Abstract
Background and Aims
Oilseed rape (Brassica napus) is one of the most important oil crops worldwide. Phosphorus (P) deficiency severely decreases the plant height and branch number of B. napus. However, the genetic bases controlling plant height and branch number in B. napus under P deficiency remain largely unknown. This study aims to mine candidate genes for plant height and branch number by genome-wide association study (GWAS) and determine low-P-tolerance haplotypes.
Methods
An association panel of B. napus was grown in the field with a low P supply (P, 0 kg ha−1) and a sufficient P supply (P, 40 kg ha−1) across 2 years and plant height and branch number were investigated. More than five million single-nucleotide polymorphisms (SNPs) were used to conduct GWAS of plant height and branch number at two contrasting P supplies.
Key Results
A total of 2127 SNPs were strongly associated (P < 6·25 × 10−07) with plant height and branch number at two P supplies. There was significant correlation between phenotypic variation and the number of favourable alleles of associated loci on chromosomes A10 (chrA10_821671) and C08 (chrC08_27999846), which will contribute to breeding improvement by aggregating these SNPs. BnaA10g09290D and BnaC08g26640D were identified to be associated with chrA10_821671 and chrC08_27999846, respectively. Candidate gene association analysis and haplotype analysis showed that the inbred lines carrying ATT at BnaA10g09290Hap1 and AAT at BnaC08g26640Hap1 had greater plant height than lines carrying other haplotype alleles at low P supply.
Conclusion
Our results demonstrate the power of GWAS in identifying genes of interest in B. napus and provided insights into the genetic basis of plant height and branch number at low P supply in B. napus. Candidate genes and favourable haplotypes may facilitate marker-based breeding efforts aimed at improving P use efficiency in B. napus.
Keywords: Oilseed rape, genome wide association study, plant height, branch number, low phosphorus supply, haplotype analysis
INTRODUCTION
Oilseed rape (Brassica napus) is one of the most important sources of vegetable oil globally. The global production of vegetable oil reached 75 million tons in 2020 (http://www.fao.org/faostat/zh/#data/QC/visualize). Oilseed rape canopy architecture is determined by plant height (PH), the length of the main inflorescence and the number (BN) and distribution of ancillary branches (Li et al., 2016). Canopy architecture indirectly influences cultivar yield potential by significantly influencing the number of siliques per plant. It has previously been shown that PH negatively correlates with siliques per plant, owing to the greater lodging risk of taller plants, and BN positively correlates with the number of siliques per plant (Qiu et al., 2006; Chen et al., 2014).
Phosphorus (P) is an essential macroelement for plant growth and development (Kochian, 2012). Oilseed rape has a high P requirement and P deficiency reduces PH and BN, and the subsequent seed yield of B. napus (Shi et al., 2013). Phosphorus in the soil is easily fixed by metal cations to form insoluble compounds or bound in organic forms, which are difficult to be directly absorbed and used by plants (Holford, 1997). Therefore, P fertilizers must be supplied to ensure the normal growth and development of crops. The use of P fertilizer increased from 34·5 million tons in 2001 to 45·4 million tons in 2017 (http://www.fao.org/faostat/zh/#data/QC/visualize). However, long-term excess P fertilizer is problematic because of its limited bioavailability and potential environmental problems such as eutrophication, and the depletion of non-renewable rock phosphate resources (Vance et al., 2003). Therefore, uncovering the genetic mechanisms of B. napus tolerance to low P (LP) and breeding P-efficient varieties is an important way to reduce P use in agricultural systems.
Linkage mapping analysis has been widely used to study the genetic basis of P tolerance in B. napus (Yang et al., 2010a, b; Ding et al., 2012; Shi et al., 2013). Sixty-two significant quantitative trait loci (QTLs) were associated with plant P uptake, total root surface area, root length, root volume and total dry weight under high and low P conditions in three experiments, explaining 12·7–31·9 % of the phenotypic variation (Yang et al., 2010a). Ding et al. (2012) detected 21 significant QTLs associated with PH and BN under high and low P conditions, explaining 8.5–19·8 % of the phenotypic variation. One hundred and fifty-five significant QTLs were associated with seed yield and yield-related traits from three different trials, and 79 QTLs detected at LP (a deficient P supply) and 76 at SP (a sufficient P supply) (Shi et al., 2013). Among them, 16 QTLs were associated with PH and 5 QTLs were associated with BN (Shi et al., 2013). However, due to the limited number of genetic markers and the frequency of recombination in mapping populations, the physical interval for these QTLs are usually large, which makes it difficult to determine the candidate genes (Xiao et al., 2017).
Compared with QTL mapping, genome wide association study (GWAS) can achieve a higher resolution of the underlying genetic loci, which greatly improves the efficiency of gene mapping (Nordborg and Weigel, 2008). GWAS has been widely applied to the genetic dissection of complex traits in crops, such as grain size in rice (Si et al., 2016), PH in maize (Zhang et al., 2019b) and silique number in B. napus (Li et al., 2020). GWAS combined with QTL analysis has been used successfully to mine candidate genes in B. napus, such as root-related traits (Wang et al., 2017) and clubroot resistance (Laperche et al., 2017) in B. napus and capsaicinoid content in Capsicum (Han et al., 2018).
GWAS of the PH and BN of B. napus under SP conditions has been conducted in several studies (Li et al., 2016; Sun et al., 2016; Zheng et al., 2017). However, the genetic bases controlling PH and BN in B. napus under P deficiency remain unknown. In this study, we investigated the PH and BN of a B. napus association panel at LP and SP across 2 years. GWAS of PH and BN was performed using high-quality single-nucleotide polymorphisms (SNPs) by whole-genome resequencing (Tang et al., 2021). We aimed to identify the (1) genetic diversity of the population, (2) significant SNPs and candidate genes associated with PH and BN at contrasting P supplies and (3) reveal the favourable haplotypes for breeding P-efficient B. napus cultivars.
MATERIALS AND METHODS
Plant materials and growth conditions
The association panel of Brassica napus comprises 403 cultivars and inbred lines, including 350 semi-winter, 44 spring, 8 winter and 1 unknown type, collected from major B. napus breeding centres across China (Supplementary Data Table S1). Of them, 361 lines originated in China, 21 from Europe, 8 from Japan, 5 from Canada, 4 from Australia, 3 from Korea and 1 unknown (Supplementary Data Table S1). The panel was grown in the field at LP supply (P, 0 kg ha−1) and SP supply (P, 40 kg ha−1) with three replications at Meichuan Town, Wuxue city, Hubei province, China (115·55° E , 29·85° N) from 2018 to 2019 (Trial 1) and from 2019 to 2020 (Trial 2). The soil was sandy loam soil. The topsoil (0–30 cm) was collected before sowing (before fertilization) for determination of the available nutrient concentrations (Table 1). We used 0·5 m NaHCO3 (pH 8·5) to measure available soil P. All the plots received basal fertilizer, and the application rate was as follows (per hectare), 108 kg of N (supplied as urea), 0 or 40 kg of P (supplied as calcium superphosphate), 87 kg of K (supplied as potassium chloride) and 6 kg of B (supplied as borax). These fertilizers were thoroughly mixed and applied in bands near the crop rows. The remaining N (72 kg ha−1) was top-dressed as urea in equal amounts at the four- to five-leaf stage. Each accession had four rows and each plot had eight plants in each row. At the mature stage, six plants in each plot were selected to measure PH and BN. the PH was the length of the plant from the base of the stem to the tip of the main inflorescence, and BN was calculated as the number of primary branches arising from the main shoot. PHr and BNr were defined as the ratio of PH_LP to PH_SP and that of BN_LP to BN_SP, respectively.
