UC patients compared with healthy controls, yet with wide heterogeneity,122 suggesting decreased IL-22 signalling regardless of IL-22 serum levels. Interestingly again, IL-22BP produced by CD4 T cells, but not DC, varies in association to response to anti-TNF treatment122 [Table 1], and therefore, frequencies of IL-22BP+ T cells might be a criterion for patient stratification.
In animal models, IL-22 attenuates colitis in a Th2-mediated chronic colitis model [TCRα KO mice].125 Moreover, anti-IL-22 antibodies significantly delay recovery from the DSS model of colitis.125 Also, a ligand for aryl hydrocarbon receptor is protective against TNBS colitis and the protective effect is partially blocked by IL-22 antibodies.126 Further, transfer of IL-22-deficient CD45RBhi CD4+ T cells into Il22-/- Rag1-/- mice causes more aggressive inflammation compared with transfer of wild-type CD45RBhi CD4+ T cells.127 Kamanaka et al. demonstrated that transfer of Foxp3-depleted CD45RBlo CD4+ T cells into Rag1-/- mice induces a Th17-type intestinal inflammation compared with Th1-type inflammation caused by transfer of CD45RBhi CD4+ T cells.128 Interestingly, transfer of IL-22 KO Foxp3-CD45RBlo CD4+ T cells caused reduced inflammation compared with transfer of WT Foxp3-CD45RBlo CD4+ T cells, demonstrating a pathological role of IL-22 in this model of intestinal inflammation in contrast to the CD45RBhi CD4+ T cell transfer model, as well as heterogeneous functions of IL-22 in IBD.128 In addition, in anti-CD40 antibody-induced acute innate colitis models in Rag1-/- mice, IL-23R-dependent IL-22 production promoted inflammation, demonstrating that IL-22 can have pathogenic effects also in innate models of colitis.129 Thus, results from studies with different animal models demonstrate high functional heterogeneity of IL-22 depending, for example, on cytokine environment.130 The mean serum IL-22 is significantly higher in CD patients with the CD-associated IL23R variant compared with patients with the variant associated with decreased CD risk, but the clinical relevance of this has yet to be ascertained.120 Overall, no clinically applicable stratification has yet been demonstrated to translate the observed heterogeneity in IL-22 functions.
3. Transcriptional stratification strategies
Stratification strategies of IBD patients based on transcriptomic profiles have been recently developed. Kugathasan et al. showed that at disease onset, paediatric CD patients who later developed stricturing disease behaviour had upregulation of extracellular matrix accumulation-associated genes in ileum, whereas patients who later developed penetrating disease had upregulation of genes associated with acute microbial immune responses131 [Table 1]. Biasci et al. validated a transcriptional signature based on 17 genes in whole-blood samples from IBD patients which was predictive of aggressive disease course.132 The clustering was based on an unbiased patient stratification according to transcriptomic profile in circulating CD8+ T-cells, which was earlier shown to predict disease course in IBD.132,133 Interestingly, both the stratification strategy in whole blood and that in CD8+ T cells were independent of the diagnosis UC or CD.132,133 However, an attempt at validation of patient stratification based on transcriptional profile in CD8+ T cells has recently reported negative results,134 highlighting the importance of validating molecular predictors of such clinical importance in independent cohorts. Of note however, in this validation cohort the CD8+ T cell profile was associated with the age of the patient, indicating that this insight might shed light on the clinical heterogeneity between childhood- and adult-onset IBD.
3.1. Heterogeneous responses to anti-TNF treatment
Heterogenereous responses to different treatment options among IBD patients have led to active search for predictive biomarkers.135 Stevens et al. performed in 2018 a literature search and found 92 articles on predictive biomarkers of response to therapies, mostly to anti-TNF agents, but no study was evaluated to have overall low risk of bias.136 However, high pre-treatment levels of oncostatin M [OSM] have been shown to predict poor response to anti-TNF treatment.137 Further, OSM was shown to have pathological significance in driving anti-TNF-resistant colitis in a murine model, and blocking OSM could attenuate colitis.137 Thus, OSM is not only a biomarker but also a potential treatment target with pathological significance.137 Further, high OSM expression is associated with a generally different transcriptomic profile,137 possibly revealing that different pathological mechanisms are relevant in anti-TNF responders and non-responders. On the other hand, OSM expression was significantly correlated with inflammation severity and macroscopic lesions, suggesting that anti-TNF resistance might reflect an escalated state of inflammation.137 However, baseline clinical Mayo scores [consisting of categories stool frequency, rectal bleeding, endoscopic findings, and physician’s global assessment, where each is scored between 0–3] were not different between anti-TNF responders and non-responders, although their OSM expression levels were significantly different.137
Developments in scRNA-seq technologies have provided novel ways for patient stratification based on cellular composition. In that way, Martin et al. found that a subset of ileal CD patients has distinct cellular profile in the inflamed ileum, with accumulation of IgG plasma cells, inflammatory mononuclear phagocytes, activated T cells, and stromal cells.138 Further, they demonstrated that presence of the cellular profile is associated with altered cytokine signalling compared with other ileal CD patients, as well as non-response to anti-TNF therapy.138 Correspondingly, Smillie et al. demonstrated a cellular profile in UC with increase in inflammatory fibroblasts that are enriched in gene expression associated with colitis, fibrosis, and cancer, and inflammatory monocytes and DC2 subsets that predicted non-response to anti-TNF treatment.139,140 Using gene expression profiling to predict disease course has great clinical potential, but the extent to which these profiles identify more aggressive inflammation in a way that is not captured by traditional endoscopic scores such as Mayo, and to what extent these profiles identify patients with immunological signatures that are specifically resistant to particular drugs, has not yet been resolved.
3.2. Molecular taxonomy of IBD
Molecular profiling will likely be central in future clinical decision making.141 For example, in patients with multifocal inflammation, finding the driving factors of inflammation with specific EIMs could allow treatment of many inflammatory manifestations simultaneously.9 However, in biomarker discovery there is the well-acknowledged problem that inter-individual variability that is not relevant for the disease itself, e.g., differences in environment and lifestyle, accounts for most of the observable data variability.142 For example, Lloyd-Price et al. followed longitudinally gut microbial constitution in UC and CD patients and controls and showed that inter-individual variability explained most of the variation between samples, surpassing even the explaining effect of the diagnosis.52 Thus, to support the field of personalised medicine, new methodologies are required to distinguish data variability that is relevant for the disease itself from random data noise.
