Abstract
Repairing DNA double-strand breaks (DSBs) with homologous chromosomes as templates is the hallmark of meiosis. The critical outcome of meiotic homologous recombination is crossovers, which ensure faithful chromosome segregation and promote genetic diversity of progenies. Crossover patterns are tightly controlled and exhibit three characteristics: obligatory crossover, crossover interference, and crossover homeostasis. Aberrant crossover patterns are the leading cause of infertility, miscarriage, and congenital disease. Crossover recombination occurs in the context of meiotic chromosomes, and it is tightly integrated with and regulated by meiotic chromosome structure both locally and globally. Meiotic chromosomes are organized in a loop-axis architecture. Diverse evidence shows that chromosome axis length determines crossover frequency. Interestingly, short chromosomes show different crossover patterns compared to long chromosomes. A high frequency of human embryos are aneuploid, primarily derived from female meiosis errors. Dramatically increased aneuploidy in older women is the well-known “maternal age effect.” However, a high frequency of aneuploidy also occurs in young women, derived from crossover maturation inefficiency in human females. In addition, frequency of human aneuploidy also shows other age-dependent alterations. Here, current advances in the understanding of these issues are reviewed, regulation of crossover patterns by meiotic chromosomes are discussed, and issues that remain to be investigated are suggested.
Keywords: aneuploidy, chromosome, crossover, crossover interference, crossover pattern, meiosis, recombination
INTRODUCTION: MEIOTIC RECOMBINATION IS INTEGRATED INTO THE DEVELOPMENT OF CHROMOSOME STRUCTURE
Meiosis, a specific type of cell division generating gametes with half DNA complements of progenitor cells, is essential for successful sexual reproduction. During meiosis, DNA is replicated only once but is followed by two successive rounds of chromosome segregation. Homologous chromosomes (homologs) are segregated during meiosis I and sister chromatids (sisters) are segregated during meiosis II.
Meiotic homologous recombination, a crucial feature of meiosis, is initiated from DNA double-strand breaks (DSBs) catalyzed by SPOrulation 11 (SPO11) transesterase.1,2,3,4,5 After SPO11-oligo complexes are removed, the DSB ends are further resected by exonuclease to yield long 3’ single-stranded tails (ssDNA). With the help of Recombinase A (Rec A) related strand-exchange proteins, Disrupted Meiotic cDNA 1 (DMC1) and RADiation sensitive 51 (RAD51), the ssDNA tails search for and invade intact homologous duplexes to form displacement loops (D-loops).6 Among a large number of DSBs, only a small subset (approximately 10% in mice) is repaired as crossovers (COs), with the reciprocal exchange of chromosome arms flanking the break site. However, the majority of DSBs are repaired as noncrossovers (NCOs) without the exchange of flanking arms (Figure 1a).2
Figure 1.
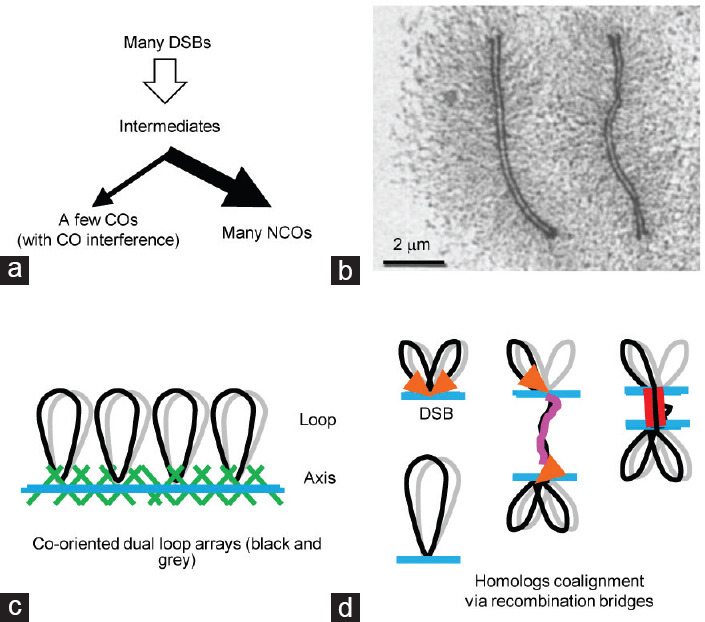
Meiotic recombination is integrated into the development of chromosome structure. (a) Meiotic recombination. Usually, many DSBs occur in a nucleus. But, only a few of them are repaired as COs (with interference) and the majority are repaired as NCOs. (b) An electron microscopy picture to show pachytene chromosomes of the moth Hyalophora columbia.149 The copyright license of reproducing this picture was received from the publisher Elsevier. (c) Cartoon for meiotic chromosome organization. (d) The tethered loop-axis complex brings DNA from loops to axes to generate DSBs and one DSB end is released to search for and bring its homolog into proximity. DSB: double-strand breaks; CO: crossover; NCO: noncrossover.
Homologous recombination occurs in meiotic chromosomes. The meiotic chromosome is proposed to be organized as a linear array of loops, and the base of these loops plus a large number of proteins compose the chromosome axis (Figure 1b and 1c).7 The density of loops along a chromosome axis is highly conserved among various organisms (approximately 20 loops per micron of axis).8 Therefore, loop size is negatively correlated with axis length.8,9
The process of homologous recombination is accompanied by elaborate meiotic chromosome dynamics and tightly integrated into meiotic chromosome structure.7,10,11,12,13,14,15 At the DNA level, DSB sites are mapped to sequences located in chromatin loops. However, cytologically, recombination complexes are observed on axes that are DSB cold spots.16,17,18,19,20 This paradox is resolved by the proposed tethered loop-axis complex (TLAC; Figure 1d).16,17,21 In this model, SPO11 complexes located on chromatin loops are recruited to axes and activated to generate DSBs. After resection, one end of a DSB is released to search for its homologous template (Figure 1d). In most organisms, most of the DSB-mediated interhomolog interactions seem to be responsible for homolog alignment. However, homolog synapsis mediated by the synaptonemal complex (SC) initiates only from some of these interactions.11,12,22 The SC also has important roles in homologous recombination, such as inhibiting excessive DSB formation and ensuring efficient recombination completion.23,24
CO PATTERNS AND THE UNDERLYING LOGIC
Besides exchanging genetic information, COs also have a specific role in ensuring faithful homolog segregation at anaphase I, through physically connecting homologs together in combination with sister chromatid cohesion (Figure 2a and 2b). COs are cytologically visualized as chiasmata after SC disassembly (Figure 2a and 2b). COs can also be observed as late/large (recombination) nodules at pachytene under electron microscopy,25 marked by specific recombination protein foci including MutL homolog 1/3 (MLH1/3), ZIPper 3 (Zip3), or Homo sapiens Enhancer of Invasion 10 (HEI10) in diverse organisms,26,27,28 and defined by genetic or DNA polymorphism analysis from progeny.29,30
Figure 2.
