Version Changes
Revised. Amendments from Version 1
- Clarified design and rationale for treatment groups used in the metabolomics experiment - Included description of methods, results and conclusions for a new experiment testing for an influence of blood glucose rhythms on the IDC - Includes new figure and table - Clarified rationale for in vitro approach to following up the metabolomics screen - Increased depth of discussion on how glucose and isoleucine might interact - Updated all figure and table numbers following the addition of the new experiment - Improved clarity of Figures 3, 4, 8 (now 9) and 9 (now 10) - Revised Figure 5 (now 6) to include negative control - Included references to newly cited literature
Abstract
Background: Rapid asexual replication of blood stage malaria parasites is responsible for the severity of disease symptoms and fuels the production of transmission forms. Here, we demonstrate that a Plasmodium chabaudi’s schedule for asexual replication can be orchestrated by isoleucine, a metabolite provided to the parasite in a periodic manner due to the host’s rhythmic intake of food.
Methods: We infect female C57BL/6 and Per1/2-null mice which have a disrupted canonical (transcription translation feedback loop, TTFL) clock with 1×10 5 red blood cells containing P. chabaudi (DK genotype). We perturb the timing of rhythms in asexual replication and host feeding-fasting cycles to identify nutrients with rhythms that match all combinations of host and parasite rhythms. We then test whether perturbing the availability of the best candidate nutrient in vitro changes the schedule for asexual development.
Results: Our large-scale metabolomics experiment and follow up experiments reveal that only one metabolite - the amino acid isoleucine – fits criteria for a time-of-day cue used by parasites to set the schedule for replication. The response to isoleucine is a parasite strategy rather than solely the consequences of a constraint imposed by host rhythms, because unlike when parasites are deprived of other essential nutrients, they suffer no apparent costs from isoleucine withdrawal.
Conclusions: Overall, our data suggest parasites can use the daily rhythmicity of blood-isoleucine concentration to synchronise asexual development with the availability of isoleucine, and potentially other resources, that arrive in the blood in a periodic manner due to the host’s daily feeding-fasting cycle. Identifying both how and why parasites keep time opens avenues for interventions; interfering with the parasite’s time-keeping mechanism may stall replication, increasing the efficacy of drugs and immune responses, and could also prevent parasites from entering dormancy to tolerate drugs.
Keywords: Plasmodium, periodicity, circadian rhythm, metabolism, metabolomics, intraerythrocytic development cycle, isoleucine, asexual replication
Introduction
Circadian rhythms are assumed to have evolved to coordinate organisms’ activities with daily rhythms in the environment ( Hozer et al., 2020; Ouyang et al., 1998; Spoelstra et al., 2016). The value of organising sleep/wake cycles according to whether it is light or dark, whether predators or prey are active, etc., is clear, but how parasites cope with rhythmicity in the within-host environment has been neglected ( Martinez-Bakker & Helm, 2015; Reece et al., 2017; Westwood et al., 2019). Many of the processes underlying interactions between hosts and parasites have a circadian basis, yet why these rhythms exist and what their consequences are for hosts and parasites remain mysterious. For example, parasites are confronted with rhythms in host behaviours and physiologies, including immune responses and metabolism. Thus, some host rhythms offer opportunities for parasites to exploit, whilst other rhythms impose constraints parasites must cope with. Some parasites use their own circadian clocks to control metabolism ( Rijo-Ferreira et al., 2017), and virulence ( Hevia et al., 2015), suggesting that host rhythms are a selective (evolutionary) driver of parasite rhythms. Host rhythms have fitness consequences for malaria ( Plasmodium) parasites ( O’Donnell et al., 2011; O’Donnell et al., 2013), whose rhythmicity in development during blood-stage replication is aligned with the timing of host feeding-fasting cycles ( Hirako et al., 2018; O’Donnell et al., 2020; Prior et al., 2018).
Explaining the timing and synchrony exhibited by malaria parasites during successive cycles of blood-stage replication has been a puzzle since the Hippocratic Era, when the duration of these cycles (24-, 48, or 72-hours, depending on the Plasmodium spp.) were used to diagnose malaria infection ( Garcia et al., 2001; Mideo et al., 2013). Blood-stage asexual replication is orchestrated by the intraerythrocytic development cycle (IDC), which is characterised by parasite invasion of red blood cells (RBC), growth, division, and finally bursting of RBC to release the next cohort of asexually replicating parasites. Given that asexual replication is responsible for the disease symptoms of malaria and fuels the production of transmission forms, explaining how and why the vast majority of Plasmodium species progress though the IDC in synchrony, and why transitions between these stages occur at particular times of day, may unlock novel interventions and improve the efficacy of existing treatments. However, such endeavours rely on identifying the precise host rhythm(s) that parasites align to and explaining why this matters for their fitness ( Prior et al., 2020).
Malaria parasites are able to keep time ( Rijo-Ferreira et al., 2020; Subudhi et al., 2020) and organise their IDC schedule to coordinate with an aspect(s) of host feeding-fasting rhythms ( Hirako et al., 2018; Prior et al., 2018), but not the act of eating itself ( Rijo-Ferreira et al., 2020), or processes directly controlled by canonical host circadian clocks ( O’Donnell et al., 2020). Coordination with host feeding-fasting rhythms may allow parasites to couple the nutritional needs of each IDC stage with circadian fluctuations in the concentrations of nutrients in the blood ( Prior et al., 2020). Whilst parasites are able to scavenge most amino acids from haemoglobin digestion, and can biosynthesise some metabolites themselves, other nutrients are essential and must be taken up from the RBCs or blood plasma. Haemoglobin digestion and biosynthesis can occur at any time-of-day but essential nutrients that both host and parasite must acquire from the host’s food (including glucose and the amino acid, isoleucine) follow circadian rhythms in the blood ( Skene et al., 2018). Hosts forage during their active phase and fast during the rest phase, which generally generates peaks in the blood concentrations of metabolites shortly after the onset of feeding and their lowest point (nadir) occurs during the fasting phase.
Nutritional needs vary across IDC stages ( Figure 1A), with for example, requirements for glycerophospholipids and amino acids to fuel biogenesis, increasing as parasites progress through the IDC, and the release of parasite progeny at the end of the IDC consumes a lot of glucose ( Déchamps et al., 2010; Olszewski et al., 2009). Whether essential nutrients are available around-the-clock at sufficient concentrations to satisfy the needs of all parasite cells within an infection is unknown, but in culture, IDC progression is affected by nutrient limitation. For example, parasites experiencing glucose limitation quickly mount starvation responses ( Daily et al., 2007) and isoleucine starvation rapidly induces dormancy ( Babbitt et al., 2012). Moreover, the nadir of blood glucose rhythms in the rest phase can be exacerbated by malaria infection ( Hirako et al., 2018). Thus, to avoid the problems of temporal resource limitation, we hypothesise that parasites match transitions between IDC stages to rhythmicity in the availability of essential nutrients in the blood, ensuring the most demanding IDC stages (trophozoites and schizonts) coincide with when resources are most abundant. Parasites could synchronise with resource rhythms by using an essential nutrient itself as a time-cue to set the schedule for IDC transitions, and/or allow the host to impose the IDC schedule by, for example, selectively killing IDC stages that are not “on time”. By getting the timing of the IDC schedule right, parasites garner fitness in two ways. They maximise asexual replication, which underpins within-host survival ( O’Donnell et al., 2011; O’Donnell et al., 2013), and benefit from coordinating the production of sexual transmission stages with the time-of-day their vectors forage for blood ( Pigeault et al., 2018; Schneider et al., 2018).
Figure 1. Rhythms in the intraerythrocytic development cycle (IDC) and host rhythms.
( A) The role of each IDC stage during cycles of asexual replication, and the resources known to be essential to each stage. RG=ring stage, ET=early stage trophozoite, MT=mid stage trophozoite, LT=late stage trophozoite, SZ=schizont. ( B), The experimental design used wild type mice housed in a 12h light: 12h dark regime (indicated by the light-dark bar) with unrestricted access to food for 12 hours either during the night-times (DF, dark feeding) or the day times (LF, light feeding) as indicated by the position of the cheeses. Per1/2-null mice were housed in continuous darkness (DD) and either experienced cycles of 12-hours with food followed by 12 h without access to food (TRF, time restricted feeding,) or given constant access to food (ALF, ad libitum feeding). The parasite IDC is rhythmic in DF, LF and TRF mice but not ALF mice ( O’Donnell et al., 2020) causing the IDC rhythm to be substantially dampened. ( C) Metabolites that were significantly rhythmic in DF, LF and TRF mice (highlighted in red), but not in ALF mice, were sought. Treatment groups colour coded throughout: DF=orange, LF=light blue, TRF=green, ALF=dark blue.
We integrate evolutionary ecology and parasitology with chronobiology to, first, undertake a hypothesis-driven screen of several metabolite classes and glucose to identify nutrients with daily fluctuations in the blood of malaria-infected hosts. Second, we determine which metabolites follow rhythms set by the timing of different perturbations of host feeding-fasting rhythms as well as matching the timing of the IDC schedules in all treatment groups. We find that the IDC schedule cannot be explained by fluctuations in blood glucose concentrations and almost all metabolites in the screen, but the experiment does reveal a single candidate; the amino acid, isoleucine. Third, we test if the timing of isoleucine provision and withdrawal affects IDC progression in the manner expected if parasites use it as a time-of-day cue to schedule their IDC or in the manner expected if the IDC schedule is imposed by starvation of mis-timed IDC stages. We capitalise on the rodent malaria P. chabaudi model system in which in vivo experiments exploit ecologically relevant host-parasite interactions and short-term in vitro studies allow within-host ecology to be probed in-depth.
Results
Metabolites that associate with host feeding rhythms and the IDC schedule
To identify metabolites whose rhythms correspond to the timing of host feeding and the IDC schedule, we compared four groups of malaria infections in mice that were either wild type (WT) C57BL/6J strain or Per1/2-null (Period 1 and Period 2) circadian clock-disrupted mice. Per1/2-null mice have an impaired canonical clock (transcription translation feedback loop, TTFL) and exhibit no circadian rhythms in physiology or behaviour when kept in constant darkness ( Bae et al., 2001; Maywood et al., 2014; O’Donnell et al., 2020). We generated three different groups of hosts whose feeding-fasting rhythms differed with respect to the light:dark cycle and whether they had an intact TTFL clock, and a 4 th group of hosts which lacked both feeding rhythms and an intact TTFL clock ( Figure 1B). Specifically, we housed 2 groups of wild type mice in light-dark cycles to entrain their SCN-driven rhythms, and applied time-restricted (TRF) feeding protocols such that one group had access to food throughout the 12 hours of dark (dark fed, DF) or light (light fed, LF), every circadian cycle. Using TRF for both groups of wild types avoided any potentially confounding effects of elevated food intake on metabolite concentrations that could occur if the DF mice were instead allowed continuous access to food. Both groups of Per1/2-null mice were housed in constant darkness and one group experienced TRF such that food was available for the same daily window as the LF mice, and the other group had continuous access to food (ad lib fed, ALF). We chose these combinations because they span the manner that feeding-fasting rhythms, but not photoperiod, impact the IDC schedule, observed by us and others ( Hirako et al., 2018; O’Donnell et al., 2020; Prior et al., 2018; Rijo-Ferreira et al., 2020).
All infections were sampled every two hours from day 5 post infection (when infections are at a low parasitaemia, ~10%, to minimise the contribution of parasite molecules to the dataset, Olszewski et al., 2009) for 26 hours. We hypothesised that a time-cue/time-dependent resource must vary or have rhythmicity across 24 hours, with a peak concentration that associates with the timing of host feeding-fasting across the three treatment groups with rhythmic feeding, yet also be arrhythmic in the 4 th group ( Figure 1B). In addition, the parasite IDC must be rhythmic in the three treatment groups with rhythmic feeding, with the same IDC stage present at the time of feeding, and be arrhythmic in the 4 th group. Thus, we identified candidate metabolites by intersecting rhythmic metabolites in the treatment groups ( Figure 1C).
