Abstract
Embryonic stem cells (ESCs) are progenitor cells that retain the ability to differentiate into various cell types and are necessary for tissue repair. Improving cell culture conditions to maintain the pluripotency of ESCs in vitro is an urgent problem in the field of regenerative medicine. Here, we reveal that Spautin-1, a specific small-molecule inhibitor of ubiquitin-specific protease (USP) family members USP10 and USP13, promotes the maintenance of self-renewal and pluripotency of mouse ESCs in vitro. Functional studies reveal that only knockdown of USP13, but not USP10, is capable of mimicking the function of Spautin-1. Mechanistically, we demonstrate that USP13 physically interacts with, deubiquitinates, and stabilizes serine/threonine kinase Raf1 and thereby sustains Raf1 protein at the posttranslational level to activate the FGF/MEK/ERK prodifferentiation signaling pathway in naïve mouse ESCs. In contrast, in primed mouse epiblast stem cells and human induced pluripotent stem cells, the addition of Spautin-1 had an inhibitory effect on Raf1 levels, but USP13 overexpression promoted self-renewal. The addition of an MEK inhibitor impaired the effect of USP13 upregulation in these cells. These findings provide new insights into the regulatory network of naïve and primed pluripotency.
Keywords: Embryonic stem cells, self-renewal, Spautin-1, USP13, Raf1
Abbreviations: AP, alkaline phosphatase; CHIR, CHIR99021; CS, CHIR99021 and Spautin-1; EpiSCs, epiblast stem cells; iPSCs, induced pluripotent stem cells; LIF, leukemia inhibitory factor; mESCs, mouse embryonic stem cells; Nanog, Nanog homeobox; Oct4, POU domain, class 5, transcription factor 1; PB, PiggyBac system; qRT-PCR, quantitative real-time PCR; Raf1, v-raf-leukemia viral oncogene 1; shRNA, the short hairpin RNA; Tfcp2l1, transcription factor CP2-like 1; USP10, ubiquitin-specific peptidase 10; USP13, ubiquitin-specific peptidase 13
Embryonic stem cells (ESCs) derived from the inner cell mass of blastocysts have the ability to maintain self-renewal and pluripotency under suitable culture conditions in vitro. In 1981, Evans and Kaufman successfully isolated mouse ESCs (mESCs) by using feeder cells cultured in serum-containing medium (1, 2). In 1988, Smith AG reported that leukemia inhibitory factor (LIF) maintains the stemness of mESCs without feeder cells (3). However, the serum composition is too complicated, which is not conducive to the future application of stem cells. The identification of serum-free culture conditions has become a research hotspot. In 2003, Ying et al. found that bone morphogenetic protein 4 (BMP4) sustains mESC self-renewal by synergizing with LIF/STAT3 signaling in serum-free medium (4). Ying et al. aimed to further optimize the culture conditions and discovered that two small-molecule inhibitors (2i), PD0325091 (PD03) and CHIR99021 (CHIR), which are specific inhibitors of mitogen-activated protein kinase kinase (MEK) and glycogen synthase kinase-3 (Gsk3), respectively, enable robust proliferation of mESCs without inducing differentiation (5). In addition, 2i is also suitable for the isolation and culture of germline competent rat ESCs in vitro (6, 7). Mouse and rat ESCs thus represent a “naïve” pluripotent state. Notably, LIF and 2i fail to retain human pluripotent stem cells, such as human ESCs and induced pluripotent stem cells (hiPSCs) (7, 8, 9), which share similar features with mouse postimplantation epiblast stem cells (mEpiSCs) (10, 11, 12). Therefore, these types of pluripotent stem cells exhibit a “primed” pluripotent state.
Naïve and primed pluripotent states have many similarities and differences. For instance, both mouse and human ESCs are capable of propagating indefinitely and require the core genes Oct4, Sox2, and Nanog to maintain pluripotency (13). However, many signaling pathways have the opposite functions in the maintenance of the undifferentiated state of mouse and human pluripotent cells, such as the Wnt/β-catenin and FGF/MEK/ERK pathways. Activation of the Wnt/β-catenin signaling pathway promotes mESC self-renewal by releasing Tcf3-suppressed pluripotent genes, while inducing human pluripotent stem cell differentiation (6, 14, 15). Therefore, the addition of IWR1, an inhibitor of the Wnt/β-catenin pathway, facilitates the maintenance of their undifferentiated state (16). In contrast to this phenotype, FGF4 triggers the transition of mESCs to lineage commitment, and inhibition of the FGF/MEK/ERK pathway is important for safeguarding naïve pluripotency (6, 17). Conversely, human pluripotent stem cells require basic fibroblast growth factor (bFGF) to maintain their identity (10). Unfortunately, the biological characteristics of many animal ESCs are similar to those of human pluripotent stem cells. New targets and regulatory mechanisms must be discovered, and novel culture conditions must be invented for ESCs in vitro to efficiently isolate and maintain the naïve pluripotent state of ESCs. As 2i/LIF conditions have been used to sustain the pluripotent state of mouse and rat ESCs, the maintenance of ESCs from different species may share a set of conserved molecular mechanisms. Additionally, a screen using small-molecule libraries is an effective method. Previously, we have found that inhibition of protein kinase D by CID755673 promotes the short-term maintenance of mouse and human ESC pluripotency in vitro (18).
Here, we continued our screen using a compound library that has not been previously tested in ESCs. The chemical compound Spautin-1 (Sp-1) is beneficial to the maintenance of mouse ESCs in vitro and supports the long-term self-renewal of mESCs in vitro in combination with CHIR while promoting hiPSC differentiation. Sp-1 functions mainly by inhibiting the activity of USP13, which associates with and deubiquitinates RAF1. These results expand our understanding of different states of pluripotency, which will facilitate the establishment and maintenance of naïve pluripotent ESCs from multiple species.
Results
Screening of small molecules that promote the self-renewal of mESCs
We incubated 46C mESCs at low density in serum-containing medium without LIF, and then 1312 small molecules were individually added into each different well to identify new conditions that can maintain mESC identity in vitro. After 8 days, we discovered that many mESCs were in an undifferentiated state in the presence of Spautin-1 (Sp-1) and exhibited strong alkaline phosphatase (AP) activity, a marker of ESCs (Fig. 1A). In addition, immunofluorescence assays and western blotting showed that Sp-1-treated mESCs expressed higher levels of the pluripotent marker genes Oct4, Sox2, and Klf4 than untreated cells (Figs. 1B and S1). Next, different concentration gradients of Sp-1, from 2 to 20 μM, were added into serum-containing medium to determine the optimal concentration of Sp-1 that promoted mESC maintenance. AP staining showed that Sp-1 promoted mESC self-renewal in a dose-dependent manner (Fig. 1C). mESCs grew best in the presence of 8 μM Sp-1 but underwent apoptosis once the concentration exceeded 8 μM (Fig. 1C). Therefore, we chose 8 μM for subsequent experiments. These cells expressed the pluripotency genes Oct4, Nanog, Tfcp2l1, and Klf4 at higher levels but exhibited lower levels of the differentiation-associated genes Gata4, Sox17, and Cdx2 (Fig. 1, D and E). Notably, this dose of Sp-1 only maintained mESC self-renewal for no more than three passages (Fig. 1F). Together, these data indicate that Sp-1 promotes the short-term self-renewal of mESCs.
Figure 1.
