Abstract
Eukaryotic translation initiation factor 2B (eIF2B) is the guanine nucleotide exchange factor for protein synthesis initiation factor 2 (eIF2). Composed of five subunits, it converts eIF2 from a GDP-bound form to the active eIF2-GTP complex. This is a regulatory step of translation initiation. In vitro, eIF2B catalytic function can be provided by the largest (epsilon) subunit alone (eIF2Bɛ). This activity is stimulated by complex formation with the other eIF2B subunits. We have analyzed the roles of different regions of eIF2Bɛ in catalysis, in eIF2B complex formation, and in binding to eIF2 by characterizing mutations in the Saccharomyces cerevisiae gene encoding eIF2Bɛ (GCD6) that impair the essential function of eIF2B. Our analysis of nonsense mutations indicates that the C terminus of eIF2Bɛ (residues 518 to 712) is required for both catalytic activity and interaction with eIF2. In addition, missense mutations within this region impair the catalytic activity of eIF2Bɛ without affecting its ability to bind eIF2. Internal, in-frame deletions within the N-terminal half of eIF2Bɛ disrupt eIF2B complex formation without affecting the nucleotide exchange activity of eIF2Bɛ alone. Finally, missense mutations identified within this region do not affect the catalytic activity of eIF2Bɛ alone or its interactions with the other eIF2B subunits or with eIF2. Instead, these missense mutations act indirectly by impairing the enhancement of the rate of nucleotide exchange that results from complex formation between eIF2Bɛ and the other eIF2B subunits. This suggests that the N-terminal region of eIF2Bɛ is an activation domain that responds to eIF2B complex formation.
Eukaryotic translation initiation factor 2B (eIF2B) is a guanine nucleotide exchange factor (GEF) that converts its substrate, eIF2, from an inactive eIF2-GDP binary complex to eIF2-GTP. This active complex binds charged initiator tRNAMet (Met-tRNAiMet) forming a ternary complex that interacts with eIF3 and the 40S ribosomal subunit. Following addition of mRNA, associated initiation factors, and the 60S ribosomal subunit, the G-protein cycle is completed by hydrolysis of eIF2-bound GTP and the release of eIF2-GDP from the ribosome (reviewed in references 14, 31, and 42). Thus, the functions of eIF2 and eIF2B are believed to be similar to those of the small GTPases and exchange factors, respectively, of the RAS superfamily (4). Recent three-dimensional structure determinations have demonstrated that while the nucleotide binding domains of the G proteins are very similar, GEF structures differ markedly from one another, each employing different amino acid motifs to drive the release of GDP (8, 41).
eIF2 and eIF2B are complex proteins of three (α to γ) and five (α to ɛ) nonidentical subunits, respectively. The subunit complexity of eIF2B reflects, at least in part, its novel mechanism of regulation. Four protein kinases, called PKR, HCR (HRI), PERK (PEK), and GCN2, specifically phosphorylate the seryl residue at position 51 of the α subunit of eIF2 (eIF2α) under different stress conditions (12, 23, 40). Phosphorylation of eIF2α at this site converts eIF2 from a substrate into an inhibitor of eIF2B (33, 38), thus inhibiting global translation initiation. In the yeast Saccharomyces cerevisiae, the protein kinase GCN2 phosphorylates eIF2α in response to amino acid or purine starvation. Under moderate amino acid starvation conditions, the level of phosphorylated eIF2 produced is not sufficient to inhibit total protein synthesis; however, it specifically enhances translation of GCN4 mRNA, which encodes a transcriptional regulator of amino acid biosynthetic genes (24). GCN4 translation is inversely coupled to ternary complex concentration and thus to eIF2B activity by the presence of inhibitory short open reading frames in the 5′ leader of its mRNA. Recently, homologues of GCN2 have been identified in Drosophila melanogaster (32) and mammals (2), indicating that this kinase may be universally conserved in eukaryotes.
By using both genetic and biochemical methods, it has been demonstrated that three subunits of S. cerevisiae eIF2B (α, β, and δ encoded by GCN3, GCD7, and GCD2, respectively) act together to mediate regulation of eIF2B activity in response to phosphorylation of its substrate, eIF2 (33, 34, 43). We also found that the ɛ subunit of eIF2B, encoded by GCD6 in yeast, is a catalytic subunit of eIF2B: the ability of extracts from yeast cells overexpressing eIF2Bɛ alone to dissociate GDP from eIF2-GDP binary complexes was higher than that of nonoverexpressing cell extracts (33). Interestingly, eIF2Bɛ catalyzed nucleotide exchange at a reduced rate compared with that of the five-subunit eIF2B complex. Others have obtained similar results expressing mammalian eIF2Bɛ cDNA in insect cells (18). In addition, we showed that the ɛ and γ subunits can form an eIF2B catalytic subcomplex in the absence of the other three subunits. This γɛ catalytic subcomplex promoted release of GDP from eIF2-GDP at a higher rate than ɛ alone and could also bind stably to eIF2 (33), but in contrast to the full five-subunit complex, nucleotide exchange and binding of this subcomplex to eIF2 were not affected by the phosphorylation of eIF2α.
In this study, we decided to follow up on our observation that eIF2Bɛ showed catalytic activity to determine what regions or residues of this polypeptide are important for its GEF activity. Examination of the primary sequence of eIF2Bɛ reveals no significant sequence identity with any other GEF. However, eIF2Bɛ does share significant sequence similarity with eIF2Bγ (5, 35) (see Fig. 1), to which it binds, forming the eIF2B catalytic subcomplex (33). In addition, eIF2Bγ and eIF2Bɛ both share extended sequence similarity with two other protein families found mainly in bacteria–nucleoside triphosphate (NTP)-hexose pyrophosphorylases and acyltransferases (see Fig. 1A). It has been proposed that the region of similarity with the bacterial NTP-hexose pyrophosphorylase family represents a nucleotide binding domain composed of a modified P-loop and magnesium ion coordinating region (28), suggesting a role for nucleotide binding by eIF2B in the guanine nucleotide exchange reaction. Finally, it has been shown recently that the sequence motif shared between the extreme C termini of eIF2Bɛ and eIF5 (a potential GTPase-activating protein for eIF2) (28) provides a binding site in both proteins for the β subunit of their common substrate eIF2 (1).
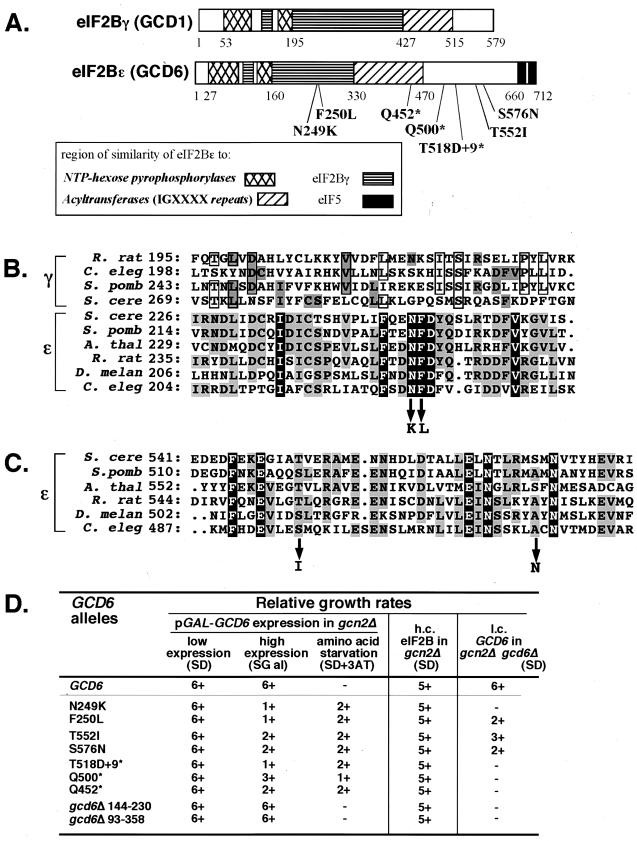
FIG. 1.
Genetic characterization of novel mutations in yeast eIF2Bɛ. (A) eIF2Bγ and eIF2Bɛ subunits encoded by yeast genes GCD1 and GCD6 are shown schematically from N to C termini. The patterns indicate regions of significant sequence similarity both between these proteins and with other protein families as shown in the key. The amino acids at the boundaries of these regions are indicated by numbers. The relative positions and nature of missense and nonsense mutations identified in GCD6 are indicated below the eIF2Bɛ schematic. (B) A segment of a multiple-sequence alignment of eIF2Bγ and eIF2Bɛ proteins from diverse organisms. The region around the mutations at N249 and F250 is shown. These mutant changes are indicated with arrows pointing down. Residues identical in all eIF2Bɛ sequences are shown in reverse type. Residues identical in at least three eIF2Bɛ sequences are shaded, as are residues in eIF2Bγ sequences that are identical to those of any eIF2Bɛ sequence. Other residues shared by three or more eIF2Bγ sequences are boxed. The sequences used are as follows (GenBank accession numbers given in brackets): S. cerevisiae (S. cere) GCD6 [Z68195] and GCD1 [Z75168], Schizosaccharomyces pombe (S. pomb) eIF2Bɛ (ɛ) [P56287] and eIF2Bγ (γ) [P56288], Arabidopsis thaliana (A. thal) ɛ [AAC12836], Rattus norvegicus (R. rat) ɛ [Q64350] and γ [P70541], Caenorhabditis elegans (C. eleg) ɛ [CAA91063.1] and γ [P80361], and D. melanogaster (D. melan) ɛ [AL021086]. The number after each sequence abbreviation indicates the position in the protein of the first residue of each sequence shown in the alignment. (C) The segment of the multiple-sequence alignment of eIF2Bɛ proteins from the region around the mutations at T552 and S576. All shading and other information are as described above for panel B. (D) Rates of growth of cells transformed with mutant alleles of GCD6. Rates of growth are scored on a linear scale from 6+ (wild type, maximal growth rate) to − (no visible growth) for growth on SD or SGal. Medium supplemented with 25 mM 3-aminotriazole (3AT) was used to assess response to amino acid starvation. Growth tests were performed in three genetic backgrounds. Columns 2 to 4 show results following transformation of strain GP3667 (gcn2Δ) with galactose-inducible GCD6-only plasmids carrying the indicated allele. Column 5 shows results of cooverexpressing mutant alleles of GCD6 with all other eIF2B subunits from high-copy-number (h.c.) plasmids transformed into strain GP3667. Column 6 shows the ability of low-copy-number (l.c.) GCD6-only plasmid-borne alleles to complement a deletion of GCD6 in strain GP3751 (gcd6Δ gcn2Δ).
