Abstract
Background
Head and neck squamous cell carcinoma (HNSCC) is an extremely heterogeneous malignant cancer with poor prognosis. Pyroptosis is defined as a novel inflammation-dependent programmed cell death. However, the pyroptosis-associated gene expression in HNSCC and their relationship with prognosis are still indistinct.
Material and Methods
We acquired the mRNA expression information of HNSCC patients from publicly available TCGA and GEO databases. We compared the tumor issues and adjacent normal tissues in terms of the gene expression for the purpose of identifying differentially expressed genes (DEGs). Based on these genes, we established a risk signature by the LASSO Cox regression in the TCGA cohort and validated the results in a GEO cohort. We also verified the levels of relevant mRNA expression in the model by RT-qPCR analysis. Eventually, functional enrichment approach was carried out to explore the potential mechanisms.
Results
Our team found a total of 18 differentially expressed genes (DEGs) between the HNSCC and healthy samples, and 4 DEGs displayed a remarkable association with the overall survival (OS) (P < 0.05). A 4-gene signature was constructed, presenting beneficial forecast power in both TCGA and GEO cohorts. Our team categorized patients into a group with high risk and another group with low risk as per the average risk value of the 4-gene feature. The individuals in the low risk group displayed a notably greater OS compared with the high risk one (P < 0.01). The Cox regression study demonstrated the independent forecast capability of the risk score. The receiver operating characteristic approach facilitated the verification of the forecast function of the gene signature. Posterior to verification, 4 genes were aberrantly expressed in the HNSCC and healthy samples. Functional study displayed that these groups presented diverse immunity conditions.
Conclusion
Pyroptosis-associated genes are pivotal for the prognosis of HNSCC and can serve as potential therapeutic targets.
Keywords: pyroptosis, head and neck squamous cell carcinoma, overall survival, risk signature, prognosis
Introduction
HNSCC marks the 6th most commonly seen malignant cancer across the globe, which originates from the nasal cavity, larynx, hypopharynx, oropharynx, nasopharynx, oral cavity, and salivary gland.1,2 Owing to the absence of effective methods in early diagnosis, most of the HNSCC patients are already at advanced stages when diagnosed, and will suffer from recurrence after treatment, with a poor survival rate of 35%.3 The multimodal strategies, such as surgery, radiotherapy and chemotherapy, are the main treatments for HNSCC. In spite of the recent progress in therapies, the improvement of survival rate is still slow.4 Therefore, novel therapeutic targets are urgently required to bring better results for HNSCC patients, and it is necessary to develop dependable new prognostic models to bring more feasibility for targeted therapies.
Pyroptosis is a cell death triggered by caspase-1/4/5/11 and results in cell swelling, chromatin fragmentation, plasma membrane cleavage, and intracellular proinflammatory release.5 More and more studies reveal that pyroptosis is vital for cancer progression. Some previous studies have reported that some key components of pyroptosis, such as inflammatory vesicles, proinflammatory cytokines, and gasdermin proteins are closely related to tumourigenesis, invasion, and metastasis.6,7 A few latest studies have confirmed that pyroptosis plays a critical role in tumour immune microenvironment and antitumour processes as well.8,9 However, it remains unclear whether pyroptosis has an effect on the development of HNSCC.
In this research, we conducted a systematic research to identify the pyroptosis-associated gene expression in healthy tissues and HNSCC tissues, and the DEGs were adopted to establish a model of prognosis so as to reveal the prognostic role of those genes. The predictive ability of the model was validated by GEO data, and the expression of mRNA of the pyroptosis-associated genes in our model was validated by RT-qPCR analysis. Eventually, our research investigated the potential mechanisms by functional enrichment method.
