Abstract
Human papillomavirus (HPV) infection is the leading cause of cervical cancer. The Papanicolaou cytology test is the usually employed type of screening for this infection; however, its sensibility is limited. Only a small percentage of women infected with high-risk HPV develop cervical cancer with an array of genetic and epigenetic modifications. Thus, it is necessary to develop rapid, reproducible and minimally invasive technologies for screening. DNA methylation has gained attention as an alternative method for molecular diagnosis and prognosis in HPV infection. The aim of the present review was to highlight the potential of DNA methylation in cervical neoplasia screening for clinical applications. It was observed that the methylation human and viral genes was correlated with high-grade lesions and cancer. Methylation biomarkers have shown a good capacity to discriminate between high-grade lesions with a transformative potential and cervical cancer, being able to detect these modifications at an early stage. With further research, the epigenetic profiles and subtypes of the tumors could be elaborated, which would aid in therapy selection by opening avenues in personalized precision medicine. Response to therapy could also be evaluated through such methods and the accessibility of liquid biopsies would allow a constant monitoring of the patient's status without invasive sampling techniques.
Keywords: DNA methylation, cervical cancer, human papillomavirus, cervical screening, epigenetics
1. Introduction
Cervical cancer is a common cause of morbidity and mortality among women worldwide, with 604,000 new diagnosed cases and ~342,000 mortalities reported in 2020 alone (1). Epidemiological data confirm that persistent infections with high-risk (hr) genotypes of human papillomavirus (HPV) are associated with cervical cancer development. hrHPVs have the potential to induce carcinogenesis driven by E6 and E7 viral oncoproteins (2). In sexually active women, acute hrHPV infections are common and the majority resolve within 1-2 years from onset without clinical consequences. A small percentage of these infections persist and develops into cancer (3). However, the progression from hrHPV-induced cervical lesion to cervical cancer is slow, taking years or even decades and occurs through well-defined cytological abnormalities. Thus, timely identification of such lesions is the goal of cervical screening (3). The long process of cervical cancer development allows periodic screening tests to detect preneoplastic cellular changes sufficiently early in order to prevent the onset of this malignancy. Cervical cancer can therefore be reduced or even prevented through screening programs that aim to diminish the incidence and mortality of this type of cancer (4). Epigenetic modifications, as hallmarks of carcinogenesis, can serve as a powerful tool for risk stratification, prognosis and treatment of hrHPV-induced cervical lesions in screening programs. For example, there are clinically used panels that assess DNA hypermethylation or hypomethylation associated with oncogene activation and genomic instability for breast cancer. Such kits can be developed and used for cervical cancer early detection, prognosis and treatment management, particularly because such epigenetic modifications are reversible (5).
2. Typical cervical screening and the necessity of designing novel efficient assays
Traditional methods are available to screen women for cervical precancer and cancer, including cytology-based screening, visual examination of the cervix, colposcopy, cervicography and HPV DNA test. Each screening test has its own strengths and limitations and the selection of test will depend on the context (lesion severity, socio-economic factors, sample availability and invasiveness).
Cytological screening
In 1927, the Romanian pathologist Aurel Babes noted the usefulness of exfoliated cells from the cervix in the detection of cervical cancer (6). Subsequently implemented by Papanicolaou and Traut under the name of the Papanicolaou (Pap) test, this method has been used since 1960s in national cervical screening programs in Europe and USA (7). In developed countries that have introduced cytology-based screening programs, the incidence and mortality associated with cervical cancer have been markedly reduced and ≤2,000 new cases of cervical cancer are prevented per month (8). At present, the majority of cervical intraepithelial neoplasia (CIN) that may lead to malignancies are detected with Pap cytology screening programs. However, applying such a screening method and preserving a high quality, particularly in developing countries, is markedly difficult (9). The data published over the last two decades reveal the high quality of Pap cytology, but they also highlight that >30% of CIN2, CIN3 and invasive carcinoma are not distinguished (10). Cytological testing has disadvantages determined by the quality of the smear and the experience of the cytopathologist, which contribute to the decrease in sensitivity of this method. The diagnosis of cervical cancer by Pap test is however limited due to its lack of sensitivity in identifying precancerous lesions and its increased specificity, which leads to over-diagnosis and over-treatment, mainly among young women (11). Therefore, alternative screening methods and algorithms to provide early, sensitive and specific information regarding precursor lesions are evaluated in the present review.
HPV testing
Based on the role of HPV infection in cervical epithelium transformation and cervical cancer development, a new approach for screening has been developed by using HPV oncogenic genotypes testing, which has already been implemented in certain developed countries, such as the USA. The increased sensitivity and limited specificity of the HPV test do not allow discrimination between women with transient, self-limiting infection and those at risk of developing neoplastic lesions. HPV DNA detection does not necessarily indicate a pre-cancerous lesion or cancer. However, since the immune system fights against the majority of HPV infections and prevents the development of cervical precursor lesions or carcinomas, the main challenge is the management of HPV-positive results in primary screening (12). Performing HPV testing as often as Pap screening in women who do not present CIN2 or CIN3 would produce more positive results than cytological screening (10). Furthermore, in a joint European cohort study, an extended screening interval to 6 years was recommended for women with a negative HPV test (13).
HPV testing has various limitations for screening, including low specificity and reproducibility, as well as high cost, considering the different types of tests used. Additionally, HPV testing as a primary screening should be accompanied by a highly specific second-line test, so that recommendations for additional examinations (colposcopy) are reduced (11). Thus, in numerous countries, screening strategies are based on co-testing (HPV and Pap testing). For instance, the American Cancer Society (ACS) and the American Society for Colposcopy and Cervical Pathology (ASCCP) recommend either co-testing every 5 years or cytology screening every 3 years for women aged 30-65 years. Since in young women (21-30 years) HPV infection is more frequent, ACS and ASCCP recommend performing cytology screening alone (14). Currently, hrHPV testing is a more effective method of screening precancerous lesions and carcinomas than cytology-based assay, as the process relies on the detection of nucleic acids rather than microscopic examination of the lesions (15).
To date, neither cytology nor HPV testing provide sufficient specificity and sensitivity as a basis for screening. Thus, there is still a need for developing a screening strategy that is based on objective, easily reproducible and accurate methods (16). In this regard, viral proteins have received attention and several commercial assays have been accordingly developed, which suggests that they may have improved diagnostic capacities and may allow an improved discrimination between productive and transforming infections. In addition, these methods may be less invasive and more cost effective than hrHPV testing (17).
New approaches in cancer diagnosis
Classic diagnostic approaches for tumor subtyping rely on tissue biopsies and histological analysis of tumor specimens. These methods can provide an incomplete image of the cellular composition of the tumor due to sampling a small subpopulation of the cell types in a tumor and often miss the aspect of heterogeneity. Neoteric epigenetic methods may use samples obtained by using such techniques, or fresh frozen or formalin-fixed paraffin-embedded (FFPE) tissue, thus providing a more comprehensive analysis of the modifications occurring at a cellular level (18). Novel epigenetic methods can also be applied to liquid biopsies, thus offering information on tumorigenesis, growth, immune-cancer interactions and cell death by analyzing circulating tumor cells and microvesicles (exosomes). Another advantage of this type of assays is their ability to analyze circulating tumor DNA (ctDNA), which can better illustrate the heterogeneity and clonality of the tumor cell populations. Since it is likely that all cells contribute to this type of cell-free DNA (cfDNA) (19). Analyzing the mutational and epigenetic profiles without special handling of the samples while using non-invasive or minimally invasive sampling techniques, further illustrates the advantages of such novel approaches in cancer diagnosis (20).
A previous study on the molecular mechanisms that could contribute to the onset and progression of cervical cancer have shown that the theory of genetic mutations alone cannot explain the development of tumors (21). Research in the field of epigenetic changes has shown that aberrant methylation of cellular DNA is a common alteration in cancer and hypermethylation of specific DNA regions during carcinogenesis could serve as a sensitive screening tool (22), particularly because distinct methylation patterns of tumor suppressor genes have been found in HPV-induced tumors (23). Epigenetic markers associated with molecular events occurring in HPV-induced oncogenesis can be used to triage women at risk of developing cervical cancer. Consequently, cervical screening is undergoing major changes with the development of new screening tests with improved diagnostic performances using the same clinical specimens as those employed for cytology and HPV testing. The specificity of methylation-based tests could be enhanced by identifying the specific pattern of each type of lesion (24). On the other hand, the performance of screening based on HPV detection could be increased through co-testing of epigenetic changes. According to Cuzick (24), due to the lack of clear indicators on whom to screen for cervical cancer, the necessity for a more accurate initial triaging test is increased. Thus, combining HPV genotyping, cytology screening and detection of p16 and methylation levels through less invasive methods may contribute to the development of more efficient approaches for cancer diagnosis (24).
3. Epigenetic alterations in human tumors
DNA methylation
DNA methylation is one of the three different epigenetic mechanisms that have been identified to date. Catalyzed by DNA methyltransferase enzymes (DNMTs), DNA methylation often occurs at CpG islands, when a methyl group is transferred from S-adenyl methionine to the fifth carbon of a cytosine residue, thus generating 5-methylcytosine (5-mC) (25). There are four members in the DNMT family, including DNMT1, DNMT3A, DNMT3B and DNMT3L. DNMT1 encodes for the maintenance methyltransferase, which is responsible for the transfer of methylation patterns to newly synthesized DNA strands following DNA replication, while DNMT3A and DNMT3B are involved in de novo methylation and maintenance in addition to DNMT1 (26,27). Furthermore, the oxidation of 5-mC leads to 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in the presence of Ten-eleven translocation (TET) enzymes (TET1, TET2 and TET3) in a step-by-step reaction, which indicates that methylation is a reversible process (28). DNA methylation that involves CpG sites in the regulatory sequences leads to gene silencing, while active genes have hypomethylated or unmethylated sites (29). In addition, in almost all types of carcinoma, the genomes of tumor cells exhibit decreased levels of 5-mC, while regulatory sites display increased levels of 5-mC (30). Additionally, the molecular effectors and enzymes involved in DNA methylation process can suffer cancer-specific alterations, such as those shown in Table I (31).
Table I.
Several DNA methylation genes altered in various cancer types.
| Cancer type | DNA methylation genes with modified expression |
|---|---|
| Breast cancer | DNMT3a, DNMT3b, DNMT3L, TET |
| Cervical cancer | DNMT3L |
| Colorectal cancer | MBD1, MBD2, MDB3, MBD4, Kaiso |
| Glioma | TET1, TET2, TET3 |
| Lung cancer | MBD1, MBD2, MDB3, Kaiso, TET |
| Ovarian cancer | DNMT1, DNMT3a |
| Pancreatic cancer | TET |
| Prostate cancer | MeCP2, MBD1, MBD2, TET |
DNMTs, DNA methyltransferase enzymes; TETs, ten-eleven-translocation enzymes; MBDs, methyl-CpG binding domain; MeCP2, methyl-CpG binding protein 2.
