Abstract
Recently, the highest wave of SARS-CoV-2 epidemic occurred since the beginning of the pandemic in Brazil was registered in Rio Grande do Sul (RS) State, Southern Brazil, considering the number of cases, deaths and hospitalization per day caused by COVID-19. In this study we described which lineages were circulating in the first quarter of 2021 in Southern Brazil to better understand the viral factors involved in the health crisis caused by SARS-CoV-2 in the region, searching also for possible additional SARS-CoV-2 sequence mutations. A total of 70 positive SARS-CoV-2 samples collected between January 28th, 2021 until April 23rd, 2021, were selected to sequencing. Whole genome sequencing of 70 SARS-CoV-2 samples showed a predominance of Gamma lineage (67%, 47/70), followed by P.2 lineage (27%, 19/70) and B.1.1.28 (6%, 4/70). Two Gamma lineage consensus sequences presented a new S:D614A mutation. Newly mutations could be emerging due the quick SARS-CoV-2 spreading. Thus, the greater understanding about immune protection and variants vigilance is essential to the better management of the health SARS-CoV-2 crisis.
Keywords: Health crisis, COVID-19, B.1.1.25, P.2 lineage, Gamma lineage
1. Introduction
In Brazil, the initially dominant lineages of SARS-CoV-2 were B.1.1.28 and B.1.1.33 (Candido et al., 2020). In late 2020 two relevant variants arose independently, the P.1 lineage (Gamma) from Manaus and P.2 lineage from Rio de Janeiro. Both lineages exhibit the E484K mutation and descendant of the B.1.1.28 strain (Faria et al., 2021; Voloch et al., 2021). Over time, the P.2 lineage was almost completely replaced by Gamma (De Almeida et al., 2021; Barbosa et al., 2021). Since few mutations can change virus fitness, the aim of this research was to investigate the lineages that were circulating in the beginning of 2021 in Rio Grande do Sul State (RS), Southern Brazil, to better understand the possible virus-related causes of this major health crisis caused by SARS-CoV-2 in the region, searching also possible additional SARS-CoV-2 mutations.
2. Material and methods
2.1. Sampling
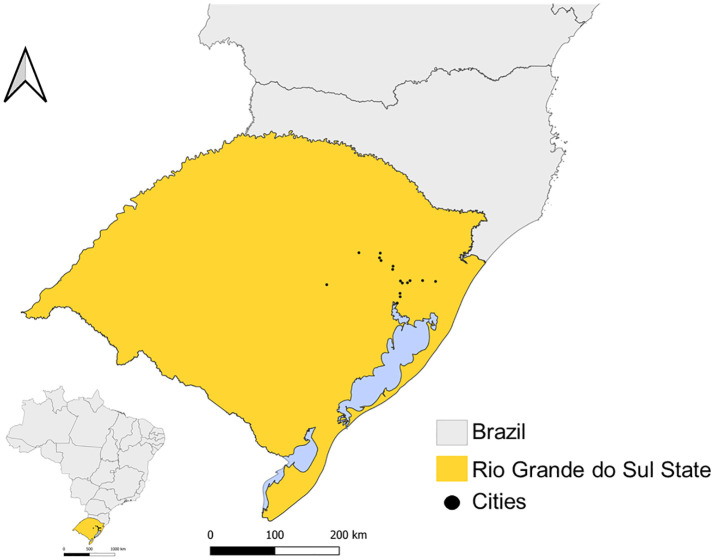
A total of 70 positive samples to SARS-CoV-2, collected between January 28, 2021, until April 23, 2021, were selected to high throughput sequencing. The samples were from 16 cities distributed in three RS regions (Metropolitan Porto Alegre, North East and Eastern center) (Fig. 1 ). The sampling is characterized by 42 males and 28 females, with ages ranging from 10 to 86 years old, but no association between the lineages and metadata information was inferred.
Fig. 1.
Rio Grande do Sul State (RS) geographic location. On the left, Brazil map (in grey) with the RS highlighted in yellow. The black dots represent the 16 cites from Metropolitan Porto Alegre, Northeast and Eastern center regions of RS. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.2. SARS-CoV-2 library preparation
Total RNA was extracted from nasopharyngeal swabs using commercial MagMAX™ CORE Nucleic Acid Purification Kit (Applied biosystems™) kit and the automated equipment KingFisher™ Duo Prime (Thermo Fisher Scientific™). After, reverse transcriptase reaction was achieved using SuperScript IV kit and the library preparation was performed according to QIAseq® SARS-CoV-2 Primer Panel Handbook kit. The sequencing occurred on an Illumina MiSeq platform using MiSeq Reagent Kit v3 (600-cycle).