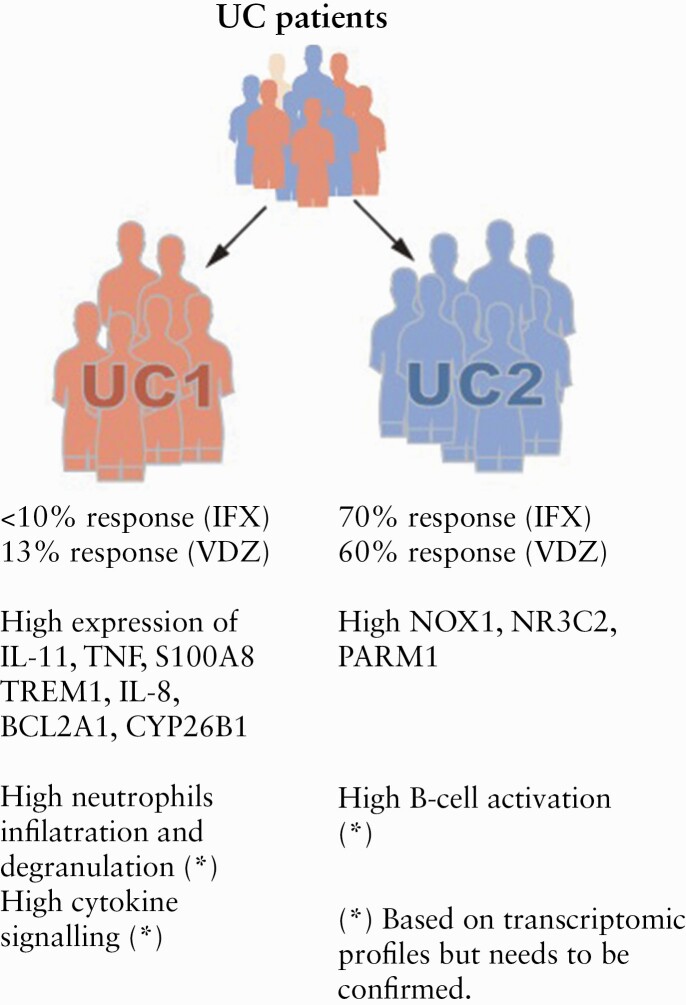
Here, as an example of one approach to this problem, we will describe a novel classification of UC patients into two molecularly distinct groups, UC1 and UC2, which differed in immune-mediated transcriptomic signatures but had similar partial Mayo score [clinical Mayo score excluding endoscopic findings]143 [Figure 3]. The classification was obtained by unsupervised human patient stratification in respect to evolutionarily conserved transcriptomic profile between mouse and human colitis,143 which is a totally new approach to study heterogeneity in IBD. Selecting genes that were differentially expressed also during the mouse colitis made it possible to create a clustering based on genes with a true biological significance during inflammation.143 Also, the fact that they are evolutionary conserved suggests that they are fundamental to the inflammatory process.
Importantly, it showed up that less than 10% of patients in cluster UC1 were responsive to infliximab [anti-TNF] therapy, whereas on average 70% of patients in cluster UC2 were responsive143 [Figure 3]. Furthermore, response to vedolizumab was an average 60% and 13% in clusters UC2 and UC1, respectively143 [Figure 3]. Since infliximab and vedolizumab have fundamentally different mechanisms of action, it is unclear how the same clustering can predict response to both drugs. However, this unbiased clustering suggests that responders and non-responders for these medications have biologically different disease types. One could speculate that UC1 patients are generally less responsive to all kinds of treatments or that they would need much higher doses compared with UC2 patients. For example, UC1 could patients leak more drug through a damaged gut barrier.144 Indeed, it has been demonstrated that infliximab primary non-responders have higher faecal concentration of infliximab during the first day after infusion compared with responders, although the same weight-adjusted dose is given.145 Also, there are data suggesting that the Fc part of infliximab is needed to obtain its therapeutic effect in colitis,146 and the Fc part is also present in vedolizumab. Further, it is possible that the UC1-UC2 stratification relates to a downstream immunological pathway common to both drugs despite their differing mechanisms of action. Whether immune function differences between UC1 and UC2 clusters are stable in a given individual or dynamic during the disease course, and how environmental factors among others affect the classification, is still to be discovered.
Among the most differentially expressed genes [DEGs] between clusters UC1 and UC2, higher expression of TNF, S100 calcium-binding protein A8 [S100A8], IL-11, triggering receptor expressed on myeloid cells 1 [TREM-1], IL8, BCL2-related protein A1 [BCL2A1] and cytochrome P450 26B1 [CYP26B1], and lower expression of NADPH oxidase 1 [NOX1], nuclear receptor subfamily 3 group c member 2 [NR3C2], and prostatic androgen-repressed message-1 [PARM-1], defined UC1 from UC2143 [Figure 3, Table 2]. Totally there were 56 DEGs that defined the UC1/UC2 clustering [Table 2]. Overall, results were partly in agreement with reports in which biased stratification based on response to therapy was made.140,147
Table 2.
All UC1 and UC2 genes. This table summarises all the genes in UC1 and UC2 clustering together with possible implications in colitis.
| Cluster | Gene | Possible functional implications in colitis |
|---|---|---|
| Reduced in UC1 relative to UC2 | PARM1 | Expression increased by inflammatory cytokines, but no found functions in colitis |
| NR3C2 | Receptor activated by cortisol and aldosterone, but no found functions in colitis | |
| NOX1 | Oxidative stress | |
| MAP2K2 | A member of mitogen-activated protein kinase family that has many pro-inflammatory effects in colitis1 | |
| Increased in UC1 relative to UC2 | COL15A1 | Collagen synthesis2 |
| COL12A1 | ||
| COL4A1 | ||
| COL4A2 | ||
| COL7A1 | ||
| MMP3 | ECM breakdown2 | |
| MMP1 | ||
| WNT5A | WNT gene family,2 important in intestinal epithelial regeneration and probably in wound healing in colitis,3 linked to many inflammatory pathways3 and might be possible activators of expression of metalloproteases4 | |
| WNT2 | ||
| WISP1 | ||
| IL6 | Cytokine and cell-to-cell signalling2 | |
| IL24 | ||
| IL11 | ||
| IL1B | ||
| IL33 | ||
| IL1R2 | ||
| IL1RN | ||
| IL7R | ||
| IL18RAP | ||
| IL13RA2 | ||
| TNF | ||
| TNFRSF11B | ||
| CSFF2RB | ||
| CSF3 | ||
| CSF3R | ||
| TGFBI | ||
| SOCS3 | ||
| TREM1 | ||
| CXCR2 | ||
| ICAM1 | ||
| ITGA5 | ||
| CXCR1 | ||
| CXCL8 | ||
| CXCL6 | ||
| CCR1 | ||
| ICAM3 | ||
| TLR1 | Innate immunity, cell migration and neutrophil degranulation2 | |
| TLR2 | ||
| CD14 | ||
| S100A4 | ||
| S100A9 | ||
| S100A8 | ||
| S100A12 | ||
| PLAU | ||
| C5AR1 | ||
| SELL | ||
| SELE | ||
| FGR | ||
| BCL2A1 | ||
| PTGS2 | ||
| CYP26B1 | ||
| FAM49A | No found functions in colitis |
UC, ulcerative colitis; ECM, extracellular matrix.
