Abstract
A dangerous infectious disease of the current century, the COVID-19 has apparently originated in a city in China and turned into a widespread pandemic within a short time. In this paper, a novel method has been presented for improving the screening and classification of COVID-19 patients based on their chest X-Ray (CXR) images. This method eliminates the severe dependence of the deep learning models on large datasets and the deep features extracted from them. In this approach, we have not only resolved the data limitation problem by combining the traditional data augmentation techniques with the generative adversarial networks (GANs), but also have enabled a deeper extraction of features by applying different filter banks such as the Sobel, Laplacian of Gaussian (LoG) and the Gabor filters. To verify the satisfactory performance of the proposed approach, it was applied on several deep transfer models and the results in each step were compared with each other. For training the entire models, we used 4560 CXR images of various patients with the viral, bacterial, fungal, and other diseases; 360 of these images are in the COVID-19 category and the rest belong to the non-COVID-19 diseases. According to the results, the Gabor filter bank achieves the highest growth in the values of the defined evaluation criteria and in just 45 epochs, it is able to elevate the accuracy by up to 32%. We then applied the proposed model on the DenseNet-201 model and compared its performance in terms of the detection accuracy with the performances of 10 existing COVID-19 detection techniques. Our approach was able to achieve an accuracy of 98.5% in the two-class classification procedure; which makes it a state-of-the-art method for detecting the COVID-19.
Keywords: COVID-19, Generative adversarial network, Classification, Gabor, Data augmentation, Deep learning
1. Introduction
In the early 2019, the people of world were astonished with the extensive proliferation of a new type of highly-contagious virus [1]. The COVID-19, a pandemic disease that causes an acute respiratory syndrome, was first detected in December 2019, in Wuhan, China, and then quickly spread to the other parts of the world [2]. In Feb. 11, 2020, this disease was officially named the novel coronavirus disease (COVID-19) by the World Health Organization (WHO) [3]. The serious threat posed by this pandemic on the world population has prompted WHO to issue a global emergency alert. Up to now, and as this paper is being written, about 220 countries and territories are grappling with this illness and, according to the official statistics, the number of people with COVID-19 and the number of deaths due to this disease have surpassed 162 m and 3 m, respectively [4].
The coronaviruses belong to an extensive family of viruses that have been discovered back in the 1960 s; therefore, the COVID-19 is not the first epidemic coronavirus-caused acute respiratory disease in the world [5]. However, due to its high transmissibility and its rapid spread in many parts of the world within a very short time span, WHO has now declared this disease as a pandemic [6]. In general, the family of coronaviruses can be divided into the following four groups [7]:
-
•
Common cold viruses
-
•
The virus causing the severe acute respiratory syndrome (SARS)
-
•
The virus causing the Middle East respiratory syndrome (MERS)
-
•
The novel Coronavirus (the new coronavirus originating in China)
Considering the similarity between the clinical signs of this disease and those of the other infectious diseases of the lungs, an accurate and rapid detection of COVID-19 is highly essential. Those suspected of catching the COVID-19 must quickly know if they are infected or not so that they can isolate themselves, start their treatment, and inform those they have been in close contact with. Generally, the coronavirus diagnostic methods can be divided into two major approaches: i) the laboratory techniques and ii) the methods based on medical imaging [8].
One of the most common laboratory techniques for diagnosing the respiratory illnesses and screening the patients is to collect their initial respiratory tract specimens. The lab techniques, which are normally based on the examination of antigens, nucleic acids and antibodies, are themselves divided into the direct and the indirect approaches. In the direct methods, the coronavirus infection is verified by detecting the viral RNA through the reverse transcriptase-polymerase chain reaction (RT-PCR) test. While in the indirect methods, the diagnosis of COVID-19 is confirmed by measuring the antiviral antibodies or the viral antigens [9], [10].
The laboratory techniques have their own advantages and disadvantages. For example, although the pathogen test in the lab is considered to be the best way of diagnosing a disease, the process is time-consuming and costly and the sampling procedures may sometimes lead to erroneous results. In addition, the sensitivity of this test is relatively low and even if the test results are negative, the existence of a disease cannot be refuted with absolute certainty.
Due to the existing problems in lab procedures (i.e., high costs, low sensitivity, likely sampling errors, and the relatively long process time), various imaging techniques are employed as an auxiliary diagnostic tool to speed up the detection and the clinical decision-making process pertaining to the COVID-19. To be clearer, while a physician is waiting for the lab results (e.g., RT-PCR) or when the lab results are negative but a patient still shows the signs of COVID-19 (false negative results), the physician can rely on the chest images to diagnose the disease.
Presently, and with regards to the quick proliferation of COVID-19, the radiologists are burdened with a heavy responsibility and they have to analyze a lot of infection cases daily. Therefore, developing intelligent and automated approaches based on deep learning for analyzing the medical images can play a crucial role in the control of this disease and reduce the working pressure of the medical personnel, and especially the radiologists.
As one of the sub-branches of artificial intelligence, machine learning has recently found extensive applications in different medical fields. With the help of machine learning, the accuracy of the physicians in diagnosing the COVID-19 disease can be improved quickly and substantially and an important step can be taken in preventing and controlling this illnesses, especially in its early stages [11], [12].
In this research, we have focused on the first application of artificial intelligence in a medical field (i.e., the processing and analysis of medical images). In this respect, first, we have reviewed the exclusive indicators of the COVID-19 disease in the chest X-Ray (CXR) images of the patients’ lungs and then presented a novel and intelligent method for processing and classifying these images. This method has been able to significantly improve the accuracy of classifying the lung images and diagnosing the COVID-19 disease.
Due to the scarcity of the sufficiently large datasets with the CXR images of the COVID-19 patients, we employed the data augmentation techniques in order to generate new data. To this end, we combined the traditional data augmentation techniques (e.g., image resizing, cropping, rotating, flipping) with the generative adversarial networks (GANs). In the next step, by applying each of these techniques on the limited initial data available, we were able to obtain the final dataset needed for the fine-tuning of our deep network. We then passed each data in the dataset through a pre-processing block consisting of the Gabor, Laplacian of Gaussian (LoG), and the Sobel filters before using them to train our deep network.
The overall contributions of this work can be summarized as follows:
-
•
Generating new training data from previous data by applying the traditional data augmentation techniques and combining these approaches with the GANs.
-
•
Passing the data produced in previous step along with the main initial data through the various processing filters.
-
•
Selecting several deep convolutional neural networks (CNNs) and applying the proposed method on them.
-
•
Classifying the patients’ lung images into the COVID-19 and the non-COVID-19 categories.
-
•
Comparing and evaluating the obtained results by applying the criteria of accuracy, precision, recall, specificity, F1-Score, and the Mathews correlation coefficient (MCC).
The rest of this paper has been organized as follows. The works related to the detection and diagnosis of the COVID-19 disease based on the image processing and deep learning techniques are reviewed in Section 2. The datasets and the proposed method are fully described in Section 3. Then in Section 4, we present our results along with the evaluation criteria and compare them with the findings of some other methods. And finally, the conclusion of the paper and the potential future works are presented in Section 5.
2. Literature review
In the last decade, the artificial intelligence methods such as the deep learning, machine learning, fuzzy logic, artificial neural network, genetic programming and the regression techniques have been employed to tackle many problems, including the classification and diagnosis of various diseases.
In this section, we briefly review the works that have been conducted on the applications of the image processing and deep learning techniques in the medical fields, and especially in the diagnosis of the respiratory and COVID-19 diseases.
2.1. Methods based on machine learning
The applications of the deep learning methods in the detection of the respiratory (and specifically, the Corona) patients can be explored from three aspects: the classification [13], segmentation [14], and the hybrid applications. The aim of the segmentation process is to extract useful information from images, such as the image edges and shapes and the characteristics of each region in an image. Segmentation is the first and the most critical step in image analysis and it can be considered as one of the most difficult aspects of image processing. This technique has a significant influence on the evaluation of image features and it can be implemented in two different ways: histogram-based [15] and clustering-based [16]. In medical applications (and especially in the detection of the COVID-19), the segmentation technique is often used to segregate the cancerous and abnormal tissues from the other types of infections in different images. The main goal of the classification process, however, is to detect and identify the Corona-related signs and infections in the medical images; which includes the steps of feature extraction, feature selection, the grouping of features, decision-making, and producing the final output. In the hybrid method, both of the abovementioned techniques are used simultaneously [17]. In general, the applications of the deep learning technique in medical imaging can be summarized as follows [18], [19], [20]:
-
•
Medical image analysis:
-
•
Helping in the diagnosis of neural conditions
-
•
Detection of the cardiovascular disorders
-
•
Cancer screening
2.1.1. Segmentation
The image segmentation process in computer vision refers to a procedure in which a digital image is divided into several segments or regions. In this technique, the output is not a selected class or a number but an image in which the boundaries of different objects have been delineated. Since in this process, the intended changes are implemented on the image pixels, we can somehow call it the “classification at the pixel level”.