Methylated genes in cancer
Previous studies have highlighted the important role served by DNA methylation in various types of cancer (32-39). Genes that perform different functions (oncogenes or tumor suppressors) are frequently targeted and, as a result, their expression profile is altered, particularly in the transformation process. A common phenomenon in cancer is the hypermethylation of promoters of tumor suppressor genes, which leads to a decrease in their expression pattern. To date, numerous methylated gene panels have been identified and validated as biomarkers for various pathologies (Table II).
Table II.
Several histone modification genes altered in various cancers.
| Authors, year | Cancer type | Methylated genes | Refs. |
|---|---|---|---|
| Reinert, 2012 | Bladder cancer | ZNF154, HOXA9, POU4F2, TWIST1, VIM | (32) |
| de Groot et al, 2014 | Breast cancer | SLC5A8, AKR1B1, ALX1, GPX7, RASSGRF2, SFRP2, TM6SF1 and TMEFF2 | (33) |
| Yi, 2021 | Colorectal cancer | TFPI2, FBN2, SEPT9, SMAP8, MLH1, CDH1, TIMP3, O6-MGMT, SFRP1, SFRP2, p16, APC, HIC1, CHFR | (34) |
| Etcheverry et al, 2010; LeBlanc and Marra, 2016 | Glioblastoma | SLC5A8, MGMT, SOX10, RUNX3, WIF1, CD133, HTATIP2, PDE4C, TES | (35,36) |
| Zhang et al, 2016 | Hepatocellular carcinoma | RASSF1A, APC, GSTP1, CDH1, p15, RUNX3, SOCS1, SFRP1, PRDM2, p14, RARβ and p73 | (37) |
| Shen et al, 2019 | Lung cancer | SLC5A8, HOXA9, KRTAP8-1, CCND1, TULP2 | (38) |
| Yang and Park, 2012 | Prostate cancer | CAV1, CDKN2A, CCND2, DAPK, HIC1, LPL, PITX2, PTGS2, RASSF1A, SLC5A8 | (39) |
Methylated genes specific to cervical cancer
There have been a number findings on a series of methylations in gene promoter sequences associated with the pathogenesis and progression of cervical cancer. In addition, epigenetic modifications have been reported to occur earlier than genetic alterations in cervical cancer. In addition, abnormal DNA methylation could occur as early as in low-grade intraepithelial lesion (LSIL), indicating the potential application of abnormal DNA methylation in the early diagnosis of cervical cancer alone or combined with the existing monitoring methods. The detection of altered DNA methylation at this stage serves an important role as LSIL can either regress or progress to higher grade lesions (40).
At present, the DNA methylation sites that have been demonstrated to have a possible association with the early pathogenesis of cervical cancer include cell adhesion molecule 1, junctional adhesion molecule B, Ras association domain family member 1 and fragile histidine triad and hypermethylation of some of them has been detected in blood, urine and exfoliative cell samples (40). Currently, the gene methylation sites that have been validated to be possibly associated with the prognosis of cervical cancer include anaphase-promoting complex subunit 1, chromodomain helicase DNA binding protein 1, voltage-dependent calcium channel subunit α-2/δ-2, Dickkopf-3 and Cyclin Dependent Kinase Inhibitor 2A (41). A previous study showed a strong correlation between host and viral genome methylation and the severity of cervical lesions (CIN2/CIN3) and cervical carcinoma, respectively (42). The most commonly reported host genes exhibiting promoter methylation include erythrocyte membrane protein band 4.1 like 3 (EPB41L3), myelin and lymphocyte protein, cell adhesion molecule (CADM), family with sequence similarity 19 [chemokine (C-C motif)-like] member A4 (FAM19A4) and microRNA (miRNA/miR) 124, as well as CpG sites in the late regions of various HPV genomes (43). Among the genes that display higher levels of methylation in higher grade lesions adenylate cyclase activating polypeptide 1, achaete-scute family BHLH transcription factor 1, CADM1, deleted in colorectal carcinoma, ATPase phospholipid transporting 10A, deleted in breast cancer 1, heparan sulfate-glucosamine 3-sulfotransferase 2, proto-oncogene serine/threonine-protein kinase mos, SRY-box transcription factor (SOX1), myogenic differentiation 1 (MYOD1), SOX17 and transmembrane protein with EGF like and two follistatin like domains 2 are included (44). In addition, LIM homeobox 8 and ST6 N-acetylgalactosaminide α-2,6-sialyltransferase 5 exhibit increased DNA methylation in cervical cancer (45). Verlaat et al (46) identified novel potential biomarkers including ankyrin repeat domain 18C pseudogene, chromosome 13 open reading frame 18, junctional adhesion molecule 3 (JAM3), zinc finger and SCAN domain containing 1, growth hormone secretagogue receptor (GHSR), somatostatin (SST), zinc family member 1 (ZIC1), phosphatase and actin regulator 3 (PHACTR3) and PR-domain containing protein 14 (PRDM14)] that display increased levels of methylation as lesions progress from precancerous to cancerous driven by all hrHPV types. In an exploratory study, EPB41L3 and JAM3 methylation had similar diagnostic accuracy in detecting CIN2+ lesions as hrHPV testing (47).
Aberrant methylation levels are also found in the promoter regions of p16INK4A, death-associated protein kinase, O-6-methylguanine-DNA methyltransferase (MGMT), cadherin 1 and retinoic acid receptor β of cervical cancer tissues, as well as calcitonin related polypeptide α, human telomerase reverse transcriptase, MYOD1, progesterone receptor and TIMP metallopeptidase inhibitor 3 in serum samples (48).
Another mechanism involved in cervical carcinogenesis is the methylation of the promoter of genes encoding miRNAs. Varghese et al (49) demonstrate that hypermethylation of the miR-434 gene's promoter is associated with the development of cervical carcinoma. Botezatu et al (50) found a significant methylation percentage in tumors vs. the healthy control group for miR-124a (90.33% vs. 13.33%), miR-34b (74.19% vs. 6.67%) and miR-203 (87.09% vs. 10.00%).
Regarding the classification of cervical cancer into molecular subtypes based on the degree of methylation of certain genes, a previous study has been conducted in silico, but has not been validated in a patient cohort. Using The Cancer Genome Atlas database, Li et al (41) identify in silico that 1,253 CpG islands were correlated with prognosis in cervical cancer. The authors built a computational model based on novel biomarkers to contribute to prognostic prediction and subtype classification at the molecular level. This model could be of further use in the clinic to guide the medical practitioner to a more personalized approach (targeted therapy based on epigenetic subtypes).
DNA hypomethylation
In types of cancer, hypomethylation of transcription regulatory regions is a phenomenon less encountered than hypermethylation of gene promoters and is usually found in the early stages of oncogenesis (51). Yin et al (52) show that DNA hypomethylation is caused by increased expression levels of the serine/threonine kinase 31 gene, which is epigenetically upregulated by E6 and E7 viral proteins. The promoter of collagen type XVII α 1 chain was found hypomethylated in tumoral vs. normal tissue and was able to predict invasiveness and patient outcome in cervical cancer (53). Varghese et al (49) show that miR-200b and miR-34c are hypomethylated during cervical cancer development. A methylation study performed in a cohort of women with precancerous lesions and cervical carcinoma highlighted a small number of hypomethylated genes: Non-SMC Condensin I Complex Subunit G, presenilin, Histone cluster 1 H3 family member H and ribonucleotide reductase regulatory subunit M2 with increased expression, particularly in CIN3 lesions and cancer (54).
Alteration in histone modification genes
Another epigenetic mechanism, histone modifications rely on post-translational modifications (PTMs) that include methylation, phosphorylation, acetylation, ubiquitylation and SUMOylating. These PTMs lead to conformational change in nucleosomes, thus facilitating a closed or open chromatin structure. Histone methylation involves the transfer of methyl groups from S-adenosyl-L-methionine to lysine or arginine residues of histone proteins by histone methyltransferases (HMTs) (55). In the cell nucleus, HMTs are often part of regulatory complexes that control and modulate DNA methylation Histone methylation affects gene expression via chromatin-dependent transcriptional repression or activation (56). Histone methyltransferases are specific for the lysine or arginine residue that they modify. SET Domain Containing 1A (SET1), SET7/9, ASH1 Like Histone Lysine Methyltransferase, acute lymphoblastic leukemia, methyltransferase mixed lineage leukemia, augmenter of liver regeneration, thioredoxin and SET and MYND domain containing 3 are histone methyltransferases that catalyze the methylation of histone H3 at lysine 4 (H3-K4) in mammalian cells, which is associated with an active transcription status (57). The methylation of both H3-K9 and H3-K27 is associated with heterochromatin formation and leads to silenced gene expression. This is achieved by the activity of the following HMTs: ERG-associated protein with SET domain, Euchromatic Histone Lysine Methyltransferase 2 (EHMT2), suppressor of variegation 3-9 homolog (SUV39-h) 1, SUV39-h2, SET domain bifurcated histone lysine methyltransferase 1, Dim-5, enhancer of zeste homolog 2 (EZH2) and EHMT2(57). Increased global H3-K27 methylation is also found to be involved in certain pathological processes such as cancer progression. The histone methylation process is reversible and methyl groups are removed by histone demethylases that belong to two families: Amino oxidase homolog lysine demethylase (KDM)1 and JmjC domain-containing histone demethylases (58).
Histone acetyl transferases (HATs) promote gene transcription, while histone deacetylases (HDACs) are responsible for removing the acetyl group from lysine, silencing gene expression. Histone modification genes exhibit alterations in tumors and each cancer type exhibits a specific molecular pattern, as summarized in Table III (31). The identification of key factors in cancer histone modification could help to develop novel epigenetic tests (59).
Table III.
Several histone modification genes that are altered in various cancer types.
| Cancer type | Histone modification genes with abnormal expression |
|---|---|
| Bladder cancer | HBO1 |
| Breast cancer | DAC4, HDAC6, SIRT3, SIRT7, p300, GCN5, HBO1, SRC1, NCOA3, ATF2, ELP3, EHMT2, EZH2, SUZ12, BMI1, NSD3, SYMD2, CARM1, Suv4-20h, LSD1, BHC110, JHDM2A, JMJD1A, TSGA, JARID1A-D |
| Cervical cancer | HDAC1 |
| Colon cancer | HDAC1, HDAC2, HDAC3, HDAC4, HDAC5, HDAC7, HDAC8, SIRT1, EHMT2, SYMD2 |
| Glioblastoma | JHDM1a, FBXL10, FBXL11 |
| Lung cancer | SETDB1, LSD1, BHC110, JMJD2A, JHDM3A, UTX, JMJD3 |
| Ovarian cancer | GCN5, EZH2, SUZ12, BMI1 |
| Prostate cancer | Tip60, GCN5, SRC1 EZH2, SUZ12, BMI1, CARM1, Suv4-20h, JHDM1a, FBXL10, FBXL11, JARID1A-D |
Histone modifications in cervical cancer
In cervical cancer, chromatin modifications induced by hrHPV infection have not been used thus far in screening strategies, but they have been documented thoroughly in order to further understand the evolution and behavior of HPV infection.
