ABSTRACT
Adult stem cells are critical for the maintenance of tissue homeostasis. However, how the proliferation and differentiation of intestinal stem cells (ISCs) are regulated remains not fully understood. Here, we find a mutant, stum 9-3, affecting the proliferation and differentiation of Drosophila adult ISCs in a forward genetic screen for factors regulating the proliferation and differentiation ISCs. stum 9-3 acts through the conserved Notch signaling pathway, upstream of the S2 cleavage of the Notch receptor. Interestingly, the phenotype of stum 9-3 mutant is not caused by disruption of stumble (stum), where the p-element is inserted. Detailed mapping, rescue experiments and mutant characterization show that stum 9-3 is a new allele of O-fucosyltransferase 1 (O-fut1). Our results indicate that unexpected mutants with interesting phenotype could be recovered in forward genetic screens using known p-element insertion stocks.
KEY WORDS: O-fut1, Intestinal stem cell, Drosophila, Notch signaling
Summary: A mutant, stum 9-3, affecting the proliferation and differentiation of Drosophila adult intestinal stem cells (ISCs) was identified in a forward genetic screen for factors regulating the proliferation and differentiation ISCs. stum 9-3 acts through the Notch signaling pathway. Detailed mapping, rescue experiments and characterization show that stum 9-3 is not a stumble mutant where the p-element is inserted, but a new allele of O-fucosyltransferase 1 (O-fut1).
INTRODUCTION
Stem cells are responsible for maintaining the homeostasis of adult tissues, where the frequently lost cells are constantly replenished by stem cell progeny. The proliferation and differentiation of adult stem cells must be tightly controlled. Disruption of this balance will lead to stem cell accumulation or stem cell depletion, eventually resulting in various diseases, such as cancer and aging (Jasper, 2020; Lin, 2008; Morrison and Spradling, 2008; Radtke and Clevers, 2005; Singh et al., 2019). Therefore, understanding of the mechanisms controlling stem cell proliferation and differentiation will provide insight into the development of therapeutics to treat human diseases.
The posterior midgut of the adult Drosophila intestine has proven to be an excellent system to study the regulation of stem cell proliferation and differentiation. Drosophila intestines show marked similarities with their mammalian counterparts in terms of cellular make-up, development and genetic control (Banerjee et al., 2019; Casali and Batlle, 2009; Gervais and Bardin, 2017; Jasper, 2020; Stainier, 2005; Wang and Hou, 2010). ISCs are distributed along the basement membrane of the Drosophila adult midgut epithelium (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Initial studies proposed that ISCs constantly undergo asymmetric divisions and produce non-dividing daughter cells, enteroblasts (EBs) (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006, 2007). Delta (Dl), one of the Notch (N) ligands, is specifically expressed in ISCs, while Notch receptor is expressed in both ISCs and EBs. ISCs signal via Dl to activate Notch signaling in EBs, which terminally differentiate into either absorptive enterocytes (ECs) or secretory enteroendocrine cells (EE) depending on their signaling environments (Beebe et al., 2010; Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006; Perdigoto et al., 2011; Yeung et al., 2011). Recent studies show that a significant proportion of ISCs divides symmetrically in response to differentiation and subsequent loss of a neighboring ISC (or vice versa) (de Navascués et al., 2012; Goulas et al., 2012; O'Brien et al., 2011). Moreover, EE cells may not be generated from EBs, but directly from ISCs or EE progenitor cells (EEPs) (Biteau and Jasper, 2014; Chen et al., 2018; Zeng et al., 2015). Interestingly, unlike other systems in which differentiated cells could be de-differentiated into stem cells, no new ISCs could be re-generated after all progenitors were ablated, indicating that fully differentiated intestinal cells could not de-differentiate into ISCs in the absence of any progenitors (Brawley and Matunis, 2004; Lu and Li, 2015a; Raff, 2003).
Previous studies have shown that the proliferation and differentiation of ISCs under physiological and stressed conditions are regulated by many signaling pathways including the Notch, Wingless (Wg), Janus Kinase/Signal Transducer and Activator of Transcription (JAK/STAT), Epidermal Growth Factor Receptor (EGFR), Hippo (Hpo), Insulin, Hedgehog (Hh), and Bone Morphogenetic Protein (BMP) pathways (see reviews by Colombani and Andersen, 2020; Gervais and Bardin, 2017; Guo et al., 2016; Jiang et al., 2016; Joly and Rousset, 2020; and references therein). The evolutionarily conserved Notch signaling pathway plays essential roles in the control of cell proliferation and specification/differentiation during animal development (Artavanis-Tsakonas and Muskavitch, 2010). Before being presented on the plasma membrane, the Notch receptor undergoes post-translational modifications such as fucosylation in the ER and Golgi apparatus where the S1 proteolytic cleavage of the Notch receptor occurs. Upon binding of its ligand (Dl/Ser) from the signal sending cell on the plasma membrane, the Notch receptor in the signal receiving cell undergoes two consecutive proteolytic cleavages (S2 and S3 cleavages), producing Notch extracellular truncate (NEXT, the S2 cleavage product) and finally releasing the intracellular domain of Notch (NICD, the S3 cleavage product) from the plasma membrane to regulate the expression of downstream genes in the nucleus (Artavanis-Tsakonas and Muskavitch, 2010; Artavanis-Tsakonas et al., 1999; Bray, 2006; Okajima and Irvine, 2002; Pandey et al., 2021). Notch signaling is essential for the proliferation and differentiation of ISCs in Drosophila, loss of Notch signaling in ISCs leads to increased ISC proliferation and progeny differentiation defects, generating ISC and/or EE tumors, while constitutive activation of Notch signaling forces direct differentiation of ISCs into ECs, resulting in ISC loss (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006, 2007).
