Abstract
The dorsal vagal complex (DVC) in the hindbrain, composed of the area postrema, nucleus of the solitary tract, and dorsal motor nucleus of the vagus, plays a critical role in modulating satiety. The incretins glucagon-like peptide 1 (GLP-1) and glucose-dependent insulinotropic polypeptide (GIP) act directly in the brain to modulate feeding, and receptors for both are expressed in the DVC. Given the impressive clinical responses to pharmacologic manipulation of incretin signaling, understanding the central mechanisms by which incretins alter metabolism and energy balance is of critical importance. Here, we review recent single-cell approaches used to detect molecular signatures of GLP-1 and GIP receptor–expressing cells in the DVC. In addition, we discuss how current advancements in single-cell transcriptomics, epigenetics, spatial transcriptomics, and circuit mapping techniques have the potential to further characterize incretin receptor circuits in the hindbrain.
Introduction
The incretins glucagon-like peptide 1 (GLP-1) and glucose-dependent insulinotropic polypeptide (GIP) are peptide hormones released by intestinal enteroendocrine cells in response to a meal (1). In addition to augmenting insulin secretion during an oral glucose load, both hormones modulate satiety (1). GLP-1 receptor agonists (GLP1RA) are approved for the treatment of obesity (2), and GLP-1 and GIP receptor coagonists show pronounced synergistic effects on weight loss in clinical trials (3). Moreover, genome-wide association studies (GWAS) for BMI (a surrogate measure to approximate adiposity) have associated coding variants in the GIP receptor (GIPR) with lowered body weight (4), further supporting a weight-modulatory role of GIPR in human obesity.
Intracerebroventricular injection of either GIP or GLP-1 reduces body weight and combined intracerebroventricular injection of GIP and GLP-1 (at subeffective doses for each individual peptide) synergistically reduces body weight in mice (5). Due to the rapid degradation of endogenous GLP-1 and GIP by dipeptidyl peptidase 4, it is unlikely that intestinal-derived incretins reach receptors in the brain. However, the brain mediates the anorexic effects induced by degradation-resistant incretin receptor agonists; for instance, knockdown of Glp1r in the central nervous system abolishes weight loss in mice treated peripherally with GLP1RA (6). Interestingly, both peripherally and centrally administered GIPR antagonists have been shown to protect against obesity in rodents and nonhuman primates (7,8), indicating that the cellular mechanisms through which GIPR signaling improves metabolic function remain undefined.
Neurons in the hindbrain and especially the dorsal vagal complex (DVC) are optimally positioned to play a key role in mediating incretin’s effects on food intake. The DVC is located in the medulla oblongata by the fourth ventricle and comprises the area postrema (AP), the nucleus of the solitary tract (NTS), and the dorsal motor nucleus of the vagus (DMV). The AP senses circulating metabolic signals (9), and the NTS processes and relays intestinal information to the brain (10). Studies in rodents and/or nonhuman primates have shown that GLP1R is expressed in the AP, NTS, and DMV (11–16) and that Gipr is expressed in the AP (15–17). In addition, the NTS houses a small population of GLP-1–producing neurons (18). Moreover, GLP1R in glutamatergic and not GABAergic neurons are required for weight loss induced by peripherally administered GLP1RA in mice (14). Indeed, Glp1r is predominantly expressed in glutamatergic neurons in the AP and NTS (14), and administration of GLP1RA in the fourth ventricle suppresses feeding (19). Furthermore, peripherally administered GLP1RA-induced transcriptional changes are particularly pronounced in the AP compared with the NTS and extra-DVC brain areas (16). However, some studies have suggested an involvement of GABAergic NTS neurons and question whether AP cells are necessary for GLP1RA to exert control on feeding (19–22), leaving the role of the DVC unclear.
Here we argue that elucidating the mechanisms by which incretins affect feeding requires a more detailed understanding of the incretin receptor circuits in the hindbrain. Whereas immunohistochemistry, in situ hybridization, flow cytometry, and traditional large-scale molecular profiling approaches can provide information on the spatial distribution and expression levels of incretin receptors, they provide limited information on the cell types expressing the receptors. Given the cellular heterogeneity of the DVC, large-scale molecular approaches operating at single-cell resolution are imperative to further map the role of hindbrain incretin signaling in body weight regulation. Here, we describe recent advancements in transcriptional characterization of GLP-1 and GIP receptor cells in the DVC and outline how single-cell transcriptomics, epigenetics, spatial transcriptomics, and circuit mapping techniques have the potential to further our understanding of incretin signaling in hindbrain circuits.
Mapping the Distribution of Incretin Receptors Using Single-Cell Transcriptomics
Due to the development of robust protocols using minimal amounts of biological input material and the introduction of easy-to-use commercial systems, RNA sequencing (RNA-seq) has become the preferred choice for single-cell profiling. Below we describe key concepts and considerations that need to be taken into account when profiling incretin receptor circuits in the hindbrain.
Hindbrain Single-Cell RNA-seq Data Generation and Analysis
The first step in generating single-cell RNA-seq (scRNA-seq) data is to dissociate tissue into a solution of single cells or single nuclei. Single-nucleus RNA-seq (snRNA-seq) has emerged as the preferred protocol for single-cell sequencing analysis of the hindbrain based on its applicability to frozen tissue samples and because nuclei are less vulnerable to mechanical stresses imposed by whole-cell dissociation protocols. Although nucleus-based approaches do not assay cytosolic RNAs, RNAs in whole cells and nuclei are similarly distributed so that nuclei-based protocols are expected to provide an equally good estimate of the cells’ transcriptional state (23). Typically, data from 2,000–3,000 nuclei can be obtained from a single mouse DVC isolated from a frozen brain sample. The RNA from each cell is captured and processed to “sequencing-ready libraries” that integrate a unique barcode (cell barcode) for all transcripts originating from a given cell and a transcript-specific unique molecular identifier to control for PCR-induced amplification biases. Single-cell level barcoding can be achieved in multiple ways, but most approaches capture individual cells in an isolated reaction environment until the cell barcode is encoded. This can be achieved through serial dilution into reaction wells, FACS, micropipetting, and valve-based, nanowell-based, or droplet-based microfluidics. An alternative approach is combinatorial indexing where cells are repeatedly mixed into different wells to attain a unique combination of barcodes. Whereas valve- and nanowell-based protocols enable complex operations such as in-well imaging and capture of full-length RNAs, allowing detection of splice isoforms (e.g., the human GLP1R and GIPR are transcribed into at least four and eight splice variants, respectively [24]), they typically have lower throughput in terms of the total number of cells as compared with droplet- and combinatorial indexing–based techniques. Due to their scalability and ease of use, microfluidic-based techniques have become a popular choice for scRNA-seq. See ref. 25 for a comparison of specific techniques.
