Abstract
Background
Ureteropelvic junction obstruction (UPJO) is one of the most common causes of hydronephrosis in children. This study explored the effects and the regulatory mechanisms of the circular RNA (circRNA) hsa_circRNA_0045861 (circRNA_0045861) in UPJO.
Methods
RNA sequencing was used to identify the differentially expressed circRNAs in UPJO. The effects of circRNA_0045861 on renal cell apoptosis was investigated by flow cytometry and Western blot analysis. Furthermore, we used bioinformatics methods to predict the possible target genes of circRNA_0045861. Fluorescence in-situ hybridization and dual-luciferase reporter assays were performed to validate the target genes of circRNA_0045861. Finally, we evaluated the effects of circRNA_0045861 target gene miR-181d-5p on UPJO-induced renal fibrosis in vivo.
Results
RNA sequencing identified 63 upregulated and 64 downregulated circRNAs in UPJO patients. The expression of circRNA_0045861 was significantly elevated in kidney damage both in vivo and in vitro. Silencing circ_0045861 inhibited transforming growth factor (TGF)-β1-induced apoptosis in vitro in human kidney 2 (HK-2) cells. Furthermore, circ_0045861 was shown to directly interact with the microRNA miR-181d-5p and regulate the expression of sirtuin 1 (SIRT1), thereby promoting the progression of apoptosis and renal injury. In addition, overexpression of miR-181d-5p inhibited cell apoptosis and renal fibrosis in a mouse model through downregulating the SIRT1/p53 pathway.
Conclusions
Circ_0045861 may be a novel candidate circRNA in the pathogenesis of UPJO by acting as a pro-apoptotic factor via the miR-181d-5p/SIRT1 axis.
Keywords: Ureteropelvic junction obstruction (UPJO), circRNA_0045861, miR-181d-5p, SIRT1, apoptosis
Introduction
Congenital ureteropelvic junction obstruction (UPJO) is the main cause of hydronephrosis in children with chronic kidney disease (1,2). The major pathological change associated with UPJO is renal fibrosis which is characterized by renal tubular epithelial cell apoptosis, podocyte inflammation, fibroblast proliferation, and excessive extracellular matrix accumulation (3-5). Although urinary obstruction can be surgically resolved, in some cases, kidney function cannot be fully restored, leading to the need for hemodialysis or kidney transplantation. Therefore, in depth understanding of the pathological mechanisms of UPJO and the identification of effective therapeutic targets is crucial for the early restoration of renal function in these children.
Sirtuin 1 (SIRT1) is a key sensor protein of deacetylases and plays a vital role in the regulation of renal diseases (6-8). SIRT1 can regulate cell proliferation, apoptosis, DNA damage repair, senescence, and metabolism (9). However, the mechanisms by which SIRT1 is activated and facilitates this process remains unclear. Circular RNAs (circRNAs) are endogenous non-coding RNAs that function predominantly as microRNA (miRNA) sponges, blocking the inhibitory effects of miRNAs on target genes (10,11). CircRNAs are involved in the regulation of genes that contribute to nervous system diseases, cardiovascular diseases, and various types of cancers, including renal clear cell carcinoma (12-16). Thus, modulation of circRNA function may be an effective therapeutic strategy for renal diseases. Therefore, this study explored the regulatory role of circRNAs in UPJO.
In this investigation, differentially expressed circRNAs in UPJO were identified by RNA sequencing. The results revealed 63 upregulated and 64 downregulated circRNAs in UPJO patients. Of these circRNAs, hsa_circRNA_0045861 (circRNA_0045861) was identified as a potential regulator of SIRT1 by using bioinformatics methods. CircRNA_0045861 was shown to promote transforming growth factor (TGF)-β1-induced apoptosis in vitro. Furthermore, circRNA_0045861 enhanced the progression of UPJO through regulation of SIRT1 expression by sponging miR‐181d‐5p. The study also demonstrated that overexpression of miR-181d-5p inhibited the progression of unilateral ureteral obstruction (UUO)-induced renal fibrosis in mice and inhibited cell apoptosis through downregulation of the SIRT1/p53 pathway. We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-5060).
Methods
Patient selection and determination of renal function
A total of 6 patients with a diagnosis of UPJO (details in Table S1) confirmed by the Pediatric Urology Department at Shengjing Hospital of China Medical University were included in this study. Glomerular filtration rate (GFR) was evaluated by renal dynamic imaging. Based on split renal function (SRF; SRF = injured kidney GFR/total GFR), the 6 patients were divided into 2 group, namely, patients with an SRF <30% and patients with SRF >40%. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Medical Research and New Technology Ethics Committee of Shengjing Hospital of China Medical University (approval No. 2012PS81K) and informed consent was taken from all the patients, and all experiments were conducted in accordance with the Good Clinical Practice recommendations of the institute.
RNA sequencing and circRNAs analysis
Total RNA was extracted from renal tissue samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The RNA samples were quantified after removal of DNA contamination. The RNA library was sequenced in the HiSeq 4000 system (Illumina, San Diego, CA, USA). CircRNAs were identified using CIRI software based on reference data from the circBase database (http://www.circbase.org/) (17). All data were uploaded to the NCBI Sequence Read Archive database under the identifier PRJNA591062. CircRNAs with a P value <0.05 and log2 fold change >1 were considered differentially expressed circRNAs. The circBase 2.0 software was used to identify all possible miRNA of the differentially expressed circRNAs and TargetScan was then used to identify all possible target mRNAs of the miRNAs of interest (18).
