Abstract
Background
The rice weevil Sitophilus oryzae is one of the most important agricultural pests, causing extensive damage to cereal in fields and to stored grains. S. oryzae has an intracellular symbiotic relationship (endosymbiosis) with the Gram-negative bacterium Sodalis pierantonius and is a valuable model to decipher host-symbiont molecular interactions.
Results
We sequenced the Sitophilus oryzae genome using a combination of short and long reads to produce the best assembly for a Curculionidae species to date. We show that S. oryzae has undergone successive bursts of transposable element (TE) amplification, representing 72% of the genome. In addition, we show that many TE families are transcriptionally active, and changes in their expression are associated with insect endosymbiotic state. S. oryzae has undergone a high gene expansion rate, when compared to other beetles. Reconstruction of host-symbiont metabolic networks revealed that, despite its recent association with cereal weevils (30 kyear), S. pierantonius relies on the host for several amino acids and nucleotides to survive and to produce vitamins and essential amino acids required for insect development and cuticle biosynthesis.
Conclusions
Here we present the genome of an agricultural pest beetle, which may act as a foundation for pest control. In addition, S. oryzae may be a useful model for endosymbiosis, and studying TE evolution and regulation, along with the impact of TEs on eukaryotic genomes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12915-021-01158-2.
Keywords: Coleoptera, Weevil, Sitophilus oryzae, Genome, Transposable elements, Endosymbiosis, Immunity, Evolution
Background
Beetles account for approximately 25% of known animals, with an estimated number of 400,000 described species [1–3]. Among them, Curculionidae (true weevils) is the largest animal family described, comprising about 70,000 species [1, 4, 5]. Despite being often associated with ecological invasion and ecosystem degradation, only three Curculionidae genomes are publicly available to date [6–8]. Among the cereal weevils, the rice weevil Sitophilus oryzae is one of the most important pests of crops of high agronomic and economic importance (wheat, maize, rice, sorghum, and barley), causing extensive quantitative and qualitative losses in field, stored grains, and grain products throughout the world [9–11]. Moreover, this insect pest is of increasing concern due to its ability to rapidly evolve resistance to insecticides such as phosphine, a fumigant used to protect stored grains from insect pests [12–14].
Like other holometabolous insects, the life cycle of S. oryzae can be divided into four stages: egg, larva, pupa, and adult (Fig. 1). Females drill a small hole in the grain, deposit a single egg, and seal it with secretions from their ovipositor. Up to six eggs can be laid daily by each female, totaling around 400 eggs over its entire lifespan [15]. Larvae develop and pupate within the grain kernel, metamorphose, and exit the grain as adults. The whole process takes on average 30 days [10]. Like many insects living on nutritionally poor diets, cereal weevils permanently associate with nutritional intracellular bacteria (endosymbionts) that supply them with nutrients that are not readily available in the grains, thereby increasing their fitness and invasive power. The endosymbiont of S. oryzae, the gamma-proteobacterium Sodalis pierantonius [16, 17], is housed within specialized host cells, named bacteriocytes, that group together into an organ, the bacteriome [18]. Contrasting with most studied symbiotic insects, the association between Sitophilus spp. and S. pierantonius was established recently (less than 30,000 years ago), probably following the replacement of the ancestor endosymbiont, Candidatus Nardonella, in the Dryophthorinae subfamily [19, 20]. As a result, contrary to long-lasting endosymbiotic associations, the genome of S. pierantonius is GC rich (56.06%), and its size is similar to that of free-living bacteria (4.5 Mbp) [16]. Moreover, it encodes genes involved in bacterial infection, including type three secretion systems (TTSS), as well as genes encoding microbial associated molecular patterns (MAMPs) that trigger Pattern Recognition Receptors (PRR) and are usually absent or reduced in bacteria involved in long-lasting associations [16, 21, 22]. Nevertheless, many features indicate that the genome of S. pierantonius is in a process of degradation, as it contains many pseudogenes (43% of the predicted protein-coding sequences) and a large number of mobile elements (18% of the genome size) [16, 23]. Finally, it is important to note that no other symbionts, with the exception of the familiar Wolbachia endosymbiont in some strains, have been described in S. oryzae.
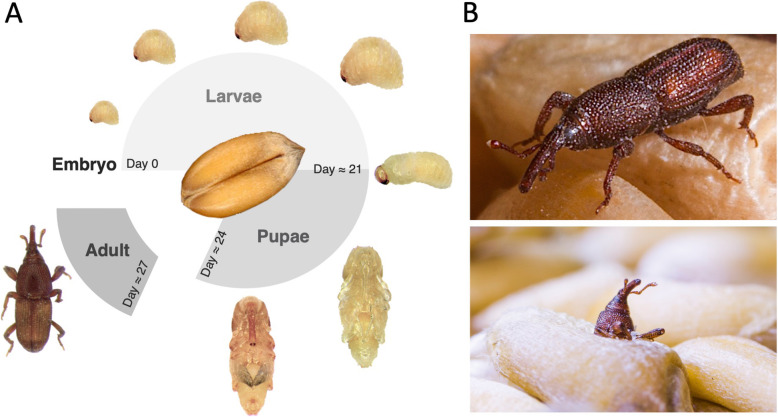
Fig. 1.
Sitophilus oryzae overview. A Life cycle of cereal weevil Sitophilus oryzae. The embryo develops into a larva and pupa, and metamorphoses into a young adult, exiting the grain around 3 days after metamorphosis completion. The developmental times indicated are from a rearing condition at 27 °C and 70% relative humidity. B Photos of adult S. oryzae. Lower panel shows an adult exiting the grain
In order to help unravel potential adaptive functions and features that could become the basis for identifying novel control strategies for weevils and other major insect pests, we have undertaken the sequencing, assembly, and annotation of the genome of S. oryzae. Strikingly, the repeated fraction of S. oryzae’s genome (repeatome), composed mostly of transposable elements (TEs), is among the largest found to date in insects. TEs, the most versatile DNA units described to date, are sequences present in multiple copies and capable of relocating or replicating within a genome. While most observed TE insertions evolve neutrally or are slightly deleterious, there are a number of documented cases where TEs may facilitate host adaptation (for reviews, see [24–26]). For instance, gene families involved in xenobiotic detoxification are enriched in TEs in Drosophila melanogaster [27], Plutella xylostella [28], a major crop pest, and Myzus persicae, another phytophagous insect causing significant agronomic losses [29]. TEs have also been frequently associated with insecticide resistance in Drosophila species [30–32]. In addition, population genetics studies suggested that more than 84 TE copies in D. melanogaster may play a positive role in fitness-related traits [33], including xenobiotic resistance [32] and immune response to Gram-negative bacteria [34].
In eukaryotes, TE content varies drastically and contributes significantly to the size and organization of the genome. From TE-rich genomes as maize (85% [35]), humans (≈45% [36]), and the recently sequenced lungfish (≈90% [37]) for instance, to TE-poor genomes, as D. melanogaster (12–15% [38]), or Arabidopsis thaliana (≈10% [39]), repeatomes thrive on a high level of diversity. These drastic variations are also observed within animal clades, such as insects, where the proportion of TE ranges from 2% in the Antarctic midge (Belgica antarctica) to 65% in the migratory locust (Locusta migratoria) [40–42] and up to 75% in morabine grasshoppers (Vandiemenella viatica species) [43]. In addition to the overall TE content, the number of different TE families (homogeneous groups of phylogenetically related TE sequences), their size (number of copies per family), and sequence diversity are also very high among insect species [44]. For instance, SINEs (short interspersed elements) are almost absent from most insect genomes, but many lepidopterans harbor these elements [44]. In flies, long terminal repeat retrotransposons (LTRs) are a staple of the Drosophila genus, but such TEs are nearly absent from other dipteran genomes (e.g., Glossina brevipalpis and Megaselia scalaris) [44]. Recent advances in sequencing have dramatically increased the level to which TEs can be studied across species and reveal that such variations can persist even within recently diverged groups, as observed within Drosophila species [45], or among Heliconius butterflies [46]. An increasing number of insect genomes are reported with large repeatomes (e.g. Aedes aegypti and Ae. albopictus 40–50% [47, 48], L. migratoria 60–65% [40, 41], Dendrolimus punctatus 56% [49], Vandiemenella viatica species 66–75% [43]).
Here we present the genome of S. oryzae, with a strong focus on the repeatome, its largest genomic compartment, spanning over ≈74% of the assembly. S. oryzae represents a model system for stored grain pests, host-TE evolutionary biology, and the study of the molecular mechanisms acting at the early steps of symbiogenesis. Moreover, the features uncovered suggest that S. oryzae and its relatives have the potential to become a platform to study the interplay between TEs, host genomes, and endosymbionts.
Results and discussion
Genome assembly and annotation
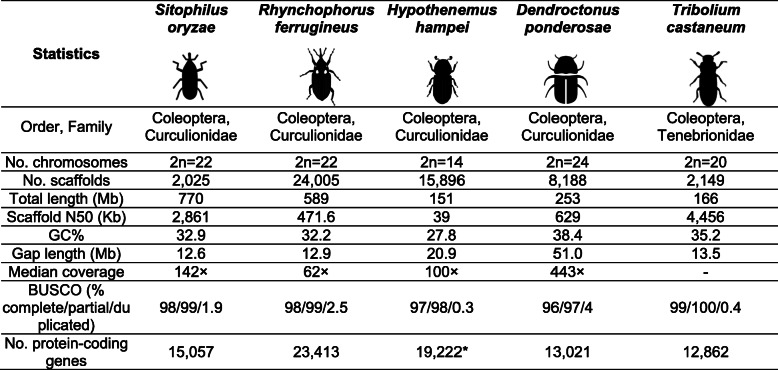
We have sequenced and assembled the genome of the rice weevil S. oryzae at a base coverage depth of 142× using a combination of short- and long-read strategies (see “Methods” and Additional file 1). The assembly pipeline was defined to optimize multiple criteria including gene completeness (BUSCO scores [50]) and reference-free metrics (number of contigs, total length, N50, number of N’s per 100 kbp and the proportion of shared 100-mers between the assembly and short reads). The karyotype of S. oryzae comprises 22 chromosomes [51], and the genome assembly consists of 2025 scaffolds spanning 770 Mbp with a N50 of 2.86 Mbp, demonstrating a high contiguity compared to other Coleopteran genomes (Table 1). The assembly size is consistent with the genome size measured through flow cytometry (769 Mbp in females and 768 Mbp in males [51]). Haploid genome size estimations based on k-mer distributions of the short reads ranged from 785 Mbp (GenomeScope [58]) over 814 Mbp (gce [59]) to 818 Mbp (findGSE [60]), in agreement with the assembly size. BUSCO scores show the assembly is complete (97.9% complete and 0.7% fragmented), with a low duplication rate (1.9%). Consistent with the low duplication rate at the gene level, no significant haplotig contamination was observed. Finally, to confirm the completeness and consensus quality of S. oryzae’s assembly, we have firstly performed a K-mer analysis (100-mers), revealing that around 92% of the 100-mers of our assembly are covered by the 100-mers from the short reads, and secondly, 98% of the short reads were able to map to the assembly. Hence, thanks to the aforementioned statistics, S. oryzae is the best assembled Curculionidae genome to date [7, 52, 61] (Table 1). The complete analysis of gene content and function can be found in Additional files 2 and 3.
Table 1.
Assembly statistics of S. oryzae’s genome in comparison to Curculionidae genomes and T. castaneum [6, 7, 51–57]
*All genes, no NCBI RefSeq annotation report available
Annotation of the Sitophilus oryzae genome
Among the different pathways we were able to decipher in the genome of S. oryzae, we present here highlights of the main annotation efforts, followed by a detailed analysis of the TE content and impact on the host genome. A comprehensive analysis for each highlight is presented as Supplemental Notes in Additional file 2.
Phylome and horizontal gene transfer
Sitophilus oryzae has a high gene expansion rate when compared to other beetles. Some of the families with the largest expansions include genes coding for proteins with DNA binding motifs, potentially regulating functions specific to this clade. Olfactory receptors, antimicrobial peptides (AMPs), and P450 cytochromes were expanded as well, probably in response to their ecological niche and lifestyle. Additionally, we noticed an expansion of plant cell wall-degrading enzymes that originated from horizontal gene transfer (HGT) events from both bacteria and fungi. Given the intimate relationship between S. oryzae and its endosymbiont, including the permanent infection of the female germline, we searched for evidence for HGT in the weevil genome possibly coming from S. pierantonius. Contrary to the genome of the tsetse fly Glossina, where at least three HGT events from Wolbachia have been reported [62], we were unable to pinpoint any HGT event from either the ancient endosymbiont Nardonella, Wolbachia, or the recently acquired S. pierantonius. A detailed description is reported in Additional file 2: Supplemental Note 1 [19, 63–86], and Note 3 for digestive enzymes [52, 87–107].
Global analysis of metabolic pathways
Using the CycADS [108] pipeline and Pathway Tools [109], we have generated BioCyc metabolism reconstruction databases for S. oryzae and its endosymbiont S. pierantonius. We compared S. oryzae metabolism to that of other arthropods available in the ArthropodaCyc [110] collection and we explored the metabolic exchanges between weevils and their endosymbionts. The metabolic reconstruction reveals that, despite its large genome for an endosymbiotic bacterium, S. pierantonius relies on its host for several central compounds, including alanine and proline, but also isocitrate, inosine monophosphate (IMP), and uridine monophosphate (UMP), to produce essential molecules to weevils, including the essential amino acids tryptophan, phenylalanine, lysine, and arginine, the vitamins pantothenate, riboflavin, and dihydropteroate as a folate precursor, and nicotinamide adenine dinucleotide (NAD) (Additional file 2: Supplemental Note 2). Among the amino acids listed above, phenylalanine, in particular, is an essential precursor for the cuticle synthesis in emerging adults [111]. In addition, several studies have shown that S. pierantonius improves host fitness, including fertility, developmental time, and flight capacity, in part by supplying the host with vitamins and improving its mitochondrial energy metabolism [112–114]. See Additional file 2: Supplemental Note 2 [19, 20, 108–110, 112, 113, 115–120] for more information.
Development
Developmental gene regulatory networks of D. melanogaster and Tribolium castaneum were used to annotate S. oryzae genes with roles in signaling, embryonic patterning, oogenesis, segmentation and segment identity, organogenesis, appendage and eye development, and insect size and developmental transitions. Overall, we observed a high level of conservation in comparison to the red flour beetle Tribolium castaneum, a model coleopteran. When compared to D. melanogaster, several key coordinate group genes are absent in T. castaneum and S. oryzae, most notably the anterior group genes bicoid and swallow and the posterior group gene oskar. Moreover, seven developmental genes with two homologs in the Drosophila genome are represented by a single ortholog in T. castaneum and S. oryzae. We also observed that homologs for signaling pathway ligands could not always be identified, which, given the presence of conserved receptors, is probably due to divergent primary sequence of the ligands. A detailed description is reported in Additional file 2: Supplemental Note 4 [52, 121–138].
Cuticle protein genes
Among the distinctive biological features of coleopterans is the ability to generate a hard and thick cuticle that protects them against dehydration and represents the first physical barrier from infections and topical insecticide penetration. The analysis of cuticle proteins (CPs) showed that S. oryzae has an average number of CPs, but with an enrichment of members of the CPAP1 family. While some members of this family are known to be involved in molting and maintaining the integrity of the cuticle in T. castaneum, most are still uncharacterized [139, 140]. Thus, these proteins might be involved in the development of specific cuticular tissues in S. oryzae or other weevils. The total number of CPs did not follow the taxonomy of beetles, suggesting instead that it might be an adaptation to their diverse lifestyles. For details, see Additional file 2: Supplemental Note 5 [139–143].
Innate immune system
The analysis of immunity-related genes revealed that the genome of S. oryzae encodes the canonical genes involved in the three main antimicrobial pathways Toll, Imd, and JAK-STAT, suggesting functional conservation of these pathways in cereal weevils. The conservation of the Imd pathway in the S. oryzae genome is of particular interest as its degradation in other symbiotic insects (Acyrthosiphon pisum [144], B. tabaci [145], or Rhodnius prolixus [146]) was initially correlated to their symbiotic status. The Imd pathway is not only present in S. oryzae, but it is also functional [147, 148], and has evolved molecular features necessary for endosymbiont control [147] and host immune homeostasis [148]. Thus, not only is the Imd pathway conserved in cereal weevils, contrary to aphids and some other hemimetabolous insects, but it seems to have been evolutionary “rewired” toward additional functions in symbiotic homeostasis [147]. A detailed description can be seen in Additional file 2: Supplemental Note 6 [41, 62, 144–193].
Detoxification and insecticide resistance
Fumigation using phosphine, hydrogen phosphide gas (PH3), is by far the most widely used treatment for the protection of stored grains against insect pests due to its ease of use, low cost, and universal acceptance as a residue-free treatment [194, 195]. However, high-level resistance to this fumigant has been reported in S. oryzae from different countries [13, 196–203]. Hence, we searched for genes associated with detoxification and resistance to insecticide and more generally to toxins, including plant allelochemicals. The S. oryzae repertoire of detoxification and insecticide resistance genes includes more than 300 candidates, similar to what is seen in other coleopteran genomes. For more details, see Additional file 2: Supplemental Note 7 [12–14, 110, 194–212].
Odorant receptors
One promising pest management strategy relies on modifying insect behavior through the use of volatile organic compounds that act on odorant receptors (ORs) [213, 214]. ORs play a significant role in many crucial behaviors in insects by mediating host-seeking behavior, mating, oviposition, and predator avoidance [215]. Interfering with the behavior of pest insects and modulating their ability to find suitable hosts and mates has been shown to reduce population numbers, notably using plants that are capable of producing attractants and repellents [216, 217]. Sitophilus spp. are known to use kairomones for host detection [218, 219], as well as aggregation pheromones [220, 221]. We annotated 100 candidate OR genes in S. oryzae (named SoryORs), including the gene encoding the co-receptor Orco. Of these genes, 46 were predicted to encode a full-length sequence. The global size of the SoryOR gene repertoire is in the range of what has been described in other species of the coleopteran suborder Polyphaga (between 46 in Agrilus planipennis and more than 300 in T. castaneum) and close to the number of OR genes annotated in the closely related species Dendroctonus ponderosae (85 genes, [222]) (Additional file 2: Supplemental Note 8 [204, 218–242]).
