Abstract
Introduction
Apolipoproteins are predictive biomarkers for cardiovascular, neoplasms and cerebrovascular diseases and are postulated as prognostic biomarkers in infectious diseases, as COVID-19. Thus, we assessed the prognosis value of apolipoproteins for COVID-19 severity and mortality.
Methods
We conducted a systematic review and meta-analysis using observational studies that reported the association between apolipoproteins and severity or mortality in COVID-19 patients. Newcastle-Ottawa was used for the quality assessment of included studies. Effects measurements were shown as odds ratios (ORs) with 95% confidence intervals (CIs), and Egger-test was developed for assessing the risk of bias publication.
Results
We analyzed 12 cohort studies (n = 3580). Patients with low ApoliproteinA1 (ApoA1) (OR 0.35; 95%CI 0.24 to 0.49; P < 0.001) and ApoliproteinB (ApoB) (OR = 0.78; 95%CI 0.69 to 0.87; P < 0.001) values had a higher risk of developing severe disease. ApoB/ApoA1 ratio showed no statistically significant association with higher odds of severity. Low ApoA1 levels were associated with higher odds of all-cause mortality (OR = 0.34; 95%CI 0.20 to 0.57; P < 0.001). ApoB values showed no statistically significant association with a high risk of all-cause mortality.
Conclusion
We suggest that adequate levels of ApoA1 and ApoB can be a protective factor for severity in COVID-19, and ApoB/ApoA1 ratio did not show predictive utility for severity.
Keywords: Sars-Cov-2, ApoA, ApoB, Prognosis, Mortality
1. Introduction
Since new coronavirus disease 2019 (COVID-19) was declared as pandemic and a global health emergency by World Health Organization (WHO) [1], clinical research has been focused on describing the natural history of disease and setting effective treatments, as well as on developing vaccines for COVID-19. As a result, it has provided evidence for diagnosis criteria, categorizing patients with a higher risk of poor outcomes, and suitable allocation of resources, especially for middle-income and low-income countries. This way, clinical research has been able for health systems to manage COVID-19 patients optimally and build an evidence-based treatment.
Nonetheless, the emergence of new SARS-CoV-2 variants with higher associated mortality and the likelihood that SARS-CoV-2 will become an endemic virus constitutes an uncertain future for the population and mainly for health staff [2,3]. Furthermore, daily COVID-19 patient care requires routine laboratory examinations and specific laboratory profiles for underlying diseases. As a result, laboratory tests can be helpful to classify patients according to their risk of progression, prognosis, treatment strategies and other objectives [4].
In this sense, several biological markers and proportions derived from them have been evaluated as indicators of severity and mortality in COVID-19 patients, such as D-dimer [[5], [6], [7]], C-reactive protein [8,9], neutrophil to lymphocyte ratio [10], apolipoproteins [11], albumin to globulin ratio [12], among others. In the case of ApoA1, ApoB and the ApoA1/ApoB ratio, its prognostic value in cardiovascular and cerebrovascular diseases and neoplasms is known. Although there are studies that suggest its prognostic value in sepsis and bacterial diseases, to our knowledge, the evidence of its prognostic value in patients hospitalized for COVID-19 was not systematized. Recently, these biological markers have been evaluated as prognosis indicators of severity in COVID-19 patients [[13], [14], [15]]. In order to keep increasing knowledge about COVID-19 and supporting clinical practise, we conducted a meta-analysis of available evidence to assess the prognosis value of apolipoproteins and ApoB/ApoA1 ratio for COVID-19 severity and mortality.
2. Methods
2.1. Report, register, study design and research question
This systematic review was registered on the International Prospective Register of Systematic Reviews (PROSPERO) with code CRD42021274326, and the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA) statement was followed for reporting [16]. The research question was based on Population, Exposure, Comparison and Outcome (PECO) strategy: Do COVID-19 patients (P) with low values of apolipoproteins (E) have more risk of severity or all-cause mortality (O) compared to normal values of apolipoproteins (C)?
2.2. Data sources and searches
The Peer Review of Electronic Search Strategies (PRESS) [17] checklist was used for building the search strategy, and no language or date restriction was applied. On August 28, 2021, a systematic search was performed for retrieving studies assessing the association between Apolipoprotein A1 (ApoA1), Apolipoprotein B (ApoB) or ApoB/ApoA1 ratio, and severity of COVID-19, through the following peer review databases: Embase, PubMed, Web of Science, Scielo, Scopus, LILACS and The Cochrane Library. In addition, a manual search was carried out in preprint databases (Medrixv, Scielo Preprints and ResearchSquare) and other sources (Wangfang Database and CNKI databases). At first, a search strategy based on MeSH and free terms was built for Pubmed, and it was adapted to the other databases (see Search Strategy in Appendix 1 of the Supplemental Information).
2.3. Study selection and data extraction
We included studies with a case-control or cohort design, conducted in patients aged more than 18 years old with a confirmed COVID-19 diagnosis, and assessed the association between ApoA1, ApoB or ApoB/ApoA1 ratio values reported at hospital admission and COVID-19 severity or mortality. Duplicates and studies without all eligibility criteria were excluded. The primary outcome was the severity of COVID-19, and mortality was a secondary outcome. COVID-19 severity was defined as meeting at least one of the following criteria: shortness of breath, respiration rate (RR) ≥ 30 times per minute, blood oxygen saturation at rest ≤93%, PaO2/FiO2 ≤ 300 mmHg or ICU admission. However, definitions for severity are diverse among studies and could be sources of heterogeneity.
Rayyan QCRI software was used for study selection and removing duplicates [18]. First, two authors (JRUB and EAHB) screened the retrieved records independently by titles and abstracts. Then, the same two authors assessed the remaining records independently by full-text. Any conflict in the screening process was resolved by two authors (VABZ, PH-A). Afterwards, two authors collected data from included studies in a preset data extraction Microsoft Excel © sheet (JRUB and EAHB). Collected data were: first author, study title, publication date, study design, study location, population baseline characteristics (number of participants, age, sex, comorbidities, stratified sample data), exposure measurements (mean with standard deviation or median with interquartile range, for ApoA1, ApoB or ApoB/ApoA1 ratio from the overall sample and according to sample stratification) outcome type (severity or mortality) and association measures (crude and adjusted).
