Abstract
Topoisomerase I cleavage complexes can be induced by a variety of DNA damages and by the anticancer drug camptothecin. We have developed a ligation-mediated PCR (LM-PCR) assay to analyze replication-mediated DNA double-strand breaks induced by topoisomerase I cleavage complexes in human colon carcinoma HT29 cells at the nucleotide level. We found that conversion of topoisomerase I cleavage complexes into replication-mediated DNA double-strand breaks was only detectable on the leading strand for DNA synthesis, which suggests an asymmetry in the way that topoisomerase I cleavage complexes are metabolized on the two arms of a replication fork. Extension by Taq DNA polymerase was not required for ligation to the LM-PCR primer, indicating that the 3′ DNA ends are extended by DNA polymerase in vivo closely to the 5′ ends of the topoisomerase I cleavage complexes. These findings suggest that the replication-mediated DNA double-strand breaks generated at topoisomerase I cleavage sites are produced by replication runoff. We also found that the 5′ ends of these DNA double-strand breaks are phosphorylated in vivo, which suggests that a DNA 5′ kinase activity acts on the double-strand ends generated by replication runoff. The replication-mediated DNA double-strand breaks were rapidly reversible after cessation of the topoisomerase I cleavage complexes, suggesting the existence of efficient repair pathways for removal of topoisomerase I-DNA covalent adducts in ribosomal DNA.
DNA topoisomerases are ubiquitous enzymes that regulate the topological state of DNA. They participate in essential cellular processes, including replication, transcription, chromosome segregation, and recombination (22, 34, 71). Eukaryotic DNA topoisomerase I (top1) acts as a monomer, and its catalytic activity can be divided into four steps (61): (i) binding of the enzyme to duplex DNA, (ii) single-stranded DNA cleavage by a transesterification reaction in which a top1 tyrosine-hydroxyl group becomes covalently linked to the 3′ phosphate of a DNA phosphodiester bond to generate a 5′-hydroxyl DNA terminus, (iii) DNA relaxation by controlled rotation around the intact DNA strand (61); and (iv) religation of the cleaved DNA by nucleophilic attack from the 5′-hydroxyl DNA end and dissociation of the top1 tyrosyl residue from the 3′ end. The topoisomerase-linked DNA breaks are commonly referred to as cleavage complexes (22, 34, 71). Under physiological conditions, they are short-lived catalytic intermediates.
A number of physiological and environmental DNA modifications can inhibit top1 by inducing top1 cleavage complexes. These include DNA mismatches or abasic sites (37, 48, 73), oxidative base damage (47), base alkylation and carcinogenic adducts (44, 66), UV photoproducts (50, 62), and DNA breaks (11, 45). Trapping of top1 cleavage complexes is also the primary mechanism of action of camptothecin (CPT), a potent anticancer agent which reversibly inhibits the religation step of the top1 catalytic cycle (25, 29, 39, 40). The cytotoxicity of top1 cleavage complexes is attested by the potent cell killing and anticancer activity of CPTs. In both human and yeast cells, cleavage complexes induce DNA damage by interference with DNA replication (16, 24, 26, 55). Studies with simian virus 40-infected cells indicate that top1 cleavage complexes generate double-stranded DNA breakage at replication forks (2, 59, 68). Persistent DNA double-strand breaks have also been detected by pulsed-field gel electrophoresis in replicating DNA of human cells treated with CPT (51, 60).
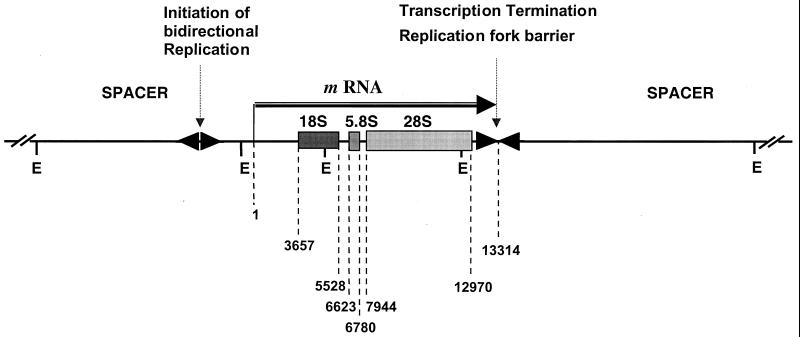
The aim of the present study was to analyze top1-linked DNA double-strand breaks at the molecular level in human cells using ligation-mediated PCR (LM-PCR). The rRNA gene cluster was chosen for this analysis for the following reasons. First, it contains a high frequency of top1 cleavage sites (14, 41, 75). Second, immunolocalization studies show that top1 is concentrated in nucleoli (3, 7, 15, 20, 31, 33). Third, each 13-kb transcribed region of the human rRNA gene complex (Fig. 1) is tandemly repeated about 40 times on each of the five acrocentric chromosomes (21, 65). Fourth, ribosomal DNA (rDNA) replication is unidirectional towards the 3′ end of the 28S gene, and rRNA is one of the few human genes whose replication origin and termination are known. Replication fork barriers have been described at the 3′ end of the 28S ribosomal gene region, which prevent fork migration in the opposite direction (19, 23, 28) (Fig. 1). The LM-PCR assay enabled us to examine replication-dependent DNA damage induced by top1 cleavage complexes and to map replication-mediated double-strand breaks at the nucleotide level. Our data suggest that top1 cleavage complexes lead to replication runoff on the leading strand with 5′-end phosphorylation and that these lesions are effectively repaired in rDNA.
FIG. 1.
Map of the human rRNA gene repeat. The human rRNA gene forms a 44-kb repeat unit, with the four segments defined by EcoRI (E) sites. Boxes show the positions of sequences coding for the 18S, 5.8S, and 28S rRNA genes. Spacer, nontranscribed region. The horizontal arrow indicates the transcribed region. Numbers for genomic positions are according to GenBank (accession no. U13369). 1 to 3656, 5′ external spacer; 3657 to 5527, 18S segment; 5528 to 6622, internal spacer I; 6623 to 6779, 5.8S segment; 6780 to 7943, internal spacer II; 7944 to 12969, 28S segment; 12970 to 13314, 3′ external spacer. Replication starts bidirectionally in the nontranscribed intergenic spacer (18, 28, 74). Unidirectional replication fork barriers are located at the 3′ end of the transcribed region (23, 28).
MATERIALS AND METHODS
Drugs, chemicals, and enzymes.
20-(S)-Camptothecin lactone (CPT) was obtained from the Drug Synthesis and Chemistry Branch, Division of Cancer Treatment, National Cancer Institute. The drug was dissolved in dimethyl sulfoxide at a concentration of 10 mM. Further dilutions were made in water immediately before use. Taq DNA polymerase was purchased from Qiagen (Santa Clarita, Calif.). T4 polynucleotide kinase (10 U/μl) was purchased from Gibco-BRL, Life Technologies (Gaithersburg, Md.), and aphidicolin was obtained from Sigma Chemical Co. (St. Louis, Mo.). T4 DNA ligase (4 U/μl) was obtained from New England Biolabs (Beverly, Mass.). [γ-32P]ATP (6,000 Ci/mmol) was purchased from New England Nuclear (Boston, Mass.).
Cell culture and drug treatment.
HT29 cells were provided by the Developmental Therapeutics Program (National Cancer Institute) and grown at 37°C in the presence of 5% CO2 in RPMI 1640 supplemented with 16% heat-inactivated fetal bovine serum (Gibco-BRL, Grand Island, N.Y.), 2 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Exponentially growing cells were, unless otherwise stated, treated with 10 μM CPT for the indicated times. Treatments at 0°C were performed after replacing the culture medium with ice-cold medium and transferring the cells on ice to a cold room. To examine the reversal of CPT-induced top1-linked DNA cleavage, treated cells were washed twice with drug-free ice-cold Hanks' balanced salt solution and allowed to grow in drug-free RPMI 1640 medium for the indicated time.