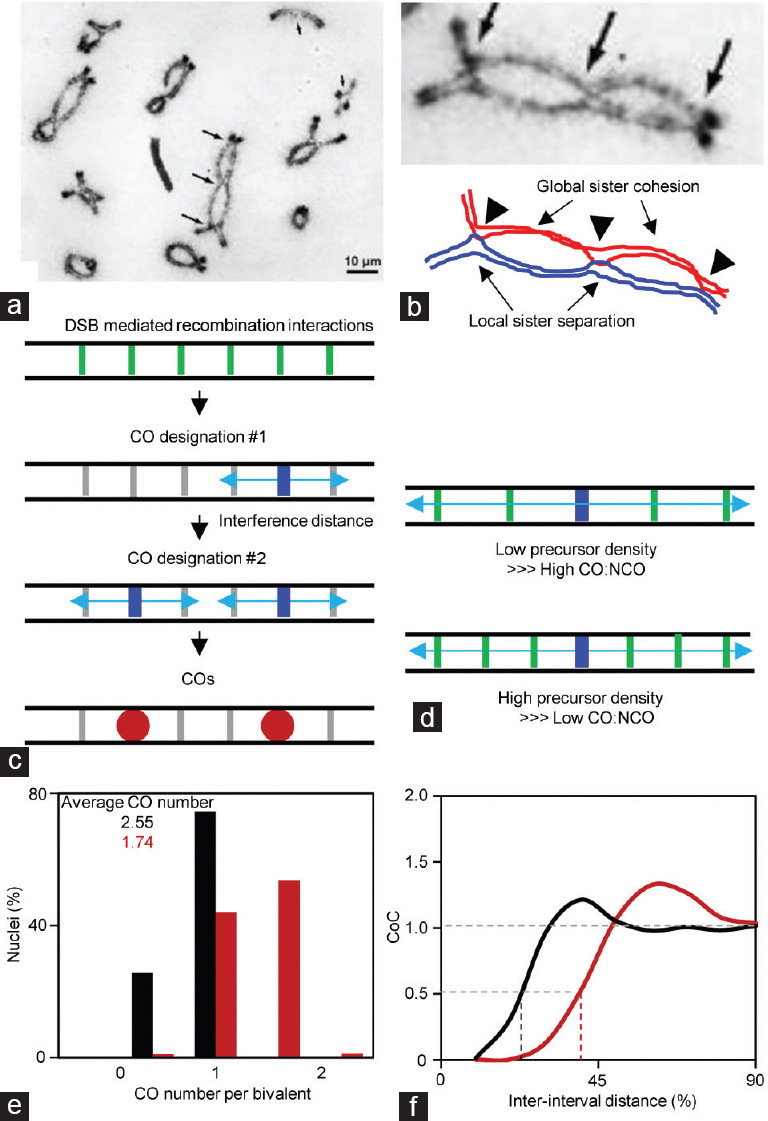
Crossover (chiasmata) and the logic of crossover patterning. (a) A spread nucleus of the locust Schistocerca gregaria to show chiasmata.32 The copyright license of reproducing this picture was received from the publisher John Wiley and Sons. Multiple chiasmata (crossovers) are evenly spaced (long arrows). (b) Homologs are linked together by chiasmata combined with global sister cohesion. Sisters are locally separated around chiasmata. (c) The logic of crossover patterning. Among a large number of crossover precursors (DSB mediated inter-homolog interactions), the most sensitive one is first designated and crossover interference spreads out to inhibit a second crossover nearby. The next crossover if occurs would occur far away from the existed ones to fill in the holes. (d) When more (less) precursors exist, there would be more (less) precursors suppressed by crossover interference, however, crossover number is maintained less altered. (e) Crossover number distribution from two artificial datasets generated by simulations. Interference distance L = 0.55 and 0.3 for chromosomes with less and more crossovers, respectively; other parameters are the same, CO designation driving force, Smax= 3.5; clamped left and right ends, cL = cR = 1; the average precursor number, N= 14; precursor distribution among chromosomes, B = 0.6; the evenness level of precursors along chromosomes, E = 0.6. (f) CoC curves from the two simulated data set in (e). DSB: double-strand breaks; CO: crossover; NCO: noncrossover; CoC: coefficient of coincidence.
CO patterns (the number and distribution of chiasmata/COs) are tightly controlled (Figure 2a and 2b).15,31,32,33,34 Aberrant CO patterns usually result in chromosome segregation errors and thus aneuploidy, which is the leading cause of infertility, abortion, and congenital birth defects in humans.35,36 The tight control of CO patterns is exhibited as three major features: obligatory CO, CO interference, and CO homeostasis.31,32,33
In most organisms, each nucleus usually has only a small number of COs. If they are randomly distributed among chromosomes, a large number of chromosomes would not get any CO. For example, each mouse spermatocyte has approximately 23 COs on 19 autosomes.37 If COs are randomly distributed among chromosomes with CO numbers proportional to axis lengths, at least one chromosome in each nucleus will fail to get even one CO (from 10 000 simulations). As expected, the longest chromosome, chromosome 2, has the lowest probability of absence of COs (approximately 16%), and the shortest chromosome, chromosome 19, has the highest probability of absence of COs (approximately 50%). However, given its essential role for accurate chromosome segregation, each pair of homologs acquires at least one CO, which is traditionally referred to as the obligatory CO.31,32 As a result, the frequency of CO absence from chromosomes is maintained at a very low level (usually <2%, except for human females at approximately 10%)9,31,35
Not only the numbers but also the positions of COs on chromosomes are not random. When two or more COs are present on a pair of homologs, these COs tend to be farther away from each other rather than, appearing to be randomly placed. This phenomenon, known as CO interference, was noted more than 100 years ago, and reflects the existence of a CO generating an interference signal to inhibit the occurrence of other COs nearby.38,39 Therefore, CO interference restricts the maximal number of COs on each pair of homologs, usually only 1–3 in most organisms31,33,40,41
Despite COs originating from DSBs, alterations in DSB numbers change CO numbers less proportionally at the cost of NCOs.42,43,44,45,46,47 This phenomenon is called CO homeostasis, which maintains the number of COs at a relatively constant level and is less affected by fluctuations of DSBs.33,42
Studies have shown that CO interference spreads along chromosomes with micron of axis as a metric.43,48,49 Consistently, various studies suggest that intact chromosome axes are required for proper CO interference.43,50,51,52,53 However, the mechanism of CO interference is unknown.
Among several models proposed to explain CO interference, the “fill-in-the-holes” model (also known as beam-film or stress model) has been widely accepted to explain the basic logic of CO patterning (Figure 2c).33,54,55,56,57,58 In this model, CO precursors, an array of DSB-mediated interhomolog interactions, are distributed along chromosomes. Among them, the most sensitive precursor is first designated to become a CO (i.e., CO designation), and simultaneously, the interference signal from this designation site propagates along the chromosome axis in both directions to inhibit further CO designations in nearby regions. The distance over which the interference signal spreads is the “interference distance,” measured as physical axis length (micron).33,43 The strength of interference dissipates with increasing distance. If a subsequent CO designation occurs, it will tend to occur far away from the existing designation sites to “fill in the holes.” Finally, multiple CO designation sites and thus COs tend to be evenly spaced along a chromosome.
The other two features of CO patterns, the obligatory CO and CO homeostasis, can be easily integrated into this logic and understood as described below. At least one CO is required for faithful chromosome segregation, and this can be ensured by a highly efficient CO designation process, which is probably regulated evolutionarily. The existence of the CO interference signal creates an inhibition zone, where other CO precursors are inhibited regardless of the increased or decreased number of precursors in that zone (Figure 2d). Therefore, the numbers of CO designations and corresponding COs are maintained at a relatively stable level, and are less affected by altered precursor numbers. Moreover, stronger CO interference results in fewer COs and stronger CO homeostasis (Figure 2d and 2e).
It is worth noting that studies based on different criteria suggest that there are two types of COs, interference-dependent and -independent COs. In most organisms, a majority of COs are sensitive and subject to CO interference, and require ZMM (including at least Zip1-3, Meiotic recombination 3 (Mer3), and MutS homolog 4/5 (MSH4/5) in budding yeast) group proteins. In both mouse and human, this type of CO accounts for 90%–95% of total COs and can be marked by MLH1 foci at pachytene. For the other 5%–10%, a minority of COs are insensitive to CO interference and require MMS and UV Sensitive 81 (MUS81) and MutS homolog 4 (MMS4)/Essential Meiotic structure-specific Endonuclease 1 (EME1), which is thought to arise in a different manner from the majority of COs.2,33,34 However, Mus81-/- mutant mouse shows an upregulated number of MLH1 foci and the normal number of chiasmata.59 Using combined fluorescence and electron microscopy, a study in tomato revealed interference between the two types of COs.25 Therefore, these studies suggest an interaction between the two types of COs. Further investigation is necessary to elucidate the mechanistic relationship.
MEASUREMENTS OF PHENOMENOLOGICAL AND MECHANISTIC CO INTERFERENCE
CO interference is originally described as the occurrence of one CO that interferes with the occurrence of another CO nearby on the same pair of homologs.38,39 CO interference is traditionally measured using the coefficient of coincidence (CoC) method. For any two intervals, the frequency of CO occurrence in each interval can be calculated, and the expected frequency of CO coincidence in both intervals can be obtained by multiplying the frequencies of CO occurrence in the two intervals, while the observed frequency of CO coincidence in both intervals can be calculated from experimental data. CoC is defined as the ratio of the frequency of observed double COs to the frequency of expected double COs (observed/expected). A more rigorous way to measure CO interference is a CoC curve analysis when multiple CO intervals are available. For this purpose, CoC values are calculated from all pairs of intervals and plotted as a function of inter-interval distance (Figure 2f). Generally, the CoC value is very small at the short inter-interval distance, reflecting strong interference, and CoC increases to approximately 1 with increasing inter-interval distance, reflecting no interference. At a particular inter-interval distance (i.e., the average distance between adjacent COs), a hump with CoC value much larger than 1 is often seen, especially for genetically short chromosomes, reflecting evenly spaced COs (Figure 2f).33,60 Therefore, the CoC (curve) method integrates both the distance and the “evenness” information.
Because COs tend to be evenly distributed along chromosomes due to CO interference, a gamma distribution, which fits the frequency distribution of inter-adjacent CO distances, has often been applied to measure the strength of CO interference.61,62 A higher value shape parameter (γ) indicates more evenly spaced COs, and thus stronger CO interference.61,62 However, the gamma shape parameter only reflects the “evenness” regardless of the absolute distance.
