Summary
MDMX is overexpressed in the vast majority of patients with acute myeloid leukemia (AML). We report that MDMX overexpression increases preleukemic stem cell (pre-LSC) number and competitive advantage. Utilizing five newly generated murine models, we found that MDMX overexpression triggers progression of multiple chronic/asymptomatic preleukemic conditions to overt AML. Transcriptomic and proteomic studies revealed that MDMX overexpression exerts this function, unexpectedly, through activation of Wnt/β-Catenin signaling in pre-LSC. Mechanistically, MDMX binds CK1α and leads to accumulation of β-Catenin in a p53-independent manner. Wnt/β-Catenin inhibitors reverse MDMX-induced pre-LSC properties, and synergize with MDMX-p53 inhibitors. Wnt/β-Catenin signaling correlates with MDMX expression in patients with preleukemic myelodysplastic syndromes and is associated with increased risk of progression to AML. Our work identifies MDMX overexpression as a pervasive preleukemic-to-AML transition mechanism in different genetically-driven disease subtypes, and reveals Wnt/β-Catenin as a non-canonical MDMX-driven pathway with therapeutic potential for progression prevention and cancer interception.
Keywords: Cancer interception, precision prevention, preleukemia, myelodysplastic syndromes, acute myeloid leukemia, MDMX, β-Catenin, CK1α, preleukemic stem cells, targeted therapy
eTOC
Using mouse models and data from MDS patients, Ueda et al. identify MDMX as a pervasive preleukemic-to-acute myeloid leukemia transition mechanism across multiple different genetic disease subtypes. MDMX physically interacts with CK1α inducing accumulation of β-Catenin. Blocking both canonical and noncanonical activity of MDMX enhances therapeutic effects against preleukemia/leukemia.
Graphical Abstract

Introduction
While mutations in the tumor suppressor p53 (Tumor Protein P53; TP53) are very common in various solid tumors (Kandoth et al., 2013; Muller and Vousden, 2013), they are relatively rare (< 10% of patients) in patients with acute myeloid leukemia (AML) (Fenaux et al., 1992; Kadia et al., 2016; Peller and Rotter, 2003; Zeisig et al., 2012). In the majority of AML with wildtype (WT) TP53, p53 protein function is inhibited by its suppressors, Murine Double Minute 2 (MDM2) and Murine double minute X (MDMX, also known as MDM4) (Wade et al., 2013). In particular, MDMX, which interacts with p53 and prevents its transactivation, is overexpressed in the vast majority of patients with AML (~ 90%) including at the stem cell level, in part due to altered splicing between a short form and more stable full length MDMX transcript (Carvajal et al., 2018; Han et al., 2016).
We and others have previously demonstrated that AML with WT TP53 is responsive to a novel MDMX/MDM2 dual inhibitor (ALRN-6924) and several clinical trials are currently ongoing (Carvajal et al., 2018; Sallman et al., 2018). However, the precise contribution and causative effects of MDMX overexpression and its functional consequences on normal and malignant hematopoiesis have not yet been studied. This is of particular significance for preleukemic aberrations and conditions. While AML is characterized by substantial genetic and epigenetic inter-patient as well as intra-patient subclonal heterogeneity (Cancer Genome Atlas Research et al., 2013; Li et al., 2016; Papaemmanuil et al., 2016), a large body of recent work has established pre-leukemic stem cells (pre-LSC) as the initial reservoir for consecutive transformation to overt leukemia (Bereshchenko et al., 2009; Busque et al., 2012; Chen et al., 2019; Genovese et al., 2014; Jaiswal et al., 2014; Jan et al., 2012; Kuo et al., 2006; Shlush et al., 2014; Shlush et al., 2017; Steidl et al., 2006; Will et al., 2012; Will et al., 2015; Xie et al., 2014). However, despite increasing investigation and more refined characterization of pre-LSC, the mechanisms that trigger the preleukemic to overt leukemic transition, i.e. that are causative for genetically aberrant pre-LSC to progress to AML rather than remaining compatible with healthy aging in the vast majority of individuals, remain largely unclear. This prompted us to systematically study MDMX overexpression in the context of different, frequently occurring preleukemic aberrations to assess its possible role as a pervasive preleukemic-to-AML transition mechanism.
Results
MDMX overexpression increases the number, proliferation and competitivity of HSCs.
To investigate the role of MDMX overexpression in normal and malignant hematopoiesis, we utilized Mdmx transgenic mice (Mdmx-Tg) which die from a variety of solid tumors or lymphomas after long latency (median of 550 days), without evidence of any myeloid lineage disorders (Xiong et al., 2010; Xiong et al., 2017).
First, we analyzed hematolgical phenotypes of Mdmx-Tg compared to WT littermates. Hematopoietic stem and progenitor cells (HSPCs; lineage− cKit+) from Mdmx-Tg animals overexpressed Mdmx 2.9±0.1 fold on average compared to WT. Mdmx expression in hematopoietic stem cells (HSCs; lineage− cKit+ Sca-1+(LSK) Flk2−) was 17.3±7.4 fold elevated compared to WT (Figure S1A, left and center). Also, we measured the ratio of oncogenic full-length Mdmx (Mdmx-FL) against short-form Mdmx (Mdmx-S; reported to suppress oncogenic activity of MDMX-FL) as previously described (Dewaele et al., 2016). The Percent Spliced In (PSI) index (Mdmx-FL/Mdmx-FL+Mdmx-S) was approximately 0.6 in WT HSCs, and nearly 1 in Mdmx-Tg HSCs suggesting that relative levels of endogenous Mdmx-S are minimal in transgenic mice (Figure S1A, right). Blood cell counts were similar between Mdmx-Tg and WT throughout the lifespan, suggesting that MDMX overexpression alone does not cause any overt change in hematopoiesis (Figure S1B). Total bone marrow (BM) cell count of 3-month-old Mdmx-Tg mice was also similar to WT controls (Figure S1C). We analyzed BM HSPCs by flow cytometry (FCM), and detected no significant differences in Mdmx-Tg compared to WT controls in the number of progenitors or LSK cells (Figure S1D). Also, spleen weight and cellularity in 3-month-old Mdmx-Tg mice were undistingishable from WT controls (Figure S1E–F). However, more detailed analysis of the BM LSK fraction revealed that phenotypic long term hematopoietic stem cells (LT-HSC; Flk2low, CD48low, CD150high LSK) were increased in Mdmx-Tg compared to WT mice (Figure 1A). We compared cell cycling of HSCs (Flk2− LSK) by in vivo BrdU assays of Mdmx-Tg compared to WT littermates. BrdU uptake in Mdmx-Tg HSCs was significantly increased, indicative of faster proliferation of HSCs (Figure 1B). In addition, Mdmx-Tg BM cells displayed significantly increased serial replating capacity in methylcellulose relative to WT BM cells (Figure 1C).
Figure 1. MDMX overexpression increases the number, proliferation, and competitiveness of HSCs.
(A) Upper: Representative flow cytometric plots of bone marrow LSK cells from a 3-month-old WT and Mdmx-Tg mouse. Lower: Ratio of Flk2low CD48low CD150high LT-HSC in LSK (%) are shown (n=6). (B) Upper: BrdU uptake in HSCs (Flk2− LSK) after 48 hours treatment with 1mg/ml BrdU in drinking water. Representative flow cytometric plots. Lower: Ratio of BrdU+ HSCs are shown (n=4). (C) Colony number from serial replating assays of 1×104 bulk bone marrow (BM) cells (n=4). (D) 1.0×106 WT or Mdmx-Tg BM cells (Ly45.2) and 1.0×106 WT competitors (Ly45.1/2) were competitively transplanted into lethally irradiated recipients (Ly45.1). Graph shows ratio of donor cells (Ly45.2) at indicated time points (months) after transplantation for each lineage (n=10 for WT, 9 for Mdmx-Tg). # (A)-(D): statistical differences were calculated by T test. Data shown as Mean±SEM. *: 0.01≤P<0.05, **: 0.001≤P<0.01, ***: P<0.001, N.S.: not significant. See also Figure S1.
To determine whether HSCs from Mdmx-Tg animals have increased functional repopulating capacity compared to WT controls in vivo, we performed competitive congenic transplantation experiments. While homing of Ly45.2+ WT and Mdmx-Tg cells to the BM right after transplantation was indistiguishable (Figure S1G), Mdmx-Tg cells significantly outcompeted WT cells over time, and in both the myeloid and lymphoid lineages (Figure 1D). Collectively, our data reveal that HSCs in Mdmx-Tg mice are increased in number, proliferative activity, in vitro serial replating capacity, and in vivo repopulating capacity, compared to HSCs from WT control mice.
MDMX overexpression acts as a preleukemic-to-AML transition mechanism in conjunction with different myeloid disease alleles
Next, we wanted to interrogate a potential cooperative effect of MDMX overexpression in the contexts of known leukemia-driving aberrations.
MDMX overexpression transforms PU.1 knockdown-induced pre-LSC
We crossbred Mdmx-Tg mice with PU.1 URE knockout mice (URE−/−), which lack a PU.1 upstream regulatory element resulting in 80% reduction of expression of PU.1 compared to WT controls and develop AML at 4 to 8 months of age (Rosenbauer et al., 2004). PU.1 downregulation is very frequent (50–70%) preleukemic event in both mouse and human (Lavallee et al., 2015; Mizuki et al., 2003; Muller and Vousden, 2013; Sive et al., 2016; Steidl et al., 2006; Steidl et al., 2007; Vangala et al., 2003; Will et al., 2015; Yoshida et al., 2007). Young URE−/− mice (2 to 4-month-old) display preleukemic characteristics including at the stem cell level and have myeloid-skewed hematopoiesis but do not give rise to overt AML upon transplantation, and can therefore be utilized as a preleukemic-to-leukemic transition model (Steidl et al., 2006). To test whether MDMX overexpression plays a role in the preleukemia-to-AML transition, we transplanted preleukemic BM cells from preleukemic 3-month-old URE−/−;Mdmx-Tg or 4-month-old URE−/−, or leukemic 4-month-old URE−/−;Mdmx-Tg animals into sublethally irradiated NOD-SCID IL2R-gamma null (NSG) mice (Figure 2A). BM cells from 4-month-old URE−/− mice, as expected, did not cause AML in recipients. However, BM cells of 4-month-old URE−/−;Mdmx-Tg animals were sufficient to precipitate overt AML in recipients (Figure 2B). Moreover, when we performed the transplantation with younger, preleukemic URE−/−;Mdmx-Tg BM cells at 3 months of age, we obtained the same result of induction of overt AML in all recipient animals, albeit at a sligthly longer latency (Figure 2B–C). These findings indicate that MDMX overexpression plays a causative role in the preleukemic-to-AML transition in the context of PU.1 inactivation. We therefore further investigated the preleukemic stage of URE−/−;Mdmx-Tg mice. Immunophenotypic Flk2− LSK cells of URE−/−;Mdmx-Tg mice had 7.1±1.2 fold higher Mdmx levels compared to URE−/− Flk2− LSK cells (pre-LSCs) (Figure S2A). Interestingly, 3-month-old URE−/−;Mdmx-Tg mice displayed neutrophilia (Figure 2D) and expanded total BM cell counts in comparison to URE−/− mice (Figure 2E), with only a minor increase in blast-like cells (Figure 2F), indicating that these mice were still in a preleukemic stage. Further, the ratio of BM cKit+ cells within non-lymphoid cells and the ratio of lineage− cKit+ cells, which includes the pre-LSC/LSC fraction (Steidl et al., 2006), were significantly increased in URE−/−;Mdmx-Tg compared to URE−/− mice (Figure 2G, Figure S2B–C). Spleens were also larger in URE−/−;Mdmx-Tg compared to URE−/− mice, and they were predominantly comprised of myeloid cells both in URE−/−;Mdmx-Tg and URE−/− mice (Figure 2H, Figure S2D). Overall, MDMX overexpression led to a significantly more aggressive disease with reduced survival of URE−/−;Mdmx-Tg primary mice (median 115 days) compared to URE−/− alone (median 178 days) (Figure S2E). Also, moribund URE−/−;Mdmx-Tg mice displayed a greatly increased blast percentage in the BM (URE−/−;Mdmx-Tg; 71.3±4.2%, compared to URE−/−: 21.3±4.2%, p<0.001) (Figure S2F). In summary, these findings support a concept of MDMX overexpression mediating the transition of pre-LSC to LSC in a model of PU.1 knockdown-induced preleukemia.
Figure 2. MDMX overexpression transforms PU.1 knockdown-induced pre-LSC.
(A) Schema of the bone marrow transplantation (BMT) assay. Bone marrow (BM) cells from 3-month-old (3M) URE−/−;Mdmx-Tg mice (preleukemic), 4-month-old (4M) URE−/− mice (preleukemic) or 4M URE−/−;Mdmx-Tg mice (leukemic) were transplanted into sublethally irradiated NSG mice. (B) Survival after BMT of 1×106 BM cells from indicated mice into NSG recipients. (n=5 for URE−/− (4M), Mdmx-Tg, and URE−/−;Mdmx-Tg (3M), n=10 for URE−/−;Mdmx-Tg (4M)). Statistical significance was calculated by log rank test. **: 0.001≤P<0.01, ***: P<0.001. (C) Left: BM smears of BMT recipients. Right: BM blast counts of the recipients of URE−/− (4M) cells and moribund recipients of URE−/−;Mdmx-Tg (3M) cells. (D) Peripheral blood cell counts from 3M primary mice (n=17 for URE−/−, 11 for URE−/−;Mdmx-Tg). WBC; white blood cell, Neu; neutrophil, Hb; hemoglobin, PLT; platelet. (E) Total BM cell number from 3M primary mice. Cell numbers were counted from crushed tibia, femur, ileum, sternum and vertebrae (n=7 for URE−/−, 8 for URE−/−;Mdmx-Tg). (F) Representative picture of bone marrow cytospins of 3M primary mice. (G) Left: Ratio of cKit positive cells in non-lymphoid BM cells of 3M primary mice (n=5 for URE−/−, 4 for URE−/−;Mdmx-Tg). Right: Ratio of live lineage− cKit+ cells (LK and LSK) in the BM. (H) Left: Representative image of spleens from 3M primary mice. The order of the genotypes is WT, URE−/−, URE−/−;Mdmx-Tg, URE−/−;Mdmx-Tg (moribund) from left to right. Right: Spleen weights of 3M primary mice (n=9 for URE−/−, 13 for URE−/−;Mdmx-Tg). # (C) (F) Arrowheads indicate blast cells. Scale bars: 40μm. # (C) (D) (E) (G) (H): Statistical significance was calculated by T test. Data shown as Mean±SEM. *: 0.01≤P<0.05, **: 0.001≤P<0.01, ***: P<0.001. See also Figure S2.
