Abstract
Pecan (Carya illinoinensis) is a tree nut crop of worldwide economic importance that is rich in health-promoting factors. However, pecan production and nut quality are greatly challenged by environmental stresses such as the outbreak of severe fungal diseases. Here, we report a high-quality, chromosome-scale genome assembly of the controlled-cross pecan cultivar ‘Pawnee’ constructed by integrating Nanopore sequencing and Hi-C technologies. Phylogenetic and evolutionary analyses reveal two whole-genome duplication (WGD) events and two paleo-subgenomes in pecan and walnut. Time estimates suggest that the recent WGD event and considerable genome rearrangements in pecan and walnut account for expansions in genome size and chromosome number after the divergence from bayberry. The two paleo-subgenomes differ in size and protein-coding gene sets. They exhibit uneven ancient gene loss, asymmetrical distribution of transposable elements (especially LTR/Copia and LTR/Gypsy), and expansions in transcription factor families (such as the extreme pecan-specific expansion in the far-red impaired response 1 family), which are likely to reflect the long evolutionary history of species in the Juglandaceae. A whole-genome scan of resequencing data from 86 pecan scab-associated core accessions identified 47 chromosome regions containing 185 putative candidate genes. Significant changes were detected in the expression of candidate genes associated with the chitin response pathway under chitin treatment in the scab-resistant and scab-susceptible cultivars ‘Excell’ and ‘Pawnee’. These findings enable us to identify key genes that may be important susceptibility factors for fungal diseases in pecan. The high-quality sequences are valuable resources for pecan breeders and will provide a foundation for the production and quality improvement of tree nut crops.
Key words: pecan, genome assembly, paleo-subgenome, pecan scab, fungal disease, population genetics
A high-quality, chromosome-scale reference genome of pecan reveals two paleo-subgenomes that show asymmetry in their features and evolution. Resequencing of pecan scab-associated core accessions identified several key genes in the chitin response pathway that may be important susceptibility factors for fungal diseases. This assembly is a valuable resource for pecan breeders and supports the production and quality improvement of tree nut crops.
Introduction
The East Asia–North Eastern American disjunctive genus Carya (hickories) are widely grown for their wood, edible nuts, and ornamental value. The most economically significant Carya species is pecan (Carya illinoinensis), whose nuts are known to be rich in health-promoting factors, such as unsaturated fatty acids, antioxidant polyphenols, and vitamins. Pecan is consumed worldwide both directly and as a primary ingredient in many foods and confectionary products or as cooking oil after pressing (Huang et al., 2019). Pecan is native to North America; its range spans tropical to temperate regions, and it is currently cultivated across six continents (Grauke et al., 2016).
Increasing consumer demands have promoted efforts toward the genetic improvement of pecan as a nut crop. Nonetheless, this work has been largely limited to the domestication and identification of varieties with good performance in yield-related traits, and the majority of cultivars have poor resistance to abiotic and biotic stresses (Goff et al., 1996; Wood et al., 2003; Thompson and Conner, 2012; Bock et al., 2020a). Among the economically damaging fungal diseases, pecan scab, caused by the phytopathogenic fungus Venturia effusa, is the most significant disease-related constraint to pecan production in the southeastern regions of the US, where severe fruit infection often results in major or complete crop loss (Bock et al., 2016, 2017, 2018, 2020a, 2020b). Although several natural scab-resistant genotypes have been selected and bred to limit yield losses, scab-susceptible cultivars still predominate in much of the existing and expanding pecan acreage (Thompson and Conner, 2012; Wells, 2014). In the last several years, control of this disease in pecan orchards in major cropping regions has relied largely on repeated and costly fungicide spraying, which also leads to fungicide resistance in the scab pathogen (Bock et al., 2020a). However, the genetic basis of scab resistance is poorly understood, and sources of genetic resistance to the pathogen are urgently needed.
As with many tree species, pecan requires a long generation time to reach full productivity and also displays sporophytic self-incompatibility (Thompson and Conner, 2012). This makes selection for many agronomically valuable traits by classical breeding approaches extremely slow, and it may take over 20 years to release a new cultivar (Thompson and Grauke, 1994; Conner, 2012). Therefore, genome-wide database resources that enable the identification and selection of many genetic loci simultaneously have huge potential to accelerate pecan research and breeding.
A draft genome assembly of the pecan cultivar ‘Pawnee’ was recently published (Huang et al., 2019). Some molecular and SSR markers have also been developed for pecan (Conner and Wood 2001; Grauke et al., 2003; Beedanagari et al., 2005; Chaney et al., 2015), and others are in progress (Jenkins et al., 2015). Although these studies provide essential resources for the identification of scab-resistant cultivars, there is still a need for a high-quality reference genome sequence of pecan to identify key candidate genes and to facilitate the development of more scab resistance-specific markers to aid in scab resistance breeding. Toward this goal, we used a whole-genome shotgun sequencing strategy that combined Oxford Nanopore long-read sequencing and Hi-C (high-throughput chromosome conformation capture) technology to construct a de novo chromosome-scale Pawnee genome assembly consisting of 16 pseudomolecules. Comparison of the pseudomolecules revealed two recent whole-genome duplication (WGD)- and genome rearrangement-related paleo-subgenomes with asymmetry in genome size, gene content, and transposable element (TE) distribution, as well as significant expansion of transcription factor families. We also used whole-genome resequencing data from 86 accessions of 36 genotypes with susceptibility or resistance to pecan scab to identify 47 resistance-related chromosome regions containing 185 putative candidate genes. The candidate gene set highlights genetic selection on putative genes involved in chitin responses, such as chitinase, MAP3K3, GLRs, and so forth, and it provides potential seedling screening markers for the development of fungal disease-resistant varieties.
Results
Chromosome-scale assembly and annotation of pecan
A chromosome-scale assembly of a grafted plant derived from the controlled-cross pecan cultivar Pawnee (Figure 1A) was produced by integration of data generated from Oxford Nanopore sequencing and Hi-C technologies. A total of 71.7 Gb high-quality, cleaned Nanopore sequencing data, representing about 104-fold coverage of the estimated 691.28 Mb genome size with a heterozygosity of 1.52%, were used for de novo assembly (supplemental Figures 1 and 2; Table 1 and supplemental Tables 1 and 2). A 636.26-Mb initial assembly with a contig N50 length of 4.20 Mb and a longest contig of 23.88 Mb was obtained by combining de novo assembly of Nanopore sequences, error correction with Illumina sequences (generated previously), and removal of redundant and bacterial contamination sequences (supplemental Tables 1 and 3). The resulting 636.41-Mb, high-quality final assembly (Cil_v. 2.0) was generated using 75.93 Gb of Hi-C paired-end sequences, and 90.51% of the contigs in the initial assembly were anchored onto 16 pseudochromosomes with lengths ranging from 20.8 to 50.7 Mb (Figure 1C and supplemental Figure 3; Table 1 and supplemental Tables 1 and 4). Completeness assessment of the assembly revealed complete coverage of 95.1% of the core eukaryotic genes in the BUSCO database (Waterhouse et al., 2018) (supplemental Table 5). At least 93.72% of the Illumina short reads could be mapped to the assembly with coverages of 4-, 10-, and 20-fold (supplemental Table 6). The scatterplots of GC distribution showed a good concentration of nearly 36% and were close to the Poisson distribution (supplemental Figure 4). These metrics indicate the high accuracy and overall completeness of the assembly.
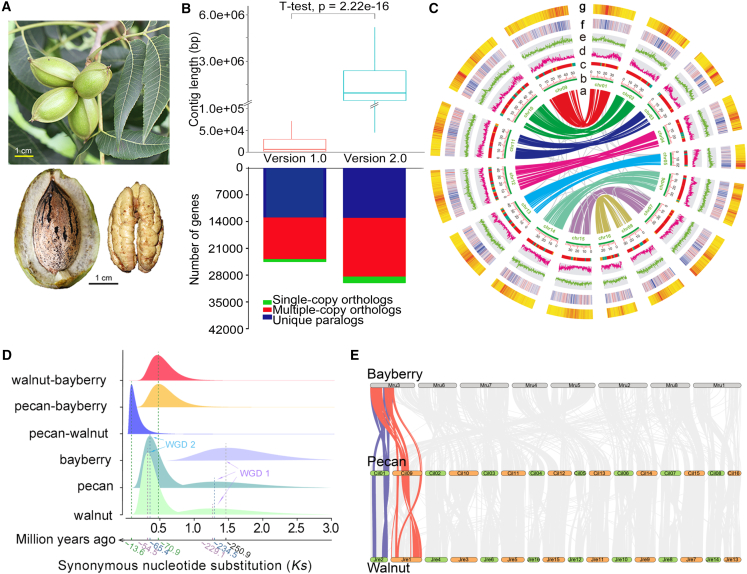
Figure 1.
Genome features of the chromosome-scale assembly and evolution of the pecan genome.
(A) Morphology of fruit, nut, and fresh kernel of Pawnee. Scale bar corresponds to 1 cm.
(B) Comparison of contig lengths and clustered gene sets between versions 1.0 and 2.0.
(C) Landscape of the chromosome-scale pecan genome assembly. (a) Synteny of gene pairs from the recent WGD (WGD 2); (b) chromosomes; (c) contigs of version 1.0 on chromosomes of version 2.0; red, green, and yellow indicate contigs >2, 1–2, and <1 Mb in length in the version 1.0 assembly; (d) protein-coding genes; (e) LTR density distribution; (f–g) distribution of Copia and Gypsy elements.
(D) WGD and divergence within and among species of pecan, walnut, and bayberry.
(E) Syntenic distribution of pecan compared with walnut and bayberry.