Genomic DNA isolation.
Following treatment, cells were lysed in 250 mM Tris-HCl–5 mM NaCl–25 mM EDTA–0.5% sodium dodecyl sulfate (pH 8.0). Cell lysates were incubated with DNase-free RNase A (final concentration, 100 μg/ml) for 1 h at 37°C and then with proteinase K (800 μg/ml) at 37°C for 3 h. The DNA was isolated by two phenol-chloroform extractions followed by one chloroform extraction. After ethanol precipitation and washing of the pellet with 75% ethanol, the DNA was dissolved in TE buffer (1 mM EDTA, 10 mM Tris-HCl [pH 7.6]).
Oligonucleotide primers.
Table 1 shows the sequences of the oligonucleotide primers used to map top1-linked DNA single- and double-strand breaks in the 18S (primers RA, RC, and RD) and 28S (primers RF and RG) segments. Oligonucleotide primers 1 were only used to sequence top1-induced DNA single-strand breaks. Primers 2 were the PCR primers, and primers 3 were used in the labeling step of the LM-PCR (see Fig. 2).
TABLE 1.
Oligonucleotides used in the present study
| Primer | 5′-end coordinatea | DNA sequence | Annealing temp (°C) |
|---|---|---|---|
| RA-1 | 5030 | 5′-CGATAACGAACGAGACTCTGG | 60.0 |
| RA-2 | 5049 | 5′-GGCATGCTAACTAGTTACGCGACCCC | 72.0 |
| RA-3 | 5059 | 5′-CTAGTTACGCGACCCCCGAGCG | 70.0 |
| RC-1 | 5541 | 5′-CCTCCGGGCTCCGTTAATG | 56.0 |
| RC-2 | 5523 | 5′-GATCCTTCCGCAGGTTCACC | 64.0 |
| RC-3 | 5517 | 5′-TCCGCAGGTTCACCTACGGAAACC | 69.0 |
| RD-1 | 5294 | 5′-CTCGTTCATGGGGAATAATTG | 54.0 |
| RD-2 | 5275 | 5′-TGCAATCCCCGATCCCCATCA | 64.0 |
| RD-3 | 5270 | 5′-TCCCCGATCCCCATCACGAATGG | 64.0 |
| RF-1 | 12064 | 5′-CGCTCCGGGGACAGTGCCAG | 62.0 |
| RF-2 | 12083 | 5′-GGTGGGGAGTTTGACTGGGGCGGTAC | 72.0 |
| RF-3 | 12091 | 5′-GTTTGACTGGGGCGGTACACCTGTC | 72.0 |
| RG-1 | 12642 | 5′-GGCGCTGCCGTATCGTTCGC | 62.0 |
| RG-2 | 12623 | 5′-CCTGGGCGGGATTCTGACTTAGAGG | 72.0 |
| RG-3 | 12616 | 5′-GGGATTCTGACTTAGAGGCGTTCAGTC | 72.0 |
| Linker | Linker | 5′-GCGGTGACCCGGGAGATCTGAATTC CTAGACTTAAG-5′ | 72.0 |
From GenBank accession no. U13369.
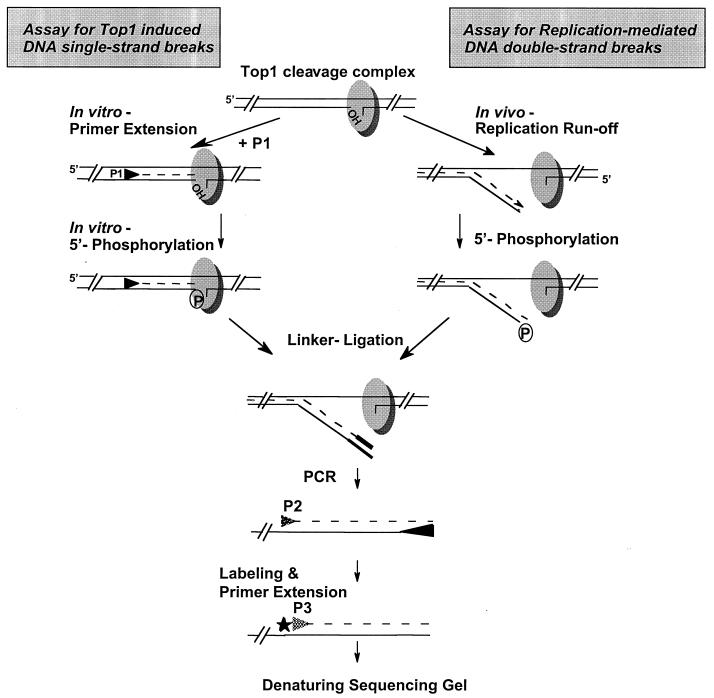
FIG. 2.
Diagram of the LM-PCR protocol to detect top1-induced DNA single-strand breaks and replication-mediated DNA double-strand breaks. Top1 is shown as a shaded oval with covalent linkage to the 3′ end of a DNA single-strand break. In the assay for top1-induced DNA single-strand breaks, top1-induced DNA single-strand breaks (i.e., top1 cleavage complexes) were detected (upper left) as described previously (41) by annealing primer 1 (P1) to denatured genomic DNA. After primer extension and in vitro phosphorylation of the 5′-OH termini with T4 polynucleotide kinase, ligation to the double-stranded linker was performed. Thereafter, rRNA gene-specific DNA fragments were amplified with Taq DNA polymerase using the linker-primer and a nested, gene-specific PCR primer. After 26 cycles of PCR, a third primer (5′ end labeled with 32P; star) was used for two primer extension cycles before the samples were separated in 7% denaturing polyacrylamide gels. In the assay for replication-mediated DNA double-strand breaks, collision between a replication fork and a top1 cleavage complex is proposed to lead to replication runoff, with generation of a DNA double-strand break (upper right). Because of in vivo 5′-end phosphorylation of replication-mediated DNA double-strand breaks, ligation to the linker could be performed without prior T4 polynucleotide kinase reaction. The following reaction steps were the same as for the detection of top1-induced DNA single-strand breaks. Note that the single-strand break assay detects both single- and double-strand breaks.
LM-PCR.
Figure 2 shows the reaction steps for the detection of replication-mediated DNA double-strand breaks. Genomic DNA (0.5 μg) from CPT-treated cells was first incubated with Taq DNA polymerase and deoxynucleoside triphosphates (dNTPs) to allow 3′-end filling. Then, a T4 polynucleotide kinase reaction was performed before ligation to the duplex oligonucleotide linker, consisting of a 26-mer annealed to an 11-mer oligonucleotide (Table 1) (32, 41). Ligation was performed overnight at 14°C. After precipitation of the DNA, rRNA gene-specific DNA fragments were amplified with Taq DNA polymerase using the 26-mer strand from the linker (Table 1) and a nested, gene-specific PCR primer (primers 2). After 26 cycles of PCR, a third internal primer (primers 3, 5′ end labeled with 32P using T4 polynucleotide kinase) was used for two cycles. Samples were extracted once with phenol-chloroform, and the amplified fragments were precipitated with ethanol and separated on 7% denaturing polyacrylamide gels. Samples were run at 60 W for 90 min. After drying, gels were exposed to a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Top1 cleavage complexes (i.e., top1-induced DNA single-strand breaks) were detected as described previously (41). Briefly, 0.5 μg of genomic DNA was denatured for 5 min, and oligonucleotide primers 1 were annealed for 30 min (annealing temperatures are given in Table 1). Primer extension was then carried out at 72°C with Taq DNA polymerase. After phosphorylation of the 5′-OH termini using T4 polynucleotide kinase and ligation to the linker, reaction steps were the same as for the detection of replication-mediated DNA double-strand breaks. Chemical DNA sequencing reactions (30) performed with genomic DNA from HT29 cells were used to provide position markers with all LM-PCRs. The genomic positions of the cleavage sites presented in the figures correspond to the nucleotide covalently linked to top1.