Cell culture and treatment
The human kidney epithelial HK-2 cell line was purchased from BeNa Culture Collection (Suzhou, China) and cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Gibco, Waltham, MA, USA). HK-2 cells were treated with TGF-β1 (10 ng/mL; R&D Systems, Minneapolis, MN, USA) or a negative control. Cells were transfected with circ_0045861 small interfering RNA (siRNA) or the corresponding scrambled negative control (NC) (GeneSeed Biotech, Guangzhou, China), miR-181d-5p mimic or the corresponding NC mimic, miR-181d-5p inhibitor or the corresponding NC inhibitor (RiboBio, Guangzhou, China), or a SIRT1 expressing plasmid or the corresponding empty plasmid (GeneChem, Shanghai, China). The cells were harvested 24 hours after transfection. Target sequences of these siRNA and microRNA were as follows: si-circ-0054861: 5'-CTCAAGAGTGGAGCTGGAC-3'; si-circ-0054861-NC: 5'-CAGCCGACAGCATCCAGCAG-3'; miR-181d-5p mimic: 5'-AACAUUCAUUGUUGUCGGUGGGU-3'; miR-181d-5p mimic NC: 5'-UUUGUACUACACAAAAGUACUG-3'; miR-181d-5p inhibitor: 5'-ACCCACCGACAACAAUGAAUGUU-3'; miR-181d-5p inhibitor NC: 5'-CAGUACUUUUGUGUAGUACAAA-3'.
In situ hybridization of circ_0045861 and miR-181d-5p
HK-2 cells were fixed, permeabilized, and rehydrated. The cells were then hybridized overnight with a biotin-labeled-circ_0045861 and miR-181d-5p probe. Samples were then placed on coverslips with Prolong Gold Antifade Reagent containing 4’,6-diamidino-2-phenylindole (DAPI; Cell Signaling Technology, Danvers, MA, USA) and observed by confocal microscopy. The sequence for the circ-0045864 label was CY3: 5'-GGTTCAGTCCAGCTCCACTCTTG-3' and the sequence for the miR-181d-5p-FITC-label was 5'-ACCCACCGACAACAATGAATGTT-3'.
Dual-luciferase reporter assay
The 3'-UTR sequence of SIRT1 or the 3'-UTR sequence of circ_0045861 was cloned into the pMIR-GLO vector (GenePharma, Shanghai, China) between the SacI and HindIII restriction sites. HK-2 cells were co-transfected with 2 µg vector plus 2 µg miR-181d-5p mimics or negative control using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA). Cells were harvested after 72 hours of incubation. Luciferase values were determined using the Dual-Luciferase Reporter Assay System (Beyotime Biotechnology, Beijing, China).
Annexin V and propidium iodide (PI) staining for flow cytometry analysis
HK-2 cells were seeded into 6-well plates. After treatment, the cells were harvested, washed with cold phosphate buffered saline (PBS), and stained with 10 µL Annexin V-FITC and 5 µL PI in 300 µL binding buffer. After incubation for 20 minutes at room temperature in the dark, the samples were diluted and filtrated. The apoptotic rate of each sample was measured by flow cytometry (Becton-Dickinson and Company, Franklin Lakes, NJ, USA).
Animals and experimental design
Animal experiments were performed under a project license (No. 2018PS229K) granted by ethics committee of Shengjing Hospital of China Medical University, in compliance with the NIH guidelines for the care and use of animals. Eight-week-old male C57BL/6 mice (20±5 g) were purchased from Changsheng Biotechnology (Shenyang, China) and housed under normal temperature (25±2 °C) and humidity (50–60%), with a 12-hour light/dark cycle. The mice were randomly divided into 4 groups of 8 mice as follows: (I) sham group; (II) UUO model group; (III) UUO + miR-181d-5p agomir-NC; and (IV) UUO + miR-181d-5p agomir group. All surgeries were performed under 1% sodium pentobarbital anesthesia (50 mg/kg intraperitoneal administration). The right ureter of the mouse was ligated to establish the UUO model. In detail, their left ureters were exposed via a lateral incision and ligated with two separate silk ties, level with the lower pole of the kidney. The model was considered successful when expansion of the renal pelvis and reduction in renal function was observed. On day 2 post-surgery, mice were given miRNA agomir (100 nmol/kg). On post-operative day 7, the animals were sacrificed and the kidneys were harvested for further analysis.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells or renal tissues using TRIzol reagent (Takara, Dalian, China). The RNA PCR Kit (TaKaRa) was used to reverse-transcribe RNA into complementary cDNA. qRT-PCR was performed using the SYBR Premix Ex Taq Kit (Takara) and the 7500 Real-time PCR System (Applied Biosystems, Foster City, CA, USA). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the endogenous control for circRNA, and U6 was used for miRNA. qRT-PCR data were measured using the 2−∆∆Ct method. Primers for circ_0045861, miR-181d-5p, GAPDH, and U6 were designed and synthesized by RiboBio (Guangzhou, China) and are listed in Table 1.
Table 1. Primer sequences for real-time polymerase chain reaction.