Materials and Methods
Data Collection
The clinical information and the mRNA expression of 528 HNSCC sufferers were obtained from the TCGA database (https://portal.gdc.cancer.gov/), and another 96 HNSCC sufferers from the GEO cohort (GSE31056) were used as the verification set. Our research was conducted without ethical examination, given that the entire data were accessible openly. Our team chose an overall 33 pyroptosis-associated genes as per past researches.7,10
Construction and Validation of a Prognostic Pyroptosis-Related Gene Signature
Our team compared the levels of pyroptosis-related gene expression of cancer and neighboring healthy samples to determine the DEGs via the “limma” R package.11 The DEGs standard registered a false discovery rate (FDR) below 0.05. The DEGs are as described below: *P < 0.05, **P < 0.01, and ***P < 0.001. A PPI network regarding the DEGs was established via STRING 11.0 (https://string-db.org/). The Cox study facilitated the capability examination of pyroptosis-associated genes to forecast the OS. Benjamini-Hochberg modulated p-scores were adopted to diminish FDR. The “glmnet” R package was applied to implement a LASSO Cox regression in order to determine whether DEGs could forecast the OS and HNSCC suffer conditions.12 The best score of the penalty parameter (λ), correlating with the lowest partial possible abnormality, was determined by cross-verification at 10 folds. Our team computed the risk score by the formula below: risk score = esum (the standardized expression level of every gene× the regression coefficient). Afterwards, our team applied the mid-value risk score as the criterion for categorizing sufferers into the two above mentioned groups. Out team described the gene distribution in these groups via implementing PCA by the “stats” R package.13 The “surv_cutpoint” function of the “survminer” R package was adopted to analyze the survival in order to determine the ideal cut-off values. Subsequently, our team leveraged the “survivalROC” R package to carry out time-dependent ROC study to assess whether the genetic feature could forecast or not.14 Afterwards, our research applied the univariable and multivariable Cox regression method to identify whether the risk score enabled the independent forecast of the prognosis on OS. For the validation, a HNSCC cohort from the GEO (GSE31056) was included in the analysis. The risk score was computed via the identical formula in the TCGA cohort. The sufferers in the GSE31056 cohort were also classified into two groups likewise on account of the mid-value risk score of TCGA, and these groups were utilized for verifying the gene signature.
Functional Enrichment Analysis
Our team finished the GO enrichment and the KEGG pathway analyses for the DEGs in these groups with the “clusterProfiler” R package and |log2 (fold-change)| ≥ 1 and FDR < 0.05 were deemed as the standards of these DEGs.15 The “gsva” R package was adopted on the ssGSEA (single-sample gene set enrichment analysis) to compute the clustering value of immune cells and pathways associated with immunity.16
Patients and Specimens
Our team chose 15 HNSCC samples and 10 healthy samples from the First Hospital of Jiaxing between 2020 and 2021. This research was approved by the Ethics Committee of the First Hospital of Jiaxing and was conducted in accordance with the Declaration of Helsinki, with all individuals offering informed consents. Each sample was put in liquid nitrogen and reserved at −80°C directly.
Quantitative Real-Time PCR
After trizol extracted the entire RNA from the samples, it was turned into cDNA by reverse transcription. PCR was implemented via TB Green® Premix Ex Taq™II kit (Takara, China). The reactive activity is demonstrated as follows: half a minute at 94°C, half a minute at 58°C, one minute at 72°C, forty times. GAPDH served as the interior control, and the comparative level of expression was identify via 2−ΔΔ CT.
Statistical Analyses
Student’s t-test was applied to contrast the gene expressions in cancer samples and neighboring healthy samples. Our team implemented the Mann–Whitney test to contrast the enrichment values of immune cells and the pathways associated with immunity in these groups. The Log rank test and Kaplan–Meier approach were utilized to describe the survival curves. Afterwards, our team carried out Cox regression study to reveal the OS-predicting factors. Our research used all statistical observation in R (Version 4.0.3). A two-tailed P value below 0.05 was important on statistics. The overall flow diagram of the study is shown in Figure 1 and graphical abstract.
Figure 1.
Workflow diagram of data analysis.
Results
Identification of DEGs Between Normal and Tumor Tissues
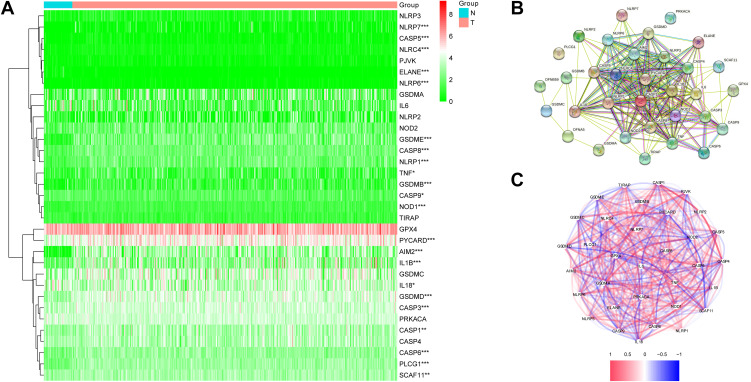
We compared the expression levels of 33 pyroptosis-associated genes between 44 healthy and 502 cancer samples from the TCGA cohort, and we found 18 differentially expressed genes (DEGs) (P < 0.05). Among them, 17 genes (AIM2, CASP1, CASP5, CASP6, CASP8, GSDMB, GSDMD, GSDME, IL1B, NLRC4, NLRP1, NLRP6, NLRP7, NOD1, PLCG1, PYCARD, TNF) were upregulated while only 1 gene (ELANE) was downregulated in the tumor group. The heatmap presented the expression levels of these genes (Figure 2A). To further investigate the interactions of these genes, we used a protein–protein interaction (PPI) method, and the outcomes are presented in Figure 2B. Figure 2C shows the associations between these pyroptosis-associated genes (red: positive association; blue: negative association).
