ABSTRACT
Recombinant adeno-associated virus (rAAV) vectors are one of the leading tools for the delivery of therapeutic genes in human gene therapy applications. For a successful transfer of their payload, the AAV vectors have to circumvent potential preexisting neutralizing host antibodies and bind to the receptors of the target cells. Both of these aspects have not been structurally analyzed for AAVrh.10. Here, cryo-electron microscopy and three-dimensional image reconstruction were used to map the binding site of sulfated N-acetyllactosamine (LacNAc; previously shown to bind AAVrh.10) and a series of four monoclonal antibodies (MAbs). LacNAc was found to bind to a pocket located on the side of the 3-fold capsid protrusion that is mostly conserved to AAV9 and equivalent to its galactose-binding site. As a result, AAVrh.10 was also shown to be able to bind to cell surface glycans with terminal galactose. For the antigenic characterization, it was observed that several anti-AAV8 MAbs cross-react with AAVrh.10. The binding sites of these antibodies were mapped to the 3-fold capsid protrusions. Based on these observations, the AAVrh.10 capsid surface was engineered to create variant capsids that escape these antibodies while maintaining infectivity.
IMPORTANCE Gene therapy vectors based on adeno-associated virus rhesus isolate 10 (AAVrh.10) have been used in several clinical trials to treat monogenetic diseases. However, compared to other AAV serotypes little is known about receptor binding and antigenicity of the AAVrh.10 capsid. Particularly, preexisting neutralizing antibodies against capsids are an important challenge that can hamper treatment efficiency. This study addresses both topics and identifies critical regions of the AAVrh.10 capsid for receptor and antibody binding. The insights gained were utilized to generate AAVrh.10 variants capable of evading known neutralizing antibodies. The findings of this study could further aid the utilization of AAVrh.10 vectors in clinical trials and help the approval of the subsequent biologics.
KEYWORDS: AAVrh.10, adeno-associated virus, antibody, capsid, cryo-EM, galactose, gene therapy, glycan, keratan sulfate, receptors
INTRODUCTION
Adeno-associated viruses (AAVs) are small nonenveloped viruses of the family Parvoviridae that package linear single-stranded DNA (ssDNA) genomes of ∼4.7 kb (1). These viruses are extensively studied due to their utilization as vectors for gene delivery applications, particularly for the treatment of monogenetic diseases. Thus far, three gene therapy biologics based on AAV vectors have been approved for commercialization: Glybera, an AAV1 vector for the treatment of lipoprotein lipase deficiency by the European Medicines Agency (EMA) in 2012 (2); Luxturna, an AAV2 vector for the treatment of Leber’s congenital amaurosis in 2018 (3); and Zolgensma, an AAV9 vector for the treatment of spinal muscular atrophy type 1 in 2019 (4), both by the Food and Drug Administration (FDA) and EMA.
The AAVs are composed of T=1 icosahedral capsids with a diameter of ∼260 Å that contain the ssDNA genome (5). The capsids are assembled from 60 overlapping viral proteins (VPs): VP1 (∼82 kDa), VP2 (∼73 kDa), and VP3 (∼61 kDa) that are incorporated in an approximate stochastic ratio of 1:1:10 (6). The VPs share a C terminus that includes the entirety of VP3. VP1 and VP2 are N-terminal extended forms of VP3 that contain a phospholipase A2 (PLA2) domain, a calcium-binding domain, and nuclear localization signals, all of which are required for infectivity (7, 8).
Thirteen human and primate AAV serotypes, and hundreds of genomic isolates from various primate and nonprimate species, have been described (9–11). The amino acid sequence identity of the VPs can vary by ∼50%. The three-dimensional (3D) structures of numerous AAV capsids have been determined by X-ray crystallography and/or cryo-electron microscopy and image reconstruction (cryo-EM) (12–18). Regardless of the method of structure determination, only the common VP3 region, with the exception of the first ∼15 amino acids, is ordered and structurally observed. Each VP consists of a core eight-stranded antiparallel β-barrel motif (βB to βI), with the BIDG β-sheet forming the capsid interior surface. An additional β-strand, βA, is situated antiparallel to the βB strand. Furthermore, all VPs have an alpha helix (αA) located between the βC and βD strands. Connecting the core β-strands large are insertion loops that exhibit sequence and structure variability that are responsible for the AAV serotype-specific exterior capsid surface features. These loops are named after their flanking β-strands (e.g., HI loop, between the βH and βI strands) and nine regions of significant structural diversity have been defined as variable regions (VR I to IX), by structural alignment comparison (19).