2.4. Evaluation of study quality and publication bias
Quality assessment was evaluated independently with the Newcastle-Ottawa Scale (NOS) [19] by two authors (JRUB and EAHB), and scores greater than or equal to six were categorized as low risk of bias. Publication bias was assessed through funnel plots, Egger's test and the trim-and-fill method [20].
2.5. Data synthesis and analysis
Statistical analysis was performed using Review Manager 5.4 (RevMan 5.4) (The Cochrane Collaboration, Copenhagen, Denmark). Continuous data reported as the median and interquartile range (IQR) were transformed into means and standard deviations (SD) according to Wan et al. [21]. In order to analyze continuous values of apolipoproteins, standardized mean differences were converted to the natural logarithm of odds ratio and its standard error following Chinn method [22].
Heterogeneity analysis was assessed using the I2 test and Cochran's Q-statistic. Test values were categorized as severe heterogeneity (>60%), moderate heterogeneity (40–60%) and mild heterogeneity (<40%). A p-value of <0.05 was considered statistically significant. Due to anticipated heterogeneity, a random-effects meta-analysis was performed. Additionally, a subgroup analysis was carried out by study location (Chinese vs non-Chinese studies), and the interaction test p-value per subgroup analysis was reported. Finally, sensitivity analyses were performed using the low risk of bias studies only.
3. Results
3.1. Study selection
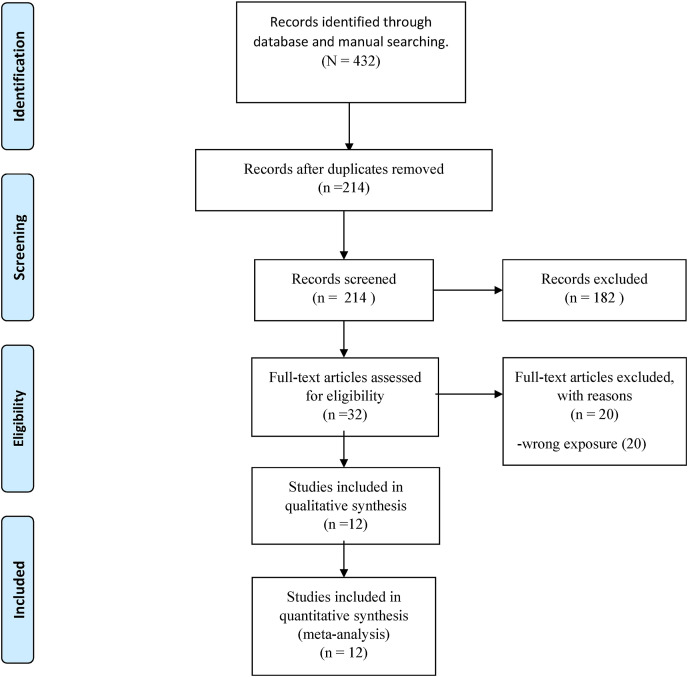
The comprehensive search strategy identified 432 articles, and 214 studies remained after removing duplicates. The screening process by titles and abstracts left 32 studies for full-text review (see Excluded articles by full-text in Supplemental Table S1). In turn, screening by full-text left 12 studies respecting all eligibility criteria [[13], [14], [15],[23], [24], [25], [26], [27], [28], [29], [30], [31]]. This process is summarised in a flow chart (Fig. 1 ).
Fig. 1.
Prisma flow diagram.
3.2. Study characteristics
Collected data from included studies are reported in Table 1 and Table 2 . A total of 12 cohort studies were included, of which nine studies analyzed severity, three studies analyzed mortality, and only one study analyzed both outcomes. In addition, eight studies were conducted in China, two in the United Kingdom, one in France and one in Belgium.
Table 1.
Characteristics of studies evaluating the association of Apolipoproteins and Severity.
| Author | Year | Location | Outcome | Participants (Male) | Median/mean Age (IQR/SD) | Apolipoprotein analyzed | Apolipoprotein mean (SD) in severe patients | Apolipoprotein mean (SD) in non-severe patients | Std Mean Difference between severe and non-severe patients | OR |
|---|---|---|---|---|---|---|---|---|---|---|
| Hilser Jet al. | 2021 | United Kingdom | Severity | 1110(NR) | 60 (14) | ApoA1 | NR | NR | NR | 0.82 (0.73–0.91) p < 0.001 ±b |
| Zhu Z et al. | 2021 | China | Severity | 142 (55) | 49 (16) | ApoA1 | 0.98(0.14) | 1.22 (0.16) | −1.51 [-2.05, −0.98] | NR |
| ApoB | 0.76(0.14) | 0.81 (0.17) | −0.30 [-0.81, 0.21] | NR | ||||||
| Shuke N et al. | 2020 | China | Severity | 97 (34) | 39 (30–60) | ApoA1 | 1.22(0.23) | 1.51 (0.14) | −1.61 [-2.18, −1.05] | NR |
| ApoB | 0.76(0.23) | 0.88 (0.25) | −0.49 [-0.99, 0.01] | NR | ||||||
| ApoB/A1 ratio | 1.64(0.6) | 1.84 (0.42) | −0.40 [-0.90, 0.09] | NR | ||||||
| Qin C et al. | 2020 | China | Severity | 248 (130) | 55 (16) | ApoA1 | 0.73 (0.18) | 0.81 (0.24) | −0.34 [-0.82, 0.14] | NR |
| ApoB | 0.72 (0,18) | 0,76 (0,25) | −0.16 [-0.64, 0.31] | NR | ||||||
| Sun JT et.al | 2020 | China | Severity Mortality |
99 (60) | 61 (42–83) | ApoA1 | 1.01(0.32) | 1.42 (0.3) | −1.32 [-1.66, −0.98] | NR |
| ApoB | 0.85(0.33) | 0.93 (0.21) | −0.29 [-0.60, 0.02] | NR | ||||||
| Dierckx T et.al (Cohort A) | 2020 | Hasselt, Belgium | Severity | 164 (84) | 58 (81) | ApoA1 | NR | NR | NR | 0.513(0.375–0.691), p < 0.001a |
| ApoB | NR | NR | NR | 0.71(0.53–0.94), p < 0.05a | ||||||
| ApoB/A1 ratio | NR | NR | NR | 1.39(0.83–2.34), p = 0.07a | ||||||
| Dierckx T et.al(Cohort B) | 2020 | Leuven, Belgium | Severity | 219 (114) | 67 (56–80) | ApoA1 | NR | NR | NR | 0.571(0.436–0.7478), p < 0.001a |
| ApoB | NR | NR | NR | 0.71(0.55–0.91), p < 0.001 | ||||||
| ApoB/A1 ratio | NR | NR | NR | 1.28(0.98–1.6718), p = 0.07a | ||||||
| Julkunen H et.al | 2021 | United Kingdom | Severity | 652 (372) | 60 (40–70) | ApoA1 | NR | NR | NR | 0.8151(0.7454–0.8913), p < 0.001 ±c |
| ApoB | NR | NR | NR | 0.8645(0.7983–0.9363) < 0.001 ±c | ||||||
| ApoB/A1 ratio | NR | NR | NR | 0.9754(0.8920–1.0667) p = 0.5857 ±c | ||||||
| Li C et.al | 2020 | China | Severity | 242 (133) | 63 (53–68) | ApoA1 | 1.02(0.22) | 1.07(0.22) | −0.23 [-0.51, 0.05] | NR |
| ApoB | 0.87(0.22) | 0.92(0.22) | −0.23 [-0.51, 0.05] | |||||||
| ApoB/A1 ratio | 0.9(0.29) | 0.82(0.37) | 0.23 [-0.05, 0.51] | NR | ||||||
| Qi J et.al | 2020 | China | Severity | 104 (47) | 42 (33–56) | ApoA1 | 0.71(0.12) | 0.92(0.14) | −1.51 [-2.19, −0.83] | NR |
| ApoB | 0.72(0.22) | 0.8(0.25) | −0.32 [-0.96, 0.31] | NR |
OR CRUDE, ±: OR Adjusted, ‡ NR: NOT REPORTED.