The term “interference” only describes the phenomenon. Both CoC (curve) analysis and gamma distribution methods measure the “phenomenon” but do not reveal the essence or the mechanism of CO interference, for example, the interference signal. Alterations in CoC or gamma shape parameter can result from an altered patterning process other than mechanistic interference, especially factors acting before interference imposition, such as the number of precursors.33,57 Additionally, gamma distribution but not CoC can also be affected by alterations after interference spreading, for example, CO maturation inefficiency.9,13,33,62,63
To distinguish the effects of different factors on observed phenomenological interference, a mathematical simulation approach based on the “fill-in-the-holes” model was developed. According to the CO patterning logic described above, the following four sets of parameters are required for this simulation: (1) the array of CO precursors; (2) the strength of CO designation and the response of precursors to CO designation; (3) the strength of CO interference, that is, the distance over which interference spreads; and (4) CO maturation efficiency, that is, the probability of a designated CO becoming a real CO. This mathematic simulation approach can quantitatively mimic the CO patterning process and is very useful to differentiate how different factors alter CO interference.13,33,43,56,58 This simulation method has very accurately captured observed CO patterns in several investigated organisms, including budding yeast, Sordaria, Arabidopsis, maize, human, and mouse.9,12,27,33,43,60,64 Moreover, it helps in identifying the first CO interference regulatory pathway, discovering human female-specific CO maturation inefficiency, clarifying the per-nucleus CO co-variation resulting from the co-variation of chromosome axis lengths, and explaining CO pattern differences between the two sexes and between long and short chromosomes.43,60,63
CO FREQUENCY IS MAINLY REGULATED BY CHROMOSOME AXIS LENGTH
Although the positions of COs are stochastic on a given chromosome, COs preferentially occur in some chromosome regions (CO hotspots) and are rare in other regions, such as the pericentromeric and rDNA regions.29,65,66 The existence of DSBs is the prerequisite for the occurrence of COs; however, the probability of a DSB becoming a CO varies significantly from locus to locus.67,68,69,70,71,72,73,74 This may be regulated at the stages of partner choice (interhomolog vs intersister), CO/NCO differentiation, or CO maturation. Differences in partner choice of DSB repair have been observed in Schizosaccharomyces pombe;75 both DSB formation near centromeres and its repair by homologs are inhibited in a distance-dependent manner;67,76 there are different CO/NCO ratios at different loci in mouse meiosis and a reduced CO/NCO ratio near telomeres in budding yeast.67,70,71 Therefore, local chromosome structures influence the occurrence of COs, and CO hot spots do not always overlap with DSB hot spots.77,78,79,80
Regardless of the local regulation of CO formation, it has long been known that there is a strong positive correlation between chromosome axis length and CO number under diverse conditions:
In one nucleus, usually a chromosome with more DNA content tends to have a longer axis and more COs. However, when two chromosomes have very similar DNA content, one chromosome axis can be longer than the other one. In this case, the chromosome with the longer axis also has more COs. Therefore, it is the axis length but not DNA content correlates with CO number. Consistent with this, the number of COs correlates with axis length better than with DNA content. For example, in mouse spermatocyte, r = 0.96 between CO number and axis length; however, r = 0.86 between CO number and DNA content (Figure 3a and 3b)
Among different nuclei, both chromosome axis lengths and the numbers of COs can vary significantly, and nuclei with longer chromosome axis tend to have more COs.40,81,82,83,84 Moreover, our recent studies have found that the numbers of COs co-vary among chromosomes at a per-nucleus basis, which results from co-variation of chromosome axis lengths (Figure 3c and 3d)13,85
In many organisms, CO frequencies are different between males and females, and the sex with longer chromosome axis also has more COs.9 For example, human females have 2-fold longer chromosome axes compared to human males, and have approximately 1.6-fold more COs as revealed by various measurements.9,29,30,86,87,88 Similarly, in mouse and zebrafish meiosis, females have approximately 20% longer chromosome axes and also approximately 20% more COs than males.49,89 However, in some other organisms including Arabidopsis and maize, males have longer meiotic chromosome axes and thus more COs48,60,64,91,92,93
For the same species, different genetic backgrounds may show different CO frequencies. Among CAST/EiJ, C3H/HeJ, and C57BL/6J mice, C57BL/6J spermatocytes have the longest chromosome axes and also the highest number of COs. However, CAST/EiJ spermatocytes have the shortest chromosome axes and also the lowest number of COs94
Several mutants are found to have altered chromosome axis lengths in diverse organisms, and these mutants also have correspondingly altered CO numbers. For example, studies from diverse organisms show that mutants of cohesin and related factors have decreased meiotic chromosome axis lengths and reduced COs.53,95,96,97,98,99,100,101,102 However, mutants, such as Hr6b-/- mouse spermatocytes, have longer axes and more COs.103
Figure 3.
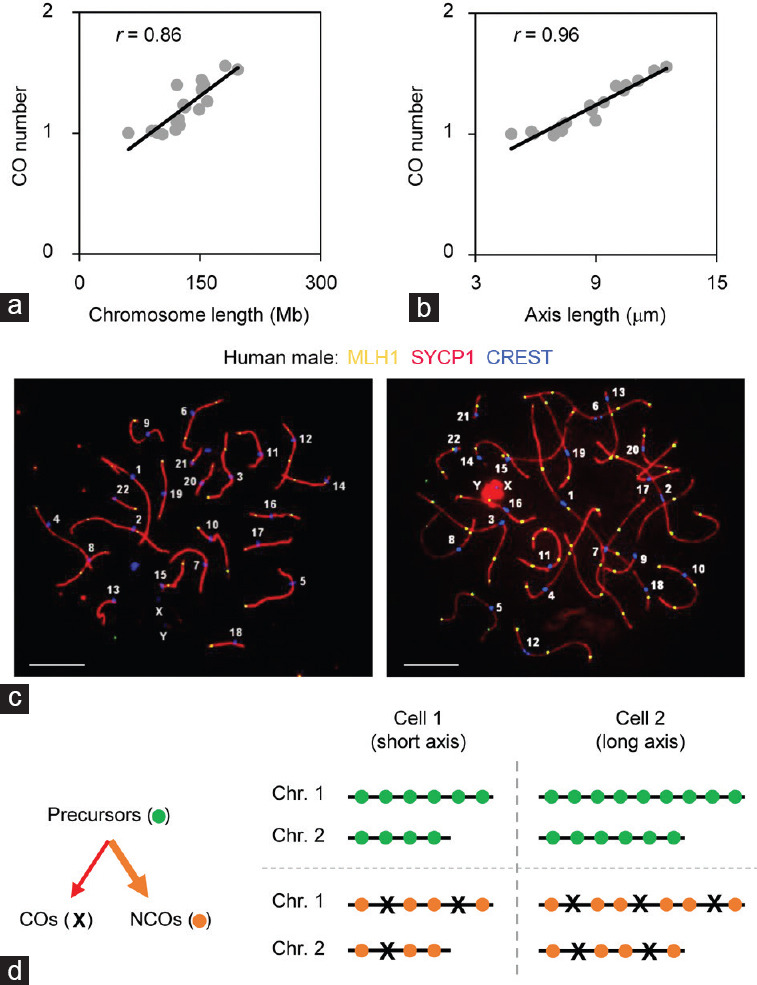
The co-variation of chromosome axis lengths results in the co-variation of crossover numbers. (a) Correlation between crossover number and chromosome DNA content in male mice. (b) Correlation between crossover number and chromosome axis length in male mice (data provided by Lorida Anderson). (c) Representative human male pachytene nuclei to show co-variation of axis lengths and covariation of crossover numbers among chromosomes within individual nuclei. Left nucleus: 37 COs; 170 micron (axis length). Right nucleus: 61 COs; 230 micron (axis length). Scales bars=5 μm. (d) Cartoon to show co-variation of axis lengths leads to co-variation of crossover numbers. CO: crossover; NCO: noncrossover; Chr: chromosome; MLH1: MutL homolog 1; SYCP1: Synaptonemal complex protein 1; CREST: Calcium-responsive transactivator.
