Abstract
The dysregulation of myeloid cell responses is increasingly demonstrated to be a major mechanism of pathogenesis for COVID-19. The pathological cellular and cytokine signatures associated with this disease point to a critical role of a hyperactivated innate immune response in driving pathology. Unique immunopathological features of COVID-19 include myeloid-cell dominant inflammation and cytokine release syndrome (CRS) alongside lymphopenia and acute respiratory distress syndrome (ARDS), all of which correlate with severe disease. Studies suggest a range of causes mediating myeloid hyperactivation, such as aberrant innate sensing, asynchronized immune cellular responses, as well as direct viral protein/host interactions. These include the recent identification of new myeloid cell receptors that bind SARS-CoV-2, which drive myeloid cell hyperinflammatory responses independently of lung epithelial cell infection via the canonical receptor, angiotensin-converting enzyme 2 (ACE2). The spectrum and nature of myeloid cell dysregulation in COVID-19 also differs from, at least to some extent, what is observed in other infectious diseases involving myeloid cell activation. While much of the therapeutic effort has focused on preventative measures with vaccines or neutralizing antibodies that block viral infection, recent clinical trials have also targeted myeloid cells and the associated cytokines as a means to resolve CRS and severe disease, with promising but thus far modest effects. In this review, we critically examine potential mechanisms driving myeloid cell dysregulation, leading to immunopathology and severe disease, and discuss potential therapeutic strategies targeting myeloid cells as a new paradigm for COVID-19 treatment.
Keywords: COVID-19, SARS-CoV-2, Pathogenesis, Immunopathology, Myeloid cell, Hyperactivation, Immunotherapy
1. Introduction
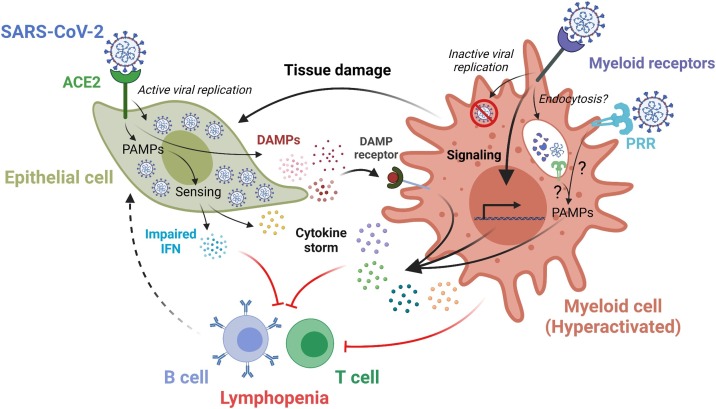
Caused by infections from the SARS-CoV-2 virus, the resulting COVID-19 pandemic has now surpassed over 250 million cases and 5 million deaths globally (https://covid19.who.int/). Though most individuals experience mild symptoms, severe disease occurs in about 5–15 % of patients [1]. Compared to other respiratory viruses such as influenza, COVID-19 has a longer clinical course (9-day average, from onset of symptoms to severe disease, compared to 4-day average in influenza) [[2], [3], [4], [5], [6]]. The disease can cause more serious symptoms requiring prolonged mechanical ventilation. It can be lethal in susceptible demographics, especially older men with comorbidities such as hypertension or diabetes [7]. Postmortem pathology reveals disease features of COVID-19 that include lymphopenia and myeloid-cell dominant inflammation alongside cytokine storm syndrome (CRS) and acute respiratory distress syndrome (ARDS) [8]. Though lymphocytes, such as B and T cells, consistently show decreased numbers in patients with severe disease, their role in orchestrating antiviral cellular immunity is more complex, with studies showing both positive and negative effects on disease progression [[8], [9], [10]]. However, the significant increase and hyperactive nature of inflammatory myeloid cells is generally regarded as a key hallmark of disease pathogenesis (Fig. 1 ) [11,12], especially given that macrophage-associated cytokines and chemokines [13], as well as other molecules such as neutrophil extracellular traps (NETs) [14,15], damage-associated molecular patterns (DAMPs) [16], and S100 alarmins [17] have shown clear prognostic value.
Fig. 1.
Myeloid dysregulation upon SARS-CoV-2 infection. ACE2-mediated SARS-CoV-2 infection leads to active viral replication and release of DAMPs and cytokines by lung epithelial cells. Sensing DAMPs by DAMP receptors on myeloid cells, direct viral engagement with myeloid receptors, or viral interaction with PRRs on the plasma membrane and/or in the endocytic compartments may lead to the hyperactivation of myeloid cells. This triggers production and release of proinflammatory cytokines, which in turn may elicit cytokine storm and ultimately lead to lymphopenia. ACE2, angiotensin-converting enzyme 2; PAMP, pathogen-associated molecular pattern; DAMP, damage-associated molecular pattern; PRR, pattern recognition receptor.
For the initial viral infection, it has been extensively demonstrated that SARS-CoV-2, an airborne virus, primarily infects specialized epithelial cells (type II pneumocytes) in the lung alveoli, nasal goblet cells, and ciliated cells in the airway or intestine by binding the host receptor angiotensin-converting enzyme 2 (ACE2) via its spike (S) protein (Fig. 2 ) [[18], [19], [20]]. While infection occurs initially in the lung, other tissues, such as kidney, small intestine, brain, heart, etc., show evidence of viral infection upon autopsy [21]. In general, viral infection in epithelial cells leads to the accumulation of viral genomes and replicative intermediates that are produced during the viral life cycle, collectively termed virus-derived pathogen-associated molecular patterns (PAMPs). These PAMPs are subsequently detected by a diverse array of host-encoded endosomal and cytosolic nucleic acid sensors, known as the pattern recognition receptors (PRRs), which are key sensing mechanisms of the host immune system. Given the nature of SARS-CoV-2 as a RNA virus, multiple cytosolic PRRs, including RIG-I [22], MDA-5 [23], and LGP2 [23], mediate recognition of SARS-CoV-2 viral components in infected lung epithelial cells. Viral infection also triggers the production of DAMPs, which are endogenous host-derived molecules that are released from damaged or dying cells. DAMPs can be sensed by PRRs as well as other innate receptors, including inflammasomes, TREM receptors, and G protein-coupled receptors (GPCRs) [24,25]. After detection of viral infection through PAMP or DAMP sensing, innate sensors rapidly trigger the production of cytokines and chemokines to initiate antiviral innate and adaptive immunity [26]. While protective in moderation, elevated cytokine levels observed in CRS in patients with severe disease have functional ties to the innate immune system and reveal an immune asynchrony involving an overheated innate immune system coupled with adaptive immunosuppression [13]. The mechanisms for lymphopenia and the suppression of adaptive immunity are still under investigation. How myeloid hyperactivation is linked to initial viral infection and disease pathogenesis remains a key open question for the basic understanding of COVID-19 and the development of related therapeutic strategies. Earlier stages of the disease currently focus on antivirals, disease prevention via vaccination, or neutralizing antibodies that block viral infection. However, taming innate hyperactivation is critical towards designing effective interventions for late stage COVID-19, for which therapies are still urgently required [10]. Thus, while COVID-19 pathophysiology is still being investigated, herein we focus on myeloid cells as a critical mediator of immunopathology, delving into potential mechanisms and outlining possible myeloid-targeted immunotherapies that may influence patient outcomes (Fig. 1).
Fig. 2.
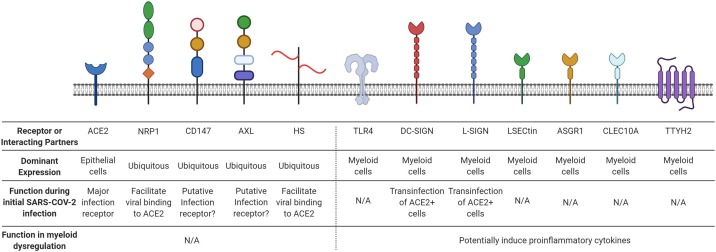
Receptor(s) or interacting partner(s) of SARS-CoV-2. ACE2, angiotensin-converting enzyme 2; NRP1, neuropilin 1; CD147, basigin; AXL, tyrosine-protein kinase receptor UFO; HS, heparan sulfate; TLR4, toll-like receptor 4; DC-SIGN, dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin; L-SIGN, C-type lectin domain family 4 member M; LSECtin, C-type lectin domain family 4 member G; ASGR1, asialoglycoprotein receptor 1; CLEC10A, C-type lectin domain family 10 member A; TTYH2, tweety family member 2. N/A, not applied.
2. Features of myeloid dysregulation in COVID-19
The lung microenvironment is normally composed of a heterogenous group of cells that maintain homeostasis while serving as a frontline barrier against particulates and pathogens [27]. However, absolute numbers and ratios of these populations, particularly those involved in immunity, change upon infection by SARS-CoV-2. Single-cell profiling of broncho-alveolar lavage samples of hospitalized patients with COVID-19 across multiple studies have shown that hyperactivated myeloid cells correlate with disease progression and severity [[28], [29], [30]]. Along with granulocytes such as neutrophils [30,31], proinflammatory monocyte-derived macrophages (MDM) are remarkably increased in lungs with severe disease compared to normal lungs across many studies. Levels of other macrophage subsets [28], such as transitioning MDM, can also be higher in COVID-19 patients, while tissue-resident alveolar macrophages (AM) are instead enriched in control and mild disease [13,30]. Similar to the phenotype within the lung, elevated levels of dysfunctional monocytes are likewise found in the peripheral blood of severe COVID-19 patients [12,32]. These monocytes are likely poor antigen presenters, potentially myeloid-derived suppressor cells (MDSCs), given their low expression of HLA-DR, in contrast to HLA-DRhi cells in patients with mild disease [12]. However, some subsets, such as transitional or nonclassical monocytes, are lower in the blood than the lung within patients with severe disease, hinting at potential trafficking and expansion from blood to the infected tissue [33]. Expansion of the inflammatory monocytic population occurs at the expense of other antiviral cell types within the myeloid lineage, such as dendritic cells (DCs) and plasmacytoid dendritic cells (pDCs), which remain at depressed levels even seven months post infection [34,35]. These two populations are critical for antigen presentation and type I interferon (IFN) production, respectively. Combined with the increased inflammatory myeloid population, their relative decrease may further escalate symptoms. However, whether the decrease is more causal or consequential in disease progression remains to be seen.