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| circ_0045861 | ACCGCTACCTCCTCAAGAGTG | TCCTTCACCGACAAGCCAC |
| miR-181d-5p | TGCGCAACATTCATTGTTGTCG | CTCAAGTGTCGTGGAGTCGGCAA |
| GAPDH | GCAAGTTCAACGGCACAG | ACGCCAGTAGACTCCACGAC |
| U6 | CTCGCTTCGGCAGCACA | GCGAGCACAGAATTAATACGAC |
GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Western blot analysis
Proteins were isolated using radioimmunoprecipitation assay buffer (RIPA) and phenylmethylsulphonyl fluoride (PMSF) in a 10:1 ratio (Beyotime Institute of Biotechnology, Shanghai, China) and quantified using the Enhanced BCA Protein Assay Kit (Beyotime Institute of Biotechnology, Shanghai, China). Protein lysates were separated by gel electrophoresis and transferred onto polyvinylidene difluoride membranes. The membranes were exposed to primary antibodies against SIRT1 (Abcam, Cambridge, United Kingdom, ab7343), p53 (Abcam, ab1101), cleaved caspase-3 (Abcam, ab2324), bax (Abcam, ab7977), bcl-2 (Abcam, ab692), alpha smooth muscle actin (Abcam, ab5694), vimentin (Abcam, ab40863), and GAPDH (Abcam, ab11256) overnight at 4 °C. Membranes were then incubated for 2 hours at 37 °C with an anti-rabbit IgG secondary antibody (Abcam). The proteins were detected using the ProtoBlot II AP System (Promega, Madison, WI, USA). GAPDH was used as the loading control. Relative protein expression was determined by densitometry analysis using the ImageJ software.
Masson’s trichrome staining and immunohistochemistry (IHC)
Kidney tissues were fixed in 10% formalin and embedded in paraffin. Sections were then stained with Masson’s trichrome dyes. The fibrotic area was assessed by outlining the blue staining. IHC was performed using the Polink-1 one-step polymer detection system (ZSGB-BIO, Beijing, China). Slides were incubated with antibodies against SIRT1, p53, cleaved caspase-3, bcl-2, or bax (Abcam) overnight. A negative control was prepared by replacing the primary antibody with phosphate-buffered saline solution. All sections were imaged by microscopy (Nikon ECLIPSE 80i).
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
Frozen tissue sections were treated with 20 µg/mL proteinase K (ZSGB-BIO, Beijing, China) and incubated in a nucleotide mixture containing fluorescein-12-dUTP and terminal deoxynucleotidyl transferase. Samples were labeled with fluorescent-conjugated affinity-purified secondary antibody, co-labeled with DAPI, and mounted with Vectashield (VectorLabs, Burlingame, CA, USA). The number of fluorescent apoptotic cells were counted in 20 random fields of view and the mean number of apoptotic cells per field was recorded.
Statistical analysis
Data are expressed as the mean ± standard deviation. All tests were repeated at least 3 times. Differences between 3 or more groups were evaluated using one-way analysis of variance. All statistical analyses were conducted using the SPSS 19.0 (IBM Corp., Armonk, NY, USA) and Prism 7.0 software (GraphPad Software, San Diego CA, USA). A P value <0.05 was considered statistically significant.
Results
Expression and characteristics of circ_0045861 in UPJO
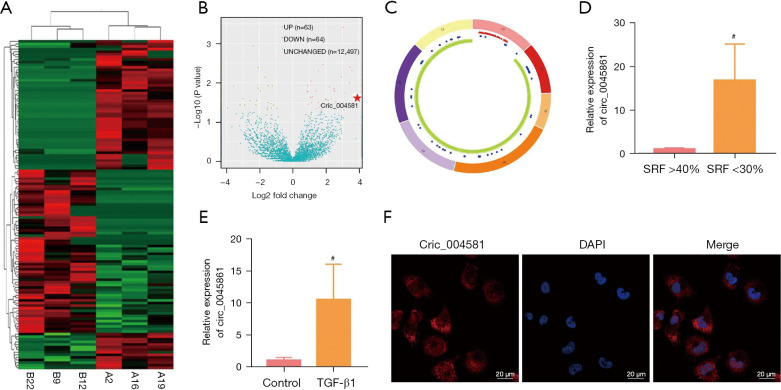
The heatmap in Figure 1A shows significant differences in the expression of circRNAs between two groups of UPJO samples. There were 63 upregulated differentially expressed circRNAs (P<0.05) and 64 downregulated circRNAs (P<0.05) in the UPJO samples (Figure 1B). Circ_0045861 was determined to be located on chromosome 17, with a total length of 14570 bp, containing 7 exons that are formed by exon circularization (Figure 1C). Furthermore, among the UPJO patients, the expression of circ_0045861 was significantly higher in patients with SRF <30% compared to patients with SRF >40% (Figure 1D).
Figure 1.
Expression and characteristics of circ_0045861 in UPJO. (A) A heatmap showing the differentially expressed circRNAs. (B) A volcano plot showing the differentially expressed circRNAs. (C) Circular structure of circ_0045861. (D) The expression levels of circ_0045861 in patients with SRF >40% and patients with SRF <30%. (E) The expression levels of circ_0045861 in TGF-β1-stimulated HK-2 cells and control cells. (F) In situ hybridization of hsa_circ_0045861 in HK-2 cells, scale bar: 20 µm. #P<0.05. UPJO, ureteropelvic junction obstruction. SRF, split renal function; TGF, transforming growth factor.