References: 1Broom OJ, Widjaya B, Troelsen J, Olsen J, Nielsen OH. Mitogen Activated Protein Kinases: A Role in Inflammatory Bowel Disease? Clin Exp Immunol 2009;158[3]:272–80. 2National Center for Biotechnology Information, U.S. National Library of Medicine. NCBI Gene. https://www.ncbi.nlm.nih.gov/gene/ Last accessed June 26, 2020.3Moparthi L, Koch S. Wnt signaling in intestinal inflammation. Differentiation 2019;108:24–32. 4Tamamura Y, Otani T, Kanatani N, et al. Developmental Regulation of Wnt/beta-catenin Signals Is Required for Growth Plate Assembly, Cartilage Integrity, and Endochondral Ossification. J Biol Chem 2005;280:19185–95.
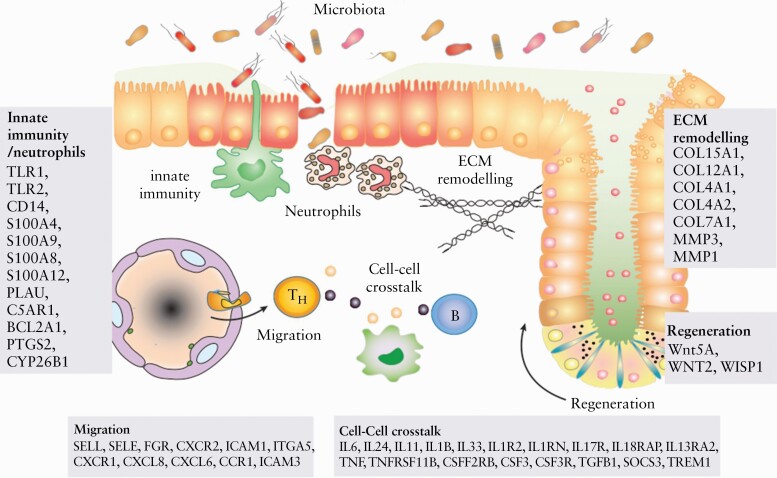
The cluster UC1 is characterised by both genes previously linked to IBD pathogenesis and genes without previous known significance in IBD [Figure 4]. Fascinatingly, the genes expressed at higher level in UC1 cover many of the functions previously identified to be involved with IBD pathogenesis, namely pro-inflammatory cytokine signalling, granulocyte maturation and degranulation, molecules effecting immune amplification, epithelial cell regeneration and gut barrier function, immune cell chemotaxis, regulatory effects of RA, cellular metabolic processes, and apoptosis. This may imply that the UC1 cluster comprehensively captures multiple dimensions of IBD pathogenesis which lead to unresponsiveness to biologic drugs infliximab and vedolizumab [Figure 2].143 In contrast, the UC2 cluster is characterised by upregulation of genes that have few specific implications in gut immune functions [Table 2].
Figure 4.
Illustration of the function of genes differentially upregulated in UC1 compared with UC2 patients in the context of gut inflammation. The genes that are differentially upregulated are related to innate immunity and neutrophil activation, migration of immune cells, cell-cell cross-talk in inflammation, tissue regeneration, and ECM remodelling.118 Thus, it may be that these functions are more highly activated in UC1 than UC2 patients. However, this hypothesis should be confirmed in future studies. UC, ulcerative colitis; Th, T helper cell; B, B cell; ECM, extracellular matrix.
It may be speculated that patients in cluster UC1 have more severe colitis, rather than a distinct inflammatory phenotype. Also, in a mouse model of colitis with adoptive transfer of CD4+CD45RBhi T cells, intestinal TREM-1 expression correlates [albeit weakly] with the severity of the inflammation.148 An earlier study has associated high baseline C-reactive protein and low baseline albumin levels, which are markers of acute severe colitis, with poor response to infliximab in the long term.149
Severe disease also predicts poor response for vedolizumab.150 However, Sjöberg et al. have shown that 50% and 54% of patients receiving infliximab as a rescue treatment for hospitalisation-requiring flare of UC had steroid-free clinical remission at 3 and 12 months, respectively, which are remarkably higher response rates compared with those seen in cluster UC1 patients in our study,143,151 supporting that the UC1 and UC2 clustering is more accurate in predicting response to infliximab than clinically observable disease activity is. Corresponding results have been reported by Jarnerot et al.152 Further, the UC1-UC2 stratification appears to provide a greater value for patient stratification than partial Mayo score, which was same between patients in clusters UC1 and UC2.143 However, among paediatric UC1 patients, total clinical Mayo and Paediatric Ulcerative Colitis Activity Index [PUCAI] scores were higher compared with UC2 patients, although no significant difference existed between UC1 and UC2 for histological score or calprotectin.143
One could also speculate that the UC1 cluster represents an earlier state of a flare, that is that UC1 and UC2 may represent different phases of the disease and as such may be observed in the same patient at different time points. Indeed, in the DSS mouse colitis model, genes associated with inflammatory responses and neutrophil degranulation were upregulated during the acute phase of DSS colitis143 but not in later phases. It may be that molecular changes towards recovery occur before clinically observable signs, explaining why UC1 and UC2 patients could not generally be distinguished with partial Mayo score. In that case, it could be also that UC1 patients have a prolonged acute phase of the inflammation and are unable to initiate the recovery phase, whereas UC2 patients may have already activated mechanisms of healing and are thus primed to respond positively to drug treatments. Hence, UC1 and UC2 classification could provide knowledge on optimal drug dose and optimal time point for the start of a biological therapy. Moreover, if UC1 and UC2 represent different phases of the patients’ progression from inflammation to healing, this would provide potential molecular targets to directly enhance the recovery and regeneration process or enhance response to biologic therapies.
