Abstract
Background
The administration of docetaxel chemotherapy is one therapeutic option to delay disease progression and increase overall survival in metastatic castration resistant prostate cancer (mCRPC). However, about 15% of patients are primary resistant to chemotherapy and hence would benefit from an alternative mCRPC treatment. Despite intensive research, there are no robust clinical validated biomarkers to predict mCRPC therapy response. Thus, the aim of the study was to determine KDM5D expression in archival radical prostatectomy specimens of patients medicated with docetaxel at time of mCRPC development in order to correlate KMD5D expression with treatment response.
Methods
We used in situ hybridization (ISH) (RNA scope 2.5 HD) to determine KDM5D expression in tissue samples of 28 prostate cancer patients. KDM5D status was correlated to chemotherapy response (PSA and radiographic response).
Results
Data revealed that KDM5D is significantly overexpressed in tumor cells (P<0.0001) but also in benign cells (P<0.02) of those patients who responded to chemotherapy compared to non-responders.
Conclusions
To summarize, KDM5D is a promising novel biomarker predicting response to docetaxel chemotherapy already at the time of localized disease and thus potentially avoiding metastatic biopsies in the mCRPC stage of disease.
Keywords: KDM5D, metastatic castration resistant prostate cancer (mCRPC), biomarker, therapy response, docetaxel chemotherapy, in situ hybridization (ISH)
Introduction
In the past years therapy for patients with metastatic castration resistant prostate cancer (mCRPC) evolved leading to the fact that currently diverse treatment options are available for the same stage of disease including taxane based chemotherapeutic agents (docetaxel, cabazitaxel), second generation androgen deprivation agents (abiraterone acetate, enzalutamide) or radium-223 in the third line setting (1,2). In addition, some months ago, also the PARP inhibitor olaparib was approved for mCRPC patients with BRCA1 or BRCA2 mutations (3). Considering the increasing therapeutic options in the mCRPC stage of disease there is urgent medical need for robust biomarkers to select patients, who benefit from a certain therapy, pursuing a personalized treatment approach (4).
Histone lysine methylation plays a pivotal role in epigenetically regulating gene expression associated with either gene activation or silencing (5,6). KDM5D is a male-specific histone-modifying enzyme and an important co-regulator of multiple transcription factors encoded at the locus Yq11. KDM5D specifically demethylates di- and trimethylated forms of lysine 4 in histone H3, thereby repressing certain genes at the level of transcription (7).
There is upcoming evidence for deregulation of KDM5D and important phenotypic consequences in various types of cancer by demonstrating lower KDM5D mRNA expression in lung, bladder and esophageal cancer compared to normal tissue (The Human Protein Atlas. https://www.proteinatlas.org/ENSG00000012817-KDM5D/pathology) (7). For example, it has been demonstrated that loss of chromosome Y leads to downregulation of KDM5D epigenetic modifiers in clear cell renal cell carcinoma (8). Furthermore, KDM5D was associated with the metastasis of gastric cancer because of its critical role in the epithelial-mesenchymal transition of gastric cancer cells (9). Moreover, KDM5 demethylases have a pivotal role in cancer cell chemo-resistance (10). Concerning prostate cancer (PCa), in vitro studies demonstrated lower KDM5D expression in metastatic and aggressive PCa compared to less aggressive primary tumors (11,12). Preclinical evidence also suggests that KDM5D plays an important role in docetaxel sensitivity by interacting with androgen receptor (AR) signaling as KDM5D downregulation in AR positive cells (LNCaP) leads to docetaxel resistance in the presence of dihydrotestosterone (13). Moreover, KMD5D regulates AR transcriptional activity and suppresses cell invasion (11).
The aim of the study was to investigate for the first time the expression of KDM5D in tissue samples of patients who underwent radical prostatectomy in localized stage of disease, progressed to mCRPC at a later time point and underwent a subsequent docetaxel therapy. In this cohort, we tested KDM5D as early biomarker for therapy response to docetaxel. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tau-20-1084).