Massive expansion of TE copies in the genome of S. oryzae
Detection and annotation of the repeatome
The repeatome represents the fraction of the genome categorized as repetitive. It encompasses TEs, satellites, tandem, and simple repeats. Eukaryotic TEs can be separated into two classes, depending on their replication mode [243]. DNA (Class II)-based elements are able to directly move within a genome and include terminal inverted repeat (TIR), Crypton, rolling circle (RC/Helitron), and large composite elements (Maverick). Conversely, retrotransposons (Class I) have an RNA intermediate and replicate through RNA retrotranscription. Retrotransposons can be further divided into long terminal repeat (LTR), and non-LTR elements, including long and short interspersed nuclear repeat elements (LINEs and SINEs). Other retrotransposons include Penelope-like (PLEs) and DIRS-like elements. Each one of these TE orders can be further classified into specific superfamilies (as for instance Copia or Gypsy LTR elements, and hAT or Tc1/Mariner TIR elements) that may encompass hundreds of TE families, each containing thousands of copies. The intrinsic diversity of TEs complicates their identification and annotation, especially in understudied species genera.
We used multiple state-of-the-art TE detection tools, including RepeatModeler2 and EDTA [244, 245], to generate consensus sequences of the TE families in S. oryzae. After an initial discovery step, more than 10,000 likely redundant TE families were identified by the dedicated programs; we combined their results using multiple sequence alignments and clustering (see “Methods” and Additional file 2: Figure S1) to reduce this number to 3399. After quality filtering (see “Methods”), the final library includes a total of 3361 sequences. Due to the evolutionary distance between S. oryzae and other known coleopterans, the consensus sequences obtained were further classified using a thorough combination of sequence homology and structure (see “Methods”). The S. oryzae genome is among the most TE-rich insect genomes to date. Comparison of TE genomic content as given by RepeatMasker using TE libraries from RepeatModeler 2.0.1, EDTA v1.7.8 or our custom pipeline shows that traditional methods miss ~ 5% of TEs in spite of harboring more complexity (more total TE consensus, Table S1). Thus, we conclude that our method is likely to improve the overall quality of the TE annotation.
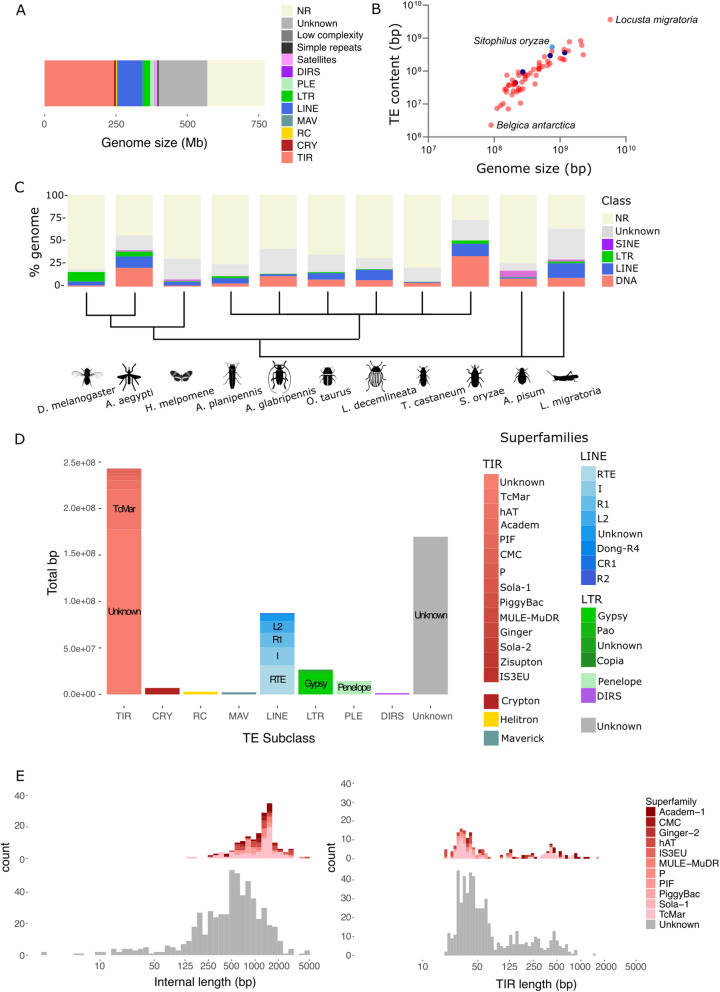
We uncovered 570 Mbp of repeat sequences, corresponding to ≈74% of the S. oryzae genome: ≈2% of satellite sequences, simple or low-complexity repeats, and ≈72% of other mobile elements, including TEs (Fig. 2A, Additional file 4). Given the limitation of the sequencing technologies, the proportion of satellites and TEs usually abundant in the heterochromatin is likely underestimated. We took advantage of a recent comparative analysis of TE content in 62 insect species [40] to contrast with the S. oryzae TE compartment. The S. oryzae genome ranks among those with the highest TE fraction observed in insects (Fig. 2B, C). Within the largest insect order, Coleoptera, very little is known regarding TE distribution and evolution. T. castaneum harbors only 6% of TEs [52] and Hypothenemus hampei contains 8.2% of TEs [6, 247], while Dichotomius schiffleri harbors 21% [248]. The species closest to S. oryzae, Rhynchophorus ferrugineus, has a TE content of 45% [8]. Therefore, while TE content has been described to follow phylogenetic relationships in most insects [44, 45], there is a large variation among the few Coleoptera species with available genomes. It is important to note that the pipeline we used to detect and annotate TEs in S. oryzae differs from the method implemented by Petersen and colleagues [40], as we incorporated 31 manually curated TE references for S. oryzae, and specifically annotated DNA/TIR elements based on their sequence structure (see “Methods”), increasing the annotation sensitivity.
Fig. 2.
A Proportion of repeat content in S. oryzae’s genome. The majority of repeats detected in S. oryzae are represented by Class II (TIR) elements, LINEs (Class I), and unclassified repeats (unknown). NR: non repetitive. B Variation of genome size and TE content in 62 insect species from [40] and S. oryzae. Coleopteran species are depicted in dark blue, and S. oryzae in light blue. S. oryzae is clearly a TE-rich genome. C TE proportion across 11 insect species, including six coleoptera. In agreement with the data used for comparison [40], PLEs are included in the LINE superfamilies, DIRS in LTRs, and RC, CRY, MAV and TIR in the DNA superfamilies. NR: non repetitive. S. oryzae harbors the largest TE content among Coleopterans and most insect species studied to date. Within Coleoptera, there is a large variation in TE content and type, with A. planipennis, L. decemlineata, and O. taurus carrying an abundant LINE content, while S. oryzae, T. castaneum, and A. glabripennis show larger DNA content. Cladogram based on [246]. D Classification of the 570 Mbs of TEs present in the S. oryzae genome. Most TIR families detected were not classified into known superfamilies. RTE LINE and Gypsy LTR elements are the most abundant superfamilies among retrotransposons. Around 21% of repeats in S. oryzae’s genome were not classified by our pipeline, and remain unknown (gray). E Distribution of TIR length sequences (right) detected by einverted and the internal region present between both TIRs (left) for complete consensus of TIR superfamilies (color) and unknown TIR families (gray)
Class II (DNA) elements dominate S. oryzae’s genome
The most striking feature of the genome of S. oryzae is the high abundance of Class II (DNA) elements (≈32% of the genome, ≈43% of the TE content) (Fig. 2A), which is the highest observed among all 62 insect species included in this analysis [40–42]. The most DNA transposon-rich genomes include mosquito Culex quinquefasciatus and Ae. aegypti, harboring 25% and 20% of DNA transposon content in their genome, amounting to 54% and 36% of the total TE compartment, respectively [6]. The TE-rich grasshopper L. migratoria repeatome comprises only 14% of DNA transposons, while LINE retroelements (Class I) amount to 25%. Morabine grasshoppers, with up to 75% of TE content, show equivalent amounts of DNA, LINE, and Helitrons [43]. Finally, among Coleoptera, a large diversity of repeatomes is observed (Fig. 2C) with A. planipennis, Leptinotarsa decemlineata, and Onthophagus taurus carrying an abundant LINE content, while S. oryzae, T. castaneum, and Anoplophora glabripennis show larger DNA transposon content.
Among the Class II elements present in S. oryzae, the majority belongs to the TIR subclass but has not been assigned a known superfamily (Fig. 2D), while Tc Mariners make up ≈6% of DNA elements. Among the consensus sequences, we were able to assemble from 5′TIR to 3′TIR (highest confidence, see “Methods”), the length distribution shows a continuum starting at a couple of hundred bases to a maximum of ≈ 5 kbp (see Fig. 2E). We hypothesize that most of the smaller TIR families observed are miniature inverted repeat elements (MITEs). MITEs are non-autonomous elements, deriving from autonomous Class II/TIR copies, comprising two TIRs flanking a unique, non-coding, region (sometimes absent) of variable length. While the TE detection pipeline used was able to detect and annotate most Class II/TIR elements based on transposase homologies, we also specifically searched for non-autonomous TIR sequences, allowing the detection of putative MITEs that lack protein-coding regions (Additional file 2: Figure S1). Among all Class II/TIR superfamilies, TIR length varies between tens of base pairs to ≈1 kbp (Fig. 2E). We identified short elements, composed mostly of their TIR sequences (Fig. 2E), typical of MITEs. Interestingly, the unknown TIR families show an average size smaller than 1 kbp, while TIRs with an annotated superfamily, show larger sizes (Additional file 2: Figure S3), suggesting that most unknown families could be indeed non-autonomous MITEs. MITE size ranges were previously described from around 100 bp to copies reaching more than 1 kbp [249]. Finally, the distribution of the proportions of TIR relative to the consensus length appears superfamily-specific (Fig. 2E and Additional file 2: Figure S3), and unknown families recapitulate these patterns. In conclusion, while most unknown TIR families seem to be composed of MITEs, we cannot exclude that our homology database is limited, likely missing some unknown protein domains. The most abundant TE family detected in the S. oryzae genome is indeed a MITE element (TE2641_SO2_FAM0704), with 10,486 genomic hits (or the equivalent of ≈4117 copies based on the consensus size), corresponding to 1.3% of the genome. Large fractions of MITEs were also reported in Class II-rich genomes, such as the aforementioned mosquitoes [48, 250] and the invasive Ae. albopictus [47], but also in many plant species such as the rice Oryza sativa [251–253]. Among Class II elements, we have also detected Crypton (0.9% of the genome), RC/Helitrons (0.4% of the genome). and Mavericks (0.3% of the genome).
LINE elements are the second most abundant TE subclass, representing ≈11% of the S. oryzae genome, among which ≈35% are assigned to RTE elements and ≈22% to I elements (Fig. 2D). No SINE families have been detected. LTRs are rather scarce, representing only ≈3% of the genome (Fig. 2D), and the vast majority belong to the Gypsy superfamily (≈30%). Another retrotransposon order detected are Penelope (PLEs), reaching nearly 2% of S. oryzae’s genome, and DIRS (tyrosine recombinase retrotransposons, 0.14% of the genome).
Finally, around 22% of the genome is composed of repeats for which our pipeline could not assign a known TE class (Fig. 2D). CDD search on peptides greater than 100 aa extracted from “Unknown” consensus found a total of 74 distinct hits (P ≤ 0.01), for a total of 50 consensus. We identified 14 unknown consensus with hits against known TE domains or viral sequences. The other 36 sequences had significant hits against Eukaryotic or Prokaryotic domains, traditionally not associated with TEs. Therefore, potential non-TE sequences within the unknown fraction represent an estimated total of 0.35% of the genome and were removed from the TE library. These unknown families highlight the wealth and diversity of TEs among insects and Coleopteran genomes in particular.
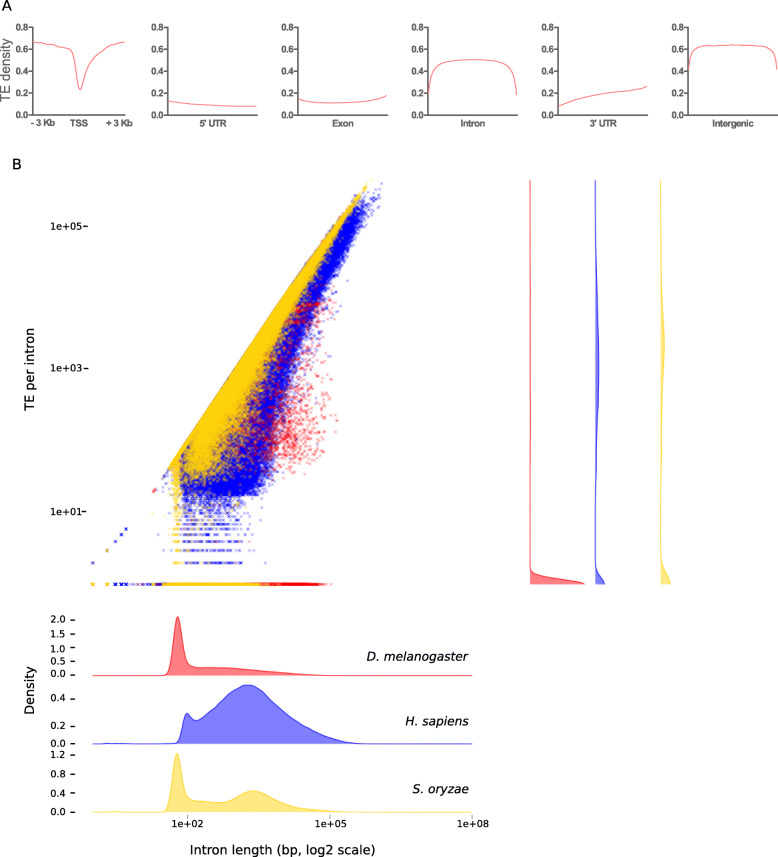
TE copies make up most of non-coding sequences of S. oryzae’s genome
TE copies are interspersed around the S. oryzae genome. TEs are less frequently found close to gene transcription start sites (TSS), 5′ and 3′ untranslated regions (5′ and 3′ UTRs) and exons (Fig. 3A), as expected. On the contrary, introns and intergenic sequences harbor the highest TE content (Fig. 3A), amounting to around 50% of TE density, close to the general TE proportion in the genome (72%), suggesting that most non-coding DNA sequences in the S. oryzae genome are virtually made of TEs. To grasp the impact of TEs on intron size, we compared intron length in S. oryzae with two very well assembled genomes: D. melanogaster with a very compact and small genome, and the large, TE-rich human genome (Fig. 3B). In D. melanogaster, introns are small and harbor few TEs, while in humans, introns are much larger potentially due to high TE accumulation [254]. S. oryzae intron sizes also seem to be due, at least partly, to TE accumulation. Interestingly, the S. oryzae genome presents a bimodal distribution, with a large proportion of small introns, as found in D. melanogaster, but also a noticeable amount of larger, TE-packed and more human-like introns. This could suggest that specific regions of the genome could be more prone to TE elimination, and be associated with high rates of recombination and/or signature of purifying selection.
Fig. 3.
TE distribution in S. oryzae’s genome. A Density of TE copies within gene regions. TE copies are the least abundant within TSSs, 5′ and 3′ UTRs and exons, while introns and intergenic regions are riddled with TEs. TSS: transcription start site, UTR: untranslated regions. B Relationship between intron length and TE per intron in D. melanogaster (red), H. sapiens (blue), and S. oryzae (yellow). S. oryzae shares characteristics of both Drosophila with short and TE-poor introns and Humans with a significant number of large, TE-packed introns
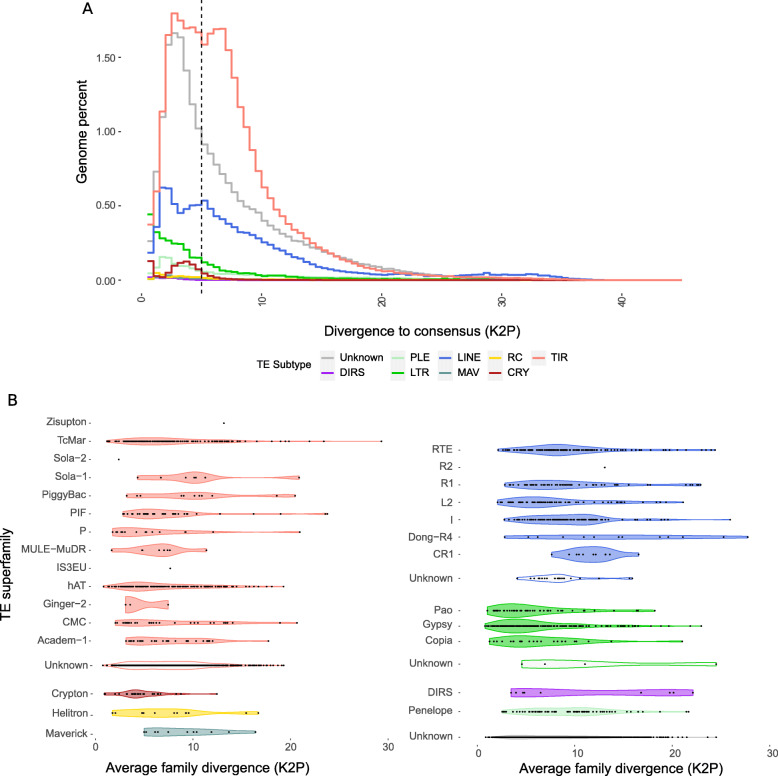
TE activity inferred by evolutionary history
Within reconstructed TE families, nucleotide substitution levels (Kimura 2 parameters, K2P) between copies and their consensus sequences allowed estimation of their relative ages and identified potentially active ones (Fig. 4A). Such “TE landscapes” are extremely helpful to pinpoint potential TE amplifications (modes in the distribution) and extinctions (valleys) within the 0–30% K2P range (beyond, the increased divergence between copies affects negatively the sensitivity of the alignments, such that TE-derived sequences are no longer recognizable). The landscape analysis revealed a heterogeneous distribution of the TE copy divergence to their consensus within and between the main TE subclasses (Fig. 4A). Most identified TE copies have a K2P divergence under 10, which is often observed in insects, and strikingly distinguishes itself from TE-rich mammalian genomes (RepeatMasker.org, [40]). While S. oryzae’s TE density and distribution evokes the architecture of mammalian genomes, this relatively younger TE landscape suggests higher deletion rates, and possibly a higher TE turnover rate, as observed in Drosophila [255, 256]. LINEs and DNA transposons have the wider spectrum of divergence levels, suggesting an aggregation of distinct dynamics for the TE families present in S. oryzae. By contrast, the rare LTR copies identified appear to be the most homogeneous within families, with only a few substitutions between copies and their consensuses, suggesting a very recent amplification in this subclass. Finally, unknown TEs share a large part of their K2P distribution with TIR elements, though relatively less divergent from their consensus sequences as a whole. A breakdown of the K2P distributions at the superfamily level reveals specific evolutionary dynamics (Fig. 4B). Diverse superfamilies, such as Tc-Mar and hAT (TIR) or RTE (LINE), show more uniform distributions, suggesting sustained activity of some of its members throughout S. oryzae’s genome evolution, though this could also indicate that these subfamilies could be subdivided further. As observed at the class level, all three identified LTR superfamilies (Pao, Gypsy, and Copia) show families within the lowest K2P range.