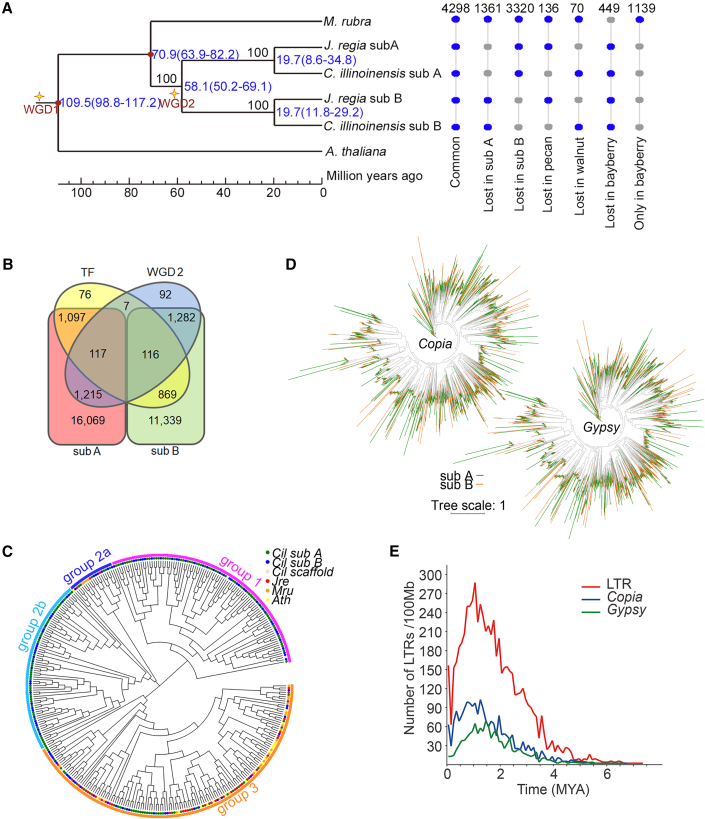
Figure 2.
The features, evolution, and divergence of the pecan subgenomes.
(A) Evolutionary analysis of genomes, subgenomes, and gene sets in pecan, walnut, and bayberry, using Arabidopsis as an outgroup. Numbers indicate the number of gene families.
(B) Venn diagram of genes in the recent WGD (WGD 2), subgenomes (sub A and sub B), and transcription factors. Numbers indicate the number of genes.
(C) Significantly expanded TF families.
(D) Distribution of Copia and Gypsy LTRs on the subgenomes.
(E) Insertion time estimates of LTRs.
Table 1.
Global statistics of the chromosome-scale pecan genome assembly (Cil_v. 2.0).
| Estimated genome size (Mb) | 691.28 |
|---|---|
| Total length of scaffolds (Mb) | 636.41 |
| Number of scaffolds and contigs | 124 & 564 |
| Longest scaffold (Mb) | 55.75 |
| N50 of scaffold and contig length (Mb) | 38.78 & 2.89 |
| Number of predicted protein-coding genes | 33 472 |
| Pseudochromosomes | 16 |
| Anchored sequence to pseudochromosome (Mb) | 608.60 |
| Protein-coding genes in pseudochromosomes | 32 104 |
| Average gene length (CDS + intron) (bp) | 5482.63 |
| Masked repeat sequence length (Mb) | 304.41 |
| Percentage of repeat sequences (%) | 47.83 |
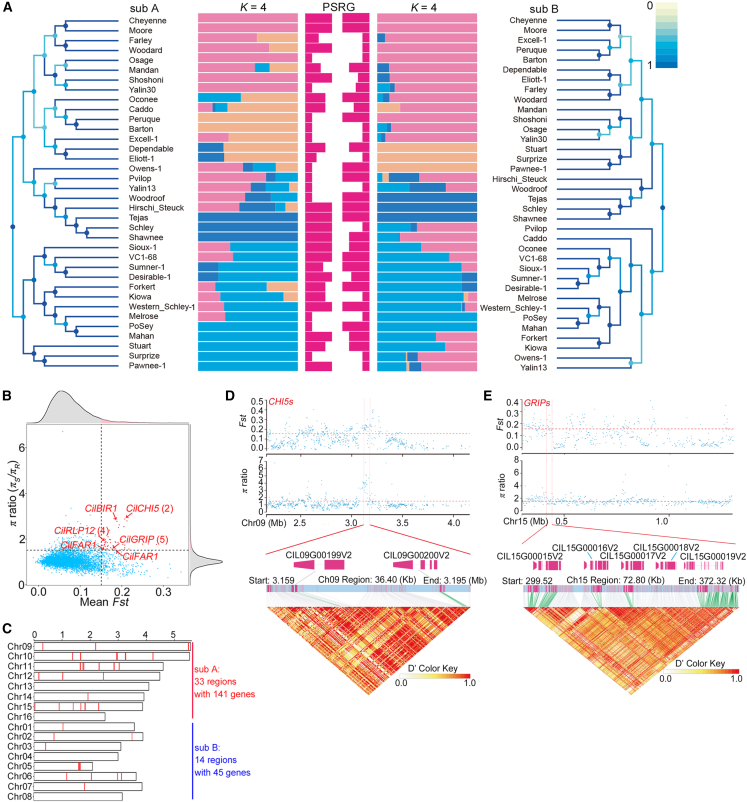
Figure 3.
Population genomics and identification of pecan scab-associated candidate genomic regions and genes under selection.
(A) NJ trees and population structures of subgenomes and scab-resistance grades of each accession. A score of 1 on the color scale bar indicates that the tree structure of the node is identical to the tree structure of its best corresponding node. K = 4, the best substructure. PSRG, pecan scab-resistance grade. The length of the red bars in the middle indicates the disease-resistance grade.
(B) Plots of the highest 5% π and Fst values in pecan scab-related accessions. Arrowheads indicate the loci of key candidate genes associated with pecan scab resistance.
(C) Locations of the selected regions of pecan scab resistance on chromosomes.
(D)Fst and π ratio (upper, coordinate diagrams), gene details (middle, color bars), and LD heatmap of the candidate region containing two putative chitinase-encoding genes.
(E)Fst and π ratio (upper, coordinate diagrams), gene details (middle, color bars), and LD heatmap of the candidate region containing five putative GRIP-encoding genes. The pairwise LD between the SNPs is indicated as D′ values, where red indicates a value of 1 and yellow indicates 0. Rose-red indicates the CDS regions of genes, and green indicates polymorphic SNP sites in the promoter and CDS regions.
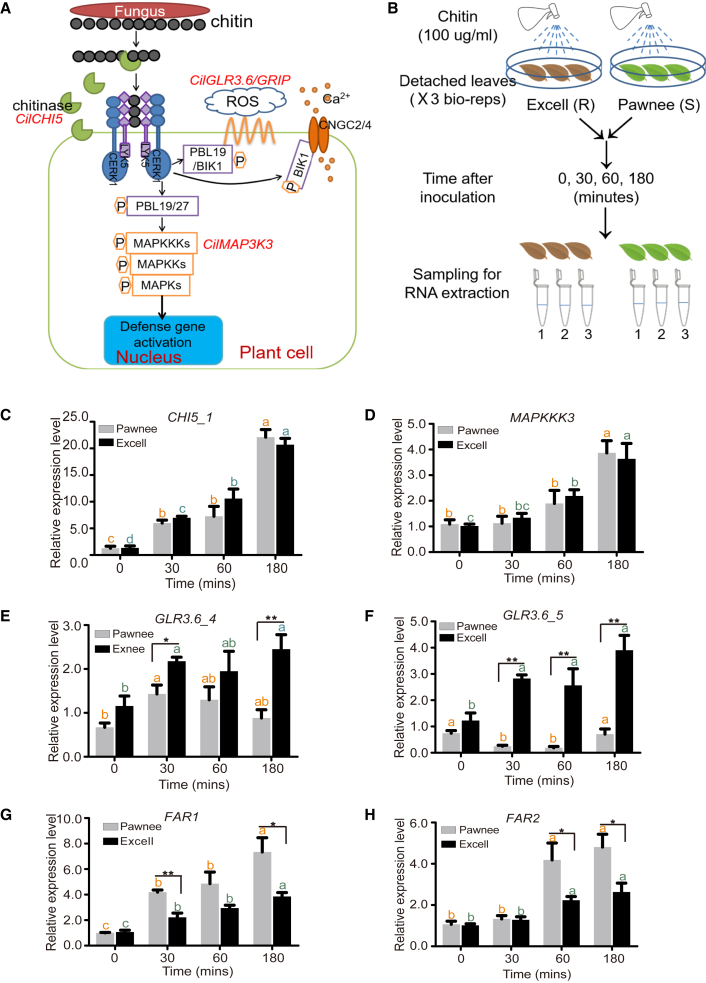
Figure 4.
Identification and expression analyses of key candidate genes involved in the chitin signaling pathway.
(A) A simplified chitin signaling pathway in plants and the key genes under selection. Genes in red are putative candidate genes selected for further analysis.
(B) Chitin treatment and sampling strategy for expression analysis by qPCR. R, pecan scab resistance; S, pecan scab sensitivity.
(C–H) Expression of selected candidate key genes involved in chitin signaling pathways in chitin-treated leaves of the pecan cultivars Excell and Pawnee. Letters show the significance of differences between time points within cultivars (P < 0.05, one-way ANOVA), and stars show the significance of differences between cultivars (∗P < 0.05, ∗∗P < 0.01). qPCR was performed using five replicate leaves from each time point and cultivar.
A combination of homology searches and de novo prediction resulted in the identification of 304.41 Mb of repetitive sequences in the Cil_v. 2.0 assembly, representing 47.83% of the pecan genome (Table 1 and supplemental Tables 7–9). The TE content of the assembly is 45.37%. LTR is the most abundant type, accounting for 35.15% of the pecan assembly (supplemental Tables 8 and 9), and the Gypsy and Copia subfamilies are the dominant subtypes (34.69% and 34.23% of TE length). Gypsy is usually enriched in the centromere regions of angiosperms, and this is also the case for Pawnee (Figure 1C; supplemental Table 9).