We show here that the C-terminal region of eIF2Bɛ is responsible for binding to the substrate eIF2 and contains the catalytic domain for GEF activity. Missense alleles in which single conserved amino acids within this region were changed dramatically reduce the GEF activity of eIF2Bɛ without affecting eIF2 binding, indicating that different residues are responsible for these two functions. In contrast, the N-terminal half of eIF2Bɛ is required for its interactions with the other eIF2B subunits. Missense alleles, where single conserved residues have been altered, in this region of the gene affect the stimulation of eIF2B activity observed upon eIF2B complex formation without detectably altering binding to eIF2. The implications of these results for eIF2B function are discussed.
MATERIALS AND METHODS
S. cerevisiae strains and genetic methods.
Standard genetic methods were used to construct yeast strains and to characterize the phenotypes conferred by the gcd6 mutations described here (20). Transformation of yeast strains with plasmids was done by the lithium acetate method (26). Plasmid shuffling employing 5-fluoro-orotic acid was done as described previously (3). Yeast strain GP3751 (MATα leu2-3 leu2-112 ura3-52::[HIS4-lacZ ura3-52] ino1 gcd6Δ gcn2Δ::hisG [GCD6 CEN6 LEU2]) was constructed by transformation of strain KAY16 (1) with pJB102 (6) followed by plasmid shuffling to lose pJB5. Yeast strain GP3667 (MATα leu2-3 leu2-112 trp1-Δ63 ura3-52 gcn2ΔGAL2+) was constructed by deletion of GCN2 in strain H1511 (19) using plasmid p1144 as previously described (15).
Plasmids.
Standard methods were used to construct all plasmids (39). pAV1427 is a 2μm URA3 plasmid derived from pEMBL-yex4 (7) and was used to overexpress hexahistidine and FLAG double-tagged eIF2Bɛ (GCD6) from a galactose-inducible GAL-CYC hybrid promoter. pAV1427 was constructed by modifying the coding region of GCD6 in plasmid pJB85 (GCD6 CEN URA3) (6) to position an MluI restriction site immediately upstream of the AUG start codon, move an NcoI site from codon 15 to the start codon, and introduce a BspEI restriction site at codon 9 without altering the sequence of the encoded protein. This was done by the ligation of a pair of annealed complementary oligonucleotides of sequence 5′-CGC GTG CCA TGC TGG AAA AAA GGG ACA AAA GAA ATC CGG ACT AGG CAA T and 5′-CAT GAT TGC CTA GTC CGG ATT TCT TTT GTC CCT TTT TTC CAG CAT GGC A between the upstream MluI site (beginning at nucleotide −213) and the NcoI site at codon 15 to generate plasmid pAV1426. This modified GCD6 was subcloned on a 2.3-kb MluI-to-SpeI fragment into plasmid pGAL-GCN2FH digested with MluI and NheI. Plasmid pGAL-GCN2FH (a gift from Jinsheng Dong and Alan Hinnebusch, National Institutes of Health [NIH], Bethesda, Md.) is pEMBL-yex4 expressing N-terminally FLAG- and hexahistidine-tagged GCN2. The subcloning replaced the GCN2 DNA with the GCD6 sequence, creating a galactose-inducible GCD6-expressing plasmid with N-terminal FLAG and hexahistidine tags.
pAV1464 was derived from pAV1427 by partial digestion with EcoRI and religation to generate an in-frame deletion between residues A+274 and G+1074 of GCD6 (corresponding to amino acids E93 and E358), termed gcd6Δ93-358. pAV1466 was derived from pAV1427 by ClaI digestion and religation. This resulted in an in-frame deletion between GCD6 residues C+429 and T+692 (corresponding to amino acids D144 and D230), termed gcd6Δ144-230. Subcloning was used to introduce gcd6 mutations (isolation described in the section below) from the original pAV1427-derived plasmid into the low-copy-number URA3 plasmid pJB85, containing GCD6 under the control of its own promoter and without epitope tags (5). The DNA in these plasmids was sequenced to confirm the presence of the mutation. Plasmids generated are pAV1514 (gcd6-T518D+9*), pAV1515 (gcd6-N249K), pAV1522 (gcd6Δ93-358), pAV1524 (gcd6-F250L), pAV1527 (gcd6Δ144-230), pAV1566 (gcd6-Q500*), pAV1582 (gcd6-T552I), pAV1586 (gcd6-S576N), and pAV1588 (gcd6-Q452*) where asterisks indicate nonsense codons.
A high-copy-number URA3 plasmid pRS426 (9) containing GCD1 with C-terminal six-histidine and two copies of the FLAG epitope, called pAV1431, was created by using complementary oligonucleotides to introduce the amino acid sequence SGDYKDDDKDITGDYKDDDKDITGHHHHHHTG immediately prior to the stop codon of GCD1. This plasmid was derived from the six-histidine-tagged GCD1 plasmids described previously (33) by adding tandem copies of oligonucleotides specifying the FLAG tag (underlined) immediately 5′ to the hexahistidine tag. The immediate parent of pAV1431 is pTK1.11, containing GCD6 and double-tagged GCD1 (constructed by Thanuja Krishnamoorthy and Alan Hinnebusch, NIH). Double-tagged GCD1 was subcloned on a 2.4-kb BamHI fragment into similarly cleaved pRS426 so that the GCD1 and URA3 genes are transcribed in the same direction in pAV1431. Plasmids coexpressing FLAG- and hexahistidine-tagged GCD1 and different GCD6 alleles were constructed by subcloning each GCD6 allele on a 2.5-kb BamHI (made blunt ended with Klenow polymerase)-to-XhoI fragment from the low-copy-number plasmids described above into SmaI- and XhoI-cut pAV1431 (GCD1 2μm URA3). The plasmids created are pAV1533 (GCD1 GCD6), pAV1535 (GCD1 gcd6-T518D+9*), pAV1539 (GCD1 gcd6-F250L), pAV1541 (GCD1 gcd6-N249K), pAV1549 (GCD1 gcd6Δ93-358), pAV1551 (GCD1 gcd6Δ144-230), pAV1574 (GCD1 gcd6-S576N), pAV1576 (GCD1 gcd6-Q500*), pAV1578 (GCD1 gcd6-Q452*), and pAV1580 (GCD1 gcd6-T552I).
pAV1494 is a high-copy-number LEU2 plasmid made by subcloning GCN3, GCD2, and GCD7 on a 8.1-kb SacII-to-SalI fragment from p1871 (43) into pRS425 (9).
Isolation of mutations in GCD6 with reduced eIF2B activity.
Plasmid pAV1427 was subjected to random mutagenesis using the error-prone bacterial strain XL-1 Red (Stratagene), as described by the supplier, to generate a pool of randomly mutated plasmid DNA termed pAV1427M. Dominant mutations in GCD6 were selected from this DNA pool. pAV1427M transformants of yeast strain GP3667 (GCD6 gcn2Δ) were selected on synthetic minimal medium containing 2% glucose (SD). Approximately 10,000 fast-growing colonies were screened for slow growth (Slg−) following replica plating to synthetic minimal medium containing 2% galactose (SGal). Slg− cells were then tested for resistance to 3-aminotriazole (3ATr) on SD plates additionally supplemented with 25 mM 3AT (Fluka). These phenotypes are dominant, as they require that the mutant allele be incorporated into the eIF2B complex in place of the chromosomally encoded wild-type allele. The Slg− and 3ATr phenotypes screened for have been associated with Gcd− alleles of eIF2B genes isolated previously (22). Plasmid DNA was recovered from 109 candidates, and of these, only 9 retransformed to generate the original phenotype. The GCD6 insert from each mutant was subjected to automated dye terminator sequencing (ABI), and seven unique alleles resulted, two being isolated twice each. The gcd6 alleles are caused by the following nucleotide (protein) changes; T+747A (N249K) in plasmid pAV1459, T+748C (F250L) in plasmid pAV1456, A+1484G (N495S) and C+1655T (T552I) in pAV1460, G+1727A (S576N) in pAV1458 and pAV1462, C+1354T (Q452*) in pAV1455, C+1498T (Q500*) in pAV1457 and pAV1462, and A+1550AA (an insertion of an adenine residue to alter the reading frame and induce a premature stop-T518D+KEKKNDVCQ* (abbreviated T518D+9*) in pAV1554. Subcloning and nucleotide sequence confirmation were used to separate the two residue changes in pAV1460, generating plasmids pAV1603 (gcd6-N495S) and pAV1605 (gcd6-T552I). The results of phenotypic and biochemical analyses of the resulting purified proteins demonstrated that the mutant phenotype was entirely due to the T552I substitution (data not shown).
