Abstract
Repetitive DNA elements frequently are precursors to chromosomal deletions in prokaryotes and lower eukaryotes. However, little is known about the relationship between repeated sequences and deletion formation in mammalian cells. We have created a novel integrated plasmid-based recombination assay to investigate repeated sequence instability in human cells. In a control cell line, the presence of direct or inverted repeats did not appreciably influence the very low deletion frequencies (2 × 10−7 to 9 × 10−7) in the region containing the repeat. Similar to what has been observed in lower eukaryotes, the majority of deletions resulted from the loss of the largest direct repeat present in the system along with the intervening sequence. Interestingly, in closely related cell lines that possess a mutant p53 gene, deletion frequencies in the control and direct-repeat plasmids were 40 to 300 times higher than in their wild-type counterparts. However, mutant p53 cells did not preferentially utilize the largest available homology in the formation of the deletion. Surprisingly, inverted repeats were approximately 10,000 times more unstable in all mutant p53 cells than in wild-type cells. Finally, several deletion junctions were marked by the addition of novel bases that were homologous to one of the preexisting DNA ends. Contrary to our expectations, only 6% of deletions in all cell lines could be classified as arising from nonhomologous recombination.
Genomic instability is a hallmark of cancer. A situation where a cell promiscuously rearranges its DNA is likely to lead to further oncogenic progression. With the human genome containing large amounts of repetitive DNA, a deregulation of stability between repeats could facilitate unchecked large-scale rearrangements. Despite the recent cloning of several putative recombination enzymes, such as Rad 51 (36) and Rad52 (28), there is no definitive evidence indicating whether homologous or nonhomologous recombination (HR or NHR) is the predominant pathway for the formation of DNA deletions. Several reports of human germ line mutations suggest that when highly homologous sequences are present, such as Alu repeats in the LDLR or CI1 gene, the majority of deletions tend to occur via a mechanism in which one repeat and the entire intervening sequence is deleted (8). Since there is a selection bias toward analyzing DNA from patients who present with mutations in the LDLR or CI1 gene, it is difficult to know whether this frequency of HR is typical of somatic cells.
In the absence of systematic studies to examine the effect of the orientation of repetitive DNA on deletions, only anecdotal reports exist to suggest that inverted repeats are exceptionally unstable DNA elements. Large DNA inverted repeats (>150 bp) lead to a unique genomic instability in prokaryotes and lower eukaryotes (12). In Escherichia coli, inverted repeats are rapidly deleted and may lead to death (39). Similarly, artificially created inverted repeats in Saccharomyces cerevisiae are approximately 3 orders of magnitude more unstable than direct repeats (20). It has been demonstrated that large inverted repeats fold back upon themselves to form a hairpin or cruciform structure in vitro and in vivo (17, 44). The formation of a cruciform structure is believed to stall DNA polymerases, and the structure may be a prime substrate for recombinases (20). When an inverted repeat is resolved in E. coli or S. cerevisiae, it is common to find part or all of one arm of the hairpin to be missing (5, 11). In both organisms, these deletions usually occur at short direct repeats, one lying within the putative hairpin and one lying outside, flanking its base. Several recombination and excision repair proteins are known to play a role in excision of the cruciform structure in S. cerevisiae (10, 13), and patients who lack the human homologues of these genes (XP-F, XP-G) are at increased risk for cancer (30, 37). On the other hand, numerous reports exist that detail the seemingly random rejoining of DNA. Unfortunately, searching a large number of endogenously mutated genes, such as hypoxanthine phosphoribosyltransferase (hprt) and aprt, to find a few sequencable breakpoints is costly and time-consuming and often results in one junction being derived from an unsequenced region of the genome. Furthermore, it is extremely difficult to engineer various repeats into the endogenous gene of interest in order to create a controlled system similar to those that have been successfully used in bacteria and lower eukaryotes.
In the absence of a system to assess deletion formation in a controlled setting, little information is available about the role that the genetic background might play in the frequency and nature of large (kilobase-sized) deletions in human cells. The p53 protein has several purported characteristics that implicate it in the maintenance of large-scale genomic instability. The p53 tumor suppressor gene is the most commonly mutated gene in human tumors (15). This protein plays a role in regulating G1 arrest (18) and apoptosis following DNA damage (21). It has been hypothesized that the loss of functional p53 may influence genomic instability through direct or indirect roles in DNA repair. Cells with mutated p53 have been observed to undergo increased gene amplification (19) and homologous recombination (3, 24) and to have an elevated frequency of chromosome abnormalities (4). However, it is not clear whether this observed genomic instability is due to the failure of heavily damaged cells to undergo programmed cell death, to the lack of functional G1 arrest, or to the fact that p53 plays a direct role in repairing (or regulating the repair of) lesions that may lead to large-scale mutations. The p53 protein possesses an exonucleolytic function (27) and can bind short DNA free ends and stimulate their renaturation (2). p53 also interacts with several proteins that are believed to play a role in recombination: replication protein A (RPA) (9), Rad51 (6, 38), and Rad52 (35).
A minigene deletion reporter gene was constructed to examine the genetic and physical factors that influence deletions in human cells. Several deletion characteristics were found that were similar to those of lower eukaryotes in a control cell line. In comparison to controls, cells possessing a mutant p53 gene exhibited a hyperdeletion phenotype and an altered involvement of homology in the deletion formation.
