Abstract
During 1994, 10 isolates of extended-spectrum β-lactamase-producing Salmonella typhimurium were recovered from children transferred to our hospital from two different centers. Two additional isolates were recovered from two nurses from one of these centers. The aim of this study was to determine if there is any relationship between these isolates. The characterization was done by phenotypic and genotypic methods: biotyping, phage typing, antibiotic susceptibility pattern determination, plasmid analysis, ribotyping (by the four endonucleases EcoRI, SmaI, BglII, and PvuII), pulsed-field gel electrophoresis (PFGE) of genome macrorestriction patterns with XbaI, and randomly amplified polymorphic DNA (RAPD) pattern determination (with the three primers 217 d2, B1, and A3). The same biotype, the same serotype, and an identical antibiotype were found. All isolates were resistant to oxyimino-β-lactams, gentamicin, tobramycin, and sulfamethoxazole-trimethoprim. All isolates showed an indistinguishable pattern by ribotyping and very similar patterns by PFGE and RAPD. The overall results indicated the spread of a closely related strain of S. typhimurium in children and nurses.
The incidence of infections caused by salmonellae other than Salmonella typhi has increased considerably in many countries (7, 44). The most common serotypes, isolated from human and animal sources, in the United States (21, 44), France (10, 27, 30, 42), and Tunisia (2, 20), are Salmonella enteritidis, Salmonella typhimurium, and Salmonella wien, respectively. The most prevalent serotypes in Casablanca, Morocco, are S. typhimurium and S. enteritidis (unpublished results). In recent years, S. typhimurium strains were responsible for outbreaks in pediatric units and were often resistant to multiple antibiotics, including aminopenicillins, gentamicin, tetracycline, chloramphenicol, and sulfonamides (9, 10, 27, 44).
From February to September 1994, 10 distinct isolates of extended-spectrum β-lactamase (ESBL)-producing S. typhimurium (S1 to S10) were isolated at the microbiology laboratory of the Ibn Rochd University Hospital, Casablanca, Morocco, from children with acute diarrhea and septicemia. These children were transferred to our hospital from two different centers (center 1 and center 2). In September 1994, two additional strains of S. typhimurium were isolated from stools of nurses from center 1 (S11 and S12) (Table 1). Because it was the first time such isolates were isolated in our laboratory and this type of resistance is rarely associated with the genus Salmonella (8, 14), and because children are transferred between the two centers, the aim of this study was to determine if these isolates belong to the same or to related clones. These isolates were characterized by phenotypic methods, including biotyping, serotyping, phage typing, and determination of antibiotic susceptibility patterns, and by genotypic techniques such as plasmid analysis, ribotyping, pulsed-field gel electrophoresis (PFGE), and randomly amplified polymorphic DNA (RAPD).
TABLE 1.
Origin and phenotypic characteristics of outbreak-related isolates of S. typhimurium
| Strain | Center no. | Date of isolation (mo/day/yr) | Origin | Phage type | Antibiotypea |
|---|---|---|---|---|---|
| From patients | |||||
| S124 | 1 | 12/11/92 | Blood | 91 | S |
| S1 | 2 | 02/22/94 | Blood | 91 | R |
| S2 | 2 | 02/26/94 | Blood | 29 | R |
| S3 | 2 | 05/26/94 | Blood | 91 | R |
| S4 | 1 | 08/24/94 | Blood | 91 | R |
| S5 | 1 | 08/24/94 | Blood | 91 | R |
| S6 | 1 | 08/26/94 | Blood | 91 | R |
| S7 | 1 | 08/26/94 | Blood | 91 | R |
| S8 | 1 | 08/26/94 | Blood | 29 | R |
| S9 | 1 | 09/30/94 | Stool | 91 | R |
| S10 | 1 | 09/30/94 | Stool | 91 | R |
| From nurses | |||||
| S11 | 1 | 09/16/94 | Stool | 91 | R |
| S12 | 1 | 09/20/94 | Stool | 91 | R |
S, susceptible to all antibiotics tested; R, resistant to ampicillin, amoxicillin-clavulanic acid, cephalothin, cefotaxime, ceftazidime, aztreonam, gentamicin, tobramycin, and trimethoprim-sulfamethoxazole, but not tetracycline, chloramphenicol, netilmicin, or amikacin.
