Abstract
Several studies of the yeast Saccharomyces cerevisiae support differential regulation of heat shock mRNA (hs mRNA) and non-hs mRNA nuclear export during stress. These include the finding that hs mRNA export at 42°C is inhibited in the absence of the nucleoporinlike protein Rip1p (also called Nup42p) (C. A. Saavedra, C. M. Hammell, C. V. Heath, and C. N. Cole, Genes Dev. 11:2845–2856, 1997; F. Stutz, J. Kantor, D. Zhang, T. McCarthy, M. Neville, and M. Rosbash, Genes Dev. 11:2857–2868, 1997). However, the results reported in this paper provide little evidence for selective non-hs mRNA retention or selective hs mRNA export under heat shock conditions. First, we do not detect a block to non-hs mRNA export at 42°C in a wild-type strain. Second, hs mRNA export appears to be mediated by the Ran system and several other factors previously reported to be important for general mRNA export. Third, the export of non-hs mRNA as well as hs mRNA is inhibited in the absence of Rip1p at 42°C. As a corollary, we find no evidence for cis-acting hs mRNA sequences that promote transport during heat shock. Taken together, our data suggest that a shift to 42°C in the absence of Rip1p impacts a late stage of transport affecting most if not all mRNA.
In eukaryotic cells, macromolecules are constantly moving between the nucleus and the cytoplasm. Transport occurs through nuclear pore complexes (NPCs), which are imbedded in the double membrane surrounding the nucleus. The NPC is 66 MDa in the yeast Saccharomyces cerevisiae and consists of about 50 different nuclear pore proteins (nucleoporins) (4, 34, 37, 53). Nucleocytoplasmic transport of all macromolecular substrates studied to date is receptor mediated, energy dependent, and saturable (reviewed in references 27 and 33).
Accumulating data on protein import and export point to common principles. To access an NPC, transport substrates need to be recognized by soluble receptors. Most well-characterized receptors belong to a family of proteins called β-importins or β-karyopherins, which recognize specific sequences within their respective substrates. The directionality of transport, nucleus to cytoplasm or cytoplasm to nucleus, is determined in large part by a predicted asymmetry in the intracellular nucleotide-bound status of a key transport player, the small GTPase Ran (Gsp1p in yeast) (17, 18, 28; reviewed in reference 9). Due to the nuclear localization of the Ran GTP exchange factor (Prp20p in yeast) and the cytoplasmic localization of the Ran GTPase-activating protein (Rna1p in yeast), nuclear Ran is mostly in the GTP-bound form, whereas cytoplasmic Ran is mostly GDP bound. Nuclear Ran-GTP promotes the assembly of several export complexes that are formed by a cooperative association between export cargo, receptor, and Ran-GTP. Ran-GTP hydrolysis in the cytoplasm promotes the dissociation of such export complexes, whereas Ran-GTP in the nucleus promotes the dissociation of import complexes.
Upon transcription and processing, mRNA becomes associated with many different RNA-binding proteins, forming heterogeneous nuclear ribonucleoprotein (hnRNP) particles (6). Some hnRNPs have been shown to remain associated with mRNA during transport to the cytoplasm, leading to the hypothesis that hnRNPs contain export signals and serve as adaptors recognized by export receptors (reviewed in references 29 and 30). This idea is also based on retroviral systems, in which RNA-binding proteins recognize both specific sequences within viral mRNA and the export receptor Crm1p (also called Xpo1p), resulting in the export of unspliced viral mRNA (reviewed in reference 50). Crm1p is a protein exporter with no major role in general mRNA export (32, 50), suggesting that Rev-like nuclear export signals (NESs) do not make a major contribution to mRNA export. Moreover, there are no known functional Rev-like NESs in any hnRNP, and there are no reported interactions between an hnRNP and a known β-karyopherin-like export receptor. Indeed, the few identified mRNA export factors do not fall into the β-karyopherin class of soluble transport receptors (reviewed in references 27 and 50). These factors (human TAP, or yeast Mex67p; human p15, or yeast Mtr2p; yeast Gle1p, or Rss1p; yeast Rip1p; and yeast Dbp5p, or Rat8p) are all characterized by at least transient association with the NPC.
Microinjection-competition experiments with Xenopus oocytes pointed to the existence of nonoverlapping export pathways for different classes of RNA molecules (19). This strategy also indicated multiple separable mRNA export pathways (17, 18, 35, 38). Previous studies of mRNA export during stress in the yeast S. cerevisiae strongly supported this notion, in that exposure to 42°C or 10% ethanol resulted in pronounced nuclear accumulation of non-hs mRNA, whereas hs mRNA was efficiently exported (39). In further support of separate transport pathways for hs and non-hs mRNAs, it was suggested that hs mRNA contains cis-acting sequences that allow its preferential export under stress conditions. hs mRNA export was also proposed not to require Ran and its auxiliary proteins, unlike the transport of non-hs mRNA. The discovery that hs mRNA export at 42°C requires the nucleoporinlike protein Rip1p supported this view of a specialized transport route (40, 47).
However, our studies indicate that hs mRNA export is mediated by the Ran system and many other factors involved in non-hs mRNA export. Furthermore, we do not observe a block to non-hs mRNA export at 42°C in a wild-type strain. Finally, we show that nuclear export of various non-hs as well as hs mRNAs is severely affected in the absence of Rip1p at 42°C. One can therefore picture mRNA transport under both normal and stress conditions as a competition among different mRNA molecules for common transport factors.
MATERIALS AND METHODS
DNA manipulations and yeast transformations were performed using standard protocols (1, 13, 26). The yeast strains used in this study are described in Table 1.