RESULTS
DNA double-strand breaks in the rRNA gene cluster in CPT-treated cells are dependent on DNA replication.
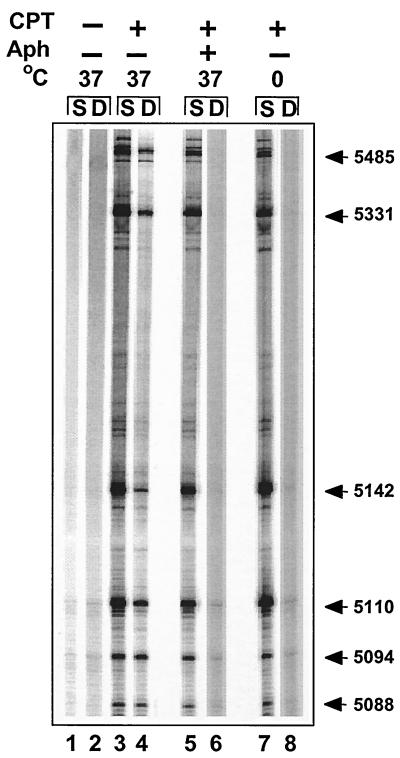
Figure 3 shows that top1-induced DNA double-strand breaks were detectable by LM-PCR in the rRNA gene from CPT-treated cells (lane 4). Previous studies suggested that inhibition of DNA replication with the DNA polymerase inhibitor aphidicolin prevents the conversion of DNA single-strand breaks into replication-mediated double-strand breaks (24, 26, 51). As expected, DNA double-strand breaks were not detectable in aphidicolin-treated cells (Fig. 3, lane 6), whereas aphidicolin had no detectable effect on top1-induced DNA single-strand breaks (Fig. 3, lanes 3, 5, and 7) (24). Conversion of top1 cleavage complexes into DNA double-strand breaks was also prevented when cells were treated at 0°C, conditions under which the top1 cleavage complexes (single-strand breaks) still form (13) (compare lanes 7 and 8). These results indicate that the top1-induced DNA double-strand breaks detected by LM-PCR are dependent on DNA replication.
FIG. 3.
Replication-mediated DNA double-strand breaks on the leading strand of the 18S human rRNA gene are prevented by the DNA synthesis inhibitor aphidicolin or treatment at 0°C. DNA single-strand breaks (S) and double-strand breaks (D) were determined in untreated HT29 cells (lanes 1 and 2) or after 1 h of treatment with CPT alone (lanes 3 and 4) or in combination with aphidicolin (Aph; 10 μM, 5-min pretreatment and 1-h cotreatment with CPT; lanes 5 and 6) or after treatment with CPT for 1 h on ice (lanes 7 and 8). Numbers correspond to genomic positions of the DNA lesions (GenBank accession no. U13369).
Replication-mediated DNA double-strand breaks are 5′ phosphorylated and coincide with top1-induced single-strand breaks.
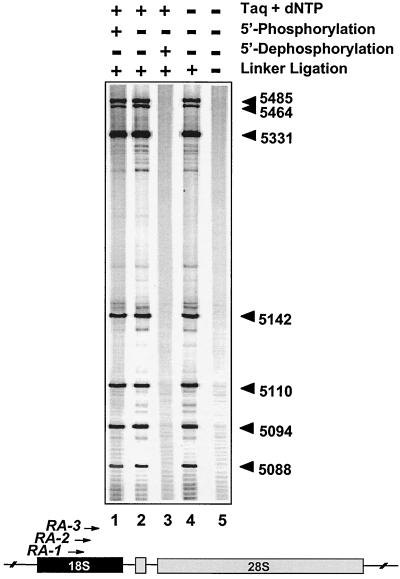
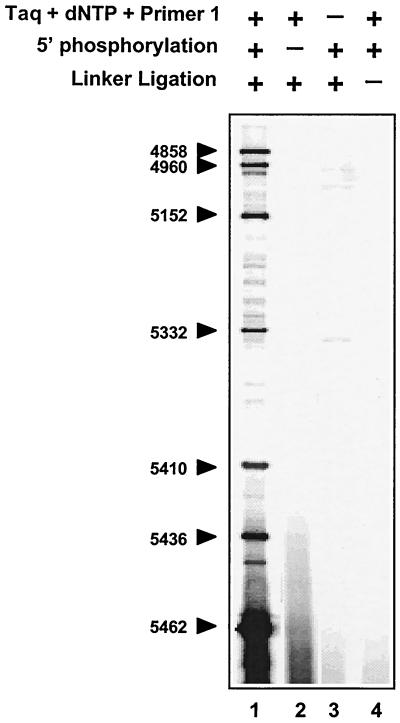
We next investigated the biochemical characteristics of the 5′ ends of the replication-mediated DNA double-strand breaks (Fig. 4). Even when T4 polynucleotide kinase was omitted (lane 2), strong signals that could be mapped to the same nucleotide positions as in the complete reaction (lane 1) were found. Conversely, we found that 5′ dephosphorylation of genomic DNA from CPT-treated cells with shrimp alkaline phosphatase prevented detection of the replication-mediated double-strand breaks (lane 3), suggesting that no ligation to the linker occurred. These results indicate that the replication-mediated DNA double-strand breaks are 5′ phosphorylated in vivo. Because 3′-end filling with Taq DNA polymerase (lane 4) was dispensable for ligation to the double-stranded linker and detection of the replication-mediated double-strand breaks, it appears that the leading strand is replicated up to the last nucleotide at the 5′ end of the top1 cleavage complex, suggesting replication runoff at top1 cleavage complexes (see Fig. 2 and 9).
FIG. 4.
Replication-mediated DNA double-strand breaks are 5′-phosphorylated and coincide with top1-induced single-strand breaks. HT29 cells were exposed to CPT for 4 h. Experiments were performed with the RA primers (Table 1). Lane 1, complete reaction (see Fig. 2); lanes 2 to 5, 5′ phosphorylation of genomic DNA with T4 polynucleotide kinase omitted; lane 3, 5′ dephosphorylation with shrimp alkaline phosphatase (USB); lanes 4 and 5, incubation with Taq DNA polymerase and dNTPs omitted; lane 5, linker ligation omitted. Numbers correspond to genomic positions of the DNA lesions (GenBank accession no. U13369).
FIG. 9.
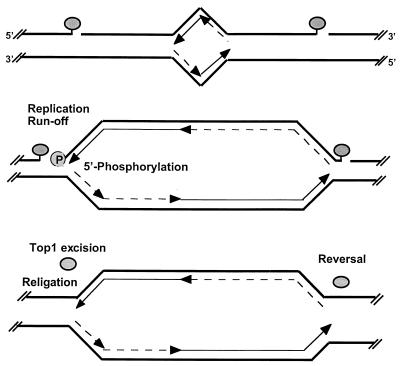
Proposed interactions of DNA replication forks with CPT-stabilized top1 cleavage complexes and hypothetical repair pathways. Two covalent top1 cleavage complexes (shaded ovals) are shown, one on each side of a growing replication bubble (top). Parental DNA strands are represented as thick lines. Leading-strand synthesis is shown as thin arrows, and Okazaki fragments are shown as broken-line arrows. The differential effect of replication fork collision into top1 cleavage complexes on the leading and lagging strands is shown in the middle panel. Our results suggest that replication-mediated DNA double-strand breaks are formed by replication fork runoff on the leading strand with phosphorylation (P) of the 5′ end of the DNA template strand. By contrast, replication-mediated DNA double-strand breaks are not detectable on the lagging strand, which suggests that the replication fork is arrested upstream from the top1 cleavage complex without bypass, that the replication complex forces the dissociation of the top1 cleavage complex, or that Okasaki fragment synthesis bypasses the top1-mediated single-strand break interruption (see Discussion). In any case, no replication runoff would occur on the replicating lagging strand. (Bottom) Hypothetical excision repair of top1 cleavage complexes on the leading strand (see text for details and references). On the lagging strand, top1 might religate the DNA template strand directly upon drug removal. Replication-mediated DNA double-strand breaks are potential targets for homologous and illegitimate recombination.
