Abstract
Studying the structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein is important to understand the infection process. The S protein is necessary in completing the virus life cycle and is responsible for the appearance of new variants and drug and vaccine resistance. Understanding the structure and dynamics of biological macromolecules is essential for understanding how they function. In this work, we investigated the effects of mutations on S protein stability and solubility through molecular dynamic (MD) simulation in a 100 ns (ns) period. We screened four variants in addition to the wild type (WT). Results show that changes on MD simulation parameters of S protein indicate fluctuations and changes in the conformation, especially in the area between 300 and 600 amino acids (aa). This provides us an image of how the virus protein can reshape itself to adapt to any changes that occur in human angiotensin-converting enzyme 2 or drugs that can target the protein region. Our results also show that the Brazil variant has high fluctuations and unstable folding at some stages compared with other variants.
Keywords: SARS-CoV-2, Variant, S protein, MD simulation
1. Introduction
At present, seven types of severe acute respiratory syndrome coronavirus (SARS-CoV) have been found to infect humans, including SARS-CoV, SARS-CoV-2, MERS-CoV, hCoV-OC43, hCoV-HKU1, hCoV-229E, and hCoV-NL63. They all cause cold-like symptoms with a difference in the degree of illness severity. However, SARS-CoV-2, SARS-CoV, and MERS-CoV have a high mortality rate. The seven members of human CoV (hCoV) share some common feature, including RNA, single-stranded, length of the genome about ∼30 kb, consist of around 16 nonstructural proteins and 10–12 structural proteins. However, four among them are extensively studied the envelope (E), membrane (M), nucleocapsid (N), and spike (S) [1].
The S protein is the main virus receptor that interacts with the receptor angiotensin-converting enzyme 2 (ACE2) in human cells. It consists of subunit S1 (receptor-binding domain (RBD) and N-terminal domain (NTD) and S2. Both subunits are targeted by vaccines. The mRNA vaccines from BioNTech/Pfizer and Moderna consist of a full-length S protein with two K986P and V987P mutations in two proline substitutions (2P). The Novavax vaccines consist of five K986P and V987P mutations in 2P, R682Q, R683Q, and R685Q in the S protein. The Janssen vaccines consist of four K986P and V987P mutations in 2P, R682S, and R685G in the S protein [2]. At present, several variants of SARS-CoV-2 have appeared. Herein, we compared the S protein structure of the wild-type (WT) and early SARS-CoV-2 variants known by the country where they were first detected (United Kingdom, South Africa, California, and Brazil). Our analyses showed that structural conformation occurred between variants and even in the same variant at different time points, special in the region where it interacts with human protein receptor. In this regard, the high flexibility of variant allows the virus to react to drugs in a variety of ways. Many questions remain to be answered. For example, do variants have the same 3D structure, or are there variations? If there are variations inside the structure, can molecular dynamic (MD) simulation and artificial intelligence determine which variant is resistant against vaccines, why some variants have higher mortality rate, and whether these variants have similar reactions to certain drugs? If the virus with one receptor can interact with different or alternate receptors, does this mean that the virus receptor can reshape itself to adapt to new organs or environment? Our work is just the first step toward answering these questions.
2. Methodology
2.1. MD simulation
The PDB file 7CWU [3] was used as WT SARS-CoV-2 S protein. The MD simulation was run on Desmond with default protocols [4]. The TIP3P model in Desmond System Builder tool was used to solvate the protein. Periodic boundary conditions with 10 Å orthorhombic box were used on the outer protein surface, and 0.15 M NaCl was used to neutralize the simulation system. The simulation was run at temperature of 310 K and pressure of 1.013 bar for 100 ns. The trajectory was analyzed by Desmond, VMD [5], and PyMOL.