Though the exact myeloid activation program differs between studies, pathways increased in severe disease include proteases such as cathepsins [28], scavenger receptors, toll-like receptor (TLR) and signaling pathways, inflammatory transcriptional regulators [29], and cytokine and chemokine programs [36]. Cytokines, released by hyperactivated myeloid cells [28,30], are of special pathogenic importance; with no strict definition, the phenomenon of CRS is instead characterized by a large release of proinflammatory cytokines that can lead to enhanced vascular hyperpermeability, triggering ARDS [37], a major cause of COVID-19-related fatality [[38], [39], [40], [41], [42]]. The exact composition of upregulated inflammatory mediators can differ between patients and studies [43]. Cytokines such as IL-6, IL-8, IL-1, S100A8/9, GM-CSF, TNF-α, IL-12, CXCL10, MCP-1, MIP1A, etc., have all been shown to be upregulated in patients with severe disease or associated with pulmonary inflammation or disease severity [2,12,[44], [45], [46], [47], [48], [49], [50]]. Though studies suggest that myeloid cells may be the dominant producers for inflammatory cytokines such as IL-6 [51] and TNF-α [52], we note that the precise cellular source of cytokines has not been determined, as other cells such as epithelial cells are likely to also be a significant producer of inflammatory cytokines.
In addition to the upregulation of inflammatory innate cytokines, a blunted IFN response is also a general immune signature for severe disease. A central paradigm of protective antiviral immunity, first informed by studying immunological responses to influenza infection, is that IFN-mediated host responses precede downstream proinflammatory ones [53]. Such multitiered and well-regulated immune responses maximize antiviral protection while avoiding collateral damage to the host. However, this does not seem to hold true for COVID-19. Through longitudinal analysis of hospitalized patients with moderate-to-severe COVID-19, it was shown that production of IFN-I and IFN-III is diminished, delayed, and induced only in a fraction of patients [54]. Meanwhile, production of proinflammatory cytokines such as TNF-α, IL-6, and IL-8 precedes IFNs in all patients and persists for a prolonged time. This is in stark contrast to cytokine responses in influenza virus-infected patients hospitalized for viral pneumonia, which are characterized by robust and early IFN production and acutely produced proinflammatory cytokines. This imbalanced response to SARS-CoV-2 infection is also recapitulated in human lung epithelium-derived cell lines and ferrets [55]. The lack of proper IFN induction could be explained by the various viral mechanisms that evade and suppress the IFN response [56,57]. Indeed, it has been extensively shown that SARS-CoV, MERS-CoV, and SARS-CoV-2 can interfere with any of the following processes in innate antiviral immunity: 1) innate sensing, 2) IFN production, 3) IFN signaling, and 4) ISG effector function [[58], [59], [60], [61], [62]]. The ability for SARS-CoV-2 to evade innate immunity is further enhanced in new variants, such as the Alpha (B.1.1.7) and Beta (B.1.351) strains [[63], [64], [65]]. This raises the possibility that emerging viral variants may cause more pronounced immunopathology through further dampening of IFN-I-based antiviral immunity.
3. Mechanisms of myeloid dysregulation
The immune response to SARS-CoV-2 infection defined by low levels of IFN juxtaposed to elevated proinflammatory cytokines and chemokines could explain the myeloid cell hyperactivation observed in COVID-19. With lessons learned from SARS-CoV, in a theoretical framework of immune asynchrony [13], diminished IFN levels lead to an insufficient ability to clear SARS-CoV-2, resulting in further viral replication. Chronic low IFN released in response to viral infection generates further myeloid cell accumulation and activation alongside a suppressed adaptive immune response [66]. Throughout this cycle, many innate cytokines are produced, further amplifying the innate hyperactivation [11,67]. Many innate cytokines that correlate with severe disease, such as TNF-α, IL-6, IL-8, GM-CSF, and CCL2, have diverse functional effects on myeloid cells, including generation, activation, and recruitment (Fig. 3 ). A classic cytokine elicited in chronic inflammation [68], TNF-α promotes hematopoietic stem cell (HSC) survival and myeloid differentiation, in addition to supporting the immunosuppressive function of myeloid derived suppressor cells [69,70]. Lung epithelial and endothelial apoptosis is also caused by TNF-α [71], contributing to the release of DAMPs and the hyperinflammatory environment. Some notable examples of DAMPs include members of the S100 protein family, in particular S100A8 and S100A9, which are alarmins that critically regulate the migration and trafficking of leukocytes and confer anti-microbial protection during bacterial infection. It has been shown that S100A8/9 becomes dramatically upregulated at mRNA and protein levels following SARS-CoV-2 infection in animal models and in patients. Inhibition of S100A8/A9 with a small-molecule inhibitor, paquinimod, has been demonstrated to alleviate COVID-19-associated inflammatory disorder in a mouse model of SARS-CoV-2 infection [72,73]. It is also worth noting that there are counter-regulatory mechanisms by the host to prevent DAMPs from triggering excessive inflammation. One such mechanism is CD24, a surface receptor that has been increasingly recognized as a negative regulator of DAMP signaling. CD24 represses DAMP-induced immunity by directly trapping DAMPs to prevent agonistic receptor binding and/or by actively suppressing downstream signaling pathways through Siglec-10/G receptor [74,75].
Fig. 3.
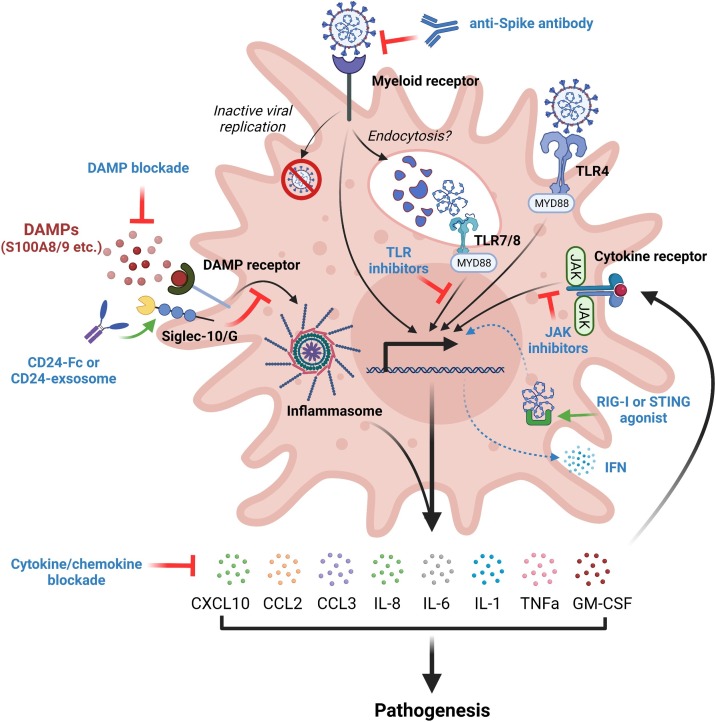
Potential immunotherapies for COVID-19 by targeting various pathways in dysregulated myeloid cells to restore immune homeostasis. These immunotherapies may include: 1) blocking spike-myeloid receptor interaction; 2) inhibiting TLR signaling pathways; 3) engaging Siglec-10/G by CD24-Fc fusion protein or exosome to suppress DAMP signaling; 4) blocking JAK signaling pathway; 5) enhancing RIG-I or STING mediated production of IFNs; and 6) blocking myeloid cell-released proinflammatory cytokines, chemokines, and DAMPs.
Outside of these DAMP-related pathways, other cytokines, such as IL-6, have been shown to induce myelopoiesis in severe infection [76]. Importantly, IL-6 plays a key role in CRS in the course of chimeric antigen-receptor T cell (CAR-T) therapy, with anti-IL-6 monoclonal antibodies being effective in ameliorating CRS symptoms [42]. Survival of myeloid cells such as neutrophils is promoted by IL-8 via inhibition of apoptosis [77]. IL-8 has been closely linked to the induction of MDSCs that potently inhibit lymphocyte activation, which can be a possible mechanism of lymphopenia. Produced by epithelial and hematopoietic cells during inflammation, GM-CSF is another important cytokine that confers a pro-inflammatory phenotype in myeloid cells characterized by the production of reactive oxygen species (ROS), cytokines, and chemokines [78]. Elevation of GM-CSF is unique to fatal COVID-19 but not influenza, suggesting GM-CSF as a disease-specific mechanism [44]. The chemoattractant cytokine CCL2 has been shown to be secreted by SARS-CoV-2 infected cardiomyocytes, promoting monocyte recruitment to the heart [79]. Although myeloid cells can limit viral replication through engulfment of infected cells upon arrival at the site of infection, an excessive production of proinflammatory cytokines, such as IL-6, TNF-α, IL-8, IL-1β, GM-CSF, IL-18, S100A8/9, etc., was observed during COVID-19. These molecular features are closely associated with dysregulated myeloid activation and can result in secondary immune recruitment, vascular leakage, and lethal respiratory dysfunction that likely underlie COVID-19 pathogenesis [42]. In the absence of appropriate regulation in normal resolution of infection, these cytokines can further activate the innate immune system, cause lymphopenia, and damage host tissue, leading to a potentially deadly feed-forward loop [42]. Altogether, these data provide compelling evidence that exposure to innate cytokines is a crucial driver of myeloid recruitment and activation [12,32].