General protein methods.
Protein concentrations were determined by the Bradford assay (Bio-Rad) using bovine serum albumin (BSA) as a standard. eIF2 and eIF2B proteins were resolved by sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis (SDS–12.5% PAGE) (39) and detected by Western blotting as described previously (16, 43) using appropriate rabbit polyclonal primary antisera (6, 10, 11) with a horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (HRP-conjugated protein A was used for the experiment shown in Fig. 4C) and the enhanced chemiluminescence system from Amersham.
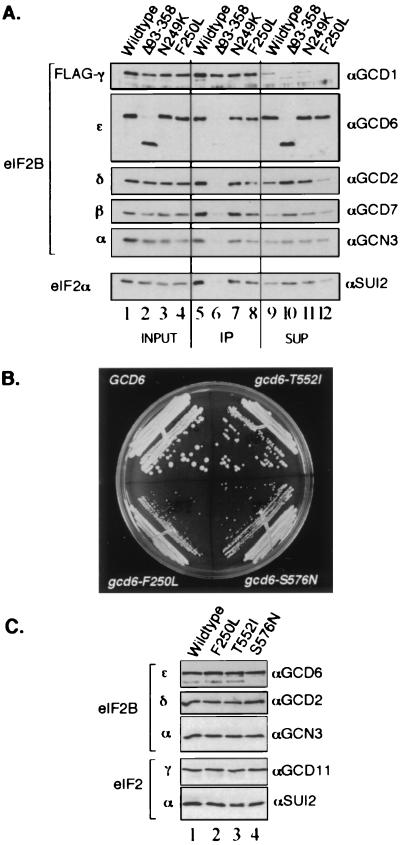
FIG. 4.
In vivo analysis of eIF2Bɛ mutants. (A) Immunoprecipitation of FLAG-tagged eIF2Bγ and associated eIF2B subunits from extracts of yeast strain GP3667 overexpressing all five subunits of wild-type or mutant eIF2B as indicated. Cell extract (10 μg) was loaded in the input lanes (lanes 1 to 4), and the equivalent of 20 μg was loaded in the immune precipitated (IP) lanes (lanes 5 to 8) and unbound supernatant (SUP) lanes (lanes 9 to 12). Proteins were visualized as described in the legend to Fig. 3. (B) Three missense mutations complement a deletion of GCD6. Low-copy-number plasmids bearing the indicated alleles of GCD6 were introduced into strain GP3751 (gcd6Δ), and plasmid shuffling was used to make the indicated alleles the only source of GCD6. Growth on rich medium YPD is shown. (C) Immunoprecipitation of eIF2Bɛ and associated eIF2B and eIF2 polypeptides from extracts of cells shown in panel B using anti-GCD6 (αGCD6) antiserum. Pellets from 300 μg of cell extract were loaded in each lane.
Protein purification.
Yeast eIF2 was purified as described previously (33). Wild-type and mutant eIF2Bɛ proteins were overexpressed in yeast and partially purified by nickel-affinity chromatography as described below. Hexahistidine- and FLAG-tagged eIF2Bɛ was expressed from a galactose-inducible promoter in plasmid pAV1427 using yeast strain GP3667. Yeast cells transformed with pAV1427 were grown overnight at 30°C in 160 ml of synthetic complete medium containing 2% glucose but without uracil to maintain plasmid selection (SC-URA). Cells were collected by centrifugation, resuspended in 1.6 liters of the same medium except that it contained a mixture of 0.4% glucose and 2% galactose as carbon sources and lacked uracil, leucine, isoleucine, and valine supplements, and grown for an additional 24 h to allow growth and induction of GCD6 expression. Cells were then collected by centrifugation, washed with water, and suspended in lysis buffer (1 M KCl, 20 mM Tris-HCl [pH 7.5], 3 mM MgCl2, 5% glycerol, 5 mM β-mercaptoethanol, 5 mM NaF, 0.1% Triton X-100, 10 mM imidazole, Complete EDTA-free protease inhibitor cocktail [Roche Molecular Biochemicals]) to twice the volume of the cell pellet. Cells were lysed at 4°C using acid-washed glass beads in 50-ml Falcon tubes with vortexing (five times for 1 min each time) and 1-min cooling intervals. Cell lysates were cleared by centrifugation, and eIF2Bɛ was purified by use of Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen). Cell lysates were incubated with Ni-NTA beads for 3 h at 4°C. Agarose beads were collected by low-speed centrifugation (2,000 × g for 2 min) and washed three times in wash buffer (same as lysis buffer but without any added Triton X-100) containing 10 or 40 mM imidazole. Ni-NTA agarose-bound proteins were then eluted by two incubations in the same buffer in the presence of 500 mM imidazole. After dialysis overnight in storage buffer (100 mM KCl, 20 mM Tris-HCl [pH 7.5], 5% glycerol, 5 mM β-mercaptoethanol, 5 mM NaF, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mg leupeptin per liter 0.7 mg of pepstatin per liter, and 1 mg of aprotinin per liter) and concentration using Centricon 30,000 concentrators (Amicon), proteins were aliquoted and stored at −80°C. Protein concentrations were assessed by the Bradford assay (Bio-Rad), and the integrity and purity of eIF2Bɛ were confirmed by SDS-PAGE, Coomassie brilliant blue staining, and Western blotting. The yield was typically ∼1 mg of eIF2Bɛ from 10 g (wet weight) of yeast cells at 75% purity. eIF2Bɛ mutants were purified in exactly the same way as and in parallel with the wild-type protein, only using strain GP3667 transformed with the appropriate plasmid derivative of pAV1427. The final purity varied for each mutant.
Purification of five-subunit eIF2B was performed essentially as described above for eIF2Bɛ proteins with the following modifications. Yeast strain GP3667 was transformed with plasmid pAV1494 (2μm LEU2 GCN3 GCD2 GCD7) and pAV1533 (2μm URA3 GCD1 GCD6) or an equivalent plasmid expressing the appropriate eIF2Bɛ mutant. These cells were grown in 2.4 liters of SC medium with dextrose but without uracil, leucine, isoleucine, and valine to A600 of 2.5 to 4.5. The yield was typically ∼1 mg of eIF2B from 10 g (wet weight) of yeast cells at ∼85% purity.
Guanine nucleotide exchange assays.
Binary complexes of yeast eIF2 and [3H]GDP (Amersham) were formed exactly as described previously (33). Displacement of [3H]GDP from binary complexes was also measured as described previously (33), except that the indicated amount of eIF2Bɛ or five-subunit eIF2B, purified as described above, was used in place of extracts from cells overexpressing eIF2B subunits.
In vitro protein-protein interaction assays.
We performed a binding assay using anti-FLAG M2 affinity resin to analyze the interactions between purified eIF2 and FLAG- and hexahistidine double-tagged wild-type and mutant eIF2Bɛ proteins, either alone or within the eIF2B complex. Purified eIF2Bɛ proteins (200 nM), 100 nM purified eIF2B complex, or an equivalent concentration of control FLAG peptide was incubated with 20 μl (wet volume) of anti-FLAG M2 affinity resin (Eastman Kodak) with rotation for 2 h at 4°C in 100 μl of buffer A (100 mM KCl, 20 mM Tris-HCl [pH 7.5], 2 mM MgCl2, 5 mM β-mercaptoethanol, 0.1% Triton X-100) in the presence of Complete EDTA-free protease inhibitor (Roche Diagnostics) and 10 μg of BSA. Beads were washed three times with 100 μl of buffer A. Purified eIF2 at concentrations of 0.625 to 40 nM (as indicated in the figure legends) was then added to the beads in 100 μl of buffer A in the presence of 10 μg of BSA and rotated for 2 h at 4°C. After three washes with 100 μl of buffer A, the beads were suspended in Laemmli sample buffer (29) and incubated at 100°C for 5 min to elute proteins remaining bound to the resin.
In vivo immune precipitations.
Anti-FLAG immune precipitations were performed from extracts of whole cells of strain GP3667 cooverexpressing all five subunits of eIF2B from two plasmids where GCD1 (eIF2Bγ) is FLAG tagged and using anti-FLAG M2 affinity gel. Cells in 100 ml of SC medium lacking uracil, leucine, isoleucine, and valine were grown to A600 of 0.5 to 0.8. Cells were harvested by centrifugation, washed in water, and resuspended in buffer A (see above) containing 1 mM PMSF, 1 μg of leupeptin per ml, 0.7 μg of pepstatin per ml, and 1 μg of aprotinin per ml. Cells were lysed using glass beads and cleared by centrifugation at 14,000 × g. The resulting extract (250 μg) was incubated with 10 μl of prewashed M2 anti-FLAG resin (IBI Kodak) overnight with rotation at 4°C. Bound immune complexes were washed three times in the same buffer and eluted into Laemmli sample buffer by incubation at 100°C for 5 min.
Anti-GCD6 immune precipitations from extracts of whole cells were done exactly as described previously (1) using derivatives of strain GP3751 (as indicated in the legend to Fig. 4).