The earlier described stratification strategy by Biasci D et al. which is based on transcriptomic profiles in whole blood does not include any genes present among the most differentially expressed genes between UC1 and UC2, demonstrating entirely distinct biological backgrounds of these two stratification strategies.132,143 Of note, OSM was not found among differentially expressed genes between UC1 and UC2, though both OSM expression levels and UC1 and UC2 stratification predict response to anti-TNF therapy.137,143 This might indicate that OSM is not biologically related to UC1 and UC2 stratification profiles and predicts anti-TNF response based on different biological grounds. However, IL-1β, IL-6, and IL-11 are upregulated in both UC1 patients and patients with high OSM,137,143 suggesting that these profiles could be partly overlapping. Smillie et al. demonstrated that inflammation-associated fibroblasts, inflammatory monocytes, and DC2 subsets are not only predictive of non-response to anti-TNF, but also that fibroblasts express OSM receptor whereas monocytes and DC2 subsets express OSM, thus likely being linked to OSM-mediated TNF resistance.140 Interestingly, they also demonstrated that UC1 genes IL-11, IL13RA2, TNFRSF11B, and CXCL6 are mostly expressed by inflammation-associated fibroblasts, TREM-1 by inflammatory monocytes and DC2s, and UC1 gene PTGS2 by all of these cell types, suggesting that these key cell types might be involved in both OSM signalling and the pathogenesis in the UC1 cluster.140,143 Further characterisation of the UC1 and UC2 clusters might provide additional clues on how immunity underlines clinical disease heterogeneity.
4. Concluding remarks
Patient stratification, including prediction of disease course, complications, and treatment responses, are currently the main goals of IBD research.153 Thus, clustering strategies like UC1 and UC2 are needed to provide the immunological foundation for understanding the observed heterogeneity of IBD. This review has mostly focused on immune functions as a base for heterogeneity in IBD, but many other intestinal cell types and pathways also have their role in observed heterogeneity. For example, the epithelial layer has a key function in intestinal homeostasis and antimicrobial defence, integrating with other intestinal cell types and luminal content.154 Indeed, integration of data of, for example, genetics, epigenetics, microbiology, immunology, and metabolism in combination with clinical data, so called -omics technology, will now facilitate the search for non-invasive and even more precise biomarkers and enhance understanding of the aetiopathogenesis of IBD.29,141,142 Also, big data and biostatistical approaches, similar to those employed to detect UC1 and UC2 clusters, provide numerous possibilities to find new approaches for precision medicine.155 Although there are still few clinically useable clustering strategies, technological developments continue apace,141 and are likely to revolutionise future clinical management of IBD.
Funding
No specific funding has been received for this work.
Conflicts of Interest
CRHH has received speaker fees from Takeda, Ferring, AbbVie, and Janssen, and consultancy fees from Pfizer, and has acted as local principal investigator for clinical trials for Janssen and GlaxoSmithKline. EJV has received research funding from F. Hoffmann-La Roche AG.
Author Contributions
The manuscript and figures were drafted by KAS and repeatedly revised for important intellectual content by CRHH and EJV.
References
- 1. Kirsner JB. Historical origins of current IBD concepts. World J Gastroenterol 2001;7:175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Subramanian S, Ekbom A, Rhodes JM. Recent advances in clinical practice: a systematic review of isolated colonic Crohn’s disease: the third IBD? Gut 2017;66:362–81. [DOI] [PubMed] [Google Scholar]
- 3. Feuerstein JD, Moss AC, Farraye FA. Ulcerative colitis. Mayo Clin Proc 2019;94:1357–73. [DOI] [PubMed] [Google Scholar]
- 4. Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn’s disease. Lancet 2017;389:1741–55. [DOI] [PubMed] [Google Scholar]
- 5. Summers JE. VII. The surgical treatment of chronic mucomembranous and ulcerative colitis, with special reference to technique. Ann Surg 1905;42:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abdalla MI, Sandler RS, Kappelman MD, et al. Prevalence and impact of inflammatory bowel disease-irritable bowel syndrome on patient-reported outcomes in CCFA partners. Inflamm Bowel Dis 2017;23:325–31. [DOI] [PubMed] [Google Scholar]
- 7. Dulai PS, Singh S, Vande Casteele N, et al. Should we divide Crohn’s disease into ileum-dominant and isolated colonic diseases? Clin Gastroenterol Hepatol 2019;17:2634–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sørensen JØ, Nielsen OH, Andersson M, et al. Inflammatory bowel disease with primary sclerosing cholangitis: a Danish population-based cohort study 1977-2011. Liver Int 2018;38:532–41. [DOI] [PubMed] [Google Scholar]
- 9. Hedin CRH, Vavricka SR, Stagg AJ, et al. The pathogenesis of extraintestinal manifestations: implications for IBD research, diagnosis, and therapy. J Crohns Colitis 2019;13:541–54. [DOI] [PubMed] [Google Scholar]
- 10. Harbord M, Annese V, Vavricka SR, et al. ; European Crohn’s and Colitis Organisation. The first European evidence-based consensus on extra-intestinal manifestations in inflammatory bowel disease. J Crohns Colitis 2016;10:239–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mitsialis V, Wall S, Liu P, et al. ; Boston Children’s Hospital Inflammatory Bowel Disease Center; Brigham and Women’s Hospital Crohn’s and Colitis Center. Single-cell analyses of colon and blood reveal distinct immune cell signatures of ulcerative colitis and Crohn’s disease. Gastroenterology 2020;159:591–608.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gower-Rousseau C, Vasseur F, Fumery M, et al. Epidemiology of inflammatory bowel diseases: new insights from a French population-based registry [EPIMAD]. Dig Liver Dis 2013;45:89–94. [DOI] [PubMed] [Google Scholar]
- 13. Guariso G, Gasparetto M, Visonà Dalla Pozza L, et al. Inflammatory bowel disease developing in paediatric and adult age. J Pediatr Gastroenterol Nutr 2010;51:698–707. [DOI] [PubMed] [Google Scholar]
- 14. Vernier-Massouille G, Balde M, Salleron J, et al. Natural history of pediatric Crohn’s disease: a population-based cohort study. Gastroenterology 2008;135:1106–13. [DOI] [PubMed] [Google Scholar]
- 15. Charpentier C, Salleron J, Savoye G, et al. Natural history of elderly-onset inflammatory bowel disease: a population-based cohort study. Gut 2014;63:423–32. [DOI] [PubMed] [Google Scholar]
- 16. Paul T, Birnbaum A, Pal DK, et al. Distinct phenotype of early childhood inflammatory bowel disease. J Clin Gastroenterol 2006;40:583–6. [DOI] [PubMed] [Google Scholar]