While the effect of SARS-CoV-2 induced cytokines on myeloid cells is relatively understood, few studies have examined whether proinflammatory myeloid activation occurs dependently or independently of epithelial SARS-CoV-2 infection. Single-cell transcriptomic studies of lung tissues from COVID-19 patients have revealed myeloid cells as the cellular compartment most enriched for SARS-CoV-2 RNA, with particular abundance of CD14highCD16high inflammatory monocytes and LDB2highOSMRhighYAP1high macrophages [29,36]. Notably, myeloid cells positive for SARS-CoV-2 RNA appear to have distinct transcriptional programs compared to cells that do not harbor viral RNA, including genes associated with chemokine and cytokine signaling and responses to IFN, TNF, intracellular pathogens and viruses. These results highlight the potential for viral RNA to directly trigger activation of myeloid cells. However, unlike epithelial cells, myeloid cells do not express canonical cellular receptors and proteases, such as ACE2 and TMPRSS2, that are utilized by SARS-CoV-2 for viral entry [80]. While SARS-CoV-2 viral antigens have been detected in myeloid cells [[81], [82], [83]], there is no evidence thus far of productive infection. Consequently, activation of myeloid cells by SARS-CoV-2 is unlikely a result of direct infection by the virus via known receptors or general viral uptake mechanisms. Here, we propose two major mechanisms linking SARS-CoV-2 infection and myeloid cells to explain how the virus can directly induce myeloid activation: 1) direct PRR stimulation in myeloid cells, and 2) modulation of myeloid activation by viral proteins.
4. Myeloid cell activation via direct innate sensing of SARS-CoV-2
The first mechanism by which myeloid cells respond to SARS-CoV-2 is through direct innate sensing. Toll-like receptor 4 (TLR4) is a cell surface PRR that is abundantly expressed on myeloid cells and responsible for detecting bacterial ligands such as lipopolysaccharides (LPS) [84]. TLR4 signaling can be activated by oxidized phospholipid (OxPL), which is produced during the respiratory virus-induced oxidative stress response, to trigger cytokine production in macrophages and acute lung injury (ALI) [85]. A recent study also found that the S protein from SARS-CoV-2 directly interacts with and activates TLR4 (Fig. 2), triggering high levels of IL1B and IL6 transcription in vitro [86]. In addition to TLR4, myeloid cells use specialized endosomal PRR, including TLR3 (receptor for double-stranded RNA (dsRNA)), TLR7 (receptor for single-stranded RNA (ssRNA)), TLR8 (receptor for ssRNA), and TLR9 (receptor for double-stranded DNA (dsDNA)), to mediate sensing of viral genomes and replication products [87]. Upon endocytosis of viruses, endosomal TLRs sense viral genomes, presumably after the envelopes and capsids are uncoated by the degradative enzymes therein, and trigger cytokine and type I IFN production. However, SARS-CoV-2 is unlikely to directly enter the endosome in non-epithelial cells through endocytosis downstream of the spike-ACE2 interaction, as the canonical cellular receptor ACE2 is not abundantly expressed in these cells. Instead, viral products of SARS-CoV-2 may be delivered into the endosomal/lysosomal compartment through phagolysosomal fusion, a process by which the phagosome formed upon phagocytosis fuses with the lysosome and acquires lysosomal contents, such as PRRs and hydrolytic enzymes [88]. By doing so, viral particles or PAMPs in which viral endocytosis does not normally occur can be delivered to and sampled by the endosomal PRRs to initiate an antiviral response. The phagocytic process can be further augmented by Fc-mediated phagocytosis of antibody-virion immune complexes [89]. However, we note that phagocytosis is a general viral uptake mechanism, and the specific features of SARS-CoV-2 infection, such as the hyperactivation of myeloid cells, have yet to be thoroughly explained by this theory.
In addition to initiating PRR activation de novo, myeloid cells can also bypass the upstream sensing and production of second messengers to directly engage downstream responses. Rather than producing second messengers from within, host cells can directly acquire these intermediate signaling molecules from the extracellular space through transport mechanisms. A salient example for this is the cGAS/STING pathway, in which cGAS detects the presence of dsDNA in the cytoplasm of infected or damaged cells [90,91]. In the context of SARS-CoV-2 infection, it was recently shown that cGAS recognizes cytoplasmic chromatin DNA derived from the nucleus due to cell-to-cell fusion caused by the infection [92]. Upon ligand recognition, cGAS then produces 2’3’-cyclic-GMP-AMP (cGAMP) to activate STING, eliciting robust IFN-I responses. Interestingly, cGAMP is not only a cell-intrinsic activator of STING, but also a paracrine mediator that orchestrates larger-scale biological responses. During viral infection, cGAMP can be exported directly to the extracellular space in a soluble, non-membrane bound form by LRRC8A:C/E heteromeric channels and imported into neighbor host cells by importers such as SLC19A1 and SLC46A2 [[93], [94], [95]]. In particular, SLC46A2 represents the major cGAMP importer in primary human monocytes and monocyte-derived macrophages [95]. In the context of COVID-19, uncontrolled infection with SARS-CoV-2 in epithelial cells leads to pervasive cell damage and death [96], a process that likely triggers cGAS/STING activation and cGAMP release. Extracellular cGAMP can then be transported into myeloid cells via LRRC8A and induce STING activation [94]. However, the importance of this pathway in SARS-CoV-2 infection awaits further investigation.
5. Myeloid cell activation via viral/host protein interactions
The second mechanism by which SARS-CoV-2 induces myeloid cell activation is mediated by functional interactions between viral and host proteins. Emerging evidence suggests that the SARS-CoV-2 virion contains numerous proteins capable of interacting with host receptors and modulating immune responses. The SARS-CoV-2 genome has six functional open reading frames (ORFs) that encode replicase (ORF1a/ORF1b), spike (S), envelope (E), membrane (M), and nucleocapsid (N). In addition to the genomic RNA, a nested set of subgenomic RNAs (sgRNAs) encoding accessory proteins are also produced as a part of the viral life cycle. Though the canonical receptor for viral infection, ACE2, is not expressed on myeloid cells, a myeloid cell receptor-focused ectopic expression screen recently identified five C-type lectin receptors (DC-SIGN [97], L-SIGN, LSECtin, ASGR1, and CLEC10A), in addition to Tweety family member 2 (TTYH2), as novel binding partners for the viral S protein (Fig. 2) [80]. S protein binding epitopes are distinct between ACE2 (mainly the receptor-binding domain (RBD) of S) and C-type lectins (mainly the N-terminal domain (NTD) and C-terminal domain (CTD) of S) but partially overlapping between ACE2 and TTYH2, though all of the binding occurs in an ACE2-independent manner. Although there is no active infection and replication downstream of SARS-CoV-2 engagement with these myeloid cell receptors, these glycan-dependent interactions trigger robust proinflammatory responses in myeloid cells, characterized by potent production of inflammatory cytokines such as IL-1α, IL-1β, IL-8, CXCL10, CCL2, and CCL3. Rationally engineered bispecific antibodies blocking these new interactions show significantly reduced proinflammatory responses, demonstrating the functional importance of targeting not only viral infection, but also inflammatory receptors. Outside of ACE2 and these novel myeloid receptors, other extracellular receptors have also been identified, such as neuropilin-1 (NRP1) [98,99], heparan sulfate [100], CD147 [101], sialic acids [102], and AXL [103]. Both NRP-1 and heparan sulfate binding appear to be ACE2 dependent [98,100], which may limit the benefit of combinatorial therapy targeting these interactions alongside anti-S/ACE2 blocking antibody. Of the ACE2 independent interactions, the reproducibility of the CD147/S protein interaction is currently under debate [101,104], while sialic acids show both promotion and prevention of SARS-CoV-2 binding [105]. AXL may bind to S protein and mediate infection in the lung independently of ACE2 and its other known ligands, GAS6 or protein S [103]. Moreover, some of these receptors, particularly DC-SIGN/L-SIGN, have been reported to elicit SARS-CoV-2 trans-infection of ACE2 positive cells and thus facilitate viral infection (Fig. 2) [106]. Overall, the functional relevance of many other viral/receptor interactions downstream of receptor engagement remains to be determined, along with the therapeutic utility of targeting these interactions in addition to ACE2.
Viral proteins other than S may also directly modulate myeloid cells. E protein from SARS-CoV-2 is sensed by TLR2, another PRR expressed on the cell surface or endosomal compartment [107]. The interaction is required for the release of inflammatory cytokines, such as TNF-α and IFN-γ [108], during Murine Hepatitis Virus (MHV) infection. Blocking TLR2 also protects against SARS-CoV-2-induced lethality in vivo. In addition, N protein has been shown to trigger NLRP3-dependent inflammasome activation, which correlates with severe COVID-19 disease [81], by directly binding to the inflammasome sensor NLRP3 [109]. This binding promotes its interaction with ASC (the adaptor molecule apoptosis-associated speck-like protein containing a CARD) and facilitates inflammasome assembly. N protein induces the production of IL-1β and IL-6, aggravates lung injury, and instigates disease in mouse models of sepsis and acute inflammation [109]. Given that inflammasomes are located in the cytoplasm, we note that inflammasome activation by N protein likely pertains to ACE2-expressing cells or cells that can be infected by SARS-CoV-2. Several accessory proteins have also been implicated in the induction of myeloid activation in COVID-19 [110]. ORF3a is the largest among all accessory proteins encoded by SARS-CoV-2, sharing 72.7 % amino acid sequence homology with SARS-CoV ORF3a. SARS-CoV ORF3a promotes inflammasome activation by inducing IL1b gene expression and IL-1β maturation [111]. Interestingly, infection of K18-hACE2 transgenic mice with an ORF3a-deficient SARS-CoV-2 mutant results in less pathology and improved survival compared to that of wild type virus [112]. Whether the observed attenuation of disease severity is mediated by reduced immunopathology remains to be determined. In addition, it was recently reported that SARS-CoV-2 ORF7a directly triggers the activation of CD14+ monocytes ex vivo, which is characterized by a marked reduction in surface HLA expression and significantly increased production of proinflammatory cytokines, including IL-6, IL-1β, IL-8, and TNF-α [113]. ORF8, the only secreted SARS-CoV-2 viral protein, also promotes the expression of proinflammatory mediators via the IL-17 signaling pathway by interacting with host IL17RA [114]. It has also been shown to modulate cytokine expression in primary human macrophages, most notably downregulation of IL-6 and IL-8 [115]. ORF9c, which encodes a small, unstable protein with a putative transmembrane domain, has also been reported to stimulate IL-6 signaling while potently repressing IFN-I responses [116]. Whether these ORFs functionally encode for and/or structurally resemble physiological ligands of PRR and inflammasome sensors, as well as their functional receptors in the myeloid cells, remains an open question [42]. The virus also does not actively replicate in myeloid cells [80,117], resulting in fewer templates for transcription and translation - the functional relevance of viral proteins requiring host machinery in myeloid cells thus awaits further study. Recently, reversal of post-translational modifications of downstream effector pathways, such as conjugation of ubiquitin-like protein ISG15 (interferon-stimulated gene 15) [118], have also been shown to correlate with macrophage hyperpolarization [119]. Exactly which substrate(s) are responsible for this functional perturbation await future investigation.