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| W303 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 canR1-100 | 48 |
| W303ΔRIP1 | MATa ΔRIP1::KANr; otherwise isogenic to W303 | 48 |
| rna1-1 | MATa ura3-52 leu2Δ1 trp1 rna1-1 | 3 |
| prp20-1 | MATα ura3-52 leu2Δ1 trp1Δ63 prp20-1 | 21 |
| prp20-7 | MATa ura3-52 his3Δ200 tyr1 ade2-1 prp20-7 | 39 |
| prp20-101 | MATa ura3-52 leu2Δ1 his3Δ200 Gal+ prp20-101 | 51 |
| ΔYRB2 | MATa ura3 leu2 ade2 trp1 YRB2::HIS3 | 51 |
| mex67-5 | MATa ade2 leu2 ura3 trp1 MEX67::HIS3 pRS314-TRP1-mex67-5 | 41 |
| mtr2-9 | MATa ade2 leu2 ura3 trp1 MTR2::HIS3 pRS315-LEU2-mtr2-9 | 41 |
| xpo1-1 | MATα ade2 his3 trp1 ura3 can1 XPO1::LEU2 pKW457-HIS3-xpo1-1 | 45 |
| PSY580 | MATa ura3-52 trp1Δ63 leu2Δ1 Gal+ | 42 |
| pse1-1 | MATa ura3-52 trp1Δ63 leu2Δ1 Gal+ pse1-1 | 42 |
| ΔKAP123 | MATα ura3-52 leu2Δ1 KAP123::HIS3 | 42 |
| ΔSXM1 | MATa ura3-52 leu2Δ1 trp1Δ63 ADE+ SXM1::HIS3 | 42 |
| rat8-2 (or dbp5-2) | MATa leu2Δ1 trp1Δ63 ura3-52 rat8-2 | 44 |
| gle1-8 | MATa ade2 his3 leu2 trp1 ura3 gle1-8 | F. Stutz |
| rat7-1 (or nup159-1) | MATa trp1Δ63 ura3-52 leu2Δ1 rat7-1 | 10 |
| ΔNUP133 | MATα ade2 his3 leu2 trp1 ura3 NUP133::HIS3 | 5 |
| ΔNUP100 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 nup100-3::TRP1 | 52 |
| ΔNUP116 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 nup116-5::HIS3 | 52 |
| ΔNUP145 | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 nup145-1::URA3 | 52 |
| NUP82Δ108 | MATa ade2 leu2 ura3 trp1 NUP82::HIS3 pRS316-URA-NUP82Δ108 | 16 |
| nup49-313 | MATα ade2 ade3 his3 leu2 ura3 NUP49::TRP1 pUN100-LEU2-nup49-313 | 5 |
| ΔNUP57 | MATa ade2 leu2 ura3 trp1 NUP57::HIS3 | E. Hurt, V. Doye |
| rpb1-1 | MATα leu2 ura3 Gal+ rpb1-1 | R. Young |
Plasmid construction. (i) pHS-GFP.
The SSA4 heat shock promoter and 5′ untranslated region (UTR) (HS) were PCR amplified from pEC702 (SSA4 gene in YEp351; a gift of E. Craig, University of Wisconsin) using primers ACGTACGGCGGCCGCCTGATACCTTCCATACTAGAAAAG and ACGTACGCTAGCCATGATTATTGTTTTGTTTATTTT; green fluorescent protein (GFP) was PCR amplified from pJK19-1 (a gift from P. Silver, Harvard University) using primers AAAAGAATTCATGGCTAGCAAAGGAGAAGAACTC and ACGTACAAGCTTTTAGCAGCCGGATCCTTTGTATAGTTC. HS and GFP were digested with NheI, ligated, PCR amplified with terminal primers, and cloned into pRS316 using NotI and HindIII to generate pHS-GFP.
(ii) pHS-GFP-3′ SSA4.
The SSA4 3′ UTR was PCR amplified from pEC702 using primers ACGTACAAGCTTATAAATACAAAGATGCGATGAAGT and ACGTACCTCGAGTATGATTGCTGTACATTTCCGAGC and cloned into pHS-GFP using HindIII and XhoI to generate pHS-GFP-3′ SSA4.
(iii) pHS-GFP*SSA4-3′ SSA4.
SSA4 was PCR amplified from pEC702 using primers ACGTACGGATCCGGCTGCTAAATGTCAAAAGCTGTTGGTATTGAT and ACGTACAAGCTTCTAATCAACCTCTTCAACCGTTGG and cloned into pHS-GFP-3′ SSA4 using BamHI and HindIII to generate pHS-GFP*SSA4-3′ SSA4.
Plasmid Gal-LacZ was identical to the previously described pLGSD5 plasmid (23).
In vivo protein labeling.
In vivo protein labeling was performed as described by Stutz et al. (48) with the following modifications. Yeast cultures were grown overnight at 25°C to an optical density at 600 nm (OD600) of 0.05 to 0.1. The cells were pelleted, resuspended in the same volume of medium lacking methionine (Met−), and grown for another 2 h at 25°C. The cultures were rapidly mixed with 1 volume of Met− medium preheated to 49 or 59°C and incubated as shaking cultures at 37 or 42°C, respectively. Control samples (25°C) were mixed with 1 volume of 25°C Met− medium. After 10 to 15 min (for single-time-point assays) or at various times after temperature shift (for a time course), 1-ml samples were withdrawn and mixed with 50 μCi of Trans35S Label (1,191 Ci/mmol; 11.02 mCi/ml; ICN Pharmaceuticals, Inc.) and incubated for an additional 20 min at the relevant temperature. Protein labeling was stopped by centrifugation at 4°C, medium removal, and immediate freezing on dry ice. The samples were resuspended in 30 μl of 1× sodium dodecyl sulfate (SDS) sample buffer, boiled for 10 min, and spun in a microcentrifuge for 5 min prior to being loaded on a 7.5% SDS-polyacrylamide gel. The gels were dried, and bands were visualized by autoradiography.
Thermotolerance assays.
Yeast cultures were grown overnight at 25°C to an OD600 of 0.05 to 0.1. Aliquots (5 ml) of each culture were rapidly mixed with an equivalent volume of medium at 25, 49, or 59°C and incubated at 25, 37, 42, or 50°C. For 42°C thermotolerance experiments, the cells were either incubated directly at 42°C for up to 6 h or pretreated at 37°C for 30 min prior to incubation at 42°C. For 50°C thermotolerance experiments, the cells were either incubated directly at 50°C for 20 min or pretreated at 42°C for 30 min prior to incubation at 50°C. Samples (1 ml) were withdrawn, chilled on ice, and serially diluted in sterile water. Eight microliters of each serial dilution was spotted on yeast extract-peptone-dextrose plates, and the plates were incubated for 2 days at 25°C.
Sample preparation for RNA and protein analyses.
Yeast cultures were grown overnight at 25°C to an OD600 of 0.05 to 0.1. Twenty-five milliliters of each culture was mixed with an equivalent volume of medium at 25, 49, or 59°C and incubated as shaking cultures at 25, 37, or 42°C, respectively. At time points after temperature shift (for a time course), two 1-ml samples were withdrawn (one for Western blot analysis and one for RNA analysis) and centrifuged at 4°C, and the cell pellets were immediately frozen on dry ice.
Cultures harboring Gal-LacZ were treated as described above with the following modifications. Cells were grown overnight in medium lacking glucose (2% lactate [pH 5.5], 2% glycerol). At 10 to 15 min after temperature shift, 20% galactose was added to a final concentration of 2%, and the incubation was continued at the relevant temperature.
RNA extractions and primer extensions.