Fig. 4.
A TE divergence landscape. Distribution of the divergence (Kimura two parameters, K2P) between TE copies and their consensus, aggregated by TE class reported in percent of the genome. The less divergent superfamilies are distributed to the left and suggest recent activity. Strikingly, most of the TE copies have less than 10% divergence to their consensus, with a large number of copies under 5% (dotted line). The distribution of the “unknown” class overlaps with the leftmost mode of the TIR distribution, suggesting that many more TIR families are yet to be described in S. oryzae. Strikingly, LTR elements are the least diverged altogether with the mode of the distribution on the 0–1% divergence bin. B Mean K2P distributions within TE superfamilies. Left panel depicts Class II families, and all Class I (retrotransposons) and unknown families are on the right panel. LTR superfamilies harbor some of the least divergent TE families, suggesting that this class may host some of the youngest TE
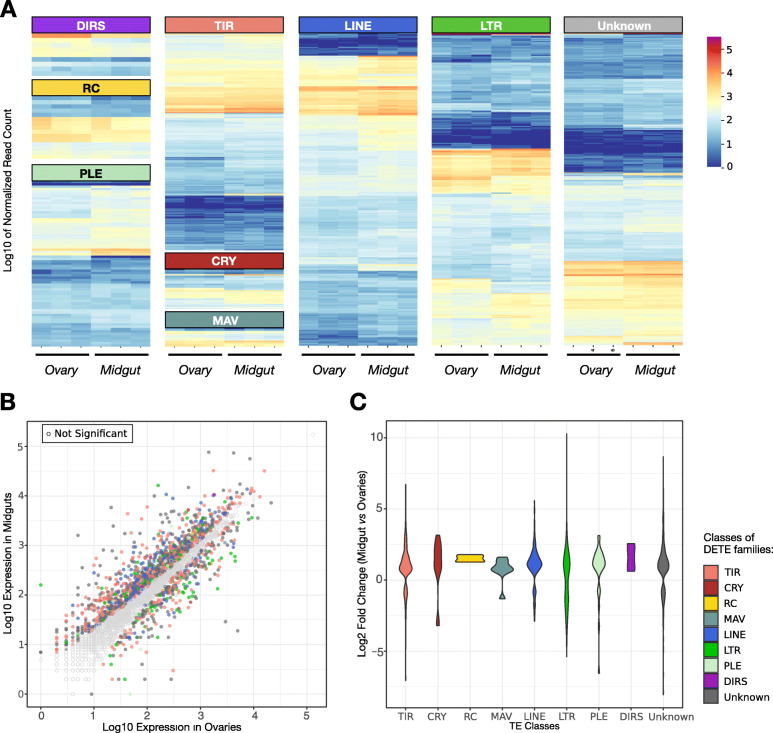
TEs are transcriptionally active in somatic and germline tissues
The TE K2P landscape suggests that LTR elements as well as some LINE families and several Class II subclasses are among the youngest, and thus potentially active. In order to estimate the transcriptional activity of S. oryzae’s TE families, we have produced somatic (midgut) and germline (ovary) transcriptomic data. While germline tissues allow identification of potential TE families capable of producing vertically transmitted new copies, TE derepression in somatic tissues represents the potential mutational burden due to TEs. The expression of TE families varied extensively within a class and the proportion of transcriptionally active/inactive TE families between classes was also distinct (Fig. 5A). In total, 1594 TE families were differentially expressed between ovary and midgut tissues (Fig. 5B, Additional file 5); of which 329 have an absolute log2 fold change higher than 2 (71 downregulated and 258 upregulated in midgut). In total, we detected 360 TE families downregulated in midgut when compared to ovaries: A much larger set of upregulated TE families was detected in midgut when compared to ovaries (1 236), illustrating the tighter regulation of TE copies in germline tissues. Moreover, the distribution of log2 fold changes were similar between TE subclasses but different for LTRs, which had a higher proportion of upregulated TE families in ovaries compared to other classes (Fig. 5C. Kruskall and Wallis rank-sum test: H = 36.18, P < 0.01; LTR vs. LINE, Class II or Unknown: Dunn’s test: P-adj < 0.01). In conclusion, the large TE compartment in S. oryzae shows abundantly expressed TE families, and tissue-specific expression patterns.
Fig. 5.
TE family expression in midguts and ovaries from S. oryzae. A Log10 normalized counts in midguts and ovaries triplicates. Normalized counts show different proportions of transcriptionally active TE families in different TE classes. B Log10 of base mean average expression of TE families in ovaries and midguts from three biological replicates. Depicted in color only TE families which had differential expression between ovary and gut tissues (padj< 0.05, |log2FC| > 2). Most TE families are upregulated in midguts compared to ovaries. C Distribution of all significant (padj< 0.05). Log2FC depicts specifically deregulated TE classes in each tissue. LTR elements are predominantly upregulated in ovaries
To estimate the TE transcriptional load imposed on S. oryzae, we computed the percentage of total RNAseq poly-A enriched reads mapping to TE consensus sequences in gut and ovaries (Additional file 2: Figure S4). Around 5% of the midgut transcriptome corresponds to TE sequences, while such reads represent only ~ 2% of ovarian transcriptomes, reinforcing the tighter regulation of TEs in germ tissues. We compared such transcriptional burden to a TE-poor (D. melanogaster, ≈12%) and a TE-rich (Ae. albopictus ≈50%) genome, using similar technology in equivalent tissues (adult midgut, see “Methods”). It is important to note that, despite being a TE-poor genome, D. melanogaster harbors many young LTR elements that have been recurrently shown to transpose [257]. We did not detect a direct correlation between genomic TE content and TE expression (Additional file 2: Figure S4). S. oryzae bears the highest proportion of RNAseq reads mapped against TE consensus sequences (≈5%), followed by D. melanogaster (≈1%) and Ae. albopictus (≈0.01%). Henceforth, not only is S. oryzae a TE-rich genome, but the transcriptional load from TEs is higher than in other TE-rich genomes (Ae. albopictus), and in genomes harboring young and active TE copies (D. melanogaster, [38, 258]).
Finally, it is important to note that while transcriptional activation of TE copies may have an impact on the host genome, it does not indicate high transposition and therefore higher mutation rates. The high transcriptional load of S. oryzae compared to other species might stem from differences in TE regulation. In insects, TEs are mainly silenced by small RNAs and repressive chromatin marks [259]. More specifically, piwi-interacting RNAs (piRNAs) are able to target post-transcriptional repression of TEs, and guide chromatin silencing complexes to TE copies [259–261]. Therefore, we have annotated genes implicated in small RNA biogenesis and found that all three pathways (piRNAs but also microRNAs and small interfering RNAs biogenesis pathways) are complete (Additional file 2: Supplemental Note 9). Genes involved in piRNA biosynthesis are expressed mainly in ovaries and testes, while somatic tissues (midgut) show smaller steady-state levels (Additional file 2: Supplemental Note 9 [41, 94, 259, 260, 262–284]), suggesting the piRNA pathway is potentially functional in S. oryzae ovaries, and could efficiently reduce transposition.
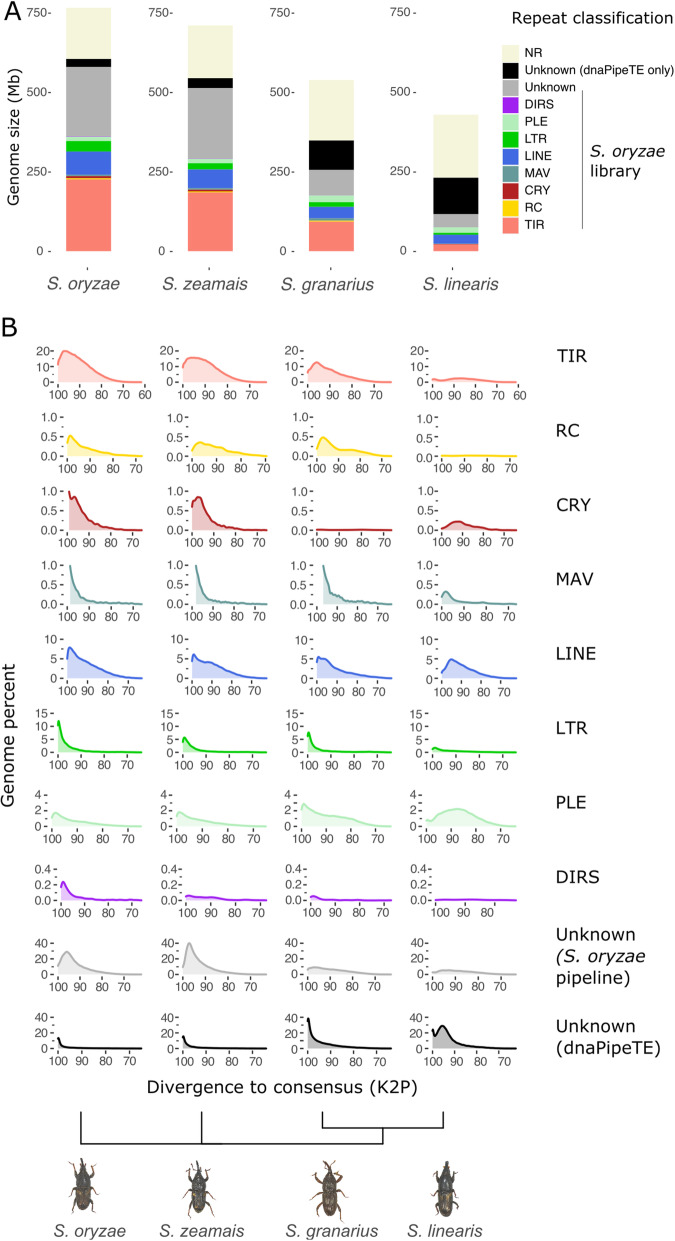
TE content is variable among Sitophilus species
Cereal weevils are part of the Dryophthoridae family that includes more than 500 species. Very little is known about genome dynamics in this massive phylogenetic group, and Sitophilus species divergence is estimated to the Neogene (2.5–25 Ma) [285] . Because of the unusual high TE copy number and landscape observed in S. oryzae, we analyzed three other closely related species namely Sitophilus zeamais, Sitophilus granarius, and Sitophilus linearis. We produced low-coverage sequencing and estimated the TE content from raw reads using our annotated S. oryzae TE library with dnaPipeTE [47]. Remarkably, among Sitophilus species, repeat content is variable (Fig. 6A), with S. linearis harboring the smaller repeat load (≈54%) compared to S. oryzae (≈80%), S. zeamais (≈79%), and S. granarius (≈65%). Most importantly, Class II (DNA) elements of S. oryzae are nearly absent from S. linearis, and no recent burst of LTR elements is observed, contrary to the other Sitophilus species, suggesting alternative TE evolutionary histories (Fig. 6B). It is important to note that our analysis is biased toward S. oryzae, as the library used to annotate the TEs in the other Sitophilus species stems from automatic and manual annotation of the S. oryzae genome.
Fig. 6.
TE landscape across Sitophilus species. A Proportion of TE per species estimated from short reads with dnaPipeTE and a custom TE library including Repbase (release 2017) and annotated TE consensus discovered in S. oryzae. S. oryzae, S. zeamais, and S. granarius harbor similar TE content, while S. granarius presents a smaller TE load, and S. linearis harbors the smallest TE content and the higher proportion of unknown repeats. The proportion of unknown repeats only found by dnaPipeTE (black) increases from S. oryzae to S. linearis with the phylogenetic distance. B Distribution of divergence values between raw reads and repeats contig assembled with dnaPipeTE (blastn) across four Sitophilus species. S. oryzae appears to share its TE landscape with S. zeamais and S. granarius, but the three species display a distinct repeatome than S. linearis, in spite of their phylogenetic proximity. SO2: S. oryzae’s TE library produced in this analysis, DPTE: DNApipeTE TE annotation (repeats only found by dnaPipeTE)
Overall, the comparison of TE content in closely related species highlights the influence of phylogenetic inertia, but reveals a possible TE turnover in the S. linearis lineage. In addition to the regulation mechanisms that strongly contribute to TE amount and variation, TE accumulation is conditioned by the drift/selection balance in populations. Indeed, effective population size has been suggested to be a major variable influencing TE content, as small, inbred, or expanding populations suffer drift, allowing detrimental insertions to stay in the gene pool and thus favor TE fixation [286]. Such hypotheses should be addressed in the future, especially on recently sequenced TE-rich but rather small (< 1 Gbp) genomes such as S. oryzae.
Endosymbionts might impact TE transcriptional regulation
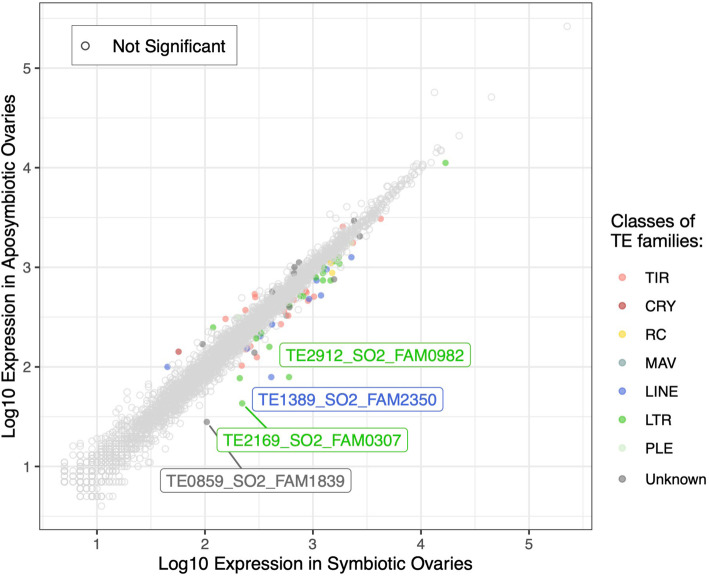
The four Sitophilus species studied have different ecologies. S. oryzae and S. zeamais infest field cereals and silos, while S. granarius is mainly observed in cereal-containing silos. S. linearis, however, lives in a richer environment, i.e., tamarind seeds. In association with their diets, the interaction of Sitophilus species with endosymbiotic bacteria differs: the cereal weevils (S. oryzae, S. zeamais, and S. granarius) harbor the intracellular gram-negative bacteria S. pierantonius, albeit at very different loads. While S. oryzae and S. zeamais show high bacterial load, S. granarius has a smaller bacterial population [111]. In contrast, S. linearis has no nutritional endosymbionts, in correlation with its richer diet. We wondered whether the presence of intracellular bacteria impacts TE regulation, and took advantage of artificially obtained aposymbiotic S. oryzae animals to search for TE families differentially expressed in symbiotic versus aposymbiotic ovaries. There were 50 TE families upregulated in symbiotic ovaries compared to artificially obtained aposymbiotic ones, while 15 families were downregulated (Fig. 7 and Additional file 5). Only three families presented an absolute log2 fold change higher than 2: one LINE and two LTR/Gypsy elements. The three of them were upregulated both in symbiotic versus aposymbiotic ovaries, and in ovaries versus midgut (Additional file 5), suggesting that such elements have tissue specificity, and their expression is modulated by the presence of intracellular bacteria. Such TE families would be ideal candidates to further study the crosstalk between host genes, intracellular bacteria and TE transcriptional regulation. It is important to note that the process used to obtain aposymbiotic insects relies in heat treatments that could impact overall transcriptional regulation, and henceforth, the TEs differentially expressed between symbiotic and aposymbiotic ovaries could stem from such treatment and not from the lack of endosymbionts. In order to confirm the link between intracellular bacteria and TE regulation, it is mandatory to deplete insects of their endosymbionts using other methods, as antibiotic treatment, and reassess TE expression in aposymbiotic individuals.
Fig. 7.
Differentially expressed TE families between symbiotic and aposymbiotic S. oryzae ovaries. Log10 of base mean average expression of TE families in symbiotic vs aposymbiotic ovaries from two biological replicates. Depicted in color only TE families which had differential expression between both ovary types (padj< 0.05, |log2FC| > 2). Two LTR elements and one LINE element are upregulated (log2FC > 2) in symbiotic ovaries
Conclusion
The success of obtaining a TE-rich genome assembly complete enough to understand genome architecture and regulatory networks relies on the use of multiple sequencing platforms [287]. Here, we describe the first assembly of the repeat-rich (74%) S. oryzae genome, based on a combination of long- and short-read sequencing, and a new assembly method, WENGAN [288]. While this first assembly reaches quality standards similar to other coleopteran species (Table 1), it is important to stress that new sequencing methods have emerged in order to improve genome assemblies, including linked reads and optical mapping [287].
We uncovered around 74% of repeated sequences in the S. oryzae genome, mostly TE families. While the TE landscape is marked by a wealth of Class II elements, especially non-autonomous MITE elements, ~ 21% of the genome is composed of unknown repeats. Large duplicated gene families can be present in such a category, but it is tempting to speculate that the majority is composed of novel Class II elements. Indeed, unknown and TIR elements share the same K2P landscapes, and many Class II elements have only been detected through an inverted repeat search for TIRs, and not proteins, excluding therefore TE copies old enough that TIRs are too divergent to be recognized. Moreover, we have shown that many TE families in S. oryzae are present in the transcriptome, suggesting that several families can be transcriptionally active. How such TE families are able to escape host silencing remains unknown. It seems obvious today that insect models such as D. melanogaster only represent a small window on the complex biology and evolution of TEs, and the sequencing and annotation of species with high TE content—while challenging [289]—is key to understanding how genomes, their size, their structure, and their function evolve. In conclusion, S. oryzae constitutes an excellent model to understand TE dynamics and regulation and the impact on genome function.
Sitophilus species not only differ in their TE landscape, but also in their ecology and as a consequence, their association with intracellular bacteria. Comparison of TE content within the Sitophilus genus shows variable TE amount and diversity. In addition, intracellular bacterium impacts transcription of specific TE families in ovaries. The molecular mechanisms behind the co-evolution between an insect, its endosymbiotic bacterium, and TEs remain unexplored. The impact of intracellular bacteria on host genomes is poorly studied, and the Sitophilus genus offers a simpler experimental setting, with a single intracellular bacterium present within specific host cells [19, 112], and a well-established knowledge of host-bacteria interaction [111, 147, 148, 193, 290].