Tissue dissociation, cell capture, barcoding, and library construction are followed by massively parallel sequencing and data analysis. Transcripts are mapped to a reference genome, and a count matrix of the captured transcripts is constructed. Following quality control steps and data normalization, cells are clustered and marker genes for each cluster are identified. Cell type labels can be automatically inferred from one of the growing numbers of DVC scRNA-seq data sets becoming available with use of data integration tools (26). Downstream analyses of hindbrain single-cell data include but are not limited to identification of differentially expressed genes between treatment groups, network-based analyses to identify sets of coregulated genes within a given cell type, integration with GWAS data to identify cell types expressing marker genes that colocalize with genetic association signals for metabolic traits, and mapping of intercellular signaling between cell types (27). See ref. 28 for a review of best-practice recommendations in single-cell data analysis.
Key Questions in Hindbrain Single-Cell Transcriptomics Studies
Four important questions arise when using single-cell experiments to study incretin receptor expression in the hindbrain.
What Should I Pay Attention to During the Study Design?
We anticipate that most hindbrain single-cell sequencing studies will be designed to investigate stimulus-induced gene expression changes, e.g., transcriptional characterization of cell types directly and indirectly responding to incretin dual agonist treatment in model organisms. Ensuring a sufficient number of biological replicates per group is key, and we advise using power calculations based on established DVC single-cell data sets to estimate that number. Next, because the induced transcriptional changes can be subtle, and despite the emergence of computational approaches, technical variation can introduce batch effects (26). Thus, to ensure preservation of biological variation, we recommend using “cell hashing” techniques that allow cells or nuclei from a greater number of samples to be pooled and processed together not only to reduce batch effects but also to minimize cell doublets and experiment costs (29).
Which Technique Should I Use?
If the aim is to map hindbrain cell types, then a single-nucleus droplet-based approach typically constitutes an appropriate choice due to its minimal tissue-handling requirements and the capability to work with frozen tissue and because it facilitates unbiased capture of activity-regulated transcription factor genes—a class of genes typically being transcribed shortly after neuron activation. If the experimental focus is on specific cell types that can be genetically labeled with fluorophores, then FACS purification followed by scRNA-seq represents an efficient way to profile the genetically defined cell type. CITE-seq (cellular indexing of transcriptomes and epitopes by sequencing) (30) and inCITE-seq (intranuclear CITE-seq) (31) are two related techniques, which provide quantitative information on cellular and intranuclear protein levels, respectively, along with transcriptome measurements. Such approaches can for instance be applied to quantify the protein abundance of activity-regulated transcription factors in characterization of cell types responding to physiological stimuli such as incretin receptor agonists.
How Many Cells Do I Need and What Sequencing Depth Should I Aim for When Profiling the Incretin Response in the Hindbrain?
Typically, the number of cells and sequencing read depth are traded off against each other (32). Heterogeneous structures like the hindbrain require a larger number of cells to be profiled than more homogeneous tissues. However, incretin receptor–expressing cells constitute a small fraction of the total number of cells in the DVC and we suggest using one of the following two sorting strategies to profile stimulus-induced expression changes in GLP1R- and GIPR-positive hindbrain cells: 1) a transgenic animal model reporter line expressing a nuclear membrane protein such as SUN1 driven by the promoter for Glp1r, Gipr, or a gene marking the focal cell type; or 2) the Probe-Seq approach (33), which relies on fluorescent in situ hybridization (FISH) to label intranuclear GLP1R or GIPR RNAs.
What Drawbacks Should I Be Aware of When Using Single-Cell Sequencing to Explore Hindbrain Incretin Receptor Circuits?
Glp1r and Gipr are expressed at relatively low levels and detected in only ∼0.7% and 1% of the cells in the mouse DVC, respectively (16). Unbiased profiling studies investigating incretin receptor signaling in response to relevant physiological stimuli will be costly unless effective sorting-based enrichment of the target cells is used; however, such sorting-based approaches can be difficult to set up and apply. Finally, single-cell sequencing results are only descriptive; identified genes will need to be validated in a relevant physiological context using reverse genetics approaches, and relevant cell types will need to be validated in vivo using direct manipulation of cell activity in response to defined physiologic stimuli.
Single-Cell Profiling of Incretin Receptor–Expressing Cell Populations in the DVC
Targeted studies combining genetic mouse models and immunohistochemistry have made valuable insights about incretin receptor expression in the hindbrain. For example, the majority of Glp1r AP and NTS neurons coexpress solute carrier family 17 member 6 (Slc17a6) and tyrosine hydroxylase (Th) (14,34), suggesting that glutamatergic and dopaminergic/noradrenergic neurotransmission may be important mediators of central GLP1RA action. However, unbiased and comprehensive transcriptional characterization of incretin receptor–expressing cells in the hindbrain is still in its infancy. Below we combine and expand on findings from two recent studies using single-cell techniques to profile DVC cell populations in mouse models.
Insights From Mouse Single-Cell Transcriptomics Studies
Zhang et al. (15) profiled the transcriptomes of 3,657 cells from the AP and proximal NTS of mice to unravel groups of AP neurons mediating nausea-associated behaviors. To understand cell populations involved in body weight control, we (Ludwig et al. [16]) characterized the transcriptomes of 72,128 cells from AP-centric DVC tissue of mice with diet-induced obesity that were 1) treated with GLP1RA peripherally, 2) given vehicle and fed ad libitum, or 3) weight matched to the GLP1RA-treated group for 7 days.
