PURPOSE
Two poly (ADP-ribose) polymerase (PARP) inhibitors (olaparib and rucaparib) are US Food and Drug Administration–approved for patients with metastatic castration-resistant prostate cancer (mCRPC) harboring BRCA1/2 mutations, but the relative efficacy of PARP inhibition in BRCA1- versus BRCA2-altered mCRPC is understudied.
METHODS
We conducted a multicenter retrospective analysis involving 12 sites. We collected genomic and clinical data from 123 patients with BRCA1/2-altered mCRPC who were treated with PARP inhibitors. The primary efficacy end point was the prostate-specific antigen (PSA) response (≥ 50% PSA decline) rate. Secondary end points were PSA progression-free survival (PSA-PFS), clinical or radiographic PFS, and overall survival. We compared clinical outcomes, and other genomic characteristics, among BRCA1- versus BRCA2-altered mCRPC.
RESULTS
A total of 123 patients (13 BRCA1 and 110 BRCA2) were included. PARP inhibitors used were olaparib (n = 116), rucaparib (n = 3), talazoparib (n = 2), and veliparib (n = 2). At diagnosis, 72% of patients had Gleason 8-10 disease. BRCA1 patients were more likely to have metastatic disease at presentation (69% v 37%; P = .04). Age, baseline PSA, metastatic distribution, and types of previous systemic therapies were similar between groups. There were equal proportions of germline mutations (51% v 46%; P = .78) in both groups. BRCA1 patients had more monoallelic (56% v 41%; P = .49) and concurrent TP53 (55% v 36%; P = .32) mutations. PSA50 responses in BRCA1- versus BRCA2-altered patients were 23% versus 63%, respectively (P = .01). BRCA2 patients achieved longer PSA-PFS (HR, 1.94; 95% CI, 0.92 to 4.09; P = .08), PFS (HR, 2.08; 95% CI, 0.99 to 4.40; P = .05), and overall survival (HR, 3.01; 95% CI, 1.32 to 6.83; P = .008). Biallelic (compared with monoallelic) mutations, truncating (compared with missense) mutations, and absence of a concurrent TP53 mutation were associated with PARP inhibitor sensitivity.
CONCLUSION
PARP inhibitor efficacy is diminished in BRCA1- versus BRCA2-altered mCRPC. This is not due to an imbalance in germline mutations but might be related to more monoallelic mutations and/or concurrent TP53 alterations in the BRCA1 group.
INTRODUCTION
Metastatic castration-resistant prostate cancer (mCRPC) remains a lethal disease with a poor prognosis.1 Germline or somatic mutations in DNA damage repair genes have been described in approximately 20%-25% of these patients, which include BRCA1 and BRCA2.2,3 Although patients with these mutations generally have more aggressive disease and higher mortality than those with proficient homologous recombination repair (HRR),4,5 they also present as a novel therapeutic opportunity. Poly (ADP-ribose) polymerase (PARP) inhibitors efficiently kill tumor cells by synthetic lethality in cancers with damaged BRCA1 or BRCA2 genes, with PARP-mediated repair pathway inhibition resulting in DNA disruption and, therefore, genomic instability causing cancer cell death.6,7
CONTEXT
Key Objective
We conducted a multicenter retrospective study to determine whether the efficacy of poly (ADP-ribose) polymerase (PARP) inhibitors differs between cancers with BRCA1 and BRCA2 mutations and to examine differences in other genomic alterations that coexist with BRCA1/2 mutations.
Knowledge Generated
We show that PARP inhibitor efficacy is diminished in BRCA1- versus BRCA2-altered metastatic castration-resistant prostate cancer. This is not due to an imbalance in germline mutations but might be related to more monoallelic mutations and/or concurrent TP53 alterations in the BRCA1 group.
Relevance
Additional therapeutic approaches are needed for patients with BRCA1-altered prostate cancer. These findings may have broad implications for other BRCA1/2-associated malignancies (breast, ovarian, and pancreatic cancers) where PARP inhibitors are used.
In May 2020, the US Food and Drug Administration approved two PARP inhibitors (olaparib and rucaparib) for men with mCRPC harboring a germline or somatic mutation in a gene associated with HRR.8 The approval of olaparib was based on the PROfound study in which men with previously treated mCRPC and HRR deficiency (mutation in one of the 14 HRR genes) who received olaparib had improved progression-free survival and overall survival relative to next-generation hormone therapy (enzalutamide and abiraterone).9,10 The approval of rucaparib was based on the TRITON2 trial, which reported prostate-specific antigen (PSA) and objective response rates of 63 and 44 percent in men with previously treated mCRPC and somatic or germline BRCA1 and BRCA2 mutations, respectively.11 One of the most interesting post hoc findings from TRITON2 study was the possible disparity in the effectiveness of rucaparib in men with BRCA1 relative to BRCA2 mutations. More precisely, the therapeutic activity of rucaparib appeared to be generally higher in BRCA2-altered compared with BRCA1-altered mCRPC tumors, although formal statistical analyses comparing outcomes by gene mutation were not conducted. Similar trends were observed in the PROfound study, which also suggested dampened activity of olaparib in the BRCA1 population.10
The activity of rucaparib was also previously explored in patients with mCRPC harboring non-BRCA1/2 HRR gene mutations and was not sufficient to merit regulatory approval for that molecular subset of patients12; similar observations have been made with olaparib in the gene-by-gene post hoc analyses from the PROfound study.9,10 However, differences in PARP inhibitor sensitivity between patients with BRCA1- and BRCA2-mutated mCRPC have not been formally examined to date. We hypothesized that patients with BRCA1 mutations would not exhibit the same responses to PARP inhibition as patients with BRCA2 mutations. Here, we describe the differential sensitivity to treatment with PARP inhibitors among men with BRCA1 versus BRCA2 mutations.
METHODS
Study Population and Design
This was a multicenter retrospective analysis of 123 consecutive patients from 12 academic sites with mCRPC who received single-agent PARP inhibitor treatment between December 2014 and July 2020. All PARP inhibitors were permitted in this analysis. Only patients harboring deleterious (somatic or germline) mutations in BRCA1 or BRCA2 were included in the study; other HRR genes were excluded. All centers participating in the study obtained local institutional review board approval before data abstraction.
Deleterious mutations in BRCA1/2 were defined as any alterations resulting in protein truncation (frameshift or nonsense mutations, canonical splice-site mutations, and truncating rearrangements) or homozygous genomic deletions. Selected missense mutations in BRCA1/2 were also classified as deleterious, but only if they were designated as pathogenic or likely pathogenic in the ClinVar and/or COSMIC databases. All other alterations were considered variants of unknown significance and were excluded. Mutation origin (germline or somatic) and zygosity status (monoallelic or biallelic) were also recorded.
Study Outcomes
Demographic, clinical, and genomic characteristics were collected for all patients. This included age, Gleason score, PSA level, stage at diagnosis, metastatic distribution, and the type and number of previous systemic therapies received. We also captured information on concurrent genomic alterations in key prostate cancer genes (namely, TP53, PTEN, RB1, SPOP, AR, and TMPRSS2-ERG).
The primary efficacy end point was the percentage of men achieving a confirmed ≥ 50% decline in PSA level from baseline at the initiation of PARP inhibitor treatment (PSA50 response). Secondary end points were PSA progression-free survival (PSA-PFS, defined as the time until a ≥ 25% increase in PSA from baseline or nadir), progression-free survival (PFS, defined as the time to investigator-assessed clinical or radiographic progression, excluding PSA progression), and overall survival (OS, defined as the time to death from any cause). These definitions are broadly consistent with the Prostate Cancer Working Group 3 guidelines.13
Statistical Analysis
The desired sample size was prospectively defined, on the basis of the primary end point. In the published literature, the prevalence of BRCA2 mutations relative to BRCA1 mutations in prostate cancer is approximately 9:1.14,15 We hypothesized that the PSA50 response rate to PARP inhibition would be 60% in BRCA2-mutated prostate cancers and 20% in BRCA1-mutated prostate cancers. To achieve 80% power to detect this difference using Fisher's exact test with a one-sided alpha of .05, we would need to collect at least a total of 120 patients (108 with a BRCA2 mutation and 12 with a BRCA1 mutation).