A total of 33472 protein-coding genes were predicted by integrating the ab initio prediction, homology search, and transcriptome assembly approaches, and 95.9% of these genes were anchored to the 16 pseudochromosomes (Table 1 and supplemental Tables 9 and 10). In total, 1349 of the predicted protein-coding genes can be completely matched with the BUSCO database (1440 genes), indicating the high completeness (93.7%) of the gene set (supplemental Table 5). Of the protein-coding genes, 31 247 (93.35%) have known functions in the SwissProt, Kyoto Encyclopedia of Genes and Genomes (KEGG), TrEMBL, InterPro, and Gene Ontology (GO) public databases (supplemental Tables 10–12). Compared with our previously reported scaffold-scale draft assembly (version 1.0), the chromosome-level assembly filled in 98.06% of the gaps with highly improved contig length, and this led to the identification of 2397 additional protein-coding genes (Figure 1B, 1C, and supplemental Figure S5; supplemental Table 13). Moreover, some agronomic trait-related genes that have been studied previously have been improved in the new genome version. For example, the copy numbers of genes encoding key components of oil accumulation and polyphenol metabolism decreased significantly in the Cil_v. 2.0 assembly (supplemental Table 14), probably because the short-read-based v. 1.0 assembly was inaccurate in the assembly of multicopy genes (Huang et al., 2019). In addition, the Pawnee assembly encodes 121 miRNAs, 565 tRNAs, 414 rRNAs, and 1318 snRNAs (supplemental Table 15).
When compared to the most recent published genomes of pecan (Lovell et al., 2021), the improved Pawnee Cil_v. 2.0 assembly is similar to the assemblies of four pecan genotypes in assembly completeness (Lovell et al., 2021). The Cil_v. 2.0 assembly is also similar to the genotypes ‘Oaxaca’, ‘Lakota’, and ‘Elliott’ in terms of genomic features, except that it has fewer scaffolds/contigs (supplemental Table 13), indicating that there are fewer gaps and missing sequences in the improved assembly. The gap-free Pawnee assembly reported by Lovell et al. (2021) shows slightly higher scores in genomic features than Cil_v. 2.0 (supplemental Table 13). Synteny analysis between the gap-free Pawnee assembly and Cil_v. 2.0 reveals high collinearity and one-to-one chromosome correspondence between the 2 assemblies, with 21 504 gene pairs (∼67%) in syntenic blocks (supplemental Figures 6–7 and supplemental Table 16). Differences outside the syntenic blocks of the two Pawnee assemblies probably result from the syntenic block cutoff (at least five genes) and haplotype selection when assembling the chromosomes, as the outbred Pawnee has a highly heterozygous genome.
Genome evolution and identification of two paleo-subgenomes
We identified orthologous gene pairs in pecan, walnut, and bayberry, estimated species divergence times based on the synonymous nucleotide substitution (Ks) sites of orthologous genes, and corrected the times using the earliest fossil records of Myricaceae and Juglandaceae (64–84 mya) (Sauquet et al., 2012; Ho and Phillips, 2009). Our analysis revealed a shallow peak that occurred about 234.5 mya and very close to the ancient WGD event (WGD 1) in walnut and bayberry (∼229.1 and ∼250.9 mya) (Figure 1D), probably reflecting the paleopolyploidy WGD (γ) event in the angiosperm lineage (Landis et al., 2017). Pecan and walnut also experienced a recent WGD event (WGD 2) at about 65.4 and 54.5 mya (Figure 1D). Estimates of divergence times between species of walnut–bayberry and pecan–bayberry indicated that the speciation event in Juglandaceae and Myricaceae occurred before the tetraploidization in the genera of Juglandaceae, whereas the divergence between pecan and walnut (∼13.6 mya) occurred after their tetraploidization (Figure 1D), suggesting a common ancestor between pecan and walnut.
The two WGD events involved a total of 3,683 orthologous gene pairs, 2,829 of which were in WGD 2, reflecting its important contribution to the protein-coding gene set. The gene pairs from WGD 2 were mapped to the 16 pseudochromosomes to visualize the detailed syntenic relationships among chromosomes in pecan. We observed eight chromosome pairs with one-to-one corresponding collinear relationships in the pecan assembly (Figure 1C). Further mapping of all identified orthologous genes in syntenic blocks between chromosomes of any two genomes (among pecan, walnut, and bayberry) revealed a considerable number of pair-to-pair relationships between the chromosomes of pecan and walnut but not between the chromosomes of pecan and bayberry (Figure 1E). Based on the results of syntenic analysis, the chromosomes of pecan, walnut, and bayberry were divided into eight groups, and one-to-one orthologous gene pairs identified in the groups (supplemental Table 17) were used to construct eight phylogenetic trees by the neighbor-joining (NJ) method (supplemental Figure 8). Two paleo-subgenomes—subgenome A (chromosomes Chr09 to Chr16) and subgenome B (chromosomes Chr01 to Chr08)—were identified based on the branch lengths of the NJ trees for both pecan and walnut (Figure 1C, 1D, and supplemental Figure 8). The walnut paleo-subgenomes were the same as those reported in the recently published walnut assembly (Zhang et al., 2020). These results reflected frequent large-scale chromosome rearrangements in the pecan and walnut comparing with bayberry genomes after the divergence of Myricaceae and Juglandaceae, as well as rare rearrangement events between pecan and walnut because of their relatively short divergence time.
Features and evolution of the two paleo-subgenomes
Comparison of the sequence similarity of the two paleo-subgenomes of pecan against the bayberry genome showed higher average identity for each chromosome in subgenome B than in subgenome A (supplemental Table 17). The overall identity distribution between the two subgenomes displayed a trend similar to that of the average identity for each chromosome (supplemental Figure 9). Selection analysis (Ka/Ks, i.e., ω) revealed that the chromosomes in subgenome B had experienced stronger positive selection than those in subgenome A, in addition to the PAIR-5 chromosomes (supplemental Figure 10; supplemental Table 17). We also detected asymmetry in lengths and protein-coding genes between the paired chromosomes of the two subgenomes. Except for the PAIR-8 chromosomes, all chromosomes in subgenome A were longer and contained more protein-coding genes (345.88 Mb and 18 498 genes) than those in subgenome B (328.41 Mb and 13 606 genes) (supplemental Table 17).
To compare the paleo-subgenome features in pecan, the values of Ka, Ks, and Ka/Ks were estimated based on 6316 orthologous gene pairs from the subgenomes. According to the values of Ks and Ka, 6253 of the analyzed orthologous gene pairs had been subjected to negative selection (Ka/Ks < 1), whereas 63 had been subjected to positive selection (Ka/Ks > 1). No genes under neutral selection were detected (Ka/Ks = 1). These results indicated that these genes may have undergone lower selection pressure, evolving at a slower evolutionary rate. In addition, we also established the relationships among Ka/Ks, Ka, and Ks in the pecan genome. We found that Ka increased gradually with increasing Ks (supplemental Figure 11), with R = 0.71, P < 2.2 × 10−16 (Spearman's rank correlation). These data were basically consistent with those in pear (R = 0.75) (Cao et al., 2019), suggesting that mechanisms that affect both Ka and Ks sites may be shared in different genomes. In addition, the Ka/Ks ratio was negatively correlated with both Ka (R = 0.34, P < 2.2 × 10−16) and Ks (R = −0.28, P < 2.2 × 10−16) (supplemental Figure 11). The correlation between Ka and Ka/Ks was greater than that between Ks and Ka/Ks, indicating that Ka may be a determining factor for the Ka/Ks ratio between the subgenomes.
For evolutionary analyses of the two paleo-subgenomes in pecan, a simplified phylogenetic tree was constructed using 1080 single-copy orthologous genes from pecan, with walnut, bayberry, and Arabidopsis as references (Figure 2A). The topology of the phylogenetic tree confirmed the close relationship between pecan and walnut, as did our previous report (Huang et al., 2019). Interestingly, the group A or B subgenomes of pecan and walnut were clustered into a terminal clade (Figure 2A). The corrected estimate for the split between subgenomes A and B in the 2 species was about 58 mya, earlier than 11.8–29.2 mya, a time representing the splits of subgenomes A or B between the species (Figure 2A) and during which time a speciation event occurred between pecan and walnut (∼13.6 mya) (Figure 1D). This analysis indicated that differentiation between the two paleo-subgenomes occurred in the common ancestor of pecan and walnut after the Juglandaceae had diverged from the Myricaceae.
Gene family statistics showed that 4298 families were common to pecan, walnut, and bayberry; 1361 or 3320 families were lost in subgenome A or subgenome B of both pecan and walnut; 136 families were lost only in pecan, and 70 were lost only in walnut; and 449 families were retained in both subgenomes of pecan and walnut (Figure 2A). The considerable alteration of gene families among species and subgenomes largely reflects genome-wide rearrangements before and after the recent WGD during the species’ evolution.
The pecan genome encoded a total of 2282 transcription factors (TFs) from 61 TF families, ∼53% (1204) of them in subgenome A and ∼43% (985) in subgenome B. Only 117 in subgenome A and 116 in subgenome B were related to the recent WGD event (WGD 2) (Figure 2B). Of the 2829 WGD 2 genes, 1332 were encoded by subgenome A, and slightly fewer were encoded by subgenome B (1398). Among the TF families, 18 were significantly over-represented in pecan and walnut, and the far-red impaired response 1 (FAR1) family showed extreme expansion, especially in pecan (supplemental Table 18). A maximum likelihood (ML) tree of 264 FAR1-encoding genes revealed three major clades representing the three groups of this family, and group 1 and group 2b FAR1s existed specifically in the pecan genome (Figure 2C). Based on the diverse biological functions of this gene family, expansion of the FAR1s may account for the enhanced light signaling in pecan life processes, including plant development, stress response, and immunity (Ma et al., 2016; Wang et al., 2016). Further chromosome mapping revealed an asymmetrical distribution of FAR1 loci between subgenomes, with more members in subgenome A (Figure 2C and supplemental Figure 12).