IDC schedules followed the expected patterns for each treatment group ( Figure 2, and O’Donnell et al., 2020). Specifically, parasites displayed opposite IDC schedules in dark (i.e. night, DF, n=18) and light (i.e. day, LF, n=17) fed WT mice because their feeding-fasting rhythms are inverted ( Figure 2, Table 1). Parasites in Per1/2-null mice (kept in constant darkness), fed during a time-restricted 12 h window (TRF mice, n=17; Table 2) that coincided with when LF mice were fed, followed the same IDC schedule as parasites in LF mice. Finally, parasites in Per1/2-null mice (kept in constant darkness) with constant access to food (ALF mice, n=16; Table 2) exhibited dampened IDC rhythms because their hosts have no feeding-fasting rhythm ( O’Donnell et al., 2020). Across the entire data set, 110 metabolites exhibited variation in their blood concentrations during the 26-hour sampling window (101 in DF, 91 in LF, 50 in TRF and one in ALF; Table 3). That only one metabolite (lysoPC a C20:3) exhibited a rhythm in ALF hosts demonstrates that host TTFL clocks and timed feeding are required to generate metabolite rhythms ( Figure 3A, Reinke & Asher, 2019). Further, that approximately half of the metabolites rhythmic in hosts with TTFL clocks (DF and LF) were also rhythmic in TRF hosts reveals that feeding rhythms alone can maintain metabolic rhythms during infection ( Figure 3A). Of all metabolites, only 42 were rhythmic in all three groups of infections with both feeding-fasting and IDC rhythms (i.e. the red area in Figure 1C), consisting of three acylcarnitines, 11 amino acids, nine biogenic amines and 19 glycerophospholipids ( Figure 3A). Next, we identified 33 metabolites that exhibited a peak concentration (timing or phase) that corresponded to the timing of the host’s feeding-fasting rhythm and the parasite’s IDC ( Figure 3B; metabolites clustering in the light blue and purple areas).
Figure 2. Intraerythrocytic development cycle (IDC) schedule and host feeding rhythms.
The proportion of parasites at ring stage (phase marker) for: ( A) DF: dark feeding WT mice food access ZT 12-24 (12 h at night) in 12h:12h light:dark, ( B) LF: light feeding WT mice food access ZT 0-12 (12 h in day) in 12 h:12 h light:dark. ( C) TRF: time restricted feeding Per1/2-null mice food access 0–12 hours (12 h at the same time (GMT) as LF mice) in constant darkness (DD). ( D) ALF: ad libitum feeding Per1/2-null mice access in constant darkness (see Figure 1 for experimental design and more details). Feeding-fasting rhythms are indicated by cheeses, the white panel denotes lights on (Zeitgeber Time=0–12 h), dark grey panel denotes lights off (Zeitgeber Time=12–24 h for DF and LF, 0–24 h for TRF and ALF). Model fits for each group are plotted on the raw data (n= 2–5 infections per time point). The fitted sine/cosine curve for DF ( A) is distorted due to a large amplitude which exceeds the bounds possible for proportion data (between 0 and 1) so is truncated for visualisation. The patterns for groups are explained better by sine/cosine curves than by a straight line (see Table 1).
Figure 3. Rhythmic metabolites that associate with intraerythrocytic development cycle (IDC) timing and host feeding-fasting cycles.
( A) The numbers of rhythmic intersecting metabolites out of a total of 109. ( B) Peak timing (phase) concentration of the 42 metabolites that are rhythmic in DF, LF and TRF infections. Top left panel (light blue) denotes peaks during the fasting phase in all groups and the bottom right panel (purple) denotes peaks in the feeding phase for all groups, yielding 33 candidates linked to the feeding-fasting cycle. The top right and bottom left regions contain metabolites that are not in phase with feeding-fasting rhythms in all groups. Metabolite classes: acylcarnitines=green circles, amino acids/biogenic amines=orange triangles, glycerophospholipids=purple squares. See Table 7 for peak times of each metabolite. Ring stages (RG), late trophozoites (LT) and schizonts (SZ) peak during the hosts feeding period (purple) when some amino acids/biogenic amines peak. Early trophozoites (ET) and mid trophozoites (MT) peak during host fasting (light blue) when glycerophospholipids and acylcarnitines peak.
Table 1. Degrees of Freedom (df), log-Likelihood (logLik), AICc, ΔAICc (AICc i ΔAICc min) and AICc w (AICc weight) for each linear model in the parasite stage proportion analysis ordered in descending fit (best-fitting model at the top).
The response variable for each model is proportion of ring stages (Ring.prop), with “sine” and “cosine” terms being the sine or cosine function of (2π × time of day)/24 with a fixed 24-hour period fitted for each treatment group (DF, LF, TRF or ALF). AICc is a form of the Akaike Information Criteria corrected for smaller sample sizes to address potential overfitting, used for model selection. DF=dark fed wild type mice, LF=light fed wild type mice, TRF=time restricted fed Per1/2-null mice, ALF=ad libitum fed Per1/2-null mice.
| Model description:
Ring.prop ~ |
df | logLik | AICc | ΔAICc | AICc w | |
|---|---|---|---|---|---|---|
| DF | sine + cosine | 4 | 13.30 | -16.9 | 0.00 | 1.000 |
| sine | 3 | -5.77 | 18.5 | 35.44 | 0.000 | |
| cosine | 3 | -8.69 | 24.3 | 41.27 | 0.000 | |
| null | 2 | -14.60 | 33.7 | 50.60 | 0.000 | |
| LF | sine + cosine | 4 | 8.57 | -7.5 | 0.00 | 0.788 |
| sine | 3 | 5.92 | -4.9 | 2.63 | 0.212 | |
| cosine | 3 | -1.03 | 9.0 | 16.52 | 0.000 | |
| null | 2 | -2.68 | 9.8 | 17.34 | 0.000 | |
| TRF | sine + cosine | 4 | 21.85 | -33.5 | 0.00 | 1.000 |
| sine | 3 | 10.98 | -14.7 | 18.77 | 0.000 | |
| cosine | 3 | -0.14 | 7.5 | 41.01 | 0.000 | |
| null | 2 | -2.54 | 9.7 | 43.15 | 0.000 | |
| ALF | sine + cosine | 4 | 32.12 | -53.6 | 0.00 | 0.995 |
| sine | 3 | 24.51 | -41.5 | 12.05 | 0.002 | |
| cosine | 3 | 24.49 | -41.5 | 12.08 | 0.002 | |
| null | 2 | 19.90 | -35.1 | 18.47 | 0.000 |
Table 2. Mouse numbers for experimental treatment groups.
A) Mice in the metabolomics experiment were sampled in blocks (A–D – see methods) with 4/5 mice per block sampled every eight hours. Totals for each treatment group: DF=18, LF=17, TRF=17, ALF=16. B) Mice in the glucose experiment were sampled every two hours. Totals for each treatment group: DF=5, LF=5, TRF=5, ALF=5. For each experiment repeated measures from mice were controlled for during the analysis. DF=dark fed wild type mice, LF=light fed wild type mice, TRF=time restricted fed Per1/2-null mice, ALF=ad libitum fed Per1/2-null mice.
| Time (ZT/hour) | 0 | 2 | 4 | 6 | 8 | 10 | 12 | 14 | 16 | 18 | 20 | 22 | 24/0 | 2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A Metabolomics experiment | ||||||||||||||
| Block | A | B | C | D | A | B | C | D | A | B | C | D | A | B |
| DF | 5 | 5 | 4 | 4 | 5 | 5 | 4 | 4 | 5 | 5 | 4 | 4 | 5 | 5 |
| LF | 5 | 4 | 4 | 4 | 5 | 4 | 4 | 4 | 5 | 4 | 4 | 4 | 5 | 4 |
| TRF | 5 | 4 | 4 | 4 | 5 | 4 | 4 | 4 | 5 | 4 | 4 | 4 | 5 | 4 |
| ALF | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| B Glucose experiment | ||||||||||||||
| DF | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| LF | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| TRF | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| ALF | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
Table 3. Metabolite numbers that significantly fluctuate every 24 hours in the mouse blood for three methods.
A) Data for all metabolites were run through three circadian programmes to find those following a 24h rhythm using ECHO (Benjamini-Hochberg (BH) adjusted p value of 0.05), CircWave (standard p value of 0.05) and JTK_Cycle (BH adjusted p value of 0.05). Metabolites that are excluded using the Surrey LC/MS assay criteria were removed from the analysis ( Figure 10). B) Rhythmic metabolites in each programme (ECHO, CircWave and JTK) were intersected, with a metabolite counted as rhythmic if it is significantly rhythmic in at least two programmes (ECHO=CW, ECHO=JTK, CW=JTK). C) The metabolites not fulfilling these criteria were analysed using ANOVA (including time-of-day as a factor), with BH adjusted p values at the 5% level. Metabolites from both methods (circadian programmes and ANOVA) were then combined to perform a final intersection between DF, LF and TRF to find common metabolites. Metabolites rhythmic in ALF mice were then removed. DF=dark fed wild type mice, LF=light fed wild type mice, TRF=time restricted fed Per1/2-null mice, ALF=ad libitum fed Per1/2-null mice.
| A | ECHO | CircWave (CW) | JTK | |||
|---|---|---|---|---|---|---|
| Rhythmic with
24h period |
Not-rhythmic with
24h period |
Rhythmic
with 24h period |
Not-rhythmic
with 24h period |
Rhythmic
with 24h period |
Not-rhythmic
with 24h period |
|
| DF | 75 | 59 | 67 | 67 | 47 | 87 |
| LF | 106 | 28 | 83 | 51 | 70 | 64 |
| TRF | 55 | 77 | 41 | 93 | 23 | 111 |
| ALF | 2 | 127 | 6 | 128 | 0 | 134 |
| B | ECHO==CW | ECHO==JTK | CW==JTK | Unique | ||
| DF | 62 | 47 | 46 | 63 | ||
| LF | 79 | 68 | 63 | 88 | ||
| TRF | 37 | 23 | 21 | 39 | ||
| ALF | 1 | 0 | 0 | 1 | ||
| C | Not-rhythmic
with 24h period |
Time-of-day effect | No time-of-
day effect |
Time-of-day
effect + rhythmic with 24h period |
Total candidates | |
| DF | 71 | 38 | 33 | 101 | 42 | |
| LF | 46 | 3 | 43 | 91 | ||
| TRF | 95 | 11 | 84 | 50 | ||
| ALF | 133 | 0 | 133 | 1 | ||
Figure 10. Median concentration of all metabolites across the time series for all groups, those included and excluded from the analysis.
The median concentration (µM) of each metabolite for each of the 4 treatment groups. In black are metabolites excluded since they failed set LC/MS assay criteria and those metabolites taken forward into the analysis are red. The numbers on the x axis correspond to the metabolites as follows: (1)Ala, (2)Arg, (3)Asn, (4)Asp, (5)Cit, (6)Gln, (7)Glu, (8)Gly, (9)His, (10)Ile, (11)Leu, (12)Lys, (13)Met, (14)Orn, (15)Phe, (16)Pro, (17)Ser, (18)Thr, (19)Trp, (20)Tyr, (21)Val, (22)Ac.Orn, (23)ADMA, (24)alpha.AAA, (25)c4.OH.Pro, (26)Carnosine, (27)Creatinine, (28)DOPA, (29)Dopamine, (30)Histamine, (31)Kynurenine, (32)Met.SO, (33)Nitro.Tyr, (34)PEA, (35)Putrescine, (36)Sarcosine, (37)SDMA, (38)Serotonin, (39)Spermidine, (40)Spermine, (41)t4.OH.Pro, (42)Taurine, (43)C0, (44)C2, (45)C3, (46)C3.DC..C4.OH., (47)C3.OH, (48)C3.1, (49)C4, (50)C4.1, (51)C5, (52)C5.DC..C6.OH., (53)C5.M.DC, (54)C5.OH..C3.DC.M., (55)C5.1, (56)C5.1.DC, (57)C6..C4.1.DC., (58)C6.1, (59)C7.DC, (60)C8, (61)C9, (62)C10, (63)C10.1, (64)C10.2, (65)C12, (66)C12.DC, (67)C12.1, (68)C14, (69)C14.1, (70)C14.1.OH, (71)C14.2, (72)C14.2.OH, (73)C16, (74)C16.OH, (75)C16.1, (76)C16.1.OH, (77)C16.2, (78)C16.2.OH, (79)C18, (80)C18.1, (81)C18.1.OH, (82)C18.2, (83)lysoPC.a.C14.0, (84)lysoPC.a.C16.0, (85)lysoPC.a.C16.1, (86)lysoPC.a.C17.0, (87)lysoPC.a.C18.0, (88)lysoPC.a.C18.1, (89)lysoPC.a.C18.2, (90)lysoPC.a.C20.3, (91)lysoPC.a.C20.4, (92)lysoPC.a.C24.0, (93)lysoPC.a.C26.0, (94)lysoPC.a.C26.1, (95)lysoPC.a.C28.0, (96)lysoPC.a.C28.1, (97)PC.aa.C24.0, (98)PC.aa.C26.0, (99)PC.aa.C28.1, (100)PC.aa.C30.0, (101)PC.aa.C32.0, (102)PC.aa.C32.1, (103)PC.aa.C32.2, (104)PC.aa.C32.3, (105)PC.aa.C34.1, (106)PC.aa.C34.2, (107)PC.aa.C34.3, (108)PC.aa.C34.4, (109)PC.aa.C36.0, (110)PC.aa.C36.1, (111)PC.aa.C36.2, (112)PC.aa.C36.3, (113)PC.aa.C36.4, (114)PC.aa.C36.5, (115)PC.aa.C36.6, (116)PC.aa.C38.0, (117)PC.aa.C38.3, (118)PC.aa.C38.4, (119)PC.aa.C38.5, (120)PC.aa.C38.6, (121)PC.aa.C40.1, (122)PC.aa.C40.2, (123)PC.aa.C40.3, (124)PC.aa.C40.4, (125)PC.aa.C40.5, (126)PC.aa.C40.6, (127)PC.aa.C42.0, (128)PC.aa.C42.1, (129)PC.aa.C42.2, (130)PC.aa.C42.4, (131)PC.aa.C42.5, (132)PC.aa.C42.6, (133)PC.ae.C30.0, (134)PC.ae.C30.1, (135)PC.ae.C30.2, (136)PC.ae.C32.1, (137)PC.ae.C32.2, (138)PC.ae.C34.0, (139)PC.ae.C34.1, (140)PC.ae.C34.2, (141)PC.ae.C34.3, (142)PC.ae.C36.0, (143)PC.ae.C36.1, (144)PC.ae.C36.2, (145)PC.ae.C36.3, (146)PC.ae.C36.4, (147)PC.ae.C36.5, (148)PC.ae.C38.0, (149)PC.ae.C38.1, (150)PC.ae.C38.2, (151)PC.ae.C38.3, (152)PC.ae.C38.4, (153)PC.ae.C38.5, (154)PC.ae.C38.6, (155)PC.ae.C40.1, (156)PC.ae.C40.2, (157)PC.ae.C40.3, (158)PC.ae.C40.4, (159)PC.ae.C40.5, (160)PC.ae.C40.6, (161)PC.ae.C42.0, (162)PC.ae.C42.1, (163)PC.ae.C42.2, (164)PC.ae.C42.3, (165)PC.ae.C42.4, (166)PC.ae.C42.5, (167)PC.ae.C44.3, (168)PC.ae.C44.4, (169)PC.ae.C44.5, (170)PC.ae.C44.6, (171)SM..OH..C14.1, (172)SM..OH..C16.1, (173)SM..OH..C22.1, (174)SM..OH..C22.2, (175)SM..OH..C24.1, (176)SM.C16.0, (177)SM.C16.1, (178)SM.C18.0, (179)SM.C18.1, (180)SM.C20.2, (181)SM.C24.0, (182)SM.C24.1, (183)SM.C26.0, (184)SM.C26.1.