PSA50 response rates were compared between BRCA1- and BRCA2-altered patients using logistics regression. Time-to-event outcomes of PSA-PFS, PFS, and OS were estimated using Kaplan-Meier method, and comparisons between groups were carried out using Cox proportional-hazards model. Multivariable logistic regression and Cox models were used to estimate odds ratio (OR) for PSA response and hazard ratios (HRs) for time-to-event outcomes and corresponding 95% CIs and to test for the association between BRCA mutation and patient outcomes, after adjusting for important clinical variables of age, Gleason sum, M stage, baseline PSA, and previous taxane treatment. Separate multivariable models were used to estimate the association between BRCA1/2 mutations and clinical outcomes adjusting for TP53 and PTEN mutations. The database was locked on August 30, 2020. Patients not meeting one or more of the time-to-event end points at the time of database lock were censored for that end point at the time of the last contact with the health system. R 4.0.1 was used for statistical analyses. All tests were two-sided, and P values of ≤ .05 were considered statistically significant; adjustments were not made for multiple statistical comparisons.
RESULTS
Cohort Characteristics
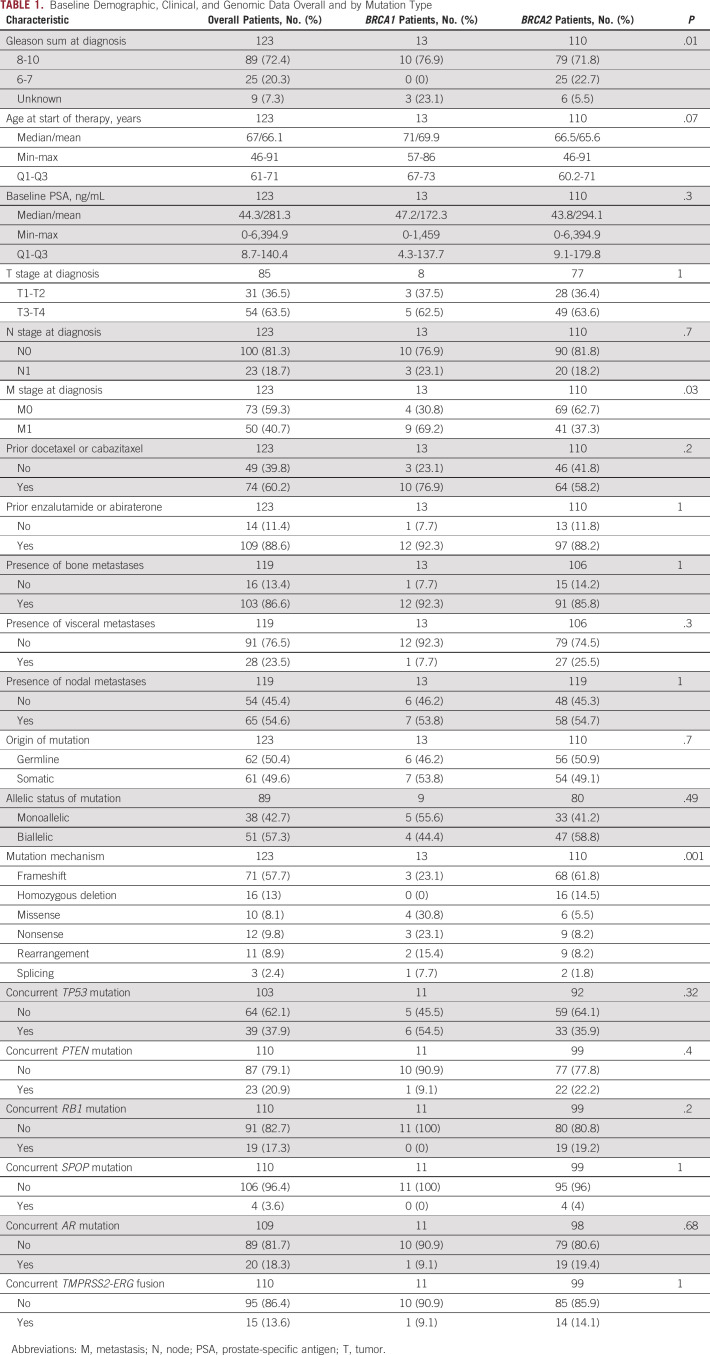
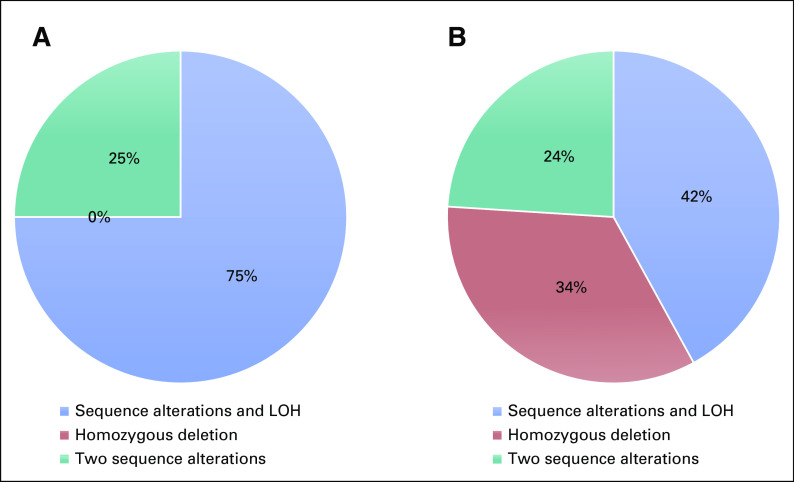
One hundred twenty-three consecutive mCRPC patients with deleterious mutations in BRCA1 (n = 13) or BRCA2 (n = 110) were included in this analysis. Most mutations were frameshift alterations (58%), followed by homozygous deletions (13%); missense alterations were found in 8.1% of cases. PARP inhibitors used were olaparib (n = 116), rucaparib (n = 3), talazoparib (n = 2), and veliparib (n = 2). Table 1 displays the baseline demographic, clinical, and genomic characteristics of these patients. Men with BRCA1 mutations did not significantly differ from those with BRCA2 mutations, except that more patients in the BRCA1 group had metastatic disease at initial diagnosis (69% v 37% P = .04). In the overall cohort, 72% of patients had Gleason 8-10 disease, the median age at the time of PARP inhibitor initiation was 67 years (interquartile range 61-71), and the median baseline PSA level was 44.3 ng/mL (interquartile range 8.7-140). Germline (compared with somatic) BRCA1/2 mutations were equally distributed in the two groups. There were numerically more monoallelic mutations (56% v 41%; P = .49) and more concurrent TP53 mutations (55% v 36%; P = .32) in the BRCA1 group, but broadly similar numbers of mutations in other key genes (PTEN, RB1, SPOP, TMPRSS2-ERG, and AR) across groups (Fig 1). Differences in the mechanisms of biallelic inactivation among the BRCA1- versus BRCA2-altered cancers are summarized in Appendix Table A1 and Figure A1.
TABLE 1.
Baseline Demographic, Clinical, and Genomic Data Overall and by Mutation Type
FIG 1.
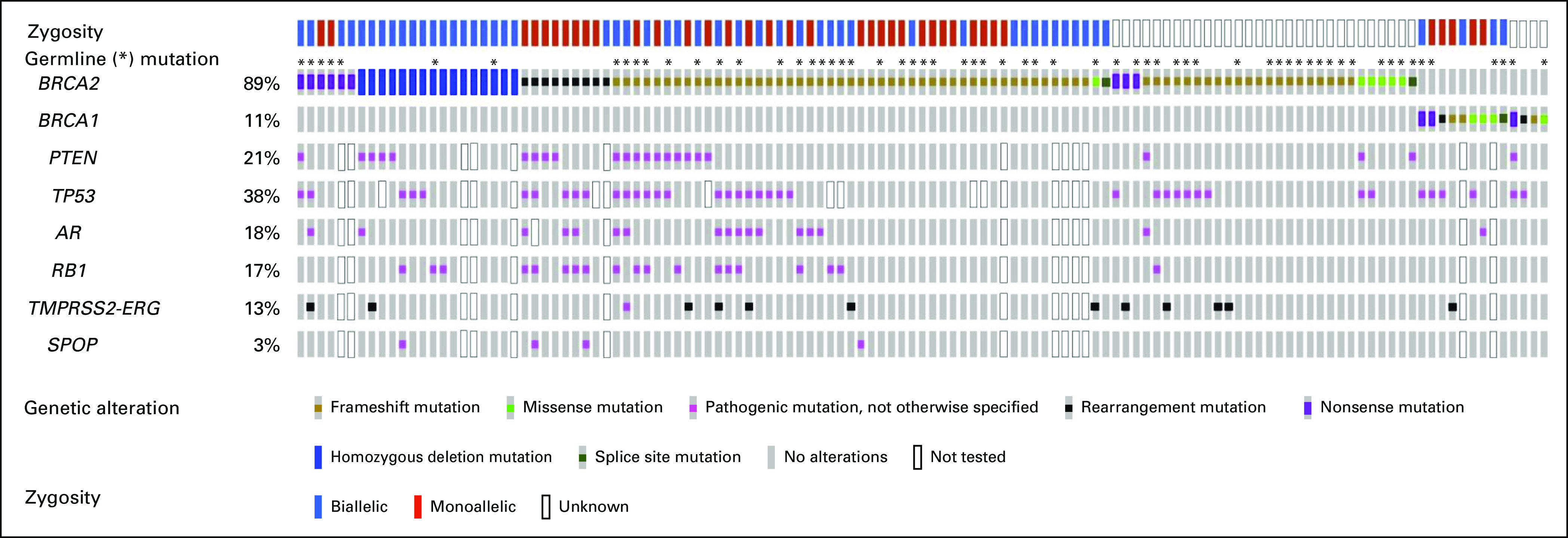
Summary of key genetic alterations identified among patients in the overall cohort (N = 123).