Table 1.
Soil physical and chemical properties in the field trials
| Trial (year) | Replications | pH (1:1 H2O) | Organic matter (g kg−1) | Total N concentration (g kg−1) | Available P concentration (g kg−1) | Total P concentration (g kg−1) | Total K concentration (g kg−1) |
|---|---|---|---|---|---|---|---|
| Trial 1 (2018–19) | 1 | 5·61 | 7·37 | 0·91 | 11·56 | 0·57 | 13·2 |
| 2 | 5·69 | 8·04 | 1·12 | 11·71 | 0·53 | 12·1 | |
| 3 | 5·65 | 8·33 | 1·30 | 12·19 | 0·46 | 14·6 | |
| Trial 2 (2019–20) | 1 | 5·70 | 8·42 | 1·19 | 12·51 | 0·63 | 11·8 |
| 2 | 5·58 | 7·88 | 1·33 | 16·95 | 0·56 | 10·5 | |
| 3 | 5·56 | 7·03 | 1·06 | 14·10 | 0·62 | 12·7 |
Genome-wide association analysis and candidate gene identification
More than 10 million high-quality SNP markers of the association panel of B. napus were derived from a previous study (Tang et al., 2021). After filtering the SNPs with minor-allele frequency (MAF) >0.05 and missing rate <0·2, we obtained 5·58 million SNPs for GWAS. GWAS for PH was carried out using general linear models (GLMs) and mixed linear models (MLMs) by Tassel 5.0 software (Bradbury et al., 2007). In order to minimize the contribution from regions of extensive strong linkage dissequilibrium (LD), we scanned the whole genome with a sliding window of 500 kb (in steps of 100 SNPs), and used Plink software to remove SNPs related to other SNPs within the window with correlation coefficient (R2) > 0·2. Finally, we obtained 497 761 SNPs. Admixture software was used to calculate the Q matrix and Tassel 5.0 software was used to calculate the K matrix. The Manhattan plot was drawn by ggplot2 (https://cran.r-project.org/web/packages/ggplot2/index.html) software and the Quantile–Quantile plot was drawn by the CMplot software (https://github.com/YinLiLin/CMplot). The threshold for the significance of associations between SNPs and traits was P < 6·25 × 10−07. The LD statistic was calculated by PopLDdecay software (Zhang et al., 2019a). According to the LD decay (238 kb) of the panel, the genes located within 300 kb upstream and downstream of the peak SNPs were considered as candidate genes. The genotypes of BnaA02g33340D, BnaA10g09290D and BnaC08g26640D in the association panel of B. napus were obtained by bcftools software (http://samtools.github.io/bcftools/bcftools.html). Candidate gene association analysis of BnaA02g33340D, BnaA10g09290D and BnaC08g26640D were performed with Tassel 5.0 software (Bradbury et al., 2007). The SNP markers from 2 kb upstream of the gene to 2 kb downstream of the gene were used to conduct association analysis with the PH of the association panel of B. napus at both P supplies.
Haplotype analysis
The haplotype analysis was predicted using the HaploView software (Barrett et al., 2005). One-way ANOVA and Student’s t-test were employed to compare the differences in PH among the haplotypes. Haplotypes containing at least 20 B. napus cultivars were used for further comparative analysis. Student’s t-test was used to compare the differences in PH among the haplotypes.
Statistical analysis of phenotypic data
The mean values of PH and BN of six plants of each replication were calculated using Excel 2007. Best linear unbiased prediction (BLUP) of PH and BN at LP and SP supply for each line was calculated using the R package ‘lme4’ (https://cran.r-project.org/web/packages/lme4/index.html). Trial 1_BLUP and Trial 2_BLUP were calculated with the phenotypic values of three replications in Trial 1 and Trial 2, respectively (Supplementary Data Table S2). The R language was used to calculate the correlation coefficients between phenotypes. The broad-sense heritability was calculated as: h2 = VG/(VG/(VG + VE)/nr), where VG is genetic variance, VE is environmental variance, n is the number of environments and r is the number of replicates.
RESULTS
Phenotypic variation for PH and BN of an association panel of B. napus at LP and SP supplies
From the seedling stage to the silique stage at LP, the old leaves turned dark and purple, and the inhibition of root and shoot growth was observed in most lines (Supplementary Data Fig. S1A–E). In addition, the PH and BN of B. napus decreased at LP compared with SP (Supplementary Data Fig. S1F–I). Extensive phenotypic variations for PH and BN were observed in the association panel of B. napus at LP and SP (Supplementary Data Figs S1F and S2, Table 2, Supplementary Data Table S2). At LP, PH varied from 57·5 to 179·3 cm (Supplementary Data Fig. S2, Table 2, Supplementary Data Table S2) and BN varied from 0 to 11 per plant across the 2 years (Supplementary Data Fig. S2, Table 2, Supplementary Data Table S2). At SP, PH varied from 84·0 to 235·0 cm (Supplementary Data Fig. S2, Table 2, Supplementary Data Table S2) and BN varied from 1 to 16 per plant across the 2 years (Supplementary Data Fig. S2, Table 2, Supplementary Data Table S2). PHr varied from 0·40 to 1·00 and BNr varied from 0·10 to 1·00 across the 2 years (Supplementary Data Fig. S2, Table 2, Supplementary Data Table S2). High h2 values were observed for all traits (Table 2). The correlation coefficients of PH_SP, BN_SP, PH_LP, BN_LP, PHr and BNr between Trial 1 and Trial 2 were 0·57, 0·28, 0·27, 0·09, 0·19 and 0·09, respectively (Supplementary Data Fig. S3). Thus, BLUP analysis was employed to deal with multi-year and field phenotypic values of PH and BN.
Table 2.