MDMX overexpression induces AML in the Tet2−/− MPN/MDS model and the Tet2+/− clonal hematopoiesis model
Our observations in the context of reduced levels of PU.1, a preleukemic driver and frequently encountered in patients with myelodysplastic syndrome (MDS) and AML, prompted us to examine other pre-leukemic and chronic-leukemic murine models for the effects of MDMX overexpression. We utilized Tet2-deficient models, as TET2 mutations represent a frequent early stage event in patients with myeloproliferative neoplasms (MPN), MDS and AML, as well as in individuals with clonal hematopoiesis (CH) (Genovese et al., 2014; Jaiswal et al., 2014). Tet2−/− mice have been reported to develop MPN/MDS-like disease with more than 1 year latency while Tet2+/− mice can serve as a CH model with many mice being asymptomatic and a few developing a mild form of MPN (Ko et al., 2011; Moran-Crusio et al., 2011). We crossbred Tet2-deficient mice (Ko et al., 2011) with Mdmx-Tg mice (Figure 3A). We first compared Tet2−/− with Tet2−/−;Mdmx-Tg mice. Tet2−/−;Mdmx-Tg animals died significantly earlier than Tet2−/− mice (Figure 3B), and strikingly, developed an agressive overt AML presenting with massive splenomegary with myeloid infiltration, BM infiltration with high blast counts and expansion of cKit-high myeloid cell and CD48+ CD150− LSK cells (Figure 3C–E). Tet2−/− mice only developed an MPN/MDS-like phenotype consistent with prior reports (Figure 3B–E). In addition, we found that out of 7 mice in our Tet2+/−;Mdmx-Tg cohort developed overt AML at 9, 14 and 17-month-old of age, while Tet2+/− mice did not (Figure 3F–H). Taken together, MDMX overexpression induces transformation to acute myeloid leukemia in both a Tet2-deficient as well as a Tet2-haploinsufficient background.
Figure 3. MDMX overexpression induces AML in the Tet2−/− MPN/MDS model and Tet2+/− clonal hematopoiesis model.
(A) Schema of the breeding strategy, resultant genotypes and phenotypes. (B) Survival of Tet2−/− and Tet2−/−;Mdmx-Tg mice (n=15 for Tet2−/−, 18 for Tet2−/−;Mdmx-Tg). **: 0.001≤P<0.01 by log-rank test. (C) Abdominal cavity of moribund Tet2−/−;Mdmx-Tg mouse and a Tet2−/− littermate reveals drastic hepatosplenomegaly in the Tet2−/−;Mdmx-Tg mouse. (D) Left: Bone marrow (BM) cytospins of a non-diseased Tet2−/− mouse at 12-months of age (12M), a moribund (MPN/MDS-like) Tet2−/− mouse at 18-months of age (18M), and a moribund (AML) Tet2−/−;Mdmx-Tg mouse at 12M. Right: percentage of blasts in the BM cells of moribund Tet2−/−;Mdmx-Tg and Tet2−/− mice. Bars indicate Mean±SEM. *: 0.01≤P<0.05 by T test. (E) Flow cytometric analysis of BM/spleen cells of a non-diseased Tet2−/− mouse (12M), a moribund (MPN/MDS-like) Tet2−/− mouse (18M), and a moribund (AML) Tet2−/−;Mdmx-Tg mouse (12M). (F) Abdominal cavity of a moribund Tet2+/−;Mdmx-Tg mouse and its littermate (Tet2+/−). (G) BM cytospins of a moribund (AML) Tet2+/−;Mdmx-Tg mouse (15-month-old; 15M) and a Tet2+/− littermate. (H) Flow cytometric analysis of BM and spleen cells of a moribund Tet2+/−;Mdmx-Tg mouse and a Tet2+/− littermate. # (D) (G) Arrows indicate morphological blasts. Scale bars: 40μm.
MDMX overexpression induces AML in the context of asymptomatic heterozygous Flt3 mutations
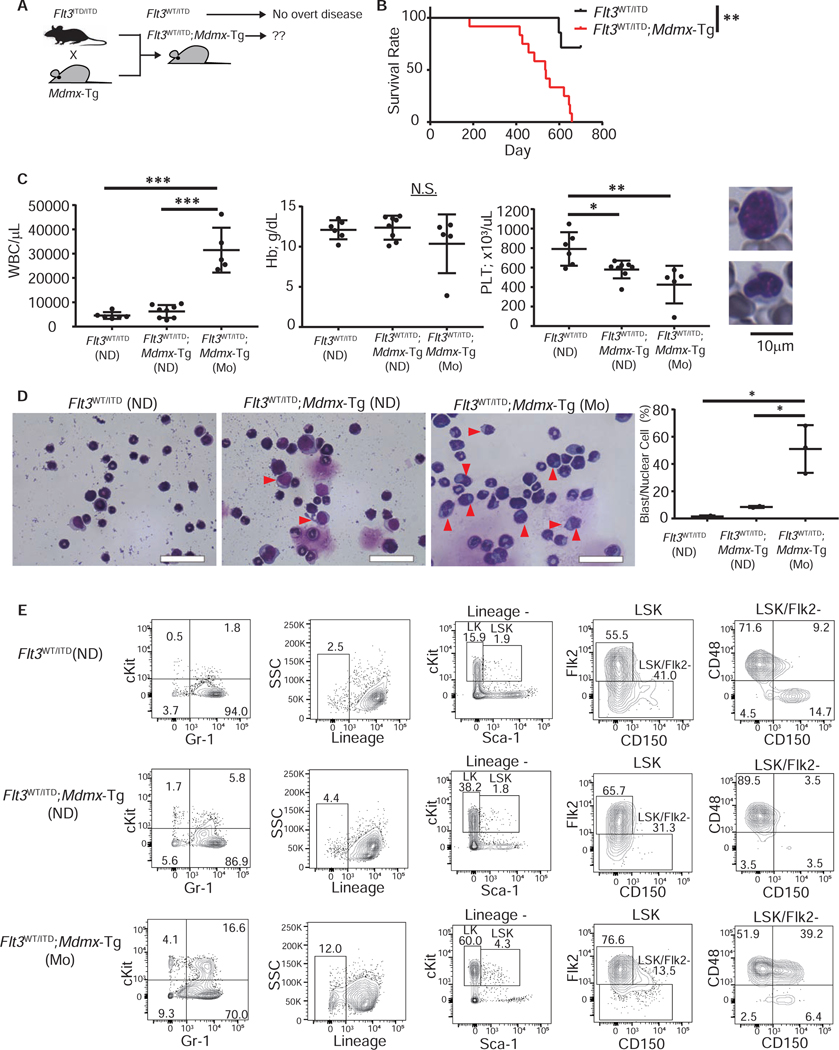
We investigated whether MDMX induces leukemic transformation in other non-leukemic genetic models. Heterozygous FLT3-activating mutations are amongst the most frequent mutations in AML patients, however, Flt3WT/ITD mice do not develop an overt leukemic phenotype and have a life-span comparable to WT mice (Lee et al., 2007). We bred Flt3ITD/ITD with Mdmx-Tg mice to generate Flt3WT/ITD;Mdmx-Tg mice which we compared to Flt3WT/ITD littermate controls (Figure 4A). Flt3WT/ITD;Mdmx-Tg mice died between 6 to 20-months of age (median: 536.5 days), while most of the Flt3WT/ITD animals lived more than 24 months (median: not reached) (Figure 4B). Moribund Flt3WT/ITD;Mdmx-Tg mice (n=5) exhibited significant increase of white blood cell counts with invasion of blasts and a moderate but significant reduction of platelets relative to age-matched Flt3WT/ITD littermates (Figure 4C). The BM of these mice was heavily infiltrated with myeloblasts, while no blast-like cells were observed in Flt3WT/ITD BM (Figure 4D). Flow cytometric analysis of BM cells revealed a significant increase in myeloid precursors (cKit+ Gr-1+) as well as more immature lineage-negative cKit-positive cells, and a increase in CD48+ CD150− LSKs in Flt3WT/ITD;Mdmx-Tg (Figure 4E). Spectral karyotyping (SKY) of lineage-negative cKit-positive AML cells revealed the presence of complex chromosomal abnormalities including deletions, duplications, and translocations, providing evidence for clonality of malignant cells (Figure S3A and S3B). Moribund mice displayed severe splenomegaly (Figure S3C) with the myeloid cell fraction in the spleen markedly increased compared to control littermates (Figure S3D). A group of Flt3WT/ITD;Mdmx-Tg mice, age-matched to moribund mice but not overtly sick yet (labeled as “non-diseased”), were also analyzed and displayed qualitatively similar, but less severe, AML-like changes in the blood, spleen and BM compared to those of moribund mice (Figure 4C–E). Taken together, these data revealed that MDMX overexpression combined with the non-penetrant Flt3WT/ITD allele triggers leukemic transformation and induces overt AML.
Figure 4. MDMX overexpression induces AML in the context of asymptomatic heterozygous Flt3 mutations.
(A) Schema of the breeding strategy of Flt3WT/ITD;Mdmx-Tg mice. (B) Survival of FLT3WT/ITD and Flt3WT/ITD;Mdmx-Tg mice (n=7 for FLT3WT/ITD, 12 for Flt3WT/ITD;Mdmx-Tg). **: 0.001≤P<0.01 by log-rank test. (C) Left: Peripheral blood counts of moribund (Mo) mice and 16-month-old non-diseased (ND) mice. (n=6 for FLT3WT/ITD (ND), 8 for Flt3WT/ITD;Mdmx-Tg (ND), 5 for Flt3WT/ITD;Mdmx-Tg (Mo)). Statistics were calculated by Tukey HSD test. Data shown as Mean±SEM. *: 0.01≤P<0.05, **: 0.001≤P<0.01, ***: P<0.001, N.S.: not significant. Right: Representative blasts in peripheral blood. (D) Left: Bone marrow cytospins from Mo or age-matched ND mice. Scale bars: 40μm. Arrowheads indicate blast cells. Right: Percentage of blasts among nuclear cells (n=2, 2, 3 respectively). Statistics were calculated by Tukey HSD test. Data shown as Mean±SEM. *: 0.01≤P<0.05. (E) Representative flow cytometric analysis of bone marrow of Mo mice and 16-month-old ND mice. See also Figures S3–S5.
Because of MDMX’s function as a p53 inhibitor, we analyzed the coexistence of FLT3 mutations with TP53 mutations in human AML cases. Using TCGA datasets, we found that mutations of FLT3 and TP53 are mutually exclusive (Figure S3E), consistent with prior reports (Hou et al., 2015; Kadia et al., 2016). On the other hand, MDMX overexpression is observed in the vast majority of AML cases regardless of co-mutations and includes FLT3-mutant patients (Carvajal et al., 2018; Han et al., 2016).This prompted us to consider the intriguing possibility of a potential p53-independent function of MDMX overexpression in driving leukemogenesis, which we will describe further below.
MDMX overexpression triggers AML in the context of Nras-G12D-driven chronic myelomonocytic disease
To further establish the contribution of MDMX overexpression to AML onset, we expanded our studies to an additional model of a slowly-progressing, chronic myeloid disease, driven by the Nras-G12D mutation. Adoptive transfer of Nras-G12D-transduced BM cells leads to a chronic myelomonocytic leukemia (CMML)-like disease at around 6 months post transplantation (Parikh et al., 2007). We retrovirally introduced the Nras-G12D mutant into cKit+ WT or Mdmx-Tg BM cells (transduction efficiency; 19±1%), followed by transplantation into lethally irradiated C57BL/6 recipients (Figure S4A). Recipients of Nras-G12D Mdmx-Tg cells (NrasG12D;Mdmx-Tg) rapidly succumbed to disease within around 2 months after transplantation (median: 67 days) while NRAS-G12D recipients (NrasG12D) developed disease at 4 to 8 months after transplantation (median: 153 days) (Figure S4B). Several of the NrasG12D;Mdmx-Tg mice we analyzed showed BM invasion of GFP+ CD11b+ Gr-1+ cKitdim AML-like cells (Figure S4C). In NrasG12D mice, there were no detectable GFP-positive tumor cells in the BM at 2 months after transplantation; however, by 5 months there was infiltration with GFP+ CD11b+ Gr-1− cKitlow monocytic cells, which is compatible with CMML-like disease and consistent with prior reports (Figure S4C) (Parikh et al., 2007; Zhang et al., 2017). Moreover, severe hepatosplenomegaly was observed in all NrasG12D;Mdmx-Tg mice at 2 months, whilst the spleen weight of NrasG12D mice was normal at the analyzed time point (Figure S5A). The GFP+ cells from G12D spleens and BM at 5 months were CD11b+ Gr-1− monocytic cells, reflecting a CMML-like phenotype (Figure S5B–C). However, the GFP+ spleen cells of NrasG12D;Mdmx-Tg mice additionally displayed acute leukemia phenotypes: either Gr-1− CD11b− CD8+ and Gr-1− CD11b+ CD8+ co-existing bulk tumor cells (i.e. CMML/AML + T-ALL) or Gr-1− CD11b+ and Gr-1+ CD11b+ cKit+ co-existing bulk tumor cells (i.e. CMML + AML type) (Figure S5C). GFP+ thymus cells from NrasG12D;Mdmx-Tg mice with the combined myeloid plus T-ALL phenotype showed CD8+ skewed lymphopoiesis, and they contained an altered CD8/CD4 double negative (DN) fraction (Figure S5D). These observations support a co-diagnosis of murine T-ALL/thymotic lymphoma in some animals (Tremblay et al., 2010), in addition to the AML phenotype present in all NrasG12D;Mdmx-Tg recipients (Figure S5E).
