Abstract
Human apoptosis-linked gene-2 interacting protein X (ALIX), a versatile adapter protein, regulates essential cellular processes by shuttling between late endosomal membranes and the cytosol, determined by its interactions with Src kinase. Here, we investigate the molecular basis of these transitions and the effects of tyrosine phosphorylation on the interplay between structure, assembly, and intramolecular and intermolecular interactions of ALIX. As evidenced by transmission electron microscopy, fluorescence and circular dichroism spectroscopy, the proline-rich domain of ALIX, which encodes binding epitopes of multiple cellular partners, formed rope-like β-sheet–rich reversible amyloid fibrils that dissolved upon Src-mediated phosphorylation and were restored on protein-tyrosine phosphatase 1B–mediated dephosphorylation of its conserved tyrosine residues. Analyses of the Bro1 domain of ALIX by solution NMR spectroscopy elucidated the conformational changes originating from its phosphorylation by Src and established that Bro1 binds to hyperphosphorylated proline-rich domain and to analogs of late endosomal membranes via its highly basic surface. These results uncover the autoinhibition mechanism that relocates ALIX to the cytosol and the diverse roles played by tyrosine phosphorylation in cellular and membrane functions of ALIX.
Keywords: NMR, paramagnetic relaxation enhancement, programmed cell death 6 interacting protein, endosomal sorting complexes required for transport, post-translational modifications
Abbreviations: ALG-2, apoptosis linked gene-2; ALIX, apoptosis-linked gene-2 interacting protein X; CD, circular dichroism; CEP55, centrosomal protein of 55 kDa; CHMP, charged multivesicular body protein; CIL, Cambridge Isotopes Laboratories; CR, congo red; DLS, dynamic light scattering; D2O, deuterium oxide; ESI, electrospray ionization; GB1, B1 domain of protein G; HSQC, heteronuclear single quantum coherence; LBPA, 2,2′ lysobisphosphatidic acid; LC, liquid chromatography; MBP, maltose-binding protein; MS, mass spectrometry; MTS, (1-acetoxy-2,2,5,5-tetramethyl-δ-3-pyrroline-3-methyl) methanethiosulfonate; MTSL, (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate; NIH, National Institutes of Health; PK, proteinase K; POPC, 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine; POPE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine; POPS, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine; PRD, proline-rich domain; PRE, paramagnetic relaxation enhancement; PTM, post-translational modification; PTP1B, protein-tyrosine phosphatase 1B; TEM, transmission electron microscopy; TEMPOL, 4-hydroxy-2,2,6,6-tetramethylpiperidine 1-oxyl; TEV, tobacco etch virus; ThT, thioflavin T; TOF, time-of-flight; TROSY, transverse relaxation optimized spectroscopy; UCSD, University of California San Diego; UEV, ubiquitin E2 variant
Human apoptosis-linked gene-2 interacting protein X (ALIX) is a conserved adapter protein involved in essential cellular processes, including endosomal protein sorting, apoptosis, exosome biogenesis, and many others (1, 2, 3, 4). ALIX also regulates budding of diverse enveloped viruses, such as HIV (5, 6) and Ebola (7). It comprises an amino (N)-terminal Bro1 domain (residues 1–359), a central coiled-coil domain termed V (residues 360–702), and a carboxy (C)-terminal proline-rich domain (PRD; residues 703–868) (Fig. 1A). ALIX, also known as programmed cell death 6 interacting protein (PDCD6IP), undergoes tyrosine phosphorylation at multiple sites, which regulates its cellular functions; it comprises 28 conserved tyrosine residues (8), among which 11 are localized in the C-terminal portion of PRD, residues 803 to 846. ALIX interacts with and is phosphorylated by Src kinase at late endosomal membranes (8, 9); ALIX–endosomal membrane interactions are governed by its Bro1 domain, which binds to negatively charged 2,2′ lysobisphosphatidic acid (2,2′ LBPA; referred to as LBPA), a phospholipid found in late endosomes (10, 11, 12). ALIX–Src interactions result in phosphorylation of the Bro1 domain and hyperphosphorylation of the tyrosine-rich portion of PRD (8, 9). These interactions promote exosome secretion and cause ALIX to relocate from a membrane-bound active conformation to a cytosolic inactive state. Interdomain interactions between PRD and the upstream globular domains, Bro1 and V, are suggested to cause ALIX autoinhibition in the cytosol (13, 14). Structural details of these interactions are unclear because of the unavailability of recombinant full-length ALIX, owing to difficulties in expressing ALIX-PRD in bacteria and its disordered nature. Intact ALIX was, however, successfully produced using a baculovirus–insect cell expression system (14). ALIX-PRD encodes binding epitopes of numerous cellular proteins (15), and its hyperphosphorylation by Src affects its cellular interactions (8). We recently discovered that the N-terminal portion of ALIX-PRD (residues 703–800) is disordered, whereas its C-terminal tyrosine-rich portion (residues 800–868) forms amyloid fibrils that disassemble on Src-mediated hyperphosphorylation (16). However, the mechanistic effects of tyrosine phosphorylation on cellular redistribution and functions of ALIX are unclear.
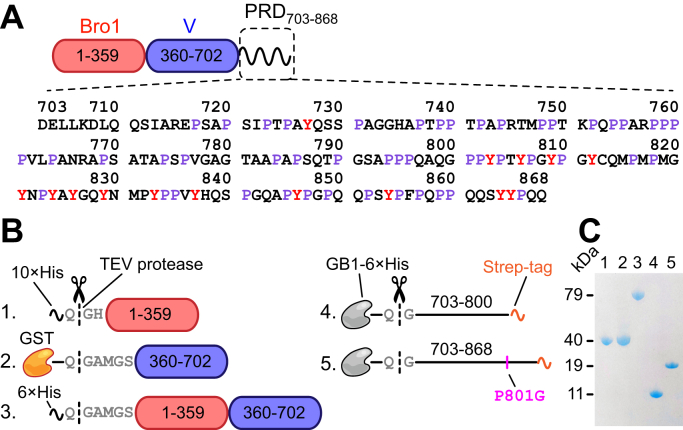
Figure 1.
ALIX domain organization and summary of recombinant ALIX constructs used in the current work.A, schematic of ALIX organization. Primary sequence of the PRD is shown. B, recombinant ALIX constructs used in the current work. Constructs 1 to 3 were obtained from Addgene; accession no. 80641 (47), 17639 (48), and 42577 (49), respectively. Construct 4 is described in our previous work (16); Addgene accession no. 141344. The positions of purification tags are marked. TEV protease cutting sites are shown. The point mutation, P801G, of is marked. C, SDS-PAGE analysis of TEV-cleaved ALIX constructs; the order of the constructs is the same as the one depicted in B. ALIX, apoptosis-linked gene-2 interacting protein X; PRD, proline-rich domain; TEV, tobacco etch virus.
Using recombinant ALIX domains and a range of biophysical methods, including NMR, fluorescence, and circular dichroism (CD) spectroscopy, mass spectrometry (MS), transmission electron microscopy (TEM), and dynamic light scattering (DLS), we elucidate how tyrosine phosphorylation affects the interplay between ALIX conformation, assembly, and function (poor bacterial expression and the overall size, ∼96 kDa, do not allow for a similar study in the context of full-length ALIX). Codon optimization coupled with a single-point mutation permitted expression of full-length ALIX-PRD in Escherichia coli. We show that PRD forms reversible β-sheet–rich fibrils modulated by post-translational modifications (PTMs), Src-mediated phosphorylation, and human protein-tyrosine phosphatase 1B (PTP1B)–mediated dephosphorylation of its conserved tyrosine residues. We establish that Bro1 binds to hyperphosphorylated PRD and to analogs of late endosomal membranes. The close correlation between the regions of Bro1 that associate with hyperphosphorylated PRD and with anionic phospholipids elucidates the underlying mechanism of PRD-mediated autoinhibition of ALIX–membrane interactions and how tyrosine phosphorylation causes redistribution of ALIX away from late endosomal membranes into the cytosol.
Results
Recombinant constructs
The current work made use of five recombinant ALIX constructs (Fig. 1, B and C). These include constructs representing the individual Bro1 and V domains (residues 1–359 and 360–702, respectively), a construct comprising both the Bro1 and V domains (residues 1–702; referred to as Bro1-V), and two constructs representing the N-terminal portion of PRD, (residues 703–800), and the entire PRD, (residues 703–868); “Strep” denotes a noncleavable C-terminal strep-affinity tag (17) that facilitated purification of intact PRD constructs from their truncated fragments using affinity chromatography. The latter are generated during heterologous expression of ALIX-PRD in E. coli because of translational arrest induced by its polyproline motifs. carries a P801G point mutation to resolve issues with protein expression in E. coli. We previously established that the 800GPPYP804 segment of ALIX-PRD is responsible for ribosomal stalling in E. coli, resulting in essentially no expression of wildtype PRD beyond residue 801 (16). Note that the effects of the P801G mutation on the cellular functions of ALIX are currently not known. For paramagnetic NMR, three cysteine variants of were engineered (see below). We also made use of full-length Src kinase and the catalytic domain of PTP1B; see Figure S1 for liquid chromatography (LC)–electrospray ionization (ESI)–time-of-flight (TOF)MS analyses of constructs used in the current study.
Reversible amyloid fibrils of
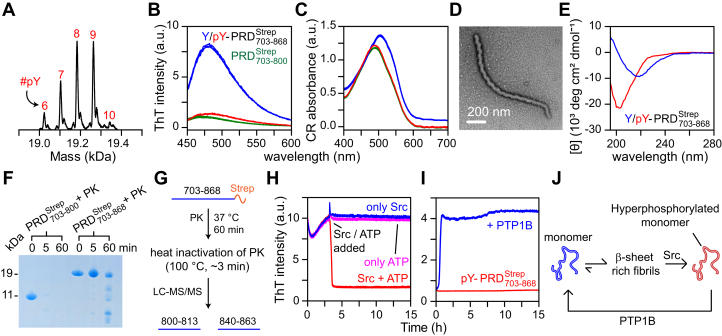
Using dye-binding assays with amyloid-specific probes, thioflavin T (ThT) and congo red (CR), and TEM, we previously established that the C-terminal tyrosine-rich fragment of ALIX-PRD (PRD800–868) forms rope-like fibrils that dissolve into monomeric units on Src-mediated hyperphosphorylation (16). Because of the poor solubility of PRD800–868 (<30 μM; pH 4.5–7), the data reported in our prior study were collected on a fusion protein with an N-terminal solubility enhancement tag comprising the B1 domain of protein G (GB1) (18). In contrast to PRD800–868, was more soluble (∼300 μM; pH 4.5–8), which allowed us to characterize its aggregation properties and its interactions with Src in the absence of a GB1 tag. LC–ESI–TOFMS analyses of Src-mediated phosphorylation revealed the formation of hyperphosphorylated with phosphorylation of six to ten (out of 15) tyrosine residues (Fig. 2A and Table S1). ThT emission assays confirmed the presence of β-sheet–rich conformations for aggregates (Fig. 2B). Spectral-shift assays carried out using CR demonstrated clear shifts toward 540 nm, further validating their amyloidogenic nature (Fig. 2C). For these assays, the soluble N-terminal fragment, (16) and hyperphosphorylated served as negative controls. NMR analysis revealed a near-perfect superimposition of 1H–15N transverse relaxation optimized spectroscopy (TROSY)–heteronuclear single quantum coherence (HSQC) spectra of these two proteins, establishing Src-mediated dissolution of aggregates into NMR-amenable soluble monomers and the complete lack of interactions between the N-terminal and C-terminal portions of PRD (Fig. S2). Backbone resonance assignments of hyperphosphorylated were not feasible owing to the complexity and heterogeneity of its phosphorylation pattern. However, a narrow chemical shift dispersion (∼7.7–8.7 ppm) suggested that the hyperphosphorylated is disordered in solution. TEM analyses revealed the presence of rope-like unbranched filaments, characteristic of amyloid fibrils (Fig. 2D). CD spectroscopic data confirmed the presence of β-sheet conformations in these fibrils (Fig. 2E), corroborating the presence of amyloid structure. The corresponding CD spectrum of hyperphosphorylated validated its random-coil conformation.
Figure 2.