The capsids are assembled via the icosahedral 2-, 3-, and 5-fold symmetry-related VP interactions (20). Several AAV capsid features have been described, including: cylindrical channels running radially to the 5-fold axis, assembled by five DE loops creating pores between interior and exterior of the capsid, depressions located at the 2-fold axis, protrusions surrounding the 3-fold axis, and raised regions between the 2- and 5-fold axes that are termed 2/5-fold walls.
While these general surface features are conserved among all AAV capsids, the precise capsid structure varies due to differences in the VR surface loop conformations. Concomitant with the amino acid and structural variation of the AAV serotype capsids are differences in binding to host cell receptors, thereby determining tissue tropism, and also differences in their antigenic profiles. For the AAV serotypes various receptors have been identified, such as heparan sulfate proteoglycan, e.g., for AAV2, AAV3, AAV6, and AAV13 (21–25); sialic acids, e.g., AAV1, AAV4, AAV5, and AAV6 (26–28); terminal galactose glycans for AAV9 (29, 30); and sulfated N-acetyllactosamine (LacNAc) for AAV rhesus isolate 10 (AAVrh.10) (31). Furthermore, a series of protein receptors have been described, e.g., AAVR (32), αvβ1 integrin (33), αvβ5 integrin (34), laminin (35), the hepatocyte growth factor receptor (36), the fibroblast growth factor receptor (37), and platelet-derived growth factor receptor (38). For some of these receptors, the binding site on the AAV capsid surface has been mapped and characterized either by substitution of critical residues required for receptor binding (39–41) or by determination of the capsid structure with the receptor bound (42–45).
Furthermore, several antibodies raised by the host immune response have been shown to bind to the capsid surfaces (46). For the AAVs the antibody seroprevalence in the human population varies for the different serotypes and has been reported to be as high as 80% in the case of AAV2 (47, 48). This is significant, since these preexisting antibodies can severely reduce the desired therapeutic effect or completely prevent a patient from receiving an AAV-based gene therapy vector. Thus, efforts to characterize the antigenic regions of the AAV capsids are invested in order to generate or identify AAV capsid variants capable of escaping preexisting neutralizing antibodies (NAbs). Previously, the binding sites of multiple monoclonal antibodies to the capsids of AAV1, AAV2, AAV4, AAV5, AAV6, AAV8, and AAV9 have been described (46, 49) that resulted in the generation of new capsid variants either by rational design or directed evolution to minimize the impact of preexisting NAbs (49–51).
Vectors based on AAVrh.10 are being used in several ongoing clinical trials (52, 53). However, many aspects with regard to AAVrh.10 receptor and antigenic interactions have not been fully analyzed. In this study, the binding site of a sulfated LacNAc on the AAVrh.10 capsid has been identified by cryo-EM and mapped to a pocket at the side of the 3-fold protrusions. This site is equivalent to the galactose-binding pocket of the AAV9 capsid, and since critical amino acids in this pocket are conserved, it was shown AAVrh.10 can also bind to terminal galactose cell surface glycans. Overall, the AAVrh.10 capsid shares high structural similarity to AAV8, and a screening of five monoclonal antibodies (MAbs) developed for AAV8 capsids showed that four cross-reacted and neutralized AAVrh.10. The binding sites of these four MAbs to the AAVrh.10 capsid were also identified by cryo-EM and mapped to the 3-fold protrusions. The amino acid contact information was used to develop AAVrh.10 variants capable of evading these NAbs.
RESULTS AND DISCUSSION
The side of the 3-fold protrusions serve as the LacNAc binding site.