Mean, maximum (max.), minimum (min.), range, heritability and coefficient of variation (CV) of PH, BN, PHr (ratio of PH_LP to PH_SP) and BNr (ratio of BN_LP to BN_SP) in an association panel of B. napus in Trial 1 and Trial 2, at LP and SP
| Trait | Year | Replication | Mean | Min. | Max. | Range | CV (%) | h 2 (%) |
|---|---|---|---|---|---|---|---|---|
| PH_LP (cm) | Trial 1 | 1 | 121·89 | 73·75 | 165·50 | 91·75 | 14·80 | 63.26 |
| 2 | 132·02 | 74·95 | 179·25 | 104·30 | 14·31 | |||
| 3 | 132·15 | 79·00 | 179·00 | 100·00 | 14·31 | |||
| Trial 2 | 1 | 128·66 | 57·50 | 176·00 | 118·50 | 18·23 | ||
| 2 | 126·93 | 60·50 | 170·50 | 110·00 | 16·42 | |||
| BN_LP (no. per plant) | Trial 1 | 1 | 2·9 | 0·0 | 8·0 | 8·0 | 53·76 | 61.01 |
| 2 | 4·2 | 0·0 | 9·5 | 9·5 | 45·80 | |||
| 3 | 4·3 | 0·0 | 10·5 | 10·5 | 46·65 | |||
| Trial 2 | 1 | 4·4 | 0·0 | 10·0 | 10·0 | 45·98 | ||
| 2 | 4·0 | 0·0 | 11·0 | 11·0 | 51·15 | |||
| PH_SP (cm) | Trial 1 | 1 | 137·43 | 90·50 | 194·00 | 103·50 | 12·72 | 88.51 |
| 2 | 151·11 | 84·00 | 207·50 | 123·50 | 12·25 | |||
| 3 | 159·85 | 97·00 | 214·00 | 117·00 | 11·02 | |||
| Trial 2 | 1 | 170·31 | 97·00 | 216·00 | 119·00 | 12·82 | ||
| 2 | 160·16 | 107·00 | 204·00 | 97·00 | 11·67 | |||
| 3 | 171·78 | 90·00 | 235·00 | 145·00 | 11·76 | |||
| BN_SP (no. per plant) | Trial 1 | 1 | 4·2 | 1·0 | 9·5 | 8·5 | 39·48 | 70.12 |
| 2 | 5·9 | 1·5 | 13·0 | 11·5 | 31·31 | |||
| 3 | 6·7 | 3·0 | 15·0 | 12·0 | 28·75 | |||
| Trial 2 | 1 | 7·2 | 1·0 | 15·0 | 14·0 | 26·85 | ||
| 2 | 6·7 | 3·0 | 14·0 | 11·0 | 26·69 | |||
| 3 | 7·0 | 2·5 | 15·5 | 13·0 | 26·77 | |||
| PHr | Trial 1 | 1 | 0·84 | 0·45 | 1·00 | 0·54 | 12·22 | 69.64 |
| 2 | 0·83 | 0·52 | 1·00 | 0·47 | 12·93 | |||
| 3 | 0·81 | 0·50 | 1·00 | 0·50 | 13·98 | |||
| Trial 2 | 1 | 0·75 | 0·40 | 1·00 | 0·60 | 16·17 | ||
| 2 | 0·73 | 0·40 | 0·97 | 0·57 | 15·93 | |||
| BNr | Trial 1 | 1 | 0·61 | 0·10 | 1·00 | 0·90 | 40·97 | 61.33 |
| 2 | 0·64 | 0·10 | 1·00 | 0·90 | 37·03 | |||
| 3 | 0·63 | 0·13 | 1·00 | 0·88 | 34·79 | |||
| Trial 2 | 1 | 0·59 | 0·11 | 1·00 | 0·89 | 35·81 | ||
| 2 | 0·59 | 0·11 | 1·00 | 0·89 | 37·74 |
Population structure, relative kinship and LD decay
A total of 5·58 million SNP markers were identified for this B. napus association panel (Supplementary Data Table S3). SNP number on each chromosome ranged from 171 159 on A08 to 475 529 on C03 (Supplementary Data Table S3). The LD decay on each chromosome ranged from 1996 kb on C07 to 45 kb on A03, when r2 was 0·1 (Supplementary Data Table S3). The LD decay on the whole genome was 238 kb (Supplementary Data Fig. S4, Table S3). A total of 497 761 SNPs were selected to assess the population structure, relative kinship and LD. The population could be divided into five subgroups based on the cross-validation errors (Supplementary Data Fig. S5). The r pairwise relative kinship is close to 0 (Supplementary Data Fig. S6, Table S4). For example, the values of the relative kinships were 0–129 976 pairs and 0·1–148 803 pairs, and the ratios to the total value were 80·03 and 91·62 %, respectively (Supplementary Data Table S4). These results showed that the genetic distance of the majority of the accessions in the association panel were large enough for the GWAS analysis.
Genome-wide association mapping of PH and BN of B. napus at LP and SP supplies
We performed GWAS with GLM and MLM approaches to identify SNPs associated with PH and BN at LP and SP in B. napus. A total of 1289 SNPs were identified to be significantly associated with PH of B. napus at LP and SP across 2 years (P < 6·25 × 10−07) (Supplementary Data Fig. S7A–D, Tables S5 and S6). Among them, 133, 685 and 471 SNPs were associated with PH_LP, PH_SP and PHr, respectively. The GLM analysis detected a total of 1275 significant SNPs at both P supplies. Among them, 131, 678 and 466 SNPs were associated with PH_LP, PH_SP and PHr, respectively, and were distributed on all chromosomes, explaining between 6·03 and 14·01 % of the phenotypic variation (Supplementary Data Table S5). Chromosome C03 had the largest number of significant SNPs (244) and chromosome A07 had the least number of significant SNPs (7 SNPs) (Supplementary Data Table S5). MLM analysis detected 106 significant SNPs on 18 of the 19 B. napus chromosomes (excluding C02) at both P supplies. Among them, 15, 74 and 17 SNPs were associated with PH_LP, PH_SP and PHr, respectively, and explained between 7·85 and 14·23 % of the phenotypic variation (Supplementary Data Table S6). Among them, 93 significant SNPs were simultaneously identified by the two models (Supplementary Data Tables S5 and S6).
For BN, the GLM analysis detected a total of 837 significant SNPs at both P supplies. Among them, 34, 770 and 33 SNPs were associated with BN_LP, BN_SP and BNr, respectively, which were distributed on all chromosomes and explained between 6·12 and 15·83 % of the phenotypic variation (Supplementary Data Fig. S7E–H, Tables S5 and S6). MLM analysis detected 93 significant SNPs at both P supplies. Among them, three and 90 SNPs were associated with BN_LP and BN_SP, respectively, which explained between 6·92 and 14·47 % of the phenotypic variation. Of them, 90 SNPs were detected by both the GLM and MLM analyses (Supplementary Data Tables S5 and S6).