Methods
DNA extraction and high-throughput sequencing
Individuals of both sexes of S. oryzae were reared on wheat grains at 27.5 °C with 70% relative humidity. The aposymbiotic strain was obtained by treating the symbiotic strain during one month at 35 °C and 90% relative humidity as previously described [291]. This strain is viable, is fertile, and was raised in the same conditions as the symbiotic strain. The aposymbiotic status was confirmed by PCR and histology. Male and female adults of S. oryzae were used for DNA extraction. Only the gonads were used to minimize DNA contamination from its diet, which could be still present in the gut. The reproductive organs were obtained from aposymbiotic adults and a DNA extraction protocol specific for Sitophilus weevils was performed. DNA extractions were performed using a STE buffer (100 mM NaCl, 1 mM Na2EDTA pH 8, 10 mM Tris HCl pH 8). Tissues were homogenized in STE buffer, then treated successively by SDS 10%, proteinase K, and RNase. Briefly, genomic DNA was purified by two successive extractions with phenol:chloroform:isoamyl alcohol (25/24/1) followed by extraction with 1 vol of chloroform:isoamyl alcohol (24/1). Genomic DNA was then precipitated by 0.7 vol isopropanol. After washing the pellet with 70% ethanol, genomic DNA was recovered in TE (1 mM EDTA, 10 mM Tris HCl pH 8) buffer. Using this protocol, we obtained six different DNA samples: four from males and two from females. Each sample corresponds to the genomic DNA from 20 individuals. Five additional DNA samples were obtained using a high molecular weight DNA extraction protocol consisting of a single phenol:chloroform:isoamyl alcohol (25/24/1) extraction step from the genomic DNA of 100 males. The DNA concentration in each of these samples was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Sequencing was performed using a combination of Illumina, PacBio, and Nanopore technologies (Additional file 1). For each sex, two Illumina libraries were generated: one paired-end library with an average fragment size of 500 bp and one mate pair library with an average fragment size of 5 kbp. The libraries were sequenced using an Illumina HiSeq 2000 platform with the V3 chemistry and a read size of 101 bp; the paired-end (PE) libraries were sequenced at the “Génomique & Microgénomique” service from ProfileXpert (Lyon, France) while the mate paired (MP) were sequenced at Macrogen (Seoul, South Korea). Two male samples were used to build (i) an Illumina library with an average fragment size of 200 bp which was sequenced on a HiSeq 2500 instrument using the V4 chemistry and a read size of 125 bp, and (ii) a PacBio library sequenced on seven SMRT cells using the P6-C4 chemistry. These two libraries were sequenced at KeyGene (Wageningen, The Netherlands). Finally, five male samples were used to build Nanopore libraries with the SQK-LSK109 kit and without DNA fragmentation step. The libraries were independently sequenced on five MinION R9.4 flow cells. These libraries were built and sequenced at the sequencing platform of the IGFL (Institut de Génomique Fonctionnelle de Lyon, Ecole Normale Supérieure de Lyon, France). Statistics and accession numbers from all the sequencing runs are listed in the Additional file 1.
Genome assembly and annotation
First, the Illumina reads were error-corrected using BFC release 181 [292]. The PacBio and Nanopore reads were error-corrected using LORDEC v0.9 [293] with the error-corrected Illumina overlapping PE reads, a k-mer size of 19 and solidity threshold of 3. Overlapping reads were then merged using FLASH2 v2.2 [294]. Based on the merged Illumina reads, a first short-read assembly was produced using a modified version of MINIA v3.2.1 [295] with a k-mer length of 211. A hybrid assembly was then performed using WENGAN v0.1 [288] on the MINIA short-read assembly and the raw Nanopore reads. The resulting assembly was polished using two rounds of PILON v1.23 [296] using the error-corrected Illumina overlapping PE reads and the --diploid option. A first scaffolding was then performed with two rounds of FAST-SG v06/2019 [297] and SCAFFMATCH v0.9 [298] with the error-corrected Illumina MP, Illumina PE, PacBio, and Nanopore libraries. The LR_GAPCLOSER algorithm v06/2019 [299] was used for the gap-filling step using the error-corrected PacBio and Nanopore libraries. An additional scaffolding step was performed using RASCAF v1.0.2 [300] with the available RNAseq libraries from the Sequence Read Archive (SRX1034967-SRX1034972 and SRX3721133-SRX3721138). The resulting scaffolds were then gap-filled using a new round of LR_GAPCLOSER as previously described followed by two rounds of SEALER v2.1.5 [301] using the error-corrected Illumina overlapping PE reads and k-mer sizes of 64 and 96. Two rounds of PILON, as previously described, were performed to produce the final assembly. During the assembly process, we assessed haplotig contamination by using purge_haplotigs [302] and purge_dups [303]. No diploid peak nor significant haplotig contamination was observed. Quality of the assembly was assessed by computing several metrics using (i) QUAST v5.0.2 [304] with a minimal contig size of 100 bp and the --large and -k options, (ii) BUSCO v4.0.5 [50] using the Insecta ODB10 database and the -geno option, and (iii) KMC v3.0.0 [305] to evaluate the percentage of shared 100-mers between the assembly and the merged Illumina reads. Genome size prediction was performed with GenomeScope v2.0 [58], findGSE v1.94.R [60], and gce v1.0.2 [59] based on 21-mer histograms generated by JellyFish v2.2.10 [306] on the R1 reads from error-corrected Illumina overlapping PE library.
Three contaminant scaffolds corresponding to the mitochondrial genome and an artifact were removed from the assembly prior to the annotation step. The “NCBI Sitophilus oryzae Annotation Release 100” was produced using the NCBI Eukaryotic Genome Annotation Pipeline v8.2.
Low-coverage genome sequencing of other Sitophilus species
Twenty pairs of ovaries were dissected from S. oryzae, S. zeamais, S. granarius, and S. linearis females. Ovaries were homogenized in 100 mM NaCl, 1 mM EDTA pH 8, and 10 mM Tris-HCl pH 8 using a small piston. Proteinase K digestion followed in the presence of SDS for 2 h at 55 °C with shaking and for 1 h at 37 °C with RNAse A. A typical phenol chloroform extraction was then performed and genomic DNA was isopropanol precipitated. Eight whole genome sequencing libraries with a median insert size of 550 bp were constructed using the Illumina TruSeq DNA PCR-free sample preparation kit (Illumina, San Diego, CA, USA), according to the manufacturer’s protocols. Briefly, 2 μg of each gDNA was sheared using a Covaris M220 Focused-ultrasonicator (Covaris, Inc. Woburn, MA, USA), end-repaired, A-tailed, and adapter ligated. Library quality control was performed using the 2100 Bioanalyzer System with the Agilent High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, CA, USA). The libraries were individually quantified via qPCR using a KAPA Library Quantification Kits (Kapa Biosystems, Wilmington, MA, USA) for Illumina platforms, then they were pooled together in equimolar quantities and sequenced in a MiSeq sequencing system. 2 × 300 paired-end reads were obtained using a MiSeq Reagent Kits (600-cycles).
TE library construction
In order to annotate the S. oryzae repeatome, we collected and combined cutting-edge bioinformatic tools to (i) create and (ii) classify a non-redundant library of repeated elements (Additional file 2: Figure S1). First, we separately ran RepeatModeler2 (v2.0.1) [244] and EDTA v1.7.8 [245] on the assembled genome. Together, these programs include most of the recent and long-trusted tools used to detect generic repeats, but also include specific modules, such as for LTR and TIR elements. Preliminary analyses of the S. oryzae genome with RepeatModeler1 [307] and dnaPipeTE (v1.3) [47] suggested a rather large fraction of Class II DNA elements with terminal inverted repeats (TIRs). Thus, MITE-Tracker [308] was incorporated in our pipeline and ran independently on the genome assembly using 1- and 2-kbp size cutoffs to detect Class II elements harboring TIRs with high sensitivity. Following this initial step, 15,510 consensus sequences obtained from RM2, EDTA, and the two runs of MITE-tracker were successively clustered using MAFFT (v6.864b) [65], Mothur (v.1.45.2) [309], and Refiner [307] to reduce redundancy in the repeat library to a total of 2754 consensus sequences (Additional file 2: Figure S1A, https://github.com/clemgoub/So2). Then, we inspected the quality of the raw library by calculating the genomic coverage of each consensus. We ran the library against the genome using RepeatMasker (v4.1.1) (52) and implemented a simple algorithm “TE-trimmer.sh” to trim or split a consensus sequence wherever the genomic support drops below 5% of the average consensus coverage (Additional file 2: Figure S1A, https://github.com/clemgoub/So2). To mitigate any redundancy generated by the splitting, the newly trimmed library was clustered before being re-quantified using RepeatMasker (v4.1.1) [307]. At this step, we removed any consensus under 200 bp and represented by less than the equivalent of two full-length copies (in total bp). In addition, TAREAN (RepeatExplorer2 v0.3.6) [310] was used to detect and quantify candidate satellite repeats. We obtained an ab initio repeat library of 3950 consensus sequences automatically generated (Additional file 2: Figure S1A).
To refine and improve the quality of the TE consensus sequences, we then turned it over to DFAM [311] who processed the ab initio library following their recent guidelines [312]. First, any sequences mostly composed of tandem repeats were removed using a custom script to remove any sequences that were greater than 80% masked and/or had a sequence less than 100 bp. To generate seed alignments for each consensus, the consensus sequences were used as a search library for RepeatMasker (v4.1.1) to collect interspersed repeats. Seed alignments in the form of Stockholm files were generated using the RepeatMasker output. To extend potentially truncated elements, the instances in the Stockholm file for each model were extended into neighboring flanking sequences until the alignment was below a threshold equivalent to ~ 3 sequences in agreement. More specifically, all sequences are extended using full dynamic programming matrices using an improved affine gap penalty (default: − 28 open, − 6 extension) and a full substitution matrix (default: 20% divergence, 43% GC background). The termination of extension occurs when the improvement by adding a further column to the multiple alignment does not exceed 27 (with default scoring system). This is equivalent to a net gain of ~ 3 sequences in agreement. Following extension, the new consensus were collected and consensus sequences greater than 80% similar for 80% of their length were considered duplicates and only one consensus was kept.
Upon completion, we used RepeatMasker to quantify the improved library. We selected the top 50 elements (by abundance in the genome) represented in each of the “LTR,” “LINE,” “Class II,” and “Unknown” classes for manual inspection (these categories represent the 4 most abundant classes of repeats in the S. oryzae genome). While most consensus sequences were correctly extended and annotated (200), we noticed some cases of over-extension with LTR (consensus doubled in size) and flagged others with non-supported fragments for further trimming (Additional file 4 | tab 1). Once our quality check completed and the sequences curated, we removed fragments with 100% identity against a previously established consensus (Additional file 4 | tab 2). The final TE library contains 3399 sequences to classify.
The classification of the final repeat library was done in successive rounds combining homology and structure methods (Additional file 2: Figure S1B). Before the final TE library was completed, we manually curated and annotated the sequences of 31 transposable elements and satellites among the most represented in S. oryzae. These 31 high-confidence consensus sequences are added to the libraries used by the annotation programs described below and Repbase v.2017 [313]. We searched for nucleotide homology using RepeatMasker (v4.1.1 [307]) with -s “-slow” search settings. Best hits were chosen based on the highest score at the superfamily level allowing non-overlapping hits of related families to contribute to the same hit. In addition, we used blastx [94] to query each consensus against a curated collection of TE proteins (available with RepeatMasker), as well as those identified in the 31 manual consensus sequences. We kept the best protein hit based on the blastx score. Based on the 200 consensus sequences manually inspected (see above), we set a hit length / consensus size threshold of 0.08 (RepeatMasker) and 0.03 (blastx) to keep a hit. In our hands, these thresholds were conservative to automate the classification. As an alternate homology-based method, we also ran RepeatClassifier (RepeatModeler2, v2.0.1). Finally, because DNA elements are often represented by non-autonomous copies (unidentifiable or absent transposase), we further used einverted to flag terminal inverted repeats located less than 100 bp of the ends of each sequence. The complete library of 3399 consensus sequences was first annotated at the subclass level (see DFAM taxonomy: https://dfam.org/classification/tree) if two out of RepeatMasker, RepeatClassifier, and blastx annotations agreed. Further, the same rule was applied for the superfamilies if possible. At this stage, consensus sequences without annotation by homology but with TIRs as flagged by einverted were classified as TIR and all other sequences classified as Unknown. We further divided the subclass “DNA” into “MAV” (Mavericks), “RC” (Rolling circle/Helitron), “CRY” (Crypton), and “TIR” (terminal inverted repeats). Finally, the classifications automatically given as “Unknown” to 16/274 manually inspected consensus sequences were replaced to match the manually reported classification.
In order to remove potential multi-copy gene families which would have made their way to the TE library, we searched for non-TE conserved protein domains using NCBI’s CDD search with all peptides ≥ 100 AA extracted from the unknown repeats [314]. Significant hits (P ≤ 0.01) against known TEs and viruses were removed and all other left consensus were removed from the TE library. In conclusion, there are 21% unknown repeats, and the number of total TE consensus sequences in S. oryzae is 3361. The data can be obtained from 10.5281/zenodo.5128603.
In order to assess the relevance of our custom TE analysis pipeline, we ran and compared the unfiltered outputs (out.tbl file) of RepeatMasker v4.1.1 using either the TE library produced by RepeatModeler 2.0.1, EDTA v1.7.8, or our final library. An optimized TE library should minimize the total number of consensus while being able to capture as much TE in the genome as possible. Thus, we compared the total number of consensus built in each library as well as the total percent of the genome masked by each respective library.
Estimation of the repeat content
The total repeat content of the S. oryzae genome was analyzed using RepeatMasker (v.4.1.1) and our classified library of 3361 consensus sequences and the following parameters: -s -gccalc -no_is -cutoff 200. The subsequent alignments were parsed with the script “parseRM.pl” [315] https://github.com/4ureliek/Parsing-RepeatMasker-Outputs) to remove hits overlap and statistically analyzed with R version 4.0.2.
Genomic distribution of TE copies
The distribution of TE copies across the S. oryzae genome was assessed using two different approaches over six different genomic regions namely TSS ± 3 kbp, 5′ UTRs, exons, introns, 3′ UTRs, and intergenic regions. Briefly, the coverage of all TE copies was computed over a sliding window of 100 bp across the whole genome sequence using the makewindows and coverage tools from the bedtools package [280] and the bedGraphToBigWig UCSC gtfToGenePred tool. Then the different genomic regions were retrieved from the S. oryzae annotation file (GFF format) using the gencode_regions script (https://github.com/saketkc/gencode_regions) and the UCSC gtfToGenePred tool (https://github.com/ENCODE-DCC/kentUtils). A matrix containing the TE coverage per genomic region was generated using the computeMatrix tool from deepTools [279] and used to generate metaplots using the plotProfile tool.
TE landscapes
The relative age of the different TE families identified in the genome assembly was drawn performing a “TE landscape” analysis on the RepeatMasker outputs. Briefly, the different copies of one TE family identified by RepeatMasker are compared to their consensus sequence, and the divergence (Kimura substitution level, CpG adjusted, see RepeatMasker webpage: http://repeatmasker.org/webrepeatmaskerhelp.html) is calculated. The TE landscape consists of the distribution of these divergence levels. In the end, the relative age of a TE family can be seen as its distribution within the landscape graph: “older” TE families tend to have wider and flatter distribution spreading to the right (higher substitution levels) than the “recent” TE families, which are found on the left of the graph and have a narrower distribution. TE landscapes were drawn from the RepeatMasker output parsed with the options -l of “parseRM.pl.” We report here the TE landscape at the level of the TE subclass (LINE, LTR, TIR, CRY, MAV, DIRS, PLE, RC, and Unknown).
dnaPipeTE comparative analysis in Sitophilus species
To compare the TE content of S. oryzae to four related species of Sitophilus (S. granarius, S. zeamais, S. linearis), we used dnaPipeTE v.1.3 [47]. dnaPipeTE allows unbiased estimation and comparison of the total repeat content across different species by assembling and quantifying TE from unassembled reads instead of a linear genome assembly. Reads for Sitophilus species were produced as described above. Using our new classified library (3 390 consensus) as TE database in dnaPipeTE, we were further able to identify the phylogenetic depth of the repeat identified in S. oryzae.
RNA sequencing and TE expression analysis
Individuals of both sexes of S. oryzae were reared on wheat grains at 27.5 °C with 70% relative humidity. Ten midguts and ovaries from 10-day-old adults were dissected in diethylpyrocarbonate-treated Buffer A (25 mM KCl, 10 mM MgCl2, 250 mM Sucrose, 35 mM Tris/HCl, pH 7.5). RNA was extracted in triplicates with RNAqueous-Micro (Qiagen), following the manufacturer recommendations. Single-indexed libraries were built using the SENSE mRNA-Seq Library Prep Kit V2 (Lexogen), following the manufacturer recommendations. Libraries were then pooled in an equimolar range and sequenced using high-throughput reagents on an Illumina NextSeq 500 (86 bp in single end). Raw sequencing reads were deposited at SRA archive BioProject PRJNA746240. Adapter sequences and low-quality reads were filtered out with Trimmomatic (v0.36) [316] and clean reads were aligned to the S. oryzae genome with STAR aligner (v2.5.4b, [317]) and featureCounts from subread package [318] to obtain gene counts. We also used the STAR aligner in single-end mode to map the clean reads against all TE copies extracted from the genome with the following options: --outFilterMultimapNmax 100 --winAnchorMultimapNmax 100 --outMultimapperOrder Random --outSAMmultNmax 1. The mapped bam files were used as input to TEtools software [319] to determine TE family expression. Genes and TE family counts were used as input for DESEq2 package (v1.30) [320] to determine differential TE expression between ovary vs gut tissues as well as ovaries from symbiotic and aposymbiotic weevils. Differentially expressed TEs were defined whenever the adjusted p value was smaller than 0.05 and log2 fold change was higher than 1 or smaller than − 1. We used the aforementioned STAR alignment parameters to map transcriptomic sequencing reads from midgut and ovaries of S. oryzae (Accession: SRX1034971, SRX1034972, and reads from this study deposited under the BioProject ID PRJNA746240), D. melanogaster (Accession: SRX029389, and SRX045361), and Ae. albopictus (Accession: SRX1512976, SRX1898481, SRX1898483, SRX1898487, SRX3939061, and SRX3939054) against the TE consensus sequences for each species.
Supplementary Information
Additional file 1: Summary of sequencing libraries produced for S. oryzae.
Additional file 2:. Supplementary notes, supplementary figures, and small tables.
Additional file 3:. Large supporting tables and datasets.
Additional file 4:. Transposable elements annotation tables.
Additional file 5:. STAR and TEtools mapping statistics.