A synthetic sulfated LacNAc tetrasaccharide, imitating natural keratan sulfate (KS) glycosaminoglycans, has been shown previously to interact with AAVrh.10 capsids using glycan array screening (31). Sulfated LacNAc (∼1 kDa) was added to purified AAVrh.10 capsids at a molecular ratio of 6,000 glycan molecules per capsid, which corresponds to a 100-fold excess of the glycan per VP binding site. Subsequently, cryo-EM data were collected on the vitrified complex sample and a 3D-image reconstruction conducted to locate the binding site of the glycan on the capsid. The capsid-glycan complex was reconstructed to 3.09-Å (FSC 0.143) resolution from 142,036 capsids (Table 1), and the resulting density map (Fig. 1A) looked very similar to the previously described AAVrh.10 capsid structure (54). At this resolution, amino acid side chains were clearly distinguishable, and the subsequent model built was nearly identical to the deposited AAVrh.10 capsid structure (PDB 6O9R) with an overall Cα-RMSD of <0.1 Å. However, additional density was clearly observed between amino acids N472 and W505 at a sigma (σ) level threshold of 1.0 (Fig. 1B) located on the side of the 3-fold protrusions (Fig. 1C).
TABLE 1.
Summary of data collection and image-processing parameters
| Parameter | AAVrh.10-7x | AAVrh.10:LacNAc |
|---|---|---|
| Data collection statistics | ||
| Total no. of micrographs | 589 | 1,437 |
| Defocus range (μm) | 0.95–3.24 | 0.99–4.05 |
| Total electron dose (e−/Å2) | 75 | 60 |
| Frames/micrograph | 50 | 43 |
| Pixel size (Å/pixel) | 1.05 | 0.97 |
| No. of capsids used for final map | 17,861 | 142,036 |
| Resolution of final map (Å) | 2.71 | 3.09 |
| Refinement statistics | ||
| Map CC | 0.870 | 0.851 |
| RMSD | ||
| Bonds (Å) | 0.01 | 0.01 |
| Angles (°) | 0.82 | 0.86 |
| All-atom clash score | 7.52 | 8.20 |
| Ramachandran plot (%) | ||
| Outliers | 0 | 0 |
| Allowed | 1.4 | 1.5 |
| Favored | 98.6 | 98.5 |
| Rotamer outliers (%) | 0 | 0 |
| C-beta deviations | 0 | 0 |
FIG 1.
Characterization of the AAVrh.10 glycan binding site. (A) Reconstructed capsid surface map colored according to radial distance (Å) from the capsid center (blue) to outer surface region (red), as indicated by the scale bar on the left. The positions of the 5-fold channel, 3- and 2-fold axes, and 2/5-fold wall are indicated. Symbols: blue square, N-acetylglucosamine; yellow circle, galactose. (B) Amino acid residues 471 to 474 and 504 to 506, including a galactose (GAL) molecule inside the density map at a sigma (σ) threshold level of 1.0, are depicted as a black mesh. The model is shown in stick representation, and the atoms are colored: C, yellow; O, red; N, blue; and S, green. Panels A and B were generated with Chimera (80). (C) Surface representation of the AAVrh.10 capsid with the positions of N472 (green) and W505 (yellow) highlighted. The positions of the 5-fold, 3-fold, and 2-fold axes are indicated. This image was generated with PyMOL (85). (D) Transduction efficiency of wtAAVrh.10, AAVrh.10-N472A, or AAVrh.10-W505A (all packaging the luciferase gene) in Pro5 and Lec2 cells. The experiments were determined by luciferase reporter gene expression and performed in triplicate and are displayed as means plus the standard deviations (SD) (n = 3). Symbols: blue square, N-acetylglucosamine; yellow circle, galactose; pink diamond, sialic acid. (E) Relative transduction efficiencies of AAV2, AAV5, AAV9, AAV8, and AAVrh.10, or variants thereof, in Pro5 and Lec2 cells, determined as for panel D.