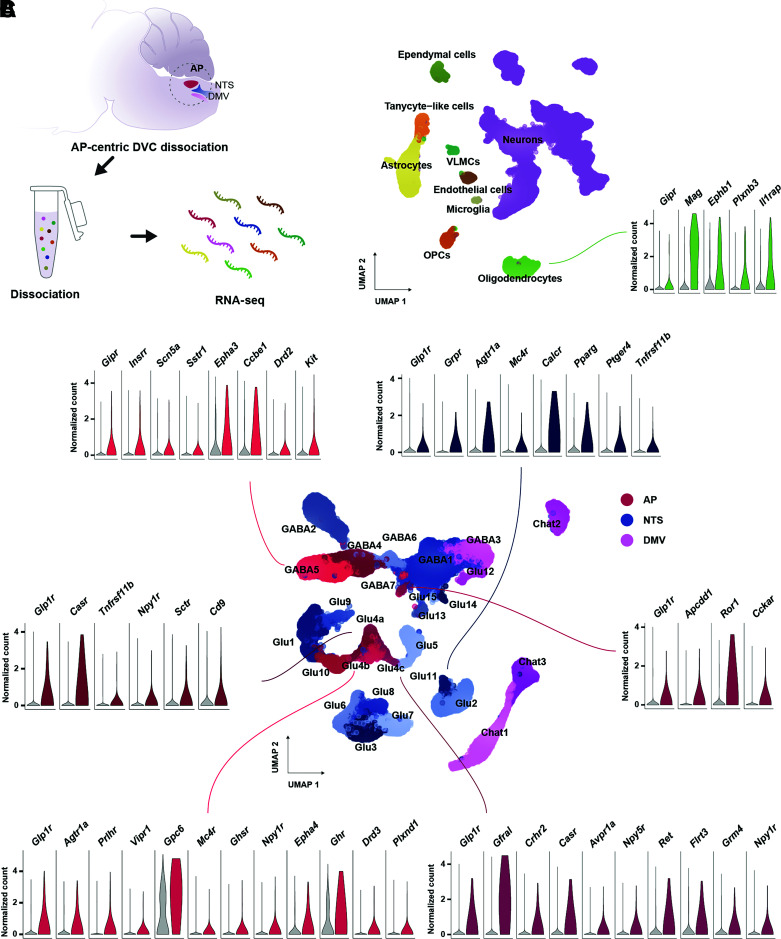
Both studies identified expression of Glp1r in glutamatergic neurons and Gipr in GABAergic neurons in the AP (15,16). In addition, Ludwig et al. detected Glp1r in GABAergic AP neurons and glutamatergic NTS neurons and Gipr in oligodendrocytes, which may have been undetected by Zhang et al. due to a less dense coverage of these cell populations. Here, we integrated cells from these two atlases into a single DVC cell population map (see https://github.com/perslab/Ludwig-Diabetes-2021/ for Methods). This atlas comprises a total of 8 glial cell types and 27 neuronal populations (Fig. 1); for a complete list of marker genes see Supplementary Table 1. We note that since the dissections in both studies are AP-focused, populations in the NTS and DMV may have been missed, such as Glp1r-expressing GABAergic NTS neurons and Glp1r-expressing DMV neurons.
Figure 1.
Combined DVC single-cell transcriptomics atlas highlights distinct groups of incretin receptor cells. A: Graphical illustration of snRNA-seq of the DVC. B: UMAP plot of 75,785 DVC cells from two independent studies (15,16). The receptors found among the top 100 most specifically expressed genes in oligodendrocytes, the only incretin receptor–expressing glial cell type, are plotted by the normalized transcript counts in the focal cell type (green) and remaining cells (gray). C: UMAP plot of 51,212 DVC neurons from the combined atlas. The cell population classifications from the atlas provided by Ludwig et al. (16) have been maintained except for Glu4 neurons, which were split into three clusters (Glu4a-c) following the classification by Zhang et al. (15). The expression of receptors being part of the top 100 most specifically expressed genes in incretin receptor–expressing neurons is depicted in terms of the normalized transcript counts in the focal neuronal population (red, AP; blue, NTS) and remaining neurons (gray). OPCs, oligodendrocyte precursor cells; VLMCs, vascular and leptomeningeal cells.
Transcriptional Signatures of Incretin Receptor Cells and Their Potential Role in Energy Balance Control
In the AP, Glp1r is expressed in a subset of glutamatergic neurons (Glu4a-c in Fig. 1C) distinct from calcitonin receptor (Calcr)-expressing and receptor activity modifying protein 3 (Ramp3)-expressing neurons (Glu10 in Fig. 1C), highlighting that GLP-1 and amylin/calcitonin activate different cell populations. Glu4a neurons were marked by solute carrier family 6 member 2 (Slc6a2), Glu4b by thymocyte selection–associated high mobility group box (Tox), and Glu4c neurons by GDNF family receptor α-like (Gfral). In all three neuronal populations, at least one of the enzymes involved in dopamine and/or noradrenaline synthesis was expressed (Th, dopamine β-hydroxylase [Dbh], and DOPA decarboxylase [Ddc]). Furthermore, Glu4b neurons expressed the melanocortin 4 receptor (Mc4r), which reduces feeding upon activation in the DVC (35) and is upregulated in the AP following GLP1RA administration (16). Glu4a and Glu4c neurons expressed the calcium-sensing receptor (Casr), which contributes to AP-mediated nausea, consistent with the finding that Slc6a2-, Gfral-, and Glp1r-expressing AP neurons activate aversive calcitonin gene–related peptide (Cgrp) neurons in the parabrachial nucleus (PBN) (15). It remains unclear whether Glp1r-expressing AP neurons also activate anorectic circuits independent of nausea. In the AP, Glp1r was also expressed in a small population of GABAergic neurons (GABA7 in Fig. 1C) that expressed the receptor for the anorexic peptide cholecystokinin (Cckar) at high specificity. This population has, however, not been confirmed using in situ hybridization or immunohistochemistry. In the NTS, Glp1r is expressed in glutamatergic neurons (Glu11 in Fig. 1C) where the gastrin-releasing peptide receptor (Grpr) was the top marker gene. These neurons expressed Th, Ddc, and Dbh and may play a role in decreased preferences for palatable foods seen after intraparenchymal NTS injection of GLP1RA (36), as DBH-immunoreactive NTS neurons project to the nucleus accumbens (a key brain area in food reward processing) (37). In contrast to the AP, Glp1r NTS neurons coexpressed Calcr, suggesting that calcitonin and GLP-1 activate partly overlapping NTS cell populations. In addition, these neurons expressed Mc4r. Following treatment with GLP1RA, genes specific to glutamatergic Glp1r-expressing AP and NTS neurons were upregulated, including Bdnf, Ptprn, and Pam, which also colocalize with GWAS-associated loci for BMI (16,38,39). In contrast, genes expressed in glial cells including astrocytes and oligodendrocytes were downregulated following GLP1RA treatment (16). The fact that Glp1r expression was not detected in glial cells in current DVC single-cell atlases suggests that glial cells respond indirectly to treatment, although other studies have described a role of Glp1r-expressing NTS astrocytes in GLP1RA-mediated feeding suppression (40). Moreover, GLP1RA administration did not alter expression of genes specific to GLP-1–producing glucagon (Gcg) neurons in the NTS (Glu14 in Fig. 1C) (16), consistent with the observation that GLP-1 neurons are not necessary for GLP1RA-induced weight loss but when activated may act synergistically to reduce food intake (41).