The asymmetry of the subgenomes was also shown in the distribution of TEs, especially in the numbers of major LTR types, the Copia (132 369 in subgenome A and 90 474 in subgenome B) and Gypsy (100 257 in subgenome A and 89 931 in subgenome B) subfamilies (Figure 2D; supplemental Table 9). Insertion time estimates show that the insertion times of total LTRs and Gypsy and Copia elements are significantly earlier in subgenome A than in subgenome B (Figure 2E).
Population phylogenetic and genetic analysis of pecan scab-associated accessions
A total of 86 pecan accessions, representing scab-associated core varieties in our collection, were selected for genome-wide analysis (supplemental Table 19). This set of germplasm includes 36 core pecan scab-associated varieties: 29 single individual varieties and 7 cloned populations. Genome resequencing of the accessions using the BGI-Seq 500 sequencer generated a total of 1624.41 Gb of sequences after trimming of low-quality reads. On average, ∼19 Gb of clean data (27× coverage of the estimated pecan genome size) were obtained for each sample (supplemental Table 20). The filtered reads from each accession were mapped to the pecan Cil_v. 2.0 assembly with an average mapping rate of 96.57%. The mapped reads covered most regions of the reference genome with a coverage ratio from 90.99% to 95.16% among the accessions. A total of 24 972 828 high-quality SNPs were detected. A further filtering step revealed 5 901 970 SNPs that were suitable for population analysis, more than half of which (3 293 001) were unique to subgenome A and 2 608 969 of which were unique to subgenome B.
NJ phylogenetic trees were built to display the phylogenetic relationships among the 36 cultivars based on the variations in each subgenome (Figure 3A). Topologies of both NJ trees clearly formed two major clades for each subgenome, but the cultivars of different major clades varied between subgenome A and subgenome B. The phylogenetic relationships of cultivars in each subclade within major clades of subgenome A or subgenome B were closely related to their genetic relationships (Figure 3A; supplemental Table 21) but showed no obvious correlation with disease resistance (Figure 3A; supplemental Table 20). Internal structure comparison between the two phylogenetic trees revealed the best correspondences of all the leaf (outer) nodes and parts of the inner nodes, with a score of 1, and relatively lower correspondences for the root nodes between the NJ trees of subgenome A and subgenome B, with a score of 0.5.
Further population structure investigation of the 36 cultivars at the subgenome level revealed that K = 4 was the best cluster number for the datasets (Figure 3A). To facilitate comparison between subgenomes, we denoted the four K numbers K1 to K4 based on the color of the visualized structure and recorded the ancestral types for each cultivar in subgenomes A and B (Figure 3A; supplemental Table 21). We found that 10 of the cultivars originated from a single ancestor and that 6 cultivars derived from 2 to 4 ancestors, and all these 16 cultivars had the same ancestral types in subgenomes A and B (Figure 3A; supplemental Table 21). The remaining 20 cultivars differed in both ancestors and ancestral types between subgenomes (Figure 3A; supplemental Table 21). These results may reflect the complex domestication history and frequent gene flow caused by natural and human selection and inter- and intra-species hybridization and admixture among the cultivars.
To evaluate the genetic diversity among the accessions, we first divided the accessions into two populations based on their pecan scab-resistance grades: the resistant population (denoted R) had grades ≤2, and the susceptible population (denoted S) had grades >2 (supplemental Table 20). We then quantified the variations in nucleotide diversity (π value) for each population and the pairwise differentiation level (Fst) between the two populations.
Identification of selected regions and candidate genes associated with pecan scab resistance
Given that disease resistance-associated regions in the R population were subjected to stronger selection pressure and therefore had lower polymorphism than corresponding regions in the S population, chromosome regions (100 kb per window) with both π ratios of πS/πR and Fst values in the top 5% were identified as selected regions associated with pecan scab resistance (Figure 3B; supplemental Table 22). The analyses revealed a total of 47 candidate regions that contained 185 putative protein-coding genes, 141 of which were located in subgenome A and 45 in subgenome B (Figure 3B; supplemental Table 23). The candidate regions were unevenly distributed on 12 chromosomes and were most abundant on chromosomes 6, 10, 11, and 15 (Figure 3C).
Of the 47 selected regions, a region of approximately 83.6 kb on chromosome 9 (between 3.1 and 3.2 Mb) displayed the highest Fst value and a relatively high π ratio (Fst = 0.208, π = 2.879), and it contained 7 putative protein-coding genes (supplemental Table 23). Two of the seven genes, Cil_09G_00199V2 and Cil_09G_00200V2, were annotated as chitinases (denoted CilCHI5_1 and CilCHI5_2) (Figure 3B–3D). They were the closest known homologs of an EP3 endochitinase in plants that has been observed to participate in the innate immune response through inhibition of fungal growth (de A. Gerhardt et al., 1997). Detailed analysis revealed two nonsynonymous nucleotide substitutions (missense variants) in CHI5_1 and four in CilCHI5_2, and CilCHI5_2 also harbored an intron variant (Figure 3D; supplemental Table 24).
An approximately 72.4-kb region on chromosome 15 with an Fst value of 0.174 and a π of 1.623 also attracted our attention because it contains 5 tandem repeat genes encoding ionotropic glutamate receptors, Cil_15G_00015V2 to Cil_15G_00019V2 (denoted CilGLR3.6/GRIP1–CilGLR3.6/GRIP5) (Figure 3B, 3C, and 3E; supplemental Table 20). Plant glutamate receptor-like (GLR) homologs have been reported to participate in many plant-specific physiological functions, such as sperm signaling, pollen tube growth, root meristem proliferation, abiotic responses, and innate immunity (Zhu, 2016; Li et al., 2019; Wudick et al., 2018). We detected a total of 183 variants in this tandem repeat region, 28 of which were synonymous substitutions and 21 of which were located in the downstream (14) or upstream (7) regions of the CilGLR genes (Figure 3E; supplemental Table 24). Most of the variants (up to 92) were located in introns, and 42 missense variants were found within the GLRs. Four variants were detected in the splice-and-intron regions of GRIP3 and GRIP5, and one was identified as a stop-gain variant in CilGRIP5 (supplemental Table 24).
In addition to two CilCHI5s and five CilGLR3.6s/GRIPs, we also identified a mitogen-activated protein kinase kinase kinase (MAP3K3/MPKKK3)-encoding gene (Cil_03G_00295V2) that has been well studied in model plant species as a key gene in the chitin-signaling cascade (Figures 3B and 4A; supplemental Table 20 [Gong et al., 2020]). We also detected eight transcription factor genes in the selected regions, including two FAR1 family members in the pecan-specific expansion (supplemental Table 23) that may contribute to pecan scab resistance in this species.
Expression patterns of candidate key genes in response to chitin treatment
To investigate the functions of candidate key genes in response to fungal disease, two cultivars with historical records of strong pecan scab resistant (Excell) and susceptible (Pawnee) phenotypes were subjected to chitin treatment for 30, 60, and 180 min (Figure 4B). Expression levels of 10 candidate key genes were examined by real-time qPCR: CilCHI5_1, CilCHI5_2, five CilGRIPs, CilMAP3K3, CilFAR1_1, and CilFAR1_2 (supplemental Table 25). Six of the genes responded to the chitin treatment, five of which were induced as early as 30 min after chitin treatment (Figure 4C–4H). CHI5_2 and MAP3K3 were induced by chitin with similar expression in both susceptible and resistant cultivars at the early response stage (Figure 4C and 4D), indicating that they may have important roles in defense against fungal pathogens at early infection stages. Two GLR3.6/GRIP homologs were significantly induced in “Excell” (Figure 4E and 4F), suggesting their close correlation with fungal pathogen resistance. By contrast, two FAR members were quickly upregulated in only the susceptible cultivar Pawnee (Figure 4G and 4H), probably reflecting their involvement in fungal disease resistance.
Discussion
Toward a reference genome for pecan
Pecan is typical of a number of important tree nut crop species with high heterozygosity and high genetic diversity due to self-incompatibility and for which limited genome data are currently available. The publicly available draft genome sequence of the pecan cultivar Pawnee makes it possible to identify most gene sequences of interest but not their chromosomal locations or the exact family members for multicopy genes. A highly continuous and complete reference genome is an essential basis for a wide range of studies on gene functions, molecular and metabolic mechanisms, population genetics, breeding, and so forth. By combining current state-of-the art technologies—Oxford Nanopore long-read (>2 kb) sequencing, Hi-C technology, and high-quality genome assemblers—we constructed a chromosome-scale genome assembly for the pecan cultivar Pawnee. The de-novo-assembled Cil_v. 2.0 Pawnee genome displays high continuity, integrity, and quality, with a contig N50 of 3.04 Mb and BUSCO assessments of 95.1% for assembly and 93.7% for protein-coding sequences. The newly assembled genome sequence is about 92% of the estimated genome size, with a total of 608.6 Mb on 16 chromosomes (95.6% of the new assembly). All of the missing sequence lengths are likely to be telomeric and centromeric repeats. The Cil_v. 2.0 assembly contains 33 472 predicted protein-coding models, of which 9516 are unique to Cil_v. 2.0 and, in total, 2397 more than the previous version (v. 1.0) (Huang et al., 2019), reflecting the higher continuity, integrity, and accuracy of the new version.