Sp-1 promotes the self-renewal of mESCs.A, morphology and AP staining of 46C mESCs cultured in serum-containing medium in the presence of absence of 5 μM Sp-1 for 8 days. Scale bar, 100 μM. B, immunofluorescence analysis for Oct4 in mESCs treated with or without Sp-1 for 8 days. Scale bar, 100 μM. C, AP staining in mESCs cultured with different concentrations of Sp-1 for 8 days. Scale bar, 100 μM. D, Western blot analysis of Oct4, Nanog, and Tfcp2l1 protein levels in 46C mESCs treated with or without 8 μM Sp-1 for 8 days. E, qRT–PCR analysis of the expression levels of Oct4, Nanog, Klf4, Tfcp2l1, Gata4, Sox17, and Cdx2 in 46C mESCs treated with or without Sp-1 for 8 days. Data represented as the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus NT. F, AP staining of 46C mESCs cultured with Sp-1-containing medium for three passages. Scale bar, 100 μM. NT, No treatment; P3, passage 3.
Sp-1 sustains mESC self-renewal and pluripotency in combination with CHIR
PD03 and CHIR were added to the cultures to examine whether Sp-1 can cooperate with other small molecules to maintain the stemness of mESCs for a long time in vitro. After one passage, Sp-1, CHIR, and PD03 alone or the combination of PD03 and Sp-1 did not maintain cell growth well (Fig. 2A). However, mESCs maintained in medium containing both CHIR and Sp-1 (named CS) exhibited a typical stem cell phenotype and stronger AP activity than untreated cells (Fig. 2A). Meanwhile, these cells were able to be continuously passaged and expressed high levels of the pluripotency markers Oct4, Nanog, Klf4, and Tfcp2l1 (Fig. 2, B and C). The same conclusion was further confirmed in another mESC line J1 (Fig. S2), suggesting that CS is capable of sustaining the long-term self-renewal of mESCs.
Figure 2.
Sp-1 cooperates with CHIR to sustain mESC self-renewal and pluripotency.A, AP staining in 46C mESCs cultured with serum-containing medium supplemented with the indicated molecules for 8 days. CS, CHIR plus Sp-1. Scale bar, 100 μM. B, immunostaining analysis of Oct4 expression in mESCs treated with or without CS for five passages. Scale bar, 100 μM. C, qRT–PCR analysis of Oct4, Nanog, Klf4, and Tfcp2l1 expression levels in mESCs maintained in serum-containing medium supplemented with or without CS. Data represent mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus CHIR. D, immunostaining analysis of Gata6, Myosin, and Tuj1 expression in EB-derived cells. Scale bar, 100 μM. E, immunostaining of female mESCs and mEpiSCs for H3K27me3 and Oct4. mEpiSCs exhibit a nuclear body indicative of the inactive X chromosome. Scale bar, 100 μM. F, generation of chimeric mice after the injection of blastocysts with CS-treated C57BL/6 mESCs.
We performed embryoid body (EB) and microinjection experiments in vitro and in vivo to further evaluate the pluripotency of CS-maintained mESCs. After cultured in CS-containing medium for ten passages, 46C mESCs were suspended in petri dishes and allowed to form EBs in serum-containing medium. Immunofluorescence staining showed that they retained the ability to generate Tuj1-positive neurons, myosin-positive myocardial cells, and Gata6-positive primitive endoderm cells (Fig. 2D). In addition, immunofluorescence staining revealed no trimethylated H3 lysine 27 (H3K27me3) nuclear foci in the female CS-treated mESCs, a diagnostic marker of a silent X chromosome, while mEpiSCs in the primed pluripotent state exhibited a prominent body of nuclear staining for H3K27me3 (Fig. 2E). CS-maintained mESCs were injected into 72 blastocysts at 3.5 days post coitum to further validate the naïve pluripotent state of these cells. These injected blastocysts were able to generate chimeric mice when put into pseudopregnant female mice (Fig. 2F). Overall, mESCs grown with CS sustain pluripotency.
Sp-1 enhances mESC self-renewal by inhibiting USP13 activity
Since a previous report documented that Sp-1 participates in the process of cell autophagy by inhibiting ubiquitin-specific proteases (USPs) USP10 and USP13 to destabilize VPS34 (19), we first wanted to investigate which USP is responsible for the effect of Sp-1 on mESCs. Lentiviruses carrying two short hairpin RNAs (shRNAs) were used to infect 46C mESCs and silence mouse USP10 expression. Compared with cells transfected with the scramble control lentivirus, stable knockdown of USP10 transcript levels by approximately 60 to 80% was observed following drug selection (Fig. 3A). Scramble and USP10 shRNA-expressing mESCs were seeded in serum-containing medium in the absence of LIF. After 8 days, all cell lines lost AP activity and expressed similar levels of the pluripotency genes Oct4, Nanog, Tfcp2l1, and Klf4 (Fig. 3, B and C), indicating that the suppression of USP10 does not replace the effect of Sp-1. Subsequently, we focused on USP13. Likewise, we designed two shRNA sequences to target and decrease the expression of USP13 transcripts in mESCs (Fig. 3D). After the withdrawal of LIF for 8 days, USP13 shRNA-expressing mESCs showed stronger AP activity and expressed higher levels of the self-renewal genes Oct4, Nanog, Tfcp2l1, and Klf4 than scramble control-expressing cells (Fig. 3, E and F). Based on these data, the suppression of USP13 but not USP10 promotes mESC self-renewal. Consistently, the addition of CHIR was sufficient to induce typical AP activity in USP13 shRNA-expressing cells but not in scramble and USP10 shRNA-expressing cells (Fig. 3, G and H). Flag-tagged USP10 and HA-tagged USP13 were overexpressed in 46C mESCs using the PiggyBac vector (PB) to further test and verify the roles of USP10 and USP13 in regulating mESC maintenance, and both USP10 and USP13 expression levels were efficiently increased (Fig. S3, A and B). As expected, forced USP13 expression caused mESCs to lose AP activity more quickly than PB- and USP10-overexpressing cells after growth in basal medium for 5 days (Fig. S3, C and D). These results suggested that Sp-1 mediates the self-renewal of mESCs mainly by inhibiting USP13.
Figure 3.
Sp-1 promotes mESC self-renewal by decreasing USP13 activity.A, qRT–PCR analysis of USP10 expression in mESCs infected with scramble or USP10 shRNA lentiviruses. The data are presented as the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus Scramble. B, qRT–PCR analysis of Oct4, Nanog, Klf4, and Tfcp2l1 expression levels in scramble and USP10 shRNA-expressing mESCs cultured in serum-containing medium without LIF. The data are presented as the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus Scramble. C, AP staining of Scramble control and USP10 shRNA-expressing mESCs cultured in serum-containing medium without LIF. Scale bar, 100 μM. D, qRT–PCR analysis of USP13 expression in mESCs infected with scramble or USP13 shRNA-expressing lentiviruses. The data are presented as the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus Scramble. E, qRT–PCR analysis of Oct4, Nanog, Klf4, and Tfcp2l1 levels in scramble and USP13 shRNA-expressing ESCs cultured with basal medium in the absence of LIF. The data are presented as the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus Scramble. F, AP staining of scramble and USP13 shRNA-expressing mESCs cultured in serum-containing medium without LIF. Scale bar, 100 μM. G, AP staining of scramble and USP10 shRNA-expressing mESCs cultured in the presence of CHIR/serum. Scale bar, 100 μM. H, AP staining of scramble and USP13 shRNA-expressing mESCs cultured in CHIR/serum-condition. Scale bar, 100 μM.