Reversible amyloids of.A, LC–ESI–TOFMS analysis of Src-mediated phosphorylation of ; the number of phosphorylated tyrosine residues is marked in red. Emission spectra of ThT, a.u., arbitrary units (B) and absorbance spectra of CR, a.u., absorbance units (C) of aggregates (blue); n = 3. (green) and hyperphosphorylated (red) were used as controls. D, negatively stained EM images of fibrils. E, CD spectra of fibrils (blue) and its hyperphosphorylated form (red). F, SDS-PAGE analysis of limited PK digestion of fibrils and monomeric . G, scheme of PK digestion and the results of LC–MS/MS analysis. H and I, the impact of tyrosine phosphorylation and dephosphorylation on aggregation kinetics of assessed using ThT assays (30 °C). H, 100 μM was incubated without Src for ∼3 h (n = 9). 10 μM Src + 1 mM ATP were added to three samples (red), whereas the remaining received either 10 μM Src (blue) or 1 mM ATP (pink). I, 100 μM hyperphosphorylated samples (n = 3) were incubated in the absence (red) and the presence (blue) of 50 nM PTP1B. J, scheme depicting potential mode of formation and dissolution of fibrils. CR, congo red; ESI, electrospray ionization; PK, proteinase K; PRD, proline-rich domain; PTP1B, protein-tyrosine phosphatase 1B; ThT, thioflavin T.
To identify the fibril core resistant to protease treatment, a limited digestion of fibrils was carried out using proteinase K (PK) (Fig. 2, F and G). Based on band intensities of the reaction mixture comprising intact protein and the proteolyzed products monitored using SDS-PAGE (Fig. 2F), we conclude that fibrils are resistant to PK digestion as the band for persisted after ∼60 min of incubation with PK, albeit at a lower intensity than the corresponding band at time 0. In contrast, the soluble was completely hydrolyzed by PK within ∼5 min of the reaction. LC–tandem MS (LC–MS/MS) analysis of PK digestion of fibrils revealed that the two most abundant fragments were localized in the C-terminal region, residues 800 to 813 and 840 to 863 (Figs. 2G and S3), indicating that these motifs likely participate in the formation of the fibril core. A limited PK digestion (incubation time of ∼5 min) was also performed on hyperphosphorylated , and the corresponding cleavage products were identified using LC–MS/MS, which permitted identification of phosphorylated tyrosine residues, namely 803, 806, 809, 812, 829, 833, 837, 846, and 854 (Fig. S4). Nothing definitive can be said about the phosphorylation status of the remaining tyrosine residues because of sequencing coverage gaps arising from problems associated with proteolytic digestion of . Note that the tyrosine-rich portion of , residues 800 to 868, carries no native lysine and arginine residues (cf., Fig. 1A) and, thus, cannot be probed by traditional trypsin-based proteomics (19). PK digestion of hyperphosphorylated was not analyzed by SDS-PAGE because of smearing of protein bands owing to phosphorylation (20).
To assess the impact of tyrosine phosphorylation on polymerization, aggregation kinetics using ThT fluorescence were monitored. exhibited sigmoidal aggregation profiles, a hallmark of fibril formation (Fig. 2H). To establish Src-mediated dissolution of fibrils, Src and ATP were added to samples upon reaching a stationary phase (at ∼3 h; Fig. 2H), which resulted in a significant loss of ThT signal with a corresponding half-life (t1/2) of ∼0.2 h. The remaining samples were mixed with either Src or ATP and displayed no drop in ThT signals, indicating that Src or ATP do not affect fibrils. Remarkably, judging by the rapid appearance of characteristic sigmoidal ThT profiles (t1/2 ∼0.5 h; Fig. 2I), dephosphorylation of hyperphosphorylated by PTP1B resulted in restoration of fibrils, whereas the hyperphosphorylated without PTP1B showed no sigmoidal ThT profile (activity of recombinant PTP1B was measured using the malachite green assay; Fig. S5. For Src, the activity was measured using the ADP-Glo assay, described in our previous work (16)). Although the exact identity of the phosphatase that works in tandem with ALIX is not known, PTP1B was suggested to regulate the endosomal sorting machinery (21). These observations establish that forms rope-like β-sheet–rich fibrils that dissolve into soluble monomers on hyperphosphorylation by Src and that the removal of phosphoryl groups via PTP1B culminates in the restoration of fibrils, making a completely reversible amyloid controlled by PTMs (Fig. 2J). In addition to PRD, Src is suggested to phosphorylate the Bro1 domain of ALIX in vivo (8). To characterize the impact of phosphorylation on Bro1, we performed detailed NMR analyses of nonphosphorylated and phosphorylated Bro1, described below.
NMR analysis of nonphosphorylated and phosphorylated Bro1
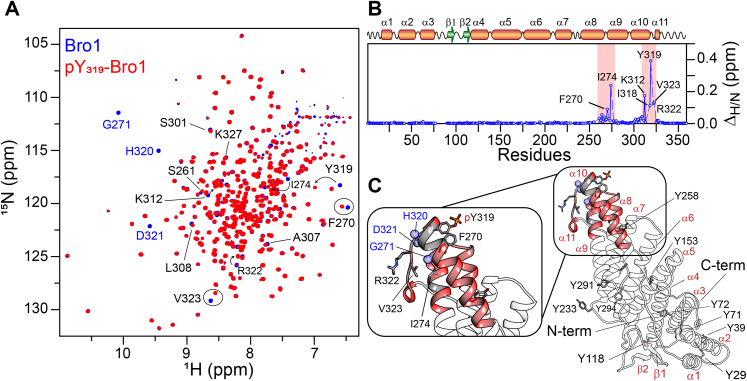
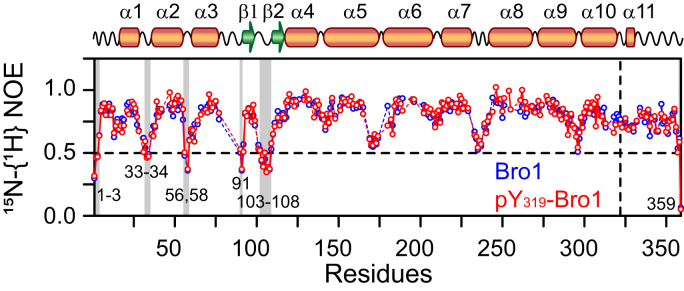
ALIX–Src interactions are suggested to proceed in a stepwise fashion in vivo, where Src first binds and phosphorylates the highly conserved region, 312KKDNDFIY319, of the Bro1 domain, followed by hyperphosphorylation of the tyrosine-rich portion of PRD, ultimately resulting in the relocation of ALIX from late endosomal membranes to the cytosol (8). Src-mediated phosphorylation of Bro1 was confirmed using Western blotting (Fig. S6A). LC–ESI–TOFMS and LC–MS/MS analyses established the formation of phosphorylated Bro1 and confirmed the phosphorylation of Y319 (Fig. S6, B and C, respectively). To explore the impact of tyrosine phosphorylation on Bro1 at atomic resolution, we conducted a detailed NMR investigation (Fig. 3). Although the overall size of Bro1 is relatively large for solution NMR studies (∼40 kDa), excellent spectral quality was obtained by perdeuteration coupled with TROSY (Fig. 3A), which allowed us to carry out nearly complete backbone resonance assignments of Bro1. Based on an excellent agreement between the secondary structure derived from the assigned backbone chemical shifts (13Cα, 13Cβ, 13C′, 15N, and 1HN) using TALOS-N (22) with that obtained from the three-dimensional crystal structure of Bro1 (Protein Data Bank entry: 5WA1 (23); Fig. S7), we conclude that recombinant Bro1 is well folded in solution. To probe structural and conformational changes that take place in Bro1 upon Src-mediated phosphorylation of Y319, 1HN/15N chemical shift perturbation mapping was utilized. NMR analyses of phosphorylated Bro1 revealed large 1HN/15N chemical shift perturbations that arise from the introduction of a negatively charged phosphoryl group (Fig. 3B). Mapping of these perturbations onto the crystal structure of Bro1 allowed identification of regions affected by tyrosine phosphorylation (Fig. 3C). All perturbations were localized in the regions surrounding Y319, which further confirmed specific phosphorylation of Bro1 by Src. The heteronuclear 15N-{1H} NOE data (Fig. 4) of nonphosphorylated and phosphorylated Bro1 indicated that the introduction of phosphoryl group did not affect the local internal mobility of Bro1. Collectively, the aforementioned data established that Src-mediated phosphorylation of Bro1 at Y319 induces local changes.
Figure 3.
NMR characterization of nonphosphorylated and phosphorylated Bro1.A, overlay of 1H–15N TROSY–HSQC spectra of nonphosphorylated (blue) and phosphorylated (red) Bro1. A few of the 1H–15N crosspeaks that undergo chemical shift changes on phosphorylation are labeled and circled. Residues that exhibit resonance line broadening upon phosphorylation, namely G271, H320, and D321, are labeled in blue. B, 1HN/15N chemical shift perturbation profile of Bro1 upon phosphorylation. Secondary structure elements are indicated above the panel. Red rectangles indicate residues (261–280 and 300–328) that exhibit large chemical shift perturbations because of phosphorylation. C, a ribbon diagram of phosphorylated Bro1 constructed using Protein Data Bank entry 5WA1 in Xplor-NIH (65); tyrosine and a few of the residues that undergo large chemical shift changes upon phosphorylation are shown in stick representation. Red ribbons represent residues that are most affected because of phosphorylation. Gray ribbons indicate residues around the phosphorylation site that could not be assigned unambiguously. Residues that undergo resonance line broadening upon phosphorylation are depicted as blue spheres. HSQC, heteronuclear single quantum coherence; NIH, National Institutes of Health; TROSY, transverse relaxation optimized spectroscopy.
Figure 4.
15N–{1H} heteronuclear NOE data for nonphosphorylated and phosphorylated Bro1. Regions with high local mobility on the nanosecond time scale, 15N–{1H} NOE <0.5, are labeled and marked with semitransparent gray rectangles. The location of Y319 is indicated by a vertical dashed line. All data were acquired with 500 μM 2H/15N labeled proteins at a spectrometer 1H frequency of 800 MHz at 30 °C.
Lack of interactions between and globular ALIX domains
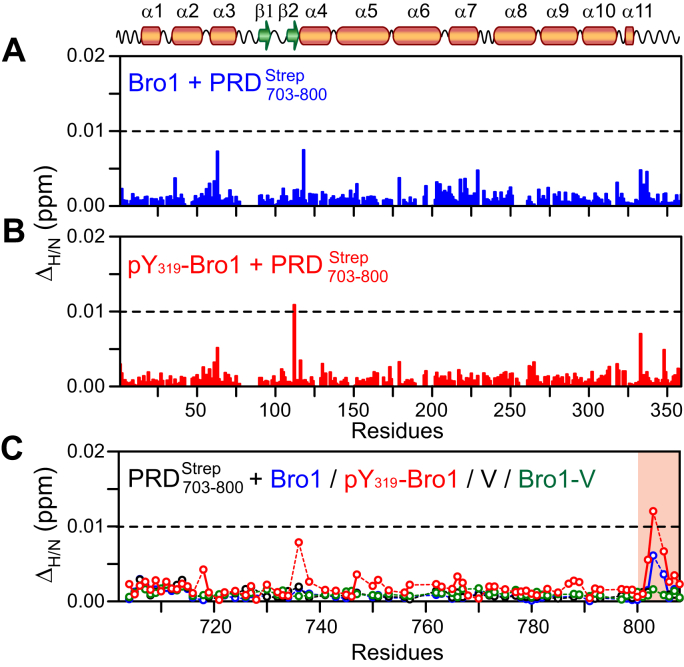
A prior study suggested that the N-terminal portion of PRD, specifically the 717PSAP720 motif that interacts with the ubiquitin E2 variant (UEV) domain of tumor-susceptibility gene 101, directly associates with the Src-binding region of Bro1 (13). PRD was also suggested to prevent the binding between the V domain and the YPXnL consensus motifs (X = any residue and n = 1–3) found in viral proteins, although molecular details of this inhibition were not determined (14). To investigate interdomain interactions of ALIX, we carried out 1HN/15N chemical shift perturbation mapping using and nonphosphorylated and phosphorylated Bro1 (Fig. 5). Negligible 1HN/15N chemical shift perturbations were observed for both nonphosphorylated and phosphorylated NMR-visible Bro1, one at a time, upon the addition of three-molar equivalents of (Fig. 5, A and B, respectively), indicating that Bro1 did not bind to . The corresponding titration experiments carried out using NMR-visible and unlabeled nonphosphorylated and phosphorylated Bro1 yielded negligible chemical shift changes (Fig. 5C), confirming the lack of Bro1– interactions. To determine the involvement of the V domain, titration experiments were carried out using NMR-visible , in the absence and presence of three-molar equivalents of V and Bro1-V constructs (one at a time). Minimal chemical shift changes indicated that V and Bro1-V constructs did not bind to (Fig. 5C). Experiments using NMR-visible V and Bro1-V constructs were not viable since both produced poor-quality NMR spectra, likely because of the open–close transitions of the V domain in solution (14). Collectively, these observations indicated that Bro1 and V domains did not interact with . These findings are consistent with the fact that unlike proline-recognition domains such as UEV (24, 25), Bro1 domains have not been known to bind to proline-rich motifs, and that ALIX-PRD carries no YPXnL consensus motif recognized by the V domain. Finally, nothing definitive can be said about the interactions of intact with Bro1 and V domains in solution because of its propensity to form fibrils. We, however, uncovered novel interactions between Bro1 and hyperphosphorylated , which are described below.