Relationship Between BRCA1/2 and TP53 Mutations
To further explore the potential relationship between BRCA1/2 mutations and concurrent TP53 mutations, we interrogated the cBioPortal for Cancer Genomics database16 that contains DNA sequencing analysis on 6,875 patients with prostate cancer. Of those, 49 (0.7%) had deleterious BRCA1 mutations or deletions and 323 (4.7%) had deleterious BRCA2 alterations. With respect to TP53, 39% of BRCA1-altered (19 of 49) and 22% of BRCA2-altered (71 of 323) prostate cancers also harbored a deleterious TP53 mutation (P for difference, .019). There were no differences with respect to concurrent PTEN alterations; 20% of BRCA1-altered (10 of 49) and 17% of BRCA2-altered (55 of 323) prostate cancers also harbored a PTEN alteration (P for difference, .548). Conversely, RB1 mutations or deletions were enriched in BRCA2-altered cases; 12% of BRCA1-altered (6 of 49) and 30% of BRCA2-altered (97 of 323) prostate cancers also harbored an RB1 alteration (P for difference, .001).
We then examined the association between BRCA1/2 mutations and concurrent TP53 mutations in the other BRCA-associated cancers, again using cBioPortal.16 Among 9,134 patients with breast cancer, 50% of BRCA1-altered (144 of 288) and 41% of BRCA2-altered (143 of 350) breast cancers also harbored deleterious TP53 mutations (P for difference, .025). Similarly, among 1,206 patients with pancreatic cancer, 56% of BRCA1-altered (14 of 25) and 44% of BRCA2-altered (7 of 16) pancreatic cancers also harbored deleterious TP53 mutations, although this did not reach significance (P for difference, .328). Conversely, among 1,668 patients with ovarian cancer, the prevalence of concurrent TP53 mutations was very high in both BRCA1-altered (94%; 77 of 82) and BRCA2-altered (94%; 80 of 85) ovarian cancers (P for difference, .604).
PSA Response Rate
The best PSA response for each patient (at any time) is depicted in Figure 2A. Overall, 72 of 123 patients (59%) achieved a PSA50 response to PARP inhibitor treatment. There were significantly fewer PSA50 responses in patients with BRCA1-altered versus BRCA2-altered mCRPC (23% v 63% respectively; OR, 0.18; 95% CI, 0.04 to 0.62; P = .01). This difference persisted after adjusting for age, Gleason sum, stage, baseline PSA, and previous taxane treatment (adjusted OR, 0.20; 95% CI, 0.04 to 0.76; P = .03) and after adjusting for concurrent TP53 and PTEN mutations (adjusted OR, 0.22; 95% CI, 0.05 to 0.86; P = .04; Table 2). Paradoxically, the median time to best PSA response (in those who achieved a response) was shorter in the BRCA1 than the BRCA2 group (6 v 17 weeks; P < .01). A forest plot showing other clinical and molecular factors that influenced PSA50 responses is depicted in Figure 3A. PSA50 responses were numerically lower in patients with somatic (compared with germline) mutations, monoallelic (compared with biallelic) mutations, missense (compared with truncating) mutations, and concurrent TP53 mutations.
FIG 2.
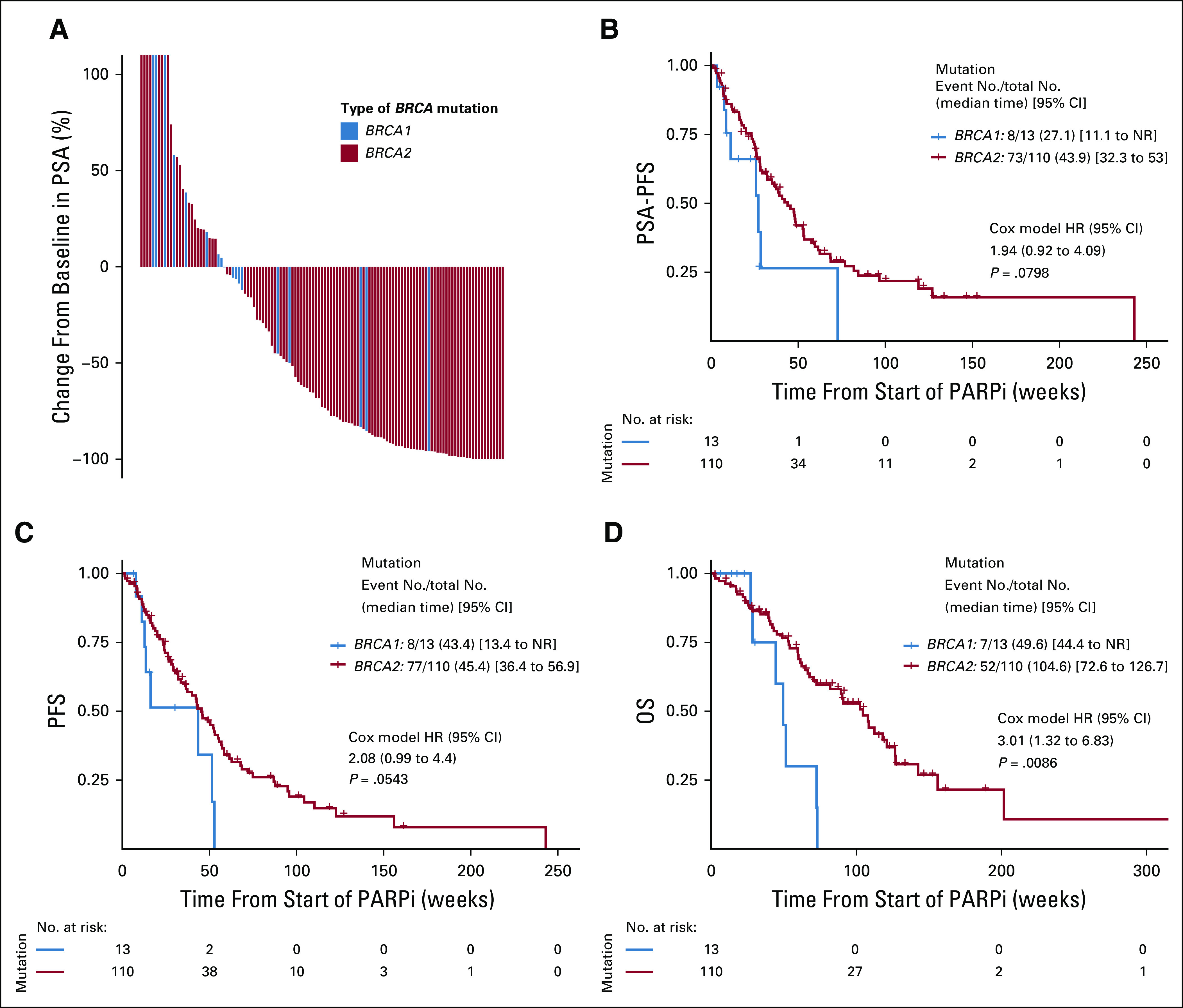
Efficacy outcomes, according to type of BRCA mutation. (A) Waterfall plot of best PSA response, comparing BRCA1- and BRCA2-altered patients. (B) Kaplan-Meier analysis of PSA-PFS, (C) PFS, and (D) OS, comparing BRCA1- and BRCA2-altered patients. HR, hazard ratio; NR, not reached; OS, overall survival; PARPi, poly (ADP-ribose) polymerase inhibitor; PFS, progression-free survival; PSA, prostate-specific antigen.