In summary, we discuss two distinct mechanisms explaining how SARS-CoV-2 infection triggers myeloid cell activation and pathogenic inflammation. First, myeloid cells can utilize a broad array of surface and endosomal PRRs to detect the presence of viral PAMPs or viral infection associated DAMPs. Second, myeloid cells could be activated by viral proteins, including but are not limited to S, N, and various ORFs. While some of these mechanisms remain speculative and await further experimental evidence, they point to the centrality of myeloid-derived cytokines and chemokines in the pathogenesis of COVID-19. Below, we further review how these insights motivate myeloid-directed immunotherapies against COVID-19 that are currently in different stages of clinical trials.
6. Potential immunotherapies
Immunomodulatory approaches have been proposed as a promising intervention for COVID-19 management given that dysregulation of the immune response, especially hyperactivated myeloid immunity, is a major contributor of systemic and lethal COVID-19 pathogenesis [8,120]. The hypothesis was supported by the results from UK-based RECOVERY and other randomized controlled trials (RCT), which indicated that the use of anti-inflammatory agents, such as corticosteroids, was associated with reduced 28-day mortality compared with the standard of care among patients receiving oxygen support [121,122]. However, delayed viral clearance and secondary infection caused by general immunosuppression from these anti-inflammatory agents warrants concern [123,124]. In this setting, exploring more specific immunomodulators, especially those targeting the hysteric innate immune response, may emerge as a promising therapeutic strategy to prevent immune-induced injury while preserving adaptive antiviral immune function.
As aforementioned, the features of hyperactivated myeloid dysregulation triggered by SARS-CoV-2 involve aberrant sensing pathways, hypersensitive alarming mechanisms, and robust innate immune cytokine release. Molecules that target myeloid dysregulation at every stage are under investigation. Previous studies suggested that critical cytokines, such as IL-6, are key mediators of myeloid overactivation and inflammatory circuit establishment. Based on these findings, targeting cytokines in this inflammation amplification stage, especially IL-6 blockade which has shown efficacy against CRS in CAR-T therapy, received early and widespread attention (Fig. 3) [125,126]. Multiple powered trials have confirmed that IL-6 blockade (tocilizumab, sarilumab and siltuximab) were associated with modest reduction of 28-day mortality among hospitalized patients, with a 3 % decrease from IL-6 blockade to standard of care (22 % vs 25 %) [[127], [128], [129], [130]]. Tocilizumab received U.S. Emergency Use Authorization for the management of hospitalized adults and children with COVID-19 in 2021. Notably, this benefit appeared more evident among patients who received non-invasive respiratory support rather than those requiring invasive mechanical ventilation, highlighting the importance of disturbing the positive-feedback cytokine network at the early stage [129]. This observation is further supported by the result from GM-CSF blockade. Intervention with lenzilumab at an early phase was shown to improve ventilation-free survival at day 28 among patients with high-risk comorbidities [131]. The successes in targeting GM-CSF may partially correlate with its critical effects on mobilizing myeloid cells and promoting the expression of IL-1, IL-6, TNF-α and other proinflammatory cytokines (Fig. 3) simultaneously [78,132,133]. However, more clinical data are required to draw conclusions regarding efficacy and safety in COVID-19 treatment. Apart from targeting cytokines themselves, inhibition of essential downstream, critical inflammatory cytokine signaling, such as the Janus kinase (JAK) pathway, has been found to strongly suppress the inflammatory cascade response [134]. Administration of orally selective JAK inhibitors, tofacitinib or baricitinib, have led to clinical improvements among hospitalized patients with COVID-19 pneumonia compared to placebo in several randomized controlled trials (RCT) [[134], [135], [136]]. Molecules that target other proinflammatory cytokines and signaling pathways associated with hyperactivated myeloid dysregulation also warrant attention. Agents which block IL-8 [137], TNF-α [138,139], IL-18 [140], and CCR2 [141], in addition to other pathways such as LIGHT (TNFSF14) [142], IL-1 [[143], [144], [145]], and IL-17 [146], have shown potential beneficial effects in preliminary data of COVID-19 management, although more evidence is still needed to identify their safety and efficacy in ongoing trials.
Correcting dysregulated myeloid activation at sensing and alarming stages also showed encouraging effects in reducing inflammatory-related injuries. In particular, targeting an impaired type I IFN response, a hallmark of severe COVID-19 and critical pathway downstream of TLR signaling [147], is becoming increasingly appreciated. In one randomized study involving 101 adults admitted to the hospital with mild-to-moderate COVID-19, patients received inhaled SNG001 for 14 days, a nebulized formulation of recombinant IFN-β. Results demonstrated a more rapid recovery and less incidence of progression to severe disease or death for those on SNG001 than placebo [148]. However, there is debate regarding optimal timing of IFN treatment for COVID-19, particularly considering that IFN treatment might exacerbate tissue destruction due to robust immune activation. Other than as monotherapy, a recent strategy to arm a RBD based vaccine fused with type I IFN offered rapid and complete protection against SARS-CoV-2 infection, which may provide a unique approach for IFN-based viral prevention [149]. Besides this, other targets, such as those involved in the PAMP signaling pathway including RIG-I agonists [150], STING agonists [151,152], and TLR inhibitors are also under investigation, which may emerge as important options for future therapeutic development. Regarding DAMP signaling, CD24, an innate immune checkpoint binding to inhibitory Siglec10/G receptor, has also been repurposed as a promising target for COVID-19 treatment (Fig. 3) [75]. In line with its encouraging results in management of graft versus host disease (GVHD), addition of CD24 fusion protein to standard of care could prevent disease progression in a pre-planned interim efficacy analysis in the SACCOVID clinical trial [153]. Unpublished data from a recent trial showed that 29 of 30 patients rapidly recovered from moderate/serious COVID-19 infections after delivering CD24-enriched exosomes to the lung, which may become a potential breakthrough for COVID-19 treatment [154]. Given the primary success of these inhaled drugs, regulating the local immune response in the lung may be a promising strategy moving forward [30,155]. Likewise, early research has found that aberrant immature neutrophils are induced during SARS-CoV-2 infection. Recent data demonstrated that alarmins S100A8/A9, which are DAMP molecules, may account for this immune disorder in severe patients. Blockade of S100A8/9 through paquinimod could rescue pneumonia with substantial reduction of SARS-CoV-2 viral loads in preclinical experiments, providing a good option for further COVID-19 treatment [72]. Collectively, these promising results provide support for the importance of correcting myeloid cell dysregulation at activation stages prior to uncontrolled, downstream cytokine storm feed forward loops (Fig. 3 and Table 1 ).
Table 1.
Key clinical trials assessing immunotherapies in the management of COVID-19.