Figure 5.
Lack of association betweenand globular ALIX domains.1HN/15N chemical shift perturbation profiles of (A) nonphosphorylated and (B) phosphorylated 2H/15N-labeled Bro1 on addition of three-molar equivalent of nonlabeled . C, 1HN/15N chemical shift perturbation profiles of 15N-labeled on addition of nonlabeled Bro1 (blue), phosphorylated Bro1 (red), V (black), and Bro-V (dark green); one at a time, to globular ALIX domains molar ratio = 1:3. Region highlighted in semitransparent orange rectangle indicates the location of the C-terminal strep purification tag (residues 801–808). ALIX, apoptosis-linked gene-2 interacting protein X; PRD, proline-rich domain.
Interactions between Bro1 and hyperphosphorylated
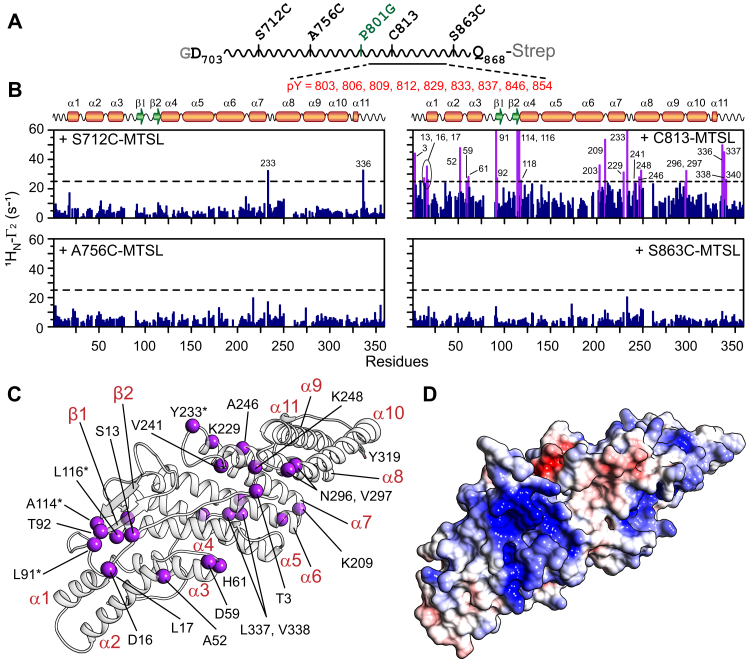
Src-mediated hyperphosphorylation of PRD relocates ALIX from late endosomal membranes to the cytosol (8), indicating a likely competition between hyperphosphorylated PRD and membranes for association with the phospholipids-binding region(s) of Bro1. To test this hypothesis and identify contacts between Bro1 and hyperphosphorylated , we made use of intermolecular paramagnetic relaxation enhancement (PRE) (26). The magnitude of the PRE effect is very large owing to the large magnetic moment of the unpaired electron of the paramagnetic label and is proportional to the <r−6> paramagnetic label—proton distance, which enables the detection of sparsely populated (as low as 0.5–1% in favorable cases) encounter complexes between binding partners (27). The paramagnetic nitroxide spin label (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate (MTSL) was introduced at four separate sites on hyperphosphorylated via disulfide linkages (Fig. S8A; also see Fig. S8B and Table S1 for LC–ESI–TOFMS analysis of MTSL conjugated products). These four sites were the native cysteine residue of , C813, and three engineered sites, S712C, A756C, and S863C (Fig. 6A; in all cysteine variants, C813 was mutated to a serine). Site C813 lies adjacent to a cluster of tyrosine residues that are phosphorylated by Src (cf., Fig. S4). Sites S712C and A756C are located near the motif that binds to tumor-susceptibility gene 101-UEV (residues 717–720; (16)) and in the highly basic region of PRD (residues 756–767; theoretical isoelectric point ∼12), respectively. Site S863C is located at the extreme C terminus of PRD and is adjacent to 864YY865 (cf., Fig. 1A); we were unable to detect the phosphorylation status of these two tyrosine residues because of sequencing coverage gaps.
Figure 6.
PRE mapping of Bro1–hyperphosphorylatedinteractions.A, schematic of ; vertical lines indicate the location of the four independent conjugation sites and of the P801G mutation (green). The location of phosphorylated tyrosine residues is shown. B, intermolecular PREs observed on 2H/15N-labeled Bro1 (200 μM) arising from MTSL label conjugated to hyperphosphorylated at four specific sites; Bro1 to molar ratio = 1:1. Conjugation of MTSL at site C813 produced a noticeable intermolecular PRE; residues with intermolecular PREs above the background are labeled and indicated by purple bars. PREs too large (>60 s−1) to be accurately measured are plotted as 60 s−1. The remaining three conjugation sites did not generate large intermolecular PREs; a few exceptions are labeled. C, ribbon representation of Bro1; residues that exhibit large PREs (≥25 s−1) with spin label at site C813 are depicted as purple spheres; residues that were completely broadened out are marked with asterisks. Y319 is shown in a stick representation. D, molecular surface of Bro1 (in the same orientation as C) color coded according to electrostatic potential; ±5 kT with blue, positive; white, neutral; and red, negative. PRD, proline-rich domain; PRE, paramagnetic relaxation enhancement; MTSL, (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate.
Intermolecular PRE (1HN–Γ2) profiles observed for NMR-visible Bro1 in the presence of paramagnetically labeled hyperphosphorylated and its cysteine variants are shown in Figure 6B. When the paramagnetic label was located at site S712C of , two lone ≥25 s−1 PREs were observed (residues 233 and 336 of Bro1), whereas no strong PRE of ≥25 s−1 were seen when the label was conjugated to site A756C of . These minimal intermolecular PRE effects indicate that interactions of Bro1 with sites S712C and A756C of are negligible. These observations validate the results of our NMR titration experiments, which demonstrated that the isolated Bro1 did not associate with the N-terminal portion of PRD and rule out transient association between the highly basic region of PRD encompassing site A756C and electronegative surface accessible regions of Bro1. When the paramagnetic label was located at site C813 of , strong PREs (≥25 s−1) were observed for several Bro1 residues, with residues 91, 114, 116, and 233 completely broadened out. These observations indicate that the spin label is in the close vicinity of the aforementioned Bro1 residues, possibly because of its proximity to phosphorylated tyrosine residues of that likely drive the interdomain electrostatic association. No strong PREs (≥25 s−1) were observed with the paramagnetic label at site S863C of , indicating minimal interactions with this site, perhaps because tyrosine residues located next to this site (864YY865) are not phosphorylated by Src. Control experiments carried out using either free water-soluble paramagnetic nitroxide label, 4-hydroxy-2,2,6,6-tetramethylpiperidine 1-oxyl (TEMPOL), or an unrelated MTSL-conjugated protein, apo maltose-binding protein carrying an E38C point mutation (MBP-E38C), showed no significant intermolecular PRE effects (Fig. S9). These results establish that the intermolecular PREs obtained for Bro1 with label at site C813 of hyperphosphorylated arise from a specific association of this site with Bro1 and not from preferential binding of the paramagnetic label to Bro1.
A quantitative interpretation of the strong intermolecular PREs obtained with paramagnetic label conjugated to site C813 is not plausible owing to the disordered nature of hyperphosphorylated and the complexity of phosphorylation pattern (cf., Fig. S8B), culminating in a highly dynamic encounter complex with Bro1. The experimental manifestation of this dynamic complex is the approximately uniform and elevated 1H–Γ2 PRE background of ∼10 to 15 s−1 (Fig. 6B; for the other three sites of , this background is relatively lower, ∼5–10 s−1 and stems from random collisions of these three sites with surface accessible sites of Bro1). Mapping these substantial PREs on to the crystal structure of Bro1, however, allowed us to identify the preferential mode of interactions between hyperphosphorylated and Bro1. Two clusters of intermolecular contacts can be discerned (Fig. 6C), both residing on the two arms of the convex face of Bro1. For comparison, the molecular surface of Bro1 color coded according to electrostatic potential calculated using APBS software (Adaptive Poisson-Boltzmann Solver) (28) is shown in Figure 6D. One contact site comprises the N-terminal portion of Bro1, residues 13, 16, 17, 52, 59, 61, 91, 92, 114, 116, and 118. Because three of these residues were completely broadened out in the presence of paramagnetically labeled hyperphosphorylated , we speculate that this cluster makes primary contact with paramagnetically labeled site C813. This cluster has a strong basic character and, therefore, can electrostatically associate with hyperphosphorylated , further validating that the interdomain interactions are governed by phosphorylated tyrosine residues of PRD. The other contact site comprises residues 203, 209, 229, 233, 241, 246, 248, 296, and 297, and residue 3, which lies in close vicinity of this cluster. Because of a less pronounced electropositive surface, we hypothesize that this Bro1 cluster serves as a secondary contact site with hyperphosphorylated . Technical difficulties prevented us from measuring intermolecular PREs on phosphorylated Bro1 owing to a significant precipitation observed upon mixing phosphorylated Bro1 with paramagnetically labeled hyperphosphorylated . However, since Y319 of Bro1 is located away from the two Bro1 regions that interact with hyperphosphorylated (Fig. 6C), we predict that Bro1 phosphorylation will have a negligible effect on the association of these two domains. Collectively, the aforementioned observations establish that site C813 of hyperphosphorylated , which lies adjacent to a group of phosphorylated tyrosine residues, transiently interacts with two electropositive regions of Bro1.
Interactions between Bro1 and analogs of late endosomal membranes
To identify the phospholipid-binding region(s) of Bro1, we made use of NMR titration experiments, where unilamellar lipid vesicles were mixed with NMR-visible Bro1. To mimic the composition of late endosomal membranes, extruded vesicles were made using 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and LBPA. We also made use of zwitterionic vesicles comprising POPC and POPE and negatively charged vesicles composed of POPC, POPE, and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS). Note that eukaryotic membranes, including those of late endosomes, are highly complex and that the vesicles used here represent an artificial system. However, the latter offer the most native-like local membrane environment and, therefore, are often considered simplified models of cellular membranes (29). The size distribution of vesicles, in the absence and presence of Bro1, was measured by DLS and found to be homogenous with mean diameters ranging from 130 to 150 nm (Fig. S10). A previous study suggested that the binding of divalent calcium to Bro1 was necessary for Bro1–membrane interactions (11). However, negligible chemical shift changes were observed for Bro1 resonances in the presence of a molar excess of calcium, which ruled out Bro1–calcium interactions (Fig. S11).