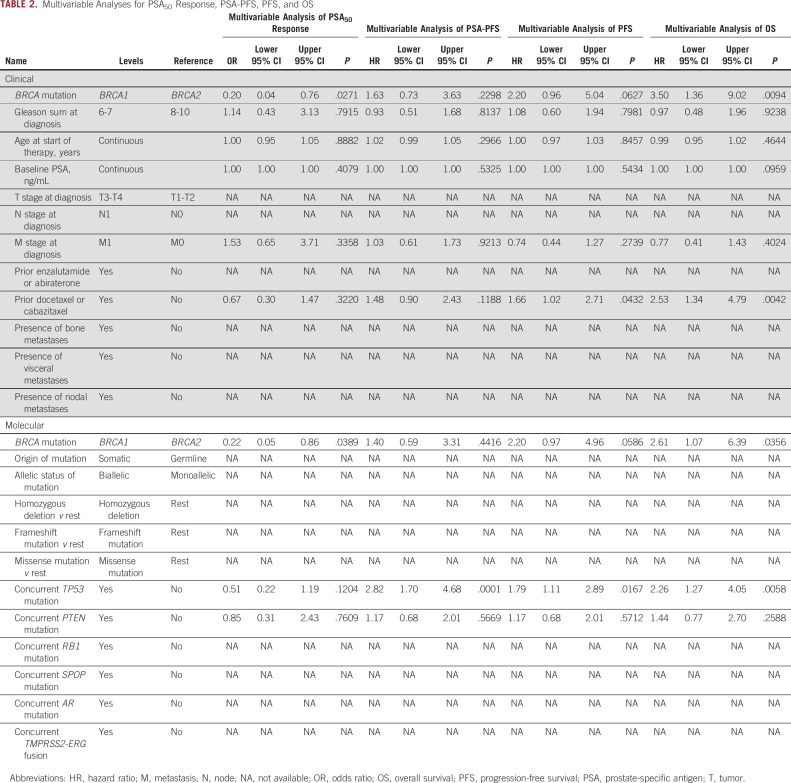
TABLE 2.
Multivariable Analyses for PSA50 Response, PSA-PFS, PFS, and OS
FIG 3.
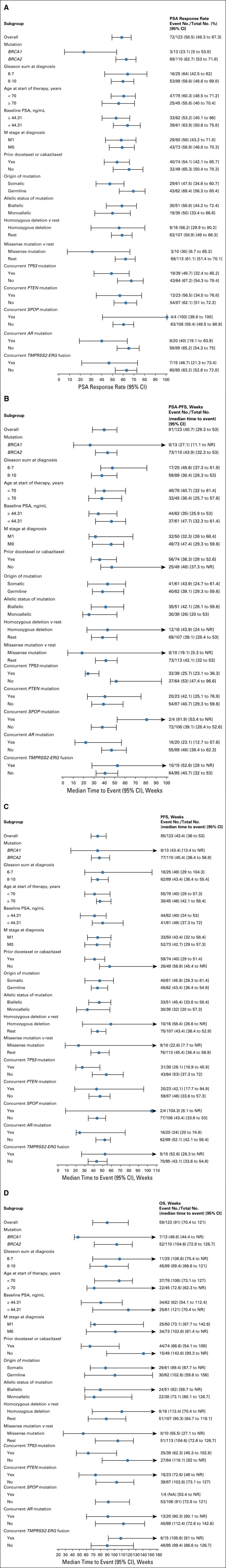
Forest plot analysis of clinical and molecular factors influencing efficacy outcomes. (A) Variables influencing PSA (> 50%) response rates. (B) Variables influencing PSA-PFS. (C) Variables influencing PFS. (D) Variables influencing OS. M, metastasis; N, node, NR, not reached; OS, overall survival; PFS, progression-free survival; PSA, prostate-specific antigen; T, tumor.
Time-to-Event Outcomes
The median PSA-PFS with PARP inhibitor treatment was 40.7 weeks (95% CI, 29.3 to 53.0) in the entire cohort. PSA-PFS was shorter in BRCA1-mutated compared with BRCA2-mutated patients (median 27.1 v 43.9 weeks; HR, 1.94; 95% CI, 0.92 to 4.09; P = .08; Fig 2B). This difference weakened after adjusting for age, Gleason sum, stage, baseline PSA, and previous taxane treatment (adjusted HR, 1.63; 95% CI, 0.73 to 3.63; P = .23) and after adjusting for concurrent TP53 and PTEN mutations (adjusted HR, 1.40; 95% CI, 0.59 to 3.31; P = .44; Table 2). PSA-PFS was numerically shorter in patients with monoallelic (compared with biallelic) mutations, missense (compared with truncating) mutations, and concurrent TP53 mutations (Fig 3B).
The median PFS was 43.4 weeks (95% CI, 36.0 to 53.0) in the overall cohort. Again, PFS was shorter in BRCA1- compared with BRCA2-mutated patients (median 43.4 v 45.4 weeks; HR, 2.08; 95% CI, 0.99 to 4.40; P = .05; Fig 2C). This difference persisted after adjusting for age, Gleason sum, stage, baseline PSA, and previous taxane treatment (adjusted HR, 2.20; 95% CI, 0.96 to 5.04; P = .06) and after adjusting for concurrent TP53 and PTEN mutations (adjusted HR, 2.20; 95% CI, 0.97 to 4.96; P = .06; Table 2). PFS was numerically shorter in patients with monoallelic mutations, missense mutations, and concurrent TP53 alterations (Fig 3C).
We also explored PSA-PFS and PFS in the responding patients only, examining BRCA1 and BRCA2 patients separately. Overall, 72 of 123 patients achieved a PSA50 response: three patients with BRCA1 and 69 patients with BRCA2 mutations. Differences in baseline characteristics between the responding and nonresponding patients are included in Appendix Tables A2 and A3. PSA-PFS was shorter in BRCA1 patients compared with BRCA2 patients (median 20 v 44 weeks; HR, 1.7; 95% CI, 0.4 to 7.3; P = .4). PFS was also shorter in BRCA1 compared with BRCA2 patients (median 40 v 60 weeks; HR, 2.1; 95% CI, 0.5 to 8.9; P = .3).
The median OS was 91.0 weeks (95% CI, 70.4 to 121) in the overall cohort. Again, OS was shorter in BRCA1- compared with BRCA2-altered patients (median 49.6 v 104.6 weeks; HR, 3.01; 95% CI, 1.32 to 6.83; P = .008; Fig 2D). This difference persisted after adjusting for age, Gleason sum, stage, baseline PSA, and previous taxane treatment (adjusted HR, 3.50; 95% CI, 1.36 to 9.02; P = .009) and after adjusting for concurrent TP53 and PTEN mutations (adjusted HR, 2.61; 95% CI, 1.07 to 6.39; P = .04; Table 2). OS was numerically shorter in patients with somatic mutations, monoallelic mutations, missense mutations, and concurrent TP53 alterations (Fig 3D).
DISCUSSION
The advent of the PARP inhibitors, olaparib and rucaparib, represents a major breakthrough in the management of advanced prostate cancer and heralds the beginning of the precision oncology era for this disease.8 However, it is becoming apparent that not all mutated HRR genes display equivalent sensitivity to PARP inhibition in prostate cancer.17,18 Previous studies have already suggested that mCRPC patients with germline or somatic non-BRCA1/2 mutations have less favorable outcomes to a variety of PARP inhibitors compared with those with BRCA1/2 alterations.12,17,19 However, less attention has been given to examining the potential differential efficacy of PARP inhibitors in BRCA1-altered versus BRCA2-altered advanced prostate cancers (potentially because of the much lower relative prevalence of BRCA1 in this disease), whereas preliminary evidence has suggested a possible blunting of PARP inhibitor sensitivity in the BRCA1 subset.20 Here, we aimed to formally examine this issue by forming a multicenter consortium to study and compare PARP inhibitor efficacy among BRCA1- versus BRCA2-associated prostate cancers.
We have found that the clinical activity of PARP inhibitors is diminished in mCRPC patients with germline or somatic BRCA1 compared with BRCA2 mutations. This decreased sensitivity did not appear to be driven by differences in baseline demographic or clinical characteristics in the two groups, and it persisted after adjusting for important clinical and genomic variables. Interestingly, when exploring the mutation characteristics of these BRCA1/2-altered patients, we observed generally improved clinical outcomes in those with biallelic (compared with monoallelic) mutations, but not in those with germline (compared with somatic) mutations. When considering BRCA1/2 mutation mechanism, we observed broadly better clinical outcomes in patients with truncating (compared with missense) alterations, but not in those with homozygous deletions compared with other alterations. Finally, the presence of concurrent TP53 mutations (which were observed in 55% and 36% of BRCA1- and BRCA2-altered cancers, respectively) were associated with numerically worse outcomes to PARP inhibitor treatment. In addition to the notion that TP53 alterations broadly portend an overall worse prognosis in many cancer types, recent reports suggest that TP53 mutations might be more permissive of the emergence of BRCA1/2 reversion mutations (that restore the open reading frame) in BRCA-altered cancers that are exposed to PARP inhibitor treatment.21,22 Such reversion mutations have been associated with secondary PARP inhibitor resistance in prostate and other BRCA-mutated cancers.