A study by Groves et al (59) shows that the distribution of interactions between the viral genome and host chromosomes are uniform between all viral genes, with a greater percentage of interactions deriving from the hrHPV E7 gene. In cervical cancer, silencing of tumor suppressor genes occurs through histone modifications as well as other epigenetic mechanisms. For example, the HPV E7 protein can block the interactions of HDACs with hypoxia-inducible factor-1α, which leads to an increase in pro-angiogenic factors (60).
HPV infection mediates modifications of cellular chromatin, leading to a differential expression profile of host genes. These changes often occur in CpG islands of gene promoters under the influence of DNMTs (40). These enzymes have been found to be considerably upregulated in hrHPV infections and these modifications can be used as biomarkers with prognostic value (40).
Among the HPV-induced epigenetic changes, cellular histone methylation status has an important role (61). E2 protein associates with a H3-K4 demethylase, lysine-specific demethylase 5C, to demethylate the HPV18 upstream regulatory region in order to downregulate early promoter activity, leading to a decreased E6/E7 expression. Another mark of HPV-mediated control is shown by a decrease in the global levels of H3-K27me3 under the regulation of KDM6A and KDM6B, which target H3-K27 specifically and their expression is increased by E7 activity, leading to a transcriptionally repressed chromatin (61). The increase in histone acetylation induced by E7 may be required to activate E2 factor (E2F)-responsive genes that favor the cell cycle re-entry (61).
Shadeo et al (62) highlight a modified expression level pattern of dihydrofolate reductase, mortality factor 4-like protein 2, Morf4 family associated protein 1, nuclear receptor corepressor 1 and SWI/SNF related, matrix associated and actin dependent regulator of chromatin subfamily c member 1; genes associated with chromatin remodeling in early cervical lesions (including CIN2), that is maintained throughout their progression, albeit at a reduced frequency in CIN3. These findings have revealed novel genes and events in the early stages of cervical dysplasia that can serve as markers in cervical screening and provide information regarding lesion progression.
Non-coding RNAs (ncRNAs)
ncRNAs include a number of categories and the best known type of ncRNAs are miRNAs, which are single-stranded RNAs with ~20 nucleotides that serve a major role in regulating gene expression. miRNAs downregulate gene expression by binding complementarily to the 3'untranslated region of mRNA. The majority of miRNAs are located within CpG islands, thus allowing miRNAs to undergo DNA methylation and/or other epigenetic modifications (63). Another category of interest in ncRNAs include long ncRNAs (lncRNAs), a family of transcripts with >200 nucleotides in length, although previous studies have shown them capable of encoding functional peptides with short open reading frames, making their mechanisms of action more elaborated than previously considered (64). Additionally, miRNAs and lncRNAs exhibit epigenetic modifications themselves and their biological functions are also affected by the methylation status (Table IV) (30,65,66).
Table IV.
Several non-coding RNAs with altered expression in various cancers.
| Cancer type | Non-coding RNAs with modified expression |
|---|---|
| Bladder cancer | miR-127, miR-205¸ MALAT1, Linc00346, NEAT |
| Breast cancer | miR-21, miR-372, miR-373, miR-155, miR-146, miR-520, miR-10b, miR-9, miR-125, miR-34a, miR-200c, miR-141, miR-429, miR-126, miR-218, miR-145, miR-101, miR-9-1, miR-205, miR-335, miR-342, LINC00922, ATB, BCAR4, PNUTS, ANRIL, miR-199a |
| Cervical cancer | miR-21, miR-127, miR-199a, miR-143, CCHE1, HOTAIR |
| Colorectal cancer | miR-124, miR-34b, miR-34c, miR-21, miR-155, miR-342, miR-145, CCAT2, PCAT-1 |
| Glioblastoma | miR-21, miR-137 |
| Lung cancer | miR-17, miR-92, miR-21, miR-155, miR-29, miR-200c, miR-141, miR-429, miR-126, miR-9, miR-218, miR-145, miR-25, miR32, miR-142, miR-124, miR183, miR-181, miR-101, LINC00922, MALAT1, HOTAIR, CCAT2, AK126698, PANDA, lncRNA-RoR, loc28519, TUG-1 |
| Ovarian cancer | miR-9, miR-9-1, miR199, miR-342, DUXAP10 |
| Pancreatic cancer | miR-146, miR-190, miR-196, lncRNA miR31HG |
| Prostate cancer | miR-21, miR-146, miR-34a, miR-146a, miR-146b, miR-145, miR-218, miR-101, miR-205, CCAT2, PCAT-1 |
miR, microRNA; Linc, long non-coding RNA; MALAT1, metastasis associated lung adenocarcinoma transcript 1 HOTAIR, HOX Transcript Antisense RNA; CCAT2, colon cancer associated transcript 2, PANDA, long non-coding RNA AK126698, P21 associated ncRNA DNA damage activated; TUG-1, taurine upregulated 1; PCAT-1, prostate cancer associated transcript 1.
ncRNAs in cervical cancer
miRNAs expression profile allows discrimination between neoplastic and normal cells. Previous studies found that miR-16, miR-25, miR-92a and miR-378 are upregulated, while miR-22, miR-27a, miR-29a and miR-100 are downregulated in cervical tissue specimens (67-69). These results correlate with cervical cancer progression. miR-21 targets programmed cell death 4 and overexpression of miR-21 can promote proliferation in HeLa cells. miR-29 has been shown to inhibit cell cycle progression (70).
A study by Gibb et al (71) found 123, 105 and 76 differentially expressed lncRNAs in mild (CIN1), moderate (CIN2) and severe (CIN3) dysplasia. Of these, 13 lncRNAs were common for all dysplasia stages. However, there is limited research on the role of aberrant lncRNA expression in cancer, although studies are currently underway to elucidate the complex mechanisms involved. In this regard, HOX Transcript Antisense RNA (HOTAIR), H19, X-inactive specific transcript, cervical carcinoma high-expressed 1, EZH2-binding lncRNA in cervical cancer, metastasis associated lung adenocarcinoma transcript 1, antisense ncRNA in the INK4 locus, lethal, nuclear paraspeckle assembly transcript 1, bladder cancer associated transcript 1, ubiquitin-fold modifier-conjugating enzyme 1, small nucleolar RNA host gene (SNHG16) and SNHG20 have been shown to serve major roles in various processes and greatly influence the mechanisms of oncogenesis (72).
Interplay between E6 and E7 viral proteins and the epigenetic machinery
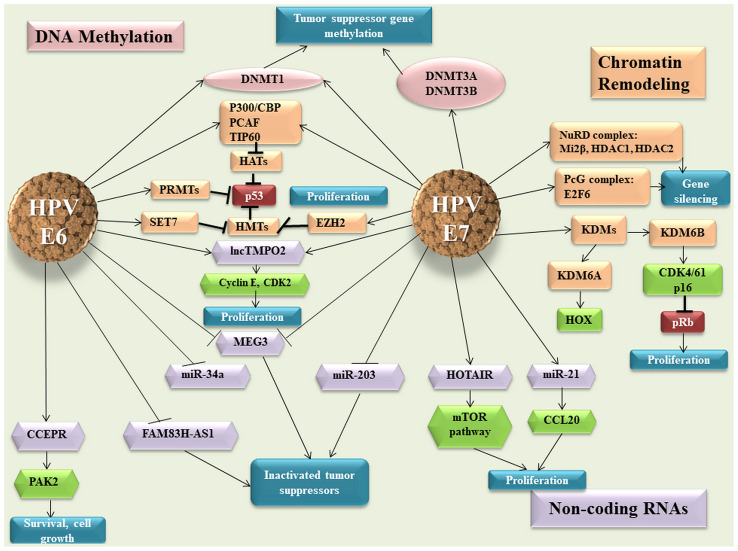
The HPV E6 and E7 oncoproteins interact with host cell factors to alter numerous cellular pathways and maintain the infected cell in a proliferative state. Viral oncoproteins act at all epigenetic levels to hijack the normal cell function to facilitate the replication of the viral genome. It has been shown that HPV16 E6 and E7 proteins increase DNMT1 activity and HPV16 E7 protein binds directly to DNMT1 through the zinc-finger conserved region 3 (CR3; Fig. 1) (73). Previous studies showed that E7 oncoproteins upregulate DNMT3A and DNMT3B at the protein level (74,75). Using this strategy, the virus ensures its viral replication by regulation of the methylation level of both the viral and host genomes (hypermethylation of tumor suppressor genes), thus promoting the oncogenesis process (75). Epigenetic modulation is accomplished not only by interaction of viral oncoproteins with different enzymes and epigenetic factors, but also by their ability to affect the transcription of these modulators (76).
Figure 1.
E6 and E7 viral oncoproteins and their effect on the host epigenetic machinery. HPV, human papillomavirus: DNMT, DNA methyltransferase enzymes; HATs, histone acetyl transferases; TIP60, Tat interactive protein 60; PRMTs, protein arginine methyltransferases; SET, SET Domain Containing 1A; EZH2, enhancer of zeste homolog 2; lnc, long non-coding RNA; MEG3, maternally expressed 3; miR, microRNA; CCEPR, cervical carcinoma expressed PCNA regulatory lncRNA; PAK2, P21 (RAC1) activated kinase 2; FAM83H-AS1, family with sequence similarity 83 member H anti-sense 1; HOTAIR, HOX Transcript Antisense RNA; PcG, polycomb group; NuRD, nucleosome remodelling and deacetylase; HDACs, histone deacetylases; KDM, lysine-specific demethylase; Transcription factor E2F6; CCL20, C-C motif chemokine ligand 20; CDK, Cyclin Dependent Kinase Inhibitor; pRB, retinoblastoma protein.
The E7 viral protein interacts with HDACs and E2F6 transcription factor, leading to the inactivation of retinoblastoma protein (pRB) in order to maintain the proliferative status of the cell. The two viral oncogenes inhibit p53 p300-CREB-binding protein-mediated acetylation, thus blocking p53 activity (77,78). Other HATs affected by viral oncoproteins are Tat interactive protein 60 (TIP60) and p300/CBP-associated factor, which promote an inhibitory effect on the function of p53 and NF-κB, thus allowing the infected cells to escape the immune response (79-81). HPVs not only modify the host genes, but also the expression of viral oncogenes (82). TIP60 targets E6 for degradation, which derepresses the HPV early promoter, thus representing an E2-independent repression mechanism (83). The E7 oncoprotein interacts not only with HATs, but also with HDAC1 and HDAC2 through Chromodomain Helicase DNA Binding Protein 4, which forms part of the nucleosome remodelling and deacetylase (NuRD) chromatin remodelling complex (84).
This interaction modulates the transcription of several genes that are necessary for the differentiation-dependent phase of the virus life cycle, but also represses IFN-β gene transcription, which leads to a suppressed cellular immune response to HPV viral infection (Fig. 1) (85,86).
Coactivator associated arginine methyltransferase 1, protein arginine methyltransferase 1 and SET7 methyltransferases are modulated by E6 protein, thus affecting p53 function (76). It has been shown that HPV E7 oncoprotein induces the expression of KDM6A and KDM6B, thereby inducing pRB degradation and the reactivation of transcription of genes that are normally repressed, such as HOX genes and p16INK4A (87).