3. Results and discussion
To our knowledge, this study is the first comprehensive MD simulation study, in which complete SARS-CoV-2 S protein mutation was compared with the WT. A total of 177 million cases and 3.8 deaths of SARS-CoV-2 have been reported worldwide (https://news.google.com/covid19/map) (December 2019 to Jun 2021). Five variants of S proteins and 26 mutations have been analyzed. To date, there are 3249 variants in North America, 23 variants in South America, 46,271 variants in Europe, 50 variants in Africa, 7581 variants in Asia, and 128 variants in Australia according to the GISAID database (https://www.gisaid.org/). Among these variants, 26 mutations were found in four countries, with the highest number detected in isolates from the United Kingdom, South Africa, California, and Brazil. At the end of December 2020, first mutant variation was reported in United Kingdom with a high mortality compared to the WT, since then, number of new variations is risen and detected across the globe. Actually, new variations can upper independently like the South Africa variation, or been modified when the variation move to new area or country through the passengers, as the result of that the genetic distinct start to be wide between the WT and the new variations, like the Brazilian variation with 15 mutations [6,7].
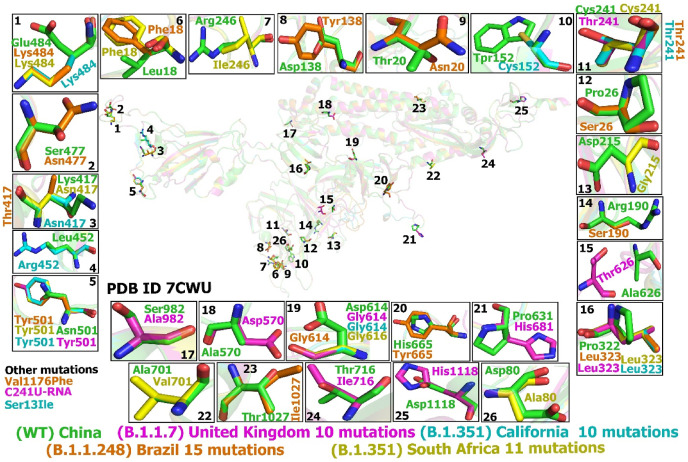
Hence, our study selected and reported the 26 mutations which mainly present in the S proteins. As the largest structural protein with 1270 aa of SARS-CoV-2, the mutant residues of the S protein have been localized at NTD, CTD, and the sequence between them. The majority of these mutations were named by the country where they were first detected, namely, United Kingdom (B.1.1.7), South Africa (B.1.351), United States also known as California (B.1.351 and BR-B.1.1.248), and Brazil (B.1.1.248) variants. These variants seem to be more transmissible and infectious than the WT [7]. Most repeated mutations that have been characterized in the United Kingdom variant include A570D, D614G, D1118H, S982A, P323L, N501Y, A626T, P681H, C241T, and T716I. while South Africa variant include D614G, P323L, C241U, N501Y, L18F, D80A, D215G, R246I, K417 N, E484K, and A701V. Those in California variant L452R, D614G, W152C, N501Y, K417 N, E484K, P323L, C241T, and S13I, and in Brazil variants include D138Y, V1176F, K417T, L18F, P26S, R190S, T20 N, T1027I, N501Y, D614G, P323L, C241T, E484K, S477 N, and H655Y (Fig. 1 ).
Fig. 1.
Location of the most common mutations in SARS-CoV-2 S proteins: (WT) China, United Kingdom, South Africa, California, and Brazil variants.
4. Root-mean-square deviation of atomic positions (RMSD) of WT and mutations
The RMSD has always been used to measure the distance between the protein atoms chains to measure the difference of two protein structures [8] during the simulation periods. A high RMSD value indicates dissimilarity, whereas a zero value indicates identical conformation structure. The RMSD graphs of S proteins of WT and mutations are shown in Fig. 2 A. All mutations appear more stable compared with the Brazil. Among all mutations, the California variant showed low RMSD value, indicating that it is more stable compared with other variants. In addition, the RMSD values of all variants increased at the first 18 ns, except the South Africa variant, which decreased after the first 5 ns and back to increase after 10 ns. No remarkable changes were observed in all variants at the last 10 ns. Desmond and VMD showed that the RMSD of WT variant increased until 50 ns (25 Å) and became almost stable from 50 ns to 100 ns (23 Å and 25 Å) (Fig. 2A). However, when we used PyMOL to calculate the RMSD, the RMSD between the first frame at 0 ns (F0) and the last frame at 100 ns (F100) reached 23.2 Å, and it was only 5.7 Å when we compared the frame at 50 ns (F50) to F100 (Fig. 3 ).
Fig. 2.
Comparison of the RMSD, RMSF, SASA, Rg, and number of HBs of WT (China), United Kingdom, South Africa, California, and Brazil variants S proteins of SARS-CoV-2 during the simulation periods. The results were calculated by Desmond and VMD.