Preparation and gradient analysis of yeast ribosomes and polysomes.
Cultures of GP3751 (gcd6Δ gcn2Δ) were transformed and plasmid shuffled to have either pJB85 (GCD6) or pAV1524 (gcd6-F250L) or pAV1586 (gcd6-S576N) as the only source of eIF2Bɛ. These cells were grown in rich medium (YEPD [yeast extract-peptone-dextrose]) at 30°C. Cycloheximide was added to 50 μg/ml, and the cells were harvested onto ice and lysed exactly as described previously (30). Gradient analysis was exactly as described previously (10, 19). Briefly, cell extracts were layered on low-salt, 7 to 47% or 15 to 35% sucrose gradients and sedimented at 39,000 rpm at 4°C in an SW41 rotor (Beckman). The gradients were scanned at 254 nm while being fractionated into 0.6-ml fractions on an ISCO gradient collector. A 20-μl portion of each fraction was analyzed by SDS-PAGE (12.5 or 15% polyacrylamide) and Western blotting with the indicated antiserum. Fractions containing 40S and 60S ribosomes were determined using a rabbit polyclonal antibody raised against RPL30 that also reacts with RPS2 (kindly supplied by Jonathan Warner).
RESULTS
eIF2Bɛ mutations with reduced activity cluster within two regions, one N terminal and one C terminal.
eIF2Bɛ in the yeast S. cerevisiae (GCD6) is the largest eIF2B subunit (81 kDa), and we have shown previously that it possesses GEF activity in vitro (33). We demonstrated higher GEF activity in extracts from yeast cells overexpressing eIF2Bɛ than in extracts from control cells. To identify regions of eIF2Bɛ that were important for catalytic activity, we used computer programs to search public databases for similarities between the primary sequence of GCD6, other known GEFs, and other proteins. These comparisons demonstrated similarities with other protein families, but not with GEFs (28) (Fig. 1A), indicating that a random mutagenesis experiment to identify residues or regions important for eIF2Bɛ function would be more rewarding.
We designed a genetic screen to identify amino acid residues of eIF2Bɛ important for catalytic activity. GCD6 is an essential gene in yeast, so we expected that mutations reducing eIF2B activity would cause slow growth or be lethal. We therefore set up a conditional expression system to highly express eIF2Bɛ from a plasmid (pAV1427) under the control of a galactose-inducible promoter (see Materials and Methods). Importantly, overexpression of GCD6 alone apparently does not increase eIF2B activity in vivo, as judged by genetic tests (43), despite its enhanced catalytic activity in vitro (33). Hence, we expected that only overexpressed mutants that compete with the chromosomally encoded eIF2Bɛ for inclusion into the eIF2B complex would be identified by our experimental approach. We screened for conditional slow-growing or conditional-lethal mutants. As a secondary screen, we used the fact that a reduction in eIF2B activity will derepress translation of GCN4 mRNA independently of the upstream activating protein kinase GCN2 (24). This will mimic the effects of amino acid starvation, derepressing expression of amino acid biosynthetic pathway enzymes to allow gcn2Δ yeast cells to grow on medium containing the 3AT, an inhibitor of histidine biosynthesis (22).
Plasmid pAV1427 was mutated at random, and the resulting pool of mutated DNA was transformed into the gcn2Δ yeast strain GP3667. Colonies that grew well on SD medium were screened for poor growth on SGal medium and for induction of GCN4 expression by growth on SD medium supplemented with 25 mM 3AT. Following further genetic tests and DNA sequencing (see Materials and Methods), seven plasmid-dependent mutant alleles were isolated: four missense mutations each changing a single amino acid residue clustered within two regions of the protein and three nonsense mutations that prematurely terminate the GCD6 open reading frame (Fig. 1A). The gcd6-N249K and gcd6-F250L mutations are within a region conserved between eIF2Bγ and eIF2Bɛ sequences. These mutations change adjacent residues that are universally conserved in all known or predicted eIF2Bɛ sequences from yeast to mammals (Fig. 1B). Interestingly, the N249F250D251 residues are the only three consecutive residues shared in all eIF2Bɛ sequences. The gcd6-T552I and gcd6-S576N mutations affect residues in a region conserved only in the eIF2Bɛ sequences (Fig. 1C). The three nonsense mutations each eliminate the region containing these C-terminal missense alleles. Growth phenotypes associated with these new eIF2Bɛ alleles are summarized in Fig. 1D. As expected from the isolation procedures, all mutations show no growth defect on glucose-containing medium when the mutated gene is poorly expressed (Fig. 1D, column 2, SD) but cause a reduced growth rate relative to the wild-type control on galactose-containing medium (column 3, SGal) and induce expression of GCN4 in the absence of the protein kinase GCN2 (column 4, SD+3AT). Interestingly, when these mutations were combined with a deletion of the gene encoding eIF2Bα (gcn3Δ), no synthetic growth phenotype was seen (data not shown). This is in contrast to other previously characterized gcd6 mutations where loss of GCN3 function exacerbated the growth phenotypes (6, 13), indicating that we have identified novel gcd6 alleles.
One unexpected result of this mutational analysis was that the mutations we had isolated were not within any of the regions of sequence similarity shared with NTP-hexose pyrophosphorylases or acyltransferases (Fig. 1A). We therefore created two additional mutant alleles defective in these regions. gcd6Δ144-230 is an in-frame internal deletion between two ClaI restriction sites at amino acid residues 144 and 230. Similarly, gcd6Δ93-358 is an in-frame internal deletion between two EcoRI restriction sites. This largest deletion removes most of the residues with sequence similarity to NTP-hexose pyrophosphorylases, a region shared with eIF2Bγ and part of the region that resembles acyltransferases. When overexpressed from the same galactose-inducible expression system as employed above, these mutants showed no detectable phenotype (Fig. 1D). These results suggested either that these regions were dispensable for eIF2Bɛ function or that the mutations were unable to compete with the endogenous wild-type GCD6 protein for eIF2B complex formation (i.e., they are recessive mutations). The results of further analysis of all the mutations presented below lead us to suggest that the regions of eIF2Bɛ that share sequence similarity with NTP-hexose pyrophosphorylases and acyltransferases are not critical for the nucleotide exchange function of eIF2Bɛ.
The eIF2Bɛ C terminus is required for catalytic activity.
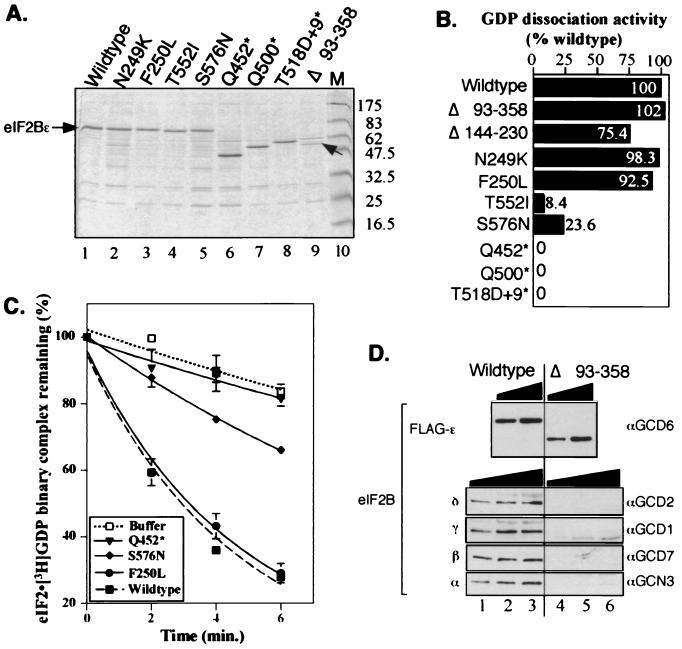
To assess directly the catalytic activity of the isolated mutations, we first purified each mutant eIF2Bɛ polypeptide in parallel with the wild-type protein (see Materials and Methods). Levels of proteins expressed were estimated by a combination of Coomassie brilliant blue-stained gels (Fig. 2A), Western blotting, and Bradford assay. Equivalent amounts of wild-type or mutant eIF2Bɛ proteins were then assayed for GEF activity in standard filter binding assays. This set of experiments demonstrated clearly that all mutations affecting the C-terminal region of eIF2Bɛ either dramatically reduced (missense mutations) or eliminated (nonsense mutations) eIF2Bɛ GEF activity (Fig. 2B and C). In contrast, mutations affecting the N terminus, including the large internal deletions, retained full in vitro activity.
FIG. 2.