Many recently published studies indicate that the CXR and CT scan images of the Corona patients’ lungs usually contain the ground-glass opacity (GGO) regions. So, for the radiologists who are trying to diagnose the COVID-19, it is important to detect the abnormal regions such as the GGOs or other anomalies in such images. One of the applications of deep learning is the automatic detection of the GGO regions in the CT images. In this regard, Chen et al. [21] have used a UNET-based architecture (which is called the UNET++ [22] and is a novel method for biomedical image segmentation) to extract the abnormal lung regions in the CT scan images of patients.
Fan et al. [23] presented a deep network, called the Inf-Net, for the automatic segmentation of the infected regions in CT images. They used a partial decoder in order to aggregate the high-level features and to produce a global map. Then, to deal with the shortage of the sufficiently-large and high-quality datasets, they presented a semi-supervised approach and also built their own exclusive dataset with a total of 1700 CT scan images (100 labeled and 1600 unlabeled images). For the first time, and in order to solve the problem of model dependency on the data with semantic labels, Yao et al. [24] proposed an unsupervised method (called the NormNet) that does not need labeled data.
In a research to detect the suspected COVID-19 patients, Shan et al. [25] used 249 CT scan images for training and 300 CT images for validation. Of these 300 samples, 97 samples showed critical conditions, and 7 patients eventually passed away. The model designed in this work is based on the Vnet and is called the Vb-Net. It includes two sections for extracting the image features and also reducing the resolution, increasing the size, and receiving the image information. Also in this model, instead of the standard 5x5x5 filters, which are used in the Vnet model, they employed a sub-network with three 1x1, 3x3 and 1x1 convolutional layer sequence and considerably increased the speed of this network relative to that of the Vnet.
2.1.2. Classification
In all the research works related to the classification of the Corona samples, usually two or three outputs are considered. In the two-output case, we have the presence or the absence of a Corona infection, and in the three-output case, the presence or the absence of the COVID-19 disease or the existence of the other types of respiratory illnesses is determined.
By combining the computer-aided detection (CAD) with the deep learning approach, and by simultaneously using the RGB superposition in the ROI, Ye et al. [26] were able to present a method for the detection of the pulmonary nodules and the GGO regions. They used the architectures of the AlexNet and the GoogleNet to detect the pulmonary nodules and the architecture of the ResNet50 to identify the GGO regions. The DenseNet-201 based deep transfer learning has been employed in [27] for the binary classification of the Corona samples. In this research, the chest CT scan images were classified as the training, testing, and the validating data with accuracies of 99.82%, 96.25% and 97.4%, respectively. In another research, Hall et al. [28] employed the deep learning techniques to classify the COVID-19 images from the chest images of patients. In this respect, by using the method of transfer learning based on the ResNet50 architecture, they were able to classify the chest images with an accuracy of 89.2%. Also, Sethy and Behea [29] combined this architecture (ResNet50) with the Support Vector Machine (SVM) and could achieve a classification accuracy of 95.38%. By presenting a model called “COVIDX-Net” in 2020, Hemdan et al. [30] were able to achieve a binary classification of COVID-19 images with an accuracy of 90%. For training their network, they used 50 CXR images, half of which belonged to the patients infected with COVID-19.
Examples of the binary and the multi-class classifications based on the transfer learning approach and the AlexNet architecture with a total of 371 chest CT scan images can be observed in [31]. It should be mentioned that multi-class classification is abundant in similar research works [32], [33]. By examining these works more closely, another classification criterion and approach that relates to the severity level of a disease can be observed (e.g., mild, medium, critical, and severely-critical). In [34], by examining the chest CT images of 176 Corona patients and by extracting and analyzing 63 features from each image, the conditions of the patients were classified as “severe” and “non-severe”. The random forest algorithm was used in this research, and the classification accuracy and the area under curve (AUC) were obtained as 87.5% and 0.91, respectively.
In another study, Li et al. [35] used the CT scan images of 78 patients and classified the severity of their Corona infections into the three levels of mild (with 30.7%), common (with 59%), and severely-critical (with 10.3%). Narin et al. [36] were able to devise a method based on the deep transfer learning approach for the binary classification of the chest images and produced the four different classes of COVID-19, normal (healthy), viral pneumonia, and bacterial pneumonia. From among the five pre-trained models in this research (ResNet50, ResNet101, ResNet152, InceptionV3, and Inception-ResNetV2), the ResNet50 was able to achieve the highest accuracy for the entire image datasets used. A model based on the Shufflenet v2 was designed by Hu et al. [37] for the classification of CT scan images. The dataset used in this work contained 521 image samples of Corona patients, 397 samples of healthy individuals, 76 samples of a bacterial disease and 48 samples of the SARS disease. This model achieved an average sensitivity of 85.71% and an average specificity of 84.88%.
2.1.3. The hybrid method
In the preceding sections, we explored the use of the classification and the segmentation models in the medical applications. However, most often, a combination of these two methods (a hybrid approach) can be employed to improve the detection accuracy. Actually, in the hybrid method, by relying on the information obtained from the classification and segmentation procedures in each model step, the precision of the model’s next step can be increased. For example, in a research in which a large dataset with the information of the disease severity levels and the pixel levels was compiled, a detection system was designed by combining the classification and the segmentation procedures. In this system, first, the classification procedure is implemented and the suspected COVID-19 cases are determined. Then, by using a segmentation encoder and combining it with a classification decoder, the infected regions in the CT images are specified. Since the designing of a new classification model has not been the goal of this research, the classification has been based on the Res2Net network. Also, this system only needs 22 s for the processing of COVID-19 images and 1 s for the non-COVID-19 images and it ultimately achieves a sensitivity of 95%, specificity of 93% and a Dice score of 78.5% in segmentation [38]. In another work, Gao et al. [39] used 1918 CT scan images of 1202 people and identified 704 patients and 498 healthy individuals. Their method achieved a final accuracy of 96.74%. The proposed model in this research has two blocks. In the first block, the Unet architecture is used to delineate the exact locations of various lung regions. In the second block, the segmentation outcomes are considered in the classification process and ultimately, a precise classification of the input images is achieved.
3. Materials and methods
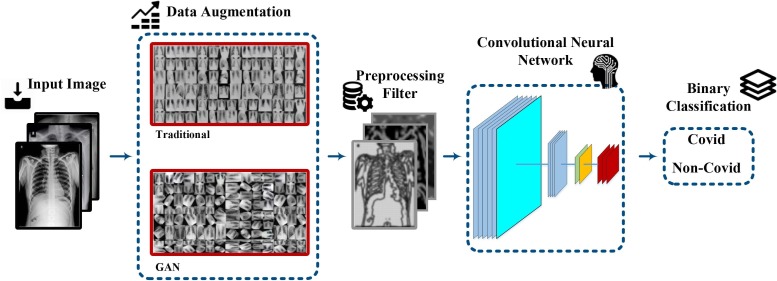
In our proposed detection system, we are going to use the CXR images of the patients. However, due to the unavailability of sufficient datasets for training, we increase the existing datasets via the data augmentation techniques. To this end, the existing data are fed to a pre-processing level consisting of different filter blocks such as the Gabor, LoG and the Sobel filters. In order to compare the network training results, we use the AlexNet, GoogleNet, VGG-19, ShuffleNet V2, DenseNet-121, and the DenseNet-201 architectures. Finally, by defining specific criteria, the classification performance of the proposed model is evaluated. The abovementioned steps and procedures in the processing of CXR images and the detection of COVID-19 lung involvement have been illustrated in Fig. 1 .
Fig. 1.
The steps and procedures of the proposed approach.