To simulate renal tubular cell injury in vitro, human proximal tubular epithelial HK-2 cells were treated with TGF-β1 to induce apoptosis. Compared to the control group, the expression level of circ_0045861 was significantly increased in HK-2 cells treated with TGF-β1 compared to control cells (Figure 1E). In situ hybridization revealed that circ_0045861 was present in both the cell nucleus and cell cytoplasm, but was significantly enriched in the cytoplasm (Figure 1F).
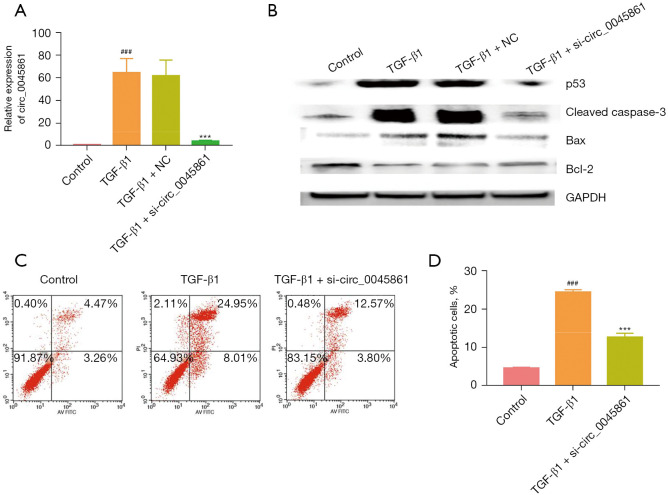
Silencing circ_0045861 inhibits TGF-β1-induced apoptosis in vitro
HK-2 cells were transfected with a targeted siRNA to silence the expression of circ_0045861. Cells transfected with si-circ_0045861 showed significantly reduced expression of circ_0045861 compared with cells treated with TGF-β1 (Figure 2A). Treatment with TGF-β1 induced the expression of proteins involved in apoptosis including p53, Bax/Bcl-2 ratio, and cleaved caspase-3. However, silencing of circ_0045861 abrogated the effects of TGF-β1 on these apoptotic proteins (Figure 2B). Furthermore, flow cytometry analysis revealed that si-circ_0045861 inhibited TGF-β1-induced apoptosis in vitro (Figure 2C,2D). Taken together, these results suggested that silencing circ_0045861 inhibited the apoptosis of HK-2 cells in vitro.
Figure 2.
Silencing circ_0045861 inhibits TGF-β1-induced apoptosis in vitro. (A) The expression of circ_0045861 in HK-2 cells treated with TGF-β1. (B) Western blot analysis of p53, cleaved caspase-3, Bax, and Bcl-2 protein expression. (C) Flow cytometry analysis showing the apoptotic effects of si-circ_0045861. (D) Statistic analysis of the percentage of apoptotic cells (n=3). ###P<0.001 vs. control group; ***P<0.001 vs. TGF-β1 group. TGF, transforming growth factor; NC, negative control; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; AV, annexin V; PI, propidium iodide.
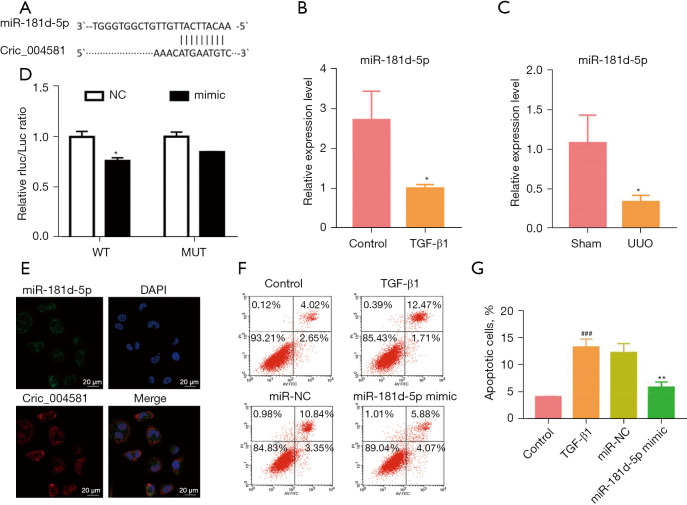
Circ_0045861 interacts with miR-181d-5p and regulates its expression
Using bioinformatics tools, miR-181d-5p was identified as a potential target of circ_0045861 (Figure 3A). Thus, the expression of miR-181d-5p was assessed in vivo and in vitro. HK-2 cells treated with TGF-β1 showed significantly decreased expression of miR-181d-5p compared to control cells (Figure 3B). The expression of miR-181d-5p was also significantly reduced in the UUO mouse model compared to sham mice (Figure 3C). Dual luciferase reporter assays were performed to verify the functional interaction between miR-181d-5p and circ_0045861. MiR-181d-5p mimics inhibited the luciferase activity of the wild-type circ_0045861 reporter in HK-2 cells but had no effect on the mutant reporter of circ_0045861 (Figure 3D). In situ hybridization analyses revealed that, similar to circ_0045861, miR-181d-5p was localized to the cell cytoplasm (Figure 3E). Furthermore, overexpression of miR-181d-5p inhibited TGF-β1-induced apoptosis in HK-2 cells (Figure 3F,3G). These findings suggested that miR-181d-5p may be a potential target of circ_0045861.