Gipr is expressed in oligodendrocytes and GABAergic AP neurons (GABA5) (Fig. 1B and C). The top marker of Gipr-expressing GABAergic neurons was the transcription factor paired box 5 (Pax5). GLP1RA administration did not alter gene expression in Gipr neurons (16). Since GABAergic neurons in the AP mainly innervate targets in the AP and proximal NTS (15), including TH-immunoreactive AP neurons (42), this suggests that GIP may presynaptically modulate the action of other metabolic peptides in the DVC. Interestingly, a single-cell transcriptomics study of the AP and NTS of fed and fasted mice published during revision of this article suggests that oligodendrocytes are particularly sensitive to changes in energy status (43), indicating a potential metabolic role for GIPR signaling in DVC oligodendrocytes.
In sum, while many questions regarding GLP-1 and GIP action in the DVC remain to be answered, single-cell transcriptomics has provided a number of important answers: a map of the transcriptional heterogeneity in DVC cell populations that express combinations of the receptors for GLP-1, GIP, amylin, calcitonin, and growth/differentiation factor 15 (GDF15) and identification of marker genes for 27 DVC neuronal populations that can be used for cell type–specific manipulations. Key issues to resolve include mapping of additional NTS and DMV cell populations, screening of incretin receptor expression in less prevalent DVC cell populations and extra-DVC hindbrain nuclei, delineation of species differences in incretin receptor circuits, and elucidation of how incretin receptor–expressing cell types interact when activated.
Mapping cis-Regulatory Factors Regulating Incretin Action in the Hindbrain
Heritability of obesity is predominantly driven by common genetic variants found mostly within intergenic, gene-regulatory regions distributed throughout the human genome (44). Consequently, insights into hindbrain circuits mediating genetic susceptibility to metabolic disease in humans can be inferred by identifying cis-regulatory elements across relevant hindbrain cell types. Below we outline some of the most popular methods to map candidate cis-regulatory elements and their target genes, and we review our recent effort to map chromatin accessibility in the DVC to identify candidate cell populations mediating genetic risk for obesity.
Single-Cell Techniques for Mapping cis-Regulatory Elements and Their Target Genes
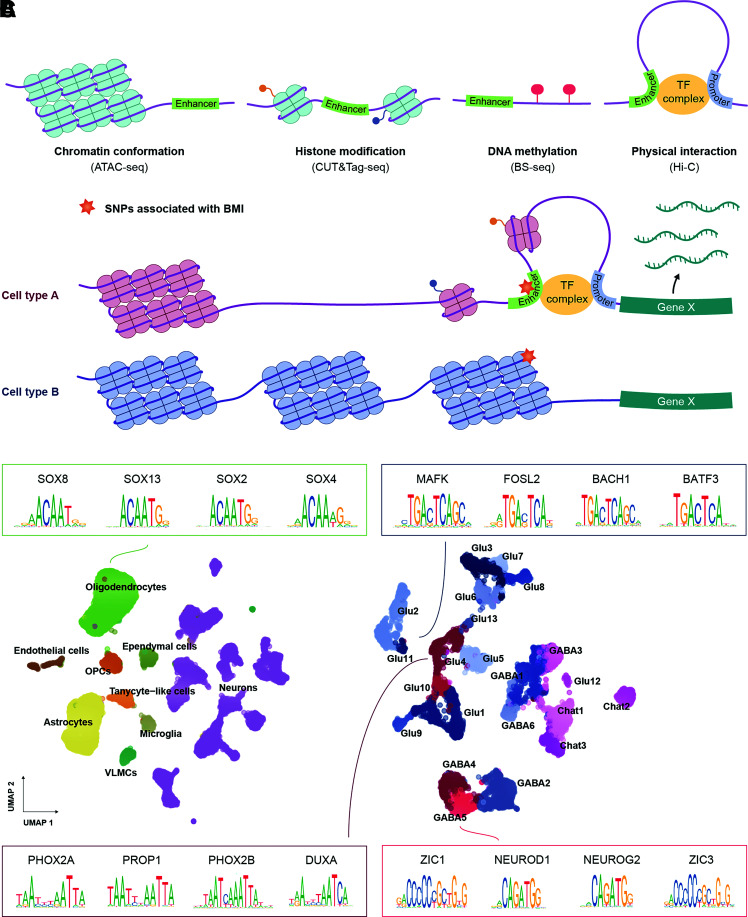
Enhancers are an important class of cis-regulatory DNA elements that bind transcription factors and regulate the expression of proximal genes. However, enhancers remain unidentified for the far majority of rodent and human cell types (45). Active enhancers typically reside in DNA stretches devoid of closed chromatin and can be characterized by high levels of active histone modifications, low levels of DNA cytosine methylation, and physical interactions with gene promoter regions (45) (Fig. 2A). Profiling of open chromatin at single-cell resolution can be accomplished by scATAC-seq (single-cell assay for transposase-accessible chromatin using sequencing) (46), which uses a hyperactive Tn5 transposase that simultaneously cuts and tags DNA at accessible regions with sequencing adaptors, enabling DNA in open chromatin to be profiled (47). Single-cell profiling of chromatin components such as acetylation of lysine 27 at the histone H3 protein (H3K27ac) (a marker for active transcription) or binding of a transcription factor can be performed using scCUT&Tag-seq (single-cell cleavage under targets & tagmentation using sequencing) (48). Here the protein of interest is bound by an antibody that recruits a Tn5 transposase–protein A fusion protein whereby sequencing adaptors are integrated at chromatin-protein binding sites. Single-cell bisulfite-sequencing (scBS-seq) can be applied to measure the DNA methylation landscape through the conversion of cytosine to uracil, leaving methylated cytosines unaffected (49). Lastly, the single-cell Hi-C technique can be used to determine chromatin interactions (such as enhancer-promoter contacts) by cross-linking and then fragmenting the DNA in a way that allows sequencing and identification of DNA that are in close proximity in the three-dimensional chromosomal organization but distant in terms of genomic distance (50).
Figure 2.
Single-cell epigenetic techniques and landscape of incretin receptor DVC cells. A: Graphical illustration of the epigenetic features regulating enhancer activity. Enhancers typically reside in DNA stretches found in open chromatin, with high levels of active histone marks, devoid of cytosine methylation and in physical proximity to gene promoter regions. These features can be measured at single-cell resolution using the indicated assays. B: Graphical illustration of how single-cell epigenetic profiling can predict cell type–specific enhancers mediating genetic risk to human obesity. Colocalization of a BMI-associated genetic variant with the active enhancer in cell type A suggests that the genetic variant exerts its effect on obesity risk by modifying expression of the associated effector gene X in cell type A (and not cell type B). C: UMAP plots of 22,545 cells and 11,651 neurons from the snATAC-seq atlas provided by Ludwig et al. (16). The top four most enriched motifs in incretin receptor–expressing cells are shown by their position weight matrices; at each position of a given motif, the relative distribution of a given nucleotide is depicted. SNP, single nucleotide polymorphism; TF, transcription factor.