As VPS34-mediated autophagy is an important event downstream of USP13 (19), we also wanted to examine whether VPS34-mediated autophagy is involved in Sp-1-mediated self-renewal using two other specific inhibitors, VPS34-PIK-Ⅲ and VPS34-IN-1. Unfortunately, both molecules failed to mimic the function of Sp-1 in sustaining AP activity in 46C mESCs in the presence of CHIR (Fig. S4), suggesting that Sp-1 promotes mESC self-renewal through a mechanism that bypasses autophagy.
Sp-1 inhibits the FGF/MEK/ERK signaling pathway by decreasing Raf1 levels
We performed high-throughput sequencing to evaluate the expression pattern regulated by CHIR or CS and to further explore the mechanism by which Sp-1 promotes mESC self-renewal. The addition of Sp-1 induced many differently expressed genes (DEGs), of which 428 genes were upregulated and 592 genes were downregulated by twofold or more (Fig. 4A). GO and KEGG analyses were carried out and identified many pluripotency associated genes induced by CS, such as Nanog, Esrrb, Tbx3, Mycn, and Otx2 (Fig. 4, B and C). Notably, the transcription of Socs3, a direct target of LIF/STAT3 signaling, was not changed. However, the expression of Egr1, a classical target of the FGF/MEK/ERK pathway, was significantly decreased and was validated by qRT-PCR (Fig. 4, B and C). This result promoted us to examine the activity of FGF/MEK/ERK signaling. As shown in Figure 4D, western blot results showed that the addition of Sp-1 did not change the total protein levels of MEK1/2 and ERK1/2, while the levels of phosphorylated MEK (P-MEK1/2) and ERK (P-ERK1/2) were reduced compared with those in the NT and CHIR groups (Fig. 4D), suggesting that the inactivation of MEK1/2 and ERK1/2 was not due to the decreased levels of the MEK1/2 and ERK1/2 proteins. Because USP13 is a deubiquitinated protein, we hypothesized that Sp-1 will accelerate the degradation of target proteins by inhibiting USP13. Hence, we focused on and detected level of the Raf1 protein, which is located upstream of MEK1/2 and controls its phosphorylation level, and found that Sp-1 significantly decreased Raf1 protein levels without significantly changing the expression of the Raf1 mRNA (Figs. 4D and S5A). Consistent with these findings, knockdown of USP13 but not USP10 exerted similar effects on levels of the Raf1 protein and phosphorylation of MEK1/2 and ERK1/2 without changing Raf1 mRNA levels (Figs. 4, E and F and S5B). Collectively, these results reveal that Sp-1 promotes mESC self-renewal by modulating Raf1 protein levels to inhibit the FGF/MEK/ERK signaling pathway.
Figure 4.
Sp-1 suppresses the FGF/MEK/ERK signaling pathway by decreasing the level of the Raf1 protein.A, volcano map showing the differentially expressed genes regulated by CHIR or CS. B, heat map shows the expression pattern of stem cell pluripotency-associated genes regulated by CHIR or CS. Genes were ranked at the level of a log2 fold change. C, qRT–PCR analysis of the expression of the indicated genes in cells exposed to CHIR or CS. The data are presented as the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus CHIR. D, Western blot analysis of Raf1, ERK1/2, P-ERK1/2, MEK1/2, and P-MEK1/2 protein levels in 46C mESCs treated with Sp-1 or/and CHIR for 24 h. E, Western blot analysis of Raf1, P-MEK1/2, MEK1/2, P-ERK1/2, ERK1/2, and Oct4 protein levels in 46C mESCs infected with scramble or USP10 shRNA lentiviruses. F, Western blot analysis of Raf1, P-MEK1/2, MEK1/2, P-ERK1/2, ERK1/2, and Oct4 protein levels in 46C mESCs infected with scramble or USP13 shRNA lentiviruses.
USP13 interacts with Raf1
A coimmunoprecipitation assay was performed in Flag-tagged Raf1 (Flag-Raf1)-overexpressing 46C mESCs carrying the HA or HA-USP13 transgene with a Flag antibody-affinity gel, which was used to bind the Flag fusion protein for immunoprecipitation analysis, to determine whether USP13 interacts with Raf1 (Fig. 5A). Ectopically expressed HA-USP13 was detected in Flag-Raf1 immunoprecipitants (Fig. 5A). Consistent with this result, endogenous USP13 was present in endogenous Raf1 immunoprecipitants (Fig. 5B). Nuclear and cytoplasmic separation and immunofluorescence staining showed that Raf1 was located in the cytoplasm, while USP13 was distributed in both the cytoplasm and nucleus (Fig. 5, C and D). Meanwhile, the results from the pull-down assay showed that purified GST-USP13 directly interacted with endogenous Raf1 in 46C ESC lysates (Fig. 5E). Together, these results suggest that USP13 and Raf1 have a direct interaction in the cytoplasm.
Figure 5.
USP13 interacts with Raf1.A, 46C cells cotransfected with Flag-Raf1 and HA-USP13 were subjected to immunoprecipitation (IP) with anti-Flag affinity gel. The immunoprecipitants were analyzed using western blot. B, endogenous USP13 was coimmunoprecipitated with a Raf1 antibody in mESCs. C, Western blot analysis of the subcellular localization of HA-USP13 and Raf1 proteins in mESCs. D, immunofluorescence staining for HA (green) and Flag (red) in mESCs transfected with HA-USP13 and Flag-Raf1. Scale bar, 100 μM. E, Raf1 proteins pulled-down by GST-USP13 were detected using immunoblotting. Purified GST or GST-USP13 was visualized using western blot analysis with the GST antibody.
USP13 deubiquitinates Raf1
We aimed to determine whether USP13 regulates the stability of the Raf1 protein via deubiquitylation, and thus 46C mESCs were treated with cycloheximide (CHX) to block protein synthesis in the presence or absence of MG132, a potent proteasome inhibitor. The half-life analysis indicated that the Raf1 protein was much more stable in MG132-treated cells (Fig. 6A). As a key target of Sp-1 and a deubiquitinating enzyme, ectopically expressed USP13 also led to an obvious increase in the stability of the endogenous Raf1 protein (Fig. 6, B and C). Moreover, the addition of MG132 further increased Raf1 levels in cells cultured with CS-containing medium (Fig. 6D), suggesting that Raf1 protein degradation occurs through the proteasomal pathway. Supporting these observations, the ubiquitylation of Flag-tagged Raf1 was markedly increased after CS supplementation (Fig. 6E), while ectopically expressed USP13 significantly reduced the level of Raf1 ubiquitination (Fig. 6F). Notably, USP13 efficiently removed the K48-linked polyubiquitylation of Raf1 (Fig. 6G). The abovementioned results indicated that USP13 deubiquitinates Raf1 to increase its stability, while Sp-1 antagonizes USP13 function and promotes Raf1 degradation.
Figure 6.