MATERIALS AND METHODS
Plasmid construction.
pBASE was constructed by cloning the PCR fragment (generated from primers ACACGTGTGAACCAACCCGC and GGAAAATTTGGCAATACCAA) that included exon 2 of hprt into the XbaI site of PNI2 (a kind gift from L. Reid). This PCR fragment splits the preexisting hprt intron 2 with a second hprt exon 2 (120 bp of homology to the first exon 2). This duplicated exon 2 is now flanked by intron A (1,088 bp) and intron B (1,174 bp), which do not possess any significant homology greater than 20 bp. pDIR and pINV were constructed by cloning the Alu sequence found in intron B into the XbaI site of intron A. The Alu sequence was cloned in both the direct-repeat orientation (pDIR) and inverted-repeat orientation (pINV), creating 369 bp of perfect homology between introns A and B. pINV-HYG was made by cloning a hygromycin cassette into the AseI site on pINV.
Cell lines.
TK6 is a human lymphoblastoid cell line derived from WIL-2 culture. It possesses a wild-type p53 gene (41). WTK-1 is a human lymphoblast cell line that is derived from the same donor as TK6 but has a mutant p53 gene (containing a mutation at Ile237) (41–43). AHH-1 is derived from a different donor from TK6 and WTK-1 and has a p53 mutation at codon 283 (26). TK6-E6 is TK6 transfected with a retroviral vector expressing the E6 gene (42). All cells were grown in RPMI 1640 supplemented with 10% horse serum (Sigma).
Transfection and characterization.
pBASE, pINV, and pDIR were transfected into hprt total-deletion lymphoblasts by electroporation and selected for integration with G418. Individual transfectants were screened for comparable expression levels by Northern blot analysis, and reverse transcription-PCR was performed to ensure that the duplicated exon was included in the mRNA and that no exon skipping was evident (data not shown). Mostly single-copy integrants (as determined by Southern blot analysis) were used for this study; however, there was no significant increase in the deletion frequency when multiple-copy integrants (all tandem arrays) were used. All candidate transfectants were initially hypoxanthine-aminopterin-thymidine (HAT) sensitive and 6-thioguanine (6-TG) resistant.
Deletion frequency.
All candidate transfectants were pretreated with 6-TG to reduce the background of cells that had rearranged the hprt minigene during transfection or passaging. The cells were simultaneously pretreated with G418 to ensure that plasmid was present. As a control of plasmid loss, cells were plated in the presence and absence of G418. No plasmid loss was detectable during 5 days of nonselective growth. After pretreatment, transfectants were allowed 5 days of growth for expression time (time for the cell to express the functional HPRT protein and turn over any dysfunctional protein). The expression time was determined empirically by treating the transfectants with UV light at 20 J/m2 and plating every day in the selective agent HAT. Day 5 was chosen because it was 1 day past the point where the deletion frequency had plateaued. For all measured deletion frequencies, cells were plated at appropriate densities in the selective agent HAT (2 × 10−4 M hypoxanthine, 10−7 M aminopterin, 8 × 10−5 M thymidine, 10−5 M deoxycytidine). The majority of transfectants used in this study were single-copy integrants as determined by Southern blot analysis (numbers of single and multiple copies were 23 and 6, respectively). There was no significant difference in the deletion frequency between single- and multiple-copy integrants. The deletion frequency for all experiments was determined by counting the number of colonies that were plated either in normal medium or in the selective agent HAT in 96-well microtiter dishes at appropriate densities such that both positive and negative wells were observed. Deletion frequencies (DF) were calculated from the Poisson distribution, using the formula DF = −ln (fraction of negative wells in HAT)/(number of cells plated per well × plating efficiency in normal medium). For all deletion frequencies, a minimum of three distinct transfectants were used to control for genomic position effect. For each transfectant, deletion frequencies were determined in triplicate and averaged.
Deletion Spectra.
Each mutant was isolated from an independent culture that had been pretreated with HAT to eliminate any previously rearranged plasmids. Genomic DNA extracted from one mutant from each experiment was amplified by PCR with primers 5′-CCCTGGCGTCGTGATTAGTG-3′ and 5′-GCCTGACCAAGGAAAGCAAA-3′. Digestion of the PCR product with restriction enzymes unique to the fragment narrowed the search for the deletion rejoining site. Sequencing was performed by cycle sequencing with appropriate primers at a contracted laboratory. Rejoining points and the ends of the deleted segments were examined for the presence of homology by visual inspection and computer analysis using Vector NTI 4.0 (Informax). Although it was hypothesized that a point mutation in the exon 2 splice acceptor site could result in exon skipping and hence in a functional HPRT protein, no representatives of this class of mutations were found.
Western blot analysis.
Western blot analyses were done as described by Xia et al. (41).
RESULTS
Development of a minigene system to detect and characterize DNA deletions.
