Abstract
SARS-CoV-2 variants of concern (VOCs) have emerged worldwide, with implications on the spread of the pandemic. Characterizing the cross-reactivity of antibodies against these VOCs is necessary to understand the humoral response of non-hospitalized individuals previously infected with SARS-CoV-2, a population that remains understudied. Thirty-two SARS-CoV-2-positive (PCR-confirmed) and non-hospitalized Canadian adults were enrolled 14–21 days post-diagnosis in 2020, before the emergence of the B.1.351 (also known as Beta), B.1.617.2 (Delta) and P.1 (Gamma) VOCs. Sera were collected 4 and 16 weeks post-diagnosis. Antibody levels and pseudo-neutralization of the ectodomain of SARS-CoV-2 spike protein/human ACE-2 receptor interaction were analyzed with native, B.1.351, B.1.617.2 and P.1 variant spike proteins. Despite a lower response observed for the variant spike proteins, we report evidence of a sustained humoral response against native, B.1.351, B.1.617.2 and P.1 variant spike proteins among non-hospitalized Canadian adults. Furthermore, this response inhibited the interaction between the spike proteins from the different VOCs and ACE-2 receptor for ≥ 16 weeks post-diagnosis, except for individuals aged 18–49 years who showed no inhibition of the interaction between B.1.617.1 or B.1.617.2 spike and ACE-2. Interestingly, the affinity (KD) measured between the spike proteins (native, B.1.351, B.1.617.2 and P.1) and antibodies elicited in sera of infected and vaccinated (BNT162b2 and ChAdOx1 nCoV-19) individuals was invariant. Relative to sera from vaccine-naïve (and previously infected) individuals, sera from vaccinated individuals had higher antibody levels (as measured with label-free SPR) and more efficiently inhibited the spike–ACE-2 interactions, even among individuals aged 18–49 years, showing the effectiveness of vaccination.
Subject terms: Antibodies, Optical sensors
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected and caused the death of millions of individuals across the globe since 20191. This RNA coronavirus of zoonotic origin has a diameter of 80–90 nm with several structural proteins, including nucleocapsid and spike2. The virus invades and replicates in the lower respiratory tract and causes pneumonia in some infected individuals, which is one of the most frequent complications of the coronavirus disease COVID-19.
The immune system fights the infection by eliciting an innate immune response3, and a B- and T-cell mediated response4,5. The humoral response against SARS-CoV-2 follows a classical pattern in which IgMs and IgAs are expressed 1–2 weeks post-diagnosis during the recovery phase and IgGs are expressed 2–4 weeks post-diagnosis during the convalescence phase6,7. IgGs may be associated with long-term humoral memory as they are detectable several months post-diagnosis8. However, antibody levels appear to be lower for asymptomatic or paucisymptomatic individuals compared to those with severe illness requiring hospitalization9–11. Given this notable difference, studies are needed to better understand the humoral response of non-hospitalized individuals, a population that remains understudied.
As an abundant surface protein with a large, accessible ectodomain, spike protein is the immunogen of SARS-CoV-2 that elicits the strongest humoral response. As a result, the ectodomain of spike protein (simply referred to as ‘spike protein’ here) forms the basis of current mRNA and viral-vector-based vaccines12,13. Spike is a trimeric glycoprotein with each monomer composed of an S1 and S2 subunit. During viral fusion with human cells, the receptor binding domain (RBD) of the S1 subunit binds to the membrane-bound angiotensin-converting enzyme 2 (ACE-2) receptor14, and the S2 subunit mediates membrane fusion. ACE-2 is particularly abundant on the surface of lower respiratory tract cells, which makes them susceptible to infection and can cause pneumonia15,16. The antibodies produced in response to current vaccines work primarily by binding to the RBD of spike protein, thus blocking its interaction with ACE-217, which is thought to mediate their effectiveness.
Over the course of the pandemic, several SARS-CoV-2 variants of concern (VOCs) have emerged in the United Kingdom (B.1.1.7, also named Alpha variant by the WHO), in South Africa (B.1.351, Beta), in Brazil (P.1, Gamma), and in India (B.1.617.1, Kappa and B.1.617.2, Delta). These VOCs are now the dominant strains worldwide and harbor multiple mutations in the spike protein18–20, which raises questions about the effectiveness of the humoral immunity of individuals who were previously infected with the native strain that originated from Wuhan, China (i.e., variant-naïve individuals), and those who were immunized by the first-generation vaccines that use the native spike protein as immunogen. These mutations may affect the ability of antibodies to bind to the virus which may thus evade neutralizing antibodies21–23. It has been reported that the N501Y and K417N mutations present in the B.1.1.7, B.1.351 and P.1 VOC spike protein reduce the activity of antibodies from convalescent and post-vaccination serum or therapeutic monoclonal antibodies24–30. It is suspected that the multiple mutations harbored by spike protein of the VOCs could lead to a conformational change in the RBD of spike and affect binding to ACE-218. Detailed investigations of the cross-reactivity of antibodies are therefore necessary to evaluate the susceptibility of individuals to infection by the VOCs31,32, particularly those infected early during the pandemic as well as those immunized, all of whom are thus presumably naïve to these variants.
Several methodological approaches are available to study the humoral response against SARS-CoV-2. Most serological assays use ELISA to assess the seroprevalence of SARS-CoV-2. These assays typically detect IgGs targeting the S1 subunit or the trimeric spike protein, which improves performance compared with assays that target the RBD33. However, detecting anti-spike IgGs alone provides an incomplete picture of the humoral response, as effective antibodies should inhibit the interaction of spike protein with ACE-234,35. Cell-based neutralization assays are the gold standard but require live viruses and thus a biosafety level 3 (BSL3) lab, which makes these assays costly, complex and lengthy to perform. Cell- and virus-free surrogate or pseudo-neutralization assays could provide valuable functional information on the inhibition of the interaction between spike protein and ACE-2, as recently demonstrated by a surrogate ELISA neutralization assay36. Sensing techniques such as surface plasmon resonance (SPR) provide complementary biochemical data to ELISA. SPR has been used to conduct serological tests37 and to measure various biochemical parameters influencing the strength of the humoral response, including the binding constant of the antibodies to spike or its subunits38, the inhibition of the spike protein:ACE-2 interaction by neutralizing antibodies39,40 and the equilibrium dissociation constant (KD) of recombinant human ACE-2 with the RBD of spike protein41.
In this longitudinal study, we assessed the cross-reactivity of antibodies produced by non-hospitalized, variant-naïve SARS-CoV-2-positive individuals against the native spike protein, as well as the B.1.351, B.1.617.2 and P.1 VOC spike proteins. Notably, an in vitro SPR pseudo-neutralization assay was developed to determine the ability of convalescent sera to inhibit the interaction between native or variant spike proteins and ACE-2, including a limited number of individuals in our cohort who were vaccinated in the late stages of the study.
Results
Cross-reactivity of antibodies with the native and B.1.351 spike proteins
Serum samples from 32 non-hospitalized individuals who tested positive for SARS-CoV-2 (PCR confirmed on average 17.25 days prior to enrollment) were collected at weeks 4 and 16 post-diagnosis. Inclusion criteria included quotas based on age, so that the cohort consisted of four age groups (18–49, 50–59, 60–69 and 70 + years; n = 8 each) that each comprised eight individuals. A second PCR test was conducted at the time of enrollment, and 7 (22%) individuals had a negative test result, indicating that some had fully recovered while most had not. The results of this second PCR-based diagnostic test did not influence eligibility to the current study. Control sera were collected from eight individuals never diagnosed with SARS-CoV-2. Of note, one individual aged 60–69 years did not report on week 16 (n = 7 for this age group).