Integration of results from these epigenetic profiling techniques into computational models enables prediction of cell type–specific cis-regulatory elements mediating genetic risk for obesity (Fig. 2B). In epigenetic single-cell data, cell clusters are typically annotated with cell type labels through correlation of epigenetic marks at promoter regions and gene bodies with single-cell transcriptomics data from the same tissue with use of label-transfer algorithms (26). A subsequent key step is to map enhancers to genes. While perturbation screens are ultimately needed to establish a causal link between an enhancer and its target gene, enhancer-gene pairs can to some extent be predicted by correlating chromatin accessibility or differentially methylated DNA at promoters with enhancer activity at less proximal sites (51,52). Fulco et al. (53) showed that by profiling multiple epigenetic layers (chromatin accessibility, H3K27ac marks, and three-dimensional chromatin conformation), enhancer-gene prediction can be substantially improved. Having mapped cell type–specific enhancers, machine learning frameworks can be used to estimate whether genetic variants impact a transcription factor’s ability to bind a given regulatory element (e.g., by using single-cell chromatin data [54]). While we anticipate that animal models will be valuable for predicting human cis-regulatory elements in incretin receptor circuits, the divergence between animal and human DNA segments must be considered. Between 65 and 80% (dependent on the sequence homology threshold) of mouse cis-regulatory elements have human orthologs, with the majority of these also being cis-regulatory elements in humans (55,56).
Mapping Chromatin Accessibility and Overlap With Obesity GWAS Signals in the DVC
As part of the above-mentioned DVC snRNA-seq study, Ludwig et al. (16) profiled the chromatin accessibility landscape of the DVC in mice with diet-induced obesity exposed to GLP1RA or control treatments for 7 days. The resulting snATAC-seq atlas comprised 253,452 accessible chromatin regions across 22,545 cells spanning 8 distinct glial and 22 neuronal cell populations (Fig. 2C). Transcription factors, potentially involved in establishing and maintaining cellular identities of incretin receptor cells, were identified from enriched transcription factor binding sites (henceforth referred to here as “motifs”) at accessible chromatin sites (the top four motifs are depicted in Fig. 2C). Glutamatergic Glp1r-expressing AP and NTS neurons contained motifs for the PHOX2A and PHOX2B transcription factors, which are known to regulate transcription of Dbh (57), consistent with the noradrenergic profile of these neurons. Upon administration of GLP1RA, the accessibility of motifs for activity-regulated transcription factor genes in glutamatergic Glp1r AP neurons increased, highlighting GLP1RA-induced reorganization of chromatin in these cells. GABAergic Gipr neurons displayed a chromatin accessibility landscape distinct from that of Glp1r neurons with an enrichment of ZIC1 and NEUROD1 motifs across the accessible regions.
To investigate whether BMI-associated genetic variants nonrandomly colocalized with cell population–specific genes and motifs, Ludwig et al. (16) separately integrated DVC snRNA-seq and snATAC-seq data with BMI GWAS data. They found that glutamatergic DVC neurons, including Glp1r-expressing AP and NTS neurons, were enriched for BMI-associated genetic variants, suggesting a role of GLP1R neurons in the predisposition to obesity.
In sum, whereas the current mouse chromatin accessibility atlas of DVC cell populations provides initial understanding of specific transcription factors maintaining cellular identity and regulating GLP1RA-induced activity, key human DVC-centric analyses remain to be done, namely, 1) mapping of active enhancers under all relevant physiological conditions, 2) identification of activity-dependent transcription factors and their target genes, and 3) identification of BMI-associated variants that perturb binding to active enhancers.
Spatial Transcriptomics and Sequencing-Based Circuit Mapping of the Hindbrain
In addition to assessing cellular identities at the transcriptional and epigenetic level, maturing single-cell spatial transcriptomics and circuit-mapping technologies have the potential to shed light on the spatial locations and physical interactions of incretin receptor cells within and outside the hindbrain. In the following, we will describe some of the currently available protocols for assessing the spatial organization of incretin receptor–expressing cell types and sequencing-based methods for tracing their connectivity.
Toward Spatial Profiling of Incretin Receptor Expression in the Hindbrain
Spatial transcriptomics facilitates the assignment of cell types to their spatial locations. The majority of these methods can be categorized as those that rely on FISH and those that use massively parallel sequencing to profile the transcriptome (58) (Table 1). FISH-based methods enable subcellular resolution (≤1 μm) and are sensitive enough to localize lowly expressed genes such as incretin receptors. Depending on the technique, anywhere from ten to a few thousand genes can be profiled in large (1 cm2) sections of tissue sections. Computational tools can adequately integrate image-based spatial transcriptomics with scRNA-seq to locate cell types (59) or impute unmeasured genes (59–61). In parallel with FISH-based spatial transcriptomics, techniques that use sequencing are rapidly improving. Currently available commercial devices and published protocols offer spatial barcoding arrays with resolution from 55 μm to 10 μm (62,63), while emerging methods push this limit down to 1 μm (64), making the resolution comparable with that offered by FISH-based imaging. A major benefit of sequencing-based spatial transcriptomics is its ability to sample the entire transcriptome without the need to define a set of genes a priori, although this comes at the cost of lower sensitivity. While sequencing-based spatial transcriptomics has been performed to construct a spatial atlas of brain areas from adult mice (65), this data set does not adequately capture the DVC. Future studies will be needed to reveal the spatial organization of incretin receptor cells in the hindbrain and how incretin treatment and other metabolic perturbations potentially alter local cell-to-cell communication.
Table 1.
Summary of spatial transcriptomics profiling technologies
| Modality | Method | Reference no. | Pixel resolution | Number of unique genes | Capture area |
|---|---|---|---|---|---|
| Imaging | RNAscope | 77 | ≤1 μm | 12 | ≥1 cm2 |
| MERFISH | 78 | ≤1 μm | 10,000 | 1 cm2 | |
| seqFISH | 79 | ≤1 μm | 10,000 | 1 cm2 | |
| CARTANA | 80 | ≤1 μm | 600 | 1 cm2 | |
| Massively parallel sequencing | 10x Genomics Visium | 81 | 55 μm | 2,500 | 0.44 cm2 |
| Slide-seqV2 | 62 | 10 μm | 2,000 | 7 mm2 | |
| DBiT-seq | 63 | 10 μm | 2,000 | 1 mm2 | |
| 25 μm | — | 6.25 mm2 | |||
| 50 μm | 3,700 | 25 mm2 | |||
| Seq-Scope | 64 | 1 μm | 500 | 2.2–5.5 mm × 125 mm |
Imaging technologies such as RNAscope, MERFISH, seqFISH, and CARTANA have large capture areas and excellent resolution while requiring the use of a predefined probe set limited to a smaller number of unique genes. Massively parallel sequencing approaches have smaller capture areas and sample from the entire transcriptome, although they capture a fraction of unique genes with reduced sensitivity compared with imaging methods. DBiT-seq, deterministic barcoding in tissue for spatial omics sequencing; MERFISH, multiplexed error-robust fluorescence in situ hybridization; seqFISH, sequential barcode fluorescence in situ hybridization; Seq-Scope, Sequence-Scope.