| Drug/Targets | Trials | Study design | Primary results | Timelines of the same or similar targets |
|---|---|---|---|---|
| Dexamethasone (Corticosteroids) | NCT04381936 | Hospitalized COVID-19 | Dexamethasone resulted in a reduction in mortality of 2.8 % for usual care (22.9 % vs. 25.7 %; p<0.001) | Hydrocortisone (NCT02735707, phase 4); Methylprednisolone (NCT04244591, phase 2/3) |
| (RECOVERY) (phase 2/3, RCT) [121] | ||||
| Tocilizumab (IL-6 receptor antagonist), EUA | NCT04381936 (RECOVERY) (phase 2/3, RCT) [130] | Hypoxia with systemic inflammation, receive SoC and either tocilizumab or SoC alone | Tocilizumab resulted in a reduction in mortality within 28 days as compared to SoC (31 % vs. 35 %; p = 0.0028) | Tocilizumab (NCT04320615, phase 3); Sarilumab (NCT04327388, phase 3); Olokizumab (NCT04380519, phase 2/3) |
| Clazakizumab (IL-6 antagonist) | NCT04363502 (phase 2, RCT) | Life-threatening COVID-19 infection, receive SoC and either clazakizumab or placebo | Not yet published | Clazakizumab (NCT04494724, phase 2); Siltuximab (NCT04322188, observational); Sirukumab (NCT04380961, phase 2) |
| Lenzilumab (GM-CSF antagonist) | NCT04351152 (phase 3, RCT) [131] | SpO2 ≤ 94 % or requiring supplemental oxygen, but not IMV, receive SoC and either lenzilumab or placebo | Lenzilumab improved the likelihood of SWOV by 54 % in the mITT population (p = 0.041) compared to placebo | Gimsilumab (NCT04351243, phase 2); Otilimab (NCT04376684, phase 2) |
| Mavrilimumab (GM-CSF receptor antagonist) | NCT04399980, NCT04463004, NCT04492514, (MASH-COVID) (phase 2, RCT) [133] | Severe COVID-19 pneumonia and systemic hyperinflammation, receive SoC and either mavrilimumab or placebo | No significant increase in the proportion of patients free of supplemental oxygen at day 14 in mavrilimumab group compared to placebo (57 % vs. 47 %; p = 0.76) | Mavrilimumab (NCT04397497, phase 2; NCT04447469, phase 2/3) |
| Baricitinib (JAK 1/2 inhibitor), EUA | NCT04421027, (COV-BARRIER) (phase 3, RCT) [136] | Hospitalized COVID-19, receive SoC and either baricitinib or placebo | Baricitinib resulted in a reduction in mortality by day 28 as compared to placebo (8 % vs. 13 %; p = 0.0018) | Tofacitinib, selective JAK1/3 inhibitor, and to lesser extent JAK2 (NCT04469114, phase 3) |
| CERC-002 (TNFSF14 antagonist) | NCT04412057 (phase 2, RCT) [142] | Mild to moderate ARDS, randomly receive a single dose of CERC-002 or placebo, in addition to standard of care that included high dose corticosteroids | CERC-002 increased the rate of survival and free of respiratory failure status through day 28 as compared to placebo (83.9 % vs. 64.5 %; p = 0.044) | Adalimumab, TNF-α antagonist, (NCT04705844, phase 3); Infliximab, TNF-α antagonist, (NCT04922827, phase 2); Etanercept, TNF-α receptor fusion protein, (pre-clinical) |
| Anakinra (IL-1 receptor antagonist) | NCT04341584 (CORIMUNO-ANA-1) (phase 2, RCT) [144] | Mild-to-moderate COVID-19 pneumonia, receive usual care plus anakinra or usual care alone | No significant difference in WHO-CPS score of >5 points at day 4 in anakinra group compared to placebo (36 % vs. 38 %) | Anakinra (NCT04680949, phase 3) |
| Canakinumab (IL-1β antagonist) | NCT04362813 (CAN-COVID) (phase 3, RCT) [145] | Patients with COVID-19 pneumonia, receive SoC and either canakinumab or placebo | No significant difference in survival rate without requiring IMV between canakinumab group and placebo group (88.8 % vs. 85.7 %, p = 0.29) | Canakinumab (NCT04476706, no longer available) |
| Secukinumab (IL-17A antagonist) | RBR-5vpyh4 (BISHOP study) (phase 2, RCT) [146] | Hospitalized COVID-19, receive SoC plus secukinumab or SoC alone | No significant difference in VFD between secukinumab group and control group (23.7 vs. 23.8 days; p = 0.62) | Ixekizumab (NCT04724629, phase 3) |
| Cenicriviroc (CCR2/CCR5 antagonist) | NCT04500418 (phase 2, RCT) | Patients with COVID-19 scoring "3" or "4" on the 7-Point Ordinal Scale, receive SoC and either cenicriviroc or placebo | Not yet published | BMS-813160 (pre-clinical) |
| SNG001 (Inhaled interferon β-1a) | NCT04385095 (phase 2, RCT) [148] | Adults admitted to hospital with COVID-19 symptoms, randomly receive SNG001 or placebo by inhalation for 14 days | Patients receiving SNG001 had greater odds of improvement on the OSCI scale on day 15 or 16 (p = 0.033) | Interferon β-1a, (NCT04350671, phase 4); Interferon β-1b (NCT04465695, phase 2); Interferon α-2b (NCT04480138, phase 2) |
| SACCOVID (CD24Fc) | NCT04317040 (SAC-COVID) (phase 3, RCT) [153] | Severe or critical COVID-19, receive best available treatment and either CD24Fc or placebo | SACCOVID improved the likelihood of clinical recovery by 60 % compared to placebo (p = 0.005). (Unpublished interim efficacy and safety analyses) | CD24-enriched exosomes (NCT04747574, phase 1; NCT04969172, phase 2) |
| BMS-986253 (IL-8 antagonist) | NCT04347226 (phase 2, RCT) | Hospitalized COVID-19, receive SoC plus BMS-986253 or SoC alone | Not yet published | ABX-IL8, IL-8 antagonist, (pre-clinical) |
| EB05 (TLR4 antagonist) | NCT04401475 (phase 2/3, RCT) | Hospitalized COVID-19, receive SoC and either EB05 or placebo | Not yet published | M5049, TLR7/8 inhibitor (NCT04448756, phase 2) |
RCT, randomized controlled trial; EUA, emergency use authorization; SoC, standard-of-care; SpO2, oxygen saturation; SWOV, survival without ventilation; mITT, modified intention-to-treat analysis; JAK, Janus kinase; WHO-CPS, World Health Organization 10-point Clinical Progression Scale; OSCI, Ordinal Scale for Clinical Improvement; IMV, invasive mechanical ventilation; ARDS, acute respiratory distress syndrome; VFD, ventilator-free days.
Strategies targeting the virus-host interaction, especially the virus-immune cell interaction, have been of great interest. A strong association between neutralization antibody levels and immune protection has been increasingly recognized [156]. Administration of neutralizing antibody, such as sotrovimab (VIR-7831), may reduce the risk of hospitalization and death in patients with mild and moderate diseases [157]. However, most of these neutralizing antibodies seem ineffective for patients with severe disease [158]. Some neutralizing antibodies may facilitate various viral functions and even worsen clinical outcomes, particularly via the antibody-dependent enhancement (ADE) effect, complicating neutralizing antibody treatment [158,159]. Of note, current antibody programs are mostly designed to block the SARS-CoV-2 S/ACE2 interaction. Given that ACE2 is minimally expressed on most human peripheral immune cells, exploring novel antibodies that target additional viral receptors are urgently required, especially those that interfere with functional interactions between immune cells and the coronavirus. Specifically, our previous research identified that SARS-CoV-2 engages immune cells through specific extracellular proteins, C-type lectins and TTYH2, which are interactions distinct from ACE2 [80]. Engineered nanobodies (Fig. 3), which can be nebulized and delivered to the lung, potently blocked all viral immune interactions along with ACE2 and reduced proinflammatory responses. Our data reveals an important therapeutic paradigm for neutralizing not only receptors for viral infection, but also for hyperinflammation [80]. We are advancing these nanobodies towards development as a new therapeutic option for patients with COVID-19, though the clinical benefits and appropriate timing for use of these agents await further evaluation.
Although the dysregulation of the immune response was artificially separated to early and late phases, key pathogenic mechanisms in the clinic can occur in parallel and synergize with one another [160]. Consistent with the benefit of combining IL-6 blockade with corticosteroids and baricitinib with remdesivir, the combinatorial usage of antiviruses and immunomodulators (or agents targeting different phases of immune dysregulation together) are rational and under further investigation, especially for patients with clear signs of hyperinflammation. However, there is still limited evidence regarding the safety and efficacy of combinatorial treatment in early COVID-19. The combinational usage of immunomodulators with molnupiravir, a recent investigational oral antiviral drug primarily based on RNA mutagenesis for outpatients [161,162], might provide the possibility controlling infection early and efficiently. However, the potential risk of accelerating the emergence of variants [163] and safety concerns of its mutagenic effect on host cells must be noted. Several genetic and metabolic defects have been recently identified in patients with life-threatening COVID-19 [164]; identification of patients with high risk of immune dysregulation at initial stages provides the possibility for early combined and personalized therapy in select patients with known defects. It is also important to explore multi-specific antibodies or cocktails to prevent virus mutational escape due to new and emerging variants [165,166]. In addition, recent research suggests that dysregulated metabolic profiles and energy generation, such as upregulation of oxidative phosphorylation and downregulation of fructose and mannose metabolism [167], may be an underlying mechanism for imbalanced immune responses [168,169]. This contributes to increased mortality in COVID-19 patients with type 2 diabetes mellitus and obesity [170,171]. These new findings offer the possibility to combine agents targeting key immune-metabolic pathways for COVID-19 treatment, though disruption of normal host metabolism should be avoided. Overall, strategies targeting dysregulated myeloid responses have improved clinical outcomes of COVID-19. Earlier immunological intervention and combination of therapies with different mechanisms carry the potential to achieve greater levels of success for COVID-19 management.
7. Conclusion
Hyperactivated myeloid cells constitute a key part of COVID-19 immunopathogenesis. In this review, we summarize the major mechanisms describing how SARS-CoV-2 dysregulates myeloid cells, their functional consequences, and potential myeloid targeted immunotherapies. We outline direct innate sensing and viral/host protein interactions as potential drivers of myeloid cell hyperactivation, discussing in particular novel myeloid cell receptors for SARS-CoV-2 S protein binding that trigger subsequent myeloid overactivation. Further investigation of myeloid biology over the course of disease progression is necessary and will yield fertile ground for the development of novel immunotherapies targeting all stages of myeloid cell activation. Combinatorial strategies with early intervention will hopefully correct myeloid dysregulation while reducing viral burden, offering patients new options to manage and treat COVID-19.
Author contributions
J.W., T.T.S., T.M., and R.G. conceived the study and wrote the manuscript, Q.L. and J.W. drafted the figures.
Declaration of Competing Interest
J.W. and Q.L. are named inventors on a patent application that describes the anti-SARS-CoV-2 spike nanobodies that block both ACE2 and myeloid cell receptor interactions. J.W. is a consultant for Lilly Asia Ventures and is on the Scientific Advisory Board of Rootpath Genomics, which is not relevant to this work. T.M. is named an inventor on a patent entitled “Compositions and Methods for Treating, Ameliorating, and/or Preventing Viral Infections”. The other authors declare no competing interests exist.
Acknowledgments
This work is supported by internal funds provided by the Office of Science & Research (OSR) and the Department of Pathology of New York University Grossman School of Medicine, and a NIH grant R21-AI163924 (to J.W.).