Glucose does not associate with the IDC schedule
Previous work suggests that blood glucose rhythms are responsible for the IDC schedule ( Hirako et al., 2018; Prior et al., 2018) but glucose could not be measured by the UPLC/MS-MS metabolomics platform. Therefore, we set up an additional set of DF, LF, TRF and ALF infections (n=5/group), which followed the same feeding-fasting and IDC schedules as infections in the metabolomics screen, Figure 2, Table 2), to quantify blood-glucose rhythms. Mean blood-glucose concentration across the time series differed between the groups, being higher in DF and TRF mice (DF=8.55±0.14 mmol/L, TRF=8.59±0.13 mmol/L) than in LF and ALF mice (LF=7.90±0.14 mmol/L, ALF=7.68±0.11 mmol/L). We found that two competitive models, including only time-of-day (Zeitgeber time, ZT/h) or both time-of-day and treatment (DF, LF, TRF and ALF) as main effects, can explain patterns of glucose concentration ( Figure 4, Table 4). Specifically, glucose concentration varied throughout the day in DF mice but much less in LF mice, and glucose was invariant in both groups of Per1/2-null mice (TRF and ALF) ( Table 4). The rhythmic IDC schedule in the DF, LF and TRF groups but the lack of significant rhythmicity in blood glucose in the TRF (and possibly LF) infections indicates that glucose does not explain the connection between feeding rhythms and the IDC schedule across all groups.
Figure 4. Blood glucose concentrations (mmol/L).
( A) DF: dark feeding WT mice food access ZT 12-24 (12 h at night) in 12h:12h light:dark, ( B) LF: light feeding WT mice food access ZT 0-12 (12 h in day) in 12 h:12 h light:dark. ( C) TRF: time restricted feeding Per1/2-null mice food access 0–12 hours (12 h at the same time (GMT) as LF mice) in constant darkness (DD). ( D) ALF: ad libitum feeding Per1/2-null mice access in constant darkness (see Figure 1 for experimental design). Feeding-fasting rhythms are indicated by cheeses, the white panel denotes lights on (Zeitgeber Time=0–12 h), dark grey panel denotes lights off (Zeitgeber Time=12–24 h for DF and LF, 0–24 h for TRF and ALF). The lines and shading are mean ± SEM at each time point and the dots are the raw data. At time points 0.5 and 2.5 for DF, TRF and ALF, and 12.5 and 14.5 for LF the points are stacked because the time course lasted ~26 hours and the data are plotted on a 0–24 hour axis.
Table 4. Degrees of Freedom (df), log-Likelihood (logLik), AICc, ΔAICc (AICc i – AICc min) and AICc w (AICc weight) for each linear model in the glucose concentration analysis ordered in descending fit (best-fitting model at the top).
The response variable for each model is glucose concentration (Glucose.conc) and the random effect is “mouseID”. “Treatment” refers to the treatment group (DF, LF, TRF or ALF) and “time” refers to the time-of day (ZT 0-24h or 0-24h) which was fitted as a factor. DF=dark fed wild type mice, LF=light fed wild type mice, TRF=time restricted fed Per1/2-null mice, ALF=ad libitum fed Per1/2-null mice.
| Model description:
Glucose.conc ~ + (1|mouseID) |
df | logLik | AICc | ΔAICc | AICc w | |
|---|---|---|---|---|---|---|
| time | 14 | -369.71 | 769.0 | 0.00 | 0.555 | |
| treatment + time | 17 | -366.55 | 769.4 | 0.44 | 0.445 | |
| treatment*time | 50 | -331.38 | 785.1 | 16.12 | 0.000 | |
| null | 3 | -390.80 | 787.7 | 18.68 | 0.000 | |
| treatment | 6 | -387.75 | 787.8 | 18.80 | 0.000 | |
| DF | time | 14 | -90.06 | 215.7 | 0.00 | 0.996 |
| null | 3 | -110.28 | 226.9 | 11.17 | 0.004 | |
| LF | null | 3 | -94.83 | 196.0 | 0.00 | 0.550 |
| time | 14 | -80.39 | 196.4 | 0.40 | 0.450 | |
| TRF | null | 3 | -85.23 | 176.8 | 0.00 | 0.999 |
| time | 14 | -77.54 | 190.7 | 13.9 | 0.001 | |
| ALF | null | 3 | -88.02 | 182.4 | 0.00 | 0.994 |
| time | 14 | -78.43 | 192.6 | 10.21 | 0.006 |
Further, we directly tested whether glucose can schedule the IDC by comparing IDC rhythms in Per1/2-null mice (housed in DD) experiencing daily transient perturbations to blood glucose concentration. At the same time each day, infected mice (n= 5 or 6 / group) experienced one of the following treatments for 6 days: intra-peritoneal injection of glucose alone (to simulate the effects of a large meal) or with insulin (to reduce blood glucose, following Crosby et al., 2019), or PBS carrier only (control). Mice were infected with synchronous ring stages from WT donors, injections began on day 1 post infection and occurred 12 hours out of phase to peak feeding in the donor mice. Thus, the only rhythm parasites were exposed to was a predictable daily rhythm in blood glucose concentration. We reasoned that if parasites responded to a spike or trough in blood glucose, the IDC schedule would diverge from the controls in the glucose and/or the glucose+insulin groups.
After 5 days we quantified blood glucose concentration and the IDC schedule every 4 hours for 24 hours, starting 1 hour post injections. We found glucose concentration increases or decreases, as expected, with the best fitting model including the effect of “time” and “treatment” plus their interaction (ΔAICc=0, Figure 5A, Table 5A). Specifically, blood glucose concentration remained stable in control mice that received daily injections of PBS (8.30±0.17 mmol/L). Whereas, compared to the control group, the glucose only treatment experienced elevated blood glucose (by 3.62 mmol/L) and the insulin + glucose group experienced reduced blood glucose (by 2.42 mmol/L). Perturbations of blood glucose were transient, returning to the levels of control mice 2–4 hours post injection. Despite regularly perturbing glucose levels over a sufficient number of days for the IDC schedule to change, we find no effect on parasite rhythms, with all groups following near identical rhythms ( Figure 5B, Table 5B). The best fitting model included only the effect of “time” (ΔAICc=0); the addition of “treatment” did not improve the model fit (ΔAICc=4.72). Furthermore, parasite or RBC density did not vary significantly between the treatment groups ( Figure 5C and 5D, Table 5C and 5D) suggesting parasite replication activities are impervious to time of day signals provided by glucose.
Figure 5. Predictable daily perturbations of blood glucose concentration do not influence the IDC schedule nor the density of parasites or red blood cells.
For 6 days following infection A) Mice were injected at Time=1h with either glucose (grey, n=5), glucose + insulin (yellow, n=6) or PBS (n=5) and their glucose concentration (mmol/L) measured in the blood every 4 hours for 24 hours over days 5–6 post infection. B) Over the same 24 hour window on days 5–6 the proportion of ring stage parasites was quantified as a marker for the IDC schedule. C) Parasite and D) red blood cell density across the 24 sampling window for each treatment. Means and ±SEM plotted.
Table 5. Degrees of Freedom (df), log-Likelihood (logLik), AICc, ∆AIC (AICci – AICcmin) and AICc w (AICc weight) for each linear model ordered in descending fit (best-fitting model at the top) for glucose perturbations.
The response variable for each model is either proportion of ring stage parasites (Ring.prop), parasite density (Parasite) or RBC density (RBC).
| Model description: | df | logLik | AICc | ∆AIC | AICc w |
|---|---|---|---|---|---|
| A) Glucose.conc ~ + (1|mouseID) | |||||
| hour * treatment | 26 | -184.802 | 435.6 | 0.00 | 1 |
| treatment | 5 | -225.597 | 461.7 | 26.05 | 0 |
| null | 3 | -230.388 | 467.0 | 31.33 | 0 |
| hour + treatment | 12 | -221.788 | 470.3 | 34.67 | 0 |
| hour | 10 | -226.544 | 475.0 | 39.34 | 0 |
| B) Ring prop ~ | |||||
| hour | 9 | 115.438 | -211.4 | 0.00 | 0.911 |
| hour + treatment | 11 | 115.456 | -206.6 | 4.72 | 0.086 |
| hour * treatment | 25 | 131.257 | -199.8 | 11.58 | 0.003 |
| null | 2 | 33.888 | -63.7 | 147.67 | 0.000 |
| treatment | 4 | 33.893 | -59.5 | 151.89 | 0.000 |
| C) Parasite ~ + (1|hour) + (1|mouseID) | |||||
| null | 4 | -7.358 | 23.0 | 0.00 | 0.98 |
| treatment | 6 | -9.053 | 30.8 | 7.76 | 0.02 |
| D) RBC ~ + (1|hour) + (1|mouseID) | |||||
| null | 4 | 224.77 | -441.2 | 0.00 | 0.999 |
| treatment | 6 | 219.69 | -426.7 | 14.54 | 0.001 |
Linking metabolites to the IDC schedule
Having ruled out a role for blood glucose, we explored whether the metabolites from our screen could explain why the IDC schedule follows feeding-fasting rhythms. The metabolites whose peak associated with both feeding and late IDC stages, are alanine, asparagine, isoleucine, leucine, methionine, phenylalanine, proline, threonine, valine, methionine-sulfoxide and serotonin. Whereas 20 acylcarnitines and glycerophospholipids (with the exception of two biogenic amines) associated with fasting and early IDC stages (ADMA, SDMA, C14.1, C16, C18.1, lysoPCaC16:1, lysoPCaC18:1, lysoPCaC18:2, PCaaC32:1, PCaaC34:4, PCaaC38:3, PCaaC38:4, PCaaC38:5, PCaaC38:6, PCaaC40:4, PCaaC40:5, PCaeC34:3, PCaeC38:0, PCaeC38:3 and PCaeC42:1; Table 6 and Table 7). We asked if these metabolites could schedule the IDC through the following non-mutually exclusive mechanisms. First, by being sufficiently limiting at a certain time-of-day to enforce a rhythm on the IDC because mis-timed IDC stages starve and die. This scenario requires that parasites are unable to overcome resource limitation by synthesising the metabolite itself or scavenging it from a source (such as haemoglobin) that is available around the clock. Second, by acting as a time-of-day cue to which certain IDC stages respond by for example, initiating the transition to the next stage (called a “just-in-time” strategy) or by acting as a Zeitgeber to entrain an endogenous circadian clock that allows parasites to anticipate when IDC transitions should occur. For a metabolite to be a reliable time-of-day cue/Zeitgeber, it should be something the parasite cannot synthesise/scavenge to avoid the challenge of differentiating between inaccurate endogenous and accurate exogenous time information leading to the risk of mistakenly responding to an endogenous signal. Of the 33 metabolites on the shortlist, only isoleucine fulfils these criteria ( Table 8, Figure 6), suggesting that parasites could use isoleucine both as a resource and a time-of-day cue.