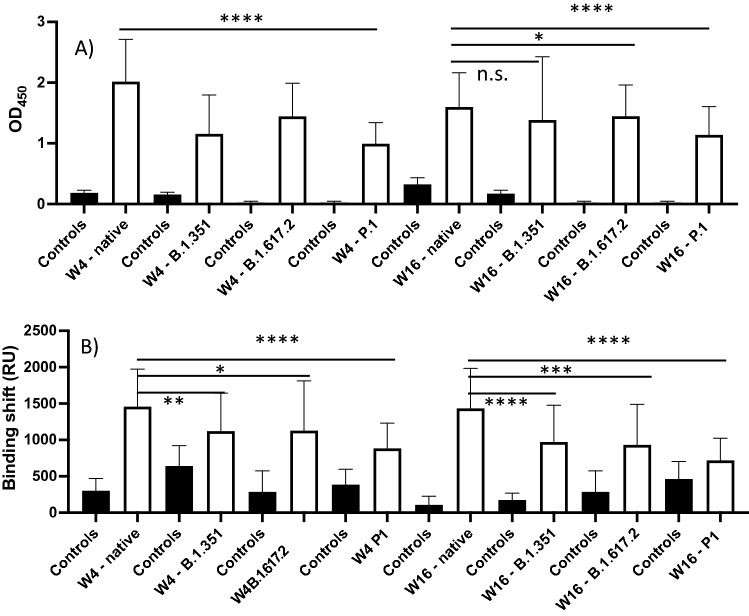
ELISA performed well to identify SARS-CoV-2-positive individuals from the negative control group based on the native spike protein (Area under the curve (AUC) = 1.00 and p < 0.0001), correctly identifying all positive and negative samples (sensitivity = 100%, specificity = 100%, Tables 1, S1, S2, Figs. 1, S1). ELISA results showed no difference among individuals with a positive test result, whether their second PCR test result was negative or positive at the time of enrollment (14–21 days post diagnosis, Fig. S2). The SPR assays also performed well for the native spike protein (AUC = 0.99 and p < 0.0001), correctly identifying all samples with the exception of one positive sample which tested negative at week 4 (sensitivity = 97%, specificity = 100%, Tables 2, S3, S4). Whereas the OD450 decreased by approximately 20% from weeks 4 to 16 for the native spike protein, the SPR binding shift did not change significantly (Fig. 1). The ELISA OD450 and the SPR shift tended to decrease for B.1.351, B.1.617.2 and P.1 at weeks 4 and 16 in comparison to the native spike protein, the greatest difference being week 4 (Figs. 1, S1). As a result, a few positive samples tested negative when SPR assays were conducted with the B.1.351 spike variant, a tendency that increased with the B.1.617.2 and P.1 spike variants (Table 2). This affected the AUCs (range: 0.75–0.99) and the ability of SPR to differentiate positive samples from the negative controls (Fig. S3). However, the performance of ELISA remained excellent (AUC: 1.00 and p < 0.0001 for all VOCs, except for B.1351 at week 16, AUC: 0.99 and p < 0.01, Table 1, Fig. S4).
Table 1.
ELISA for the detection of IgG targeting the native, B.1.351, B.1.617.2, and P.1 spike proteins using serum from variant-naïve, SARS-CoV-2-positive individuals at weeks 4 and 16 post-diagnosis.
| Native spike | B.1.351 spike | B.1.617.2 spike | P.1 spike | ||||||
|---|---|---|---|---|---|---|---|---|---|
| W4 | W16 | W4 | W16 | W4 | W16 | W4 | W16 | ||
| OD450 (A.U) | Pos | 2.0 ± 0.7 | 1.6 ± 0.6 | 1.4 ± 0.7 | 1.5 ± 0.8 | 1.4 ± 0.5 | 1.4 ± 0.5 | 1.0 ± 0.3 | 1.1 ± 0.5 |
| OD450 (A.U) | Neg | 0.18 ± 0.04 | 0.32 ± 0.11 | 0.16 ± 0.04 | 0.17 ± 0.05 | 0.02 ± 0.03 | 0.02 ± 0.03 | 0.02 ± 0.03 | 0.02 ± 0.03 |
| Threshold (A.U) | 0.4 | 0.6 | 0.25 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | |
| COVID + sera | # Pos | 32 | 31 | 32 | 31 | 30 | 30 | 31 | 31 |
| # Neg | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| Control sera | # Pos | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| # Neg | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | |
| AUC | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | |
| p value | < 0.0001 | < 0.0001 | < 0.0001 | < 0.01 | < 0.0001 | < 0.0001 | < 0.0001 | < 0.0001 | |
| Sensitivity | 100 | 100 | 100 | 96 | 100 | 100 | 100 | 100 | |
| Specificity | 100 | 100 | 100 | 88 | 100 | 100 | 100 | 100 | |
Figure 1.
Average ELISA OD450 (A) and SPR binding shifts (B) for the detection of anti-spike IgG in SARS-CoV-2-positive sera (n = 32) at week 4 (W4) and week 16 (W16) post-diagnosis for the native, B.1.351, B.1.617.2, and P.1 spike proteins. Controls are sera from SARS-CoV-2-negative individuals who were never diagnosed with SARS-CoV-2 (n = 8). Error bars represent one standard deviation. n.s., not statistically significant, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Table 2.
SPR assay for the detection of human IgG targeting the native, B.1.351, B.1.617.2, and P.1 spike proteins using serum from variant-naïve, SARS-CoV-2-positive individuals at weeks 4 and 16 post-diagnosis.
| Native spike | B.1.351 spike | B.1.617.2 spike | P.1 spike | ||||||
|---|---|---|---|---|---|---|---|---|---|
| W4 | W16 | W4 | W16 | W4 | W16 | W4 | W16 | ||
| Shift (kRU) | Pos | 1.5 ± 0.5 | 1.4 ± 0.6 | 1.1 ± 0.5 | 1.0 ± 0.5 | 1.1 ± 0.7 | 0.9 ± 0.5 | 0.8 ± 0.3 | 0.7 ± 0.3 |
| Shift (kRU) | Neg | 0.3 ± 0.2 | 0.1 ± 0.1 | 0.6 ± 0.3 | 0.2 ± 0.1 | 0.3 ± 0.3 | 0.3 ± 0.3 | 0.4 ± 0.2 | 0.5 ± 0.2 |
| Threshold (kRU) | 0.6 | 0.35 | 0.7 | 0.35 | 0.7 | 0.7 | 0.6 | 0.7 | |
| Positive sera | # Pos | 31 | 31 | 26 | 27 | 22 | 17 | 27 | 16 |
| # Neg | 1 | 0 | 6 | 4 | 9 | 14 | 5 | 15 | |
| Control sera | # Pos | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 |
| # Neg | 8 | 8 | 7 | 8 | 7 | 7 | 6 | 6 | |
| AUC | 0.99 | 0.99 | 0.82 | 0.93 | 0.87 | 0.83 | 0.90 | 0.75 | |
| P value | < 0.0001 | < 0.0001 | < 0.05 | < 0.0001 | < 0.01 | < 0.01 | < 0.001 | < 0.05 | |
| Sensitivity | 97 | 100 | 81 | 87 | 71 | 55 | 84 | 51 | |
| Specificity | 100 | 100 | 88 | 100 | 88 | 88 | 86 | 86 | |
ELISA and SPR experiments were then repeated with the RBD of native and B.1.351 spike. However, the ability of the RBD assays to discriminate between positive to negative samples was generally lower than that obtained using full-length spike protein (Figs. S5–S7 and Tables S5–S8, compared to Tables 1, 2), in agreement with previous reports in which using the S1 subunit or trimeric spike ectodomain protein improved assay performance2,33,42. Therefore, the full-length spike protein was used for the remaining experiments. Taken together, these results suggest that the sera of variant-naïve, SARS-CoV-2-positive individuals contain antibodies that cross-react with the B.1.351, B.1.617.2 and P.1 spike variants during at least 16 weeks post-diagnosis, although antibody binding appeared reduced for the variants compared with the native protein (Fig. 1), in agreement with a previous report43.
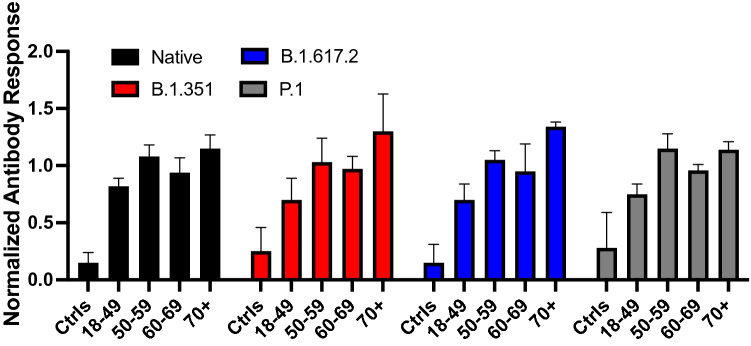
When stratified by age, antibody binding increased with age for native spike. This trend was even more evident for data acquired with the VOCs spike proteins (Fig. 2). On average, individuals in the 70 + years group exhibited ELISA and SPR responses 15% (native) to 30% (VOCs) above the mean of the overall cohort, those aged 50–59 or 60–69 years exhibited responses within 10% of the mean, and those aged 18–49 years exhibited responses 18% (native) to 30% (VOCs) below the mean.
Figure 2.
Average normalized antibody binding stratified by age group for human anti-IgGs targeting the native (black), B.1.351 (red), B.1.617.2 (blue) and P.1 (gray) spike proteins among variant-naïve, non-hospitalized individuals. SPR and ELISA data from weeks 4 and 16 post-diagnosis were normalized to the mean of the overall cohort and pooled. A normalized antibody binding of 1 thus refers to a response equivalent to the average of all SARS-CoV-2-positive individuals (n = 8 for each data set) and the error bars correspond to one standard deviation). Ctrls: SARS-CoV-2-negative sera (n = 8).