Fig. 3.
RMSD structure superimposed at 0, 25, and 50 ns simulation periods of S proteins of SARS-CoV-2. The WT (China) has high RMSD value compared with the United Kingdom, South African, Californian, and Brazil variants. The RMSD was calculated by PyMOL. The cycle refers to RBD (the area where the virus binding to human protein).
The RMSD of the South African variant showed fluctuations at the beginning of the simulation and reached 25 Å and became almost stable until the end of the simulation according to Desmond and VMD (Fig. 2A). However, the RMSD between F50 and F100 was 7.07 Å, and that between F0 and F100 was 19.7 Å according to PyMOL (Fig. 3). The RMSD of the Californian variant showed fluctuations between 17 ns and 80 ns around 12 to 18 Å after 80 ns until to end of simulation the range of fluctuations became narrow about 15 Å according to Desmond and VMD (Fig. 2A). However, the RMSD was 11.6 Å when we compared F0 to F50 and 4.2 Å when we compared F50 and F100 according to PyMOL (Fig. 3). The RMSD of the United Kingdom variant showed fluctuations between 18 ns and 58 ns around 17 to 22.5 Å, while between 60 and 80 ns the RMSD fluctuations between 20 and 22.5 Å, after 80 ns until to end of simulation the range of fluctuations became narrow and stable about 22.5 Å according to Desmond and VMD (Fig. 2A). However, the RMSD was 4.3 Å when we compared F50 to F100 and 21.8 Å when we compared F0 to F100 according to PyMOL (Fig. 3). The RMSD of the Brazil variant showed instability value compare to another variant can be categorize to three-time point early stage of the simulation 0–18 ns process around 5 to 19 Å, the second time point 18–48 ns got RMSD range 12–17.5 Å, the third time point between 48 ns to the end of the simulation, the RMSD increase gradually from 17.5 to 36 Å according to Desmond and VMD (Fig. 2A). However, the RMSD was 17.4 Å when we compared F50 to F100 and 23.1 Å when we compared F0 to F100 according to PyMOL (Fig. 3).
5. Root mean square fluctuation (RMSF) of WT and mutations
The RMSF is used to measure the flexibility of each residue and how much the residue moves or fluctuates over a simulation period. Practically, the structure refers to which amino acids in the protein sequence responsible for more structure motion. In general, the Brazil variant significantly has higher RMSF value compared with other variants, and the WT and South African variants have a higher RMSF value compared with the United Kingdom and Californian variants (Fig. 2B).
Variations in the RMSF were observed at some positions of WT and mutations (Fig. 2B). The protein sequence of the WT with 2 range has high RMSF value, first range between the aa 200 to 280 exhibited 17.5 Å, and the second range between aa 400 to 500 exhibited 31 Å. The WT and South African range between aa 700 to 790 and aa 800 to 880 and 910 to 1110 got the same value or RMSF 17, 16 and 19 Å, respectively. Brazil variant showed a high RMSF value compared to other mutant variants, protein sequence of Brazil variant consist 4 range has high RMSF value first range between the aa 500 to 600 exhibited 22 Å and, the second range between aa 700 to 790 exhibited 27 Å. the third range between aa 800 to 880 exhibited 25 Å. the fourth range between aa 910 to 1110 exhibited 35 Å. The RMSF at the position of aa 50 to 200 and 700 to 1110 exhibited difference between Californian variant and other variants. In fact, Californian variant got the lost RMSF value compared to other variants, the United Kingdom and South African variants RMSF value close to each other at the N terminal of the sequence while South African variant got high RMSF value at the C terminal of the sequence compared to United Kingdom variant.