Adjusted to age, sex, obesity, hypertension, type 2 diabetes, and coronary artery disease.
Adjusted to adjusted for age, sex, and assessment centre.
Table 2.
Characteristics of studies evaluating the association of Apolipoproteins and mortality.
| Author | Year | Location | Outcome | Participants (Male) | Median/mean Age (IQR/SD) | Apolipoprotein analyzed | Apolipoprotein mean (SD) in deceased | Apolipoprotein mean (SD) in survivors | Std Mean Difference between deceased and survivors |
|---|---|---|---|---|---|---|---|---|---|
| Ressaire Q et al. | 2020 | France | Mortality | 31 (24) | 63 (60–68) | ApoA1 | 0.65 (0.2) | 0.72 (0.24) | −0.29 [-1.14, 0.55] |
| ApoB | 0.58 (0.17) | 0.8 (0.2) | −1.10 [-2.00, −0.21] | ||||||
| Sun JT et.al | 2020 | China | Mortality Severity |
99 (60) | 61 (42–83) | ApoA1 | 0.87 (0.4) | 1.02 (0.33) | −0.42 [-1.03, 0.19] |
| ApoB | 0.78 (0.3) | 0.89 (0.3) | −0.36 [-0.97, 0.25] | ||||||
| Li Yiet.al | 2021 | China | Mortality | 424 (220) | 61 (12) | ApoA1 | 0.67 (0.07) | 0.82 (0.22) | −0.71 [-1.06, −0.35] |
| ApoB | 0.95 (0.29) | 0.97 (0.22) | −0.09 [-0.44, 0.26] | ||||||
| Yue J et al. | 2021 | China | Mortality | 48 (32) | 68 (62–78) | ApoB | 0.83 (0.28) | 0.82 (0.37) | 0.03 [-0.56, 0.61] |
The included studies were conducted between 2020 and 2021, with 3580 patients hospitalized for COVID-19, of which only 1305 are male. The age range among the total participants ranged from 33 to 83 years.
According to the quality assessment of included studies by NOS, seven studies were at low risk of bias, while the remaining five were at moderate risk of bias (Supplemental Table S2).
3.3. Association of apolipoproteins with severity in hospitalized COVID-19 patients
3.3.1. Apolipoprotein A1
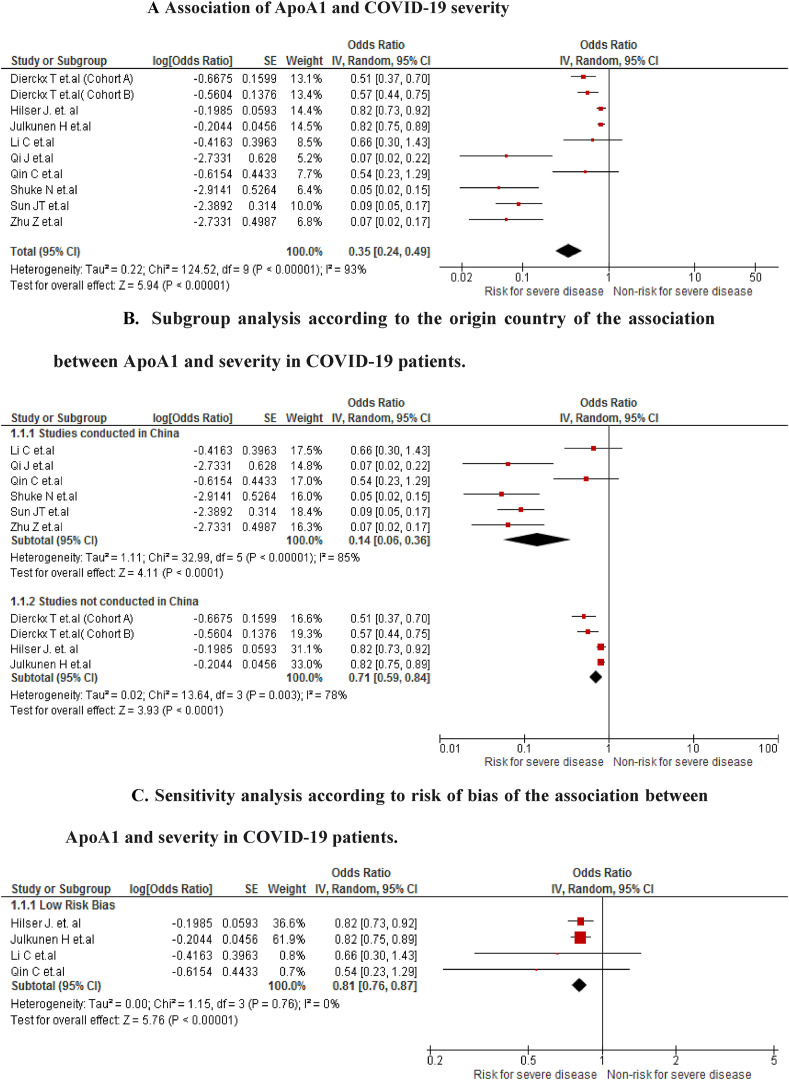
The association was found in 10 studies (n = 3077). We found that COVID-19 patients with low ApoA1 values have a higher risk of developing severe disease (OR 0.35; 95% CI 0.24 to 0.49; P < 0.001) with severe heterogeneity (I2 = 93%) (Fig. 2 A). In the analysis of subgroups by study location, differences were found between Chinese studies (OR 0.14; 95% CI 0.06 to 0.36; P < 0.001) and non-Chinese studies (OR 0.71; 95% CI 0.59 to 0.84; P < 0.001) (Fig. 2B). In the sensitivity analysis for including only articles with a low risk of bias (Fig. 2C), it was found that the association between ApoA1 and the risk of developing severity is still present (OR 0.81; 95% CI 0.76 to 0.87; P < 0.001), but with null heterogeneity (I2 = 0%)
Fig. 2.