In summary, our studies demonstrate that MDMX overexpression leads to a transition from non-penetrant, or slowly progessing chronic myeloid disease to overt aute myeloid leukemia in five different genetic models including PU.1 URE−/−;Mdmx-Tg, Tet2−/−;Mdmx-Tg, Tet2+/−;Mdmx-Tg, Flt3WT/ITD;Mdmx-Tg, and NrasG12D;Mdmx-Tg.
MDMX overexpression leads to upregulation of Wnt/β-Catenin signaling in pre-LSC, an effect that is mediated by physical interaction of MDMX with CK1α
Our finding of cooperativity of MDMX overexpression in the context of Flt3-mutation, combined with the clinical observation of mutual exclusivity of FTL3 and TP53 mutations, raised the interesting possibility of p53-independent mechanisms playing a role in the AML-triggering effects of MDMX overexpression. To study the possible underlying molecular mechanisms, we performed both RNA sequencing (RNA-seq) as well as immunoprecipitation followed by mass spectrometry (LC-MS/MS) experiments.
HSCs (Flk2- LSK) from WT and Mdmx-Tg mice, and pre-LSCs (Flk2− LSK) from URE−/− and URE−/−;Mdmx-Tg mice were transcriptionally interrogated via RNA-seq (Table S1, S2). As expected, expression of p53 targets was repressed in Mdmx-Tg HSCs compared to WT HSCs, however, only to a relatively modest extent (NES: −1.15, P=0.25) (Figure 5A). Interestingly, using Ingenuity Pathway Analysis (IPA), we found that Wnt/β-Catenin was the most significantly upregulated canonical pathway (Z-score: 1.6, P=0.006) (Figure 5B). Gene set enrichment analysis also revealed a Ctnnb1 (β-Catenin) oncogenic signature to be upregulated in Mdmx-Tg HSCs (NES: 1.52, P=0.04) (Figure 5C). RNA-seq comparison of URE−/− versus URE−/−;Mdmx-Tg pre-LSC also exhibited upregulated Ctnnb1 oncogenic (NES: 1.36, P=0.059) and Wnt/β-Catenin signaling signatures (NES: 1.41, P=0.072) (Figure 5E). In addition to upregulation of the Wnt/β-Catenin pathway, the suppression of p53 targets and apoptosis pathways was evident in URE−/−;Mdmx-Tg pre-LSC (NES: −1.68, P=0.004, and NES: −1.40, P=0.016, respectively) (Figure 5D). We confirmed increased levels of β-Catenin protein by immunofluorescence (IF) staining in Mdmx-Tg, URE−/−;Mdmx-Tg, and Tet2+/−;Mdmx-Tg HSC/pre-LSC (Figure 5F, S6A).
Figure 5. MDMX overexpression leads to upregulation of Wnt/β-Catenin signaling in pre-LSC, an effect that is mediated by physical interaction of MDMX with CK1α.
(A) RNA sequencing data of WT and Mdmx-Tg HSCs analyzed by Gene set enrichment analysis (GSEA) for p53 targets. (n=3) (B) Canonical pathways representing the top 10 Z-scores by Ingenuity Pathway Analysis (IPA) of the RNA sequencing data. (C) RNA sequencing data of WT and Mdmx-Tg HSCs were analyzed by GSEA for oncogenic β-Catenin signature. (D) RNA sequencing of pre-LSCs from URE−/− and URE−/−;Mdmx-Tg was performed (n=3), including GSEA analysis of p53 targets and apoptosis. (E) GSEA analysis for β-Catenin oncogenic signature and hallmark Wnt/β-Catenin signaling for URE−/− and URE−/−;Mdmx-Tg. (F) Left: Protein expression of β-Catenin (green) by immunofluorescence (IF) staining with DAPI (blue) counterstain in HSCs of WT and Mdmx-Tg mice. Representative pictures are shown (scale bars: 1.85μm). Center: Signal of total cell staining of β-Catenin. Right: Nuclear signal of β-Catenin with DAPI counterstain. Statistics were calculated by T test. ***: P<0.001. (G) Schema of experimental strategy to detect MDMX interacting proteins. (H) Whole cell lysate (WCL), cytoplasmic and nuclear proteins were extracted from 32D cells transduced with HA-tagged Mdmx or Empty control lentivirus. Immunoprecipitation (IP) with anti-HA beads was performed for WCL. Samples were blotted for indicated antibodies. Molecular weight (kDa) was indicated in the figure. (I) Relative expression of Ctnnb1 RNA in 32D cells transduced with Mdmx or Empty control lentivirus (n=3). Statistical significance was calculated by T test. N.S.: not significant. Data shown as Mean±SEM. See also Figure S6 and Tables S1–S3.
Next, we set out to identify proteins that interact with MDMX in myeloid cells. We transduced the p53-WT/MDMX-low murine AML cell line 32D with an HA-tagged murine MDMX-expressing lentiviral vector (Figure 5G). We extracted total cellular protein and performed MDMX-interactome screening using immunoprecipitation against the HA-tag followed by mass-spectrometry (LC-MS/MS). Among all interacting proteins, casein kinase CK1α (Csnk1a1) was the top, non-structural/non-housekeeping MDMX-interacting protein in 32D AML cells (Table S3, Figure S6B). This appeared to be of particular interest to us as previous studies had reported that CK1α can bind and regulated both MDMX and β-Catenin in other cell types (Chen et al., 2005; Liu et al., 2002; Wu et al., 2012). We therefore focused on the possible interaction of MDMX with CK1α and its downstream effects on Wnt/β-Catenin signaling in leukemia/preleukemia cells. Using co-immunoprecipitation (IP) assays followed by western blotting (WB), we validated the LC-MS/MS results and found that MDMX indeed interacts with CK1α in AML cells (Figure 5H). We further hypothesized that this interaction may prevent CK1α binding to β-Catenin upon MDMX overexpression. In agreement with this hypothesis, we found that total protein expression as well as nuclear import of β-Catenin was increased in 32D cells upon MDMX overexpression (Figure 5H); at the same time, RNA expression levels of Ctnnb1 remained unchanged (Figure 5I), indicating that β-Catenin is regulated at the protein level. Also, overexpression of CK1α in MDMX overexpressing 32D cells reduced expression and nuclear transportation of β-Catenin (Figure S6C), indicating that CK1α abundance plays a causative role in the MDMX-mediated elevation of β-Catenin levels. Collectively, our results indicate that MDMX binds to and reduces cellular abundance of CK1α in AML cells, and as a consequence, leads to increased nuclear levels of β-Catenin.
Wnt/β-Catenin inhibition or elevation of CK1α levels rescue MDMX-overexpression-induced functional properties of pre-LSC.
We next investigated whether the observed elevation of β-Catenin signaling and sequestration of CK1α are functionally relevant in MDMX overexpressing pre-LSCs. For this purpose, we first utilized two pharmacological inhibitors of Wnt/β-Catenin, WNT974 and PNU74654. WNT974 is a porcupine inhibitor that prevents the secretion of Wnt, hence broadly inhibits Wnt/β-Catenin signaling (Liu et al., 2013), while PNU74654 inhibits the interaction of nuclear β-Catenin with transcription factors and suppresses canonical Wnt/β-Catenin signaling (Trosset et al., 2006). Colony forming assays showed that WT HSCs tolerated Wnt/β-Catenin inhibition well (IC50s: 13.7 μM for WNT974, >20μM for PNU74654) except at very high concentrations of WNT974 (Figure 6A), in line with a previous study demonstrating that canonical Wnt/β-Catenin signaling is largely dispensable for adult hematopoiesis (Cobas et al., 2004). On the other hand, Mdmx-Tg HSCs were significantly more sensitive (IC50s: 0.1μM for WNT974, 0.5μM for PNU74654) to both the Wnt/β-Catenin inhibitors including at nanomolar concentrations (Figure 6A).
Figure 6. Wnt/β-Catenin inhibition or elevation of CK1α levels rescue MDMX-overexpression-induced functional properties of pre-LSC.
(A) IC50 determination in colony formation assays of 1000 WT or Mdmx-Tg HSCs with indicated concentration of Wnt/β-Catenin inhibitors (n=2). (B) Colony numbers from 5000 WT or Mdmx-Tg cKit+ bone marrow cells transduced with Csnk1a1 (CK1α) or empty vector (Emp) (n=2). Cells were serially replated for 3 times. (C) Colony number from 10000 URE−/− or URE−/−;Mdmx-Tg pre-LSCs with indicated concentration of Wnt/β-Catenin inhibitors (n=2). (D) Colony number from 5000 URE−/− or URE−/−;Mdmx-Tg lineage- cKit+ bone marrow (BM) cells transformed with Csnk1a1 (CK1α) or empty (Epm) vector (n=2). Cells were serially replated for 3 times. (E) IC50 assays of lineage− cKit+ BM cells from WT, URE−/− and URE−/−;Mdmx-Tg preleukemic mice cultured with the indicated concentrations of PNU74654, ALRN-6924, or both, for 24hours. # (A) (C): Statistical significance was calculated by T test for each drug concentration. Data shown as Mean±SEM. *: 0.01≤P<0.05, **: 0.001≤P<0.01, ***: P<0.001. NR: not reached. # (B) (D): Statistical significance was calculated by Tukey HSD test. Data shown as Mean±SEM. *: 0.01≤P<0.05, **: 0.001≤P<0.01, ***: P<0.001. See also Figure S7, S8.
Next, we investigated whether retroviral overexpression of CK1α in cKit+ hematopoietic stem/progenitor cells (HSPCs) also impacted colony forming ability of MDMX-overexpressing cells in a similar manner. We transduced WT or Mdmx-Tg HSPCs with an MSCV-CK1α(Csnk1a1)-IRES-GFP (Jaras et al., 2014) or empty vector and performed serial replating assays. Strikingly, CK1α overexpression indeed resulted in complete suppression of colony formation of Mdmx-Tg HSPCs after the third replating, while empty vector-transduced Mdmx-Tg HSPCs maintained high colony-forming capacity (Figure 6B). Moreover, we did not observe any significant change in serial replating capacity of WT HSPCs upon CK1α overexpression.
Similarly, URE−/−;Mdmx-Tg and Flt3WT/ITD;Mdmx-Tg pre-LSCs were sensitive to Wnt/β-Catenin inhibition by WNT974 and PNU74654, while URE−/− and Flt3WT/ITD pre-LSCs were not (Figure 6C, Figure S7A). Likewise, CK1α overexpression rescued the increased colony forming ability of URE−/−;Mdmx-Tg as well as Flt3WT/ITD;Mdmx-Tg, whereas URE−/− and Flt3WT/ITD pre-LSCs were not significantly affected (Figure 6D, Figure S7B). Also, knock-down of β-Catenin (Ctnnb1) in LSCs (lineage− cKit+ cells in URE−/−;Mdmx-Tg leukemic mice) remarkably reduced colony formation, while reduction of colony by Ctnnb1 knock-down was limited in LSCs of URE−/− mice (Figure S7C).
Altogether, these findings using different model systems demonstrate that increased Wnt/β-Catenin signaling is, at least in part, mediating the increase in colony forming capacity of MDMX-overexpressing pre-LSC/LSC. Furthermore, our data demonstrate that CK1α overexpression can abrogate the increased serial replating capacity of MDMX overexpression-induced pre-LSC.
In a next step, we explored the potential utility of our findings for possible combinatorial therapy. Specifically, we investigated whether combination of a Wnt/β-Catenin inhibitor (PNU74654) and ALRN-6924, which re-activates p53 by inhibiting interaction of MDMX with TP53 protein (Carvajal et al., 2018), would be beneficial. We found that URE−/−;Mdmx-Tg preleukemic lineage− cKit+ cells were only modestly sensitive to single drug treatment by either PNU74654 or ALRN-6924. However, the combination of PNU74654 and ALRN-6924 showed strong synergy in eradicating URE−/−;Mdmx-Tg pre-LSCs (Figure 6E, left), and the combination index (CI) indicated substantially stronger synergism in URE−/−;Mdmx-Tg cells (CI=0.12) compared to WT (CI=0.59) or URE−/− (CI=0.56) controls (Figure 6E, right). We next tested the combination treatment in vivo, and injected ALRN-6924 and BC2059 (a Wnt/β-Catenin inhibitor suitable for in vivo studies) into recipient mice of 3-month-old URE−/−;Mdmx-Tg cells (which lead to AML in recipients). While single drug treatment with either ALRN-6924 or BC2059 slightly prolonged survival, combination treatment with both drugs was significantly more effective (Figure S7D). Moreover, we found that treatment with Wnt/β-Catenin inhibitor sensitized a previously reported ALRN-6924-resistant human AML cell line, OCI-AML3 (p53-wildtype), to ALRN-6924 treatment (Combination index: 0.17) (Figure S7E). Collectively, these data suggest that the MDMX/CK1α/β-catenin axis in pre-LSC, which our mechanistic studies revealed, is therapeutically targetable.
MDMX-driven HSC expansion and Wnt/β-Catenin upregulation is p53 independent.