The nucleotide level resolution of the LM-PCR enabled us to further compare the distribution and kinetics of the top1 cleavage complexes (i.e., the DNA single-strand breaks) and the replication-mediated DNA double-strand breaks. Top1 cleavage complexes (i.e., DNA single-strand breaks) appeared within 30 min of exposure to CPT (Fig. 5, lanes 0 in left panel, and data not shown), which is consistent with quantitation of top1 cleavage complexes at the overall genome level (13). Replication-mediated DNA double-strand breaks were also detectable within 30 min of CPT exposure, and they tended to increase in intensity from 2 to 4 h of ongoing CPT treatment (data not shown), which is consistent with the requirement for replication fork collision (replication runoff) for the generation of the DNA double-strand breaks.
FIG. 5.
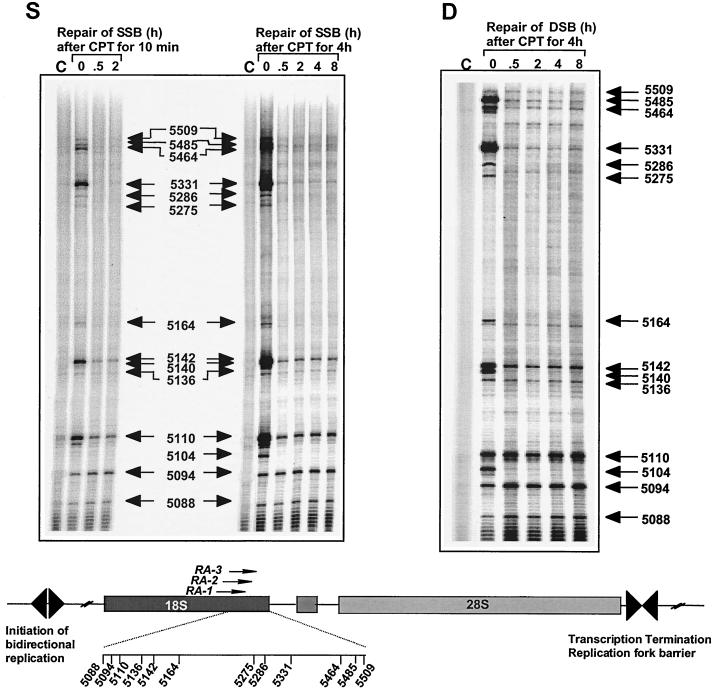
Repair of replication-mediated DNA double-strand breaks (panel D) induced by top1 cleavage complexes (panel S) on the leading strand of the 18S human rRNA gene. HT29 cells were treated with CPT for either 10 min or 4 h. Top1 cleavage complexes (i.e., DNA single-strand breaks [SSB], panel S) and replication-mediated DNA double-strand breaks (DSB, panel D) were studied at the indicated times (in hours after CPT removal). Numbers correspond to genomic positions of the DNA lesions (GenBank accession no. U13369). Lanes C, control DNA from untreated cells.
Reversal kinetics of replication-mediated DNA double-strand breaks suggests the existence of effective mechanisms to remove top1 covalent complexes in rDNA.
Prior studies have suggested that persistent DNA double-strand breaks in replicating DNA determine the cellular response to CPT through either cell cycle arrest or apoptosis (5, 51). The LM-PCR approach enabled us to monitor the site-specific persistence of replication-mediated DNA double-strand breaks. Figure 5 compares the kinetics of disappearance of the top1 cleavage complexes and the replication-mediated DNA double-strand breaks after CPT washout from the tissue culture medium. Most top1 cleavage complexes (panel S) induced by 10-min or 4-h CPT treatments were reversed after 30 min (e.g., at positions 5509, 5485, 5464, 5331, 5286, 5275, 5164, 5140, and 5104).
We next examined whether a relationship existed between the kinetics of disappearance of the top1-induced cleavage complexes (DNA single-strand breaks) and of the replication-mediated DNA double-strand breaks (Fig. 5, panel D). Unexpectedly (51), DNA double-strand breaks disappeared (almost) completely and rapidly at a number of sites (e.g., at positions 5485, 5464, 5331, 5286, 5275, 5164, 5140, and 5104), whereas removal of top1 covalent complexes was incomplete or not detectable at other sites (e.g., at positions 5110, 5094, and 5088). Differential persistence was observed for DNA double-strand breaks very close to each other (e.g., at position 5110 or 5094 versus 5104), indicating that removal of replication-mediated DNA double-strand breaks in this genomic region did not reveal consistent directionality towards either end of the rRNA gene. Differential reversal of top1-linked DNA breaks in vitro has previously been reported to depend on the local DNA sequence (42, 67). It is also possible that top1-induced single-strand breaks located closely and simultaneously on both strands of the DNA duplex (compare Fig. 5 and 6) can lead to the formation of double-strand breaks independent of DNA replication. Such double-strand breaks might be more persistent (72) than the replication-mediated double-strand breaks. Taken together, these results suggest that repair of replication-mediated DNA double-strand breaks can be remarkably fast in rDNA.
FIG. 6.
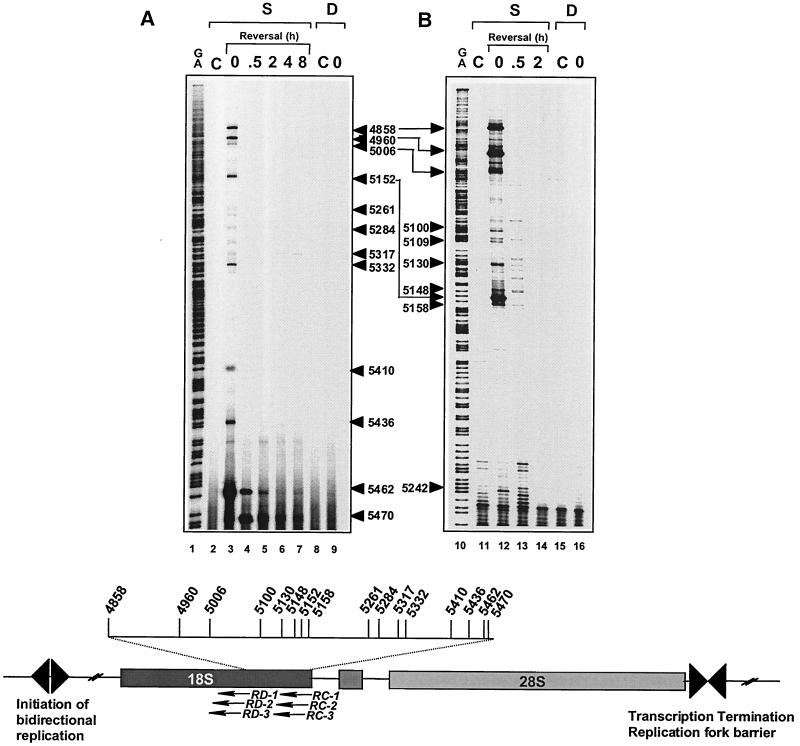
Replication-mediated DNA double-strand breaks are not detectable on the lagging strand of the 18S human rRNA gene. HT29 cells were treated with CPT for 4 h. DNA was extracted immediately after drug treatment at time 0 (lanes 3, 9, 12, and 16) or at various times after CPT removal. Time in drug-free medium (in hours) is indicated above lanes 4 to 7, 13, and 14. Lanes C, control (untreated) cells; lanes S, detection of top1 cleavage complexes (i.e., DNA single-strand breaks); lanes D, detection of replication-mediated DNA double-strand breaks. The scheme at the bottom illustrates the positions of the primers used with the landmarks of the rRNA gene. Data were obtained with primer set RC (panel A) and primer set RD (panel B). Lanes G+A, Maxam-Gilbert sequencing reactions. Numbers correspond to genomic positions of the DNA lesions (GenBank accession no. U13369).