The 12 isolates of S. typhimurium were identified by Gram stain, by determining biochemical characteristics with the API 20E system (Biomérieux), and by serological identification of somatic (O) and flagellar (H) antigens with commercial antisera (Sanofi Diagnostics Pasteur) according to the Kauffman-White serotyping scheme (25). All strains were stored frozen at −70°C in 20% glycerol and in nutrient agar stab cultures at room temperature. The type strain, ATCC 43971, and one nonrelated S. typhimurium strain, S124, were studied for comparison.
Antibiotic susceptibility testing was performed by a disk diffusion method on Mueller-Hinton agar and interpreted in accordance with criteria of the National Committee for Clinical Laboratory Standards (34). The strains were screened for their resistance to the following antibiotics (Sanofi Diagnostics Pasteur): ampicillin, amoxicillin-clavulanic acid, cephalothin, imipenem, cefotaxime, ceftazidime, aztreonam, gentamicin, amikacin, netilmicin, tobramycin, chloramphenicol, tetracycline, and trimethoprim-sulfamethoxazole. The double-disk synergy test was performed with cefotaxime, ceftazidime, aztreonam, and clavulanic acid plus amoxicillin on Mueller-Hinton agar (24). Escherichia coli ATCC 25922 was used as a reference strain.
Conjugation experiments were carried out in Luria broth supplemented with 0.5% sucrose by mixing equal volumes (1 ml) of exponentially growing cultures of donors (S. typhimurium) and the recipient E. coli K-12 J53-2 resistant to rifampin. After incubation at 37°C overnight with slow shaking (3), transconjugants of E. coli were selected on MacConkey agar supplemented with cefotaxime (1 μg/ml) and rifampin (100 μg/ml). Extended-spectrum β-lactamase production was confirmed in the transconjugants by the double-disk diffusion test (24).
Phage typing was done, as previously described, at the French National Center for Enteric Molecular Typing (Pasteur Institute, Paris, France) (15).
Bacterial strains were screened for plasmid DNA by a modification of the Birnboim-Doly and Ish-Horowicz Bruke extraction procedure (40). Extracted plasmid DNA was electrophoresed on an 0.7% horizontal agarose gel containing 0.5 μg of ethidium bromide solution per ml and analyzed under UV illumination.
For ribotyping, total S. typhimurium DNA was extracted as described by Picard-Pasquier (36). DNA (2 to 5 μg) was digested with four different endonucleases: PvuII, BglII, SmaI, and EcoRI (Boehringer GmbH, Mannheim, Germany) and analyzed by electrophoresis on submarine ethidium bromide-containing 0.8% agarose gels. Genomic restriction digests were subjected to Southern blotting on Hybond-N nylon membranes (Amersham) by the classical procedure of Southern (43). Ribosomal 16+23S RNA from E. coli (Boehringer) was used as a probe (16) and was cold-labeled by random oligopriming with a mixture of hexanucleotides (Pharmacia, Uppsala, Sweden) and cloned Moloney murine leukemia virus reverse transcriptase (Bethesda Research Laboratories, Gaithersburg, Md.) in the presence of 0.35 mM DiG-II-dUTP (digoxigenin-II–deoxyuridine 5′-11 triphosphate; Boehringer). Chemiluminescence detection procedures were done as described by the manufacturer (Boehringer) by incubating the membranes in the presence of an antidigoxigenin antibody linked to alkaline phosphatase and its substrate, chemiluminescence substrate phenyl-phosphate disodium (CSPD; Boehringer). Filters were autoradiographed by exposure to X-Omat AR 5 film (Kodak) for 3 h at room temperature. Isolates which differed by one fragment were considered to be different strains. Each distinct combination of patterns was used to define a ribotype.