Next, we asked whether β-Catenin upregulation and resultant functional effects in an MDMX overexpression background are p53 dependent or not. We therefore generated Mdmx-Tg mice in a p53 null background (Trp53−/−;Mdmx-Tg). HSCs of Trp53−/−;Mdmx-Tg mice overexpressed Mdmx 20.3±2.4 fold compared to Trp53−/− (Figure 7A), and we found that Trp53−/−;Mdmx-Tg BM cells had significantly higher serial replating capacity compared to Trp53−/− (Figure 7B). We then transplanted Trp53−/− or Trp53−/−;Mdmx-Tg (Ly45.2) BM cells together with WT competitor cells (Ly45.1/2) into lethally irradiated C57BL/6 recipients (Ly45.1). Trp53−/−;Mdmx-Tg cells reconstituted the myeloid lineage to a significantly greater extent than Trp53−/− cells (Figure 7C); however, unlike the Mdmx-Tg versus WT competitive transplantation assay (Figure 1D), reconstitution of the lymphoid lineage did not differ between Trp53−/− and Trp53−/−;Mdmx-Tg. In addition, mice who received Trp53−/−;Mdmx-Tg cells displayed a higher proportion of LSK cells than Trp53−/− recipients (Figure 7C). We concluded that MDMX overexpression enhances competitiveness of HSCs and specifically myeloid reconstitution independent of its functions involving p53. We also found protein levels of β-Catenin (both total as well as nuclear) and RNA expression of its major targets, Ccnd1 and c-Myc, to be significantly increased in Trp53−/−;Mdmx-Tg HSCs (Figure 7D–E). In line with this, we found the colony forming ability of Trp53−/−;Mdmx-Tg HSCs to be impaired by treatment with the canonical Wnt/β-Catenin inhibitor PNU74654, while Trp53−/− HSCs were unaffected (Figure 7F). Lastly, we overexpressed FLAG-tagged MDMX in the p53-null human AML cell line HL-60. While RNA expression of CTNNB1 was not different between empty vector and MDMX transduced cells (Figure 7G), we found increased expression of β-Catenin protein in whole cell lysates as well as cytoplasmic and nuclear extracts from MDMX overexpressing HL-60 cells (Figure 7H). Furthermore, we detected interaction of MDMX and CK1α in the p53-null context (Figure 7H). In summary, these data reveal that MDMX overexpression leads to expansion of the HSC compartment, and specifically enhances their reconstitution of the myeloid lineage, in a p53-independent manner. At the mechanistic level, the physical interaction of MDMX with CK1α is also not dependent on p53.
Figure 7. MDMX-driven HSC expansion and Wnt/β-Catenin upregulation is p53 independent.
(A) Mdmx relative transcript expression in HSCs (Flk2- LSK) (n=3). (B) Colony formation assay from 1×104 Trp53−/− or Trp53−/−;Mdmx-Tg bone marrow cells. (n=2). (C) 0.5×106 Trp53−/− or Trp53−/−;Mdmx-Tg bone marrow cells (Ly45.2) with 2.0×106 WT competitors (Ly45.1/2) were competitively transplanted into lethally irradiated recipients (Ly45.1). Ratio of donor cells (Ly45.2) at each time point (months) from transplantation (n=7) and ratio of donor cells (Ly45.2) in the LSK population at 4 months after transplantation (n=7). (D) Immunofluorescence (IF) staining of β-Catenin (green) with DAPI (blue) in HSC of Trp53−/− and Trp53−/−;Mdmx-Tg mice. Top; Representative pictures (Scale bars; 1.85μm). Bottom; Left: Signal of total cell staining of β-Catenin. Right: Nuclear signal of β-Catenin with DAPI counterstain. (E) Relative transcript levels of the β-Catenin target genes Ccnd1 and c-Myc. (F) Colony number from 1000 Trp53−/− or Trp53−/−;Mdmx-Tg HSCs with indicated concentration of Wnt/β-Catenin inhibitor PNU74654 (n=2). NR: not reached. (G) Relative expression of CTNNB1 (β-Catenin) RNA in HL-60 cells transduced with MDMX or Empty control lentivirus (n=3). (H) Whole cell lysate (WCL), cytoplasmic and nuclear extracts from HL-60 cells transduced by FLAG-tagged MDMX (MDMX) or Empty control lentivirus (Empty). Immunoprecipitation (IP) for WCL by anti-FLAG beads was performed. WCL, IP, cytoplasmic and nuclear extract were blotted by indicated antibodies. Molecular weight (KDa) is indicated in the figure. # (A)-(G): statistical significance was calculated by T test. Data shown as Mean±SEM. N.S.: not significant. *: 0.01≤P<0.05, **: 0.001≤P<0.01, ***: P<0.001.
Clinical-correlative data indicate relevance of the MDMX-Wnt/β-Catenin axis in patients with MDS
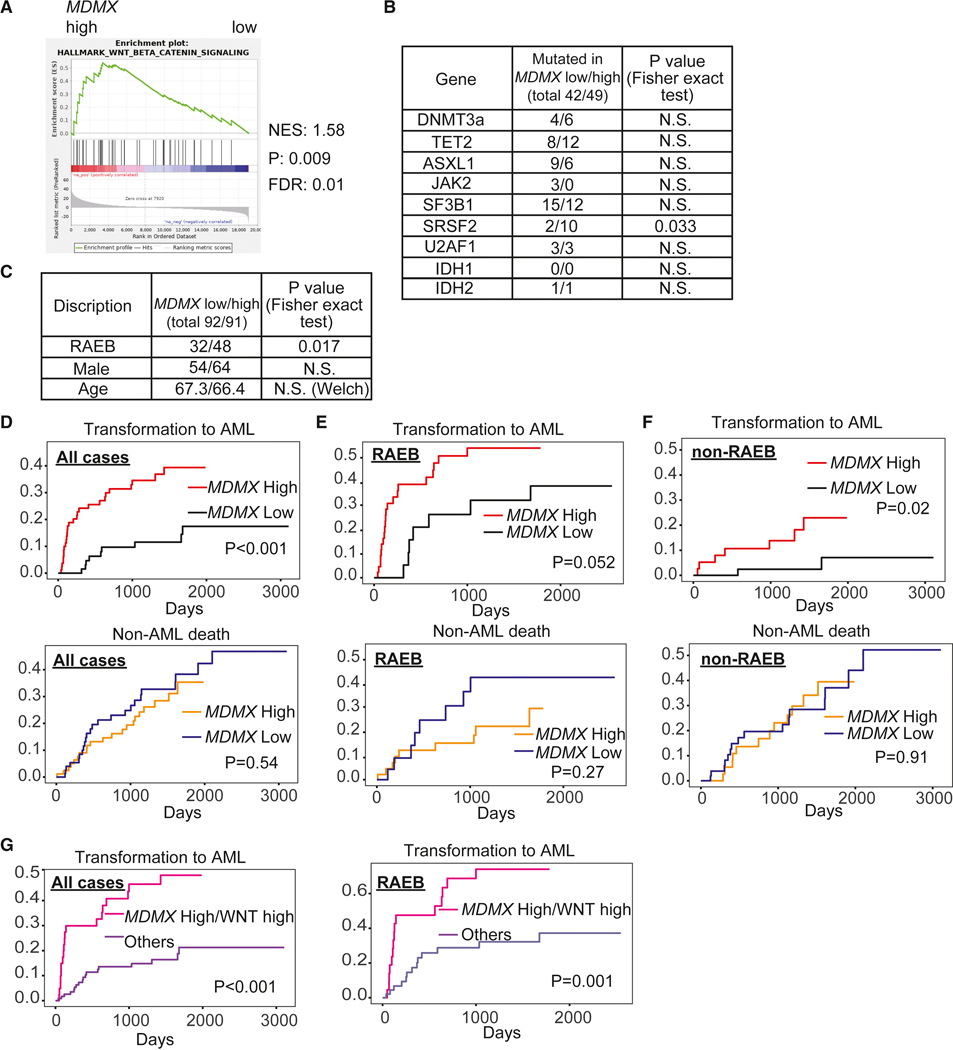
We analyzed large published patient cohorts to examine the potential clinical relevance of the MDMX/β-Catenin axis. As our data suggests that Wnt/β-Catenin upregulation by MDMX overexpression is important for leukemic transformation from a preleukemic stage, we focused on a cohort of patients with non-treated myelodysplastic syndrome (MDS) for which both molecular as well as clinical time-to-event data was available (GSE19429). We dichotomized patients into MDMX-high and MDMX-low expressers, and found enrichment of a Wnt/β-Catenin signature in patients with high MDMX expression (Figure 8A). The mutation status of CH-related genes was comparable in both the MDMX-high and MDMX-low groups, except for SRSF2 (Figure 8B). Interestingly, MDMX-high patients included more Refractory Anemia with Excess Blasts (RAEB) cases which is a more severe subtype of MDS (Figure 8C). In a next step, we evaluated cumulative incidence of transformation to AML treating non-leukemic death as competing risk. Strikingly, MDS patients with high MDMX expression displayed a higher transformation rate to AML in the entire cohort compared to patients with low MDMX expression (Figure 8D), but also when we analyzed patients with RAEB (Figure 8E) and patients with non-RAEB subtype (Figure 8F) separately, while non-leukemic death was not significantly different in all subgroups. Moreover, patients who were simultaneously MDMX-high as well as WNT score (Bhagat et al., 2017) high showed very rapid progression to AML, and this transformation rate was highly significantly different compared to all other patients (Figure 8G). Patients with RAEB showed no significant difference in blast percentage between the MDMX low versus high groups (10.25±4.00%, and 12.16±4.60%, respectively). We next investigated primary BM samples of MDS. About half of MDS RAEB cases had elevated MDMX, about 5- to 6-fold increased levels compared to normal (Figure S8A), and HSPCs (Lineage−/CD34+) from MDMX high patients were significantly more sensitive to treatment with β-Catenin inhibitor (Figure S8B), consistent with our previous findings in murine models. Taken together, these data suggest that the MDMX-Wnt/β-Catenin axis is functionally important in patients with MDS and relevant for the clinical course of preleukemic disease.
Figure 8. Clinical-correlative data indicate relevance of the MDMX-Wnt/β-Catenin axis in patients with MDS.
Analyses of GSE19429. (A) Gene set enrichment analysis (GSEA) of MDMX high versus low patients reveals highly significant enrichment of Wnt/β-Catenin signature. N.S.; not significant. (B) Mutation status of clonal hematopoiesis related genes between MDMX low and high patients. P values were calculated by Fisher exact test. N.S.; not significant. (C) Disease subtype, sex and age between MDMX low and high patients. P values were calculated by Fisher exact test for subtype and sex, and Welch’s test for age. (D) Cumulative incidence plots of transformation to AML and non-leukemic death treating each other as competing risk (MDMX low versus high, all cases), (E) patients with RAEB subtype, and (F) patients with MDS subtypes other than RAEB. P values were calculated by Gray’s test. (G) Cumulative incidence plots of transformation to AML treating non-leukemic death as competing risk (MDMX high, WNT score high versus all other cases) (Left: all patients, right: patients with RAEB subtype). P values were calculated by Gray’s test. See also Figure S8.
Discussion
Our studies of MDMX overexpression in murine hematopoiesis revealed an important and pervasive role of MDMX in the induction of AML in the context of several different molecular aberrations frequently occurring in human leukemia. MDMX overexpression is detected in almost 90% of AML patients irrespective of mutational subtypes (Carvajal et al., 2018; Han et al., 2016; Quintas-Cardama et al., 2017). The different preleukemic models we combined with Mdmx transgenic overexpression act via distinct molecular mechanisms and reflect aberrations that occur frequently in patients. PU.1 inactivation has been previously classified as a so-called ‘class II’ (i.e. differentiation blocking) aberration which is caused by various genetic, epigenetic, and posttranslational mechanisms and is encountered in 60–70% of all AML patients (Gilliland and Tallman, 2002; Sive et al., 2016; Tenen, 2003; Will et al., 2015). TET2 loss-of-function mutations are frequent pre-leukemic alterations including in individuals with CH (~10%) (Genovese et al., 2014; Jaiswal et al., 2014). Heterozygous FLT3 or NRAS mutations are two of the most frequent genetic aberrations in AML, and are thought to primarily act as ‘proliferative hits’ and at a later stage in many patients (Bacher et al., 2006; Papaemmanuil et al., 2016) (i.e. class I aberrations) (Bacher et al., 2006; Gilliland and Tallman, 2002; Kiyoi et al., 1999; Nakao et al., 1996; Sive et al., 2016; Staber et al., 2014). Of note, heterozygous Tet2 and Flt3 mutations are not sufficient to induce hematopoietic malignancy and Nras mutations only lead to a smoldering MDS/CMML-like phenotype in mice. Furthermore, MDMX overexpression occurs in the majority of patients with TET2, FLT3 or NRAS-G12D heterozygous mutations (Cancer Genome Atlas Research et al., 2013). Overall, our new MDMX-driven models and findings cover and are applicable to a range of different leukemogenic genetic contexts and mechanisms that are frequently encountered in patients.
Notably, each of the murine models we tested do not develop overt AML as a stand-alone and without MDMX overexpression, while representing classical “pre-leukemic or CH-like conditions (Tet2 (haplo)insufficiency and PU.1low, respectively) or changes that are associated with myeloproliferative conditions (Tet2 (homozygous) deficiency, heterozygous Flt3-ITD mutations and Nras mutations). In either situation, MDMX was causative in the induction of an acute leukemia phenotype. Since our models do not allow the timely control of induction of the different aberrations, it will be interesting to test in future studies whether the exact timing of MDMX overexpression is critical (early versus late), and may depend on the specific cooperating aberrations.
Our cooperativity models with overexpression of the Mdmx transgene showed marked molecular and phenotypic differences from previously studied combinatorial phenotypes with p53 mutants (Dumble et al., 2007). Most importantly, we observed greater competitive advantage of Trp53−/−;Mdmx-Tg compared to Trp53−/− cells, indicating that MDMX exerts p53-independent mechanisms of clonal expansion. Furthermore, we found only modest suppression of p53 target genes in MDMX-overexpressing HSCs, which is in contrast to the strong suppression of p53 targets including cellular quiescence related genes that has been previously reported in Trp53−/− HSCs (Liu et al., 2009).
Using approach RNA-seq and proteomic studies, we found, unexpectedly, that upregulation of Wnt/β-Catenin signaling in Mdmx-Tg mice plays an important role in the expansion of HSCs. We also observed activation of Wnt/β-Catenin signaling in MDMX overexpressing pre-LSCs, as well as clinical-correlative evidence that MDMX expression levels correlate with Wnt/β-Catenin signature in patients with MDS. Wnt/β-Catenin signaling has previously been reported to be implicated in various cancers, including AML (Gruszka et al., 2019; Zhan et al., 2017). On the other hand, canonical Wnt/β-Catenin signaling is dispensable in adult hematopoiesis (Cobas et al., 2004), but its constitutive activation leads to increased cell cycling of HSCs (Scheller et al., 2006), which is consistent with our findings in the MDMX overexpression setting.