Research shows that, the amino acid between aa 300 to 600 involves in the interaction between the S proteins and human ACE2. We believe this give us a good image how the virus protein can reshape itself to adapt with any change occur in human ACE2 or any drug can target the region in protein. At the same time, the RMSF of this region can be used as an indicator of virus pathogenicity. Thus, pharmaceutical companies should avoid these regions to increase the drug efficiency and stability. According to the Annette B the variant which consists one of these mutation K417 N or N501Y had some resistant against currently vaccine. In our study, all four variants had an N501Y mutation, and the South African variant had K417 N and N501Y mutations, indicating that it might be more resistant against current vaccines compared with other variants (Fig. 2B). MD simulations provide insights into the dynamic fluctuations at the molecular level, which may lead to some difficulty during experimental work. Previous studies have determined that the RMSF is correlated with protein function [9,10]. Generally speaking, when two proteins have an RMSD less the 2 Å that mean the two proteins have the same structure. However, an increase in the RMSD value indicates conformational structural changes. We can say that all S proteins of SARS-CoV-2 variants can be changed at any time and will allow the virus to hide from our immune system or even infect various types of cells. This is one of the reasons why SARS-CoV-2 has a high infectivity.
6. Solvent-accessible surface area (SASA) of WT and mutations
SASA is used to measure the surface area of proteins that are accessible to a solvent [11,12]. Our results revealed that the SASA of all variants dramatically decreased during the simulation period (Fig. 2C). The United Kingdom and WT got the lost SASA value compared with other variants and this result totally agree with Rg result. This result indicates that the United Kingdom has a small surface to bind with the human protein compared with other variants. While, In the Brazil variant a high value of SASA was obtained from 30 ns to 80 ns because this variant had a wide packing and folding amount at the mid of the simulation. In other side, Californian variant go the lost SASA value at the same period of time. At last 12 ns of simulation period Californian, Brazil and South African variant overlap the SASA value. At the end simulation rang of SASA value for all SARS-CoV-2 variants in our study confined between 55000 and 59000 nm2.
7. Radius of gyration (Rg) of WT and mutations
Rg is the RMS distance of all electrons from their center of gravity [13], used to measure of the elastic stability of a cross-section [14]. The Rg values of WT and mutations throughout the simulation time period were examined simultaneously, and variations in folding state were observed. A low Rg value indicates tight packing through the energy interaction and the conformational entropy between residues [15]. In all of our target simulations, United Kingdom variant proteins exhibiting an obvious decrease in Rg might be due to the decomposition of some α-helix and conversion to random coil-like structure. The WT and United Kingdom variant showed a dramatic decrease in Rg value during the first 58 ns compared with other variants. But after that WT increase and stable around 46 nm2 up to the end of the simulation. while United Kingdom variant continue decrease until 75 ns after that increased slightly around 40 nm2. Moreover, United Kingdom variant has lost Rg value compared to other variants indicating that minimum tightness packing value compared to other variants, but it did not reach to aggregating stage because the Rg value got stable at last 25ns. We can say that the WT and United Kingdom variants almost have close folding state compared with other variants at the beginning of the simulation until the mid of the simulation, while from the mid of the simulation until the end WT and Californian variant got a close folding state compared with other variants, the range of Californian variant Rg between 44 and 48 nm2 is the closest value of Rg compared to other variants.
The Rg of South African variant immediately decreased and in less than 5 ns back to increase to reach to 52 nm2 after that decreased sharply to reach to 43 nm2. From 48 ns the Rg value it same to be stable around 48 nm2 up to the end of the simulation It is not clear why the Rg of the Brazil variant dramatically increased and fluctuated several times during the simulation and reach to 60 nm2 before it decreases slightly at end of the simulation. It seems that, the fluctuations due to unstable folding sate. We as assume that, Brazil variant has wide packing value compared to other variants depend on the Rg result. Moreover, The Rg plotted for all SARS-CoV-2 S proteins shows remarkable stability than the Brazil variant, and the variants have a less Rg fluctuation average value than the Brazil variant. When protein folding increases, the accessible surface area for solvent or any type of binding (small molecule or interaction with other proteins) decreases. Thus, The Brazil and South African variants have greater surface area compared with the WT and other variants. However, the Californian variant has a more stable folding sate compared with the South African variant given that the latter has more fluctuations than the former. In general, as the protein RMSD or RMSF or Rg is sensitive indication and any changing on this parameter leading to in the chain in the conformation, and that will reflect in the protein function and activity, also as the number of the research in molecular docking and simulation is rising we have to suggests here to when study drug or new molecule it will be better to do that with different variant of the protein when it is available, that will help us to understand more about the thermodynamic interactions of the protein and many of effective defense strategies the virus have.