A Association of ApoA1 and COVID-19 severity
Fig. 2B. Subgroup analysis according to the origin country of the association between ApoA1 and severity in COVID-19 patients.
Fig. 2C. Sensitivity analysis according to risk of bias of the association between ApoA1 and severity in COVID-19 patients.
3.3.2. Apolipoprotein B
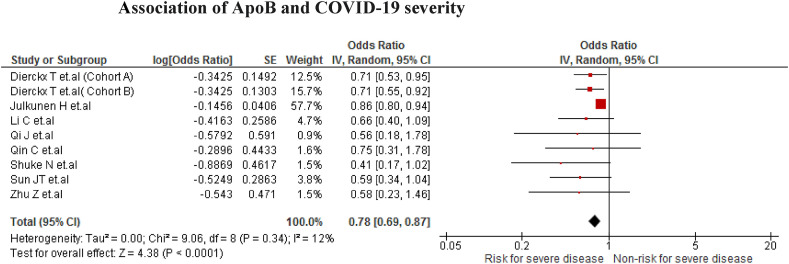
The association was found in nine studies (n = 1375). We found that COVID-19 patients with low ApoB values have a higher risk of developing severe disease (OR = 0.78; 95% CI 0.69 to 0.87; P < 0.001) with mild heterogeneity (I2 = 12%) (Fig. 3 ).
Fig. 3.
Association of ApoB and COVID-19 severity.
3.3.3. ApoB/ApoA1 ratio
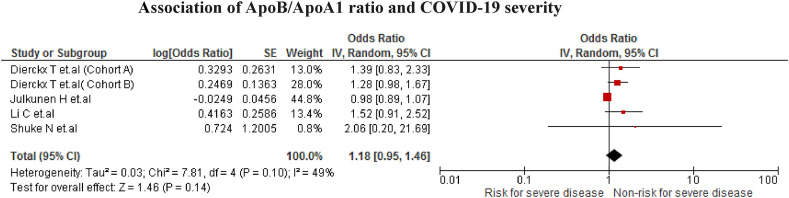
The association was found in five studies (n = 1374). However, no statistically significant association was found when low ApoB/ApoA1 values increased the risk of developing the severe disease due to COVID-19 (OR = 1.18; 95% CI 0.95 to 1.46; p = 0.14) with moderate heterogeneity (I2 = 49%) (Fig. 4 ).
Fig. 4.
Association of ApoB/ApoA1 ratio and COVID-19 severity.
3.4. Association of apolipoproteins with mortality in hospitalized COVID-19 patients
3.4.1. ApolipoproteinA1
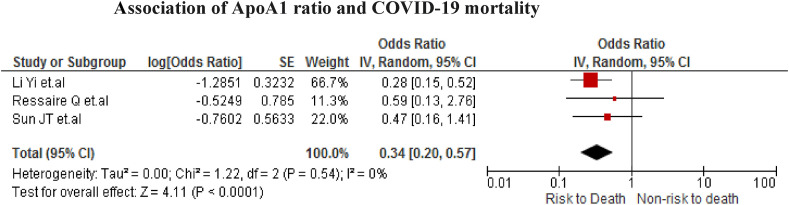
The association was found in three studies (n = 554). We found that COVID-19 patients with low ApoA1 values have a higher risk of all-cause mortality (OR = 0.34; 95% CI 0.20 to 0.57; P < 0.001) with no heterogeneity (I2 = 0%) (Fig. 5 ).
Fig. 5.
Association of ApoA1 ratio and COVID-19 mortality.
3.4.2. Apolipoprotein B
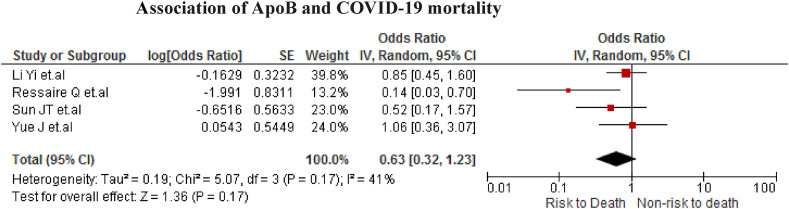
The association was found in four studies (n = 602). No statistically significant association was found when low ApoB values increase the risk of death of COVID-19 patients (OR = 0.63; 95% CI 0.32 to 1.23; p = 0.17) with moderate heterogeneity (I2 = 41%) (Fig. 6 ).
Fig. 6.
Association of ApoB and COVID-19 mortality.
3.5. Publication bias
In the association between ApoA1 and ApoB with disease severity, publication bias was found (Egger test <0.1). We corrected the publication bias using the trim-and-fill method (OR = 0.39; 95% CI 0.28 to 0.56 and OR: 0.82; 95% CI: 0.74 to 0.90, respectively) (supplemental Figures S1A and S1·B).
4. Discussion
The present systematic review found evidence to recommend low levels of ApoA1 and Apo B as predictors of disease severity and low levels of ApoA1 as a predictor of mortality in patients hospitalized for COVID-19.