Acknowledgements
The authors acknowledge supercomputing resources made available by the Rhône-Alpes Bioinformatics center (PRABI-AMSB, http://www.prabi.fr) to perform the NGS data analyses. The authors would like to thank S. Robin, F. Legeai, and A. Bretaudeau at the INRAE BioInformatics Platform for Agro-ecosystems Arthropods (BIPAA) (https://bipaa.genouest.org) for the S. oryzae genome integration. The authors would like to thank F. Barbet, S. Croze, and N. Nazaret from profileXpert for Illumina sequencing of Sitophilus oryzae libraries. The authors thank the network REacTION and its coordinators (N. Ponts, G. Le Trionnaire, I. Fudal, M. Jubault), funded by INRAE-SPE, for organizing a Bisulfite-seq workshop that allowed the production of the data presented here. We also thank J.-Y. Rasplus for collecting Sitophilus linearis individuals from Niger. The authors would like to thank C. Feschotte at Cornell University for the support, insights, and the occasional use of bioinformatic resources.
Abbreviations
- AMPs
Antimicrobial peptides
- CPs
Cuticle proteins
- HGT
Horizontal gene transfer
- IMP
Inosine monophosphate
- K2P
Kimura 2 parameters
- LINE
Long-interspersed element
- LTR
Long terminal repeat
- MAMPs
Microbial associated molecular patterns
- MITEs
Miniature inverted repeat elements
- ORs
Odorant receptors
- PRR
Pattern recognition receptors
- PLE
Penelope-like
- RC
Rolling circle
- SINE
Short interspersed element
- TIR
Terminal inverted repeat
- TSS
Transcription start sites
- TEs
Transposable elements
- TTSS
Type three secretion systems
- UTR
Untranslated regions
- UMP
Uridine monophosphate
Authors’ contributions
AH and AL conceived the original sequencing project and were joined by NP, RR, CG, CVC, AM, and CV who participated in the coordination of the project. AV1, CVM, EDA, JM, FM, and AV2 reared the inbred lines and AV1 extracted genomic DNA and RNA that was used for library construction and sequencing. BG and SH produced and sequenced the Nanopore libraries. CG, AV1, MB, NB, CV, AG, and ATRDV produced and sequenced the low-coverage Illumina libraries. NP, CVC, ADG, and MFS performed the genome assembly and automated gene prediction. CVC, MMH, and TG analyzed and wrote the phylome and horizontal gene transfer note. PBP, GF, SC, HC, and FC analyzed and wrote the global analysis of metabolic pathways note. NP analyzed and wrote the digestive enzymes and the detoxification and insecticide resistance notes. PC analyzed and wrote the development note. CV-C analyzed and wrote the cuticle protein genes note. CVM, CVC, NP, JM, LB, AB, WZ, FM, AV2, and AZR analyzed and wrote the innate immune system note. NM, CM, ADSB, and EJJ analyzed and wrote the odorant receptors note. TC, CB, AV1, and RR produced the data for the epigenetic pathways note. TC, CB, AV1, GR, CVC, CV, and RR analyzed and wrote the epigenetic pathways note. MGF, CG, EDA, RR, SB, GF, NM, CVM, and NP produced the figures. CG, RR, JMS, JR, RH, and AFAS annotated and analyzed the TE content while MGF analyzed the TE RNAseq data. NP, CVC, CG, CV, RR, AL, and AH wrote the manuscript. All authors read and approved the final manuscript.
Authors’ information
Twitter handles: @digenoma (Alex Di Genova), @atrvlncc (Ana Tereza R Vasconcelos), @AnnaZaidmanRemy (Anna Zaidman-Rémy), @CamilleBurgh (Camille Meslin), @VargasChavezC (Carlos Vargas-Chavez), @cmonegat (Carole Vincent-Monegat), @clementgoubert (Clément Goubert), @Cosmicomica (Elisa Dell'Aglio), @EmmaJoly8 (Emmanuelle Jacquin-Joly), @flmasson (Florent Masson), @Justin_Maire (Justin Maire), @MGFerrarini (Mariana Galvao Ferrarini), @niparisot (Nicolas Parisot), @rita_rebollo (Rita Rebollo), @StevaTravaggio (Stefano Colella), @Toni_Gabaldon (Toni Gabaldón).
Funding
Funding for this project was provided by the Fondation de l’Institut National des Sciences Appliquées-Lyon (INSA-Lyon), the research direction of INSA-Lyon, the Santé des Plantes et Environnement (SPE) department at the Institut National de Recherche pour l'Agriculture, l'Alimentation et l'Environnement (INRAE), the French ANR-10-BLAN-1701 (ImmunSymbArt), the French ANR-13-BSV7-0016-01 (IMetSym), the French ANR-17_CE20_0031_01 (GREEN), and a grant from la Région Rhône-Alpes (France) to AH. RR received funding from the French ANR-17-CE20-0015 (UNLEASH) and the IDEX-Lyon PALSE IMPULSION initiative. The project was also funded by European Regional Development Fund (ERDF) and Ministerio de Ciencia, Innovación y Universidades (Spain) PGC2018-099344-B-I00 to AL, and PID2019-105969GB-I00 to AM and Conselleria d’Educació, Generalitat Valenciana (Spain), grant number PROMETEO/2018/133 to AM. CV-C was a recipient of a fellowship from the Ministerio de Economía y Competitividad (Spain) and a grant from la Région Rhône-Alpes (France).
Availability of data and materials
Data generated and analyzed during this study are included in the published article, its additional files and publicly available repositories. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession PPTJ00000000 [321]. The version described in this paper is version PPTJ02000000. The assembly can be visualized, along with gene models and supporting data, on a dedicated genome browser (https://bipaa.genouest.org/sp/sitophilus_oryzae/). Raw reads from low-coverage genome sequencing of S. zeamais, S. granarius, and S. linearis have been deposited at NCBI Sequence Read Archive (SRA) under the BioProject accessions PRJNA647530 [322], PRJNA647520 [323], and PRJNA647347 [324] respectively. TE annotation (GFF) and consensus sequences can be found at 10.5281/zenodo.4570415 [325]. RNAseq reads obtained in this manuscript were deposited under the BioProject accession PRJNA746240 [326]. Bisulfite-seq reads have been deposited at NCBI SRA, under the BioProject accession PRJNA681724 [327].
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nicolas Parisot, Carlos Vargas-Chavez and Clément Goubert contributed equally to this work.
Contributor Information
Cristina Vieira, Email: cristina.vieira@univ-lyon1.fr.
Rita Rebollo, Email: rita.rebollo@inrae.fr.
Amparo Latorre, Email: amparo.latorre@uv.es.
Abdelaziz Heddi, Email: abdelaziz.heddi@insa-lyon.fr.
References
- 1.Hunt T, Bergsten J, Levkanicova Z, Papadopoulou A, John OS, Wild R, et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 2007;318:1913–1916. doi: 10.1126/science.1146954. [DOI] [PubMed] [Google Scholar]
- 2.Stork NE, McBroom J, Gely C, Hamilton AJ. New approaches narrow global species estimates for beetles, insects, and terrestrial arthropods. Proc Natl Acad Sci U S A. 2015;112:7519–7523. doi: 10.1073/pnas.1502408112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammond P. Species inventory. In: Groombridge B, editor. Global biodiversity: status of the Earth’s living resources. London: Chapman and Hall; 1992. pp. 17–39. [Google Scholar]
- 4.McKenna DD, Sequeira AS, Marvaldi AE, Farrell BD. Temporal lags and overlap in the diversification of weevils and flowering plants. Proc Natl Acad Sci U S A. 2009;106:7083–7088. doi: 10.1073/pnas.0810618106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberprieler RG, Marvaldi AE, Anderson RS. Weevils, weevils, weevils everywhere*. Zootaxa. 2007;1668:491–520. doi: 10.11646/zootaxa.1668.1.24. [DOI] [Google Scholar]
- 6.Vega FE, Brown SM, Chen H, Shen E, Nair MB, Ceja-Navarro JA, et al. Draft genome of the most devastating insect pest of coffee worldwide: the coffee berry borer, Hypothenemus hampei. Sci Rep. 2015;5:12525. doi: 10.1038/srep12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keeling CI, Yuen MM, Liao NY, Roderick Docking T, Chan SK, Taylor GA, et al. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 2013;14:R27. doi: 10.1186/gb-2013-14-3-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazzouri KM, Sudalaimuthuasari N, Kundu B, Nelson D, Al-Deeb MA, Le Mansour A, et al. The genome of pest Rhynchophorus ferrugineus reveals gene families important at the plant-beetle interface. Commun Biol. 2020;3:1–14. doi: 10.1038/s42003-020-1060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zunjare R, Hossain F, Muthusamy V, Jha SK, Kumar P, Sekhar JC, et al. Genetic variability among exotic and indigenous maize inbreds for resistance to stored grain weevil (Sitophilus oryzae L.) infestation. Cogent Food Agric 2016;2:1137156. 10.1080/23311932.2015.1137156.
- 10.Longstaff BC. Biology of the grain pest species of the genus Sitophilus (Coleoptera: Curculionidae): a critical review. Prot Ecol. 1981;3:83–130. [Google Scholar]
- 11.Grenier A-M, Mbaiguinam M, Delobel B. Genetical analysis of the ability of the rice weevil Sitophilus oryzae (Coleoptera, Curculionidae) to breed on split peas. Heredity. 1997;79:15–23. doi: 10.1038/hdy.1997.118. [DOI] [Google Scholar]
- 12.Champ BR, Dyte CE. FAO global survey of pesticide susceptibility of stored grain pests. FAO Plant Protec Bull. 1977;25(2):49–67. [Google Scholar]
- 13.Nguyen TT, Collins PJ, Ebert PR. Inheritance and characterization of strong resistance to phosphine in Sitophilus oryzae (L.) PLoS One. 2015;10:e0124335. doi: 10.1371/journal.pone.0124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills KA. Phosphine resistance: where to now? In: Donahaye EJ, Navarro S, Leesch JG, editors. Proceeding international conference on controlled atmosphere and fumigation in stored products; 2000 Oct 29-Nov 3. USA: Fresno; 2000. pp. 583–591. [Google Scholar]
- 15.Campbell JF. Fitness consequences of multiple mating on female Sitophilus oryzae L. (Coleoptera: Curculionidae) Environ Entomol. 2005;34:833–843. doi: 10.1603/0046-225X-34.4.833. [DOI] [Google Scholar]
- 16.Oakeson KF, Gil R, Clayton AL, Dunn DM, von Niederhausern AC, Hamil C, et al. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol. 2014;6:76–93. doi: 10.1093/gbe/evt210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heddi A, Charles H, Khatchadourian C, Bonnot G, Nardon P. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G + C content of an endocytobiotic DNA. J Mol Evol. 1998;47:52–61. doi: 10.1007/pl00006362. [DOI] [PubMed] [Google Scholar]
- 18.Heddi A, Charles H, Khatchadourian C. Intracellular bacterial symbiosis in the genus Sitophilus: the ‘biological individual’ concept revisited. Res Microbiol. 2001;152:431–437. doi: 10.1016/S0923-2508(01)01216-5. [DOI] [PubMed] [Google Scholar]
- 19.Lefèvre C, Charles H, Vallier A, Delobel B, Farrell B, Heddi A. Endosymbiont phylogenesis in the Dryophthoridae weevils: evidence for bacterial replacement. Mol Biol Evol. 2004;21:965–973. doi: 10.1093/molbev/msh063. [DOI] [PubMed] [Google Scholar]
- 20.Clayton AL, Oakeson KF, Gutin M, Pontes A, Dunn DM, von Niederhausern AC, et al. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect–bacterial symbioses. PLoS Genet. 2012;8:e1002990. doi: 10.1371/journal.pgen.1002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002;32:402–407. doi: 10.1038/ng986. [DOI] [PubMed] [Google Scholar]
- 22.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature, 2000;407:81–6. 10.1038/35024074. [DOI] [PubMed]
- 23.Gil R, Belda E, Gosalbes MJ, Delaye L, Vallier A, Vincent-Monégat C, et al. Massive presence of insertion sequences in the genome of SOPE, the primary endosymbiont of the rice weevil Sitophilus oryzae. Int Microbiol Off J Span Soc Microbiol. 2008;11:41–48. [PubMed] [Google Scholar]
- 24.Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- 25.Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, et al. Ten things you should know about transposable elements. Genome Biol. 2018;19:199. doi: 10.1186/s13059-018-1577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, Li X. Transposable elements are enriched within or in close proximity to xenobiotic-metabolizing cytochrome P450 genes. BMC Evol Biol. 2007;7:46. doi: 10.1186/1471-2148-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.You M, Yue Z, He W, Yang X, Yang G, Xie M, et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet. 2013;45:220–225. doi: 10.1038/ng.2524. [DOI] [PubMed] [Google Scholar]
- 29.Singh KS, Troczka BJ, Duarte A, Balabanidou V, Trissi N, Paladino LZC, et al. The genetic architecture of a host shift: an adaptive walk protected an aphid and its endosymbiont from plant chemical defenses. Sci Adv. 2020;6:eaba1070. doi: 10.1126/sciadv.aba1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carareto CMA, Hernandez EH, Vieira C. Genomic regions harboring insecticide resistance-associated Cyp genes are enriched by transposable element fragments carrying putative transcription factor binding sites in two sibling Drosophila species. Gene. 2014;537:93–9. 10.1016/j.gene.2013.11.080. [DOI] [PubMed]
- 31.Rostant WG, Wedell N, Hosken DJ. Chapter 2 - Transposable Elements and Insecticide Resistance. In: Goodwin SF, Friedmann T, Dunlap JC, editors. Adv. Genet., vol. 78, Academic Press; 2012, p. 169–201. 10.1016/B978-0-12-394394-1.00002-X. [DOI] [PubMed]
- 32.Mateo L, Ullastres A, González J. A transposable element insertion confers xenobiotic resistance in Drosophila. PLoS Genet. 2014;10:e1004560. 10.1371/journal.pgen.1004560. [DOI] [PMC free article] [PubMed]
- 33.Rech GE, Bogaerts-Márquez M, Barrón MG, Merenciano M, Villanueva-Cañas JL, Horváth V, et al. Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila. PLoS Genet. 2019;15:e1007900. 10.1371/journal.pgen.1007900. [DOI] [PMC free article] [PubMed]
- 34.Ullastres A, Merenciano M, González J. Regulatory regions in natural transposable element insertions drive interindividual differences in response to immune challenges in Drosophila. Genome Biol. 2021;22:265. 10.1186/s13059-021-02471-3. [DOI] [PMC free article] [PubMed]
- 35.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 36.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 37.Meyer A, Schloissnig S, Franchini P, Du K, Woltering JM, Irisarri I, et al. Giant lungfish genome elucidates the conquest of land by vertebrates. Nature. 2021:1–6. 10.1038/s41586-021-03198-8. [DOI] [PMC free article] [PubMed]
- 38.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 39.The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 40.Petersen M, Armisén D, Gibbs RA, Hering L, Khila A, Mayer G, et al. Diversity and evolution of the transposable element repertoire in arthropods with particular reference to insects. BMC Evol Biol. 2019;19:11. doi: 10.1186/s12862-018-1324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Fang X, Yang P, Jiang X, Jiang F, Zhao D, et al. The locust genome provides insight into swarm formation and long-distance flight. Nat Commun. 2014;5:2957. doi: 10.1038/ncomms3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley JL, Peyton JT, Fiston-Lavier A-S, Teets NM, Yee M-C, Johnston JS, et al. Compact genome of the Antarctic midge is likely an adaptation to an extreme environment. Nat Commun. 2014;5:4611. doi: 10.1038/ncomms5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palacios-Gimenez OM, Koelman J, Palmada-Flores M, Bradford TM, Jones KK, Cooper SJB, et al. Comparative analysis of morabine grasshopper genomes reveals highly abundant transposable elements and rapidly proliferating satellite DNA repeats. BMC Biol. 2020;18:199. doi: 10.1186/s12915-020-00925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilbert C, Peccoud J, Cordaux R. Transposable elements and the evolution of insects. Annu Rev Entomol. 2021;66:355–372. doi: 10.1146/annurev-ento-070720-074650. [DOI] [PubMed] [Google Scholar]
- 45.Sessegolo C, Burlet N, Haudry A. Strong phylogenetic inertia on genome size and transposable element content among 26 species of flies. Biol Lett. 2016;12:20160407. doi: 10.1098/rsbl.2016.0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ray DA, Grimshaw JR, Halsey MK, Korstian JM, Osmanski AB, Sullivan KAM, et al. Simultaneous TE analysis of 19 heliconiine butterflies yields novel insights into rapid TE-based genome diversification and multiple SINE births and deaths. Genome Biol Evol. 2019;11:2162–2177. doi: 10.1093/gbe/evz125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goubert C, Modolo L, Vieira C, Valiente Moro C, Mavingui P, Boulesteix M. De novo assembly and annotation of the Asian tiger mosquito (Aedes albopictus) repeatome with dnaPipeTE from raw genomic reads and comparative analysis with the yellow fever mosquito (Aedes aegypti) Genome Biol Evol. 2015;7:1192–1205. doi: 10.1093/gbe/evv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu Z (Jake), et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S, Shen S, Peng J, Zhou X, Kong X, Ren P, et al. Chromosome-level genome assembly of an important pine defoliator, Dendrolimus punctatus (Lepidoptera; Lasiocampidae) Mol Ecol Resour. 2020;20:1023–1037. doi: 10.1111/1755-0998.13169. [DOI] [PubMed] [Google Scholar]
- 50.Seppey M, Manni M, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness. In: Kollmar M, editor. Gene Prediction. Methods Mol Biol. 2019;1962. p. 227–45. 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed]
- 51.Silva AA, Braga LS, Corrêa AS, Holmes VR, Johnston JS, Oppert B, et al. Comparative cytogenetics and derived phylogenic relationship among Sitophilus grain weevils (Coleoptera, Curculionidae, Dryophthorinae) Comp Cytogenet. 2018;12:223–245. doi: 10.3897/CompCytogen.v12i2.26412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tribolium Genome Sequencing Consortium. Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 53.Dias GB, Altammami MA, El-Shafie HAF, Alhoshani FM, Al-Fageeh MB, Bergman CM, et al. Haplotype-resolved genome assembly enables gene discovery in the red palm weevil Rhynchophorus ferrugineus. Sci Rep. 2021;11:9987. doi: 10.1038/s41598-021-89091-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-Qahtani AH, Al-Khalifa MS, Al-Saleh AA. Karyotype, meiosis and sperm formation in the red palm weevil Rhynchophorus ferrugineus. Cytologia. 2014;79:235–242. doi: 10.1508/cytologia.79.235. [DOI] [Google Scholar]
- 55.Brun LO, Stuart J, Gaudichon V, Aronstein K, French-Constant RH. Functional haplodiploidy: a mechanism for the spread of insecticide resistance in an important international insect pest. Proc Natl Acad Sci U S A. 1995;92:9861–9865. doi: 10.1073/pnas.92.21.9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lanier GN, Wood DL. Controlled mating, karyology, morphology, and sex-ratio in the Dendroctonus ponderosae complex. Ann Entomol Soc Am. 1968;61:517–526. doi: 10.1093/aesa/61.2.517. [DOI] [Google Scholar]
- 57.Stuart JJ, Mocelin G. Cytogenetics of chromosome rearrangements in Tribolium castaneum. Genome. 1995. 10.1139/g95-085. [DOI] [PubMed]
- 58.Vurture GW, Sedlazeck FJ, Nattestad M, Underwood CJ, Fang H, Gurtowski J, et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics. 2017;33:2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu B, Shi Y, Yuan J, Hu X, Zhang H, Li N, et al. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. ArXiv13082012 Q-Bio 2020.