USP13 stabilizes the Raf1 protein.A, Western blot analysis of Raf1 protein levels in mESCs treated with 50 μM cycloheximide (CHX) in the presence or absence of 20 μM MG132. B, Western blot analysis of Raf1 protein levels in mESCs transfected with HA or HA-USP13 in the presence of CHX. C, the half-life curves of the Raf1 protein corresponding to Figure 6B. The data are presented as the mean ± SD (N = 3 biological replicates). ∗∗p < 0.01 versus HA. D, Western blot analysis of Raf1 protein levels in 46C mESCs treated with CS in the presence or absence of MG132 for 24 h. E, mESCs cotransfected with Flag-Raf1 and ubiquitin B (Ubb) were treated with or without CS for 24 h. Flag-Raf1 was immunoprecipitated with an anti-Flag affinity gel and assessed using an anti-Ub antibody. F, flag-Raf1 and Ubb were cotransfected in mESCs overexpressing HA or HA-USP13. These cells were subjected to IP with an anti-Flag affinity gel. The immunoprecipitants were analyzed by western blot with an anti-Ub antibody. G, the Raf1 ubiquitylation linkage was analyzed in 46C mESCs transfected with Raf1, USP13, and the ubiquitin Lys 48-only plasmid in which all sites of Lys were mutated but not site 48.
Raf1 is capable of mediating the self-renewal-promoting effect of Sp-1
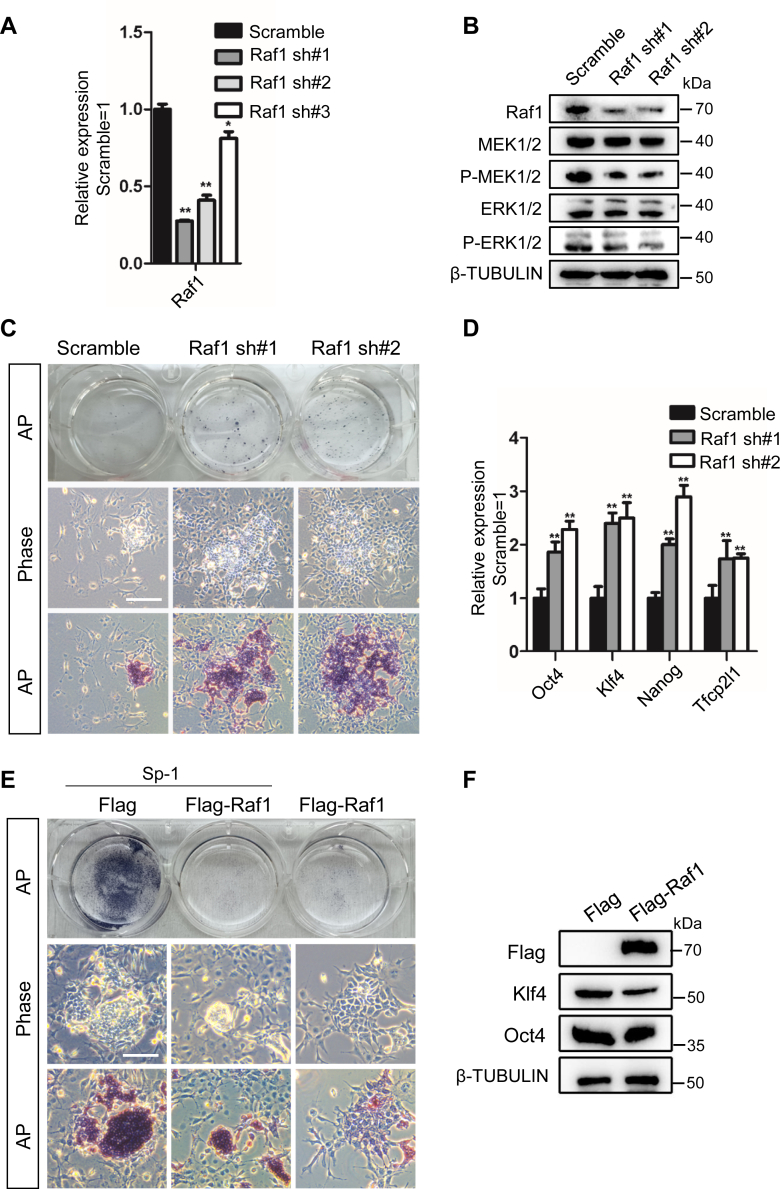
We first infected 46C ESCs with a scramble control shRNA or three shRNAs designed to decrease endogenous Raf1 expression (Raf1 sh#1, sh#2, and sh#3) to ascertain whether Raf1 is indispensable for Sp-1-mediated mESC self-renewal. Raf1 expression was reduced by 60%–70%, as measured using qRT–PCR in Raf1 sh#1 and Raf1 sh#2-expressing cells (Fig. 7A). As expected, Raf1 knockdown inhibited the phosphorylation of MEK and ERK (Fig. 7B). Moreover, Raf1 knockdown efficiently delayed the differentiation of mESCs cultured in serum-containing medium in the absence of LIF (Fig. 7C). After 8 days, they displayed stronger AP activity and higher levels of the self-renewal markers Oct4, Nanog, Klf4, and Tfcp2l1 (Fig. 7, C and D). Next, Flag-tagged Raf1 was overexpressed in 46C mESCs with the PB system (Flag-Raf1). In the presence of Sp-1, Flag-Raf1-expressing mESCs generated fewer AP-positive colonies than empty vector control (Flag)-transfected cells (Fig. 7E). They thus expressed lower levels of the pluripotency markers Oct4 and Klf4 than Flag-transfected cells (Fig. 7F), indicating that Raf1 overexpression impairs the function of Sp-1 in promoting mESC self-renewal.
Figure 7.
Raf1 is required for Sp-1 to promote mESC self-renewal.A, qRT–PCR analysis of Raf1 expression levels in scramble control and Raf1 shRNA-expressing mESCs cultured with serum-containing medium in the presence of LIF. The data are presented as the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus scramble. B, Western blot analysis of Raf1, P-MEK1/2, MEK1/2, P-ERK1/2, and ERK1/2 protein levels in 46C mESCs infected with scramble or Raf1 shRNA lentiviruses. C, morphology and AP staining of scramble and Raf1 shRNA-expressing cells cultured in serum-containing medium in the absence of LIF for 8 days. Scale bar, 100 μM. D, qRT–PCR analysis of the expression of the Oct4, Nanog, Klf4, and Tfcp2l1 in scramble and Raf1 shRNA-expressing cells without LIF treatment. The data are presented as the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus scramble. E, morphology and AP staining of empty vector control or Raf1 overexpressing cells treated with or without Sp-1 for 8 days. Scale bar, 100 μM. F, Western blot analysis of Flag, Oct4, and Klf4 levels in empty vector or Raf1 overexpressing cells.
Sp-1 reduces mESC proliferation
Although Sp-1 was beneficial to the maintenance of the stemness of mESCs in vitro, we also noticed that the proliferation rate of the cells slowed down (Fig. S6, A and B). Flow cytometry was then applied to analyze the distribution of the cell cycle and indicated that the addition of Sp-1 prolonged G1 phase from 20.84% to 53.74% (Fig. S6C). In fact, a KEGG analysis of the RNA-sequencing data showed that 65 DEGs regulated by Sp-1 were enriched in the cell cycle and 42 DEGs were closely associated with the P53 signaling pathway, both of which are essential for the regulation of cell cycle arrest (Fig. S6D). qRT–PCR validation also showed that the expression of some genes that positively modulate the G1 phase was reduced, such as CDK1, CDK4, and c-Myc (Fig. S6E), whereas genes that restrict cell proliferation were increased, such as Gadd45b and P53 (Fig. S6E). Their protein levels also changed consistently (Fig. S6F). Collectively, these results imply that Sp-1 limits mESC proliferation through tight regulation of cell-cycle-associated genes.