Label-free response and affinity of antibodies produced by variant-naïve individuals against native and variant spike proteins
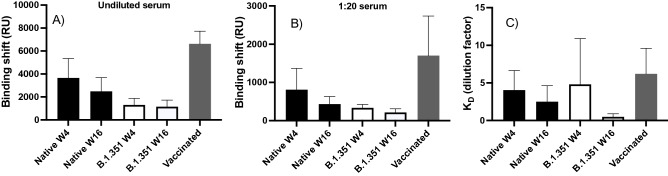
SPR sensing is ideally suited for the measurement of antibodies and proteins in moderately diluted (i.e., 1:10) to undiluted serum44. Because SPR is a label-free method, it can be used to study biochemical properties of protein–protein interactions, such as the KD. Prior studies have successfully used SPR to study the affinity of antibodies produced following SARS-CoV-2 vaccination45 or COVID-19 infection38. Hence, we adapted the SPR assay to measure antibody binding and affinity for the native spike protein based on the sera of four SARS-CoV-2-positive individuals in each of the four age groups; we also determined affinity for the B.1.351 variant spike protein for the individuals aged 18–49 years (Fig. S8). As a result of vaccination roll-out in late 2020/early 2021, a limited number of the SARS-CoV-2-positive individuals in our cohort were vaccinated in the late stages of the study (Table S12). Their sera samples were acquired 24 weeks post-diagnosis and at least 2 weeks post-vaccination. Analysis of five of these allowed for a first glance into the impact of vaccination on affinity of their antibody development.
The SPR binding shift for the undiluted sera decreased slightly between weeks 4 and 16 for the native spike protein (p = 0.03) and remained constant for B.1.351 (p = 0.73) spike protein (Fig. 3). The SPR binding shift decreased from weeks 4 to 16 when using a 1:20 dilution for the native spike protein (p = 0.02) and tended to decrease for the B.1.351 (p = 0.11) spike protein. As shown earlier, a similar decrease was obtained using ELISA (Fig. 1), which was performed using samples at a greater dilution (1:50). For the B.1.351 variant, the SPR signal appeared modestly lower (although not statistically significant at week 4) than that observed for native spike at week 4 (p = 0.11) and week 16 (p = 0.05; Fig. 3). In comparison, vaccinated individuals exhibited a twofold higher SPR response for both 1:20 dilution (p = 0.02) and undiluted samples (p < 0.01).
Figure 3.
SPR binding shift for undiluted serum (A) and serum at a 1:20 dilution (B) from SARS-CoV-2-positive individuals at weeks 4 (W4) and 16 (W16) post-diagnosis for the native (black, n = 16, 4 age groups) and B.1.351 (white, n = 4, 18–49 years) spike proteins, and for vaccinated individuals (gray, n = 5) for the native spike protein. (C) A dissociation constant was estimated from the dilution series assuming a 1:1 binding model according to the data presented in panels (A) and (B). The dilution series used in SPR data acquisition consisted of the following: 1:40, 1:20, 1:10, 1:5, 1:2.5 and undiluted serum. In all panels, error bars represent one standard deviation.
The label-free SPR assay revealed that binding shift increased almost linearly with serum dilution (Fig. S8—left panel), and KDs were extracted by fitting a 1:1 binding site Langmuir isotherm (Fig. S8—right panel). Because the serum concentration is expressed as a dilution factor (i.e., a high dilution factor corresponds to low antibody levels), a high KD corresponds to a high affinity of the antibodies for spike. Attempts to extract the association (kon) and dissociation (koff) rates were unsuccessful since sera contain a mixture of polyclonal antibodies. The estimated KD appeared to decrease with time for the two spike proteins, but the difference was not statistically significant (native: p = 0.08 and B.1.351: p = 0.29) between weeks 4 to 16 (Fig. 3C). No significant difference in KD was observed between native and B.1.351 spike proteins at week 4 (p = 0.71), week 16 (p = 0.13) or with vaccinated individuals (p = 0.15).
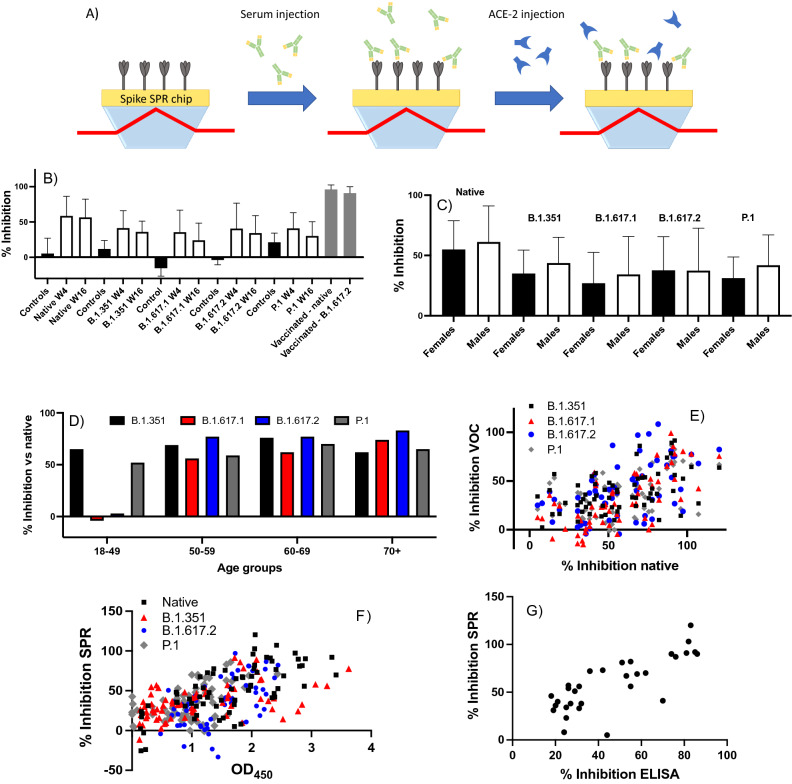
Pseudo-neutralization SPR inhibition assay
To better understand the potential functional implications of antibody cross-reactivity, we adapted a previously reported in vitro pseudo-neutralization assay39,46 to measure how antibody binding to spike protein affects its interaction with ACE-2 (details in supporting information, Schemes S1 and S2, Figs. S9–S11 and Tables S9, S10). The difference in inhibition of the spike–ACE-2 interaction between weeks 4 and 16 post-diagnosis was not statistically significant for the native (mean inhibition %: week 4 = 59%, week 16 = 57%, p = 0.67), B.1.351 (week 4 = 41%, week 16 = 36%, p = 0.1), and B.1.617.2 spike proteins (week 4 = 41%, week 16 = 34%, p = 0.1), while statistically significant reductions were observed for the B.1.617.1 (week 4 = 36%, week 16 = 24%, p < 0.01) and P.1 spike proteins (week 4 = 41%, week 16 = 30%, p < 0.01; Table S11). Sera from variant-naïve SARS-CoV-2-positive individuals inhibited the spike–ACE-2 interaction significantly less for all variants compared to the native spike protein at week 4 (B.1.351, p < 0.0001; B.1.617.1, p < 0.0001; B.1.617.2, p < 0.01; P.1, p < 0.0001; Fig. 4 and Table S11) and at week 16 (B.1.351, p < 0.001; B.1.617.1, p < 0.0001; B.1.617.2, p < 0.001; P.1, p < 0.0001; Fig. 4 and Table S11). No significant difference in inhibition was observed between female and male individuals for the native (females = 55%, males = 61%, p = 0.37), B.1.351 (females = 35%, males = 44%, p = 0.10), B.1.617.1 (females = 27%, males = 34%, p = 0.31), B.1.617.2 (females = 38%, males = 37%, p = 0.98), and P.1 (females = 31%, males = 42%, p = 0.051) spike proteins (Fig. 4C). The normalized inhibition for the variants relative to the native spike protein was relatively constant (range: 56–83%, Fig. 4D) for the different age groups, except for the 18–49 age group which showed no inhibition (normalized inhibition of − 4% and 3%) for the two variants in the B.1.617 lineage. The percent inhibition observed for each individual showed only a mild correlation between native and B.1.351 spike proteins (r = 0.46; Fig. 4E). However, the percent inhibition correlated well with the ELISA OD450 results of the native spike proteins (r = 0.70) and moderately well with the VOCs (B.1.351: r = 0.52, B.1.351: r = 0.54, and P.1: r = 0.51), suggesting that anti-spike IgG concentration is a predictor of the inter-individual variation in the ability of sera to inhibit the interaction between spike protein and ACE-2 (Fig. 4F). The percent inhibition obtained with SPR also correlated well (r = 0.78) with the results of a similar pseudo-neutralization assay performed with ELISA (Fig. 4G), adapted from a recently reported protocol47. Finally, the cohort of nine vaccinated individuals (BNT162b2 n = 8 and ChAdOx1 nCoV-19 n = 1) showed significantly higher percent inhibition to the native (96% ± 6%, p < 0.001 vs infected only) and B.1.617.2 (91% ± 9%, p < 0.001 vs infected only) spike proteins in comparison to vaccine-naïve infected individuals (Fig. 4B, Table S12).