Strand specificity of replication-mediated DNA double-strand breaks suggests absence of replication runoff on the lagging strand.
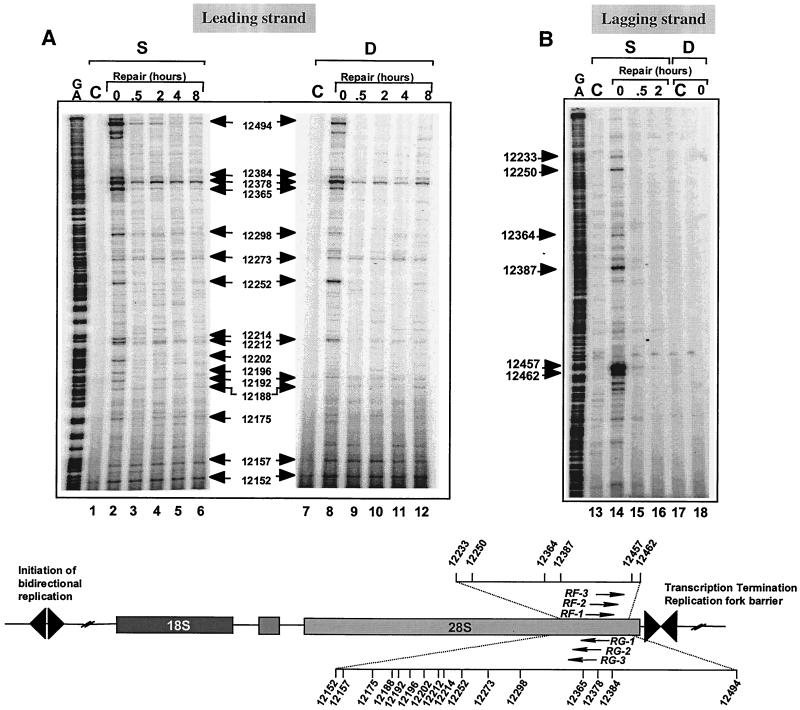
DNA lesions affect replication differently depending on their location on the leading or lagging strand. For example, replication forks are able to bypass UV-induced DNA damage on the lagging strand but not on the leading strand (64). The known directionality of DNA replication in the rRNA genes (19, 23, 28) (Fig. 1) allowed us to examine whether the location of top1 cleavage complexes on the leading or lagging strand affected replication-mediated DNA damage. The data shown in Fig. 3 to 5 demonstrated the formation of replication-mediated double-strand breaks on the leading strand for DNA replication. The experiments shown in Fig. 6 investigated whether Okazaki fragment synthesis could also generate detectable double-strand breaks. Although reversible top1 cleavage complexes (single-strand breaks) were readily apparent (lanes 3 and 12) and were formed independently of DNA replication (in aphidicolin-treated cells; data not shown), replication-mediated DNA double-strand breaks were not detectable on the lagging strand (lanes 9 and 16, Fig. 6A). This observation was confirmed using different sets of oligonucleotide primers (Fig. 6A and B). We next extended these studies to the 28S region of the rRNA gene (Figure 7). Consistent with the observations in the 18S region (Fig. 5), replication-mediated DNA double-strand breaks on the leading strand (Fig. 7A, positions 12494, 12378, and 12252) were readily detectable and coincided with the top1 cleavage complexes. By contrast, on the lagging strand (Fig. 7B), replication-mediated DNA double-strand breaks were not detectable. Thus, it appears that replication fork collision and replication runoff are not detectable on the lagging strand for DNA synthesis (see Fig. 9).
FIG. 7.
Top1 cleavage complexes occur on both strands of the human 28S rRNA gene, whereas replication-mediated DNA double-strand breaks are detectable only on the leading strand. The scheme at the bottom illustrates the positions of the primers used with the landmarks of the rRNA gene. Experiments were performed with primer set RF for the leading DNA strand (panel A) and primer set RG for the lagging strand (Table 1) (panel B). HT29 cells were treated with CPT for 4 h. DNA was extracted immediately after drug treatment at time 0 (lanes 2, 8, 14, and 18) or at the indicated times after CPT removal. Time in drug-free medium is indicated above lanes 3 to 6, 9 to 12, 15, and 16. Lanes C, control (untreated) cells; lanes S, detection of DNA single-strand breaks; lanes D, detection of double-strand breaks; lanes G+A, Maxam-Gilbert sequencing reactions. Numbers correspond to genomic positions of the DNA lesions (GenBank accession no. U13369).
In vivo 5′ phosphorylation is not detectable on the lagging strand for DNA synthesis.
Because in vivo 5′ phosphorylation was associated on the leading strand with the production of DNA double-strand breaks, we analyzed top1 cleavage complexes on the lagging strand for in vivo 5′ phosphorylation (Fig. 8). After denaturation, genomic DNA from CPT-treated cells was annealed to oligonucleotide primers 1 and extended with Taq DNA polymerase and dNTPs. After phosphorylation of the 5′ termini with T4 polynucleotide kinase, the oligonucleotide linker could be ligated, and subsequent PCR allowed sequencing of top1 cleavage sites (Fig. 8, lane 1). However, omission of T4 polynucleotide kinase prevented the detection of top1-induced DNA cleavage (lane 2), suggesting lack of in vivo 5′ phosphorylation on the lagging strand. This observation suggests that 5′ phosphorylation is dependent on prior replication runoff, which is detectable only on the leading strand.
FIG. 8.
In vivo 5′ phosphorylation is not detectable on the lagging strand for DNA replication, suggesting that 5′-end phosphorylation is only detectable at replication-mediated DNA double-strand breaks. HT29 cells were exposed to CPT for 4 h. Experiments were performed with primers RC for the lagging DNA strand. Lane 1, complete reaction as described before (41), using primer 1; lane 2, incubation with T4 polynucleotide kinase omitted; lane 3, annealing and extension of primer 1 omitted; lane 4, linker ligation omitted. Numbers correspond to genomic positions of the DNA lesions (GenBank accession no. U13369).
DISCUSSION
Formation of top1-induced DNA double-strand breaks by replication fork runoff on the leading strand for DNA synthesis.
The present study provides novel insights into the replication-mediated DNA double-strand breaks induced by top1 cleavage complexes, which have been proposed to be the primary cytotoxic DNA lesions produced by this type of covalent protein-DNA adduct (24, 26, 51, 56). Using LM-PCR, we found that in HT29 cells treated with CPT, replication-dependent DNA double-strand breaks are readily detectable on the leading strand of the rRNA genes.
The 5′ end of the replication-mediated DNA double-strand breaks matched, at the nucleotide level, the sites of CPT-stabilized top1 cleavage complexes. This coincidence is remarkable considering that in cleavage complexes, top1 binds not only to the 3′-phosphate end of the cleaved DNA strand but also noncovalently to at least nine nucleotides around the actual cleavage site on the scissile strand and to five additional nucleotides on the noncleaved strand (49, 63). Thus, it would have been conceivable that replication-associated DNA polymerase should have been blocked before reaching the top1 cleavage site. However, this would result in a double-strand break with a 5′ protruding end of at least seven nucleotides (63), which should not be ligated to the blunt-ended double-stranded linker by T4 DNA ligase (12) in the LM-PCR experiments performed in the absence of Taq DNA polymerase (Fig. 4). Alternatively, it would have been conceivable that the 5′ protruding end might have been digested in vivo by an exonuclease, resulting in a blunt end. However, in this case, the 5′ ends of the replication-mediated DNA double-strand breaks should not have matched the 5′ ends of the top1 cleavage complexes. Because we found that the 5′ ends of the replication-mediated DNA double-strand breaks coincided with those of the top1 cleavage complexes, we interpret our results as indicating that top1-induced replication-mediated DNA double-strand breaks result from replication fork runoff at the top1 cleavage complex sites on the leading strand for DNA replication (Fig. 2 and 9).