For PFGE, chromosomal DNA was prepared by using the Chef Genomic DNA Plug kit (Bio-Rad Laboratories, Hercules, Calif.). Chromosomal DNA was digested overnight at 37°C with 30 U of XbaI in a 250-μl reaction volume. The resulting restriction fragments were then analyzed on 14- by 20-cm 0.8% agarose gels (CHEF Mapper electrophoresis system; Bio-Rad Laboratories), stained with ethidium bromide, and visualized by UV transillumination. Isolates which differed by no more than three restriction fragment positions were considered to represent subtypes of a common epidemic strain (45).
Bacterial DNA was also studied by a RAPD procedure, which was adapted from the method of Williams et al. (51) by using the in-house-synthesized PCR primers 217 d2 (5′GCCCCCAGGGGCACAGT 3′), A3 (5′AGTCAGCCAC 3′), and B1 (5′GTTTCGTCC 3′). The reaction took place in 50 μl of 100 mM Tris-HCI buffer (pH 8.3) containing 50 mM KCl, 4 mM MgCl2, 0.4 mM deoxynucleoside triphosphate, 3 μM primer, 50 ng of DNA, and 2.5 U of Taq DNA polymerase (Beckman, Fullerton, Calif.). Amplification was performed in a DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.) programmed for 35 cycles of 1 min at 94°C, 1 min at 36°C, and 2 min at 72°C. Amplification products were resolved by electrophoresis in a 2% agarose gel and were detected by staining with ethidium bromide. Isolates which differed by two or more prominent bands were considered sufficiently divergent to warrant separate strain designations. Profiles differing from one another by only one major band or by one or two weak bands were considered minor variant types representing subtypes of a common epidemic strain (5, 26).
In the present study, the enzymatic resistance to oxyimino-β-lactam antibiotics was reported among isolates of S. typhimurium for the first time in our laboratory. The production of ESBL is rarely associated with the genus Salmonella (8, 14). The first such strains were detected in France in 1984 and 1987 (S. typhimurium), in Tunisia in 1988 (Salmonella wien), in Algeria in 1990 (Salmonella mbandaka), and in Argentina in 1991 (S. typhimurium) (1, 10, 20, 37). The most frequent types of ESBLs found in Salmonella species were SHV-2, CTX-2, CTX-M2, TEM-27, CTX-M5, and PER-1 (1, 8, 20, 31, 37, 49).
The combined results of antigenic, biochemical typing and antibiotyping demonstrated the existence of the same Salmonella strain with API profile 6704552, serotype 4,5,12:i-1,2, and the same antibiotype characterized by the production of ESBL and resistance to gentamicin and trimethoprim-sulfamethoxazole but susceptibility to chloramphenicol, tetracycline, and quinolones (Table 1). In other countries, the resistance of Salmonella to several antibiotics was more worrisome. In the United States, 32% of the 282 human S. typhimurium isolates tested at the Centers for Disease Control in 1996 were multidrug resistant, including isolates with a recently emerged resistance to quinolones (23). In England and Wales, in 1995, 27% of human S. typhimurium isolates were multidrug resistant and 6% were also resistant to ciprofloxacin (48).
Another powerful phenotypic typing technique for Salmonella species is phage typing (13, 44, 50). It has been reported that this technique was the most useful marker for distinguishing clonal groups of S. typhimurium when compared to plasmid analysis, biotyping, and antibiotic susceptibility pattern (29). In our study, phage typing discriminated two groups (Table 1). For most isolates (10 of 12), phage typing correlated with biotyping and antibiotyping. However, phage typing may be problematic in ruling out reinfection because of the high prevalence of one or a few phage types of S. typhimurium in a community. Phage type may also be modified by type phage-determining plasmids because acquisition of a plasmid may partially restrict the susceptibility to the typing bacteriophage (13). Furthermore, the use of this technique is limited to a few specialized centers. Of the traditional techniques most accessible to clinical laboratories, i.e., biotyping, serotyping, and antibiograms, we found that antibiograms worked well in discriminating between strain S124, the unrelated strain isolated in 1992, and the 12 outbreak-related isolates of S. typhimurium, so an antibiogram can be used as an initial screen to determine strain relatedness.