3.1. Datasets
In this paper, we have used a total of 4560 CXR images as the input data. 360 of these images are related to the corona patients, and 4200 images belong to the patients with the other types of pulmonary diseases and non-pulmonary illnesses. All these images have been extracted from the following open-source datasets and they have been classified only into the binary classes of COVID-19 and non-COVID-19. These four datasets, which include two types of medical images (CT-Scan and CXR) taken from different angles (mostly, frontal view), belong to more than 64,000 patients with different viral, bacterial, fungal, lipoid aspiration, unknown, and other diseases. It should be mentioned that each of the 4560 images used in this study has been selected under the direct supervision of a physician and that all the factors involved in the patient physiology indexes (e.g., age, gender, geographical region, etc.) have been taken into consideration in image selection [40]. 50% of these images belong to the male patients and 50% to the female patients, and they cover the age groups of 8–85. We have also tried to use the CXR images of the patients from various geographical locations such as China, United States, South Korea, Spain, Saudi Arabia, and others.
-
•
Kaggle: This dataset contains 5863 CXR images in two categories (normal and pneumonia) [41].
-
•
IEEE8023: This dataset covers five different categories (viral, bacterial, fungal, lipoid aspiration, and unknown), with 506, 46, 26, 9 and 59 images, respectively [42].
-
•
CheXpert-v1.0-small: The limited edition of this dataset has a training section and a validation section, which respectively contain 64,540 and 200 front-view CXR images of different patients. We should mention that this dataset does not include the COVID-19 patients and that we have used it simply as the chest images of non-Corona patients [43].
-
•
Radiology assistant: A limited number of patients have been considered in this dataset on a case by case basis. Also, the infection percentages and scores have been reported [44].
3.2. Data augmentation
The supervised machine learning models are severely data-dependent and their performance is based, to a large extent, on the size of the training data available. In many cases, it is difficult to create sufficiently large training datasets. This is also true about the existing data and the images related to the COVID-19 disease. Due to the quick spread of this virus in a short period, unfortunately, there hasn’t been enough time so far to collect sufficient data on this disease on a large scale. Of course, there are some different solutions to this problem, including the data augmentation techniques and the use of the pre-trained networks (i.e., the transfer learning method).
In this section, we focus on the data augmentation process and divide it into the traditional data augmentation techniques and the GANs. We then generate new data on the basis of these two approaches and subsequently use the produced datasets as the input data for the pre-processing block.
3.2.1. Traditional data augmentation
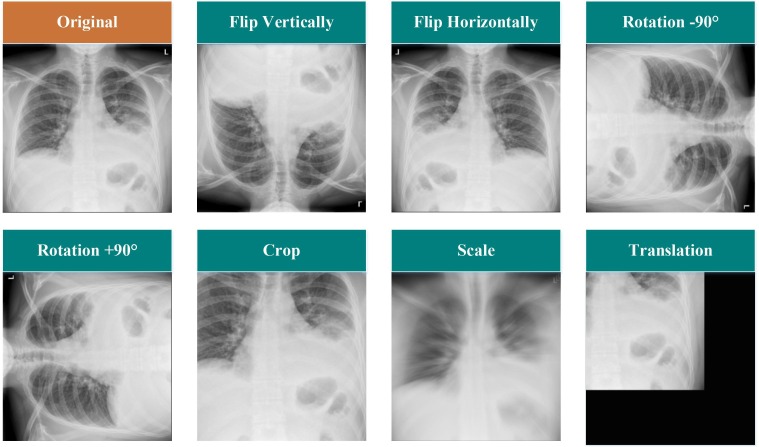
In the field of machine learning, the first and simplest method of increasing the size of an existing dataset is to apply the traditional data augmentation procedures. In this way, new images can be produced by implementing slight changes to the old ones. In this paper, we have applied five data augmentation techniques (horizontal and vertical flips, rotation, scale, translation, and crop) on the CXR images. Several examples of these modifications have been illustrated in Fig. 2 .
Fig. 2.
Sample modifications made to the traditional data augmentation process.
3.2.2. Generative adversarial network (GAN)
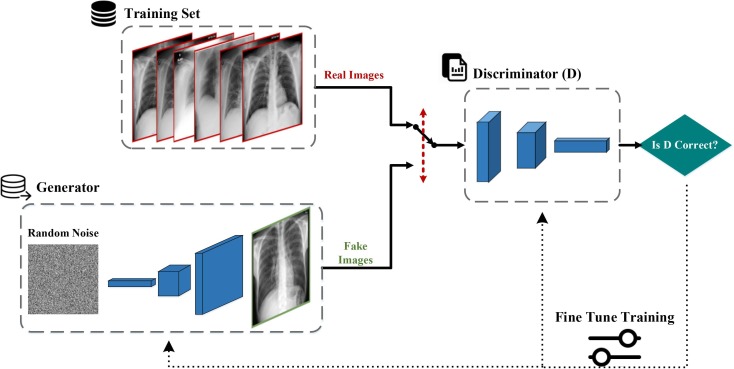
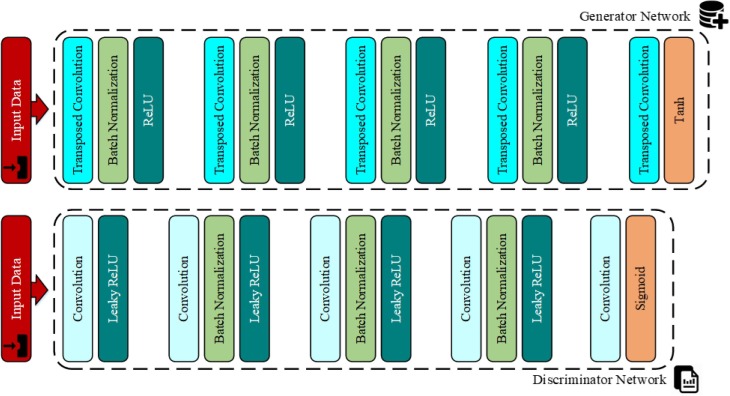
Another technique for producing new data is to use the GANs. The GANs, a group of machine learning systems, were first developed by Ian Goodfellow et al. [45] in 2014. Using this method, we can generate artificial and fake data (sounds and images) which are similar to the input data. This technique involves a game (as it exists in the game theory concept [46]) in which two neural network models (a generator and a discriminator) compete with each other. The duty of the generator model is to produce content (images) and the task of the rival model (discriminator) is to differentiate the real images from the fake ones. At first, the rival artificial intelligence model is able to easily distinguish the fake images from the real ones. However, as time passes, the accuracy and the performance of the generator artificial intelligence model improves so much that it becomes very hard for the rival discriminator model to differentiate between the fake and the real contents. The procedures of this process and the arrangement of the existing layers in each model have been illustrated in Fig. 3, Fig. 4 , respectively. The architectural details and the layers of each of the two models designed in this paper are as follows [47]:
Fig. 3.
The structure of the GANs.
Fig. 4.
The arrangement of the layers in the generator and the discriminator models.
The generator network: This network includes 5 transposed convolutional layers with a window size of 4x4 pixels along with 64 filters, 4 ReLU activation functions, 4 batch normalization layers, and the Tanh activation function.
The discriminator network: This network also consists of 5 transposed convolutional layers with a window size of 4x4 pixels along with 64 filters, 4 Leaky ReLU activation functions, and 3 batch normalization layers and the Sigmoid activation function.
The details of the designed GAN and the specifications of every existing layer and the values of the hyper parameters considered in each of the generator and the discriminator models have been listed in Table 1 . As was mentioned before, with this technique, the data volume can be increased considerably (e.g., up to 20-fold). In this paper, we have used this method exclusively to increase the number of images related to the COVID-19 infections. Fig. 5 shows several examples of the fake images produced by the GANs from the real images containing the COVID-19 infections.
Table 1.
The hyper parameters of the GAN.
| Operation | Kernel | Strides | Feature maps | BN | Activation |
|---|---|---|---|---|---|
| Generator | |||||
| Transposed Convolution | 44 | 11 | 512 | ✓ | ReLU |
| Transposed Convolution | 44 | 22 | 256 | ✓ | ReLU |
| Transposed Convolution | 44 | 22 | 128 | ✓ | ReLU |
| Transposed Convolution | 44 | 22 | 256 | ✓ | ReLU |
| Transposed Convolution | 44 | 22 | 3 | – | Tanh |
| Discriminator | |||||
| Convolution | 44 | 22 | 64 | – | Leaky ReLU |
| Convolution | 44 | 22 | 128 | ✓ | Leaky ReLU |
| Convolution | 44 | 22 | 256 | ✓ | Leaky ReLU |
| Convolution | 44 | 22 | 512 | ✓ | Leaky ReLU |
| Convolution | 44 | 11 | 1 | – | Sigmoid |
| Frame Work | PyTorch | ||||
| Batch size for real data | 128 | ||||
| Batch size for generator | 128 | ||||
| Number of worker | 2 | ||||
| Number of epochs | 400 | ||||
| Optimizer | Adam () | ||||
| Learning rate | 0.0002 | ||||
| Leaky ReLU slope | 0.2 | ||||
| Loss function | Binary Cross Entropy |
Fig. 5.