Regarding ncRNAs, cervical carcinoma expressed PCNA regulatory lncRNA is upregulated in response to HPV16 E6 expression, enhancing P21 (RAC1) activated kinase 2 expression through miR-922 sponging (88). In silico analysis predicts that HPV16 E7 may bind HOTAIR and this was validated by E7 immunoprecipitation followed by reverse transcription-quantitative (RT-q) PCR analysis (89). This interaction may impede the ability of HOTAIR to interact with polycomb repressive complex 2 and/or KDM1A, thus causing the de-repression of polycomb-regulated genes (87). In cervical cancer, maternally expressed 3 (MEG3) presents a decreased expression as a direct consequence of HPV E6/E7 expression (90). In vitro ectopic MEG3 expression inhibits cell proliferation, increases apoptosis and reduces tumorigenicity in xenograft models (90).
E6 and E7 hrHPV are found to regulate the expression of lncRNA thymopoietin pseudogene 2 (TMPOP2) in a feedback-positive manner, leading to an increased expression of this lncRNA (91). The molecular mechanism involves the tumor suppressor p53, which represses the transcription of lncRNA TMPOP2 through direct interaction (91). The same study shows that TMPOP2 has a role in cell proliferation, which suggests that this lncRNA could be a potential diagnostic marker and therapeutic target of cervical cancer (Fig. 1) (91).
It has been demonstrated that certain miRNAs with a tumor suppressor role present a downregulated expression in cervical cancer, which is associated with the expression of viral oncogenes. Decreased miR-34a expression is associated with the early productive phase and is correlated with E6 expression (92). Melar-New and Laimins (93) show that miR-203 exhibits an inhibitory effect upon HPV amplification and HPV proteins act to suppress the expression of this miRNA to allow viral replication in differentiating cells (Fig. 1). miR-21 is another miRNA that is associated with HPV infection and is upregulated in cervical cancer (94). The same study suggests that chemokine ligand 20 expression is negatively correlated with miR-21 expression and promotes cell proliferation (94).
Viral genes modifications
The HPV genome is subjected to epigenetic regulation, besides DNA methylation, at the level of PTMs of histones, including acetylation, phosphorylation and methylation. These processes control the viral gene expression, serving a main part in the life cycle pf the virus (61).
In addition to host genes methylation, the methylation of viral genes exhibits high levels, particularly in capsid genes and late open reading frames. This has been associated with the progression of lesions from CIN2/3 to invasive cancer (95,96). In a study on HPV-infected Asian women with different grade of lesions, the methylation status of CpG islands in HPV16, 18, 52 and 58 was investigated. A correlation was observed between the methylation status of the Papillomavirus Major Capsid Protein 1 (L1) gene and the disease severity and progression (96). The methylation status of the L1 gene of HPV16 and HPV18 can discriminate between normal tissue and CIN1 lesions. In HPV52 infection, it can differentiate between <CIN2 and ≥CIN2 lesions and between <CIN3 and ≥CIN3 lesions. Furthermore, improved cervical lesion diagnosis was achieved by combining host and HPV genome methylation analysis. Thus, a positive correlation between HPV16 and HPV18 methylation status and paired box 1 (PAX1) and SOX1 was observed. On the other hand, a negative correlation was observed between HPV52 methylation and the same host genes (96). This led to the development of a biomarker panel called S5, using the DNA methylation levels of HPV16 L1, HPV16 Minor Capsid Protein (L2), HPV18 L2, HPV31 L1, HPV33 L2 and EPB41L3 tumor suppressor gene. The S5 methylation assay provided a 90% sensitivity and a 49% specificity for detecting high-grade cervical lesions in a cohort of women from United Kingdom (97). In a Mexican study, the S5 classifier was able to detect all cancer cases, reducing the number of colposcopies required by 30-50% as opposed to triage by cytology and HPV16/18 genotyping (42) Considering that only a small percentage of lesions progress to cervical cancer and the process is rather slow, it is important to identify accordingly the women who are at risk of developing a malignancy and avoid unnecessary expenses and treatment for the women who are most likely to overcome naturally the infection (42,98).
4. DNA methylation for cervical screening
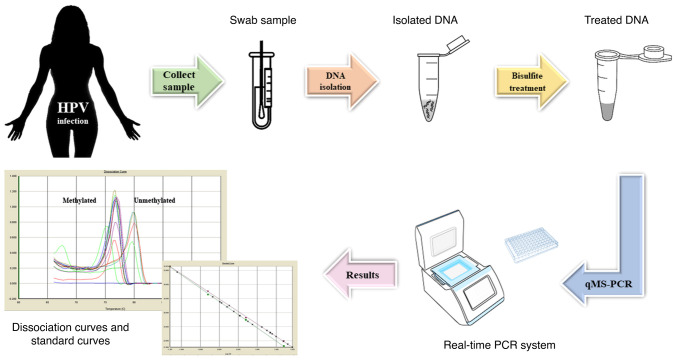
Methylation markers can be used in cervical cancer screening programs, with studies demonstrating their increased specificity compared with that of testing for HPV and immunohistochemistry (p16/Ki-67) (99). Previous studies that have shown a correlation between the expression of HPV E6 and E7 oncogenes and the activity of DNMTs support the validation of DNA hypermethylation as a common event during cervical carcinogenesis (100-103). Certain studies have demonstrated that methylation assays are a useful tool for triaging women with advanced HPV infections, although their performance is rather poor in on-going hrHPV infections that have not progressed yet to CIN3 or cervical cancer (97,104-106). In addition, with advancements in technology, HPV DNA testing could be replaced by DNA methylation screening (95). At present, methylation testing is based on RT-qPCR to estimate the percentage of methylated DNA molecules in the sample for the targeted region (Fig. 2). Currently, the most important application of novel biomarkers in cervical carcinoma is the triage of women with positive cytology and/or HPV tests.
Figure 2.
Workflow of methylation testing. qMS, quantitative methylation-specific.
Commercial screening kits based on DNA methylation
The QIAsure Methylation Test (Qiagen GmbH) is a multiplex quantitative methylation-specific PCR (qMSP)-based assay that amplifies the methylated promoter regions of the tumor suppressor genes family with sequence similarity 19 [chemokine (C-C motif)-like) member A4] and Homo sapiens-miR-124-2, as well as a methylation-unspecific fragment of a reference gene, actin β, that acts as a quality control marker. The QIAsure Methylation Test has a sensitivity of 20.5% for identifying ≤CIN1 infections, 38.1% for CIN2 infections, 67% for CIN3 infections, 90% for advanced transforming CIN and 100% for cervical cancer in hrHPV-positive samples (107).
GynTect® (Oncgnostics GmbH) is another MSP-based assay that distinguishes between cervical lesion types by investigating the methylation status of the promoter regions of six genes (astrotactin1, distal-less homeobox 1, integrin subunit α4, relaxin family peptide receptor 3, SOX17 and zinc finger protein 671) and uses two quality control markers (iduronate 2-sulfatase-M and acetylcholinesterase). Data from clinical trials show that GynTect® markers are positive prior to any histological finding of CIN1, CIN2 or CIN3 in a number of patients (108).
The QIAsure Methylation Test records slightly higher detection rates than GynTect® among CIN samples, but no significant differences have been found. Instead, a significant difference is observed in the detection rates of negative for intraepithelial lesion or malignancy samples, irrespective of the hrHPV status. Also, a notable difference was noticed in the specificity of CIN2+ and CIN3+ cases. The tests scored positive predictive values of 64.3% for GynTect® and 51.4% for the QIAsure Methylation Test and their negative predictive values were 85.5 and 88.1%, respectively. The two kits showed a markedly high sensitivity for the detection of cancer cases, recognizing all cancer and carcinoma in situ samples. Another study found similar results for the detection rates of different stages of CIN, ranging from 13.3-20% for CIN1, 33.3-44.4% for CIN2 and 60-61.2% for CIN3 for the GynTect® assay (109). For the QIAsure Methylation Test, the detection rates were 26.7% for CIN1, 27.8% for CIN2 and 74.3% for CIN3. With the exception of CIN2 cases, these positivity rates are in agreement with those from previous studies (CIN1, 27.7%; CIN2, 44.3%; and CIN3, 75.8%) (24,108,109). The differences between the capabilities of these two tests are illustrated in Table V.
Table V.
The differences between two methylation assays.
| QIAsure Methylation Test | Sensitivity, % | Specificity, % |
|---|---|---|
| CIN 2+ | 67.9 | 66.2 |
| CIN 3+ | 73.3 | 65.9 |
| Cancer | 100.0 | - |
| GynTect | - | - |
| CIN 2+ | 60.4 | 88.2 |
| CIN 3+ | 66.7 | 86.9 |
| Cancer | 100.0 | - |
CIN 2+, cervical intraepithelial neoplasia grade 2; CIN 3+, cervical intraepithelial neoplasia grade 3; -, not available.
The CONFIDENCE™ assay developed by Neumann Diagnostics Kft. detects HPV DNA (CONFIDENCE HPV™) via a TaqManVR-based L1 region-specific multiplex qPCR assay and it also contains a human epigenetic biomarker test (CONFIDENCE Marker™) that measures the methylation level of CpG sites in the promoter region of POU class 4 homeobox 3 by qMSP. As an internal reference for normalizing the methylation level, a quantitative measurement of type II collagen is used. The CONFIDENCE Marker™ test was compared with cytology-based methods for triaging hrHPV-positive cases and it was found to be more sensitive and to have a comparable specificity, scoring for CIN2+ and CIN3+ a relative sensitivity of 1.67 and 1.74, respectively, with a relative specificity of 1.01 for CIN2+ and 0.98 for CIN3+ (110).
PAX1 DNA Detection kit and ZNF582 DNA Detection kit (manufactured by iStat Biomedical Co. Ltd.) are based on qPCR technology and target the PAX1 and zinc finger protein 582 (ZNF582) genes. PAX1 exhibits biomarker potential in a study on 443 patient samples. The study revealed that CIN3+ detection sensitivity was 92%, while the specificity was 83% (111). Another study that included 449 patients reveals that the presence of HPV16/18 and the methylation status of PAX1 and ZNF582 genes is correlated with CIN3+ dysplasia (110). HPV 16/18 detection combined with PAX1 displayed a sensitivity of 89.2% and a specificity of 76.0%, while HPV 16/18 detection combined with ZNF582 displayed a sensitivity and specificity of 85.4 and 80.1%, respectively (111,112).
Methylation-based assays have been developed and used successfully for other types of cancer, demonstrating that this type of triaging/diagnostic approach is feasible and less invasive than traditional methods. Each test includes cancer-specific genes and they aim at recurrence, survival and early detection.