Several studies have shown the stability of plasmid profile analysis of Salmonella species. Thus, plasmid analysis appears to be the more effective method for grouping strains with the same serotype obtained from a single outbreak (7, 49). Holmberg et al. (22) compared plasmid profiles, phage types, and antibiotypes in the investigation of 20 outbreaks of S. typhimurium infections. In 17 of these 20 outbreaks, a correlation was found between these three techniques. The most discriminatory method was plasmid profile analysis in two outbreaks and phage typing in one outbreak (22). Several investigators reported that resistance to different antimicrobial agents was mediated by a large plasmid (2, 7, 13, 20, 31). This plasmid was found in all our strains (data not shown); the only difference in the plasmid profiles was the absence of one small plasmid in the isolates from nurses. However, this may not exclude an epidemiological relationship between all isolates because plasmids are unstable genetic elements that can be readily lost or acquired.
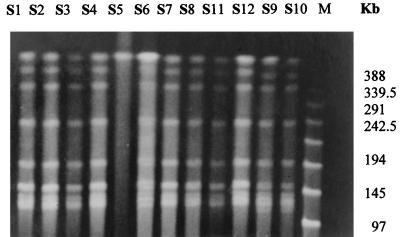
Ribotyping has been used for the study of many bacterial species responsible for nosocomial infections (6, 11) and also for different species of the genus Salmonella (12, 19, 30, 35). In our study, ribotyping revealed an identical pattern (Fig. 1; Table 2) for all isolates, including the unrelated strain, S124, by four different restriction endonucleases, including EcoRI, an enzyme which has been suggested to be the most discriminative and as having the most easily defined banding distribution (12, 49). These results suggest that ribotyping is of limited value in the epidemiological analysis of these Salmonella species. However, ribotyping with hybridization with the IS200 probe was more sensitive than phage typing or ribotyping for discriminating between S. typhimurium isolates because of the wide diversity of IS200 profiles among S. typhimurium isolates (30). Our findings also suggest, as reported by others researchers (12, 33, 35), that ribotyping should be used in parallel with phage typing, antibiotyping, and plasmid analysis.
FIG. 1.
Ribotyping profiles after digestion by EcoRI of S. typhimurium isolates S124, S1 to S12, and type strain ATCC 43971. Lane M contains molecular size markers.
TABLE 2.
Genotypic characteristics of outbreak-related isolates of S. typhimurium and the type strain
| Strain | rDN RFLP pattern
|
Ribotype | PFGE XbaIa | RAPD results
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EcoRI | PvuII | BglII | SmaI | With primer
|
Overall pattern | |||||
| 217 d2 | A3 | B1 | ||||||||
| ATCC 43971 | E1 | P1 | B1 | S1 | 1 | a | a | a | A | |
| From patients | ||||||||||
| S124 | E2 | P2 | B2 | S2 | 2 | b | b | a | B | |
| S1 | E2 | P2 | B2 | S2 | 2 | X1 | c | b | b | C |
| S2 | E2 | P2 | B2 | S2 | 2 | X1 | c | b | b | C |
| S3 | E2 | P2 | B2 | S2 | 2 | X1 | c | b | b | C |
| S4 | E2 | P2 | B2 | S2 | 2 | X1 | c | b | b | C |
| S5 | E2 | P2 | B2 | S2 | 2 | c | b | b | C | |
| S6 | E2 | P2 | B2 | S2 | 2 | X2 | c | b | b | C |
| S7 | E2 | P2 | B2 | S2 | 2 | X1 | c | b | b | C |
| S8 | E2 | P2 | B2 | S2 | 2 | X1 | c | b | b | C |
| S9 | E2 | P2 | B2 | S2 | 2 | X2 | c | b | b | C |
| S10 | E2 | P2 | B2 | S2 | 2 | X2 | c | b | b | C |
| From nurses | ||||||||||
| S11 | E2 | P2 | B2 | S2 | 2 | X1 | c | b | b | C |
| S12 | E2 | P2 | B2 | S2 | 2 | X1 | c | b | b | C |
The macrorestriction genotype was determined by PFGE after digestion with XbaI. The two patterns were designated X1 and X2.