Figure 6. Isoleucine is rhythmic and coincides with the intraerythrocytic development cycle (IDC) schedule and host feeding-fasting cycles.
Model fit (best fit line and 95% prediction interval) for DF, LF and TRF infections, from the time since feeding commences, with feeding-fasting windows overlaid. In the ALF group, a flat line with an intercept of 77.7 μM (illustrated as a dotted line) is the best fit to describe isoleucine concentration across the sampling window.
Table 6. Highlighted in green are the metabolites rhythmic in each treatment group (according to the circadian programmes and ANOVA).
DF: 101 y, 33 n; LF: 91 y, 43 n; TRF: 50 y, 84 n; ALF: 1 y, 133 n.
| Metabolite | Class | DF | LF | TRF | ALF |
|---|---|---|---|---|---|
| Ala | Amino.acid | ||||
| Arg | Amino.acid | ||||
| Asn | Amino.acid | ||||
| Asp | Amino.acid | ||||
| Cit | Amino.acid | ||||
| Gln | Amino.acid | ||||
| Glu | Amino.acid | ||||
| Gly | Amino.acid | ||||
| His | Amino.acid | ||||
| Ile | Amino.acid | ||||
| Leu | Amino.acid | ||||
| Lys | Amino.acid | ||||
| Met | Amino.acid | ||||
| Orn | Amino.acid | ||||
| Phe | Amino.acid | ||||
| Pro | Amino.acid | ||||
| Ser | Amino.acid | ||||
| Thr | Amino.acid | ||||
| Trp | Amino.acid | ||||
| Tyr | Amino.acid | ||||
| Val | Amino.acid | ||||
| ADMA | Biogenic_amine | ||||
| alpha.AAA | Biogenic_amine | ||||
| Carnosine | Biogenic_amine | ||||
| Histamine | Biogenic_amine | ||||
| Kynurenine | Biogenic_amine | ||||
| Met.SO | Biogenic_amine | ||||
| Putrescine | Biogenic_amine | ||||
| Sarcosine | Biogenic_amine | ||||
| SDMA | Biogenic_amine | ||||
| Serotonin | Biogenic_amine | ||||
| Spermidine | Biogenic_amine | ||||
| t4.OH.Pro | Biogenic_amine | ||||
| Taurine | Biogenic_amine | ||||
| C0 | Acylcarnitines | ||||
| C2 | Acylcarnitines | ||||
| C3 | Acylcarnitines | ||||
| C4 | Acylcarnitines | ||||
| C14.1 | Acylcarnitines | ||||
| C16 | Acylcarnitines | ||||
| C18.1 | Acylcarnitines | ||||
| lysoPC.a.C16.0 | Glycerophospholipids | ||||
| lysoPC.a.C16.1 | Glycerophospholipids | ||||
| lysoPC.a.C17.0 | Glycerophospholipids | ||||
| lysoPC.a.C18.0 | Glycerophospholipids | ||||
| lysoPC.a.C18.1 | Glycerophospholipids | ||||
| lysoPC.a.C18.2 | Glycerophospholipids | ||||
| lysoPC.a.C20.3 | Glycerophospholipids | ||||
| lysoPC.a.C20.4 | Glycerophospholipids | ||||
| lysoPC.a.C24.0 | Glycerophospholipids | ||||
| lysoPC.a.C26.0 | Glycerophospholipids | ||||
| lysoPC.a.C26.1 | Glycerophospholipids | ||||
| lysoPC.a.C28.0 | Glycerophospholipids | ||||
| lysoPC.a.C28.1 | Glycerophospholipids | ||||
| PC.aa.C24.0 | Glycerophospholipids | ||||
| PC.aa.C28.1 | Glycerophospholipids | ||||
| PC.aa.C30.0 | Glycerophospholipids | ||||
| PC.aa.C32.0 | Glycerophospholipids | ||||
| PC.aa.C32.1 | Glycerophospholipids | ||||
| PC.aa.C32.2 | Glycerophospholipids | ||||
| PC.aa.C32.3 | Glycerophospholipids | ||||
| PC.aa.C34.1 | Glycerophospholipids | ||||
| PC.aa.C34.2 | Glycerophospholipids | ||||
| PC.aa.C34.3 | Glycerophospholipids | ||||
| PC.aa.C34.4 | Glycerophospholipids | ||||
| PC.aa.C36.1 | Glycerophospholipids | ||||
| PC.aa.C36.2 | Glycerophospholipids | ||||
| PC.aa.C36.3 | Glycerophospholipids | ||||
| PC.aa.C36.4 | Glycerophospholipids | ||||
| PC.aa.C36.5 | Glycerophospholipids | ||||
| PC.aa.C36.6 | Glycerophospholipids | ||||
| PC.aa.C38.0 | Glycerophospholipids | ||||
| PC.aa.C38.3 | Glycerophospholipids | ||||
| PC.aa.C38.4 | Glycerophospholipids | ||||
| PC.aa.C38.5 | Glycerophospholipids | ||||
| PC.aa.C38.6 | Glycerophospholipids | ||||
| PC.aa.C40.2 | Glycerophospholipids | ||||
| PC.aa.C40.3 | Glycerophospholipids | ||||
| PC.aa.C40.4 | Glycerophospholipids | ||||
| PC.aa.C40.5 | Glycerophospholipids | ||||
| PC.aa.C40.6 | Glycerophospholipids | ||||
| PC.aa.C42.0 | Glycerophospholipids | ||||
| PC.aa.C42.1 | Glycerophospholipids | ||||
| PC.aa.C42.2 | Glycerophospholipids | ||||
| PC.aa.C42.4 | Glycerophospholipids | ||||
| PC.aa.C42.5 | Glycerophospholipids | ||||
| PC.aa.C42.6 | Glycerophospholipids | ||||
| PC.ae.C30.1 | Glycerophospholipids | ||||
| PC.ae.C30.2 | Glycerophospholipids | ||||
| PC.ae.C32.1 | Glycerophospholipids | ||||
| PC.ae.C32.2 | Glycerophospholipids | ||||
| PC.ae.C34.0 | Glycerophospholipids | ||||
| PC.ae.C34.1 | Glycerophospholipids | ||||
| PC.ae.C34.2 | Glycerophospholipids | ||||
| PC.ae.C34.3 | Glycerophospholipids | ||||
| PC.ae.C36.0 | Glycerophospholipids | ||||
| PC.ae.C36.1 | Glycerophospholipids | ||||
| PC.ae.C36.2 | Glycerophospholipids | ||||
| PC.ae.C36.3 | Glycerophospholipids | ||||
| PC.ae.C36.4 | Glycerophospholipids | ||||
| PC.ae.C36.5 | Glycerophospholipids | ||||
| PC.ae.C38.0 | Glycerophospholipids | ||||
| PC.ae.C38.1 | Glycerophospholipids | ||||
| PC.ae.C38.2 | Glycerophospholipids | ||||
| PC.ae.C38.3 | Glycerophospholipids | ||||
| PC.ae.C38.4 | Glycerophospholipids | ||||
| PC.ae.C38.5 | Glycerophospholipids | ||||
| PC.ae.C38.6 | Glycerophospholipids | ||||
| PC.ae.C40.1 | Glycerophospholipids | ||||
| PC.ae.C40.2 | Glycerophospholipids | ||||
| PC.ae.C40.3 | Glycerophospholipids | ||||
| PC.ae.C40.4 | Glycerophospholipids | ||||
| PC.ae.C40.5 | Glycerophospholipids | ||||
| PC.ae.C40.6 | Glycerophospholipids | ||||
| PC.ae.C42.1 | Glycerophospholipids | ||||
| PC.ae.C42.2 | Glycerophospholipids | ||||
| PC.ae.C42.3 | Glycerophospholipids | ||||
| PC.ae.C44.3 | Glycerophospholipids | ||||
| PC.ae.C44.5 | Glycerophospholipids | ||||
| PC.ae.C44.6 | Glycerophospholipids | ||||
| SM..OH..C14.1 | Sphingolipids | ||||
| SM..OH..C16.1 | Sphingolipids | ||||
| SM..OH..C22.1 | Sphingolipids | ||||
| SM..OH..C22.2 | Sphingolipids | ||||
| SM..OH..C24.1 | Sphingolipids | ||||
| SM.C16.0 | Sphingolipids | ||||
| SM.C16.1 | Sphingolipids | ||||
| SM.C18.0 | Sphingolipids | ||||
| SM.C18.1 | Sphingolipids | ||||
| SM.C20.2 | Sphingolipids | ||||
| SM.C24.0 | Sphingolipids | ||||
| SM.C24.1 | Sphingolipids | ||||
| SM.C26.0 | Sphingolipids | ||||
| SM.C26.1 | Sphingolipids |
Table 7. Timing of peak of each final candidate metabolite in the blood for each treatment group.
For LF and TRF groups ZT 0/ 0 hours is the start of the feeding window and the time of lights on for LF, while for DF ZT0 is the start of the fasting window and the time of lights on. LF and DF groups are in 12h:12h light:dark although feeding in the day and night, respectively. TRF are in constant darkness although feed for the same 12h window (0–12 hours) as the LF group (same experimenter time). DF=dark fed wild type mice, LF=light fed wild type mice, TRF=time restricted fed Per1/2-null mice.
| Metabolite | Class | DF (ZT, hours) | LF (ZT, hours) | TRF (hours) |
|---|---|---|---|---|
| C14.1 | Acylcarnitines | 7.12 | 20.85 | 19.13 |
| C16 | Acylcarnitines | 6.83 | 19.97 | 20.44 |
| C18.1 | Acylcarnitines | 6.65 | 19.92 | 20.18 |
| Ala | Amino.acid | 19.17 | 7.99 | 7.38 |
| Asn | Amino.acid | 18.47 | 6.39 | 6.03 |
| Asp | Amino.acid | 17.46 | 20.42 | 18.01 |
| Glu | Amino.acid | 17.59 | 20.08 | 18.38 |
| Ile | Amino.acid | 21.03 | 7.70 | 0.40 |
| Leu | Amino.acid | 20.36 | 6.65 | 2.85 |
| Met | Amino.acid | 19.90 | 6.20 | 5.90 |
| Phe | Amino.acid | 19.78 | 6.99 | 5.91 |
| Pro | Amino.acid | 19.08 | 6.95 | 6.34 |
| Thr | Amino.acid | 20.06 | 7.93 | 6.33 |
| Val | Amino.acid | 20.50 | 7.95 | 5.05 |
| ADMA | Biogenic_amine | 9.74 | 20.85 | 19.43 |
| Carnosine | Biogenic_amine | 16.39 | 20.41 | 19.03 |
| Histamine | Biogenic_amine | 15.27 | 19.06 | 19.21 |
| Met.SO | Biogenic_amine | 19.37 | 8.04 | 6.54 |
| Putrescine | Biogenic_amine | 15.80 | 19.05 | 16.13 |
| SDMA | Biogenic_amine | 9.83 | 20.78 | 20.54 |
| Serotonin | Biogenic_amine | 13.31 | 6.29 | 18.74 |
| Spermidine | Biogenic_amine | 16.54 | 19.78 | 19.13 |
| Taurine | Biogenic_amine | 16.07 | 20.22 | 19.60 |
| lysoPCaC16:1 | Glycerophospholipids | 6.50 | 18.17 | 18.20 |
| lysoPCaC18:1 | Glycerophospholipids | 6.97 | 18.57 | 16.64 |
| lysoPCaC18:2 | Glycerophospholipids | 5.90 | 15.27 | 12.05 |
| PCaaC32:1 | Glycerophospholipids | 1.41 | 16.26 | 18.89 |
| PCaaC32:2 | Glycerophospholipids | 23.87 | 16.15 | 18.57 |
| PCaaC34:4 | Glycerophospholipids | 6.71 | 17.92 | 0.54 |
| PCaaC38:3 | Glycerophospholipids | 7.48 | 19.01 | 22.51 |
| PCaaC38:4 | Glycerophospholipids | 8.18 | 21.07 | 1.18 |
| PCaaC38:5 | Glycerophospholipids | 7.70 | 20.03 | 0.89 |
| PCaaC38:6 | Glycerophospholipids | 6.81 | 20.36 | 23.97 |
| PCaaC40:4 | Glycerophospholipids | 5.80 | 18.63 | 23.57 |
| PCaaC40:5 | Glycerophospholipids | 7.01 | 19.68 | 23.88 |
| PCaeC34:1 | Glycerophospholipids | 23.59 | 16.45 | 18.99 |
| PCaeC34:3 | Glycerophospholipids | 0.48 | 15.22 | 14.65 |
| PCaeC36:2 | Glycerophospholipids | 22.36 | 12.23 | 10.04 |
| PCaeC38:0 | Glycerophospholipids | 7.80 | 19.97 | 0.71 |
| PCaeC38:2 | Glycerophospholipids | 20.45 | 12.78 | 9.58 |
| PCaeC38:3 | Glycerophospholipids | 2.18 | 16.49 | 19.91 |
| PCaeC42:1 | Glycerophospholipids | 8.78 | 20.70 | 2.72 |
Table 8. Potential for acylcarnitines, glycerophospholipids and amino acids/amines candidates that associate with the feeding-fasting cycle to explain the intraerythrocytic development cycle (IDC) schedule.