Our findings revealed the mechanism of Wnt/β-Catenin signaling upregulation by MDMX overexpression to be the result of physical interaction with and reduced abundance of CK1α, which phosphorylates β-Catenin. CK1α is known to act as a tumor suppressor by phosphorylating β-Catenin on Ser45. This phosphorylation is required for further phosphorylation by GSK3β and subsequent protease-mediated degradation of β-Catenin in the cytoplasm (Liu et al., 2002). Our data indicate that MDMX binds CK1α, thereby preventing its function to phosphorylate β-Catenin and subsequent degradation. CK1α overexpression counteracted the β-Catenin upregulation in MDMX overexpressing cells and rescued the repopulating phenotypes of Mdmx-Tg HSCs/pre-LSCs. Our findings are in line with several reports in other cell types. It has been reported that during the DNA damage response in ES cells, reduced abundance of CK1α activates Wnt/β-Catenin signaling via increased β-Catenin protein levels (Carreras Puigvert et al., 2013); further, CK1α agonists have shown activity against Wnt/β-Catenin dependent solid tumors via enhanced degradation of β-Catenin (Li et al., 2017). Although CK1α harbors multiple functions (Jiang et al., 2018a), our findings demonstrate that regulation of β-Catenin is a critical role of CK1α in preleukemic and leukemic cells; our data demonstrate a novel mechanism of reduced CK1α availability caused by MDMX overexpression in hematopoietic cells. Moreover, since we observed upregulation of Wnt/β-Catenin signaling upon MDMX overexpression even in the absence of p53, we conclude that reduced abundance of CK1α and subsequent Wnt/β-Catenin upregulation is a p53-independent, principal mechanism of MDMX.
With regards to potential translational utility of our findings, we found that high levels of MDMX correlated with upregulation of Wnt/β-Catenin signaling and higher leukemic transformation rate in a large cohort of MDS patients suggesting relevance of the uncovered mechanisms for human disease. Furthermore, we tested whether Wnt/β-Catenin can be a therapeutic target in AML and preleukemic disease. Although Wnt/β-Catenin has been reported to be functionally relevant in LSCs (Wang et al., 2010), and preclinical studies revealed that Wnt/β-Catenin inhibitors are active against AML in vitro and in vivo (Fiskus et al., 2015; Li et al., 2014), there have been no published clinical trials on the use of Wnt/β-Catenin inhibitors/modulators in AML/high-risk MDS (Gruszka et al., 2019). Our data therefore suggest that Wnt/β-Catenin inhibitors might be particularly effective in MDMX-overexpressing patients, and specifically at the preleukemic stage rather than completely transformed AML, or that they may sensitize preleukemic/leukemic cells to other drugs (Heidel et al., 2012; Jiang et al., 2018b). In this regard, we tested a combination of Wnt/β-Catenin inhibitor and the clinical drug ALRN-6924 which inhibits the MDMX/TP53 interaction. This combinatorial treatment showed promise in that it revealed significant synergism and led to complete elimination of MDMX-overexpressing pre-LSCs and MDMX/p53 inhibitor-resistant AML cells. As MDMX overexpression is common in AML and we showed overexpression of MDMX and Wnt/β-Catenin is associated with poor outcome of preleukemic patients, this combination could be a promising therapeutic approach for patients with MDS and AML. Currently, there are no clinical options available for the treatment of preleukemic conditions (e.g. clonal hematopoiesis in progression) or to specifically prevent leukemic transformation (e.g. of high-risk MDS). Our data suggest that Wnt/β-Catenin inhibitors in combination with MDMX/p53 inhibitors may offer such a targeted anti-pre-LSC strategy in patients with elevated MDMX and should be considered for further testing.
Overall, our findings highlight an important role of MDMX overexpression in preleukemic to acute leukemic progression, and reveal CK1α/Wnt/β-Catenin as a therapeutically targetable pathway and as a novel p53-independent non-canonical mechanism of MDMX overexpression. Combinatorial targeting of these pathways may provide a possible approach for precision prevention and cancer interception in the early stages of the pathogenesis of myeloid and possibly other MDMX-driven malignancies.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requestst for resources and reagent should be directed to and will be fulfilled by the Lead Contact, Ulrich Steidl (Ulrich.steidl@einsteinmed.org).
Materials Availability
Plasmids are detailed in the Key Resources Table and available upon request. The sequence of oligos are detailed in Table S4. Mouse models used in this study are detailed in the Key Resources Table and available upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-Human CD2 (PE/Cyamine5) | Thermo Fisher Scientific | Cat# 15-0029-73, RRID:AB_468687 |
| Mouse Anti-Human CD3 (PE/Cyamine5) | Thermo Fisher Scientific | Cat# 15-0038-42, RRID:AB_10598354 |
| Mouse Anti-Human CD4 (Tri-Color) | Thermo Fisher Scientific | Cat# MHCD0406, RRID:AB_1473737 |
| Mouse Anti-Human CD7 (Tri-Color) | Thermo Fisher Scientific | Cat# MHCD0706, RRID:AB_1482844 |
| Mouse Anti-Human CD8 (Tri-Color) | Thermo Fisher Scientific | Cat# MHCD0806, RRID:AB_10372207 |
| Mouse Anti-Human CD10 (PE/Cyamine5) | Thermo Fisher Scientific | Cat# 15-0106-73, RRID:AB_657520 |
| Mouse Anti-Human CD14 (Tri-Color) | Thermo Fisher Scientific | Cat# MHCD1406, RRID:AB_10373566 |
| Mouse Anti-Human CD19 (PE/Cyamine5) | Thermo Fisher Scientific | Cat# 15-0199-42, RRID:AB_10853658 |
| Mouse Anti-Human CD20 (PE/Cyamine5) | Thermo Fisher Scientific | Cat# 15-0209-42, RRID:AB_10548510 |
| Mouse Anti-Human CD235a (PE/Cyamine5) | BD Biosciences | Cat# 559944, RRID:AB_397387 |
| Mouse Anti-Human CD56 (Tri-Color) | Thermo Fisher Scientific | Cat# MHCD5606, RRID:AB_10372520 |
| Mouse Anti-Human CD33 (APC) | Thermo Fisher Scientific | Cat# A15727, RRID:AB_2534507 |
| Mouse Anti-Human CD34 (Pacific Blue) | BioLegend | Cat# 343512, RRID:AB_1877197 |
| Rat Anti-Mouse CD3e (Biotin) | Thermo Fisher Scientific | Cat# 13-0032-82, RRID:AB_2572762 |
| Rat Anti-Mouse CD3e (eFluor450) | Thermo Fisher Scientific | Cat# 48-0031-82, RRID:AB_10735092 |
| Rat Anti-Mouse CD4 (Biotin) | Thermo Fisher Scientific | Cat# 13-0041-82, RRID:AB_466325 |
| Rat Anti-Mouse CD4 (eFluor450) | Thermo Fisher Scientific | Cat# 48-0041-82, RRID:AB_10718983 |
| Rat Anti-Mouse CD4 (PE-Cyamine7) | Thermo Fisher Scientific | Cat# 25-0041-82, RRID:AB_469576 |
| Rat Anti-Mouse CD8a (Biotin) | Thermo Fisher Scientific | Cat# 13-0081-82, RRID:AB_466346 |
| Rat Anti-Mouse CD8a (eFluor450) | Thermo Fisher Scientific | Cat# 48-0081-82, RRID:AB_1272198 |
| Rat Anti-Mouse CD8a (PE-Cyamine7) | Thermo Fisher Scientific | Cat# 25-0081-82, RRID:AB_469584 |
| Rat Anti-Mouse CD11b (APC-eFluor780) | Thermo Fisher Scientific | Cat# 47-0112-82, RRID:AB_1603193 |
| Rat Anti-Mouse CD11b (Biotin) | Thermo Fisher Scientific | Cat# 13-0112-82, RRID:AB_466359 |
| Rat Anti-Mouse CD11b (eFluor450) | Thermo Fisher Scientific | Cat# 48-0112-82, RRID:AB_1582236 |
| Rat Anti-Mouse CD11b (PE) | Thermo Fisher Scientific | Cat# 12-0112-83, RRID:AB_2734870 |
| Rat Anti-Mouse CD16/32 (PE/Cyamine7) | Thermo Fisher Scientific | Cat# 25-0161-82, RRID:AB_469598 |
| Rat Anti-Mouse CD16/32 TruStain fcX (Blocking-Ab) | BioLegend | Cat# 101320, RRID:AB_1574975 |
| Rat Anti-Mouse CD19 (Biotin) | Thermo Fisher Scientific | Cat# 13-0193-82, RRID:AB_657656 |
| Rat Anti-Mouse CD19 (eFluor450) | Thermo Fisher Scientific | Cat# 48-0193-82, RRID:AB_2734905 |
| Rat Anti-Mouse CD25 (eFluor450) | BD Biosciences | Cat# 553866, RRID:AB_395101 |
| Rat Anti-Mouse CD34 (eFluor450) | Thermo Fisher Scientific | Cat# 48-0341-82, RRID:AB_2043837 |
| Rat Anti-Mouse CD34 (FITC) | Thermo Fisher Scientific | Cat# 11-0341-82, RRID:AB_465021 |
| Rat Anti-Human/Mouse CD44 (eFluor450) | Thermo Fisher Scientific | Cat# 48-0441-82, RRID:AB_1272246 |
| Rat Anti-Human/Mouse CD45.1 (FITC) | Thermo Fisher Scientific | Cat# 11-0453-82, RRID:AB_465058 |
| Rat Anti-Human/Mouse CD45.2 (APC-Cyamine7) | Thermo Fisher Scientific | Cat# 47-0454-82, RRID:AB_1272175 |
| Rat Anti-Human/Mouse CD45.2 (PE) | Thermo Fisher Scientific | Cat# 12-0454-82, RRID:AB_465678 |
| Rat Anti-Human/Mouse CD45R (B220) (Biotin) | Thermo Fisher Scientific | Cat# 13-0452-82, RRID:AB_466449 |
| Rat Anti-Human/Mouse CD45R (B220) (eFluor450) | Thermo Fisher Scientific | Cat# 48-0452-82, RRID:AB_1548761 |
| Rat Anti-Mouse CD48 (Alexa-Fluor488) | BioLegend | Cat# 103414, RRID:AB_571979 |
| Rat Anti-Mouse CD48 (PE) | BioLegend | Cat# 103405, RRID:AB_313020 |
| Rat Anti-Mouse cKit (CD117) (APC) | Thermo Fisher Scientific | Cat# 17-1171-82, RRID:AB_469430 |
| Rat Anti-Mouse CD127 (Biotin) | Thermo Fisher Scientific | Cat# 13-1271-82, RRID:AB_466588 |
| Rat Anti-Mouse CD127 (eFluor450) | Thermo Fisher Scientific | Cat# 48-1271-82, RRID:AB_2016698 |
| Rat Anti-Mouse CD135 (Flt3/Flk2) (PE) | Thermo Fisher Scientific | Cat# 12-1351-82, RRID:AB_465859 |
| Rat Anti-Mouse CD150 (PE-Cyamine7) | BioLegend | Cat# 115914, RRID:AB_439797 |
| Rat Anti-Mouse F4/80 (eFluor450) | Thermo Fisher Scientific | Cat# 48-4801-82, RRID:AB_1548747 |
| Rat Anti-Mouse Ly-6A/E (Sca-1) (APC-Cyamine7) | BD Biosciences | Cat# 560654, RRID:AB_1727552 |
| Rat Anti-Mouse Ly-6G (Gr-1) (Biotin) | Thermo Fisher Scientific | Cat# 13-5931-82, RRID:AB_466800 |
| Rat Anti-Mouse Ly-6G (Gr-1) (eFluor450) | Thermo Fisher Scientific | Cat# 48-5931-82, RRID:AB_1548788 |
| Rat Anti-Mouse Ly-6G (Gr-1) (PE) | Thermo Fisher Scientific | Cat# 12-5931-81, RRID:AB_466044 |
| Rat Anti-Mouse-Ter119 (Biotin) | Thermo Fisher Scientific | Cat# 13-5921-82, RRID:AB_466797 |
| Rat Anti-Mouse-Ter119 (eFluor450) | Thermo Fisher Scientific | Cat# 48-5921-82, RRID:AB_1518808 |
| Streptavidin eFluor450 | Thermo Fisher Scientific | Cat# 48-4317-82, RRID:AB_10359737 |
| Streptavidin Pacific Orange | Thermo Fisher Scientific | Cat# S32365 |
| Streptavidin PE-Cyamine7 | Thermo Fisher Scientific | Cat# 25-4317-82, RRID:AB_10116480 |
| Rabbit Anti-Actin (polyclonal) | Sigma Aldrich | Cat# A2066, RRID:AB_476693 |
| Rabbit Anti-β-Catenin (CAT-15) (polyclonal) | Thermo Fisher Scientific | Cat# 71-2700, RRID:AB_2533982 |
| Rabbit Anti-β-Catenin (D10A8) (monoclonal) | Cell Signaling Technology | Cat# 8480 RRID:AB 11127855 |
| Rabbit Anti-β-Tubulin (polyclonal) | Cell Signaling Technology | Cat# 2146, RRID:AB_2210545 |
| Rabbit Anti-CK1α (EPR1961(2)) (monoclonal) | Abcam | Cat# ab108296, RRID:AB_10864123 |
| Rabbit Anti-FLAG(DYKDDDDK)-Tag (polyclonal) | Cell Signaling Technology | Cat# 2368, RRID:AB_2217020 |
| Rabbit Anti-HA-Tag (C29F4) (monoclonal) | Cell Signaling Technology | Cat# 3724, RRID:AB_1549585 |
| Rabbit Anti-LaminB1 (polyclonal) | Abcam | Cat# ab16048, RRID:AB_443298 |
| Mouse Anti-MDMX (MDMX_82) (monoclonal) | Abcam | Cat# ab49993, RRID:AB_880928 |
| Rabbit Anti-phospho-β-Catenin (Thr41/Ser45) (polyclonal) | Cell Signaling Technology | Cat# 9565, RRID:AB_331731 |
| Horse Anti-Mouse-IgG, HRP-linked (secondary antibody) | Cell Signaling Technology | Cat# 7076, RRID:AB_330924 |
| Horse Anti-Rabbit-IgG, HRP-linked (secondary antibody) | Cell Signaling Technology | Cat# 7074, RRID:AB_2099233 |
| Goat Anti-Rabbit-IgG, Alexa Fluor 488 conjugated | Abcam | Cat# ab150077, RRID:AB 2630356 |
| Mouse TrueBlot ULTRA: Anti-Mouse Ig HRP (secondary antibody) | Rockland | Cat# 18-8817-30, RRID:AB_2610849 |
| Bacterial and Virus Strains | ||
| FUW-IRES-GFP lentiviral vector | (Ito et al., 2019) | N/A |
| MSCV-IRES-GFP retroviral vector (MIG) | Addgene | 20672 |
| PAX2 (lentiviral helper) | Addgene | 35002 |
| pCAD-IRES-GFP lentiviral vector | (Kawahara et al., 2012) | Originally from Dr. Bruce Torbett |
| Phi-Eco (retroviral helper) | Gift (Kira Gritsman) | N/A |
| pMD2.G (lentiviral helper) | Addgene | 12259 |
| Biological Samples | ||
| N/A | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Murine recombinant IL-3 | GEMINI | 300-324P |
| Murine recombinant IL-6 | GEMINI | 300-327P |