Figure 4.
(A) Schematic illustrating the principle of the SPR pseudo-neutralization assay to quantify the inhibition of the interaction between spike protein and ACE-2 in the presence of SARS-CoV-2-positive sera. Serum from an individual who tested positive for SARS-CoV-2 is injected onto an SPR chip that has been functionalized with spike protein, and a preparation of recombinant human ACE-2 is subsequently injected. A reduced SPR response (and thus higher percent inhibition) is observed if the serum antibodies inhibit the spike–ACE-2 interaction. (B) Observed inhibition of the interaction of ACE-2 with native, B.1.351, B.1.617.2, and P.1 spike proteins by variant-naïve SARS-CoV-2-positive sera (n = 32). Controls: SARS-CoV-2-negative sera (n = 8). Error bars represent one standard deviation. (C) Observed inhibition of the interaction of ACE-2 with native, B.1.351, B.1.617.2, and P.1 spike proteins by variant-naïve SARS-CoV-2-positive sera as a function of sex (females: n = 18; males: n = 14). (D) Normalized inhibition for the B.1.351, B.1.617.1, B.1.617.2 and P.1 variants expressed as a percentage of inhibition compared to the native spike protein for the same age group (n = 8 for each age group). Data from weeks 4 and 16 are pooled. (E) Correlation between the percent inhibition (for weeks 4 and 16 post-diagnosis combined) observed for native and for the B.1.351, B.1.617.1, B.1.617.2 and P.1 spike proteins; each dot corresponds to an average of 2 replicates for a single individual (n = 31 or 32 depending on the data sets). (F) Correlation between the ELISA OD450 results (Fig. 1 and Tables S1, S2) and pseudo-neutralization results for weeks 4 and 16 combined (n = 31 or 32 depending on the data sets). (G) Correlation between the ELISA and SPR pseudo-neutralization results obtained with the native spike protein for the samples at week 4 (n = 31).
Discussion
The protein–protein interaction data reported in this study show cross-reactivity of antibodies produced by variant-naïve, PCR-confirmed SARS-CoV-2-positive individuals against the B.1.351, B.1.617.2, and P.1 VOCs. Notably, none of the SARS-CoV-2-positive individuals included in this study experienced severe symptoms that required hospitalization; this population remains understudied in the existing literature.
Age correlated well with antibody binding: individuals aged 70 + years generally had higher than average ELISA OD450 and SPR responses, those younger than 49 years exhibited below average responses, and those aged 50–69 years exhibited intermediate responses (Fig. 2). This was exacerbated with the Delta variant (B.1.617.2) and its closely-related B.1.617.1 variant. Convalescent sera from individuals younger than 49 years were not able to inhibit the interaction of the spike protein of those two variants with ACE-2, a result in agreement with a recent study showing that the B.1.617.2 variant was less sensitive to sera from naturally immunized individuals20. Other studies also found that age is positively correlated with antibody levels as measured by ELISA among individuals hospitalized for COVID-1948 or any cause (whether it be COVID-19 or any other condition)49. Our study builds on this evidence by showing a similar correlation for the spike proteins of the B.1.351, B.1.617.2 and P.1 VOCs.
The higher ELISA and SPR signals observed against native spike protein than spike proteins for VOCs (Fig. 1, Tables 1, 2) are consistent with the period of our sample acquisition, prior to reports of any VOC and were thus presumably infected with the native strain. This agrees with a report suggesting that antibodies produced by variant-naïve individuals cross-reacted with the B.1.351 spike protein, albeit at lower titer50.
Furthermore, KDs (expressed in units of dilution) appeared to be higher (therefore tighter binding) at week 4 than week 16 for native and B.1.351 spike proteins, but the difference was not statistically different. Further data would need to be acquired to validate if there is presence of greater affinity of antibodies to all the spike proteins tested earlier post-diagnosis. Taken together, these results suggest that the affinity of antibodies wanes between weeks 4 and 16 post-diagnosis, and that antibody binding levels were lower in sera of individuals 16 weeks post-diagnosis. Our confirmation that the antibodies elicited in variant-naïve, SARS-CoV-2-positive individuals effectively cross-react with B.1.351, B.1.617.2 and P.1 spike protein up until at least 16 weeks post-diagnosis agrees with other reports8,51–55.
While informative, biochemical data on antibody binding provide only a partial picture of the humoral response. The pseudo-neutralization SPR assay showed that convalescent sera effectively inhibited the interaction between ACE-2 and the native or B.1.351, B.1.617.1, B.1.617.2 and P.1 spike proteins. The decrease in inhibition for the VOCs in comparison to the native spike protein agrees with a recent report on vaccinated individuals which used a lentiviral spike pseudotyped virus assay56. Furthermore, the degree of inhibition correlated with the observed ELISA response for anti-spike protein IgGs (Fig. 4), suggesting that antibody levels explain a large proportion of the inter-individual variation in the ability of sera to inhibit the spike–ACE-2 interaction. Consistent with the ELISA response being higher for the native protein, inhibition of this interaction was more important for the native spike protein. Other reports found that the levels of neutralizing antibodies decreased within 3 months post-diagnosis52,57–59, which was not seen here in non-hospitalized individuals. One of those reports associated IgMs with improved neutralization early post-diagnosis57. While such association with antibody isotype cannot be established with our data, the antibody levels and the apparent decrease in KD from weeks 4 to 16 post-diagnosis tend to support that antibody affinity is more efficient shortly after infection. These results suggest that antibodies from convalescent sera can inhibit the interaction of spike protein with ACE-2, although the degree of inhibition was larger for native spike protein given that included individuals were presumably infected with the native strain of SARS-CoV-2.
Label-free SPR measurements indicated that vaccination led to an increase in antibody levels in previously infected individuals. However, higher antibody levels did not appear to improve the KD of the antibodies in vaccinated individuals. Vaccination improved inhibition of the spike–ACE-2 interaction, seeing > 90% percent inhibition in sera of vaccinated individuals compared to < 60% for vaccine-naïve individuals. Interestingly, one previously infected individual received two doses of vaccine over the 24-week follow-up period (i.e. at weeks 5 and 20), allowing for the assessment of the humoral response before and after vaccination. Of note, this individual was aged 18–49 years, an age group that showed a relatively poor inhibition of the spike–ACE-2 interaction (especially that involving the B.1.617.2 spike protein). At weeks 2 and 4 (i.e. before vaccination, post-infection), the percent inhibition of this individual was < 20% for the native and B.1.617.2 spike proteins (Fig. S12), in agreement with the low percent inhibition observed in that age group (Fig. 4D, Table S11). After week 4 (i.e. post-vaccination), the percent inhibition increased > 80% and was sustained for at least 20 weeks following vaccination for both the native and B.1.617.2 spike proteins (Fig. S12). These data provide further evidence that vaccination is beneficial to elicit neutralizing antibodies to the B.1.617.2 variant, which is currently dominant across the world.
This study has some limitations, including the limited number of enrollees and the small number of time points assessed post-diagnosis. Increasing sample size should improve the representativeness for some of the experiments. Furthermore, it remains unclear whether ELISA and SPR data may be used to predict the effectiveness of an individual’s humoral response to prevent a new infection, whether from the native strain or from a variant strain, since clinical outcomes following a subsequent SARS-CoV-2 exposure were not collected. As vaccination progresses, this question will also apply to immunized individuals. Despite the in vitro nature of these experiments, cell-based assays should yield largely similar results since the results of another in vitro, surrogate neutralization assay were found to strongly correlate with those of a cell-based neutralization assay36,60,61.
In conclusion, antibodies in sera of SARS-CoV-2-positive, variant-naïve individuals cross-reacted with the spike protein of the B.1.351, B.1.617.2 and P.1 VOCs, albeit with a decrease in antibody binding as measured by ELISA and SPR. Antibody levels decreased from weeks 4 to 16 and were positively correlated with age. SPR results suggested that the higher affinity and higher concentration antibodies wane from weeks 4 to 16 post-diagnosis. However, remaining antibodies effectively inhibited the interaction between native and variant spike proteins and recombinant human ACE-2. ELISA results correlated with the degree of inhibition, suggesting that high antibody levels are needed for optimal pseudo-neutralization. Vaccination increased the antibody levels and strongly improved the percent inhibition of the spike–ACE-2 interaction. Taken together, these results suggest that variant-naïve, non-hospitalized, SARS-CoV-2-positive individuals have sustained humoral immunity at later times post-diagnosis and that vaccination improves the humoral response to SARS-CoV-2.