Requirements for these metabolites is likely to increase as each parasite cell progresses through its IDC.
| Metabolite class &
general function |
Role in the IDC? | Putative time-cue? |
|---|---|---|
|
Acylcarnitines
• Energy metabolism • Mitochondrial function • Fatty acid transport |
No evidence a lack of acylcarnitines affects either the IDC schedule or replication rate. Peak during fasting when parasites are in their least energetically/ metabolically demanding stages. |
Very unlikely. |
|
Glycerophospholipids
• Cellular signalling and trafficking • Membrane neogenesis • Haemozoin formation |
Phospholipid composition of the host RBC increases during infection, especially oleic acid (18:1)
( Déchamps et al., 2010). Parasites scavenge fatty acids and also lysoPCs from host plasma, which compete with each other as a source of the acyl components required for lipid synthesis. Lysophosphatidylcholine a C18:1 (lysoPC a 18:1), which contains an oleic acid side chain, is associated with IDC progression. Majority peak during host fasting when parasites are in their least energetically/ metabolically demanding stages. Thus, to be a time-of day cue |
Unlikely. Not essential; unlike the liver
stage, IDC stages can synthesise fatty acids de novo via type II FA synthase (FASII), ( Tarun et al., 2009). |
|
Biogenic amines
and amino acids: • Nucleic acid and protein synthesis |
Parasites must scavenge several amino acids from the host, including six host-‘essential’ (isoleucine,
leucine, methionine, phenylalanine, threonine and valine) and one host-‘non-essential’ (alanine) amino acid ( Payne & Loomis, 2006) that are rhythmic. Parasites can scavenge most amino acids from catabolism of host haemoglobin ( Babbitt et al., 2012; Liu et al., 2006; Martin & Kirk, 2007). Isoleucine is the only amino acid absent from human haemoglobin and is one of the least abundant amino acids in rodent haemoglobin (1-3%, Figure 9) yet makes up 9% of both P. falciparum’s and P. chabaudi’s amino acids ( Yadav & Swati, 2012). The response of P. falciparum to isoleucine withdrawal is not influenced by whether they were previously cultured in a high or low isoleucine concentration environment and results in dormancy that can be broken upon isoleucine replenishment ( Babbitt et al., 2012). Coinciding with the rise in isoleucine concentration during the feeding window, trophozoites transition to schizonts before bursting and beginning development as ring stages at the end of the feeding window ( Figure 6, Figure 8). |
Likely. Isoleucine is the only essential
amino acid for P. falciparum, it cannot be stored, and dormancy has no apparent ill-effects on subsequent development. P. falciparum deprived of other amino acids develop as normal ( Liu et al., 2006). |
Figure 9.
Frequency of amino acids in alpha ( A) and beta ( B) chains of human (orange) and mouse (blue) haemoglobin. Amino acid codes: A - alanine, C - cysteine, D - aspartic acid, E - glutamic acid, F - phenylalanine, G - glycine, H - histidine, I - isoleucine, K - lysine, L - leucine, M - methionine, N - asparagine, P - proline, Q - glutamine, R - arginine, S - serine, T - threonine, V - valine, W - tryptophan, Y - tyrosine.
Figure 8. Schematic for isoleucine’s role in the intraerythrocytic development cycle (IDC) schedule.
( A) In the absence of isoleucine (Ile), but in the presence of all other essential components of culture media, P. chabaudi’s IDC progression stops or continues very slowly, as observed for P. falciparum ( Liu et al., 2006 and Babbitt et al., 2012). This observation is consistent with isoleucine being an essential resource and/or a time-cue. This means the time-of-day cue may also be a direct selective agent (i.e. the reason why a timing strategy has evolved). In such situations it is very difficult to unravel the consequences of cue loss. Here, the challenge is differentiating to what extent the absence of isoleucine (i) creates a non-permissive environment, imposing a constraint on parasites that simply forces IDC progression to stop, versus (ii) parasites responding in a strategic manner and ‘deciding’ to stop development. Under scenario (i) parasites experience resource limitation, and starvation by definition should have negative consequences. For example, the longer parasites are starved, the more die, or experience increasingly lower or slower rates of recovery upon isoleucine replenishment. Under scenario (ii) there are no such costs because parasites avoid starvation by stopping/slowing development and waiting for a signal that it is time to expect resources to be replenished. That parasite number is not affected by isoleucine withdrawal, even for prolonged periods, and that parasites recover IDC completion at the same rate regardless of the duration of withdrawal, is consistent with scenario (ii) not scenario (i). ( B) It is now well established that P. chabaudi’s IDC schedule is synchronised to host feeding-fasting cycles. Our findings suggest this coordination is achieved by parasites responding to daily rhythmicity in the blood concentration of isoleucine for the purpose of maximising exploitation of the host’s isoleucine, and potentially other nutrients, they can more easily acquire from the host’s digestion of its food than by biosynthesis or scavenging from haemoglobin.
Timing and completion of the parasite IDC depends on the availability of isoleucine
That malaria parasites rely on exogenous isoleucine as both a resource and time-cue to complete the IDC is supported by previous observations across species of Plasmodium. First, when the human malaria P. falciparum is deprived of isoleucine, parasites immediately and dramatically slow cell cycle progression akin to dormancy ( Liu et al., 2006) yet are able to recover even after long periods of starvation (~4 IDCs) ( Babbitt et al., 2012). Second, in contrast to isoleucine, if other amino acids are removed from culture media, parasites switch to scavenging them from haemoglobin with only minor effects on IDC completion ( Babbitt et al., 2012). Third, not only is isoleucine crucial for the development of P. knowlesi in culture, parasites only incorporate isoleucine for part of the IDC (until the point that schizogony starts ( Butcher & Cohen, 1971; Polet & Conrad, 1968; Polet & Conrad, 1969; Sherman, 1979)). Fourth, isoleucine is one of few amino acids that exhibits a daily rhythm in the blood of mice and humans. Specifically, isoleucine was inverted with a 12-hour shift in simulated day and night shift working humans (a phase difference of 11:49 ± 02:10 h between the day and night shift conditions) and follows the timing of food intake ( Skene et al., 2018). Therefore, our next experiments tested whether an exogenous supply of isoleucine is capable of scheduling the IDC.
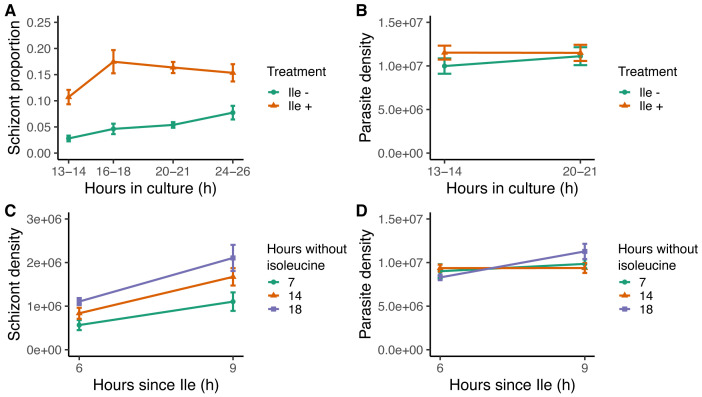
We carried out two experiments in parallel to quantify how P. chabaudi’s IDC progression is affected when deprived of isoleucine, and whether the IDC is then completed (defined as the proportion of parasites that reach the schizont stage) when isoleucine is restored. We conducted these experiments in vitro where isoleucine levels and timing can be controlled more successfully than by attempting to perturb blood isoleucine concentration in vivo. The challenges of an in vivo manipulation include that isoleucine is essential for mice so hosts cannot be maintained in isoleucine-free conditions without negative health consequences, particularly via its roles in the immune system and response to anaemia ( Mao et al., 2018; Murata & Moriyama, 2007; Rivas-Santiago et al., 2011). These potential off-target effects could confound conclusions about how the IDC behaves in vivo. First, parasites cultured in the absence of isoleucine (n=32 cultures from the blood of eight mice, which were split equally across both treatments) develop extremely slowly with approximately three-fold fewer completing the IDC compared to parasites with isoleucine (50 mg/L, which is the same concentration as standard culture media; RPMI 1640) (n=32 cultures) ( Figure 7A). The best fitting model contained only “treatment” (parasites cultured with or without isoleucine) as a main effect (ΔAICc=0, Table 9A). The reduction in schizonts in isoleucine-free conditions was not due to a higher death rate because the density of parasites remains constant during the experiment and did not differ between the treatments ( Figure 7B) (best fitting model is the null model ΔAICc=0, Table 9B). Further, incorporating either “treatment” or “hours in culture” into the model did not improve the model fit (treatment: ΔAICc=4.16, hours in culture: ΔAICc=5.50, Table 9B). This experiment extends previous findings for P. falciparum ( Babbitt et al., 2012; Liu et al., 2006; McLean & Jacobs-Lorena, 2020) across species as well as provides greater resolution on the parasite phenotypes involved.
Figure 7. Isoleucine provision and withdrawal drives intraerythrocytic development cycle (IDC) completion but does affect parasite mortality.
( A) IDC completion defined as the proportion of parasites that are schizonts, in cultures with (orange triangles, Ile +, 50 mg/L) or without (green circles, Ile -) isoleucine. ( B) Density of all parasite stages when parasites are cultured with or without isoleucine. Density of ( C) schizonts and ( D) all parasite stages, after the addition of isoleucine into cultures following isoleucine deprivation for seven (green circles), 14 (orange triangles) and 18 hours (purple squares). The proportion of schizonts in the blood seeding the cultures was ~0.005.
Table 9. Degrees of Freedom (df), log-Likelihood (logLik), AICc, ΔAICc (AICc i – AICc min) and AICc w (AICc weight) for each linear model in the schizont/parasite proportion/density analysis ordered in descending fit (best-fitting model at the top).