The AAVrh.10 sulfated LacNAc binding site is located in the analogous position to the reported galactose binding site of AAV9 (40) that has both the asparagine and tryptophan amino acids in structurally equivalent positions. In addition, since the terminal saccharide of the LacNAc glycan is also a galactose (Fig. 1A), these observations suggested that the density observed between N472 and W505 could be the terminal galactose (Fig. 1B). No other interpretable density was observed, most likely due to the flexibility of the tetrasaccharide linkage causing the structure to be disordered. While amino acid W505 is conserved in almost all AAV serotypes, except for AAV4, −5, −11, and −12, and amino acid N472 is only present in AAV9 and AAVrh.10. Amino acid substitutions at either position 472 or position 505 to alanine in AAV9 have been shown previously to reduce the virus transduction efficiency by 50 to 70% in the case of N470A (AAV9 numbering) or >90% in the case of W503A (AAV9 numbering), preventing binding of the terminal galactose (40). Similarly, amino substitutions of the tryptophan in nongalactose binders such as AAV1, AAV2, or AAV6 also significantly reduced the virus transduction efficiency (44, 55). In the case of AAV1 and AAV6, the tryptophan participates in sialic acid binding (44) and also in adeno-associated virus receptor (AAVR) binding for most serotypes except AAV4, AAV5, and likely AAV11 and AAV12 (42, 43, 56).
In order to determine the importance of these residues for AAVrh.10, both amino acids were changed to alanine, and the infectivity of AAVrh.10 and the variants was studied in Chinese hamster ovary (CHO) Pro5 and Lec2 cells that display alternative cell surface glycan profiles. While Pro5 cells display glycans with terminal sialic acids, the Lec2 cells have a mutation in a sialic acid transporter causing cell surface glycans to terminate in galactoses (Fig. 1D) (57). When comparing the transduction level of AAVrh.10 vectors in both cell lines, the transduction efficiency was slightly higher in Lec2 cells by a factor of ca. 1.5- to 2-fold. The observed increase in transduction in Lec2 cells could indicate that AAVrh.10 utilizes terminal galactoses for cell attachment. However, the enhanced transduction efficiency in Lec2 cells is lower compared to AAV9, with an ∼4-fold-higher transduction efficiency in Lec2 cells versus Pro5 (Fig. 1E). Nonetheless, galactose binding of AAVrh.10 is further supported by the N472A variant that resulted in an ∼50% reduction of transduction efficiency in Pro5 cells but showed comparable transduction levels of the variant in Lec2 cells (Fig. 1D). While the change of the asparagine to alanine probably abrogated galactose binding, the moderate decrease in transduction efficiency also indicated the presence of an alternative receptor. In contrast, the AAVrh.10 W505A variant resulted in a severe loss of transduction ability in both cell lines (Fig. 1D). Although the substitution of the tryptophan might impact galactose binding, it is highly likely that this amino acid substitution affects other functions of the capsid such as AAVR binding since the equivalent tryptophan to alanine variant in other non-galactose-binding AAV serotypes, including AAV8 (Fig. 1E), also led to reduced transduction efficiencies (44, 55).
AAV8 is structurally very similar to AAVrh.10 (54) but did not bind to the sulfated LacNAc on the glycan array (31) and showed no significant transduction differences in Pro5 and Lec2 cells, similar to AAV2 (Fig. 1E). This implies the utilization of an alternate cell surface receptor of these AAV serotypes, such as HSPG in the case of AAV2 (21). Interestingly, compared to AAVrh.10 and AAV9, AAV8 has a threonine at position 472, but introduction of an asparagine (T472N) to AAV8 did not result in a gain of function or variation of transduction compared to the wild-type capsid (Fig. 1E). Similarly, the transduction efficiency of the AAV8 T472A variant is largely unaffected.
Keratan sulfate affects AAVrh.10 transduction.
A previous study that analyzed the transduction efficiency of various AAV capsids in mouse cornea showed enhanced transduction of AAVrh.10 compared to most of AAV serotypes tested (58). Particularly, high transduction in keratocytes that highly express keratocan was observed. Keratocan is a proteoglycan that displays keratan sulfate chains. The fact that AAVrh.10 also bound to sulfated LacNAc on a glycan array (31) was a strong indicator that keratan sulfate was a potential receptor for the capsid.