RESULTS AND DISCUSSION
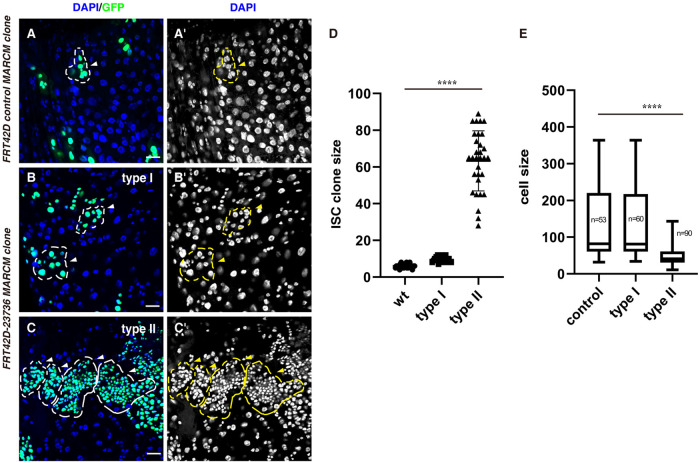
To identify factors regulating the proliferation and differentiation of ISCs, we carried out a small-scale forward genetic screen using some p-element insertions by MARCM technique in the posterior midgut (data not shown) (Lee and Luo, 2001). We were surprised to find that there are two types of ISC MARCAM clones generated upon induction of stumMB01421 mutant (Fig. 1A,B). A p-element is inserted in the coding region of stumble (stum, stumMB01421), which is required for locomotion and mechanical sensing in proprioceptive neurons by transducing dendrite stretching into cellular responses (Desai et al., 2014). The type I stumMB01421 ISC MARCM clones are similar as those of wild-type (WT) control (Fig. 1B,D,E). While the type II stumMB01421 ISC MARCM clones grow much faster than those of WT clones, and the cell size in these clones is tiny and quite uniform compared to the varied cell size of control and type I clones, suggesting that ISC proliferation and/or differentiation is affected in these clones (Fig. 1C–E). We purified the mutant for the type II clones by consecutive backcrosses and named this mutant as stum 9-3 (data not shown).
Fig. 1.
Two type of ISC clones are observed in FRT42D-23736 mutant. (A) FRT42D control ISC MARCM clones (green) (white arrowhead). One ISC MARCM clone is labeled with dotted lines. DAPI staining for the nucleus is showed separately. (B) Type I ISC MARCM clones from FRT42D-23736 mutant (green) (white arrowheads). Please note that the size of type I clone is larger than that of control clone. DAPI staining for the nucleus is showed separately. (C) Type II ISC MARCM clones from FRT42D-23736 mutant (green) (white arrowheads). Please note that the size of type II clone is much larger than that of control and type I clones and the cells in these clones are small and quite uniform in size. DAPI staining for the nucleus is showed separately. (D) Quantification of the size of ISC MARCM clones indicated. Mean±s.d. is shown. n=30–35. ****P<0.0001. (E) Quantification of the cell size of ISC MARCM clones indicated. Mean±s.d. is shown. ****P<0.0001. Please note that the type II clones are deformed, preventing accurate quantification of ISC MARCM clone size. Scale bars: 20 μm.
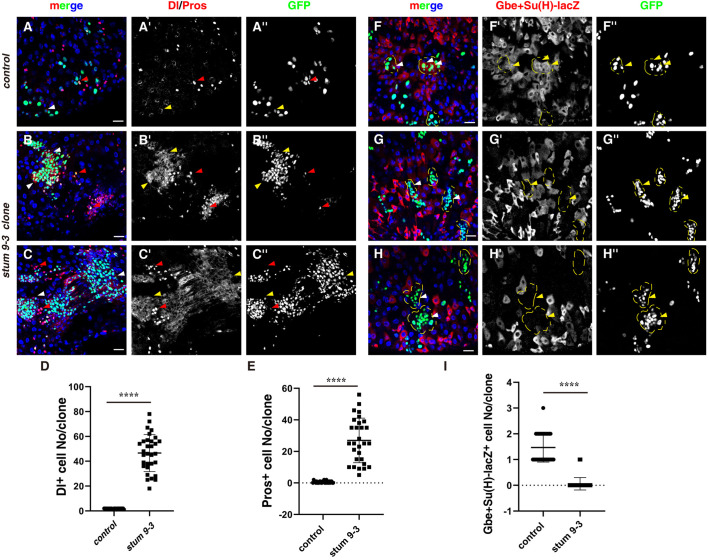
We examined the cell identity in stum 9-3 mutant clones. One to two Dl+ cells could be observed in control clones, while the number of Dl+ cells is dramatically increased in stum 9-3 mutant clones, suggesting that stum 9-3 mutant affects ISC proliferation and/or differentiation (Fig. 2A–D). Meanwhile, EE cell (Pros+) could be observed in control clones (Fig. 2A). On the contrary, two types of stum 9-3 mutant clones are observed: the number of EE cells is significantly increased and increased number of EE cells could be observed adjacent to some clones; while the majority of the other type clones are EE cells (Fig. 2B,C,E). These data suggest that stum 9-3 mutant likely affects ISC proliferation and differentiation. The phenotype of stum 9-3 mutant is reminiscent of Notch loss of function (Fig. S1) (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006, 2007). We then examined whether Notch signaling is affected in stum 9-3 mutant ISC MARCM clones. Notch signaling is activated [by Gbe+Su(H)-lacZ] in control clones (Fig. 2F) (Furriols and Bray, 2001). However, no Gbe+Su(H)-lacZ+ cells are observed in stum 9-3 ISC clones, indicating that Notch signaling is abolished in stum 9-3 mutant (Fig. 2G–I). In supporting of this, the expression of Notch downstream target Cut during wing development is also abolished in stum 9-3 mutant clones, indicating that stum 9-3 mutant is generally required for the activation of Notch signaling (Fig. S2). Furthermore, we find that both NECD and NICD are highly accumulated in stum 9-3 ISC MARCM clones, suggesting that the full-length Notch receptor is accumulated in stum 9-3 mutant clones (Fig. S3). Altogether, these data indicate that stum 9-3 mutant affects the activation of Notch signaling.