- 60.Sun H, Ding J, Piednoël M, Schneeberger K. findGSE: estimating genome size variation within human and Arabidopsis using k-mer frequencies. Bioinformatics. 2018;34:550–557. doi: 10.1093/bioinformatics/btx637. [DOI] [PubMed] [Google Scholar]
- 61.McKenna DD, Scully ED, Pauchet Y, Hoover K, Kirsch R, Geib SM, et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle-plant interface. Genome Biol. 2016;17:227. doi: 10.1186/s13059-016-1088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Initiative IGG. Genome sequence of the tsetse fly (Glossina morsitans): vector of African trypanosomiasis. Science. 2014;344:380–386. doi: 10.1126/science.1249656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 64.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lassmann T, Sonnhammer ELL. Kalign--an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wallace IM, O’Sullivan O, Higgins DG, Notredame C. M-coffee: combining multiple sequence alignment methods with T-coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009;25:1972–1973. 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed]
- 69.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huerta-Cepas J, Capella-Gutiérrez S, Pryszcz LP, Marcet-Houben M, Gabaldón T. PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 2014;42:D897–D902. doi: 10.1093/nar/gkt1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wehe A, Bansal MS, Burleigh JG, Eulenstein O. DupTree: a program for large-scale phylogenetic analyses using gene tree parsimony. Bioinformatics. 2008;24:1540–1541. doi: 10.1093/bioinformatics/btn230. [DOI] [PubMed] [Google Scholar]
- 72.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 73.Al-Shahrour F, Díaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of gene ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 74.Huerta-Cepas J, Serra F, Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 76.R Core Team . R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 77.Chung H-R, Schäfer U, Jäckle H, Böhm S. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila. EMBO Rep. 2002;3:1158–62. 10.1093/embo-reports/kvf243. [DOI] [PMC free article] [PubMed]
- 78.Chung H-R, Löhr U, Jäckle H. Lineage-specific expansion of the zinc finger associated domain ZAD. Mol Biol Evol. 2007;24:1934–1943. doi: 10.1093/molbev/msm121. [DOI] [PubMed] [Google Scholar]
- 79.Masson F, Vallier A, Vigneron A, Balmand S, Vincent-Monégat C, Zaidman-Rémy A, et al. Systemic infection generates a local-like immune response of the bacteriome organ in insect symbiosis. J Innate Immun. 2015;7:290–301. doi: 10.1159/000368928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reid WR, Sun H, Becnel JJ, Clark AG, Scott JG. Overexpression of a glutathione S-transferase (Mdgst) and a galactosyltransferase-like gene (Mdgt1) is responsible for imidacloprid resistance in house flies. Pest Manag Sci. 2019;75:37–44. doi: 10.1002/ps.5125. [DOI] [PubMed] [Google Scholar]
- 81.Altincicek B, Knorr E, Vilcinskas A. Beetle immunity: identification of immune-inducible genes from the model insect Tribolium castaneum. Dev Comp Immunol. 2008;32:585–595. doi: 10.1016/j.dci.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 82.Podell S, Gaasterland T. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol. 2007;8:R16. doi: 10.1186/gb-2007-8-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nguyen M, Ekstrom A, Li X, Yin Y. HGT-finder: a new tool for horizontal gene transfer finding and application to Aspergillus genomes. Toxins. 2015;7:4035–53. 10.3390/toxins7104035. [DOI] [PMC free article] [PubMed]
- 84.Nakabachi A. Horizontal gene transfers in insects. Curr Opin Insect Sci. 2015;7:24–29. doi: 10.1016/j.cois.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 85.Brelsfoard C, Tsiamis G, Falchetto M, Gomulski LM, Telleria E, Alam U, et al. Presence of extensive Wolbachia symbiont insertions discovered in the genome of its host Glossina morsitans morsitans. PLoS Negl Trop Dis. 2014;8:e2728. 10.1371/journal.pntd.0002728. [DOI] [PMC free article] [PubMed]
- 86.Nikoh N, Nakabachi A. Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol. 2009;7:12. doi: 10.1186/1741-7007-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pauchet Y, Wilkinson P, Chauhan R, Ffrench-Constant RH. Diversity of beetle genes encoding novel plant cell wall degrading enzymes. PLoS One. 2010;5:e15635. doi: 10.1371/journal.pone.0015635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Terra WR, Cristofoletti PT. Midgut proteinases in three divergent species of Coleoptera. Comp Biochem Physiol B Biochem Mol Biol. 1996;113:725–730. doi: 10.1016/0305-0491(95)02037-3. [DOI] [Google Scholar]
- 90.Murdock LL, Brookhart G, Dunn PE, Foard DE, Kelley S, Kitch L, et al. Cysteine digestive proteinases in Coleoptera. Comp Biochem Physiol Part B Comp Biochem. 1987;87:783–787. doi: 10.1016/0305-0491(87)90388-9. [DOI] [Google Scholar]
- 91.Liang C, Brookhart G, Feng GH, Reeck GR, Kramer KJ. Inhibition of digestive proteinases of stored grain Coleoptera by oryzacystatin, a cysteine proteinase inhibitor from rice seed. FEBS Lett. 1991;278:139–142. doi: 10.1016/0014-5793(91)80102-9. [DOI] [PubMed] [Google Scholar]
- 92.Mossé J. Acides aminés de 16 céréales et protéagineux : variations et clés du calcul de la composition en fonction du taux d’azote des grain(e)s. Conséquences nutritionnelles INRA Prod Anim 1990;3:103–19.
- 93.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martynov AG, Elpidina EN, Perkin L, Oppert B. Functional analysis of C1 family cysteine peptidases in the larval gut of Тenebrio molitor and Tribolium castaneum. BMC Genomics. 2015;16:75. doi: 10.1186/s12864-015-1306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schoville SD, Chen YH, Andersson MN, Benoit JB, Bhandari A, Bowsher JH, et al. A model species for agricultural pest genomics: the genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae) Sci Rep. 2018;8:1931. doi: 10.1038/s41598-018-20154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jongsma MA, Bolter C. The adaptation of insects to plant protease inhibitors. J Insect Physiol. 1997;43:885–895. doi: 10.1016/S0022-1910(97)00040-1. [DOI] [PubMed] [Google Scholar]
- 99.Ryan CA. Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu Rev Phytopathol. 1990;28:425–449. doi: 10.1146/annurev.py.28.090190.002233. [DOI] [Google Scholar]
- 100.Sosulski FW, Minja LA, Christensen DA. Trypsin inhibitors and nutritive value in cereals. Plant Foods Hum Nutr. 1988;38:23–34. doi: 10.1007/BF01092307. [DOI] [PubMed] [Google Scholar]
- 101.Feng GH, Richardson M, Chen MS, Kramer KJ, Morgan TD, Reeck GR. α-Amylase inhibitors from wheat: amino acid sequences and patterns of inhibition of insect and human α-amylases. Insect Biochem Mol Biol. 1996;26:419–426. doi: 10.1016/0965-1748(95)00087-9. [DOI] [PubMed] [Google Scholar]
- 102.Yetter MA, Saunders RM, Boles HP. Alpha-amylase inhibitors from wheat kernels as factors in resistance to postharvest insects. Cereal Chem. 1979;56:243–244. [Google Scholar]
- 103.Agrawal S, Kelkenberg M, Begum K, Steinfeld L, Williams CE, Kramer KJ, et al. Two essential peritrophic matrix proteins mediate matrix barrier functions in the insect midgut. Insect Biochem Mol Biol. 2014;49:24–34. doi: 10.1016/j.ibmb.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 104.Tellam RL, Wijffels G, Willadsen P. Peritrophic matrix proteins. Insect Biochem Mol Biol. 1999;29:87–101. doi: 10.1016/s0965-1748(98)00123-4. [DOI] [PubMed] [Google Scholar]
- 105.McKenna DD, Shin S, Ahrens D, Balke M, Beza-Beza C, Clarke DJ, et al. The evolution and genomic basis of beetle diversity. Proc Natl Acad Sci U S A. 2019;116:24729–24737. doi: 10.1073/pnas.1909655116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kirsch R, Gramzow L, Theißen G, Siegfried BD, Ffrench-Constant RH, Heckel DG, et al. Horizontal gene transfer and functional diversification of plant cell wall degrading polygalacturonases: key events in the evolution of herbivory in beetles. Insect Biochem Mol Biol. 2014;52:33–50. doi: 10.1016/j.ibmb.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 107.Shen Z, Denton M, Mutti N, Pappan K, Kanost MR, Reese JC, et al. Polygalacturonase from Sitophilus oryzae: possible horizontal transfer of a pectinase gene from fungi to weevils. J Insect Sci Online. 2003;3:24. doi: 10.1093/jis/3.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vellozo AF, Véron AS, Baa-Puyoulet P, Huerta-Cepas J, Cottret L, Febvay G, et al. CycADS: an annotation database system to ease the development and update of BioCyc databases. Database. 2011;2011:bar008. doi: 10.1093/database/bar008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Karp PD, Midford PE, Billington R, Kothari A, Krummenacker M, Latendresse M, et al. Pathway tools version 23.0 update: software for pathway/genome informatics and systems biology. Brief Bioinform. 2019. 10.1093/bib/bbz104. [DOI] [PMC free article] [PubMed]
- 110.Baa-Puyoulet P, Parisot N, Febvay G, Huerta-Cepas J, Vellozo AF, Gabaldón T, et al. ArthropodaCyc: a CycADS powered collection of BioCyc databases to analyse and compare metabolism of arthropods. Database. 2016;2016:baw081. doi: 10.1093/database/baw081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vigneron A, Masson F, Vallier A, Balmand S, Rey M, Vincent-Monégat C, et al. Insects recycle endosymbionts when the benefit is over. Curr Biol. 2014;24:2267–2273. doi: 10.1016/j.cub.2014.07.065. [DOI] [PubMed] [Google Scholar]
- 112.Heddi A, Grenier A-M, Khatchadourian C, Charles H, Nardon P. Four intracellular genomes direct weevil biology: nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc Natl Acad Sci U S A. 1999;96:6814–9. 10.1073/pnas.96.12.6814. [DOI] [PMC free article] [PubMed]
- 113.Grenier AM, Nardon C, Nardon P. The role of symbiotes in flight activity of Sitophilus weevils. Entomol Exp Appl. 1994;70:201–208. doi: 10.1111/j.1570-7458.1994.tb00748.x. [DOI] [Google Scholar]
- 114.Rio RVM, Lefevre C, Heddi A, Aksoy S. Comparative genomics of insect-symbiotic bacteria: influence of host environment on microbial genome composition. Appl Environ Microbiol. 2003;69:6825–6832. doi: 10.1128/aem.69.11.6825-6832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baker JE, Woo SM. Purification, partial characterization, and postembryonic levels of amylases from Sitophilus oryzae and Sitophilus granarius. Arch Insect Biochem Physiol. 1985;2:415–428. doi: 10.1002/arch.940020409. [DOI] [Google Scholar]
- 116.Wicker C. Differential vitamin and choline requirements of symbiotic and aposymbiotic S. oryzae (Coleoptera: Curculionidae) Comp Biochem Physiol A Physiol. 1983;76:177–182. doi: 10.1016/0300-9629(83)90311-0. [DOI] [Google Scholar]
- 117.Heddi A, Lestienne P, Wallace DC, Stepien G. Steady state levels of mitochondrial and nuclear oxidative phosphorylation transcripts in Kearns-Sayre syndrome. Biochim Biophys Acta. 1994;1226:206–212. doi: 10.1016/0925-4439(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 118.Hernández L, Afonso D, Rodríguez EM, Díaz C. Phenolic compounds in wheat grain cultivars. Plant Foods Hum Nutr Dordr Neth. 2011;66:408–415. doi: 10.1007/s11130-011-0261-1. [DOI] [PubMed] [Google Scholar]
- 119.Panfili G, Fratianni A, Irano M. Improved normal-phase high-performance liquid chromatography procedure for the determination of carotenoids in cereals. J Agric Food Chem. 2004;52:6373–6377. doi: 10.1021/jf0402025. [DOI] [PubMed] [Google Scholar]
- 120.Hall RJ, Thorpe S, Thomas GH, Wood AJ. Simulating the evolutionary trajectories of metabolic pathways for insect symbionts in the genus Sodalis. Microb Genomics. 2020;6:e000378. doi: 10.1099/mgen.0.000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wieschaus E, Nüsslein-Volhard C. The Heidelberg screen for pattern mutants of Drosophila: a personal account. Annu Rev Cell Dev Biol. 2016;32:1–46. 10.1146/annurev-cellbio-113015-023138. [DOI] [PubMed]
- 122.Schmidt-Ott U, Lynch JA. Emerging developmental genetic model systems in holometabolous insects. Curr Opin Genet Dev. 2016;39:116–128. doi: 10.1016/j.gde.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 123.Schmitt-Engel C, Schultheis D, Schwirz J, Ströhlein N, Troelenberg N, Majumdar U, et al. The iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology. Nat Commun. 2015;6:7822. doi: 10.1038/ncomms8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Peel AD. The evolution of developmental gene networks: lessons from comparative studies on holometabolous insects. Philos Trans R Soc Lond Ser B Biol Sci. 2008;363:1539–1547. doi: 10.1098/rstb.2007.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Herndon N, Shelton J, Gerischer L, Ioannidis P, Ninova M, Dönitz J, et al. Enhanced genome assembly and a new official gene set for Tribolium castaneum. BMC Genomics. 2020;21:47. doi: 10.1186/s12864-019-6394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tiegs OW, Murray FV. Memoirs: the embryonic development of Calandra oryzae. J Cell Sci. 1938;s2-80:159–273. doi: 10.1242/jcs.s2-80.318.159. [DOI] [Google Scholar]
- 127.Duncan EJ, Benton MA, Dearden PK. Canonical terminal patterning is an evolutionary novelty. Dev Biol. 2013;377:245–261. doi: 10.1016/j.ydbio.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 128.Sano H, Renault AD, Lehmann R. Control of lateral migration and germ cell elimination by the Drosophila melanogaster lipid phosphate phosphatases Wunen and Wunen 2. J Cell Biol. 2005;171:675–683. doi: 10.1083/jcb.200506038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Savard J, Marques-Souza H, Aranda M, Tautz D. A segmentation gene in Tribolium produces a polycistronic mRNA that codes for multiple conserved peptides. Cell. 2006;126:559–569. doi: 10.1016/j.cell.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 130.Angelini DR, Kaufman TC. Comparative developmental genetics and the evolution of arthropod body plans. Annu Rev Genet. 2005;39:95–119. doi: 10.1146/annurev.genet.39.073003.112310. [DOI] [PubMed] [Google Scholar]
- 131.Shippy TD, Brown SJ, Denell RE. maxillopedia is the Tribolium ortholog of proboscipedia. Evol Dev. 2000;2:145–51. 10.1046/j.1525-142x.2000.00055.x. [DOI] [PubMed]
- 132.Angelini DR, Smith FW, Jockusch EL. Extent with modification: leg patterning in the beetle Tribolium castaneum and the evolution of serial homologs. G3. 2012;2:235–48. 10.1534/g3.111.001537. [DOI] [PMC free article] [PubMed]
- 133.Ober KA, Jockusch EL. The roles of wingless and decapentaplegic in axis and appendage development in the red flour beetle, Tribolium castaneum. Dev Biol. 2006;294:391–405. doi: 10.1016/j.ydbio.2006.02.053. [DOI] [PubMed] [Google Scholar]
- 134.Mirth CK, Anthony Frankino W, Shingleton AW. Allometry and size control: what can studies of body size regulation teach us about the evolution of morphological scaling relationships? Curr Opin Insect Sci. 2016;13:93–98. doi: 10.1016/j.cois.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 135.Nijhout HF, Callier V. Developmental mechanisms of body size and wing-body scaling in insects. Annu Rev Entomol. 2015;60:141–156. doi: 10.1146/annurev-ento-010814-020841. [DOI] [PubMed] [Google Scholar]
- 136.Huybrechts J, Bonhomme J, Minoli S, Prunier-Leterme N, Dombrovsky A, Abdel-Latief M, et al. Neuropeptide and neurohormone precursors in the pea aphid, Acyrthosiphon pisum. Insect Mol Biol. 2010;19(Suppl 2):87–95. doi: 10.1111/j.1365-2583.2009.00951.x. [DOI] [PubMed] [Google Scholar]