Interestingly, numerically better outcomes were observed in patients with concurrent SPOP mutations, although this represented a very small subset (4%) of the total population. The presence of SPOP mutations has also been associated with improved outcomes to a number of hormonal therapies as well.23,24 In addition, studies have shown that SPOP mutations may increase genomic instability, potentially explaining the higher PARP inhibitor sensitivity in these cancers.25 These hypothesis-generating findings require further validation.
Why might patients with BRCA1-altered mCRPC respond less favorably to PARP inhibitor treatment than BRCA2-altered patients? Our current data, taken together with previous findings, do not support the idea that there are differences in mutational origins (germline v somatic) when comparing BRCA1- and BRCA2-related cancers; the two genes are affected by germline alterations at equal frequencies.14,21 Nor are there experimental or clinical data to support the notion that biallelic BRCA1 inactivation in prostate cancer is associated with less HRR deficiency (and hence less synthetic lethality with PARP inhibition)6 than biallelic BRCA2 inactivation; in fact, HRR function might be impaired more significantly from BRCA1 deficiency.14,21 Therefore, one might predict that patients with mCRPC harboring biallelic BRCA1 or BRCA2 mutations would respond equally to PARP inhibitor treatment. However, our data and those of other investigators suggest that biallelic mutations are less common in BRCA1- compared with BRCA2-associated prostate cancers11,14,26; this appears to be one of the key differences potentially driving better responses to PARP inhibitors among the latter. Interestingly, after adjusting for zygosity status, there were no differences in outcomes among patients with biallelic BRCA1- versus BRCA2-mutated mCRPC (HR for PFS: 0.85, 95% CI, 0.20 to 3.60, P = .29; HR for OS: 1.80, 95% CI, 0.41 to 7.87, P = .77). Notably, a recent pan-cancer analysis of BRCA1/2-mutated cancers also suggested a greater amount of genome-wide loss of heterozygosity among cancers with biallelic versus monoallelic mutations, predicting enhanced PARP inhibitor sensitivity.14 Finally, another important observation (that has also been confirmed in other studies) is the greater coexistence of TP53 mutations in BRCA1- versus BRCA2-mutated cancers11,20; the presence of concurrent TP53 mutations may blunt PARP inhibitor efficacy in prostate cancer and possibly other cancers. Notably, after adjusting for TP53 status, there were no differences in outcomes among patients with BRCA1- versus BRCA2-mutated mCRPC (HR for PFS: 1.22, 95% CI, 0.36 to 4.13, P = .20; HR for OS: 1.75, 95% CI, 0.51 to 6.02, P = .25).
This study had not only some strengths but also several limitations. The main shortcoming was that a variety of germline and somatic DNA sequencing platforms were used for the interrogation of BRCA1/2 mutations. Furthermore, the tissue types used for tumor mutation analyses varied (sometimes involving archival primary tumor tissue, sometimes involving metastatic biopsies, and sometimes relying on ctDNA samples). Although the use of archival tissues is not likely to significantly alter the prevalence of BRCA1/2 (or SPOP or TMPRSS2-ERG) mutations, which are generally thought to be truncal events,27,28 archival samples would certainly underestimate the true prevalence of PTEN, RB1, and AR mutations.29 However, the strict definition of BRCA1/2 pathogenicity was a strength of our study, minimizing the inclusion of patients with variants of unknown significance alterations in this analysis. In addition, because of the inclusion of patients from 12 centers, it was not possible to centrally adjudicate clinical or radiographic progression events, and we relied on investigators' assessment of these end points in this retrospective study. We were also not able to harmonize the frequency of PSA assessments or radiographic assessments across the 12 sites. Thus, the most reliable clinical end point in this study is OS, whereas PSA-PFS and PFS estimates should be interpreted with caution. Some strengths of this study include the prospective determination of the sample size, the relatively large number of BRCA1/2-altered patients included (eg, larger than the PARP inhibitor–treated BRCA1/2 populations in either the PROfound or TRITON2 studies), and the collection and annotation of additional molecular data allowing us to formulate hypotheses to explain the observed clinical differences.
In conclusion, to our knowledge, this is the first dedicated study to examine the potential differential effect of PARP inhibitor efficacy in BRCA1- versus BRCA2-associated prostate cancers. We show that PARP inhibitor activity is attenuated in mCRPC patients with BRCA1 mutations and that sensitivity is highest in those with BRCA2 mutations. The diminished efficacy among BRCA1-altered patients is not due to differences in mutation origin (germline v somatic) but rather appears to be associated with a greater number of monoallelic (rather than biallelic) mutations and a higher prevalence of concurrent TP53 alterations in the BRCA1 group. Thus, additional treatment options are needed for patients with BRCA1-altered mCRPC. These findings may have broad implications for other BRCA1/2-associated malignancies (breast, ovarian, and pancreatic cancers) where PARP inhibitors are used.
APPENDIX
FIG A1.
Pie charts of the biallelic inactivation mechanisms by BRCA mutation type: (A) BRCA1 and (B) BRCA2. LOH, loss of heterozygosity.
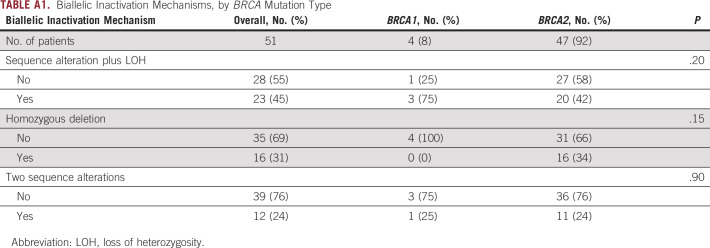
TABLE A1.
Biallelic Inactivation Mechanisms, by BRCA Mutation Type
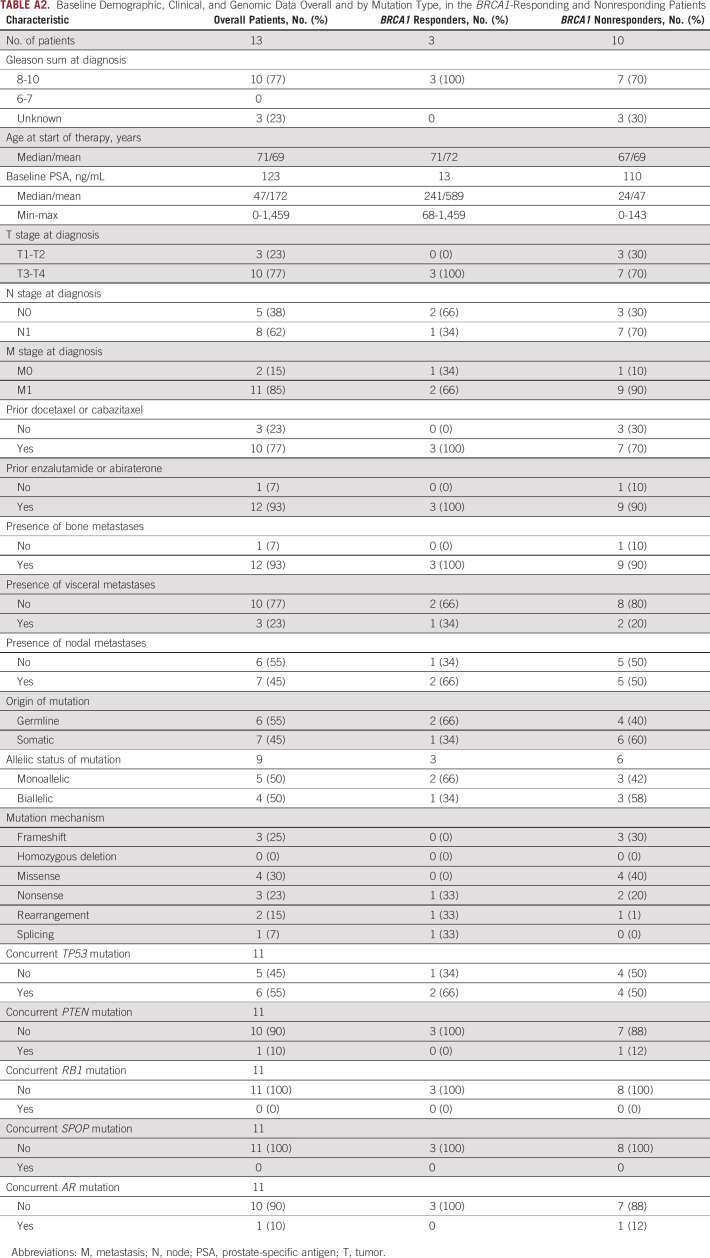
TABLE A2.