RNA extractions and primer extensions were performed as described previously (36) using three different oligonucleotide primers. Oligonucleotide primer DT320 (CACCAGTGAGACGGGC) is complementary to positions 27 to 42 of the initiation codon for the β-galactosidase coding sequence in pLGSD5 (23). Oligonucleotide primer IV99 (GGTAGCTTCCCAGTAGTGC) is complementary to positions 167 to 186 of the initiation codon in the GFP coding sequence. Oligonucleotide primer DT58 (GCCAAAAAATGTGTATTGTAA), which is complementary to U2 snRNA and gives an approximately 120-base primer extended product, was used as an internal control for loading. Samples were loaded on 5 to 7% polyacrylamide denaturing gels, and bands were visualized by autoradiography.
Western blot analysis.
Frozen cell pellets were resuspended in 30 μl of 1× SDS sample buffer, boiled for 10 min, and spun in a microcentrifuge for 5 min prior to being loaded on a 7% (for LacZ) or 10% (for GFP) SDS-polyacrylamide gel. Transfer to nitrocellulose filters was performed by standard protocols (1). The filters were incubated for 2 h at room temperature with either rabbit α-GFP polyclonal antibody (1:100 dilution; Clontech) or mouse α-β-galactosidase monoclonal antibody (1:2,000; Boehringer Mannheim). Immunoreactive bands were detected by enhanced chemiluminescence (Amersham Life Science, Inc.).
In situ hybridization assays with Cy3 fluorochrome-conjugated oligonucleotides.
Briefly, cells were grown at 30°C to an OD600 of 0.2 and shifted to the appropriate temperature for 1 h prior to fixation. In the case of 42°C cultures, the cells were diluted with an equal volume of 54°C medium to ensure a rapid temperature shift. In situ hybridization to detect poly(A)+ RNA and SSA4 mRNA was performed as described previously (24). A mixture of two oligonucleotides complementary to the SSA4 3′ UTR (GTT*AAGAGGGAAAACT*AAGAAATTCGAT*GCTGCTACTT*CATCGCATCTT*TG and GAGAACGT*ACAAATAGTAGT*CATTTGCTAAT*TACTGATTGT*GTATCTTATAT*AT) was used to localize SSA4 mRNA. T* represents 5′-dimethoxytrityl-S-[N-(trifluoroacetylaminohexyl)-3-acrylimido]-2′-deoxyuridine, 3′-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite (Amino-Modifier dT; Glen Research), that was subsequently coupled to Cy3 fluorochrome (Amersham Pharmacia Biotech). PolyA+ RNA was localized using an oligo(dT)70, including seven T* residues spaced approximately 10 nucleotides apart.
RESULTS
A ΔRIP1 strain is deficient in hs mRNA nuclear export as determined by thermotolerance assays.
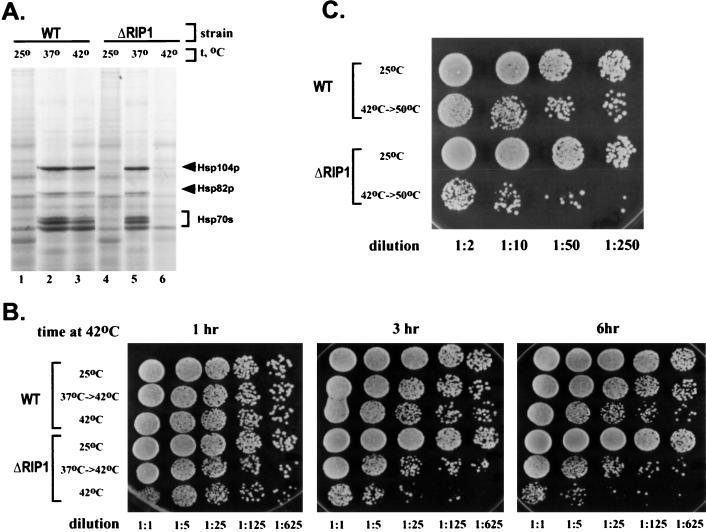
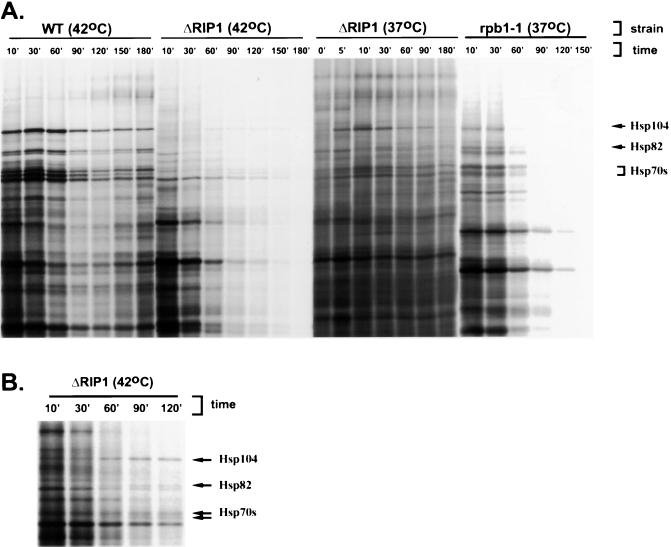
It has been previously shown that hs mRNA nuclear export is severely inhibited in a ΔRIP1 strain, which carries a deletion of the gene encoding the nucleoporinlike protein Rip1p (40, 48). hs mRNA export in the ΔRIP1 strain was first assayed indirectly, by pulse-labeling with [35S]methionine followed by SDS-polyacrylamide gel electrophoresis (PAGE) after transcriptional induction at high temperatures (48) (Fig. 1A). Consistent with previous results, labeled bands corresponding to major heat shock proteins were absent from the ΔRIP1 strain at 42°C but present at 37°C, indicating that the mutant phenotype of the ΔRIP1 strain is observed only at temperatures higher than 37°C (Fig. 1A, lanes 5 and 6).
FIG. 1.
A ΔRIP1 strain is deficient in nuclear export of hs mRNA at 42°C. (A) SDS-PAGE of total cellular proteins labeled with [35S]methionine in wild-type (WT) and ΔRIP1 strains treated for 30 min at 25, 37, or 42°C. The positions of the stress-inducible yeast heat shock proteins Hsp104p, Hsp82p, and Hsp70s are shown by arrows. (B and C) Thermotolerance assays. Each row represents serial dilutions of the same culture. The dilution factor is indicated at the bottom of each column. The number of colonies corresponds to the number of cells that have survived the treatment. (B) 42°C thermotolerance. Wild-type and ΔRIP1 strains were either maintained at 25°C or incubated at 42°C for various times, with or without pretreatment at 37°C prior to the 42°C incubation. The three plates show the number of cells that survived the 42°C treatment for 1, 3, and 6 h, respectively. (C) 50°C thermotolerance. Wild-type and ΔRIP1 strains were either maintained at 25°C or pretreated for 30 min at 42°C prior to a 20-min 50°C treatment.