The above evidence suggests that CO number and meiotic chromosome axis length are tightly linked. Moreover, alterations in chromosome axis length and corresponding alterations in DSBs or DSB-mediated interhomolog interactions are also observed, between males and females in both human and mouse, and among mice in different genetic backgrounds.86,87,94 These results suggest that it is axis length that determines the number of COs but not the other way around, which is also supported by the following evidence. (1) Absence of or alterations in DSBs and/or DSB-mediated interhomolog interactions do not have an obvious effect on chromosome axis length in various organisms including mouse.12,33,43,44,104,105 In some organisms, chromosome axes are formed before DSB formation and thus before the recombination process.23 (2) CO formation has little effect on overall chromosome axis length alteration. For example, studies on Caenorhabditis elegans and Sordaria show that the occurrence of a CO (designation) only locally alters chromosome axis length/structure; however, this change is subtle in terms of the overall axis length.52,104
UNIQUE CO PATTERNS ON SHORT CHROMOSOMES
During meiosis, short chromosomes behave differently from long chromosomes in several aspects. In budding yeast, short chromosomes complete homolog pairing late.106 However, in other organisms, short chromosomes seem to complete homolog pairing earlier than long chromosomes.107 Structurally, short chromosomes are organized with a longer axis and smaller loops than the genomic average as observed in budding yeast.33,108 In both budding yeast and mouse, short chromosomes tend to have higher DSB density, which is attributed to multiple effects including early DNA replication, centromere and telomere effects, and an “intrinsic boost.”21,80,109,110
A special case is mammal XY chromosomes with a pretty short homologous region (pseudoautosomal region [PAR]), where DSB formation mediated by Spo11α and homolog pairing are late relative to autosomes. Although PAR is very short (<1 Mb in mouse), it is organized with an extremely long axis and small loops (approximately 1 μm of axis per Mb compared with approximately 0.1 μm of axis per Mb for autosomes), and obtains 10- to 20-fold higher DSB density than the genomic average.111,112,113,114
Studies on meiotic recombination have revealed that short chromosomes have different CO configurations in various organisms including human and mouse (Figure 4).9,37,83,115 (1) A high frequency of short chromosomes does not have the obligatory CO. (2) Short chromosomes tend to have higher CO density (CO number per micron of chromosome axis), which is usually interpreted as the obligatory CO effect. The higher DSB density on short chromosomes may also make a small, but not a large, contribution given the existence of CO homeostasis.33,79 (3) COs tend to be located more distally and the average inter-adjacent-CO distance takes up a larger proportion of the chromosome axis. However, the absolute distance between adjacent COs is shorter.9,37,83,115 (4) The distribution of inter-CO distances tends to be more even as indicated by a bigger gamma shape parameter. Sometimes, this is interpreted as short chromosomes having higher CO interference; however, this is not true.115,116 At least some of these features (e.g., chromosomes without COs) contribute to the observed high frequency of chromosome mis-segregation.9,12,33 However, it is unclear whether these special CO configurations are just because short chromosomes have short axes or because they have other special features.
Figure 4.
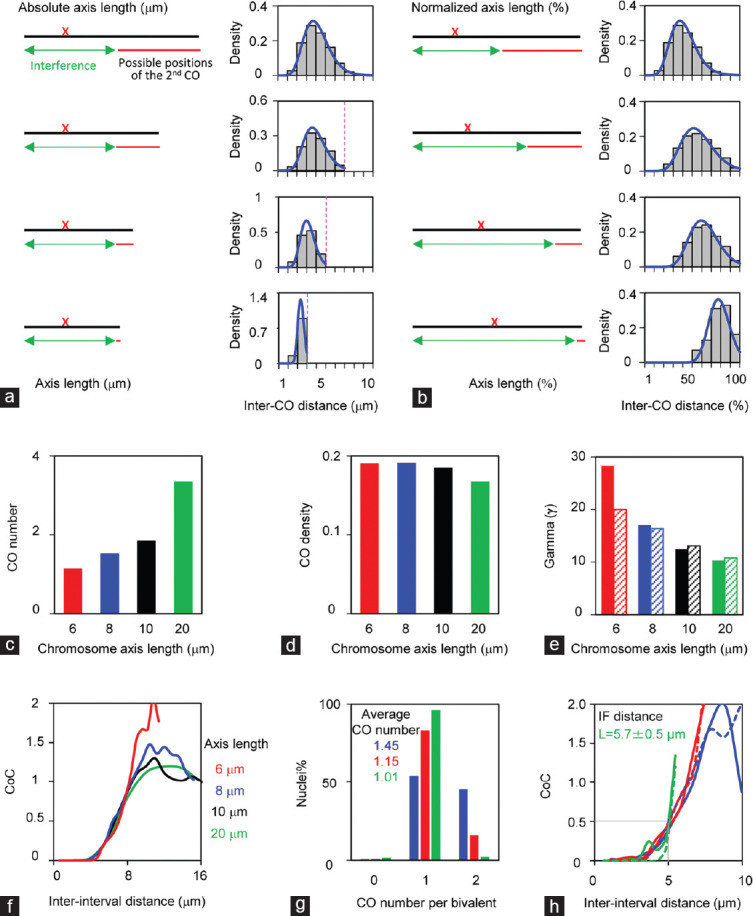
Chromosome axis length regulates crossover patterns. (a–f) Simulated data from four artificial chromosomes with different axis lengths (20 μm, 10 μm, 8 μm, and 6 μm) to illustrate the effects of chromosome lengths on crossover patterns. The same set of parameters used for all chromosomes: CO designation driving force, Smax= 4.1; clamped left and right ends, cL = cR = 1; the evenness level of precursors along chromosomes, E = 0.6; precursor distribution among chromosomes, B = 0.6; interference distance, L = 5 μm; precursor number N is adjusted according to axis length (N= 20, 10, 8, and 6, separately). (a and b) Illustration of how short chromosomes alter crossover number and distribution. Chromosome axis lengths are (a) shown as microns or (b) normalized to “1.” (c–e) Short chromosomes have (c) fewer crossovers, (d) higher crossover density, and (e) higher gamma values estimated from gamma distribution (solid bars) or modified gamma distribution (hatched bars). (f) CoC curves show that all chromosomes have the same CO interference. (g) Crossover numbers and (h) CoC curves of male mice (data provided by Lorida Anderson). Mouse autosomes are divided into three groups according to axis lengths (long chromosomes: chromosomes 1, 2, 4, 5, 7, and 11; medium chromosomes: chromosomes 3, 6, 8, 9, 10, 12, 13, and 14; short chromosomes: chromosomes 15–19). CoC analyses show that all chromosomes have the same crossover interference (curves with solid lines) as confirmed by simulations (curves with dot lines). Smax= 3.5, L = 6 μm, cL = 0.6, cR = 1, E = 0.6, B = 0.6, N is adjusted according to axis length (14, 10, and 8 for long, medium, and short chromosomes, respectively). CO: crossover; CoC: coefficient of coincidence.
Analysis of CO interference in diverse organisms based on gamma distribution and related methods raises the question of whether short chromosomes have stronger CO interference.63,115,116,117,118,119,120 Gamma distribution is used for the analysis of distances between events along an infinite axis. However, a chromosome axis is finite and only chromosomes with two or more COs are included in this analysis, which introduces bias and results in inappropriately fitted gamma distributions, especially for short chromosomes.62,63 This is illustrated by a set of “artificial” data generated by the beam-film application using the same set of parameters except for different chromosome axis lengths (Figure 4a–f). To resolve the above problem, a modified gamma distribution analysis has also been applied. In this analysis, all chromosomes, including chromosomes with only one or zero CO, and also the distances from chromosome ends to the nearest CO, are included. However, this modified gamma distribution analysis only partially improves the fitting (Figure 4e), solid bars from gamma distribution vs dashed bars from modified gamma distribution). The CoC analyses show that all these chromosomes have the same CO interference, and CoC curves of shorter chromosomes increase rapidly (Figure 4f). The CoC analysis of male mouse MLH1 data also shows that all chromosomes have the same CO interference, which is further confirmed by beam-film simulations (Figure 4g and 4h, dashed vs solid lines). Therefore, it seems like short chromosomes have the same CO interference as long chromosomes. Similarly, CoC analyses in human and mouse also show that both males and females have the same/similar CO interference, although females have longer axis and thus more COs than males (Figure 4h).9,49,119,121 The same CO interference in both sexes is also confirmed in Arabidopsis, in which male meiosis has longer axes and more COs.48,60
CO PATTERNS AND THE HIGH FREQUENCY OF HUMAN ANEUPLOIDIES
The frequency of human embryo aneuploidy increases with increasing maternal age, and for women close to their end of the reproductive lifespan, the frequency of embryo aneuploidy can even reach 50% or more, which is known as the “maternal age effect.”35,122,123,124,125 Compared with other organisms, even in young women, the frequency of embryo aneuploidy is still very high (approximately 10%).35,123,124 A recent study reveals that human female-specific CO maturation inefficiency underlies the high aneuploidy frequency.9 In addition, several other age-related CO alterations in humans are noted.