Exchangeable Apolipoproteins, including Apo As, Apo E, and ApoCs, are constituents of HDL and triglyceride-rich lipoproteins such as VLDL. The best-studied family members are Apo A-I, the most significant HDL protein, in which an anti-atherogenic effect has been documented [32]. In contrast, non-exchangeable Apolipoproteins, such as Apo B, share a similar sequence and structure and can be reversibly associated with lipid surfaces [33]. Apo A is primarily bound to low-density lipoprotein (LDL) in subjects with average triglyceride values. However, Apo A can also bind to APOB100 or triglyceride particles in dyslipidemic states, called very low and intermediate-density lipoproteins [33].
Due to their potential effects and prominence in different pathologies, apolipoproteins have been studied as predictors of clinical outcomes in some diseases. For example, various systematic reviews associated the Apo E with ischemic and hemorrhagic stroke and a higher risk of worse outcomes in patients with traumatic brain disease [[34], [35], [36], [37], [38]]. Similarly, Apo C was associated with the risk of ischemic stroke, although a systematic review found no evidence of this association [39].
In the case of Apo A1, probably due to its anti-atherogenic effect, some systematic reviews and meta-analyses sought its association with cardiovascular outcomes. Haji Aghajani M et al., in a review of seventeen case-control studies, found an association between Apo A 1 levels and premature coronary artery disease. However, the authors note the lack of good quality prospective cohort studies [40]. Erqou S et al., in a systematic review of thirty-six studies, found that people with smaller Apo A isoforms have an approximately 2-fold higher risk of coronary heart disease or ischemic stroke than those with larger proteins [41]. As with cardiovascular outcomes, other systematic reviews found evidence of Apo A1 as a diagnostic marker for bladder cancer [42], a poor prognosis of multiple cancers [43,44], and it was found at lower levels in patients with Alzheimer's disease [45].
To the best of our knowledge, no systematic reviews have been published on the association between Apolipoprotein values as a prognostic factor in patients with COVID-19 or some other infectious disease; however, our results are not surprising. Apo-I's presence characterizes High-density lipoproteins (HDL), and their ability to transport cholesterol from peripheral tissues back to the liver gives it a cardioprotective function [46]. Similarly, it has antioxidant, anti-apoptotic, anti-thrombotic, anti-inflammatory or anti-infectious functions and decreases rapidly in patients with sepsis, which could explain our findings [46]. A study in pediatric patients in intensive care for sepsis found that Apo A5 serum levels were significantly lower in patients who died than survivors. Similarly, Apo A5 serum levels were significantly correlated with multiple organ failure, shock, acute kidney injury, acute liver injury, and gastrointestinal dysfunction, although not respiratory failure [47]. In adults, an association was also found between low levels of Apolipoproteins and a poor prognosis in patients with sepsis. Although the mechanisms are not well understood, it is suggested that the association is explained due to increased platelet activation and monocyte activation [48,49]. In addition, the low levels of Apo A are related to high levels of inflammation [50], and this being a prognostic marker in patients infected by COVID-19 [51], its role in the binding and neutralization of lipopolysaccharides in bacterial infections is known [52].
In patients with virus infections, changes in plasma HDL-C levels were reported during infections, where the viruses would take advantage of the HDL lipid transfer activity in host cells [53]. Although the best evidence is in patients with hepatitis C virus and acquired immunodeficiency virus, in the case of patients with COVID-19 infection, a similar theory is suggested [54]. Therefore, the HDL lipid transfer activity mechanism could explain our results as the relationship between viral load and worse prognosis in patients with COVID-19 is known [55]. Similarly, the hypothesis of the relationship between lipoproteins and inflammation and thrombosis was raised. In this way, our findings could explain since the association between thrombosis and the prognosis are known [56].
Finally, due to its known association with brain and cardiovascular disease, it is possible that in patients with COVID-19, the prognostic value of ApoA1 is mediated by the occurrence of these diseases. Indeed, complications including myocarditis, acute myocardial infarction, heart failure, arrhythmias and venous thromboembolic events are described in these patients [57,58]. Similarly, concerning cerebrovascular complications, episodes of stroke, necrotizing hemorrhagic encephalitis, among others, were reported [59,60].
Our study is the first systematic review to evaluate the prognostic value of Apo A1, ApoB and the ratio of both in patients with an infectious disease. In addition, our study used the NOS to assess the risk of bias of the included articles, which allowed sensitivity analyses when the association between Apo A1 and Apo B with the severity of the disease of patients hospitalized for COVID-19 was analyzed. Our findings allow us to suggest a potential low-cost prognostic marker in patients hospitalized for COVID-19 that will allow health personnel to prioritize or individualize management strategies in patients with low values of these markers.
4.1. Limitations
The main limitation is the clinical and methodological heterogeneity in the studies analyzed, which we assumed a priori. However, heterogeneity was addressed and explained mainly by studies with a high risk of bias and, to a lesser extent, by studies done in China. Also, the small number of participants in some studies could be overrepresented in their weights in the meta-analysis. In addition, we found publication bias, which was addressed using the trim and fill method, which did not change the direction of the effect found in the meta-analysis. Likewise, the studies in this meta-analysis do not evaluate the effect that some sociodemographic and clinic variables may have on Apo A1 and Apo B. Indeed, some studies find that Apo AI was significantly higher and Apo B levels were significantly lower among women in general, but, between women, Apo B levels were higher in post-versus premenopausal women [61]. Other studies show that some parameters present a large interindividual variability of response, which is significantly influenced by cofactors, such as weight or BMI, for apo B and apo E [62]. We did not find studies reported in African, American or Oceanic countries, so the generalizability of the results should be taken with caution. Therefore, cohort studies are necessary for various populations to establish Apolipoproteins' generalizability in the severity prognosis of COVID-19. Finally, the heterogeneity of the quantitative assays of plasma Apolipoproteins was not considered, which has been suggested as a problem previously [63,64] and still represents a challenge since tools are needed for the characterization and accurate quantification of apolipoproteins, including their diverse array of variant forms, are required to understand their salutary and disease-related roles [65].
5. Conclusion
We conclude that adequate levels of Apolipoproteins are a protective factor for severity in COVID-19. In contrast, only adequate levels of Apo A1 evidenced a protective effect against mortality in patients hospitalized for COVID-19. Furthermore, our findings showed that the ApoA1/ApoB ratio did not show more excellent predictive utility for severity. Therefore, apolipoproteins could be included in the clinical assessment of hospitalized patients with COVID-19. However, primary studies are necessary to define the optimal cut-off point for Apolipoproteins according to the profile of the hospitalized COVID-19 patient.