Genomic macrorestriction fragment analysis by PFGE has been used successfully for many bacterial species (18, 38, 39). This is also true for S. typhi (47) and S. enteritidis (46). However, PFGE analysis of S. typhimurium was rarely done (32, 41). In the present study, an identical pattern, X1, was found for most isolates (in 9 of 12 isolates); the second pattern, X2, was very similar to X1, differing only by one weak band, and with genetic methods, such a difference is not reliable proof for concluding that isolates represent different strains (17, 28, 45) (Fig. 2; Table 2). The disadvantages of PFGE are time-consuming DNA preparation and electrophoresis, costly reagents, and requirement of specialized equipment.
FIG. 2.
PFGE patterns of XbaI-digested genomic DNA obtained from S. typhimurium isolates S1, S2, S3, S4, S5, S6, S7, S8, S11, S12, S9, and S10. Lane M contains molecular size markers.
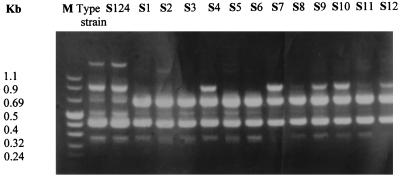
RAPD is another powerful typing method. It has the advantages of speed and simplicity. Its stability and reproducibility have been recently reported (4). Its discriminatory power relies on the primer sequences chosen. Its usefulness for S. typhimurium typing has not been well documented. In this study, we selected primers which have been shown to differentiate clones of members of the family Enterobacteriaceae. With primer A3, all isolates (S1 to S12) and S124 yielded identical patterns, whereas with primers 217 d2 and B1, the isolates were highly related or identical, differing by only one band, and were genetically unrelated to S124 and the ATCC type strain (Fig. 3; Table 2).
FIG. 3.
RAPD patterns of S. typhimurium isolates with primer B1 (5′GTTCGCC3′). Strains include type strain ATCC 43971, S124, and S1 to S12. Lane M contains molecular size markers.
Among phenotypic methods, plasmid analysis and antibiotyping remain interesting for use in the study of S. typhimurium. Among recent genetic methods, RAPD typing seems well adapted to situations in which a rapid comparison of bacterial strains is needed. PFGE is more discriminatory and can be used as a confirmatory method.
REFERENCES
- 1.Bauernfeind A, Casellas J M, Goldberg M, Holley M, Jungwirth R, Mangold P, Rohnisch T, Schweighart S, Wilhelm R. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection. 1992;20:158–163. doi: 10.1007/BF01704610. [DOI] [PubMed] [Google Scholar]
- 2.Ben Hassen A, Bejaoui M, Lakhoua M R, Ben Redjeb S. Profil èpidemiologique de la résistance de 153 souches de Salmonella (S. typhi exclues) isolées en milieu pédiatrique tunisien de 1985 á 1990. Pathol Biol. 1994;41:706–712. [PubMed] [Google Scholar]
- 3.Bergquist P L. Incompatibility. In: Hardy K G, editor. Plasmids, a practical approach. New York, N.Y: Oxford University Press; 1987. p. 37. [Google Scholar]
- 4.Bingen E, Boissinot C, Desjardin P, Cave H, Brahimi N, Lambert-Zechovsky N, Denamur E, Blot P, Elion J. Arbitrarily primed polymerase chain reaction provides rapid differentiation of Proteus mirabilis isolates from a pediatric hospital. J Clin Microbiol. 1993;31:1055–1059. doi: 10.1128/jcm.31.5.1055-1059.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bingen E, Bonacorsi S, Rohrlich P, Duval M, Lhopital S, Brahimi N, Vilmer E, Goering R V. Molecular epidemiology provides evidence of genotypic heterogeneity of multidrug-resistant Pseudomonas aeruginosa serotype O: 12 outbreak isolates from a pediatric hospital. J Clin Microbiol. 1996;34:3226–3229. doi: 10.1128/jcm.34.12.3226-3229.