Maternal age effect
Maternal age effect is proposed to be mainly caused by age-dependent loss of sister chromatid cohesion.35,126,127,128,129,130,131 Sister cohesion is mainly mediated by the cohesin complex, which is loaded before or during DNA replication and not replenished.132,133 Therefore, gradual deterioration of cohesin over time results in loss of sister cohesion. First, decreased peri-centromeric cohesion impairs the function of centromeres and directly results in chromosome segregation errors. Second, decreased arm cohesion between two adjacent COs and/or between the distal CO and chromosome end weakens the function of the chiasma in connecting homologs.85
Besides sister cohesion, many other factors including the states of kinetochore, spindle, checkpoint, environment, and recurrence of DNA damage also have important contributions to chromosome mis-segregation and thus maternal age effect.134,135,136,137,138,139,140
CO maturation inefficiency in human oocytes
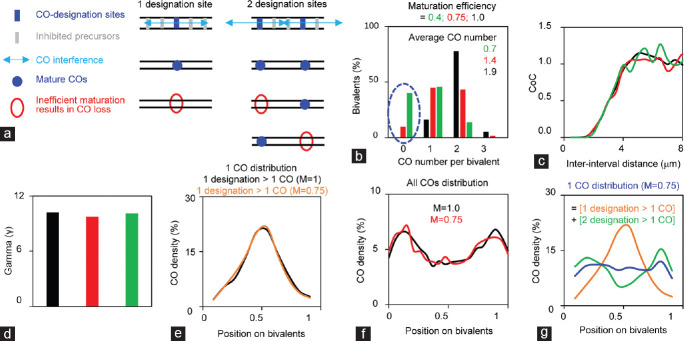
The maternal age effect is an important factor for human embryo aneuploidy. However, even in young women, the frequency of aneuploidy is still approximately 10%.35,122 Most aneuploidy (approximately 90%) results from oocyte meiosis errors, especially homologous chromosome mis-segregation during meiosis I.35 A recent study comparing meiotic recombination between human males and females, reveals that human females have CO maturation inefficiency, which leads to a fraction of CO designations failing to become mature COs (Figure 5).9,13
Figure 5.
Crossover maturation inefficiency generates aberrant crossover configurations, especially on short chromosomes. (a) Cartoon to show the effects of crossover maturation inefficiency. (b) Crossover maturation inefficiency decreases crossover number. (c and d) Crossover maturation inefficiency does not affect crossover interference as revealed by (c) CoC and (d) gamma distribution analysis. Maturation efficiency, M = 0.4 (green), 0.75 (red), or 1 (black). Crossover maturation inefficiency does not alter (e) relative crossover density distribution for chromosome with only one crossover designation, or (f) overall crossover density distribution. (g) Crossover density distribution of chromosomes with one crossover altered by crossover maturation inefficiency. Data used from simulations: CO designation driving force, Smax= 3.8; clamped left and right ends, cL = cR = 1.1; the evenness level of precursors along chromosomes, E = 0.6; precursor distribution among chromosomes, B = 0.6; precursor number, N = 7; interference distance, L = 0.43; M = 0.4, 0.75 or 1 as indicated.
CO maturation inefficiency significantly alters CO patterns to give rise to a high frequency of chromosomes with error-prone CO configurations (Figure 5).13,85 First, CO maturation inefficiency decreases the number of COs on all chromosomes, probably in proportion to the chromosome axis length (Figure 5a and 5b). Second, CO maturation inefficiency has more severe effects on short chromosomes than on long chromosomes, by generating high levels of chromosomes with error-prone CO configurations, for example, without the obligatory CO, or with distal-only or proximal-only COs (Figure 5). Aberrant COs cause improper tensions at metaphase I, which leads to homolog segregation errors and thus aneuploidy. Third, different from human males, the completion of meiotic recombination and CO maturation inefficiency occur in the human female fetal stage and before the maternal age effect occurs. Therefore, CO maturation inefficiency is a major basis for human female aneuploidy. With increasing age, the maternal age effect interacts with aberrant COs caused by CO maturation inefficiency, to significantly elevate the frequency of chromosome mis-segregation and thus aneuploidy.
Other age-related alterations in CO and aneuploidy frequency
Several other age-related alterations in COs and human aneuploidy have also been recognized but less studied.
Younger parents tend to produce a higher frequency of aneuploid embryos.35,123,124,141,142 However, the reason is not known. Cole and colleagues have found that compared to adult males, juvenile males tend to have longer meiotic chromosome axes, less proportionally elevated CO number, and thus decreased CO density (CO number per micron of axis).142 This similarity between juvenile males and adult females (compared with adult males) raises the possibility that CO maturation inefficiency may work in juvenile males. Studies in mouse show a similar recombination process in juvenile and adult spermatocytes, however, in juveniles, a fraction of recombination intermediates is processed to be NCOs by structure-selective nucleases and alternative complexes.142 As a result, decreased CO frequency and CO density is observed in juvenile spermatocytes.142,143 Intriguingly, younger women also tend to have a high frequency of aneuploid pregnancies than adults.35,122 Further investigation is required to know why younger mothers have increased incidence of aneuploid pregnancies and whether the mechanism found in juvenile mouse spermatocytes also works in younger men and women. Detailed analysis of alterations in CO patterns will help to illustrate whether they have CO maturation inefficiency
Compared with the dramatic effect of maternal age, a mild paternal age effect is also observed in some studies.144,145 Although analysis of parent-child pairs does not detect increased CO frequency in old fathers,68,146 studies in old mice detect a small increase in CO frequency and SC/axis length.143 Increased meiotic errors with increasing paternal age are also observed in old mice, but these aberrant nuclei are probably eliminated and thus do not give rise to sperm.143 It is unknown whether this also exists in men, and examination of CO frequency in spermatocytes of old men will provide valuable information
Several studies have shown that older women tend to have children with higher CO frequency than young women.29,68,146,147,148 This is sometimes explained as old mothers tend to have more interference-insensitive COs based on two-pathway gamma model analysis.148 However, CO recombination is completed at the meiotic pachytene stage during the human female fetal stage and it is less likely that additional COs occur during the long arrest period (dictyate). Moreover, CO frequency is maintained constantly in oocytes at different ages of mothers.94 A more reasonable explanation is that oocytes with more COs have a higher probability to keep at least one chiasma (the loss of sister cohesion results in chiasma loss), and thus a higher probability to have euploid eggs and children.85 These nuclei have more COs likely because they have either relatively longer axes or more sensitive precursors.13,33,85
CONCLUSION
Crossover recombination is essential for meiosis, which not only ensures faithful chromosome segregation but also promotes the genetic diversity of progenies for evolutionary adaption. The process of crossover recombination is tightly integrated into the meiotic chromosome structure, which is essential in regulating crossover patterns. Chromosomes with aberrant crossover configuration are subject to mis-segregation, which is the primary cause of aneuploidy and thus infertility, abortion, stillbirth, and congenital birth defects in humans. Therefore, crossover recombination remains at the forefront of biological and reproductive medicine research and attracts the interest of different research fields. Although significant advances have been made in recent years, many outstanding questions need to be further investigated. For example, identifying the CO interference signal and how it is regulated; how and when CO/NCO differentiation is determined, and what factors are involved; how meiotic chromosome loop/axis forms; and how CO maturation inefficiency is regulated and identifying its evolutionary advantages.
AUTHOR CONTRIBUTIONS
SXW and LRZ drew figures and wrote the draft. All authors discussed and edited the manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declared no competing interests.
ACKNOWLEDGMENTS
Many thanks to Lorida Anderson (Colorado State University) who kindly provided male mouse MLH1 data, Fei Sun (Nanchang University) who kindly provided the pictures in Figure 3c. This work is supported by grants from the National Key R&D Program of China (2018YFC1003700, 2018YFC1003400) and National Natural Science Foundation of China (31671293, 31801203, and 31890782).