Authors contributions
Juan R. Ulloque-Badaracco: Conceptualization, Methodology, Investigation, Formal analysis and Writing - Original Draft. Enrique A. Hernandez-Bustamante: Methodology, Investigation, Formal analysis and Writing - Original Draft. Percy Herrera-Añazco: Investigation, Writing - Original Draft, Visualization and Supervision. Vicente A. Benites-Zapata: Conceptualization, Methodology, Writing - Review & Editing, Visualization and Supervision.
Funding
This research has been funded by the Universidad Peruana de Ciencias Aplicadas through grant IP008-2016.
Declaration of competing interest
The authors do not have conflicts of interest.
Acknowledgements
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tmaid.2021.102200.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.World Health Organization WHO Director-General’s opening remarks at the media briefing on COVID-19 - 11 March 2020 2020. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 (accessed October 7, 2021)
- 2.Phillips N. The coronavirus is here to stay - here's what that means. Nature. 2021;590:382–384. doi: 10.1038/D41586-021-00396-2. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization Tracking SARS-CoV-2 variants. 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- 4.Jutzeler C.R., Bourguignon L., Weis C.v., Tong B., Wong C., Rieck B., et al. Comorbidities, clinical signs and symptoms, laboratory findings, imaging features, treatment strategies, and outcomes in adult and pediatric patients with COVID-19: a systematic review and meta-analysis. Trav Med Infect Dis. 2020;37:1–32. doi: 10.1016/J.TMAID.2020.101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu H.-H., Qin C., Chen M., Wang W., Tian D.-S. D-dimer level is associated with the severity of COVID-19. Thromb Res. 2020;195:219–225. doi: 10.1016/J.THROMRES.2020.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Düz M.E., Balci A., Menekşe E. D-dimer levels and covid-19 severity: systematic Review and Meta-analysis. Tuberk Toraks. 2020;68:353–360. doi: 10.5578/TT.70351. [DOI] [PubMed] [Google Scholar]
- 7.Paliogiannis P., Mangoni A.A., Dettori P., Nasrallah G.K., Pintus G., Zinellu A. D-dimer concentrations and COVID-19 severity: a systematic review and meta-analysis. Frontiers in Public Health. 2020;8:1–7. doi: 10.3389/FPUBH.2020.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahu B.R., Kampa R.K., Padhi A., Panda A.K. C-reactive protein: a promising biomarker for poor prognosis in COVID-19 infection. Clin Chim Acta. 2020;509:91–94. doi: 10.1016/J.CCA.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erika P., Domenica Z., Paolo I., Luca R., Giulia L., Alessandro D., et al. Lactate dehydrogenase and C-reactive protein as predictors of respiratory failure in CoVID-19 patients. Clin Chim Acta. 2020 doi: 10.1016/j.cca.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulloque-Badaracco J.R., Salas-Tello W.I., Al-kassab-Córdova A., Alarcón-Braga E.A., Benites-Zapata V.A., Maguiña J.L., et al. Prognostic value of neutrophil-to-lymphocyte ratio in COVID-19 patients: a systematic review and meta-analysis. Int J Clin Pract. 2021:1–16. doi: 10.1111/ijcp.14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poynard T., Deckmyn O., Rudler M., Peta V., Ngo Y., Vautier M., et al. Performance of serum apolipoprotein-A1 as a sentinel of Covid-19. PLoS One. 2020;15 doi: 10.1371/JOURNAL.PONE.0242306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feketea G.M., Vlacha V. The diagnostic significance of usual biochemical parameters in coronavirus disease 19 (COVID-19): albumin to globulin ratio and CRP to albumin ratio. Front Med. 2020;7:1–3. doi: 10.3389/FMED.2020.566591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin C., Minghan H., Ziwen Z., Yukun L. Alteration of lipid profile and value of lipids in the prediction of the length of hospital stay in COVID-19 pneumonia patients. Food Sci Nutr. 2020;8:6144–6152. doi: 10.1002/FSN3.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z., Yang Y., Fan L., Ye S., Lou K., Hua X., et al. Low serum level of apolipoprotein A1 may predict the severity of COVID-19: a retrospective study. J Clin Lab Anal. 2021;35 doi: 10.1002/JCLA.23911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilser J.R., Han Y., Biswas S., Gukasyan J., Cai Z., Zhu R., et al. Association of serum HDL-cholesterol and apolipoprotein A1 levels with risk of severe SARS-CoV-2 infection. JLR (J Lipid Res) 2021;62:100061. doi: 10.1016/J.JLR.2021.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liberati A., Altman D.G., Tetzlaff J., Mulrow C., Gøtzsche P.C., Ioannidis J.P.A., et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339 doi: 10.1136/BMJ.B2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGowan J., Sampson M., Salzwedel D., Cogo E., Foerster V., Lefebvre C. PRESS peer review of electronic search strategies: 2015 guideline statement. J Clin Epidemiol. 2016;75:40–46. doi: 10.1016/J.JCLINEPI.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 18.Ouzzani M., Hammady H., Fedorowicz Z., Elmagarmid A. Rayyan—a web and mobile app for systematic reviews. Syst Rev. 2016;5:1–10. doi: 10.1186/S13643-016-0384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wells G, Shea B, O'Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analyses n.d. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (accessed September 1, 2021).