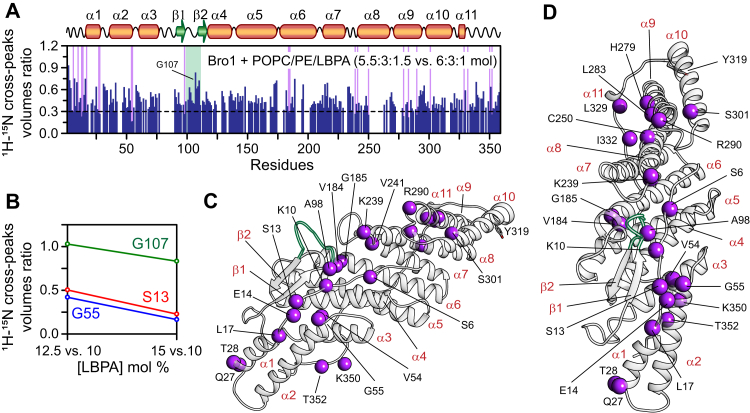
On addition of POPC/POPE/LBPA vesicles (molar ratio of 6:3:1) to 100 μM NMR-visible Bro1 (protein to lipid molar ratio: 1:30), ∼20 to 30% resonance broadening was observed for a majority of 1H–15N cross peaks of Bro1 (Fig. S12A; no chemical shift perturbations and new resonances were observed upon addition of vesicles to Bro1). When LBPA concentration was increased in a stepwise fashion (10, 12.5, and 15 mol% of LBPA; Fig. S12, A–C), crosspeaks of Bro1 were progressively attenuated. These results indicate the formation of a high–molecular weight complex between Bro1 and lipid vesicles and an increase in the vesicle-bound population of Bro1 as a function of LBPA concentration; all NMR measurements were carried out using the same protein to lipid molar ratio of 1:30. The reductions in 1H–15N cross-peak volumes of Bro1 as a function of increasing LBPA concentration (Bro1 + POPC/POPE/LBPA vesicles, molar ratio 5.5:3:1.5 versus Bro1 + POPC/POPE/LBPA vesicles, molar ratio 6:3:1) are plotted in Figure 7A, which permitted identification of the most affected residues that likely associate directly with LBPA-enriched particles. The solvent-exposed motif, residues 101KGSLFGGSVK110, of the extended loop that connects the two β-strands of Bro1 was predicted to insert into LBPA-enriched membranes (11). Although reduction of cross-peak volumes was observed for residues 104 to 106, the other residues of this exposed motif were comparatively less affected in the presence of lipid particles, with G107 being the least affected residue (Figs. 7A and S12). A quantitative analysis of the attenuation of 1H–15N cross-peak volumes of G107 as a function of increasing LBPA concentration revealed the ratios of ∼1 and ∼0.8 between 12.5 versus 10 and 15 versus 10 mol% of LBPA, respectively (∼20% attenuation; Fig. 7B). On the other hand, the most affected Bro1 residues, S13 and G55, exhibited ∼60% attenuation in their cross-peak volumes as a function of increasing LBPA concentration. These observations rule out the insertion of this exposed loop into the vesicles since in the event of such insertion, the corresponding residues would exhibit a significantly greater resonance line broadening. Mapping of the most affected Bro1 residues in the presence of LBPA-enriched particles onto the crystal structure of Bro1 revealed two clusters, localized on two arms of the convex face of Bro1, which likely make direct contacts with lipid particles (Fig. 7, C and D). Cluster 1 comprised residues 10, 13, 14, 17, 54, 55, 98, 184, and 185, whereas residues 6, 239, 241, 250, 279, 283, 290, 301, 329, and 322 formed cluster 2. Residues 27 to 28 and two extreme C-terminal residues, 352 to 353, were outliers, likely because of nonspecific association with the lipid particles. Y319 of Bro1 is located away from both clusters that interact with lipid particles. A near-perfect match was observed with the pattern and the magnitude of attenuation of resonances of nonphosphorylated and phosphorylated Bro1 (one at a time) in the presence of LPBA-enriched particles, indicating that Src-mediated phosphorylation of Bro1 plays no major role in interactions with phospholipids (Fig. S13). A close agreement between the two clusters that interact with lipid vesicles and the electropositive regions of Bro1 (cf., Figs. 6C and 7, C and D) indicated that interactions between Bro1 and lipid particles were primarily governed by the negative charge of LBPA. To confirm the electrostatic nature of these interactions, similar titration experiments were carried out using Bro1, in the absence and presence of zwitterionic and negatively charged vesicles (POPC/POPE and POPC/POPE/POPS, one at a time, Fig. S14, A and B, respectively). Negligible signal reductions were observed with zwitterionic vesicles (POPC/POPE molar ratio 7:3). These results also rule out the role of viscosity on attenuation of Bro1 resonances in the presence of vesicles. Negatively charged POPC/POPE/POPS particles (molar ratio of 5.5:3:1.5) produced a very similar attenuation profile as that of Bro1 + POPC/POPE/LBPA vesicles, indicating that ionic interactions drive the association between Bro1 and lipid particles. Finally, we note that despite similarities, the magnitude of cross-peak attenuation is greater with LBPA than POPS particles (cf., Figs. S12C and S14B, respectively), indicating a stronger association of Bro1 with LBPA-enriched particles.
Figure 7.
Interaction of Bro1 with LBPA enriched lipid vesicles.A, reduction in cross-peak volumes of 2H/15N-labeled Bro1 (100 μM) as a function of increasing amount of LBPA; Bro1 + POPC/POPE/LBPA vesicles (5.5:3:1.5 versus 6:3:1), the numbers in parenthesis represent the molar ratio of each phospholipid in vesicles. Bro1 to lipid molar ratio = 1:30. Semitransparent purple rectangles indicate residues that exhibit significant attenuation (≤0.3) in the presence of increasing concentrations of LBPA. A semitransparent green rectangle marks the location of the flexible loop connecting the two β-strands; G107 is marked. B, relative changes in cross-peak volumes of three representative residues as a function of increasing concentration of LBPA. The ratios of cross-peak volumes were obtained by recording three 1H–15N TROSY–HSQC spectra of Bro1 in the presence of vesicles containing increasing amounts of LBPA. The following vesicle compositions were used: POPC/POPE/LBPA molar ratios 6:3:1, 5.75:3:1.25, and 5.5:3:1.5. C and D, ribbon representation of Bro1 in two different orientations; the most affected residues upon association with LBPA-enriched vesicles are depicted as purple spheres. Y319 is shown in a stick representation. The extended loop connecting the two β-strands is marked in green. HSQC, heteronuclear single quantum coherence; LBPA, 2,2′ lysobisphosphatidic acid; POPC, 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine; POPE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine; TROSY, transverse relaxation optimized spectroscopy.
Discussion
In summary, we establish that tyrosine phosphorylation of ALIX-PRD plays a critical role in its polymerization and that hyperphosphorylated PRD likely competes with anionic phospholipids for binding to the Bro1 domain. These data elucidate the underlying mechanism by which tyrosine phosphorylation triggers the cellular redistribution of ALIX and its impact on intramolecular and intermolecular associations that dictate the broad functional repertoire of ALIX in cell signaling.
exhibited remarkable aggregation properties and formed β-sheet–rich amyloid fibrils that dissolved on Src kinase–mediated phosphorylation and reassembled on PTP1B-mediated dephosphorylation of its conserved tyrosine residues. LC–MS/MS analyses of PK digestion of fibrils revealed two protected fragments, residues 800 to 813 and 840 to 863, that likely form the fibril core. The motif encompassing residues 800 to 813 of PRD encodes epitopes of two binding partners, centrosomal protein of 55 kDa (CEP55), which is a mitotic phosphoprotein involved in cytokinetic abscission (30), and apoptosis linked gene-2 (ALG-2), which is a calcium-binding protein necessary for cell death (31). We speculate that the formation of ALIX fibrils provides a stringent regulatory control as the binding epitopes required for the recruitment of CEP55 and ALG-2 will be unavailable because of their involvement in fibril formation, whereas Src-mediated disassembly of ALIX fibrils will allow for the subsequent recruitment of CEP55 and ALG-2 by ALIX. In line with these hypotheses, a cellular study uncovered that inhibition of the focal adhesion kinase–Src signaling pathway affected the timing of CEP55 recruitment at the midbody and blocked cytokinetic abscission (32). Src-mediated hyperphosphorylation of PRD results in a heterogenous phosphorylation pattern in vitro. This complexity may provide additional regulatory control for ALIX-mediated ALG2 recruitment and subsequent cell death. This is because the tyrosine residues of the PRD motif, residues 800 to 813, were shown to be critical for ALG2 association (33). The C-terminal residues of PRD (residues 832–868, especially motif 852PSYP855) were shown to be essential for ALIX multimerization in vivo (30). These observations are consistent with our in vitro findings that residues 840 to 863 of PRD are involved in fibril formation. The filament-forming components of the endosomal sorting complexes required for transport pathway known as charged multivesicular body proteins (CHMPs) recruit ALIX through its Bro1 domain to endosomal membranes (34, 35). Conversely, ALIX-Bro1 recruits and nucleates CHMPs (36), which are the main drivers of endosomal sorting complexes required for transport-mediated membrane remodeling. We predict that the formation of PRD fibrils will likely result in a multiplicative increase in the binding affinity between Bro1 and CHMPs, culminating in nucleation and polymerization of CHMPs. These observations make a strong case for the existence of PRD-mediated ALIX assemblies in vivo and suggest a vital role of PTMs in regulating the timing of the recruitment of signaling partners of ALIX.
The structural characteristics and dynamics of the Bro1 domain were assessed using NMR spectroscopy. Conformational changes stemming from Src-mediated phosphorylation of residue Y319 of Bro1 were found to be localized around this tyrosine. NMR titration experiments ruled out the interdomain association between and Bro1. These results are contradictory to a previous study, which indicated that the N-terminal portion of the PRD of intact ALIX associates with the Bro1 domain and inhibits its interaction with CHMPs (13). Our results agree with a previous biophysical study that used full-length ALIX produced in insect cells and found that the presence of PRD did not influence the interactions between intact ALIX and peptide analogs of CHMPs (14). The discrepancy between these two studies was attributed to the use of pure monomeric ALIX in the biophysical investigation (14) versus the use of crude extracts in the cellular study (13) and the likely involvement of additional factors such as avidity effects because of ALIX and CHMP oligomerization influencing their association. Because highly sensitive PRE experiments also ruled out the association between Bro1 and the N-terminal portion of PRD at sites S712C and A756C, we argue that the cellular results of intramolecular association between the N-terminal portion of PRD and Bro1 and the corresponding prevention of Bro1 binding to CHMPs (13) could have been influenced by other adapter proteins present in crude cell extracts. Note that fibril formation was not observed in full-length ALIX made using insect cells (14), likely because of potential unaccounted PTMs such as hyperphosphorylation of PRD or because the globular domains of ALIX increase the critical concentration required for its fibrillization.
Paramagnetic NMR measurements revealed transient electrostatic interactions between Bro1 and hyperphosphorylated PRD. In intact ALIX, these interdomain interactions will likely be substantially stronger owing to their intramolecular nature. These interactions stem from a specific association between site C813 of hyperphosphorylated PRD and the basic surface of Bro1 and are governed by a patch of phosphorylated tyrosine residues encompassing residue C813. Since the same motif, residues 800 to 813, is likely involved in fibril formation, these intramolecular interactions will not be plausible in PRD-mediated ALIX assemblies. The biophysical study mentioned previously uncovered that the presence of PRD results in inhibition of the binding of the V domain to YPXnL motifs in monomeric full-length ALIX (14). Although NMR experiments rule out the association between the N-terminal portion of PRD and the V domain, interdomain association between Bro1 and hyperphosphorylated PRD in full-length ALIX can likely interfere with the interactions of the V domain with its binding partners. NMR titration experiments between Bro1 and LBPA-enriched lipid vesicles revealed that these electrostatic interactions localized on the same basic surface of Bro1 that comes into direct contact with hyperphosphorylated PRD. Competition between intramolecular and intermolecular interactions is, thus, likely to redistribute ALIX away from late endosomal membranes to the cytosol. These observations are consistent with a recent in vivo study that demonstrated that deletion of PRD facilitated ALIX membrane association (37). Phosphorylation of Bro1 neither was found to play a major role in interactions between Bro1 and anionic lipid vesicles nor is it likely involved in the interdomain interactions with hyperphosphorylated PRD. Even though the Src-binding motif represents the most conserved set of residues in mammalian Bro1 domains, mutation of Y319 of the yeast Bro1 ortholog had no effect on multivesicular body sorting (38). Our results are consistent with this observation and indicate that the phosphorylation of this residue is linked to other functions in mammalian cells that do not involve membrane associations.