2.3. Data analysis
FASTQ reads were imported to Geneious Prime, trimmed using BBDuk 37.25 and also analyzed using ARTIC Illumina pipeline. Consensus sequences were assembled mapping the reads against the reference sequence hCoV-19/Wuhan/WIV04/2019 (EPI_ISL_402124) available in EpiCoV database from GISAID (https://www.gisaid.org/ 45) using Geneious Prime software. The sequences were first classified using the Pangolin (https://pangolin.cog-uk.io/) and a phylogenetic tree was inferred. The newly assembled genomes and other SARS-CoV-2 Brazilian sequences (Supplementary material) retrieved from GISAID were aligned using Clustal Omega, with the reference sequence from Wuhan as an outgroup. The Maximum Likelihood phylogenetic analysis under the General Time Reversible model allowing for a proportion of invariable sites and substitution rates were inferred empirically in IQ-TREE v2.1.2 webserver (Nguyen et al., 2015) applying 200 replicates and 1000 bootstrap. The putative structures of SARS-CoV-2 S protein harboring mutations S:D614G and S:D614A were generated by homology modeling the Phyre2 (Kelley et al., 2015) web server. The obtained models with optimal levels of quality were structurally aligned using MUSTANG v3.2.3 (Konagurthu et al., 2006) and compared for conformational changes in topology and dynamics using WEBnm@ v2.0 server (Tiwari et al., 2014). The models and comparisons were finally visualized with Pymol v2.3.0 (https://pymol.org/2/).
3. Results
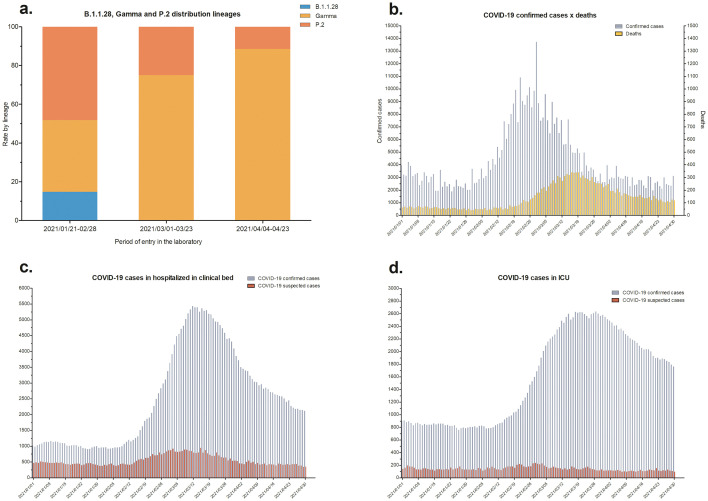
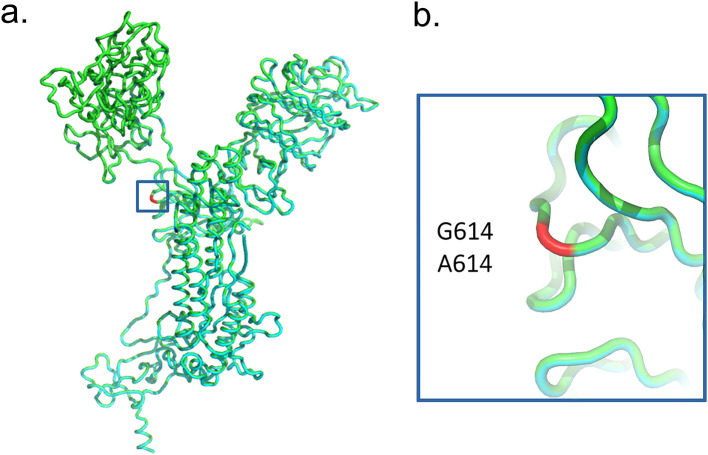
SARS-CoV-2 high-quality complete genomes were retrieved with a mean coverage depth ranging between 517 and 18,904 and breadth of coverage 99% considering the totality of coding regions sequences. The sequences show a predominance of Gamma lineage (67%, 47/70), followed by P.2 lineage (27%, 19/70) and B.1.1.28 (6%, 4/70) (Fig. 2 ). The Gamma sequences generated herein presented the classical mutational signatures in spike glycoprotein L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I and V1176F, with some exceptions, as in the sample hCoV-19/Brazil/LMM53965 and hCoV-19/Brazil/LMM54004 that change a guanine for an adenine in 614 position (D614G → D614A) and the hCoV-19/Brazil/LMM54029 that change a histidine for a glutamic acid in 655 position (H655Y → H655E). P.2 sequences presented E484K, D614G e V1176F and the B.1.1.28 sequences, D614G and V1176F spike mutations. The distribution rate of lineages by period were the following 2021/01/28 – 02/28: 15% of B.1.1.28, 37% of Gamma and 48% of P.2; 2021/03/01 – 03/23: none B.1.1.28, 25% of P.2 and 75% of Gamma; 2021/04/08 – 04/23: none B.1.1.28, 11% of P.2 and 89% of Gamma (Fig. 2). Homology modeling and dynamics analysis revealed no conformational changes or other remarkable differences in the proximal site of Spike protein when comparing models generated from G614 or A614 mutant deduced amino acid sequences (Fig. 3 ).