Purification and GEF activity of eIF2Bɛ mutants. (A) SDS–12.5% polyacrylamide gel of the indicated nickel affinity gel-purified eIF2Bɛ polypeptides (lanes 1 to 9) stained with Coomassie brilliant blue. eIF2Bɛ polypeptides were added to the lanes of the gel as follows; 2.5 μg was loaded in lanes 2, 5, and 6, while 1.25 μg was loaded in lanes 1, 3, 4, 7, 8, and 9. In lane 9, the polypeptide corresponding to eIF2BɛΔ93-358 is indicated with an arrow. All proteins were purified with Triton X-100 (0.1%) added to the buffer, except for proteins shown in lanes 2 and 7. Lane 10 contains prestained molecular mass markers (M) (New England BioLabs) with the approximate masses (in kilodaltons) indicated to the right. (B) The initial rates of nucleotide exchange for the mutant polypeptides are shown as a percentage of the wild-type protein activity. Initial rates of [3H]GDP release were determined from exponential curves fitted to the data using a computer program (Cricketgraph 3.0) of time course nucleotide exchange assays performed using a standard filter binding assay. The eIF2BɛΔ144-230 mutant was expressed very poorly, resulting in high copurification of contaminating proteins. Partially purified cell extract (15 μg) was used for its assay. It is likely that this mutant retains full activity. (C) Nucleotide exchange assay results for selected purified proteins. Some of the primary data used in panel B is shown. In these experiments, 2.5-μg samples of nickel-purified extract were used, except for the Q452* mutant where 5 μg was used. The wild-type and buffer-only control curves are shown as broken lines, and the mutant curves are shown as solid lines. Experiments were done in duplicate and replicated two to eight times. Typical data are shown with error bars indicating the standard deviation (ς3) where ς3 ≤ 5.65 for each time point. (D) Western blot of eIF2B subunits in purified fractions from the wild type (lanes 1 to 3) and gcd6Δ93-358 mutant (lanes 4 to 6). Blots were probed with the antisera indicated to the right of each panel to detect the eIF2B subunits indicated to the left. For eIF2Bɛ, 62.5 ng (lanes 2 and 4) and 125 ng (lanes 3 and 5) were loaded. For detection of eIF2Bδ and -γ, 2.5 μg (lanes 1 and 4), 5 μg (lanes 2 and 5), and 7.5 μg (lanes 3 and 6) were loaded. For detection of eIF2Bβ and -α, 1 μg (lanes 1 and 4), 1.5 μg (lanes 2 and 5), and 2 μg (lanes 3 and 6) were loaded.
As the mutants were dominant when overexpressed, we expected them to interact and copurify, at low level, with the other eIF2B subunits that had not been overexpressed. Western blotting of each mutant with antisera to each eIF2B subunit confirmed this (data not shown). In contrast to these results, we did find that the purified proteins with large N-terminal internal deletions were free of detectable amounts of other eIF2B subunits (Fig. 2D and data not shown) and still retained full GEF activity (Fig. 2B). The findings shown in Fig. 2 demonstrate that, in a purified system, eIF2Bɛ alone is a catalytic subunit of eIF2B. They also show that residues between 93 and 358 are not required for eIF2Bɛ catalytic activity, while residues between amino acids 518 and 712 are necessary for this function.
The eIF2Bɛ C terminus is required for interaction with eIF2 in vitro.
We wished to determine whether the eIF2Bɛ mutants had an altered affinity for eIF2. We set up a binding assay using the N-terminal FLAG epitope tags on purified eIF2Bɛ proteins to measure the relative binding affinities between eIF2Bɛ and purified eIF2 in immune precipitation assays with anti-FLAG M2 affinity gel. First, we examined the binding between a fixed concentration of eIF2Bɛ (∼200 nM) or FLAG peptide as a control (200 nM) and various concentrations of eIF2. We detected concentration-dependent binding between eIF2 and eIF2Bɛ that saturated the detection system (Western blotting) at 20 to 40 nM eIF2 (Fig. 3A). Next, we used saturating amounts of eIF2 (20 nM) to examine binding with our panel of mutants (Fig. 3B). We found that all three nonsense mutants showed dramatic reductions in stable binding to eIF2, with the shortest polypeptide, eIF2Bɛ-Q452*, exhibiting the most defective binding (Fig. 3B, lane 11). In contrast, all the missense mutants and the eIF2Bɛ-Δ93-358 mutant bound eIF2 as the wild type did. By using more limiting concentrations of eIF2, lower-affinity interactions could result in reduced steady-state binding. However, even when a lower concentration (5 nM) of eIF2 was used (Fig. 3C), these mutants bound eIF2 in a manner indistinguishable from that of wild-type eIF2Bɛ. These data strongly suggest that the nonsense mutants are defective for nucleotide exchange (Fig. 2B), because they fail to bind to eIF2 (Fig. 3B). However, the T552I and S576N mutants bind to eIF2 as well as wild-type eIF2Bɛ did in this assay but nonetheless are defective for nucleotide exchange activity, implying that these mutations directly impair the catalytic function of eIF2Bɛ and that these residues may be directly involved in catalysis.
FIG. 3.
In vitro binding between eIF2 and eIF2Bɛ proteins. (A) Titration of interaction between a fixed concentration (200 nM) of FLAG-tagged eIF2Bɛ (even-numbered lanes) or 200 nM FLAG peptide as a control (odd-numbered lanes) and the indicated concentration of eIF2. Proteins remaining bound after washing were identified by SDS-PAGE and Western blotting. Subunits indicated to the left of each panel were identified with the antisera shown to the right. Pellet fractions (33%) were loaded for probing with eIF2 antibodies, and 10% was used for eIF2Bɛ. (B) Binding of mutants at saturating concentrations of eIF2. Binding reactions were performed with a 200 nM concentration of the indicated FLAG-tagged eIF2Bɛ protein (lanes 6 to 14) or 200 nM control FLAG peptide (lane 5) and 20 nM eIF2. Proteins were visualized as described above for panel A. Lanes 1 to 4 show decreasing concentrations of input eIF2 (equivalent 20, 10, 5, and 2.5% of the 20 nM used in the reaction mixtures) and a single concentration input eIF2Bɛ (5%) (lane 4). (C) Binding of mutants at limiting concentrations of eIF2. Binding reactions were performed with a 200 nM concentration of the indicated FLAG-tagged eIF2Bɛ protein (lanes 2 to 7) or 200 nM control FLAG peptide (lane 1) and 5 nM eIF2. Proteins were visualized as described above for panel A.
The eIF2Bɛ N terminus is important for interactions with other eIF2Bɛ subunits.
Having accounted for the mutant phenotypes affecting the C terminus of eIF2Bɛ, our attention turned to the gcd6-N249K and gcd6-F250L mutants. These mutations affect adjacent, absolutely conserved residues (Fig. 1B), suggesting that they each impair the same function of eIF2B. However, these mutations cause a reduction in eIF2B function in vivo, as implicated by a slow-growth phenotype, without affecting the in vitro biochemical functions of the epsilon subunit (substrate binding and GEF activity [Fig. 2 and 3]). Similarly, the Δ93-358 mutant with a large region of the protein deleted retains catalytic activity and eIF2 binding. However, this mutant protein failed to copurify with the other eIF2B subunits (Fig. 2D). This suggested that the gcd6-N249K and gcd6-F250L alleles might more subtly affect the stability of the eIF2B complex to cause a mutant phenotype, so we tested this idea. However, in the experiments described in the next section, we could not detect any differences in association of these mutants either with subunits of eIF2B or with eIF2 in vivo.
First, we subcloned the eIF2Bɛ mutants onto high-copy-number plasmids under the control of the normal GCD6 promoter. These plasmids cooverexpressed hexahistidine and FLAG epitope-tagged eIF2γ. When cotransformed into a yeast strain with a plasmid overexpressing the eIF2Bα, -β, and -δ subunits, all mutants grew as well as the wild type did (Fig. 1D, column 5). This confirmed that eIF2B function was no longer limiting when all five subunits were overexpressed. Next, anti-FLAG immune precipitation reactions were performed using extracts from these eIF2B-overexpressing cells. Western blotting showed that similar amounts of eIF2B subunits and eIF2α coimmune precipitated with the FLAG-eIF2Bγ from wild-type cells and eIF2BɛN249K- and eIF2BɛF250L-overexpressing cells (Fig. 4A, compare lane 5 with lanes 7 and 8). In contrast to these results but in agreement with the protein purification experiments, eIF2BɛΔ93-358 failed to associate with FLAG-eIF2Bγ (Fig. 4A, lane 6). It is noteworthy that the eIF2B regulatory subcomplex of α, β, and δ subunits also failed to interact with FLAG-eIF2Bγ here. This indicates that the N terminus of eIF2Bɛ is required to stabilize the interaction between eIF2Bγ and the regulatory subcomplex to form the intact eIF2B complex.
We next subcloned the eIF2Bɛ mutants onto yeast low-copy-number plasmids and plasmid shuffled them into a yeast strain with the chromosomal GCD6 gene deleted. We found that only three mutants could support growth (F250L, T552I, and S576N) but at reduced rates (Fig. 1D, column 6, and 4B). Because in these cells no genes are tagged or overexpressed, we performed an experiment to precipitate eIF2B and eIF2 proteins from whole-cell extracts using antisera directed against eIF2Bɛ (as previously described [1]). Consistent with all previous experiments using tagged or overexpressed proteins, we found no differences in the expression levels or complex-forming abilities of these three mutants in vivo that could account for their phenotypes (Fig. 4C).
The F250L mutation impairs translation initiation.