REFERENCES
- 1.Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu Rev Genet. 2014;48:187–214. doi: 10.1146/annurev-genet-120213-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol. 2015;7:a016618. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–99. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 4.Lichten M, de Massy B. The impressionistic landscape of meiotic recombination. Cell. 2011;147:267–70. doi: 10.1016/j.cell.2011.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paigen K, Petkov PM. PRDM9 and its role in genetic recombination. Trends Genet. 2018;34:291–300. doi: 10.1016/j.tig.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown MS, Bishop DK. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb Perspect Biol. 2014;7:a016659. doi: 10.1101/cshperspect.a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 8.Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–94. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Hassold T, Hunt P, White MA, Zickler D, et al. Inefficient crossover maturation underlies elevated aneuploidy in human female meiosis. Cell. 2017;168:977–89. doi: 10.1016/j.cell.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borde V, de Massy B. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr Opin Genet Dev. 2013;23:147–55. doi: 10.1016/j.gde.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Zickler D, Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol. 2015;7:a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Espagne E, de Muyt A, Zickler D, Kleckner NE. Interference-mediated synaptonemal complex formation with embedded crossover designation. Proc Natl Acad Sci U S A. 2014;111:E5059–68. doi: 10.1073/pnas.1416411111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, Veller C, Sun F, Ruiz-Herrera A, Shang Y, et al. Per-nucleus crossover covariation and implications for evolution. Cell. 2019;177:326–38. doi: 10.1016/j.cell.2019.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolcun-Filas E, Handel MA. Meiosis: the chromosomal foundation of reproduction. Biol Reprod. 2018;99:112–26. doi: 10.1093/biolre/ioy021. [DOI] [PubMed] [Google Scholar]
- 15.Smith GR, Nambiar M. New solutions to old problems: molecular mechanisms of meiotic crossover control. Trends Genet. 2020;36:337–46. doi: 10.1016/j.tig.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 17.Ito M, Kugou K, Fawcett JA, Mura S, Ikeda S, et al. Meiotic recombination cold spots in chromosomal cohesion sites. Genes Cells. 2014;19:359–73. doi: 10.1111/gtc.12138. [DOI] [PubMed] [Google Scholar]
- 18.Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, et al. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell. 2008;14:263–74. doi: 10.1016/j.devcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Kugou K, Fukuda T, Yamada S, Ito M, Sasanuma H, et al. Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol Biol Cell. 2009;20:3064–76. doi: 10.1091/mbc.E08-12-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, et al. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol. 2011;13:599–610. doi: 10.1038/ncb2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–83. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Gruhn JR, Al-Asmar N, Fasnacht R, Maylor-Hagen H, Peinado V, et al. Correlations between synaptic initiation and meiotic recombination: a study of humans and mice. Am J Hum Genet. 2016;98:102–15. doi: 10.1016/j.ajhg.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Colaiácovo MP. Zipping and unzipping: protein modifications regulating synaptonemal complex dynamics. Trends Genet. 2018;34:232–45. doi: 10.1016/j.tig.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu X, Keeney S. Zip it up to shut it down. Cell Cycle. 2014;13:2157–8. doi: 10.4161/cc.29747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson LK, Lohmiller LD, Tang X, Hammond DB, Javernick L, et al. Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers. Proc Natl Acad Sci U S A. 2014;111:13415–20. doi: 10.1073/pnas.1406846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcon E, Moens P. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics. 2003;165:2283–7. doi: 10.1093/genetics/165.4.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Muyt A, Zhang L, Piolot T, Kleckner N, Espagne E, et al. E3 ligase Hei10: a multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes Dev. 2014;28:1111–23. doi: 10.1101/gad.240408.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–55. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- 29.Kong A, Thorleifsson G, Frigge ML, Masson G, Gudbjartsson DF, et al. Common and low-frequency variants associated with genome-wide recombination rate. Nat Genet. 2014;46:11–6. doi: 10.1038/ng.2833. [DOI] [PubMed] [Google Scholar]
- 30.Lu S, Zong C, Fan W, Yang M, Li J, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–30. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones GH. The control of chiasma distribution. Symp Soc Exp Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- 32.Jones GH, Franklin FC. Meiotic crossing-over: obligation and interference. Cell. 2006;126:246–8. doi: 10.1016/j.cell.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Liang Z, Hutchinson J, Kleckner N. Crossover patterning by the beam-film model: analysis and implications. PLoS Genet. 2014;10:e1004042. doi: 10.1371/journal.pgen.1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray S, Cohen PE. Control of meiotic crossovers: from double-strand break formation to designation. Annu Rev Genet. 2016;50:175–210. doi: 10.1146/annurev-genet-120215-035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet. 2012;13:493–504. doi: 10.1038/nrg3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ottolini CS, Newnham L, Capalbo A, Natesan SA, Joshi HA, et al. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet. 2015;47:727–35. doi: 10.1038/ng.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Froenicke L, Anderson LK, Wienberg J, Ashley T. Male mouse recombination maps for each autosome identified by chromosome painting. Am J Hum Genet. 2002;71:1353–68. doi: 10.1086/344714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sturtevant AH. The behavior of the chromosomes as studied through linkage. Z Indukt Abstamm-u VererbLehre. 1915;13:234–87. [Google Scholar]
- 39.Muller HJ. The mechanism of crossing over. Am Nat. 1916;50:193–434. [Google Scholar]
- 40.Ruiz-Herrera A, Vozdova M, Fernández J, Sebestova H, Capilla L, et al. Recombination correlates with synaptonemal complex length and chromatin loop size in bovids-insights into mammalian meiotic chromosomal organization. Chromosoma. 2017;126:615–31. doi: 10.1007/s00412-016-0624-3. [DOI] [PubMed] [Google Scholar]
- 41.Stapley J, Feulner PG, Johnston SE, Santure AW, Smadja CM. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160455. doi: 10.1098/rstb.2016.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–95. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Wang S, Yin S, Hong S, Kim KP, et al. Topoisomerase II mediates meiotic crossover interference. Nature. 2014;511:551–6. doi: 10.1038/nature13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole F, Kauppi L, Lange J, Roig I, Wang R, et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol. 2012;14:424–30. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosu S, Libuda DE, Villeneuve AM. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science. 2011;334:1286–9. doi: 10.1126/science.1212424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, et al. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012;149:75–87. doi: 10.1016/j.cell.2012.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Globus ST, Keeney S. The joy of six: how to control your crossovers. Cell. 2012;149:11–2. doi: 10.1016/j.cell.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 48.Drouaud J, Mercier R, Chelysheva L, Berard A, Falque M, et al. Sex specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet. 2007;3:e106. doi: 10.1371/journal.pgen.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petkov PM, Broman KW, Szatkiewicz JP, Paigen K. Crossover interference underlies sex differences in recombination rates. Trends Genet. 2007;23:539–42. doi: 10.1016/j.tig.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 50.Hillers KJ, Villeneuve AM. Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol. 2003;13:1641–7. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 51.Nabeshima K, Villeneuve AM, Hillers KJ. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics. 2004;168:1275–92. doi: 10.1534/genetics.104.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Libuda DE, Uzawa S, Meyer BJ, Villeneuve AM. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature. 2013;502:703–6. doi: 10.1038/nature12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Boer E, Dietrich AJ, Höög C, Stam P, Heyting C. Meiotic interference among MLH1 foci requires neither an intact axial element structure nor full synapsis. J Cell Sci. 2007;120:731–6. doi: 10.1242/jcs.003186. [DOI] [PubMed] [Google Scholar]