Top1-induced replication-mediated DNA double-strand breaks are 5′ phosphorylated.
Because the replication-mediated DNA double-strand breaks did not require addition of T4 polynucleotide kinase in the LM-PCRs, we conclude that replication-mediated DNA double-strand breaks resulting from top1 cleavage complexes are 5′ phosphorylated. This finding contrasts with the biochemical properties of the DNA termini in the top1 cleavage complexes, which bear a 5′-hydroxyl and a 3′-phosphotyrosyl top1 covalent linkage (22, 34, 71). The 5′-hydroxyl terminus enables the reversibility of the top1 cleavage complexes. Thus, 5′ phosphorylation should prevent religation by top1 itself (11, 45). Because our data indicate that many of the replication-mediated DNA double-strand breaks are readily reversible in rDNA, it appears that the 3′ termini, including the covalently linked top1 protein, are processed or repaired in vivo. The simplest scheme would involve excision of the top1 protein with or without covalently linked nucleotides (Fig. 9). Recently, a eukaryotic tyrosyl-DNA-phosphodiesterase has been identified that specifically hydrolyzes the chemical bond between the active-site tyrosine of top1 and the 3′ end of DNA (43). Alternatively, nucleotide excision repair might excise the top1-DNA conjugates by incising the DNA strand 5′ from the top1 cleavage complex (53). The repair of top1 cleavage complexes remains, however, generally poorly defined (40). Based on the hypersensitivity of Rad52-deficient yeast strains to CPT (17), recombination repair has been invoked. Replication-mediated DNA double-strand breaks that are no longer tethered by proteins could become potential targets for illegitimate recombination (10, 11, 35, 52, 54, 69). Ligation between nascent and parental strands at the arrested replication fork might also be mediated by top1 itself, since purified top1 is able to ligate heterologous DNA in vitro (8, 9, 11, 36, 57, 58). Recombination repair involving nascent and parental strands is consistent with the induction of sister chromatid exchanges in CPT-treated cells (1).
Stabilized top1 cleavage complexes do not lead to replication runoff on the lagging strand for DNA replication.
Our results demonstrate an asymmetry between the leading and lagging strands for DNA replication. Because replication-mediated DNA double-strand breaks were not detectable on the lagging strand, a first possibility is that lagging-strand DNA synthesis cannot bypass top1 cleavage complexes (Fig. 9). Westergaard and coworkers previously reported that a top1 cleavage complex could arrest RNA polymerase in vitro (4). Such a replication block by top1 cleavage complexes on the lagging strand would be in contrast to the effects of bulky DNA adducts and UV-induced DNA lesions, which fail to arrest fork progression when the DNA modification is on the lagging strand (64, 70). A replication fork block in the case of the top1 cleavage complexes on the lagging strand could be related to the size of the protein (100 kDa)-DNA adducts. Inhibition of the DNA-relaxing activity of top1 might also affect lagging-strand replication by preventing the chromatin structural changes that might be required for coordinated synthesis of both leading and lagging strands (6). Consistently, we observed that Okasaki fragment synthesis was altered by CPT-induced top1 cleavage complexes (M. Smith, R. Hickey, and L. Malkas, unpublished data). Two other possible mechanisms might explain the lack of detectable replication runoff on the lagging strand. First, replication fork progression might destabilize the top1 cleavage complexes on the lagging strand and force their reversal (68). Destabilization of top1 cleavage complexes by a DNA helix-tracking protein, such as simian virus 40 large T antigen, has been observed (38). Finally, if lagging-strand synthesis could be initiated past the top1 cleavage complexes, it is possible that nascent Okasaki fragment synthesis might bypass the top1-associated single-stranded DNA interruptions on the lagging strand (no experimental data are yet available on this issue), which would most likely result in a double-stranded DNA end with top1 covalently attached to the protruding 3′ end. Such lesions would not be detectable in our double-strand break assay. Further analyses using different assays are needed to establish unambiguously the effects of top1 cleavage complexes on DNA synthesis on the lagging strand.
Cellular implications of top1-mediated DNA damage in replicating DNA.
Considering the frequency of top1 cleavage complexes in genomic DNA and the growing number of pharmacological (39, 40, 46, 66) as well as physiological and environmental DNA modifications (11, 27, 37, 44, 45, 47, 48, 62, 73) that can trap top1 cleavage complexes, it is important to consider the cellular consequences of top1 cleavage complexes. We recently demonstrated that replication-mediated DNA double-strand breaks can recruit the DNA end-binding Ku proteins, activate DNA-dependent protein kinase, and induce phosphorylation of RPA2 (55). It is therefore possible that the replication-mediated DNA double-strand breaks activate the S-phase checkpoint by this DNA-dependent protein kinase-dependent RPA2 pathway (55).
The ability to sequence DNA strand-specific replication-mediated DNA damage at the nucleotide level and to study the processing of such DNA lesions in specific gene regions of human cells raises a number of interesting and novel issues, including the basis for the leading- versus lagging-strand asymmetry in the production of double-strand breaks, the mechanism(s) of formation and removal of replication-associated top1-DNA adducts (which appears to be rapid at many sites in rDNA) in different genomic regions, and the identity of the kinase(s) that phosphorylates the 5′ end of the replication-mediated DNA double-strand breaks on the leading strand for DNA synthesis.
ACKNOWLEDGMENTS
We thank M. L. DePamphilis and K. W. Kohn for helpful discussions.