- 17. Heyman MB, Kirschner BS, Gold BD, et al. Children with early-onset inflammatory bowel disease [IBD]: analysis of a pediatric IBD consortium registry. J Pediatr 2005;146:35–40. [DOI] [PubMed] [Google Scholar]
- 18. Ruel J, Ruane D, Mehandru S, Gower-Rousseau C, Colombel JF. IBD across the age spectrum: is it the same disease? Nat Rev Gastroenterol Hepatol 2014;11:88–98. [DOI] [PubMed] [Google Scholar]
- 19. Conte MP, Schippa S, Zamboni I, et al. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 2006;55:1760–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Cruz P, Prideaux L, Wagner J, et al. Characterization of the gastrointestinal microbiota in health and inflammatory bowel disease. Inflamm Bowel Dis 2012;18:372–90. [DOI] [PubMed] [Google Scholar]
- 21. Kelsen JR, Baldassano RN. The role of monogenic disease in children with very early onset inflammatory bowel disease. Curr Opin Pediatr 2017;29:566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bianco AM, Girardelli M, Tommasini A. Genetics of inflammatory bowel disease from multifactorial to monogenic forms. World J Gastroenterol 2015;21:12296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tegtmeyer D, Seidl M, Gerner P, Baumann U, Klemann C. Inflammatory bowel disease caused by primary immunodeficiencies ‐ clinical presentations, review of literature, and proposal of a rational diagnostic algorithm. Pediatr Allergy Immunol 2017;28:412–29. [DOI] [PubMed] [Google Scholar]
- 24. Uhlig HH, Muise AM. Clinical genomics in inflammatory bowel disease. Trends Genet 2017;33:629–41. [DOI] [PubMed] [Google Scholar]
- 25. de Valles-Ibáñez G, Esteve-Solé A, Piquer M, et al. Evaluating the genetics of common variable immunodeficiency: monogenetic model and beyond. Front Immunol 2018;9:636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fischer A, Notarangelo LD, Neven B, Cavazzana M, Puck JM. Severe combined immunodeficiencies and related disorders. Nat Rev Dis Primers 2015;1:15061. [DOI] [PubMed] [Google Scholar]
- 27. McGovern DP, Kugathasan S, Cho JH. Genetics of Inflammatory Bowel Diseases. Gastroenterology 2015;149:1163–76.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prescott NJ, Lehne B, Stone K, et al. ; UK IBD Genetics Consortium. Pooled sequencing of 531 genes in inflammatory bowel disease identifies an associated rare variant in BTNL2 and implicates other immune related genes. PLoS Genet 2015;11:e1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stylianou E. Recent advances in the etiopathogenesis of inflammatory bowel disease: the role of omics. Mol Diagn Ther 2018;22:11–23. [DOI] [PubMed] [Google Scholar]
- 30. Furey TS, Sethupathy P, Sheikh SZ. Redefining the IBDs using genome-scale molecular phenotyping. Nat Rev Gastroenterol Hepatol 2019;16:296–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Verstockt B, Smith KG, Lee JC. Genome-wide association studies in Crohn’s disease: past, present and future. Clin Transl Immunology 2018;7:e1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu JZ, van Sommeren S, Huang H, et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013;45:1238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marigorta UM, Denson LA, Hyams JS, et al. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn’s disease. Nat Genet 2017;49:1517–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee JC, Biasci D, Roberts R, et al. ; UK IBD Genetics Consortium. Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat Genet 2017;49:262–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dragasevic S, Stankovic B, Milosavljevic T, et al. Genetic and environmental factors significant for the presentation and development of inflammatory bowel disease. Eur J Gastroenterol Hepatol 2017;29:909–15. [DOI] [PubMed] [Google Scholar]
- 37. Diaz OE, Morales RA, Srustidhar D, Villablanca EJ. Experimental models of intestinal inflammation: lessons from mouse and zebrafish. In: Hedin C, Rioux JD, D’Amato M, editors. Molecular Genetics of Inflammatory Bowel Disease. New York, NY: Springer; 2019. [Google Scholar]
- 38. Bauer C, Duewell P, Mayer C, et al. Colitis induced in mice with dextran sulfate sodium [DSS] is mediated by the NLRP3 inflammasome. Gut 2010;59:1192–9. [DOI] [PubMed] [Google Scholar]
- 39. Holgersen K, Kutlu B, Fox B, et al. High-resolution gene expression profiling using RNA sequencing in patients with inflammatory bowel disease and in mouse models of colitis. J Crohns Colitis 2015;9:492–506. [DOI] [PubMed] [Google Scholar]
- 40. Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, Strober W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity 2002;17:629–38. [DOI] [PubMed] [Google Scholar]
- 41. Meroni E, Stakenborg N, Gomez-Pinilla PJ, et al. Functional characterization of oxazolone-induced colitis and survival improvement by vagus nerve stimulation. PLoS One 2018;13:e0197487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ostanin DV, Bao J, Koboziev I, et al. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol 2009;296:G135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Powrie F, Leach MW, Mauze S, Menon S, Caddle LB, Coffman RL. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity 1994;1:553–62. [DOI] [PubMed] [Google Scholar]
- 44. Keubler LM, Buettner M, Häger C, Bleich A. A multihit model: colitis lessons from the interleukin-10-deficient mouse. Inflamm Bowel Dis 2015;21:1967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Berg DJ, Davidson N, Kühn R, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4[+] TH1-like responses. J Clin Invest 1996;98:1010–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yen D, Cheung J, Scheerens H, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest 2006;116:1310–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilson MS, Ramalingam TR, Rivollier A, et al. Colitis and intestinal inflammation in IL10-/- mice results from IL-13Rα2-mediated attenuation of IL-13 activity. Gastroenterology 2011;140:254–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kozik AJ, Nakatsu CH, Chun H, Jones-Hall YL. Age, sex, and TNF associated differences in the gut microbiota of mice and their impact on acute TNBS colitis. Exp Mol Pathol 2017;103:311–9. [DOI] [PubMed] [Google Scholar]
- 49. Roy U, Gálvez EJC, Iljazovic A, et al. Distinct microbial communities trigger colitis development upon intestinal barrier damage via innate or adaptive immune cells. Cell Rep 2017;21:994–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Halfvarson J, Brislawn CJ, Lamendella R, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017;2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. ; IBDMDB Investigators. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. ; IBDMDB Investigators. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol 2018;11:1–10. [DOI] [PubMed] [Google Scholar]