Asymmetry in the evolution and features of the pecan paleo-subgenomes
An ancient WGD event, i.e., the γ triplication event, has been widely reported in many angiosperms, including Fagales (Tuskan et al., 2006; Jaillon et al., 2007; Huang et al., 2009, 2019; Luo et al., 2015). Our analysis confirmed these ancient WGD events (WGD 1) and a recent WGD (WGD 2) in the pecan genome (Figure 1D). Based on the syntenic relationships of homologous gene pairs in WGD 2, we divided the 16 pecan chromosomes into 8 pairs of homoeologs and further divided them into 2 paleo-subgenomes based on the phylogenetic relationships among pecan, walnut, and bayberry (Figures 1C, 1E, and 2), which were similar to those reported in walnut (Luo et al., 2015; Zhang et al., 2020). Previous studies suggested that the γ triplication generated a genome with n = 21, indicating that the 8 homoeologous chromosome pairs in haploid genomes of Juglandaceae evolved from n = 8 by a recent WGD rather than n = 21 by dysploid reduction (Salse, 2012; Luo et al., 2015). Our analysis of the pecan genome was consistent with this conclusion (Figures 3D and 4B). One hypothesis of x = 8 as the ancestral state was supported by the presence of chromosome number n = 8 in the genera Roipterlea and Myrica, which are closely related to Juglandaceae (Luo et al., 2015). Timing inference in this study revealed that the recent WGD events in pecan and walnut (65.4 and 54.5 mya) happened in the “juglandoid” WGD (56–66 mya) near the Cretaceous–Tertiary (K–T) boundary about 66 mya (Manchester, 1989; Luo et al., 2015), after the divergence from bayberry (Figures 3D and 4B). The divergence between the paleo-subgenomes preceded the split between pecan and walnut, and the divergence time between paleo-subgenomes followed and/or coincided with the “juglandoid” WGD events and was accompanied by extensive genome rearrangements, probably reflecting rapid genome evolution and adaptive evolution to survive the adverse environmental conditions associated with the K–T boundary (Fawcett et al., 2009; Soltis and Burleigh, 2009; Van de Peer et al., 2009; Luo et al., 2015). The relatively lower syntenic relationship of bayberry with pecan or walnut in this study provided solid evidence for this conclusion (Figure 1E). Moreover, the asymmetry between paleo-subgenome features, such as genome size, number of gene models, TE distribution, and pecan-specific gene family expansion, all strongly supported the inference of a “juglandoid” WGD and a large-scale genome rearrangement-associated evolutionary trajectory during the K–T boundary in the Juglandaceae. Nonetheless, further evidence is still needed to uncover the sources of the “juglandoid” WGD, i.e., data derived from parental ancestral hybridization or from duplication of one ancestral species.
Genome-based insights into breeding targets for fungal disease resistance
Fungal pathogens constitute major threats to land plants and pose growing challenges to global crop production; they have led to losses of approximately 30% in annual global crop production before and after harvest (Gong et al., 2020). In plants, the first layer of innate immunity relies on the perception of conserved pathogen-associated molecular patterns (PAMPs). This perception is mediated by pattern recognition receptors located at the cell surface, including membrane-localized receptor-like kinases and receptor-like proteins, which elicit PAMP-triggered immunity (Wang et al., 2017; Dodds and Rathjen, 2010; Tena et al., 2011). Chitin, an insoluble polymer of β-1,4-linked N-acetylglucosamine, is a highly conserved building block of fungal cell walls and a broadly effective elicitor of plant immunity. Invasion by fungal pathogens can induce the secretion of plant chitinases into the apoplast to hydrolyze fungal cell walls and release chitin oligomers. PAMP recognition then rapidly initiates a series of early immune responses, including the activation of mitogen-activated protein kinase (MAPK) cascades and the production of reactive oxygen species (ROS) to combat pathogen infection.
To date, many key genes involved in chitin perception and signaling pathways have been identified in model plants such as Arabidopsis and rice (Liu et al., 2016; Wang et al., 2017; Gong et al., 2020). In this study, genome-based population genetic diversity enabled us to identify 47 selected regions containing 185 putative candidate genes associated with scab resistance in pecan (Figure 3). Fourteen of them were annotated as receptor(-like) proteins, including one bacterial flagellin receptor-like protein (FLS2) and five proteins with ionotropic GLR activity (supplemental Table 23). FLS2 has been extensively reported to function in bacterial-derived PAMPs but not in fungal-derived PAMPs (Wang et al., 2017), and its identification here may reflect its important role in fungal disease resistance of pecan. However, the chitin receptor kinase CERK homolog did not appear to be under selection in this study, possibly because of the small population sample. GLRs in plants participate in diverse and important biological processes, such as photosynthesis, cellular C/N balance, plant organ development, abiotic stress response, plant-pathogen interactions, calcium-mediated signal transduction, and so forth (Kang and Turano, 2003; Kang et al., 2004, 2006; Singh et al., 2006; Li et al., 2013; Manzoor et al., 2013; Cheng et al., 2016). The relatively high Fst values of the five GLR genes detected in pecan scab-resistant cultivars probably suggest enhanced chitin-induced fungal resistance. Recognition of chitin is known to trigger the intracellular activation of MAPK cascades and the rapid production of ROS (Tsutomu et al., 2017). The activation of MAPK cascades is the core step in chitin-induced immune responses (Yamada et al., 2017), and a homolog of MAP3K3 (Cil_03G_00295V2) was identified and shown to be induced by chitin treatment in scab-resistant pecan cultivars, implying that it may have an important role in the fungal defense response of pecan. Our results also highlighted two putative EP3 endochitinase-like genes, CHI5s, which have been reported to function in innate immune responses by degrading the fungal cell wall to inhibit fungal growth in plants (de A. Gerhardt et al., 1997). One of the two detected CilCHI5 genes was strongly induced in both resistant and susceptible cultivars at the early stages of chitin treatment, indicating its important role in early defense against fungal pathogens. The expression levels of CHI5_1 showed no obvious differences between the cultivars, indicating that the defense mechanism of resistant varieties may involve downstream signal transduction and corresponding processes. Also, the specific induction of GLR3.6_4 and GLR3.6_5 expression in resistant cultivars (Figure 4E and 4F) may serve as a potential marker for the screening of fungal pathogen-resistant varieties at the seedling stage after further experimental validation. Our findings provide important clues and potential targets for uncovering the intrinsic mechanisms of fungal disease resistance and breeding in the future.
Materials and methods
Plant materials
The widely planted cultivar Pawnee, which was released in 1985 as the progeny of a controlled cross between ‘Mohawk’ × ‘Starking Hardy Giant’ performed in 1963 (Thompson and Hunter, 1985), was selected for whole-genome sequencing. Fresh young leaves were collected from a grafted plant growing in the plantation of Zhejiang A&F University, Lin'an District, Hangzhou, China in April 2018. To investigate genome-wide associations with scab resistance, fresh young leaves from 86 accessions representing 36 genotypes, including 7 cloned populations, were collected from April to May 2019 (supplemental Table 19). All the collected samples were frozen and transported in liquid nitrogen and stored at −80°C in a freezer before use.
Preparation of genomic DNA and Nanopore sequencing
High-molecular-weight genomic DNA was extracted from young Pawnee leaves using the DNeasy Plant Mini Kit (QIAGEN, Germany) for use in v. 2.0 genome assembly. DNA quality and quantity were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA), Qubit dsDNA HS Assay Kits, and a Qubit 2.0 fluorometer (Invitrogen, USA). Genomic DNA of over 2 kb in length was purified using a BluePippin automatic nucleic acid electrophoresis and fragment recovery system (Sage Science, USA). The recovered DNA was used to construct libraries for whole-genome sequencing using the Nanopore PromethION platform (Oxford Nanopore Technologies, UK) at Biomarker Technologies, Beijing, China.
Hi-C library preparation and sequencing
To enable a high-quality, chromosome-level assembly of the pecan reference genome, fresh young leaves were collected from the tree and used for whole-genome sequencing. Leaf samples frozen in liquid nitrogen were fixed with 2% formaldehyde solution in PBS buffer for 30 min, and the reaction was terminated using 2.5 M glycine for 5 min. The fixed samples were sent to BGI-Qingdao (Qingdao, China) for Hi-C library construction using the DNA restriction endonuclease DpnII, according to the standard library preparation protocol (Burton et al., 2013). The BGISEQ-500 platform (BGI-Shenzhen, China) was used for library preparation and sequencing.
Genome survey
Approximately 168 Gb of Illumina data that had been used for scaffold-level assembly of the version 1.0 pecan genome (Huang et al., 2019) were used for a genome survey in the present study. We first used SOAPnuke software (Chen et al., 2018) to remove low-quality paired-end raw reads and then used GenomeScope (Vurture et al., 2017) to estimate the genome size, heterozygosity, and repeat rate based on the 17-mer depth frequency distribution.
Genome assembly
The quality-filtered Nanopore data were assembled using CANU v. 1.6 software (Koren et al., 2017) with optimized parameters (genomeSize=700m minReadLength=500 -correctedErrorRate=0.20 -fast). The accuracy of the initial assembly was then improved three times using Pilon v. 1.22 (Walker et al., 2014), and redundant contigs were removed using Purge_Haplotigs in the CANU package. To remove possible contamination by bacterial sequences identified by GC depth analysis, NT Blast was launched to scan all assembled contigs and eliminate those contigs with best hits to bacterial sequences. Next, all contigs were mapped to the pecan chloroplast genomes deposited in GenBank (accessions MW410238, MH909600, and MH909599) to remove chloroplast sequences. To evaluate the consistency and integrity of the initial polished assembly, Illumina short reads were blast-searched against the genome assembly using BWA v. 0.7.12 (Li and Durbin, 2009), and BUSCO v. 4.1.2 (Simᾶo et al., 2015) analysis was performed to further evaluate the assembly.
To generate a chromosome-level assembly, Hi-C paired-end reads were subjected to quality control using HiC-Pro v. 2.8.0 (Servant et al., 2015). Low-quality bases and adapter sequences were then removed using Bowtie 2 v. 2.2.5 (Langmead et al., 2009), and Juicer v. 1.5 (Durand et al., 2016) was used to analyze the Hi-C datasets. Finally, a 3D de novo assembly (3D-DNA, v. 170123) pipeline (Dudchenko et al., 2017) was used to scaffold the assembly onto pseudochromosomes.