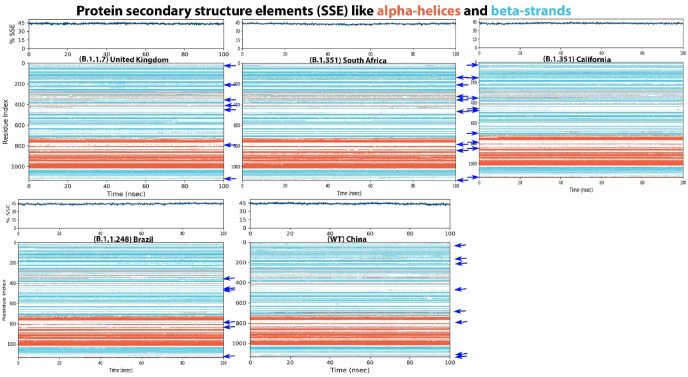
8. Protein secondary structure elements (SSE)
SSE is used to determine the changes that occur in the 3D structure of the protein during the simulation period for each frame in the trajectory. The SSE index shows the percentage that alpha-helices (α) and beta-strands (β) occur during the simulation period by residue. Overall, the SSE composition of the South African variant consists of less amounts of β compared with other variants, while the WT and Brazil variant has high amount of β compared with other variants (Table 1 and Fig. 4, Fig. 5 ). According to this result the S proteins of SARS-CoV-2 if very fixable and by using this way the virus maybe be able to hid from the immune system, also these maybe one of the techniques the virus used to reinfect people or even some of the vaccinate people.
Table 1.
Percentage of α and β in the 3D structure during the simulation period.
| % Alpha | % Beta | % Total SSE | |
|---|---|---|---|
| WT | 17.31 | 26.25 | 43.56 |
| United Kingdom | 16.99 | 26.53 | 43.53 |
| South Africa | 17.48 | 25.18 | 42.66 |
| California | 17.14 | 25.27 | 42.42 |
| Brazil | 17.39 | 26.52 | 43.91 |
Fig. 4.
Protein SSE such as alpha-helices and beta-strands are monitored throughout the simulation periods of S proteins of SARS-CoV-2. The blue arrow indicates the difference. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
Changes in the residue index in the formation of protein SSE such as alpha-helices and beta-strands are characterized during the simulation times of S proteins of SARS-CoV-2. The blue arrow indicates the difference. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
9. Number of hydrogen bonds (HB) of WT and mutants
The HB, hydrophobic, ionic, van der Waals, and electrostatic forces and water bridges are all involved in protein folding, stability, and flexibility. Among them, HB is the most important as it allows the binding between the reside to form the 3D structure in the shape of α and β. HB mainly occurs between the carbonyl oxygen and amide nitrogen in the protein chain. Fig. 2F shows that the WT and Brazil variant consists of fewer numbers of HB compared with other mutations during the simulation. The number of HB in the Californian variant increased between 32 and 62 ns. While The number of HB in the South African variant increased between 70 and 80 ns otherwise South African and United Kingdom got almost the same number of HB.
10. Discussion
The SARS-CoV-2 S protein is the first target of vaccine developers. Several studies have explained the complex between the S protein and neutralizing antibodies. Two published data have pinpointed the variability and flexibility of S protein with structural conformation particularly RBDs aa 300 to 600 which known as RBD up-open when it is binding to hACE2 and RBD down-closed it is not yet clear [16,17]. The suggested severity risk of infection scale in COVID-19 patients is dependent on hACE2 receptor and SARS-CoV-2 spike protein composition alterations. The COVID-19 illness severity has been classified into five zones based on the primary cause (very weak, weak, moderate, strong and very strong). The wild hACE2 receptor and the SARS-CoV-2 spike protein have a tight binding relationship [18]. A recent study showed that changes in the pH value of 6.5, 7.4, and 8.0 create several conformation states [19]. A previous work showed that 50% of S proteins have an open conformation, while 41% have an undefined state [20]. Actually, it is seeming the flexibility of S protein component RBD, NTD and S2 is relativity and each one of them has a certain degree of independent confirmation, this means open/closed-confirmation is gradient state, here one question will upper what is max distance can be found between RBD and NTD in open confirmation state. The interaction between C840 and C851 through the disulfide bond controls the open and closed conformations of S protein [20]. In fact, the S protein WT crystal structure consists of 14 disulfide bonds. This amount remains the same as no mutation occurs in the amino acids involved in disulfide bonds at least in our four variants. According to a previous study, amino acids such as D830, Y837, D839, D843, A846, and L849 add more stability to S protein open/closed conformation [20]. How these amino acids, which are located at S2 far from RBD and NTD, interfere with open/closed conformation remains unknown. However, our four variants did not carry mutation in these amino acids. Other uncharacterized amino acids may be able to interfere with the open/closed conformation. Pramanick et al. found that the packing variability of amino acids affects the solvent accessibility depending on the environment, which is consistent with our findings [19]. With all of these conformation state (it can be name as conformational masking strategy), rearrangement and continuous movement what is human antibodies capability to catch that, to which level the strength of binding with virus can be reach, might lead to failure of the antibodies and our immune system.