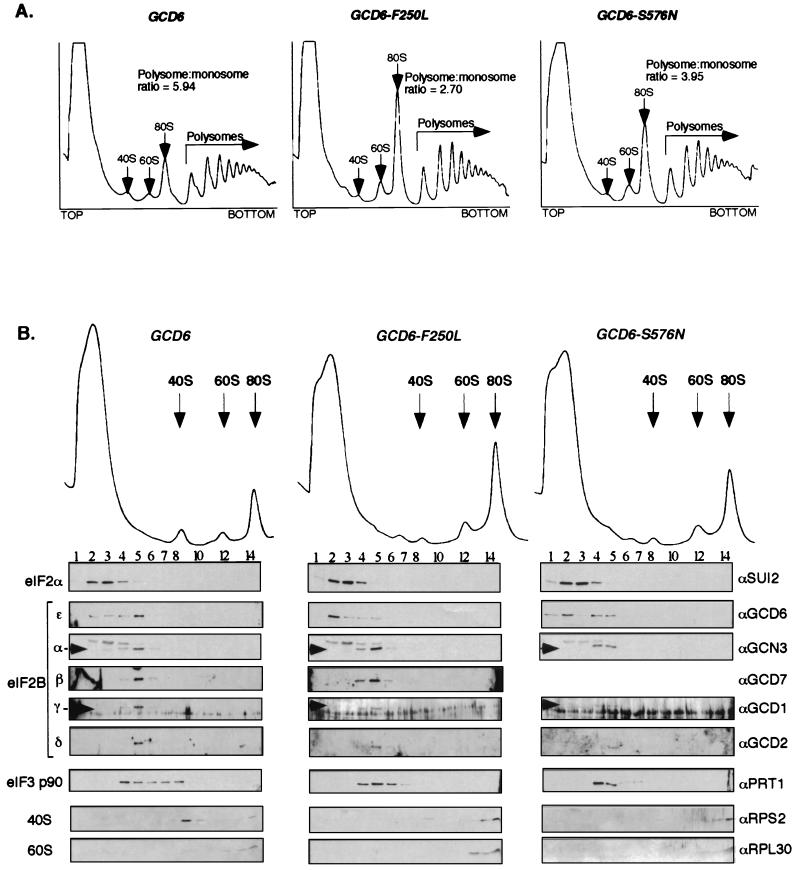
To determine whether the gcd6-F250L mutation resulted in a defect in translation initiation or some other (unknown) function of eIF2B, we performed low-salt 7 to 47% sucrose density gradient centrifugation to resolve ribosomal and polyribosomal fractions. We used extracts of cells containing this mutant and compared the resulting pattern to the patterns seen for wild-type cells and cells containing the gcd6-S576N mutation. We chose the gcd6-S576N strain as a control, because its rate of growth was almost identical to that for the isogenic strain with the gcd6-F250L mutation (Fig. 4B) and because we had determined that its eIF2B activity was impaired (Fig. 2B and C). Figure 5A shows that extracts from both the gcd6-F250L and gcd6-S576N mutant strains (center and right panels) each display increased 80S monosome peaks and reduced polysome size when compared with the wild-type gradient control (left panel). These features are indicative of a translation initiation defect as seen before for other mutants affecting translation initiation factors including eIF2B subunits (10, 19) and confirm that the F250L mutation does impair a function of eIF2B in translation initiation.
FIG. 5.
Analysis of polysome profiles from gcd6 mutant yeast strains using low-salt sucrose density gradient centrifugation. (A) Extracts prepared from cells grown in YPD medium at 30°C were centrifuged on low-salt 7 to 47% sucrose gradients. Gradients were fractionated while scanning at 254 nm, and the resulting profiles are shown. The positions of ribosomal subunits, 80S monosomes, and polysomes are indicated. The ratio of polysomes to 80S monosomes was determined by measuring the area under the peaks using NIH Image software. (B) Extracts from the same yeast strains were centrifuged on low-salt 15 to 35% sucrose gradients. This provides greater separation of the top portion of the gradient. Proteins collected in fractions from the gradients were analyzed by SDS-PAGE and Western blotting using the antisera indicated to the right.
We next went on to examine the association of different translation factors with these fractions. This was achieved by repeating the sucrose density gradients using 15 to 35% sucrose to better separate the 40S, 60S, and 80S region. eIF2, as monitored by antibodies to eIF2α, migrated mainly as a ribosome-free complex and concentrated in fractions 2 to 4 (Fig. 5B, left panel). This migration pattern has been observed previously for yeast eIF2 in wild-type cells (6). Similarly, eIF2B migrated as a ribosome-free complex primarily in fraction 5. The behavior of eIF2α was unaffected by the eIF2Bɛ mutations. However, in both mutants tested, but not in the wild-type gradients, eIF2Bɛ itself appeared to dissociate partially from the other eIF2B subunits so that a significant portion of eIF2Bɛ was localized to fraction 2 (Fig. 5B, center and right panels). This suggests that during gradient centrifugation the mutant eIF2B complexes partially dissociate, although the significance of this is unclear. Consistent with the reduction in translation initiation deduced from the profiles, the fraction of eIF3p90 apparently associated with 40S subunits was reduced in both mutants compared with the wild type. This analysis revealed no differences between the two eIF2B mutants examined, despite marked differences in GEF activity in vitro. This suggested to us that although the catalytic activity of the ɛ subunit alone remained intact in the F250L mutant, the activity of the five-subunit eIF2B complex might be impaired.
The N249K and F250L mutations impair nucleotide exchange activity of the five-subunit eIF2B complex.
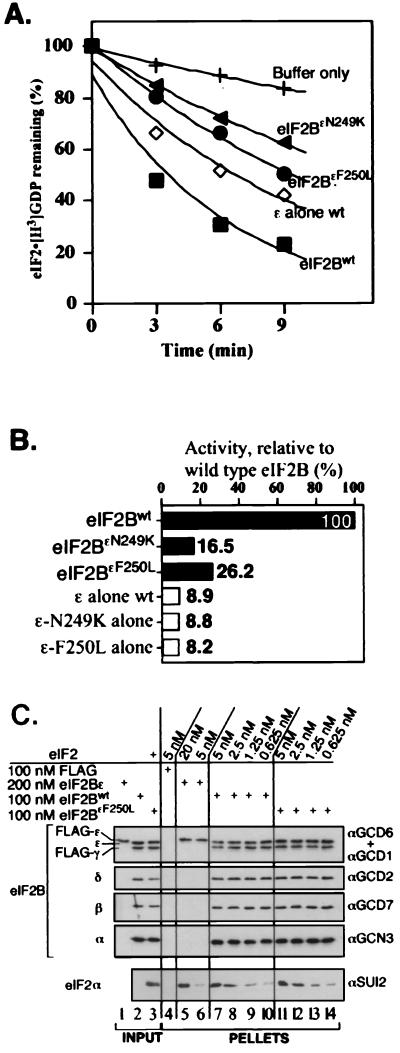
From the results of the experiments described in the section above (Fig. 5), it seemed most likely that the gcd6-N249K and gcd6-F250L mutations impair the GEF activity of eIF2B, rather than some other novel eIF2B function. To test this idea directly, we purified five-subunit eIF2B (wild-type and both eIF2BɛN249K and eIF2BɛF250L mutant forms) from cells overexpressing all five subunits from high-copy-number plasmids and assayed GEF activity in vitro. We used our plasmids encoding FLAG and hexahistidine epitope-tagged eIF2Bγ and partially purified eIF2B by nickel affinity chromatography (see Materials and Methods). Using just one purification step, eIF2B was purified to ∼85% homogeneity, as assessed by Coomassie brilliant blue staining, and free from contaminating eIF2 as judged by Western blotting (data not shown). Consistent with previous experiments using cell extracts as a source of eIF2B, we found that the five-subunit eIF2B complex promoted nucleotide exchange at a higher rate than that for the epsilon subunit alone (Fig. 6A). When compared on a molar basis, the wild-type eIF2B complex was found to be 11-fold more active than eIF2Bɛ alone (Fig. 6B). However, greatly reduced rates of nucleotide exchange relative to that for wild-type eIF2B were observed when the eIF2BɛN249K and eIF2BɛF250L mutant complexes were assayed (6- and 4-fold reductions, respectively) (Fig. 6A and B), demonstrating that these mutants are defective for eIF2B activity within the full five-subunit complex.
FIG. 6.
In vitro analysis of purified mutant eIF2B five-subunit complexes. (A) Nucleotide exchange assays comparing activities for mutant eIF2B complexes containing N249K (filled triangle) and F250L (filled circle) alleles of eIF2Bɛ with wild-type eIF2B (eIF2Bwt) (filled square) (1 μg each) and eIF2Bɛ alone (2.5 μg, open diamond). Assays were done in triplicate, with a standard deviation of less than 2.5 for each time point. (B) Analysis of rates of nucleotide exchange activity for mutant eIF2B complexes and isolated eIF2Bɛ subunits relative to wild-type eIF2B activity (percent initial activity). Analysis was performed as described in the legend to Fig. 2. (C) Analysis of binding between the indicated concentration of purified eIF2 and FLAG-tagged wild-type eIF2B complex (lanes 7 to 10) or eIF2BɛF250L (lanes 11 to 14). Also shown are control lanes using FLAG peptide (lane 4) or wild-type eIF2Bɛ alone (lanes 5 and 6) in place of eIF2B. Lanes 1 to 3 contain inputs; 5% of each eIF2B preparation used in the reaction mixtures was loaded and 6.25 ng of eIF2 was also loaded in lane 3 (representing 10% of 5 nM used in the reaction mixtures). Detection as described in the legend to Fig. 3, with 33% of each reaction pellet loaded to probe for eIF2 and 10% loaded to probe for each eIF2B subunit.
