Abstract
Twelve Salmonella typhimurium strains resistant to broad-spectrum cephalosporins were isolated from cases of gastroenteritis during 1996 to 1998 in Russia, Hungary, and Greece. Resistance was due to the production of CTX-M-type extended-spectrum β-lactamases encoded by similar 12-kb plasmids. By pulsed-field gel electrophoresis, all strains shared the same chromosomal type. These data suggest that an S. typhimurium clone resistant to broad-spectrum cephalosporins is present in at least three European countries.
Over the years, an increasing proportion of Salmonella isolates have been acquiring resistance to various “older” antimicrobial drugs (4, 10, 19). Lately, sporadic appearance of nontyphoid salmonella isolates that are resistant to broad-spectrum cephalosporins due to production of various plasmid-mediated β-lactamases, including the CTX-M-type extended-spectrum (ES) enzymes, has been noted (summarized in reference 15). We have reported previously the emergence of resistance to cephalosporins in Salmonella typhimurium. Strains from Russia and Greece displayed an unusual phenotype, being resistant to cefotaxime, ceftriaxone, and aztreonam, but susceptible to ceftazidime (7, 20). These strains possessed plasmids encoding CTX-M-type β-lactamases (8, 9). In the meantime, additional S. typhimurium strains with similar resistance phenotypes were isolated in Russia and Hungary. In the present study, we examined the possibility of the older and newer strains being clonally related and analyzed the resistance mechanisms to β-lactams of the recent isolates.
Twelve cefotaxime-resistant S. typhimurium strains were included in the study. They were identified by the API 20E system and were serotyped with respect to cell wall (O) and flagellar (H) antigens (17). Six strains were isolated from an equal number of patients during an outbreak of gastroenteritis in a psychiatric institution in St. Petersburg, Russia, in 1997 (R-strains). Strain S-661 was representative of an outbreak of gastroenteritis that had occurred in St. Petersburg in 1996 (7). Three strains were isolated from sporadic cases of gastroenteritis in Budapest, Hungary, during 1998 (H-strains). The two Greek strains, AS30 and AS31, were also epidemiologically unrelated (20).
Genomic DNA was extracted and pulsed-field gel electrophoresis (PFGE) was performed as described previously (17). Cell lysis with lysozyme was followed by a proteinase K treatment and DNA digestion with XbaI. Electrophoresis through 1% agarose–0.5× (wt/vol) Tris-borate-EDTA was performed by using a CHEF DRIII apparatus (Bio-Rad). Isolates with electrophoretic patterns differing by four or more DNA fragments were assigned to distinct types (16, 18). Following visual inspection, PFGE banding patterns were also analyzed with GelCompar software (Applied Maths).
Conjugal transfer was carried out in broth with E. coli 26R793 (Rifr Lac−) as the recipient (21). Transconjugant clones were selected on nutrient agar containing cefotaxime (10 μg/ml) and rifampin (200 μg/ml).
Susceptibility to β-lactam antibiotics was assessed by an agar dilution method (12). Susceptibility to other antibiotics was assessed by a disk diffusion assay (13).
Isolation of plasmids was performed by an alkaline lysis procedure (11). Agarose-purified plasmid DNA was digested with SacII or HaeIII restriction endonucleases and subjected to agarose gel electrophoresis.
β-Lactamase extracts were obtained after ultrasonic treatment of mid-log-phase cultures in tryptone-soy broth. The lysates were clarified by ultracentrifugation and dialyzed against phosphate buffer (50 mM, pH 7.0). Isoelectric focusing (IEF) of β-lactamases was performed in polyacrylamide gels containing ampholytes (pH range 3.5 to 9.5) (Pharmacia-LKB). β-Lactamase bands were visualized with nitrocefin (Oxoid).