Baseline Demographic, Clinical, and Genomic Data Overall and by Mutation Type, in the BRCA1-Responding and Nonresponding Patients
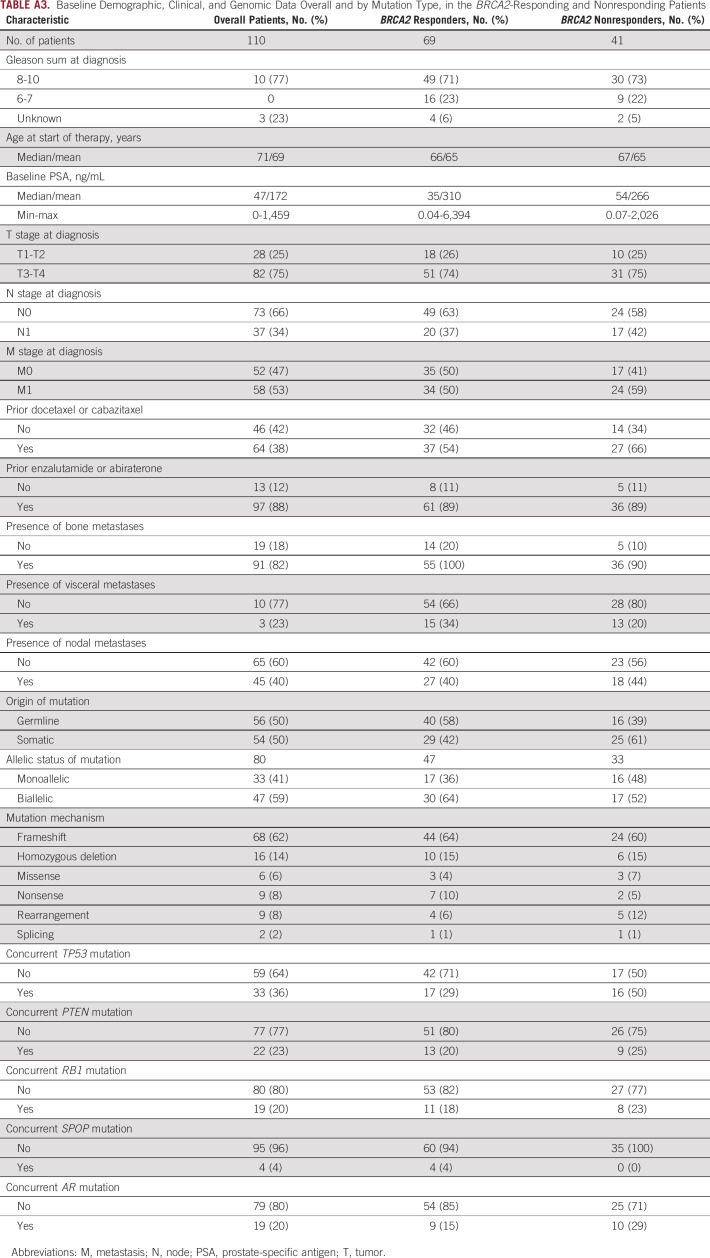
TABLE A3.
Baseline Demographic, Clinical, and Genomic Data Overall and by Mutation Type, in the BRCA2-Responding and Nonresponding Patients
Albert E. Holler
Stock and Other Ownership Interests: CareDX
Nabil Adra
Consulting or Advisory Role: Astellas Pharma, Bristol Myers Squibb Foundation, Merck
Research Funding: Merck, Genentech, Exelixis
Costantine Albany
Stock and Other Ownership Interests: Advaxis
Honoraria: Sanofi, AstraZeneca, Seattle Genetics
Consulting or Advisory Role: Seattle Genetics, AstraZeneca/MedImmune
Speakers' Bureau: Sanofi
Research Funding: Astex Pharmaceuticals, Merck, Bristol Myers Squibb, Lilly, Bayer
Travel, Accommodations, Expenses: Sanofi
Heather H. Cheng
Consulting or Advisory Role: AstraZeneca
Research Funding: Sanofi, Janssen, Clovis Oncology, Color Genomics Foundation, Fred Hutchinson Cancer Research Center
Neeraj Agarwal
Consulting or Advisory Role: Pfizer, Medivation/Astellas, Bristol Myers Squibb, AstraZeneca, Nektar, Lilly, Bayer, Pharmacyclics, Foundation Medicine, Astellas Pharma, Exelixis, Pfizer, Merck, Novartis, Eisai, Seattle Genetics, EMD Serono, Janssen Oncology, AVEO, Calithera Biosciences, MEI Pharma, Genentech, Gilead Sciences
Research Funding: Bayer, Bristol Myers Squibb, Takeda, Pfizer, Exelixis, Amgen, AstraZeneca, Calithera Biosciences, Celldex, Eisai, Genentech, Immunomedics, Janssen, Merck, Lilly, Nektar, ORIC Pharmaceuticals, CRISPR therapeutics, Arvinas
Alan Bryce
Honoraria: Astellas Pharma, Bayer
Travel, Accommodations, Expenses: Clovis Oncology, Phosplatin Therapeutics
Pedro Barata
Consulting or Advisory Role: Bayer, Bristol Myers Squibb, Pfizer, EMD Serono, Eisai, Caris Life Sciences, Clovis Oncology, Dendreon
Research Funding: Blue Earth Diagnostics
A. Oliver Sartor
Stock and Other Ownership Interests: Lilly, GlaxoSmithKline, AbbVie, Cardinal Health, United Health Group, PSMA Therapeutics, Clarity Pharmaceuticals, Noria Therapeutics, Clovis Oncology
Consulting or Advisory Role: Bayer, Sanofi, AstraZeneca, Dendreon , Constellation Pharmaceuticals, Advanced Accelerator Applications, Pfizer, Bristol Myers Squibb, Bavarian Nordic, EMD Serono, Astellas Pharma, Progenics, Blue Earth Diagnostics, Myovant Sciences, Myriad Genetics, Novartis, Clarity Pharmaceuticals, Fusion Pharmaceuticals, Isotopen Technologien, Janssen, Noxopharm, Clovis Oncology, Noria Therapeutics, Point Biopharma, TeneoBio, Telix Pharmaceuticals, Theragnostics
Research Funding: Bayer, Sanofi, Endocyte, Merck, InVitae, Constellation Pharmaceuticals, Advanced Accelerator Applications, AstraZeneca, Dendreon, SOTIO, Janssen, Progenics
Expert Testimony: Sanofi
Travel, Accommodations, Expenses: Bayer, Johnson & Johnson, Sanofi, AstraZeneca, Progenics
Diogo Bastos
Honoraria: MSD, Roche, Bristol Myers Squibb, Janssen-Cilag, Astellas Pharma, AstraZeneca, Bayer
Consulting or Advisory Role: Roche, Bayer, Janssen-Cilag, MSD Oncology
Research Funding: Janssen-Cilag, Pfizer, Astellas Pharma
Travel, Accommodations, Expenses: Janssen-Cilag, Bayer
Oren Smaletz
Stock and Other Ownership Interests: AstraZeneca, GlaxoSmithKline, Novartis, Roche, Sanofi
Honoraria: AstraZeneca, Astellas Pharma
Consulting or Advisory Role: Astellas Pharma, Pfizer, Ipsen, Lilly, Janssen
Research Funding: Janssen, Bristol Myers Squibb
Travel, Accommodations, Expenses: Bristol Myers Squibb, Ipsen
Jacob E. Berchuck
Employment: Cityblock Health
Stock and Other Ownership Interests: Genome Medical, Cityblock Health
Consulting or Advisory Role: Genome Medical
Patents, Royalties, Other Intellectual Property: Institutional patent filed on methods for detecting prostate cancer resistance using cell-free DNA
Mary-Ellen Taplin
Honoraria: Janssen-Ortho, Clovis Oncology, Astellas Pharma, Incyte, UpToDate, Research to Practice, Pfizer, Bayer, Amgen, AstraZeneca, Progenics, Guidepoint Global, Celgene, Merck, GlaxoSmithKline, Myovant Sciences, Roivant, AbbVie, Arcus Biosciences, Constellation Pharmaceuticals
Consulting or Advisory Role: Janssen-Ortho, Bayer, Guidepoint Global, Best Doctors Inc, UpToDate, Clovis Oncology, Research to Practice, Myovant Sciences, Incyte, Pfizer, AstraZeneca, Arcus Ventures
Research Funding: Janssen-Ortho, Medivation, Bayer, Pfizer
Travel, Accommodations, Expenses: Medivation, Janssen Oncology, Tokai Pharmaceuticals, Astellas Pharma, Incyte, Pfizer, Clovis Oncology, Bayer
Rahul Aggarwal
Honoraria: Clovis Oncology
Consulting or Advisory Role: AstraZeneca, Dendreon, Advanced Accelerator Applications, Clovis Oncology, Axiom Biotechnologies
Research Funding: Zenith Epigenetics, Novartis, Xynomic Pharma, Cancer Targeted Technology, Janssen, Merck, AbbVie, Amgen, AstraZeneca, BioXcel Therapeutics
Cora N. Sternberg
Consulting or Advisory Role: Bayer, MSD, Pfizer, Roche, Incyte, AstraZeneca, Merck, Medscape, UroToday, Astellas Pharma, Genzyme, Immunomedics, Foundation Medicine
Ajjai S. Alva
Consulting or Advisory Role: AstraZeneca, Merck, Pfizer, Bristol Myers Squibb
Research Funding: Genentech, Bristol Myers Squibb, Merck Sharp & Dohme, Prometheus, Mirati Therapeutics, AstraZeneca, Roche, Bayer, Progenics, Astellas Pharma, Arcus Biosciences, Harpoon Therapeutics, Celgene, Janssen
Travel, Accommodations, Expenses: Merck, Bristol Myers Squibb
Catherine H. Marshall
Consulting or Advisory Role: McGraw-Hill Education, Dendreon, Bayer
Research Funding: Conquer Cancer Foundation/Bristol Myers Squibb
Travel, Accommodations, Expenses: Dava Oncology, Bayer
Emmanuel S. Antonarakis
Honoraria: Sanofi, Dendreon, Medivation, Janssen Biotech, ESSA, Astellas Pharma, Merck, AstraZeneca, Clovis Oncology
Consulting or Advisory Role: Sanofi, Dendreon, Janssen Biotech, ESSA, Merck, AstraZeneca, Clovis Oncology, Lilly, Bayer
Research Funding: Janssen Biotech, Johnson & Johnson, Sanofi, Dendreon, Aragon Pharmaceuticals, Exelixis, Millennium, Genentech, Novartis, Astellas Pharma, Tokai Pharmaceuticals, Merck, AstraZeneca, Clovis Oncology, Constellation Pharmaceuticals
Patents, Royalties, Other Intellectual Property: Coinventor of a biomarker technology that has been licensed to Qiagen
Travel, Accommodations, Expenses: Sanofi, Dendreon, Medivation
No other potential conflicts of interest were reported.