Figure 2.
Expressions of 33 pyroptosis-related genes and interactions between them. (A) Heatmap of pyroptosis-related genes between tumor and normal tissues. P values were showed as: *P < 0.05; **P< 0.01; ***P < 0.001. (B) The PPI network showing the interactions of the pyroptosis-related genes. (C) Correlation network of pyroptosis-related genes.
Construction of a Prognostic Gene Signature in the TCGA Cohort
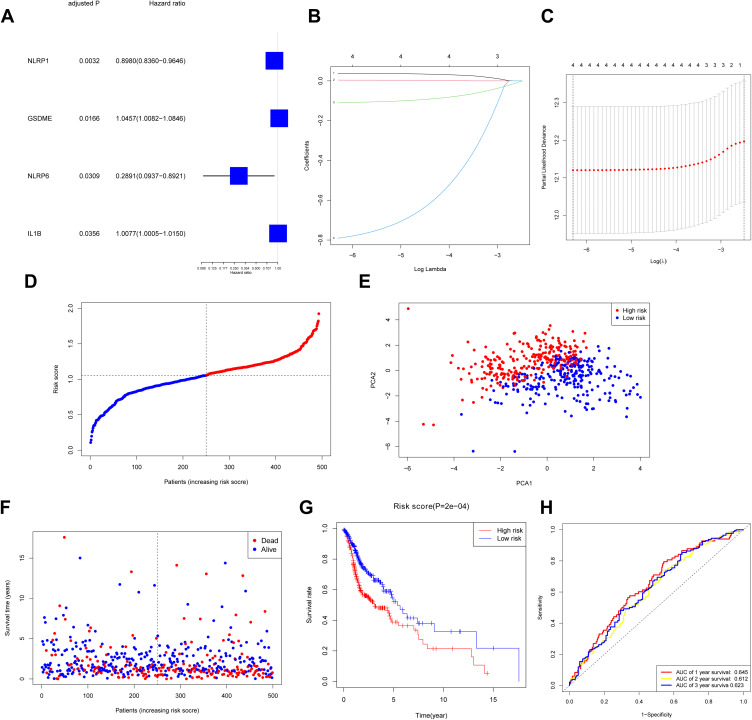
Our team conducted the univariate Cox regression observation using the DEGs to screen out the survival-related genes, and finally, 4 genes (GSDME, NLRP1, NLRP6, IL1B) were tightly related to OS (P < 0.05) (Figure 3A). The aforementioned 4 genes were used for the purpose of establishing a model of prognosis via LASSO Cox regression method. A 4-gene signature was then established in the light of the best score of λ (Figure 3B and C). Our research adopted the formula below to compute the risk score: e(0.8938 * the NLRP1 expression level + 1. 0402 * the GSDME expression level + 0.4383* the level of expression of NLRP6 +1. 0041 * the IL1B expression level). Our study applied the mid-value risk score as the criterion for separating cases into low-risk and high-risk groups (Figure 3D). The PCA study presented that there were two diverse distribution of these HNSCC cases (Figure 3E). People in the high-risk group displayed a greater death ratio versus the other group (Figure 3F). Likewise, the Kaplan–Meier curve revealed that the low-risk group exhibited a notably greater OS versus the high-risk group (Figure 3G, P < 0.001). Figure 3H unveils the OS forecast role of the risk score, with the region below the curve (AUC) being 0.645, 0.612, and 0.623 of 1, 2, 3 years, separately.
Figure 3.
Construction of risk signature in TCGA cohort. (A) Screen out of OS-related genes with univariate cox regression analysis. (B) LASSO regression of 4 OS-related genes. (C) Cross-validation for tuning parameter selection in LASSO regression. (D) Distribution of HNSCC patients based on risk score. (E) PCA plot for HNSCCs based on risk score. (F) Survival status for each patient. (G) Kaplan–Meier curves for OS of patients in high- and low-risk groups. (H) The ROC curves demonstrated the predictive efficiency of the risk score.