- 54.Fowler KR, Hyppa RW, Cromie GA, Smith GR. Physical basis for long-distance communication along meiotic chromosomes. Proc Natl Acad Sci U S A. 2018;115:E9333–42. doi: 10.1073/pnas.1801920115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zickler D, Kleckner N. A few of our favorite things: pairing, the bouquet, crossover interference and evolution of meiosis. Semin Cell Dev Biol. 2016;54:135–48. doi: 10.1016/j.semcdb.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, et al. A mechanical basis for chromosome function. Proc Natl Acad Sci U S A. 2004;101:12592–7. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S, Zickler D, Kleckner N, Zhang L. Meiotic crossover patterns: obligatory crossover, interference and homeostasis in a single process. Cell Cycle. 2015;14:305–14. doi: 10.4161/15384101.2014.991185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White MA, Wang S, Zhang L, Kleckner N. Quantitative modeling and automated analysis of meiotic recombination. Methods Mol Biol. 2017;1471:305–23. doi: 10.1007/978-1-4939-6340-9_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 2008;4:e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lloyd A, Jenczewski E. Modelling sex-specific crossover patterning in Arabidopsis. Genetics. 2019;211:847–59. doi: 10.1534/genetics.118.301838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Boer E, Stam P, Dietrich AJ, Pastink A, Heyting C. Two levels of interference in mouse meiotic recombination. Proc Natl Acad Sci U S A. 2006;103:9607–12. doi: 10.1073/pnas.0600418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Boer E, Lhuissier FG, Heyting C. Cytological analysis of interference in mouse meiosis. Methods Mol Biol. 2009;558:355–82. doi: 10.1007/978-1-60761-103-5_21. [DOI] [PubMed] [Google Scholar]
- 63.Housworth EA, Stahl FW. Is there variation in crossover interference levels among chromosomes from human males? Genetics. 2009;183:403–5. doi: 10.1534/genetics.109.103853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo C, Li X, Zhang Q, Yan J. Single gametophyte sequencing reveals that crossover events differ between sexes in maize. Nat Commun. 2019;10:785. doi: 10.1038/s41467-019-08786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:11383–91. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Talbert PB, Henikoff S. Centromeres convert but don’t cross. PLoS Biol. 2010;8:e1000326. doi: 10.1371/journal.pbio.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen SY, Tsubouchi T, Rockmill B, Sandler J, Richards D, et al. Global analysis of the meiotic crossover landscape. Dev Cell. 2008;15:401–15. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Halldorsson BV, Palsson G, Stefansson OA, Blanshard R, Capalbo A, et al. Characterizing mutagenic effects of recombination through a sequence-level genetic map. Science. 2019;363:eaau1043. doi: 10.1126/science.aau1043. [DOI] [PubMed] [Google Scholar]
- 69.Chen Y, Lyu R, Rong B, Zheng Y, Lin Z, et al. Refined spatial temporal epigenomic profiling reveals intrinsic connection between PRDM9-mediated H3K4me3 and the fate of double-stranded breaks. Cell Res. 2020;30:256–68. doi: 10.1038/s41422-020-0281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cole F, Keeney S, Jasin M. Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol Cell. 2010;39:700–10. doi: 10.1016/j.molcel.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Boer E, Jasin M, Keeney S. Local and sex-specific biases in crossover vs. noncrossover outcomes at meiotic recombination hot spots in mice. Genes Dev. 2015;29:1721–33. doi: 10.1101/gad.265561.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Medhi D, Goldman AS, Lichten M. Local chromosome context is a major determinant of crossover pathway biochemistry during budding yeast meiosis. Elife. 2016;5:e19669. doi: 10.7554/eLife.19669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shodhan A, Medhi D, Lichten M. Noncanonical contributions of Mutlγ to VDE-initiated crossovers during Saccharomyces cerevisiae meiosis. G3 (Bethesda) 2019;9:1647–54. doi: 10.1534/g3.119.400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang T, Yuan S, Gao L, Li M, Yu X, et al. The histone modification reader ZCWPW1 links histone methylation to PRDM9-induced double strand break repair. Elife. 2020;9:e53459. doi: 10.7554/eLife.53459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hyppa RW, Smith GR. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell. 2010;142:243–55. doi: 10.1016/j.cell.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vincenten N, Kuhl LM, Lam I, Oke A, Kerr AR, et al. The kinetochore prevents centromere-proximal crossover recombination during meiosis. Elife. 2015;4:e10850. doi: 10.7554/eLife.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–85. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Serrentino ME, Borde V. The spatial regulation of meiotic recombination hotspots: are all DSB hotspots crossover hotspots? Exp Cell Res. 2012;318:1347–52. doi: 10.1016/j.yexcr.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 79.Serrentino ME, Chaplais E, Sommermeyer V, Borde V. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet. 2013;9:e1003416. doi: 10.1371/journal.pgen.1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lange J, Yamada S, Tischfield SE, Pan J, Kim S, et al. The landscape of mouse meiotic double-strand break formation, processing, and repair. Cell. 2016;167:695–708. doi: 10.1016/j.cell.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, et al. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science. 2002;296:2222–5. doi: 10.1126/science.1071220. [DOI] [PubMed] [Google Scholar]
- 82.Kleckner N, Storlazzi A, Zickler D. Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet. 2003;19:623–8. doi: 10.1016/j.tig.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 83.Sun F, Turek P, Greene C, Ko E, Rademaker A, et al. Abnormal progression through meiosis in men with nonobstructive azoospermia. Fertil Steril. 2007;87:565–71. doi: 10.1016/j.fertnstert.2006.07.1531. [DOI] [PubMed] [Google Scholar]
- 84.Shi Q, Martin R. Spontaneous frequencies of aneuploid and diploid sperm in 10 normal Chinese men: assessed by multicolor fluorescence in situ hybridization. Cytogenet Cell Genet. 2000;91:79–83. doi: 10.1159/000015668. [DOI] [PubMed] [Google Scholar]
- 85.Wang S, Liu Y, Shang Y, Zhai B, Yang X, et al. Crossover interference, crossover maturation, and human aneuploidy. Bioessays. 2019;41:e1800221. doi: 10.1002/bies.201800221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng EY, Hunt PA, Naluai-Cecchini TA, Fligner CL, Fujimoto VY, et al. Meiotic recombination in human oocytes. PLoS Genet. 2009;5:e1000661. doi: 10.1371/journal.pgen.1000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gruhn JR, Rubio C, Broman KW, Hunt PA, Hassold T. Cytological studies of human meiosis: sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PLoS One. 2013;8:e85075. doi: 10.1371/journal.pone.0085075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hou Y, Fan W, Yan L, Li R, Lian Y, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 89.Kochakpour N, Moens PB. Sex-specific crossover patterns in Zebrafish (Danio rerio) Heredity (Edinb) 2008;100:489–95. doi: 10.1038/sj.hdy.6801091. [DOI] [PubMed] [Google Scholar]
- 90.Falque M, Anderson LK, Stack SM, Gauthier F, Martin OC. Two types of meiotic crossovers coexist in maize. Plant Cell. 2009;21:3915–25. doi: 10.1105/tpc.109.071514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.do Vale Martins L, Yu F, Zhao H, Dennison T, Lauter N, et al. Meiotic crossovers characterized by haplotype-specific chromosome painting in maize. Nat Commun. 2019;10:4604. doi: 10.1038/s41467-019-12646-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kianian PM, Wang M, Simons K, Ghavami F, He Y, et al. High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nat Commun. 2018;9:2370. doi: 10.1038/s41467-018-04562-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sidhu GK, Fang C, Olson MA, Falque M, Martin OC, et al. Recombination patterns in maize reveal limits to crossover homeostasis. Proc Natl Acad Sci U S A. 2015;112:15982–7. doi: 10.1073/pnas.1514265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baier B, Hunt P, Broman KW, Hassold T. Variation in genome-wide levels of meiotic recombination is established at the onset of prophase in mammalian males. PLoS Genet. 2014;10:e1004125. doi: 10.1371/journal.pgen.1004125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Biswas U, Hempel K, Llano E, Pendás A, Jessberger R. Distinct roles of meiosis-specific cohesin complexes in mammalian spermatogenesis. PLoS Genet. 2016;12:e1006389. doi: 10.1371/journal.pgen.1006389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, et al. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 2004;6:555–62. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- 97.Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, et al. Cohesin Smc1beta determines meiotic chromatin axis loop organization. J Cell Biol. 2008;180:83–90. doi: 10.1083/jcb.200706136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ding DQ, Sakurai N, Katou Y, Itoh T, Shirahige K, et al. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J Cell Biol. 2006;174:499–508. doi: 10.1083/jcb.200605074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ding DQ, Matsuda A, Okamasa K, Nagahama Y, Haraguchi T, et al. Meiotic cohesin-based chromosome structure is essential for homologous chromosome pairing in Schizosaccharomyces pombe. Chromosoma. 2016;125:205–14. doi: 10.1007/s00412-015-0551-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jin H, Guacci V, Yu HG. Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J Cell Biol. 2009;186:713–25. doi: 10.1083/jcb.200810107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Viera A, Berenguer I, Ruiz-Torres M, Gómez R, Guajardo A, et al. PDS5 proteins regulate the length of axial elements and telomere integrity during male mouse meiosis. EMBO Rep. 2020;21:e49273. doi: 10.15252/embr.201949273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hong S, Joo JH, Yun H, Kleckner N, Kim KP. Recruitment of Rec8, Pds5 and Rad61/Wapl to meiotic homolog pairing, recombination, axis formation and S-phase. Nucleic Acids Res. 2019;47:11691–708. doi: 10.1093/nar/gkz903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baarends WM, Wassenaar E, Hoogerbrugge JW, van Cappellen G, Roest HP, et al. Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase. Mol Cell Biol. 2003;23:1151–62. doi: 10.1128/MCB.23.4.1151-1162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Storlazzi A, Tessé S, Gargano S, James F, Kleckner N, et al. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 2003;17:2675–87. doi: 10.1101/gad.275203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tessé S, Bourbon HM, Debuchy R, Budin K, Dubois E, et al. Asy2/Mer2: an evolutionarily conserved mediator of meiotic recombination, pairing, and global chromosome compaction. Genes Dev. 2017;31:1880–93. doi: 10.1101/gad.304543.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee CY, Conrad MN, Dresser ME. Meiotic chromosome pairing is promoted by telomere-led chromosome movements independent of bouquet formation. PLoS Genet. 2012;8:e1002730. doi: 10.1371/journal.pgen.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Scherthan H, Schonborn I. Asynchronous chromosome pairing in male meiosis of the rat (Rattus norvegicus) Chromosome Res. 2001;9:273–82. doi: 10.1023/a:1016642528981. [DOI] [PubMed] [Google Scholar]