To determine whether soluble keratan sulfate, derived and purified from chicken egg white (59), affects the transduction of AAVrh.10 and other AAV serotypes, competition experiments with keratan sulfate were performed. Surprisingly, up to an ∼3-fold enhancement of AAVrh.10 transduction was observed with increasing concentration of keratan sulfate (Fig. 2). At the same time, AAV8 transduction was also enhanced, whereas AAV5 transduction seemed to be largely unaffected by the glycan. In contrast, transduction of AAV2 was ∼70% reduced in the presence of high concentrations of keratan sulfate (Fig. 2). This observation was not surprising since other glucosamine glycans, such as dermatan sulfate and dextran sulfate, were previously also able to partially inhibit AAV2 transduction (21). However, the unexpected enhancement is more challenging to explain. Keratan sulfate, which is most abundant in the human cornea and the brain, has multiple functions and interacts with many proteins, including membrane proteins (60–62). It is possible that the keratan sulfate binds to the cells and capsid simultaneously, thereby facilitating cellular attachment of these viruses.
FIG 2.
Analysis of the effect of keratan sulfate on AAV transduction. The relative transduction efficiencies of AAV2, AAV5, AAV8, and AAVrh.10 (all packaging the luciferase gene) were determined in the presence of increasing concentrations of keratan sulfate. The experiments were determined from luciferase reporter gene expression in HEK293 cells, performed in triplicate, and are displayed as means plus the SD (n = 3). Symbols: blue square, N-acetylglucosamine; yellow circle, galactose.
Similar to the LacNAc tetrasaccharide, AAVrh.10 capsids were complexed with the long-chain keratan sulfate, and cryo-EM data were collected on the sample. The 3D reconstructed map of this complex at a resolution of 3.15 Å also showed some density between N472 and W505 but with significantly more structural disorder (data not shown). This implies that keratan sulfate might be a weak binder to AAVrh.10 capsids.
AAV8 and AAVrh.10 share antigenic properties.
AAVrh.10 was originally isolated from nonhuman primates. However, analyses of human sera have shown that up to ∼60% possess antibodies against AAVrh.10 (63). This high percentage can likely be explained by the existence of very closely related AAV variants of clade E that have been found in human tissues, such as AAVhu.6 that differs by only six amino acids from the AAVrh.10 VP1 protein sequence (9). In addition, AAV8, the prototype member of clade E, differs by 48 amino acids from AAVrh.10, and their capsid structures have been shown to be similar (54).
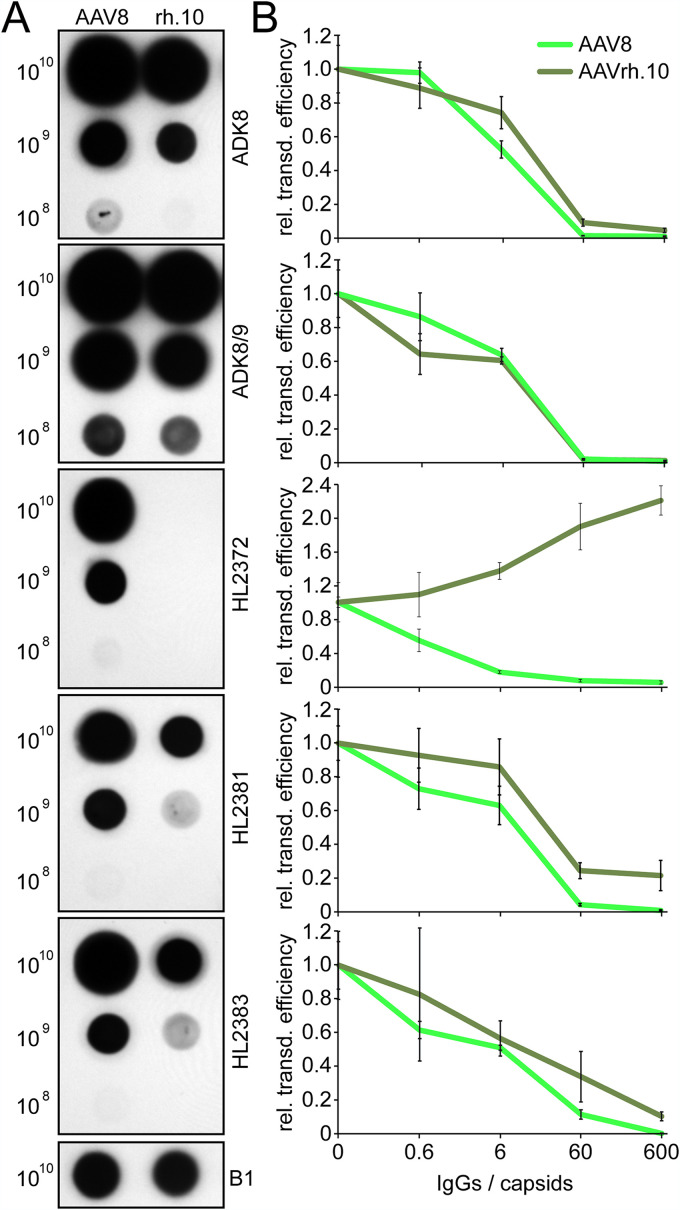
For the AAV8 capsid a series of MAbs (ADK8, ADK8/9, HL2372, HL2381, and HL2383) have been developed (64, 65), and their binding epitopes have been mapped (49). Hence, their cross-reactivity to the AAVrh.10 capsid was evaluated by native dot blot analysis. Four of five AAV8 MAbs showed reactivity to AAVrh.10, while HL2372 did not (Fig. 3A). This antibody was shown previously to bind to the 5-fold region of the AAV8 capsid (49). Despite the absence of structural differences around the 5-fold channel, the different amino acid compositions of the DE and HI loop are likely responsible for the differential binding observed for the two capsids. The remaining four antibodies did not just bind to the AAVrh.10 capsid but were also able to neutralize AAVrh.10 vectors in a cell transduction assay (Fig. 3B). For all of these antibodies, significant levels of neutralization were observed at a concentration equivalent to 60 IgGs and higher per capsid.