Candidate genes for PH and BN at LP supply in B. napus
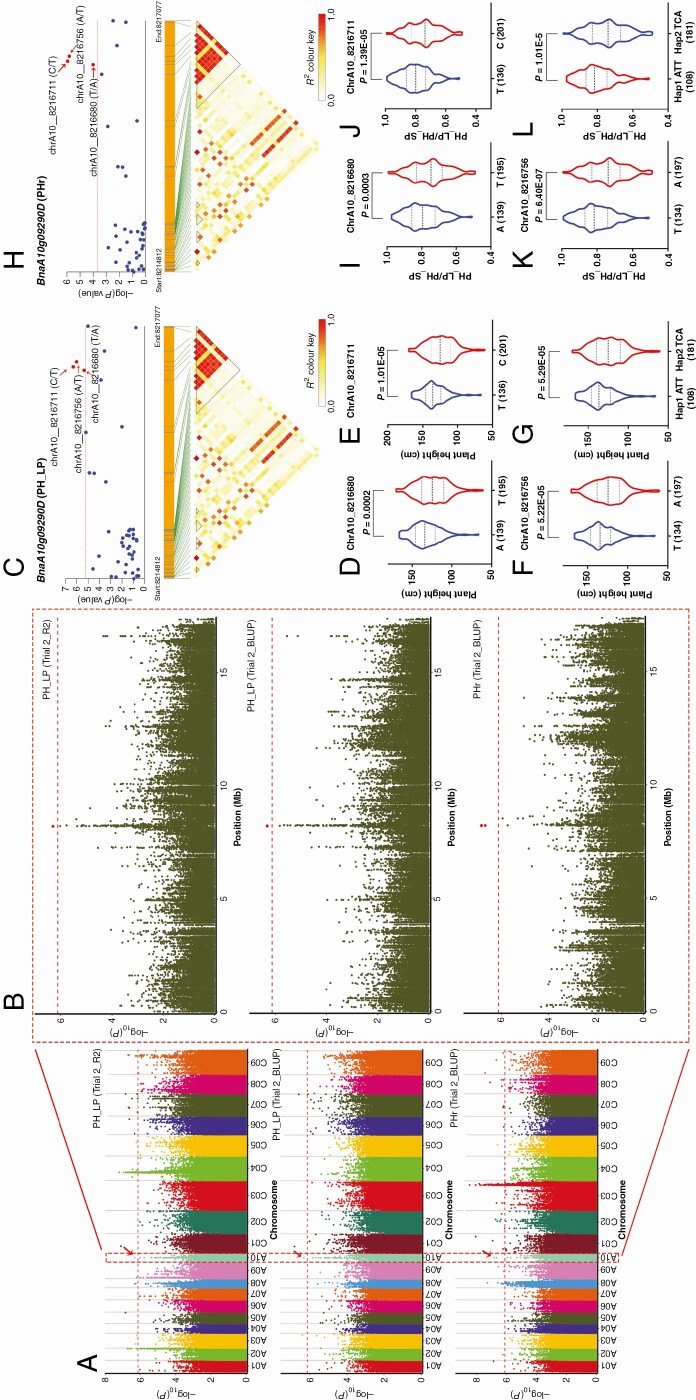
We identified 1289 significant SNPs significantly associated with PH_LP and PHr. Two, 19, 3, 9, 88, 5 and 5 significant SNPs were associated with PH_LP in Trial 1_R1, Trial 1_R2, Trial 1_R3, Trial 1_BLUP, Trial 2_R1, Trial 2_R2 and Trial 2_BLUP, respectively (Supplementary Data Fig. S7A–D, Tables S5 and S6). Fifteen, 17, 12, 23, 250, 1 and 158 significant SNPs were associated with PHr in Trial 1_R1, Trial 1_R2, Trial 1_R3, Trial 1_BLUP, Trial 2_R1, Trial 2_R2 and Trial 2_BLUP, respectively (Supplementary Data Fig. S7A–D, Tables S5 and S6). The SNP of chrA10_8216711 on chromosome A10 was associated with PH_LP and PHr in both Trial 2_R2 and Trial 2_BLUP (Fig. 1A, B). In this study, the LD decay was 238 kb for this association panel (Supplementary Data Table S3). Based on the LD decay, 300 kb up/downstream of the significant SNPs were selected to identify candidate genes on A10 and 76 candidate genes were detected. The significant SNP associated with PH_LP and PHr identified at the 8 216 711 bp position on chromosome A10 was found located in the genetic region of BnaA10g09290D, whose function was unknown. Candidate gene association analysis of BnaA10g09290D with SNP markers from the 2-kb promoter region and the entire coding region showed that three SNPs in BnaA10g09290D were significantly associated with the PH_LP and PHr, and had strong LD with each other (Fig. 1C, H). Further analysis demonstrated that the A allele of chrA10_8216680, T allele of chrA10_8216711 and T allele of chrA10_8216756 were the P-deficiency-tolerant alleles (Fig. 1). Two major haplotypes based on the three significant SNPs were detected in this association panel, among which BnaA10g09290Hap2 (TCA) was P deficiency sensitive, while BnaA10g09290Hap1 (ATT) was P deficiency tolerant (Fig. 1G, L). Notably, chrA10_8216680 (T/A) located in the exon region of the gene BnaA10g09290D and resulted in amino acid changes from isoleucine to asparagine and might contribute to the phenotypic difference in PH_LP and PHr (Table 3).
Fig. 1.
The co-localized locus and haplotypes on chromosome A10 associated with PH_LP and PHr of B. napus. (A) Manhattan plot of co-localized locus for PH_LP and PHr in Trial 2_R2 and Trial 2_BLUP. (B) Significant SNPs associated with PH_LP and PHr on chromosome A10. The big red dots represent the significant SNPs. (C) Candidate gene association analysis of BnaA10g09290D with PH_LP. Association of the three alleles in chrA10_8216680 (D), chrA10_8216711 (E) and chrA10_8216756 (F) with PH_LP, respectively. (G) Two haplotypes of BnaA10g09290D. (H) Candidate gene association analysis of BnaA10g09290D with PHr. Association of the three alleles in chrA10_8216680 (I), chrA10_8216711 (J) and chrA10_8216756 (K) with PHr, respectively. (L) Two haplotypes of BnaA10g09290D. The number of inbred lines harbouring the corresponding allele is shown in brackets at the bottom.
Table 3.
List of synonymous and non-synonymous SNP variants identified in the candidate genes of BnaA02g33090D, BnaA10g09290D and BnaC08g26640D
| Gene | SNP | Major allele |
Minor allele |
SNP location | SNP type | Amino acid changes |
|---|---|---|---|---|---|---|
| BnaA02g33090D | chrA02_23899623 | G | A | Intron | – | – |
| chrA02_23899654 | G | T | Intron | – | – | |
| chrA02_23899669 | T | A | Intron | – | – | |
| chrA02_23899686 | C | T | Exon | Synonymous | – | |
| chrA02_23899688 | A | C | Exon | Non-synonymous | Histidine to proline | |
| BnaA10g09290D | chrA10_8216680 | T | A | Exon | Non-synonymous | Isoleucine to asparagine |
| chrA10_8216711 | C | T | Intron | – | ||
| chrA10_8216756 | A | T | Exon | Synonymous | ||
| BnaC08g26640D | chrC08_27999709 | A | T | Intron | – | – |
| chrC08_27999778 | A | T | Exon | Non-synonymous | Isoleucine to asparagine | |
| chrC08_27999846 | T | C | Exon | Synonymous | – |
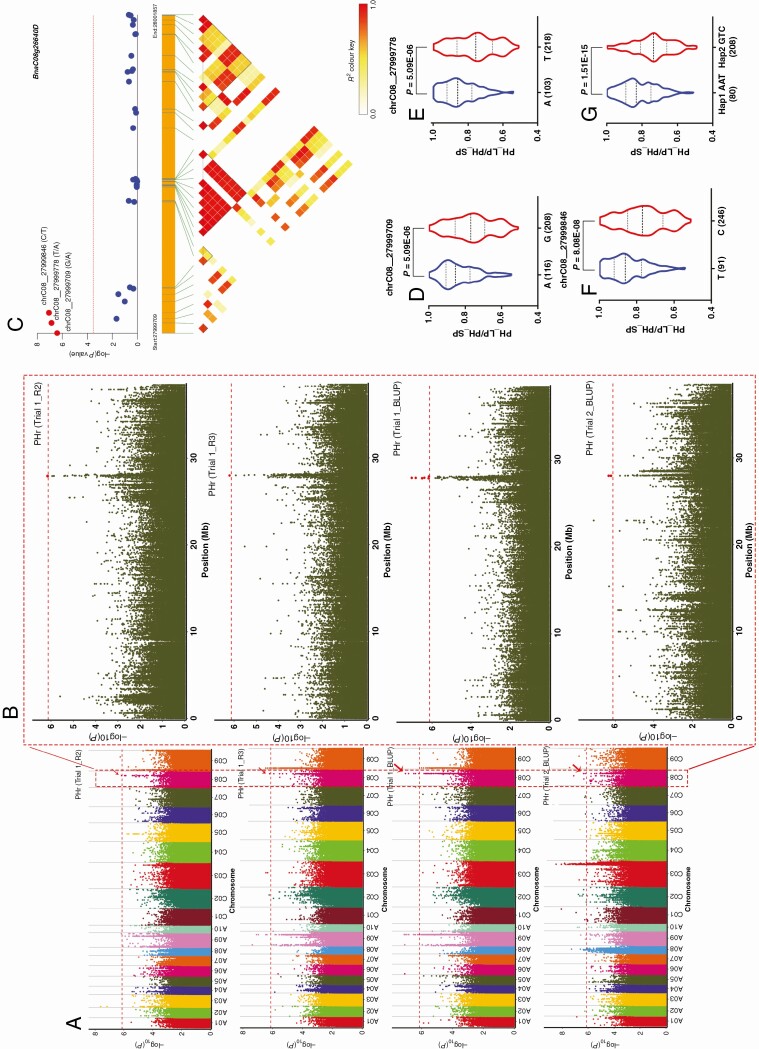
The region significantly associated with PHr in both Trial 1_R2, Trial 1_R3, Trial 1_BLUP and Trial 2_BLUP on chromosome C08 ranged from 27·95 to 28·13 Mb (Fig. 2A, B). Based on the LD decay (238 kb), 300 kb upstream and downstream regions of the significant SNP (chrC08_27999846, P = 8·91E-08, phenotypic variance explained = 12.46 %) were selected and found to contain 101 genes. The lead SNP of chrC08_27999846 was located within BnaC08g26640D. Three SNPs in BnaC08g26640D were detected to be significantly associated with the PHr (Fig. 2C), and the A allele of chrC08_27999709, A allele of chrC08_27999778 and T allele of chrC08_27999846 were the P-deficiency-tolerant alleles (Fig. 2D–F). These three significant SNPs revealed two major haplotypes, and BnaC08g26640Hap1 (AAT) had significantly greater PHr value than BnaC08g26640Hap2 (GTC) (P = 1·51E-15) (Fig. 2G). The SNP chrC08_27999709 (A/G) was located in the intron region, and did not result in any amino acid change (Table 3). The SNP of chrC08_27999846 (T/C) was in the exon region, and it was a synonymous mutation that did not result in any amino acid change (Table 3). The SNP of chrC08_27999778 (A/T) located in the exon region of the gene BnaC08g26640D and resulted in amino acid changes from isoleucine to asparagine and might contribute to the phenotypic difference in PHr (Table 3).