For bladder cancer, the necessity of early detection and the monitoring of recurrence led to the development and approval of different types of DNA methylation based-assays. The following assays are all based on DNA extracted from patients' urine samples. Bladder EpiCheck® (Nucleix), a non-muscle-invasive bladder cancer (NMIBC) diagnosis assay, is based on 15 methylation biomarkers and the results are compiled as an EpiScore ranging from 0 to 100, where scores >60 indicate a positive result for recurrence (113-115). A similar assay is Bladder CARE™ (Pangaea Laboratories, Ltd.), which is used in NMIBC recurrence and is based on a three-gene panel: SOX1, interleukin 1 receptor associated kinase 3 and long interspersed nuclear element 1(116). By contrast, AssureMDx™ for Bladder Cancer (MDxHealth) is an assay based on a negative result that excludes bladder cancer by targeting the orthodenticle homeobox 1, one cut homeobox 2 and Twist family BHLH transcription factor 1 genes in via MSP (117). Similarly, UroMark (University College London) has the capacity to detect the methylation status of 150 loci across the genome. The two assays are promising options in patients presenting hematuria (118).
For breast cancer detection, the current DNA methylation-based assays are limited and the only available predictive test is therascreen® PITX2 RQG test developed by Qiagen GmbH/Therawis Pharma GmbH. The assay is based on a qPCR assay that establishes the ratio between methylated and unmethylated DNA in tumoral samples. The ratio is also an indicator of survival when standard therapy is associated with anthracyclines (119).
For the diagnosis of primary colorectal cancer (CRC), two DNA methylation assays with Food and Drug Administration approval are available: Epi proColon® (Epigenomics AG) and Cologuard® (Exact Sciences Corporation). The former is a blood-based assay that targets the methylation changes of the septin 9 (SEPT9) gene promoter in cfDNA. Similar, Cologuard® is a multi-target stool DNA-based test used in screening that targets the methylated promoter of the bone morphogenetic protein 3 and N-myc downstream-regulated gene 4 genes and seven mutations in the KRAS gene. The two assays are available for CRC screening of patients who are ≥50 years of age, with re-testing recommended every 3 years (120).
The current detection method based on DNA methylation in patients with cirrhosis is the HCCBloodTest (Epigenomics AG). This assay is designed to quantify the methylation levels of the SEPT9 gene in blood samples using RT-qPCR technology (121). Another test, OncoguardTM Liver (Exact Sciences Corp.), for detection of early hepatocellular carcinoma (HCC) was developed in association with Mayo Clinic Healthcare. The panel test is blood based and targets four genes (disabled homolog 2-interacting protein (DAB2IP), empty spiracles homeobox 1 (EMX1), homeobox A1 (HOXA1) and testis-specific Y-encoded-like protein 5 (TSPYL5)) and two protein markers [α fetoprotein (AFP) and lectin-bound AFP] using immunochemical methods and QuARTS™ technology (122). Another promising assay is IvyGene Dx Liver Cancer Test (Laboratory for Advanced Medicine Inc.), which detects the methylation levels of genes in cfDNA isolated from plasma. The test validates the presence of HCC by using next-generation sequencing technology with 95% sensitivity and 97.5% specificity in detection (123).
For glioblastoma diagnosis, MGMT methylation detection status represents the target for different assays. Of these assays, four include CE-marked (Therascreen® MGMT Pyro® kit from Qiagen GmbH, Human MGMT Gene Methylation Detection kit from Xiamen Spacegen Co. Ltd., MGMT Promoter Methylation Detection kit from EntroGen Inc. and EntroGen MGMT Promoter Methylation Detection kit from EntroGen Inc.). These tests use different molecular methods (pyrosequencing technology, amplification refractive mutation system and pyrophosphorolysis-activated polymerization reaction and semi-quantitative RT-PCR with fluorescent hydrolysis probes (124). Another assay is PredictMDx (MdxHealth), which is a laboratory developed test (LDT) based on qMSP technology for detecting MGMT methylation in DNA extracted from FFPE biopsies (125).
At present, the only assay available for early detection in prostate cancer is an LDT. ConfirmMDx (MDxHealth) is a biopsy tissue-based assay design to target glutathione S-transferase π 1, Ras association domain family member 1 and adenomatous polyposis coli genes that uses DNA qMSP technology (126).
Currently, Epi proLung® (Epigenomics AG) is the only CE-marked test available for lung cancer diagnosis in patients at increased risk of the disease. The assay is based on a triplex RT-qPCR with fluorescent hydrolysis probe that targets two genes, short stature homeobox 2 and prostaglandin E receptor 4, which are detected in cfDNA from plasma. The test also uses a third gene, β-actin, as a housekeeping gene (127).
For the detection of multiple tumors, two types of assay have been developed. The first, IvyGene® Cancer Blood Test (Laboratory for Advanced Medicine Inc.) is designed to detect the presence of breast, colon, liver and lung tumors from blood samples by evaluating the methylation levels of cfDNA by using a panel of 46 markers (128). The second, EPICUP™ (Merete GmbH) is a tissue-based assay that can use either freshly frozen or FFPE biopsies. To classify the type of Cancer of Unknown Primary, the test uses bead array technology from Illumina, Inc. (129).
5. Conclusions and perspectives
HPV diagnostic and triaging assays need to advance in terms of capacities and affordability, particularly in low-to-middle income countries where the prevalence of this type of infection remains markedly high. One area that shows promise is associated with DNA methylation classifiers, which can be improved by identifying improved gene combinations and by automating the process.
A study by Snoek et al (130) found a correlation coefficient varying from 0.691 [PR/SET domain 14 (PRDM14)] to 0.9 (SST) for six DNA methylation markers (FAM19A4, GHSR, PHACTR3, PRDM14, SST and ZIC1) in samples obtained from urine sediments and native urine. In the same study, a correlation coefficient varying from 0.508 (PRDM14) to 0.717 (PHACTR3) was observed between urine sediments and cervical scrapes, although the DNA methylation levels in cervical scrapes were considerably higher than those in urine. A significant correlation was found in DNA methylation markers in patients with cervical cancer compared with those in controls. The study showed that DNA methylation marker testing is feasible for detecting cervical cancer and all markers have a high discriminatory power in urine sediments.
Another study by Clarke et al (18) shows that HPV type-specific DNA methylation performs well for risk stratification and has an improved performance compared with that of HPV 16/18 genotyping and Pap cytology. The association between increased methylation with CIN3 or adenocarcinoma in situ has been evaluated across 12 carcinogenic HPV types and is shown to have a strong correlation, allowing for an improved observation of the transition from HPV lesions to cancer (42).
DNA methylation modifications and chromatin changes have been demonstrated to be hallmarks of epigenetic reprogramming in malignant disease and should be investigated in order to further understand the precise role of chromatin remodeling and protein-DNA interactions in HPV-induced carcinogenesis (131). The findings could provide novel, more suitable markers for triaging HPV-infected women.
DNA methylation tests present numerous advantages. They can be used not only in tissue samples, but in any body fluid (liquid biopsy) (18). In cervical cancer, this type of assay may be combined with traditional screening for an improved triage and therapy management, adding valuable information regarding epigenetic profiles (24).
Such novel approaches would provide a clearer primary diagnosis, whereas a number of classic tests would be inconclusive and would help triaging via a minimally invasive screening method. With further research, the epigenetic profiles and subtypes of the tumors could be elaborated, which would aid in therapy selection by opening avenues in personalized precision medicine. Response to therapy could also be evaluated through such methods and the accessibility of liquid biopsies would allow a constant monitoring of the patient's status without invasive sampling techniques.
In addition, tissue biopsies sample only a subpopulation of all the cell types found within a tumor and could potentially miss the aspect of heterogeneity and clonality of the entire tumor, thus providing a skewed image of the cellular phenotype (18).
According to previous studies, ctDNA may offer a more accurate picture regarding the molecular composition of the tumor and precursor lesions in HPV infection since all cell types are likely to contribute (18,132,133). Other sources of information regarding the composition of the tumor include cfDNA and nucleic acids from exosomes. Exosomes are found in both physiological and pathological conditions in the majority of body fluids, including blood, urine, cerebrospinal fluid, saliva, serum, amniotic fluid, breast milk and cervical lavage (132). An important feature of exosomes is that they represent with high fidelity a ‘snapshot’ of the cell they originate from and their molecular cargo can be transferred to other cells. It has been noticed that, in the oncogenic process, the number of exosomes increases significantly and tumors cells release a high number of exosomes that contain different markers that could promote cancer progression and metastasis (133).
Furthermore, there is increasing evidence that supports the involvement of ncRNA species, such as lncRNAs, in cancer pathogenesis. The data show a different lncRNA profile of expression in cervical lesions and cancer and a large number of the identified molecules have the potential to be used as markers for diagnostic and prognosis (134). Therefore, exosomal lncRNAs from cervical lavage and urine samples could be used as cancer biomarkers and could serve to develop a novel, non-invasive array for early detection/triage of precursor lesions and cervical cancer (135).
Acknowledgements
Not applicable.
Funding Statement
Funding: The present study was funded by UEFISCDI (grant no. PN-III-P1-1.1-TE 39/2020).
Availability of data and materials
Not applicable.