Figure 3.
Circ_0045861 interacts with miR-181d-5p and regulates its expression. (A) The binding site between circ_0045861 and miR-181d-5p. (B) The expression of miR-181d-5p in HK-2 cells treated with TGF-β1. (C) The expression of miR-181d-5p in UUO mice and sham animals. (D) Dual-luciferase reporter assay showing the relationship between miR-181d-5p and circ_0045861 in HK-2 cells. (E) In situ hybridization of circ_0045861 (red) and miR-181d-5p (green) in HK-2 cells, scale bar: 20 µm. (F) Flow cytometry analysis showing the effects of miR-181d-5p mimics on apoptosis in HK-2 cells. (G) Statistic analysis of the percentage of apoptotic cells (n=3). *P<0.05; ###P<0.001 vs. control group; **P<0.01 vs. TGF-β1 group. TGF, transforming growth factor; UUO, unilateral ureteral obstruction; NC, negative control; AV, annexin V; PI, propidium iodide.
Circ_0045861 regulates SIRT1 via miR-181d-5p
Prediction algorithms for miRNA targets identified SIRT1 as a potential target gene of miR-181d-5p (Figure 4A). Western blot analysis showed significantly increased expression of SIRT1 protein in HK-2 cells treated with TGF-β1 compared to control cells (Figure 4B). Similarly, the protein expression of SIRT1 was elevated 2.43-fold in UUO mice compared to sham mice (Figure 4C). Furthermore, dual-luciferase reporter assays demonstrated that miR-181d-5p overexpression inhibited SIRT1 3'-UTR-WT sequence reporter, but no inhibition was observed in the SIRT1 3'-UTR-MUT sequence (Figure 4D). Silencing circ_0045861 reduced SIRT1 expression in HK-2 cells, while miR-181d-5p inhibitors reversed this effect (Figure 4E). Finally, the results showed that miR-181d-5p mimics reduced the protein expression level of SIRT1, while miR-181d-5p inhibitor increased SIRT1 protein expression (Figure 4F). These results suggested that miR-181d-5p functions as a downstream effector of circ_0045861 to regulate SIRT1.
Figure 4.
Circ_0045861 regulates SIRT1 via miR-181d-5p. (A) Possible binding site between miR-181d-5p and the 3’ noncoding region of SIRT1. (B) The protein expression of SIRT1 in HK-2 cells treated with TGF-β1. (C) The protein expression of SIRT1 in the UUO mouse model. (D) Dual-luciferase reporter assay showing the relationship between miR-181d-5p and SIRT1 in HK-2 cells. (E) The mRNA expression of SIRT1 in HK-2 cells transfected with si-circ_0045861. (F) Western blot analysis showing the protein expression of SIRT1 in HK-2 cells treated with the miR-181d-5p mimics or the miR-181d-5p inhibitor. #P<0.05, ##P<0.01 vs. control group; *P<0.05, **P<0.01 vs.TGF-β1 group. SIRT1, sirtuin 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; TGF, transforming growth factor; UUO, unilateral ureteral obstruction; WT, wildtype; MUT, mutant; NC, negative control.
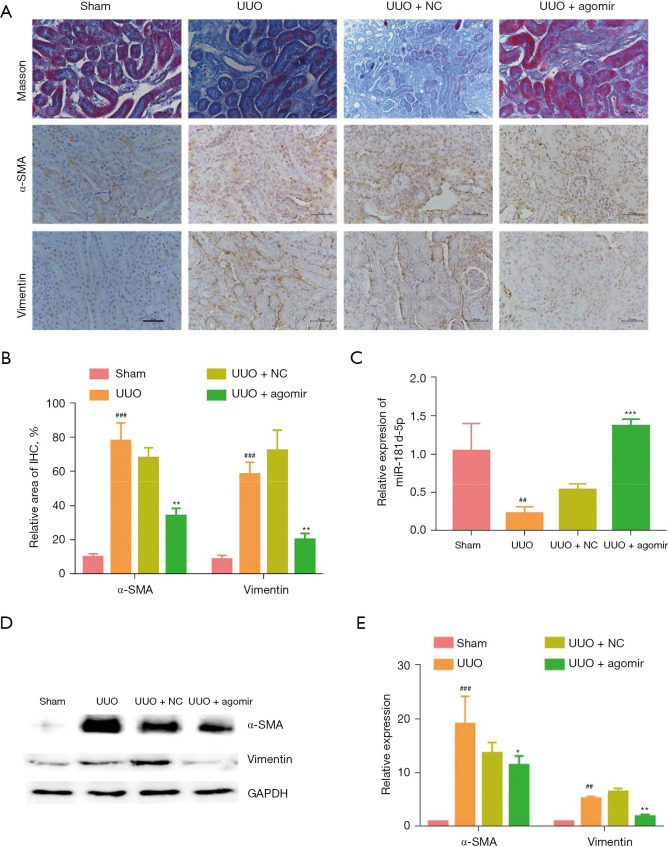
Overexpression of miR-181d-5p inhibits the progression of unilateral ureteral obstruction-induced renal fibrosis in vivo
Since the homologous gene of circ_0045861 was not found in mice, we verified the function of miR-181d-5p in vivo. Tubulointerstitial fibrosis of the renal parenchyma occurred in UUO mice at day 7 after surgery. Masson’s trichrome staining showed that miR-181d-5p agomir treatment reduced renal fibrosis in UUO mice (Figure 5A). Additionally, IHC results revealed that overexpression of miR-181d-5p significantly decreased the expression of α-smooth muscle actin (α-SMA) and vimentin in vivo (Figure 5A,5B). Furthermore, the expression of miR-181d-5p was significantly depressed in UUO mice, and this was elevated upon treatment with the miR-181d-5p agomir to levels higher than that observed in the sham mice (Figure 5C). Western blot analysis confirmed that the protein expression of α-SMA and vimentin was significantly elevated in UUO mice, but the miR-181d-5p agomir significantly decreased the expression of both α-SMA and vimentin (Figure 5D,5E). These results suggested that overexpression of miR-181d-5p inhibits the progression of UUO-induced renal injury in vivo.