The response variable for each model is either schizont proportion (Schizont.prop), parasite density (Parasite.dens) or schizont density (Schizont.dens) and the random effect is “mouseID”. “Treatment” refers to the treatment group (DF, LF, TRF or ALF), “hours in culture” refers to the number of hours spent in culture since being extracted from the mice, and “hours since isoleucine” refers to the number of hours since isoleucine was added to the cultures. Treatment, hours in culture and hours since isoleucine were all fitted as factors. DF=dark fed wild type mice, LF=light fed wild type mice, TRF=time restricted fed Per1/2-null mice, ALF=ad libitum fed Per1/2-null mice.
| Model description:
A) Schizont.prop ~ + (1|mouseID) |
df | logLik | AICc | ΔAICc | AICc w |
|---|---|---|---|---|---|
| treatment | 4 | 106.26 | -203.8 | 0.00 | 0.996 |
| treatment + hours in culture | 7 | 104.29 | -192.6 | 11.26 | 0.004 |
| treatment * hours in culture | 10 | 98.81 | -173.5 | 30.37 | 0.000 |
| null | 3 | 80.96 | -155.5 | 48.31 | 0.000 |
| hours in culture | 6 | 74.89 | -136.3 | 67.54 | 0.000 |
| B) Parasite.dens ~ + (1|mouseID) | |||||
| null | 3 | 0.14 | 6.6 | 0.00 | 0.836 |
| treatment | 4 | -0.63 | 10.7 | 4.16 | 0.104 |
| hours in culture | 4 | -1.30 | 12.1 | 5.50 | 0.053 |
| treatment + hours in culture | 5 | -2.07 | 16.4 | 9.88 | 0.006 |
| treatment * hours in culture | 6 | -2.79 | 20.9 | 14.36 | 0.001 |
| C) Schizont.dens ~ + (1|mouseID) | |||||
| treatment + hours since isoleucine | 6 | 69.67 | -125.3 | 0.00 | 0.887 |
| hours since isoleucine | 4 | 65.04 | -121.2 | 4.14 | 0.112 |
| treatment * hours since isoleucine | 8 | 65.67 | -111.6 | 13.65 | 0.001 |
| null | 3 | 56.87 | -107.2 | 18.10 | 0.000 |
| treatment | 5 | 56.67 | -101.9 | 23.39 | 0.000 |
| D) Parasite.dens ~ + (1|mouseID) | |||||
| hours since isoleucine | 4 | 12.96 | -17.0 | 0.00 | 0.568 |
| null | 3 | 11.49 | -16.4 | 0.56 | 0.430 |
| treatment + hours since isoleucine | 6 | 9.10 | -4.1 | 12.85 | 0.001 |
| treatment | 5 | 7.76 | -4.1 | 12.90 | 0.001 |
| treatment * hours since isoleucine | 8 | 10.47 | -1.2 | 15.75 | 0.000 |
The substantial slowing of IDC progression in the absence of isoleucine is further supported by our second experiment. This experiment revealed that like for P. falciparum, P. chabaudi completes IDC development when isoleucine deprivation ends. Parasites (~10 7 per culture) were added to isoleucine-free media and incubated for seven, 14, or 18 hours, after which isoleucine (50 mg/L) was added to their cultures (n=16 cultures per treatment). Parasites completed development when isoleucine became available, regardless of the duration of deprivation (seven, 14, or 18 hours), with the best fitting model containing main effects of “treatment” and “hours since isoleucine added” (ΔAICc=0, Table 9C). Importantly, including the interaction did not improve the model fit (ΔAICc=13.65, Table 9C), demonstrating that IDC completion proceeds at the same rate despite different durations of isoleucine starvation. Specifically, the rate of IDC completion in the 6–9 hours following isoleucine replenishment was approximately 50% for all groups ( Figure 7C). Again, higher death rates in cultures deprived of isoleucine for the longest time period did not give the appearance of equal IDC rates because the model incorporating “hours since isoleucine” was competitive with the null model (ΔAICc=0.56, Figure 7D, Table 9D), revealing parasites were still as viable after 18 hours of deprivation as those deprived for seven and 14 hours. Furthermore, cultures deprived the longest achieved the most schizonts (18 hours, mean±SEM: 1.61×10 6 ±0.20 Figure 7C), while the fewest schizonts were observed in cultures deprived for the shortest period (seven hours, mean±SEM: 0.83×10 6 ±0.14 Figure 7C). Unlike previous studies, by comparing rates of completion across treatments (i.e. statistically testing the roles of the duration of deprivation and its interaction with treatment), our experiment demonstrates that isoleucine deprivation is not costly to parasites ( Figure 8A). The variation in the intercepts of Figure 7C (<1% schizonts after seven hours, 3% after 14 hours and 5% after 18 hours deprivation) is likely explained by the 18 hour deprivation cultures accumulating a higher proportion of schizonts at the time of isoleucine provision simply as a product of developing very slowly during a longer window of deprivation (in line with Liu et al., 2006 and Babbitt et al., 2012). The lack of costs and ability to recover development when isoleucine is replenished - which differs from the response to the loss of other essential nutrients - is consistent with isoleucine being sufficient to act as a time-cue as well as being an essential resource ( Figure 8B).
Discussion
Our large-scale metabolomics screening experiment ( Figure 3, Figure 4, Figure 6, Table 8) and follow-up experiments ( Figure 5, Figure 7), demonstrate that isoleucine is sufficient to explain how malaria parasites schedule their IDC in line with host feeding-fasting rhythms. A key challenge was differentiating to what extent the absence of isoleucine creates a non-permissive environment, imposing a constraint on parasites that simply forces IDC progression to stop, versus parasites responding in a strategic manner and ‘deciding’ to stop development ( Figure 8A). There are several reasons why our findings are not a case of revealing the obvious, that parasites cannot develop when resource-limited. First, when deprived of other essential resources such as glucose, P. falciparum rapidly displays stress responses at the transcriptional level yet when deprived of isoleucine, its normal transcriptional pattern persists, albeit very slowly ( Babbitt et al., 2012), suggesting it is waiting for a signal to proceed (a “gate”). Second, parasites cannot cope with deprivation of other essential resources, for example dying within hours of glucose deprivation ( Babbitt et al., 2012), yet the IDC restarts with no apparent ill-effects when isoleucine is replenished ( Figure 7). Third, that parasites cannot create isoleucine stores, nor generate it endogenously, and mount rapid transcriptional responses to changes in exogenous isoleucine concentration ( Babbitt et al., 2012), are all hallmarks of an informative cue and a sensitive response system. Thus, we propose that either parasites are so well adapted to isoleucine starvation they cope just fine without it, unlike their ability to cope with other forms of starvation, or that isoleucine exerts its effects on IDC progression as both a time-cue and resource ( Figure 8B).
Most studies of isoleucine uptake and use in malaria parasites focus on P. falciparum. This parasite uses several channels and receptors (both parasite- and host-derived) to acquire resources from the host. Uninfected human RBC take up both isoleucine and methionine via the saturable L-system ( Cobbold et al., 2011), which supplies 20% of the necessary isoleucine ( Martin & Kirk, 2007). When parasitised, there is a five-fold increase in isoleucine entering RBC which is attributable to new permeability pathways (NPPs) introduced into the RBC membrane by the parasite ( Martin & Kirk, 2007; McLean & Jacobs-Lorena, 2020). NPPs supply 80% of the necessary isoleucine ( Martin & Kirk, 2007) and are active only in the host membrane at the trophozoite and schizont stages of the IDC, suggesting this influx of isoleucine occurs only at certain times of day (e.g. after host feeding) ( Kutner et al., 1985). Assuming P. chabaudi has analogous mechanisms, we propose that elevated isoleucine is used by the parasite as a marker for a sufficiently nutrient-rich environment to traverse cell cycle checkpoints and complete the IDC ( McLean & Jacobs-Lorena, 2020; O’Neill et al., 2020).
Glucose has previously been suggested as a time-cue or scheduling force for the IDC ( Hirako et al., 2018; Prior et al., 2018). Our experiment - which is the first to quantify glucose and IDC rhythms across different feeding-fasting rhythms and expose parasites to artificial glucose rhythms in arrhythmic hosts - does not support a direct effect of glucose but it may be indirectly involved. Parasites that are glucose-limited fail to concentrate isoleucine ( Martin & Kirk, 2007), likely due to a lack of glycolysis and ATP production needed to operate isoleucine transporters. High concentrations of isoleucine in the blood are also associated with uptake of glucose by tissues, potentially contributing to the hypoglycaemia associated with TNF-stimulation of immune cells during malaria infection ( Elased & Playfair, 1994; Hirako et al., 2018). Additionally, rodent models and humans with obesity and type 2 diabetes-like pathologies have elevated levels of isoleucine and dampened glucose rhythms in the blood ( Lynch & Adams, 2014; Isherwood et al., 2017). Thus, if glucose limitation or elevation interferes with the parasite’s ability to acquire time-of-day information from isoleucine, the IDC schedule will be disrupted, as observed in diabetic mice ( Hirako et al., 2018). Connections between isoleucine and glucose might also explain why the parasite protein kinase ‘KIN’ is involved in nutrient sensing ( Mancio-Silva et al., 2017). Whilst isoleucine appears to be the main driver of the IDC schedule, glucose likely mediates isoleucine’s impact via multiple, non-mutually exclusive, activities.
Given that daily variation in the concentration of isoleucine in the blood of infected mice appears modest (55 μM to 80 μM from nadir to peak; Figure 6), we suggest that like P. falciparum, P. chabaudi is very sensitive to changes in isoleucine levels. This observation could be interpreted in two non-mutually exclusive ways. Perhaps this seemingly damp rhythm means that sufficient isoleucine is available around-the-clock to meet the parasites resource-needs (from the blood and scavenging from the very low level (<3%) of isoleucine in rodent haemoglobin; Figure 9), so isoleucine acts on the IDC schedule only as a time-cue (to align with resources that are limited at certain times of day) rather than as a developmental-rate limiting resource as well. For instance, the appearance of isoleucine may signal a window during which vitamins, cofactors, purines, folic acid, pantothenic acid, and glucose, that are also required for successful replication, are available ( Müller & Kappes, 2007; Müller et al., 2010; Sherman, 1979). Furthermore, isoleucine is also a reliable cue for the timing of other nutrients that are perhaps easier for parasites to acquire from the host’s digestion of food than from biosynthesis or scavenging from haemoglobin. In particular, that the expression of genes associated with translation are the most commonly disrupted when P. chabaudi’s rhythms are perturbed away from the host’s rhythms ( Subudhi et al., 2020) suggests parasites align the IDC schedule with the resources required for proteins. However, if isoleucine is not a limiting resource at certain times of day, this may only be the case early infections.
Like blood glucose concentration, the isoleucine rhythm may become exacerbated as infections develop high parasite burdens and hosts become sick. In this case, by aligning the IDC schedule correctly early in infections, parasites minimise the costs of resource limitation later on.
Our findings offer a route into identifying the molecular pathways involved in transducing environmental signals to the IDC schedule. To date, the only gene known to be involved in the IDC schedule is SR10, which modulates IDC duration in response to perturbations of host time-of-day ( Subudhi et al., 2020). Identifying KIN ( Mancio-Silva et al., 2017) regulated pathways and whether they, along with pathways associated with SR10 ( Subudhi et al., 2020), and PK7/MORC (which is involved in IDC stage transitions; Singh et al., 2021), are sensitive to isoleucine might reveal how the parasites’ time-keeping mechanism operates. Whilst by no means conclusive, our results feed the debate about whether malaria parasites keep time by an endogenous circadian oscillator or a simpler mechanism. The hallmarks of an endogenous circadian clock are (i) temperature compensation, (ii) free running in constant conditions, and (iii) entrainment to a Zeitgeber ( Pittendrigh, 1960). Recent observations are consistent with (ii) ( Rijo-Ferreira et al., 2020; Smith et al., 2020; Subudhi et al., 2020) and our results now allow entrainment to isoleucine to be tested for (iii), as well as free-running in isoleucine-constant conditions (ii). However, an endogenous clock may exist to allow parasites to schedule activities other than the IDC. Overall, the rapid stop-start responses to isoleucine withdrawal and replenishment that we observe, and that is known for P. falciparum ( Babbitt et al., 2012; Liu et al., 2006; McLean & Jacobs-Lorena, 2020) , appear more parsimoniously explained by a “just-in-time reactionary strategy” than an endogenous oscillator. This is because clocks generally require several cycles to catch up with a large change in the phase (timing) of their Zeitgeber (which is why jet lag occurs). Thus, in the absence of isoleucine or a phase-shift in its availability, if the ticking of an endogenous clock cannot be readily de-coupled from IDC progression, development would have continued as best as possible, instead of stopping. Furthermore, most clocks are set by a Zeitgeber (usually light) that is a good proxy for something else (e.g. predation risk) that is the ultimate driver of selection for a timing mechanism. Yet, isoleucine appears to be both cue and selective driver, further suggesting a just-in-time response. This strategy has the advantage of cueing in to the most accurate information about resource availability, and providing a more fast-responding strategy than a clock when the bulk of host foraging could occur anytime within the broad window of the active phase, and whose timing may be disrupted if hosts become severely ill.
By whatever mechanism the IDC is scheduled, coordinating development according to the availability of the resources needed to produce progeny intuitively seems like a good strategy to maximise fitness. Yet, the costs/benefits of the IDC schedule demonstrated by parasites may be mediated by parasite density. At low parasite densities (e.g. at the start of infection), resources may be sufficient to support IDC completion of all parasites at any time-of-day, but at intermediate densities, parasites may need to align their IDC needs with rhythms in resource availability. Finally, at very high densities and/or when hosts become sick, resources could be limiting so a synchronous IDC leads to deleterious competition between related parasites. Quantifying whether the costs and benefits of a synchronous IDC vary during infections in line with parasite density and resource availability is complicated by the connection between the IDC schedule and the timing of transmission stages. By aligning the IDC schedule with the feeding-fasting rhythm which is set by light-dark rhythms in the environment, parasites benefit from the knock-on alignment of transmission stage maturation with vector biting activity ( O’Donnell et al., 2011; Pigeault et al., 2018; Schneider et al., 2018). Therefore, a key question is to what extent does the IDC schedule contribute to parasite fitness via transmission benefits versus variation during infections in the benefits and costs of aligning asexual replication with rhythmic resources in the blood? More broadly, by integrating evolutionary and mechanistic insight, it may be possible to improve antimalarial treatments. For example, impairing the parasites ability to detect or respond to isoleucine may stall the IDC, reducing virulence and buy time for host immune responses to clear the infection, as well as preventing ring stage dormancy from providing tolerance to antimalarials ( Codd et al., 2011).