Methods
Patients
Twenty-eight mCRPC patients treated with 75 mg/m2 docetaxel (median 6 cycles, range 3–10 cycles) who previously underwent a radical prostatectomy at our department were identified retrospectively. The study was approved by the local ethics committee (No. 1148/2017) of Medical University Innsbruck (Austria). All patients enrolled completed the informed consent form. Only patients who underwent docetaxel chemotherapy as first systemic therapy in the mCRPC setting were included, patients with docetaxel chemotherapy in the hormone naïve setting or those with docetaxel in later mCRPC lines were excluded from the study. In addition, the availability of paraffin-embedded tissue stored in our prostate cancer biobank was an inclusion criterion. The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Immunohistochemistry and in situ hybridization
Formalin-fixed paraffin-embedded (FFPE) primary tumor specimens were obtained from previously untreated patients who had undergone radical prostatectomy at the Department of Urology, Innsbruck Medical University, KDM5D in situ hybridization (ISH) were performed using the RNAscope 2.5 HD Red kit according to the manufacturer’s instruction (Advanced Cell Diagnostics, Inc. Newark, CA). Positive (PPIB) and negative (DapB) control probes were hybridized in parallel for all experiments. For characterization of cancer areas and discrimination from benign tissue, hematoxylin/eosin staining and AMACR/p63 immunohistochemistry (IHC) double staining (Monoclonal Rabbit Anti-Human AMACR, clone 13H4, Dako, Code M361601-2, 1:100, CC1 and Ventana Anti-p63 (4A4) Mouse Anti-Human Monoclonal, Catalog Number: 790-4509, BM, CC1) were used. KDM5D expression was determined by evaluation of ISH dot intensities in cancer and benign areas. The corresponding proliferation indices were determined using Ki-67 immunohistochemistry (Monoclonal Mouse Anti-Human Ki-67 Antigen, clone MIB-1, Dako Code M724001-2, 1:100, CC1).
Stained slides, which were done on consecutive paraffin sections, were digitally scanned by a Pannoramic 250 Flash III scanning system (3DHISTECH, Hungary) and for each case, ISH and IHC slides were placed side by side and aligned using CaseViewer digital microscopy software (3DHISTECH, Hungary) for systematic analysis by the uro-pathologist (G.S). For quantifying KDM5D expression levels an established semi-quantitative “quick score” system combining the proportion of positive cells and the average staining intensity based on the established method first described by Detre et al. was used (14) . Briefly, quick score categories were based on both the proportion (denoted category A) and intensity (denoted category B) of positively stained cells. The proportion of positive cells (category A) was stratified into 4 groups (0: negative, 1: ≤30%, 2: 30–60%, 3 : ≥60%). Average staining intensity (category B) corresponding to the presence of negative, weak, intermediate, and strong staining was given a score from 0 to 3, respectively. An average multiplicative quick score (category A × category B) was subsequently obtained from 3 different benign and malignant tissue cores for each case. Ki-67 proliferation indices were defined for the same areas by the percentage of positive nuclear stained tumor cells among total number of tumor cells counted for each core.
Statistics
Numerical data are presented as mean ± SEM from at least three independent experiments using independent donors. Statistical evaluation was performed using a one-way ANOVA followed by Tukey’s post-hoc test (ns, not significant; *, P<0.05; **, P<0.01, ***, P<0.001) using GraphPad Prism version 7 (La Jolla, CA). For nonparametric distributed data, Mann-Whitney U was used. All demographic and baseline characteristics were analyzed descriptively [absolute and relative frequency for qualitative data and mean and standard deviation (SD) for quantitative data].
Results
Patient characteristics and histopathological findings at the time of radical prostatectomy are presented in Tables S1,S2. Median time from surgery until development of mCRPC was 86 months (range, 8–220 months), median PSA value at chemotherapy start was 10.4 ng/mL (range, 0.5–987.8 ng/mL), with a median PSA nadir of 2.18 ng/mL (range, 0.0–431.7 ng/mL) during chemotherapy.
Patients characteristics at the time of chemotherapy start as well as chemotherapy related information are illustrated in Table 1. Tables 2,3 provide an overview about treatment response to chemotherapy which was correlated to KDM5D expression. Summarizing, 10/21 patients (47.6%) were classified as non-responders while 11/21 patients (52.4%) responded to therapy. Response stratification was based on PSA progression according to PCWG2 2007 (PSA increase ≥50% from nadir, PSA ≥5 ng/mL) and median time to radiographic progression (appearance of ≥2 new lesions according to RECIST 1.1). Furthermore, information on consecutive therapy lines after progression to docetaxel treatment has been elucidated (Table 4).
Table 1. Overview about patient characteristics at the time of chemotherapy (n=28).
| Parameter | Mean | Median | Range |
|---|---|---|---|
| Age (years) | 70.7 | 72 | 48–82 |
| PSA start CTX (ng/mL) | 73.3 | 10.4 | 0.5–978.8 |
| Time surgery: mCRPC (months) | 131 | 86 | 8–220 |
| Number of CTX cycles | 7.5 | 6 | 3–10 |
| PSA nadir during CTX (ng/mL) | 31.9 | 2.18 | 0.0–431.7 |
mCRPC, metastatic castration resistant prostate cancer; CTX, chemotherapy; PSA, prostate specific antigen.