Fig. 2.
The co-localized locus and haplotypes on chromosome C08 associated with PHr of B. napus. (A) Manhattan plot of co-localized locus for PHr in Trial 1_R2, Trial 1_R3, Trial 1_BLUP and Trial 2_BLUP. (B) Significant SNP associated with PHr on chromosome C08. The big red dots represent the significant SNPs. (C) Candidate gene association analysis of BnaC08g26640D with PHr. Association of the three alleles in chrC08_27999709 (D), chrC08_27999778 (E) and chrC08_27999846 (F) with PHr, respectively. (G) Two haplotypes of BnaC08g26640D. The number of inbred lines harbouring the corresponding allele is shown in brackets at the bottom.
Three SNPs associated with BN_LP, chrA01_13846343, chrA03_20898013 and chrC07_521008 were detected simultaneously by GLM and MLM (Supplementary Data Tables S5 and S6). On chromosome A01, 40 candidate genes were detected underlying the region around chrA01_13846343 (Supplementary Data Table S7). The ATP phosphoribosyl transferase 1 (BnaA01g21560D) and low-temperature- and salt-responsive protein (BnaA01g21470D) were identified in the candidate region. On chromosome A03, the significant SNP chrA03_20898013 was also detected by GLM and MLM model simultaneously, with P = 4·97E-07 and R2 = 9·21 % (Supplementary Data Tables S5 and S6). Two genes encoding heat shock transcription factor A7A (BnaA03g41540D and BnaA03g41550D) were identified in the upstream region from chrA03_20898013, and their homologue in Arabidopsis responds to abiotic stresses (Lin et al., 2018). In addition, a phosphatidic acid phosphatase family protein (BnaA03g41050D) and a pyruvate orthophosphate dikinase (BnaA03g41960D) were identified in the downstream region of chrA03_20898013. On chromosome C07, a total of 62 genes were detected underlying the candidate region around the SNP chrC07_521008 (P = 5·95E-07). Among these candidate genes was an EXP1 gene (BnaC07g00140D), which is involved in unidimensional cell growth and plant-type cell wall loosening, and may be involved in BN determination at LP (Supplementary Data Table S8).
Comparison of the significant SNPs with QTLs for P efficiency
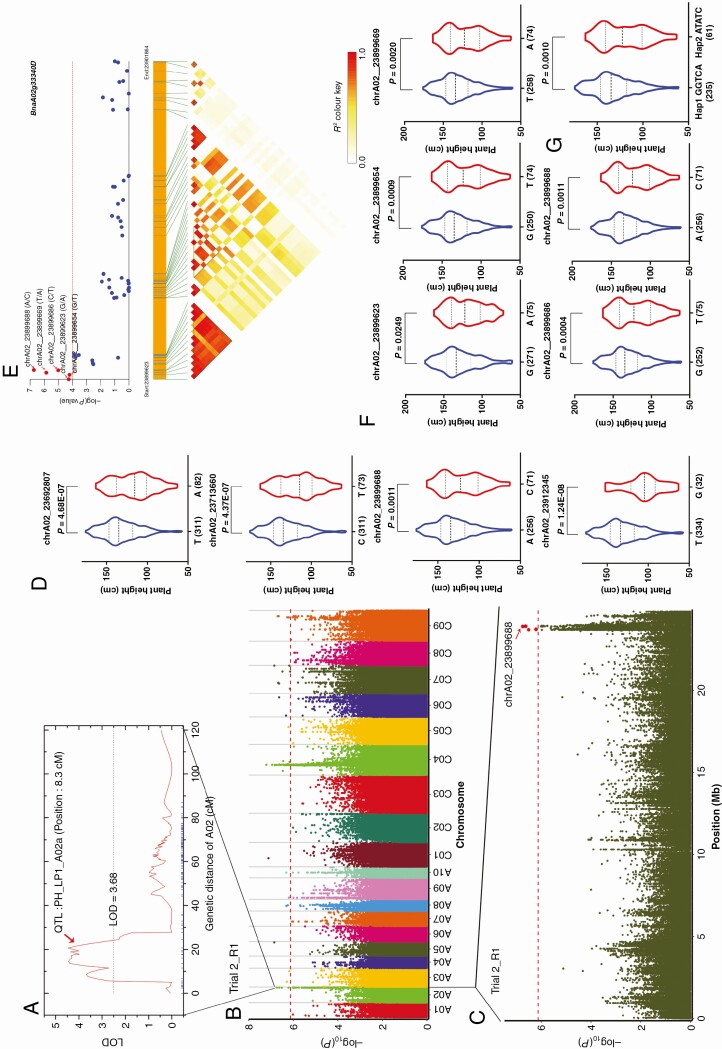
Based on the Darmor-bzh reference genome, we analysed the co-localization of the significant SNPs detected in our study and the QTLs detected by previous study (Shi et al., 2013). Thirty-three significant SNPs detected by GWAS co-localized with the intervals of the QTLs for the same traits in a previous linkage analyses of the BnaTNDH population (Shi et al., 2013), including 4 SNPs on A02, 7 SNPs on A09, 7 SNPs on C06 and 15 SNPs on C07 (Supplementary Data Table S9). Among them, four co-located SNPs on A02 chromosome associated with PH_LP (Fig. 3A–C), including the P-tolerant T allele of chrA02_23692807, C allele of chrA02_23713660, A allele of chrA02_23899688 and T allele of chrA02_23912345 (Fig. 3D). A total of 152 candidate genes were within the QTL (PH_LP1_A02a; Shi et al., 2013) confidence interval on A02 (Fig. 3A, Supplementary Data Table S10). Among the four co-localized SNPs, the lead SNP chrA02_23899688 (P = 1·85E-07) was located in the CDS region of BnaA02g33340D (Supplementary Data Table 3). Since BnaA02g33340D was identified by association with PH_LP, we examined the association of sequence variation in BnaA02g33340D with the PH_LP of the association panel. Five SNPs in BnaA02g33340D were detected to be significantly associated with the PH_LP (Fig. 3E). Further analyses showed that the G allele of chrA02_23899623, G allele of chrA02_23899654, T allele of chrA02_23899669, C allele of chrA02_23899686 and A allele of chrA02_23899688 were P-tolerant alleles (Fig. 3F). There were two major haplotypes associated with PH and Hap1 (GGTCA) and had higher PH at low P than Hap2 (ATATC) with a P-value of 0·0010, suggesting that the variation in BnaA02g33340D was associated with PH_LP (Fig. 3G). Further analysis indicated that chrA02_23899623 (G/A), chrA02_23899654 (G/T) and chrA02_23899669 (T/A) were located in the intron region, and the SNP ‘chrA02_23899686 (C/T)’ was located in the exon region and was a synonymous mutation, which was unlikely to affect the function of the BnaA02g33340D protein (Table 3). The SNP ‘chrA02_23899688 (A/C)’ was located in the exon region of the gene BnaA02g33340D and resulted in amino acid changes from histidine to proline and may be causative of the difference in PH_LP (Table 3). These observations demonstrated that BnaA02g33340D is most likely to be the candidate gene for the locus represented by lead SNP chrA02_23899688.