- 20.Duval S., Tweedie R. A nonparametric “trim and fill” method of accounting for publication. Bias in Meta-Analysis. 2012;95:89–98. doi: 10.1080/01621459.2000.10473905. [DOI] [Google Scholar]
- 21.Wan X., Wang W., Liu J., Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. 2014;14(1):1–13. doi: 10.1186/1471-2288-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chinn S. A simple method for converting an odds ratio to effect size for use in meta-analysis. Stat Med. 2000;19:3127–3131. doi: 10.1002/1097-0258(20001130)19:22<3127::aid-sim784>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 23.Nie S., Zhao X., Zhao K., Zhang Z., Zhang Z., Zhang Z. Metabolic disturbances and inflammatory dysfunction predict severity of coronavirus disease 2019 (COVID-19): a retrospective study. MedRxiv. 2020:1–28. doi: 10.1101/2020.03.24.20042283. [DOI] [Google Scholar]
- 24.Ressaire Q., Dudoignon E., Moreno N., Coutrot M., Dépret F. Low total cholesterol blood level is correlated with pulmonary severity in COVID-19 critical ill patients. Anaesthesia, Critical Care & Pain Medicine. 2020;39:733. doi: 10.1016/J.ACCPM.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Zhang Y., Lu R., Dai M., Shen M., Zhang J., et al. Lipid metabolism changes in patients with severe COVID-19. Clin Chim Acta. 2021;517:66–73. doi: 10.1016/J.CCA.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue J., Xu H., Zhou Y., Liu W., Han X., Mao Q., et al. Dyslipidemia is related to mortality in critical patients with coronavirus disease 2019: a retrospective study. Front Endocrinol. 2021:618. doi: 10.3389/FENDO.2021.611526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J.T., Chen Z., Nie P., Ge H., Shen L., Yang F., et al. Lipid profile features and their associations with disease severity and mortality in patients with COVID-19. Frontiers in Cardiovascular Medicine. 2020:290. doi: 10.3389/fcvm.2020.584987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dierckx T., Elslande J van, Salmela H., Decru B., Wauters E., Gunst J., et al. The metabolic fingerprint of COVID-19 severity. MedRxiv. 2020 doi: 10.1101/2020.11.09.20228221. 11.09.20228221. [DOI] [Google Scholar]
- 29.Julkunen H., Cichońska A., Slagboom P.E., Würtz P. Metabolic biomarker profiling for identification of susceptibility to severe pneumonia and COVID-19 in the general population. ELife. 2021;10:1–20. doi: 10.7554/ELIFE.63033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C., Zhang W., Xu C., Tan H., Cao G., Li L., Sun Q., Wu G., Hu M., Wu S., Li Q., Wang G., Zhang X., Zeng C. Coronavirus disease 2019 induced inflammatory response are associated with changes in lipid profiles. Research Square. 2020 doi: 10.21203/rs.3.rs-64766/v1. [DOI] [Google Scholar]
- 31.Qi J., He D., Yang D., Wang M., Ma W., Cui H., et al. Severity-associated markers and assessment model for predicting the severity of COVID-19: a retrospective study in Hangzhou, China. BMC Infect Dis. 2021;21(1):1–10. doi: 10.1186/S12879-021-06509-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gursky O. Apolipoprotein structure dynamics. Curr Opin Lipidol. 2005;16:287–294. doi: 10.1097/01.MOL.0000169348.61191.AC. [DOI] [PubMed] [Google Scholar]
- 33.Scanu A., Nakajima K., Edelstein C. Apolipoprotein(a): structure and biology. Front Biosci. 2001;6:546–554. doi: 10.2741/SCANU. [DOI] [PubMed] [Google Scholar]
- 34.Khan T.A., Shah T., Prieto D., Zhang W., Price J., Fowkes G.R., et al. Apolipoprotein E genotype, cardiovascular biomarkers and risk of stroke: systematic review and meta-analysis of 14 015 stroke cases and pooled analysis of primary biomarker data from up to 60 883 individuals. Int J Epidemiol. 2013;42:475–492. doi: 10.1093/IJE/DYT034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schilling S., DeStefano A.L., Sachdev P.S., Choi S.H., Mather K.A., DeCarli C.D., et al. APOE genotype and MRI markers of cerebrovascular disease: systematic review and meta-analysis. Neurology. 2013;81:292–300. doi: 10.1212/WNL.0B013E31829BFDA4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nie H., Hu Y., Liu N., Zhang P., Li G., Li Y., et al. Apolipoprotein E gene polymorphisms are risk factors for spontaneous intracerebral hemorrhage: a systematic review and meta-analysis. Current Medical Science. 2019;39:111–117. doi: 10.1007/S11596-019-2007-5. [DOI] [PubMed] [Google Scholar]
- 37.Talha K.A., Selina F., Nasir M., Kausar A., Islam T., Perveen R.A. Systematic review on apolipoprotein E: a strong genetic cause of hemorrhagic stroke. Mymensingh Med J. 2020;29:1026–1032. [PubMed] [Google Scholar]
- 38.McFadyen C.A., Zeiler F.A., Newcombe V., Synnot A., Steyerberg E., Gruen R.L., et al. Apolipoprotein E4 polymorphism and outcomes from traumatic brain injury: a living systematic review and meta-analysis. J Neurotrauma. 2021;38:1124–1136. doi: 10.1089/NEU.2018.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ballmoos MCW von, Haring B., Sacks F.M. The risk of cardiovascular events with increased apolipoprotein CIII: a systematic review and meta-analysis. Journal of Clinical Lipidology. 2015;9:498–510. doi: 10.1016/J.JACL.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Aghajani M.H., Neishaboori A.M., Ahmadzadeh K., Toloui A., Yousefifard M. The association between apolipoprotein A-1 plasma level and premature coronary artery disease: a systematic review and meta-analysis. Int J Clin Pract. 2021 doi: 10.1111/ijcp.14578. [DOI] [PubMed] [Google Scholar]
- 41.Erqou S., Thompson A., di Angelantonio E., Saleheen D., Kaptoge S., Marcovina S., et al. Apolipoprotein(a) isoforms and the risk of vascular disease: systematic review of 40 studies involving 58,000 participants. J Am Coll Cardiol. 2010;55:2160–2167. doi: 10.1016/J.JACC.2009.10.080. [DOI] [PubMed] [Google Scholar]