FIG 3.
AAV8 MAbs cross-react with AAVrh.10 capsids. (A) Native dot blot analysis of AAV8 and AAVrh.10. The amounts of loaded capsids are given on the left side, and the those for the utilized antibody are given on the right side. The B1 antibody was used on a blot with denatured capsids as a positive control. (B) Neutralization assay of AAV8 and AAVrh.10 in HEK293 cells. Preincubation was done with increasing amounts of indicated antibodies relative to the capsids as indicated. The experiments were determined using luciferase reporter gene expression, and the results are shown relative to the luciferase expression in the absence of the antibodies. The experiments were performed in triplicate and represent means plus the SD (n = 3).
The AAV8 MAbs bind to the 3-fold protrusions of the AAVrh.10 capsid.
In order to determine the binding sites of the AAV8 MAbs to the AAVrh.10 capsid, cryo-EM and image reconstruction were used. For this purpose, purified capsids were mixed with purified Fabs (fragment antigen-binding) at a molecular ratio of 1:120 (capsid to antibody) and cryo-EM images collected of the complexes. For the ADK8, ADK8/9, HL2381, and HL2383 AAVrh.10 complexes, maps were reconstructed from ∼3,500 to ∼6,900 individual capsid-Fab complexes to a approximately 6.2- to 7.1-Å resolution (Fig. 4 and Table 2). The binding footprints of these antibodies had been previously mapped for the AAV8 capsid (49). While ADK8, HL2381, and HL2383 were shown to recognize the top of the protrusions surrounding the 3-fold axis, ADK8/9 was shown to bind at the side of the 3-fold protrusions and the 2/5-fold wall toward the 2-fold axis. As expected, the AAV8 MAbs bound to the AAVrh.10 capsid in a similar manner (Fig. 4A). Pseudo-atomic models of the Fabs were fitted into the complex density maps resulting in correlation coefficients ranging from 0.72 to 0.89 (Table 1). ADK8, HL2381, and HL2383 bound identically to the 3-fold protrusions of AAVrh.10 with contacts to VR-IV, -V, and -VIII (Fig. 4B). Previously, amino acid substitutions in these surface loops confirmed that residues in VR-VIII are responsible for ADK8 binding and not VR-IV or VR-V (66). Furthermore, an AAV8 capsid variant, Hum8, was generated that was engineered through structure-guided evolution, which, among others, contains amino acid substitutions in VR-VIII and thus enabled an escape phenotype from the neutralizing antibodies (49, 67). This variant was also shown to escape ADK8/9 neutralization, which showed potential contact residues in VR-I, -V, and -VIII (Fig. 4B). Although all of the MAbs neutralize AAVrh.10 transduction (Fig. 3B), none of the MAb epitopes overlap the LacNAc binding pocket (Fig. 1B and C). Thus, these MAbs might not block receptor binding of AAVrh.10 to the cells and instead affect a postentry step. A similar antibody, MAb A20, binds at the 2/5-fold wall of the AAV2 capsid apart from its heparin-binding site and neutralization of viral transduction occurs at a postentry step (39, 68, 69).