USP13 facilitates mEpiSC and hiPSC self-renewal
Unlike mESCs, mEpiSCs and human pluripotent stem cells rely on the FGF/MEK/ERK signaling pathway to maintain an undifferentiated state (20). As USP13 increases the stability of the Raf1 protein, we inferred that USP13 may favor the maintenance of primed pluripotency. We first overexpressed the HA-tagged mouse USP13 gene in mEpiSCs and cultured these cells in serum-containing medium without Activin A and bFGF to validate this hypothesis (Fig. S7A). Upregulation of USP13 delayed the differentiation of mEpiSCs and sustained the expression of the pluripotency genes Oct4, Sox2, and Nanog (Fig. S7, B–D). However, the addition of the MEK inhibitor PD03 substantially eliminated the effect of USP13 and rapidly induced cell differentiation (Fig. S7, B–D). Thus, USP13 promotes mEpiSC self-renewal in an MEK-dependent manner. Then, we wanted to examine transgene-free hiPSCs. First, Sp-1 was added to routine hiPSC culture medium. After 10 days, hiPSCs exhibited a robust self-renewal ability in the absence of Sp-1, while Sp-1-treated hiPSCs became flat and lost AP activity (Fig. 8A). Next, FLAG tagged human USP13 (FLAG-hUSP13) was overexpressed efficiently in hiPSCs with PB (Fig. 8B). Enforced expression of hUSP13 increased the levels of P-MEK1/2 and P-ERK1/2 (Fig. 8C). After seeding in hiPSC basal medium without Activin A and bFGF, USP13 overexpressing hiPSCs expressed OCT4, PRDM14, SOX2, and NANOG at higher levels (Fig. 8, D and E), but exhibited a slower differentiation rate than PB control cells (Fig. 8F). Finally, we determined whether USP13 associates with and deubiquitinates human RAF1. HA-labeled human RAF1 (HA-hRAF1) and FLAG-hUSP13 were overexpressed in hiPSCs together. Co-IP was performed and showed that RAF1 interacted with USP13 in hiPSCs (Fig. 8G). Finally, the ubiquitination of HA-hRAF1 was examined in FLAG-hUSP13-expressing hiPSCs and the western blot results showed that USP13 suppressed RAF1 ubiquitination (Fig. 8H). Taken together, Sp-1 inhibits mEpiSC and hiPSC self-renewal, while USP13, an Sp-1 substrate, exerts the opposite function.
Figure 8.
USP13 overexpression promotes hiPSC self-renewal.A, morphology and AP staining of hiPSCs cultured with ncTarget medium, in the presence or absence of 8 μM Sp-1 for 10 days. Scale bar, 100 μM. B, Western blot analysis of human RAF1 and FLAG levels in FLAG and FLAG-hUSP13-expressing hiPSCs. C, Western blot analysis of MEK1/2, P-MEK1/2, P-ERK1/2, and ERK1/2 protein levels in FLAG and FLAG-hUSP13-expressing hiPSCs cultured in hiPSC routine medium. D, immunofluorescence staining for OCT4 in FLAG and FLAG-hUSP13-expressing hiPSCs cultured in N2B27/KSR medium for 10 days. Scale bar, 100 μM. E, qRT–PCR analysis of the expression of the OCT4, PRDM14, SOX2, and NANOG in FLAG and FLAG-hUSP13-expressing hiPSCs cultured in hiPSC basal medium. The data are presented as the mean ± SD (N = 3 biological replicates). ∗p < 0.05, ∗∗p < 0.01 versus FLAG. F, morphology and AP staining of FLAG and FLAG-hUSP13-expressing hiPSCs cultured in N2B27/KSR medium for 10 days. Scale bar, 100 μM. G, HA-hRAF1 hiPSCs overexpressed FLAG-hUSP13. Co-IP of FLAG-hUSP13 using the anti-FLAG affinity gel. The association of hUSP13 and hRAF1 was analyzed by western blot using an HA antibody. H, Co-IP of HA-hRAF1 in Ubb/HA-hRAF1-expressing hiPSCs transfected with FLAG or FLAG-hUSP13. The ubiquitylated hRAF1 protein was detected using the Ub antibody.
Discussion
Our research has identified that Sp-1 promotes the self-renewal of mESCs but induces mEpiSC and human iPSC differentiation. Sp-1 mainly functions by inhibiting the deubiquitinating protein USP13, which interacts with and thereby deubiquitinates the Raf1 protein in the cells; the latter is a key component in the activation of MEK/ERK signaling (Fig. 9). Therefore, Sp-1 plays different roles in the naïve and primed pluripotent states by inducing the degradation of the Raf1 protein to repress the FGF/MEK/ERK signaling pathway.
Figure 9.
Diagram of the working model of Sp-1 functions in pluripotent stem cells. Schematic diagram of Sp-1 input to the naïve and primed pluripotency network. Sp-1 integrates USP13 into the FGF/MEK/ERK signal pathway to regulate the self-renewal and differentiation of mouse ESCs, mEpiSCs and hiPSCs through the posttranslational modification of Raf1 protein stability.
Protein ubiquitination is a reversible process catalyzed by ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin-protein ligase (E3), while ubiquitination is removed from the ubiquitinated substrates by deubiquitinating enzymes (DUBs) (21). The most important discovery of our project is the identification of USP13 as a modulator of ESC identity (Figs. 1, 2 and 8). Many DUB proteins are expressed in ESCs, and they are important for balancing self-renewal and differentiation. For example, USP21 is critical for maintaining the undifferentiated state of mESCs by deubiquitinating Nanog and facilitates chromatin remodeling by Nanog to stimulate gene transcription (22). The specific deubiquitylation site of USP21 may be lys48, which links the polyubiquitination chains to Nanog (23). Depletion of USP21 induces Nanog degradation and mESC differentiation (22). USP4 also removes inhibitory monoubiquitylation from SMAD4 to enhance BMP signaling, which is beneficial for mESC self-renewal (4, 24). In addition, USP8 binds to the coiled-coil domain of EPG5 and directly removes nonclassical K63-linked ubiquitin chains from EPG5 at lysine 252 to preserve autophagy flux in ESCs and maintain stemness (25). On the other hand, USP22 promotes the mouse and human ESC transition from self-renewal to differentiation by repressing Sox2 transcription through deubiquitylation of histone H2B at the Sox2 locus (26). Monoubiquitylation of H2B on lysine 120 (H2Bub1) increases during mouse and human ESC differentiation and is critical for the occurrence of this process (27), while USP44 is a negative regulator of H2B ubiquitylation, and its downregulation during ESC differentiation promotes H2Bub1 upregulation (27). Here, we revealed the function of another DUB member, USP13, in mouse ESCs and human iPSCs (Figs. 1 and 8). These results enriched the regulatory network between DUB and ESCs. USP13 is mainly involved in autophagy. It forms a deubiquitylation complex with NEDD4-1 to decrease the K48-linked ubiquitination of VPS34 and PIK3C3 and promote autophagy flux (28, 29). We also showed that USP13 directly removes classical K48-linked ubiquitin chains from the Raf1 protein (Fig. 6G), while inhibition of USP13 by Sp-1 promotes mESC self-renewal in an autophagy-independent manner (Fig. S4). Additionally, USP13 enhances the protein stability of many mESCs-associated factors in vivo, such as c-Myc, Mcl-1, and PTEN, to control tumorigenesis (30, 31, 32). It stabilizes c-Myc by antagonizing FBXL14-mediated ubiquitination to maintain glioma stem cell self-renewal and tumorigenic potential (33). Similarly, USP13 represents as a potential oncogene in cervical cancer that functions to deubiquitinate and stabilize the prosurvival protein Mcl-1 (34). In contrast, USP13 directly binds and deubiquitylates PTEN to suppress tumorigenesis and glycolysis in PTEN-positive breast cancer cells (35). Studies investigating whether these substrates are engaged in Sp-1-mediated ESC self-renewal and whether USP13-mediated Raf1 deubiquitylation functions in these biological processes rather than in ESC maintenance will be interesting.