Methods
Ethics statement
All procedures complied with the UK Home Office regulations (Animals Scientific Procedures Act 1986; project 483 licence number 70/8546) and were approved by the University of Edinburgh. All efforts were made to ameliorate harm to animals through monitoring of animal health by twice daily visual inspections and daily routine health checks, such as weight and RBC count.
Experimental designs and terminology
The same four perturbations of host and parasite rhythms were used in the metabolomics experiment and the first glucose monitoring experiment. The Per1/2-null mice (non-functional proteins Period 1 & 2, backcrossed onto a C57BL/6 background for over 10 generations) were donated by Michael Hastings (MRC Laboratory of Molecular Biology, Cambridge, UK) and generated by David Weaver (UMass Medical School, Massachusetts, USA). Wild type C57BL/6 mice were housed in a 12h light: 12h dark regime (12 hours of light followed by 12 hours of darkness) and the Per1/2-null mice housed in constant darkness (DD). All mice acclimated to their photo schedule and feeding treatments for two weeks prior to the start of infections and conditions were maintained throughout sampling. We randomly separated mice into their treatment groups. We refer to time-of-day using Zeitgeber Time (ZT) for mice housed under entrained conditions (light:dark cycles) and hours when Per1/2-null mice are housed under constant conditions (dark:dark). WT mice either had access to food at night (dark feeding, DF) or in the day (light feeding, LF). Per1/2-null mice either had access to food for 12 hours (time restricted feeding, TRF) or constant access to food ( ad libitum feeding, ALF). Every 12 hours, food was added/removed accordingly from the DF, LF and TRF cages and the cages were checked for evidence of hoarding, which was never observed. ALF cages were also disturbed during food removal/provision of the other groups. Investigators could not be blinded to the mouse strain due the difference in coat colour, or the lighting and feeding regime due to the need to add and remove food manually.
Sampling. All mice were infected intravenously with P. chabaudi DK genotype by injection of 1×10 5 infected RBC at ring stage. Sampling started on day 5 post infection and occurred every two hours for both the metabolomics and glucose experiments. We made a thin blood smear each time a mouse was sampled to quantify parasite rhythms by counting ~100 parasites per blood smear using microscopy. Smears were read without knowledge of the treatment group, and excluded where smear quality was too poor to count RBCs. Following Prior et al. (2018); O’Donnell et al. (2020) and Rijo-Ferreira et al. (2020), we used the proportion of ring stages as a phase marker (an estimate of the timing of parasite development in the blood) of parasite rhythms. We calculated amplitude and time-of-day of peak for each treatment group using sine and cosine curves in a linear model to confirm the IDC schedules for each group as used in O’Donnell et al. (2020).
Metabolomics experiment. We infected 68 eight-week-old female mice: 35 C57BL/6 wild type animals (DF and LF mice) and 33 Per1/2-null TTFL clock-disrupted mice (TRF and ALF) ( Table 2). Sample sizes were chosen based on previous data collected from our lab using our study system. We sampled mice in blocks (A–D), meaning each individual mouse was sampled every eight hours during the 26-hour sampling window, with 14 time points in total. We did not sample each mouse at each sampling point to minimise the total volume of blood being taken. At each sampling point for each designated host, 20 µl blood was taken from the tail vein to provide 10 µl blood plasma for snap freezing using dry ice.
Glucose monitoring. We infected 20 eight-week-old male mice: 10 C57BL/6 wild type animals and 10 Per1/2-null circadian clock-disrupted mice (as described above, Table 2). Sample sizes were chosen based on previous data collected using our study system. We recorded blood glucose concentration from all mice every two hours by taking 1 µl blood using an Accu-Chek Performa Nano glucometer.
Glucose perturbations. We infected 16 eight-to-ten week old male Per1/2/-null mice (housed in DD to ensure arrhythmicity) with 1×10 5 infected RBC at ring stage (note, a 10 fold higher density than all the other experiments). From day 1 post infection, intra-peritoneal injections of 100µl glucose (3g/kg of glucose, n=5), glucose + insulin (4.5IU/kg of insulin + 3g/kg of glucose, n=6) or PBS (n=5) occurred every 24h up to and including day 6 post infection. Injections occurred at the same time every day, at 12 hours out of phase with the IDC schedule of the parasites at the point of infection (i.e. 12 hours out of phase with peak feeding in the donor mice). Mice were sampled on days 5-6 at 0h, injections administered at 1h, then sampled again at 2h and 4h after which all mice were sampled every 4 hours up to and including 24h after the first sample. Samples consisted of a thin blood smear to stage parasites as well as calculate parasite density, blood glucose concentration ( Accu-Chek Performa Nano glucometer), and RBC density (Beckman Coulter).
Targeted metabolomics
We quantified metabolites by analysing plasma samples using the AbsoluteIDQ p180 targeted metabolomics kit (Biocrates Life Sciences AG, Innsbruck, Austria) and a Waters Xevo TQ-S mass spectrometer coupled to an Acquity UPLC system (Waters Corporation, Milford, MA, USA) ( Isherwood et al., 2017; Skene et al., 2018). We prepared the plasma samples (10 µl) according to the manufacturer’s instructions, adding several stable isotope–labelled standards to the samples prior to the derivatization and extraction steps. Using UPLC/MS (ultra performance liquid chromatography/mass spectrometry), we quantified 185 metabolites from five different compound classes (acylcarnitines, amino acids, biogenic amines, glycerophospholipids, and sphingolipids). We ran the samples on two 96-well plates, randomised the sample order and ran three levels of quality control (QC) on each plate. We normalised the data between the plates using the results of quality control level 2 (QC2) repeats across the plate (n=4) and between plates (n=2) using Biocrates METIDQ software (QC2 correction). Metabolites were excluded if the CV% of QC2 was >30% or if all four groups contained >25% of samples that were below the limit of detection, below the lower limit of quantification, or above the limit of quantification or blank out of range. The remaining 134 quantified metabolites comprised of seven acylcarnitines, 19 amino acids, 15 biogenic amines, 79 glycerophospholipids and 14 sphingolipids (see Figure 10).
Isoleucine response experiments
To test the effect of the amino acid isoleucine on the IDC, we compared parasite developmental progression in cultures with and without isoleucine (50 mg/L), as well as after different durations (seven, 14, 18 hours) of isoleucine starvation. We set up N = 112 cultures (from eight mice) so that for each time point within each treatment, an independent culture was sampled, avoiding any bias associated with repeat-sampling individual cultures. We used eight-week-old wild type female mice, MF1 strain, housed in a 12h:12h light:dark regime before and during infection. We infected mice intraperitoneally with 1×10 6 P. chabaudi genotype DK infected red blood cells at ring stage and terminally bled them on day 6 post infection, when the parasitaemia was around 15%. Mice were bled at ZT4, when parasites were late rings/ early trophozoites (see Prior et al., 2018). Approximately 1 ml of blood was collected from each mouse, which was then split equally across cultures in all five treatment groups.
Culturing. We washed infected blood twice with buffered RPMI containing no amino acids (following Spence et al., 2011), before being reconstituted in the RPMI medium corresponding to each treatment (containing isoleucine, or not) ( dx.doi.org/10.17504/protocols.io.buuqnwvw). We cultured parasites in 96-well round bottom plates with total culture volumes of 200–250 µl, at ~3% haematocrit and kept the culture plates inside a gas chamber which was gassed upon closing with 88% nitrogen 7% carbon dioxide and 5% oxygen, and then placed inside a 37°C incubator. The culture medium was custom made RPMI from Cell Culture Technologies, Switzerland.
Sampling. We sampled parasites in the first experiment (comparing IDC completion in isoleucine rich versus isoleucine free media) at 13–14, 16–18, 20–21, 24–26 and 27 hours after culture initiation. We also sampled parasites that had been isoleucine deprived for seven, 14 or 18 hours at six and nine hours after isoleucine addition. Samples consisted of a thin blood smear from each culture fixed with methanol and Giemsa stained. We measured the proportion of parasites in the schizont stage (as an indicator of parasites having completed their IDC) by counting ~300 parasites per blood smear. Smears were read without knowledge of the treatment group, and excluded where smear quality was too poor to count RBCs.
Statistical analysis
We defined daily rhythmicity in the concentrations of the 134 metabolites detected by the UPLC/MS-MS platform as the detection of rhythmicity in at least two of the following circadian analysis programmes: ECHO, CircWave and JTK_Cycle. For metabolites not found to be rhythmic by at least two of the circadian analysis programmes, we also carried out Analysis of Variance to identify metabolites that varied across the day but without a detectable 24-hour rhythm ( Table 3 for breakdown of which candidate metabolites were rhythmic in each programme). To calculate the acrophase (timing of peak), we used linear mixed-effects regression models containing sine and cosine terms on all metabolites that varied across the day. Metabolites whose acrophase fell in the same 12-hour feeding or fasting window (ZT0-12 or ZT12-24) in LF and TRF infections but fell in the opposite window for the DF group were shortlisted as potential candidates to connect host feeding rhythms and the IDC schedule. We compared schizont proportion and the density of combined IDC stages using linear regression models. To avoid overfitting due to small sample sizes, “Akaike information criterion-corrected” (AICc) values were calculated for each model, and a change in two AICc (ΔAICc = 2) was chosen to select the most parsimonious model ( Brewer et al., 2016).
An earlier version of this article can be found on bioRxiv (doi: https://doi.org/10.1101/2020.08.24.264689).
Data availability
Underlying data
Figshare: Plasmodium chabaudi genotype DK parasite stages during time series. https://doi.org/10.6084/m9.figshare.14842695.v1 ( Prior et al., 2021a).
Figshare: Targeted metabolomics on malaria-infected mouse blood. https://doi.org/10.6084/m9.figshare.14842638.v1 ( Prior et al., 2021b).
Figshare: Plasmodium schizont proportion in culture grown with and without isoleucine. https://doi.org/10.6084/m9.figshare.14842713.v1 ( Prior et al., 2021c).
Figshare: Plasmodium schizont densities in culture grown with and without isoleucine, as well as isoleucine starvation for different durations of time. https://doi.org/10.6084/m9.figshare.14842716.v1 ( Prior et al., 2021d).
Figshare: Blood glucose concentration time series in mice infected with malaria parasites https://doi.org/10.6084/m9.figshare.14912709.v1 ( Prior et al., 2021e).
Reporting guidelines
Figshare: ARRIVE checklist for “Synchrony between daily rhythms of malaria parasites and hosts is driven by an essential amino acid”. https://doi.org/10.6084/m9.figshare.14912703.v1 ( Prior et al., 2021f).
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Acknowledgements
We thank Daan van der Veen, Sam Rund, Petra Schneider, Alejandra Herbert-Mainero and Ronnie Mooney for practical help and discussion.
Funding Statement
This work was supported by the Wellcome Trust [202769, <a href=https://doi.org/10.35802/202769>https://doi.org/10.35802/202769</a>]; 108905; FC001043] and the Royal Society [UF110155; NF140517].