Table 2. Overview about responses to chemotherapy (n=28).
| Responses rates to chemotherapy | Number (%) |
|---|---|
| No PSA declines | 3 (10.7) |
| PSA decline <30% | 3 (10.7) |
| PSA decline ≥30% | 1 (3.6) |
| PSA decline ≥50% | 5 (17.9) |
| PSA decline ≥90% | 14 (50) |
| Unknown | 2 (7.1) |
| Radiographic complete response | 4 (14.3) |
| Radiographic partial response | 11 (39) |
| Stable disease | 2 (7.1) |
| Radiographic progression | 9 (32.1) |
| Unknown | 2 (7.1) |
PSA, prostate specific antigen.
Table 3. Overview about progressive patients during chemotherapy (n=9).
| Patient | Progressive metastatic lesions | PSA response % |
|---|---|---|
| #1 | Osseous; lung; local | ↓ >90% |
| #2 | Osseous | ↓ <30% |
| #3 | Osseous | ↓ >50% |
| #4 | Osseous; liver | ↓ >50% |
| #5 | Osseous; lymph nodes | ↓ >50% |
| #6 | Osseous; lymph nodes; lung | No PSA response |
| #7 | Osseous; lung | ↓ >90% |
| #8 | Osseous | ↓ <30% |
| #9 | Lymph nodes; penis; local | No PSA response |
PSA, prostate specific antigen.
Table 4. Consecutive therapies after docetaxel relapse (n=28).
| Patient number | 1. Subsequent therapy | 2. Subsequent therapy | 3. Subsequent therapy | 4. Subsequent therapy |
|---|---|---|---|---|
| #1 | Radium-223 | |||
| #2 | Abiraterone | |||
| #3 | Enzalutamide | Abiraterone | Cabazitaxel | |
| #4 | Enzalutamide | Cabazitaxel | PSMA Lutetium | |
| #5 | Radium-223 | Enzalutamide | ||
| #6 | Enzalutamide | |||
| #7 | Abiraterone | |||
| #8 | Radium-223 | |||
| #9 | – | |||
| #10 | Radiation | |||
| #11 | Radium-223 | PSMA Lutetium | ||
| #12 | Abiraterone | |||
| #13 | Enzalutamide | Radium-223 | ||
| #14 | – | |||
| #15 | – | |||
| #16 | Abiraterone | Cabazitaxel | PSMA Lutetium | |
| #17 | unknown | |||
| #18 | – | |||
| #19 | Enzalutamide | Abiraterone | ||
| #20 | Abiraterone | |||
| #21 | Abiraterone | |||
| #22 | Enzalutamide | Cabazitaxel | ||
| #23 | Enzalutamide | Abiraterone | Cabazitaxel | PSMA Lutetium |
| #24 | – | |||
| #25 | – | |||
| #26 | – | |||
| #27 | Enzalutamide | Cabazitaxel | ||
| #28 | Radium-223 | Cabazitaxel | Abiraterone |
Concerning KDM5D expression in PCa tissue, we initially aimed to use IHC, however, the production of IHC recommended HPA060807 anti-KDM5D antibody (with manufacturer approved use for IHC) was discontinued, alternatively we tested three different primary antibodies (Affinity Biosciences DF2548, Proteintech 22739-1-AP and Sigma-Aldrich SAB1300510), but they showed either insufficient reactivity (also on recommended control tissue/kidney) or immunohistochemically positive reaction also in PC3 cells, which we used as negative control, since PC3 cells are known to have no KDM5D locus (12).
Thus, we decided to use ISH for further analyses. Generally, ISH is a technique that detects and localizes the presence of specific NA sequence within tissue samples by binding of labeled DNA or RNA probe by complementary base pairing. In this study, RNAscope® Technology was performed, an ISH assay for detection of target RNA within intact cells, which amplifies target-specific signals. RNAscope® combines a highly specific probe strategy with multiple cycles of signal amplification and is commercially available from Advanced Cell Diagnostics. ISH was performed in 26/28 patients among them 21 cases were evaluable for final analyses.