Another key discovery of our study is that we revealed for the first time the opposite function of USP13 in mouse and human ESC maintenance by deubiquitinating Raf1. Interestingly, USP10 overexpression promotes an increase in Raf1 protein levels through deubiquitylation to promote the proliferation and migration of endometrial stromal cells (36). Furthermore, the deubiquitylation of the pluripotency proteins c-Myc and Klf4 is also induced by USP10 in human cancer cells (37, 38, 39). Sp-1 simultaneously inhibits the activity of USP10 and USP13 (19). However, knockdown of USP10 had no effect on the Raf1 protein or the total protein and phosphorylation levels of MEK and ERK (Fig. 4E), suggesting that USP10 may not function similarly to USP13 in mESCs and hiPSCs (Figs. 3H and 8, D–F). USP13 knockdown mimicked the function of Sp-1 in mESCs by promoting Raf1 protein degradation (Fig. 4, D and F). These differences may be due to the use of different experimental animal models and cell types. Raf1 is a critical bridge that transmits external signals to the cytoplasm and nucleus via the FGF/MEK/ERK pathway. Upon FGF cytokines binding to FGF receptor, Grb2-SOS complexes are recruited and then fix Ras protein to the cell membrane; the latter is activated and interacts with Raf1. The catalytic domain of Raf1 appears to activate the MEK/ERK axis (40). Raf1 determines the hierarchical regulation of the FGF/MEK/ERK signaling pathway (40), which is one of the key pathways that distinguishes and establishes naïve and primed pluripotency (13, 41). FGF4 produced by mESCs is important to initiate differentiation by increasing ERK1/ERK2 signaling (17). Inhibition of FGF/MEK/ERK signaling using small molecules thus increases mESC stability and stemness (6, 17). In contrast, activation of this signaling pathway is necessary for maintenance of the undifferentiated state of mEpiSCs, human ESCs, and hiPSCs (9, 10, 11, 12). This information also explains why we observed that USP13 is not conducive to the maintenance of mESC stemness but promotes the self-renewal of mEpiSCs and hiPSCs (Figure 3, Figure 8 and S7). Notably, although both Sp-1 and PD0325901 inhibited the activity of the FGF/MEK/ERK signaling pathway (Fig. 4D) (5), a difference was observed between the CS and 2i conditions in maintaining ESC stemness, as USP13 has multiple substrates in addition to Raf1, and their sublocalization in the cells is not exactly the same (Fig. 5D). Moreover, Sp-1 functions by inhibiting the activity of USP10 and USP13, while PD0325901 is a specific inhibitor of MEK (5, 19). USP10 and USP13 participate in the biological regulation of autophagy (19), which has been reported to be involved in regulating ESC identity by modulating metabolism and epigenetic events (25, 42). Therefore, the growth rate of cells under CS culture conditions was slow (Figs. 2A and S6). Further examination of the advantages and disadvantages of using Sp-1 or the MEK inhibitor PD0325901 in naïve ESCs is required.
In summary, we reported that Sp-1 promotes the maintenance of naïve but not primed pluripotency in vitro and further identified USP13 as a novel deubiquitinase for Raf1. These results not only revealed a new regulatory network in ESCs, enabling us optimize the current stem cell culture conditions for different pluripotent states, but also provided new insights into developmental biology and tumor biology research because Raf1 is an evolutionarily conserved protein.
Experimental procedures
Cell culture
46C mESCs, provided by Qi-Long Ying (University of Southern California, USA), were routinely cultured in DMEM supplemented with 10% FBS (FND500, ExCell Bio), 1nonessential amino acids (N1250, Solarbio), 0.1 mM β-mercaptoethanol (M3148, Sigma), and 1000 U/ml LIF (made in house). HEK293 T cells were also cultured in DMEM medium supplemented with 10% FBS (FB25015, CLARK Bioscience). Human transgene-free iPSCs were kindly provided by NuwaCell Ltd (ZSSY-001) and were cultured in ncTarget medium (RP01020, NuwaCell.Ltd). CD1 mouse EpiSCs were cultured in mESC basal medium supplemented with bFGF (AF-100-18B, Peprotech), Activin A (120-14E, Peprotech), and 4 μM IWR-1 (HY-12238, MedChemExpress).
Plasmid construction
The full-length code sequences of USP10, USP13, Raf1, and Ubb were subcloned into PiggyBac (PB) transposon vectors. The target sequences of shRNA were synthesized, annealed, and inserted into the pLKO.1-TRC vector (Addgene plasmid #10878). The target sequences are listed in Table S1.
Alkaline phosphatase staining
Alkaline phosphatase staining was carried out by using the alkaline phosphatase kit (C3206, Beyotime Biotechnology). First, cells were fixed in 4% paraformaldehyde for 2 min. Subsequently, alkaline phosphatase mixed solution was added to the cells. After 30 min, the alkaline phosphatase-positive colonies were observed under a Leica DMI8 microscope.
EB differentiation
Spontaneous differentiation was induced by embryoid body (EB) formation for 8 days and further plating of EBs on gelatin-coated plates and cultured for an additional 8 days in mESC media without LIF to examine the pluripotency of mESCs sustained by CS conditions.
Western blot
The cells were lysed in ice-clod RIPA cell buffer (P0013B, Beyotime Biotechnology). The total protein was separated by 10% SDS-PAGE and was transferred into PVDF membrane. The primary antibodies were incubated for overnight at 4 °C. They were USP13 (R26051, ZENBIO, 1:500), Raf1 (251817, ZENBIO, 1:1000), Phospho-MEK1/2 (Ser217/221) (310050, ZENBIO, 1:1000), MEK1/2 (380797, ZENBIO, 1:1000), Phospho-ERK1 (Thr202/Tyr204)/ERK2 (Thr185/Tyr187) (301245, ZENBIO, 1:500), Klf4 (381633, ZENBIO, 1:1000), c-Myc (382809, ZENBIO, 1:1000), HA (3724, Cell Signaling Technology, 1:1000), Flag (SG110-26, GNI, 1:1000), Ub (SC-8017, Santa Cruz, 1:1000), Oct4 (SC-5279, Santa Cruz, 1:1000), Nanog (14295-1-AP, Proteintech, 1:1000), Sox2 (66411-1-Ig, Proteintech, 1:1000), P53 (345567, ZENBIO, 1:1000), CDK4 (11026-1-AP, Proteintech, 1:1000), β-TUBULIN (200608, ZENBIO, 1:2000), and CDK1 (19532-1-AP, Proteintech, 1:1000). The bands were analyzed with High-sig ECL kit (180-501, Tanon).