References
- 1.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasselli G., Zangrillo A., Zanella A., Antonelli M., Cabrini L., Castelli A., et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong S.B., Choi E.Y., Kim S.H., Suh G.Y., Park M.S., Lee M.G., et al. Epidemiological analysis of critically ill adult patients with pandemic influenza A(H1N1) in South Korea. Epidemiol. Infect. 2013;141:1070–1079. doi: 10.1017/S0950268812001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nin N., Soto L., Hurtado J., Lorente J.A., Buroni M., Arancibia F., et al. Clinical characteristics and outcomes of patients with 2009 influenza A(H1N1) virus infection with respiratory failure requiring mechanical ventilation. J. Crit. Care. 2011;26:186–192. doi: 10.1016/j.jcrc.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 7.Sanyaolu A., Okorie C., Marinkovic A., Patidar R., Younis K., Desai P., et al. Comorbidity and its impact on patients with COVID-19. SN Compr. Clin. Med. 2020:1–8. doi: 10.1007/s42399-020-00363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Candia P., Prattichizzo F., Garavelli S., Matarese G. T cells: warriors of SARS-CoV-2 infection. Trends Immunol. 2021;42:18–30. doi: 10.1016/j.it.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y., Fu B., Zheng X., Wang D., Zhao C., Qi Y., et al. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Sci. Rev. 2020;7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulte-Schrepping J., Reusch N., Paclik D., Bassler K., Schlickeiser S., Zhang B., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440. doi: 10.1016/j.cell.2020.08.001. e1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou T., Su T.T., Mudianto T., Wang J. Immune asynchrony in COVID-19 pathogenesis and potential immunotherapies. J. Exp. Med. 2020:217. doi: 10.1084/jem.20200674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuo Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J.A., et al. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5 doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodríguez-Espinosa O., Rojas-Espinosa O., Moreno-Altamirano M.M., López-Villegas E.O., Sánchez-García F.J. Metabolic requirements for neutrophil extracellular traps formation. Immunology. 2015;145:213–224. doi: 10.1111/imm.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huckriede J., Anderberg S.B., Morales A., de Vries F., Hultström M., Bergqvist A., et al. Evolution of NETosis markers and DAMPs have prognostic value in critically ill COVID-19 patients. Sci. Rep. 2021;11:15701. doi: 10.1038/s41598-021-95209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biji A., Khatun O., Swaraj S., Narayan R., Rajmani R.S., Sardar R., et al. Identification of COVID-19 prognostic markers and therapeutic targets through meta-analysis and validation of Omics data from nasopharyngeal samples. EBioMedicine. 2021;70 doi: 10.1016/j.ebiom.2021.103525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flerlage T., Boyd D.F., Meliopoulos V., Thomas P.G., Schultz-Cherry S. Influenza virus and SARS-CoV-2: pathogenesis and host responses in the respiratory tract. Nat. Rev. Microbiol. 2021;19:425–441. doi: 10.1038/s41579-021-00542-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J., Li Y., Liu Q., Yao Q., Wang X., Zhang H., et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021;7:17. doi: 10.1038/s41421-021-00249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada T., Sato S., Sotoyama Y., Orba Y., Sawa H., Yamauchi H., et al. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021;22:820–828. doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- 23.Yin X., Riva L., Pu Y., Martin-Sancho L., Kanamune J., Yamamoto Y., et al. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swanson K.V., Deng M., Ting J.P.Y. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019;19:477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong T., Liu L., Jiang W., Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2020;20:95–112. doi: 10.1038/s41577-019-0215-7. [DOI] [PubMed] [Google Scholar]
- 26.Iwasaki A., Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altorki N.K., Markowitz G.J., Gao D., Port J.L., Saxena A., Stiles B., et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat. Rev. Cancer. 2019;19:9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melms J.C., Biermann J., Huang H., Wang Y., Nair A., Tagore S., et al. A molecular single-cell lung atlas of lethal COVID-19. Nature. 2021;595:114–119. doi: 10.1038/s41586-021-03569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delorey T.M., Ziegler C.G.K., Heimberg G., Normand R., Yang Y., Segerstolpe A., et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595:107–113. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 31.Vitte J., Diallo A.B., Boumaza A., Lopez A., Michel M., Allardet-Servent J., et al. A granulocytic signature identifies COVID-19 and its severity. J. Infect. Dis. 2020;222:1985–1996. doi: 10.1093/infdis/jiaa591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Cerrillo I., Landete P., Aldave B., Sanchez-Alonso S., Sanchez-Azofra A., Marcos-Jimenez A., et al. COVID-19 severity associates with pulmonary redistribution of CD1c+ DCs and inflammatory transitional and nonclassical monocytes. J. Clin. Invest. 2020;130:6290–6300. doi: 10.1172/JCI140335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez-Gómez A., Vitallé J., Gasca-Capote C., Gutierrez-Valencia A., Trujillo-Rodriguez M., Serna-Gallego A., et al. Dendritic cell deficiencies persist seven months after SARS-CoV-2 infection. Cell. Mol. Immunol. 2021;18:2128–2139. doi: 10.1038/s41423-021-00728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grant R.A., Morales-Nebreda L., Markov N.S., Swaminathan S., Querrey M., Guzman E.R., et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 2020;8:e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasan S.S., Capstick T., Ahmed R., Kow C.S., Mazhar F., Merchant H.A., et al. Mortality in COVID-19 patients with acute respiratory distress syndrome and corticosteroids use: a systematic review and meta-analysis. Expert Rev. Respir. Med. 2020;14:1149–1163. doi: 10.1080/17476348.2020.1804365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinha P., Matthay M.A., Calfee C.S. Is a “cytokine storm” relevant to COVID-19? JAMA Intern. Med. 2020;180:1152–1154. doi: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 40.Behrens E.M., Koretzky G.A. Review: cytokine storm syndrome: looking toward the precision medicine era . 2017;69:1135–1143. doi: 10.1002/art.40071. [DOI] [PubMed] [Google Scholar]
- 41.Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012;76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangalmurti N., Hunter C.A. Cytokine storms: understanding COVID-19. Immunity. 2020;53:19–25. doi: 10.1016/j.immuni.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buszko M., Nita-Lazar A., Park J.H., Schwartzberg P.L., Verthelyi D., Young H.A., et al. Lessons learned: new insights on the role of cytokines in COVID-19. Nat. Immunol. 2021;22:404–411. doi: 10.1038/s41590-021-00901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thwaites R.S., Sanchez Sevilla Uruchurtu A., Siggins M.K., Liew F., Russell C.D., Moore S.C., et al. Inflammatory profiles across the spectrum of disease reveal a distinct role for GM-CSF in severe COVID-19. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abg9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jontvedt Jorgensen M., Holter J.C., Christensen E.E., Schjalm C., Tonby K., Pischke S.E., et al. Increased interleukin-6 and macrophage chemoattractant protein-1 are associated with respiratory failure in COVID-19. Sci. Rep. 2020;10:21697. doi: 10.1038/s41598-020-78710-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silvin A., Chapuis N., Dunsmore G., Goubet A.G., Dubuisson A., Derosa L., et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418. doi: 10.1016/j.cell.2020.08.002. e1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danlos F.X., Ackermann F., Rohmer J., Roumier M., Marabelle A., Michot J.M. High levels of TNFalpha in patients with COVID-19 refractory to tocilizumab. Eur. J. Cancer. 2021;149:102–104. doi: 10.1016/j.ejca.2021.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coperchini F., Chiovato L., Rotondi M. Interleukin-6, CXCL10 and infiltrating macrophages in COVID-19-related cytokine storm: not one for all but all for one! Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.668507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 50.Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo C., Li B., Ma H., Wang X., Cai P., Yu Q., et al. Single-cell analysis of two severe COVID-19 patients reveals a monocyte-associated and tocilizumab-responding cytokine storm. Nat. Commun. 2020;11:3924. doi: 10.1038/s41467-020-17834-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pelaia C., Tinello C., Vatrella A., De Sarro G., Pelaia G. Lung under attack by COVID-19-induced cytokine storm: pathogenic mechanisms and therapeutic implications. Ther. Adv. Respir. Dis. 2020;14 doi: 10.1177/1753466620933508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwasaki A., Pillai P.S. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galani I.E., Rovina N., Lampropoulou V., Triantafyllia V., Manioudaki M., Pavlos E., et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021;22:32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- 55.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park A., Iwasaki A. Type I and Type III interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020;27:870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sa Ribero M., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., et al. Evasion of Type I interferon by SARS-CoV-2. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsu J.C., Laurent-Rolle M., Pawlak J.B., Wilen C.B., Cresswell P. Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2101161118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schroeder S., Pott F., Niemeyer D., Veith T., Richter A., Muth D., et al. Interferon antagonism by SARS-CoV-2: a functional study using reverse genetics. Lancet Microbe. 2021;2:e210–e218. doi: 10.1016/S2666-5247(21)00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thorne L.G., Bouhaddou M., Reuschl A.K., Zuliani-Alvarez L., Polacco B., Pelin A., et al. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant. bioRxiv. 2021 [Google Scholar]
- 64.Guo K., Barrett B.S., Mickens K.L., Hasenkrug K.J., Santiago M.L. Interferon resistance of emerging SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1073/pnas.2203760119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perlman S., Dandekar A.A. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Popa C., Netea M.G., van Riel P.L., van der Meer J.W., Stalenhoef A.F. The role of TNF-alpha in chronic inflammatory conditions, intermediary metabolism, and cardiovascular risk. J. Lipid Res. 2007;48:751–762. doi: 10.1194/jlr.R600021-JLR200. [DOI] [PubMed] [Google Scholar]