Fig. 3.
Co-localized locus on chromosome A02 for PH of B. napus at LP. (A) QTL detected for PH in the BnaTNDH population linkage analysis (Shi et al., 2013). (B) Manhattan plot of PH_LP (Trial 2_R1). (C) Peak SNP for PH_LP (Trial 2_R1). (D) Association of the co-localized locus with PH at LP. (E) Candidate gene association analysis of BnaA02g33340D. (F) Association of the five alleles in chrA02_23899623, chrA02_23899654, chrA02_23899669, chrA02_23899686 and chrA02_23899688 with PH at LP, respectively. (G) Two haplotypes of BnaA02g33340D. The number of inbred lines harbouring the corresponding allele is shown in brackets at the bottom. R1, replication 1.
DISCUSSION
Significant differences in the PH and BN in the association panel of B. napus at LP supply
In this study, the available soil P concentration in three fields ranged from 11·56 to 16·95 mg kg−1, which are between P deficient and slightly P deficient (Zou et al., 2009). Some typical P-deficiency symptoms were observed in most lines in the field, such as greater anthocyanin visibility in old leaves, restricted shoot growth at the seedling stage (Supplementary Data Fig. S1A–C), and reduced PH and BN at the mature stage (Supplementary Data Fig. S1D–F). Plant height and BN are essential components of plant canopy architecture in B. napus, which are closely correlated with its yield (Zheng et al., 2020). Compared with SP, PH and BN of ‘Eyou Changjia’ (a P-efficient variety) decreased by 11 and 25·4 %, respectively; and PH and BN of B104-2’ (a P-inefficient variety) decreased by 23 and 46·9 %, respectively (Ding et al., 2012). In addition, the PH and BN of ‘Eyou Changjia’ were higher than those of ‘B104-2’ at LP in two field trials (Ding et al., 2012). Cultivars ‘Tapidor’ and ‘Ningyou 7’ are parental cultivars of the BnaTNDH population and PH and BN at LP were also less than those at SP (Shi et al., 2013). Compared with the control (no P application), the PH of B. napus increased by 11·2, 16·7 and 19·4 %, respectively, and the BN of B. napus increased by 25·5, 36·2 and 40·4 % in the P application rates of 103·0, 206·0 and 309·0 kg hm−2 (Lu et al., 2005).
Extensive phenotypic variations for PH and BN are observed in several natural populations of B. napus at SP (Li et al., 2016; Sun et al., 2016; Luo et al., 2017; Zheng et al., 2017). For example, the PH varied from 48·3 to 228·3 cm among 496 B. napus accessions, with an average ranging from 133·5 to 187·3 cm across six environments (Sun et al., 2016), and varied from 86.2 to 206·0 cm among 333 B. napus accessions across 2 years, with 1·6- to 2·4-fold variations across the 2 years (Zheng et al., 2017). In this study, we measured PH and BN in a panel of 403 B. napus accessions across 2 years under both LP and SP conditions. At SP, the PH varied from 84.0 to 235·0 cm and BN varied from 1 to 15 per plant (Table 2; Supplementary Data Table S1 and Fig. S2). Compared with the association panel with 496 B. napus accessions (Sun et al., 2016), the natural population in this study has more extensive genetic variation at SP. At LP conditions, the PH varied from 57·5 to 179·2 cm and BN varied from 0 to 11 per plant (Table 2; Supplementary Data Table S1 and Fig. S2). These data show that P deficiency significantly reduced the PH and BN of B. napus compared with those grown at SP in our study and others. In addition, extensive phenotypic variations for PH and BN at LP would be useful for screening for P-efficient cultivars that have greater potential to give higher yields at LP.
Candidate genes associated with significant SNPs for PH and BN of B. napus at LP
GWAS has been successfully applied to the genetic dissection of low P tolerance in different crops, such as in maize (Xu et al., 2018; Luo et al., 2019), rice (Wissuwa et al., 2015), wheat (Liu et al., 2015), soybean (Zhang et al., 2014) and oilseed rape (Wang et al., 2017). However, up to now there has been no report on the GWAS of B. napus PH and BN at LP, major determinants of seed yield and yield in B. napus. A total of 2127 significant SNPs were detected for PH and BN and 33 SNPs were identified co-localized with previously identified QTLs (Supplementary Data Tables S5, S6, S9). Closer examination of allelic frequency of the four co-located SNPs on chromosome A02 among this association panel indicated that the inbred lines carrying a minor allele have lower PH than inbred lines carrying a major allele at LP (Fig. 3D). Interestingly, the PH of the accessions carrying the minor allele and major allele did not show a significant difference at SP supply (Supplementary Data Fig. S8). The four co-localized SNPs decrease the PH at LP and might affect the seed yield. ChrA10_8216711 for PH_LP and PHr in both Trial 2_R2 and Trial 2_BLUP was located in the genetic region of a gene with unknown function, BnaA10g09290D. ChrC08_27999846 for PHr was located in the exon of BnaC08g26640D. We performed candidate gene association analysis and haplotype analysis for BnaA10g09290D and BnaC08g26640D, and high-P-efficiency haplotypes of BnaA10g09290Hap1 and BnaC08g26640Hap1, respectively, were identified (Figs 1G, L and 2G).