- 108.Schalbetter SA, Fudenberg G, Baxter J, Pollard KS, Neale MJ. Principles of meiotic chromosome assembly revealed in S. cerevisiae. Nat Commun. 2019;10:4795. doi: 10.1038/s41467-019-12629-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Murakami H, Lam I, Huang P, Song J, van Overbeek M. Multilayered mechanisms ensure that short chromosomes recombine in meiosis. Nature. 2020;582:124–8. doi: 10.1038/s41586-020-2248-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Subramanian VV, Zhu X, Markowitz TE, Vale-Silva LA, San-Segundo PA, et al. Persistent DNA-break potential near telomeres increases initiation of meiotic recombination on short chromosomes. Nat Commun. 2019;10:970. doi: 10.1038/s41467-019-08875-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kauppi L, Barchi M, Baudat F, Romanienko PJ, Keeney S, et al. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science. 2011;331:916–20. doi: 10.1126/science.1195774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kauppi L, Jasin M, Keeney S. The tricky path to recombining X and Y chromosomes in meiosis. Ann N Y Acad Sci. 2012;1267:18–23. doi: 10.1111/j.1749-6632.2012.06593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boekhout M, Karasu ME, Wang J, Acquaviva L, Pratto F, et al. REC114 partner ANKRD31 controls number, timing, and location of meiotic DNA breaks. Mol Cell. 2019;74:1053–68. doi: 10.1016/j.molcel.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Papanikos F, Clément JA, Testa E, Ravindranathan R, Grey C, et al. Mouse ANKRD31 regulates spatiotemporal patterning of meiotic recombination initiation and ensures recombination between X and Y sex chromosomes. Mol Cell. 2019;74:1069–85. doi: 10.1016/j.molcel.2019.03.022. [DOI] [PubMed] [Google Scholar]
- 115.Lian J, Yin Y, Oliver-Bonet M, Liehr T, Ko E, et al. Martin RH. Variation in crossover interference levels on individual chromosomes from human males. Hum Mol Genet. 2008;17:2583–94. doi: 10.1093/hmg/ddn158. [DOI] [PubMed] [Google Scholar]
- 116.Borodin PM, Karamysheva TV, Belonogova NM, Torgasheva AA, Rubtsov NB, et al. Recombination map of the common shrew, Sorex araneus (Eulipotyphla, Mammalia) Genetics. 2008;178:621–32. doi: 10.1534/genetics.107.079665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Barchi M, Roig I, Di Giacomo M, de Rooij DG, Keeney S, et al. ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet. 2008;4:e1000076. doi: 10.1371/journal.pgen.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Basheva EA, Bidau CJ, Borodin PM. General pattern of meiotic recombination in male dogs estimated by MLH1 and RAD51 immunolocalization. Chromosome Res. 2008;16:709–19. doi: 10.1007/s10577-008-1221-y. [DOI] [PubMed] [Google Scholar]
- 119.Housworth EA, Stahl FW. Crossover interference in humans. Am J Hum Genet. 2003;73:188–97. doi: 10.1086/376610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Z, Shen B, Jiang J, Li J, Ma L. Effect of sex, age and genetics on crossover interference in cattle. Sci Rep. 2016;6:37698. doi: 10.1038/srep37698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Billings T, Sargent EE, Szatkiewicz JP, Leahy N, Kwak IY, et al. Patterns of recombination activity on mouse chromosome 11 revealed by high resolution mapping. PLoS One. 2010;5:e15340. doi: 10.1371/journal.pone.0015340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–91. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 123.Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, et al. Aneuploidy across individual chromosomes at the embryonic level in trophectoderm biopsies: changes with patient age and chromosome structure. J Assist Reprod Genet. 2014;31:1501–9. doi: 10.1007/s10815-014-0333-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, et al. The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril. 2014;101:656–63.e1. doi: 10.1016/j.fertnstert.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 125.Gruhn JR, Zielinska AP, Shukla V, Blanshard R, Capalbo A, et al. Chromosome errors in human eggs shape natural fertility over reproductive life span. Science. 2019;365:1466–9. doi: 10.1126/science.aav7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wolstenholme J, Angell RR. Maternal age and trisomy – a unifying mechanism of formation. Chromosoma. 2000;109:435–8. doi: 10.1007/s004120000088. [DOI] [PubMed] [Google Scholar]
- 127.Storlazzi A, Tesse S, Ruprich-Robert G, Gargano S, Pöggeler S, et al. Coupling meiotic chromosome axis integrity to recombination. Genes Dev. 2008;22:796–809. doi: 10.1101/gad.459308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jessberger R. Age-related aneuploidy through cohesion exhaustion. EMBO Rep. 2012;13:539–46. doi: 10.1038/embor.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Handyside AH. Molecular origin of female meiotic aneuploidies. Biochim Biophys Acta. 2012;1822:1913–20. doi: 10.1016/j.bbadis.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 130.Chiang T, Schultz RM, Lampson MA. Meiotic origins of maternal age-related aneuploidy. Biol Reprod. 2012;86:1–7. doi: 10.1095/biolreprod.111.094367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cheng JM, Liu YX. Age-related loss of cohesion: causes and effects. Int J Mol Sci. 2017;18:1578. doi: 10.3390/ijms18071578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–62. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- 134.Zielinska AP, Holubcova Z, Blayney M, Elder K, Schuh M. Sister kinetochore splitting and precocious disintegration of bivalents could explain the maternal age effect. eLife. 2015;4:e11389. doi: 10.7554/eLife.11389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zielinska AP, Bellou E, Sharma N, Frombach AS, Seres KB, et al. Meiotic kinetochores fragment into multiple lobes upon cohesin loss in aging eggs. Curr Biol. 2019;29:3749–65. doi: 10.1016/j.cub.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Perkins AT, Greig MM, Sontakke AA, Peloquin AS, McPeek MA, et al. Increased levels of superoxide dismutase suppress meiotic segregation errors in aging oocytes. Chromosoma. 2019;128:215–22. doi: 10.1007/s00412-019-00702-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gheldof A, Mackay DJ, Cheong Y, Verpoest W. Genetic diagnosis of subfertility: the impact of meiosis and maternal effects. J Med Genet. 2019;56:271–82. doi: 10.1136/jmedgenet-2018-105513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Capalbo A, Hoffmann ER, Cimadomo D, Ubaldi FM, Rienzi L. Human female meiosis revised: new insights into the mechanisms of chromosome segregation and aneuploidies from advanced genomics and time-lapse imaging. Hum Reprod Update. 2017;23:706–22. doi: 10.1093/humupd/dmx026. [DOI] [PubMed] [Google Scholar]
- 139.Lu YQ, He XC, Zheng P. Decrease in expression of maternal effect gene mater is associated with maternal ageing in mice. Mol Hum Reprod. 2016;22:252–60. doi: 10.1093/molehr/gaw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rémillard-Labrosse G, Dean NL, Allais A, Mihajlović AI, Jin SG, et al. Human oocytes harboring damaged DNA can complete meiosis I. Fertil Steril. 2020;113:1080–9.e2. doi: 10.1016/j.fertnstert.2019.12.029. [DOI] [PubMed] [Google Scholar]
- 141.Steiner B, Masood R, Rufibach K, Niedrist D, Kundert O, et al. An unexpected finding: younger fathers have a higher risk for offspring with chromosomal aneuploidies. Eur J Hum Genet. 2015;23:466–72. doi: 10.1038/ejhg.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zelazowski MJ, Sandoval M, Paniker L, Hamilton HM, Han J, et al. Age-dependent alterations in meiotic recombination cause chromosome segregation errors in spermatocytes. Cell. 2017;171:601–14. doi: 10.1016/j.cell.2017.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vrooman LA, Nagaoka SI, Hassold TJ, Hunt PA. Evidence for paternal age-related alterations in meiotic chromosome dynamics in the mouse. Genetics. 2014;196:385–96. doi: 10.1534/genetics.113.158782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Martin RH. Meiotic errors in human oogenesis and spermatogenesis. Reprod Biomed Online. 2008;16:523–31. doi: 10.1016/s1472-6483(10)60459-2. [DOI] [PubMed] [Google Scholar]
- 145.Fonseka KGL, Griffin DK. Is there a paternal age effect for aneuploidy? Cytogenet Genome Res. 2011;133:280–91. doi: 10.1159/000322816. [DOI] [PubMed] [Google Scholar]
- 146.Campbell CL, Furlotte NA, Eriksson N, Hinds D, Auton A. Escape from crossover interference increases with maternal age. Nat Commun. 2015;6:6260. doi: 10.1038/ncomms7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Martin HC, Christ R, Hussin JG, O’Connell J, Gordon S, et al. Multicohort analysis of the maternal age effect on recombination. Nat Commun. 2015;6:7846. doi: 10.1038/ncomms8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–8. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 149.Moens PB, Pearlman RE. Chromatin organization at meiosis. Bioessays. 1988;9:151–3. doi: 10.1002/bies.950090503. [DOI] [PubMed] [Google Scholar]