Fig. 2.
Distribution lineages and Rio Grande do Sul state COVID-19 general data between 2021 January and April. a) B.1.1.28, P.2 and Gamma distribution lineages demonstrated by lineages rate by period of entry in the Microbiology Molecular Laboratory. The first bar (2021/01/28-02/28) presents a total of 27 sequences, the second bar (2021/03/01-03/23) presents 8 sequences and the third bar (2021-04-08-04/23) represents 35 sequences. b) COVID-19 confirmed cases and deaths. c) COVID-19 confirmed and suspected cases in clinical bed hospitalizations. d) COVID-19 confirmed and suspected cases in ICU hospitalizations.
Fig. 3.
Structure-based alignment between homology models of the spike proteins from a local Gamma G614 sample (cyan) and one of the A614 mutants (green). The S:614 residue is colored in red both in the overall view of the models (a) and in detail (b) showing complete superposition between models and no conformational changes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
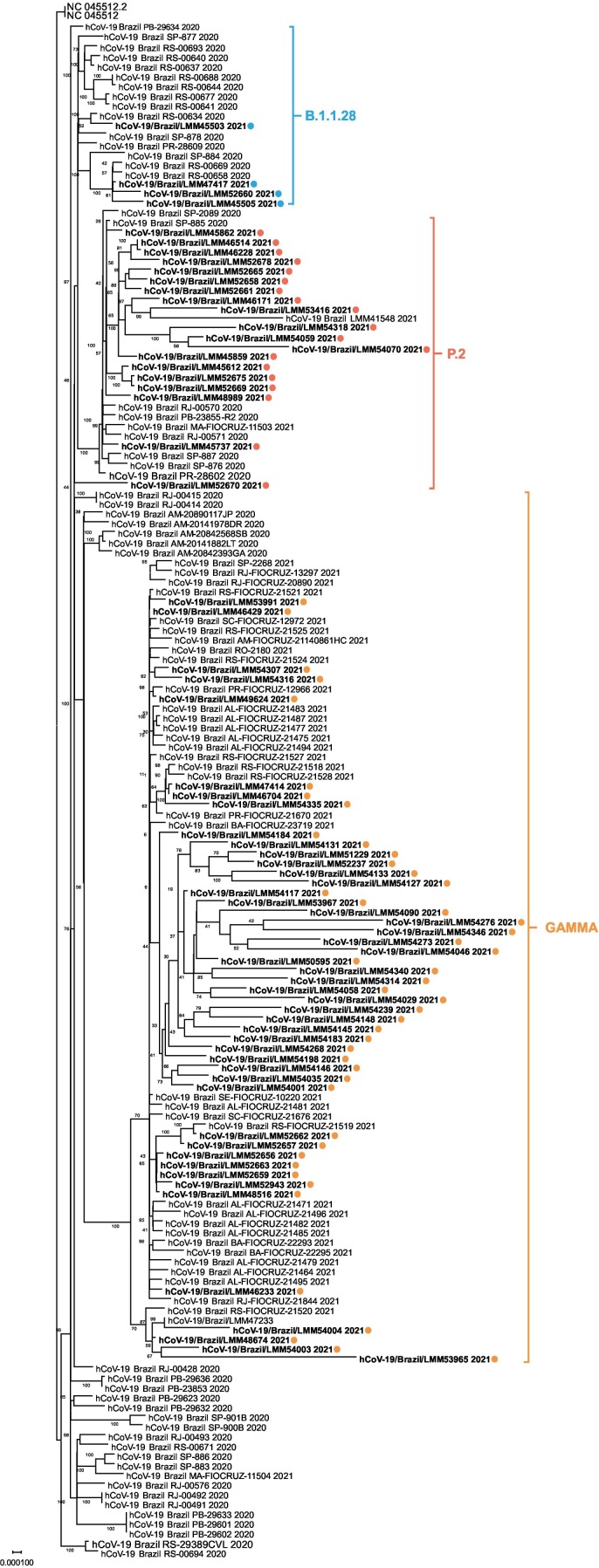
The phylogenetic analysis from samples between January 28, 2021 to April 23 period showed that the majority of the sequences belong to Gamma lineage. P.2 lineage presented a medium frequency and the B.1.1.28 lineage in a lower frequency (Fig. 4 ). At the beginning of 2021, the fast Gamma lineage spread, and dominance was also seen in other Brazilian States. In São Paulo State on March 2021, the Gamma variant was observed in a higher frequency, where the detection rates were 78.6% (first week) and 91.7% (second week). At the same time, other variants as P.2, B.1.1.7 and B.1.1.28 presented a low frequency (Barbosa et al., 2021). In the RJ state a fast replacement of P.2 by Gamma lineage was observed between 24 and 28 of March 2021 (De Almeida et al., 2021). In March and April, this happened here, the Gamma lineage representing 75% and 89% of all sequences, respectively.
Fig. 4.
SARS-CoV-2 complete genome phylogenetic tree. Samples from this study were highlighted in bold followed by a colored dot. In blue the B.1.1.28 lineage sequences, in orange the P.2 lineage sequences and in yellow the Gamma lineage sequences. The Maximum Likelihood phylogenetic analysis was performed in IQ-TREE v2.1.2 web server applying 200 replicates and 1000 bootstrap. The long branches observed in some sequences could be related to a lack primers annealing in some ORF1ab and ORF8 short regions. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Between the end of January until the end of February 2021 our results showed the presence of Gamma and P.2 lineages in the approximate proportion rate. However, in March, the P.2 lineage represented only a third of sequences and, in April, an even higher prevalence of Gamma lineage was identified. This scenario was concomitant with the increase in the numbers of cases (13,723), deaths (342), and hospitalization per day (5435 in clinical hospital beds and 2634 in ICU) caused by COVID-19. Those numbers marked the highest SARS- CoV-2 epidemic wave during the pandemic in this region (date observed until July 2021) (Fig. 2). It has already been described that Gamma lineage can escape of the neutralization antibodies acquired by infection or immunization with previously circulating viral variants (de Souza et al., 2021). As well, it was suggested that the Gamma is more transmissible than previously circulating lineages (Naveca et al., 2021). These facts, in addition to the lack an efficient social distancing and other prevention measures probably caused the high numbers observed in this big SARS-CoV-2 wave.
In this research, two genomes presented the change of a guanine for an adenine in amino acid 614 of spike protein. In Gamma lineage, this mutation was identified for the first time. Besides those sequenced, 11 sequences presented the D614A mutation in GISAID, however, they belong to A.24 lineage from Asia (6 sequences), B.1.1.7 lineage from Europe (4 sequences) and B.1.2 lineage from North American (1 sequence). The D614G is a relevant Spike mutation observed in early March 2020 (Korber et al., 2020). This mutation presents high viral viability probably attributable to minuscule changes in partitioning energy (Laha et al., 2020). Prior to March, the D614G was present in 10% of global sequences, from 1st to March 31, 2020, the detection rate already reached 67% of sequences and, from 1st April to May 18, 2020, the D614G mutations have already been observed in 78% of the sequences (Korber et al., 2020). This rapid spread and dominancy of sequences with D614G mutation globally, probably is related with the advantage observed in replication efficiency when comparing to D614, which can increase the probability of human-to-human transmission (Plante et al., 2021; Yurkovetskiy et al., 2020). The D614G for D614A change has already been identified in July 2020 in three individuals who returning to Korea from Uzbekistan (Park et al., 2020). Nevertheless, the lesser frequency of D614A mutation may be related to lower transmission efficiency or less adaptation in the environment, although no conformational changes were seen when comparing the modeled Spike structures from the original G614 viruses and those A614 mutants.