Quantitative RT-PCR (qRT-PCR)
The total RNA in the cells was isolated with the EZ-10 RNA extraction kit (B610583, BBI), and cDNA was synthesized using the with Reverse Transcription (with dsDNase) (BL699A, Biosharp) according to the manufacturer's instructions. Rapid amplification of cDNA by a two-step method using Hieff qPCR SYBR Green Master Mix (11201ES03, YEASEN) in a PikoReal Real-Time PCR machine (Thermo Scientific). The primers were listed in the Table S2.
Coimmunoprecipitation assay
Cells were lysed in NP-40 lysis buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, and protease inhibitors). The cell lysate (500 μl) was centrifuged at 12,000 rpm for 15 min at 4 °C. The supernatant was immunoprecipitated with 10 μl of anti-FLAG affinity gel (450-FG, GNI) or anti-HA affinity gel (450-HA, GNI) overnight at 4 °C. For the coimmunoprecipitation of endogenous proteins, the specific antibody of Raf1 was used and followed by adding Protein A/G. The precipitate was washed with PBS for three times and then was resolved in 1 × SDS loading buffer.
Ubiquitylation assay
Flag tagged Raf1, HA tagged USP13, and Ubb were cotransfected into cells. The target proteins were purified by immunoprecipitation with an anti-Flag affinity gel. The purified protein was analyzed by western blotting using an anti-ubiquitin antibody.
Immunofluorescence staining
The cells were fixed with 4% paraformaldehyde for 20 min and were blocked in blocking solution containing 5% BSA and 0.02% Triton X-100 for 2 h at 37 °C. The primary antibodies were Flag (SG110-26, GNI, 1:500), HA (3724, Cell Signaling Technology, 1:500), Oct4 (SC-5279, Santa Cruz, 1:500), H3K27me3 (R26242, ZENBIO, 1:200), Gata6 (5851T, Cell Signaling Technology, 1:300), Myosin (ab50967, Abcam, 1:100), and Tuj1 (MAB1195, R&D, 1:100). The nuclei were stained with Hoechst 33342 (H3570, Invitrogen, 1:10,000).
Nuclear and cytoplasmic separation
A total of 1 × 106 cells were grown in a 6 cm dish and digested with trypsin. After centrifugation at 1000 rpm for 3 min, the supernatant was discarded and resuspended in precooled PBS. Nuclear and cytoplasmic extraction reagents (78833, Thermo Scientific) were used to prepare nuclear and cytoplasmic extracts according to the manufacturer's instructions.
Cell proliferation assay
The cells were seeded in a 96-well plate in serum-containing medium. The rate of cell growth was detected every 12 h with Cell Counting Kit-8 reagent (K1085, APE x BIO). In total, 10 μl of CCK8 reagent was added to each well. The OD450nm value was measured using a microplate reader (Molecular Devices, SpectraMax).
Cell cycle assays
Cells were prepared according to the instructions of the cell cycle and apoptosis kit (C1052, Beyotime Biotechnology) to detect the cell cycle distribution. Cells were washed twice with ice-cold PBS. After fixation with ice-cold 70% ethanol overnight, the cells were washed with ice-cold PBS and stained with propidium iodide for 30 min. The cell cycle was analyzed using flow cytometry.
GST pull-down assay
GST-tagged mouse USP13 was inserted into the vector PGEX-4T, which was transformed into E. coli BL21, and protein expression was induced with 0.6 mM isopropyl-β-D-thiogalactopyranoside and then purified using glutathione Sepharose 4B. The purified proteins were validated using western blot. A GST pull-down assay was performed using a GST Protein Interaction Pull-Down Kit (21516, Pierce) according to the manufacturer’s instructions. Briefly, GST or the recombinant GST-USP13 protein was conjugated to glutathione agarose resin, followed by incubation with 46C mESC lysates. The immobilized proteins were then washed, eluted, and subjected to western blot analysis with GST and Raf1 antibodies.
Chimera generation
The method of microinjection was described in our previous report (18). Briefly, 12 to 15 single cells were injected into 3.5 dpc blastocysts collected from B6(Cg)-Tyrc-2J/J mice (The Jackson Laboratory, 000058). The injected blastocysts were then transferred into the uterine horns of pseudopregnant female mice. The chimeric offspring were identified by coat color.
Accession number
Our Microarray data set has been deposited in the GEO database under ID number GSE172469.
Statistical analysis
All data are reported as the mean ± SD. Student's t test is used to determine the significance of the comparison difference by using GraphPad Prism 8 software. Values with p < 0.05 are considered statistically significant.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank everyone in the Ye lab for their technical help. All animal experiments were conducted in accordance with an approved protocol and carried out according to the institutional animal welfare guidelines of the Anhui University (Protocol No: 2021-029). This work was supported by the Natural Science Foundation of Anhui Province [1908085J13], the Anhui Provincial Key Research and Development Plan [202104b11020026], the University Synergy Innovation Program of Anhui Province [GXXT-2020-064], the Open Fund for Discipline Construction, Institute of Physical Science and Information Technology, Anhui University [S01003106], and the Funding supported by the Department of Education of Anhui Province and the Department of Human Resources and Social Security of Anhui Province [gxyqZD2020001 and 2020H210].
Author contributions
Xin Wang, X. Z., and S.-D. Y. conceptualization; Y. L., M. Z., and J. J. formal analysis; Xin Wang, Xiaoxiao Wang, X. Z., Y. Z., Z. Z., and Y. Y. investigation; Xiaoxiao Wang, Y. Z., Z. Z., and Y. Y. methodology; S.-D. Y. supervision; Y. L., M. Z., and J. J. validation; Xiaoxiao Wang, Z. Z., and Y. Y. visualization; Xin Wang, writing–original draft; S.-D. Y. writing–review and editing.