- 54. Gevers D, Kugathasan S, Knights D, Kostic AD, Knight R, Xavier RJ. A microbiome foundation for the study of Crohn’s disease. Cell Host Microbe 2017;21:301–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Plichta DR, Graham DB, Subramanian S, Xavier RJ. Therapeutic opportunities in inflammatory bowel disease: mechanistic dissection of host-microbiome relationships. Cell 2019;178:1041–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schirmer M, Garner A, Vlamakis H, Xavier RJ. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol 2019;17:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Scanlan PD, Shanahan F, O’Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol 2006;44:3980–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol 2006;44:4136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vich Vila A, Imhann F, Collij V, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med 2018;10:eaap8914. [DOI] [PubMed] [Google Scholar]
- 61. Yilmaz B, Juillerat P, Øyås O, et al. ; Swiss IBD Cohort Investigators. Microbial network disturbances in relapsing refractory Crohn’s disease. Nat Med 2019;25:323–36. [DOI] [PubMed] [Google Scholar]
- 62. Dovrolis N, Michalopoulos G, Theodoropoulos GE, et al. The interplay between mucosal microbiota composition and host gene-expression is linked with infliximab response in inflammatory bowel diseases. Microorganisms 2020;8:438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rajca S, Grondin V, Louis E, et al. Alterations in the intestinal microbiome [dysbiosis] as a predictor of relapse after infliximab withdrawal in Crohn’s disease. Inflamm Bowel Dis 2014;20:978–86. [DOI] [PubMed] [Google Scholar]
- 64. Strömbeck A, Lasson A, Strid H, et al. Fecal microbiota composition is linked to the postoperative disease course in patients with Crohn’s disease. BMC Gastroenterol 2020;20:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Moller FT, Knudsen L, Harbord M, et al. Danish cohort of monozygotic inflammatory bowel disease twins: clinical characteristics and inflammatory activity. World J Gastroenterol 2016;22:5050–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Spehlmann ME, Begun AZ, Burghardt J, Lepage P, Raedler A, Schreiber S. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm Bowel Dis 2008;14:968–76. [DOI] [PubMed] [Google Scholar]
- 68. Ananthakrishnan AN, Bernstein CN, Iliopoulos D, et al. Environmental triggers in IBD: a review of progress and evidence. Nat Rev Gastroenterol Hepatol 2018;15:39–49. [DOI] [PubMed] [Google Scholar]
- 69. Khalili H, Chan SSM, Lochhead P, Ananthakrishnan AN, Hart AR, Chan AT. The role of diet in the aetiopathogenesis of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2018;15:525–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Castro F, de Souza HSP. Dietary composition and effects in inflammatory bowel disease. Nutrients 2019;11:1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 2010;107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science 2008;320:1647–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Costea I, Mack DR, Lemaitre RN, et al. Interactions between the dietary polyunsaturated fatty acid ratio and genetic factors determine susceptibility to pediatric Crohn’s disease. Gastroenterology 2014;146:929–31. [DOI] [PubMed] [Google Scholar]
- 74. Ananthakrishnan AN, Khalili H, Song M, et al. Genetic polymorphisms in fatty acid metabolism modify the association between dietary n3: n6 intake and risk of ulcerative colitis: a prospective cohort study. Inflamm Bowel Dis 2017;23:1898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Khalili H, Malik S, Ananthakrishnan AN, et al. Identification and characterization of a novel association between dietary potassium and risk of Crohn’s disease and ulcerative colitis. Front Immunol 2016;7:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Khalili H, de Silva PS, Ananthakrishnan AN, et al. Dietary iron and heme iron consumption, genetic susceptibility, and risk of Crohn’s disease and ulcerative Colitis. Inflamm Bowel Dis 2017;23:1088–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kmieć Z, Cyman M, Ślebioda TJ. Cells of the innate and adaptive immunity and their interactions in inflammatory bowel disease. Adv Med Sci 2017;62:1–16. [DOI] [PubMed] [Google Scholar]
- 78. Agace WW, Persson EK. How vitamin A metabolizing dendritic cells are generated in the gut mucosa. Trends Immunol 2012;33:42–8. [DOI] [PubMed] [Google Scholar]
- 79. Jaensson-Gyllenbäck E, Kotarsky K, Zapata F, et al. Bile retinoids imprint intestinal CD103+ dendritic cells with the ability to generate gut-tropic T cells. Mucosal Immunol 2011;4:438–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Villablanca EJ, Wang S, de Calisto J, et al. MyD88 and retinoic acid signaling pathways interact to modulate gastrointestinal activities of dendritic cells. Gastroenterology 2011;141:176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cassani B, Villablanca EJ, Quintana FJ, et al. Gut-tropic T cells that express integrin α4β7 and CCR9 are required for induction of oral immune tolerance in mice. Gastroenterology 2011;141:2109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hadis U, Wahl B, Schulz O, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 2011;34:237–46. [DOI] [PubMed] [Google Scholar]
- 83. Bakdash G, Vogelpoel LT, van Capel TM, Kapsenberg ML, de Jong EC. Retinoic acid primes human dendritic cells to induce gut-homing, IL-10-producing regulatory T cells. Mucosal Immunol 2015;8:265–78. [DOI] [PubMed] [Google Scholar]
- 84. Magnusson MK, Strid H, Sapnara M, et al. Anti-TNF therapy response in patients with ulcerative colitis is associated with colonic antimicrobial peptide expression and microbiota composition. J Crohns Colitis 2016;10:943–52. [DOI] [PubMed] [Google Scholar]
- 85. Dige A, Magnusson MK, Öhman L, et al. Reduced numbers of mucosal DR[int] macrophages and increased numbers of CD103[+] dendritic cells during anti-TNF-α treatment in patients with Crohn’s disease. Scand J Gastroenterol 2016;51:692–9. [DOI] [PubMed] [Google Scholar]
- 86. Hart AL, Al-Hassi HO, Rigby RJ, et al. Characteristics of intestinal dendritic cells in inflammatory bowel diseases. Gastroenterology 2005;129:50–65. [DOI] [PubMed] [Google Scholar]
- 87. Platt AM, Bain CC, Bordon Y, Sester DP, Mowat AM. An independent subset of TLR expressing CCR2-dependent macrophages promotes colonic inflammation. J Immunol 2010;184:6843–54. [DOI] [PubMed] [Google Scholar]
- 88. Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells: a new paradigm in immunology. Science 2015;348:aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Vivier E, Artis D, Colonna M, et al. Innate lymphoid cells: 10 years on. Cell 2018;174:1054–66. [DOI] [PubMed] [Google Scholar]
- 90. Fuchs A, Vermi W, Lee JS, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 2013;38:769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bernink JH, Peters CP, Munneke M, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol 2013;14:221–9. [DOI] [PubMed] [Google Scholar]