Examples of the unreal images produced by the GANs.
3.3. Image processing
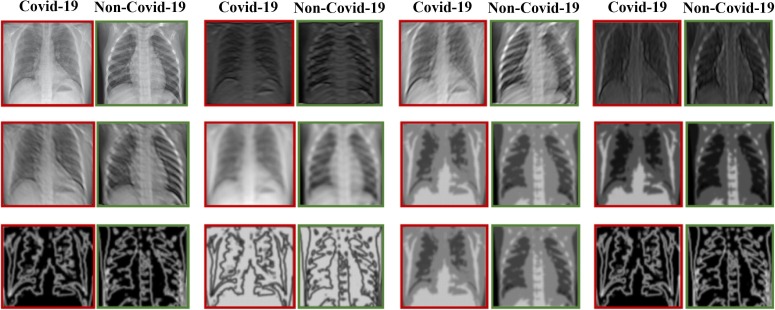
Our main goal in this research is to improve the performance of the classifiers by means of the preprocessing blocks. To this end, after examining the existing filter banks, we have finally selected the Gabor filter and applied it on the data which had been augmented in the preceding steps. We have then considered the outputs of this filter along with the original data (before the application of the Gabor filter) as the new data and used them to train our network. Furthermore, to prove the superiority of this filter over the other filters, we have performed the above steps once again using the LoG and the Sobel filters and compared the outputs of each filter in the Results section.
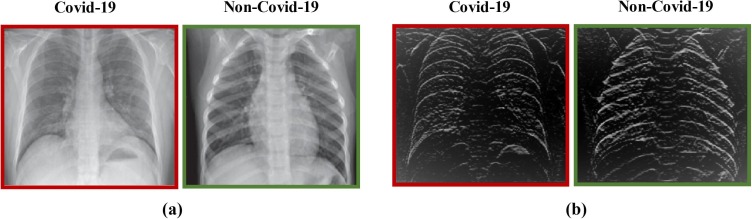
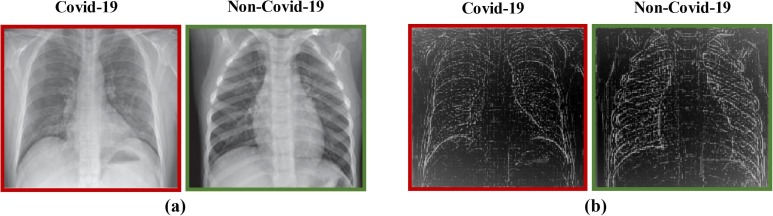
The LoG filter [48] is a common edge finding method based on the second derivative. First, an image is made smooth with a Gaussian filter and then edges are obtained by computing the second derivative (e.g., Laplacian). Conversely, the Sobel filter uses the first derivative to find the edges. In this filter, two convolution kernels and , which are known as the Sobel edge filters, are employed to find the vertical and the horizontal edges of an image, respectively [49], [50]. Some examples of applying the Sobel and LoG filters on the CXR images can be observed in Fig. 6, Fig. 7 , respectively.
Fig. 6.
The outcome of applying the Sobel filter on a data; (a) the original data, (b) the data obtained after applying the Sobel filter.
Fig. 7.
The outcome of applying the LoG filter on a data; (a) the original data, (b) the data after applying the LoG filter.
3.3.1. The Gabor filter
The Gabor filter, named after Denis Gabor, is a type of linear filter which is used for texture analysis in image processing tasks. This filter basically checks to see if a specific frequency exists in an image along a certain orientation at a local zone around a point of interest or a region of analysis [51]. Many contemporary researchers claim that the frequencies and orientations presented by the Gabor filters are similar to those of the human vision system; although this view has not been corroborated by any experimental evidence or practical rationality. Nevertheless, due to the multifaceted differentiating and resolving capabilities of the Gabor transformation function in the position and frequency domains, it has become a very useful tool for extracting and analyzing the texture features and also for detecting the image edges. In an actual texture analysis process, a Gabor filter bank with filters of different orientations and frequencies is normally used to obtain the features of the examined images. The complex form of a two-dimensional Gabor filter in the spatial domain can be expressed as [51]
| (1) |
where
where, λ is the wavelength of the sinusoidal component, θ is the orientation of the normal to the parallel stripes of the Gabor function, ψ is the phase offset, σ is the standard deviation of the Gaussian envelope, and γ is the spatial aspect ratio which specifies the elliptically of the support of the Gabor function.
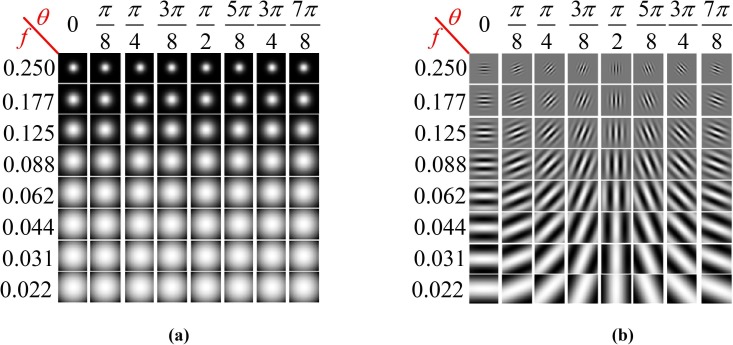
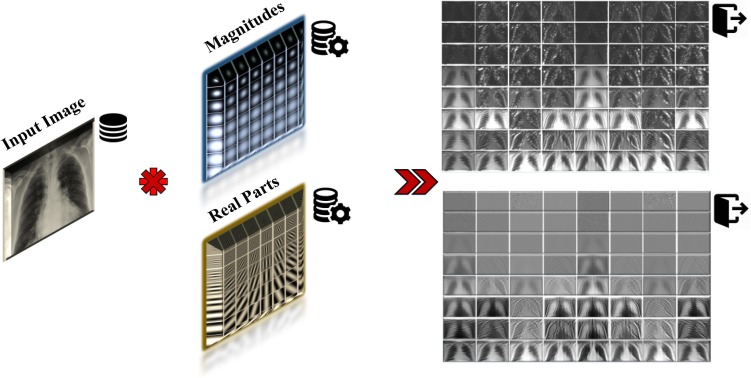
To obtain diverse information with regards to different scales and orientations, we have selected a set of Gabor filters with 8 orientations and 8 scales in order to extract the image features from the existing CXR images. Therefore, the total number of filters used in our experiments is 64. In most cases, it is sufficient to use 8 different orientations to cover the local directions of image features. The values of these parameters have been given in Table 2 . In addition, the real parts and magnitudes of these 64 filters, in 8 different scales and 8 orientations, as well as their convolution with the sample image have been illustrated in Fig. 8 and Fig. 9 , respectively. Fig. 10 shows the output CXR images obtained after applying the Gabor filters.
Table 2.
The values of the Gabor filter parameters.
| Parameters | Values |
|---|---|
| Number of scales | 8 |
| Number of orientations | 8 |
| Wavelength () | |
| Orientation () | |
| Maximum frequency | 0.250 |
| Frequency () | |
| Sigma () | |
| Phase offset () | 0.1 |
| Spatial aspect ratio () | |
| Envelope | Gaussian |
Fig. 8.
The magnitudes (a) and the real parts (b) of the 64 filters used in our experiments in 8 scales and 8 orientations.
Fig. 9.
The convolution of magnitudes and real parts of 64 filters with the sample image.
Fig. 10.
The outcome of applying the Gabor filter.
3.4. Network training
As was mentioned earlier, the use of the transfer learning method is another way of dealing with data scarcity. In applying the deep learning approach to the classification of objects, two methods are generally employed: 1) training from scratch, and 2) transfer learning. For training a deep network from scratch, we need to have a huge set of classified data. Furthermore, in designing a neural network, the deep learning model should be trained by means of these classified data. However, because of the immensity of the existing data and the prolonged training process, this method is used less often. On the other hand, the transfer learning process involves the tuning of a pre-trained model. Hence, we can start with a ready neural network such as the AlexNet or the GoogleNet and tune it with new data. The advantage of the transfer learning technique is that it requires fewer data and, thus, needs less time to train; therefore, it is frequently used in many deep learning programs. Here, we are going to use the DenseNet-121, DenseNet-201, ShuffleNet V2, VGG-19, AlexNet, and the GoogleNet architectures to train the data after applying the intended changes on them. The main network specifications and parameters, including the learning rate, the type of the optimizer, and the batch size, have been listed in Table 3 .