| Murine recombinant Flt3-Ligand | GEMINI | 300-306P |
| Murine recombinant SCF | GEMINI | 300-348P |
| Murine recombinant TPO | GEMINI | 300-351P |
| ALRN-6924 | Aileron Therapeutics | N/A |
| BC2059 | Targetmol | T5642 |
| BrdU (5-Bromo-2’-deoxiuridin) | Sigma-Aldrich | 19160 |
| MG132, Ready Made Solution | Sigma-Aldrich | M7449 |
| PNU75654 | Selleckchem | S8429 |
| Puromycin dihydrochloride | Sigma-Aldrich | P8833 |
| WNT974 (LGK974) | InvivoChem | V1353 |
| Critical Commercial Assays | ||
| CD117 (cKit) MicroBeads, mouse | Miltenyi Biotec | 130-091-224 |
| CellTiter-Blue Cell Viability Assay | Promega | G8081 |
| cOmplete Protease Inhibitor Cocktail (IP/WB) | Millipore Sigma | 11697498001 |
| Dynabeads Untouched Mouse T cells (negative depletion) | Thermo Fisher Scientific | 11413D |
| FITC BrdU Flow Kit | BD Pharmingen | 51-2354AK |
| iScript cDNA Synthesis Kit | BIO-RAD | 1708890 |
| Mouse methylcellulose complete media | R&D systems | HSC007 |
| Nucleospin Plasmid (cloning) | Takara Bio | 740588 |
| Pierce Anti-DYKDDDDK(FLAG) magnetic beads | Thermo Scientific | A36797 |
| Pierce Anti-HA magnetic beads | Thermo Scientific | 88836 |
| Phosphatase inhibitor cocktail 2 (IP/WB) | Sigma-Aldrich | P5726 |
| Phusion High-Fidelity DNA Polymerase (cloning) | Thermo Scientific | F530 |
| Phusion Site-Directed Mutagenesis Kit (cloning) | Thermo Scientific | F541 |
| Polybrene (virus transduction) | Santa Cruz Biotechnology | Sc-134220 |
| Power SYBR Green PCR Mster Mix | Applied biosystems | 4367659 |
| Primocin | InvivoGen | ant-pm |
| QIAquick Gel Extraction Kit (cloning) | QIAGEN | 28704 |
| QIAquick PCR Purification Kit (cloning) | QIAGEN | 28106 |
| RetroNectin (virus transduction) | Takara Bio | T100 |
| RNeasy Micro Kit (RNA extraction) | QIAGEN | 74004 |
| StemSpan SFEM | STEMCELLS | 09600 |
| Deposited Data | ||
| All RNA sequencing data | This paper | GSE164838 |
| Gene expression data of MDS cohort | (Bhagat et al., 2017) | GSE19429 |
| Experimental Models: Cell Lines | ||
| Human: HEK293T cell line | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| Human: HL-60 | ATCC | Cat# CCL-240 RRID: CVCL_0002 |
| Human: OCI-AML3 | Leibniz Institute | Cat# ACC-582 RRID: CVCL_1844 |
| Mouse: 32D (clone3) | ATCC | Cat# CRL-11346 RRID: CVCL_0119 |
| Experimental Models: Organisms/Strains | ||
| Mouse: B6.129-Flt3tm1Dgg/J (Flt3WT/ITD) | Jackson (Lee et al., 2007) |
011112 |
| Mouse: B6.129S2-Trp53tm1Tyj/J (Trp53−/−) | Jackson | 002101 |
| Mouse: B6.SJL-Ptprca Pepcb/BoyJ (Ly45.1) | Jackson | 002014 |
| Mouse: C57BL/6J | Jackson | 000664 |
| Mouse: Mdmx-Tg (Tg-15) | (Xiong et al., 2010) | N/A |
| Mouse: NOD.Cg-Prkdcscidll2rgtm1Wjl/SzJ (NSG) | Jackson | 005557 |
| Mouse: PU.1 URE−/− | (Rosenbauer et al., 2004) | N/A |
| Oligonucleotides | ||
| See table S5 | ||
| Recombinant DNA | ||
| pCMV3.(Human)MDM4-Flag | Sino Biological | HG15395-CF |
| FUW.(Human)MDMX.IRES.Puro lentiviral vector | This paper | N/A |
| FUW.(Mouse)Mdmx.IRES.Puro lentiviral vector | This Paper | N/A |
| MSCV.NRAS-G12D.IRES.GFP retroviral vector | (Parikh et al., 2007) | N/A |
| MSCV.(Mouse)Csnk1a1.IRES.GFP retroviral vector | (Jaras et al., 2014) | N/A |
| pCAD.(Mouse)Mdmx.IRES.GFP lentiviral vector | This paper | N/A |
| pGFP-C-shLenti control shRNA | Origene | TR30021 |
| pGFP-C-shLenti shRNA for Ctnnb1-A | Origene | TL500280A |
| pGFP-C-shLenti shRNA for Ctnnb1-B | Origene | TL500280B |
| Software and Algorithms | ||
| CompuSyn ver.1.0 | Compusyn Inc, (Chou, 2006) |
http://www.combosyn.com/ |
| DESeq2 | (Love et al., 2014) | http://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| FastQC | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| FlowJo_10.6.1_CL | FlowJo | https://www.flowjo.com/ |
| GSEA 4.0.2 | Broad institute | https://software.broadinstitute.org/gsea/index.jsp |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| IPA v01.13 | Qiagen | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis |
| Volocity Quantitation | Quorum Technologies | https://quorumtechnologies.com/volocity/volocity/quantitation |
| R v3.6.0 | R Core Team | www.r-project.org |
| Salmon v0.11.3 | (Patro et al., 2017) | https://salmon.readthedocs.io/en/latest/salmon.html |
| Other | ||
Data and Code Availability
RNA sequencing data sets of murine HSC/pre-LSC were deposited (GSE164838). Publicly available gene expression data sets of a large MDS cohort (GSE19429) were used for the analysis.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
Human HL-60 cells were cultured in IMDM medium supplemented with 20% Fetal Bovine Serum (FBS) and 1% penicillin streptomycin. FUW-MDMX-IRES-Puro or empty vector (FUW-IRES-Puro) was transduced into HL-60 as previously described (Ito et al., 2019) to generate MDMX-overexpressing HL-60 and control. Human OCI-AML3 cells were cultured in αMEM medium supplemented with 20% FBS and 1% penicillin streptomycin. Mouse 32D leukemia cells were cultured in IMDM medium supplemented with 10% Fetal Bovine Serum (FBS), 10% WEHI conditioned media (including murine IL3) and 1% penicillin streptomycin. FUW-Mdmx-IRES-Puro, pCAD-Mdmx-IRES-GFP, MSCV-Csnk1a1-IRES-GFP or empty vectors were transduced into 32D as previously described (Kawahara et al., 2012) to generate MDMX and/or CSNK1A1 (CK1α) overexpressing 32D and controls. FUW-MDMX-IRES-Puro or empty vectors were transduced into HL-60 to generate MDMX overexpressed HL-60.
Animal Studies
All animal experiments were performed in compliance with institutional guidelines and approved by the Animal Institute Committee of the Albert Einstein College of Medicine (#00001099). The origin of murine models used in the study are detailed in the Key Resources Table. For competitive transplantation assays and Nras-G12D transplantation assays, Mdmx-Tg were backcrossed to C57BL/6J at least 8 generations. For other analyses, mice were used in Sv129 and C57BL/6J mixed background. Mdmx-Tg were bred with PU.1 URE+/− or Flt3ITD/ITD, for generation of generated URE−/−;Mdmx-Tg and Flt3WT/ITD;Mdmx-Tg animals with controls. 10 to 14-week-old male mice were used for the experiments otherwise specified.
Clinical Samples
Bone marrow samples from patients with MDS were obtained after written informed consent, from Montefiore Medical Center / Albert Einstein Cancer Center (IRB# 11–02-060E).
METHOD DETAILS
Analyses of Murine Models
Peripheral blood was counted using FORCYTE or Genesis Hematology System (Oxford Science) instruments. Spleen and thymus were weighed, and cells were dissociated on 70μm filters. BM cells were collected by crushing bones (tibia, femur, iliac, sternum and vertebrae). 1×105 cells of spleen and BM were used for cytospin, and cells were stained by standard Wright-Giemsa staining. For further analysis of nuclear cells, red blood cells were lysed with lysis buffer (150mM NH4Cl, 1mM KHCO3 and 0.1mM EDTA).
Flow Cytometry (FCM)
1×106 nuclear cells of spleen and thymus or 5×106 nuclear cells of BM were stained (see Table S4) for FCM analysis using FACS Aria2 (BD Biosciences). Definition of HSC/HSPC fractions was described previously (Kondo et al., 1997; Mayle et al., 2013; Pietras et al., 2015). Thymus cells were analyzed as previously described (Tremblay et al., 2010).
In Vivo BrdU Assays
BrdU assay was performed as previously described with minor modifications (Liu et al., 2009). Briefly, BrdU was administered by mixing into drinking water (1mg/ml). After 48 hours, mice were sacrificed and BM cells were stained by biotin-conjugated lineage markers (see Table S5). 2.5×105 Lineage negative cells were selected by Magnetic-activated cell sorting system (MACS; Miltenyi Biotec), and stained for additional FCM antibodies (see Table S5). Cells were fixed, permeabilized and stained with FITC-conjugated anti-BrdU antibody and 7AAD (BD Pharmingen), and analyzed using a FACS Aria II instrument.
In Vitro Colony Forming Assays
Bulk BM cells or sorted HSPCs/HSCs/pre-LSCs were spread in 1mL cytokine-containing methylcellulose media (HSC007 for mouse, HSC003 for human, R&D systems), and plated in 35mm culture dish. For murine cells, colonies were counted and replated after 7 days.
Competitive Transplantation Assays
1×106 donor cells (Ly45.2) and 1×106 WT competitors (Ly45.1/2 double positive) were transplanted into lethally irradiated (12Gy split dose) recipients (Ly45.1). 20μl of peripheral blood was collected monthly, lysed with lysis buffer (150mM NH4Cl, 1mM KHCO3 and 0.1mM EDTA) and stained (see Table S5) to measure ratio of Ly45.2 using a FACS Aria II. Also, p53−/− and p53−/−;Mdmx-Tg recipients were sacrificed 4 months after transplantation, and BM cells were collected. 5×106 cells were stained (see Table S4) to measure the ratio of CD45.2 using a FACS Aria II instrument. For homing assay, ratio of Ly45.2 in BM was measured at 1 week from transplantation.
Transplantation Assay of Preleukemic/Leukemic BM Cells
1×106 BM cells/recipient from preleukemic/leukemic URE−/− or URE−/−;Mdmx-Tg mice were transplanted into sublethally (2Gy) irradiated NSG mice. 1×105 sorted GFP+ BM cells from Nras-G12D leukemic recipients were serially transplanted into lethally (12Gy split dose) irradiated recipients (Ly45.1).
Spectral karyotyping (SKY)
Lineage-negative cKit-positive BM cells were sorted from Flt3WT/ITD;Mdmx-Tg AML mice, and cultured overnight. Cells were stained and analyzed for mitotic phase cells as previously described (Montagna et al., 2003; Padilla-Nash et al., 2006).
RNA sequencing (RNA-seq)
1×104 HSCs (WT and Mdmx-Tg) or pre-LSCs (URE−/− and URE−/−;Mdmx-Tg) were sorted using a FACS Aria II instrument (see Table S4). Total RNA was isolated using RNeasy Micro Kit. Total RNA was subjected to mRNA enrichment, fragmentation, cDNA synthesis, adapter ligation, and PCR amplification for library construction, with incorporation of the DNA nanoball technology as previously described (Zhu et al., 2018). RNA-seq was performed by BGI Americas using the BGISEQ-500 platform, paired-end 100-bp read length (Xu et al., 2019). Cleaned and trimmed reads were analyzed by FastQC for base sequence quality, GC content, N content, and sequence duplication levels. Sequencing reads passing quality control criteria were aligned to the Mus Musculus (mm10/GRCm38) transcriptome and quantified using Salmon (Patro et al., 2017). Raw counts were normalized and analyzed for differential expression in R using the Bioconductor package DESeq2 (Love et al., 2014). For GSEA analysis, pre-ranked gene lists were generated using the negative logarithm of the adjusted p-value multiplied by the sign of the fold change for each gene (equation 1), and input into GSEA Preranked to calculate the enrichment score for each gene set (Subramanian et al., 2005). Pre-ranked gene lists were queried against standard MSigDB gene sets, including hallmark and c1–7 curated gene sets, as well as selected p53 and β-catenin gene lists. In parallel, DESeq2 results were input into IPA for further pathway enrichment analysis (https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis).
| Equation 1: |
Immunofluorescence (IF) staining
Sorted primary Flk2- LSK cells were plated on retronectin coated coverslips and incubated at 4 degree for 30 min prior to fixation. After microscopic confirmation of cell attachment, cells were fixed with 3.2% PFA in PBS for 10 min at room temperature followed by permeabilization in 0.2% Triton X-100 in PBS for 10 min at room temperature. Blocking was done in blocking buffer (1% BSA/0.1%TritonX-100/1XPBS) for 30 min at room temperature. Cells were then incubated overnight at 4 degrees with 1:500 dilution of rabbit anti-β-Catenin (Cell Signaling) in blocking buffer. Cells were washed 3 times in PBS and incubated for 2 hours at room temperature in 1:1000 dilution of AF488-conjugated goat anti-rabbit IgG (abcom) in blocking buffer. Slides were then washed 3 times in PBS and mounted using ProLong Diamond Antifade Mountant with DAPI (Invitrogen, P36962). Cells were imaged on a Leica SP8 Upright Confocal Microscope (Leica, with 63X objective). Images were processed and analyzed using Volocity® Quantitation software.