In addition, some genes reported to be associated with P uptake and homeostasis were also detected in our association analysis (Supplementary Data Table S11). PHOSPHATE1 (PHO1), a gene that encodes a membrane protein consisting of SPX, plays an important role in loading P into xylem in roots (Hamburger et al., 2002; Pacak et al., 2016; Che et al., 2020). The peak SNP chrA07_6641098 on A07 was identified to be located within 280 kb of B. napus BnaA07.PHO1 (Supplementary Data Table S11). Many plant proteins with SPX domains are involved in Pi signalling (Wang et al., 2004; Duan et al., 2008). In this study, BnaC03g65110D, a gene with an SPX (SYG1/Pho81/XPR1) domain-containing protein, was located within the interval of the SNP chrC03_54249584 for the trait of PHr (Supplementary Data Table S11). BnaC05g27910D was a homologous gene of BnaC03g65110D, also containing an SPX (SYG1/Pho81/XPR1) domain, and was located within the interval of the leading SNP chrC05_25541489 for the trait of PHr (Supplementary Data Table S11). In Arabidopsis, AtUBP14 is involved in the response of root systems to P deficiency (Lee et al., 2008). In this study, BnaA08g08470D (BnaA08.UBP20), a gene that contains ubiquitin-specific protease activity, was located within the interval of the SNP chrA08_8488725 and linked with the trait for PHr (Supplementary Data Table S11). The PHT1 family encode plant Pi transporters to transport P into plants (Huang et al., 2019). Recently, BnaPht1;4 was found to participate in the uptake and transportation of P, and promotes seed germination and seedling growth of B. napus by regulating the biosynthesis of ABA and GA (Huang et al., 2019). In this study, we identified the BnaA04g22280D (BnaA04.PHT11;4) by SNP chrA04_16843321 for BN_LP, which was likely to affect the growth and development of BN at LP (Supplementary Data Table S11). In addition, we identified BnaA09g16430D (BnaA09.Pht1;6) by SNP chrA09_9605643 for BN_LP, which is located 170 kb downstream of the SNP chrA09_9605643 (Supplementary Data Table S11). Purple acid phosphatases (PAPs) are a family of binuclear metalloenzymes (Olczak et al., 2003). Overexpression of OsPAP10c (purple acid phosphatase 10c) enhances the utilization of phytate-P, root growth and yield at LP in rice (Deng et al., 2020). BnaA03g41050D (BnaA03.PAP2) was located 310 kb downstream of the SNP chrA03_20898013 and linked with the trait for BN_LP (Supplementary Data Table S11). These candidate genes might play important roles in the development of PH and BN in B. napus at a deficient P supply.
Co-location of the significant SNPs for root system architecture traits and PH and BN of B. napus at a deficient P supply
Previously, 285 SNPs were detected associated with root-related traits of B. napus at LP and a P-efficient haplotype, BnA03Hap, on chromosome A03 was identified (Wang et al., 2017). Fifty-three significant SNPs in this study were adjacent to previously published significant SNPs associated with root system architecture at LP, which may control both the root system and PH or BN at LP (Supplementary Data Table S12). For example, SNP chrA08_10449680 was associated with PHr in this study and chrA08_10481443 was associated with primary root length at LP (PRL_LP) in Wang et al. (2017). BnaA08g11400D was detected by chrA08_10449680 and linked with the trait for PHr and located 106 kb upstream of chrA08_10449680 (Supplementary Data Table S12). BnaA08g11400D was homologous to Arabidopsis root hair defective 6-like 2 (RSL2). RSL2 encodes a basic helix–loop–helix transcription factor that promotes root hair initiation, growth and elongation (Yi et al., 2010; Han et al., 2017; Mangano et al., 2018). In addition, mutants lacking RSL2 have disrupted root hair formation under LP (Lan et al., 2012; Bhosale et al., 2018). BnaA08g11400D may affect PH by regulating root hair development at LP or have pleiotropic effects on shoot growth and development in B. napus.
Conclusions
Taken together, the PH and BN of a panel of 403 B. napus accessions collected worldwide were investigated at LP and SP, with 2127 significant SNPs associated with these traits identified by GWAS. Thirty-three SNPs co-localized with previous QTLs, including four at LP and 22 at SP. In addition, candidate gene association and haplotype analysis of BnaA02g33340D, BnaA10g09290D and BnaC08g26640D revealed several P-tolerant haplotypes. These significant SNP loci, favourable alleles and haplotypes, and the accessions carrying these desired alleles and haplotypes will be useful for breeding for low-P-tolerance B. napus cultivars.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: shoot and root growth of an association panel of B. napus in the field at LP and SP. Figure S2: frequency distribution of PH and BN of an association panel of B. napus in Trial 1and Trial 2 at LP and SP. Figure S3: correlation coefficients of PH and BN of B. napus between Trial 1 and Trial 2 at LP and SP. Figure S4: LD decay of an association panel of B. napus. Figure S5: population structure of an association panel of B. napus with K from 2 to 8. Figure S6: kinship of an association panel of 403 B. napus accessions. Figure S7: GWAS of PH and BN in an association panel of B. napus at LP and SP supplies. Figure S8: association of alleles for PH_SP. Table S1: list of 403 accessions of B. napus used in the study. Table S2: PH and BN at LP and SP supplies in an association panel of B. napus. Table S3: number of SNPs and LD decay on the 19 chromosomes of B. napus in this study. Table S4: kinship of an association panel of B. napus. Table S5: significant SNP loci for PH and BN of B. napus by GWAS in Trial 1 and Trial 2 at LP and SP by GLM. Table S6: significant SNP loci for PH and BN of B. napus by GWAS in Trial 1 and Trial 2 under LP and SP by MLM. Table S7: gene ontology (GO) enrichment analysis of candidate genes within 300 kb upstream and downstream of the lead SNP of chrA01__13846343 on A01 chromosome for BN_LP. Table S8: GO enrichment analysis of candidate genes within 300 kb upstream and downstream of the lead SNP of chrC07__521008 on C07 chromosome for BN_LP. Table S9: co-location of SNPs associated with plant height detected by GWAS with an association panel of B. napus and QTLs for PH detected in the BnaTNDH mapping population at LP and SP. Table S10: GO enrichment analysis of candidate genes located in the QTL linkage disequilibrium intervals on A02 chromosome for PH_LP. Table S11: candidate genes associated with phosphorus uptake and utilization in this study. Table S12: Comparison of SNPs detected by PH_LP and BN_LP in this study with previously identified SNPs for root and shoot traits at LP.
FUNDING
This work was supported by the National Nature Science Foundation of China (Grant No. 31972498). We also acknowledge the Applied Basic Research Fronts Program of Wuhan city (Grant No. 2018020401011302), the National Key R & D Program of China (Grant No. 2017YFD0200200) and the Natural and Fundamental Research Funds for the Central Universities of China (Grant No. 2662019PY013).