- 92. Geremia A, Arancibia-Cárcamo CV, Fleming MP, et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med 2011;208:1127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Takayama T, Kamada N, Chinen H, et al. Imbalance of NKp44[+]NKp46[-] and NKp44[-]NKp46[+] natural killer cells in the intestinal mucosa of patients with Crohn’s disease. Gastroenterology 2010;139:882–92. [DOI] [PubMed] [Google Scholar]
- 94. Buonocore S, Ahern PP, Uhlig HH, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 2010;464:1371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pearson C, Thornton EE, McKenzie B, et al. ILC3 GM-CSF production and mobilisation orchestrate acute intestinal inflammation. Elife 2016;5:e10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gwela A, Siddhanathi P, Chapman RW, et al. Th1 and innate lymphoid cells accumulate in primary sclerosing cholangitis-associated inflammatory bowel disease. J Crohns Colitis 2017;11:1124–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Panda SK, Colonna M. Innate lymphoid cells in mucosal immunity. Front Immunol 2019;10:861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Forkel M, van Tol S, Höög C, Michaëlsson J, Almer S, Mjösberg J. Distinct alterations in the composition of mucosal innate lymphoid cells in newly diagnosed and established Crohn’s disease and ulcerative colitis. J Crohns Colitis 2019;13:67–78. [DOI] [PubMed] [Google Scholar]
- 99. Roncarolo MG, Gregori S, Bacchetta R, Battaglia M, Gagliani N. The biology of T regulatory type 1 cells and their therapeutic application in immune-mediated diseases. Immunity 2018;49:1004–19. [DOI] [PubMed] [Google Scholar]
- 100. Mohr A, Atif M, Balderas R, Gorochov G, Miyara M. The role of FOXP3+ regulatory T cells in human autoimmune and inflammatory diseases. Clin Exp Immunol 2019;197:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Saruta M, Yu QT, Fleshner PR, et al. Characterization of FOXP3+CD4+ regulatory T cells in Crohn’s disease. Clin Immunol 2007;125:281–90. [DOI] [PubMed] [Google Scholar]
- 102. Holmén N, Lundgren A, Lundin S, et al. Functional CD4+CD25high regulatory T cells are enriched in the colonic mucosa of patients with active ulcerative colitis and increase with disease activity. Inflamm Bowel Dis 2006;12:447–56. [DOI] [PubMed] [Google Scholar]
- 103. Reikvam DH, Perminow G, Lyckander LG, et al. Increase of regulatory T cells in ileal mucosa of untreated pediatric Crohn’s disease patients. Scand J Gastroenterol 2011;46:550–60. [DOI] [PubMed] [Google Scholar]
- 104. Ueno A, Jijon H, Chan R, et al. Increased prevalence of circulating novel IL-17 secreting Foxp3 expressing CD4+ T cells and defective suppressive function of circulating Foxp3+ regulatory cells support plasticity between Th17 and regulatory T cells in inflammatory bowel disease patients. Inflamm Bowel Dis 2013;19:2522–34. [DOI] [PubMed] [Google Scholar]
- 105. Veltkamp C, Anstaett M, Wahl K, et al. Apoptosis of regulatory T lymphocytes is increased in chronic inflammatory bowel disease and reversed by anti-TNFα treatment. Gut 2011;60:1345–53. [DOI] [PubMed] [Google Scholar]
- 106. Gagliani N, Magnani CF, Huber S, et al. Coexpression of CD49b and LAG-3 identifies human and mouse T regulatory type 1 cells. Nat Med 2013;19:739–46. [DOI] [PubMed] [Google Scholar]
- 107. Brockmann L, Soukou S, Steglich B, et al. Molecular and functional heterogeneity of IL-10-producing CD4+ T cells. Nat Commun 2018;9:5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Iboshi Y, Nakamura K, Ihara E, et al. Multigene analysis unveils distinctive expression profiles of helper T-cell-related genes in the intestinal mucosa that discriminate between ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis 2014;20:967–77. [DOI] [PubMed] [Google Scholar]
- 109. Butera A, Di Paola M, Vitali F, et al. IL-13 mRNA tissue content identifies two subsets of adult ulcerative colitis patients with different clinical and mucosa-associated microbiota profiles. J Crohns Colitis 2020;14:369–80. [DOI] [PubMed] [Google Scholar]
- 110. Ogino T, Nishimura J, Barman S, et al. Increased Th17-inducing activity of CD14+ CD163 low myeloid cells in intestinal lamina propria of patients with Crohn’s disease. Gastroenterology 2013;145:1380–91.e1. [DOI] [PubMed] [Google Scholar]
- 111. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium [IIBDGC]. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Moschen AR, Tilg H, Raine T. IL-12, IL-23 and IL-17 in IBD: immunobiology and therapeutic targeting. Nat Rev Gastroenterol Hepatol 2019;16:185–96. [DOI] [PubMed] [Google Scholar]
- 113. Sandborn WJ, Gasink C, Gao LL, et al. ; CERTIFI Study Group. Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med 2012;367:1519–28. [DOI] [PubMed] [Google Scholar]
- 114. Sandborn WJ, Feagan BG, Fedorak RN, et al. ; Ustekinumab Crohn’s Disease Study Group. A randomized trial of Ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with moderate-to-severe Crohn’s disease. Gastroenterology 2008;135:1130–41. [DOI] [PubMed] [Google Scholar]
- 115. Fuss IJ, Becker C, Yang Z, et al. Both IL-12p70 and IL-23 are synthesized during active Crohn’s disease and are down-regulated by treatment with anti-IL-12 p40 monoclonal antibody. Inflamm Bowel Dis 2006;12:9–15. [DOI] [PubMed] [Google Scholar]
- 116. Neurath MF, Fuss I, Kelsall BL, Stüber E, Strober W. Antibodies to interleukin 12 abrogate established experimental colitis in mice. J Exp Med 1995;182:1281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Matsuoka K, Inoue N, Sato T, et al. T-bet upregulation and subsequent interleukin 12 stimulation are essential for induction of Th1 mediated immunopathology in Crohn’s disease. Gut 2004;53:1303–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Strober W, Fuss IJ. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology 2011;140:1756–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dudakov JA, Hanash AM, van den Brink MR. Interleukin-22: immunobiology and pathology. Annu Rev Immunol 2015;33:747–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Schmechel S, Konrad A, Diegelmann J, et al. Linking genetic susceptibility to Crohn’s disease with Th17 cell function: IL-22 serum levels are increased in Crohn’s disease and correlate with disease activity and IL23R genotype status. Inflamm Bowel Dis 2008;14:204–12. [DOI] [PubMed] [Google Scholar]
- 121. Arj A, Razavizadeh M, Mohammadi H, Nikoueinejad H, Akbari H. The correlation between the numerical status of Th22 cells and serum level of IL-22 with severity of ulcerative colitis. Iran J Allergy Asthma Immunol 2018;17:78–84. [PubMed] [Google Scholar]
- 122. Pelczar P, Witkowski M, Perez LG, et al. A pathogenic role for T cell-derived IL-22BP in inflammatory bowel disease. Science 2016;354:358–62. [DOI] [PubMed] [Google Scholar]