FIG 4.
AAVrh.10-MAb complex interactions. (A) 3D reconstruction maps of AAVrh.10 (uncomplexed) and of AAVrh.10:ADK8, ADK8/9, HL2381, and HL2383 complexes. The maps are radially colored according to distance to the capsid center (blue to red, as indicated by the scale bar on the left). The icosahedral 2-, 3-, and 5-fold axes, the 2/5-fold wall, and AVB are indicated on the uncomplexed AAVrh.10 capsid map. (B) The AAVrh.10 surface loops (dark green) and Fab models (orange) are shown as ribbons inside their black mesh density maps. The VRs involved in the capsid-Fab interaction are indicated.
TABLE 2.
Summary of data collection for AAVrh.10-Fab complexes
| Parameter | AAVrh.10: ADK8 | AAVrh.10: ADK8/9 | AAVrh.10: HL2381 | AAVrh.10: HL2383 |
|---|---|---|---|---|
| No. of micrographs | 50 | 50 | 50 | 50 |
| Defocus range (μm) | 1.00–4.50 | 1.40–4.69 | 1.27–4.40 | 1.00–4.50 |
| No. of particles | 6,536 | 3,496 | 4,337 | 6,870 |
| Pixel size (Å/pixel) | 1.83 | 1.83 | 1.83 | 1.83 |
| Resolution (Å) | 6.60 | 6.45 | 6.21 | 7.10 |
| CC of Fab in map | 0.72 | 0.89 | 0.78 | 0.76 |
Generation of AAVrh.10 antibody-escape variants.
In order to generate AAVrh.10 variants capable of escaping the AAV8 MAbs a rational design approach was conducted. As previously shown, the AAV8 MAbs, with the exception of ADK8/9, also cross-reacted with AAV3 and AAV7 (64, 70) (Fig. 5A). A selection of other antibodies detecting conformational epitopes on the capsid surface in AAV9 (ADK9) (65) and in AAV2, AAV3, and AAV13 (A20) (68) did not cross-react with AAVrh.10. Due to the known importance of VR-VIII for the binding of this set of AAV8 antibodies, the amino acid sequences of the binding AAV serotypes were analyzed. Furthermore, VR-VIII displays generally high structural similarity among most AAV serotypes (12, 14). Thus, it is highly likely that the antibodies bind comparably to these serotypes. Among the ADK8-binding AAV serotypes, an asparagine in position 590 and an alanine in position 592 (AAVrh.10 numbering) appeared to be conserved (Fig. 5B). Substituting these residues with a serine or glutamine, respectively, significantly reduced the ability of ADK8, HL2381, and HL2383 to bind these AAVrh.10 variants, confirming their importance in the binding of the antibodies (Fig. 5A). The combination of both amino acid substitutions further reduced the binding affinity of these MAbs. It is worth noting that ADK8, HL2381, and HL2383 were all developed independently of each other (64, 65), but yet they bind identically. This indicated that the VR-VIII loop is a “hot spot” for antibody binding recognition for AAV8 and AAVrh.10.
FIG 5.
Generation of AAVrh.10 escape variants. (A) Immune-dot blot analysis of native rAAV capsids of the indicated AAV serotypes or variants thereof spotted on a nitrocellulose membrane. The membranes were incubated with a panel of MAbs, as indicated to the right of the membrane. MAb B1 served as an internal loading control with denatured capsids. (B) Sequence alignment for the VR-VIII loop of a selection of AAV serotypes. Amino acids highlighted in yellow indicate sequence identity among all of the AAVs. In addition, amino acids in green indicate common residues among the ADK8-binding AAV serotypes in and residues in dark green among the ADK8/9-binding AAVs. (C) Amino acid substitutions of the 3x and 7x variant of AAVrh.10. (D) Transduction efficiency of the different AAVrh.10 variants in HEK293 cells. (E) Relative transduction efficiency of the AAVrh.10 variants in the presence of increasing concentrations of the different MAbs. The experimental results displayed in panels D and E were obtained from studies to determine luciferase reporter gene expression that were performed in triplicate; the data indicate means plus the SD (n = 3).