The DNA sequences of blaCTX-M genes were determined directly from the wild-type plasmids by the dideoxy chain termination method with the Sequenase 2.0 kit (United States Biochemicals) and a set of blaCTX-M-specific oligonucleotide primers based on the blaCTX-M-4 sequence (8).
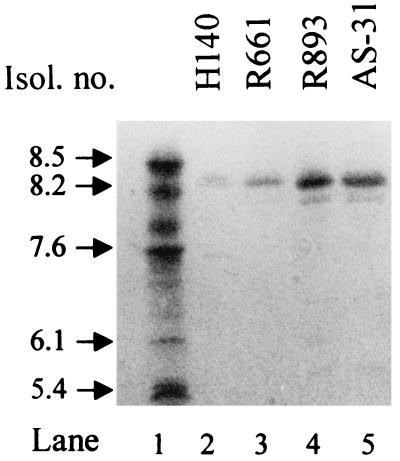
All cefotaxime-resistant S. typhimurium isolates exhibited similar resistance phenotypes to β-lactams, displaying at most 1-dilution differences in MIC; hence, results for only two representative strains are shown in Table 1. They were resistant to penicillins, cefotaxime, ceftriaxone, and aztreonam, but susceptible to ceftazidime. Piperacillin-tazobactam was highly active, while the combinations of clavulanate with amoxicillin or ticarcillin were less active. In IEF experiments, the isolates produced a single β-lactamase species that focused at 8.4 (Fig. 1). The resistance phenotype and pI of the enzymes were indicative of a CTX-M-type β-lactamase. Resistance phenotypes to various non-β-lactam antibiotics were also similar (Table 1).
TABLE 1.
Antibiotic susceptibilities of representative S. typhimurium isolates and E. coli transconjugant clones
| Strain | MIC of β-lactam(s) (μg/ml)
|
Resistance to other antibioticsb | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | Piperacillin | Amoxicillin-clavulanate (2:1) | Ticarcillin-clavulanatea | Piperacillin-tazobactama | Cefotaxime | Ceftriaxone | Ceftazidime | Aztreonam | Cefepime | ||
| S. typhimurium R-893 | >256 | >256 | 32 | 32 | 4 | >128 | >128 | 8 | 64 | 32 | Su, Tm, Te, C, Gm, Tb |
| E. coli (type 1) | >256 | >256 | 32 | 16 | 2 | >128 | >128 | 2 | 32 | 16 | Su, Tm, Te, C, Gm, Tb |
| E. coli (type 2) | >256 | >256 | 16 | 16 | 2 | >128 | >128 | 2 | 64 | 16 | None |
| S. typhimurium H-140 | >256 | >256 | 32 | 64 | 8 | >128 | >128 | 16 | 128 | 64 | Su, Tm, Te, C, Gm, Tb |
| E. coli (type 1) | >256 | >256 | 32 | 64 | 4 | >128 | >128 | 4 | 64 | 32 | Su, Tm, Te, C, Gm, Tb |
| E. coli (type 2) | >256 | >256 | 32 | 64 | 4 | >128 | >128 | 4 | 64 | 32 | None |
| E. coli 26R793 | 2 | 1 | 2 | 2 | 1 | 0.12 | 0.12 | 0.24 | 0.06 | 0.06 | |
Inhibitor fixed at 4 μg/ml.
Su, sulfonamide; Tm, trimethoprim; Te, tetracycline; C, chloramphenicol; Gm, gentamicin; Tb, tobramycin.
FIG. 1.
IEF of β-lactamase preparations from the indicated cefotaxime-resistant isolates (Isol.). β-Lactamases of the indicated pIs are in lane 1.