The hs mRNA export defect of the ΔRIP1 strain results in a lower thermotolerance. Thus, the viability of the ΔRIP1 strain is compromised after incubation at 42°C for more than 1 h (Fig. 1B). Since the absence of Rip1p does not affect heat shock protein synthesis at 37°C, a 30-min pretreatment at 37°C restores the viability of the ΔRIP1 strain at 42°C almost to wild-type levels (Fig. 1B). When wild-type and ΔRIP1 strains were treated for 20 min at the lethal temperature of 50°C, the survival rates of the two strains were similar and very low (data not shown). However, when these strains were pretreated for 30 min at 42°C prior to the 50°C shift, wild-type survival was significantly increased whereas the survival of the ΔRIP1 strain was virtually unchanged (Fig. 1C and data not shown). On the basis of multiple experiments, we estimate that under the latter conditions, the thermotolerance of the wild type is at least 30 times higher than that of the ΔRIP1 strain.
Nuclear export of hs mRNA is mediated by Ran and other factors involved in non-hs mRNA export.
Based on in situ hybridization with oligo(dT)- and hs mRNA-specific probes, it has previously been suggested that hs mRNA is exported from the nucleus via a different pathway than non-hs mRNA (39). However, hs mRNA nuclear export may still contain features in common with other export routes. Therefore, we investigated hs mRNA export in various temperature-sensitive yeast mutants previously shown to produce nuclear accumulation of non-hs mRNA at nonpermissive temperatures. We used a combination of protein labeling and thermotolerance assays to determine if these factors are involved in hs mRNA nuclear export.
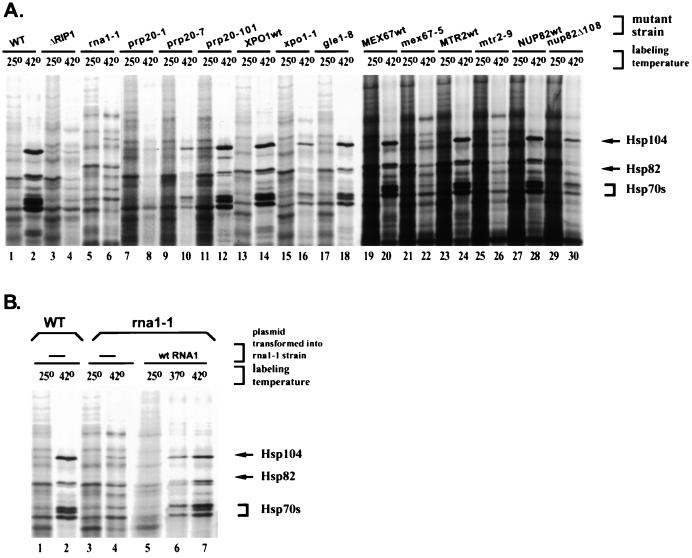
Surprisingly, many of the mutants had a pronounced defect in heat shock protein production, as determined by 35S labeling at 42°C (Fig. 2A). Most importantly, heat shock protein bands were completely absent in rna1-1 and prp20-1 strains after 30 min at 42°C (Fig. 2A, lanes 6 and 8). The wild-type protein-labeling pattern was restored by transformation with the corresponding wild-type genes (Fig. 2B, lanes 4 and 7, and data not shown). The rna1-1 and prp20-1 strains have mutations in the Ran GTPase-activating protein and the Ran GTP exchange factor, respectively, indicating that the Ran system is involved in hs mRNA export. There was a clear thermotolerance defect in rna1-1 and prp20-1 strains which was reversed by transformation with the wild-type genes (Fig. 3). We analyzed three different PRP20 mutants (prp20-1, prp20-7, and prp20-101) and observed a correlation between the severity of the heat shock protein synthesis defect and the severity of the growth phenotypes in thermotolerance assays (Fig. 2A, lanes 8, 10, and 12, and data not shown).
FIG. 2.
Several poly(A)+ RNA transport mutants show a defect in heat shock protein synthesis at 42°C. SDS-PAGE of total cellular proteins labeled with [35S]-methionine in wild-type (WT) and various mutant strains treated for 30 min at 25 or 42°C is shown. The positions of the stress-inducible yeast heat shock proteins Hsp104p, Hsp82p, and Hsp70s are shown by arrows. (B) Wild-type heat shock protein labeling pattern is restored upon transformation of rna1-1 with a plasmid carrying the wild-type RNA1 gene.
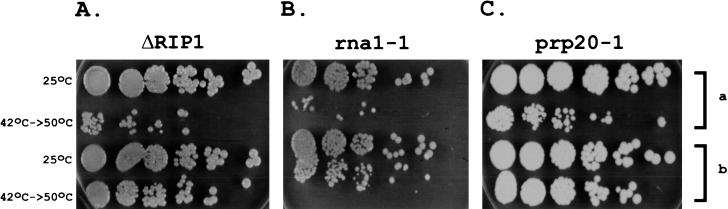
FIG. 3.
A block in hs mRNA export in ΔRIP1 (A), rna1-1 (B), and prp20-1 (C) strains is reflected in lowered thermotolerance. Yeast cultures were either maintained at 25°C or pretreated at 42°C for 30 min prior to a 20-min 50°C incubation. Each row represents serial dilutions of the same culture (the dilution factors are the same as in Fig. 1C). The number of growing colonies corresponds to the number of cells that survived the treatment. The thermotolerance defect of the strains transformed with empty vector (a) can be reversed (b) upon transformation with pRIPHi (A), pRNA1 (B), or pPRP20 (C).
In addition to Ran system mutants, we observed inhibition of hs mRNA export in mex67-5 and mtr2-9 strains (Fig. 2A, lanes 22 and 26), which carry mutations in factors shown to play an important role in the nuclear export non-hs mRNA (20, 41, 43). We also investigated the 42°C labeling pattern in the xpo1-1 strain. This strain carries a temperature-sensitive mutation in the β-karyopherinlike protein Crm1p, implicated in NES-mediated export (45). The xpo1-1 strain showed a lower level of heat shock protein labeling, but the overall pattern was identical to that of either the wild-type or xpo1-1 strain transformed with a vector carrying wild-type XPO1 (Fig. 2A, lanes 2, 14, and 16). This suggests that Crm1p is not a major export receptor for hs mRNA. It has recently been proposed that Crm1p is also not a major export receptor for non-hs mRNA (32).