Western Blotting (WB)
Whole cell extracts were prepared using lysis buffer (150mM NaCl, 50mM Tris-Cl, 5mM EDTA, 1% NP-40, 1% Phosphatase inhibitor cocktail, 1X Protease Inhibitor Cocktail, 1mM PMSF, 10% Glycerol). SDS-PAGE was performed with equal amounts of protein per sample and transferred to PVDF membranes for further immunoblotting using primary antibodies (1:5000 for Actin, 1:2000 for LaminB and β-Tublin, and 1:1000 for others; See Key Resource Table) followed by HRP-conjugated secondary antibodies (1:5000; See Key Resource Table). Imaging of western blots was performed using chemiluminescent ECL substrate on a LI-COR Odyssey Fc imager.
Immunoprecipitation and mass-spectrometry (IP and LC-MS/MS)
For MDMX interactome screening, equal numbers of cells were lysed in mild lysis buffer (1XPBS, 1%NP-40, 0.1% TritonX-100, 1% Phosphatase inhibitor cocktail, 1X Protease Inhibitor Cocktail, 1mM PMSF) followed by IP with HA-conjugated magnetic Dynabeads and eluates were submitted for mass-spectrometry analysis to MS Bioworks. Briefly, cells were incubated in lysis buffer with intermittent vortexing on ice for 45 minutes. The lysates were centrifuged at 13,000g for 15 minutes and supernatant was diluted 5 times with 1X PBS to effectively reduce the concentration of NP-40 to 0.2%. Immunoprecipitation was performed using anti-HA or anti-FLAG magnetic beads and bound proteins were eluted using 0.1M glycine pH2.0. Eluate was neutralized by neutralization buffer (1M Tris, pH8.5). Mass spectrometry data was analyzed using Scaffold Proteome software and significantly detected proteins were shortlisted for further analysis.
Subcellular fractionation
Subcellular fractionation was performed using the REAP method as described previously (Nabbi and Riabowol, 2015). Briefly, equal numbers of cells per condition were resuspended in low NP-40 buffer (0.1%NP-40 in 1XPBS, 1mM PMSF, 1% Phosphatase inhibitor cocktail, 1X Protease Inhibitor Cocktail) and rotated at 4°C for 10 minutes followed by centrifugation at 10,000g for 60 seconds. Obtained supernatant was designated as cytoplasmic extract. Pellets were washed with PBS twice to remove residual cytoplasmic contaminants and further sonicated for 5 minutes in buffer (1%NP-40 and 0.1%TritonX-100 in 1XPBS, 1mM PMSF, 1% Phosphatase inhibitor cocktail, 1X Protease Inhibitor Cocktail). Sonicated nuclear pellets were centrifuged at 13,000g for 15 minutes and supernatant was transferred and labelled as nuclear extract.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR)
RNA was isolated from 5.0×105 32D or HL-60 cells using RNeasy Micro Kit. Reverse transcriptase reaction (RT) was performed with 1ug of total RNA using the iScript cDNA Synthesize Kit followed by polymerase chain reaction (qPCR) using Power SYBR Green Master Mix on a ViiA7 Real Time PCR System (ThermoFisher). Expression levels were normalized by GAPDH. Primer sequences are available in Table S5.
Cell Viability Assay (IC50)
For primary pre-LSC model, lineage-negative cKit-positive cells from BM of preleukemic URE−/−;Mdmx-Tg mice were sorted using a FACS-Aria II instrument (BD Biosciences). 2×104 Cells were cultured in 100μl of StemSpan SFEM supplemented with 5% FBS, 1% glutamine, 100μg/ml Primocin, 50ng/ml recombinant mouse SCF, TPO, FLT3-L, IL3 and IL6 with various concentrations of PNU74654 and/or ALRN-6924 for 16 hours. For leukemia model, 1×104 OCI-AML3 cells were cultured as described above (see “Cell Lines”) with various concentrations of drugs for 24 hours. All cells were cultured in 96-well flat bottom culture dishes by triplicate for each condition. After culture, 20ul of CellTiter-Blue Viability Assay was added, and resazurin fluorescence (560EX/590EM) was measured using an FLUOstar Omega instrument (BMG Labtech). IC50 and combination-index (CI) were calculated by CompuSym software (Chou, 2006).
Plasmid Construction
For cloning of human MDMX, we purchased pCMV3-MDM4(MDMX)-Flag and the insert was amplified with primers harboring BamH1 sites at 5’-UTR and 3’UTR (see Table S5). The amplified DNA was cloned into the FUW-IRES-Puro (Ito et al., 2019) lentiviral vector using the BamH1 site. For murine Mdmx, we amplified the insert of a murine Mdmx target vector (Xiong et al., 2010) using primers harboring EcoR1 sites at 5’-UTR and 3’UTR (see Table S5). The amplified DNA was sequenced for validation and cloned into pCAD-IRES-GFP lentiviral vector (Kawahara et al., 2012), and an HA-tag was added using Phusion Site-Directed Mutagenesis Kit. In addition, HA-tagged murine Mdmx was cloned into the FUW-IRES-Puro lentiviral vector using EcoR1 site. Other plasmids are detailed in the Key Resources Table.
Virus Production and Transduction
15μg Lentiviral/retroviral vectors were transfected to 293T cells spread in 100mm culture dish with helpers (7.5μg PAX2 and 2.5μg pMD2.G for lentivirus, 7.5μg Phi-Eco for retrovirus) using Polyethyleneimine as previously described (Reed et al., 2006). Virus-containing supernatant was collected and filtered at 48 hours and 72 hours from transfection. For cell lines, virus was transduced into cells by centrifuging at 1000g, 37 Celsius, 1 hour with 10μg/ml polybrene. For primary cells, virus was attached on retronectin-coated 24 well dishes by centrifuging 1000g, 37 Celsius, 2 hours and cells were centrifuged at 1000g on virus-attached dish for 1 hour.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses except for Figure 8 were performed by GraphPad PRISM 8. Student t-test or Wilcoxon-test were used for the comparisons of the means of two groups. The Tukey HSD-test was used for the comparisons of the means of more than two groups. The Log-rank test was used for the survival curve analyses. N represents biological replicates in cell culture experiments and the number of mice in animal studies. For Figure 8, R fundamentals and package “cmprsk” were used.
Supplementary Material
RNA sequencing data of WT vs Mdmx-Tg. Related to Figure 5.
RNA sequencing data of URE−/− vs URE−/−;Mdmx-Tg. Related to Figure 5.
Highlights.
MDMX overexpression induces preleukemic-to-AML transition in multiple mouse models.
MDMX causes preleukemic-to-AML transition by p53-independent activation of β-Catenin.
MDMX binds to CK1α and prevents CK1α−dependent degradation of β-Catenin.
MDMX overexpression correlates with progression to AML in patients with MDS.
Acknowledgments
We thank M. Dawlaty at the Albert Einstein College of Medicine for sharing FUW-IRES-puro lentiviral vector, K. Gritsman at the Albert Einstein college of Medicine for sharing Phi-Eco retroviral helper vector, R. Levine at Memorial Sloan Kettering Cancer Center for sharing MSCV-NRAS-G12D-IRES-GFP retroviral vector, S. Martinez-Hoyer at Erasmus Medical Centre and B. Ebert at Dana-Farber Cancer Institute for sharing MSCV-Csnk1a1-IRES-GFP retroviral vector. Expert technical supported was provided by the Stem Cell Isolation and Xenotransplantation Core Facility (funded by NYSTEM grant #C029154) of the Gottesman Institute for Stem Cell and Regenerative Medicine Research, Analytical Imaging Facility (Andrea Briceno) and Genetics core (David Raynolds) of the Albert Einstein College of Medicine. This work was supported by Jane A. and Myles P. Dempsey, NIH grants (U.S.: R01CA217092, R01HL150832; A.V.: R01HL139487, R01HL150832; G.L.: CA47296), as well as the Albert Einstein Cancer Center Support grant (P30CA013330), the Taub Foundation Grants Program for MDS Research (to U.S.), EvansMDS (to U.S. and A.V.), the V Foundation for Cancer Research (to U.S. and A.V.), the Albert Einstein Medical Scientist Training Program (J.C.W, and E.S.; T32GM007288), the Albert Einstein Training Program in Stem Cell Biology (T.I.T.; NYSTEM #N14I-006), a Ruth L. Kirschstein National Research Service Award (J.C.W.; F30GM122308), an NYSCF Druckenmiller fellowship (S.J.T.), and postdoctoral fellowships of the Leukemia & Lymphoma Society (R.K., and S.J.T.). U.S. is the Diane and Arthur B. Belfer Scholar in Cancer Research of the Albert Einstein College of Medicine.
Footnotes
Declaration of interests
L.A.C. has been a past employee of Aileron Therapeutics. J.C. is currently an employee of Stelexis Therapeutics. ALRN-6924 was provided to U.S. from Aileron Therapeutics. U.S. has received research funding from GlaxoSmithKline, Bayer Healthcare, Aileron Therapeutics, and Novartis; has received compensation for consultancy services and for serving on scientific advisory boards from GlaxoSmithKline, Bayer Healthcare, Celgene, Aileron Therapeutics, Stelexis Therapeutics, and Pieris Pharmaceuticals; and has equity ownership in and is serving on the board of directors of Stelexis Therapeutics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bacher U, Haferlach T, Schoch C, Kern W. & Schnittger S. (2006). Implications of NRAS mutations in AML: a study of 2502 patients. Blood. 107, 3847–3853. [DOI] [PubMed] [Google Scholar]
- Bereshchenko O, Mancini E, Moore S, Bilbao D, Mansson R, Luc S, Grover A, Jacobsen SE, Bryder D. & Nerlov C. (2009). Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPalpha mutant AML. Cancer Cell. 16, 390–400. [DOI] [PubMed] [Google Scholar]
- Bhagat TD, Chen S, Bartenstein M, Barlowe AT, Von Ahrens D, Choudhary GS, Tivnan P, Amin E, Marcondes AM, Sanders MA, et al. (2017). Epigenetically Aberrant Stroma in MDS Propagates Disease via Wnt/beta-Catenin Activation. Cancer Res. 77, 4846–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, et al. (2012). Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 44, 1179–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N., Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ Jr., Laird PW, et al. (2013). Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 368, 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras Puigvert J, Von Stechow L, Siddappa R, Pines A, Bahjat M, Haazen LC, Olsen JV, Vrieling H, Meerman JH, Mullenders LH, et al. (2013). Systems biology approach identifies the kinase Csnk1a1 as a regulator of the DNA damage response in embryonic stem cells. Sci Signal. 6, ra5. [DOI] [PubMed] [Google Scholar]
- Carvajal LA, Neriah DB, Senecal A, Benard L, Thiruthuvanathan V, Yatsenko T, Narayanagari SR, Wheat JC, Todorova TI, Mitchell K, et al. (2018). Dual inhibition of MDMX and MDM2 as a therapeutic strategy in leukemia. Sci Transl Med. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kao YR, Sun D, Todorova TI, Reynolds D, Narayanagari SR, Montagna C, Will B, Verma A. & Steidl U. (2019). Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat Med. 25, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li C, Pan Y. & Chen J. (2005). Regulation of p53-MDMX interaction by casein kinase 1 alpha. Mol Cell Biol. 25, 6509–6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC (2006). Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 58, 621–681. [DOI] [PubMed] [Google Scholar]
- Cobas M, Wilson A, Ernst B, Mancini SJ, Macdonald HR, Kemler R. & Radtke F. (2004). Beta-catenin is dispensable for hematopoiesis and lymphopoiesis. J Exp Med. 199, 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewaele M, Tabaglio T, Willekens K, Bezzi M, Teo SX, Low DH, Koh CM, Rambow F, Fiers M, Rogiers A, et al. (2016). Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest. 126, 68–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumble M, Moore L, Chambers SM, Geiger H, Van Zant G, Goodell MA & Donehower LA (2007). The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood. 109, 1736–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenaux P, Preudhomme C, Quiquandon I, Jonveaux P, Lai JL, Vanrumbeke M, Loucheux-Lefebvre MH, Bauters F, Berger R. & Kerckaert JP (1992). Mutations of the P53 gene in acute myeloid leukaemia. Br J Haematol. 80, 178–183. [DOI] [PubMed] [Google Scholar]
- Fiskus W, Sharma S, Saha S, Shah B, Devaraj SG, Sun B, Horrigan S, Leveque C, Zu Y, Iyer S, et al. (2015). Pre-clinical efficacy of combined therapy with novel beta-catenin antagonist BC2059 and histone deacetylase inhibitor against AML cells. Leukemia. 29, 1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. (2014). Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 371, 2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland DG & Tallman MS (2002). Focus on acute leukemias. Cancer Cell. 1, 417–420. [DOI] [PubMed] [Google Scholar]
- Gruszka AM, Valli D. & Alcalay M. (2019). Wnt Signalling in Acute Myeloid Leukaemia. Cells. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Medeiros LJ, Zhang YH, You MJ, Andreeff M, Konopleva M. & Bueso-Ramos CE (2016). High Expression of Human Homologue of Murine Double Minute 4 and the Short Splicing Variant, HDM4-S, in Bone Marrow in Patients With Acute Myeloid Leukemia or Myelodysplastic Syndrome. Clin Lymphoma Myeloma Leuk. 16 Suppl, S30–38. [DOI] [PubMed] [Google Scholar]