Table 3.
The network specifications.
| Parameters | Type / Value |
|---|---|
| Framework | PyTorch |
| Learning rate | 0.001 |
| Learning rate decay interval | 30 |
| Epochs | 45 |
| Batch size | 32 |
| Regularization | Dropout |
| Optimizer | Adam |
| Loss function | Cross entropy |
| LR Scheduler | StepLR |
The network training process in this section can be divided into five parts:
Step 1 (changing the size of all the collected images): In this step, all the images of different sizes are normalized and changed to a single correct size.
Step 2 (creating two main classes): In this step, after applying the data augmentation techniques and passing through the pre-processing block, all the existing data in the dataset are divided into just two main classes: healthy (non-COVID-19) and COVID-19. We should mention that in this case, all the viral and bacterial pneumonia data will be grouped in the healthy category.
Step 3 (dividing the data): In this step, the existing data (CXR images) are divided into two portions; 60% of the data (4800 CXRs) are used for training and 40% (3200 CXRs) for testing and validation.
Step 4 (creating the main models): In this step, the CNNs with the DenseNet-121, DenseNet-201, ShuffleNet V2, VGG-19, AlexNet, and the GoogleNet architectures are created for training the available data.
Step 5 (training the data): In this step, by considering appropriate weights, 4800 CXR images are trained by the created CNNs. This step is a kind of backtrack to Step 4, and it will be repeated as necessary. In the current work, 45 training epochs have been considered.
Before presenting the obtained results and evaluating our approach, we outline the complete steps of the proposed CNN in Algorithm (1).
4. The experimental results
Considering the type of dataset images used, there are two classes in this scheme: unhealthy (COVID-19) and healthy (non-COVID-19). At the end, the output of the model will specify the type of class or disease. The presented model has been coded in the Python environment and with the NVIDIA RTX 2060 Super GPU. We have used 25% of the whole generated images for testing and 15% for validation. The validation dataset is usually used to check and monitor the quality of the neural network model being trained and to determine the learning termination condition for the training process; while the test dataset is independently used to determine the ultimate quality of the trained network in terms of precision and the generalization capabilities of the main system.
Algorithm (1). —
The introduced algorithm
| Algorithm 1: Pseudo Code of Proposed Method | ||
|---|---|---|
| Require: number of covid-19 images n; number of healthy images m; number of GAN epochs p; transfer model T= {AlexNet, GoogleNet, VGG-19, Shufflenet V2, DenseNet-121, DenseNet-201}; number of transfer epochs q. | ||
| Input: Chest X-ray Images | ||
| Output: Binary Classifier | ||
| 1 | First Step // Preparing Data | |
| 2 | Split I into two subfolders (), where and represent covid-19 and non-covid-19 folders, respectively. | |
| 3 | Split data directory into training, validation, and test sets | |
| 4 | Second Step // Data Augmentation | |
| 5 | fordo // Traditional Augmentation | |
| 6 | Rotate () | |
| 7 | Crop () | |
| 8 | Flip () | |
| 9 | Scale () | |
| 10 | Translate () | |
| 11 | end for | |
| 12 | ||
| 13 | fordo // Generative adversarial networks | |
| 14 | Update discriminator network | |
| 15 | Train with real batch | |
| 16 | Calculate loss on real batch | |
| 17 | Calculate gradients for discriminator | |
| 18 | Train with fake batch | |
| 19 | Generate fake image batch with generator | |
| 20 | Classify all fake batch with discriminator | |
| 21 | Calculate discriminator’s loss on the fake batch | |
| 22 | Update and Calculate gradients for generator | |
| 23 | end for | |
| 24 | Third Step // Applying Block Filter | |
| 25 | fordo | |
| 26 | Convert from RGB to Gray scale | |
| 27 | imfilter ( , fspecial ('LoG')) // apply Laplacian of Gaussian filter | |
| 28 | imfilter ( , fspecial ('Sobel')) // apply Sobel filter | |
| 29 | globalgaborfeatures (, gaborfilters, Output); // apply Gabor filter | |
| 30 | end for | |
| 31 | Fourth Step // Training Model | |
| 32 | Loading all X-ray training data | |
| 33 | Resize Images | |
| 34 | for epochs = 1 to qdo | |
| 35 | Train the AlexNet model | |
| 36 | Train the GoogleNet model | |
| 37 | Train the VGG-19 model | |
| 38 | Train the ShuffleNet V2 model | |
| 39 | Train the DenseNet-121 model | |
| 40 | Train the DenseNet-201 model | |
| 41 | end for | |
| 42 | Test and evaluate model | |
| 43 | Compare results | |
4.1. Validation and testing accuracy
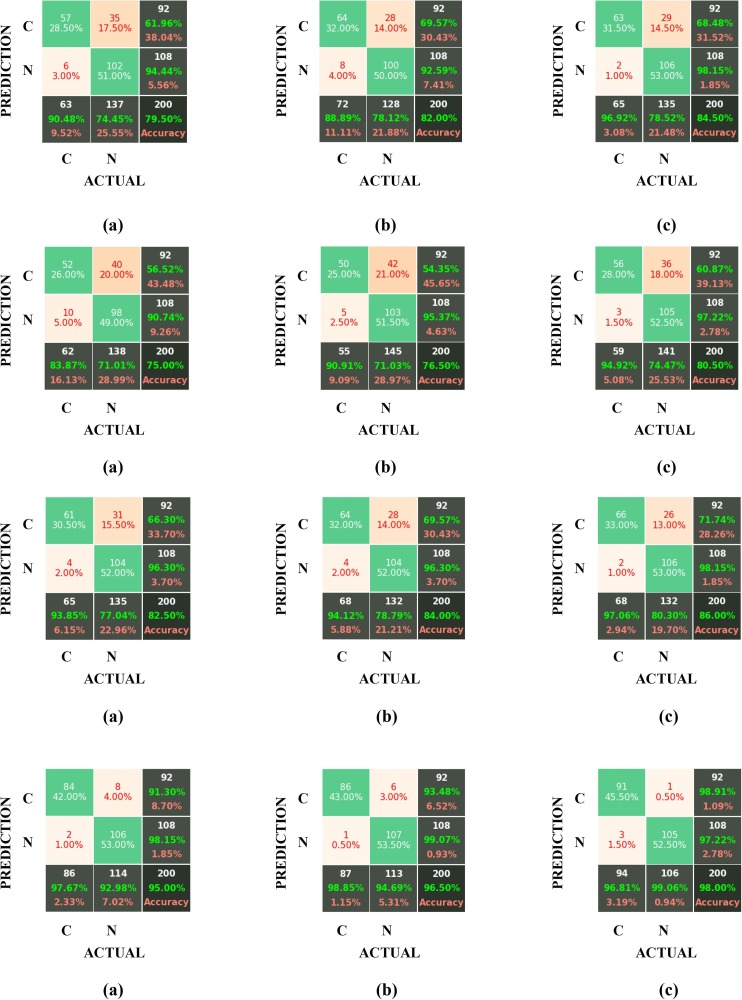
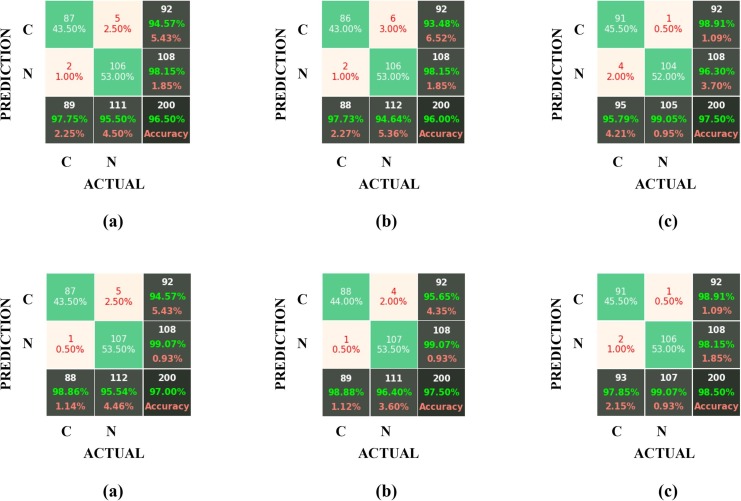
We can always use a Confusion Matrix for validating and evaluating the network model and obtaining its parameters (accuracy, sensitivity, specificity, etc.). This matrix provides the needed information regarding the classification accuracies of the two considered classes (COVID-19 and non-COVID-19) in the “real” and “predicted” categories. In this section, to confirm the superiority of the Gabor filter over the Sobel and the LoG filters, the results obtained by applying these three filters on the datasets developed by GAN and by traditional data augmentation methods have been compared in the form of confusion matrices for six different architectures (AlexNet, GoogleNet, VGG-19, ShuffleNet V2, DenseNet-121 and DenseNet-201). For checking the test accuracies, the above confusion matrices have been displayed in Fig. 11 , where the labels “C” and “N” represent the COVID-19 and non-COVID-19 (healthy) cases, respectively.