- 137.Gurska D, Vargas Jentzsch IM, Panfilio KA. Unexpected mutual regulation underlies paralogue functional diversification and promotes epithelial tissue maturation in Tribolium. Commun Biol. 2020;3:552. doi: 10.1038/s42003-020-01250-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dönitz J, Schmitt-Engel C, Grossmann D, Gerischer L, Tech M, Schoppmeier M, et al. iBeetle-base: a database for RNAi phenotypes in the red flour beetle Tribolium castaneum. Nucleic Acids Res. 2015;43:D720–D725. doi: 10.1093/nar/gku1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jasrapuria S, Arakane Y, Osman G, Kramer KJ, Beeman RW, Muthukrishnan S. Genes encoding proteins with peritrophin A-type chitin-binding domains in Tribolium castaneum are grouped into three distinct families based on phylogeny, expression and function. Insect Biochem Mol Biol. 2010;40:214–227. doi: 10.1016/j.ibmb.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 140.Jasrapuria S, Specht CA, Kramer KJ, Beeman RW, Muthukrishnan S. Gene families of cuticular proteins analogous to peritrophins (CPAPs) in Tribolium castaneum have diverse functions. PLoS One. 2012;7:e49844. doi: 10.1371/journal.pone.0049844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Balabanidou V, Kefi M, Aivaliotis M, Koidou V, Girotti JR, Mijailovsky SJ, et al. Mosquitoes cloak their legs to resist insecticides. Proc Biol Sci. 2019;286:20191091. doi: 10.1098/rspb.2019.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Arakane Y, Lomakin J, Gehrke SH, Hiromasa Y, Tomich JM, Muthukrishnan S, et al. Formation of rigid, non-flight forewings (elytra) of a beetle requires two major cuticular proteins. PLoS Genet. 2012;8:e1002682. doi: 10.1371/journal.pgen.1002682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ioannidou ZS, Theodoropoulou MC, Papandreou NC, Willis JH, Hamodrakas SJ. CutProtFam-Pred: detection and classification of putative structural cuticular proteins from sequence alone, based on profile hidden Markov models. Insect Biochem Mol Biol. 2014;52:51–59. doi: 10.1016/j.ibmb.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gerardo NM, Altincicek B, Anselme C, Atamian H, Barribeau SM, de Vos M, et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010;11:R21. doi: 10.1186/gb-2010-11-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang C-R, Zhang S, Xia J, Li F-F, Xia W-Q, Liu S-S, et al. The immune strategy and stress response of the mediterranean species of the Bemisia tabaci complex to an orally delivered bacterial pathogen. PLoS One. 2014;9:e94477. doi: 10.1371/journal.pone.0094477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Salcedo-Porras N, Guarneri A, Oliveira PL, Lowenberger C. Rhodnius prolixus: identification of missing components of the IMD immune signaling pathway and functional characterization of its role in eliminating bacteria. PLoS One. 2019;14:e0214794. doi: 10.1371/journal.pone.0214794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Maire J, Vincent-Monégat C, Masson F, Zaidman-Rémy A, Heddi A. An IMD-like pathway mediates both endosymbiont control and host immunity in the cereal weevil Sitophilus spp. Microbiome. 2018;6:6. doi: 10.1186/s40168-017-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Maire J, Vincent-Monégat C, Balmand S, Vallier A, Hervé M, Masson F, et al. Weevil pgrp-lb prevents endosymbiont TCT dissemination and chronic host systemic immune activation. Proc Natl Acad Sci U S A. 2019;116:5623–32. 10.1073/pnas.1821806116. [DOI] [PMC free article] [PubMed]
- 149.Sheehan G, Garvey A, Croke M, Kavanagh K. Innate humoral immune defences in mammals and insects: the same, with differences ? Virulence. 2018;9:1625–1639. doi: 10.1080/21505594.2018.1526531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Strand MR. The insect cellular immune response. Insect Sci. 2008;15:1–14. doi: 10.1111/j.1744-7917.2008.00183.x. [DOI] [Google Scholar]
- 151.He Y, Cao X, Li K, Hu Y, Chen Y, Blissard G, et al. A genome-wide analysis of antimicrobial effector genes and their transcription patterns in Manduca sexta. Insect Biochem Mol Biol. 2015;62:23–37. doi: 10.1016/j.ibmb.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 153.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–5. 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed]
- 154.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, et al. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 2007;8:R177. doi: 10.1186/gb-2007-8-8-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cao X, He Y, Hu Y, Wang Y, Chen Y-R, Bryant B, et al. The immune signaling pathways of Manduca sexta. Insect Biochem Mol Biol. 2015;62:64–74. doi: 10.1016/j.ibmb.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Arp AP, Hunter WB, Pelz-Stelinski KS. Annotation of the Asian citrus psyllid genome reveals a reduced innate immune system. Front Physiol. 2016;7. 10.3389/fphys.2016.00570. [DOI] [PMC free article] [PubMed]
- 158.Kang X, Dong F, Shi C, Liu S, Sun J, Chen J, et al. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci Data. 2019;6:148. doi: 10.1038/s41597-019-0154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Palmer WJ, Jiggins FM. Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol Biol Evol. 2015;32:2111–2129. doi: 10.1093/molbev/msv093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Smith CA. Structure, function and dynamics in the mur family of bacterial cell wall ligases. J Mol Biol. 2006;362:640–655. doi: 10.1016/j.jmb.2006.07.066. [DOI] [PubMed] [Google Scholar]
- 161.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, et al. The Drosophila immune response against gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–4. 10.1038/nature734. [DOI] [PubMed]
- 162.Choe K-M, Werner T, Stöven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–62. 10.1126/science.1070216. [DOI] [PubMed]
- 163.Kleino A, Silverman N. The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol. 2014;42. 10.1016/j.dci.2013.05.014. [DOI] [PMC free article] [PubMed]
- 164.Park JT. Why does Escherichia coli recycle its cell wall peptides? Mol Microbiol. 1995;17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 165.Johnson JW, Fisher JF, Mobashery S. Bacterial cell-wall recycling. Ann N Y Acad Sci. 2013;1277:54–75. doi: 10.1111/j.1749-6632.2012.06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kaneko T, Yano T, Aggarwal K, Lim J-H, Ueda K, Oshima Y, et al. PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol. 2006;7:715–23. 10.1038/ni1356. [DOI] [PubMed]
- 167.Bosco-Drayon V, Poidevin M, Boneca IG, Narbonne-Reveau K, Royet J, Charroux B. Peptidoglycan sensing by the receptor PGRP-LE in the Drosophila gut induces immune responses to infectious bacteria and tolerance to microbiota. Cell Host Microbe. 2012;12:153–65. 10.1016/j.chom.2012.06.002. [DOI] [PubMed]
- 168.Neyen C, Poidevin M, Roussel A, Lemaitre B. Tissue- and ligand-specific sensing of Gram-negative infection in Drosophila by PGRP-LC isoforms and PGRP-LE. J Immunol Baltim Md 1950. 2012;189:1886–97. 10.4049/jimmunol.1201022. [DOI] [PubMed]
- 169.Tindwa H, Patnaik BB, Kim DH, Mun S, Jo YH, Lee BL, et al. Cloning, characterization and effect of TmPGRP-LE gene silencing on survival of Tenebrio molitor against Listeria monocytogenes infection. Int J Mol Sci. 2013;14:22462–82. 10.3390/ijms141122462. [DOI] [PMC free article] [PubMed]
- 170.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–9. 10.1038/414756a. [DOI] [PubMed]
- 171.Wang J, Song X, Wang M. Peptidoglycan recognition proteins in hematophagous arthropods. Dev Comp Immunol. 2018;83:89–95. doi: 10.1016/j.dci.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chowdhury M, Li C-F, He Z, Lu Y, Liu X-S, Wang Y-F, et al. Toll family members bind multiple Spätzle proteins and activate antimicrobial peptide gene expression in Drosophila. J Biol Chem. 2019;294:10172–81. 10.1074/jbc.RA118.006804. [DOI] [PMC free article] [PubMed]
- 173.Valanne S, Wang J-H, Rämet M. The Drosophila Toll signaling pathway. J Immunol Baltim Md 1950. 2011;186:649–56. 10.4049/jimmunol.1002302. [DOI] [PubMed]
- 174.Muhammad A, Habineza P, Wang X, Xiao R, Ji T, Hou Y, et al. Spätzle homolog-mediated toll-like pathway regulates innate immune responses to maintain the homeostasis of gut microbiota in the red palm weevil, Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae) Front Microbiol. 2020;11:846. doi: 10.3389/fmicb.2020.00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gupta SK, Kupper M, Ratzka C, Feldhaar H, Vilcinskas A, Gross R, et al. Scrutinizing the immune defence inventory of Camponotus floridanus applying total transcriptome sequencing. BMC Genomics. 2015;16:540. doi: 10.1186/s12864-015-1748-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bang IS. JAK/STAT signaling in insect innate immunity. Entomol Res. 2019;49:339–353. doi: 10.1111/1748-5967.12384. [DOI] [Google Scholar]
- 177.Wu Q, Patočka J, Kuča K. Insect antimicrobial peptides, a mini review. Toxins. 2018;10. 10.3390/toxins10110461. [DOI] [PMC free article] [PubMed]
- 178.Callewaert L, Michiels CW. Lysozymes in the animal kingdom. J Biosci. 2010;35:127–160. doi: 10.1007/s12038-010-0015-5. [DOI] [PubMed] [Google Scholar]
- 179.Mohrig W, Messner B. Lysozyme as antibacterial agent in honey and bees venom. Acta Biol Med Ger. 1968;21:85–95. [PubMed] [Google Scholar]
- 180.Hultmark D. Insect lysozymes. EXS. 1996;75:87–102. doi: 10.1007/978-3-0348-9225-4_6. [DOI] [PubMed] [Google Scholar]
- 181.Beckert A, Wiesner J, Baumann A, Pöppel A-K, Vogel H, Vilcinskas A. Two c-type lysozymes boost the innate immune system of the invasive ladybird Harmonia axyridis. Dev Comp Immunol. 2015;49:303–312. doi: 10.1016/j.dci.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 182.Beckert A, Wiesner J, Schmidtberg H, Lehmann R, Baumann A, Vogel H, et al. Expression and characterization of a recombinant i-type lysozyme from the harlequin ladybird beetle Harmonia axyridis. Insect Mol Biol. 2016;25:202–215. doi: 10.1111/imb.12213. [DOI] [PubMed] [Google Scholar]
- 183.Brandazza A, Angeli S, Tegoni M, Cambillau C, Pelosi P. Plant stress proteins of the thaumatin-like family discovered in animals. FEBS Lett. 2004;572:3–7. doi: 10.1016/j.febslet.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 184.Anselme C, Pérez-Brocal V, Vallier A, Vincent-Monegat C, Charif D, Latorre A, et al. Identification of the weevil immune genes and their expression in the bacteriome tissue. BMC Biol. 2008;6:43. doi: 10.1186/1741-7007-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Masson F, Moné Y, Vigneron A, Vallier A, Parisot N, Vincent-Monégat C, et al. Weevil endosymbiont dynamics is associated with a clamping of immunity. BMC Genomics. 2015;16:819. doi: 10.1186/s12864-015-2048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chung KT, Ourth DD. Viresin. A novel antibacterial protein from immune hemolymph of Heliothis virescens pupae. Eur J Biochem. 2000;267:677–683. doi: 10.1046/j.1432-1327.2000.01034.x. [DOI] [PubMed] [Google Scholar]
- 187.Benoit JB, Adelman ZN, Reinhardt K, Dolan A, Poelchau M, Jennings EC, et al. Unique features of a global human ectoparasite identified through sequencing of the bed bug genome. Nat Commun. 2016;7:10165. doi: 10.1038/ncomms10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A. 2010;107:12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pachebat JA, van Keulen G, Whitten MMA, Girdwood S, Del Sol R, Dyson PJ, et al. Draft genome sequence of Rhodococcus rhodnii strain LMG5362, a symbiont of Rhodnius prolixus (Hemiptera, Reduviidae, Triatominae), the principle vector of Trypanosoma cruzi. Genome Announc. 2013;1. 10.1128/genomeA.00329-13. [DOI] [PMC free article] [PubMed]
- 190.Rispe C, Legeai F, Nabity PD, Fernández R, Arora AK, Baa-Puyoulet P, et al. The genome sequence of the grape Phylloxera provides insights into the evolution, adaptation, and invasion routes of an iconic pest. BMC Biol. 2020;18:90. doi: 10.1186/s12915-020-00820-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Nishide Y, Kageyama D, Yokoi K, Jouraku A, Tanaka H, Futahashi R, et al. Functional crosstalk across IMD and Toll pathways: insight into the evolution of incomplete immune cascades. Proc R Soc B Biol Sci. 2019;286:20182207. 10.1098/rspb.2018.2207. [DOI] [PMC free article] [PubMed]
- 192.Matetovici I, De Vooght L, Van Den Abbeele J. Innate immunity in the tsetse fly (Glossina), vector of African trypanosomes. Dev Comp Immunol. 2019;98:181–188. doi: 10.1016/j.dci.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 193.Login FH, Balmand S, Vallier A, Vincent-Monégat C, Vigneron A, Weiss-Gayet M, et al. Antimicrobial peptides keep insect endosymbionts under control. Science. 2011;334:362–365. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 194.Chaudhry MQ. Phosphine resistance. Pestic Outlook. 2000;11:88–91. doi: 10.1039/B006348G. [DOI] [Google Scholar]
- 195.Chaudhry MQ. A review of the mechanisms involved in the action of phosphine as an insecticide and phosphine resistance in stored-product insects. Pestic Sci. 1997;49:213–228. doi: 10.1002/(SICI)1096-9063(199703)49:3<213::AID-PS516>3.0.CO;2-#. [DOI] [Google Scholar]
- 196.Athié I, Gomes RAR, Bolonhezi S, Valentini SRT, De Castro MFPM. Effects of carbon dioxide and phosphine mixtures on resistant populations of stored-grain insects. J Stored Prod Res. 1998;34:27–32. doi: 10.1016/S0022-474X(97)00026-X. [DOI] [Google Scholar]
- 197.Rajendran S. Phosphine resistance in stored grain insect pests in India. Proc. 7th Int. Work. Conf. Stored-Prod. Prot., 1998, p. 14–9.
- 198.Zeng L. Development and countermeasures of phosphine resistance in stored grain insects in Guangdong, China, 642–647. Proc. Seventh Int. Work. Conf. Stored-Prod. Prot. Eds J Zuxun Quan Yongsheng T Xianchang G Lianghua14–19 Oct. 1998 Beijing China Sichuan Publ. House Sci. Technol. Chengdu China, 1999.
- 199.Benhalima H, Chaudhry MQ, Mills KA, Price NR. Phosphine resistance in stored-product insects collected from various grain storage facilities in Morocco. J Stored Prod Res. 2004;40:241–249. doi: 10.1016/S0022-474X(03)00012-2. [DOI] [Google Scholar]
- 200.Pimentel MAG, Faroni LRD, da Silva FH, Batista MD, Guedes RNC. Spread of phosphine resistance among brazilian populations of three species of stored product insects. Neotrop Entomol. 2010;39:101–107. doi: 10.1590/S1519-566X2010000100014. [DOI] [PubMed] [Google Scholar]
- 201.Nguyen TT, Collins PJ, Duong TM, Schlipalius DI, Ebert PR. Genetic conservation of phosphine resistance in the rice weevil Sitophilus oryzae (L.) J Hered. 2016;107:228–237. doi: 10.1093/jhered/esw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Holloway JC, Falk MG, Emery RN, Collins PJ, Nayak MK. Resistance to phosphine in Sitophilus oryzae in Australia: a national analysis of trends and frequencies over time and geographical spread. J Stored Prod Res. 2016;69:129–137. doi: 10.1016/j.jspr.2016.07.004. [DOI] [Google Scholar]
- 203.Agrafioti P, Athanassiou CG, Nayak MK. Detection of phosphine resistance in major stored-product insects in Greece and evaluation of a field resistance test kit. J Stored Prod Res. 2019;82:40–47. doi: 10.1016/j.jspr.2019.02.004. [DOI] [Google Scholar]
- 204.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019;47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sigrist CJA, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, et al. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Scott JG, Wen Z. Cytochromes P450 of insects: the tip of the iceberg. Pest Manag Sci. 2001;57:958–967. doi: 10.1002/ps.354. [DOI] [PubMed] [Google Scholar]
- 208.Hu F, Ye K, Tu X-F, Lu Y-J, Thakur K, Jiang L, et al. Identification and expression profiles of twenty-six glutathione S-transferase genes from rice weevil, Sitophilus oryzae (Coleoptera: Curculionidae) Int J Biol Macromol. 2018;120:1063–1071. doi: 10.1016/j.ijbiomac.2018.08.185. [DOI] [PubMed] [Google Scholar]
- 209.Kim K, Yang JO, Sung J-Y, Lee J-Y, Park JS, Lee H-S, et al. Minimization of energy transduction confers resistance to phosphine in the rice weevil, Sitophilus oryzae. Sci Rep. 2019;9:14605. doi: 10.1038/s41598-019-50972-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Schlipalius DI, Tuck AG, Jagadeesan R, Nguyen T, Kaur R, Subramanian S, et al. Variant linkage analysis using de novo transcriptome sequencing identifies a conserved phosphine resistance gene in insects. Genetics. 2018;209:281–290. doi: 10.1534/genetics.118.300688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Haddi K, Valbon WR, Viteri Jumbo LO, de Oliveira LO, Guedes RNC, Oliveira EE. Diversity and convergence of mechanisms involved in pyrethroid resistance in the stored grain weevils, Sitophilus spp. Sci Rep. 2018;8:16361. doi: 10.1038/s41598-018-34513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Blanton AG, Peterson BF. Symbiont-mediated insecticide detoxification as an emerging problem in insect pests. Front Microbiol. 2020;11. 10.3389/fmicb.2020.547108. [DOI] [PMC free article] [PubMed]
- 213.Carey AF, Carlson JR. Insect olfaction from model systems to disease control. Proc Natl Acad Sci U S A. 2011;108:12987–12995. doi: 10.1073/pnas.1103472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Andersson MN, Newcomb RD. Pest control compounds targeting insect chemoreceptors: another silent spring? Front Ecol Evol. 2017;5. 10.3389/fevo.2017.00005.