It should be noted that some other nuclear export mutant (gle1-8, dbp5-2, NUP82Δ108, ΔNUP57, ΔNUP145, ΔYRB2, nup49-313, ΔNUP133, pse1-1, ΔSXM1, and ΔKAP123) strains produced a wild-type labeling pattern (Fig. 2A, lanes 18 and 30, and data not shown). As certain alleles of these genes have been previously demonstrated to accumulate nuclear non-hs mRNA at nonpermissive temperatures, the absence of an effect in our experiments might be due to the singular experimental protocol: for example, a 10- to 15-min preincubation at 42°C may not be sufficient to induce a mutant phenotype. These strains were not investigated in more detail. Taken together, the data indicate that hs mRNA nuclear export is mediated by Ran and other factors involved in the export of non-hs mRNA.
Nuclear export of several non-hs mRNAs is severely inhibited in a ΔRIP1 strain at 42°C.
Previous observations suggested that Rip1p is an NPC-associated factor specialized in mediating hs mRNA export under stress conditions (40, 48). We therefore decided to test whether Rip1p is indeed specific for the hs mRNA export pathway or whether the export of other mRNAs is also affected in the ΔRIP1 strain. To directly visualize mRNA, we performed in situ hybridization to detect the general poly(A)+ RNA population (Fig. 4).
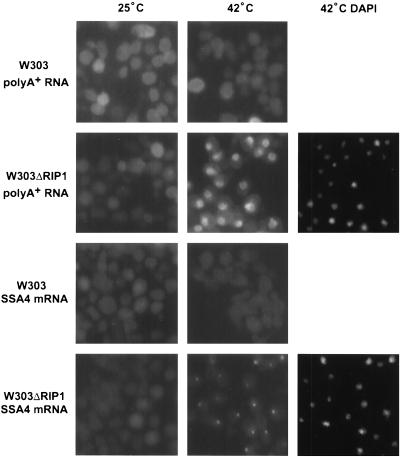
FIG. 4.
Stress-induced nuclear accumulation of poly(A)+ RNA and SSA4 mRNA occurs only in the absence of Rip1p. Cy3 in situ hybridization was performed to localize poly(A)+ RNA and SSA4 mRNA in wild-type and ΔRIP1 strains. The cells were maintained at 25 or 42°C for 1 h. DAPI (4′,6′-diamidino-2-phenylindole) staining for the ΔRIP1 strain at 42°C is shown on the right.
Similar to previous observations, poly(A)+ RNA was mostly cytoplasmic in both the wild-type and ΔRIP1 strains at 25°C, although high cell autofluorescence obscures the signal. However, we detected little or no stress-induced nuclear accumulation of poly(A)+ RNA in the wild-type strain, in contrast to previous reports (40; compare Fig. 4). Similar observations have been made elsewhere (F. Stutz, personal communication). The apparent discrepancy with the previous report might well result from a difference in strain background. Importantly, however, nuclear accumulation of poly(A)+ RNA in the ΔRIP1 strain was dramatic. The poly(A)+ RNA accumulation in the ΔRIP1 strain may reflect in large part the defect in hs mRNA export, as hs mRNA may represent a large fraction of newly transcribed mRNA and therefore much of the nuclear poly(A)+ signal at 42°C. SSA4 mRNA also shows a Rip1p-dependent nuclear accumulation at 42°C, although the accumulation is morphologically distinct from that of poly(A)+ RNA (Fig. 4).
To further address the involvement of Rip1p in the export of non-hs mRNA, we used protein-labeling assays. The wild-type and ΔRIP1 strains were incubated for various times at 37 or 42°C, followed by a 5-min labeling with [35S]methionine to give a representation of cytoplasmic mRNA levels (Fig. 5A). As a control, we used an rpb1-1 strain that carries a temperature-sensitive RNA polymerase II mutation. Inhibition of transcription at 37°C in the rpb1-1 strain leads to a decline in mRNA abundance and a decline in the protein-labeling intensity, with most of the bands disappearing by 90 min. As previously described, this decline is similar to that caused by a complete block to mRNA export (32). In contrast, when the wild-type strain was incubated at 37 or 42°C, or when the ΔRIP1 strain was incubated at 37°C, virtually no decline in protein labeling was observed for at least 3 h. The fact that the RIP1 deletion has no effect on cell growth at 37°C is consistent with the lack of an export defect at this temperature. Incubation of the ΔRIP1 strain at 42°C, however, resulted in a gradual decline in protein labeling. The decline was not as dramatic as in the case of the rpb1-1 strain, as many bands were still visible after 2 h at 42°C. This suggests a less-than-complete mRNA export block in the ΔRIP1 strain at 42°C.
FIG. 5.
Incubation of a ΔRIP1 strain at 42°C causes a decline in the protein labeling pattern. SDS-PAGE of total cellular proteins labeled with [35S]methionine is shown. The positions of the stress-inducible yeast heat shock proteins Hsp104p, Hsp82p, and Hsp70s are shown by arrows. (A) Wild-type, ΔRIP1, and rpb1-1 strains were incubated for various times at 42 or 37°C, followed by a 5-min labeling with [35S]methionine. (B) Enlargement of a portion of the gel in panel A, showing the delayed appearance of heat shock protein bands in the ΔRIP1 strain at 42°C.
Taken together, our data strongly suggest that the export of many different mRNAs is severely, but not completely, inhibited in the absence of Rip1p at 42°C. This conclusion is further supported by the appearance of heat shock protein bands in the ΔRIP1 strain after an hour of incubation at 42°C (Fig. 5B). This implies that hs mRNA export is also incompletely inhibited in the ΔRIP1 strain at 42°C.
Nuclear export of specific transcripts is inhibited in a ΔRIP1 strain at 42°C.