D. Strumberg was supported by the Deutsche Forschungsgemeinschaft (grant Str 527/1-1), Bonn, Germany.
REFERENCES
- 1.Anderson R D, Berger N A. Mutagenicity and carcinogenicity of topoisomerase-interactive drugs. Mutat Res. 1994;309:109–142. doi: 10.1016/0027-5107(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 2.Avemann K, Knippers R, Koller T, Sogo J M. Camptothecin, a specific inhibitor of type I DNA topoisomerase, induces DNA breakage at replication forks. Mol Cell Biol. 1988;8:3026–3034. doi: 10.1128/mcb.8.8.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker S D, Wadkins R M, Stewart C F, Beck W T, Danks M K. Cell cycle analysis of amount and distribution of nuclear DNA topoisomerase I as determined by fluorescence digital imaging microscopy. Cytometry. 1995;19:134–145. doi: 10.1002/cyto.990190208. [DOI] [PubMed] [Google Scholar]
- 4.Bendixen C, Thomsen B, Alsner J, Westergaard O. Camptothecin-stabilized topoisomerase I-DNA adducts cause premature termination of transcription. Biochemistry. 1990;29:5613–5619. doi: 10.1021/bi00475a028. [DOI] [PubMed] [Google Scholar]
- 5.Bennett C B, Westmoreland T J, Snipe J R, Resnick M A. A double-strand break within a yeast artificial chromosome (YAC) containing human DNA can result in YAC loss, deletion, or cell lethality. Mol Cell Biol. 1996;16:4414–4425. doi: 10.1128/mcb.16.8.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brush G S, Kelly T J. Mechanisms for replicating DNA. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 7.Buckwalter C A, Lin A H, Tanizawa A, Pommier Y G, Cheng Y C, Kaufmann S H. RNA synthesis inhibitors alter the subnuclear distribution of DNA topoisomerase I. Cancer Res. 1996;56:1674–1681. [PubMed] [Google Scholar]
- 8.Bullock P, Champoux J J, Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985;230:954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- 9.Champoux J J, McCoubrey W K, Jr, Been M D. DNA structural features that lead to strand breakage by eukaryotic type-I topoisomerase. Cold Spring Harbor Symp Quant Biol. 1984;49:435–442. doi: 10.1101/sqb.1984.049.01.049. [DOI] [PubMed] [Google Scholar]
- 10.Chen C L, Fuscoe J C, Liu Q, Pui C H, Mahmoud H H, Relling M V. Relationship between cytotoxicity and site-specific DNA recombination after in vitro exposure of leukemia cells to etoposide. J Natl Cancer Inst. 1996;88:1840–1847. doi: 10.1093/jnci/88.24.1840. [DOI] [PubMed] [Google Scholar]
- 11.Christiansen K, Svejstrup A B, Andersen A H, Westergaard O. Eukaryotic topoisomerase I-mediated cleavage requires bipartite DNA interaction. Cleavage of DNA substrates containing strand interruptions implicates a role for topoisomerase I in illegitimate recombination. J Biol Chem. 1993;268:9690–9701. [PubMed] [Google Scholar]
- 12.Cimmino C, Santori F, Donini P. Ligation of nonmatching DNA molecule ends. Plasmid. 1995;34:1–10. doi: 10.1006/plas.1995.1028. [DOI] [PubMed] [Google Scholar]
- 13.Covey J M, Jaxel C, Kohn K W, Pommier Y. Protein-linked DNA strand breaks induced in mammalian cells by camptothecin, an inhibitor of topoisomerase I. Cancer Res. 1989;49:5016–5022. [PubMed] [Google Scholar]
- 14.Culotta V, Sollner-Webb B. Sites of topoisomerase I action on X. laevis ribosomal chromatin: transcriptionally active rDNA has an approximately 200 bp repeating structure. Cell. 1988;52:585–597. doi: 10.1016/0092-8674(88)90471-0. [DOI] [PubMed] [Google Scholar]
- 15.Danks M K, Garrett K E, Marion R C, Whipple D O. Subcellular redistribution of DNA topoisomerase I in anaplastic astrocytoma cells treated with topotecan. Cancer Res. 1996;56:1664–1673. [PubMed] [Google Scholar]
- 16.D'Arpa P, Beardmore C, Liu L F. Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res. 1990;50:6919–6924. [PubMed] [Google Scholar]
- 17.Eng W K, Faucette L, Johnson R K, Sternglanz R. Evidence that DNA topoisomerase I is necessary for the cytotoxic effects of camptothecin. Mol Pharmacol. 1988;34:755–760. [PubMed] [Google Scholar]
- 18.Gencheva M, Anachkova B, Russev G. Mapping the sites of initiation of DNA replication in rat and human rRNA genes. J Biol Chem. 1996;271:2608–2614. doi: 10.1074/jbc.271.5.2608. [DOI] [PubMed] [Google Scholar]
- 19.Gerber J K, Gogel E, Berger C, Wallisch M, Muller F, Grummt I, Grummt F. Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell. 1997;90:559–567. doi: 10.1016/s0092-8674(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 20.Gilmour D S, Pflugfelder G, Wang J C, Lis J T. Topoisomerase I interacts with transcribed regions in Drosophila cells. Cell. 1986;44:401–407. doi: 10.1016/0092-8674(86)90461-7. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez I L, Sylvester J E. Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics. 1995;27:320–328. doi: 10.1006/geno.1995.1049. [DOI] [PubMed] [Google Scholar]
- 22.Gupta M, Fujimori A, Pommier Y. Eukaryotic DNA topoisomerases I. Biochim Biophys Acta. 1995;1262:1–14. doi: 10.1016/0167-4781(95)00029-g. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez P, Martin-Parras L, Martinez-Robles M L, Schvartzman J B. Conserved features in the mode of replication of eukaryotic ribosomal RNA genes. EMBO J. 1993;12:1475–1485. doi: 10.1002/j.1460-2075.1993.tb05791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm C, Covey J M, Kerrigan D, Pommier Y. Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res. 1989;49:6365–6368. [PubMed] [Google Scholar]
- 25.Hsiang Y H, Hertzberg R, Hecht S, Liu L F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J Biol Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 26.Hsiang Y H, Lihou M G, Liu L F. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 27.Lanza A, Tornaletti S, Rodolfo C, Scanavini M C, Pedrini A M. Human DNA topoisomerase I-mediated cleavages stimulated by ultraviolet light-induced DNA damage. J Biol Chem. 1996;271:6978–6986. doi: 10.1074/jbc.271.12.6978. [DOI] [PubMed] [Google Scholar]
- 28.Little R D, Platt T H, Schildkraut C L. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L F, Duann P, Lin C T, D'Arpa P, Wu J. Mechanism of action of camptothecin. Ann NY Acad Sci. 1996;803:44–49. doi: 10.1111/j.1749-6632.1996.tb26375.x. [DOI] [PubMed] [Google Scholar]
- 30.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 31.Meyer K N, Kjeldsen E, Straub T, Knudsen B R, Hickson I D, Kikuchi A, Kreipe H, Boege F. Cell cycle-coupled relocation of type I and II topoisomerases and modulation of catalytic enzyme activities. J Cell Biol. 1997;136:775–788. doi: 10.1083/jcb.136.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mueller P R, Garrity P A, Wold B. Ligation-mediated PCR. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seideman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1992. pp. 15.5.1–15.5.26. [Google Scholar]
- 33.Muller M T, Pfund W P, Mehta V B, Trask D K. Eukaryotic type I topoisomerase is enriched in the nucleolus and catalytically active on ribosomal DNA. EMBO J. 1985;4:1237–1243. doi: 10.1002/j.1460-2075.1985.tb03766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nitiss J L. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim Biophys Acta. 1998;1400:63–81. doi: 10.1016/s0167-4781(98)00128-6. [DOI] [PubMed] [Google Scholar]
- 35.Osman F, Fortunato E A, Subramani S. Double-strand break-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Genetics. 1996;142:341–357. doi: 10.1093/genetics/142.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pommier Y, Jenkins J, Kohlhagen G, Leteurtre F. DNA recombinase activity of eukaryotic DNA topoisomerase I: effects of camptothecin and other inhibitors. Mutat Res. 1995;337:135–145. doi: 10.1016/0921-8777(95)00019-g. [DOI] [PubMed] [Google Scholar]
- 37.Pommier Y, Kohlhagen G, Pourquier P, Sayer J M, Kroth H, Jerina D M. Benzo[a]pyrene epoxide adducts in DNA are potent inhibitors of a normal topoisomerase I cleavage site and powerful inducers of other topoisomerase I cleavages. Proc Natl Acad Sci USA. 2000;10:2040–2045. doi: 10.1073/pnas.040397497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pommier Y, Kohlhagen G, Wu C, Simmons D T. Mammalian DNA topoisomerase I activity and poisoning by camptothecin are inhibited by simian virus 40 large T antigen. Biochemistry. 1998;37:3818–3823. doi: 10.1021/bi972067d. [DOI] [PubMed] [Google Scholar]