Conjugal transfer of cefotaxime resistance was attempted by using R-893 and H-140 as representative donors. Cefotaxime-resistant E. coli transconjugants were readily obtained from both isolates at a frequency of 10−4. Based on their resistance phenotypes, they were divided into two types. In type 1, which included the majority of transconjugants, all resistance characters of the donor strain had been transferred. The remaining transconjugants (type 2) were resistant only to β-lactams (Table 1). The latter clones had acquired relatively small plasmids (12 kb) and produced a β-lactamase of pI 8.4 (data not shown). In type 1 clones, the other resistance determinants were most likely transferred by larger plasmids (60 to 80 kb) which could be detected along with the small CTX-M-encoding plasmids. Therefore, the genes encoding the cefotaxime-hydrolyzing β-lactamase resided in the small plasmids observed in both transconjugant types. In subsequent conjugation experiments, however, type 2 clones were unable to transfer resistance to β-lactams to another E. coli recipient strain. When the 12-kb plasmid DNA from different isolates was digested with restriction endonuclease HaeIII or SacII, the patterns obtained were indistinguishable, with the exception of plasmids derived from the Hungarian isolates, which differed in one band of approximately 0.6 kb (data not shown).
Nucleotide sequencing with purified plasmid preparations from R-893 and H-140 confirmed the presence of blaCTX-M genes. The coding and promoter regions of the blaCTX-M gene from R-893 were identical to those of the previously described blaCTX-M-4 gene found in isolate S-661 (8). Sequencing of the coding region of the blaCTX-M gene from H-140 showed that its deduced amino acid sequence differed from that of CTX-M-4 only at position 211, where a leucine had been replaced by an isoleucine.
PFGE can successfully identify epidemiological and clonal relationships among S. typhimurium isolates, either in concordance with or with higher discrimination than phage typing (1, 5, 14). This method showed that all cefotaxime-resistant S. typhimurium isolates were also highly related at the chromosomal level. Their PFGE patterns differed by three bands at most, thus classifying them in the same type, D, clearly distinguishable from PFGE types A, B and C, obtained with cefotaxime-susceptible control isolates (Fig. 2). Types A and B are the dominant types in Greek S. typhimurium (unpublished data). Type D, on the other hand, was observed for the first time with AS-30 and AS-31. The macrorestriction pattern of the Greek strain AS-30 was 94% identical to those of the Russian or Hungarian cefotaxime-resistant isolates, while that of AS-31 was 84% identical to the rest of the cluster.
FIG. 2.
(A) PFGE of cefotaxime-resistant and -susceptible S. typhimurium isolates. The sizes (in kilobase pairs) of bacteriophage λ concatamers are indicated on the right. All lanes are from the same gel. (B) Dendrogram based on the similarity of the PFGE patterns shown in panel A. Isolates are indicated on the right, and a percentage similarity scale is shown at the top.
S. typhimurium strains producing CTX-M-type β-lactamases have also been isolated in Argentina (2) and Latvia (3). These plasmid-mediated class A enzymes are related to the species-specific β-lactamases of Klebsiella oxytoca (6) and constitute a small but rapidly expanding group of ES β-lactamases. CTX-M β-lactamases preferentially hydrolyze cefotaxime and ceftriaxone, but, unlike most ES TEM and SHV enzymes, they spare ceftazidime. They are inhibited by low concentrations of tazobactam, while clavulanic acid exerts a less potent inhibitory activity (3, 8).
Based on all assays performed, the cefotaxime-resistant isolates should be considered as clonally related. However, it was not possible to demonstrate a clear epidemiological relationship among them. The R-strain isolates might be connected with the outbreak isolates represented by S-661 (7), both outbreaks having occurred in the region of St. Petersburg within 1 year. Interestingly, an ongoing epidemic of cefotaxime-resistant S. typhimurium producing plasmid-mediated CTX-M-type β-lactamases that may be related to those described here, has been reported in nearby Latvia (3). As reported previously, strains AS-30 and AS-31 may have been acquired in Eastern Europe (20). The available patients' data did not reveal any epidemiological association of the Hungarian isolates with the rest of the cluster.