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; peer review: 2 approved]
References
- Babbitt SE, Altenhofen L, Cobbold SA, et al. : Plasmodium falciparum responds to amino acid starvation by entering into a hibernatory state. Proc Natl Acad Sci U S A. 2012;109(47):E3278–E3287. 10.1073/pnas.1209823109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, et al. : Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30(2):525–536. 10.1016/s0896-6273(01)00302-6 [DOI] [PubMed] [Google Scholar]
- Brewer MJ, Butler A, Cooksley SL: The relative performance of AIC, AIC c and BIC in the presence of unobserved heterogeneity. Methods Ecol Evol. 2016;7(6):679–692. 10.1111/2041-210X.12541 [DOI] [Google Scholar]
- Butcher GA, Cohen S: Short-term culture of Plasmodium knowlesi. Parasitology. 1971;62(2):309–320. 10.1017/s0031182000071547 [DOI] [PubMed] [Google Scholar]
- Cobbold SA, Martin RE, Kirk K: Methionine transport in the malaria parasite Plasmodium falciparum. Int J Parasitol. 2011;41(1):125–135. 10.1016/j.ijpara.2010.09.001 [DOI] [PubMed] [Google Scholar]
- Codd A, Teuscher F, Kyle DE, et al. : Artemisinin-induced parasite dormancy: a plausible mechanism for treatment failure. Malar J. 2011;10:56. 10.1186/1475-2875-10-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby P, Hamnett R, Putker M, et al. : Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell. 2019;177(4):896–909.e20. 10.1016/j.cell.2019.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily JP, Scanfeld D, Pochet N, et al. : Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007;450(7172):1091–1095. 10.1038/nature06311 [DOI] [PubMed] [Google Scholar]
- Déchamps S, Shastri S, Wengelnik K, et al. : Glycerophospholipid acquisition in Plasmodium - a puzzling assembly of biosynthetic pathways. Int J Parasitol. 2010;40(12):1347–1365. 10.1016/j.ijpara.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Elased K, Playfair JH: Hypoglycemia and hyperinsulinemia in rodent models of severe malaria infection. Infect Immun. 1994;62(11):5157–5160. 10.1128/iai.62.11.5157-5160.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia CR, Markus RP, Madeira L: Tertian and quartan fevers: temporal regulation in malarial infection. J Biol Rhythms. 2001;16(5):436–443. 10.1177/074873001129002114 [DOI] [PubMed] [Google Scholar]
- Hevia MA, Canessa P, Müller-Esparza H, et al. : A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana. Proc Natl Acad Sci USA. 2015;112(28):8744–8749. 10.1073/pnas.1508432112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirako IC, Assis PA, Hojo-Souza NS, et al. : Daily Rhythms of TNFα Expression and Food Intake Regulate Synchrony of Plasmodium Stages with the Host Circadian Cycle. Cell Host Microbe. 2018;23(6):796–808.e6. 10.1016/j.chom.2018.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozer C, Perret M, Parvard S, et al. : Survival is reduced when endogenous period deviates from 24 h in a non‐human primate, supporting the circadian resonance theory. Sci Rep. 2020;10(1):18002. 10.1038/s41598-020-75068-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isherwood CM, van der Veen DR, Johnston JD, et al. : Twenty-four-hour rhythmicity of circulating metabolites: effect of body mass and type 2 diabetes. FASEB J. 2017;31(12):5557–5567. 10.1096/fj.201700323R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutner S, Breuer WV, Ginsbururg H, et al. : Characterization of permeation pathways in the plasma membrane of human erythrocytes infected with early stages of Plasmodium falciparum: association with parasite development. J Cell Physiol. 1985;125(3):521–527. 10.1002/jcp.1041250323 [DOI] [PubMed] [Google Scholar]
- Liu J, Istvan ES, Gluzman IY, et al. : Plasmodium falciparum ensures its amino acid supply with multiple acquisition pathways and redundant proteolytic enzyme systems. Proc Natl Acad Sci U S A. 2006;103(23):8840–8845. 10.1073/pnas.0601876103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch CJ, Adams SH: Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10(12):723–736. 10.1038/nrendo.2014.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancio-Silva L, Slavic K, Ruivo MTG, et al. : Nutrient sensing modulates malaria parasite virulence. Nature. 2017;547(7662):213–216. 10.1038/nature23009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Gu C, Ren M, et al. : l-Isoleucine Administration Alleviates Rotavirus Infection and Immune Response in the Weaned Piglet Model. Front Immunol. 2018;9:1654. 10.3389/fimmu.2018.01654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RE, Kirk K: Transport of the essential nutrient isoleucine in human erythrocytes infected with the malaria parasite Plasmodium falciparum. Blood. 2007;109(5):2217–2224. 10.1182/blood-2005-11-026963 [DOI] [PubMed] [Google Scholar]
- Martinez-Bakker M, Helm B: The influence of biological rhythms on host-parasite interactions. Trends Ecol Evol. 2015;30(6):314–26. 10.1016/j.tree.2015.03.012 [DOI] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, Smyllie NJ, et al. : The Tau mutation of casein kinase 1ε sets the period of the mammalian pacemaker via regulation of Period1 or Period2 clock proteins. J Biol Rhythms. 2014;29(2):110–118. 10.1177/0748730414520663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean KJ, Jacobs-Lorena M: The response of Plasmodium falciparum to isoleucine withdrawal is dependent on the stage of progression through the intraerythrocytic cell cycle. Malar J. 2020;19(1):147. 10.1186/s12936-020-03220-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mideo N, Reece SE, Smith AL, et al. : The Cinderella syndrome: why do malaria-infected cells burst at midnight? Trends Parasitol. 2013;29(1):10–16. 10.1016/j.pt.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller IB, Hyde JE, Wrenger C: Vitamin B metabolism in Plasmodium falciparum as a source of drug targets. Trends Parasitol. 2010;26(1):35–43. 10.1016/j.pt.2009.10.006 [DOI] [PubMed] [Google Scholar]
- Müller S, Kappes B: Vitamin and cofactor biosynthesis pathways in Plasmodium and other apicomplexan parasites. Trends Parasitol. 2007;23(3):112–121. 10.1016/j.pt.2007.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Moriyama M: Isoleucine, an Essential Amino Acid, Prevents Liver Metastases of Colon Cancer by Antiangiogenesis. Cancer Res. 2007;67(7):3263–8. 10.1158/0008-5472.CAN-06-3739 [DOI] [PubMed] [Google Scholar]
- O’Donnell AJ, Mideo N, Reece SE: Disrupting rhythms in Plasmodium chabaudi: costs accrue quickly and independently of how infections are initiated. Malar J. 2013;12:372. 10.1186/1475-2875-12-372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell AJ, Prior KF, Reece SE: Host circadian clocks do not set the schedule for the within-host replication of malaria parasites. Proc Biol Sci. 2020;287(1932):20200347. 10.1098/rspb.2020.0347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell AJ, Schneider P, McWatters HG, et al. : Fitness costs of disrupting circadian rhythms in malaria parasites. Proc Biol Sci. 2011;278(1717):2429–2436. 10.1098/rspb.2010.2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, Hoyle NP, Robertson JB, et al. : Eukaryotic cell biology is temporally coordinated to support the energetic demands of protein homeostasis. Nat Commun. 2020;11(1):4706. 10.1038/s41467-020-18330-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski KL, Morrisey JM, Wilinski D, et al. : Host-parasite interactions revealed by Plasmodium falciparum metabolomics. Cell Host Microbe. 2009;5(2):191–199. 10.1016/j.chom.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, et al. : Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95(15):8660–8664. 10.1073/pnas.95.15.8660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne SH, Loomis WF: Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Eukaryotic Cell. 2006;5(2):272–276. 10.1128/EC.5.2.272-276.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigeault R, Caudron Q, Nicot A, et al. : Timing malaria transmission with mosquito fluctuations. Evol Lett. 2018;2(4):378–389. 10.1002/evl3.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS: Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol. 1960;25:159–184. 10.1101/sqb.1960.025.01.015 [DOI] [PubMed] [Google Scholar]
- Polet H, Conrad ME: Malaria: extracellular amino acid requirements for in vitro growth of erythrocytic forms of Plasmodium knowlesi. Proc Soc Exp Biol Med. 1968;127(1):251–253. 10.3181/00379727-127-32666 [DOI] [PubMed] [Google Scholar]
- Polet H, Conrad ME: The influence of three analogs of isoleucine on in vitro growth and protein synthesis of erythrocytic forms of Plasmodium knowlesi. Proc Soc Exp Biol Med. 1969;130(2):581–586. 10.3181/00379727-130-33612 [DOI] [PubMed] [Google Scholar]
- Prior K, Middleton B, Owolabi A, et al. : Plasmodium chabaudi genotype DK parasite stages during time series. figshare. Dataset.2021a. 10.6084/m9.figshare.14842695.v1 [DOI] [Google Scholar]
- Prior K, Middleton B, Owolabi A, et al. : Targeted metabolomics on malaria-infected mouse blood. figshare. Dataset.2021b. 10.6084/m9.figshare.14842638.v1 [DOI] [Google Scholar]
- Prior K, Middleton B, Owolabi A, et al. : Plasmodium schizont proportion in culture grown with and without isoleucine. figshare. Dataset.2021c. 10.6084/m9.figshare.14842713.v1 [DOI] [Google Scholar]
- Prior K, Middleton B, Owolabi A, et al. : Plasmodium schizont densities in culture grown with and without isoleucine, as well as isoleucine starvation for different durations of time. figshare. Dataset.2021d. 10.6084/m9.figshare.14842716.v1 [DOI] [Google Scholar]
- Prior K, Middleton B, Owolabi A, et al. : Blood glucose concentration time series in mice infected with malaria parasites. figshare. Dataset.2021e. 10.6084/m9.figshare.14912709.v1 [DOI] [Google Scholar]
- Prior K, Middleton B, Owolabi A, et al. : ARRIVE Checklist.pdf. figshare. Journal contribution.2021f. 10.6084/m9.figshare.14912703.v1 [DOI] [Google Scholar]
- Prior KF, Rijo-Ferreira F, Assis PA, et al. : Periodic parasites and daily host rhythms. Cell Host Microbe. 2020;27(2):176–187. 10.1016/j.chom.2020.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior KF, van der Veen DR, O’Donnell AJ, et al. : Timing of host feeding drives rhythms in parasite replication. PLoS Pathog. 2018;14(2):e1006900. 10.1371/journal.ppat.1006900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece SE, Prior KF, Mideo N: The Life and Times of Parasites: Rhythms in Strategies for Within-host Survival and Between-host Transmission. J Biol Rhythms. 2017;32(6):516–533. 10.1177/0748730417718904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Asher G: Crosstalk between metabolism and circadian clocks. Nat Rev Mol Cell Biol. 2019;20(4):227–241. 10.1038/s41580-018-0096-9 [DOI] [PubMed] [Google Scholar]
- Rijo-Ferreira F, Pinto-Neves D, Barbosa-Morais NL, et al. : Trypanosoma brucei metabolism is under circadian control. Nat Microbiol. 2017;2:17032. 10.1038/nmicrobiol.2017.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijo-Ferreira F, Acosta-Rodriguez VA, Abel JH, et al. : The malaria parasite has an intrinsic clock. Science. 2020;368(6492):746–753. 10.1126/science.aba2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-Santiago CE, Rivas-Santiago B, León DA, et al. : Induction of β-defensins by l-isoleucine as novel immunotherapy in experimental murine tuberculosis. Clin Exp Immunol. 2011;164(1):80–9. 10.1111/j.1365-2249.2010.04313.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider P, Rund SSC, Smith NL, et al. : Adaptive periodicity in the infectivity of malaria gametocytes to mosquitoes. Proc Biol Sci. 2018;285(1888):20181876. 10.1098/rspb.2018.1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman IW: Biochemistry of Plasmodium (malarial parasites). Microbiol Rev. 1979;43(4):453–495. 10.1128/mr.43.4.453-495.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Tessarin-Almeida G, Dias BKM, et al. : A nuclear protein, PfMORC confers melatonin dependent synchrony of the human malaria parasite P. falciparum in the asexual stage. Sci Rep. 2021;11(1):2057. 10.1038/s41598-021-81235-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene DJ, Skornyakov E, Chowdhury NR, et al. : Separation of circadian- and behavior-driven metabolite rhythms in humans provides a window on peripheral oscillators and metabolism. Proc Natl Acad Sci U S A. 2018;115(30):7825–7830. 10.1073/pnas.1801183115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Motta FC, Chopra G, et al. : An intrinsic oscillator drives the blood stage cycle of the malaria parasite Plasmodium falciparum . Science. 2020;368(6492):754–759. 10.1126/science.aba4357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence PJ, Cunningham D, Jarra W, et al. : Transformation of the rodent malaria parasite Plasmodium chabaudi. Nat Protoc. 2011;6(4):553–561. 10.1038/nprot.2011.313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoelstra K, Wikelski M, Daan S, et al. : Natural selection against a circadian clock gene mutation in mice. Proc Natl Acad Sci USA. 2016;113(3):686–691. 10.1073/pnas.1516442113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subudhi AK, O’Donnell AJ, Ramaprasad A, et al. : Malaria parasites regulate intra-erythrocytic development duration via serpentine receptor 10 to coordinate with host rhythms. Nat Commun. 2020;11(1):2763. 10.1038/s41467-020-16593-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun AS, Vaughan AM, Kappe SHI: Redefining the role of de novo fatty acid synthesis in Plasmodium parasites. Trends Parasitol. 2009;25(12):545–550. 10.1016/j.pt.2009.09.002 [DOI] [PubMed] [Google Scholar]
- Westwood ML, O'Donnell AJ, de Bekker C, et al. : The evolutionary ecology of circadian rhythms in infection. Nat Ecol Evol. 2019;3(4):552–560. 10.1038/s41559-019-0831-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav MJ, Swati D: Comparative genome analysis of six malarial parasites using codon usage bias based tools. Bioinformation. 2012;8(24):1230–1239. 10.6026/97320630081230 [DOI] [PMC free article] [PubMed] [Google Scholar]