Consequently, various forms at the virus's surface can be generated to bind cognate receptors while altering the stability and flexibility of S proteins. To enhance efficiency, a new medication with a novel physics idea, in addition to the present chemical and biological concepts, is undoubtedly required. One of the weaknesses in virus studies is that we always study the virus in optimum conditions. This means that the copy variant is almost same. However, in real infection, the virus faces many challenging and different environments that force it to adapt and change dramatically. Thus, we need new techniques such as artificial intelligence with old componential biology to obtain good results. As difficult will be to cure infection with mix variant, this can develop new super variant and acquire new traits. From the available biochemical and structural data, there is a gap in our knowledge about SARS-CoV-2 infectivity vs. new variants. From a therapeutic angle SARS-CoV-2 is host/multifaceted pathogen with the appreciation complexity of the infection process. Hence, research in needing to more combination clinical, structural and biochemical studies to stop SARS-CoV-2 entry.
11. Conclusion
Here, we presented the stability and flexibility changes of S proteins in WT and variants of SARS-CoV-2 by using MD simulation. The homologous region between the WT and variants is high in S protein. However, the interaction patterns showed variation. As a result, all variant proteins have a high degree of flexibility to improve the effect of function and catalytic activities. Understanding the flexibility of the main protein of SARS-CoV-2 and its interaction with human cell receptors can help to improve the design and development of vaccines. According to our findings, physical factors are essential as well as biochemical factors to strengthen the virus variants and allow them to enter the host cell. Notably, in our analysis, some conformations were noticed such as α4β1, α4β7, and α9β1 to make the S protein as heterodimers.
12. Future prospects
Coronaviruses are a global threat, and our understanding of their proteins is still lacking. Thus, the transition paths and conformational changes of S proteins should be studied. In addition, the S protein is very large, and we need large amounts of computational power and time to study it in the monomer form. In the future, we should study the S protein in tetrameric and heterodimer forms.
Ethical approval
Not applicable.
Consent to participate
All authors participated in this work.
Consent to publish
All authors agree to publish this work.
Author contribution
MA wrote the manuscript and designed the study, WAE, AAE, KS and XJ contributed to manuscript revision. All authors approved the final version.
Funding
This work was supported by Shandong Provincial Key Research and Development Program (Major Scientific and Technological Innovation Project) (2019JZZY021013) Shandong Provincial Key Research and Development Program (2019GSF108080), Funds for Youth Interdisciplinary and Innovation Research Groups of Shandong University (2020QNQT003) and Shandong University postdoctoral fellowship to Mohnad Abdalla.
Declaration of competing interest
The authors declare no competing interests.