Experimental section
Materials
N-Ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride, (EDC, crystalline, cat. no. E6383), N-hydroxysuccinimide (NHS, 98%, cat. no. 130672), ethanolamine hydrochloride (≥ 99.0%, cat. no. E6133), glycine hydrochloride (≥ 99.0%, cat. no. G2879), bovine serum albumin (≥ 98.0%, cat. no. 5470), Tween20 (cat. no. P.1379), and human AB serum (cat. no. H4522) were obtained from Sigma Aldrich. The running buffer was composed of phosphate buffer saline 1X (VWR, cat. no. L0119), 0.1% BSA, and 0.005% Tween20. Goat anti-human IgG (Jackson Immunoresearch, cat. no. 109-005-003) and human recombinant ACE-2 (Sino Biologicals, cat. no. 10108-H08H) were obtained from commercial sources. Native (PRO1-429 (SmT1-162), B.1.351 (PRO6429-1 (SmT1 (SA))), B.1.617.1 (PRO7109-1 (SmT1 (B.1.617.1))), B.1.617.2 (PRO7176-1 (SmT1 (B.1.617.2))) and P.1 (PRO6875-2 (SmT1v3 (BR))) spike proteins and biotin-ACE-2 (PRO5436-5 (SH6F-ACE2-BAP)) were obtained from the National Research Council of Canada and expressed based on the protocols reported elsewhere63.
ELISA assays
Semi-quantitative ELISA was performed based on the protocols of Krammer and of Finzi and Bazin11,64,65, as recently reported37. The sera were heat inactivated for 60 min at 55 °C in a heating block and diluted 1:50 before use. ELISA operates at a greater dilution factor (1:50) compared to SPR (1:5). Therefore, ELISA preferentially measures high concentration and high affinity (high KD) antibodies, whereas SPR measures both high and low concentration or high and low affinity antibodies.
The relevant SARS-CoV-2 antigenic spike protein was diluted in PBS at a concentration of 2.5 μg/mL. Immulon 1B 96-well plates (Thermo Fischer Scientific) were coated with 100 μL of diluted antigen and incubated at 4 °C overnight. In parallel, clinical samples were inactivated for 1 h at 56 °C in a heating block, then kept at 4 °C overnight. The following day, plates were washed 4 times with PBS-T using a 50 TS Microplate Washer (Biotek) automated plate washer followed by addition of 300 μL of blocking solution (PBS-T + 3% (w/v) milk powder) to each well. After 1 h of incubation at RT, plates were washed 4 times with PBS-T. Serum samples were diluted 1:50 in PBS with 0.1% Tween20 and 1% milk powder, in a 96-well polystyrene dilution plate and 100 μL was added to each well. Plates were incubated for 1 h at RT and washed 4 times with PBS-T. The secondary antibody (100 μL of 1:10,000 dilution Anti-Human IgG (gamma-chain specific)-Peroxidase antibody produced in goat, Sigma-Aldrich, cat. no. A6029-1ML) was added. Plates were incubated for 1 h at RT then washed 4´ with PBS-T. Addition of 100 μL of TMB (3,3',5,5'-tetramethylbenzidine, Sigma-Aldrich) to each well was followed by a 20 min incubation at RT. Color development was initiated by addition of 100 μL of 2 M HCl. Absorbance was immediately recorded at 450 nm and 595 nm (plate background, subtracted from absorbance at 450 nm) in a FLUOstar Optima microplate reader (BMG Labtech).
SPR measurements for IgG detection
The methods for the SPR detection of human IgG antibodies were described recently37 using a portable SPR instrument66 (Affinité Instruments, Canada). In brief, after NHS and EDC activation for 2 min, the spike proteins or RBD for the native strain of SARS-CoV-2 or the B.1.351 variants were immobilized at a concentration of 20 µg/mL for 20 min in pH 4.5 acetate buffer and the sensors were washed with 1 M ethanolamine pH 8.5 (10 min) and running buffer composed of pH 7.4 PBS (137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, pH 7.4) supplemented with 0.1% bovine serum albumin (BSA) and 0.005% Tween 20 as described previously. Serum samples were diluted 1:5 in the running buffer and injected for 10 min. Following a quick wash with running buffer, secondary detection was performed for 10 min with a 40 µg/mL solution for the spike protein and 20 µg/mL for RBD of AffiniPure goat anti-human IgG (H + L). The surface was regenerated with 10 mM glycine pH 2.2 solution for a few seconds and washed with running buffer before the next set of sera were injected. Experiments were performed with the SPR instrument inside a laminar flow cabinet in a biosafety Level 2 (BSL2) laboratory. The SPR instrument had 4 independent channels on the fluidic cell allowing the measurement of up to 4 samples in a single run.
Antibody binding affinity
For affinity measurements using SPR, sera were diluted in the running buffer with dilutions ranging from 1:40 to no dilution (undiluted serum). The sera were then injected sequentially from the greatest dilution to the undiluted sera. Samples were measured in triplicate and the reference channel was used for the background response correction due to nonspecific binding of negative sera at the same dilution factors. In this case, the fluidic cell of the SPR system was identical to that previously described by Zhao et al.66. Data collected at equilibrium were fit to a single binding site Langmuir model to extract a KD in dilution titer.
Surrogate inhibition assay
A SPR assay was designed to serve as an in vitro surrogate to cell-based neutralization assay. The spike proteins for the native strain of SARS-CoV-2 or the B.1.351 variant were immobilized on the SPR chip as described above. Following the passivation of the surface with ethanolamine, the SPR sensor was equilibrated in a commercial serum exempt of anti-spike antibody diluted 1:5 in running buffer for typically 10–15 min until a stable baseline. Then, different samples were injected on the four channels of the SPR instrument. On two channels, sera from an individual in the negative control group or sera from a PCR-confirmed SARS-CoV-2 infected individual (4- and 16-weeks post infection were tested) diluted 1:5 in running buffer were reacted with the spike protein for 10 min. Following a quick wash with running buffer for a few seconds, human recombinant ACE-2 was then injected at a concentration of 5 µg/mL for 10 min. If antibodies blocking the interaction of human recombinant ACE-2 with the spike protein were present, a lower SPR response was recorded; signals were compared to a positive and a negative control run in the other two channels. The positive control consisted of an identical SPR chip but using a SARS-COV-2-negative, pooled commercial serum and for which human recombinant ACE-2 was then injected to obtain the maximum SPR signal from the human recombinant ACE-2–spike interaction. The negative control consisted in the injection of the same serum sample diluted 1:5 with running buffer as in the measurement channels, but with the injection of running buffer (no human recombinant ACE-2) in otherwise identical conditions; the background response was subtracted from the measurements. No heat treatment was applied to the sera to ensure native conditions for the measurement of pseudo-neutralization.
Clinical samples
Adult volunteers were recruited after written informed consent at the Centre Hospitalier Universitaire de Québec—Université Laval (CHUL, approved by the “Comité d’éthique de la recherche du CHU de Québec-UL”, Registration Number 2021-5241) in Quebec City, Canada. All experiments were performed in accordance with relevant guidelines and regulations. Sera from the same cohort of individuals as previously reported were used in this study37. Blood samples were collected and processed to obtain the sera as previously described37. All nasopharyngeal samples (outpatients and inpatients) were PCR-tested for COVID-19 at a reference laboratory early during the pandemic, but prior to enrollment. The database storing these results was searched, and the patients who had a positive nasopharyngeal PCR test result for COVID-19 (index PCR) were sent a letter explaining the study and giving contact information if they were interested to participate. Interested participants were enrolled if they satisfied the following criteria: aged > 18 years, and neither hospitalized nor admitted to intensive care unit at the time of enrollment. Individuals were recruited in four age group: 18–49, 50–59, 60–69 and 70 + years of age at the time of enrollment; they had received a positive PCR diagnosis (index PCR) for COVID-19 four weeks prior to serum collection and had blood drawn at 16 weeks post infection. Of these, eight samples were randomly selected in each age group for a total of 32 participants, 14 males and 18 females. In one case, a volunteer in the 60–69 age group did not provide a sample on week 16, resulting in n = 7 for that data point. One control nasopharyngeal PCR was done 14–21 days post index PCR (average of 17.25 days); 78% of the participants (25/32) still had a positive PCR. Participants were enrolled regardless of the result of this control PCR. One participant had no symptom related to COVID-19. All other volunteers31 were considered mildly symptomatic with an average of 3 symptoms among fever, myalgia, headache, sore throat, new olfactory or taste disorder, cough or difficulty breathing. Negative controls were collected from eight individuals (age range: 20–55 and median: 47.5 years of age, 7 females and 1 male) having never received a COVID-positive test. Enrollment was completed prior to October 1st 2020. According to the Institut National de Santé Publique du Québec (INSPQ, Quebec National Public Health Institute), the local public health authority where the study was conducted, the first cases of the B.1.351 VOC was reported in early February 202167, while the other variants (B.1.617.1, B.1.617.2 and P.1) were reported even later. Hence, all individuals in the current study were infected with the native SARS-CoV-2 strain originating from Wuhan. In all cases, sera from 8 individuals in the following age groups (18–49, 50–59, 60–69 and 70 + years old) were compared to 8 negative controls from individuals with no confirmed exposure to SARS-CoV-2. In addition to the vaccine-naïve individuals reported above, nine previously infected individuals were vaccinated during the study with either the ChAdOx1 nCoV-19 or the BNT162b2 vaccines and subsequently provided study-related post-vaccination samples. One of these individuals provided six samples between weeks 2 to 24 following a PCR positive SARS-CoV-2 tests, where vaccination occurred on weeks 5 and 20 post diagnosis.