SUPPORT
Supported by National Cancer Institute (NCI) SPORE Grant No. CA097186, Grant No. P30 CA015704, Congressional Designated Medical Research Program (CDMRP) Grant No. W81XWH-17-2-0043 all to H.H.C.
AUTHOR CONTRIBUTIONS
Conception and design: Fadi Taza, Costantine Albany, Neeraj Agarwal, A. Oliver Sartor, Cora N. Sternberg, Catherine H. Marshall, Emmanuel S. Antonarakis
Administrative support: Fadi Taza, Emmanuel S. Antonarakis
Provision of study materials or patients: Fadi Taza, Heather H. Cheng, Neeraj Agarwal, Alan Bryce, A. Oliver Sartor, Oren Smaletz, Mary-Ellen Taplin, Cora N. Sternberg, Panagiotis J. Vlachostergios, Ajjai S. Alva, Emmanuel S. Antonarakis
Collection and assembly of data: Fadi Taza, Albert E. Holler, Nabil Adra, Costantine Albany, Ryan Ashkar, Heather H. Cheng, Alexandra O. Sokolova, Neeraj Agarwal, Adam Kessel, Alan Bryce, Nellie Nafissi, A. Oliver Sartor, Diogo Bastos, Oren Smaletz, Jacob E. Berchuck, Mary-Ellen Taplin, Rahul Aggarwal, Cora N. Sternberg, Panagiotis J. Vlachostergios, Ajjai S. Alva, Catherine H. Marshall, Emmanuel S. Antonarakis
Data analysis and interpretation: Fadi Taza, Albert E. Holler, Wei Fu, Hao Wang, Nabil Adra, Costantine Albany, Alexandra O. Sokolova, Neeraj Agarwal, Alan Bryce, Pedro Barata, A. Oliver Sartor, Diogo Bastos, Mary-Ellen Taplin, Cora N. Sternberg, Ajjai S. Alva, Christopher Su, Catherine H. Marshall, Emmanuel S. Antonarakis
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by the authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Albert E. Holler
Stock and Other Ownership Interests: CareDX
Nabil Adra
Consulting or Advisory Role: Astellas Pharma, Bristol Myers Squibb Foundation, Merck
Research Funding: Merck, Genentech, Exelixis
Costantine Albany
Stock and Other Ownership Interests: Advaxis
Honoraria: Sanofi, AstraZeneca, Seattle Genetics
Consulting or Advisory Role: Seattle Genetics, AstraZeneca/MedImmune
Speakers' Bureau: Sanofi
Research Funding: Astex Pharmaceuticals, Merck, Bristol Myers Squibb, Lilly, Bayer
Travel, Accommodations, Expenses: Sanofi
Heather H. Cheng
Consulting or Advisory Role: AstraZeneca
Research Funding: Sanofi, Janssen, Clovis Oncology, Color Genomics Foundation, Fred Hutchinson Cancer Research Center
Neeraj Agarwal
Consulting or Advisory Role: Pfizer, Medivation/Astellas, Bristol Myers Squibb, AstraZeneca, Nektar, Lilly, Bayer, Pharmacyclics, Foundation Medicine, Astellas Pharma, Exelixis, Pfizer, Merck, Novartis, Eisai, Seattle Genetics, EMD Serono, Janssen Oncology, AVEO, Calithera Biosciences, MEI Pharma, Genentech, Gilead Sciences
Research Funding: Bayer, Bristol Myers Squibb, Takeda, Pfizer, Exelixis, Amgen, AstraZeneca, Calithera Biosciences, Celldex, Eisai, Genentech, Immunomedics, Janssen, Merck, Lilly, Nektar, ORIC Pharmaceuticals, CRISPR therapeutics, Arvinas
Alan Bryce
Honoraria: Astellas Pharma, Bayer
Travel, Accommodations, Expenses: Clovis Oncology, Phosplatin Therapeutics
Pedro Barata
Consulting or Advisory Role: Bayer, Bristol Myers Squibb, Pfizer, EMD Serono, Eisai, Caris Life Sciences, Clovis Oncology, Dendreon
Research Funding: Blue Earth Diagnostics
A. Oliver Sartor
Stock and Other Ownership Interests: Lilly, GlaxoSmithKline, AbbVie, Cardinal Health, United Health Group, PSMA Therapeutics, Clarity Pharmaceuticals, Noria Therapeutics, Clovis Oncology
Consulting or Advisory Role: Bayer, Sanofi, AstraZeneca, Dendreon , Constellation Pharmaceuticals, Advanced Accelerator Applications, Pfizer, Bristol Myers Squibb, Bavarian Nordic, EMD Serono, Astellas Pharma, Progenics, Blue Earth Diagnostics, Myovant Sciences, Myriad Genetics, Novartis, Clarity Pharmaceuticals, Fusion Pharmaceuticals, Isotopen Technologien, Janssen, Noxopharm, Clovis Oncology, Noria Therapeutics, Point Biopharma, TeneoBio, Telix Pharmaceuticals, Theragnostics
Research Funding: Bayer, Sanofi, Endocyte, Merck, InVitae, Constellation Pharmaceuticals, Advanced Accelerator Applications, AstraZeneca, Dendreon, SOTIO, Janssen, Progenics
Expert Testimony: Sanofi
Travel, Accommodations, Expenses: Bayer, Johnson & Johnson, Sanofi, AstraZeneca, Progenics
Diogo Bastos
Honoraria: MSD, Roche, Bristol Myers Squibb, Janssen-Cilag, Astellas Pharma, AstraZeneca, Bayer
Consulting or Advisory Role: Roche, Bayer, Janssen-Cilag, MSD Oncology
Research Funding: Janssen-Cilag, Pfizer, Astellas Pharma
Travel, Accommodations, Expenses: Janssen-Cilag, Bayer
Oren Smaletz
Stock and Other Ownership Interests: AstraZeneca, GlaxoSmithKline, Novartis, Roche, Sanofi
Honoraria: AstraZeneca, Astellas Pharma
Consulting or Advisory Role: Astellas Pharma, Pfizer, Ipsen, Lilly, Janssen
Research Funding: Janssen, Bristol Myers Squibb
Travel, Accommodations, Expenses: Bristol Myers Squibb, Ipsen
Jacob E. Berchuck
Employment: Cityblock Health
Stock and Other Ownership Interests: Genome Medical, Cityblock Health
Consulting or Advisory Role: Genome Medical
Patents, Royalties, Other Intellectual Property: Institutional patent filed on methods for detecting prostate cancer resistance using cell-free DNA
Mary-Ellen Taplin
Honoraria: Janssen-Ortho, Clovis Oncology, Astellas Pharma, Incyte, UpToDate, Research to Practice, Pfizer, Bayer, Amgen, AstraZeneca, Progenics, Guidepoint Global, Celgene, Merck, GlaxoSmithKline, Myovant Sciences, Roivant, AbbVie, Arcus Biosciences, Constellation Pharmaceuticals
Consulting or Advisory Role: Janssen-Ortho, Bayer, Guidepoint Global, Best Doctors Inc, UpToDate, Clovis Oncology, Research to Practice, Myovant Sciences, Incyte, Pfizer, AstraZeneca, Arcus Ventures
Research Funding: Janssen-Ortho, Medivation, Bayer, Pfizer
Travel, Accommodations, Expenses: Medivation, Janssen Oncology, Tokai Pharmaceuticals, Astellas Pharma, Incyte, Pfizer, Clovis Oncology, Bayer
Rahul Aggarwal
Honoraria: Clovis Oncology
Consulting or Advisory Role: AstraZeneca, Dendreon, Advanced Accelerator Applications, Clovis Oncology, Axiom Biotechnologies
Research Funding: Zenith Epigenetics, Novartis, Xynomic Pharma, Cancer Targeted Technology, Janssen, Merck, AbbVie, Amgen, AstraZeneca, BioXcel Therapeutics
Cora N. Sternberg
Consulting or Advisory Role: Bayer, MSD, Pfizer, Roche, Incyte, AstraZeneca, Merck, Medscape, UroToday, Astellas Pharma, Genzyme, Immunomedics, Foundation Medicine
Ajjai S. Alva
Consulting or Advisory Role: AstraZeneca, Merck, Pfizer, Bristol Myers Squibb
Research Funding: Genentech, Bristol Myers Squibb, Merck Sharp & Dohme, Prometheus, Mirati Therapeutics, AstraZeneca, Roche, Bayer, Progenics, Astellas Pharma, Arcus Biosciences, Harpoon Therapeutics, Celgene, Janssen
Travel, Accommodations, Expenses: Merck, Bristol Myers Squibb
Catherine H. Marshall