To analyze further how eIF2B complex formation enhances eIF2B activity, we examined the binding between purified eIF2 and purified five-subunit eIF2B complexes in vitro using our anti-FLAG immunoprecipitation assay. This analysis shows that five-subunit eIF2B has a greatly enhanced binding affinity for eIF2 than that for eIF2Bɛ alone (compare lanes 5 and 6 with lanes 7 to 10 in Fig. 6C). One explanation for the reduced GEF activity of the eIF2BɛN249K and eIF2BɛF250L mutant complexes could be that binding of mutant eIF2B to eIF2 is impaired. However, in accord with our previous in vivo results (Fig. 4C), we found no defect in eIF2 binding for the eIF2BɛF250L mutant (Fig. 6C, compare lanes 7 to 10 with lanes 11 to 14) or for eIF2BɛN249K (data not shown). These results suggest that eIF2B complex formation both enhances eIF2 binding and stimulates the rate of nucleotide exchange and that only the latter function is impaired by the N249K and F250L mutations in eIF2Bɛ.
DISCUSSION
eIF2B is a complex GEF composed of five subunits that is required to promote and regulate protein synthesis initiation in eukaryotes. To gain insight into its function, we have examined the roles of eIF2Bɛ in catalysis of guanine nucleotide exchange and in the formation of complexes with other eIF2B subunits and with its substrate, eIF2. Our analysis here involved a directed genetic screen to isolate mutations in eIF2Bɛ with reduced catalytic activity. Following purification of mutant eIF2Bɛ proteins, each was analyzed for guanine nucleotide exchange function and protein-protein interaction properties. These studies have established the likely biochemical defect of the mutant phenotypes observed. We find that regions within the C terminus of eIF2Bɛ are responsible for both substrate (eIF2) binding and nucleotide exchange activity, while regions within the N-terminal half are necessary for interactions with the other eIF2B subunits. We show by using our purified system, and in agreement with previous results (18, 33), that complex formation enhances eIF2B activity. We find that this increased activity may in part be caused by an increased affinity for the substrate, eIF2. However, missense mutations at universally conserved residues within the N-terminal half of eIF2B (gcd6-N249K and gcd6-F250L) retain the high binding affinity for eIF2 mediated by eIF2B complex formation without enhancing GEF activity. The implications of these results for the function of eIF2B are discussed below.
eIF2Bɛ is the principal catalytic subunit of eIF2B, and its N-terminal region is required for interactions with other eIF2B subunits.
Previously, we used extracts from yeast cells overexpressing different combinations of eIF2B subunits as a source of eIF2B in our in vitro nucleotide exchange assays (33). From this analysis we concluded that eIF2Bɛ was the principal catalytic subunit. To prove this, we set out to purify the epsilon subunit free from other contaminating eIF2B subunits. In our initial experiments, recombinant yeast eIF2Bɛ protein expressed and purified from Escherichia coli was catalytically inactive (our unpublished observations), so we set up the yeast expression system described in Materials and Methods. In this system, the other eIF2B subunits always copurified with wild-type hexahistidine-tagged eIF2Bɛ at a low level through several different chromatographic columns (Fig. 2D and data not shown). As the activity we measured for our purified eIF2Bɛ was approximately 11-fold lower than that for the purified eIF2B complex (Fig. 6B), this raised doubts as to whether eIF2Bɛ was indeed the catalytic subunit. Was the activity we were measuring due to the low-level copurifying eIF2B complex? Two mutants we constructed with large deletions of the N terminus of eIF2Bɛ (gcd6Δ93-358 and gcd6Δ144-230) allowed us to answer this question. These N-terminal deletion mutants failed to associate stably with the other eIF2B subunits in vivo (Fig. 4A) and were purified by a single step free from the other eIF2B subunits, as determined by Western blotting (Fig. 2D). The largest deletion, eIF2BɛΔ93-358, exhibited the same activity as purified wild-type eIF2Bɛ in our in vitro assay (Fig. 2B), demonstrating that eIF2Bɛ is indeed a catalytic subunit and that a large section of the N terminus of this polypeptide is not required for this activity.
This analysis further shows that elements within the deleted N-terminal region, residues 93 to 358, are necessary for assembly of eIF2Bɛ (Fig. 2D) and eIF2Bγ (Fig. 4A) into the eIF2B complex. Both deletions in the N terminus remove some of the region of similarity with NTP-hexose pyrophosphorylases (residues 27 to 159) and either part (gcd6Δ144-230) or the entire region (gcd6Δ93-358) that shows similarity with eIF2Bγ (residues 160 to 330). In addition, the deletion in gcd6Δ93-358 removes some IGXXXX repeat sequences also found in acyltransferases (residues 330 to 470) (36). The region homologous to NTP-hexose pyrophosphorylases was proposed to contain a potential nucleotide binding domain composed of a modified P-loop and Mg2+ binding site (28), so we thought that this region in eIF2Bɛ might be important for binding GDP and/or GTP to promote catalysis of guanine nucleotide exchange. Following our analysis described here, a direct role for this region in the catalysis of nucleotide exchange now seems unlikely. Instead, our analysis of the gcd6Δ93-358 mutation suggests that one or more of these regions of similarity in eIF2Bɛ and eIF2Bγ are important for mediating the protein-protein interactions required for eIF2B complex formation.
The C terminus of eIF2Bɛ contains the catalytic domain and regions for interaction with eIF2.
In agreement with the finding that the N-terminal region of eIF2Bɛ between residues 93 and 358 is dispensable for catalytic activity, we found that the C terminus is necessary for this function. All three nonsense mutants that prematurely terminate the GCD6 open reading frame show a dramatic reduction in binding to eIF2, with the binding of the shortest polypeptide (Q452*) being almost undetectable (Fig. 3B, lane 11). Not surprisingly, given this dramatic binding defect, all three mutants show no guanine nucleotide exchange function in vitro (Fig. 2B) and fail to complement a deletion of GCD6 (gcd6Δ) in vivo (Fig. 1D). These results suggest that sequences C terminal to residue 517, the last unchanged residue in our longest nonsense mutant gcd6-T518D+9*, are essential for GEF activity.
Asano et al. (1) showed recently that a motif in the extreme C terminus of eIF2Bɛ, called the AA-box as it is rich in aromatic and acidic residues that is shared with eIF5 (the potential GTPase-activating protein for eIF2) was important in both factors for mediating binding to eIF2. By changing all seven conserved residues of this motif between residues 696 and 706 (out of 712 residues in the protein) to alanine residues, the gcd6-7A allele was made. This mutant showed reduced binding to eIF2 in several assays but impaired the guanine nucleotide exchange activity only modestly in vivo (sufficient to induce translation of GCN4) (1). The activity of this mutant was not impaired in in vitro assays (G. D. Pavitt, K. Asano, and A. G. Hinnebusch, unpublished observations). The results of Asano et al. (1) are in agreement with those presented here. Both studies show that the C terminus of eIF2Bɛ is an important determinant for eIF2B binding to eIF2.
We isolated and characterized two missense mutations that change single residues within the region between residues 518 to 696 essential for eIF2B GEF function. Our analysis of these mutants suggests they may be directly involved in the catalytic mechanism of nucleotide exchange. Both the T552I and S576N mutations do not detectably affect eIF2Bɛ binding to eIF2 in vitro (Fig. 3B, lanes 9 and 10, and C, lanes 5 and 6) or in vivo (Fig. 4C, lanes 3 and 4). However, they do dramatically reduce but not eliminate the guanine nucleotide exchange activity of the purified mutant epsilon subunit (Fig. 2B). These mutations fall within a 44-amino-acid region between residues 543 and 586 that has been well conserved through evolution among eIF2Bɛ subunits. Five residues in this region are invariant in the eIF2Bɛ sequences shown in Fig. 1C, and several other residues are highly conserved. Three-dimensional structures for other GEFs complexed with their G-protein partner show direct interactions between residues surrounding the nucleotide binding pocket of the G protein and residues of the GEF that alter the structure of this pocket. These interactions are believed to displace the bound nucleotide (GDP), thus forming a nucleotide-free complex that can then bind GTP (reviewed in references 8 and 41). If these observations also apply to eIF2 and eIF2B interactions, this implies a direct interaction between eIF2γ (GDP- or GTP-binding subunit) (17) and eIF2Bɛ.
Missense mutations in the N-terminal region reveal an activation domain that responds to eIF2B complex formation.
The observations that the missense mutations N249K and F250L do not affect the intrinsic activity of the isolated epsilon subunit (Fig. 2B and C) but do eliminate the enhancement of GEF activity observed with the intact five-subunit complex (Fig. 6A and B) is intriguing and was quite unexpected. These mutations do not alter the affinity between eIF2B and eIF2 either in vivo (Fig. 4) or in vitro (Fig. 6C and data not shown for eIF2BɛN249K), suggesting that the effects of the mutations are not exerted through changes in eIF2 binding. Instead, the results suggest that these mutations affect residues critical for enhancing further the intrinsic GEF function of eIF2Bɛ upon complex formation and further suggest that this activation is an important consequence of eIF2B complex formation. This may be a major reason that the genes encoding three of the four other subunits of eIF2B are essential in yeast (GCD7, GCD1, and GCD2 encoding the β to δ subunits, respectively).