- Heidel FH, Bullinger L, Feng Z, Wang Z, Neff TA, Stein L, Kalaitzidis D, Lane SW & Armstrong SA (2012). Genetic and pharmacologic inhibition of beta-catenin targets imatinib-resistant leukemia stem cells in CML. Cell Stem Cell. 10, 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou HA, Chou WC, Kuo YY, Liu CY, Lin LI, Tseng MH, Chiang YC, Liu MC, Liu CW, Tang JL, et al. (2015). TP53 mutations in de novo acute myeloid leukemia patients: longitudinal follow-ups show the mutation is stable during disease evolution. Blood Cancer J. 5, e331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Lee J, Chrysanthou S, Zhao Y, Josephs K, Sato H, Teruya-Feldstein J, Zheng D, Dawlaty MM & Ito K. (2019). Non-catalytic Roles of Tet2 Are Essential to Regulate Hematopoietic Stem and Progenitor Cell Homeostasis. Cell Rep. 28, 2480–2490 e2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. (2014). Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 371, 2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR & Majeti R. (2012). Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 4, 149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaras M, Miller PG, Chu LP, Puram RV, Fink EC, Schneider RK, Al-Shahrour F, Pena P, Breyfogle LJ, Hartwell KA, et al. (2014). Csnk1a1 inhibition has p53-dependent therapeutic efficacy in acute myeloid leukemia. J Exp Med. 211, 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Zhang M, Sun J. & Yang X. (2018a). Casein kinase 1alpha: biological mechanisms and theranostic potential. Cell Commun Signal. 16, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Mak PY, Mu H, Tao W, Mak DH, Kornblau S, Zhang Q, Ruvolo P, Burks JK, Zhang W, et al. (2018b). Disruption of Wnt/beta-Catenin Exerts Antileukemia Activity and Synergizes with FLT3 Inhibition in FLT3-Mutant Acute Myeloid Leukemia. Clin Cancer Res. 24, 2417–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadia TM, Jain P, Ravandi F, Garcia-Manero G, Andreef M, Takahashi K, Borthakur G, Jabbour E, Konopleva M, Daver NG, et al. (2016). TP53 mutations in newly diagnosed acute myeloid leukemia: Clinicomolecular characteristics, response to therapy, and outcomes. Cancer. 122, 3484–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, Mclellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, Mcmichael JF, Wyczalkowski MA, et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature. 502, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara M, Pandolfi A, Bartholdy B, Barreyro L, Will B, Roth M, Okoye-Okafor UC, Todorova TI, Figueroa ME, Melnick A, et al. (2012). H2.0-like homeobox regulates early hematopoiesis and promotes acute myeloid leukemia. Cancer Cell. 22, 194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyoi H, Naoe T, Nakano Y, Yokota S, Minami S, Miyawaki S, Asou N, Kuriyama K, Jinnai I, Shimazaki C, et al. (1999). Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 93, 3074–3080. [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB & Rao A. (2011). Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 108, 14566–14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Weissman IL & Akashi K. (1997). Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91, 661–672. [DOI] [PubMed] [Google Scholar]
- Kuo YH, Landrette SF, Heilman SA, Perrat PN, Garrett L, Liu PP, Le Beau MM, Kogan SC & Castilla LH (2006). Cbf beta-SMMHC induces distinct abnormal myeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 9, 57–68. [DOI] [PubMed] [Google Scholar]
- Lavallee VP, Baccelli I, Krosl J, Wilhelm B, Barabe F, Gendron P, Boucher G, Lemieux S, Marinier A, Meloche S, et al. (2015). The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet. 47, 1030–1037. [DOI] [PubMed] [Google Scholar]
- Lee BH, Tothova Z, Levine RL, Anderson K, Buza-Vidas N, Cullen DE, Mcdowell EP, Adelsperger J, Frohling S, Huntly BJ, et al. (2007). FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer Cell. 12, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Orton D, Neitzel LR, Astudillo L, Shen C, Long J, Chen X, Kirkbride KC, Doundoulakis T, Guerra ML, et al. (2017). Differential abundance of CK1alpha provides selectivity for pharmacological CK1alpha activators to target WNT-dependent tumors. Sci Signal. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Hu C, Mei C, Ren Z, Vera JC, Zhuang Z, Jin J. & Tong H. (2014). Sequential combination of decitabine and idarubicin synergistically enhances anti-leukemia effect followed by demethylating Wnt pathway inhibitor promoters and downregulating Wnt pathway nuclear target. J Transl Med. 12, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Garrett-Bakelman FE, Chung SS, Sanders MA, Hricik T, Rapaport F, Patel J, Dillon R, Vijay P, Brown AL, et al. (2016). Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med. 22, 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X. & He X. (2002). Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 108, 837–847. [DOI] [PubMed] [Google Scholar]
- Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T, Kasibhatla S, Schuller AG, Li AG, Cheng D, et al. (2013). Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci U S A. 110, 20224–20229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, et al. (2009). p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 4, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W. & Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle A, Luo M, Jeong M. & Goodell MA (2013). Flow cytometry analysis of murine hematopoietic stem cells. Cytometry A. 83, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuki M, Schwable J, Steur C, Choudhary C, Agrawal S, Sargin B, Steffen B, Matsumura I, Kanakura Y, Bohmer FD, et al. (2003). Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 101, 3164–3173. [DOI] [PubMed] [Google Scholar]
- Montagna C, Lyu MS, Hunter K, Lukes L, Lowther W, Reppert T, Hissong B, Weaver Z. & Ried T. (2003). The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 63, 2179–2187. [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. (2011). Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 20, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA & Vousden KH (2013). p53 mutations in cancer. Nat Cell Biol. 15, 2–8. [DOI] [PubMed] [Google Scholar]
- Nabbi A. & Riabowol K. (2015). Rapid Isolation of Nuclei from Cells In Vitro. Cold Spring Harb Protoc. 2015, 769–772. [DOI] [PubMed] [Google Scholar]
- Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K, Sonoda Y, Fujimoto T. & Misawa S. (1996). Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 10, 1911–1918. [PubMed] [Google Scholar]
- Padilla-Nash HM, Barenboim-Stapleton L, Difilippantonio MJ & Ried T. (2006). Spectral karyotyping analysis of human and mouse chromosomes. Nat Protoc. 1, 3129–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Dohner H. & Campbell PJ (2016). Genomic Classification in Acute Myeloid Leukemia. N Engl J Med. 375, 900–901. [DOI] [PubMed] [Google Scholar]
- Parikh C, Subrahmanyam R. & Ren R. (2007). Oncogenic NRAS, KRAS, and HRAS exhibit different leukemogenic potentials in mice. Cancer Res. 67, 7139–7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peller S. & Rotter V. (2003). TP53 in hematological cancer: low incidence of mutations with significant clinical relevance. Hum Mutat. 21, 277–284. [DOI] [PubMed] [Google Scholar]
- Pietras EM, Reynaud D, Kang YA, Carlin D, Calero-Nieto FJ, Leavitt AD, Stuart JM, Gottgens B. & Passegue E. (2015). Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell. 17, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintas-Cardama A, Hu C, Qutub A, Qiu YH, Zhang X, Post SM, Zhang N, Coombes K. & Kornblau SM (2017). p53 pathway dysfunction is highly prevalent in acute myeloid leukemia independent of TP53 mutational status. Leukemia. 31, 1296–1305. [DOI] [PubMed] [Google Scholar]
- Reed SE, Staley EM, Mayginnes JP, Pintel DJ & Tullis GE (2006). Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J Virol Methods. 138, 85–98. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Wagner K, Kutok JL, Iwasaki H, Le Beau MM, Okuno Y, Akashi K, Fiering S. & Tenen DG (2004). Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 36, 624–630. [DOI] [PubMed] [Google Scholar]
- Sallman DA, Borate U, Cull EH, Donnellan WB, Komrokji RS, Steidl U, Corvez MM, Payton M, Annis DA, Pinchasik D, et al. (2018). Phase 1/1b Study of the Stapled Peptide ALRN-6924, a Dual Inhibitor of MDMX and MDM2, As Monotherapy or in Combination with Cytarabine for the Treatment of Relapsed/Refractory AML and Advanced MDS with TP53 Wild-Type. Blood (meeting abstruct). 132 (Supplement1), 4066. [Google Scholar]
- Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, Tenen DG & Leutz A. (2006). Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nat Immunol. 7, 1037–1047. [DOI] [PubMed] [Google Scholar]
- Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, Medeiros JJF, Rao-Bhatia A, Jaciw-Zurakowsky I, Marke R, et al. (2017). Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 547, 104–108. [DOI] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, et al. (2014). Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 506, 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive JI, Basilico S, Hannah R, Kinston SJ, Calero-Nieto FJ & Gottgens B. (2016). Genome-scale definition of the transcriptional programme associated with compromised PU.1 activity in acute myeloid leukaemia. Leukemia. 30, 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staber PB, Zhang P, Ye M, Welner RS, Levantini E, Di Ruscio A, Ebralidze AK, Bach C, Zhang H, Zhang J, et al. (2014). The Runx-PU.1 pathway preserves normal and AML/ETO9a leukemic stem cells. Blood. 124, 2391–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidl U, Rosenbauer F, Verhaak RG, Gu X, Ebralidze A, Otu HH, Klippel S, Steidl C, Bruns I, Costa DB, et al. (2006). Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet. 38, 1269–1277. [DOI] [PubMed] [Google Scholar]
- Steidl U, Steidl C, Ebralidze A, Chapuy B, Han HJ, Will B, Rosenbauer F, Becker A, Wagner K, Koschmieder S, et al. (2007). A distal single nucleotide polymorphism alters long-range regulation of the PU.1 gene in acute myeloid leukemia. J Clin Invest. 117, 2611–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenen DG (2003). Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 3, 89–101. [DOI] [PubMed] [Google Scholar]
- Tremblay M, Tremblay CS, Herblot S, Aplan PD, Hebert J, Perreault C. & Hoang T. (2010). Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev. 24, 1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trosset JY, Dalvit C, Knapp S, Fasolini M, Veronesi M, Mantegani S, Gianellini LM, Catana C, Sundstrom M, Stouten PF, et al. (2006). Inhibition of protein-protein interactions: the discovery of druglike beta-catenin inhibitors by combining virtual and biophysical screening. Proteins. 64, 60–67. [DOI] [PubMed] [Google Scholar]
- Vangala RK, Heiss-Neumann MS, Rangatia JS, Singh SM, Schoch C, Tenen DG, Hiddemann W. & Behre G. (2003). The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 101, 270–277. [DOI] [PubMed] [Google Scholar]
- Wade M, Li YC & Wahl GM (2013). MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 13, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI & Armstrong SA (2010). The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 327, 1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will B, Vogler TO, Narayanagari S, Bartholdy B, Todorova TI, Da Silva Ferreira M, Chen J, Yu Y, Mayer J, Barreyro L, et al. (2015). Minimal PU.1 reduction induces a preleukemic state and promotes development of acute myeloid leukemia. Nat Med. 21, 1172–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will B, Zhou L, Vogler TO, Ben-Neriah S, Schinke C, Tamari R, Yu Y, Bhagat TD, Bhattacharyya S, Barreyro L, et al. (2012). Stem and progenitor cells in myelodysplastic syndromes show aberrant stage-specific expansion and harbor genetic and epigenetic alterations. Blood. 120, 2076–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Chen L, Becker A, Schonbrunn E. & Chen J. (2012). Casein kinase 1alpha regulates an MDMX intramolecular interaction to stimulate p53 binding. Mol Cell Biol. 32, 4821–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, Mclellan MD, Johnson KJ, Wendl MC, Mcmichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. (2014). Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 20, 1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S, Pant V, Suh YA, Van Pelt CS, Wang Y, Valentin-Vega YA, Post SM & Lozano G. (2010). Spontaneous tumorigenesis in mice overexpressing the p53-negative regulator Mdm4. Cancer Res. 70, 7148–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S, Pant V, Zhang Y, Aryal NK, You MJ, Kusewitt D. & Lozano G. (2017). The p53 inhibitor Mdm4 cooperates with multiple genetic lesions in tumourigenesis. J Pathol. 241, 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lin Z, Tang C, Tang Y, Cai Y, Zhong H, Wang X, Zhang W, Xu C, Wang J, et al. (2019). A new massively parallel nanoball sequencing platform for whole exome research. BMC Bioinformatics. 20, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Ichikawa H, Tagata Y, Katsumoto T, Ohnishi K, Akao Y, Naoe T, Pandolfi PP & Kitabayashi I. (2007). PML-retinoic acid receptor alpha inhibits PML IV enhancement of PU.1-induced C/EBPepsilon expression in myeloid differentiation. Mol Cell Biol. 27, 5819–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisig BB, Kulasekararaj AG, Mufti GJ & So CW (2012). SnapShot: Acute myeloid leukemia. Cancer Cell. 22, 698–698 e691. [DOI] [PubMed] [Google Scholar]
- Zhan T, Rindtorff N. & Boutros M. (2017). Wnt signaling in cancer. Oncogene. 36, 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kong G, Rajagopalan A, Lu L, Song J, Hussaini M, Zhang X, Ranheim EA, Liu Y, Wang J, et al. (2017). p53−/− synergizes with enhanced NrasG12D signaling to transform megakaryocyte-erythroid progenitors in acute myeloid leukemia. Blood. 129, 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FY, Chen MX, Ye NH, Qiao WM, Gao B, Law WK, Tian Y, Zhang D, Zhang D, Liu TY, et al. (2018). Comparative performance of the BGISEQ-500 and Illumina HiSeq4000 sequencing platforms for transcriptome analysis in plants. Plant Methods. 14, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA sequencing data of WT vs Mdmx-Tg. Related to Figure 5.
RNA sequencing data of URE−/− vs URE−/−;Mdmx-Tg. Related to Figure 5.
Data Availability Statement
RNA sequencing data sets of murine HSC/pre-LSC were deposited (GSE164838). Publicly available gene expression data sets of a large MDS cohort (GSE19429) were used for the analysis.