Authors' contributions
AA, AF, AB and GA conceived and designed the review. AA, AF, AB, IVI, AP and GA performed the literature research. AA, AF, AB, IVI, AP and GA were involved in the interpretation of the findings in the literature. AA, AF, AB and GA were involved in the writing of the manuscript. Data authentication if not applicable. All authors have read and approved the final manuscript for publication.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Durzynska J, Lesniewicz K, Poreba E. Human papillomaviruses in epigenetic regulations. Mutat Res Rev Mutat Res. 2017;772:36–50. doi: 10.1016/j.mrrev.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Sawaya GF, Brown AD, Washington AE, Garber AM. Current approaches to cervical-cancer screening. N Engl J Med. 2001;344:1603–1607. doi: 10.1056/NEJM200105243442107. [DOI] [PubMed] [Google Scholar]
- 4.Sawaya GF, Smith-McCune K, Kuppermann M. Cervical cancer screening: More choices in 2019. JAMA. 2019;321:2018–2019. doi: 10.1001/jama.2019.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botezatu A, Iancu IV, Plesa A, Manda D, Popa O, Bostan M, Mihaila M, Albulescu A, Fudulu A, Vladoiu SV, et al. Methylation of tumour suppressor genes associated with thyroid cancer. Cancer Biomark. 2019;25:53–65. doi: 10.3233/CBM-182265. [DOI] [PubMed] [Google Scholar]
- 6.Tasca L, Ostör AG, Babeş V. XII. Aurel Babeş. Int J Gynecol Pathol. 2002;21:198–202. doi: 10.1097/00004347-200204000-00016. [DOI] [PubMed] [Google Scholar]
- 7.Tan SY, Tatsumura Y. George Papanicolaou (1883-1962): Discoverer of the Pap Smear. Singapore Med J. 2015;56:586–587. doi: 10.11622/smedj.2015155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petry KU, Woermann B, Schneider A. Benefits and risks of cervical cancer screening. Oncol Res Treat. 2014;37 (Suppl 3):S48–S57. doi: 10.1159/000365059. [DOI] [PubMed] [Google Scholar]
- 9.Lazcano-Ponce E, Lorincz AT, Cruz-Valdez A, Salmeron J, Uribe P, Velasco-Mondragón E, Nevarez PH, Acosta RD, Hernandez-Avila M. Self-collection of vaginal specimens for human papillomavirus testing in cervical cancer prevention (MARCH): A community-based randomised controlled trial. Lancet. 2011;378:1868–1873. doi: 10.1016/S0140-6736(11)61522-5. [DOI] [PubMed] [Google Scholar]
- 10.Koliopoulos G, Nyaga VN, Santesso N, Bryant P, Martin-Hirsch P, Mustafa RA, Schünemann H, Paraskevaidis E, Arbyn M. Cytology vs. HPV testing for cervical cancer screening in the general population. Cochrane Database Syst Rev. 2017;8(CD008587) doi: 10.1002/14651858.CD008587.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz M, Eichelkraut K, Schmidt D, Zeiser I, Hilal Z, Tettenborn Z, Hansel A, Ikenberg H. Performance of a DNA methylation marker panel using liquid-based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer. 2018;18(1197) doi: 10.1186/s12885-018-5125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wentzensen N, Schiffman M, Palmer T, Arbyn M. Triage of HPV positive women in cervical cancer screening. J Clin Virol. 2016;76 (Suppl 1):S49–S55. doi: 10.1016/j.jcv.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillner J, Rebolj M, Birembaut P, Petry KU, Szarewski A, Munk C, de Sanjose S, Naucler P, Lloveras B, Kjaer S, et al. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: Joint European cohort study. BMJ. 2008;337(a1754) doi: 10.1136/bmj.a1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saslow D, Solomon D, Lawson HW, Killackey M, Kulasingum SL, Cain J, Garcia FAR, Moriarty AT, Waxman AG, Wilbur DC, et al. American Cancer Society, American Society for Colposcopy and Cervical Pathology and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. CA Cancer J Clin. 2012;62:147–172. doi: 10.3322/caac.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bian M, Cheng J, Ma L, Cong X, Liu J, Chen Y, Chen X. Evaluation of the detection of 14 high-risk human papillomaviruses with HPV 16 and HPV 18 genotyping for cervical cancer screening. Exp Ther Med. 2013;6:1332–1336. doi: 10.3892/etm.2013.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian Y, Wu NYY, Liou YL, Yeh CT, Cao L, Kang YN, Wang HJ, Li Y, Chu TY, Li W, et al. Utility of gene methylation analysis, cytological examination and HPV-16/18 genotyping in triage of high-risk human papilloma virus-positive women. Oncotarget. 2017;8:62274–62285. doi: 10.18632/oncotarget.19459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fudulu A, Albulescu A, Anton G. Human papillomaviruses' proteins with clinical utility. J Immunoassay Immunochem. 2013;40:81–90. doi: 10.1080/15321819.2018.1553790. [DOI] [PubMed] [Google Scholar]
- 18.Clarke MA, Gradissimo A, Schiffman M, Lam J, Sollecito CC, Fetterman B, Lorey T, Poitras N, Raine-Bennett TR, Castle PE, et al. Human papillomavirus DNA methylation as a biomarker for cervical precancer: Consistency across 12 genotypes and potential impact on management of HPV-positive women. Clin Cancer Res. 2018;24:2194–2202. doi: 10.1158/1078-0432.CCR-17-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gai W, Sun K. Epigenetic biomarkers in cell-free DNA and applications in liquid biopsy. Genes (Basel) 2019;10(32) doi: 10.3390/genes10010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 21.Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8(a019505) doi: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lleras RA, Smith RV, Adrien LR, Schlecht NF, Burk RD, Harris TM, Childs G, Prystowsky MB, Belbin TJ. Unique DNA methylation loci distinguish anatomic site and HPV status in head and neck squamous cell carcinoma. Clin Cancer Res. 2013;19:5444–5455. doi: 10.1158/1078-0432.CCR-12-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: Appraisal of the state-of-the-science. Gynecol Oncol. 2009;112:293–299. doi: 10.1016/j.ygyno.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorincz AT. Virtues and weaknesses of DNA methylation as a test for cervical cancer prevention. Acta Cytol. 2016;60:501–512. doi: 10.1159/000450595. [DOI] [PubMed] [Google Scholar]
- 25.Plesa A, Iancu IV, Botezatu A, Huica I, Stoian M, Anton G. The involvement of epigenetic mechanisms in HPV-induced cervical cancer, Rajamanickam Rajkumar, human papillomavirus-research in a global perspective, IntechOpen, London, 191-239, 2016. https://www.intechopen.com/chapters/50425. Accessed June, 16, 2021. [Google Scholar]
- 26.Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin B, Robertson KD. DNA methyltransferases, DNA damage repair and cancer. Adv Exp Med Biol. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 30.Sina AA, Carrascosa LG, Liang Z, Grewal SY, Wardiana A, Shiddiky MJA, Gardiner RA, Samaratunga H, Gandhi MK, Scott RJ, et al. Epigenetically reprogrammed methylation landscape drives the DNA self-assembly and serves as a universal cancer biomarker. Nat Commun. 2018;9(4915) doi: 10.1038/s41467-018-07214-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanwal R, Gupta K, Gupta S. Cancer epigenetics: An introduction. Methods Mol Biol. 2015;1238:3–25. doi: 10.1007/978-1-4939-1804-1_1. [DOI] [PubMed] [Google Scholar]
- 32.Reinert T. Methylation markers for urine-based detection of bladder cancer: The next generation of urinary markers for diagnosis and surveillance of bladder cancer. Adv Urol. 2012;2012(503271) doi: 10.1155/2012/503271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Groot JS, Pan X, Meeldijk J, van der Wall E, van Diest PJ, Moelans CB. Validation of DNA promoter hypermethylation biomarkers in breast cancer-a short report. Cell Oncol (Dordr) 2014;37:297–303. doi: 10.1007/s13402-014-0189-1. [DOI] [PubMed] [Google Scholar]
- 34.Yi JM. DNA Methylation change profiling of colorectal disease: Screening towards clinical use. Life (Basel) 2021;11(412) doi: 10.3390/life11050412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Etcheverry A, Aubry M, de Tayrac M, Vauleon E, Boniface R, Guenot F, Saikali S, Hamlat A, Riffaud L, Menei P, et al. DNA methylation in glioblastoma: Impact on gene expression and clinical outcome. BMC Genomics. 2010;11(701) doi: 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LeBlanc VG, Marra MA. DNA methylation in adult diffuse gliomas. Brief Funct Genomics. 2016;15:491–500. doi: 10.1093/bfgp/elw019. [DOI] [PubMed] [Google Scholar]
- 37.Zhang C, Li J, Huang T, Duan S, Dai D, Jiang D, Sui X, Li D, Chen Y, Ding F, et al. Meta-analysis of DNA methylation biomarkers in hepatocellular carcinoma. Oncotarget. 2016;7:81255–81267. doi: 10.18632/oncotarget.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen N, Du J, Zhou H, Chen N, Pan Y, Hoheisel JD, Jiang Z, Xiao L, Tao Y, Mo X. A diagnostic panel of DNA methylation biomarkers for lung adenocarcinoma. Front Oncol. 2019;9(1281) doi: 10.3389/fonc.2019.01281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang M, Park JY. DNA methylation in promoter region as biomarkers in prostate cancer. Methods Mol Biol. 2012;863:67–109. doi: 10.1007/978-1-61779-612-8_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: From pathogenesis to therapy. Tumour Biol. 2014;35:5083–5093. doi: 10.1007/s13277-014-1737-z. [DOI] [PubMed] [Google Scholar]
- 41.Li C, Ke J, Liu J, Su J. DNA methylation data-based molecular subtype classification related to the prognosis of patients with cervical cancer. J Cell Biochem. 2020;121:2713–2724. doi: 10.1002/jcb.29491. [DOI] [PubMed] [Google Scholar]
- 42.Hernández-López R, Lorincz AT, Torres-Ibarra L, Reuter C, Scibior-Bentkowska D, Warman R, Nedjai B, Mendiola-Pastrana I, León-Maldonado L, Rivera-Paredez B, et al. Methylation estimates the risk of precancer in HPV-infected women with discrepant results between cytology and HPV16/18 genotyping. Clin Epigenetics. 2019;11(140) doi: 10.1186/s13148-019-0743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorincz AT. Cancer diagnostic classifiers based on quantitative DNA methylation. Expert Rev Mol Diagn. 2016;14:293–305. doi: 10.1586/14737159.2014.897610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke MA, Luhn P, Gage JC, Bodelon C, Dunn ST, Walker J, Zuna R, Hewitt S, Killian JK, Yan L, et al. Discovery and validation of candidate host DNA methylation markers for detection of cervical precancer and cancer. Int J Cancer. 2017;141:701–710. doi: 10.1002/ijc.30781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kremer WW, Van Zummeren M, Novianti PW, Richter KL, Verlaat W, Snijders PJF, Heideman DAM, Steenbergen RDM, Dreyer G, Meijer CJ. Detection of hypermethylated genes as markers for cervical screening in women living with HIV. J Int AIDS Soc. 2018;21(e25165) doi: 10.1002/jia2.25165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verlaat W, Van Leeuwen RW, Novianti PW, Schuuring E, Meijer CJLM, Van Der Zee AGJ, Snijders PJF, Heideman DAM, Steenbergen RDM, Wisman GBA. Host-cell DNA methylation patterns during high-risk HPV-induced carcinogenesis reveal a heterogeneous nature of cervical pre-cancer. Epigenetics. 2018;13:769–778. doi: 10.1080/15592294.2018.1507197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong L, Wang L, Wang Z, Xiao X, Xu T, Wu H, Wu M, Liu P, Li L. DNA methylation for cervical cancer screening: A training set in China. Clin Epigenetics. 2020;12(91) doi: 10.1186/s13148-020-00885-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shivapurkar N, Gazdar AF. DNA methylation-based biomarkers in non-invasive cancer screening. Curr Mol Med. 2010;10:123–132. doi: 10.2174/156652410790963303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varghese VK, Shukla V, Kabekkodu SP, Pandey D, Satyamoorthy K. DNA methylation regulated microRNAs in human cervical cancer. Mol Carcinog. 2018;57:370–382. doi: 10.1002/mc.22761. [DOI] [PubMed] [Google Scholar]