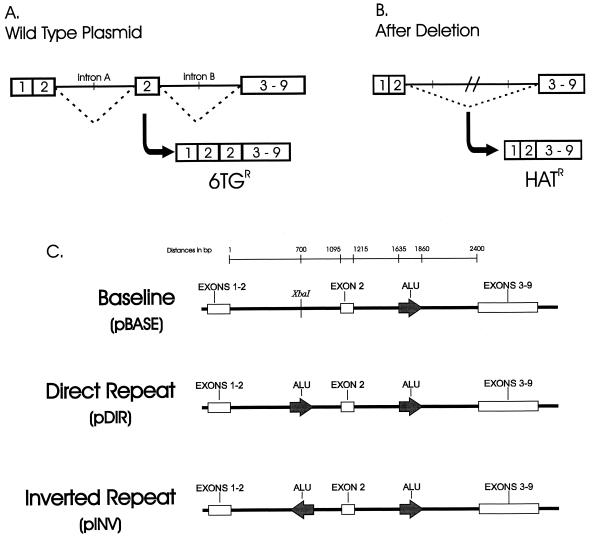
To determine the role that p53 plays in large-scale genomic instability, a plasmid-based system was designed that can quantitatively and qualitatively assess deletion formation in any human cell line. The system consists of an hprt minigene that must delete a duplicated exon 2 in order to code for a functional HPRT protein and hence confer resistance to the selective agent HAT (Fig. 1A and B). Three variants of the plasmid were created (Fig. 1C). pBASE has no significant homology (>19 bp) between intron A and B. The pDIR plasmid has two Alu repeats of 369 bases in direct-repeat orientation, while pINV has the same Alu fragment in inverted-repeat orientation. The plasmids were individually transfected into recipient hprt− (total deletion) human lymphoblast lines and selected with G418 for integration. At least three independent transfectants from each cell line were used to measure deletion frequencies and generate individual mutants for sequence analysis.
FIG. 1.
Deletion system and plasmid substrates. (A) Functional schematic of the deletion system. The hprt minigene consists of fused hprt exons 1 and 2, intron A, duplicated exon 2, intron B, and fused exons 3 to 9. When incorporated into a hprt mutant cell line, the hprt minigene incorporates the duplicated exon 2 into its mRNA. This creates a 105-bp insertion and therefore codes for a dysfunctional protein. (B) A deletion that eliminates the duplicated exon 2 restores hprt function and thus confers HAT resistance (HATR). (C) Plasmid variants. pDIR and pINV were created by cloning the 369-bp Alu sequence from intron B in an XbaI site in intron A in both orientations. All plasmids were sequenced to ensure proper sequence identity.
Quantitative study of deletions in a wild-type cell line.
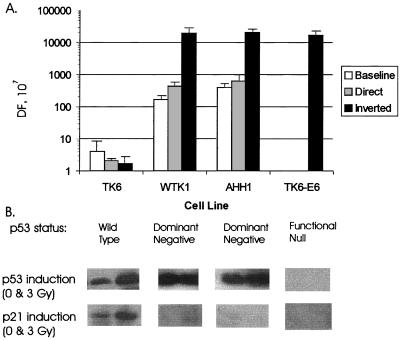
The human lymphoblast cell line TK6 contains a wild-type p53 gene (19, 30, 31). Frequencies of spontaneous deletion in the baseline plasmid averaged 4 × 10−7 (range, 2 × 10−7 to 9 × 10−7). This deletion frequency is of the same order as has been observed in endogenous genes (31). Interestingly, the addition of 369 bp of flanking homology (Alu) did not alter the observed deletion frequencies (Fig. 2). pDir and pINV exhibited average deletion frequencies of 2.1 × 10−7 and 1.8 × 10−7 (ranges, 2.0 × 10−7 to 2.1 × 10−7 and 1.1 × 10−7 to 2.5 × 10−7), respectively.
FIG. 2.
Spontaneous deletion frequencies (DF) in different p53 backgrounds. (A) Deletion frequencies. For each determination of deletion frequency, at least three individual transfectants of each of the three plasmid variants were pretreated with 6-TG for 2 days to reduce the background. Deletion frequencies were determined from the formula described in Materials and Methods. (B) p53 status of human lymphoblasts. Western blot analyses of p21 and p53 protein levels were performed as described previously (41).
Quantitative study of deletions in mutant p53 cell lines.
In contrast to the observed stability of repeats in p53 wild-type cells, when the plasmid variants were integrated into two independent p53 mutant lymphoblasts, striking differences emerged (Fig. 2A). First, the mutant p53 status markedly increased the deletion frequency for all plasmid vectors. The deletion frequency for the baseline plasmid was 42 and 155 times higher in WTK-1 and AHH-1, respectively, than in TK6; the deletion frequencies were 166 × 10−7 and 400 × 10−7, respectively (Fig. 2A). In WTK-1 and AHH-1, pDIR was also about 200 to 300 times more unstable than in the control cell line, with deletion frequencies of 432 × 10−7 to 620 × 10−7. This elevated instability is in agreement with other reports of increased instability in p53 mutants with homologous recombination substrates. Surprisingly, the inverted repeat was ∼10,000 times more unstable in both mutant p53 cell lines. The deletion frequency averaged 19,200 × 10−7 in WTK-1 cells and 20,000 × 10−7 in AHH-1 cells. To confirm that p53 was responsible for the elevated deletion frequencies seen in WTK-1 cells, further studies were conducted with TK6-E6, which possesses E6 gene and thus is a functional p53-null cell line (42); in these cells, the pINV plasmid deletion frequency averaged 17,000 × 10−7.
Deletion spectra in wild-type cells.