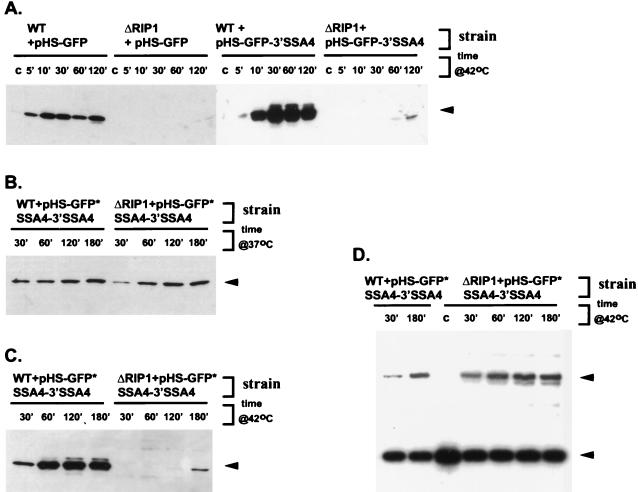
We next examined three heat-shock-inducible GFP constructs, all of which have a GFP open reading frame (ORF) cloned downstream of the SSA4 promoter and the SSA4 5′ UTR. pHS-GFP contained only the GFP ORF; pHS-GFP-3′ SSA4 contained the GFP ORF followed by the SSA4 3′ UTR, and pHS-GFP*SSA4-3′ SSA4 contained the GFP ORF followed by the complete SSA4 ORF and SSA4 3′ UTR. All three constructs were transformed into the wild-type and ΔRIP1 strains, and mRNA export after heat shock was monitored indirectly by Western blotting with α-GFP antibody. The time course of mRNA induction was verified by primer extension using a GFP mRNA-specific primer.
None of the constructs produced any GFP mRNA or protein under non-heat-shock conditions (Fig. 6A and data not shown). However, all three hybrid mRNAs and their protein products were readily detectable after a 5-min incubation of the wild-type strain at 37 or 42°C or after a 5-min incubation of the ΔRIP1 strain at 37°C (Fig. 6A to C and data not shown). The protein and mRNA accumulation continued for 60 min at 42°C, by which time it reached saturation. That induction of all three hybrid mRNAs resulted in GFP synthesis at both 37 and 42°C indicates that these mRNAs are successfully exported from the nucleus in wild-type cells under stress conditions (Fig. 6A to D). For all three mRNAs, the time courses of mRNA and protein induction were similar, suggesting that mRNA export kinetics under heat shock conditions does not depend on any cis-acting sequences contained within the SSA4 ORF and the SSA4 3′ UTR. In contrast to the situation in the wild type, none of the three constructs produced detectable levels of GFP in the ΔRIP1 strain incubated at 42°C for up to 1 h (Fig. 6A and C). As the level and timing of the hybrid mRNA induction in the ΔRIP1 strain is similar to that in the wild-type strain (Fig. 6D), the absence of detectable protein product is likely the result of an mRNA export block. The fact that the block takes place at 42 but not at 37°C is consistent with observations described earlier and strengthens the idea that Rip1p plays an important role in the nuclear export of most mRNA, but only at temperatures higher than 37°C. For all three constructs, we observed low levels of GFP in the ΔRIP1 strain after a 2- to 3-h incubation at 42°C (Fig. 6A and C and data not shown). This resembles the late appearance of labeled heat shock protein bands in the labeling time course experiment (Fig. 5B) and probably reflects the incomplete inhibition of mRNA export in the ΔRIP1 strain at 42°C (see Discussion).
FIG. 6.
Three heat-shock-inducible SSA4-GFP hybrid mRNAs are efficiently exported in a wild-type strain but not a ΔRIP1 strain at 42°C. (A, B, and C) Analysis of GFP synthesis by immunoblotting with α-GFP antibody. The position corresponding to GFP is marked by the arrowhead. Lanes marked c are from cultures maintained at 25°C. (A) Time course of GFP expression upon induction of HS-GFP and HS-GFP-3′ SSA4 mRNAs at 42°C in wild-type (WT) and ΔRIP1 strains. (B) Time course of GFP expression upon induction of HS-GFP*SSA4-3′ SSA4 mRNA at 37°C in wild-type and ΔRIP1 strains. (C) Time course of GFP expression upon induction of HS-GFP*SSA4-3′ SSA4 mRNA at 42°C in wild-type and ΔRIP1 strains. (D) Primer extension analysis of HS-GFP*SSA4-3′ SSA4 mRNA induction at 42°C in wild-type and ΔRIP1 strains (the same experiment as in panel C). Denaturing PAGE analysis of primer extension products with GFP- and U2-specific primers is shown. Positions corresponding to HS-GFP*SSA4-3′ SSA4 mRNA and U2 RNA are marked by the upper and lower arrowheads, respectively.
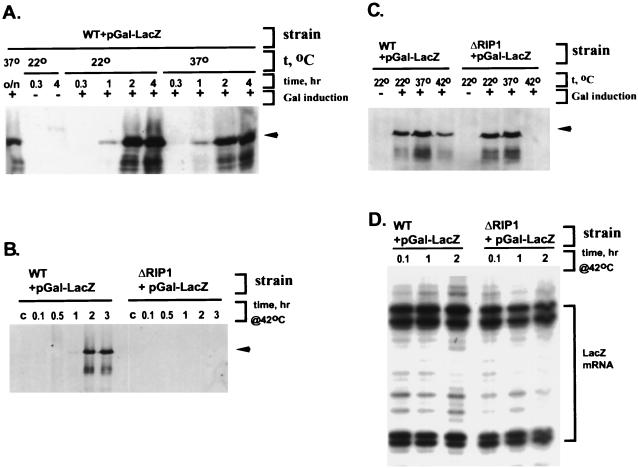
Using the experimental approach described above, we also examined the nuclear export of a fully non-hs mRNA, transcribed from a construct containing a bacterial β-galactosidase gene under the control of a galactose promoter (pGal-LacZ). After a 10- to 15-min preincubation at the relevant temperature, lacZ transcription was initiated by galactose, and the incubation was continued at the same temperature. In the wild- type strain, the time courses of LacZ induction at 22, 37, and 42°C were similar (Fig. 7A and B), with protein amounts decreasing slightly at 42°C (Fig. 7C). In the ΔRIP1 strain at 22 and 37°C, LacZ was induced with the same kinetics and to the same levels as in the wild type (Fig. 7C and data not shown). For the ΔRIP1 strain at 42°C, however, no LacZ was detected even at 3 h after the addition of galactose (Fig. 7B and C). Like the SSA4-GFP chimeric mRNAs, the level and timing of LacZ mRNA induction in the ΔRIP1 strain at 42°C was similar to that in the wild type (Fig. 7D). Therefore, the absence of detectable protein in the ΔRIP1 strain is likely due to a block in non-hs mRNA export at 42°C.
FIG. 7.