Figure 5.
Overexpression of miR-181d-5p inhibits the progression of UUO-induced tubulointerstitial fibrosis in vivo. (A) Masson staining and immunohistochemistry staining of α-SMA and vimentin in kidney tissues, scale bar: 50 µm. (B) Relative positive area of immunohistochemistry staining analysis. (C) The expression of miR-181d-5p in UUO mice after treatment with miR-181d-5p agomir. (D) Western blot analysis showing the protein level of α-SMA and vimentin in the UUO mice. (E) Relative expression of α-SMA and vimentin in the UUO mice. ##P<0.01, ###P<0.001 vs. control group; *P<0.05, **P<0.01, ***P<0.001 vs. UUO group. UUO, unilateral ureteral obstruction; SMA, smooth muscle actin; NC, negative control; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Overexpression of miR-181d-5p prevents unilateral ureteral obstruction-induced apoptosis through downregulation of SIRT1
The effects of miR-181d-5p on UUO-induced apoptosis were assessed in mice. The TUNEL staining clearly showed a significant increase in TUNEL-positive cells in the UUO model group compared to the sham group, which was partially reduced upon treatment with the miR-181d-5p agomir (Figure 6A,6B). Immunohistochemical staining revealed positive expression of apoptosis-related molecules including Bax, cleaved caspase-3, p53, and SIRT1 in UUO mice compared to sham animal. However, treatment with the miR-181d-5p agomir inhibited SIRT1 upregulation and prevented the expression of p53, Bax, and cleaved caspase-3 in the UUO mice (Figure 6C). Western blot analysis also confirmed that miR-181d-5p agomir treatment downregulated the expression of SIRT1 and altered the ratio of Bax/Bcl2, and reduced the expression of cleaved caspase-3 (Figure 6D,6E). These results suggested that miR-181d-5p overexpression prevents UUO-induced apoptosis through downregulation of SIRT1.
Figure 6.
Overexpression of miR-181d-5p prevents unilateral ureteral obstruction-induced apoptosis through downregulation of SIRT1. (A) TUNEL stained kidney sections from UUO mice and sham mice. The green fluorescence indicates positive cells, scale bar: 100 µm. (B) Statistical analysis of the relative proportion of TUNEL-positive cells (n=3). (C) Immunohistochemistry staining of bax, cleaved caspase-3, p53, and SIRT1, scale bar: 50 µm. (D) Western blot analysis of SIRT1, p53, bax, bcl2, and cleaved caspase-3. (E) Densitometric analysis of the Western blots. #P<0.05, ##P<0.01 vs. sham group, *P<0.05, **P<0.01 vs. UUO group. UUO, unilateral ureteral obstruction; NC, negative control; SIRT1, sirtuin 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Discussion
This report analyzed the renal circRNA expression profile of children with UPJO. The RNA sequencing analysis identified 63 upregulated circRNAs and 64 downregulated circRNAs in UPJO patients. Of these differential circRNAs, the expression of circ_0045861 was significantly elevated in injured renal cells both in vivo and in vitro. Furthermore, circ_0045861 was shown to promote TGF-β1-induced apoptosis in HK-2 cells in vitro, likely through sponging miR-181d-5p to reduce the inhibitory effects of this microRNA on SIRT1. Indeed, miR-181d-5p inhibited the apoptosis of renal cells through the SIRT1/p53 signaling axis. Therefore, the circ_0045861/miR-181d-5p/SIRT1 signaling axis promotes the apoptosis of kidney cells and this may represent a potential therapeutic target for UPJO patients.
CircRNAs can act as sponges for miRNAs to prevent the regulation of target genes, thereby exerting a competitive endogenous RNA mechanism (19). Previously, several studies have shown that circRNAs are closely related to chronic kidney disease and renal cell carcinoma (20-22). Hsa_circ_001895 can promote the development and progression of renal cell carcinomas by increasing the adsorption of miR-296-5p and the expression of sex-determining region Y (SRY)-box 12 (23). In addition, studies have reported that circ-AKT3 promotes ischemia-reperfusion (I/R)-induced acute kidney injury progression via regulating oxidative stress through sponging miR-144-5p to activate the Wnt/β-catenin signaling pathway (24). At present, the pathological mechanisms of UPJO in children are still unclear, and in particular, the regulatory mechanisms of circRNAs remain to be fully elucidated. In this study, RNA sequencing was used to identify the differentially expressed circRNAs and the expression of circ_0045861 was found to be significantly increased in children with UPJO. While circ_0045861 was detected in both the cell nucleus and cell cytoplasm, it was significantly enriched in the cytoplasm. Furthermore, silencing circ_0045861 inhibited TGF-β1-induced apoptosis in HK-2 cell in vitro. These results suggested that circ_0045861 may play an important role in the progression of UPJO.