Statistics
Statistical values (AUC and p values) were calculated with GraphPad Prism version 9.1.0. Means were compared with paired or unpaired two-tailed t-tests when appropriate. The thresholds were generally established as the response from the mean of the controls plus two standard deviations.
Supplementary Information
Acknowledgements
The authors acknowledge financial support from the Canadian Institutes of Health Research (CIHR), the Natural Sciences and Engineering Research Council of Canada (NSERC), the Pandemic Response Challenge Program of the National Research Council Canada and the Canadian Foundation for Innovation (CFI).
Author contributions
Experiments and experimental analysis was done by M.H.J., A.D., P.R., E.L., S.C.-G., J.C., M.-F.P., F.D., M.S., C.G., M.-P.C, M.-J.dG, and Da.B.. Methodology was conceived by Y.D., S.T., De.B., J.N.P. and J.-F.M. Funding was acquired by S.T., De.B., J.N.P. and J.-F.M. Writing of the original draft was done by J.-F.M, S.R.., M.-P.C, M.-J.dG, and Da.B., and was revised and edited by J.N.P, F.C., De.B., Y.D. and S.T.. All authors reviewed the manuscript.
Competing interests
J.-F.M. and J.N.P. have financial interest in Affinité Instruments. The authors declare that they have no other competing interests.
Footnotes
The original online version of this Article was revised: The original version of this Article contained an error in the spelling of the author Stella Cellier-Goetghebeur which was incorrectly given as Stella Cellier-Goethebeur.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Maryam Hojjat Jodaylami, Abdelhadi Djaïleb and Pierre Ricard.
Change history
11/19/2021
A Correction to this paper has been published: 10.1038/s41598-021-02484-9
Contributor Information
Joelle N. Pelletier, Email: joelle.pelletier@umontreal.ca
Jean-Francois Masson, Email: jf.masson@umontreal.ca.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-00844-z.
References
- 1.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huergo MAC, Thanh NTK. Current advances in the detection of COVID-19 and evaluation of the humoral response. Analyst. 2021;146:382–402. doi: 10.1039/d0an01686a. [DOI] [PubMed] [Google Scholar]
- 3.McKechnie JL, Blish CA. The innate immune system: Fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe. 2020;27:863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J, Liang B, Chen C, Wang H, Fang Y, Shen S, Yang X, Wang B, Chen L, Chen Q, Wu Y, Liu J, Yang X, Li W, Zhu B, Zhou W, Wang H, Li S, Lu S, Liu D, Li H, Krawczyk A, Lu M, Yang D, Deng F, Dittmer U, Trilling M, Zheng X. SARS-CoV-2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID-19. Nat. Commun. 2021;12:1813. doi: 10.1038/s41467-021-22034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox RJ, Brokstad KA. Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nat. Rev. Immunol. 2020;20:581–582. doi: 10.1038/s41577-020-00436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galipeau Y, Greig M, Liu G, Driedger M, Langlois M-A. Humoral responses and serological assays in SARS-CoV-2 infections. Front. Immunol. 2020;11:3382. doi: 10.3389/fimmu.2020.610688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, Dela Cruz CS, Wang Y, Wu C, Xiao Y, Zhang L, Han L, Dang S, Xu Y, Yang Q-W, Xu S-Y, Zhu H-D, Xu Y-C, Jin Q, Sharma L, Wang L, Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anand SP, Prévost J, Nayrac M, Beaudoin-Bussières G, Benlarbi M, Gasser R, Brassard N, Laumaea A, Gong SY, Bourassa C, Brunet-Ratnasingham E, Medjahed H, Gendron-Lepage G, Goyette G, Gokool L, Morrisseau C, Bégin P, Martel-Laferrière V, Tremblay C, Richard J, Bazin R, Duerr R, Kaufmann DE, Finzi A, et al. Longitudinal analysis of humoral immunity against SARS-CoV-2 Spike in convalescent individuals up to 8 months post-symptom onset. Cell Rep. Med. 2021;2:100290. doi: 10.1016/j.xcrm.2021.100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long Q-X, Tang X-J, Shi Q-L, Li Q, Deng H-J, Yuan J, Hu J-L, Xu W, Zhang Y, Lv F-J, Su K, Zhang F, Gong J, Wu B, Liu X-M, Li J-J, Qiu J-F, Chen J, Huang A-L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 10.Ravichandran S, Lee Y, Grubbs G, Coyle EM, Klenow L, Akasaka O, Koga M, Adachi E, Saito M, Nakachi I, Ogura T, Baba R, Ito M, Kiso M, Yasuhara A, Yamada S, Sakai-Tagawa Y, Iwatsuki-Horimoto K, Imai M, Yamayoshi S, Yotsuyanagi H, Kawaoka Y, Khurana S. Longitudinal antibody repertoire in “mild” versus “severe” COVID-19 patients reveals immune markers associated with disease severity and resolution. Sci. Adv. 2021;7:eabf2467. doi: 10.1126/sciadv.abf2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prévost J, Gasser R, Beaudoin-Bussières G, Richard J, Duerr R, Laumaea A, Anand SP, Goyette G, Benlarbi M, Ding S, Medjahed H, Lewin A, Perreault J, Tremblay T, Gendron-Lepage G, Gauthier N, Carrier M, Marcoux D, Piché A, Lavoie M, Benoit A, Loungnarath V, Brochu G, Haddad E, Stacey HD, Miller MS, Desforges M, Talbot PJ, Maule GTG, Côté M, Therrien C, Serhir B, Bazin R, Roger M, Finzi A. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep. Med. 2020;1:100126. doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schäfer A, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Martinez DR, Williamson LE, Chen EC, Jones T, Day S, Myers L, Hassan AO, Kafai NM, Winkler ES, Fox JM, Shrihari S, Mueller BK, Meiler J, Chandrashekar A, Mercado NB, Steinhardt JJ, Ren K, Loo Y-M, Kallewaard NL, McCune BT, Keeler SP, Holtzman MJ, Barouch DH, Gralinski LE, Baric RS, Thackray LB, Diamond MS, Carnahan RH, Crowe JE. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snijder EJ, Limpens RWAL, de Wilde AH, de Jong AWM, Zevenhoven-Dobbe JC, Maier HJ, Faas FFGA, Koster AJ, Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alanagreh LA, Alzoughool F, Atoum M. The human coronavirus disease COVID-19: Its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020;9:331. doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nel AE, Miller JF. Nano-enabled COVID-19 vaccines: Meeting the challenges of durable antibody plus cellular immunity and immune escape. ACS Nano. 2021;5:5793–5818. doi: 10.1021/acsnano.1c01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson G, Buzko O, Spilman P, Niazi K, Rabizadeh S, Soon-Shiong P. Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. bioRxiv. 2021 doi: 10.1101/2021.01.13.426558. [DOI] [Google Scholar]