Toward Mapping of Incretin Receptor–Mediated Neuronal Circuits
In contrast to GABAergic AP neurons, which mostly innervate cells within the AP and proximal NTS (15), glutamatergic AP and NTS neurons project to targets outside the DVC (15,66). Thus, in addition to determination of the molecular signatures and spatial locations of incretin receptor–expressing cells and their response to agonists, there is a need to identify the circuits in which these cells reside. Conventional mapping techniques using cell filling fluorophores, antigenic tags, or viral expression of fluorescent proteins have provided critical information regarding the connectivity of the DVC and its subregions (67,68). The development of Cre-dependent viral tracing tools has further refined these approaches by allowing cell-specific anterograde tracing from genetically defined DVC cell populations to their projection targets (66). Monosynaptic rabies tracing technologies have also allowed investigators to map afferent inputs to defined cell types within the brain (69,70).
Recent approaches have harnessed the power of single-cell transcriptomics for better understanding of connectivity in defined neural circuits. MAPseq (multiplexed analysis of projections by sequencing) takes advantage of the ability to virally express a library of short random barcoded RNAs in a defined brain region and then harvest the injected region and projection terminal areas for sequence analysis (71). As each neuron is labeled with a unique barcode, the presence of barcoded RNAs can be used to rapidly construct a terminal field map with single-neuron resolution (71). BARseq (barcoded anatomy resolved by sequencing) combines MAPseq with in situ sequencing to preserve spatial organization of the terminal neurons (72). MAPseq and BARseq are not replacements for conventional tracing approaches but, rather, serve as complementary methods that can provide a level of specificity that is not easily achievable by conventional techniques. retro-seq combines scRNA-seq with recently developed retrograde viral vectors (73). In this approach, injection of a neuronal projection area with a virus that has the capacity to travel in a retrograde fashion results in “tagging” the cell body of interest with expression of a transgene, allowing for subsequent identification and downstream processing (73). For example, to catalog the specific AP and NTS neuronal populations that project to the PBN, a retrograde virus expressing a robust fluorophore could be injected into the PBN. After allowing for retrograde transport and transgene expression, AP and NTS cells that project to the PBN will express the virally encoded fluorophore and would then be amenable to isolation and sequencing. These approaches will also require additional reagents/molecular tools that will allow simultaneous yet independent manipulation of neighboring cell types. Moreover, establishing a local circuit diagram for incretin receptor-expressing cells in the AP and NTS will benefit from the application of additional functional techniques including electrophysiology and channelrhodopsin-assisted circuit mapping. In sum, integration of conventional mapping techniques with cell-specific labeling approaches and sequencing has the promise to deepen our understanding of the neural circuits in which incretin receptor–expressing cells reside and operate.
Future Avenues
We are just in the beginning of understanding how hindbrain incretin-sensing circuits control satiety and respond to physiological stimuli. Although we have described single-cell sequencing as measures of only one modality, single-cell approaches for profiling different layers of information in the same cell are starting to emerge and have great potential to improve our understanding of cellular mechanisms controlling energy balance. Several single-cell and spatial methods have the capacity for simultaneous assessment of gene expression with chromatin accessibility, histone modifications, or DNA methylation (74–76), allowing one to directly link the epigenome with gene expression abundance. Likewise, it is possible to jointly profile multiple layers of epigenetic marks at single-cell resolution (76). We anticipate that with the emergence of multimodal profiling techniques and sophisticated machine learning prediction models all relevant enhancer-gene pairs across the hindbrain will gradually be mapped.
Once enhancers have been mapped they can be overlaid with human GWAS data; cell types with enriched colocalization of active enhancers and fine-mapped genetic association signals will provide important starting points to identify causal genes and hindbrain circuits mediating genetic susceptibility to human obesity. Whereas induced pluripotent stem cells appear as the most promising and efficient model systems, we envision that future approaches will use combinations of animal models, induced pluripotent stem cells, and postmortem brain setups to map enhancers and their target genes. With the growing number of transgenic mouse models, the ability to resolve molecular heterogeneity at the single cell level, and the increasing number of genetic variants associated with obesity, there is an unprecedented opportunity to further understand the role of incretin-responsive circuits in metabolic health and disease.
Article Information
Acknowledgments. The authors thank the reviewers for their valuable and constructive feedback. The authors also acknowledge the Single-Cell Omics Platform at the Novo Nordisk Foundation Center for Basic Metabolic Research for technical expertise and support.
Funding. Novo Nordisk Foundation Center for Basic Metabolic Research is an independent research center, based at the University of Copenhagen, and partially funded by an unconditional donation from the Novo Nordisk Foundation (https://www.cbmr.ku.dk/) (grant NNF18CC0034900). The authors acknowledge the Novo Nordisk Foundation (grant NNF16OC0021496 to T.H.P.) and the Lundbeck Foundation (grant R190-2014-3904 to T.H.P.). Furthermore, this work was supported by a research grant from the Danish Diabetes Academy, which is funded by the Novo Nordisk Foundation (grant NNF17SA0031406). Finally, this work has been supported by the National Institute of Diabetes and Digestive and Kidney Diseases (grants R01 DK104999 and P01 DK117821).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.Q.L., P.V.T., K.L.E., D.P.O., and T.H.P. wrote the manuscript. T.H.P. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this work were presented at the 81st Scientific Sessions of the American Diabetes Association, virtual meeting, 25–29 June 2021.
Footnotes
This article contains supplementary material online at https://doi.org/10.2337/figshare.14748642.