- 215.Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 216.Hassanali A, Herren H, Khan Z, Pickett J, Woodcock C. Integrated pest management: the push-pull approach for controlling insect pests and weeds of cereals, and its potential for other agricultural systems including animal husbandry. Philos Trans R Soc Lond Ser B Biol Sci. 2008;363:611–621. doi: 10.1098/rstb.2007.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hatano E, Saveer AM, Borrero-Echeverry F, Strauch M, Zakir A, Bengtsson M, et al. A herbivore-induced plant volatile interferes with host plant and mate location in moths through suppression of olfactory signalling pathways. BMC Biol. 2015;13:75. doi: 10.1186/s12915-015-0188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ukeh DA, Woodcock CM, Pickett JA, Birkett MA. Identification of host kairomones from maize, Zea mays, for the maize weevil, Sitophilus zeamais. J Chem Ecol. 2012;38:1402–1409. doi: 10.1007/s10886-012-0191-x. [DOI] [PubMed] [Google Scholar]
- 219.Germinara GS, De Cristofaro A, Rotundo G. Behavioral responses of adult Sitophilus granarius to individual cereal volatiles. J Chem Ecol. 2008;34:523–529. doi: 10.1007/s10886-008-9454-y. [DOI] [PubMed] [Google Scholar]
- 220.Phillips JK, Walgenbach CA, Klein JA, Burkholder WE, Schmuff NR, Fales HM. (R (*),S (*))-5-hydroxy-4-methyl-3-heptanone male-produced aggregation pheromone of Sitophilus oryzae (L.) and S. zeamais Motsch. J Chem Ecol. 1985;11:1263–1274. doi: 10.1007/BF01024114. [DOI] [PubMed] [Google Scholar]
- 221.Schmuff NR, Phillips JK, Burkholder WE, Fales HM, Chen C-W, Roller PP, et al. The chemical identification of the rice weevil and maize weevil aggregation pheromone. Tetrahedron Lett. 1984;25:1533–1534. doi: 10.1016/S0040-4039(01)90002-4. [DOI] [Google Scholar]
- 222.Mitchell RF, Schneider TM, Schwartz AM, Andersson MN, McKenna DD. The diversity and evolution of odorant receptors in beetles (Coleoptera) Insect Mol Biol. 2020;29:77–91. doi: 10.1111/imb.12611. [DOI] [PubMed] [Google Scholar]
- 223.de Bruyne M, Baker TC. Odor detection in insects: volatile codes. J Chem Ecol. 2008;34:882–897. doi: 10.1007/s10886-008-9485-4. [DOI] [PubMed] [Google Scholar]
- 224.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed]
- 225.Brand P, Robertson HM, Lin W, Pothula R, Klingeman WE, Jurat-Fuentes JL, et al. The origin of the odorant receptor gene family in insects. ELife. 2018;7. 10.7554/eLife.38340. [DOI] [PMC free article] [PubMed]
- 226.Montagné N, de Fouchier A, Newcomb RD, Jacquin-Joly E. Advances in the identification and characterization of olfactory receptors in insects. Prog Mol Biol Transl Sci. 2015;130:55–80. doi: 10.1016/bs.pmbts.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 227.Mansourian S, Stensmyr MC. The chemical ecology of the fly. Curr Opin Neurobiol. 2015;34:95–102. doi: 10.1016/j.conb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 228.Carey AF, Wang G, Su C-Y, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464:66–71. doi: 10.1038/nature08834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2010;107:4418–4423. doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.de Fouchier A, Walker WB, Montagné N, Steiner C, Binyameen M, Schlyter F, et al. Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat Commun. 2017;8:15709. doi: 10.1038/ncomms15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Guo M, Du L, Chen Q, Feng Y, Zhang J, Zhang X, et al. Odorant receptors for detecting flowering plant cues are functionally conserved across moths and butterflies. Mol Biol Evol. 2020. 10.1093/molbev/msaa300. [DOI] [PMC free article] [PubMed]
- 232.Pask GM, Slone JD, Millar JG, Das P, Moreira JA, Zhou X, et al. Specialized odorant receptors in social insects that detect cuticular hydrocarbon cues and candidate pheromones. Nat Commun. 2017;8:297. doi: 10.1038/s41467-017-00099-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Slone JD, Pask GM, Ferguson ST, Millar JG, Berger SL, Reinberg D, et al. Functional characterization of odorant receptors in the ponerine ant, Harpegnathos saltator. Proc Natl Acad Sci U S A. 2017;114:8586–8591. doi: 10.1073/pnas.1704647114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Mitchell RF, Hughes DT, Luetje CW, Millar JG, Soriano-Agatón F, Hanks LM, et al. Sequencing and characterizing odorant receptors of the cerambycid beetle Megacyllene caryae. Insect Biochem Mol Biol. 2012;42:499–505. doi: 10.1016/j.ibmb.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yuvaraj J, Roberts R, Sonntag Y, Hou X, Grosse-Wilde E, Machara A, et al. Putative ligand binding sites of two functionally characterized bark beetle odorant receptors. BMC Biol. 2021;19:16. doi: 10.1101/2020.03.07.980797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Antony B, Johny J, Montagné N, Jacquin-Joly E, Capoduro R, Cali K, et al. Pheromone receptor of the globally invasive quarantine pest of the palm tree, the red palm weevil (Rhynchophorus ferrugineus) Mol Ecol. 2021;30(9):2025–2039. doi: 10.1111/mec.15874. [DOI] [PubMed] [Google Scholar]
- 237.Keller O, Odronitz F, Stanke M, Kollmar M, Waack S. Scipio: using protein sequences to determine the precise exon/intron structures of genes and their orthologs in closely related species. BMC Bioinformatics. 2008;9:278. doi: 10.1186/1471-2105-9-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 241.Lefort V, Longueville J-E, Gascuel O. SMS: smart model selection in PhyML. Mol Biol Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 243.Makałowski W, Gotea V, Pande A, Makałowska I. Transposable elements: Classification, identification, and their use as a tool for comparative genomics. In: Anisimova M, editor. Evol. Genomics Stat. Comput. Methods, New York, NY: Springer; 2019, p. 177–207. 10.1007/978-1-4939-9074-0_6. [DOI] [PubMed]
- 244.Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ou S, Su W, Liao Y, Chougule K, Agda JRA, Hellinga AJ, et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019;20:275. doi: 10.1186/s13059-019-1905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346:763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 247.Hernandez-Hernandez EM, Fernández-Medina RD, Navarro-Escalante L, Nuñez J, Benavides-Machado P, Carareto CMA. Genome-wide analysis of transposable elements in the coffee berry borer Hypothenemus hampei (Coleoptera: Curculionidae): description of novel families. Mol Gen Genomics. 2017;292:565–583. doi: 10.1007/s00438-017-1291-7. [DOI] [PubMed] [Google Scholar]
- 248.Amorin I, Melo E, Moura R, Wallau G. Diverse mobilome of Dichotomius (Luederwaldtinia) schiffleri (Coleoptera: Scarabaeidae) reveals long-range horizontal transfer events of DNA transposons. Mol Gen Genomics. 2020. 10.1007/s00438-020-01703-8. [DOI] [PubMed]
- 249.Feschotte C, Zhang X, Wessler SR. Miniature inverted-repeat transposable elements and their relationship to established DNA transposons. Mob DNA II. 2002:1147–58. 10.1128/9781555817954.ch50.
- 250.Feschotte C, Mouchès C. Recent amplification of miniature inverted-repeat transposable elements in the vector mosquito Culex pipiens: characterization of the Mimo family. Gene. 2000;250:109–116. doi: 10.1016/S0378-1119(00)00187-6. [DOI] [PubMed] [Google Scholar]
- 251.Feschotte C, Swamy L, Wessler SR. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs) Genetics. 2003;163:747–758. doi: 10.1093/genetics/163.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Lu C, Chen J, Zhang Y, Hu Q, Su W, Kuang H. Miniature inverted-repeat transposable elements (MITEs) have been accumulated through amplification bursts and play important roles in gene expression and species diversity in Oryza sativa. Mol Biol Evol. 2012;29:1005–1017. doi: 10.1093/molbev/msr282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Feng Y. Plant MITEs: useful tools for plant genetics and genomics. Genomics Proteomics Bioinformatics. 2003;1:90–100. doi: 10.1016/S1672-0229(03)01013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sela N, Kim E, Ast G. The role of transposable elements in the evolution of non-mammalian vertebrates and invertebrates. Genome Biol. 2010;11:R59. doi: 10.1186/gb-2010-11-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Petrov DA. DNA loss and evolution of genome size in Drosophila. Genetica. 2002 May;115(1):81–91. [DOI] [PubMed]
- 256.Petrov DA, Hartl DL. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol Biol Evol. 1998;15:293–302. doi: 10.1093/oxfordjournals.molbev.a025926. [DOI] [PubMed] [Google Scholar]
- 257.Pasyukova EG, Nuzhdin SV. Doc and copia instability in an isogenic Drosophila melanogaster stock. Mol Gen Genet MGG. 1993;240:302–306. doi: 10.1007/BF00277071. [DOI] [PubMed] [Google Scholar]
- 258.Ashburner M, Bergman CM. Drosophila melanogaster: a case study of a model genomic sequence and its consequences. Genome Res. 2005;15:1661–1667. doi: 10.1101/gr.3726705. [DOI] [PubMed] [Google Scholar]
- 259.Czech B, Hannon GJ. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci. 2016;41:324–337. doi: 10.1016/j.tibs.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–80. 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed]
- 261.Andersen PR, Tirian L, Vunjak M, Brennecke J. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature. 2017;549:54–59. doi: 10.1038/nature23482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 263.Bewick AJ, Vogel KJ, Moore AJ, Schmitz RJ. Evolution of DNA methylation across insects. Mol Biol Evol. 2017;34:654–665. doi: 10.1093/molbev/msw264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ninova M, Griffiths-Jones S, Ronshaugen M. Abundant expression of somatic transposon-derived piRNAs throughout Tribolium castaneum embryogenesis. Genome Biol. 2017;18:184. doi: 10.1186/s13059-017-1304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Mongelli V, Saleh M-C. Bugs are not to be silenced: small RNA pathways and antiviral responses in insects. Annu Rev Virol. 2016;3:573–589. doi: 10.1146/annurev-virology-110615-042447. [DOI] [PubMed] [Google Scholar]
- 266.Chambeyron S, Seitz H. Insect small non-coding RNA involved in epigenetic regulations. Curr Opin Insect Sci. 2014;1:1–9. doi: 10.1016/j.cois.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 267.Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lewis SH, Quarles KA, Yang Y, Tanguy M, Frézal L, Smith SA, et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol. 2018;2:174–181. doi: 10.1038/s41559-017-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Guan D-L, Ding R-R, Hu X-Y, Yang X-R, Xu S-Q, Gu W, et al. Cadmium-induced genome-wide DNA methylation changes in growth and oxidative metabolism in Drosophila melanogaster. BMC Genomics. 2019;20:356. doi: 10.1186/s12864-019-5688-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Provataris P, Meusemann K, Niehuis O, Grath S, Misof B. Signatures of DNA methylation across insects suggest reduced DNA methylation levels in Holometabola. Genome Biol Evol. 2018;10:1185–1197. doi: 10.1093/gbe/evy066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Cunningham CB, Ji L, Wiberg RAW, Shelton J, McKinney EC, Parker DJ, et al. The genome and methylome of a beetle with complex social behavior, Nicrophorus vespilloides (Coleoptera: Silphidae) Genome Biol Evol. 2015;7:3383–3396. doi: 10.1093/gbe/evv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Yao B, Li Y, Wang Z, Chen L, Poidevin M, Zhang C, et al. Active N6-methyladenine demethylation by DMAD regulates gene expression by coordinating with Polycomb protein in neurons. Mol Cell. 2018;71:848–857.e6. doi: 10.1016/j.molcel.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Bestor TH, Holliday R, Monk M, Pugh JE. DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos Trans R Soc Lond Ser B Biol Sci. 1990;326:179–187. doi: 10.1098/rstb.1990.0002. [DOI] [PubMed] [Google Scholar]
- 275.Martienssen R. Transposons, DNA methylation and gene control. Trends Genet. 1998;14:263–264. doi: 10.1016/S0168-9525(98)01518-2. [DOI] [PubMed] [Google Scholar]
- 276.Zamudio N, Bourc’his D. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity. 2010;105:92–104. doi: 10.1038/hdy.2010.53. [DOI] [PubMed] [Google Scholar]
- 277.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hill PWS, Amouroux R, Hajkova P. DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: an emerging complex story. Genomics. 2014;104:324–333. doi: 10.1016/j.ygeno.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 282.Wojciechowski M, Rafalski D, Kucharski R, Misztal K, Maleszka J, Bochtler M, et al. Insights into DNA hydroxymethylation in the honeybee from in-depth analyses of TET dioxygenase. Open Biol. 2014;4. 10.1098/rsob.140110. [DOI] [PMC free article] [PubMed]
- 283.Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, et al. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161:893–906. 10.1016/j.cell.2015.04.018. [DOI] [PubMed]
- 284.Li-Byarlay H. The function of DNA methylation marks in social insects. Front Ecol Evol. 2016;4. 10.3389/fevo.2016.00057.
- 285.Chamorro ML, de Medeiros BAS, Farrell BD. First phylogenetic analysis of Dryophthorinae (Coleoptera, Curculionidae) based on structural alignment of ribosomal DNA reveals Cenozoic diversification. Ecol Evol. 2021;11:1984–1998. doi: 10.1002/ece3.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 287.Peona V, Blom MPK, Xu L, Burri R, Sullivan S, Bunikis I, et al. Identifying the causes and consequences of assembly gaps using a multiplatform genome assembly of a bird-of-paradise. Mol Ecol Resour. 2021;21(1):263–286. doi: 10.1111/1755-0998.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Di Genova A, Buena-Atienza E, Ossowski S, Sagot M-F. Efficient hybrid de novo assembly of human genomes with WENGAN. Nat Biotechnol. 2020:1–9. 10.1038/s41587-020-00747-w. [DOI] [PMC free article] [PubMed]
- 289.Platt RN, II, Blanco-Berdugo L, Ray DA. Accurate transposable element annotation is vital when analyzing new genome assemblies. Genome Biol Evol. 2016;8:403–410. doi: 10.1093/gbe/evw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Maire J, Parisot N, Galvao Ferrarini M, Vallier A, Gillet B, Hughes S, et al. Spatial and morphological reorganization of endosymbiosis during metamorphosis accommodates adult metabolic requirements in a weevil. Proc Natl Acad Sci U S A. 2020;117:19347–19358. doi: 10.1073/pnas.2007151117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Nardon P. Obtention d’une souche asymbiotique chez le charançon Sitophilus sasakii Tak: différentes méthodes d’obtention et comparaison avec la souche symbiotique d’origine. CR Acad Sci Paris D. 1973;277:981–984. [Google Scholar]
- 292.Li H. BFC: correcting Illumina sequencing errors. Bioinformatics. 2015;31:2885–2887. doi: 10.1093/bioinformatics/btv290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Salmela L, Rivals E. LoRDEC: accurate and efficient long read error correction. Bioinformatics. 2014;30:3506–3514. doi: 10.1093/bioinformatics/btu538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Chikhi R, Rizk G. Space-efficient and exact de Bruijn graph representation based on a bloom filter. Algorithms Mol Biol. 2013;8(1):22. doi: 10.1186/1748-7188-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Di Genova A, Ruz GA, Sagot M-F, Maass A. Fast-SG: an alignment-free algorithm for hybrid assembly. GigaScience. 2018;7. 10.1093/gigascience/giy048. [DOI] [PMC free article] [PubMed]
- 298.Mandric I, Zelikovsky A. ScaffMatch: scaffolding algorithm based on maximum weight matching. Bioinformatics. 2015;31:2632–2638. doi: 10.1093/bioinformatics/btv211. [DOI] [PubMed] [Google Scholar]
- 299.Xu G-C, Xu T-J, Zhu R, Zhang Y, Li S-Q, Wang H-W, et al. LR_Gapcloser: a tiling path-based gap closer that uses long reads to complete genome assembly. GigaScience. 2019;8. 10.1093/gigascience/giy157. [DOI] [PMC free article] [PubMed]
- 300.Song L, Shankar DS, Florea L. Rascaf: improving genome assembly with RNA sequencing data. Plant Genome. 2016;9:1–12. doi: 10.3835/plantgenome2016.03.0027. [DOI] [PubMed] [Google Scholar]
- 301.Paulino D, Warren RL, Vandervalk BP, Raymond A, Jackman SD, Birol I. Sealer: a scalable gap-closing application for finishing draft genomes. BMC Bioinformatics. 2015;16:230. doi: 10.1186/s12859-015-0663-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Roach MJ, Schmidt SA, Borneman AR. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 2018;19:460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Guan D, McCarthy SA, Wood J, Howe K, Wang Y, Durbin R. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics. 2020;36:2896–2898. doi: 10.1093/bioinformatics/btaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics. 2018;34:i142–i150. doi: 10.1093/bioinformatics/bty266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kokot M, Długosz M, Deorowicz S. KMC 3: counting and manipulating k-mer statistics. Bioinformatics. 2017;33:2759–2761. doi: 10.1093/bioinformatics/btx304. [DOI] [PubMed] [Google Scholar]
- 306.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Smit AF, Hubley R, Green P. RepeatMasker Open-4.0. 2013. http://repeatmasker.org/faq.html#faq3.
- 308.Crescente JM, Zavallo D, Helguera M, Vanzetti LS. MITE tracker: an accurate approach to identify miniature inverted-repeat transposable elements in large genomes. BMC Bioinformatics. 2018;19:348. doi: 10.1186/s12859-018-2376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Novák P, Ávila Robledillo L, Koblížková A, Vrbová I, Neumann P, Macas J. TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017;45:e111. doi: 10.1093/nar/gkx257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Storer J, Hubley R, Rosen J, Wheeler TJ, Smit AF. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob DNA. 2021;12:2. doi: 10.1186/s13100-020-00230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Storer JM, Hubley R, Rosen J, Smit AFA. Curation guidelines for de novo generated transposable element families. Curr Protoc. 2021;1:e154. doi: 10.1002/cpz1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Kapusta A, Suh A. Evolution of bird genomes—a transposon’s-eye view. Ann N Y Acad Sci. 2017;1389:164–185. doi: 10.1111/nyas.13295. [DOI] [PubMed] [Google Scholar]
- 316.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 319.Lerat E, Fablet M, Modolo L, Lopez-Maestre H, Vieira C. TEtools facilitates big data expression analysis of transposable elements and reveals an antagonism between their activity and that of piRNA genes. Nucleic Acids Res. 2017;45:e17. doi: 10.1093/nar/gkw953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Sitophilus oryzae breed Bouriz, whole genome shotgun sequencing project. 2019. https://www.ncbi.nlm.nih.gov/bioproject/566109.
- 322.Low coverage genome sequencing of maize weevil Sitophilus zeamais to analyse the repeatome. 2021. https://www.ncbi.nlm.nih.gov/bioproject/647530.
- 323.Low coverage genome sequencing of granary weevil Sitophilus granarius. 2021. https://www.ncbi.nlm.nih.gov/bioproject/647520.
- 324.Low coverage genome sequencing of the tamarind weevil Sitophilus linearis. 2021. https://www.ncbi.nlm.nih.gov/bioproject/647347.
- 325.Rebollo R, Goubert C. Transposable element annotation of Sitophilus oryzae. 2021. [Google Scholar]
- 326.RNAseq of midgut and ovaries of Day 10 Sitophilus oryzae females. 2021. https://www.ncbi.nlm.nih.gov/bioproject/746240.
- 327.DNA methylation in Sitophilus oryzae ovaries. 2021. https://www.ncbi.nlm.nih.gov/bioproject/681724.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Summary of sequencing libraries produced for S. oryzae.
Additional file 2:. Supplementary notes, supplementary figures, and small tables.
Additional file 3:. Large supporting tables and datasets.
Additional file 4:. Transposable elements annotation tables.
Additional file 5:. STAR and TEtools mapping statistics.
Data Availability Statement
Data generated and analyzed during this study are included in the published article, its additional files and publicly available repositories. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession PPTJ00000000 [321]. The version described in this paper is version PPTJ02000000. The assembly can be visualized, along with gene models and supporting data, on a dedicated genome browser (https://bipaa.genouest.org/sp/sitophilus_oryzae/). Raw reads from low-coverage genome sequencing of S. zeamais, S. granarius, and S. linearis have been deposited at NCBI Sequence Read Archive (SRA) under the BioProject accessions PRJNA647530 [322], PRJNA647520 [323], and PRJNA647347 [324] respectively. TE annotation (GFF) and consensus sequences can be found at 10.5281/zenodo.4570415 [325]. RNAseq reads obtained in this manuscript were deposited under the BioProject accession PRJNA746240 [326]. Bisulfite-seq reads have been deposited at NCBI SRA, under the BioProject accession PRJNA681724 [327].