Nuclear export of galactose-inducible β-galactosidase (Gal-LacZ) mRNA is inhibited at 42°C in a ΔRIP1 strain but not a wild-type strain. (A, B, and C) LacZ protein was analyzed by immunoblotting it with α-β-galactosidase antibody. The position corresponding to the LacZ band is marked by an arrowhead. (A) Time course of LacZ protein expression upon induction at 22 and 37°C in the wild type (WT). The lanes marked c are from cultures maintained at 22°C in the absence of galactose induction. (B) Time course of LacZ protein expression upon induction at 42°C in wild-type and ΔRIP1 strains. The lanes marked c are from cultures treated at 42°C but in the absence of galactose induction. (C) LacZ protein expression after 2-h incubation of wild-type and ΔRIP1 strains at 22, 37, or 42°C, with or without galactose induction. (D) Time course of induction of Gal-LacZ mRNA at 42°C in wild-type and ΔRIP1 strains (the same experiment as in panel B). Denaturing PAGE analysis of extension products with a LacZ-specific primer is shown. The area occupied by multiple extension products is marked by a square bracket.
DISCUSSION
The original studies of mRNA export during stress in S. cerevisiae suggested that hs mRNA is exported from the nucleus via a unique route (39). Using in situ hybridization assays, it was shown that exposure of yeast to 42°C or 10% ethanol resulted in nuclear accumulation of non-hs mRNA, whereas hs mRNA was efficiently exported to the cytoplasm. Hybrid mRNAs identified two independent cis-acting sequences in SSA4 mRNA that promote export of an Hsp70-like mRNA under stress conditions (39). It was further shown that hs mRNA export at 42°C is affected by deletion of the RIP1 gene and is not mediated by the GTPase Ran and its auxiliary proteins (39, 40).
We originally set out to define additional factors mediating this selective export of hs mRNA. However, we obtained data indicating that hs mRNA export and non-hs mRNA export are similar processes, a conclusion that contradicts some previous conclusions. Protein labeling and thermotolerance assays showed that both export pathways are similarly inhibited at 42°C in rna1-1, prp20-1, mex67-5, mtr2-9, rat7-1, and ΔNUP116 strains. The results suggest that hs mRNA export, like non-hs mRNA export, is Ran mediated. However, both rna1-1 and prp20-1 have rapid and profound effects on many different aspects of nuclear metabolism (2, 8), and the mRNA export block might be indirect. Consistent with this possibility, a dominant-negative Ran mutant has little effect on hs mRNA export as determined by protein labeling (data not shown). The interpretation notwithstanding, we suggest that the rna1-1 and prp20-1 mutations cause equally strong blocks to hs mRNA export and to non-hs mRNA export.
The discrepancy between this and previous observations (39, 40) can be explained at least in part by differences in the experimental protocols. For example, the penetrance of the ΔRIP1 mutant phenotype is very sensitive to growth conditions, reflecting an induction of heat shock protein synthesis at moderate cell densities. In addition, many of the previous studies used 10% ethanol to induce the stress response. We have observed some differences between heat shock protein synthesis induced by a shift to 42°C and that induced by ethanol addition (data not shown), indicating that this might also impact differences in heat shock protein synthesis regulation.
In light of the rna1-1 and prp20-1 data, we decided to address the issue of whether the nucleoporinlike protein Rip1p is specific for the hs mRNA export pathway or whether the export of non-hs mRNA is also affected in the ΔRIP1 strain. A comparison of protein synthesis profiles in the wild-type and ΔRIP1 strains indicates that nuclear export of both hs and non-hs mRNA is severely affected in the absence of Rip1p at 42°C. The original inference, that the ΔRIP1 strain is defective only in hs mRNA export, is probably due to the experimental design. In the earlier experiments, cells were examined within 10 to 30 min after the shift to the nonpermissive temperature. At these times, only the absence of heat shock proteins produced from newly transcribed hs mRNA is easily detected; any block in non-hs mRNA export is masked by preexisting cytoplasmic mRNA. At longer incubation times, the cytoplasmic mRNA turns over, and the export inhibition is visible. The relatively slow decrease in the protein labeling pattern in the ΔRIP1 strain at 42°C (Fig. 5A), as well as the appearance of heat shock and hs promoter-derived bands at late times during a 42°C incubation (Fig. 5B and 6C), suggests that the ΔRIP1 mRNA export block is incomplete. Although there may be some modest difference between hs and non-hs mRNA export efficiency in the ΔRIP1 strain at 42°C, we suggest that the ΔRIP1 42°C block to mRNA export affects most if not all mRNAs similarly. This conclusion fits well with the genetic interactions between RIP1 and other genes (GLE1, DBP5, and NUP85) involved in the export of non-hs mRNA (46, 48). A very recent study reached a similar conclusion for the mRNA export factor Mex67p (15).
It should be noted, however, that the idea of a general role of Rip1p in mRNA export does not contradict previously described observations of competition between hs mRNA and the human immunodeficiency virus type 1 protein Rev for nuclear exit (40). Rev is exported from the nucleus via interactions with the β-karyopherin-like receptor Crm1p, which has been shown to interact with Rip1p (7, 31, 32). The normal heat shock protein labeling pattern in the xpo1-1 strain at 42°C (Fig. 2A), as well another recent study (32), argues that Crm1p plays no prominent direct role in the export of either hs or non-hs mRNA. Nevertheless, it is conceivable that different nuclear export pathways converge below the level of Crm1p, so that different export complexes compete for binding sites on Rip1p or on other relevant NPC components (e.g., Nup159). We still do not known whether Rip1p is a bona fide NPC component, but we favor the notion that it is a transport factor with a more transient pore association (46).
Our data more generally suggest that mRNA nuclear export relies on many common factors, including the Ran system, Mex67p, Mtr2p, and Rip1p. A competition between different mRNA molecules for common transport factors may lead to the preferential export of more abundant mRNAs (i.e., hs mRNA under stress conditions) or mRNAs that interact more efficiently with generic transport machinery components. There may also be message-specific factors or cis-acting RNA elements that enhance or inhibit export of specific mRNAs in a constitutive or regulated fashion. For example, it has recently been suggested that the yeast hnRNP protein Np13p becomes dissociated from non-hs mRNA upon stress, leading to abnormal RNP formation and inefficient export of these mRNAs (22). There are many other examples of factors and cis-acting sequences that contribute to the regulation of mRNA export. These include the retroviral proteins Rev and Rex, the constitutive transport element of D-type retroviral mRNAs, the Caenorhabditis elegans Zn finger protein TRA-1, the intronless mRNA export elements within the mouse histone H2a mRNA, elements within herpes simplex virus thymidine kinase mRNA and hepatitis B virus RNA, and a retention element within C. elegans splicing factor U2AF mRNA (11, 12, 14, 25, 49). In the case of hs mRNA export under stress conditions, however, the evidence in favor of positively acting transport factors and elements is uncertain at best. We have been unable to detect a contribution of the SSA4 ORF or the SSA4 3′ UTR to RNA export. In addition, we have examined a set of GFP constructs with no SSA4 sequences: addition of the SSA4 5′ UTR, with or without additional SSA4 sequences, has no effect (data not shown). Of course, potent export of the basal GFP construct might obscure a positive but more modest contribution of SSA4 mRNA sequences.