- 69.Sade-Feldman M., Kanterman J., Ish-Shalom E., Elnekave M., Horwitz E., Baniyash M. Tumor necrosis factor-alpha blocks differentiation and enhances suppressive activity of immature myeloid cells during chronic inflammation. Immunity. 2013;38:541–554. doi: 10.1016/j.immuni.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 70.Yamashita M., Passegue E. TNF-alpha coordinates hematopoietic stem cell survival and myeloid regeneration. Cell Stem Cell. 2019;25:357–372. doi: 10.1016/j.stem.2019.05.019. e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo Q., Zhao Y., Li J., Liu J., Yang X., Guo X., et al. Induction of alarmin S100A8/A9 mediates activation of aberrant neutrophils in the pathogenesis of COVID-19. Cell Host Microbe. 2021;29:222–235. doi: 10.1016/j.chom.2020.12.016. e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen L., Long X., Xu Q., Tan J., Wang G., Cao Y., et al. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:992–994. doi: 10.1038/s41423-020-0492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen G.Y., Tang J., Zheng P., Liu Y. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science. 2009;323:1722–1725. doi: 10.1126/science.1168988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Y., Chen G.-Y., Zheng P. CD24-Siglec G/10 discriminates danger- from pathogen-associated molecular patterns. Trends Immunol. 2009;30:557–561. doi: 10.1016/j.it.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reyes M., Filbin M.R., Bhattacharyya R.P., Sonny A., Mehta A., Billman K., et al. Plasma from patients with bacterial sepsis or severe COVID-19 induces suppressive myeloid cell production from hematopoietic progenitors in vitro. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abe9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Butt Y., Kurdowska A., Allen T.C. Acute lung injury: a clinical and molecular review. Arch. Pathol. Lab. Med. 2016;140:345–350. doi: 10.5858/arpa.2015-0519-RA. [DOI] [PubMed] [Google Scholar]
- 78.Lang F.M., Lee K.M.C., Teijaro J.R., Becher B., Hamilton J.A. GM-CSF-based treatments in COVID-19: reconciling opposing therapeutic approaches. Nat. Rev. Immunol. 2020;20:507–514. doi: 10.1038/s41577-020-0357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang L., Nilsson-Payant B.E., Han Y., Jaffré F., Zhu J., Wang P., et al. Cardiomyocytes recruit monocytes upon SARS-CoV-2 infection by secreting CCL2. Stem Cell Rep. 2021;16:2274–2288. doi: 10.1016/j.stemcr.2021.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu Q., Liu J., Zhao S., Gomez Castro M.F., Laurent-Rolle M., Dong J., et al. SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2. Immunity. 2021;54:1304–1319. doi: 10.1016/j.immuni.2021.05.006. e1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rodrigues T.S., de Sa K.S.G., Ishimoto A.Y., Becerra A., Oliveira S., Almeida L., et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021;218 doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boumaza A., Gay L., Mezouar S., Bestion E., Diallo A.B., Michel M., et al. Monocytes and macrophages, targets of severe acute respiratory syndrome coronavirus 2: the clue for coronavirus disease 2019 immunoparalysis. J. Infect. Dis. 2021;224:395–406. doi: 10.1093/infdis/jiab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lv J., Wang Z., Qu Y., Zhu H., Zhu Q., Tong W., et al. Distinct uptake, amplification, and release of SARS-CoV-2 by M1 and M2 alveolar macrophages. Cell Discov. 2021;7:24. doi: 10.1038/s41421-021-00258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Medzhitov R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 85.Imai Y., Kuba K., Neely G.G., Yaghubian-Malhami R., Perkmann T., van Loo G., et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao Y., Kuang M., Li J., Zhu L., Jia Z., Guo X., et al. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 2021;31:818–820. doi: 10.1038/s41422-021-00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iwasaki A. A virological view of innate immune recognition. Annu. Rev. Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luzio J.P., Pryor P.R., Bright N.A. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 89.Swiecki M., Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 2015;15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Motwani M., Pesiridis S., Fitzgerald K.A. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 2019;20:657–674. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- 91.Hopfner K.P., Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 2020;21:501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 92.Zhuo Z., Xinyi Z., Xiaobo L., Xia X., Tao J., Ruiyi M., et al. 2021. Research Square. [Google Scholar]
- 93.Ritchie C., Cordova A.F., Hess G.T., Bassik M.C., Li L. SLC19A1 is an importer of the immunotransmitter cGAMP. Mol. Cell. 2019;75:372–381. doi: 10.1016/j.molcel.2019.05.006. e375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lahey L.J., Mardjuki R.E., Wen X., Hess G.T., Ritchie C., Carozza J.A., et al. LRRC8A:C/E heteromeric channels are ubiquitous transporters of cGAMP. Mol. Cell. 2020;80:578–591. doi: 10.1016/j.molcel.2020.10.021. e575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cordova A.F., Ritchie C., Bohnert V., Li L. Human SLC46A2 is the dominant cGAMP importer in extracellular cGAMP-Sensing macrophages and monocytes. ACS Cent. Sci. 2021;7:1073–1088. doi: 10.1021/acscentsci.1c00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bussani R., Schneider E., Zentilin L., Collesi C., Ali H., Braga L., et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang S., Song R., Wang Z., Jing Z., Wang S., Ma J. S100A8/A9 in inflammation. Front. Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Anton-Plagaro C., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057. doi: 10.1016/j.cell.2020.09.033. e1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang K., Chen W., Zhang Z., Deng Y., Lian J.Q., Du P., et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020;5:283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Milanetti E., Miotto M., Di Rienzo L., Nagaraj M., Monti M., Golbek T.W., et al. In-silico evidence for a two receptor based strategy of SARS-CoV-2. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.690655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shilts J., Crozier T.W.M., Greenwood E.J.D., Lehner P.J., Wright G.J. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci. Rep. 2021;11:413. doi: 10.1038/s41598-020-80464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sun X.L. The role of cell surface sialic acids for SARS-CoV-2 infection. Glycobiology. 2021 doi: 10.1093/glycob/cwab032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lempp F.A., Soriaga L.B., Montiel-Ruiz M., Benigni F., Noack J., Park Y.-J., et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature. 2021;598:342–347. doi: 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- 107.Zheng M., Karki R., Williams E.P., Yang D., Fitzpatrick E., Vogel P., et al. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021;22:829–838. doi: 10.1038/s41590-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., et al. Synergism of TNF-alpha and IFN-gamma triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168. doi: 10.1016/j.cell.2020.11.025. e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pan P., Shen M., Yu Z., Ge W., Chen K., Tian M., et al. SARS-CoV-2 N protein promotes NLRP3 inflammasome activation to induce hyperinflammation. Nat. Commun. 2021;12:4664. doi: 10.1038/s41467-021-25015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Redondo N., Zaldivar-Lopez S., Garrido J.J., Montoya M. SARS-CoV-2 accessory proteins in viral pathogenesis: knowns and unknowns. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.708264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Siu K.L., Yuen K.S., Castano-Rodriguez C., Ye Z.W., Yeung M.L., Fung S.Y., et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Silvas J.A., Vasquez D.M., Park J.G., Chiem K., Allue-Guardia A., Garcia-Vilanova A., et al. Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 human ACE2 transgenic mice. J. Virol. 2021;95 doi: 10.1128/JVI.00402-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhou Z., Huang C., Zhou Z., Huang Z., Su L., Kang S., et al. Structural insight reveals SARS-CoV-2 ORF7a as an immunomodulating factor for human CD14(+) monocytes. iScience. 2021;24 doi: 10.1016/j.isci.2021.102187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lin X., Fu B., Yin S., Li Z., Liu H., Zhang H., et al. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. iScience. 2021;24 doi: 10.1016/j.isci.2021.102293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kriplani N., Clohisey S., Fonseca S., Fletcher S., Lee H.-M., Ashworth J., et al. Secreted SARS-CoV-2 ORF8 modulates the cytokine expression profile of human macrophages. bioRxiv. 2021;2021 2008.2013.456266. [Google Scholar]
- 116.Dominguez Andres A., Feng Y., Campos A.R., Yin J., Yang C.C., James B., et al. SARS-CoV-2 ORF9c is a membrane-associated protein that suppresses antiviral responses in cells. bioRxiv. 2020 [Google Scholar]
- 117.Boumaza A., Gay L., Mezouar S., Bestion E., Diallo A.B., Michel M., et al. Monocytes and macrophages, targets of severe acute respiratory syndrome coronavirus 2: the clue for coronavirus disease 2019 immunoparalysis. J. Infect. Dis. 2021;224:395–406. doi: 10.1093/infdis/jiab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Perng Y.-C., Lenschow D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018;16:423–439. doi: 10.1038/s41579-018-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Munnur D., Teo Q., Eggermont D., Lee H.H.Y., Thery F., Ho J., et al. Altered ISGylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat. Immunol. 2021;22:1416–1427. doi: 10.1038/s41590-021-01035-8. [DOI] [PubMed] [Google Scholar]
- 120.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Group R.C., Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., et al. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.WHOREAfC-TW Group, Sterne J.A.C., Murthy S., Diaz J.V., Slutsky A.S., Villar J., et al. Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19: a meta-analysis. JAMA. 2020;324:1330–1341. doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li S., Hu Z., Song X. High-dose but not low-dose corticosteroids potentially delay viral shedding of patients with COVID-19. Clin. Infect. Dis. 2021;72:1297–1298. doi: 10.1093/cid/ciaa829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tang X., Feng Y.M., Ni J.X., Zhang J.Y., Liu L.M., Hu K., et al. Early use of corticosteroid may prolong SARS-CoV-2 shedding in non-intensive care unit patients with COVID-19 pneumonia: a multicenter, single-blind, randomized control trial. Respiration. 2021;100:116–126. doi: 10.1159/000512063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kotch C., Barrett D., Teachey D.T. Tocilizumab for the treatment of chimeric antigen receptor T cell-induced cytokine release syndrome. Expert Rev. Clin. Immunol. 2019;15:813–822. doi: 10.1080/1744666X.2019.1629904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee D.W., Gardner R., Porter D.L., Louis C.U., Ahmed N., Jensen M., et al. Current concepts in the diagnosis and management of cytokine release syndrome. Blood. 2014;124:188–195. doi: 10.1182/blood-2014-05-552729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Matthay M.A., Luetkemeyer A.F. IL-6 receptor antagonist therapy for patients hospitalized for COVID-19: who, when, and how? JAMA. 2021;326:483–485. doi: 10.1001/jama.2021.11121. [DOI] [PubMed] [Google Scholar]