- 39.Pommier Y, Pourquier P, Fan Y, Strumberg D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim Biophys Acta. 1998;1400:83–105. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 40.Pommier Y, Pourquier P, Urasaki Y, Wu J, Laco G. Topoisomerase I inhibitors: selectivity and cellular resistance. Drug Resistance Update. 1999;2:307–318. doi: 10.1054/drup.1999.0102. [DOI] [PubMed] [Google Scholar]
- 41.Pondarré C, Strumberg D, Fujimori A, Torres-Leon R, Pommier Y. In vivo sequencing of camptothecin-induced topoisomerase I cleavage sites in human colon carcinoma cells. Nucleic Acids Res. 1997;25:4111–4116. doi: 10.1093/nar/25.20.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porter S E, Champoux J J. The basis for camptothecin enhancement of DNA breakage by eukaryotic topoisomerase I. Nucleic Acids Res. 1989;17:8521–8532. doi: 10.1093/nar/17.21.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pouliot J J, Yao K C, Robertson C A, Nash H A. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topo I covalent complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 44.Pourquier P, Bjornsti M A, Pommier Y. Induction of topoisomerase I cleavage complexes by the vinyl chloride adduct 1,N6-ethenoadenine. J Biol Chem. 1998;273:27245–27249. doi: 10.1074/jbc.273.42.27245. [DOI] [PubMed] [Google Scholar]
- 45.Pourquier P, Pilon A A, Kohlhagen G, Mazumder A, Sharma A, Pommier Y. Trapping of mammalian topoisomerase I and recombinations induced by damaged DNA containing nicks or gaps. Importance of DNA end phosphorylation and camptothecin effects. J Biol Chem. 1997;272:26441–26447. doi: 10.1074/jbc.272.42.26441. [DOI] [PubMed] [Google Scholar]
- 46.Pourquier P, Takebayashi Y, Urasaki Y, Gioffre C, Kohlhagen G, Pommier Y. Induction of topoisomerase I cleavage complexes by 1-β-d-arabinofuranosylcytosine (Ara-C) in vitro and in ara-C-treated cells. Proc Natl Acad Sci USA. 2000;97:1885–1890. doi: 10.1073/pnas.97.4.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pourquier P, Ueng L M, Fertala J, Wang D, Park H J, Essigmann J M, Bjornsti M A, Pommier Y. Induction of reversible complexes between eukaryotic DNA topoisomerase I and DNA-containing oxidative base damages: 7,8-dihydro-8-oxoguanine and 5-hydroxycytosine. J Biol Chem. 1999;274:8516–8523. doi: 10.1074/jbc.274.13.8516. [DOI] [PubMed] [Google Scholar]
- 48.Pourquier P, Ueng L M, Kohlhagen G, Mazumder A, Gupta M, Kohn K W, Pommier Y. Effects of uracil incorporation, DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I. J Biol Chem. 1997;272:7792–7796. doi: 10.1074/jbc.272.12.7792. [DOI] [PubMed] [Google Scholar]
- 49.Redinbo M R, Stewart L, Kuhn P, Champoux J J, Hol W G. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 50.Rosenstein B S, Subramanian D, Muller M T. The involvement of topoisomerase I in the induction of DNA-protein crosslinks and DNA single-strand breaks in cells of ultraviolet-irradiated human and frog cell lines. Radiat Res. 1997;148:575–579. [PubMed] [Google Scholar]
- 51.Ryan A J, Squires S, Strutt H L, Johnson R T. Camptothecin cytotoxicity in mammalian cells is associated with the induction of persistent double strand breaks in replicating DNA. Nucleic Acids Res. 1991;19:3295–3300. doi: 10.1093/nar/19.12.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sargent R G, Brenneman M A, Wilson J H. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sastry S, Ross B M. Mechanisms for the processing of a frozen topoisomerase-DNA conjugate by human cell-free extracts. J Biol Chem. 1998;273:9942–9950. doi: 10.1074/jbc.273.16.9942. [DOI] [PubMed] [Google Scholar]
- 54.Shan Q, Cox M M. On the mechanism of RecA-mediated repair of double-strand breaks: no role for four-strand DNA pairing intermediates. Mol Cell. 1998;1:309–317. doi: 10.1016/s1097-2765(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 55.Shao R-G, Cao C-X, Zhang H, Kohn K W, Wold M S, Pommier Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin C G, Snapka R M. Exposure to camptothecin breaks leading and lagging strand simian virus 40 DNA replication forks. Biochem Biophys Res Commun. 1990;168:135–140. doi: 10.1016/0006-291x(90)91684-k. [DOI] [PubMed] [Google Scholar]
- 57.Shuman S. DNA strand transfer reactions catalyzed by vaccinia topoisomerase I. J Biol Chem. 1992;267:8620–8627. [PubMed] [Google Scholar]
- 58.Shuman S. Two classes of DNA end-joining reactions catalyzed by vaccinia topoisomerase I. J Biol Chem. 1992;267:16755–16758. [PubMed] [Google Scholar]
- 59.Snapka R M, Permana P A. SV40 DNA replication intermediates: analysis of drugs which target mammalian DNA replication. Bioessays. 1993;15:121–127. doi: 10.1002/bies.950150208. [DOI] [PubMed] [Google Scholar]
- 60.Squires S, Ryan A J, Strutt H L, Johnson R T. Hypersensitivity of Cockayne's syndrome cells to camptothecin is associated with the generation of abnormally high levels of double strand breaks in nascent DNA. Cancer Res. 1993;53:2012–2019. [PubMed] [Google Scholar]
- 61.Stewart L, Redinbo M R, Qiu X, Hol W G, Champoux J J. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 62.Subramanian D, Rosenstein B S, Muller M T. Ultraviolet-induced DNA damage stimulates topoisomerase I-DNA complex formation in vivo: possible relationship with DNA repair. Cancer Res. 1998;58:976–984. [PubMed] [Google Scholar]
- 63.Svejstrup J Q, Christiansen K, Andersen A H, Lund K, Westergaard O. Minimal DNA duplex requirements for topoisomerase I-mediated cleavage in vitro. J Biol Chem. 1990;265:12529–12535. [PubMed] [Google Scholar]
- 64.Svoboda D L, Vos J M. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation. Proc Natl Acad Sci USA. 1995;92:11975–11979. doi: 10.1073/pnas.92.26.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sylvester J E, Petersen R, Schmickel R D. Human ribosomal DNA: novel sequence organization in a 4.5-kb region upstream from the promoter. Gene. 1989;84:193–196. doi: 10.1016/0378-1119(89)90155-8. [DOI] [PubMed] [Google Scholar]
- 66.Takebayashi Y, Pourquier P, Yoshida A, Kohlhagen G, Pommier Y. Poisoning of human DNA topoisomerase I by ecteinascidin 743, an anticancer drug that selectively alkylates DNA in the minor groove. Proc Natl Acad Sci USA. 1999;96:7196–7201. doi: 10.1073/pnas.96.13.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanizawa A, Kohn K W, Kohlhagen G, Leteurtre F, Pommier Y. Differential stabilization of eukaryotic DNA topoisomerase I cleavable complexes by camptothecin derivatives. Biochemistry. 1995;34:7200–7206. doi: 10.1021/bi00021a035. [DOI] [PubMed] [Google Scholar]
- 68.Tsao Y P, Russo A, Nyamuswa G, Silber R, Liu L F. Interaction between replication forks and topoisomerase I-DNA cleavable complexes: studies in a cell-free SV40 DNA replication system. Cancer Res. 1993;53:5908–5914. [PubMed] [Google Scholar]
- 69.Tsukamoto Y, Ikeda H. Double-strand break repair mediated by DNA end-joining. Genes Cells. 1998;3:135–144. doi: 10.1046/j.1365-2443.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- 70.Veaute X, Sarasin A. Differential replication of a single N-2-acetylaminofluorene lesion in the leading or lagging strand DNA in a human cell extract. J Biol Chem. 1997;272:15351–15357. doi: 10.1074/jbc.272.24.15351. [DOI] [PubMed] [Google Scholar]
- 71.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 72.Wu J, Liu L F. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 1997;25:4181–4186. doi: 10.1093/nar/25.21.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yeh Y C, Liu H F, Ellis C A, Lu A L. Mammalian topoisomerase I has base mismatch nicking activity. J Biol Chem. 1994;269:15498–15504. [PubMed] [Google Scholar]
- 74.Yoon Y, Sanchez J A, Brun C, Huberman J A. Mapping of replication initiation sites in human ribosomal DNA by nascent-strand abundance analysis. Mol Cell Biol. 1995;15:2482–2489. doi: 10.1128/mcb.15.5.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H, Wang J C, Liu L F. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc Natl Acad Sci USA. 1988;85:1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]