References
- 1.Ziebuhr J. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 2004;7:412–419. doi: 10.1016/j.mib.2004.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdalla M., El-Arabey A.A., Jiang X. Progress in research on the S protein as the target of COVID-19 vaccines. Expert Rev. Vaccines. 2021;20:769–772. doi: 10.1080/14760584.2021.1918003. [DOI] [PubMed] [Google Scholar]
- 3.Wang N., Sun Y., Feng R., Wang Y., Guo Y., Zhang L., Deng Y.Q., Wang L., Cui Z., Cao L., Zhang Y.J., Li W., Zhu F.C., Qin C.F., Wang X. Structure-based development of human antibody cocktails against SARS-CoV-2. Cell Res. 2021;31:101–103. doi: 10.1038/s41422-020-00446-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowers K.J., Chow E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., Klepeis J.L., Kolossvary I., Moraes M.A., Sacerdoti F.D., Salmon J.K., Shan Y., Shaw D.E. Proc. 2006 ACM/IEEE Conf. Supercomput. SC’06. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters; p. 84. [DOI] [Google Scholar]
- 5.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 6.Walensky R.P., Walke H.T., Fauci A.S. SARS-CoV-2 variants of concern in the United States-challenges and opportunities. JAMA - J. Am. Med. Assoc. 2021;325:1037–1038. doi: 10.1001/jama.2021.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdalla M., El-Arabey A.A., Jiang X. Are the new SARS-CoV-2 variants resistant against the vaccine? Hum. Vaccines Immunother. 2021;17:3489–3490. doi: 10.1080/21645515.2021.1925503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reva B.A., Finkelstein A.V., Skolnick J. What is the probability of a chance prediction of a protein structure with an rmsd of 6 Å? Folding Des. 1998;3:141–147. doi: 10.1016/S1359-0278(98)00019-4. [DOI] [PubMed] [Google Scholar]
- 9.Berhanu W.M., Masunov A.E. Molecular dynamic simulation of wild type and mutants of the polymorphic amyloid NNQNTF segments of elk prion: structural stability and thermodynamic of association. Biopolymers. 2011;95:573–590. doi: 10.1002/bip.21611. [DOI] [PubMed] [Google Scholar]
- 10.Bavi R., Kumar R., Choi L., Lee K.W. Exploration of novel inhibitors for bruton's tyrosine kinase by 3D QSAR modeling and molecular dynamics simulation. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaytan A.K., Shaitan K.V., Khokhlov A.R. Solvent accessible surface area of amino acid residues in globular proteins: correlation of apparent transfer free energies with experimental hydrophobicity scales. Biomacromolecules. 2009;10:1224–1237. doi: 10.1021/bm8015169. [DOI] [PubMed] [Google Scholar]
- 12.Bogatyreva N.S., Ivankov D.N. The relationship between the solvent-accessible surface area of a protein and the number of native contacts in its structure. Mol. Biol. 2008;42:932–938. doi: 10.1134/S0026893308060150. [DOI] [PubMed] [Google Scholar]
- 13.Kratky O., Laggner P. In: Encycl. Phys. Sci. Technol. R.A.B.T.-E. of P.S., T. (Third E. Meyers), editors. Academic Press; New York: 2003. X-ray small-angle scattering; pp. 939–988. [DOI] [Google Scholar]
- 14.Anwar N., Najam F.A. In: Struct. Cross Sect. Anwar N., Najam F.A.B.T.-S.C.S., editors. Butterworth-Heinemann; 2017. Understanding cross-sections; pp. 39–136. [DOI] [Google Scholar]
- 15.Lobanov M.Y., Bogatyreva N.S., Galzitskaya O.V. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 2008;42:623–628. doi: 10.1134/S0026893308040195. [DOI] [PubMed] [Google Scholar]
- 16.Hwang S.S., Lim J., Yu Z., Kong P., Sefik E., Xu H., Harman C.C.D., Kim L.K., Lee G.R., Li H.B., Flavell R.A. MRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science (80-. ) 2020;367:1255–1260. doi: 10.1126/science.abb2507. [DOI] [PubMed] [Google Scholar]
- 17.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.AlGhamdi N.A., Alsuwat H.S., Borgio J.F., AbdulAzeez S. Emerging of composition variations of SARS-CoV-2 spike protein and human ACE2 contribute to the level of infection: in silico approaches. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1841032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pramanick I., Sengupta N., Mishra S., Pandey S., Girish N., Das A., Dutta S. Conformational flexibility and structural variability of SARS-CoV2 S protein. Structure. 2021;29:834–845. doi: 10.1016/j.str.2021.04.006. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.Y., Katsamba P.S., Sampson J.M., Schön A., Bimela J., Boyington J.C., Nazzari A., Olia A.S., Shi W., Sastry M., Stephens T., Stuckey J., Teng I.T., Wang P., Wang S., Zhang B., Friesner R.A., Ho D.D., Mascola J.R., Shapiro L., Kwong P.D. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe. 2020;28:867–879. doi: 10.1016/j.chom.2020.11.004. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]