Altogether, the aforementioned results indicate that the PTM-mediated formation and dissolution of ALIX amyloids will provide a strict spatiotemporal control on the recruitment of its binding partners involved in cytokinesis and apoptosis and will likely control nucleation of CHMPs and the corresponding endosomal membrane remodeling. ALIX–Src interactions at late endosomal membranes will result in phosphorylation of Bro1 and PRD of ALIX and shift the equilibrium toward monomeric ALIX. The hyperphosphorylated PRD will then compete against late endosomal membranes for binding to Bro1, culminating in a redistribution of ALIX back to the cytosol. Finally, we note that amyloids are commonly associated with proteinopathies, including Alzheimer's and Parkinson's diseases (39). Recent discoveries of functional amyloids have challenged this perception by elucidating the physiological roles of amyloids and their ubiquitous distribution in lower order and higher order organisms (40, 41, 42, 43). These discoveries have led to the emergence of a new narrative, which posits that amyloidogenesis represents a functional phase transition, similar to the one found in nature between gas, liquid, and solid phases, and suggests that cells may harness many useful characteristics of amyloids, including extreme stability, avidity, prion-like replication, and many others. Exactly how cells generate physiological amyloids without succumbing to pathology is not known. One suggested mechanism is the dissolution of amyloids, often through chaperones or changes in pH. The modulation of amyloids via PTMs described here, specifically a rapid disassembly and assembly of fibrils through tyrosine phosphorylation and dephosphorylation, respectively, represents another elegant mechanism with which cells may control the associated cytotoxicity. Given the prevalence of phosphorylation in eukaryotes (44) and that many proteins are prone to aggregation (45, 46), we hypothesize that such regulatory control on amyloid production is widespread, allowing eukaryotic cells to escape the cytotoxic effects of functional amyloids.
Experimental procedures
Materials
IPTG and arabinose were purchased from Sigma–Aldrich (catalog no. 420322 and A3256, respectively). ThT was purchased from Thermo Fisher Scientific (catalog no. AC211760050). CR was purchased from Sigma–Aldrich (catalog no. C6277). Buffered ATP solution (pH 7.5) was obtained from Thermo Fisher Scientific (catalog no. R1441). Phosphotyrosine mouse monoclonal antibody was obtained from Cell Signaling Technology (catalog no. 9411). Secondary antibody, goat antimouse IgG, was obtained from Thermo Fisher Scientific (catalog no. G-21040). The malachite green phosphate detection kit and PTP1B substrate were purchased from R&D Systems, Inc (catalog no. DY996 and ES006, respectively). PK solution (20 mg/ml) was purchased from Thermo Fisher Scientific (catalog no. 25530049). Reagents for NMR isotopic enrichment were obtained from Cambridge Isotopes Laboratories (CIL) and Sigma–Aldrich. Paramagnetic MTSL and its diamagnetic counterpart, (1-acetoxy-2,2,5,5-tetramethyl-δ-3-pyrroline-3-methyl) methanethiosulfonate (MTS), were purchased from Toronto Research Chemicals, Inc (catalog no. O875000 and A167900, respectively). TEMPOL was purchased from Sigma–Aldrich (catalog no. 176141). Phospholipids, POPC, POPE, and POPS were purchased from Avanti Polar Lipids, Inc (catalog no. 850457C, 850757C, 840034C, respectively). LBPA was purchased from Thermo Fisher Scientific (catalog no. 50720538).
Recombinant protein expression and purification
Codon-optimized (Genewiz, Inc) ALIX-PRD constructs (UniProt accession no. Q8WUM4), and , were subcloned in pET11a (Novagen, EMD Millipore) and expressed in BL21(DE3) competent cells (Agilent; catalog no. 200131). Cysteine variants of used for paramagnetic NMR measurements were generated using site-directed mutagenesis. The recombinant constructs representing globular ALIX domains, namely Bro1 (residues 1–359), V (residues 359–702), and Bro1-V (residues 1–702), were obtained from Addgene, accession no. 80641 (47), 17639 (48), and 42577 (49), respectively. The human Src kinase construct (UniProt accession no. P12931) comprising a C-terminal tobacco etch virus (TEV)-cleavable polyhistidine tag, pEX-Src-C-His, was a generous gift from the van der Vliet Group (University of Vermont) and was expressed in BL21-AI cells (Thermo Fisher Scientific; catalog no. C607003). The construct representing the catalytic domain of human PTP1B (residues 1–301; UniProt accession no. P18031) was obtained from Addgene, accession no. 102719 (50) and expressed in BL21(DE3) competent cells. The TEV protease construct was a generous gift from David S. Waugh (National Cancer Institute, the National Institutes of Health [NIH]). The construct of MBP-E38C (UniProt accession no. P0AEX9) was a generous gift from G. Marius Clore (National Institute of Diabetes and Digestive and Kidney Diseases, the NIH). MBP-E38C was expressed in BL21-CodonPlus (DE3)-RIPL competent cells (Agilent; catalog no. 230280).
ALIX-PRD constructs, and its cysteine variants and were expressed at 16 °C. Briefly, cells were grown at 37 °C in 1 L LB medium (MP Biomedicals; catalog no. 3002-036) at natural isotopic abundance or minimal M9 medium for isotopic labeling. The latter contained 1 g/l 15NH4Cl (CIL) for 15N labeling. About 30 min prior to induction, the temperature of cell culture was reduced to 16 °C. Cells were induced with 1 mM IPTG at an absorbance of ∼0.8 at 600 nm and harvested after ∼48 h. Src kinase was expressed according to our previous protocol (16). Briefly, cells were grown at 37 °C in 1 L terrific broth medium (Thermo Fisher Scientific; catalog no. BP9728) at natural isotopic abundance. About 30 min prior to induction, the temperature of cell culture was reduced to 16 °C. Cells were induced with 0.2% w/v arabinose and 1 mM IPTG at an absorbance of ∼0.8 at 600 nm and harvested ∼24 h after induction. The catalytic domain of PTP1B was expressed at 16 °C. Cells were grown at 37 °C in 1 L LB medium and ∼30 min prior to induction, and the temperature of cell culture was reduced to 16 °C. Cells were induced with 1 mM IPTG at an absorbance of ∼0.8 at 600 nm and harvested ∼24 h after induction. For TEV protease, cells were grown at 37 °C in 1 L LB medium and harvested ∼4 h after induction with 1 mM IPTG. The recombinant constructs of globular ALIX domains, namely Bro1, V, and Bro1-V, were expressed at 16 °C. Cells were grown at 37 °C in 1 L LB medium at natural isotopic abundance or in the case of Bro1, in M9 medium for isotopic labeling. The latter contained 0.3 g/l 2H/15N/13C Isogro (Sigma–Aldrich), 99.9% (v/v) deuterium oxide (D2O; CIL), 1 g/l 15NH4Cl (CIL), and 3 g/l 2H7,13C6-d-glucose (Sigma–Aldrich) for 2H/15N/13C labeling and 0.3 g/l 2H/15N Isogro (Sigma–Aldrich), 99.9% (v/v) deuterium oxide (CIL), 1 g/l 15NH4Cl (CIL), and 3 g/l 2H7,12C6-d-glucose (CIL) for 2H/15N labeling. About 30 min prior to induction, the temperature of cell culture was reduced to 16 °C. Cells were induced with 1 mM IPTG at an absorbance of ∼0.8 at 600 nm and harvested after ∼24 h.
PRD constructs were purified using a combination of affinity chromatography (ÄKTA Pure and Start Protein Purification Systems; GE Healthcare) and reverse-phase HPLC (1260 Infinity II Liquid Chromatography System; Agilent Technologies). The purification protocol for is described in our previous work (16). Briefly, the cells were resuspended in a lysis buffer containing 50 mM Tris(hydroxymethyl)aminomethane (Tris), pH 8.0, 2 mM EDTA, and 250 mM NaCl. Cells were lysed by heat shock (80 °C, ∼5 min) and cleared by centrifugation (48,380g, 25 min). The cell lysate was loaded onto a Streptactin Sepharose column (GE Healthcare), pre-equilibrated with a running buffer containing 50 mM Tris, pH 8.0, 2 mM EDTA, and 250 mM NaCl, and eluted in the same buffer containing 2.5 mM d-desthiobiotin (Sigma–Aldrich). The eluted protein was mixed with recombinant TEV protease (molar ratio of 50:1) in the presence of 1 mM DTT (Sigma–Aldrich) to hydrolyze the N-terminal GB1 solubility tag. Proteolysis was allowed to proceed at room temperature for ∼12 h and was assessed for completion by SDS-PAGE; Bolt 4–12% bis–Tris gel (Thermo Fisher Scientific). The hydrolyzed product, , was further purified using reverse-phase HPLC (Jupiter 10 μm C18 300 Å column; Phenomenex, catalog no. 00G-4055-N0) with a 5 to 55% acetonitrile gradient comprising 0.05% TFA (Sigma–Aldrich). The eluted protein was freeze dried (Labconco −84 °C Benchtop Freeze Dryer) and stored at −80 °C before use. For and its cysteine variants, the cells were resuspended in a denaturing buffer containing 50 mM Tris, pH 8.0, and 6 M guanidine hydrochloride (Sigma–Aldrich). Cells were lysed using a homogenizer, EmulsiFlex-C3 (Avestin) and cleared by centrifugation. The cell lysate was filtered through a 0.45 μm vacuum-driven filtration device (Stericup; Sigma–Aldrich) and loaded onto a HisTrap column (GE Healthcare), pre-equilibrated with a denaturing buffer containing 50 mM Tris, pH 8.0, and 6 M guanidine hydrochloride. The bound protein was washed with ten column volumes of refolding buffer containing 50 mM Tris, pH 8.0, and 250 mM NaCl and eluted in the same buffer containing 0.5 M imidazole (Sigma–Aldrich). The eluted protein was loaded onto a Streptactin Sepharose column, pre-equilibrated with a running buffer containing 50 mM Tris, pH 8.0, 2 mM EDTA, and 250 mM NaCl, and eluted in the same buffer containing 2.5 mM d-desthiobiotin. The resultant elution was mixed with recombinant TEV protease (molar ratio of 100:1) in the presence of 1 mM DTT to hydrolyze the N-terminal GB1 solubility tag. The proteolysis reaction was carried out at room temperature (∼12 h) and assessed for completion by SDS-PAGE. The hydrolyzed product, , was further purified using reverse-phase HPLC (Jupiter 10 μm C18 300 Å column) using a 25 to 37% acetonitrile gradient comprising 0.1% TFA. The eluted fractions were pooled, lyophilized, and stored at −80 °C.
The purification protocol for Src kinase is described in our previous work (16). Briefly, the cells were resuspended in a lysis buffer containing 50 mM Tris, pH 8.0, and 250 mM NaCl. Cells were lysed using an EmulsiFlex-C3 and cleared by centrifugation. The cell lysate was loaded onto a HisTrap column (GE Healthcare) with a 0 to 1 M imidazole gradient containing 50 mM Tris, pH 8.0, and 250 mM NaCl. The eluted protein was loaded onto a Q Sepharose HP column (GE Healthcare) with a 0 to 1 M NaCl gradient in a buffer containing 50 mM Tris, pH 8.0, and 5 mM β-mercaptoethanol. The eluted fractions were pooled, concentrated (Amicon Ultra-15; cutoff of 30 kDa), and loaded onto a HiLoad 26/600 Superdex 75 pg column (GE Healthcare) pre-equilibrated with 50 mM Tris, pH 7.5, 250 mM NaCl, and 5 mM β-mercaptoethanol. Relevant fractions were pooled and concentrated to ∼1 mg/ml (Amicon Ultra-15; cutoff of 30 kDa). Samples were aliquoted, flash frozen, and stored at −80 °C.
For the catalytic domain of PTP1B, the cells were resuspended in a lysis buffer containing 50 mM Tris, pH 8.0, and 250 mM NaCl. Cells were lysed using an EmulsiFlex-C3 and cleared by centrifugation. The cell lysate was loaded onto a HisTrap column (GE Healthcare) with a 0 to 1 M imidazole gradient containing 50 mM Tris, pH 8.0, and 250 mM NaCl. The eluted protein was concentrated (Amicon Ultra-15; cutoff of 10 kDa) and loaded onto a HiLoad 26/600 Superdex 75 pg column (GE Healthcare) pre-equilibrated with 50 mM Tris, pH 8, and 250 mM NaCl. Relevant fractions were pooled and mixed with recombinant TEV protease (molar ratio of 50:1) in the presence of 1 mM DTT to hydrolyze the N-terminal polyhistidine (6× His) affinity tag. The proteolysis reaction was carried out at room temperature (∼12 h) and assessed for completion by SDS-PAGE. The reaction mixture was loaded back onto a HisTrap column (GE Healthcare). The resultant flow-through fractions of hydrolyzed PTP1B were pooled, concentrated, and further purified by size exclusion chromatography (HiLoad 26/600 Superdex 75 pg column; GE Healthcare). Relevant fractions were concentrated (∼3 mg/ml; Amicon Ultra-15; cutoff of 10 kDa), flash frozen, and stored at −80 °C.