Fig. 2.
stum 9-3 affects Notch signaling. (A) Dl (ISC marker) and Pros (EE marker) (red) in intestines with control MARCM clones (white and red arrowheads). Dl/Pros and GFP channels are showed separately in black-white. Please note that EE cell could be observed in some control ISC clones (red arrowhead). (B,C) Dl and Pros (red) in intestines with stum 9-3 ISC MARCM clones (white and red arrowheads). Please note that the number of Dl+ and Pros+ cells is dramatically increased. Two types of clones are observed: ISC clones and EE clones. (D) Quantification of the number of Dl+ cells per clone in control and stum 9-3 intestines. n=25–35. Mean±s.d. is shown, ****P<0.0001. Please note that stum 9-3 ISC clones are deformed, preventing accurate quantification of Dl+ cells per clone. (E) Quantification of the number of EE cells per clone in control and stum 9-3 intestines. n=25–35. Mean±s.d. is shown, ****P<0.0001. Please note that stum 9-3 ISC clones are deformed, preventing accurate quantification of EE cells per clone. (F) Gbe+Su(H)-lacZ cells (Notch signaling reporter, EBs, red) in control ISC MARCM clones (labeled with yellow dotted lines, white arrowheads). Please note that these clones contain several Gbe+Su(H)-lacZ+ cells, indicating that Notch signaling is activated in these clones. Gbe+Su(H)-lacZ and GFP channels are showed separately in black-white. (G,H) Gbe+Su(H)-lacZ cells (EBs, red) in stum 9-3 ISC MARCM clones (with yellow dotted lines, white arrowheads). Please note that no Gbe+Su(H)-lacZ+ cells are observed in these clones, indicative of defective Notch signaling. (I) Quantification of the number of Gbe+Su(H)-lacZ+ cells per clone in control and stum 9-3 intestines. n=25–35. Mean±s.d. is shown, ****P<0.0001. GFP is in green, blue indicates DAPI staining for DNA. Scale bars: 20 μm. Yellow arrows correspond to the white arrows, pointing to the cells that the white arrows pointed to.
We then examined which step of Notch receptor processing that stum 9-3 mutant is required for. We performed rescue experiments by expressing full-length Notch (S1 cleavage product), NEXT (the S2 cleavage product) and NICD (the S3 cleavage product) in stum 9-3 mutant clones, respectively. We found that expression of full-length Notch could not rescue defects observed in stum 9-3 mutant clones (Fig. 3A,B; Fig. S4). While expression of either NEXT or NICD dramatically reduced the size of stum 9-3 mutant clones, likely causing precocious ISC differentiation due to ectopic activation of Notch signaling (Fig. 2C–E; Fig. S4) (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). These data suggest that stum 9-3 mutant affects Notch signaling upstream of the S2 cleavage of the Notch receptor, with accumulation of full-length Notch receptors in stum 9-3 mutant clones. The retromer complex is reported to directly regulate Notch receptor retrograde trafficking in Drosophila neuroblast lineages to prevent aberrant Notch signaling activation (Li et al., 2018). However, depleting the retromer complex could not rescue defects observed in stum 9-3 mutant clones, indicating that stum 9-3 mutant may act upstream of endocytic and retrograde trafficking of the Notch receptor (Fig. S5). Taken together, these data show that stum 9-3 mutant is involved in Notch signaling upstream of the S2 cleavage of the Notch receptor.
Fig. 3.