In addition to the nearly identical PGFE patterns, the similarity of plasmids encoding the CTX-M-β-lactamase variants further supported the clonal origin of the cefotaxime-resistant isolates. These plasmids were probably not self-transmissible but were mobilized by coexisting conjugative plasmids. Under such circumstances, further spread of the blaCTX-M genes is likely to occur. The CTX-M-5-encoding plasmids found in S. typhimurium isolates from Latvia were also small (10 kb) and non-self-transferable (3).
In this study, we showed that an oximino-cephalosporin-resistant clone of S. typhimurium is present in geographically distinct areas across Eastern and Southeastern Europe. By its resistance phenotype, its mechanism of resistance to β-lactams, and its phage type, DT193 (22), this clone is not related to the widespread multidrug-resistant clone of S. typhimurium DT104.
To date, it has not been possible to obtain information as to the presence of S. typhimurium isolates with similar phenotypes of resistance from other parts of Eastern Europe. The scarcity of relevant reports may indicate that such strains have not spread widely yet. The unusual resistance to β-lactams of this CTX-M-producing clone, together with its multidrug resistance, are traits that can hardly pass unnoticed during susceptibility testing. Nevertheless, the establishment and spread of an S. typhimurium clone that is resistant to therapeutically important broad-spectrum β-lactams are causes for concern.
Acknowledgments
We thank Linda R. Ward (Laboratory of Enteric Pathogens, Central Public Health Laboratory, London, United Kingdom) for kindly phage typing isolate AS-31 and Zannina Sarandopoulou for excellent technical assistance with PFGE. We are grateful to Jenny Kourea-Kremastinou and Maria Lambiri (National Reference Centre for Salmonella and Shigella, National School of Public Health, Athens, Greece) for providing the Greek control strains and Antonios Markogiannakis for their initial characterization.
ADDENDUM IN PROOF
Phage typing performed in the laboratory of W. Rabsch (National Reference Center for Salmonella and other enterobacteria, Robert Koch-Institute, Wernigerode, Germany) revealed that all cefotaxime-resistant isolates from the three countries belonged to DT193 (14a).
REFERENCES
- 1.Baggesen D L, Skov M N, Brown D J, Bisgaard M. Separation of Salmonella typhimurium DT2 and DT135: molecular characterization of isolates of avian origin. Eur J Epidemiol. 1997;13:347–352. doi: 10.1023/a:1007302205271. [DOI] [PubMed] [Google Scholar]
- 2.Bauernfeind A, Casellas J M, Goldberg M, Holey M, Jungwirth R, Mangold P, Rohnisch T, Schweighart S, Wilhelm R. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection. 1992;20:158–163. doi: 10.1007/BF01704610. [DOI] [PubMed] [Google Scholar]
- 3.Bradford P A, Yang Y, Sahm D, Grope I, Gardovska D, Storch G. CTX-M-5, a novel cefotaxime-hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob Agents Chemother. 1998;42:1980–1984. doi: 10.1128/aac.42.8.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casin I, Breuil J, Brisabois A, Moury F, Grimont F, Collatz E. Multidrug-resistant human and animal Salmonella typhimurium isolates in France belong predominantly to a DT104 clone with the chromosome- and integron-encoded beta-lactamase PSE-1. J Infect Dis. 1999;179:1173–1182. doi: 10.1086/314733. [DOI] [PubMed] [Google Scholar]
- 5.Corbett-Feeney G, Riain U N. The use of pulsed-field gel electrophoresis for subdivision of Salmonella typhimurium in an outbreak situation. J Infect. 1998;36:175–177. doi: 10.1016/s0163-4453(98)80009-1. [DOI] [PubMed] [Google Scholar]