References
- 1. Baggio LL, Drucker DJ. Biology of incretins: GLP-1 and GIP. Gastroenterology 2007;132:2131–2157 [DOI] [PubMed] [Google Scholar]
- 2. Srivastava G, Apovian CM. Current pharmacotherapy for obesity. Nat Rev Endocrinol 2018;14:12–24 [DOI] [PubMed] [Google Scholar]
- 3. Frias JP, Nauck MA, Van J, et al. Efficacy and safety of LY3298176, a novel dual GIP and GLP-1 receptor agonist, in patients with type 2 diabetes: a randomised, placebo-controlled and active comparator-controlled phase 2 trial. Lancet 2018;392:2180–2193 [DOI] [PubMed] [Google Scholar]
- 4. Turcot V, Lu Y, Highland HM, et al.; CHD Exome+ Consortium; EPIC-CVD Consortium; ExomeBP Consortium; Global Lipids Genetic Consortium; GoT2D Genes Consortium; EPIC InterAct Consortium; INTERVAL Study; ReproGen Consortium; T2D-Genes Consortium; MAGIC Investigators; Understanding Society Scientific Group . Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet 2018;50:26–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. NamKoong C, Kim MS, Jang BT, Lee YH, Cho YM, Choi HJ. Central administration of GLP-1 and GIP decreases feeding in mice. Biochem Biophys Res Commun 2017;490:247–252 [DOI] [PubMed] [Google Scholar]
- 6. Sisley S, Gutierrez-Aguilar R, Scott M, D’Alessio DA, Sandoval DA, Seeley RJ. Neuronal GLP1R mediates liraglutide’s anorectic but not glucose-lowering effect. J Clin Invest 2014;124:2456–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Killion EA, Wang J, Yie J, et al. Anti-obesity effects of GIPR antagonists alone and in combination with GLP-1R agonists in preclinical models. Sci Transl Med 2018;10:eaat3392. [DOI] [PubMed] [Google Scholar]
- 8. Kaneko K, Fu Y, Lin H-Y, et al. Gut-derived GIP activates central Rap1 to impair neural leptin sensitivity during overnutrition. J Clin Invest 2019;129:3786–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gabery S, Salinas CG, Paulsen SJ, et al. Semaglutide lowers body weight in rodents via distributed neural pathways. JCI Insight 2020;5:133429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grill HJ, Hayes MR. Hindbrain neurons as an essential hub in the neuroanatomically distributed control of energy balance. Cell Metab 2012;16:296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Merchenthaler I, Lane M, Shughrue P. Distribution of pre-pro-glucagon and glucagon-like peptide-1 receptor messenger RNAs in the rat central nervous system. J Comp Neurol 1999;403:261–280 [DOI] [PubMed] [Google Scholar]
- 12. Heppner KM, Kirigiti M, Secher A, et al. Expression and distribution of glucagon-like peptide-1 receptor mRNA, protein and binding in the male nonhuman primate (Macaca mulatta) brain. Endocrinology 2015;156:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jensen CB, Pyke C, Rasch MG, Dahl AB, Knudsen LB, Secher A. Characterization of the glucagonlike peptide-1 receptor in male mouse brain using a novel antibody and in situ hybridization. Endocrinology 2018;159:665–675 [DOI] [PubMed] [Google Scholar]
- 14. Adams JM, Pei H, Sandoval DA, et al. Liraglutide modulates appetite and body weight through glucagon-like peptide 1 receptor–expressing glutamatergic neurons. Diabetes 2018;67:1538–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang C, Kaye JA, Cai Z, Wang Y, Prescott SL, Liberles SD. Area postrema cell types that mediate nausea-associated behaviors. Neuron 2021;109:461–472.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ludwig MQ, Cheng W, Gordian D, et al. A genetic map of the mouse dorsal vagal complex and its role in obesity. Nat Metab 2021;3:530–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adriaenssens AE, Biggs EK, Darwish T, et al. Glucose-dependent insulinotropic polypeptide receptor-expressing cells in the hypothalamus regulate food intake. Cell Metab 2019;30:987–996.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holt MK, Richards JE, Cook DR, et al. Preproglucagon neurons in the nucleus of the solitary tract are the main source of brain GLP-1, mediate stress-induced hypophagia, and limit unusually large intakes of food. Diabetes 2019;68:21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hayes MR, Leichner TM, Zhao S, et al. Intracellular signals mediating the food intake-suppressive effects of hindbrain glucagon-like peptide-1 receptor activation. Cell Metab 2011;13:320–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Baraboi E-D, Smith P, Ferguson AV, Richard D. Lesions of area postrema and subfornical organ alter exendin-4-induced brain activation without preventing the hypophagic effect of the GLP-1 receptor agonist. Am J Physiol Regul Integr Comp Physiol 2010;298:R1098–R1110 [DOI] [PubMed] [Google Scholar]
- 21. Secher A, Jelsing J, Baquero AF, et al. The arcuate nucleus mediates GLP-1 receptor agonist liraglutide-dependent weight loss. J Clin Invest 2014;124:4473–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fortin SM, Lipsky RK, Lhamo R, et al. GABA neurons in the nucleus tractus solitarius express GLP-1 receptors and mediate anorectic effects of liraglutide in rats. Sci Transl Med 2020;12:eaay8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bakken TE, Hodge RD, Miller JA, et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One 2018;13:e0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol 2006;7(Suppl. 1):S12.1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ziegenhain C, Vieth B, Parekh S, et al. Comparative analysis of single-cell RNA sequencing methods. Mol Cell 2017;65:631–643.e4 [DOI] [PubMed] [Google Scholar]
- 26. Luecken MD, Büttner M, Chaichoompu K, et al. Benchmarking atlas-level data integration in single-cell genomics. 27 May 2020 [preprint]. bioRxiv:2020.05.22.111161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Armingol E, Officer A, Harismendy O, Lewis NE. Deciphering cell-cell interactions and communication from gene expression. Nat Rev Genet 2021;22:71–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol 2019;15:e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gaublomme JT, Li B, McCabe C, et al. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. Nat Commun 2019;10:2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14:865–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chung H, Parkhurst CN, Magee EM, et al. Simultaneous single cell measurements of intranuclear proteins and gene expression. bioRxiv. 2021. Jan 19;2021.01.18.427139. [Google Scholar]
- 32. Zhang MJ, Ntranos V, Tse D. Determining sequencing depth in a single-cell RNA-seq experiment. Nat Commun 2020;11:774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amamoto R, Garcia MD, West ER, et al. Probe-Seq enables transcriptional profiling of specific cell types from heterogeneous tissue by RNA-based isolation. ELife 2019;8:e51452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cork SC, Richards JE, Holt MK, Gribble FM, Reimann F, Trapp S. Distribution and characterisation of glucagon-like peptide-1 receptor expressing cells in the mouse brain. Mol Metab 2015;4:718–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williams DL, Kaplan JM, Grill HJ. The role of the dorsal vagal complex and the vagus nerve in feeding effects of melanocortin-3/4 receptor stimulation. Endocrinology 2000;141:1332–1337 [DOI] [PubMed] [Google Scholar]
- 36. Richard JE, Anderberg RH, Göteson A, Gribble FM, Reimann F, Skibicka KP. Activation of the GLP-1 receptors in the nucleus of the solitary tract reduces food reward behavior and targets the mesolimbic system. PLoS One 2015;10:e0119034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olson VG, Heusner CL, Bland RJ, During MJ, Weinshenker D, Palmiter RD. Role of noradrenergic signaling by the nucleus tractus solitarius in mediating opiate reward. Science 2006;311:1017–1020 [DOI] [PubMed] [Google Scholar]