Bioinformatics tools identified miR-181d-5p as a potential target of circ_0045861. Recently, miR-181d-5p has been reported to be involved in the development of a variety of tumors (25-28). MiR-181d-5p-containing exosomes derived from cancer-associated fibroblasts can promote epithelial-mesenchymal transition by regulating CDX2/HOXA5 in breast cancer cells (29). In addition, miR-181d-5p improves ischemia/reperfusion-induced acute renal injury through regulating apoptosis and inflammation by targeting KLF6 (30). In our current study, the expression of miR-181d-5p was significantly decreased in vivo in UUO mice and in vitro in HK-2 cells treated with TGF-β1. Dual luciferase reporter assay demonstrated an interaction between miR-181d-5p and circ_0045861. In addition, in situ hybridization analysis revealed that both miR-181d-5p and circ_0045861 were localized to the cell cytoplasm. Furthermore, overexpression of miR-181d-5p inhibited TGF-β1-induced HK-2 cell apoptosis. These results suggested that circ_0045861 interacts with miR-181d-5p and regulates its expression.
SIRT1 is known to promote mitochondria-dependent apoptosis by preventing the nuclear translocation of p53, resulting in its accumulation in the cytoplasm and its transport to the mitochondria (31,32). Bioinformatics analysis revealed that SIRT1 may be a potential target gene of miR-181d-5p and this was verified by dual luciferase reporter experiments. Furthermore, miR-181d-5p mimics reduced the protein expression level of SIRT1, while miR-181d-5p inhibitors increased SIRT1 protein expression. These results suggested that SIRT1 is a target gene of miR-181d-5p. Cleaved caspase-3 and Bax/Bcl-2 are key apoptosis-related proteins involved in mitochondria-dependent apoptosis. Furthermore, renal fibrosis in UUO mice could be reversed by treatment with miR-181d-5p agomir, and the expression of SIRT1, p53, Bax/bcl-2, and cleaved caspase-3 in the kidney tissues of these mice were also significantly decreased. This suggested that miR-181d-5p may inhibit apoptosis and renal injury by blocking SIRT1/p53-dependent apoptosis.
There were several potential limitations to this study. The homologous gene of circ_0045861 could not be identified in mice, and thus, the function of this circRNA could not be verified in the mouse model. Despite our findings, circ_0045861 may regulate apoptosis through multiple mechanisms, and further studies are warranted. Additionally, miR-181d-5p may have other target genes and further research is required to identify these genes.
The results of this investigation suggested that circ_0045861 contributes to the pathogenesis of UPJO as a competitive endogenous RNA to regulate SIRT1 expression by sponging miR‐181d‐5p. Therefore, the circ_0045861/miR-181d-5p/SIRT1 signaling axis may represent a potential therapeutic target for the treatment of UPJO patients.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (No. 81571514).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Medical Research and New Technology Ethics Committee of Shengjing Hospital of China Medical University (approval No. 2012PS81K) and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. 2018PS229K) granted by ethics committee of Shengjing Hospital of China Medical University, in compliance with the NIH guidelines for the care and use of animals.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-5060
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-5060
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-5060). The authors have no conflicts of interest to declare.
(English Language Editor: J. Teoh)
References
- 1.Ek S, Lidefeldt KJ, Varricio L. Fetal hydronephrosis; prevalence, natural history and postnatal consequences in an unselected population. Acta Obstet Gynecol Scand 2007;86:1463-6. 10.1080/00016340701714802 [DOI] [PubMed] [Google Scholar]
- 2.Vemulakonda VM. Ureteropelvic junction obstruction: diagnosis and management. Curr Opin Pediatr 2021;33:227-34. 10.1097/MOP.0000000000000994 [DOI] [PubMed] [Google Scholar]
- 3.Dudley JA, Haworth JM, McGraw ME, et al. Clinical relevance and implications of antenatal hydronephrosis. Arch Dis Child Fetal Neonatal Ed 1997;76:F31-4. 10.1136/fn.76.1.F31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberti C. Congenital ureteropelvic junction obstruction: physiopathology, decoupling of tout court pelvic dilatation-obstruction semantic connection, biomarkers to predict renal damage evolution. Eur Rev Med Pharmacol Sci 2012;16:213-9. [PubMed] [Google Scholar]
- 5.Higgins CE, Tang J, Higgins SP, et al. The Genomic Response to TGF-β1 Dictates Failed Repair and Progression of Fibrotic Disease in the Obstructed Kidney. Front Cell Dev Biol 2021;9:678524. 10.3389/fcell.2021.678524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Gao L, Yu ZH, et al. LncRNA HOTAIR promotes renal interstitial fibrosis by regulating Notch1 pathway via the modulation of miR-124. Nephrology (Carlton) 2019;24:472-80. 10.1111/nep.13394 [DOI] [PubMed] [Google Scholar]