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bingen E, Denamur E, Elion J. Use of ribotyping in epidemiological surveillance of nosocomial outbreaks. Clin Microbiol Rev. 1994;7:311–327. doi: 10.1128/cmr.7.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrego J J, Castro D, Jimenez-Notario M, Luque A, Martinez-Manzanares E, Rodriguez-Avial C, Picazo J J. Comparison of epidemiological markers of Salmonella strains isolated from different sources in Spain. J Clin Microbiol. 1992;30:3058–3064. doi: 10.1128/jcm.30.12.3058-3064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford P A, Yang Y, Sahm D, Grop I, Gardovska D, Storch G. CTX-M5, a novel cefotaxime hydrolysing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob Agents Chemother. 1998;42:1980–1984. doi: 10.1128/aac.42.8.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breuil J, Berger N, Dublanchet A the College BVH. Sensibilité aux antibiotiques de 2800 souches de Salmonelles et Shigelles isolées en France en 1994. Med Mal Infect. 1996;26:420–425. doi: 10.1016/s0399-077x(96)80186-7. [DOI] [PubMed] [Google Scholar]
- 10.Casin I, Brisabois A, Berger N, Breuil J, Collatz E. Phénotypes et Genotypes de résistance de 182 souches de Salmonella serotype typhimurium résistantes á l'ampicilline d'origine humaine et animale. Med Mal Infect. 1996;26:426–430. doi: 10.1016/s0399-077x(96)80187-9. [DOI] [PubMed] [Google Scholar]
- 11.Chetoui H, Delhalle E, Osterrieth P, Rousseaux D. Ribotyping for use in studying molecular epidemiology of Serratia marcescens: comparison with biotyping. J Clin Microbiol. 1995;33:2637–2642. doi: 10.1128/jcm.33.10.2637-2642.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteban E, Sniles K, Hird D, Kasten R, Kinde H. Use of ribotyping for characterization of Salmonella serotypes. J Clin Microbiol. 1993;31:233–237. doi: 10.1128/jcm.31.2.233-237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fica A E, Horowitz H W, Lior H, Cabello F C. Demonstration of persistence of Salmonella typhimurium in an AIDS patient by molecular methods. J Clin Microbiol. 1994;32:2327–2330. doi: 10.1128/jcm.32.9.2327-2330.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gazouli M, Sidorenko S V, Tzelepi E, Kozlova N S, Gladin D P, Tzouvelekis L S. A plasmid-mediated β-lactamase conferring resistance to cefotaxime in Salmonella typhimurium clone found in St. Petersburg, Russia. J Antimicrob Chemother. 1998;41:119–121. doi: 10.1093/jac/41.1.119. [DOI] [PubMed] [Google Scholar]
- 15.Grimont F. Les marqueurs épidemiologiques. II. Lysolypie, bactériocinotypie, ribotypie. In: Freney J, Renaud F, Hansen W, Bollet C, editors. Manuel de Bactériologie clinique. 2nd ed. Paris, France: Elsevier; 1995. pp. 83–90. [Google Scholar]
- 16.Grimont F, Grimont P A D. Ribosomal ribonucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986;137B:165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- 17.Grimont P A D, Grimont F. Apport de la biologie moléculaire au typage: données méthodologiques. Méd Mal Infect. 1996;26:379–385. doi: 10.1016/s0399-077x(96)80178-8. [DOI] [PubMed] [Google Scholar]
- 18.Grundmann H, Schneider C, Hartung D, Daschnes F D, Pitt T L. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J Clin Microbiol. 1995;33:528–534. doi: 10.1128/jcm.33.3.528-534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerra B, Landeras E, Gonzalez-Heria M A, Mendoza M C. A three-way ribotyping scheme for Salmonella serotype typhimurium and its usefulness for phylogenetic and epidemiological purposes. J Med Microbiol. 1997;46:307–313. doi: 10.1099/00222615-46-4-307. [DOI] [PubMed] [Google Scholar]