Genome annotation
Repetitive sequences were predicted in the pecan genome using homology-based searches combined with ab initio approaches. TRF (v. 4.07b; Benson, 1999) was used to identify tandem repeats. RepeatMasker and RepeatProteinMask (v. 3.3.0; http://www.repeatmasker.org/) were used to search for known TEs against the Repbase library and the TE protein database (Jurka et al., 2005). Then, a de novo repeat library was built using RepeatModeler software (v. 2.0; http://www.repeatmasker.org) with default parameters, and all TEs were classified using RepeatMasker (http://www.repeatmasker.org/).
The de novo prediction of protein-coding genes was performed using AUGUSTUS (v. 3.1; Stanke et al., 2004) and Genscan (v. 1.0; Aggarwal and Ramaswamy, 2002). GeneWise (v. 2.4.1; Birney et al., 2004) was used for homologous annotation against protein datasets from nine species (supplemental Table 8) downloaded from the National Center for Biotechnology Information (NCBI) database. To assist with gene model prediction, paired-end RNA sequencing reads from leaf, epicarp, embryo, and stem tissues in our previous study (Huang et al., 2019) were assembled de novo using Trinity (v. 2.8.5; Grabherr et al., 2013), followed by gene model prediction of transcripts using PASA (v. 2.3.3; Campbell et al., 2006). Gene models from these different approaches were integrated into a non-redundant set of gene structures using GLEAN (v. 1.0; Elsik et al., 2007) with default parameters. The final pecan gene set was assessed for completeness of the annotated protein-coding genes.
Functional annotation of protein-coding genes was achieved by homolog searches against the TrEMBL (UniProtKB), SwissProt (Bairoch and Apweiler, 2000), KEGG (Kanehisa and Goto, 2000), GO (Consortium, 2004), and InterProScan (Jones et al., 2014) databases with an E value cutoff of 1 × 10−7. Non-coding RNAs, including rRNAs, tRNAs, snRNAs, and miRNAs, were identified by searching against various RNA libraries. tRNAscan-SE v. 1.3.1 software (Lowe and Eddy, 1996) was run with eukaryote parameters to identify tRNA genes. The rRNA sequences were annotated based on homology to previously published rRNA sequences in plants. The snRNAs and miRNAs were predicted using the “cmsearch” program in Infernal v. 1.1 (Nawrocki and Eddy, 2013) to search against Rfam v. 13.0 (Kalvari et al., 2018) with an E value cutoff of 0.01.
WGD and synteny analyses
To estimate the timing of WGD events, wgd software (Zwaenepoel and Peer, 2019) was used to calculate the Ks distribution of orthologs from pecan, walnut, and bayberry, and then the Gaussian mixture model (GMM) was used to fit a curve for the Ks distribution of each species. In the same way, wgd software was also used to estimate the divergence between any two species among pecan, walnut, and bayberry by calculating the Ks values of one-versus-one ortholog pairs between two species and fitting curves using the GMM model. The divergence times of WGD events within species and the divergences between species were estimated by the formula Ks 1/time 1 = Ks 2/time 2 (Ks 1, divergence value of ortholog pairs between species; Ks 2, WGD peak; time 1, divergence time between species; time 2, WGD time), and corrected based on the earliest fossil records of Myricaceae and Juglandaceae (64–84 mya) (Ho and Phillips, 2009; Sauquet et al., 2012). Gene pairs associated with the recent WGD event in pecan were used for circos mapping among chromosomes (Krzywinski et al., 2009).
To obtain the syntenic relationships between pecan and walnut or bayberry, Blast v. 2.2.6 (Boratyn et al., 2012) was used to identify the syntenic gene pairs between species with “-e 1e-6” and other default parameters. The results were then used for syntenic mapping with MCScanX (Wang et al., 2012) using default parameters.
Insertion time estimates of all LTRs and Gypsy and Copia elements were obtained as described by Huang et al. (2019) with the model T = K/2r (r = 1.3 × 10−8 per site and per year).
Defining the two paleo-subgenomes
To define the subgenomes of pecan, protein sequences from the v. 2.0 pecan assembly and from the bayberry and walnut genomes were used to generate clusters for gene families. All protein sequences of bayberry and walnut were downloaded from the NCBI database. Orthologous genes with ratios of 2:2:1 in pecan, walnut, and bayberry were selected, and orthologous gene pairs located on two different chromosomes with syntenic relationships in pecan and walnut were filtered out and connected into super sequences according to their chromosomes. The super sequences were aligned with MUSCLE v. 3.8.31 (Edgar, 2004). Regions with gaps were removed using Gblocks v. 0.91b (Talavera and Castresana, 2007) to generate eight chromosome groups (including two chromosomes from pecan, two from walnut, and one from bayberry), and an ML tree was constructed for each group using FastTree (Price et al., 2010) and displayed using MEGA7 (Kumar et al., 2016). Subgenomes A and B and the chromosome numbers of pecan were defined based on evolutionary distance.
Subgenome features
Orthologous gene pairs of sub A and sub B were determined by bilateral Blast searches against the bayberry gene set, and orthologs with the best identity to bayberry genes in the sub A or sub B genome were selected. A set of 6316 orthologous gene pairs with the best identities was obtained, and MUSCLE v. 3.8.31 (Edgar, 2004) was used for alignment of the gene pairs with codons. KaKs_calculator (v. 2.0) software was used to estimate the Ka, Ks, and Ka/Ks values using the NG method (Wang et al., 2010). Frequency distribution histograms and scatterplots of the Ka, Ks, and Ka/Ks values were displayed using the ggplot2 package in the R language, and curve fitting of the scatterplots was performed using the “lm” method (Wickham, 2016).
Comparative genome analyses and phylogenetics
To investigate the evolutionary status of subgenomes in the Juglandaceae, protein sequences from the Cil_v. 2.0 pecan assembly and from three reference species (Arabidopsis, bayberry, and walnut) were used to generate clusters for gene families. All protein sequences of the three species were downloaded from the NCBI database. Genes with frame shifts that encoded fewer than 30 amino acids and redundant copies in each species were removed, and only the longest transcripts for each gene were selected for further analysis to ensure the analysis quality. To compare orthologous genes from the references with the protein-coding genes from the current pecan assembly (v. 2.0), all one-to-one orthologous gene sets were identified by BLASTP (Altschul et al., 1990) with an E value cutoff of 1 × 10−5, and similar genes were clustered into families using hcluster, a hierarchical clustering algorithm in the TreeFam v. 0.50 pipeline (Li et al., 2006). All the gene families were aligned with the multi-sequence alignment software MUSCLE v. 3.8.31 (Edgar, 2004). To consider sequence conservation, the aligned single-copy genes from different gene families were further concatenated into super long sequences for the subgenomes of each species using a perl script. An ML phylogenetic tree was constructed using RAxML v. 8.2.4 (Stamatakis, 2006) with the PROTGAMMAAUTO option to automatically determine the optimal amino acid substitution site model; Arabidopsis was used as an outgroup, and branch confidence settings were based on 100 bootstrap replicates. The ML tree was used as a starting tree to infer the divergence times between species or subgenomes using the MCMCTree program in the PAML package (Yang, 1997). The calibration times for the divergence between Arabidopsis and walnut (98–117 mya) and between walnut and bayberry (43–74 mya) were obtained from the TimeTree database (http://timetree.org/). The divergence time between bayberry and walnut was calibrated based on the earliest fossil records of Myricaceae and Juglandaceae (64–84 mya) (Ho and Phillips, 2009; Sauquet et al., 2012). The common and lost genes among and/or within species were determined based on the results of homologous alignment.
Transcription factors in the A and B subgenomes were predicted by combining homologous searches against Arabidopsis, walnut, and bayberry transcription factors in the PlantTFDB v. 3.0 database (http://planttfdb.gao-lab.org/) based on the core domain structure using a hidden Markov model with further manual correction. The visualized heatmap of TF quantity distribution was generated by R script homogenization processing.
Genome resequencing, SNP calling, quality control, and validation
Genomic DNA was extracted from 86 individuals using CTAB methods. One microgram of high-quality DNA from each sample was used for genome resequencing library construction with the MGIEasy DNA Rapid Library Prep Kit (BGI, catalog no. 1000006985), and libraries were sequenced on the BGISEQ-500 platform following the manufacturer’s protocol. The raw data (PE100) were filtered using SOAPnuke v. 1.5.6 (Chen et al., 2018) to remove reads with adapters or poly Ns and low-quality reads (reads in which >30% bases had Phred quality ≤25). The quality-controlled reads were then aligned to the Cil_v. 2.0 pecan assembly for SNP and indel calling by Sentieon, a pipeline that integrates BWA (Burrows–Wheeler Aligner) and GATK (Genome Analysis Tool Kit) (Kendig et al., 2019). The Haplotype Caller module of GATK was used for variant calling in the two subgenomes, and the concordance variants were filtered with the parameters “QD < 2.0 || MQ < 40.0 || MQRankSum < -12.5 || ReadPosRankSum < -8.0 || FS>60.0 || SOR>3.0”. The indels were further filtered with “QD < 2.0 || ReadPosRankSum < -20.0 || FS > 200.0 || SOR>10.0”.
Genetics and diversity analysis of the pecan scab-resistant and susceptible accessions
SNPs in subgenome A, subgenome B, and the whole genome of each sample were used to build NJ phylogenetic trees with 1000 bootstrap replicates using TreeBeST (http://treesoft.sourceforge.net/treebest.shtml), and the trees were visualized using the iTOL online tool (Letunic and Bork, 2019; http://itol.embl.de). To obtain a better alignment result, we did not split the subgenomes into two parts for processing. The internal structures of the two phylogenetic trees were compared using the phylo.io online tool (http://phylo.io/index.html). Structures of the accessions on the subgenome scale were analyzed with ADMIXTURE (Alexander and Lange, 2011).