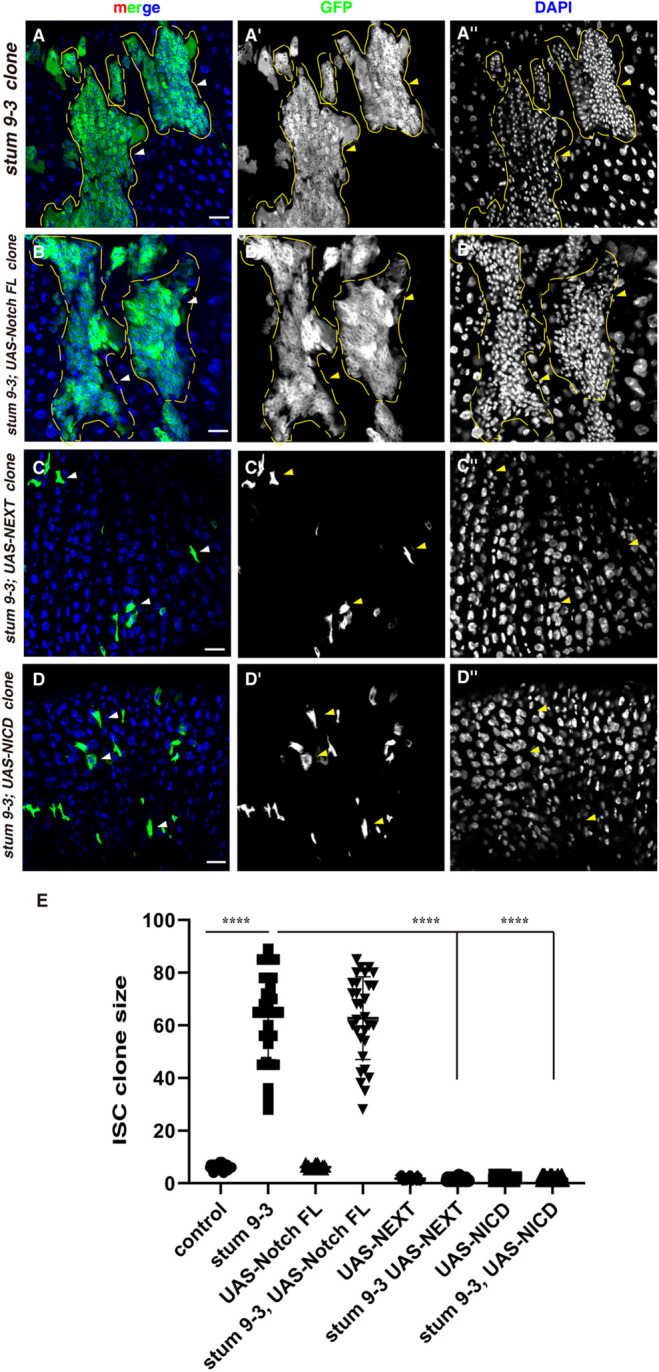
stum 9-3 functions upstream of the S2 cleavage of the Notch receptor. (A) stum 9-3 ISC MARCM clones (green) (labeled with yellow dotted lines, white arrowheads). GFP and DAPI channels are showed separately in black-white. (B) Expression of full-length Notch could not rescue defects observed in stum 9-3 ISC MARCM clones (green) (labeled with yellow dotted lines, white arrowheads). (C,D) Expression of NEXT (C) or NICD (D) could completely suppress ISC over-proliferation observed in stum 9-3 ISC MARCM clones (green) (labeled with yellow dotted lines, white arrowheads). (E) Quantification of the size of ISC MARCM clones in different genotypes indicated. n=25–35. Mean±s.d. is shown, ****P<0.0001. Please note that stum 9-3 ISC clones are deformed, preventing accurate quantification of clone size. In all panels except graphs, GFP is in green, blue indicates DAPI staining for DNA. Scale bars: 20 μm. Yellow arrows correspond to the white arrows, pointing to the cells that the white arrows pointed to.
As stum 9-3 mutant is derived from stumMB01421 mutant, which disrupts stum function, we then examined whether the defects observed in stum 9-3 mutant are resulted from stum loss of function. However, we found that stum is not expressed in the intestines (by stumGal4) (Fig. S6A–B′) (Desai et al., 2014). Furthermore, no obvious defects were detected in the adult intestines of homozygous stum null mutants (Fig. S6C,D) (Desai et al., 2014). Moreover, GFP-Stum is localized in the cell periphery when expressed in the progenitors and expression of GFP-stum could not rescue defects observed in stum 9-3 mutant (Fig. S6E–J) (Desai et al., 2014). Altogether, these data demonstrate that the defects observed in stum 9-3 mutant are not caused by stum loss of function.
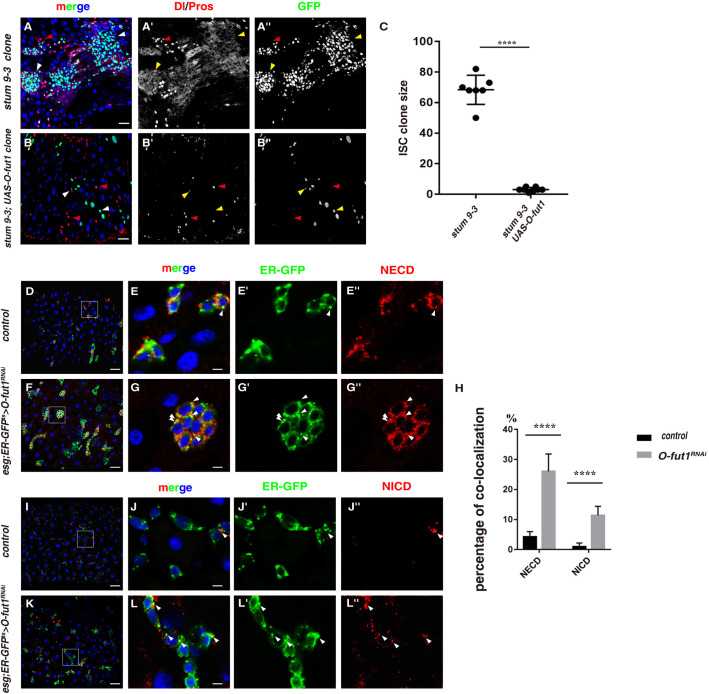
We went on to identify the gene responsible for the observed defects observed in stum 9-3 mutant. Initial examinations showed that stum 9-3 mutant is not allele of known genes involved in the Notch signaling pathway located on the right arm of the 2nd chromosome (2R), like presenilin enhancer (pen-2, encoding the key subunit of the γ-secretase responsible for the S3 cleavage of the Notch receptor) and Caf1-p105 [encoding the subunit of the chromatin assembly factor 1 (CAF-1)] (Fig. S7 and data not shown). We then carried out detailed mapping of stum 9-3 mutant using different deficiency kits for 2R and other mutants (Table S1). The lesion was finally mapped to the region of O-fut1, which is responsible for catalyzing the reaction that attaches fucose through an O-glycosidic linkage to a conserved serine or threonine residue found in the consensus sequence of EGF domains of the Notch receptor (Table S1) (Okajima and Irvine, 2002). To further confirm that the defects observed in 9-3 mutant is caused by mutations of O-fut1, we performed rescue experiments. Restoring O-fut1 function by either over-expressing O-fut1 or endogenous O-fut1 under its own promoter (O-fut1+t3.8) completely rescued defects observed in 9-3 mutant (Fig. 4A–C and data not shown) (Okajima and Irvine, 2002). These data indicate that mutation of O-fut1 in stum 9-3 mutant is responsible for the defects observed. We further examined the nature of lesion in O-fut1 region. We found that the lesion occurs in the 2nd coding exon of O-fut1, probably caused by insertion of an unknown p-element or DNA fragment (Fig. S8). We thus named this O-fut1 allele as O-fut1stum9-3.