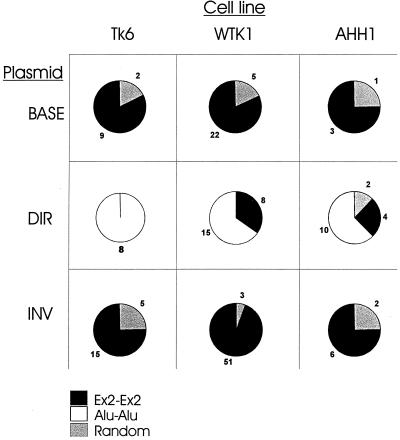
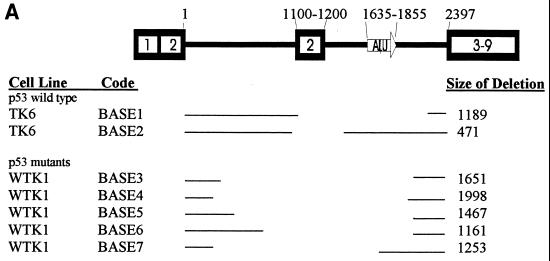
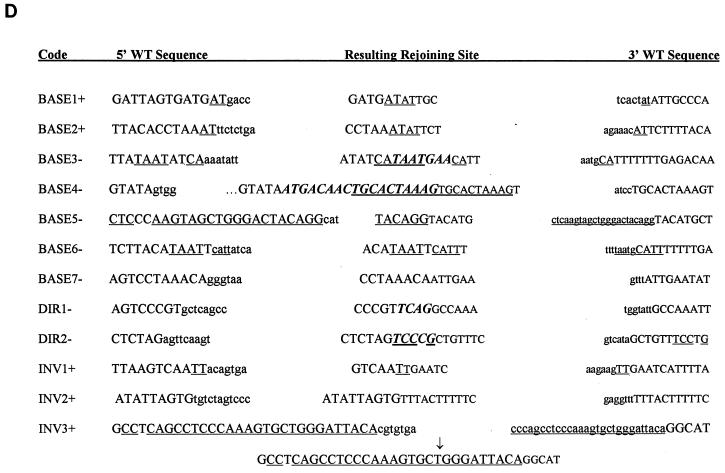
Sequence analysis of the TK6 deletion mutants revealed several interesting trends. First, 9 of 11 (82%) pBASE deletion mutants had a precise deletion of one copy of the repeated exon 2 and the intervening sequence (this class of deletion is termed Ex2-Ex2 [Fig. 3]). The remaining two deletions were termed “random” and are shown in Fig. 4A as BASE1 and BASE2. Sequencing of the rejoining sites of both of these deletion mutants revealed that both deletions occurred at repeated AT locations but that no other significant homology was present at the breakpoint (Fig. 4D).
FIG. 3.
Deletion spectrum. The pie charts show the three different classes of deletions. The Ex2-Ex2 class represents a precise deletion of one copy of exon 2 and the intervening sequence. The Alu-Alu class represents a precise deletion of one copy of the Alu sequence and the intervening sequence. All deletions that could not be classified are termed “random.”
FIG. 4.
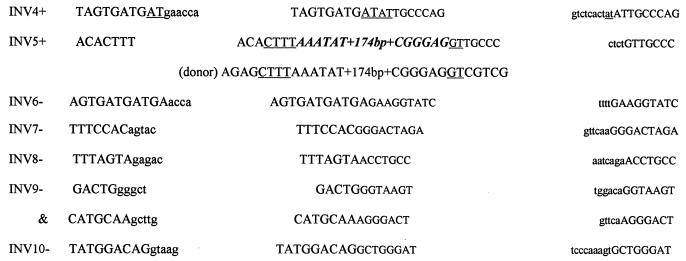
(A to C) Schematic diagrams of the DNA deletions investigated in this study. Horizontal lines represent the retained DNA. (D) Sequence and microhomology present at deletion rejoining sites. The 5′ end of the region flanking the deletion is shown in normal-sized type, while the 3′ end is shown in smaller type. Capital letters denote the retained DNA. Lowercase letters denote the sequence deleted from the wild-type intron. Underlined bases represent areas of homology between either the wild-type sequences or novel bases (shown in bold italics).
In contrast to the pBASE mutants, all TK6 pDIR deletion mutants (n = 8) had a precise deletion of one of the directly repeated Alu motifs and the entire intervening sequence (Fig. 3). By sequencing the ends of the remaining Alu motif, it was determined that in all cases the 5′ end of the Alu motif was derived from the Alu originally found in intron A (data not shown). Similarly, the 3′ end of the remaining Alu motif was derived from the Alu originally in intron B. Further resolution of the “hybrid” Alu motif is impossible due to the 100% homology of the internal Alu sequence.