- 20.Hammami A, Arlet G, BenRedjeb S, Grimont F, Ben Hassen A, Philippon A. Nosocomial outbreak of acute gastroenteritis in neonatal intensive care unit in Tunisia caused by multiply drug resistant Salmonella wien producing SHV-2 bêta-lactamase. Eur J Clin Microbiol Infect Dis. 1991;10:641–646. doi: 10.1007/BF01975816. [DOI] [PubMed] [Google Scholar]
- 21.Hedberg C W, David M J, White K E, MacDonald K L, Osterholm M T. Role of egg consumption in sporadic Salmonella enteritidis and Salmonella typhimurium infections in Minnesota. J Infect Dis. 1993;167:107–111. doi: 10.1093/infdis/167.1.107. [DOI] [PubMed] [Google Scholar]
- 22.Holmberg S D, Wachsmuth I K, Brenner F W H, Blake P A, Cohen M L. Comparison of plasmid profile analysis, phage typing, and antimicrobial susceptibility testing in characterizing Salmonella typhimurium isolates from outbreaks. J Clin Microbiol. 1981;19:100–104. doi: 10.1128/jcm.19.2.100-104.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosek G, Leschinsky D, Irons S, Safranek T J. Multidrug-resistant Salmonella serotype typhimurium in United States, 1996. Morbid Mortal Weekly Rep. 1997;46:308–310. [PubMed] [Google Scholar]
- 24.Jarlier V, Nicolas M-H, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 25.Kauffman F. Serological diagnosis of Salmonella species. Copenhagen, Denmark: Munksgaard; 1972. [Google Scholar]
- 26.Kersulyte D, Struelens M J, Deplano A, Berg D E. Comparison of arbitrarily primed PCR and macrorestriction (pulsed-field gel electrophoresis) typing of Pseudomonas aeruginosa strains from cystic fibrosis patients. J Clin Microbiol. 1995;33:2216–2219. doi: 10.1128/jcm.33.8.2216-2219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martel J L, Chaslus-Dancla E, Coudert M, Lafont J P. Evolution de la sensibilité aux antibiotiques des Salmonelles d'origine bovine en France. Med Mal Infect. 1996;26:415–419. doi: 10.1016/s0399-077x(96)80185-5. [DOI] [PubMed] [Google Scholar]
- 28.Maslow J N, Mulligan M E, Arbeit R D. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–164. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 29.McDonough P L, Timoney J F, Jacobson R H, Khakhria R. Clonal groups of Salmonella typhimurium in New York State. J Clin Microbiol. 1989;27:622–627. doi: 10.1128/jcm.27.4.622-627.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Millemann Y, Lesage M-C, Chaslus-Dancla E, Lafont J-P. Value of plasmid profiling, ribotyping, and detection of IS200 for tracing avian isolates of Salmonella typhimurium and Salmonella enteritidis. J Clin Microbiol. 1995;33:173–179. doi: 10.1128/jcm.33.1.173-179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morosini M I, Canton R, Martinez-Beltran J, Negri M C, Perez-Diaz J C, Baquero F, Blazquez J. New extended-spectrum TEM-type β-lactamase from Salmonella enterica subsp. enterica isolated in a nosocomial outbreak. Antimicrob Agents Chemother. 1995;39:458–461. doi: 10.1128/aac.39.2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murase T, Okitsu T, Suzuki R, Morozumi H, Matsushima A, Nakamura A, Yamai S. Evaluation of DNA fingerprinting by PFGE as an epidemiologic tool for Salmonella infections. Microbiol Immunol. 1995;39:673–676. doi: 10.1111/j.1348-0421.1995.tb03255.x. [DOI] [PubMed] [Google Scholar]
- 33.Nastasi A, Mammina C, Villafrate M R. DNA fingerprinting as a tool in epidemiological analysis of Salmonella typhi in infections. Epidemiol Infect. 1991;107:565–576. doi: 10.1017/s0950268800049268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing, fifth international supplement. Document M100-S5. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1994. [Google Scholar]