The purification protocol for TEV protease is described in our previous works (16, 51). Briefly, the cells were resuspended in a lysis buffer containing 50 mM Tris, pH 8.0, and 250 mM NaCl. Cells were lysed using an EmulsiFlex-C3 and cleared by centrifugation. The cell lysate was loaded onto a HisTrap column (GE Healthcare) with a 0 to 1 M imidazole gradient containing 50 mM Tris, pH 8.0, and 250 mM NaCl. The eluted protein was concentrated (Amicon Ultra-15; cutoff of 10 kDa) and loaded onto a HiLoad 26/600 Superdex 75 pg column (GE Healthcare) pre-equilibrated with 50 mM Tris, pH 8, 250 mM NaCl, and 1 mM DTT. Relevant fractions were pooled and concentrated to ∼1 mg/ml (Amicon Ultra-15; cutoff of 10 kDa). The samples were aliquoted, flash frozen, and stored at −80 °C.
For the Bro1 domain, the cells were resuspended in a lysis buffer containing 50 mM Tris, pH 8.0, and 250 mM NaCl. Cells were lysed using EmulsiFlex-C3 and cleared by centrifugation. The cell lysate was loaded onto a HisTrap column (GE Healthcare) with a 0 to 1 M imidazole gradient containing 50 mM Tris, pH 8.0, and 250 mM NaCl. The eluted protein was concentrated (Amicon Ultra-15; cutoff of 10 kDa) and loaded onto a HiLoad 26/600 Superdex 75 pg column (GE Healthcare) pre-equilibrated with 50 mM Tris, pH 8, 250 mM NaCl, and 1 mM DTT. Relevant fractions were pooled and mixed with recombinant TEV protease to cleave off the N-terminal 10× His affinity tag (molar ratio of 5:1). Proteolysis was carried out at room temperature (∼48 h) and assessed for completion using SDS-PAGE. The reaction mixture was loaded back onto a HisTrap column (GE Healthcare). The resultant flow-through fractions of hydrolyzed Bro1 were pooled, concentrated, and further purified by size exclusion chromatography (HiLoad 26/600 Superdex 75 pg column; GE Healthcare). Relevant Bro1 fractions were pooled and stored at −80 °C.
For the V domain, the cells were resuspended in a lysis buffer containing 50 mM Tris, pH 8.0, and 250 mM NaCl. Cells were lysed using an EmulsiFlex-C3 and cleared by centrifugation. Cell lysate was loaded onto a Glutathione Sepharose 4 Fast Flow column (GE Healthcare) with a 0 to 10 mM l-glutathione (Sigma–Aldrich) gradient containing 50 mM Tris, pH 8.0, and 250 mM NaCl. The eluted protein was concentrated (Amicon Ultra-15, cutoff of 10 kDa) and loaded onto a HiLoad 26/600 Superdex 75 pg column (GE Healthcare) pre-equilibrated with 50 mM Tris, pH 8, 250 mM NaCl, and 1 mM DTT. Relevant fractions were pooled and mixed with recombinant TEV protease to cleave off the N-terminal glutathione-S-transferase affinity tag (molar ratio of 50:1). Proteolysis was carried out at room temperature (∼12 h) and assessed for completion using SDS-PAGE. The reaction mixture was loaded back onto a Glutathione Sepharose 4 Fast Flow column (GE Healthcare). The resultant flow-through fractions were pooled, concentrated, and further purified by size exclusion chromatography (HiLoad 26/600 Superdex 75 pg column; GE Healthcare). Relevant fractions were pooled, concentrated (∼10 mg/ml; Amicon Ultra-15; cutoff of 10 kDa), and stored at −80 °C.
For the Bro1-V construct, the cells were resuspended in a lysis buffer containing 50 mM Tris, pH 8.0, and 250 mM NaCl. Cells were lysed using an EmulsiFlex-C3 and cleared by centrifugation. The cell lysate was loaded onto a HisTrap column (GE Healthcare) with a 0 to 1 M imidazole gradient containing 50 mM Tris, pH 8.0, and 250 mM NaCl. The eluted protein was concentrated (Amicon Ultra-15; cutoff of 30 kDa) and loaded onto a HiLoad 26/600 Superdex 200 pg column (GE Healthcare) pre-equilibrated with 50 mM Tris, pH 8, 250 mM NaCl, and 1 mM DTT. Relevant fractions were pooled and mixed with recombinant TEV protease to cleave off the N-terminal 6× His affinity tag (molar ratio of 5:1). Proteolysis was carried out at room temperature (∼48 h) and assessed for completion using SDS-PAGE. The reaction mixture was loaded back onto the HisTrap column (GE Healthcare). The resultant flow-through fractions were pooled, concentrated, and further purified by size exclusion chromatography (HiLoad 26/600 Superdex 200 pg column; GE Healthcare). Relevant fractions were pooled and stored at −80 °C.
For apo MBP-E38C, the cells were resuspended in a lysis buffer containing 50 mM Tris, pH 8.0, and 250 mM NaCl. Cells were lysed using an EmulsiFlex-C3 and cleared by centrifugation. Cell lysate was loaded onto an Amylose resin column (New England BioLabs, Inc) with a 0 to 10 mM maltose gradient containing 50 mM Tris, pH 8.0, 1 mM DTT, and 250 mM NaCl. The eluted protein was concentrated (Amicon Ultra-15; cutoff of 30 kDa) and loaded onto a HiLoad 26/600 Superdex 75 pg column (GE Healthcare) pre-equilibrated with 50 mM Tris, pH 8, 1 mM DTT, and 250 mM NaCl. Relevant fractions were pooled and concentrated to ∼10 mg/ml (Amicon Ultra-15; cutoff of 30 kDa). The samples were aliquoted and flash frozen and stored at −80 °C.
The aforementioned purification schemes resulted in ≥99% pure proteins with the following yields: (∼40 mg/l), (∼25 mg/l), Src (∼5 mg/l), PTP1B (∼45 mg/l), Bro1 (∼50 mg/l), V (∼45 mg/l), Bro1-V (∼40 mg/l), and MBP-E38C (∼10 mg/l).
MS analyses
All protein constructs, the end products of in vitro phosphorylation, and site-specific spin labeling reactions (see below) were verified by MS using our previously published protocols (16, 51). Briefly, an Agilent 6230 TOF-mass spectrometer with Jet Stream ESI was used for LC–ESI–TOFMS analysis. The Jet Stream ESI source was operated under positive ion mode with the following parameters: VCap = 3500 V, fragmentor voltage = 175 V, drying gas temperature = 325 °C, sheath gas temperature = 325 °C, drying gas flow rate = 10 l/min, sheath gas flow rate = 10 l/min, and nebulizer pressure = 40 psi. The chromatographic separation was performed at room temperature on a Phenomenex Aeris Widepore XB-C18 column (2.1 mm inner diameter × 50 mm length, 3.6 μm particle size). HPLC-grade water and acetonitrile were used as mobile phases A and B, respectively. Each phase also carried 0.1% TFA. MassHunter software (Agilent) was used for data acquisition and analysis, and MagTran software (Amgen, Inc) was used for mass spectrum deconvolution (52).
PK digestion
Fibrils of and soluble (∼40 μM each) were incubated with PK (1 μg/ml) in a buffer comprising 50 mM Tris, pH 8, 1 mM DTT, and 0.5 mM EDTA (37 °C, ∼60 min). Aliquots were taken at regular intervals and heated at 100 °C for ∼3 min to inactivate PK. The reactions were assessed using SDS-PAGE, visualized using PageBlue staining solution (Thermo Fisher Scientific; catalog no. 24620). Reactions were also analyzed by LC–MS/MS using our previously published protocol (16). Briefly, LC–MS/MS analysis was carried out by the nanoLC-Orbitrap XL spectrometer. A fused silica capillary LC column (pulled to a tip with a Sutter P-2000 laser capillary puller) was packed with Agilent Zorbax resin (C18; particle size of 5 μm). The inner diameter of the capillary was 100 μm, and the stationary phase was packed with a pressure device to a length of 70 mm. The column was equilibrated using an Agilent 1100 HPLC pump, solvent A = 100% HPLC-grade water with 0.1% formic acid and solvent B = 100% acetonitrile with several 10 to 90% solvent B step gradients. The PK-digested sample of fibrils was diluted by a factor of 100 with 2% acetonitrile and 0.1% formic acid and loaded on the column by a pressure device (2.1 μl; ∼2 pmol on column). The LC gradient program was 1 to 34% solvent B in 66 min, followed by 6 min of 90% B, followed by 1% B at 95 min. Data acquisition method parameters were as follows: the capillary LC was positioned in the Thermo nanoelectrospray interface with a 1.55 kV source voltage, 48 V capillary voltage, 85 V tube lens, and 165 °C capillary temperature. For the ion trap, three micro scans and the FT spectra, two micro scans were averaged. There were seven scan events per cycle (6.5 s), one FT scan (resolution of 30,000) from 200 to 1600 m/z, followed by six ion trap, data-dependent collision-induced dissociation MS/MS scans. Dynamic exclusion was enabled with duration of 40 s and a repeat count of 2. The ion trap collision-induced dissociation scans had an isolation width of 2.0 m/z and a normalized collision energy of 35. The data analysis method was as follows: peptide sequence matches were found using the OpenMS workflow (53) with the MS-GF + peptide sequence match search algorithm (54). The false discovery rate was set to 5%. The enzyme parameter was set to “Unspecific cleavage.” The search parameters used for LC–MS/MS analyses are tabulated in Table S2, whereas Table S3 lists all peptides identified by these analyses.
Tyrosine phosphorylation
Lyophilized was reconstituted in a buffer comprising 50 mM Tris, pH 7.5, 5 mM MgCl2, 2 mM DTT, and 0.5 mM EDTA. The resultant solution (50 μM) was mixed with 1 mM ATP and ∼5 μM Src. The phosphorylation reaction was allowed to proceed for ∼12 h at 30 °C. Src was then removed from the reaction mixture using a HisTrap column (GE Healthcare) pre-equilibrated with 50 mM Tris, pH 8, and 250 mM NaCl. The resultant flow-through fractions of phosphorylated were pooled, and excess ATP/ADP were removed using a HiPrep 26/10 Desalting column (GE Healthcare) pre-equilibrated with 20 mM sodium phosphate, pH 6.5, 50 mM NaCl, 2 mM EDTA, and 1 mM Tris(2-carboxyethyl)phosphine (Sigma–Aldrich).
The eluted fractions of Bro1 and Src from a sizing column were mixed; Bro1 to Src molar ratio of ∼5:1. The resultant mixture was concentrated to a final volume of ∼7 ml (Amicon Ultra-15; cutoff of 30 kDa) and dialyzed against a buffer containing 50 mM Tris, pH 8, 2 mM ATP, 5 mM MgCl2, 2 mM DTT, and 0.5 mM EDTA (Slide-A-Lyzer G2 dialysis cassettes; Thermo Fisher Scientific). The reaction was allowed to proceed for ∼18 h at room temperature with one buffer change at a time point of ∼4 h in the dialysis. Phosphorylated Bro1 was purified from the mixture using the procedure mentioned previously. The status of phosphorylation was assessed using LC–ESI–TOFMS, LC–MS/MS, and NMR. For LC–MS/MS, phosphorylated Bro1 was digested using trypsin (Bro1 to trypsin molar ratio of 50:1; incubation time of ∼90 min, and room temperature). Hyperphosphorylated samples were digested using PK ( to PK molar ratio of 1000:1; incubation time of ∼5 min, 37 °C). LC–MS/MS analyses were carried out using a similar procedure as the one used to identify products of PK digestion of fibrils. Up to two PTMs per peptide were allowed for the search. Phosphorylation of tyrosine was the most prevalent PTM observed (also see Tables S2 and S3). For Bro1, phosphorylation was also assessed using Western blotting. Bro1 + Src reaction mixture separated using electrophoresis was transferred onto a 0.45 μm nitrocellulose membrane (Thermo Fisher Scientific; catalog no. LC2001) using a wet-transfer Mini Blot Module (Thermo Fisher Scientific; catalog no. B1000). Procedures for blotting and subsequent product visualization are described previously (16).