Evolutionary theory predicts that viruses mutations may be selectively neutral or advantageous to viral fitness (Volz et al., 2021). In SARS-CoV-2 some mutations were correlated with advantages, as the increase of rate of transmission (example: D614G mutation and Gamma variant) and decrease neutralizing antibodies (Naveca et al., 2021; Yang and Du, 2021). In Brazil, there are still many SARS-CoV-2 cases per day and low vaccination coverage. Thus, mutations might be emerging because of the quick spreading of SARS-CoV-2, and a greater understanding of immune protection and variants vigilance is essential to the better management of the health crisis caused by SARS-CoV-2.
Ethics declarations
Project approved by the Research Ethics Committee (CEP) at Feevale University. Process number: CAAE: 33202820.7.1001.5348.
Authors' contributions
Meriane Demoliner: Conceptualization, Formal analysis, Investigation, Writing - Original Draft.: Mariana Soares da Silva: Conceptualization, Formal analysis, Investigation, Writing - Original Draft.: Juliana Schons Gularte: Conceptualization, Formal analysis, Investigation.: Alana Witt Hansen: Formal analysis, Investigation.: Paula Rodrigues de Almeida: Conceptualization, Investigation.: Matheus Nunes Weber: Conceptualization, Investigation.: Fágner Henrique Heldt: Investigation.: Flávio Silveira: Investigation.: Micheli Filippi: Investigation.: Vyctoria Malayhka de Abreu Góes Pereira: Investigation.: Francini Pereira da Silva: Investigation.: Larissa Mallmann: Investigation; Pietra Fink: Investigation.: Andréia Rosane de Moura Valim: Investigation.: Lia Gonçalves Possuelo: Investigation.: Juliane Deise Fleck: Writing - Review & Editing, Supervision.: Fernando Rosado Spilki: Conceptualization, Resources, Formal analysis, Writing - Review & Editing Supervision, Funding acquisition.
Acknowledgments
Acknowledgments
This work is an initiative of RedeCorona-ômica BR MCTI/FINEP affiliated to RedeVírus/MCTI (FINEP = 01.20.0029.000462/20, CNPq = 404096/2020-4). The authors also are grateful for the financial CAPES, CNPq and Research Support Foundation of the State of Rio Grande do Sul (FAPERGS grant 21/2551–0000081-3). See Supplementary acknowledgments.
Conflict of interests
The authors of “Predominance of SARS-CoV-2 P.1 (Gamma) lineage inducing the recent wave in southern Brazil and the finding of an additional S: D614A mutation” have no conflict of interests to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.105134.
Appendix A. Supplementary data
Supplementary material: Supplementary GISAID acknowledgments.