Consulting or Advisory Role: McGraw-Hill Education, Dendreon, Bayer
Research Funding: Conquer Cancer Foundation/Bristol Myers Squibb
Travel, Accommodations, Expenses: Dava Oncology, Bayer
Emmanuel S. Antonarakis
Honoraria: Sanofi, Dendreon, Medivation, Janssen Biotech, ESSA, Astellas Pharma, Merck, AstraZeneca, Clovis Oncology
Consulting or Advisory Role: Sanofi, Dendreon, Janssen Biotech, ESSA, Merck, AstraZeneca, Clovis Oncology, Lilly, Bayer
Research Funding: Janssen Biotech, Johnson & Johnson, Sanofi, Dendreon, Aragon Pharmaceuticals, Exelixis, Millennium, Genentech, Novartis, Astellas Pharma, Tokai Pharmaceuticals, Merck, AstraZeneca, Clovis Oncology, Constellation Pharmaceuticals
Patents, Royalties, Other Intellectual Property: Coinventor of a biomarker technology that has been licensed to Qiagen
Travel, Accommodations, Expenses: Sanofi, Dendreon, Medivation
No other potential conflicts of interest were reported.
REFERENCES
- 1.Gillessen S, Attard G, Beer TM, et al. : Management of patients with advanced prostate cancer: Report of the advanced prostate cancer Consensus Conference 2019. Eur Urol 77:508-547, 2020 [DOI] [PubMed] [Google Scholar]
- 2.Pritchard CC, Mateo J, Walsh MF, et al. : Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 375:443-453, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung JH, Dewal N, Sokol E, et al. : Prospective comprehensive genomic profiling of primary and metastatic prostate tumors. JCO Precis Oncol 3:1-23, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castro E, Romero-Laorden N, del Pozo A, et al. : PROREPAIR-B: A prospective cohort study of the impact of germline DNA repair mutations on the outcomes of patients with metastatic castration-resistant prostate cancer. J Clin Oncol 37:490-503, 2019 [DOI] [PubMed] [Google Scholar]
- 5.Annala M, Vandekerkhove G, Khalaf D, et al. : Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov 8:444-457, 2018 [DOI] [PubMed] [Google Scholar]
- 6.Farmer H, McCabe N, Lord CJ, et al. : Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434:917-921, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Dhawan M, Ryan CJ, Ashworth A: DNA repair deficiency is common in advanced prostate cancer: New therapeutic opportunities. Oncologist 21:940-945, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonarakis ES, Gomella LG, Petrylak DP: When and how to use PARP inhibitors in prostate cancer: A systematic review of the literature with an update on on-going trials. Eur Urol Oncol 3:594-611, 2020 [DOI] [PubMed] [Google Scholar]
- 9.Hussain M, Mateo J, Fizazi K, et al. : Survival with olaparib in metastatic castration-resistant prostate cancer. N Engl J Med 383:2345-2357, 2020 [DOI] [PubMed] [Google Scholar]
- 10.de Bono J, Mateo J, Fizazi K, et al. : Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 382:2091-2102, 2020 [DOI] [PubMed] [Google Scholar]
- 11.Abida W, Patnaik A, Campbell D, et al. : Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol 38:3763-3772, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abida W, Campbell D, Patnaik A, et al. : Non-BRCA DNA damage repair gene alterations and response to the PARP inhibitor rucaparib in metastatic castration-resistant prostate cancer: Analysis from the phase II TRITON2 study. Clin Cancer Res 26:2487-2496, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scher HI, Morris MJ, Stadler WM, et al. : Trial design and objectives for castration-resistant prostate cancer: Updated recommendations from the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol 34:1402-1418, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokol ES, Pavlick D, Khiabanian H, et al. : Pan-cancer analysis of BRCA1 and BRCA2 genomic alterations and their association with genomic instability as measured by genome-wide loss of heterozygosity. JCO Precis Oncol 4:442-465, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abida W, Cyrta J, Heller G, et al. : Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci 116:11428-11436, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.cBioPortal for Cancer Genomics Database: www.cbioportal.org [Google Scholar]
- 17.Marshall CH, Sokolova AO, McNatty AL, et al. : Differential response to olaparib treatment among men with metastatic castration-resistant prostate cancer harboring BRCA1 or BRCA2 versus ATM mutations. Eur Urol 76:452-458, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo J, Antonarakis ES: PARP inhibition—Not all gene mutations are created equal. Nat Rev Urol 16:4-6, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonarakis ES: Olaparib for DNA repair-deficient prostate cancer—One for all, or all for one? Nat Rev Clin Oncol 17:455-456, 2020 [DOI] [PubMed] [Google Scholar]
- 20.Markowski MC, Antonarakis ES: BRCA1 versus BRCA2 and PARP inhibitor sensitivity in prostate cancer: More different than alike? J Clin Oncol 38:3735-3739, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsson P, Bandlamudi C, Cheng ML, et al. : Tumour lineage shapes BRCA-mediated phenotypes. Nature 571:576-579, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettitt SJ, Frankum JR, Punta M, et al. : Clinical BRCA1/2 reversion analysis Identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Discov 10:1475-1488, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boysen G, Rodrigues DN, Rescigno P, et al. : SPOP-mutated/CHD1-deleted lethal prostate cancer and abiraterone sensitivity. Clin Cancer Res 24:5585-5593, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swami U, Isaacsson Velho P, Nussenzveig R, et al. : Association of SPOP mutations with outcomes in men with de novo metastatic castration-sensitive prostate cancer. Eur Urol 78:652-656, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shenoy TR, Boysen G, Wang MY, et al. : CHD1 loss sensitizes prostate cancer to DNA damaging therapy by promoting error-prone double-strand break repair. Ann Oncol 28:1495-1507, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughley R, Karlic R, Joshi H, et al. : Etiologic index—A case-only measure of BRCA1/2—Associated cancer risk. N Engl J Med 383:286-288, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abida W, Armenia J, Gopalan A, et al. : Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol 2017:1-16, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mateo J, Seed G, Bertan C, et al. : Genomics of lethal prostate cancer at diagnosis and castration resistance. J Clin Invest 130:1743-1751, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armenia J, Wankowicz SAM, Liu D, et al. : The long tail of oncogenic drivers in prostate cancer. Nat Genet 50:645-651, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]