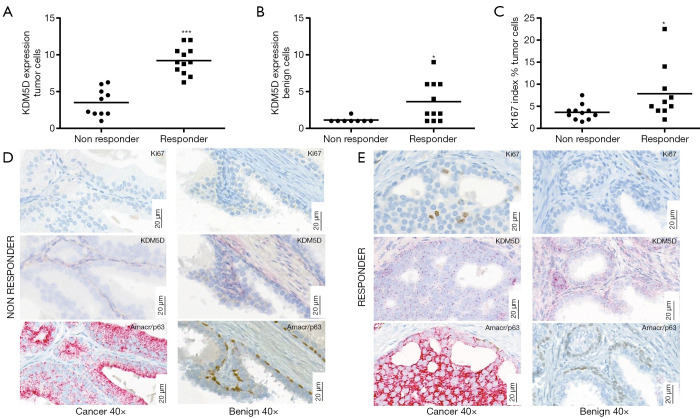
Interestingly, ISH analyses revealed significantly higher KDM5D expression in tumor cells in patients who responded to chemotherapy compared to non-responders (P<0.0001) (Figure 1A). Higher KDM5D expression was also found in non-cancerous areas of patients who were classified as responders (P=0.02) (Figure 1B). In addition, we determined the proliferation rate of cancer cells measured by Ki-67 expression and found that Ki-67 index was increased in responders compared to non-responders (P=0.041) also predicting therapy response (Figure 1C). Representative pictures of corresponding staining are illustrated in Figure 1D,E.
Figure 1.
Comparison of KDM5D expression in tumor cells (A) and benign cells (B) in radical prostatectomy specimens of docetaxel responders and non-responders. (C) Absolute number of Ki-67 index (% of positive cells in correlation to total cell number of evaluated area) in docetaxel responders and non-responders. Data presented as mean ± SEM, statistics: unpaired t-test, *, P<0.05; **, P<0.01; ***, P<0.001 (n=21); (D,E) Representative pictures of corresponding stainings (AMACR/p63 IHC-cancer: AMACR positive/p63 negative; benign: AMACR negative/p63 positive, KDM5D ISH and Ki-67 IHC) of a non-responder (D) with low KDM5D expression in cancer and benign and low Ki-67 in cancer and a responding patient (E) with high KDM5D expression in cancer and benign and high Ki-67 in cancer. Magnification ×40 (scale bar 20 µm). IHC, immunohistochemistry; ISH, in situ hybridization.
Discussion
In the past years, treatment of both primary metastatic PCa but also mCRPC expanded, in addition new therapeutic approaches like AKT- or cell cycle inhibitors demonstrated promising results in early clinical trials (www.clinicaltrias.gov). To effectively implement precision medicine in mCRPC treatment, predictive biomarkers are of urgent need.
Preclinical findings suggest that KDM5D might be involved in docetaxel chemotherapy response as KDM5D dysregulates the AR transcriptional activity in the nucleus resulting in altered docetaxel sensitivity up to resistance (13). In the present study, we described for the first time on patient tissue that KDM5D expression level might be a novel biomarker for predicting response to docetaxel therapy. We used tissue material of patients who underwent a radical prostatectomy due to localized disease and who became castration resistant after a median time of 86 months. Interestingly KDM5D is already higher expressed in radical prostatectomy tissue of those patients who responded to chemotherapy in mCRPC stage speculating that KDM5D might be used as biomarker for docetaxel response also in primary metastatic hormone sensitive disease.
Translating our findings into clinical practice, patients with low KDMD5 expression should undergo an alternative treatment than chemotherapy like ATR inhibitors, as there is a preclinical study demonstrating, that in KDM5D-deficient cells, blocking of ATR activity with an ATR inhibitor enhances DNA damage, subsequently leading to apoptosis (12). In addition, we hypothesize that patients with loss of KDM5D expression benefit more from a hormonal therapy (enzalutamide or abiraterone acetate) instead of docetaxel chemotherapy.
Of notice, in patients with higher expression of KDM5D also Ki-67 was elevated in both cancerous and benign areas. Generally, Ki-67 is a nuclear antigen expressed in the growth and synthesis phases of the cell cycle, but not in the resting phase. Furthermore Ki-67 index predicts response to neoadjuvant chemotherapy in cancer patients (15). Also, in our patient collective high Ki-67 expression correlated with therapy response.
We used ISH rather than IHC for our analyses since none of our tested primary antibodies showed a sufficient result in IHC which could be used for the evaluation of KDM5D expression on protein level. As alternative molecular analysis method KDM5D RNA ISH enabled precise microscopic localization of KDM5D on RNA level by target specific hybridization, amplification and labeling with chromogene. So cell specific expression information of KDM5D can be evaluated in a morphological context including the possibility to combine KDM5D ISH expression with AMACR/p63 double-staining IHC for characterization of cancer and benign areas and Ki-67 IHC for proliferation activity.