- 7.Ponnusamy M, Zhou X, Yan Y, et al. Blocking sirtuin 1 and 2 inhibits renal interstitial fibroblast activation and attenuates renal interstitial fibrosis in obstructive nephropathy. J Pharmacol Exp Ther 2014;350:243-56. 10.1124/jpet.113.212076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong YA, Kim JE, Jo M, et al. The Role of Sirtuins in Kidney Diseases. Int J Mol Sci 2020;21:6686. 10.3390/ijms21186686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab 2014;25:138-45. 10.1016/j.tem.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma Y, Ouyang J, Wei J, et al. Involvement of Host Non-Coding RNAs in the Pathogenesis of the Influenza Virus. Int J Mol Sci 2016;18:39. 10.3390/ijms18010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol 2015;12:381-8. 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma N, Zhang W, Wan J. Research Progress on circRNA in Nervous System Diseases. Curr Alzheimer Res 2020;17:687-97. 10.2174/1567205017666201111114928 [DOI] [PubMed] [Google Scholar]
- 13.Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 2015;444:132-6. 10.1016/j.cca.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 14.Lu D, Xu AD. Mini Review: Circular RNAs as Potential Clinical Biomarkers for Disorders in the Central Nervous System. Front Genet 2016;7:53. 10.3389/fgene.2016.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontemaggi G, Turco C, Esposito G, et al. New Molecular Mechanisms and Clinical Impact of circRNAs in Human Cancer. Cancers (Basel) 2021;13:3154. 10.3390/cancers13133154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Heng B, Ouyang P, et al. Comprehensive profiling of circRNAs and the tumor suppressor function of circHIPK3 in clear cell renal carcinoma. J Mol Histol 2020;51:317-27. 10.1007/s10735-020-09882-9 [DOI] [PubMed] [Google Scholar]
- 17.Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014;20:1666-70. 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015. doi: . 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Zhang Y, Wang P, et al. Circular RNAs in renal cell carcinoma: implications for tumorigenesis, diagnosis, and therapy. Mol Cancer 2020;19:149. 10.1186/s12943-020-01266-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin J, Sun H, Shi C, et al. Circular RNA in renal diseases. J Cell Mol Med 2020;24:6523-33. 10.1111/jcmm.15295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng F, Gong W, Li S, et al. circRNA_010383 Acts as a Sponge for miR-135a and its Downregulated Expression Contributes to Renal Fibrosis in Diabetic Nephropathy. Diabetes 2020. [Epub ahead of print]. doi: . 10.2337/db200203 [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, Xiao K, Chen S, et al. Circular RNA hsa_circ_001895 serves as a sponge of microRNA-296-5p to promote clear cell renal cell carcinoma progression by regulating SOX12. Cancer Sci 2020;111:713-26. 10.1111/cas.14261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Jiang W, Zhong L, et al. circ-AKT3 aggravates renal ischaemia-reperfusion injury via regulating miR-144-5p /Wnt/β-catenin pathway and oxidative stress. J Cell Mol Med 2020. [Epub ahead of print]. doi: . 10.1111/jcmm.16072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao LM, Zheng Y, Wang P, et al. Tumor-suppressive effects of microRNA-181d-5p on non-small-cell lung cancer through the CDKN3-mediated Akt signaling pathway in vivo and in vitro. Am J Physiol Lung Cell Mol Physiol 2019;316:L918-33. 10.1152/ajplung.00334.2018 [DOI] [PubMed] [Google Scholar]
- 26.Kim S, Bae WJ, Ahn JM, et al. MicroRNA signatures associated with lymph node metastasis in intramucosal gastric cancer. Mod Pathol 2021;34:672-83. 10.1038/s41379-020-00681-x [DOI] [PubMed] [Google Scholar]
- 27.Dong X, Liu Y, Deng X, et al. C1GALT1, Negatively Regulated by miR-181d-5p, Promotes Tumor Progression via Upregulating RAC1 in Lung Adenocarcinoma. Front Cell Dev Biol 2021;9:707970. 10.3389/fcell.2021.707970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei H, Wang J, Xu Z, et al. Hepatoma Cell-Derived Extracellular Vesicles Promote Liver Cancer Metastasis by Inducing the Differentiation of Bone Marrow Stem Cells Through microRNA-181d-5p and the FAK/Src Pathway. Front Cell Dev Biol 2021;9:607001. 10.3389/fcell.2021.607001 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Wang H, Wei H, Wang J, et al. MicroRNA-181d-5p-Containing Exosomes Derived from CAFs Promote EMT by Regulating CDX2/HOXA5 in Breast Cancer. Mol Ther Nucleic Acids 2020;19:654-67. 10.1016/j.omtn.2019.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Li C, Guan C, et al. MiR-181d-5p Targets KLF6 to Improve Ischemia/Reperfusion-Induced AKI Through Effects on Renal Function, Apoptosis, and Inflammation. Front Physiol 2020;11:510. 10.3389/fphys.2020.00510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ong ALC, Ramasamy TS. Role of Sirtuin1-p53 regulatory axis in aging, cancer and cellular reprogramming. Ageing Res Rev 2018;43:64-80. 10.1016/j.arr.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 32.Verdin E, Hirschey MD, Finley LW, et al. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci 2010;35:669-75. 10.1016/j.tibs.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as