- 19.Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DS, Mishra S, Crispim MAE, Sales FCS, Hawryluk I, McCrone JT, Hulswit RJG, Franco LAM, Ramundo MS, de Jesus JG, Andrade PS, Coletti TM, Ferreira GM, Silva CAM, Manuli ER, Pereira RHM, Peixoto PS, Kraemer MUG, Gaburo N, Camilo CC, Hoeltgebaum H, Souza WM, Rocha EC, de Souza LM, de Pinho MC, Araujo LJT, Malta FSV, de Lima AB, Silva JP, Zauli DAG, Ferreira ACS, Schnekenberg RP, Laydon DJ, Walker PGT, Schlüter HM, dos Santos ALP, Vidal MS, Del Caro VS, Filho RMF, dos Santos HM, Aguiar RS, Proença-Modena JL, Nelson B, Hay JA, Monod M, Miscouridou X, Coupland H, Sonabend R, Vollmer M, Gandy A, Prete CA, Nascimento VH, Suchard MA, Bowden TA, Pond SLK, Wu C-H, Ratmann O, Ferguson NM, Dye C, Loman NJ, Lemey P, Rambaut A, Fraiji NA, Carvalho MPSS, Pybus OG, Flaxman S, Bhatt S, Sabino EC. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 21.Saunders KO, Lee E, Parks R, Martinez DR, Li D, Chen H, Edwards RJ, Gobeil S, Barr M, Mansouri K, Alam SM, Sutherland LL, Cai F, Sanzone AM, Berry M, Manne K, Bock KW, Minai M, Nagata BM, Kapingidza AB, Azoitei M, Tse LV, Scobey TD, Spreng RL, Rountree RW, DeMarco CT, Denny TN, Woods CW, Petzold EW, Tang J, Oguin TH, Sempowski GD, Gagne M, Douek DC, Tomai MA, Fox CB, Seder R, Wiehe K, Weissman D, Pardi N, Golding H, Khurana S, Acharya P, Andersen H, Lewis MG, Moore IN, Montefiori DC, Baric RS, Haynes BF. Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature. 2021;594:553–559. doi: 10.1038/s41586-021-03594-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abu-Raddad LJ, Chemaitelly H, Butt AA. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N. Engl. J. Med. 2021;385:187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdool Karim SS, de Oliveira T. New SARS-CoV-2 variants: Clinical, public health, and vaccine implications. N. Engl. J. Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, Stowe J, Tessier E, Groves N, Dabrera G, Myers R, Campbell CNJ, Amirthalingam G, Edmunds M, Zambon M, Brown KE, Hopkins S, Chand M, Ramsay M. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe JE, Bloom JD. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e49. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M, Wei Y, Atwal GS, Murphy AJ, Stahl N, Yancopoulos GD, Kyratsous CA. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, Schaefer-Babajew D, Cipolla M, Gaebler C, Lieberman JA, Oliveira TY, Yang Z, Abernathy ME, Huey-Tubman KE, Hurley A, Turroja M, West KA, Gordon K, Millard KG, Ramos V, Da Silva J, Xu J, Colbert RA, Patel R, Dizon J, Unson-O’Brien C, Shimeliovich I, Gazumyan A, Caskey M, Bjorkman PJ, Casellas R, Hatziioannou T, Bieniasz PD, Nussenzweig MC. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, Lambson BE, de Oliveira T, Vermeulen M, van der Berg K, Rossouw T, Boswell M, Ueckermann V, Meiring S, von Gottberg A, Cohen C, Morris L, Bhiman JN, Moore PL. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 29.Wang P, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, Liu L, Kwong PD, Huang Y, Shapiro L, Ho DD. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751.e744. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cele S, Gazy I, Jackson L, Hwa S-H, Tegally H, Lustig G, Giandhari J, Pillay S, Wilkinson E, Naidoo Y, Karim F, Ganga Y, Khan K, Bernstein M, Balazs AB, Gosnell BI, Hanekom W, Moosa M-YS, Abrahams S, Alcantara LCJ, Alisoltani-Dehkordi A, Allam M, Bhiman JN, Davies M-A, Doolabh D, Engelbrecht S, Fonseca V, Giovanetti M, Glass AJ, Godzik A, Goedhals D, Hardie D, Hsiao M, Iranzadeh A, Ismail A, Korsman S, Pond SLK, Laguda-Akingba O, Lourenco J, Marais G, Martin D, Maslo C, Mlisana K, Mohale T, Msomi N, Mudau I, Petruccione F, Preiser W, San EJ, Sewell BT, Tyers L, Van Zyl G, von Gottberg A, Walaza S, Weaver S, Wibmer CK, Williamson C, York D, Archary M, Dullabh KJ, Goulder P, Harilall S, Harling G, Harrichandparsad R, Herbst K, Jeena P, Khoza T, Klein N, Kløverpris H, Leslie A, Madansein R, Marakalala M, Mazibuko M, Moshabela M, Mthabela N, Naidoo K, Ndhlovu Z, Ndung’u T, Nyamande K, Padayatchi N, Patel V, Ramjit D, Rodel H, Smit T, Steyn A, Wong E, Lessells RJ, de Oliveira T, Sigal A, Network for Genomic Surveillance in South, C.-K. Team Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheikh A, McMenamin J, Taylor B, Robertson C. SARS-CoV-2 Delta VOC in Scotland: Demographics, risk of hospital admission, and vaccine effectiveness. The Lancet. 2021;397:2461–2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lustig Y, Nemet I, Kliker L, Zuckerman N, Yishai R, Alroy-Preis S, Mendelson E, Mandelboim M. Neutralizing response against variants after SARS-CoV-2 infection and one dose of BNT162b2. N. Engl. J. Med. 2021;384:2453–2454. doi: 10.1056/NEJMc2104036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian Y, Lian C, Chen Y, Wei D, Zhang X, Ling Y, Wang Y, Yeap L-S. Sensitivity and specificity of SARS-CoV-2 S1 subunit in COVID-19 serology assays. Cell Discov. 2020;6:75. doi: 10.1038/s41421-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, Luo Y, Chan JFW, Sahi V, Figueroa A, Guo XV, Cerutti G, Bimela J, Gorman J, Zhou T, Chen Z, Yuen K-Y, Kwong PD, Sodroski JG, Yin MT, Sheng Z, Huang Y, Shapiro L, Ho DD. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 35.Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, Zhang Z, Fan P, Dong Y, Yang Y, Chen Z, Guo Y, Zhang J, Li Y, Song X, Chen Y, Xia L, Fu L, Hou L, Xu J, Yu C, Li J, Zhou Q, Chen W. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan CW, Chia WN, Qin X, Liu P, Chen MIC, Tiu C, Hu Z, Chen VC-W, Young BE, Sia WR, Tan Y-J, Foo R, Yi Y, Lye DC, Anderson DE, Wang L-F. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 37.Djaileb A, Jodaylami MH, Coutu J, Ricard P, Lamarre M, Rochet L, Cellier-Goetghebeur DMS, Charron B, Thibault V, Stevenson K, Forest S, Live LS, Abonnenc N, Guedon A, Quessy P, Lemay J-F, Farnos O, Kamen AA, Stuible M, Gervais C, Durocher Y, Cholette F, Mesa C, Kim J, Cayer M-P, De Grandmont M-J, Brouard D, Trottier S, Boudreau D, Pelletier JN, Masson J-F. Cross-validation of ELISA and a portable surface plasmon resonance instrument for IgG antibodies serology with SARS-CoV-2 positive individuals. ChemRXiv. 2021 doi: 10.26434/chemrxiv.14347013.v14347011. [DOI] [PubMed] [Google Scholar]
- 38.Schasfoort RBM, van Weperen J, van Amsterdam M, Parisot J, Hendriks J, Koerselman M, Karperien M, Mentink A, Bennink M, Krabbe H, Terstappen LWMM, Mulder AHL. Presence and strength of binding of IgM, IgG and IgA antibodies against SARS-CoV-2 during CoViD-19 infection. Biosens. Bioelectron. 2021;183:113165. doi: 10.1016/j.bios.2021.113165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker SN, Chokkalingam N, Reuschel EL, Purwar M, Xu Z, Gary EN, Kim KY, Helble M, Schultheis K, Walters J, Ramos S, Muthumani K, Smith TRF, Broderick KE, Tebas P, Patel A, Weiner DB, Kulp DW. SARS-CoV-2 assays to detect functional antibody responses that block ACE2 recognition in vaccinated animals and infected patients. J. Clin. Microbiol. 2020;58:e01533–e1520. doi: 10.1128/JCM.01533-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravichandran S, Coyle EM, Klenow L, Tang J, Grubbs G, Liu S, Wang T, Golding H, Khurana S. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci. Transl. Med. 2020;12:eabc3539. doi: 10.1126/scitranslmed.abc3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian F, Tong B, Sun L, Shi S, Zheng B, Wang Z, Dong X, Zheng P. Mutation N501Y in RBD of spike protein strengthens the interaction between COVID-19 and its receptor ACE2. eLife. 2021;10:e69091. doi: 10.7554/eLife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Arunkumar GA, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer DS, Lugo LA, Kojic EM, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, García-Sastre A, Caplivski D, Cheng AC, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, Krammer F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang P, et al. Increased resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7 to antibody neutralization. bioRxiv. 2021 doi: 10.1101/2021.01.25.428137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masson JF. Surface plasmon resonance clinical biosensors for medical diagnostics. ACS Sens. 2017;2:16–30. doi: 10.1021/acssensors.6b00763. [DOI] [PubMed] [Google Scholar]