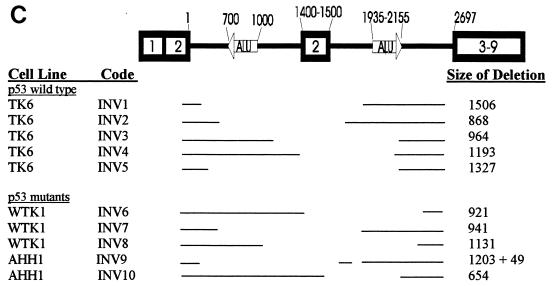
The deletion spectrum of TK6 pINV (Fig. 3) was composed of a mixture of 75% Ex2-Ex2 and 25% random mutants. Figure 4C shows the location of the random mutants. Their size ranged from 868 to 1,506 bp and averaged 1,171 bp. The sequence at the breakpoints revealed that mutants INV1, INV2, and INV4 had no significant homology (<2 bp) at the rejoining site (Fig. 4D). As shown in Fig. 4D, while the 5′ retained end of INV3 did not have any significant homology to the 3′ retained end, it had 27 of 28 bp of homology to the 3′ end of the deleted segment. Mutant INV5 represents a translocation. Sequencing the deletion uncovered a 1,327-bp deletion coupled with a 186-bp insertion. The sequence of the 186-bp insertion was 100% identical to a segment of the neo gene (found 5310 bp downstream). As shown in Fig. 4D, the donor and acceptor had 4 bp of homology at the 5′ end and 2 bp at the 3′ end. It is not known if this is a reciprocal translocation.
Deletion spectra in mutant p53 cells.
A total of 27 pBASE mutants from WTK-1 and 4 from AHH-1 were analyzed. Similar to what was seen in TK6, 82% (22 of 27) of the WTK-1 pBASE mutants fell into the Ex2-Ex2 category, as did 75% (3 of 4) of the AHH-1 pBASE mutants (Fig. 3). The remaining deletions were classified as random and are shown graphically in Fig. 4A. BASE3 had a deletion of 1,651 bp, but had an additional 7 bp (Fig. 4D). Interestingly, 4 of the 7 bp have homology to sequences that flank the 5′ end of the deletion junction site. Similarly, BASE4 added 18 bp, the last 10 of which are perfectly homologous to sequences that flank the 3′ end of the deletion junction site. Whereas BASE7 had no significant homology between the retained and deleted segments, both retained ends of BASE6 had 4 bp of homology to the opposite ends to the deleted segment. In a similar fashion, the 5′ retained end of BASE5 had 22 of 24 bp of homology to the 3′ end of the deleted segment. In total, four of five WTK-1 pBASE mutants had microhomology present, or originally present, at their rejoining sites.
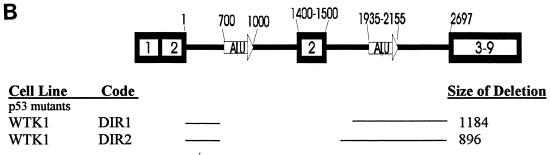
In contrast to the deletion spectrum of TK6 pDIR (100% Alu-Alu class), the mutant p53 pDIR spectrum showed greater heterogeneity. Only 62.5% of AHH-1 and 65% of WTK-1 pDIR mutants had precise deletions of one of the Alu motifs and the intervening sequence (Fig. 3). The Ex2-Ex2 class comprised 25% of AHH-1 and 35% of WTK-1 mutants. There were 2 of 16 random mutants in the AHH-1 spectrum (Fig. 4B). Both of these random deletion mutants had novel base pairs added to the ends. DIR2 added 5 bp, 4 of which were homologous to the 3′ retained end (Fig. 4D).
The WTK-1 and AHH-1 pINV deletion spectra were similar to that of TK6 and were largely composed of deletions that fell into the Ex2-Ex2 class (Fig. 3). Only 3 of 54 WTK-1, and 2 of 8 AHH1 pINV mutants could be classified as random. The sizes of these random deletions are shown in Fig. 4C. Interestingly, none of these mutants had any significant homology at their rejoining sites. One mutant, INV9 had a complex deletion in which 1,203 bp were deleted (Fig. 4C), followed by 60 bp of a retained sequence and another deletion (of 49 bp). None of the ends of this double deletion show any significant homology (INV9).
DISCUSSION
On the basis of examinations of the mutation frequencies and spectra of endogenous mutated genes, it originally was postulated that the deletion frequency of the minigene in human cells would be around 5 × 10−7. The actual frequency of ∼2 × 10−7 closely matched this estimate. It was noted that there was no difference in the deletion frequency between pBASE and pDIR transfectants, despite the addition of 369 bp of homology of the repeated Alu in wild-type cells. This would be logical if the deletions arose during repair of spontaneous double-strand breaks in the region; in that case, one would expect the addition of the longer tract of homology to shift the spectrum of events without affecting the frequency, since the level of damage would probably be similar. It was previously reported that approximately 250 bp of perfect homology was required to catalyze efficient recombination (34). Our results suggest that 120 bp of homology is sufficient to facilitate efficient recombination. The increase in the deletion frequency in the p53 mutants is in agreement with previous findings by Mekeel et al. (24) and Bertrand et al. (3). Both groups have reported that HR is elevated approximately to the same extent as what we observed in our cells (40- to 100-fold). The laboratory of S. Meyn reported that HR was elevated to a similar extent in ataxia-telangiectasia (AT)-mutant cells (25). Since AT is upstream of p53 and AT cells share some characteristics of large-scale genomic instability with p53 mutant cells, it is interesting to speculate that this pathway is involved in maintaining the integrity of the genome even under nonstressed conditions. It is possible that both genes play a role in sensing DNA damage as well as propagating the signal for repair. Several potential mechanisms for the potential role of p53 in DNA damage sensing and/or repair are discussed below.