- 38. Locke AE, Kahali B, Berndt SI, et al.; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium . Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kichaev G, Bhatia G, Loh P-R, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet 2019;104:65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reiner DJ, Mietlicki-Baase EG, McGrath LE, et al. Astrocytes regulate GLP-1 receptor-mediated effects on energy balance. J Neurosci 2016;36:3531–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brierley DI, Holt MK, Singh A, et al. Central and peripheral GLP-1 systems independently suppress eating. Nat Metab 2021;3:258–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guan JL, Wang QP, Shioda S, Ochiai H, Nakai Y. GABAergic synaptic innervation of catecholaminergic neurons in the area postrema of the rat. Acta Anat (Basel) 1996;156:46–52 [DOI] [PubMed] [Google Scholar]
- 43. Dowsett GKC, Lam BYH, Tadross JA, et al. A survey of the mouse hindbrain in the fed and fasted states using single-nucleus RNA sequencing. Mol Metab 2021;53:101240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gusev A, Lee SH, Trynka G, et al.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; SWE-SCZ Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; SWE-SCZ Consortium . Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet 2014;95:535–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nord AS, West AE. Neurobiological functions of transcriptional enhancers. Nat Neurosci 2020;23:5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 2013;10:1213–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 2019;20:207–220 [DOI] [PubMed] [Google Scholar]
- 48. Kaya-Okur HS, Wu SJ, Codomo CA, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 2019;10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smallwood SA, Lee HJ, Angermueller C, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods 2014;11:817–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nagano T, Lubling Y, Stevens TJ, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013;502:59–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cusanovich DA, Hill AJ, Aghamirzaie D, et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 2018;174:1309–1324.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu H, Zhou J, Tian W, et al. DNA methylation atlas of the mouse brain at single-cell resolution. 30 April 2020 [preprint]. bioRxiv:2020.04.30.069377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fulco CP, Nasser J, Jones TR, et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat Genet 2019;51:1664–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Corces MR, Shcherbina A, Kundu S, et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat Genet 2020;52:1158–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Moore JE, Purcaro MJ, Pratt HE, et al.; ENCODE Project Consortium . Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020;583:699–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yue F, Cheng Y, Breschi A, et al.; Mouse ENCODE Consortium . A comparative encyclopedia of DNA elements in the mouse genome. Nature 2014;515:355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang C, Kim HS, Seo H, Kim CH, Brunet JF, Kim KS. Paired-like homeodomain proteins, Phox2a and Phox2b, are responsible for noradrenergic cell-specific transcription of the dopamine beta-hydroxylase gene. J Neurochem 1998;71:1813–1826 [DOI] [PubMed] [Google Scholar]
- 58. Waylen LN, Nim HT, Martelotto LG, Ramialison M. From whole-mount to single-cell spatial assessment of gene expression in 3D. Commun Biol 2020;3:602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–1902.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nitzan M, Karaiskos N, Friedman N, Rajewsky N. Gene expression cartography. Nature 2019;576:132–137 [DOI] [PubMed] [Google Scholar]
- 61. Abdelaal T, Mourragui S, Mahfouz A, Reinders MJT. SpaGE: Spatial Gene Enhancement using scRNA-seq. Nucleic Acids Res 2020;48:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stickels RR, Murray E, Kumar P, et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat Biotechnol 2021;39:313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu Y, Yang M, Deng Y, et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 2020;183:1665–1681.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cho C-S, Xi J, Park S-R, et al. Seq-Scope: submicrometer-resolution spatial transcriptomics for single cell and subcellular studies. 27 January 2021 [preprint]. bioRxiv:2021.01.25.427807 [Google Scholar]
- 65. Ortiz C, Navarro JF, Jurek A, Märtin A, Lundeberg J, Meletis K. Molecular atlas of the adult mouse brain. Sci Adv 2020;6:eabb3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cheng W, Gonzalez I, Pan W, et al. Calcitonin receptor neurons in the mouse nucleus tractus solitarius control energy balance via the non-aversive suppression of feeding. Cell Metab 2020;31:301–312.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Han W, Tellez LA, Perkins MH, et al. A neural circuit for gut-induced reward. Cell 2018;175:665–678.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Holt MK, Pomeranz LE, Beier KT, Reimann F, Gribble FM, Rinaman L. Synaptic inputs to the mouse dorsal vagal complex and its resident preproglucagon neurons. J Neurosci 2019;39:9767–9781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Callaway EM, Luo L. Monosynaptic circuit tracing with glycoprotein-deleted rabies viruses. J Neurosci 2015;35:8979–8985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lavin TK, Jin L, Wickersham IR. Monosynaptic tracing: a step-by-step protocol. J Chem Neuroanat 2019;102:101661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kebschull JM, Garcia da Silva P, Reid AP, Peikon ID, Albeanu DF, Zador AM. High-throughput mapping of single-neuron projections by sequencing of barcoded RNA. Neuron 2016;91:975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen X, Sun Y-C, Zhan H, et al. High-throughput mapping of long-range neuronal projection using in situ sequencing. Cell 2019;179:772–786.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tasic B, Yao Z, Graybuck LT, et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 2018;563:72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kashima Y, Sakamoto Y, Kaneko K, Seki M, Suzuki Y, Suzuki A. Single-cell sequencing techniques from individual to multiomics analyses. Exp Mol Med 2020;52:1419–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhu C, Zhang Y, Li YE, Lucero J, Behrens MM, Ren B. Joint profiling of histone modifications and transcriptome in single cells from mouse brain. Nat Methods 2021;18:283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Luo C, Liu H, Xie F, et al. Single nucleus multi-omics links human cortical cell regulatory genome diversity to disease risk variants. 12 December 2019 [preprint]. bioRxiv:2019.12.11.873398 [Google Scholar]
- 77. Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2012;14:22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Moffitt JR, Zhuang X. RNA imaging with multiplexed error-robust fluorescence in situ hybridization (MERFISH). Methods Enzymol 2016;572:1–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shah S, Lubeck E, Zhou W, Cai L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 2016;92:342–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hernández I, Qian X, Laláková J, Verheyen T, Hilscher M, Kühnemund M. Application note: mapping brain cell types with CARTANA in situ sequencing on the Nikon Ti2-E microscope. Accessed 19 May 2021. Available from https://www.nature.com/articles/d42473-019-00264-8
- 81. Ståhl PL, Salmén F, Vickovic S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016;353:78–82 [DOI] [PubMed] [Google Scholar]