- 50.Botezatu A, Goia-Rusanu CD, Iancu IV, Huica I, Plesa A, Socolov D, Ungureanu C, Anton G. Quantitative analysis of the relationship between microRNA-124a,-34b and-203 gene methylation and cervical oncogenesis. Mol Med Rep. 2011;4:121–128. doi: 10.3892/mmr.2010.394. [DOI] [PubMed] [Google Scholar]
- 51.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin FF, Wang N, Bi XN, Yu X, Xu XH, Wang YL, Zhao CQ, Luo B, Wang YK. Serine/threonine kinases 31(STK31) may be a novel cellular target gene for the HPV16 oncogene E7 with potential as a DNA hypomethylation biomarker in cervical cancer. Virol J. 2016;13(60) doi: 10.1186/s12985-016-0515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thangavelu PU, Krenács T, Dray E, Duijf PHG. In epithelial cancers, aberrant COL17A1 promoter methylation predicts its misexpression and increased invasion. Clin Epigenet. 2016;8(120) doi: 10.1186/s13148-016-0290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.García AD, Abba MC, Briceño I, Aristizabal FA, Arregui AC. DNA methylation pattern in high-grade cervical intraepithelial neoplasia and cancer revealed by genomewide methylation analysis of cervical DNA. Integr Mol Med. 2017;4:1–13. doi: 10.15761/IMM.1000309. [Google Scholar]
- 55.Miller JL, Grant PA. The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell Biochem. 2013;61:289–317. doi: 10.1007/978-94-007-4525-4_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greer EL, Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D'Oto A, Tian QW, Davidoff AM, Yang J. Histone demethylases and their roles in cancer epigenetics. J Med Oncol Ther. 2016;1:34–40. [PMC free article] [PubMed] [Google Scholar]
- 59.Groves IJ, Drane ELA, Michalski M, Monahan JM, Scarpini CG, Smith SP, Bussotti G, Várnai C, Schoenfelder S, Fraser P, et al. doi: 10.1371/journal.ppat.1009875. Three-dimensional interactions between integrated HPV genomes and cellular chromatin dysregulate host gene expression in early cervical carcinogenesis. bioRxiv: Feb. 3, 2021 2021 (Epub ahead of print). doi: 10.1101/2021.02.03.429496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sen P, Ganguly P, Ganguly N. Modulation of DNA methylation by human papillomavirus E6 and E7 oncoproteins in cervical cancer. Oncol Lett. 2018;15:11–22. doi: 10.3892/ol.2017.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mac M, Moody CA. Epigenetic regulation of the human papillomavirus life cycle. Pathogens. 2020;9(483) doi: 10.3390/pathogens9060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shadeo A, Chari R, Lonergan KM, Pusic A, Miller D, Ehlen T, Van Niekerk D, Matisic J, Richards-Kortum R, Follen M, et al. Up regulation in gene expression of chromatin remodelling factors in cervical intraepithelial neoplasia. BMC Genomics. 2008;9(64) doi: 10.1186/1471-2164-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanwal R, Gupta S. Epigenetic modifications in cancer. Clin Genet. 2012;81:303–311. doi: 10.1111/j.1399-0004.2011.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu Y, Chan YT, Tan HY, Li S, Wang N, Feng Y. Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol Cancer. 2020;19(79) doi: 10.1186/s12943-020-01197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng C, Dong J, Chang W, Cui M, Xu T. The progress of methylation regulation in gene expression of cervical cancer. Int J Genomics. 2018;2018(8260652) doi: 10.1155/2018/8260652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rathinasamy B, Velmurugan BK. Role of lncRNAs in the cancer development and progression and their regulation by various phytochemicals. Biomed Pharmacother. 2018;102:242–248. doi: 10.1016/j.biopha.2018.03.077. [DOI] [PubMed] [Google Scholar]
- 67.Wang X, Wang HK, Li Y, Hafner M, Banerjee NS, Tang S, Briskin D, Meyers C, Chow LT, Xie X, et al. microRNAs are biomarkers of oncogenic human papillomavirus infections. Proc Natl Acad Sci USA. 2014;111:4262–4267. doi: 10.1073/pnas.1401430111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jia W, Wu Y, Zhang Q, Gao GE, Zhang C, Xiang Y. Expression profile of circulating microRNAs as a promising fingerprint for cervical cancer diagnosis and monitoring. Mol Clin Oncol. 2015;3:851–858. doi: 10.3892/mco.2015.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kong Q, Tang Z, Xiang F, Jiang J, Yue H, Wu R, Kang X. Diagnostic value of Serum hsa-mir-92a in patients with cervical cancer. Clin Lab. 2017;63:335–340. doi: 10.7754/Clin.Lab.2016.160610. [DOI] [PubMed] [Google Scholar]
- 70.Laengsri V, Kerdpin U, Plabplueng C, Treeratanapiboon L, Nuchnoi P. Cervical cancer markers: Epigenetics and microRNAs. Lab Med. 2018;49:97–111. doi: 10.1093/labmed/lmx080. [DOI] [PubMed] [Google Scholar]
- 71.Gibb EA, Becker-Santos DD, Enfield KS, Guillaud M, Niekerk Dv, Matisic JP, Macaulay CE, Lam WL. Aberrant expression of long noncoding RNAs in cervical intraepithelial neoplasia. Int J Gynecol Cancer. 2012;22:1557–1563. doi: 10.1097/IGC.0b013e318272f2c9. [DOI] [PubMed] [Google Scholar]
- 72.Aalijahan H, Ghorbian S. Long non-coding RNAs and cervical cancer. Exp Mol Pathol. 2019;106:7–16. doi: 10.1016/j.yexmp.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 73.Burgers WA, Blanchon L, Pradhan S, de Launoit Y, Kouzarides T, Fuks F. Viral oncoproteins target the DNA methyltransferases. Oncogene. 2007;26:1650–1655. doi: 10.1038/sj.onc.1209950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laurson J, Khan S, Chung R, Cross K, Raj K. Epigenetic repression of E-cadherin by human papillomavirus 16 E7 protein. Carcinogenesis. 2010;31:918–926. doi: 10.1093/carcin/bgq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leonard SM, Wei W, Collins SI, Pereira M, Diyaf A, Constandinou-Williams C, Young LS, Roberts S, Woodman CB. Oncogenic human papillomavirus imposes an instructive pattern of DNA methylation changes which parallel the natural history of cervical HPV infection in young women. Carcinogenesis. 2012;33:1286–1293. doi: 10.1093/carcin/bgs157. [DOI] [PubMed] [Google Scholar]
- 76.Hsu CH, Peng KL, Jhang HC, Lin CH, Wu SY, Chiang CM, Lee SC, Yu WC, Juan LJ. The HPV E6 oncoprotein targets histone methyltransferases for modulating specific gene transcription. Oncogene. 2012;31:2335–2349. doi: 10.1038/onc.2011.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bernat A, Avvakumov N, Mymryk JS, Banks L. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene. 2003;22:7871–7881. doi: 10.1038/sj.onc.1206896. [DOI] [PubMed] [Google Scholar]
- 79.Thomas MC, Chiang CM. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol Cell. 2005;17:251–264. doi: 10.1016/j.molcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 80.Avvakumov N, Torchia J, Mymryk JS. Interaction of the HPV E7 proteins with the pCAF acetyltransferase. Oncogene. 2003;22:3833–3841. doi: 10.1038/sj.onc.1206562. [DOI] [PubMed] [Google Scholar]
- 81.Huang SM, McCance DJ. Down regulation of the interleukin-8 promoter by human papillomavirus type 16 E6 and E7 through effects on CREB binding protein/p300 and P/CAF. J Virol. 2002;76:8710–8721. doi: 10.1128/jvi.76.17.8710-8721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Münger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, Huh K. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jha S, Vande Pol S, Banerjee NS, Dutta AB, Chow LT, Dutta A. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol Cell. 2010;38:700–711. doi: 10.1016/j.molcel.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brehm A, Nielsen SJ, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999;18:2449–2458. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Longworth MS, Laimins LA. The binding of histone deacetylases and the integrity of zinc finger-like motifs of the E7 protein are essential for the life cycle of human papillomavirus type 31. J Virol. 2004;78:3533–3541. doi: 10.1128/jvi.78.7.3533-3541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Longworth MS, Wilson R, Laimins LA. HPV31 E7 facilitates replication by activating E2F2 transcription through its interaction with HDACs. EMBO J. 2005;24:1821–1830. doi: 10.1038/sj.emboj.7600651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McLaughlin-Drubin ME, Crum CP, Münger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci USA. 2011;108:2130–2135. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y, Tong J, Lin H, Ma L, Xu Y. CCHE1 accelerated the initiation of oral squamous cell carcinoma through enhancing PAK2 expression by sponging miR-922. J Oral Pathol Med. 2020;49:636–644. doi: 10.1111/jop.12995. [DOI] [PubMed] [Google Scholar]
- 89.Sharma S, Mandal P, Sadhukhan T, Roy Chowdhury R, Ranjan Mondal N, Chakravarty B, Chatterjee T, Roy S, Sengupta S. Bridging links between long noncoding RNA HOTAIR and HPV oncoprotein E7 in cervical cancer pathogenesis. Sci Rep. 2015;5(11724) doi: 10.1038/srep11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang J, Gao Y. Long non-coding RNA MEG3 inhibits cervical cancer cell growth by promoting degradation of P-STAT3 protein via ubiquitination. Cancer Cell Int. 2019;19(175) doi: 10.1186/s12935-019-0893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.He H, Liu X, Liu Y, Zhang M, Lai Y, Hao Y, Wang Q, Shi D, Wang N, Luo XG, et al. Human papillomavirus E6/E7 and long noncoding RNA TMPOP2 mutually upregulated gene expression in cervical cancer cells. J Virol. 2019;93:e01808–18. doi: 10.1128/JVI.01808-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X, Wang HK, McCoy JP, Banerjee NS, Rader JS, Broker TR, Meyers C, Chow LT, Zheng ZM. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15:637–647. doi: 10.1261/rna.1442309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Melar-New M, Laimins LA. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J Virol. 2010;84:5212–5221. doi: 10.1128/JVI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yao T, Lin Z. MiR-21 is involved in cervical squamous cell tumorigenesis and regulates CCL20. Biochim Biophys Acta. 2012;1822:248–260. doi: 10.1016/j.bbadis.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 95.Kottaridi C, Leventakou D, Pouliakis A, Pergialiotis V, Chrelias G, Patsouri E, Zacharatou A, Panopoulou E, Damaskou V, Sioulas V, et al. Searching HPV genome for methylation sites involved in molecular progression to cervical precancer. J Cancer. 2019;10:4588–4595. doi: 10.7150/jca.30081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsu YW, Huang RL, Su PH, Chen YC, Wang HC, Liao CC, Lai HC. Genotype-specific methylation of HPV in cervical intraepithelial neoplasia. J Gynecol Oncol. 2017;28(e56) doi: 10.3802/jgo.2017.28.e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lorincz AT, Brentnall AR, Scibior-Bentkowska D, Reuter C, Banwait R, Cadman L, Austin J, Cuzick J, Vasiljević N. Validation of a DNA methylation HPV triage classifier in a screening sample. Int J Cancer. 2016;138:2745–2751. doi: 10.1002/ijc.30008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gu YY, Zhou GN, Wang Q, Ding JX, Hua KQ. Evaluation of a methylation classifier for predicting pre-cancer lesion among women with abnormal results between HPV16/18 and cytology. Clin Epigenetics. 2020;12(57) doi: 10.1186/s13148-020-00849-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sahasrabuddhe VV, Luhn P, Wentzensen N. Human papillomavirus and cervical cancer: Biomarkers for improved prevention efforts. Future Microbiol. 2011;6:1083–1098. doi: 10.2217/fmb.11.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wilting SM, Steenbergen RDM. Molecular events leading to HPV-induced high grade neoplasia. Papillomavirus Res. 2016;2:85–88. doi: 10.1016/j.pvr.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Au Yeung CL, Tsang WP, Tsang TY, Co NN, Yau PL, Kwok TT. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol Rep. 2010;24:1599–1604. doi: 10.3892/or_00001023. [DOI] [PubMed] [Google Scholar]