Plasmid pINV was ∼10,000-fold more unstable in mutant p53 cells than in wild-type cells. This profound difference could be due to several factors. First, p53 plays a direct role in sensing abnormal secondary structure and signaling for its repair. It has been reported that inverted repeats may form hairpins that induce double-strand breaks in E. coli and yeast (reviewed in reference 12). The p53 protein is readily activated by double-strand break-inducing agents (such as X rays) (22). Hence, an inverted repeat may be a hot spot for p53 action. In humans, hairpin cleavage may occur via the human RAD 1/10 homologues (XP-F and ERCC1) or the Mre11-Rad50 complex, creating a double-strand break (7, 16). p53 has been shown to bind DNA free ends and reanneal short stretches of unpaired strands (2). In this model, mutant p53 may leave the breaks unrepaired, which may serve as a potent signal for recombination. In an alternative model, proteins that interact with p53 may not be properly regulated in response to this abnormal secondary structure. As discussed above, the p53 protein directly interacts with several proteins, such as RPA, Rad51, and Rad52, that are involved in recombination in lower eukaryotes. In support of this theory, p53 has been reported to preferentially bind to Holliday junctions and facilitate their cleavage (14). Its presence at the center of the four-strand junction would position it in a prime location to either coordinate the resolution of the abnormal secondary structure back to a wild-type conformation or signal for its repair. Hence, a cell with mutant p53 could suffer from unregulated recombination, which could predominantly result in a deletion.
The deletion spectra of the pBASE mutants were similar for p53 mutant and wild-type cells. There were no obvious differences in the sizes of the deletions. It is interesting that with only 120 bp of homology between the duplicated exon 2's, a deletion that resembled HR was found to occur 75 to 82% of the time in all cell lines. Both the exon 2-exon 2 and Alu-Alu deletion mutants are similar to products of a single-strand annealing model (10, 23, 29), in which one direct repeat is retained while the other repeat is lost along with the intervening sequence. The minority of “random” mutants observed suggests that when a cell is given the option of a deletion occurring almost anywhere in >2,000 bp of nonhomologous DNA (the introns), it preferentially utilizes the 120 bp of homology of the duplicated exons. This result indicates that intrachromosomal HR may be a common event but has been observed in only a few instances in human cells due to the small sample size of sequenced deletions. Additionally, the degeneracy of repetitive sequences may prove to be a barrier to recombination (32). For example, although Alu sequences are involved in several deletions in genes with highly homologous Alu sequences (e.g., LDLR and C1I (8), the majority of Alu homology in the genome ranges from 72 to 99% (8). Alu homologies are particularly degenerate in the hprt gene, where Alu-Alu recombination is rare (8).
The loss of a gene that regulates the threshold for recombination between different lengths of homology may lead to promiscuous rearrangements and hence may result in a mutator phenotype. The pDIR deletion mutants showed a striking difference in their deletion spectrum. Whereas all TK6 deletions occurred as a precise Alu-Alu recombination, the p53 mutants frequently used the shorter homology of the duplicated exon 2's and only occasionally generated random deletions. The direct and indirect DNA repair functions ascribed to p53 (as described above) could lead to a model where p53 is responsible for the identification and alignment of homologous sequences to facilitate the repair. Alternatively, when this altered deletion spectrum is coupled to the elevated deletion frequency, it is possible that the formation of an abnormal secondary structure as a result of slippage during replication is regulated by p53. In fact, p53 is believed to interact with proteins that are involved in both replication and repair, such as RPA, proliferating-cell nuclear antigen (which, in turn, interacts with Flap endonuclease 1 [40]). Mutant p53 cells, in an attempt to resolve the impediment to replication, may signal for recombinational repair to remove the abnormal secondary structure at the cost of illegitimate recombination. We cannot yet distinguish between misreplication and DNA repair. Future experiments are planned that increase the intrarepeat distance (Alu or exon 2), which may address this question. The addition of novel bases was unique to mutant p53 cells. Of the four deletions that added bases, three created sequence homology to one of the retained ends. One instance of this phenomenon was reported by Morris and Thacker (26), but no mention of the p53 status of the cells used in the study was made in that report. Roth and Wilson observed this phenomenon in 4 of 17 sequenced junctions when examining rejoining sites of a linearized simian virus 40 genome that was transfected into simian cells (33). The p53 status of the simian cell line used in that study is not clear. It is possible that to rejoin nonhomologous DNA ends, bases may be added in a templated fashion in an attempt to make a patch of homology that would act as an adhesive for subsequent rejoining. Although p53 is believed to bind and remove up to 30 nucleotides from free DNA ends (1), it is unclear how this ability would result in the addition of novel bases.