- 128.Investigators R.-C., Gordon A.C., Mouncey P.R., Al-Beidh F., Rowan K.M., Nichol A.D., et al. Interleukin-6 receptor antagonists in critically ill patients with Covid-19. N. Engl. J. Med. 2021;384:1491–1502. doi: 10.1056/NEJMoa2100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Group WHOREAfC-TW, Shankar-Hari M., Vale C.L., Godolphin P.J., Fisher D., Higgins J.P.T., et al. Association between administration of IL-6 antagonists and mortality among patients hospitalized for COVID-19: a meta-analysis. JAMA. 2021;326:499–518. doi: 10.1001/jama.2021.11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Group RC Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2021;397:1637–1645. doi: 10.1016/S0140-6736(21)00676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Temesgen Z., Burger C.D., Baker J., Polk C., Libertin C., Kelley C., et al. Lenzilumab efficacy and safety in newly hospitalized Covid-19 subjects: results from the live-air phase 3 randomized double-blind placebo-controlled trial. medRxiv. 2021 [Google Scholar]
- 132.Shiomi A., Usui T. Pivotal roles of GM-CSF in autoimmunity and inflammation. Mediators Inflamm. 2015;2015 doi: 10.1155/2015/568543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cremer P.C., Abbate A., Hudock K., McWilliams C., Mehta J., Chang S.Y., et al. Mavrilimumab in patients with severe COVID-19 pneumonia and systemic hyperinflammation (MASH-COVID): an investigator initiated, multicentre, double-blind, randomised, placebo-controlled trial. Lancet Rheumatol. 2021;3:e410–e418. doi: 10.1016/S2665-9913(21)00070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Guimaraes P.O., Quirk D., Furtado R.H., Maia L.N., Saraiva J.F., Antunes M.O., et al. Tofacitinib in patients hospitalized with Covid-19 pneumonia. N. Engl. J. Med. 2021;385:406–415. doi: 10.1056/NEJMoa2101643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kalil A.C., Patterson T.F., Mehta A.K., Tomashek K.M., Wolfe C.R., Ghazaryan V., et al. Baricitinib plus remdesivir for hospitalized adults with Covid-19. N. Engl. J. Med. 2021;384:795–807. doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Marconi V.C., Ramanan A.V., de Bono S., Kartman C.E., Krishnan V., Liao R., et al. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): a randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir. Med. 2021 doi: 10.1016/S2213-2600(21)00331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li L., Li J., Gao M., Fan H., Wang Y., Xu X., et al. Interleukin-8 as a biomarker for disease prognosis of coronavirus disease-2019 patients. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.602395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen X.Y., Yan B.X., Man X.Y. TNFα inhibitor may be effective for severe COVID-19: learning from toxic epidermal necrolysis. Ther. Adv. Respir. Dis. 2020;14 doi: 10.1177/1753466620926800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stallmach A., Kortgen A., Gonnert F., Coldewey S.M., Reuken P., Bauer M. Infliximab against severe COVID-19-induced cytokine storm syndrome with organ failure-a cautionary case series. Crit. Care. 2020;24:444. doi: 10.1186/s13054-020-03158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Satış H., Özger H.S., Aysert Yıldız P., Hızel K., Gulbahar Ö, Erbaş G., et al. Prognostic value of interleukin-18 and its association with other inflammatory markers and disease severity in COVID-19. Cytokine. 2021;137 doi: 10.1016/j.cyto.2020.155302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 142.Perlin D.S., Neil G.A., Anderson C., Zafir-Lavie I., Roadcap L., Raines S., et al. CERC-002, a human anti-LIGHT mAb reduces respiratory failure and death in hospitalized COVID-19 ARDS patients. medRxiv. 2021;2021 2004.2003.21254748. [Google Scholar]
- 143.Kooistra E.J., Waalders N.J.B., Grondman I., Janssen N.A.F., de Nooijer A.H., Netea M.G., et al. Anakinra treatment in critically ill COVID-19 patients: a prospective cohort study. Crit. Care. 2020;24:688. doi: 10.1186/s13054-020-03364-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.group C-C Effect of anakinra versus usual care in adults in hospital with COVID-19 and mild-to-moderate pneumonia (CORIMUNO-ANA-1): a randomised controlled trial. Lancet Respir. Med. 2021;9:295–304. doi: 10.1016/S2213-2600(20)30556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Caricchio R., Abbate A., Gordeev I., Meng J., Hsue P.Y., Neogi T., et al. Effect of Canakinumab vs placebo on survival without invasive mechanical ventilation in patients hospitalized with severe COVID-19: a randomized clinical trial. JAMA. 2021;326:230–239. doi: 10.1001/jama.2021.9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Resende G.G., da Cruz Lage R., Lobê S.Q., Medeiros A.F., Costa e Silva A.D., Nogueira Sá A.T., et al. Blockade of Interleukin Seventeen (IL-17A) with Secukinumab in Hospitalized COVID-19 patients – the BISHOP study. medRxiv. 2021;2021 doi: 10.1080/23744235.2022.2066171. 2007.2021.21260963. [DOI] [PubMed] [Google Scholar]
- 147.Khanmohammadi S., Rezaei N. Role of Toll-like receptors in the pathogenesis of COVID-19. J. Med. Virol. 2021;93:2735–2739. doi: 10.1002/jmv.26826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Monk P.D., Marsden R.J., Tear V.J., Brookes J., Batten T.N., Mankowski M., et al. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir. Med. 2021;9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun S., Cai Y., Song T.Z., Pu Y., Cheng L., Xu H., et al. Interferon-armed RBD dimer enhances the immunogenicity of RBD for sterilizing immunity against SARS-CoV-2. Cell Res. 2021 doi: 10.1038/s41422-021-00531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mao T., Israelow B., Lucas C., Vogels C.B.F., Fedorova O., Breban M.I., et al. A stem-loop RNA RIG-I agonist confers prophylactic and therapeutic protection against acute and chronic SARS-CoV-2 infection in mice. bioRxiv. 2021;2021 doi: 10.1084/jem.20211818. 2006.2016.448754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Humphries F., Shmuel-Galia L., Jiang Z., Wilson R., Landis P., Ng S.-L., et al. A diamidobenzimidazole STING agonist protects against SARS-CoV-2 infection. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abi9002. eabi9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li M., Ferretti M., Ying B., Descamps H., Lee E., Dittmar M., et al. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abi9007. eabi9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Devenport M. 2020. OncoImmune’s SACCOVID™ (CD24Fc) Exhibits Superb Therapeutic Efficacy—A Potential Breakthrough in Treating Severe and Critical COVID-19. [Google Scholar]
- 154.Staff T. 2021. New Israeli Drug Cured 29 of 30 moderate/serious COVID Cases in Days — Hospital. [Google Scholar]
- 155.Xu G., Qi F., Li H., Yang Q., Wang H., Wang X., et al. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov. 2020;6:73. doi: 10.1038/s41421-020-00225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 157.Gupta A., Gonzalez-Rojas Y., Juarez E., Casal M.C., Moya J., Falci D.R., et al. Early Covid-19 treatment with SARS-CoV-2 neutralizing antibody sotrovimab. medRxiv. 2021;2021 doi: 10.1056/NEJMoa2107934. 2005.2027.21257096. [DOI] [PubMed] [Google Scholar]
- 158.Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Beck D.B., Aksentijevich I. Susceptibility to severe COVID-19. Science. 2020;370:404–405. doi: 10.1126/science.abe7591. [DOI] [PubMed] [Google Scholar]
- 160.Nissen C.B., Sciascia S., de Andrade D., Atsumi T., Bruce I.N., Cron R.Q., et al. The role of antirheumatics in patients with COVID-19. Lancet Rheumatol. 2021;3:e447–e459. doi: 10.1016/S2665-9913(21)00062-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gordon C.J., Tchesnokov E.P., Schinazi R.F., Götte M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Merck . 2021. Merck and Ridgeback’s Investigational Oral Antiviral Molnupiravir Reduced the Risk of Hospitalization or Death by Approximately 50 Percent Compared to Placebo for Patients With Mild or Moderate COVID-19 in Positive Interim Analysis of Phase 3 Study. [Google Scholar]
- 163.Malek R.J., Bill C.A., Vines C.M. Clinical drug therapies and biologicals currently used or in clinical trial to treat COVID-19. Biomed. Pharmacother. 2021;144 doi: 10.1016/j.biopha.2021.112276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McCallum M., Bassi J., Marco A., Chen A., Walls A.C., Iulio J.D., et al. SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. bioRxiv. 2021 [Google Scholar]
- 166.Wang L., Zhou T., Zhang Y., Yang E.S., Schramm C.A., Shi W., et al. Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science. 2021;373 doi: 10.1126/science.abh1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gardinassi L.G., Souza C.O.S., Sales-Campos H., Fonseca S.G. Immune and metabolic signatures of COVID-19 revealed by transcriptomics data reuse. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Batabyal R., Freishtat N., Hill E., Rehman M., Freishtat R., Koutroulis I. Metabolic dysfunction and immunometabolism in COVID-19 pathophysiology and therapeutics. Int. J. Obes. 2021;45:1163–1169. doi: 10.1038/s41366-021-00804-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pålsson-McDermott E.M., O’Neill L.A.J. Targeting immunometabolism as an anti-inflammatory strategy. Cell Res. 2020;30:300–314. doi: 10.1038/s41422-020-0291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Palaiodimos L., Kokkinidis D.G., Li W., Karamanis D., Ognibene J., Arora S., et al. Severe obesity, increasing age and male sex are independently associated with worse in-hospital outcomes, and higher in-hospital mortality, in a cohort of patients with COVID-19 in the Bronx, New York. Metabolism. 2020;108 doi: 10.1016/j.metabol.2020.154262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Corrao S., Pinelli K., Vacca M., Raspanti M., Argano C. Type 2 diabetes mellitus and COVID-19: a narrative review. Front. Endocrinol. (Lausanne) 2021;12:609470. doi: 10.3389/fendo.2021.609470. [DOI] [PMC free article] [PubMed] [Google Scholar]