Validation of the Risk Signature in the GEO Cohort
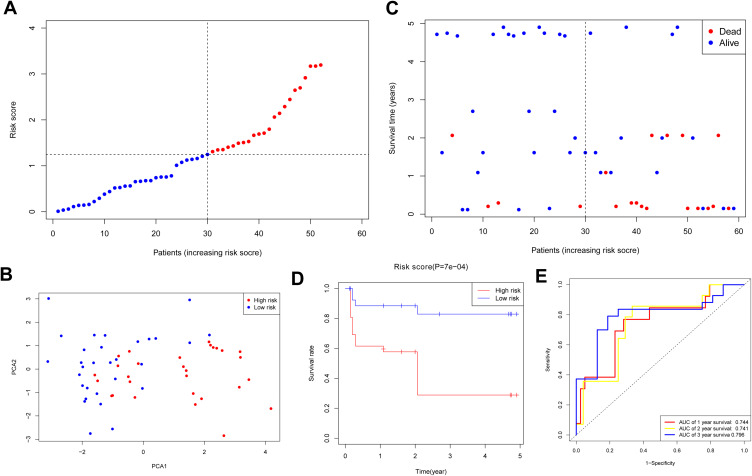
A total of 96 HNSCC sufferers from the Gene Expression Omnibus (GEO) cohort (GSE31056) were used as the verification set. Our team divided the patients into two groups as mentioned above on the foundation of the mid-value risk score of the TCGA cohort (Figure 4A). The PCA displayed acceptable separation between the two groups (Figure 4B). The individuals in the low-risk group displayed a lower mortality rate versus the other group (Figure 4C). Moreover, the Kaplan–Meier curve also demonstrated that the group with low risk exhibited a remarkably higher OS compared to the group with high risk (Figure 4D, P < 0.001). The time-dependent ROC analysis of the GEO cohort suggests that the forecast ability of the model is satisfactory, with the AUC being 0.744, 0.741, and 0.796 of 1, 2, 3 years, separately (Figure 4E).
Figure 4.
Validation of risk model in GEO cohort. (A) Distribution of HNSCC patients in GEO cohort based on risk score in TCGA cohort. (B) PCA plot for HNSCCs. (C) Survival status for each patient. (D) Kaplan–Meier curves for OS of patients in high- and low-risk groups. (E) The ROC curves demonstrated the predictive efficiency of the risk score.
Independent Prognostic Value of the Risk Signature
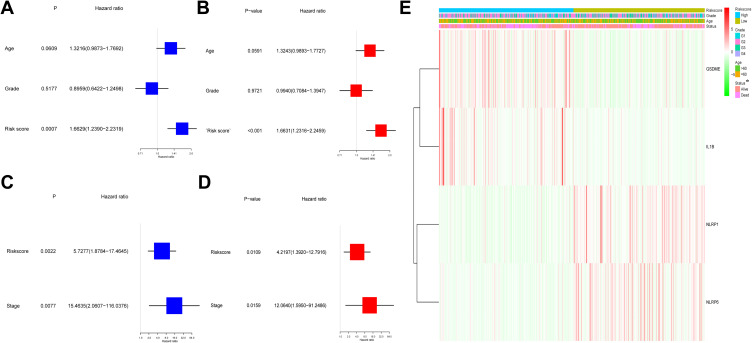
Cox regression study was employed to verify whether the risk score could independently forecast the OS. As discovered via the univariable Cox regression study, the risk score presented a tight relevance with the OS in both TCGA and GEO cohorts (HR = 1.6629, 95% CI = 1.2390–2.2319, P = 0.0007 and HR = 5.7277, 95% CI = 1.8784–17.4645, P = 0.0022) (Figure 5A and C). Likewise, the multivariable Cox regression study revealed a remarkable correlation between the OS and risk score in both cohorts (HR = 1.6631, 95% CI = 1.2316–2.2459, P < 0.001 and HR = 4.2197, 95% CI = 1.3920–12.7916, P = 0.0109) (Figure 5B and D). Moreover, our team created a heatmap of risk score and the clinical characteristics in the TCGA cohort, and the results indicated that the survival status of patients was distinctly diverse between the two groups (Figure 5E).
Figure 5.
Univariate and multivariate Cox regression analyses for risk score in TCGA and GEO cohorts. (A) Univariate analysis for TCGA cohort. (B) Multivariate analysis for TCGA cohort. (C) Univariate analysis for GEO cohort. (D) Multivariate analysis for GEO cohort. (E) The heatmap for the connections between clinicopathologic features and the risk groups (*P < 0.05).