- 102.Yeo-Teh NSL, Ito Y, Jha S. High-risk human papillomaviral oncogenes E6 and E7 target key cellular pathways to achieve oncogenesis. Int J Mol Sci. 2018;19(1706) doi: 10.3390/ijms19061706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leonard SM, Wei W, Collins SI, Pereira M, Diyaf A, Constandinou-Williams C, Young LS, Roberts S, Woodman CB. Oncogenic human papillomavirus imposes an instructive pattern of DNA methylation changes which parallel the natural history of cervical HPV infection in young women. Carcinogenesis. 2012;33:1286–1293. doi: 10.1093/carcin/bgs157. [DOI] [PubMed] [Google Scholar]
- 104.Kalantari M, Osann K, Calleja-Macias IE, Kim S, Yan B, Jordan S, Chase DM, Tewari KS, Bernard HU. Methylation of human papillomavirus 16, 18, 31, and 45 L2 and L1 genes and the cellular DAPK gene: Considerations for use as biomarkers of the progression of cervical neoplasia. Virology. 2014;448:314–321. doi: 10.1016/j.virol.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brentnall AR, Vasiljević N, Scibior-Bentkowska D, Cadman L, Austin J, Szarewski A, Cuzick J, Lorincz AT. A DNA methylation classifier of cervical precancer based on human papillomavirus and human genes. Int J Cancer. 2014;135:1425–1432. doi: 10.1002/ijc.28790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brentnall AR, Vasiljevic N, Scibior-Bentkowska D, Cadman L, Austin J, Cuzick J, Lorincz AT. DNA methylation assay for HPV33 contributes independent triage information to HPV16, HPV18, HPV31, and EPB41L3 for detecting cervical pre-cancer. Cancer Biomark. 2015;15:669–675. doi: 10.3233/CBM-150507. [DOI] [PubMed] [Google Scholar]
- 107. Qiagen: Products: Cervical Cancer Screening. https://www.qiagen.com/be/products/diagnostics-and-clinical-research/sexual-reproductive-health/cervical-cancer-screening/qiasure-methylation-test-kit-eu/#productdetails. Accessed June 16, 2021. [Google Scholar]
- 108.Dippmann C, Schmitz M, Wunsch K, Schütze S, Beer K, Greinke C, Ikenberg H, Hoyer H, Runnebaum IB, Hansel A, Dürst M. Triage of hrHPV-positive women: Comparison of two commercial methylation-specific PCR assays. Clin Epigenetics. 2020;12(171) doi: 10.1186/s13148-020-00963-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schmitz M, Eichelkraut K, Schmidt D, Zeiser I, Hilal Z, Tettenborn Z, Hansel A, Ikenberg H. Performance of a DNA methylation marker panel using liquid-based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer. 2018;18(1197) doi: 10.1186/s12885-018-5125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kocsis A, Takács T, Jeney C, Schaff Z, Koiss R, Járay B, Sobel G, Pap K, Székely I, Ferenci T, et al. Performance of a new HPV and biomarker assay in the management of hrHPV positive women: Subanalysis of the ongoing multicenter TRACE clinical trial (n > 6,000) to evaluate POU4F3 methylation as a potential biomarker of cervical precancer and cancer. Int J Cancer. 2017;140:1119–1133. doi: 10.1002/ijc.30534. [DOI] [PubMed] [Google Scholar]
- 111.Kan YY, Liou YL, Wang HJ, Chen CY, Sung LC, Chang CF, Liao CI. PAX1 methylation as a potential biomarker for cervical cancer screening. Int J Gynecol Cancer. 2014;24:928–934. doi: 10.1097/IGC.0000000000000155. [DOI] [PubMed] [Google Scholar]
- 112.Liou YL, Zhang TL, Yan T, Yeh CT, Kang YN, Cao L, Wu N, Chang CF, Wang HJ, et al. Combined clinical and genetic testing algorithm for cervical cancer diagnosis. Clin Epigenetics. 2016;8(66) doi: 10.1186/s13148-016-0232-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Witjes JA, Morote J, Cornel EB, Gakis G, van Valenberg FJP, Lozano F, Sternberg IA, Willemsen E, Hegemann ML, Paitan Y, Leibovitch I. Performance of the Bladder EpiCheck™ Methylation Test for Patients Under Surveillance for Non-muscle-invasive Bladder Cancer: Results of a Multicenter, Prospective, Blinded Clinical Trial. Eur Urol Oncol. 2018;1:307–313. doi: 10.1016/j.euo.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 114.Wasserstrom A, Frumkin D, Dotan Z, Bukin E, Gadish T, Hanuka S, Knirsh R, Darawsha AE, Leibovitch I. MP13-15 molecular urine cytology-bladder epicheck is a novel molecular diagnostic tool for monitoring of bladder cancer patients. J Urol. 2016;195(e140) [Google Scholar]
- 115.D'Andrea D, Soria F, Zehetmayer S, Gust KM, Korn S, Witjes JA, Shariat SF. Diagnostic accuracy, clinical utility and influence on decision-making of a methylation urine biomarker test in the surveillance of non-muscle-invasive bladder cancer. BJU Int. 2019;123:959–967. doi: 10.1111/bju.14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Su SF, de Castro Abreu AL, Chihara Y, Tsai Y, Andreu-Vieyra C, Daneshmand S, Skinner EC, Jones PA, Siegmund KD, Liang G. A panel of three markers hyper- and hypomethylated in urine sediments accurately predicts bladder cancer recurrence. Clin Cancer Res. 2014;20:1978–1989. doi: 10.1158/1078-0432.CCR-13-2637. [DOI] [PubMed] [Google Scholar]
- 117.Van Kessel KE, Beukers W, Lurkin I, Ziel-van der Made A, van der Keur KA, Boormans JL, Dyrskjøt L, Márquez M, Ørntoft TF, Real FX, et al. Validation of a DNA Methylation-Mutation Urine Assay to Select Patients with Hematuria for Cystoscopy. J Urol. 2017;197:590–595. doi: 10.1016/j.juro.2016.09.118. [DOI] [PubMed] [Google Scholar]
- 118.Feber A, Dhami P, Dong L, de Winter P, Tan WS, Martínez-Fernández M, Paul DS, Hynes-Allen A, Rezaee S, Gurung P, et al. UroMark-a urinary biomarker assay for the detection of bladder cancer. Clin Epigenetics. 2017;9(8) doi: 10.1186/s13148-016-0303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Maier S, Nimmrich I, Koenig T, Eppenberger-Castori S, Bohlmann I, Paradiso A, Spyratos F, Thomssen C, Mueller V, Nährig J, et al. DNA-methylation of the homeodomain transcription factor PITX2 reliably predicts risk of distant disease recurrence in tamoxifen-treated, node-negative breast cancer patients-Technical and clinical validation in a multi-centre setting in collaboration with the European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group. Eur J Cancer. 2007;43:1679–1686. doi: 10.1016/j.ejca.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 120.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 121.Oussalah A, Rischer S, Bensenane M, Conroy G, Filhine-Tresarrieu P, Debard R, Forest-Tramoy D, Josse T, Reinicke D, Garcia M, et al. Plasma mSEPT9: A novel circulating cell-free DNA-based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine. 2018;30:138–147. doi: 10.1016/j.ebiom.2018.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chalasani NP, Ramasubramanian T, Bruinsma JJ, Allawi HT, Olson M, Roberts LR, Kisiel J, Reddy KR, Lidgard GP, et al. Combined methylated DNA and protein markers: An accurate blood-based test for early-stage detection of Hepatocellular carcinoma. Hepatology. 2019;70 (Suppl 1)(109) [Google Scholar]
- 123.Taggart D, Roy D, Li G, Liu D, Zheng L, Zhang K. DNA methylation biomarkers for noninvassive detection of hepatocellular carcinoma [abstract]. SITC 2018 Abstracts 263, 2018. https://sitc.sitcancer.org/2018/abstracts/titles/poster/. [Google Scholar]
- 124.Johannessen LE, Brandal P, Myklebust T, Heim S, Micci F, Panagopoulos I. MGMT Gene Promoter methylation status-Assessment of two pyrosequencing kits and three methylation-specific PCR methods for their predictive capacity in glioblastomas. Cancer Genomics Proteomics. 2018;15:437–448. doi: 10.21873/cgp.20102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Waterhouse RL Jr, Van Neste L, Moses KA, Barnswell C, Silberstein JL, Jalkut M, Tutrone R, Sylora J, Anglade R, Murdock M, et al. Evaluation of an epigenetic assay for predicting repeat prostate biopsy outcome in African American men. Urology. 2019;128:62–65. doi: 10.1016/j.urology.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Weiss G, Schlegel A, Kottwitz D, König T, Tetzner R. Validation of the SHOX2/PTGER4 DNA Methylation Marker Panel for Plasma-Based Discrimination between Patients with Malignant and Nonmalignant Lung Disease. J Thorac Oncol. 2017;12:77–84. doi: 10.1016/j.jtho.2016.08.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hao YX, Fu Q, Guo YY, Ye M, Zhao HX, Wang Q, Peng XM, Li QW, Wang RL, Xiao WH. Effectiveness of circulating tumor DNA for detection of KRAS gene mutations in colorectal cancer patients: A meta-analysis. Onco Targets Ther. 2017;10:945–953. doi: 10.2147/OTT.S123954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moran S, Martínez-Cardús A, Sayols S, Musulén E, Balañá C, Estival-Gonzalez A, Moutinho C, Heyn H, Diaz-Lagares A, de Moura MC, et al. Epigenetic profiling to classify cancer of unknown primary: A multicentre, retrospective analysis. Lancet Oncol. 2016;17:1386–1395. doi: 10.1016/S1470-2045(16)30297-2. [DOI] [PubMed] [Google Scholar]
- 130.Snoek BC, Splunter APV, Bleeker MCG, Ruiten MC, Heideman DAM, Rurup WF, Verlaat W, Schotman H, Gent MV, Trommel NEV, Steenbergen RDM. Cervical cancer detection by DNA methylation analysis in urine. Sci Rep. 2019;9(3088) doi: 10.1038/s41598-019-39275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Darwiche N. Epigenetic mechanisms and the hallmarks of cancer: An intimate affair. Am J Cancer Res. 2020;10:1954–1978. [PMC free article] [PubMed] [Google Scholar]
- 132.Locke WJ, Guanzon D, Ma C, Liew YJ, Duesing KR, Fung KYC, Ross JP. DNA methylation cancer biomarkers: Translation to the clinic. Front Genet. 2019;10(1150) doi: 10.3389/fgene.2019.01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9(86) doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Meng Y, Sun J, Wang X, Hu T, Ma Y, Kong C, Piao H, Yu T, Zhang G. Exosomes: A promising avenue for the diagnosis of breast cancer. Technol Cancer Res Treat. 2019;18(1533033818821421) doi: 10.1177/1533033818821421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bolha L, Ravnik-Glavač M, Glavač D. Long Noncoding RNAs as biomarkers in cancer. Dis Markers. 2017;2017(7243968) doi: 10.1155/2017/7243968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.