- 45.Khurana S, Coyle EM, Dimitrova M, Castellino F, Nicholson K, Del Giudice G, Golding H. Heterologous prime-boost vaccination with MF59-adjuvanted H5 vaccines promotes antibody affinity maturation towards the hemagglutinin HA1 domain and broad H5N1 cross-clade neutralization. PLoS ONE. 2014;9:e95496. doi: 10.1371/journal.pone.0095496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J, Lan J, Yuan J, Wang H, Zhao J, Zhang S, Wang Y, Shi X, Liu L, Zhao J, Wang X, Zhang Z, Zhang L. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 47.Abe KT, Li Z, Samson R, Samavarchi-Tehrani P, Valcourt EJ, Wood H, Budylowski P, Dupuis AP, II, Girardin RC, Rathod B, Wang JH, Barrios-Rodiles M, Colwill K, McGeer AJ, Mubareka S, Gommerman JL, Durocher Y, Ostrowski M, McDonough KA, Drebot MA, Drews SJ, Rini JM, Gingras A-C. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight. 2020;5:e142362. doi: 10.1172/jci.insight.142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu F, Liu M, Wang A, Lu L, Wang Q, Gu C, Chen J, Wu Y, Xia S, Ling Y, Zhang Y, Xun J, Zhang R, Xie Y, Jiang S, Zhu T, Lu H, Wen Y, Huang J. Evaluating the association of clinical characteristics with neutralizing antibody levels in patients who have recovered from mild COVID-19 in Shanghai, China. JAMA Intern. Med. 2020;180:1356–1362. doi: 10.1001/jamainternmed.2020.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang HS, Costa V, Racine-Brzostek SE, Acker KP, Yee J, Chen Z, Karbaschi M, Zuk R, Rand S, Sukhu A, Klasse PJ, Cushing MM, Chadburn A, Zhao Z. Association of age with SARS-CoV-2 antibody response. JAMA Netw. Open. 2021;4:e214302. doi: 10.1001/jamanetworkopen.2021.4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatatos L, Czartoski J, Wan Y-H, Homad LJ, Rubin V, Glantz H, Neradilek M, Seydoux E, Jennewein MF, MacCamy A, Feng J, Mize G, De Rosa SC, Finzi A, Lemos MP, Cohen KW, Moodie Z, McElrath MJ, McGuire A. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021;372:1413. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghaffari A, Meurant R, Ardakani A. COVID-19 serological tests: How Well do they actually perform? Diagnostics. 2020;10:453. doi: 10.3390/diagnostics10070453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, Cho A, Jankovic M, Schaefer-Babajew D, Oliveira TY, Cipolla M, Viant C, Barnes CO, Bram Y, Breton G, Hägglöf T, Mendoza P, Hurley A, Turroja M, Gordon K, Millard KG, Ramos V, Schmidt F, Weisblum Y, Jha D, Tankelevich M, Martinez-Delgado G, Yee J, Patel R, Dizon J, Unson-O’Brien C, Shimeliovich I, Robbiani DF, Zhao Z, Gazumyan A, Schwartz RE, Hatziioannou T, Bjorkman PJ, Mehandru S, Bieniasz PD, Caskey M, Nussenzweig MC. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Isho B, Abe KT, Zuo M, Jamal AJ, Rathod B, Wang JH, Li Z, Chao G, Rojas OL, Bang YM, Pu A, Christie-Holmes N, Gervais C, Ceccarelli D, Samavarchi-Tehrani P, Guvenc F, Budylowski P, Li A, Paterson A, Yue FY, Marin LM, Caldwell L, Wrana JL, Colwill K, Sicheri F, Mubareka S, Gray-Owen SD, Drews SJ, Siqueira WL, Barrios-Rodiles M, Ostrowski M, Rini JM, Durocher Y, McGeer AJ, Gommerman JL, Gingras A-C. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5:eabe5511. doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, McMahon M, Meade P, Mendu DR, Muellers K, Stadlbauer D, Stone K, Strohmeier S, Simon V, Aberg J, Reich DL, Krammer F, Cordon-Cardo C. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu C, Yu X, Gao C, Zhang L, Zhai H, Hu Y, Liu E, Wang Q, Gao Y, Wei D, Zhang D, Han Y, Zhang X. Characterization of antibody responses to SARS-CoV-2 in convalescent COVID-19 patients. J. Med. Virol. 2021;93:2227–2233. doi: 10.1002/jmv.26646. [DOI] [PubMed] [Google Scholar]
- 56.Abe KT, et al. Neutralizing antibody responses to SARS-CoV-2 variants in vaccinated Ontario long-term care home residents and workers. medRxiv. 2021 doi: 10.1101/2021.08.06.21261721. [DOI] [Google Scholar]
- 57.Gasser R, Cloutier M, Prévost J, Fink C, Ducas É, Ding S, Dussault N, Landry P, Tremblay T, Laforce-Lavoie A, Lewin A, Beaudoin-Bussières G, Laumaea A, Medjahed H, Larochelle C, Richard J, Dekaban GA, Dikeakos JD, Bazin R, Finzi A. Major role of IgM in the neutralizing activity of convalescent plasma against SARS-CoV-2. Cell Rep. 2021;34:108790. doi: 10.1016/j.celrep.2021.108790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, Hemmings O, O’Byrne A, Kouphou N, Galao RP, Betancor G, Wilson HD, Signell AW, Winstone H, Kerridge C, Huettner I, Jimenez-Guardeño JM, Lista MJ, Temperton N, Snell LB, Bisnauthsing K, Moore A, Green A, Martinez L, Stokes B, Honey J, Izquierdo-Barras A, Arbane G, Patel A, Tan MKI, O’Connell L, O’Hara G, MacMahon E, Douthwaite S, Nebbia G, Batra R, Martinez-Nunez R, Shankar-Hari M, Edgeworth JD, Neil SJD, Malim MH, Doores KJ. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grzelak L, et al. Sex differences in the decline of neutralizing antibodies to SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.11.12.20230466. [DOI] [Google Scholar]
- 60.Harvala H, Robb ML, Watkins N, Ijaz S, Dicks S, Patel M, Supasa P, Wanwisa D, Liu C, Mongkolsapaya J, Bown A, Bailey D, Vipond R, Grayson N, Temperton N, Gupta S, Ploeg RJ, Bolton J, Fyfe A, Gopal R, Simmonds P, Screaton G, Thompson C, Brooks T, Zambon M, Miflin G, Roberts DJ. Convalescent plasma therapy for the treatment of patients with COVID-19: Assessment of methods available for antibody detection and their correlation with neutralising antibody levels. Transfus. Med. 2020;31:167–175. doi: 10.1111/tme.12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.GeurtsvanKessel CH, et al. Towards the next phase: evaluation of serological assays for diagnostics and exposure assessment. medRxiv. 2020 doi: 10.1101/2020.04.23.20077156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.https://nrc-digital-repository.canada.ca/eng/view/object/?id=cc208042-4c50-4aa8-8e30-3c30f4dfc1f8.
- 63.Stuible M, Gervais C, Lord-Dufour S, Perret S, L’Abbé D, Schrag J, St-Laurent G, Durocher Y. Rapid, high-yield production of full-length SARS-CoV-2 spike ectodomain by transient gene expression in CHO cells. J. Biotechnol. 2021;326:21–27. doi: 10.1016/j.jbiotec.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perreault J, Tremblay T, Fournier MJ, Drouin M, Beaudoin-Bussieres G, Prevost J, Lewin A, Begin P, Finzi A, Bazin R. Waning of SARS-CoV-2 RBD antibodies in longitudinal convalescent plasma samples within 4 months after symptom onset. Blood. 2020;136:2588–2591. doi: 10.1182/blood.2020008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stadlbauer D, Amanat F, Chromikova V, Jiang K, Strohmeier S, Arunkumar GA, Tan J, Bhavsar D, Capuano C, Kirkpatrick E, Meade P, Brito RN, Teo C, McMahon M, Simon V, Krammer F. SARS-CoV-2 seroconversion in humans: A detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao SS, Bukar N, Toulouse JL, Pelechacz D, Robitaille R, Pelletier JN, Masson JF. Miniature multi-channel SPR instrument for methotrexate monitoring in clinical samples. Biosens. Bioelectron. 2015;64:664–670. doi: 10.1016/j.bios.2014.09.082. [DOI] [PubMed] [Google Scholar]
- 67.https://www.inspq.qc.ca/nouvelles/detection-quebec-du-variant-b1351-du-sras-cov-2-ayant-emerge-en-afrique-du-sud.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.