Fig. 4.
stum 9-3 is a new allele of O-fut1. (A) Dl and Pros (red) in stum 9-3 ISC MARCM clones (white and red arrowheads). Dl/Pros and GFP channels are showed separately in black-white. (B) Expression of UAS-O-fut1 could completely rescue defects observed in stum 9-3 ISC MARCM clones (white and red arrowheads). (C) Quantification of the size of ISC MARCM clones in different genotypes indicated. Mean±s.d. is shown. n=5–10 intestines. ****P<0.0001. Please note that stum 9-3 ISC clones are deformed, preventing accurate quantification of clone size. (D–E″) NECD (red) is rarely detected in the ER (green, by KDEL-GFP) in progenitors (white arrowhead). Split channels of ER-GFP and NECD are showed separately. The boxed region in D is showed enlarged in E. (F–G″) Increased NECD puncta (red) are detected in the ER (green) in progenitors of esgts>O-fut1RNAi intestines (white arrowheads). The boxed region in F is showed enlarged in G. (H) Quantification of the percentage of NECD or NICE within the ER in progenitors in control and esgts>O-fut1RNAi intestines. (I–J″) NICD (red) is rarely detected in the ER (green, by KDEL-GFP) in progenitors (white arrowhead). Split channels of ER-GFP and NICD are showed separately. The boxed region in I is showed enlarged in J. (K-L″) Increased NICD puncta (red) are detected in the ER (green) in progenitors of esgts>O-fut1RNAi intestines (white arrowheads). The boxed region in K is showed enlarged in L. In all panels except graphs, GFP is in green, blue indicates DAPI staining for DNA. Scale bars: 20 μm (A,B,D,F,I,K) and 5 μm (E,G,J,L). Yellow arrows correspond to the white arrows, pointing to the cells that the white arrows pointed to.
Depletion of O-fut1 in progenitors resulted in identical defects as depletion of the Notch receptor (Fig. S9A–G). We further examined the consequences of depleting O-fut1 in different types of intestinal cells using cell-type specific drivers [ISC: DlGal4, EB: Gbe+Su(H)Gal4 and EE: ProsGal4]. Depletion of O-fut1 in ISCs results the same defects as that of Notch depletion, generating clusters of ISCs and EE cells, indicating that Notch signaling is required in ISCs for their proliferation and differentiation (Fig. S9H–K). Unexpectedly, no defects were observed upon depletion of either O-fut1 or Notch in EBs, in contrast to the observation that Notch signaling is activated in EBs for progeny differentiation (Fig. S9L–O). The discrepancy is likely caused by the nature of the EB driver used. The Gal4 used is directed by Gbe+Su(H), the responsive element of Notch signaling activation, in EBs (Furriols and Bray, 2001; Lu and Li, 2015b). Thus genes begin to be depleted only after the expression of this EB driver, i.e. after the activation of Notch signaling in EBs. Moreover, EB cell is in a transient differentiating state. Therefore, these facts impede the detection of the requirement of genes in EBs. Interestingly, depletion of both O-fut1 and Notch in EE cells produced no obvious defects (Fig. S9P–S). O-fut1 attaches fucose through an O-glycosidic linkage to the EGF domains of the Notch receptor using GDP-fucose as donor substrate in the ER (Okajima and Irvine, 2002; Okajima et al., 2005). Consistently, both NECD and NICD are accumulated in the ER upon depletion of O-fut1 in progenitors (Fig. 4D–H). Taken together, our data show that the defects observed in O-fut1stum9-3 mutant are caused by defective O-fucosylation of the full-length Notch receptor.
Two classic methods are generally deployed to identify factors involved in specific developmental process or signaling pathways: forward genetic screen and reverse genetic screen. Both methods have advantages and shortcomings. Factors involved in specific processes can be identified unbiasedly through forward genetic screen; however, the pinpoint of the lesion responsible for the defects observed is often tedious and time consuming, especially when the genetic background of the mutant is not clean, or the required deficiency (Df) lines are not available, or some of the Df lines are not correctly annotated, and it will be quite disappointing to find out that the identified mutants with interesting phenotype are new alleles of known genes after tedious mapping. The mapping of stum 9-3 mutant is a good example for these drawbacks as we found that the genetic background of the p-element line obtained is not clean and carries multiple lethal hits, and some Df lines are not correctly annotated (Table S1). The genes responsible for specific processes can be easily identified from reverse genetic screens; however, the off-target effects of RNAi (RNA interference), especially for library generated by dsRNA (double strand RNA), must be carefully excluded. Thus, a combination of different experiments should be carried out to confirm that the identified gene is responsible for the defects observed.