Functional Analyses Based on the Risk Model
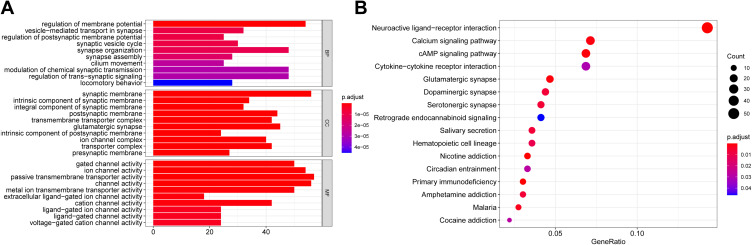
In order to reveal the biological roles and pathways associated with the risk score, we firstly used the “limma”R package to find out the DEGs between the two groups. The DEGs standard was a FDR below 0.05 and |log2FC | ≥ 1. A total of 1025 DEGs were detected between the two groups in the TCGA cohort. Subsequently, for these DEGs, our team carried out the GO enrichment and KEGG pathway assay. Figure 6A shows the top 10 MF, CC, and BP terms. The prime enriched Go terms were gated channel activity, postsynaptic membrane, integral and intrinsic components of synaptic membrane, and synaptic membrane. Figure 6B shows the main KEGG pathways, including Nicotine addiction, Primary immunodeficiency, Glutamatergic synapse, and cAMP signaling pathway.
Figure 6.
Functional analysis based on the DEGs between the two-risk group in the TCGA cohort. (A) Barplot graph for GO enrichment. (B) Bubble graph for KEGG pathways.
Comparison of the Immune Activity
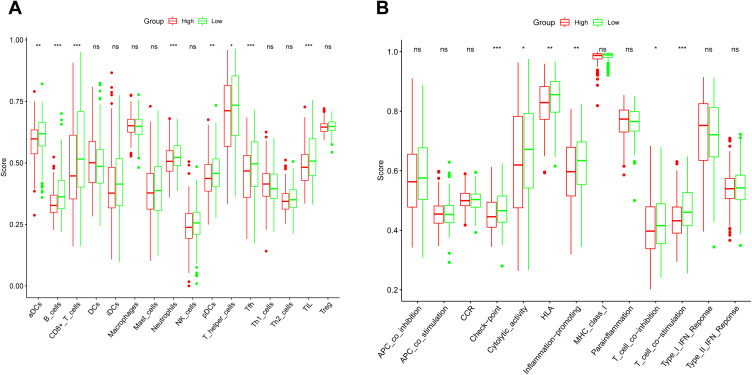
The immunity condition was calculated via ssGSEA by virtue of the enrichment score and its correlation with the risk score was studied. By comparison, the enrichment scores of TIL, Tfh, T helper cells, pDCs, Neutrophils, CD8+ T cells, B cells, and aDCs were remarkably diverse (adjusted P < 0.05, Figure 7A). These groups demonstrated a notable diversity on the values of T cell co−activation, T cell co−suppression, Inflammation promotion, HLA, Cytolytic activity, and Check point (adjusted P < 0.05, Figure 7B).
Figure 7.
The comparison of the ssGSEA scores for the immune cells and immune pathways in the TCGA cohort. (A) The comparison of the enrichment scores of 16 types of immune cells between the high- and low-risk groups. (B) The comparison of the enrichment scores of 13 immune-related pathways between the high- and low-risk groups. Adjusted P values were showed as: *P < 0.05; **P < 0.01; ***P < 0.001.
Abbreviation: ns, not significant.