- 35.Olsen J E, Brown D J, Baggesen D L, Bisgaard M. Biochemical and molecular characterization of Salmonella enterica serovar berta, and comparison of methods for typing. Epidemiol Infect. 1992;107:565–576. doi: 10.1017/s0950268800049724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Picard-Pasquier N, Ouagued M, Picard B, Goullet P, Krishnamoorty R. A simple sensitive method of analyzing bacterial ribosomal DNA polymorphism. Electrophoresis. 1989;10:186–189. doi: 10.1002/elps.1150100306. [DOI] [PubMed] [Google Scholar]
- 37.Poupart M C, Chanal C, Sirot D, Labia R, Sirot J. Identification of CTX-2, a novel cefotaximase from a Salmonella mbandaka isolate. Antimicrob Agents Chemother. 1991;35:1498–1500. doi: 10.1128/aac.35.7.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prévost G, Piemont Y, Monteil H. Intérêt de l'electrophorèse en champ pulsé en épidémiologie moléculaire. Lett Infectiol. 1993;8:279–282. [Google Scholar]
- 39.Sader H S, Pfaller M A, Tenover F C, Hollis R J, Jones R N. Evaluation and characterization of multiresistant Enterococcus faecium from 12 U.S. medical centers. J Clin Microbiol. 1994;32:2840–2848. doi: 10.1128/jcm.32.11.2840-2842.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1987. [Google Scholar]
- 41.Schwarz S, Liebish B. Pulsed-field gel electrophoretic identification of Salmonella enterica serovar typhimurium live vaccine strain Zoosaloral H. Lett Appl Microbiol. 1994;19:469–472. doi: 10.1111/j.1472-765x.1994.tb00984.x. [DOI] [PubMed] [Google Scholar]
- 42.Simonin C, Bayle A, Zurlinden A, Triozon F. Epidémiologie des Salmonella isolées au CHG de Mâcon de 1980 à 1990. Med Mal Infect. 1992;22:674–677. [Google Scholar]
- 43.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 44.Stubbs A D, Hickman-Brenner F W, Cameron D N, Farmer J J., III Differentiation of Salmonella enteritidis phage type 8 strains: evaluation of three additional phage typing systems, plasmid profiles, antibiotic susceptibility patterns, and biotyping. J Clin Microbiol. 1994;32:199–201. doi: 10.1128/jcm.32.1.199-201.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tenover F C, Arbeit R D, Goering R V, Nickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thong K-L, Ngeow Y-F, Altwegg M, Navaratnam P, Pang T. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol. 1995;33:1070–1074. doi: 10.1128/jcm.33.5.1070-1074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thong K-L, Cheong Y-M, Puthucheary S, Kott C-L, Pang T. Epidemiologic analysis of sporadic Salmonella typhi isolates and those from outbreaks by pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32:1135–1141. doi: 10.1128/jcm.32.5.1135-1141.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Threlfall E J, Hampton M D, Schofield S L, Ward L R, Frost J A, Rowe B. Epidemiological application of differentiating multiresistant Salmonella typhimurium DT 104 by plasmid profile. Communicable Dis Rep CDR Rev. 1996;6:155–158. [PubMed] [Google Scholar]
- 49.Vahaboglu H, Dodanli S, Eroglu C, Ozturk R, Soyletir G, Yildirim I, Avkan V. Characterization of multiple-antibiotic-resistant Salmonella typhimurium strains: molecular epidemiology of PER-1-producing isolates and evidence for nosocomial plasmid exchange by a clone. J Clin Microbiol. 1996;34:2942–2946. doi: 10.1128/jcm.34.12.2942-2946.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vieu J F, Jean Jean S, Tournier B, Klein B. Application d'une série unique de bactériophages à la lysotypie de Salmonella serovar Dublin et de Salmonella serovar enteritidis. Méd Mal Infect. 1990;20:229–233. [Google Scholar]
- 51.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]