Fig. 11.
The confusion matrixes for the test accuracy of the AlexNet, GoogleNet, VGG-19, ShuffleNet V2, DenseNet-121 and DenseNet-201 architectures (from top to down) for (a) Sobel, (b) LoG, and (c) the Gabor filter block.
Also, the results of each model for the two cases of “test set” and “validation set” have been listed separately in Table 4 .
Table 4.
The confusion table in the proposed approach for different filter blocks.
| Filter | Accuracy | AlexNet | GoogleNet | VGG-19 | ShuffleNet V2 | DenseNet-121 | DenseNet-201 |
|---|---|---|---|---|---|---|---|
| Sobel | Validation Set | 81.00 | 78.25 | 87.00 | 96.80 | 97.50 | 98.50 |
| Test Set | 79.50 | 75.00 | 82.50 | 95.00 | 96.50 | 97.00 | |
| LoG | Validation Set | 86.50 | 79.80 | 86.50 | 97.25 | 97.20 | 98.75 |
| Test Set | 82.00 | 76.50 | 84.00 | 96.50 | 96.00 | 97.50 | |
| Gabor | Validation Set | 86.75 | 81.50 | 88.25 | 99.00 | 98.00 | 99.25 |
| Test Set | 84.50 | 80.50 | 86.00 | 98.00 | 97.50 | 98.50 |
4.2. Performance evaluation
Although the criterion of “Accuracy” is the most common, most fundamental, and the simplest measure of a classifier’s performance quality, its main drawback is that it does not distinguish between the “False Negative” and “False Positive” errors. In fact, this criterion considers all the errors to be the same. Therefore, in this section, we introduce the other performance evaluation measures and compare them. The results have been shown in Table 5 .
-
•
The Precision criterion shows the ratio of the number of correctly classified cases from a specific class to the total number of classified cases in that particular class (whether classified correctly or falsely).
| (2) |
-
•
The Recall or Sensitivity criterion, which is also called the “True Positive Rate”, is the ratio of the positive cases which have been correctly diagnosed as positive. This measure actually shows to what extent the classifier has succeeded in detecting all the infected individuals. Therefore, the number of the healthy individuals that have been falsely diagnosed as patients by the classifier will have no effect on the calculation of this parameter. Thus,
| (3) |
-
•
The Specificity criterion, which is also called the “True Negative Rate”, is the opposite measure to the Sensitivity parameter and it is used in the cases in which the accuracy of the negative detections is important. This parameter indicates the ratio of the negative cases which have been correctly diagnosed as negative.
| (4) |
-
•
The F1-Score criterion achieves a balance between the Accuracy and the Precision criteria and it is used in the cases in which the False Positive and False Negative detections have different costs. This measure is expressed as
| (5) |
-
•
The Matthews correlation coefficient (MCC) indicates the classification quality for a binary set. The expected values for this coefficient are in the range of –1 to + 1. This parameter can be calculated as follows:
| (6) |
Table 5.
Results obtained after applying the proposed scheme.
| Architecture | Filter | Accuracy | Precision | Recall | Specificity | F1-Score | MCC |
|---|---|---|---|---|---|---|---|
| AlexNet | Sobel | 79.50 | 61.96 | 90.48 | 74.45 | 73.55 | 60.51 |
| LoG | 82.00 | 69.57 | 88.89 | 78.12 | 78.05 | 64.54 | |
| Gabor | 84.50 | 68.48 | 96.92 | 78.52 | 80.25 | 70.90 | |
| GoogleNet | Sobel | 75.00 | 56.52 | 83.87 | 71.01 | 67.53 | 50.93 |
| LoG | 76.50 | 54.35 | 90.91 | 71.03 | 68.03 | 55.50 | |
| Gabor | 80.50 | 60.87 | 94.92 | 74.47 | 74.17 | 63.49 | |
| VGG-19 | Sobel | 82.50 | 66.30 | 93.85 | 77.04 | 77.71 | 66.61 |
| LoG | 84.00 | 69.57 | 94.12 | 78.79 | 80.00 | 69.29 | |
| Gabor | 86.00 | 71.74 | 97.06 | 80.30 | 82.50 | 73.53 | |
| ShuffleNet | Sobel | 95.00 | 91.30 | 97.67 | 92.98 | 94.38 | 90.05 |
| LoG | 96.50 | 93.48 | 98.85 | 94.69 | 96.09 | 93.05 | |
| Gabor | 98.00 | 98.91 | 96.81 | 99.06 | 97.85 | 96.00 | |
| DenseNet-121 | Sobel | 96.50 | 94.57 | 97.75 | 95.50 | 96.13 | 92.98 |
| LoG | 96.00 | 93.48 | 97.73 | 94.64 | 95.56 | 92.00 | |
| Gabor | 97.50 | 98.91 | 95.79 | 99.05 | 97.33 | 95.02 | |
| DenseNet-201 | Sobel | 97.00 | 94.57 | 98.86 | 95.54 | 96.67 | 94.02 |
| LoG | 97.50 | 95.65 | 98.88 | 96.40 | 97.24 | 95.00 | |
| Gabor | 98.50 | 98.91 | 97.85 | 99.07 | 98.38 | 96.99 |
* The bold numbers indicate the maximum values in the considered architectures.
The GANs have also been employed in earlier works [52] to generate new training images in the classification-related tasks; and the efficacy of the GANs, as a novel data augmentation approach, has already been confirmed. So in order to investigate the effect of GAN on the proposed method, we trained the mentioned architectures (AlexNet, GoogleNet, VGG-19, ShuffleNet V2, DenseNet-121 and DenseNet-201) once again with and without the augmented input images and compared the outcomes. The baseline (without data augmentation and filter banks) and GAN-based results with the AUC values have been presented in Table 6, Table 7 , respectively. The comparison of metrics in achieved classifier have been summarized in Table 8 and Fig. 12 . As the results in Table 7, Table 8 indicate, the method of data augmentation with GAN has been able to respond to the severe need of the networks for more data and, thus, solve the problem of overfitting on the training data sets and improve the network performance. For example, in the baseline case, and in the AlexNet architecture, the false discovery rate and the negative predictive value are equal to 1 and the value of AUC is 0.5; this means that the classifier cannot distinguish between the examined classes. However, after applying the method of data augmentation with GAN, the model’s performance has improved considerably and an AUC value of 0.89 has been achieved.
Table 6.
Evolution metrics in baseline approach.
| Architecture | AUC |
% Evolution metrics |
|||||
|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | Specificity | F1-Score | MCC | ||
| AlexNet | 0.50 | 54.00 | 0.00 | – | 54.04 | 0.00 | – |
| GoogleNet | 0.63 | 54.00 | 1.09 | 50.00 | 54.04 | 2.13 | 0.81 |
| VGG-19 | 0.65 | 54.00 | 2.17 | 50.00 | 54.08 | 4.17 | 1.15 |
| ShuffleNet | 0.85 | 72.00 | 40.22 | 97.37 | 66.05 | 56.92 | 49.92 |
| DenseNet-121 | 0.86 | 77.00 | 58.70 | 87.10 | 72.46 | 70.13 | 55.27 |
| DenseNet-201 | 0.89 | 77.00 | 96.00 | 52.17 | 98.15 | 67.61 | 57.92 |
Table 7.