MATERIALS AND METHODS
Fly lines and cultures
Flies were maintained on standard media at 25°C. Crosses were raised at 18°C in humidity controlled incubators, or as otherwise noted. Flies hatched in 18°C incubators (2–3 days old) were picked and transferred to 29°C incubator, unless otherwise specified. Flies were transferred to new vials with fresh food every day, and dissected at time points specified in the text. In all experiments, only the female posterior midgut was analyzed. Information for alleles and transgenes used in this study can be found either in FlyBase or as noted: stumMB01421 (BL23736), stum204 BL58774), stum4487 (BL58775), stumGal4 (BL58776 and BL58776), Cyo, 2XTb (gift from A. Zhu) (BL36336), UAS-GFP-stum (BL58778) (Desai et al., 2014), pen-2MI02639 (BL36019), UAS-NotchFL (BL26820), UAS-NICD (Cooper and Bray, 2000), Ote+t5.9 (OteP, gift from Chen DH) (Jiang et al., 2008), Df(2R) deficiency kit collections (gift from J. Pastor and from Bloomington), O-fut1SH2260 (BL51666), O-fut1+t3.8 (BL44244), UAS-O-fut1 (BL9376) (Okajima and Irvine, 2002), esgGal4, UAS-GFP, tubGal80ts (esgts, gift from N. Perrimon), DlGal4, UAS-GFP, tubGal80ts (Dlts, gift from S. Hou and R. Xi) (Zeng et al., 2010), GBE+Su(H)Gal4, UAS-GFP, tubGal80ts (GBE+Su(H)ts) (Lu and Li, 2015b), vps26RNAi (THU3819/HMS01769), vps35RNAi (THU3886/HMS01858), NotchRNAi (BL33611), O-fut1RNAi (THU2167/JF02052), FRT19A-Notch264-39, Gbe+Su(H)-lacZ (gift from S. Bray) (Furriols and Bray, 2001), Caf1-p10536 (gift from R. Jiao) (Yu et al., 2013), hsFlp, ActGal4, UAS-GFP; FRT42D-tubGal80 (for MARCM clonal analysis), UAS- GFP.KDEL (BL9898).
Immunostainings and fluorescence microscopy
For standard immunostaining, intestines were dissected in 1 X PBS (10 mmol/l NaH2PO4/Na2HPO4, 175 mmol/l NaCl, pH7.4), and fixed in 4% paraformaldehyde for 25 min at room temperature. Samples were rinsed, washed with 1 X PBT (0.1% Triton X-100 in 1 X PBS) and blocked in 5% horse serum in 1 X PBT for 45 min. Embryos were fixed and stained following standard protocol. Primary antibodies were added to the samples and incubated at 4°C overnight. The following primary antibodies were used: mouse mAb anti-Dl [C594.9B, 1:50, developed by S. Artavanis-Tsakonas, Developmental Studies Hybridoma Bank (DSHB)], mouse mAb anti-Prospero (MR1A, 1:100, developed by C.Q. Doe, DSHB), mouse mAb anti-Arm (N2 7A1, 1:100, developed by E. Wieschaus, DSHB), mouse mAb anti-Cut (2B10, 1:50, developed by G.M. Rubin, DSHB), mouse mAb anti-NECD (C458.2H, 1:30, developed by S. Artavanis-Tsakonas, DSHB), mouse mAb anti-NICD (C17.9C6, 1:30, developed by S. Artavanis-Tsakonas, DSHB), rabbit anti-β-glactosidase (Cappel, 1:5000), mouse anti-β-glactosidase (Cell Signaling Technology, 1:1000). The primary antibodies were detected by fluorescent-conjugated secondary antibodies from Jackson ImmunoResearch Laboratories. Secondary antibodies were incubated for 2 h at room temperature. DAPI (Sigma-Aldrich, 0.1 μg/ml) was added after secondary antibody staining. The samples were mounted in mounting medium (70% glycerol containing 2.5% DABCO). All images were captured by a Zeiss LSM780 inverted confocal microscope, and were processed in Adobe Photoshop and Illustrator.
MARCM ISC clone analysis
The clonal analyses were achieved using the MARCM system (Lee and Luo, 2001). The ISC clones were induced by heat shocking 3–5 day-old adult flies at 37°C for 60 min. The flies were maintained at 25°C incubator and transferred to new vials with fresh food every day. The sizes of the marked clones were assayed at 6–8 days after clone induction (6–8D ACI, clones from at least ten midguts for each genotype were assayed).
Generation of UAS-pen-2-V5 and UAS-NEXT transgenic lines
The genomic coding region of pen-2 (with stop codon omitted) was cloned into the XhoI and XbaI sites of a pUAST-cV5 vector to be fused with a V5 tag at its C-terminus. The N-terminus of Notch protein (1-160 aa, containing the signal peptide) was fused to the NEXT part of Notch protein (the product after S2 cleavage, 1714-2703 aa, deleting the extracellular domain) and was cloned into a pUAST vector. Transgenic flies were obtained by standard P-element-mediated germline transformation.
Data analysis
The size of ISC clones was determined using Image-Pro Plus software from each confocal image. The number of specific cells per clone was determined manually from each confocal image. The cell size in MARCM clones was determined using Image-Pro Plus software. Confocal images of 40×lens/1.0 zoom from a defined posterior midgut region between the hindgut and the copper cells of different genotypes indicated were acquired. The number of intestines scored is indicated in the text. Statistical analysis was done using the Student's t-test. PEMS 3.1 software was used for s.d. analyses and Sigma Plot software for graph generation. The graphs were further modified using Adobe Photoshop and Illustrator. ****P<0.0001.