LITERATURE CITED
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- Bhosale R, Giri J, Pandey BK, et al. . 2018. A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nature Communications 9: 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635. [DOI] [PubMed] [Google Scholar]
- Che J, Yamaji N, Miyaji T, et al. . 2020. Node-localized transporters of phosphorus essential for seed development in rice. Plant & Cell Physiology 61: 1387–1398. [DOI] [PubMed] [Google Scholar]
- Chen B, Xu K, Li J, et al. . 2014. Evaluation of yield and agronomic traits and their genetic variation in 488 global collections of Brassica napus L. Genetic Resources and Crop Evolution 61: 979–999. [Google Scholar]
- Deng S, Lu L, Li J, et al. . 2020. Purple acid phosphatase 10c encodes a major acid phosphatase that regulates plant growth under phosphate-deficient conditions in rice. Journal of Experimental Botany 71: 4321–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding GD, Zhao ZK, Liao Y, et al. . 2012. Quantitative trait loci for seed yield and yield-related traits, and their responses to reduced phosphorus supply in Brassica napus. Annals of Botany 109: 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. 2008. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant Journal 54: 965–975. [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y. 2002. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HM, Wang HF, Han Y, et al. . 2017. Altered expression of the TaRSL2 gene contributed to variation in root hair length during allopolyploid wheat evolution. Planta 246: 1019–1028. [DOI] [PubMed] [Google Scholar]
- Han K, Lee HY, Ro NY, et al. . 2018. QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in Capsicum. Plant Biotechnology Journal 16: 1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holford ICR. 1997. Soil phosphorus: its measurement and its uptake by plants. Australian Journal of Soil Research 35: 227–39. [Google Scholar]
- Huang KL, Wang H, Wei YL, et al. . 2019. The high-affinity transporter BnPHT1;4 is involved in phosphorus acquisition and mobilization for facilitating seed germination and early seedling growth of Brassica napus. BMC Plant Biology 19: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV. 2012. Plant nutrition: rooting for more phosphorus. Nature 488: 466–467. [DOI] [PubMed] [Google Scholar]
- Lan P, Li W, Wen TN, Schmidt W. 2012. Quantitative phosphoproteome profiling of iron-deficient Arabidopsis roots. Plant Physiology 159: 403–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laperche A, Aigu Y, Jubault M, et al. . 2017. Clubroot resistance QTL are modulated by nitrogen input in Brassica napus. Theoretical and Applied Genetics 130: 669–684. [DOI] [PubMed] [Google Scholar]
- Lee YS, Huang KX, Florante AQ, et al. . 2008. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nature Chemical Biology 4: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chen B, Xu K, et al. . 2016. A genome-wide association study of plant height and primary branch number in rapeseed (Brassica napus). Plant Science 242: 169–177. [DOI] [PubMed] [Google Scholar]
- Li S, Zhu Y, Varshney RK, et al. . 2020. A systematic dissection of the mechanisms underlying the natural variation of silique number in rapeseed (Brassica napus L.) germplasm. Plant Biotechnology Journal 18: 568–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KF, Tsai MY, Lu CA, Wu SJ, Yeh CH. 2018. The roles of Arabidopsis HSFA2, HSFA4a, and HSFA7a in the heat shock response and cytosolic protein response. Botanical Studies 59: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang L, Deng M, et al. . 2015. Genome-wide association study of phosphorus-deficiency-tolerance traits in Aegilops tauschii. Theoretical and Applied Genetics 128: 2203–2212. [DOI] [PubMed] [Google Scholar]
- Lu JW, Chen F, Zhang ZQ, et al. . 2005. Effect of phosphor application rate on rapeseed yield, nutrient absorption and profit. Chinese Journal of Oil Crop Sciences 27: 73–76. [in Chinese with English abstract]. [Google Scholar]
- Luo X, Ding Y, Zhang L, et al. . 2017. Genomic prediction of genotypic effects with epistasis and environment interactions for yield-related traits of rapeseed (Brassica napus L.). Frontiers in Genetics 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Ma P, Nie Z, et al. . 2019. Metabolite profiling and genome-wide association studies reveal response mechanisms of phosphorus deficiency in maize seedling. Plant Journal 97: 947–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangano S, Denita-Juarez SP, Marzol E, Borassi C, Estevez JM. 2018. High auxin and high phosphate impact on RSL2 expression and ROS-homeostasis linked to root hair growth in Arabidopsis thaliana. Frontiers in Plant Science 9: 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Weigel D. 2008. Next-generation genetics in plants. Nature 456: 720–723. [DOI] [PubMed] [Google Scholar]
- Olczak M, Morawiecka B, Watorek W. 2003. Plant purple acid phosphatases - genes, structures and biological function. Acta Biochimica Polonica 50: 1245–1256. [PubMed] [Google Scholar]
- Pacak A, Barciszewska-Pacak M, Swida-Barteczka A, et al. . 2016. Heat stress affects Pi-related genes expression and inorganic phosphate deposition/accumulation in barley. Frontiers in Plant Science 7: 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Morgan C, Shi J, et al. . 2006. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theoretical and Applied Genetics 114: 67–80. [DOI] [PubMed] [Google Scholar]
- Shi T, Li R, Zhao Z, et al. . 2013. QTL for yield traits and their association with functional genes in response to phosphorus deficiency in Brassica napus. PLoS ONE 8: e54559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si LZ, Chen JY, Huang XH, et al. . 2016. OsSPL13 controls grain size in cultivated rice. Nature Genetics 48: 447–456. [DOI] [PubMed] [Google Scholar]
- Sun CM, Wang BQ, Yan L, et al. . 2016. Genome-wide association study provides insight into the genetic control of plant height in rapeseed (Brassica napus L.). Frontiers in Plant Science 7: 1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Zhao H, Lu S, et al. . 2021. Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Molecular Plant 14: 470–487. [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. 2003. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytologist 157: 423–447. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ribot C, Rezzonico E, Poirier Y. 2004. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiology 135: 400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Xh, Chen Yl, Thomas CL, et al. . 2017. Genetic variants associated with the root system architecture of oilseed rape (Brassica napus L.) under contrasting phosphate supply. DNA Research 24: 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissuwa M, Kondo K, Fukuda T, et al. . 2015. Unmasking novel loci for internal phosphorus utilization efficiency in rice germplasm through genome-wide association analysis. PLoS ONE 10: e0124215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YJ, Liu HJ, Wu LJ, et al. . 2017. Genome-wide association studies in maize: praise and stargaze. Molecular Plant 10: 359–374. [DOI] [PubMed] [Google Scholar]
- Xu C, Zhang H, Sun J, et al. . 2018. Genome-wide association study dissects yield components associated with low-phosphorus stress tolerance in maize. Theoretical and Applied Genetics 131: 1699–1714. [DOI] [PubMed] [Google Scholar]
- Yang M, Ding G, Shi L, Feng J, Xu F, Meng J. 2010a. Quantitative trait loci for root morphology in response to low phosphorus stress in Brassica napus. Theoretical and Applied Genetics 121: 181–193. [DOI] [PubMed] [Google Scholar]
- Yang M, Ding GD, Shi L, et al. . 2010. b. Detection of QTL for phosphorus efficiency at vegetative stage in Brassica napus. Plant and Soil 339: 97–111. [Google Scholar]
- Yi K, Menand B, Bell E, Dolan L. 2010. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nature Genetics 42: 264–267. [DOI] [PubMed] [Google Scholar]
- Zhang D, Song H, Cheng H, et al. . 2014. The acid phosphatase-encoding gene GmACP1 contributes to soybean tolerance to low-phosphorus stress. PLoS Genetics 10: e1004061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Dong SS, Xu JY, He WM, Yang TL. 2019a. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatic 35: 1786–1788. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wan J, He L, et al. . 2019. b. Genome-wide association analysis of plant height using the maize F1 population. Plants (Basel) 8: 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Peng C, Liu H, et al. . 2017. Genome-wide association study reveals candidate genes for control of plant height, branch initiation height and branch number in rapeseed (Brassica napus L.). Frontiers in Plant Science 8: 1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Zhang L, Tang M, et al. . 2020. Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnology Journal 18: 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Lu JW, Chen F, et al. . 2009. Study on abundance and deficiency indices of soil available P, K and B for winter rapeseed in Yangtze River Valley based on ASI method. Scientia Agricultura Sinica 42: 2028–2033 [in Chinese with English abstract]. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.