We found previously that cooverexpression of eIF2Bγ with eIF2Bɛ led to the formation of a subcomplex of these two subunits and that extracts prepared from these cells had higher GEF activity than that of extracts from cells overexpressing eIF2Bɛ alone (33). The simplest interpretation of these experiments was that eIF2Bγ acted to enhance the activity of eIF2Bɛ in this system, as eIF2Bγ had no activity alone, and the subcomplex was termed the catalytic subcomplex. A speculative extension of this idea is to predict that the N249K and F250L mutations in eIF2Bɛ disrupt the stimulatory function of eIF2Bγ. While this may be true, we were unable to confirm this idea experimentally. We tried to extend our earlier results using a purified system as we had done for both eIF2Bɛ alone and five-subunit eIF2B. Using purified eIF2Bγ, we confirmed that this subunit has no GEF activity (data not shown). We then purified the eIF2Bγ and eIF2Bɛ subcomplex and analyzed its GEF activity. Activity of the subcomplex was identical to that with eIF2Bɛ alone, i.e., not enhanced by eIF2Bγ. At this time, we are unable to explain the difference observed between the GEF activity of cell extracts overexpressing the catalytic subcomplex and that of the purified subcomplex. Therefore, we cannot conclude whether the enhanced activity of the purified five-subunit eIF2B complex is due to the action of eIF2Bγ alone or whether other subunits (i.e. eIF2Bβ and/or -δ) are also required.
Because complex formation between eIF2B subunits enhances binding of eIF2Bɛ to eIF2 and stimulates its catalytic activity, it could be that association of eIF2 with eIF2B is a rate-limiting step in the nucleotide exchange reaction. However, the eIF2BɛN249K and eIF2BɛF250L mutant complexes show the same enhanced binding affinity for eIF2 as wild-type eIF2B does but without conferring the same increased rate of nucleotide exchange. This implies, therefore, that a step after the association of eIF2B with eIF2 is rate limiting for nucleotide exchange. This interpretation is in agreement with kinetic studies performed for other G proteins and their GEFs. For example, Klebe et al. (27) showed that the release of bound nucleotide from a Ran-GDP-RCC1 ternary complex was rate limiting. The same step was proposed to be rate limiting for yeast RAS2 and CDC25 (21). However, for the bacterial translation elongation factor EF-Tu and its exchange factor EF-Ts, the rate-limiting step was shown to be the dissociation of EF-Ts from the EF-Tu–GTP–EF-Ts ternary complex (25, 37). The tools developed here will be useful for the further structure-function and kinetic studies required for a more detailed understanding of the function of this important regulatory molecule in translation initiation.
ACKNOWLEDGMENTS
We are grateful to Jigna Patel for technical assistance in mutant isolation; Katsura Asano, Jinsheng Dong, Thanuja Krishnamoorthy, and Alan Hinnebusch (NIH) for providing plasmids and yeast strains; Ernie Hannig (Dallas, Tex.) and Jonathan Warner (Einstein, New York) for antibodies. We thank Christopher Proud and Gert Scheper for critical reading of the manuscript.
This work was supported by an MRC career development award to G.D.P.
REFERENCES
- 1.Asano K, Krishnamoorthy T, Phan L, Pavitt G D, Hinnebusch A G. Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berlanga J J, Santoyo J, De Haro C. Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2alpha kinase. Eur J Biochem. 1999;265:754–762. doi: 10.1046/j.1432-1327.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 3.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 4.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 5.Bushman J L, Asuru A I, Matts R L, Hinnebusch A G. Evidence that GCD6 and GCD7, translational regulators of GCN4 are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1920–1932. doi: 10.1128/mcb.13.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushman J L, Foiani M, Cigan A M, Paddon C J, Hinnebusch A G. Guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae: interactions between the essential subunits GCD2, GCD6, and GCD7 and the regulatory subunit GCN3. Mol Cell Biol. 1993;13:4618–4631. doi: 10.1128/mcb.13.8.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesareni G, Murray J A H. Plasmid vectors carrying the replication origin of filamentous single-stranded phages. Genetic Eng. 1987;9:135–154. [Google Scholar]
- 8.Cherfils J, Chardin P. GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem Sci. 1999;24:306–311. doi: 10.1016/s0968-0004(99)01429-2. [DOI] [PubMed] [Google Scholar]
- 9.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 10.Cigan A M, Foiani M, Hannig E M, Hinnebusch A G. Complex formation by positive and negative translational regulators of GCN4. Mol Cell Biol. 1991;11:3217–3228. doi: 10.1128/mcb.11.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cigan A M, Pabich E K, Feng L, Donahue T F. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc Natl Acad Sci USA. 1989;86:2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemens M J. Protein kinases that phosphorylate eIF2 and eIF2B, and their role in eukaryotic cell translational control. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 139–172. [Google Scholar]
- 13.Cuesta R, Hinnebusch A G, Tamame M. Identification of GCD14 and GCD15, novel genes required for translational repression of GCN4 mRNA in Saccharomyces cerevisiae. Genetics. 1998;148:1007–1020. doi: 10.1093/genetics/148.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dever T E. Translation initiation: adept at adapting. Trends Biochem Sci. 1999;24:398–403. doi: 10.1016/s0968-0004(99)01457-7. [DOI] [PubMed] [Google Scholar]
- 15.Dever T E, Feng L, Wek R C, Cigan A M, Donahue T F, Hinnebusch A G. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- 16.Dever T E, Yang W, Åström S, Byström A S, Hinnebusch A G. Modulation of tRNAiMet, eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2 · GTP · Met-tRNAiMet ternary complexes. Mol Cell Giol. 1995;15:6351–6363. doi: 10.1128/mcb.15.11.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson F L, Hannig E M. Ligand interactions with eukaryotic translation initiation factor 2: role of the γ-subunit. EMBO J. 1996;15:6311–6320. [PMC free article] [PubMed] [Google Scholar]
- 18.Fabian J R, Kimball S R, Heinzinger N K, Jefferson L S. Subunit assembly and guanine nucleotide exchange activity of eukaryotic initiation factor-2B expressed in Sf9 cells. J Biol Chem. 1997;272:12359–12365. doi: 10.1074/jbc.272.19.12359. [DOI] [PubMed] [Google Scholar]
- 19.Foiani M, Cigan A M, Paddon C J, Harashima S, Hinnebusch A G. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [PubMed] [Google Scholar]
- 21.Haney S A, Broach J A. Cdc25p, the guanine nucleotide exchange factor for the ras proteins of Saccharomyces cerevisiae, promotes exchange by stabilizing Ras in a nucleotide-free state. J Biol Chem. 1994;269:16541–16548. [PubMed] [Google Scholar]
- 22.Harashima S, Hinnebusch A G. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3990–3998. doi: 10.1128/mcb.6.11.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harding H P, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 24.Hinnebusch A G. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y W, Miller D L. A study of the kinetic mechanism of elongation factor Ts. J Biol Chem. 1985;260:11498–11502. [PubMed] [Google Scholar]
- 26.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klebe C, Prinz H, Wittinghofer A, Goody R S. The kinetic mechanism of Ran-nucleotide exchange catalysed by RCC1. Biochemistry. 1995;34:12543–12552. doi: 10.1021/bi00039a008. [DOI] [PubMed] [Google Scholar]
- 28.Koonin E V. Multidomain organization of eukaryotic guanine nucleotide exchange translation initiation factor eIF-2B subunits revealed by analysis of conserved sequence motifs. Protein Sci. 1995;4:1608–1617. doi: 10.1002/pro.5560040819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Marton M J, Vazquez de Aldana C R, Qiu H, Chakraburtty K, Hinnebusch A G. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2α kinase GCN2. Mol Cell Biol. 1997;17:4474–4489. doi: 10.1128/mcb.17.8.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrick W C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsen D E, Jordan B, Chen D, Wek R C, Cavener D R. Isolation of the gene encoding the Drosophila melanogaster homolog of the Saccharomyces cerevisiae GCN2 eIF-2α kinase. Genetics. 1998;149:1495–1509. doi: 10.1093/genetics/149.3.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavitt G D, Ramaiah K V, Kimball S R, Hinnebusch A G. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998;12:514–526. doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavitt G D, Yang W, Hinnebusch A G. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol Cell Biol. 1997;17:1298–1313. doi: 10.1128/mcb.17.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price N T, Kimball S R, Jefferson L S, Proud C G. Cloning of cDNA for the gamma-subunit of mammalian translation initiation factor 2B, the guanine nucleotide-exchange factor for eukaryotic initiation factor 2. Biochem J. 1996;318:631–636. doi: 10.1042/bj3180631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raetz C R H, Roderick S L. A left-handed parallel β helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 37.Romero G, Chau V, Biltonen R L. Kinetics and thermodynamics of the interaction of elongation factor Tu with elongation factor Ts, guanine nucleotides, and aminoacyl-tRNA. J Biol Chem. 1985;260:6167–6174. [PubMed] [Google Scholar]
- 38.Rowlands A G, Panniers R, Henshaw E C. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem. 1988;263:5526–5533. [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Shi Y, Vattem K M, Sood R, An J, Liang J, Stramm L, Wek R C. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sprang S R, Coleman D E. Invasion of the nucleotide snatchers: structural insights into the mechanism of G protein GEFs. Cell. 1998;95:155–158. doi: 10.1016/s0092-8674(00)81746-8. [DOI] [PubMed] [Google Scholar]
- 42.Trachsel H. Binding of initiator methionyl-tRNA to ribosomes. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 113–138. [Google Scholar]
- 43.Yang W, Hinnebusch A G. Identification of a regulatory subcomplex in the guanine nucleotide exchange factor eIF2B that mediates inhibition by phosphorylated eIF2. Mol Cell Biol. 1996;16:6603–6616. doi: 10.1128/mcb.16.11.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]