The sampled accessions were divided into two groups based on their pecan scab-resistance grades: the scab-resistant group (R, grade number ≤ 2) and the scab-susceptible group (S, grade number > 2). To determine the pairwise genetic diversity Pi (π) and the fixation index Fst of the R and S groups, vcftools software (Danecek et al., 2011; http://vcftools.sourceforge.net/) was used with a 100-kb sliding window. Chromosome regions whose values of Pi ratio (πS/πR) and Fst were both in the highest 5% were selected for further analysis.
Hot-block linkage disequilibrium (LD) mapping of the chromosome regions of interest above was visualized using LDBlockShow (Dong et al., 2020) with default parameters.
Chitin treatment and real-time qPCR validation
To validate our results, fully expanded leaves from the scab-resistant cultivar Excell and the scab-susceptible cultivar Pawnee were collected and subjected to chitin (100 μg/ml) treatment for 0, 30, 60, and 180 min (Figure 4B). At least three biological replicates of each sample type were collected for RNA extraction using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China), and cDNA was obtained using the SuperScript III First-Strand Synthesis System (Takara, Dalian, China). Eleven putative genes that were involved in the chitin signaling pathway and ROS elimination were selected for expression analysis, and the 18S rRNA gene was used as the internal control (Mattison et al., 2017). Real-time qPCR was performed on an ABI 7500 Real-Time PCR System (Foster City, CA, USA) with three technical replicates for each gene in each sample. Gene expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). The genes and primers are listed in supplemental Table 25. Significant differences in relative gene expression levels between samples were determined using the SPSS program (Kretzschmar, 2000).
Data availability
Genome sequences, assembly, and annotation data have been deposited at NCBI GenBank under BioProject/BioSample numbers PRJNA727440/SAMN19020793. Resequencing reads for the 86 individuals have been deposited in the Sequence Read Archive (SRA) under BioProject/BioSample numbers PRJNA735040/SAMN19554720–19554805.
Funding
This work was supported by grants from the Natural Science Foundation of Zhejiang Province, China (grant no. Z20C160001), the State Key Laboratory of Subtropical Silviculture at Zhejiang A&F University (grant no. ZY20180202), and the Research and Development Fund of Zhejiang A&F University (grant no. 2018FR002).
Author contributions
L.X. and G.F. designed and supervised the research. L.X. wrote the paper. L.X., M.Y., R.Z., H.G., X.G., H.Z., T.D., and G.F. performed the genome assembly, annotation, and evolution analysis. J.H. and G.F. performed the population structure and genetic diversity analysis. L.X., Y.Z., J.W., S.L., and X.L. collected and prepared all samples and performed the experiments. J.H. provided valuable suggestions for the project.
Acknowledgments
The authors declare no competing interests.
Published: September 24, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information can be found online at Plant Communications Online.
Contributor Information
Lihong Xiao, Email: xiaolh@zafu.edu.cn.
Guangyi Fan, Email: fanguangyi@genomics.cn.
Supplemental information
References
- de A. Gerhardt L.B., Sachetto-Martins G., Contarini M.G., Sandroni M., de P. Ferreira R., de Lima V.M., Cordeiro M.C., de Oliveira D.E., Margis-Pinheiro M. Arabidopsis thaliana class IV chitinase is early induced during the interaction with Xanthomonas campestris. FEFS Lett. 1997;419:69–75. doi: 10.1016/s0014-5793(97)01332-x. [DOI] [PubMed] [Google Scholar]
- Aggarwal G., Ramaswamy R. Ab initio gene identification: prokaryote genome annotation with GeneScan and GLIMMER. J. Biosci. 2002;27:7–14. doi: 10.1007/BF02703679. [DOI] [PubMed] [Google Scholar]
- Alexander D.H., Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics. 2011;12:246. doi: 10.1186/1471-2105-12-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bairoch A., Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beedanagari S.R., Dove S.K., Wood B.W., Conner P.J. A first linkage map of pecan cultivars based on RAPD and AFLP markers. Theor. Appl. Genet. 2005;110:1127–1137. doi: 10.1007/s00122-005-1944-5. [DOI] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Clamp M., Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C.H., Grauke L.J., Conner P., Burrell S.L., Hotchkiss M.W., Boykin D., Wood B.W. Scab susceptibility of a provenance collection of pecan in three different seasons in the southeastern United States. Plant Dis. 2016;100:1937–1945. doi: 10.1094/PDIS-12-15-1398-RE. [DOI] [PubMed] [Google Scholar]
- Bock C.H., Hotchkiss M.W., Young C.A., Charlton N.D., Chakradhar M., Stevenson K.L., Wood B.W. Population genetic structure of Venturia effusa, cause of pecan scab, in the southeastern United States. Phytopathology. 2017;107:607–619. doi: 10.1094/PHYTO-10-16-0376-R. [DOI] [PubMed] [Google Scholar]
- Bock C.H., Young C.A., Stevenson K.L., Charlton N.D. Fine-scale population genetic structure and within-tree distribution of mating types of Venturia effusa, cause of pecan scab in the United States. Phytopathology. 2018;108:1326–1336. doi: 10.1094/PHYTO-02-18-0068-R. [DOI] [PubMed] [Google Scholar]
- Bock C.H., Alarcon Y., Conner P.J., Young C.A., Randall J.J., Pisani C., Grauke L.J., Wang X., Monteros M.J. Foliage and fruit susceptibility of pecan provenance collection to scab, caused by Vernturia effusa. CABI Agric. Biosci. 2020;1:19. [Google Scholar]
- Bock C.H., Barbedo J.G.A., Del Ponte E.M., Bohnenkamp D., Mahlein A.-K. From visual estimates to fully automated sensor-based measurements of plant disease severity: status and challenges for improving accuracy. Phytopathol Res. 2020;2:9. [Google Scholar]
- Boratyn G.M., Schäffer A.A., Agarwala R., Altschul S.F., Lipman D.J., Madden T.L. Domain enhanced lookup time accelerated BLAST. Biol. Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton J.N., Adey A., Patwardhan R.P., Qiu R., Kitzman J.O., Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M.A., Hass B.J., Hamilton J.P., Mount S.M., Buell C.B. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Jiang L., Wang L., Cai Y. Evolutionary rate heterogeneity and functional divergence of orthologous genes in Pyrus. Biomolecules. 2019;9:490. doi: 10.3390/biom9090490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney W., Han Y., Rohla C., Monteros M.J., Grauke L.J. Developing molecular marker resources for pecan. Acta Hort. 2015;1070:127–132. [Google Scholar]
- Chen Y., Chen Y., Shi C., Huang Z., Zhang Y., Li S., Li Y., Ye J., Yu C., Li Z. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018;7:1–6. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Tian Q.Y., Zhang W.H. Glutamate receptors are involved in mitigating effects of amino acids on seed germination of Arabidopsis thaliana under salt stress. Environ. Exp. Bot. 2016;130:68–78. [Google Scholar]
- Conner P.J. Pecan breeding review. Pecan South. 2012;45:34–44. [Google Scholar]
- Conner P.J., Wood B.W. Identification of pecan cultivars and their genetic relatedness as determined by randomly amplified polymorphic DNA analysis. J. Am. Soc. Hort. Sci. 2001;126:474–480. [Google Scholar]
- Consortium G.O. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Dong S., He W., Ji J., Zhang C., Guo Y., Yang T. LDBlockShow: a fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief. Bioinformatics. 2020;22:bbaa227. doi: 10.1093/bib/bbaa227. [DOI] [PubMed] [Google Scholar]
- Dudchenko O., Batra S.S., Omer A.D., Nyquist S.K., Hoeger M., Durand N.C., Shamim M.S., Machol I., Lander E.S., Aiden A.P. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand N.C., Shamim M.S., Machol I., Rao S.S., Huntley M.H., Lander E.S., Aiden E.L. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik C.G., Mackey A.J., Reese J.T., Milshina N.V., Roos D.S., Weinstock G.M. Creating a honey bee consensus gene set. Genome Biol. 2007;8:R13. doi: 10.1186/gb-2007-8-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett J.A., Maere S., Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. U S A. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff W.D., McVay J.R., Gazaway W.S. Alabama Cooperative Extension System Circular ANR-459. University; Auburn: 1996. Pecan Production in the Southeast; p. 222. [Google Scholar]
- Gong B.-Q., Wang F.-Z., Li J.-F. Hide-and-Seek: chitin-triggered plant immunity and fungal counterstrategies. Trends Plant Sci. 2020;25:805–816. doi: 10.1016/j.tplants.2020.03.006. [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., Haas B.J., Yassour M., Levin Z.J., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q. Trinity: reconstructing a full-length transcriptome without a genome from RNA-seq data. Nat. Biotechnol. 2013;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauke L.J., Iqbal M.J., Reddy A.S., Thompson T.E. Developing microsatellite DNA markers in pecan. J. Am. Soc. Hortic. Sci. 2003;128:374–380. [Google Scholar]
- Grauke L.J., Wood B.W., Harris M. Crop vulnerability: Carya. HortScience. 2016;51:653–663. [Google Scholar]
- Ho S.Y.W., Phillips M.J. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst. Biol. 2009;58:367–380. doi: 10.1093/sysbio/syp035. [DOI] [PubMed] [Google Scholar]
- Huang S.W., Li R.Q., Zhang Z.H., Li L., Gu X.F., Fan W., Lucas W.J., Wang X.W., Xie B.Y., Ni P.X. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- Huang Y., Xiao L., Zhang Z., Zhang R., Wang Z., Huang C., Huang R., Luan Y., Fan T., Wang J. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. Gigascience. 2019;8:giz036. doi: 10.1093/gigascience/giz036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., Aury J.-M., Noel B., Policriti A., Clepet C., Casagrande A., Nathalie C., Sébastien A., Nicola V., Claire J. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jenkins J., Wilson B., Grimwood J., Schmutz J., Grauke L.J. Towards a reference pecan genome sequence. Acta Hort. 2015;1070:101–108. [Google Scholar]
- Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A.L., Nuka G. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kalvari I., Argasinska J., Quinones-Olvera N., Nawrocki E.P., Rivas E., Eddy S.E., Bateman A., Finn R.D., Petrov A.I. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nacleic Acids Res. 2018;46:D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Turano F.J. The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U S A. 2003;100:6872–6877. doi: 10.1073/pnas.1030961100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Sohum M., Turano F.J. The putative glutamate receptor 1.1 (AtGLR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol. 2004;45:1380–1389. doi: 10.1093/pcp/pch159. [DOI] [PubMed] [Google Scholar]
- Kang S., Kim H.B., Lee H., Choi J.Y., Heu S., Oh C.J., Kwon S.I., An C.S. Overexpression in Arabidopsis of a plasma membrane-targeting glutamate receptor from small radish increases glutamate-mediated Ca2+ influx and delays fungal infection. Mol. Cell. 2006;21:418–427. [PubMed] [Google Scholar]
- Kendig K.I., Baheti S., Bockol M.A., Drucker T.M., Hart S.N., Heldenbrand J.R., Hernaez M., Hudson M.E., Kalmbach M.T., Klee E.W. Sentieon DNASeq variant calling workflow demonstrates strong computational performance and accuracy. Front. Genet. 2019;10:736. doi: 10.3389/fgene.2019.00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar W.A. SPSS student version 9.0 for Windows[J] J. Engl. Linguist. 2000;28:311–313. [Google Scholar]
- Krzywinski M., Schein J.E., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis J.B., Soltis D.E., Li Z., Marx H.E., Barker M.S., Tank D.C., Soltis P.S. Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 2017;105:348–363. doi: 10.1002/ajb2.1060. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Coghlan A., Ruan J., Coin L.J.M., Hériché J.K., Osmotherly L., Li R., Liu T., Zhang Z., Bolund L. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wang J., Ma C., Zhao Y., Wang Y., Hasi A., Qi Z. Glutamate receptor-like channel3.3 is involved in mediating glutathione-triggered cytosolic calcium transients, transcriptional changes, and innate immunity responses in Arabidopsis. Plant Physiol. 2013;162:1497–1509. doi: 10.1104/pp.113.217208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Jiang X., Lv X., Ahammed G.J., Guo Z., Yu J., Zhou Y. Tomato GLR3.3 and GLR3.5 mediate cold acclimation-induced chilling tolerance by regulating apoplastic H2O2 production and redox homeostasis. Plant Cell Environ. 2019;42:3326–3339. doi: 10.1111/pce.13623. [DOI] [PubMed] [Google Scholar]
- Liu Y., Huang X., Li M., Zhang Y. Loss-of-function of Arabidopsis receptor-like kinase BIR1-activates cell death and defense responses mediated by BAK1 and DOBIR1. New Phytol. 2016;212:637–645. doi: 10.1111/nph.14072. [DOI] [PubMed] [Google Scholar]
- Lovell J.T., Bentley N.B., Bhattarai G., Jenkins J.W., Sreedasyam A., Alarcon Y., Bock C., Boston L.B., Carlson J., Cervantes K. Four chromosome scale genomes and a pan-genome annotation to accelerate pecan tree breeding. Nat. Commun. 2021;12:4125. doi: 10.1038/s41467-021-24328-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1996;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M.-C., You F.M., Li P., Wang J.-R., Zhu T., Dandekar A.M., Leslie C.A., Aradhya M., McGuire P.E., Dvorak J. Synteny analysis in Rosids with a walnut physical map reveals slow genome evolution in long-lived woody perennials. BMC Genomics. 2015;16:707. doi: 10.1186/s12864-015-1906-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Tian t., Lin R., Deng X., Wang H., Li G. Arabidopsis FHY3 and FAR1 regulate light-induced myo-inositol biosynthesis and oxidative stress responses by transcriptional activation of MIPS1. Mol. Plant. 2016;9:541–557. doi: 10.1016/j.molp.2015.12.013. [DOI] [PubMed] [Google Scholar]
- Manchester S.R. Early history of the Juglandaceae. Plant Syst. Evol. 1989;162:231–250. [Google Scholar]
- Manzoor H., Kelloniemi J., Chiltz A., Wendehenne D., Pugin A., Poinssot B., Garcia-Brugger A. Involvement of the glutamate receptor AtGLR3.3 in plant defense signaling and resistance to Hyaloperonospora arabidopsidis. Plant J. 2013;76:466–480. doi: 10.1111/tpj.12311. [DOI] [PubMed] [Google Scholar]
- Mattison C.P., Rai R., Settlage R.E., Hinchliffe D.J., Madison C., Bland J.M., Brashear S., Graham C.J., Tarver M.R., Florane C. RNA-seq analysis of developing pecan (Carya illinoinensis) embryos reveals parallel expression patterns among allergen and lipid metabolism genes. J. Agric. Food Chem. 2017;65:1443–1455. doi: 10.1021/acs.jafc.6b04199. [DOI] [PubMed] [Google Scholar]
- Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y., Fawcett J.A., Proost S., Sterck L., Vandepoele K. The flowering world: a tale of duplications. Trends Plant Sci. 2009;14:680–688. doi: 10.1016/j.tplants.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J. In silico archeogenomics unveils modern plant genome organisation, regulation and evolution. Curr. Opin. Plant Biol. 2012;15:122–130. doi: 10.1016/j.pbi.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Sauquet H., Ho S.Y.W., Gandolfo M.A., Jordan G.J., Wilf P., Cantrill D.J., Bayly M.J., Bromham L., Brown G.K., Carpenter R.J. Testing the impact of calibration on molecular divergence times using a fossil-rich group: the case of Nothofagus (Fagales) Syst. Biol. 2012;61:289–313. doi: 10.1093/sysbio/syr116. [DOI] [PubMed] [Google Scholar]
- Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P., Heard E., Dekker J., Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simᾶo F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Singh S.K., Chien C.T., Chang I.F. The Arabidopsis glutamate receptor-like gene GLR3.6 controls root development by repressing the kip-related protein gene KRP4. J. Exp. Bot. 2006;67:1853–1869. doi: 10.1093/jxb/erv576. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Burleigh J.G. Surviving the K-T mass extinction: new perspectives of polyploidization in angiosperms. Proc. Natl. Acad. Sci. U S A. 2009;106:5455–5456. doi: 10.1073/pnas.0901994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stanke M., Steinkamp R., Waack S., Morgensternet B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:309–312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G., Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tena G., Boudsocq M., Sheen J. Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 2011;14:519–529. doi: 10.1016/j.pbi.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson T.E., Conner P.J. Vol. 8. Springer); USA: 2012. Pecan; pp. 771–801. (Fruit Breeding, Handbook of Plant Breeding). [Google Scholar]
- Thompson T.E., Grauke L.J. Genetic resistance to scab disease in pecan. HortScience. 1994;29:1078–1080. [Google Scholar]
- Thompson T.E., Hunter R.E. Pawnee pecan. Horts. 1985;20:776. [Google Scholar]
- Tsutomu K., Yamada K., Yoshimura S., Yamaguchi K. Chitin receptor-mediated activation of MAP kinases and ROS production in rice and Arabidopsis. Plant Signal. Behav. 2017;12:e1361076. doi: 10.1080/15592324.2017.1361076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G.A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Vurture G.W., Sedlazeck F.J., Nattestad M., Underwood C.J., Fang H., Gurtowski J., Schatz M.C. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics. 2017;34:2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B.J., Abeel T., Shea T., Priest M., Abouellie A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang G., Zhang C., Zhu P., Dai H., Yu .N., He Z., Xu L., Wang E. OsCERK1-mediated chitin perception and immune signaling requires receptor-like cytoplasmic kinase 185 to activate an MAPK cascade in rice. Mol. Plant. 2017;10:619–633. doi: 10.1016/j.molp.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Wang D., Zhang Y., Zhang Z., Zhu J., Yu J. Kaks_calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Tang W., Ma T., Niu D., Jin J., Wang H., Lin R. A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J. Integr. Plant Biol. 2016;58:91–103. doi: 10.1111/jipb.12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R.M., Seppey M., Simão F.A., Manni M., Ioannidis P., Klioutchnikov G., Kriventseva E.V., Zdobnov E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L. Pecan planting trends in Georgia. HortTechnology. 2014;24:475–479. [Google Scholar]
- Wickham H. 2nd edn. Springer; USA: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wood B.W., Conner P.J., Worley R.E. Relationship of alternate bearing intensity in pecan to fruit and canopy characteristics. HortScience. 2003;38:361–366. [Google Scholar]
- Wudick M.M., Michard E., Nunes C.O., Feijό J.A. Comparing plant and animal glutamate receptors: common traits but different fates? J. Exp. Bot. 2018;69:4151–4163. doi: 10.1093/jxb/ery153. [DOI] [PubMed] [Google Scholar]
- Yamada K., Yamaguchi K., Yashiura S., Terauchi A., Kawasaki T. Conservation of chitin-induced MAPK signaling pathways in rice and Arabidopsis. Plant Cell Physiol. 2017;58:993–1002. doi: 10.1093/pcp/pcx042. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Bioinformatics. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang W., Ji F., Qiu J., Song X., Bu D., Pan G., Ma Q., Chen J., Huang R. A high-quality walnut genome assembly reveals extensive gene expression divergences after whole-genome duplication. Plant Biotechnol. J. 2020;18:1848–1850. doi: 10.1111/pbi.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-K. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel A., Peer Y.V.de. wgd-simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics. 2019;35:2153–2155. doi: 10.1093/bioinformatics/bty915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequences, assembly, and annotation data have been deposited at NCBI GenBank under BioProject/BioSample numbers PRJNA727440/SAMN19020793. Resequencing reads for the 86 individuals have been deposited in the Sequence Read Archive (SRA) under BioProject/BioSample numbers PRJNA735040/SAMN19554720–19554805.