Edited by George DeMartino
Supporting information
References
- 1.Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U. S. A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith A.G., Heath J.K., Donaldson D.D., Wong G.G., Moreau J., Stahl M., Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 4.Ying Q.L., Nichols J., Chambers I., Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 5.Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., McLay R., Hall J., Ying Q.L., Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Li P., Tong C., Mehrian-Shai R., Jia L., Wu N., Yan Y., Maxson R.E., Schulze E.N., Song H., Hsieh C.L., Pera M.F., Ying Q.L. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daheron L., Opitz S.L., Zaehres H., Lensch M.W., Andrews P.W., Itskovitz-Eldor J., Daley G.Q. LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells. 2004;22:770–778. doi: 10.1634/stemcells.22-5-770. [DOI] [PubMed] [Google Scholar]
- 9.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., Slukvin I.I., Thomson J.A. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 10.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 11.Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 12.Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Richter L., Pedersen R.A., Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 13.Huang G., Ye S., Zhou X., Liu D., Ying Q.L. Molecular basis of embryonic stem cell self-renewal: From signaling pathways to pluripotency network. Cell Mol. Life Sci. 2015;72:1741–1757. doi: 10.1007/s00018-015-1833-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Gottgens B., Niwa H., Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson K.C., Adams A.M., Goodson J.M., McDonald C.E., Potter J.C., Berndt J.D., Biechele T.L., Taylor R.J., Moon R.T. Wnt/beta-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4485–4490. doi: 10.1073/pnas.1118777109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H., Wu J., Ye S., Tai C.I., Zhou X., Yan H., Li P., Pera M., Ying Q.L. Modulation of beta-catenin function maintains mouse epiblast stem cell and human embryonic stem cell self-renewal. Nat. Commun. 2013;4:2403. doi: 10.1038/ncomms3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunath T., Saba-El-Leil M.K., Almousailleakh M., Wray J., Meloche S., Smith A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Z., Zhang Y., Wang X., Wang X., Ye S.D. Inhibition of protein kinase D by CID755673 promotes maintenance of the pluripotency of embryonic stem cells. Development. 2020 doi: 10.1242/dev.185264. [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Xia H., Kim M., Xu L., Li Y., Zhang L., Cai Y., Norberg H.V., Zhang T., Furuya T., Jin M., Zhu Z., Wang H., Yu J., Li Y. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greber B., Wu G., Bernemann C., Joo J.Y., Han D.W., Ko K., Tapia N., Sabour D., Sterneckert J., Tesar P., Scholer H.R. Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell. 2010;6:215–226. doi: 10.1016/j.stem.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 21.Mevissen T.E.T., Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 22.Jin J., Liu J., Chen C., Liu Z., Jiang C., Chu H., Pan W., Wang X., Zhang L., Li B., Jiang C., Ge X., Xie X., Wang P. The deubiquitinase USP21 maintains the stemness of mouse embryonic stem cells via stabilization of Nanog. Nat. Commun. 2016;7:13594. doi: 10.1038/ncomms13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon S.K., Lee D.H., Kim S.Y., Park J.H., Choi J., Baek K.H. Ubiquitin-specific protease 21 regulating the K48-linked polyubiquitination of NANOG. Biochem. Biophys. Res. Commun. 2017;482:1443–1448. doi: 10.1016/j.bbrc.2016.12.055. [DOI] [PubMed] [Google Scholar]
- 24.Zhou F., Xie F., Jin K., Zhang Z., Clerici M., Gao R., van Dinther M., Sixma T.K., Huang H., Zhang L., Ten Dijke P. USP4 inhibits SMAD4 monoubiquitination and promotes activin and BMP signaling. EMBO J. 2017;36:1623–1639. doi: 10.15252/embj.201695372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu H., Shi X., Liu C., Wang C., Sui N., Zhao Y., Gong J., Wang F., Zhang H., Li W., Zhao T. USP8 maintains embryonic stem cell stemness via deubiquitination of EPG5. Nat. Commun. 2019;10:1465. doi: 10.1038/s41467-019-09430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sussman R.T., Stanek T.J., Esteso P., Gearhart J.D., Knudsen K.E., McMahon S.B. The epigenetic modifier ubiquitin-specific protease 22 (USP22) regulates embryonic stem cell differentiation via transcriptional repression of sex-determining region Y-box 2 (SOX2) J. Biol. Chem. 2013;288:24234–24246. doi: 10.1074/jbc.M113.469783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchs G., Shema E., Vesterman R., Kotler E., Wolchinsky Z., Wilder S., Golomb L., Pribluda A., Zhang F., Haj-Yahya M., Feldmesser E., Brik A., Yu X., Hanna J., Aberdam D. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol. Cell. 2012;46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie W., Jin S., Wu Y., Xian H., Tian S., Liu D.A., Guo Z., Cui J. Auto-ubiquitination of NEDD4-1 recruits USP13 to facilitate autophagy through deubiquitinating VPS34. Cell Rep. 2020;30:2807–2819e4. doi: 10.1016/j.celrep.2020.01.088. [DOI] [PubMed] [Google Scholar]
- 29.Xie W., Jin S., Cui J. The NEDD4-USP13 axis facilitates autophagy via deubiquitinating PIK3C3. Autophagy. 2020;16:1150–1151. doi: 10.1080/15548627.2020.1743071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cartwright P., McLean C., Sheppard A., Rivett D., Jones K., Dalton S. LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development. 2005;132:885–896. doi: 10.1242/dev.01670. [DOI] [PubMed] [Google Scholar]
- 31.Huskey N.E., Guo T., Evason K.J., Momcilovic O., Pardo D., Creasman K.J., Judson R.L., Blelloch R., Oakes S.A., Hebrok M., Goga A. CDK1 inhibition targets the p53-NOXA-MCL1 axis, selectively kills embryonic stem cells, and prevents teratoma formation. Stem Cell Rep. 2015;4:374–389. doi: 10.1016/j.stemcr.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W., Lu G., Su X., Tang C., Li H., Xiong Z., Leung C.K., Wong M.S., Liu H., Ma J.L., Cheung H.H., Kung H.F., Chen Z.J., Chan W.Y. Pten-mediated Gsk3beta modulates the naive pluripotency maintenance in embryonic stem cells. Cell Death Dis. 2020;11:107. doi: 10.1038/s41419-020-2271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang X., Zhou W., Wu Q., Huang Z., Shi Y., Yang K., Chen C., Xie Q., Mack S.C., Wang X., Carcaboso A.M., Sloan A.E., Ouyang G., McLendon R.E., Bian X.W. Deubiquitinase USP13 maintains glioblastoma stem cells by antagonizing FBXL14-mediated Myc ubiquitination. J. Exp. Med. 2017;214:245–267. doi: 10.1084/jem.20151673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan E.L., Patterson M.R., Barba-Moreno D., Scarth J.A., Wilson A., Macdonald A. The deubiquitinase (DUB) USP13 promotes Mcl-1 stabilisation in cervical cancer. Oncogene. 2021;40:2112–2129. doi: 10.1038/s41388-021-01679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Zhang P., Wei Y., Piao H.L., Wang W., Maddika S., Wang M., Chen D., Sun Y., Hung M.C., Chen J., Ma L. Deubiquitylation and stabilization of PTEN by USP13. Nat. Cell Biol. 2013;15:1486–1494. doi: 10.1038/ncb2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Q., Hang Y., Zhang T., Tan L., Li S., Jin Y. USP10 promotes proliferation and migration and inhibits apoptosis of endometrial stromal cells in endometriosis through activating the Raf-1/MEK/ERK pathway. Am. J. Physiol. Cell Physiol. 2018;315:C863–C872. doi: 10.1152/ajpcell.00272.2018. [DOI] [PubMed] [Google Scholar]
- 37.Ko A., Han S.Y., Choi C.H., Cho H., Lee M.S., Kim S.Y., Song J.S., Hong K.M., Lee H.W., Hewitt S.M., Chung J.Y., Song J. Oncogene-induced senescence mediated by c-Myc requires USP10 dependent deubiquitination and stabilization of p14ARF. Cell Death Differ. 2018;25:1050–1062. doi: 10.1038/s41418-018-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Z., Yang H., Tan C., Li J., Liu Z., Quan Q., Kong S., Ye J., Gao B., Fang D. USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep. 2013;5:1639–1649. doi: 10.1016/j.celrep.2013.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., Xia S., Li H., Wang X., Li C., Chao Y., Zhang L., Han C. The deubiquitinase USP10 regulates KLF4 stability and suppresses lung tumorigenesis. Cell Death Differ. 2020;27:1747–1764. doi: 10.1038/s41418-019-0458-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dailey L., Ambrosetti D., Mansukhani A., Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Ye S., Liu D., Ying Q.L. Signaling pathways in induced naive pluripotency. Curr. Opin. Genet. Dev. 2014;28:10–15. doi: 10.1016/j.gde.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Y., Zhang Y., Garcia-Canaveras J.C., Guo L., Kan M., Yu S., Blair I.A., Rabinowitz J.D., Yang X. Chaperone-mediated autophagy regulates the pluripotency of embryonic stem cells. Science. 2020;369:397–403. doi: 10.1126/science.abb4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.