The deletion spectrum of cells containing the pINV plasmid was almost identical to that of the pBASE mutants. Inverted repeats in bacteria and lower eukaryotes are generally resolved by deleting part or all of the arm of the putative hairpin. Generally, the resulting deletion occurs between two small direct repeats, one within the hairpin and one directly outside. Although there is no evidence that our inverted repeats form a hairpin in vivo, the deletion spectrum is identical to what has been observed in bacteria and lower eukaryotes. The majority of pINV deletions in all cell lines fell into the Ex2-Ex2 class. This class involves a deletion of the 5′ arm of the putative hairpin, using the exon 2 homology as the internal and external direct repeats. Similarly, deletion mutant INV3 used an internal and external 26-bp direct repeat to excise the 3′ arm of the hairpin. Interestingly, although there is a large quantitative difference between the pINV deletion frequencies of p53 wild-type and mutant cells, there is no difference in the spectrum. This would suggest that some aspect of the function of p53 is regulatory, possibly in ironing out abnormal secondary structure before additional recombination enzymes are called upon to irreversibly act on the DNA. An alternative model would place p53 as a governor of the enzymes responsible for clipping off the arm of the hairpin. In this model, the loss of p53 would allow repair enzymes to resolve secondary structures by recombination. It is of note that this is the situation observed in lower eukaryotes, where large inverted repeats are rapidly excised to minimize abnormal secondary structures that may interfere with replication and transcription.
Unlike lower eukaryotes, the human genome contains many repetitive sequences that are prime candidates to generate abnormal secondary structures, yet it is relatively stable. The data reported here suggest that not only are there conserved mechanisms to resolve this secondary structure but also these putative hairpins are well tolerated in p53 wild-type cells. It can be speculated that the loss of p53 function may result in a profound mutator phenotype due to the lack of tolerance to the misalignment of repetitive DNA, which results in resolution via deleterious repair. Hence, the preponderance of mutated p53 in cancer cells not only may be a result of the inability of the cells to arrest after DNA damage, or apoptose, but also may be a consequence of uncontrolled, evolutionarily conserved mechanisms to handle problematic repetitive DNA in the genome.
ACKNOWLEDGMENTS
We thank L. Reid for the kind gift of PNI2EX2, and we thank J. Haber and J. Nickoloff for helpful advice during the course of these experiments. Thanks are also due to J. B. Little, J. Carbon, and K. K. Hancock.
This work was supported by NIH grant CA49696. D.G. was supported by NIH training grant CA09078.
REFERENCES
- 1.Bakalkin G, Selivanova G, Yakovleva T, Kiseleva E, Kashuba E, Magnusson K P, Szekely L, Klein G, Terenius L, Wiman K G. p53 binds single-stranded DNA ends through the C-terminal domain and internal DNA segments via the middle domain. Nucleic Acids Res. 1995;23:362–369. doi: 10.1093/nar/23.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakalkin G, Yakovleva T, Selivanova G, Magnusson K P, Szekely L, Kiseleva E, Klein G, Terenius L, Wiman K G. p53 binds single-stranded DNA ends and catalyzes DNA renaturation and strand transfer. Proc Natl Acad Sci USA. 1994;91:413–417. doi: 10.1073/pnas.91.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertrand P, Rouillard D, Boulet A, Levalois C, Soussi T, Lopez B S. Increase of spontaneous intrachromosomal homologous recombination in mammalian cells expressing a mutant p53 protein. Oncogene. 1997;14:1117–1122. doi: 10.1038/sj.onc.1200931. [DOI] [PubMed] [Google Scholar]
- 4.Bouffler S D, Kemp C J, Balmain A, Cox R. Spontaneous and ionizing radiation-induced chromosomal abnormalities in p53-deficient mice. Cancer Res. 1995;55:3883–3889. [PubMed] [Google Scholar]
- 5.Brunier D, Michel B, Ehrlich S D. Copy choice illegitimate DNA recombination. Cell. 1988;52:883–892. doi: 10.1016/0092-8674(88)90430-8. . (Retracted by S. D. Ehrlich and B. Michel, 62:409, 1990.) [DOI] [PubMed] [Google Scholar]
- 6.Buchhop S, Gibson M K, Wang X W, Wagner P, Sturzbecher H W, Harris C C. Interaction of p53 with the human Rad51 protein. Nucleic Acids Res. 1997;25:3868–3874. doi: 10.1093/nar/25.19.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Kolodner R D. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 8.Cooper D N. Human gene mutation. Oxford, United Kingdom: BIOS; 1993. [Google Scholar]
- 9.Dutta A, Ruppert J M, Aster J C, Winchester E. Inhibition of DNA replication factor RPA by p53. Nature. 1993;365:79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- 10.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 11.Gordenin D A, Lobachev K S, Degtyareva N P, Malkova A L, Perkins E, Resnick M A. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol Cell Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordenin D A, Resnick M A. Yeast ARMs (DNA at-risk motifs) can reveal sources of genome instability. Mutat Res. 1998;400:45–58. doi: 10.1016/s0027-5107(98)00047-5. [DOI] [PubMed] [Google Scholar]
- 13.Habraken Y, Sung P, Prakash L, Prakash S. Holliday junction cleavage by yeast Rad1 protein. Nature. 1994;371:531–534. doi: 10.1038/371531a0. [DOI] [PubMed] [Google Scholar]
- 14.Lee S, Cavallo L, Griffith J. Human p53 binds Holliday junctions strongly and facilitates their cleavage. J Biol Chem. 1997;272:7532–7539. doi: 10.1074/jbc.272.11.7532. [DOI] [PubMed] [Google Scholar]