Large unilamellar lipid vesicles
Lipid vesicles were prepared according to the protocols published by Jiang et al. (55). Briefly, lipids were stored in chloroform/methanol (2:1) stock solutions at −20 °C. Lipid films were prepared by drying the appropriate amounts of stock solutions under a stream of dry nitrogen followed by vacuum desiccation (vacuum oven; VWR) at room temperature for ∼2 h to remove traces of organic solvents. The resultant lipid films were hydrated using appropriate volumes of buffer containing 20 mM sodium phosphate, pH 6.5, 50 mM NaCl, 1 mM EDTA, and 1 mM TCEP and subjected to vortex mixing (approximately three times, 60 s each). The multilamellar vesicle solutions were extruded through a 100 nm diameter polycarbonate membrane (Whatman, GE Healthcare) using an extrusion kit (Avanti Polar Lipids, Inc) and used immediately for NMR and size measurements.
DLS
The size distribution of lipid vesicles was determined by DLS at 25 °C using 25 μM vesicles, with and without 25 μM Bro1. Measurements were performed using the Zetasizer Nano ZS (Malvern Instruments) instrument operating at a wavelength of 633 nm. The measurements were repeated 15 times after a 1 min temperature equilibration. A cumulant fit and a sphere model were used to obtain the average hydrodynamic radius (130–150 nm).
Site-specific spin labeling
MTSL and MTS were reconstituted in ethanol (Sigma–Aldrich) to a final concentration of ∼100 mM and mixed with the protein of interest at a molar ratio of 10:1 (spin label versus protein). The spin labeling reaction was allowed to proceed in the dark for ∼12 h at room temperature and assessed for completion using LC–ESI–TOFMS. The excess of unreacted spin label was removed using the HiPrep 26/10 Desalting column (GE Healthcare) pre-equilibrated with 20 mM sodium phosphate, pH 6.5, 50 mM NaCl, and 2 mM EDTA. The resultant protein fractions were pooled, concentrated, and immediately used for NMR measurements.
NMR sample preparation
Samples of 15N-labeled , 15N-labeled hyperphosphorylated , and 15N/2H- or 15N/13C/2H-labeled Bro1 were prepared in a buffer comprising 20 mM sodium phosphate, pH 6.5, 50 mM NaCl, 2 mM EDTA, and 1 mM TCEP. For paramagnetic NMR, the samples were prepared in an identical buffer without TCEP. To assess the interactions between calcium and Bro1, samples were prepared using 20 mM MES, pH 6.5, 50 mM NaCl, 2 mM EDTA, and 1 mM TCEP. All NMR samples contained 7% v/v D2O.
NMR spectroscopy
All NMR experiments were carried out at 30 °C on Bruker 600 and 800 MHz spectrometers equipped with z-gradient triple resonance cryoprobes. Spectra were processed using NMRPipe (56) and analyzed using the CCPN software suite (57). Sequential 1H, 15N, and 13C backbone resonance assignments of nonphosphorylated and phosphorylated Bro1 samples were carried out using conventional TROSY-based three-dimensional triple resonance experiments (58). TROSY-based 15N{1H} NOE measurements (59) were carried out on 2H/15N-labeled nonphosphorylated and phosphorylated Bro1 samples at 800 MHz. The 15N-{1H} NOE and reference spectra were recorded with a 6 s saturation time for the NOE measurement and an equivalent recovery time for the reference measurement in an interleaved manner, each preceded by an additional recovery time of 1 s. Chemical shift perturbation experiments were carried out using 15N-labeled and nonlabeled Bro1, V, and Bro1-V (one at a time). The following protein concentrations were used: 100 μM and 300 μM Bro1 and V (one at a time) and 25 μM and 75 μM Bro1-V (the latter is due to the limited solubility of Bro1-V construct). Similar measurements were carried out using uniformly 2H/15N-labeled 100 μM nonphosphorylated and phosphorylated Bro1 and nonlabeled 300 μM . Perturbations were calculated as follows: ΔH/N = {(ΔδHN)2+ (0.154 × ΔδN)2}1/2, where ΔδHN and ΔδN are the 1HN and 15N chemical shift differences in parts per million, respectively, between free and bound states. To determine the effect of calcium on Bro1, titration experiments were carried out using 2H/15N-labeled 100 μM nonphosphorylated Bro1, in the absence and presence of 5 mM CaCl2. Intermolecular transverse 1HN–Γ2 PRE rates were obtained by mixing 200 μM 2H/15N-labeled Bro1 with 200 μM MTSL/MTS-labeled hyperphosphorylated and its cysteine variants (note that the corresponding PRE measurements on phosphorylated Bro1 were not feasible because of the limited solubility of phosphorylated Bro1 and MTSL-tagged hyperphosphorylated mixture). Transverse 1HN–Γ2 PRE rates were measured from the differences in the transverse 1HN–R2 relaxation rates between the paramagnetic and diamagnetic samples as described previously (26, 27). Two time points (separated by 30 ms) were used for the measurements of 1HN–R2 rates, and the errors in the 1HN–Γ2 PRE rates were calculated as described previously (26). Negative control experiments were carried out using 200 μM 2H-/15N-labeled Bro1 and 2 mM TEMPOL and 200 μM MTSL-labeled MBP-E38C. For these experiments, 200 μM 2H/15N-labeled nonphosphorylated Bro1 served as a diamagnetic control. NMR titration experiments between 2H-/15N-labeled nonphosphorylated and phosphorylated Bro1 and neutral and negatively charged lipid vesicles were carried out using 100 μM protein (protein to lipid molar ratio of 1:30). TROSY–HSQC correlation spectra were recorded on nonphosphorylated and phosphorylated Bro1 in the absence and presence of unilamellar lipid vesicles. The following vesicle compositions were used; the numbers in parenthesis represent the molar ratios of phospholipids: POPC/POPE (7:3), POPC/POPE/POPS (5.5:3:1.5), POPC/POPE/LBPA (6:3:1), POPC/POPE/LBPA (5.75:3:1.25), and POPC/POPE/LBPA (5.5:3:1.5).
CR assay
CR was dissolved in MilliQ water (MilliQ IQ 7000 purification system; Millipore–Sigma). CR stock solution (0.2% w/v) was filtered through a 0.22 μm filter and used immediately. About 100 μM solutions of nonphosphorylated and phosphorylated and were incubated overnight at room temperature and mixed with CR stock solution in the morning (100:1 dilution; protein versus CR). The mixtures were incubated at room temperature for ∼45 min. Absorption spectra were measured using an Agilent Cary 50 Bio UV–Vis spectrophotometer (quartz cuvette of 1 cm).
Fibril formation and dissolution kinetics
Aggregation and dissolution experiments were performed at 30 °C on samples in sealed 96-well flat bottom plates (Corning; catalog no. 3370) containing 100 μl sample per well (three replicates of each sample [n = 3] were placed in separate wells, and ThT fluorescence was measured through the top and bottom of the plate). Measurements were carried out with continuous linear shaking (3.5 mm, 411.3 rpm) using a microplate reader (Infinite M Plex; Tecan). ThT (10 μM) fluorescence was recorded as a function of time; excitation and emission wavelengths were 415 and 480 nm, respectively. For aggregation experiments, ThT signals as a function of time were recorded on 100 μM hyperphosphorylated samples, in the absence and presence of 50 nM PTP1B. The buffer conditions were as follows: 20 mM Hepes, pH 7.5, 1 mM DTT, and 1 mM EDTA. For Src-mediated fibril dissolution experiments, lyophilized was reconstituted in a buffer containing 50 mM Tris, pH 7.5, 5 mM MgCl2, 2 mM DTT, and 0.5 mM EDTA ( = 100 μM). ThT signals as a function of time were recorded till the samples (n = 9) reached a stationary phase; total incubation time of ∼3 h, whereupon 1 μM of recombinant Src and 1 mM ATP were added to three samples, whereas the others were mixed with either 1 μM Src or 1 mM ATP. Fibril formation of was also assessed using ThT emission spectra recorded on Agilent Cary Eclipse Fluorescence spectrophotometer. ThT fluorescence was recorded of 3 × 100 μl of 100 μM samples at 30 °C after ∼2 h incubation (20 mM Hepes, pH 7.5, 1 mM DTT, and 1 mM EDTA; micro quartz cuvettes). Similar measurements were recorded on hyperphosphorylated and the soluble N-terminal fragment, , which served as negative controls.
TEM
samples, ∼1.2 mg/ml, were incubated at 30 °C in 20 mM sodium phosphate, pH 7.0, 50 mM NaCl, 1 mM TCEP, and 1 mM EDTA. Aggregated samples were subjected to sonication (∼10 min; Elmasonic P ultrasonic bath) and diluted to ∼0.2 mg/ml immediately before application to the TEM grids (400-mesh formvar and carbon-coated copper; Electron Microscopy Sciences; catalog no. FCF400-Cu). About 1 min after deposition, the sample solution was wicked with filter paper, followed by a quick wash with 3 μl of water and the addition of three to five drops of 2% w/v aqueous uranyl acetate solution. The uranyl acetate was wicked immediately with a filter paper, and the grids were air dried at room temperature. TEM images were acquired using a JEOL JEM-1400Plus transmission electron microscope (JEOL) and recorded on a Gatan OneView digital camera (Gatan).
CD spectroscopy
CD measurements (178–280 nm, 1 nm data pitch, continuous scanning with 1 nm bandwidth, 60 nm/min, and ten accumulations) with 0.1 mg/ml nonphosphorylated and phosphorylated were carried out in 2 mm quartz cuvettes (Starna Cells, Inc) using an Aviv model 215 spectrometer. The molar ellipticity was calculated using the equation: [θ]molar = 100 ∗ MW ∗ θ/cl, where MW = molecular weight, θ = CD signal (mdeg), c = concentration (mg/ml), and l = path length (cm). The buffer and experimental conditions were as follows: 10 mM sodium phosphate, pH 7.0, 1 mM TCEP, 1 mM EDTA, and 25 °C.
Data availability
Plasmids of full-length ALIX-PRD, , and its cysteine variants, namely , , and , have been deposited in the Addgene repository, https://www.addgene.org/ (accession nos. 164444, 164445, 164446, and 164447, respectively). The chemical shift assignments of Bro1 have been deposited in the Biological Magnetic Resonance Bank, https://bmrb.io/ (accession no. 50707). LC–MS/MS data were deposited to the MassIVE data repository, https://massive.ucsd.edu/ (accession no. MSV000088057).
Supporting information
This article contains supporting information (16, 22, 23, 26, 27, 60, 61, 62, 63, 64).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Z. Jiang, R. Ghirlando, P. A. Jennings, and I. Budin for useful discussions; X. Huang (University of California San Diego [UCSD] Biomolecular NMR facility), L. Gross, and Y. Su (UCSD Molecular Mass Spectrometry Facility) and G. Castillon and T. Meerloo (UCSD Cellular and Molecular Medicine Electron Microscopy Core Facility) for technical support. We thank A. van der Vliet (University of Vermont), D. S. Waugh (National Cancer Institute, NIH), and G. Marius Clore (National Institute of Diabetes and Digestive and Kidney Diseases, NIH) for Src kinase, TEV protease, and apo-MBP-E38C constructs, respectively. We acknowledge the use of facilities and instrumentation supported by the National Science Foundation through the UCSD Materials Research Science and Engineering Center, DMR-2011924, and by the NIH for the UCSD Cellular and Molecular Medicine EM Core Facility; S10OD023527, and the UCSD Molecular Mass Spectrometry Facility; 1S10RR25636-1A1.
Author contributions
R. D. E, B. R., and L. D. methodology; R. D. E, B. R., and L. D. investigation; R. D. E, B. R., and L. D. formal analysis; R. D. E and L. D. writing–original draft.