Validation of the 4 Pyroptosis-Related Genes in HNSCC Tissues
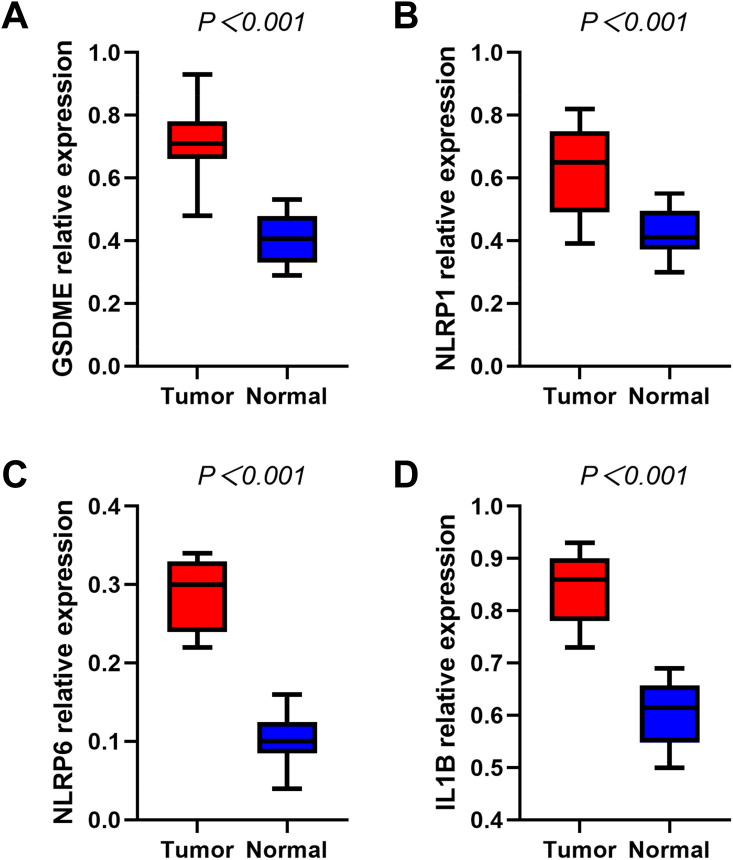
Our team verified the levels of mRNA expression as to the 4 pyroptosis-associated genes in the HNSCC samples and healthy samples via RT-qPCR, and the outcomes revealed that GSDME (Figure 8A), NLRP1 (Figure 8B), NLRP6 (Figure 8C), and IL1B (Figure 8D) were all distinctly greater in the HNSCC samples versus the healthy samples (P < 0.001).
Figure 8.
RT-qPCR detecting the mRNA expression levels of GSDME (A), NLRP1 (B), NLRP6 (C), IL1B (D) in the HNSCC and normal tissues.
Discussion
Pyroptosis is defined as a novel type of programmed cell death characterized by inflammatory response and is reported to play a dual role in cancer progress.17 Signaling pathways and inflammatory mediators in the process of pyroptosis are closely associated with the invasion, proliferation, and metastasis of cancer.18,19 Nevertheless, the induction of tumor cell pyroptosis could be a novel treatment.20 However, whether pyroptosis-related genes are related to the outcomes of HNSCC patients and how they act in HNSCC remain unknown.
In our research, we systematically analyzed the levels of mRNA expression of the 33 pyroptosis-associated genes in the HNSCC and healthy samples. The outcomes showed that more than half of the pyroptosis-related genes (55%, 18/33) were DEGs by comparing the cancer and neighboring healthy samples and four of them were associated with the OS, which hinted that pyroptosis participated in HNSCC and exhibited a potential forecast role of relevant genes. In order to explore more on the forecast power of those genes, we built a 4-gene signature through LASSO Cox regression analysis, and then the signature was validated to perform satisfactorily in a GEO cohort. We further validated the mRNA levels of these 4 genes in the HNSCC and healthy tissues via RT-qPCR analysis, and the outcomes coincided with those in the TCGA cohort. The functional analyses indicated a different immune status between the two groups.
Four pyroptosis-related genes (GSDME, NLRP1, NLRP6, IL1B), which functioned via different mechanisms, were used to establish a forecast model.21,22 GSDME, also known as DFNA5, is from the gasdermin family which is the executor of pyroptosis. The activated caspase-3 could realize the cleavage of GSDME to release a N-terminal fragment and then promote pyroptosis.23 GSDME was recognized as a tumor suppressor gene due to its inactivation and the methylation of DNA promoter region was found in many tumors, including breast cancer,24 hepatocellular carcinoma25 and so on, however, it was reported to be overexpressed in nonsmall-cell lung cancer26 and esophageal squamous cell cancer.27 Therefore, GSDME may play a specific role in different tumors. Intriguingly, GSDME seemed to be an oncogenic gene, as it was upregulated in tumor tissues. Due to the limited data and the conflicting results in different tumors, our results about GSDME provide a new direction for future research. NLRP1 is one of the NALP family proteins consisting of a pyrin domain, a CARD domain and an NBD-LRR domain which can induce pyroptosis.28 Previous studies29 reported that NLRP1 was regulated downward in colorectal cancer samples relative to healthy samples. Nevertheless, in our research, the expression of NLRP1 was distinctly higher in the HNSCC specimens versus the healthy ones, and was related to a shorter survival time. The relationship between NLRP1-mediated pyroptosis and HNSCC is still unknown and needs further study. NLRP6 is a new type of inflammasome which play critical roles in the development of cancer. The knockdown of NLRP6 encourages gastric tumor cells to proliferate, migrate, and invade.30 However, our results demonstrated that higher NLRP6 expression was identified in the tumor samples versus the normal ones. Similarly, IL1B is another type of inflammasome which is released by caspase-1 and can induce pyroptosis. A recent study has presented that the IL 1B expression is lower in hepatocellular carcinoma samples.31 On the contrary, cervical cancer cells release more IL 1B than normal cervical cells.32 We found that IL 1B was upregulated in the tumor tissues and its overexpression could predict poor outcomes, which revealed that it was a cancer-promoting gene in our research. Further research should highlight how IL 1B acts in the process of pyroptosis and tumor development. In summary, all 4 genes (GSDME, NLRP1, NLRP6, IL1B) were regulated upward in the HNSCC sufferers and were associated with unfavourable clinical results. Nevertheless, whether those genes influenced the HNSCC prognosis by virtue of modulating pyroptosis needs more studies.