The fact that Rip1p is essential only at temperatures higher than 37°C raises the intriguing possibility that the structure and/or composition of the NPC-associated transport machinery changes under conditions of more acute stress. We performed protein labeling experiments with the ΔRIP1 strain at various temperatures and ethanol concentrations and observed a gradual decline in heat shock protein labeling with increasing stress. At 42°C, the absence of Rip1p may adversely affect the activities of other essential transport factors that normally interact with it. Under conditions of mild stress, such as incubation at 37°C, this destabilization may not be very severe and/or the function of Rip1p is compensated for by other nucleoporinlike proteins. It is also conceivable that a Rip1p-dependent regulatory mechanism results in a modification of the mRNA export machinery only under severe stress conditions. Future studies will focus on understanding the role of Rip1p and its associated proteins in maintaining mRNA export under stress conditions.
ACKNOWLEDGMENTS
We thank C. Cole, E. Craig, V. Doye, P. Silver, F. Stutz, K. Weis, and S. Wente for mutant strains and plasmids and C. Hammell for help with in situ hybridization assays. We are grateful to F. Stutz for initiating this project, for advice, and for communicating data prior to publication. We thank T. H. Jensen and M. Neville for helpful discussions and for critical reading of the manuscript and C. Guthrie for comments on the manuscript. We thank L.-A. Coolege and A. Phillips for secretarial assistance and E. Dougherty for help with figures.
This work was supported by NIH grant GM 23549.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1994. [Google Scholar]
- 2.Carazo-Salas R E, Guarguaglini G, Gruss O J, Segref A, Karsenti E, Mattaj I W. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- 3.Corbett A H, Koepp D M, Schlenstedt G, Lee M S, Hopper A K, Silver P A. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 5.Doye V, Wepf R, Hurt E C. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreyfuss G, Matunis M J, Piñol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 7.Floer M, Blobel G. Putative reaction intermediates in Crm1-mediated nuclear protein export. J Biol Cell. 1999;274:16279–16286. doi: 10.1074/jbc.274.23.16279. [DOI] [PubMed] [Google Scholar]
- 8.Forrester W, Stutz F, Rosbash M, Wickens M. Defects in mRNA 3′-end formation, transcription initiation, and mRNA transport associated with the yeast mutation prp20: possible coupling of mRNA processing and chromatin structure. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- 9.Gorlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorsch L C, Dockendorff T C, Cole C N. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graves L E, Segal S, Goodwin E B. TRA-1 regulates the cellular distribution of the tra-2 mRNA in C. Elegans. Nature. 1999;399:802–805. doi: 10.1038/21682. [DOI] [PubMed] [Google Scholar]
- 12.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 13.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:389–398. [PubMed] [Google Scholar]
- 14.Huang Y, Wilmer K M, Carmichael G G. Intronless mRNA transport elements may affect multiple steps in pre-mRNA processing. EMBO J. 1998;18:1642–1652. doi: 10.1093/emboj/18.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurt E, Sträßer K, Segref A, Bailer S, Schlaich N, Presutti C, Tollervey D, Jansen R. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J Biol Chem. 2000;275:8361–8368. doi: 10.1074/jbc.275.12.8361. [DOI] [PubMed] [Google Scholar]
- 16.Hurwitz M E, Strambio-de-Castillia C, Blobel G. Two yeast nuclear pore complex proteins involved in mRNA export form a cytoplasmically orientated subcomplex. Proc Natl Acad Sci USA. 1998;95:11241–11245. doi: 10.1073/pnas.95.19.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katahira J, Strasser K, Podtelenikov A, Mann M, Jung J U, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koepp D M, Wong D H, Corbett A H, Silver P A. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krebber H, Taura T, Lee M S, Silver P A. Uncoupling of the hnRNP Np13p from mRNA during the stress-induced block in mRNA export. Genes Dev. 1999;13:1994–2004. doi: 10.1101/gad.13.15.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 24.Long R M, Singer R H, Meng X, Gonzalez I, Nasmyth K, Jansen R. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 2000;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 25.MacMorris M A, Zorio D A, Blumenthal T. An exon that prevents transport of a mature RNA. Proc Natl Acad Sci USA. 1999;7:3813–3818. doi: 10.1073/pnas.96.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 27.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 28.Nachury M V, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci USA. 1999;96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakielny S, Dreyfuss G. Nuclear export of proteins and RNAs. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- 30.Nakielny S, Fischer W M, Dreyfuss G. RNA Transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- 31.Neville M, Lee L, Stutz F, Davis L I, Rosbash M. Evidence that the importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export in S. cerevisiae. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 32.Neville M, Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 34.Ohno M, Fornerod M, Mattaj I W. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 35.Pasquinelli A E, Powers M A, Forbes D, Dahlberg J E. Inhibition of mRNA export in vertebrate cells by nuclear export signal conjugates. Proc Natl Acad Sci USA. 1997;94:14394–14399. doi: 10.1073/pnas.94.26.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pikielny C W, Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985;41:119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- 37.Rout M P, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 39.Saavedra C, Tung K-S, Amberg D C, Hopper A K, Cole C N. Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev. 1996;10:1608–1620. doi: 10.1101/gad.10.13.1608. [DOI] [PubMed] [Google Scholar]
- 40.Saavedra C A, Hammell C M, Heath C V, Cole C N. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev. 1997;11:2845–2856. doi: 10.1101/gad.11.21.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Pante N, Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seedorf M, Silver P A. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snay-Hodge C A, Colot H V, Goldstein A L, Cole C N. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 46.Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, Kantor J, Rosbash M, Stutz F. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr255p. EMBO J. 1999;18:5761–5777. doi: 10.1093/emboj/18.20.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stutz F, Izaurralde E, Mattaj I W, Rosbash M. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol Cell Biol. 1996;16:7144–7150. doi: 10.1128/mcb.16.12.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stutz F, Kantor J, Zhang D, McCarthy T, Neville M, Rosbash M. The yeast nucleoporin Rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Genes Dev. 1997;11:2857–2868. doi: 10.1101/gad.11.21.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 50.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 51.Taura T, Schlenstedt G, Silver P A. Yrb2b is a nuclear protein that interacts with Prp20p. J Biol Chem. 1997;272:31877–31884. doi: 10.1074/jbc.272.50.31877. [DOI] [PubMed] [Google Scholar]
- 52.Wente S, Blobel G. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol. 1994;125:955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Q, Rout M P, Akey C W. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]