Supplementary Material
Acknowledgements
We are grateful to Jose Pastor, Norbert Perrimon, Sarah Bray, Steven Hou, Rongwen Xi, Dahua Chen, Alan Zhu, Yu Cai and Renjie Jiao for generous gifts of reagents, the Bloomington Stock Center, VDRC, NIG-FLY Center, TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) and Tsinghua Fly Center (THFC) for fly stocks, and DSHB for antibodies. We apologize to the colleagues whose work could not be cited because of space restrictions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Zhouhua Li; Methodology: H.Z., R.M.; Validation: L.S.; Formal analysis: L.S., Zhouhua Li; Investigation: L.S., R.K., Zhengran Li, H.Z., R.M., G.B., J.L., Zhouhua Li; Resources: R.K., Zhouhua Li; Data curation: L.S., Zhouhua Li; Writing - original draft: L.S., Zhouhua Li; Writing - review & editing: L.S., Zhouhua Li; Visualization: Zhouhua Li; Supervision: Zhouhua Li; Project administration: Zhouhua Li; Funding acquisition: Zhouhua Li.
Funding
This work is supported by grants from the National Natural Science Foundation of China (numbers: 92054109, 31972893 and 31471384) and Beijing Natural Science Foundation (numbers: KZ201910028040 and 5162004).
Data availability
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
References
- Artavanis-Tsakonas, S., Rand, M. D. and Lake, R. J. (1999). Notch signaling: cell fate control and signal integration in development. Science 284, 770-776. 10.1126/science.284.5415.770 [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S. and Muskavitch, M. A. T. (2010). Notch: the past, the present, and the future. Curr. Top. Dev. Biol. 92, 1-29. 10.1016/S0070-2153(10)92001-2 [DOI] [PubMed] [Google Scholar]
- Banerjee, U., Girard, J. R., Goins, L. M. and Spratford, C. M. (2019). Drosophila as a genetic model for hematopoiesis. Genetics 211, 367-417. 10.1534/genetics.118.300223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe, K., Lee, W.-C. and Micchelli, C. A. (2010). JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev. Biol. 338, 28-37. 10.1016/j.ydbio.2009.10.045 [DOI] [PubMed] [Google Scholar]
- Biteau, B. and Jasper, H. (2014). Slit/robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila. Cell Rep. 7, 1867-1875. 10.1016/j.celrep.2014.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley, C. and Matunis, E. (2004). Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science 304, 1331-1334. 10.1126/science.1097676 [DOI] [PubMed] [Google Scholar]
- Bray, S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678-689. 10.1038/nrm2009 [DOI] [PubMed] [Google Scholar]
- Casali, A. and Batlle, E. (2009). Intestinal stem cells in mammals and Drosophila. Cell Stem Cell 4, 124-127. 10.1016/j.stem.2009.01.009 [DOI] [PubMed] [Google Scholar]
- Chen, J., Xu, N., Wang, C., Huang, P., Huang, H., Jin, Z., Yu, Z., Cai, T., Jiao, R. and Xi, R. (2018). Transient Scute activation via a self-stimulatory loop directs enteroendocrine cell pair specification from self-renewing intestinal stem cells. Nat. Cell Biol. 20, 152-161. 10.1038/s41556-017-0020-0 [DOI] [PubMed] [Google Scholar]
- Colombani, J. and Andersen, D. S. (2020). The Drosophila gut: a gatekeeper and coordinator of organism fitness and physiology. WIREs Dev. Biol. 9, e378. 10.1002/wdev.378 [DOI] [PubMed] [Google Scholar]
- Cooper, M. T. D. and Bray, S. J. (2000). R7 photoreceptor specification requires Notch activity. Curr. Biol. 10, 1507-1510. 10.1016/S0960-9822(00)00826-5 [DOI] [PubMed] [Google Scholar]
- de Navascués, J., Perdigoto, C. N., Bian, Y., Schneider, M. H., Bardin, A. J., Martínez-Arias, A. and Simons, B. D. (2012). Drosophila midgut homeostasis involves neutral competition between symmetrically dividing intestinal stem cells. EMBO J. 31, 2473-2485. 10.1038/emboj.2012.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, B. S., Chadha, A. and Cook, B. (2014). The stum gene is essential for mechanical sensing in proprioceptive neurons. Science 343, 1256-1259. 10.1126/science.1247761 [DOI] [PubMed] [Google Scholar]
- Furriols, M. and Bray, S. (2001). A model Notch response element detects suppressor of hairless-dependent molecular switch. Curr. Biol. 11, 60-64. 10.1016/S0960-9822(00)00044-0 [DOI] [PubMed] [Google Scholar]
- Gervais, L. and Bardin, A. J. (2017). Tissue homeostasis and aging: new insight from the fly intestine. Curr. Opin. Cell Biol. 48, 97-105. 10.1016/j.ceb.2017.06.005 [DOI] [PubMed] [Google Scholar]
- Goulas, S., Conder, R. and Knoblich, J. A. (2012). The Par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell 11, 529-540. 10.1016/j.stem.2012.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z., Lucchetta, E., Rafel, N. and Ohlstein, B. (2016). Maintenance of the adult Drosophila intestine: all roads lead to homeostasis. Curr. Opin. Genet. Dev. 40, 81-86. 10.1016/j.gde.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper, H. (2020). Intestinal stem cell aging: origins and interventions. Annu. Rev. Physiol. 82, 203-226. 10.1146/annurev-physiol-021119-034359 [DOI] [PubMed] [Google Scholar]
- Jiang, X., Xia, L., Chen, D., Yang, Y., Huang, H., Yang, L., Zhao, Q., Shen, L., Wang, J. and Chen, D. (2008). Otefin, a nuclear membrane protein, determines the fate of germline stem cells in Drosophila via interaction with Smad complexes. Dev. Cell 14, 494-506. 10.1016/j.devcel.2008.02.018 [DOI] [PubMed] [Google Scholar]