We analysed the DEGs between the two groups divided by risk scores and discovered that the DEGs primarily emerged in immune and inflammatory responses. The outcomes of GO and KEGG methods displayed that pyroptosis might play an important role in tumor immune microenvironment. The enrichment scores of the crucial anti-tumor immune cells and immune-associated pathways were lower in the high-risk group in contrast to the other one, suggesting that the unfavourable clinical outcomes in the high-risk group might be induced by the damaged anti-cancer immune activity.
At present, the underlying mechanism of pyroptosis is less studied, especially in HNSCC. Our study identified 4 genes which acted as the regulators of pyroptosis, and then our team carried out a preliminary research on the prognostic value of these pyroptosis-associated genes and offered a new direction for future investigation. Nevertheless, whether these regulators play the same role in pyroptosis-pathway in HNSCC is still elusive, and deserves further in-depth research. In addition, there were limitations in our model of prognosis due to the openly available data. It is pivotal to conduct further in vitro and in vivo researches to validate the clinical significance of this model.
Conclusion
This research found that pyroptosis was tightly associated with HNSCC, as more than half of the pyroptosis-associated genes were differently expressed by comparing the tumor and healthy samples. Furthermore, we constructed a novel forecast model by 4 pyroptosis-associated genes. The risk score according to the signature was able to independently forecast the OS of HNSCC in both TCGA and GEO cohorts. The potential mechanism regarding the correlation of between the pyroptosis-associated genes and HNSCC is still vague and deserves more in-depth researches.
Acknowledgments
All authors acknowledge the contributions from the TCGA and GEO project. Xiaoyu Qian and Jian Tang are co-first authors for this study.
Funding Statement
This study was funded by the Science and Technology Project of Jiaxing City (2019AD32259); the Natural Science Foundation of Zhejiang Province (LQ20H160059); the Project of the First Hospital of Jiaxing (2021-YA-041).
Ethics Statement
The research was approved by the Ethics Committee of the First Hospital of Jiaxing and all patients offered written informed consent prior to the enrollment for research and biospecimens.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Cohen E, Bell RB, Bifulco CB, et al. The society for immunotherapy of cancer consensus statement on immunotherapy for the treatment of squamous cell carcinoma of the head and neck (HNSCC). J Immunother Cancer. 2019;7(1):184. doi: 10.1186/s40425-019-0662-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu D, Ruan X, Huang J, et al. Comprehensive analysis of competitive endogenous RNAs network, being associated with esophageal squamous cell carcinoma and its emerging role in head and neck squamous cell carcinoma. Front Oncol. 2019;9:1474. doi: 10.3389/fonc.2019.01474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferlay J, Colombet M, Soerjomataram I, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021;149:778–789. doi: 10.1002/ijc.33588 [DOI] [PubMed] [Google Scholar]
- 4.Yokota T, Homma A, Kiyota N, et al. Immunotherapy for squamous cell carcinoma of the head and neck. Jpn J Clin Oncol. 2020;50(10):1089–1096. doi: 10.1093/jjco/hyaa139 [DOI] [PubMed] [Google Scholar]
- 5.Fang Y, Tian S, Pan Y, et al. Pyroptosis: a new frontier in cancer. Biomed Pharmacother. 2020;121:109595. doi: 10.1016/j.biopha.2019.109595 [DOI] [PubMed] [Google Scholar]
- 6.Kolb R, Liu GH, Janowski AM, Sutterwala FS, Zhang W. Inflammasomes in cancer: a double-edged sword. Protein Cell. 2014;5(1):12–20. doi: 10.1007/s13238-013-0001-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye Y, Dai Q, Qi H. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Discov. 2021;7(1):71. doi: 10.1038/s41420-021-00451-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang R, Xu J, Zhang B, et al. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol. 2020;13(1):110. doi: 10.1186/s13045-020-00946-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Zhang Y, Xia S, et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579(7799):415–420. doi: 10.1038/s41586-020-2071-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia X, Wang X, Cheng Z, et al. The role of pyroptosis in cancer: pro-cancer or pro-”host”. Cell Death Dis. 2019;10(9):650. doi: 10.1038/s41419-019-1883-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon N, Friedman JH, Hastie T, Tibshirani R. Regularization paths for cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39(05):1–13. doi: 10.18637/jss.v039.i05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saadatpour A, Lai S, Guo G, Yuan GC. Single-cell analysis in cancer genomics. Trends Genet. 2015;31(10):576–586. doi: 10.1016/j.tig.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32(30):5381–5397. doi: 10.1002/sim.5958 [DOI] [PubMed] [Google Scholar]
- 15.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong W, Shi Y, Ren J. Research progresses of molecular mechanism of pyroptosis and its related diseases. Immunobiology. 2020;225(2):151884. doi: 10.1016/j.imbio.2019.11.019 [DOI] [PubMed] [Google Scholar]
- 18.Derangère V, Chevriaux A, Courtaut F, et al. Liver X receptor β activation induces pyroptosis of human and murine colon cancer cells. Cell Death Differ. 2014;21(12):1914–1924. doi: 10.1038/cdd.2014.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi J, Gao W, Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci. 2017;42(4):245–254. doi: 10.1016/j.tibs.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 20.Karki R, Kanneganti TD. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat Rev Cancer. 2019;19(4):197–214. doi: 10.1038/s41568-019-0123-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Y, Chen Q, Li X, et al. Pyroptosis: a new paradigm of cell death for fighting against cancer. J Exp Clin Cancer Res. 2021;40(1):153. doi: 10.1186/s13046-021-01959-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang YY, Liu XP. [Research process of Gasdermin E in inducing cell pyroptosis]. Zhonghua Bing Li Xue Za Zhi. 2021;50(4):421–424. Chinese. doi: 10.3760/cma.j.cn112151-20200724-00589 [DOI] [PubMed] [Google Scholar]
- 23.Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8:14128. doi: 10.1038/ncomms14128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croes L, Beyens M, Fransen E, et al. Large-scale analysis of DFNA5 methylation reveals its potential as biomarker for breast cancer. Clin Epigenetics. 2018;10:51. doi: 10.1186/s13148-018-0479-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang CJ, Tang L, Shen DW, et al. The expression and regulation of DFNA5 in human hepatocellular carcinoma DFNA5 in hepatocellular carcinoma. Mol Biol Rep. 2013;40(12):6525–6531. doi: 10.1007/s11033-013-2581-8 [DOI] [PubMed] [Google Scholar]
- 26.Choubey D. Absent in melanoma 2 proteins in the development of cancer. Cell Mol Life Sci. 2016;73(23):4383–4395. doi: 10.1007/s00018-016-2296-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu M, Wang Y, Yang D, et al. A PLK1 kinase inhibitor enhances the chemosensitivity of cisplatin by inducing pyroptosis in oesophageal squamous cell carcinoma. EBioMedicine. 2019;41:244–255. doi: 10.1016/j.ebiom.2019.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Wang B, Sun J, et al. DAC can restore expression of NALP1 to suppress tumor growth in colon cancer. Cell Death Dis. 2015;6:e1602. doi: 10.1038/cddis.2014.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams TM, Leeth RA, Rothschild DE, et al. The NLRP1 inflammasome attenuates colitis and colitis-associated tumorigenesis. J Immunol. 2015;194(7):3369–3380. doi: 10.4049/jimmunol.1402098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Wang C, Chen J. NLRP6, decreased in gastric cancer, suppresses tumorigenicity of gastric cancer cells. Cancer Manag Res. 2018;10:6431–6444. doi: 10.2147/CMAR.S182980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu Q, Jiang Y, Zhang W, et al. Pyroptosis is involved in the pathogenesis of human hepatocellular carcinoma. Oncotarget. 2016;7(51):84658–84665. doi: 10.18632/oncotarget.12384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Tahhan MA, Etewa RL, El Behery MM. Association between circulating interleukin-1 beta (IL-1β) levels and IL-1β C-511T polymorphism with cervical cancer risk in Egyptian women. Mol Cell Biochem. 2011;353(1–2):159–165. doi: 10.1007/s11010-011-0782-9 [DOI] [PubMed] [Google Scholar]