- 42.Dardeer K.T., Mohammed K.A., Hussein T.D., Elsheemy M.S. Apolipoprotein A1 as a novel urinary biomarker for diagnosis of bladder cancer: a systematic review and meta-analysis. Indian J Urol. 2021;37:217–225. doi: 10.4103/IJU.IJU_69_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Yang X. Prognostic significance of pretreatment apolipoprotein A-I as a noninvasive biomarker in cancer survivors: a meta-analysis. Dis Markers. 2018;1–9 doi: 10.1155/2018/1034037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu J., Zhang C., Zhang G., Wang Y., Zhang Z., Su W., et al. Association between pretreatment serum apolipoprotein A1 and prognosis of solid tumors in Chinese population: a systematic review and meta-analysis. Cell Physiol Biochem. 2018;51:575–588. doi: 10.1159/000495277. [DOI] [PubMed] [Google Scholar]
- 45.Zuin M., Cervellati C., Trentini A., Passaro A., Rosta V., Zimetti F., et al. Association between serum concentrations of apolipoprotein A-I (ApoA-I) and alzheimer's disease: systematic review and meta-analysis. Diagnostics. 2021;11 doi: 10.3390/DIAGNOSTICS11060984. [9] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka S., Couret D., Tran-Dinh A., Duranteau J., Montravers P., Schwendeman A., et al. High-density lipoproteins during sepsis: from bench to bedside. Crit Care. 2020;24:1–11. doi: 10.1186/S13054-020-02860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C., Cui Y., Miao H., Xiong X., Dou J., Shao L., et al. Apolipoprotein A-V is a novel diagnostic and prognostic predictor in pediatric patients with sepsis: a prospective pilot study in PICU. Mediat Inflamm. 2020;2020:1–9. doi: 10.1155/2020/8052954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barlage S., Gnewuch C., Liebisch G., Wolf Z., Audebert F.-X., Glück T., et al. Changes in HDL-associated apolipoproteins relate to mortality in human sepsis and correlate to monocyte and platelet activation. Intensive Care Med. 2009;35 doi: 10.1007/S00134-009-1609-Y. 11 2009:1877–85. [DOI] [PubMed] [Google Scholar]
- 49.Berbée J.F.P., van der Hoogt C.C., de Haas C.J.C., van Kessel K.P.M., Dallinga-Thie G.M., Romijn J.A., et al. Plasma apolipoprotein CI correlates with increased survival in patients with severe sepsis. Intensive Care Med. 2008;34:907–911. doi: 10.1007/S00134-008-1006-Y. [DOI] [PubMed] [Google Scholar]
- 50.Tietge U.J.F., Maugeais C., Lund-Katz S., Grass D., deBeer F.C., Rader D.J. Human secretory phospholipase A2 mediates decreased plasma levels of HDL cholesterol and ApoA-I in response to inflammation in human ApoA-I transgenic mice. Arterioscler Thromb Vasc Biol. 2002;22:1213–1218. doi: 10.1161/01.ATV.0000023228.90866.29. [DOI] [PubMed] [Google Scholar]
- 51.Terpos E., Ntanasis-Stathopoulos I., Elalamy I., Kastritis E., Sergentanis T.N., Politou M., et al. Hematological findings and complications of COVID-19. Am J Hematol. 2020;95:834–847. doi: 10.1002/AJH.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.biao Wei X., Chen X., Li Y., Huang J., Chen X., Yu D., et al. Apolipoprotein A-I: a favorable prognostic marker in infective endocarditis. Journal of Clinical Lipidology. 2018;1–8 doi: 10.1016/j.jacl.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 53.Pirillo A., Catapano A.L., Norata G.D. vol. 224. Springer; Cham: 2015. pp. 483–508. (HDL in infectious diseases and sepsis high density lipoproteins: handbook of experimental pharmacology). [DOI] [PubMed] [Google Scholar]
- 54.Kočar E., Režen T., Rozman D. Cholesterol, lipoproteins, and COVID-19: basic concepts and clinical applications. Molecular and Cell Biology of Lipids. 2021:1866. doi: 10.1016/J.BBALIP.2020.158849. [7] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanner A.R., Phan H., Brendish N.J., Borca F., Beard K.R., Poole S., et al. SARS-CoV-2 viral load at presentation to hospital is independently associated with the risk of death. J Infect. 2021;83:458–466. doi: 10.1016/J.JINF.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gómez-Mesa J.E., Galindo-Coral S., Montes M.C., Muñoz Martin A.J. Thrombosis and coagulopathy in COVID-19. Curr Probl Cardiol. 2021;46:100742. doi: 10.1016/J.CPCARDIOL.2020.100742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Babapoor-Farrokhran S., Gill D., Walker J., Rasekhi R.T., Bozorgnia B., Amanullah A. Myocardial injury and COVID-19: possible mechanisms. Life Sci. 2020;253:1–5. doi: 10.1016/J.LFS.2020.117723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Long B., Brady W.J., Koyfman A., Gottlieb M. Cardiovascular complications in COVID-19. Am J Emerg Med. 2020 doi: 10.1016/j.ajem.2020.04.048. [4] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bridwell R., Long B., Gottlieb M. Neurologic complications of COVID-19. AJEM (Am J Emerg Med) 2020;38:e3–7. doi: 10.1016/J.AJEM.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nannoni S., de Groot R., Bell S., Markus H.S. Stroke in COVID-19: a systematic review and meta-analysis. Int J Stroke. 2021;16:137–149. doi: 10.1177/1747493020972922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gardner C.D., Tribble D.L., Young D.R., Ahn D., Fortmann S.P. Population frequency distributions of HDL, HDL2, and HDL3 cholesterol and apolipoproteins A-I and B in healthy men and women and associations with age, gender, hormonal status, and sex hormone use: the stanford five city project. Prev Med. 2000;31(4):335–345. doi: 10.1006/PMED.2000.0715. [DOI] [PubMed] [Google Scholar]
- 62.Regis-Bailly A., Visvikis S., Steinmetz J., Fournier B., Gueguen R., Siest G. Effects of apo B and apo E gene polymorphisms on lipid and apolipoprotein concentrations after a test meal. Clin Chim Acta. 1996;253(1–2):127–143. doi: 10.1016/0009-8981(96)06364-4. [DOI] [PubMed] [Google Scholar]
- 63.Grafnetter D., Molinari E., Lonsky L. International study on the comparability of Apo A-1 and Apo B methods. Clin Chim Acta. 1990;189(1):55–68. doi: 10.1016/0009-8981(90)90235-K. [DOI] [PubMed] [Google Scholar]
- 64.Ozdemir B., Selamoglu Z., Braidy N. Absolute quantification of plasma apolipoproteins for cardiovascular disease risk prediction. Methods Mol Biol. 2020;2138:373–379. doi: 10.1007/978-1-0716-0471-7_27. [DOI] [PubMed] [Google Scholar]
- 65.Poljak A., Duncan M.W., Jayasena T., Sachdev P.S. Quantitative assays of plasma apolipoproteins. Methods Mol Biol. 2020;2138:49–81. doi: 10.1007/978-1-0716-0471-7_3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.