- 15.Levine A J, Momand J, Finlay C A. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 16.Lewis L K, Westmoreland J W, Resnick M A. Repair of endonuclease-induced double-strand breaks in Saccharomyces cerevisiae. Essential role for genes associated with nonhomologous end-joining. Genetics. 1999;152:1513–1529. doi: 10.1093/genetics/152.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lilley D M. In vivo consequences of plasmid topology. Nature. 1981;292:380–382. doi: 10.1038/292380a0. [DOI] [PubMed] [Google Scholar]
- 18.Lin D, Shields M T, Ullrich S J, Appella E, Mercer W E. Growth arrest induced by wild-type p53 protein blocks cells prior to or near the restriction point in late G1 phase. Proc Natl Acad Sci USA. 1992;89:9210–9214. doi: 10.1073/pnas.89.19.9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 20.Lobachev K S, Shor B M, Tran H T, Taylor W, Keen J D, Resnick M A, Gordenin D A. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics. 1998;148:1507–1524. doi: 10.1093/genetics/148.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 22.Lu X, Lane D P. Differential induction of transcriptionally active p53 following UV or ionizing radiation: defects in chromosome instability syndromes? Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 23.Maryon E, Carroll D. Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: evidence for a nonconservative mechanism of homologous recombination. Mol Cell Biol. 1991;11:3278–3287. doi: 10.1128/mcb.11.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mekeel K L, Tang W, Kachnic L A, Luo C M, DeFrank J S, Powell S N. Inactivation of p53 results in high rates of homologous recombination. Oncogene. 1997;14:1847–1857. doi: 10.1038/sj.onc.1201143. [DOI] [PubMed] [Google Scholar]
- 25.Meyn M S. High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science. 1993;260:1327–1330. doi: 10.1126/science.8493577. [DOI] [PubMed] [Google Scholar]
- 26.Morris T, Thacker J. Formation of large deletions by illegitimate recombination in the HPRT gene of primary human fibroblasts. Proc Natl Acad Sci USA. 1993;90:1392–1396. doi: 10.1073/pnas.90.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mummenbrauer T, Janus F, Muller B, Wiesmuller L, Deppert W, Grosse F. p53 Protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85:1089–1099. doi: 10.1016/s0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- 28.Muris D F, Bezzubova O, Buerstedde J M, Vreeken K, Balajee A S, Osgood C J, Troelstra C, Hoeijmakers J H, Ostermann K, Schmidt H, et al. Cloning of human and mouse genes homologous to RAD52, a yeast gene involved in DNA repair and recombination. Mutat Res. 1994;315:295–305. doi: 10.1016/0921-8777(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 29.Ozenberger B A, Roeder G S. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol. 1991;11:1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park C H, Sancar A. Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision repair proteins. Proc Natl Acad Sci USA. 1994;91:5017–5021. doi: 10.1073/pnas.91.11.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips E N, Xia F, Kelsey K T, Liber H L. Spectra of spontaneous and X-ray-induced mutations at the hprt locus in related human lymphoblast cell lines that express wild-type or mutant p53. Radiat Res. 1995;143:255–262. [PubMed] [Google Scholar]
- 32.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 33.Roth D B, Wilson J H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubnitz J, Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984;4:2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Z, Pardington-Purtymun P E, Comeaux J C, Moyzis R K, Chen D J. Associations of UBE2I with RAD52, UBL1, p53, and RAD51 proteins in a yeast two-hybrid system. Genomics. 1996;37:183–186. doi: 10.1006/geno.1996.0540. [DOI] [PubMed] [Google Scholar]
- 36.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. . (Erratum, 5:312, 1993.) [DOI] [PubMed] [Google Scholar]
- 37.Sijbers A M, de Laat W L, Ariza R R, Biggerstaff M, Wei Y F, Moggs J G, Carter K C, Shell B K, Evans E, de Jong M C, Rademakers S, de Rooij J, Jaspers N G, Hoeijmakers J H, Wood R D. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 38.Sturzbecher H W, Donzelmann B, Henning W, Knippschild U, Buchhop S. p53 is linked directly to homologous recombination processes via RAD51/RecA protein interaction. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 39.Warren G J, Green R L. Comparison of physical and genetic properties of palindromic DNA sequences. J Bacteriol. 1985;161:1103–1111. doi: 10.1128/jb.161.3.1103-1111.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X, Lieber M R. Protein-protein and protein-DNA interaction regions within the DNA end-binding protein Ku70-Ku86. Mol Cell Biol. 1996;16:5186–5193. doi: 10.1128/mcb.16.9.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia F, Wang X, Wang Y H, Tsang N M, Yandell D W, Kelsey K T, Liber H L. Altered p53 status correlates with differences in sensitivity to radiation-induced mutation and apoptosis in two closely related human lymphoblast lines. Cancer Res. 1995;55:12–15. [PubMed] [Google Scholar]
- 42.Yu Y, Li C Y, Little J B. Abrogation of p53 function by HPV16 E6 gene delays apoptosis and enhances mutagenesis but does not alter radiosensitivity in TK6 human lymphoblast cells. Oncogene. 1997;14:1661–1667. doi: 10.1038/sj.onc.1201026. [DOI] [PubMed] [Google Scholar]
- 43.Zhan Q, Bae I, Kastan M B, Fornace A J., Jr The p53-dependent gamma-ray response of GADD45. Cancer Res. 1994;54:2755–2760. [PubMed] [Google Scholar]
- 44.Zheng G X, Sinden R R. Effect of base composition at the center of inverted repeated DNA sequences on cruciform transitions in DNA. J Biol Chem. 1988;263:5356–5361. [PubMed] [Google Scholar]