Evolution metrics in data augmentation approach.
| Architecture | AUC |
% Evolution metrics |
|||||
|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | Specificity | F1-Score | MCC | ||
| AlexNet | 0.89 | 78.50 | 58.70 | 91.53 | 73.05 | 71.52 | 59.09 |
| GoogleNet | 0.78 | 72.00 | 53.26 | 79.03 | 68.84 | 63.64 | 44.42 |
| VGG-19 | 0.92 | 82.00 | 66.30 | 92.42 | 76.87 | 77.22 | 65.37 |
| ShuffleNet | 0.98 | 96.00 | 93.48 | 97.73 | 94.46 | 95.56 | 92.00 |
| DenseNet-121 | 0.98 | 96.00 | 98.84 | 92.39 | 99.07 | 95.51 | 92.08 |
| DenseNet-201 | 0.99 | 97.50 | 100.00 | 94.57 | 100.00 | 97.21 | 95.07 |
Table 8.
Evaluation metrics improvements (%).
 |
Fig. 12.
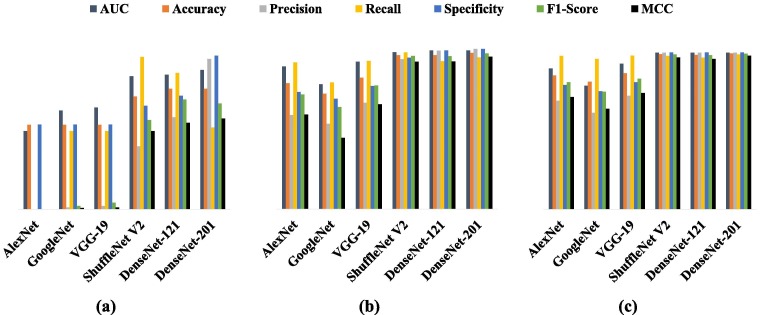
Comparing the classification metrics of the proposed algorithm in 3 cases: (a) baseline (b) with GAN (c) GAN with Gabor filter.
According to the results reported in Table 5, the highest values in the defined metrics, and for all the considered architectures, belong to the Gabor filter; which proves the superiority of this filter over the other two filters. The superiority of the proposed method is based on two facts: 1) the application of the Gabor filter banks at different orientations and scales detects different and distinct information from the input images, and by combining these data, we can create much better images and images with more prominent features and details than the original ones, 2) combining the responses from each of the 64 types of filters with the original images leads to a significant increase in the number of images needed for network training; and this can be considered as a type of data augmentation technique. We also compared the results of these three filters before and after applying them on the datasets and reported their differences in Table 8. For example, it was shown that the application of the Gabor filter bank on a dataset and the training of the GoogleNet model with these data increase the measures of classification accuracy, precision, and recall by 26.5, 59.78 and 44.92%, respectively, relative to the case in which this filter has not been applied. While the application of the other filters, such as Sobel, results in a negligible increase in the values of some parameters, and in some cases, even reduces the values of these parameters (e.g., the measure of Specificity in the DenseNet-201 model). Accordingly, instead of using deeper networks or collecting more data, the algorithm presented in this paper can be employed to significantly enhance the performance of any classification model without altering its structure and layers and with just a small number of data. Not to mention that this technique can also be applied to multiclass classifiers.
On the other hand, it is observed that the proposed scheme achieves its best performance in the DenseNet-201 architecture than in the other models; so the DenseNet-201 can be selected as the main architecture in this approach. Therefore, the application of the algorithm presented in this paper on the DenseNet-201 architecture can be introduced as a state-of-the-art deep learning method for improving the performance of image classifiers. In conclusion, we have compared the results of our model (with the DenseNet-201 architecture) with the approaches outlined in 10 of the most recent research works [53], [54], [55] on Corona and have presented these findings in Table 9 . It should be mentioned that the compared models in this table contain various images from different data sources; while our proposed model has been trained with a combination of images from different data sources.
Table 9.
Comparing the accuracies of the proposed algorithm and the existing models.
| Models | Accuracy (%) | Architecture | Imaging Type | Number of Images(Covid-19, Non-Covid-19) |
|---|---|---|---|---|
| Jaiswal et al. [27] | 96.25 | DenseNet-201 | CT | (1262, 1230) |
| Hall et al. [28] | 89.2 | ResNet-50 | CXR | (135, 320) |
| Sethy and Behr [29] | 95.38 | ResNet-50 + SVM | CXR | (25, 25) |
| Hemdan et al. [30] | 90.0 | COVIDX-Net | CXR | (25, 25) |
| Xu et al. [32] | 86.7 | ResNet + Location Attention | CT | (219, 224) |
| Apostolopoulos and Mpesiana. [33] | 96.78 | MobileNet V2 | CXR | (224, 700) |
| Hu et al. [37] | 91.21 | ShuffleNet V2 | CT | (521, 397) |
| Zheng et al. [53] | 90.8 | DeCoVNet | CT | (313, 229) |
| Ozturk et al. [54] | 98.08 | DarkCovidNet | CXR | (125, 500) |
| Wang et al. [55] | 82.9 | M−Inception | CT | (195, 258) |
| Our method | 98.50 | DenseNet-201 | CXR | (360, 4200) |
5. Conclusion
In the medical world of today, the data associated with the patients and the symptoms of various diseases are very extensive and complex; and sometimes it is impossible for a single specialist to consider and analyze all the factors involved in a particular disease. Also, the prolonged work periods and the fatigue, stress and the mental pressures endured by the medical and the laboratory personnel may have adverse and sometimes detrimental effects on the lab results. Therefore, it is essential to have an intelligent system that can help with the prediction and diagnosis of various illnesses.
In this paper, we employed a deep CNN to analyze the CXR images of patients’ lungs and presented a novel approach for improving the performance of classifying these images into the healthy (non-COVID-19) and unhealthy (COVID-19) categories. We demonstrated that the performance of the classifiers can be improved significantly by combining the convolutional deep learning techniques with the different filter banks such as the Gabor and the LoG filter. Also, another challenge in the research works on the COVID-19 disease is the data scarcity and, specifically, the shortage of adequate CXRs needed for training a network; which can adversely affect the network’s performance. To deal with this shortcoming, we generated new data by employing the traditional data augmentation techniques along with the GANs. By defining and applying several performance evaluation criteria, we showed that, depending on the type of the network architecture used, our proposed technique is able to increase the measures of accuracy, precision, recall, specificity, F1-Score, and the MCC by up to 32, 69.57, 96.92, 26.59, 80.25 and 72.38%, respectively.
In the forthcoming works, we want to study the effect of using the filters based on the deep learning techniques such as the style transfer in the more precise detection of the COVID-19 infections and in the segmentation of the infected regions in the CT and CXR images of patients and also to enhance the model accuracy by employing the attention module in the network structure.
CRediT authorship contribution statement
Amir Hossein Barshooi: Investigation, Software, Data curation, Writing – original draft, Formal analysis, Validation. Abdollah Amirkhani: Conceptualization, Software, Methodology, Writing – original draft, Formal analysis, Validation, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q.i., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q.i., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G.e., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B.o., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox LeAnne, Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. novel coronavirus in the United States”. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.’worldometer’, COVID-19 CORONAVIRUS / CASES; June, 2021, Available at: https://www.worldometers.info/coronavirus/.
- 5.M.A. Jahangir, A. Muheem, M.F. Rizvi, Coronavirus (COVID-19): history, current knowledge and pipeline medications, Int. J. Pharm. Pharmacol. 2020; 4 140. doi: 10.31531/2581, vol. 3080, no. 2, 2020.
- 6.X. Jiang, M. Coffee, A. Bari, J. Wang, X. Jiang, et al., Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity, Comput. Mater. Contin., vol. 63, no. 1, pp. 537–551, 2020.
- 7.Guarner J. Oxford University Press US; 2020. Three emerging coronaviruses in two decades: the story of SARS, MERS, and now COVID-19. [Google Scholar]
- 8.Loeffelholz M.J., Tang Y.-W. Laboratory diagnosis of emerging human coronavirus infections–the state of the art. Emerg. Microbes Infect. 2020;9(1):747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., et al. Remdesivir for the treatment of Covid-19—preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2022236. [DOI] [PubMed] [Google Scholar]
- 10.A.B. Abdulkareem, N.S. Sani, M.A. Awadallah, et al., Predicting COVID-19 based on environmental factors with machine learning, Intell. Autom. SOFT Comput., vol. 28, no. 2, pp. 305–320, 2021.