Funding and additional information
This work was funded in part by a 2018 Development Award (to L. D.) from the San Diego Center for AIDS Research (an NIH-funded program; P30 AI036214) and by Collaborative Development Awards (to L. D.) from the Cellular Host Elements in Egress, Trafficking, and Assembly of HIV (CHEETAH Center, University of Utah; an NIH-funded program; P50 AI150464) and HIV Interactions in Viral Evolution (HIVE Center, the Scripps research institute; an NIH-funded program; U54 AI150472), and a National Science Foundation Graduate Research Fellowship under grant no. DGE-1650112 (to R. D. E.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Wolfgang Peti
Supporting information
References
- 1.Odorizzi G. The multiple personalities of Alix. J. Cell Sci. 2006;119:3025–3032. doi: 10.1242/jcs.03072. [DOI] [PubMed] [Google Scholar]
- 2.Sadoul R. Do Alix and ALG-2 really control endosomes for better or for worse? Biol. Cell. 2006;98:69–77. doi: 10.1042/BC20050007. [DOI] [PubMed] [Google Scholar]
- 3.Hurley J.H., Odorizzi G. Get on the exosome bus with ALIX. Nat. Cell Biol. 2012;14:654–655. doi: 10.1038/ncb2530. [DOI] [PubMed] [Google Scholar]
- 4.Bissig C., Gruenberg J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol. 2014;24:19–25. doi: 10.1016/j.tcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Strack B., Calistri A., Craig S., Popova E., Gottlinger H.G. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 6.von Schwedler U.K., Stuchell M., Muller B., Ward D.M., Chung H.Y., Morita E., Wang H.E., Davis T., He G.P., Cimbora D.M., Scott A., Krausslich H.G., Kaplan J., Morham S.G., Sundquist W.I. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 7.Han Z., Madara J.J., Liu Y., Liu W., Ruthel G., Freedman B.D., Harty R.N. ALIX rescues budding of a double PTAP/PPEY L-domain deletion mutant of ebola VP40: A role for ALIX in ebola virus egress. J. Infect. Dis. 2015;212 Suppl 2:S138–S145. doi: 10.1093/infdis/jiu838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt M.H.H., Dikic I., Bogler O. Src phosphorylation of Alix/AIP1 modulates its interaction with binding partners and antagonizes its activities. J. Biol. Chem. 2005;280:3414–3425. doi: 10.1074/jbc.M409839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hikita T., Kuwahara A., Watanabe R., Miyata M., Oneyama C. Src in endosomal membranes promotes exosome secretion and tumor progression. Sci. Rep. 2019;9:3265. doi: 10.1038/s41598-019-39882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuo H., Chevallier J., Mayran N., Le Blanc I., Ferguson C., Fauré J., Blanc N.S., Matile S., Dubochet J., Sadoul R., Parton R.G., Vilbois F., Gruenberg J. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 11.Bissig C., Lenoir M., Velluz M.C., Kufareva I., Abagyan R., Overduin M., Gruenberg J. Viral infection controlled by a calcium-dependent lipid-binding module in ALIX. Dev. Cell. 2013;25:364–373. doi: 10.1016/j.devcel.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi T., Stang E., Fang K.S., de Moerloose P., Parton R.G., Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X., Si J., Corvera J., Gallick G.E., Kuang J. Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. Biochem. J. 2010;432:525–534. doi: 10.1042/BJ20100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai Q., Landesman M.B., Chung H.-Y., Dierkers A., Jeffries C.M., Trewhella J., Hill C.P., Sundquist W.I. Activation of the retroviral budding factor ALIX. J. Virol. 2011;85:9222–9226. doi: 10.1128/JVI.02653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren X., Hurley J.H. Proline-rich regions and motifs in trafficking: From ESCRT interaction to viral exploitation. Traffic. 2011;12:1282–1290. doi: 10.1111/j.1600-0854.2011.01208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elias R.D., Ma W., Ghirlando R., Schwieters C.D., Reddy V.S., Deshmukh L. Proline-rich domain of human ALIX contains multiple TSG101-UEV interaction sites and forms phosphorylation-mediated reversible amyloids. Proc. Natl. Acad. Sci. U. S. A. 2020;117:24274–24284. doi: 10.1073/pnas.2010635117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt T.G., Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2007;2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 18.Huth J.R., Bewley C.A., Jackson B.M., Hinnebusch A.G., Clore G.M., Gronenborn A.M. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 1997;6:2359–2364. doi: 10.1002/pro.5560061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsiatsiani L., Heck A.J. Proteomics beyond trypsin. FEBS J. 2015;282:2612–2626. doi: 10.1111/febs.13287. [DOI] [PubMed] [Google Scholar]
- 20.Baudier J., Cole R.D. Phosphorylation of tau proteins to a state like that in Alzheimer's brain is catalyzed by a calcium/calmodulin-dependent kinase and modulated by phospholipids. J. Biol. Chem. 1987;262:17577–17583. [PubMed] [Google Scholar]
- 21.Stuible M., Abella J.V., Feldhammer M., Nossov M., Sangwan V., Blagoev B., Park M., Tremblay M.L. PTP1B targets the endosomal sorting machinery: Dephosphorylation of regulatory sites on the endosomal sorting complex required for transport component STAM2. J. Biol. Chem. 2010;285:23899–23907. doi: 10.1074/jbc.M110.115295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y., Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadler J.B.A., Wenzel D.M., Williams L.K., Guindo-Martínez M., Alam S.L., Mercader J.M., Torrents D., Ullman K.S., Sundquist W.I., Martin-Serrano J. A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E8900–E8908. doi: 10.1073/pnas.1805504115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kay B.K., Williamson M.P., Sudol M. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 25.Zarrinpar A., Bhattacharyya R.P., Lim W.A. The structure and function of proline recognition domains. Sci. STKE. 2003;2003 doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]
- 26.Clore G.M., Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deshmukh L., Louis J.M., Ghirlando R., Clore G.M. Transient HIV-1 Gag-protease interactions revealed by paramagnetic NMR suggest origins of compensatory drug resistance mutations. Proc. Natl. Acad. Sci. U. S. A. 2016;113:12456–12461. doi: 10.1073/pnas.1615342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rideau E., Dimova R., Schwille P., Wurm F.R., Landfester K. Liposomes and polymersomes: A comparative review towards cell mimicking. Chem. Soc. Rev. 2018;47:8572–8610. doi: 10.1039/c8cs00162f. [DOI] [PubMed] [Google Scholar]
- 30.Carlton J.G., Agromayor M., Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trioulier Y., Torch S., Blot B., Cristina N., Chatellard-Causse C., Verna J.M., Sadoul R. Alix, a protein regulating endosomal trafficking, is involved in neuronal death. J. Biol. Chem. 2004;279:2046–2052. doi: 10.1074/jbc.M309243200. [DOI] [PubMed] [Google Scholar]
- 32.Kamranvar S.A., Gupta D.K., Huang Y., Gupta R.K., Johansson S. Integrin signaling via FAK-Src controls cytokinetic abscission by decelerating PLK1 degradation and subsequent recruitment of CEP55 at the midbody. Oncotarget. 2016;7:30820–30830. doi: 10.18632/oncotarget.9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibata H., Yamada K., Mizuno T., Yorikawa C., Takahashi H., Satoh H., Kitaura Y., Maki M. The penta-EF-hand protein ALG-2 interacts with a region containing PxY repeats in Alix/AIP1, which is required for the subcellular punctate distribution of the amino-terminal truncation form of Alix/AIP1. J. Biochem. 2004;135:117–128. doi: 10.1093/jb/mvh014. [DOI] [PubMed] [Google Scholar]
- 34.Schöneberg J., Lee I.-H., Iwasa J.H., Hurley J.H. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 2017;18:5–17. doi: 10.1038/nrm.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vietri M., Radulovic M., Stenmark H. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 2020;21:25–42. doi: 10.1038/s41580-019-0177-4. [DOI] [PubMed] [Google Scholar]
- 36.Christ L., Wenzel E.M., Liestøl K., Raiborg C., Campsteijn C., Stenmark H. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J. Cell Biol. 2016;212:499–513. doi: 10.1083/jcb.201507009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larios J., Mercier V., Roux A., Gruenberg J. ALIX- and ESCRT-III–dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020;219 doi: 10.1083/jcb.201904113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J., Sitaraman S., Hierro A., Beach B.M., Odorizzi G., Hurley J.H. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiti F., Dobson C.M. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- 40.Hammer N.D., Wang X., McGuffie B.A., Chapman M.R. Amyloids: Friend or foe? J. Alzheimers Dis. 2008;13:407–419. doi: 10.3233/jad-2008-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson M.P., Hewitt E.W. Why are functional amyloids non-toxic in humans? Biomolecules. 2017;7:71. doi: 10.3390/biom7040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang M., Audas T.E., Lee S. Disentangling a bad reputation: Changing perceptions of amyloids. Trends Cell Biol. 2017;27:465–467. doi: 10.1016/j.tcb.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otzen D., Riek R. Functional amyloids. Cold Spring Harb. Perspect. Biol. 2019;11 doi: 10.1101/cshperspect.a033860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khoury G.A., Baliban R.C., Floudas C.A. Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Sci. Rep. 2011;1:90. doi: 10.1038/srep00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 46.Schnabel J. Protein folding: The dark side of proteins. Nature. 2010;464:828–829. doi: 10.1038/464828a. [DOI] [PubMed] [Google Scholar]
- 47.McCullough J., Fisher R.D., Whitby F.G., Sundquist W.I., Hill C.P. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee S., Joshi A., Nagashima K., Freed E.O., Hurley J.H. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlson L.A., Hurley J.H. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16928–16933. doi: 10.1073/pnas.1211759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choy M.S., Li Y., Machado L., Kunze M.B.A., Connors C.R., Wei X., Lindorff-Larsen K., Page R., Peti W. Conformational rigidity and protein dynamics at distinct timescales rgulate PTP1B activity and allostery. Mol. Cell. 2017;65:644–658.e645. doi: 10.1016/j.molcel.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramaraju B., Nelson S.L., Zheng W., Ghirlando R., Deshmukh L. Quantitative NMR study of insulin-degrading enzyme using amyloid-β and HIV-1 p6 elucidates its chaperone activity. Biochemistry. 2021;60:2519–2523. doi: 10.1021/acs.biochem.1c00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z., Marshall A.G. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 53.Rost H.L., Sachsenberg T., Aiche S., Bielow C., Weisser H., Aicheler F., Andreotti S., Ehrlich H.C., Gutenbrunner P., Kenar E., Liang X., Nahnsen S., Nilse L., Pfeuffer J., Rosenberger G. OpenMS: A flexible open-source software platform for mass spectrometry data analysis. Nat. Methods. 2016;13:741–748. doi: 10.1038/nmeth.3959. [DOI] [PubMed] [Google Scholar]
- 54.Kim S., Pevzner P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang Z., de Messieres M., Lee J.C. Membrane remodeling by α-synuclein and effects on amyloid formation. J. Am. Chem. Soc. 2013;135:15970–15973. doi: 10.1021/ja405993r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 57.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., Laue E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 58.Clore G.M., Gronenborn A.M. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 1998;16:22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 59.Lakomek N.A., Ying J., Bax A. Measurement of 15N relaxation rates in perdeuterated proteins by TROSY-based methods. J. Biomol. NMR. 2012;53:209–221. doi: 10.1007/s10858-012-9626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roepstorff P., Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 61.Brademan D.R., Riley N.M., Kwiecien N.W., Coon J.J. Interactive peptide spectral annotator: A versatile web-based tool for proteomic applications. Mol. Cell. Proteomics. 2019;18:S193–S201. doi: 10.1074/mcp.TIR118.001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Z.Y., Thieme-Sefler A.M., Maclean D., McNamara D.J., Dobrusin E.M., Sawyer T.K., Dixon J.E. Substrate specificity of the protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U. S. A. 1993;90:4446–4450. doi: 10.1073/pnas.90.10.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duan X., Hall J.A., Nikaido H., Quiocho F.A. Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: Flexibility of tertiary structure and ligand binding. J. Mol. Biol. 2001;306:1115–1126. doi: 10.1006/jmbi.2001.4456. [DOI] [PubMed] [Google Scholar]
- 64.Ferreira C.M.H., Pinto I.S.S., Soares E.V., Soares H.M.V.M. (Un)suitability of the use of pH buffers in biological, biochemical and environmental studies and their interaction with metal ions – a review. RSC Adv. 2015;5:30989–31003. [Google Scholar]
- 65.Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Plasmids of full-length ALIX-PRD, , and its cysteine variants, namely , , and , have been deposited in the Addgene repository, https://www.addgene.org/ (accession nos. 164444, 164445, 164446, and 164447, respectively). The chemical shift assignments of Bro1 have been deposited in the Biological Magnetic Resonance Bank, https://bmrb.io/ (accession no. 50707). LC–MS/MS data were deposited to the MassIVE data repository, https://massive.ucsd.edu/ (accession no. MSV000088057).