- 123. Dumoutier L, Lejeune D, Colau D, Renauld JC. Cloning and characterization of IL-22 binding protein, a natural antagonist of IL-10-related T cell-derived inducible factor/IL-22. J Immunol 2001;166:7090–5. [DOI] [PubMed] [Google Scholar]
- 124. Kotenko SV, Izotova LS, Mirochnitchenko OV, et al. Identification, cloning, and characterization of a novel soluble receptor that binds IL-22 and neutralizes its activity. J Immunol 2001;166:7096–103. [DOI] [PubMed] [Google Scholar]
- 125. Sugimoto K, Ogawa A, Mizoguchi E, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest 2008;118:534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Monteleone I, Rizzo A, Sarra M, et al. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 2011;141:237–48. [DOI] [PubMed] [Google Scholar]
- 127. Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity 2008;29:947–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Kamanaka M, Huber S, Zenewicz LA, et al. Memory/effector [CD45RB[lo]] CD4 T cells are controlled directly by IL-10 and cause IL-22-dependent intestinal pathology. J Exp Med 2011;208:1027–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Eken A, Singh AK, Treuting PM, Oukka M. IL-23R+ innate lymphoid cells induce colitis via interleukin-22-dependent mechanism. Mucosal Immunol 2014;7:143–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Mizoguchi A, Yano A, Himuro H, Ezaki Y, Sadanaga T, Mizoguchi E. Clinical importance of IL-22 cascade in IBD. J Gastroenterol 2018;53:465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kugathasan S, Denson LA, Walters TD, et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 2017;389:1710–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Biasci D, Lee JC, Noor NM, et al. A blood-based prognostic biomarker in IBD. Gut 2019;68:1386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Lee JC, Lyons PA, McKinney EF, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest 2011;121:4170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Gasparetto M, Payne F, Nayak K, et al. Transcription and DNA methylation patterns of blood-derived CD8+ T cells are associated with age and inflammatory bowel disease but do not predict prognosis. Gastroenterology 2021;160:232–44.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Wang C, Baer HM, Gaya DR, Nibbs RJB, Milling S. Can molecular stratification improve the treatment of inflammatory bowel disease? Pharmacol Res 2019;148:104442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Stevens TW, Matheeuwsen M, Lönnkvist MH, et al. Systematic review: predictive biomarkers of therapeutic response in inflammatory bowel disease-personalised medicine in its infancy. Aliment Pharmacol Ther 2018;48:1213–31. [DOI] [PubMed] [Google Scholar]
- 137. West NR, Hegazy AN, Owens BMJ, et al. ; Oxford IBD Cohort Investigators. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med 2017;23:579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Martin JC, Chang C, Boschetti G, et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 2019;178:1493–508.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Perez-Jeldres T, Alvarez-Lobos M, Rivera Nieves J, Villablanca EJ. The cell circuitry of ulcerative colitis, a new view for a highly complex disease. Gastroenterology 2020;158:1506–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Smillie CS, Biton M, Ordovas-Montanes J, et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 2019;178:714–30.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Lee HS, Cleynen I. Molecular profiling of inflammatory bowel disease: is it ready for use in clinical decision-making? Cells 2019;8:535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Metwaly A, Haller D. Multi-omics in IBD biomarker discovery: the missing links. Nat Rev Gastroenterol Hepatol 2019;16:587–8. [DOI] [PubMed] [Google Scholar]
- 143. Czarnewski P, Parigi SM, Sorini C, et al. Conserved transcriptomic profile between mouse and human colitis allows unsupervised patient stratification. Nat Commun 2019;10:2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Berends SE, Strik AS, Löwenberg M, D’Haens GR, Mathôt RAA. Clinical pharmacokinetic and pharmacodynamic considerations in the treatment of ulcerative colitis. Clin Pharmacokinet 2019;58:15–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Brandse JF, van den Brink GR, Wildenberg ME, et al. Loss of infliximab into feces is associated with lack of response to therapy in patients with severe ulcerative colitis. Gastroenterology 2015;149:350–5 e2. [DOI] [PubMed] [Google Scholar]
- 146. McRae BL, Levin AD, Wildenberg ME, et al. Fc receptor-mediated effector function contributes to the therapeutic response of anti-TNF monoclonal antibodies in a mouse model of inflammatory bowel disease. J Crohns Colitis 2016;10:69–76. [DOI] [PubMed] [Google Scholar]
- 147. Arijs I, Li K, Toedter G, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 2009;58:1612–9. [DOI] [PubMed] [Google Scholar]
- 148. Saurer L, Rihs S, Birrer M, Saxer-Seculic N, Radsak M, Mueller C; Swiss IBD Cohort Study . Elevated levels of serum-soluble triggering receptor expressed on myeloid cells-1 in patients with IBD do not correlate with intestinal TREM-1 mRNA expression and endoscopic disease activity. J Crohns Colitis 2012;6:913–23. [DOI] [PubMed] [Google Scholar]
- 149. Arias MT, Vande Casteele N, Vermeire S, et al. A panel to predict long-term outcome of infliximab therapy for patients with ulcerative colitis. Clin Gastroenterol Hepatol 2015;13:531–8. [DOI] [PubMed] [Google Scholar]
- 150. Barré A, Colombel JF, Ungaro R. Review article: predictors of response to vedolizumab and ustekinumab in inflammatory bowel disease. Aliment Pharmacol Ther 2018;47:896–905. [DOI] [PubMed] [Google Scholar]
- 151. Sjöberg M, Magnuson A, Björk J, et al. ; Swedish Organization for the study of Inflammatory Bowel Disease [SOIBD]. Infliximab as rescue therapy in hospitalised patients with steroid-refractory acute ulcerative colitis: a long-term follow-up of 211 Swedish patients. Aliment Pharmacol Ther 2013;38:377–87. [DOI] [PubMed] [Google Scholar]
- 152. Järnerot G, Hertervig E, Friis-Liby I, et al. Infliximab as rescue therapy in severe to moderately severe ulcerative colitis: a randomized, placebo-controlled study. Gastroenterology 2005;128:1805–11. [DOI] [PubMed] [Google Scholar]
- 153. Denson LA, Curran M, McGovern DPB, et al. Challenges in IBD research: precision medicine. Inflamm Bowel Dis 2019;25:31–9. [DOI] [PubMed] [Google Scholar]
- 154. Sánchez de Medina F, Romero-Calvo I, Mascaraque C, Martínez-Augustin O. Intestinal inflammation and mucosal barrier function. Inflamm Bowel Dis 2014;20:2394–404. [DOI] [PubMed] [Google Scholar]
- 155. Olivera P, Danese S, Jay N, Natoli G, Peyrin-Biroulet L. Big data in IBD: a look into the future. Nat Rev Gastroenterol Hepatol 2019;16:312–21. [DOI] [PubMed] [Google Scholar]