Limitations of this study are the small unicentric patient collective, thus evaluating our hypothesis in a larger multicenter cohort is of urgent need. In addition, a consecutive study has to be performed analyzing KDM5D expression in tissue specimens of primary tumor and/or metastatic lesions at the time of castration resistance to exclude a transformation of expression levels during tumor progression.
Conclusions
To summarize, we were able to identify KMD5D as reliable biomarker for docetaxel response. Further prospective clinical studies with larger patient numbers are needed to introduce this novel biomarker into daily routine.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the local ethics committee (1148/2017) of Medical University Innsbruck (Austria). All patients enrolled completed the informed consent form. The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and the Harmonized Tripartite Guideline for Good Clinical Practice from the International Conference on Harmonization.
Footnotes
Provenance and Peer Review: This article was commissioned by the Guest Editor (Tilman Todenhöfer) for the series “Management of Advanced Genitourinary Malignancies” published in Translational Andrology and Urology. The article has undergone external peer review.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tau-20-1084
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tau-20-1084
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tau-20-1084). The series “Management of Advanced Genitourinary Malignancies” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
References
- 1.von Amsberg G, Merseburger AS. Treatment of metastatic, castration-resistant prostate cancer. Urologe A 2020;59:673-9. 10.1007/s00120-020-01187-9 [DOI] [PubMed] [Google Scholar]
- 2.Gillessen S, Attard G, Beer TM, et al. Management of Patients with Advanced Prostate Cancer: The Report of the Advanced Prostate Cancer Consensus Conference APCCC 2017. Eur Urol 2018;73:178-211. 10.1016/j.eururo.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 3.de Bono J, Mateo J, Fizazi K, et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med 2020;382:2091-102. 10.1056/NEJMoa1911440 [DOI] [PubMed] [Google Scholar]
- 4.Kafka M, Eder IE, Klocker H, et al. Emerging promising biomarkers for treatment decision in metastatic castration-resistant prostate cancer. Urol Oncol 2020;38:801-15. 10.1016/j.urolonc.2020.05.025 [DOI] [PubMed] [Google Scholar]
- 5.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007;129:823-37. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 6.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 2012;13:297-311. 10.1038/nrm3327 [DOI] [PubMed] [Google Scholar]
- 7.Blair LP, Cao J, Zou MR, et al. Epigenetic Regulation by Lysine Demethylase 5 (KDM5) Enzymes in Cancer. Cancers (Basel) 2011;3:1383-404. 10.3390/cancers3011383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar A, Kumari N, Nallabelli N, et al. Expression profile of H3K4 demethylases with their clinical and pathological correlation in patients with clear cell renal cell carcinoma. Gene 2020;739:144498. 10.1016/j.gene.2020.144498 [DOI] [PubMed] [Google Scholar]
- 9.Shen X, Hu K, Cheng G, et al. KDM5D inhibit epithelial-mesenchymal transition of gastric cancer through demethylation in the promoter of Cul4A in male. J Cell Biochem 2019;120:12247-58. 10.1002/jcb.27308 [DOI] [PubMed] [Google Scholar]
- 10.Plch J, Hrabeta J, Eckschlager T. KDM5 demethylases and their role in cancer cell chemoresistance. Int J Cancer 2019;144:221-31. 10.1002/ijc.31881 [DOI] [PubMed] [Google Scholar]
- 11.Li N, Dhar SS, Chen TY, et al. JARID1D Is a Suppressor and Prognostic Marker of Prostate Cancer Invasion and Metastasis. Cancer Res 2016;76:831-43. 10.1158/0008-5472.CAN-15-0906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komura K, Yoshikawa Y, Shimamura T, et al. ATR inhibition controls aggressive prostate tumors deficient in Y-linked histone demethylase KDM5D. J Clin Invest 2018;128:2979-95. 10.1172/JCI96769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komura K, Jeong SH, Hinohara K, et al. Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression. Proc Natl Acad Sci U S A 2016;113:6259-64. 10.1073/pnas.1600420113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Detre S, Saclani Jotti G, Dowsett M. A. "quickscore" method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol 1995;48:876-8. 10.1136/jcp.48.9.876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim KI, Lee KH, Kim TR, et al. Ki-67 as a predictor of response to neoadjuvant chemotherapy in breast cancer patients. J Breast Cancer 2014;17:40-6. 10.4048/jbc.2014.17.1.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as