References
- Barbosa G.R., Moreira L.V.L., Justo A.F.O., Perosa A.H., de Souza Luna L.K., Chaves A.P.C., Bueno M.S., Conte D.D., Carvalho J.M.A., Prates J., Dantas P.S., Faico-Filho K.S., Camargo C., Resende P.C., Siqueira M.M., Bellei N. Rapid spread and high impact of the variant of concern P.1 in the largest city of Brazil. J. Infect. 2021;83(1):119–145. doi: 10.1016/j.jinf.2021.04.008. [DOI] [PubMed] [Google Scholar]
- Candido D.S., Claro I.M., de Jesus J.G., Souza W.M., Moreira F.R.R., Dellicour S., Mellan T.A., du Plessis L., Pereira R.H.M., Sales F.C.S., Manuli E.R., Thézé J., Almeida L., Menezes M.T., Voloch C.M., Fumagalli M.J., Coletti T.M., da Silva C.A.M., Ramundo M.S.…Faria N.R. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369(6508):1255–1260. doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Almeida L.G.P., Lamarca A.P., Jr., R Da S.F., Cavalcante L., Gerber A.L., Guimarães A.P. De C., Machado D.T., Alves C., Mariani D., Cruz T.F., Ribeiro M.S., Carvalho S., Da Silva F.D., Garcia M.H. De O., De Souza L.M., Da Silva C.G., Ribeiro C.L.P., Cavalcanti A.C., De Mello C.M.B.…Vasconcelos A.T.R. Genomic Surveillance of SARS-CoV-2 in the State of Rio de Janeiro, Brazil: technical briefing. Virological. 2021 https://virological.org/t/genomic-surveillance-of-sars-cov-2-in-the-state-of-rio-de-janeiro-brazil-technical-briefing/683 [Google Scholar]
- De Souza W.M., Amorim M.R., Sesti-Costa R., Coimbra L.D., De Toledo-Teixeira D.A., Parise P.L., Barbosa P.P., Bispo-dos-Santos K., Mofatto L.S., Simeoni C.L., Brunetti N.S., Claro I.M., Duarte A.S.S., Coletti T.M., Zangirolami A.B., Costa-Lima C., Gomes A.B.S.P., Buscaratti L.I., Sales F.C.…Proenca-Modena J.L. Levels of SARS-CoV-2 lineage P.1 neutralization by antibodies elicited after natural infection and vaccination. SSRN Electr. J. 2021 doi: 10.2139/ssrn.3793486. [DOI] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D. Da S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., Hulswit R.J.G., Franco L.A.M., Ramundo M.S., De Jesus J.G., Andrade P.S., Coletti T.M., Ferreira G.M., Silva C.A.M., Manuli E.R.…Sabino E.C. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372(6544):815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konagurthu A.S., Whisstock J.C., Stuckey P.J., Lesk A.M. MUSTANG: a multiple structural alignment algorithm. Proteins Struct. Funct. Genet. 2006;64(3):559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., McDanal C., Perez L.G., Tang H.…Wyles M.D. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha S., Chakraborty J., Das S., Manna S.K., Biswas S., Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect. Genet. Evol. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveca F.G., Nascimento V., de Souza V.C., Corado A. De L., Nascimento F., Silva G., Costa Á., Duarte D., Pessoa K., Mejía M., Brandão M.J., Jesus M., Gonçalves L., Da Costa C.F., Sampaio V., Barros D., Silva M., Mattos T., Pontes G.…Bello G. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat. Med. 2021 doi: 10.1038/s41591-021-01378-7. [DOI] [PubMed] [Google Scholar]
- Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A.K., No J.S., Shin E., Chung Y.-S., Kim I.-H., Kang B.-H., Rhie G.-E., Kim H.M., Kim J.-M., Park Y.E., Kim M.J., Park J.S., Yoo C.-K., Kim J. Emergence of the D614A mutation in the spike protein of SARS-CoV-2: imported cases to the South Korea. MedRxiv. 2020 http://medrxiv.org/content/early/2020/09/07/2020.09.04.20184721.abstract 2020.09.04.20184721. [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X.…Shi P.-Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592(7852):116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.P., Fuglebakk E., Hollup S.M., Skjærven L., Cragnolini T., Grindhaug S.H.…Reuter N. WEBnm@ v2. 0: web server and services for comparing protein flexibility. BMC Bioinform. 2014;15(1):1–12. doi: 10.1186/s12859-014-0427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloch C.M., da Silva Francisco R., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L., Guimarães A.P. De C., Mariani D., Da Costa R.M., Ferreira O.C., Cavalcanti A.C., Frauches T.S., De Mello C.M.B., Leitão I. De C., Galliez R.M., Faffe D.S., Castiñeiras T.M.P.P., Tanuri A., De Vasconcelos A.T.R. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021;95(10) doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K.…Neaverson A.S. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(1):64–75. doi: 10.1016/j.cell.2020.11.020. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Du L. SARS-CoV-2 spike protein: a key target for eliciting persistent neutralizing antibodies. Signal Transduct. Target Ther. 2021;6(1):95. doi: 10.1038/s41392-021-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., Veinotte K., Egri S.B., Schaffner S.F., Lemieux J.E., Munro J.B., Rafique A., Barve A., Sabeti P.C., Kyratsous C.A.…Luban J. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183(3):739–751. doi: 10.1016/j.cell.2020.09.032. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material: Supplementary GISAID acknowledgments.