- 6.Fournier B, Roy P H, Lagrange P H, Philippon A. Chromosomal β-lactamase genes of Klebsiella oxytoca are divided into two main groups, blaOXY-1 and blaOXY-2. Antimicrob Agents Chemother. 1996;40:454–459. doi: 10.1128/aac.40.2.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gazouli M, Sidorenko S V, Tzelepi E, Kozlova N S, Gladin D P, Tzouvelekis L S. A plasmid-mediated β-lactamase conferring resistance to cefotaxime in a Salmonella typhimurium clone found in St. Petersburg, Russia. J Antimicrob Chemother. 1998;41:119–121. doi: 10.1093/jac/41.1.119. [DOI] [PubMed] [Google Scholar]
- 8.Gazouli M, Tzelepi E, Sidorenko S V, Tzouvelekis L S. Sequence of the gene encoding a plasmid-mediated cefotaxime-hydrolyzing class A β-lactamase (CTX-M-4): involvement of serine 237 in cephalosporin hydrolysis. Antimicrob Agents Chemother. 1998;42:1259–1262. doi: 10.1128/aac.42.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazouli M, Tzelepi E, Markogiannakis A, Legakis N J, Tzouvelekis L S. Two novel plasmid-mediated cefotaxime-hydrolyzing β-lactamases (CTX-M-5 and CTX-M-6) from Salmonella typhimurium. FEMS Microbiol Lett. 1998;165:289–293. doi: 10.1111/j.1574-6968.1998.tb13159.x. [DOI] [PubMed] [Google Scholar]
- 10.Glynn M K, Bopp C, Dewitt W, Dabney P, Mokhtal M, Angulo F J. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–1337. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- 11.Kado C I, Liu S-T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A2. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 14.Olsen J E, Skov M N, Angen O, Threlfall J E, Bisgaard M. Genomic relationships between selected phage-types of Salmonella enterica subsp. enterica serotype Typhimurium defined by ribotyping IS200 typing and PFGE. Microbiology. 1997;143:1471–1479. doi: 10.1099/00221287-143-4-1471. [DOI] [PubMed] [Google Scholar]
- 14a.Rabsch, W. Personal communication.
- 15.Shannon K, French G. Multiple-antibiotic-resistant salmonella. Lancet. 1998;352:490–491. doi: 10.1016/S0140-6736(05)79231-X. [DOI] [PubMed] [Google Scholar]
- 16.Struelens M J, Schwam V, Deplano A, Baran D. Genome macrorestriction analysis of diversity and variability of Pseudomonas aeruginosa strains infecting cystic fibrosis patients. J Clin Microbiol. 1993;31:2320–2326. doi: 10.1128/jcm.31.9.2320-2326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tassios P T, Markogiannakis A, Vatopoulos A C, Katsanikou E, Velonakis E N, Kourea-Kremastinou J, Legakis N J. Molecular epidemiology of antibiotic resistance of Salmonella enteritidis during a 7-year period in Greece. J Clin Microbiol. 1997;35:1316–1321. doi: 10.1128/jcm.35.6.1316-1321.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Threlfall J E, Ward L R, Skinner J A, Rowe B. Increase in multiple antibiotic resistance in nontyphoidal salmonellas from humans in England and Wales. Microb Drug Resist. 1997;3:263–266. doi: 10.1089/mdr.1997.3.263. [DOI] [PubMed] [Google Scholar]
- 20.Tzouvelekis L S, Gazouli M, Markogiannakis A, Paraskaki E, Legakis N J, Tzelepi E. Emergence of third-generation cephalosporin-resistant Salmonella typhimurium in Greece: report of the first three cases. J Antimicrob Chemother. 1998;42:273–275. doi: 10.1093/jac/42.2.273. [DOI] [PubMed] [Google Scholar]
- 21.Vatopoulos A C, Mainas E, Balis E, Threlfall E J, Kanelopoulou M, Kalapothaki V, Malamou-Lada H, Legakis N J. Molecular epidemiology of ampicillin-resistant clinical isolates of Salmonella enteritidis. J Clin Microbiol. 1994;32:1322–1325. doi: 10.1128/jcm.32.5.1322-1325.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward, L. R. Personal communication.