- 11.Jothi N., Rashid N.A., Husain W. Data mining in healthcare–a review. Procedia Comput. Sci. 2015;72:306–313. [Google Scholar]
- 12.Maroco J., Silva D., Rodrigues A., Guerreiro M., Santana I., de Mendonça A. Data mining methods in the prediction of Dementia: A real-data comparison of the accuracy, sensitivity and specificity of linear discriminant analysis, logistic regression, neural networks, support vector machines, classification trees and random forests. BMC Res. Notes. 2011;4(1):1–14. doi: 10.1186/1756-0500-4-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotsiantis S.B., Zaharakis I., Pintelas P. Supervised machine learning: A review of classification techniques. Emerg. Artif. Intell. Appl. Comput. Eng. 2007;160(1):3–24. [Google Scholar]
- 14.Pal N.R., Pal S.K. A review on image segmentation techniques. Pattern Recognit. 1993;26(9):1277–1294. [Google Scholar]
- 15.Coltuc D., Bolon P., Chassery J.-M. Exact histogram specification. IEEE Trans. Image Process. 2006;15(5):1143–1152. doi: 10.1109/tip.2005.864170. [DOI] [PubMed] [Google Scholar]
- 16.Dhanachandra N., Manglem K., Chanu Y.J. Image segmentation using K-means clustering algorithm and subtractive clustering algorithm. Procedia Comput. Sci. 2015;54:764–771. [Google Scholar]
- 17.Al-Betar M.A., Alyasseri Z.A.A., Awadallah M.A., Doush I.A. Coronavirus herd immunity optimizer (CHIO) Neural Comput. Appl. 2021;33(10):5011–5042. doi: 10.1007/s00521-020-05296-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen D., Wu G., Suk H.-I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017;19(1):221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J.-G., Jun S., Cho Y.-W., Lee H., Kim G.B., Seo J.B., Kim N. Deep learning in medical imaging: general overview. Korean J. Radiol. 2017;18(4):570. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki K. Overview of deep learning in medical imaging. Radiol. Phys. Technol. 2017;10(3):257–273. doi: 10.1007/s12194-017-0406-5. [DOI] [PubMed] [Google Scholar]
- 21.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Chen Q., Huang S., Yang M., Yang X., Hu S., Wang Y., Hu X., Zheng B., Zhang K., Wu H., Dong Z., Xu Y., Zhu Y., Chen X.i., Zhang M., Yu L., Cheng F., Yu H. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Z. Zhou, M.M.R. Siddiquee, N. Tajbakhsh, J. Liang, Unet++: A nested u-net architecture for medical image segmentation, in Deep learning in medical image analysis and multimodal learning for clinical decision support, Springer, 2018, pp. 3–11. [DOI] [PMC free article] [PubMed]
- 23.Fan D.P., Zhou T., Ji G.P., Zhou Y., Chen G., et al. Inf-net: Automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 24.Yao Q., Xiao L.i., Liu P., Zhou S.K. Label-free segmentation of covid-19 lesions in lung ct. IEEE Trans. Med. Imaging. 2021;40(10):2808–2819. doi: 10.1109/TMI.2021.3066161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shan F., Yaozong G., Jun W., Weiva S., Nannan S., et al. Abnormal lung quantification in chest CT images of COVID-19 patients with deep learning and its application to severity prediction. Med. Phys. 2021;48(4):1633–1645. doi: 10.1002/mp.14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye W., Gu W., Guo X., Yi P., Meng Y., Han F., Yu L., Chen Y.i., Zhang G., Wang X. Detection of pulmonary ground-glass opacity based on deep learning computer artificial intelligence. Biomed. Eng. Online. 2019;18(1) doi: 10.1186/s12938-019-0627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2021;39(15):5682–5689. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 28.L.O. Hall, R. Paul, D.B. Goldgof, G.M. Goldgof, Finding covid-19 from chest x-rays using deep learning on a small dataset, 2020, arXiv:2004.02060.
- 29.P.K. Sethy, S.K. Behera, Detection of coronavirus disease (COVID-19) based on deep features. Preprints; 2020. doi: 1020944/preprints2020 03030 0v1 . 2020 03030 0.
- 30.E.E.D. Hemdan, M.A. Shouman, M.E. Karar, COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in x-ray images, arXiv preprint arXiv: 200311055, 2020.
- 31.Ibrahim A.U., Ozsoz M., Serte S., Al-Turjman F., Yakoi P.S. Pneumonia classification using deep learning from chest X-ray images during COVID-19. Cognit. Comput. 2021:1–13. doi: 10.1007/s12559-020-09787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X., Jiang X., Ma C., et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Z. Tang, W. Zhao, X. Xie, Z. Zhong, F. Shi, et al., Severity assessment of coronavirus disease 2019 (COVID-19) using quantitative features from chest CT images, arXiv Prepr. arXiv2003.11988, 2020.
- 35.Li K., Fang Y., Li W., Pan C., Qin P., et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19) Eur. Radiol. 2020;30(8):4407–4416. doi: 10.1007/s00330-020-06817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021;24(3):1207–1220. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R. Hu, G. Ruan, S. Xiang, M. Huang, Q. Liang, J. Li, Automated diagnosis of covid-19 using deep learning and data augmentation on chest ct, medRxiv, 2020.
- 38.Wu Y.H., Gao S.H., Mei J., Xu J., Fan D.P., et al. Jcs: An explainable covid-19 diagnosis system by joint classification and segmentation. IEEE Trans. Image Process. 2021;30:3113–3126. doi: 10.1109/TIP.2021.3058783. [DOI] [PubMed] [Google Scholar]
- 39.Gao K., Su J., Jiang Z., Zeng L.L., Feng Z., et al. Dual-branch combination network (DCN): Towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Med. Image Anal. 2021;67 doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Z.A.A. Alyasseri, M.A. Al‐Betar, I.A. Doush, et al., Review on COVID‐19 diagnosis models based on machine learning and deep learning approaches, Expert Syst., p. e12759, 2021. doi: 10.1111/exsy.12759. [DOI] [PMC free article] [PubMed]
- 41.https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia.
- 42.J.P. Cohen, P. Morrison, L. Dao, K. Roth, T.Q. Duong, M. Ghassemi, Covid-19 image data collection: Prospective predictions are the future, arXiv Prepr. arXiv2006.11988, 2020.
- 43.Irvin J., Rajpurkar P., Ko M., Yu Y., Ciurea-Ilcus S., Chute C., Marklund H., Haghgoo B., Ball R., Shpanskaya K., Seekins J., Mong D.A., Halabi S.S., Sandberg J.K., Jones R., Larson D.B., Langlotz C.P., Patel B.N., Lungren M.P., Ng A.Y. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. Proceedings of the AAAI Conference on Artificial Intelligence. 2019;33:590–597. [Google Scholar]
- 44.https://radiologyassistant.nl/chest/covid-19/covid19-imaging-findings.
- 45.Goodfellow I.J., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., et al. Generative adversarial networks. Proc. Advances Neural Inf. Process. Syst. 2014:2672–2680. [Google Scholar]
- 46.Myerson R.B. Harvard University Press; 2013. Game theory. [Google Scholar]
- 47.Loey M., Smarandache F., Khalifa N.E.M. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry (Basel) 2020;12(4):651. [Google Scholar]
- 48.Marr D., Hildreth E. Theory of edge detection. Proc. R. Soc. London. Ser. B. Biol. Sci. 1980;207(1167):187–217. doi: 10.1098/rspb.1980.0020. [DOI] [PubMed] [Google Scholar]
- 49.Kanopoulos N., Vasanthavada N., Baker R.L. Design of an image edge detection filter using the Sobel operator. IEEE J. Solid-State Circuits. 1988;23(2):358–367. [Google Scholar]
- 50.Uçar M., Uçar E. Computer-aided detection of lung nodules in chest X-rays using deep convolutional neural networks. Sakarya University J. Comput. Inf. Sci. 2019;2(1):41–52. [Google Scholar]
- 51.Gabor D. Theory of communication. Part 1: The analysis of information. J. Inst. Electr. Eng. III Radio Commun. Eng. 1946;93(26):429–441. [Google Scholar]
- 52.A. Antoniou, A. Storkey, H. Edwards, Data augmentation generative adversarial networks, arXiv Prepr. arXiv1711.04340, 2017.
- 53.C. Zheng, X. Deng, Q. Fu, Q. Zhou, et al., Deep learning-based detection for COVID-19 from chest CT using weak label, medRxiv, 2020 .
- 54.Ozturk T., Talo M., Yildirim E.A., et al. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S., Kang B.o., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X., Xu B.o. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19) Eur. Radiol. 2021;31(8):6096–6104. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]