- Jiang, H., Tian, A. and Jiang, J. (2016). Intestinal stem cell response to injury: lessons from Drosophila. Cell Mol. Life Sci. 73, 3337-3349. 10.1007/s00018-016-2235-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly, A. and Rousset, R. (2020). Tissue adaptation to environmental cues by symmetric and asymmetric division modes of intestinal stem cells. Int. J. Mol. Sci. 21, 6362. 10.3390/ijms21176362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T. and Luo, L. (2001). Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251-254. 10.1016/S0166-2236(00)01791-4 [DOI] [PubMed] [Google Scholar]
- Li, B., Wong, C., Gao, S. M., Zhang, R., Sun, R., Li, Y. and Song, Y. (2018). The retromer complex safeguards against neural progenitor-derived tumorigenesis by regulating Notch receptor trafficking. eLife 7, e38181. 10.7554/eLife.38181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. (2008). Cell biology of stem cells: an enigma of asymmetry and self-renewal. J. Cell Biol. 180, 257-260. 10.1083/jcb.200712159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. and Li, Z. (2015a). No intestinal stem cell regeneration after complete progenitor ablation in Drosophila adult midgut. J. Genet. Genomics 42, 83-86. 10.1016/j.jgg.2014.10.002 [DOI] [PubMed] [Google Scholar]
- Lu, Y. and Li, Z. (2015b). Notch signaling downstream target E(spl)mbeta is dispensable for adult midgut homeostasis in Drosophila. Gene 560, 89-95. 10.1016/j.gene.2015.01.053 [DOI] [PubMed] [Google Scholar]
- Micchelli, C. A. and Perrimon, N. (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475-479. 10.1038/nature04371 [DOI] [PubMed] [Google Scholar]
- Morrison, S. J. and Spradling, A. C. (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598-611. 10.1016/j.cell.2008.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, L. E., Soliman, S. S., Li, X. and Bilder, D. (2011). Altered modes of stem cell division drive adaptive intestinal growth. Cell 147, 603-614. 10.1016/j.cell.2011.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein, B. and Spradling, A. (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470-474. 10.1038/nature04333 [DOI] [PubMed] [Google Scholar]
- Ohlstein, B. and Spradling, A. (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315, 988-992. 10.1126/science.1136606 [DOI] [PubMed] [Google Scholar]
- Okajima, T. and Irvine, K. D. (2002). Regulation of Notch signaling by O-linked fucose. Cell 111, 893-904. 10.1016/S0092-8674(02)01114-5 [DOI] [PubMed] [Google Scholar]
- Okajima, T., Xu, A., Lei, L. and Irvine, K. D. (2005). Chaperone activity of protein O-fucosyltransferase 1 promotes notch receptor folding. Science 307, 1599-1603. 10.1126/science.1108995 [DOI] [PubMed] [Google Scholar]
- Pandey, A., Niknejad, N. and Jafar-Nejad, H. (2021). Multifaceted regulation of Notch signaling by glycosylation. Glycobiology 31, 8-28. 10.1093/glycob/cwaa049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdigoto, C. N., Schweisguth, F. and Bardin, A. J. (2011). Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development 138, 4585-4595. 10.1242/dev.065292 [DOI] [PubMed] [Google Scholar]
- Radtke, F. and Clevers, H. (2005). Self-renewal and cancer of the gut: two sides of a coin. Science 307, 1904-1909. 10.1126/science.1104815 [DOI] [PubMed] [Google Scholar]
- Raff, M. (2003). Adult stem cell plasticity: fact or artifact? Annu. Rev. Cell Dev. Biol. 19, 1-22. 10.1146/annurev.cellbio.19.111301.143037 [DOI] [PubMed] [Google Scholar]
- Singh, S. R., Aggarwal, P. and Hou, S. X. (2019). Cancer stem cells and stem cell tumors in Drosophila. Adv. Exp. Med. Biol. 1167, 175-190. 10.1007/978-3-030-23629-8_10 [DOI] [PubMed] [Google Scholar]
- Stainier, D. Y. R. (2005). No organ left behind: tales of gut development and evolution. Science 307, 1902-1904. 10.1126/science.1108709 [DOI] [PubMed] [Google Scholar]
- Wang, P. and Hou, S. X. (2010). Regulation of intestinal stem cells in mammals and Drosophila. J. Cell. Physiol. 222, 33-37. 10.1002/jcp.21928 [DOI] [PubMed] [Google Scholar]
- Yeung, T. M., Chia, L. A., Kosinski, C. M. and Kuo, C. J. (2011). Regulation of self-renewal and differentiation by the intestinal stem cell niche. Cell. Mol. Life Sci. 68, 2513-2523. 10.1007/s00018-011-0687-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z., Wu, H., Chen, H., Wang, R., Liang, X., Liu, J., Li, C., Deng, W. M. and Jiao, R. (2013). CAF-1 promotes Notch signaling through epigenetic control of target gene expression during Drosophila development. Development 140, 3635-3644. 10.1242/dev.094599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., Chauhan, C. and Hou, S. X. (2010). Characterization of midgut stem cell- and enteroblast-specific Gal4 lines in Drosophila. Genesis 48, 607-611. 10.1002/dvg.20661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., Han, L., Singh, S. R., Liu, H., Neumüller, R. A., Yan, D., Hu, Y., Liu, Y., Liu, W., Lin, X.et al. (2015). Genome-wide RNAi screen identifies networks involved in intestinal stem cell regulation in Drosophila. Cell Rep. 10, 1226-1238. 10.1016/j.celrep.2015.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.