For the ADK8/9 antibody binding footprint, a conserved residue between AAV8 and AAVrh.10 is Q589. Substituting this residue to an asparagine did not prevent ADK8/9 binding by itself. However, adding this amino acid substitution in the context of the previous N590S/A592Q variant (i.e., the 3x variant, Fig. 5C) resulted in the inability of ADK8/9 to bind this variant (Fig. 5A). Interestingly, none of the individual amino acid substitutions or the N590S/A592Q variant was able to prevent ADK8/9 binding, indicating that the antibody potentially makes multiple contacts to this region.
While all of the AAV8 MAbs bound to VR-VIII of AAVrh.10, other antibodies might have binding sites to other regions of the capsid surface. Thus, additional compatible variants resulting in infectious AAVrh.10 particles were introduced into known antigenic regions of the capsid surface loops (46). The resulting AAV variant harbors a total of seven amino acid substitutions in comparison to the wild-type capsid (i.e., the 7x variant, Fig. 5C). The substitutions are located in VR-III (S387A), in VR-IV (ΔS453), in VR-VII (S559A), and in VR-IX (T719V). The 7x variant did not display any additional benefit when screened against the MAbs on the native dot blot (Fig. 5A). The transduction efficiencies of the 3x and 7x variants were compared to AAVrh.10-WT to confirm that they retained their infectivity. While the 3x variant displayed a comparable transduction efficiency to AAVrh.10-WT, the 7x variant maintained ∼70% of the infectivity of the WT capsid (Fig. 5D).
In order to confirm that the newly generated AAVrh.10 variants escaped the panel of antibodies tested and were capable of infecting cells in their presence, in vitro neutralization assays were performed. For this purpose, purified capsids were preincubated with various amounts of purified IgGs prior to infection. As seen before (Fig. 3B), AAVrh.10 vectors were nearly or completely neutralized in the presence of 60 IgGs for each of the four antibodies tested (Fig. 5E). The 3x variant escaped ADK8, HL2381, and HL2383 completely, even in the presence of 600 IgGs per capsids. For ADK8/9 neutralization of the 3x variant was observed in the presence of the highest concentration of ADK8/9 tested (Fig. 5E). However, compared to the wild-type AAVrh.10, a significantly higher concentration of antibodies was needed for neutralization since no measurable reduction of transduction was observed with 60 IgGs per capsid. This indicates a residual, weak ADK8/9 binding affinity of the 3x variant capsid. The 7x variant behaved similarly to the 3x variant but maintained ∼66% of its transduction efficiency in the presence of the highest ADK8/9 concentration tested (Fig. 5E). Interestingly, a slight enhancement of transduction is often seen at high antibody concentrations if these are unable to neutralize the capsids (Fig. 3B and 5E). While the reason for this observation is unknown, it is likely an unspecific effect since this enhancement has also been observed with other AAV serotypes and various antibodies (49, 50).
The 3x and 7x AAVrh.10 variants display different capsid melting temperatures.
The newly generated AAVrh.10 variants showed production efficiencies comparable to that of the wild-type capsid (data not shown) and can be readily purified on AVB columns (Fig. 6A), since the amino acid substitutions do not affect the 5-fold region where the nanobody of the affinity ligand binds to the capsid (71). For further characterization of the different capsid variants, their melting temperatures (Tm) were determined by differential scanning fluorimetry (DSF). Capsids of AAVrh.10 and AAV5 showed Tm values of 76.0 and 89.0°C, respectively (Fig. 6B), which are identical or very similar to the previously reported values for these AAV capsids (72). In contrast to the wtAAVrh.10 capsid, the 3x and 7x variants showed reduced Tms of 73.5 and 71.0°C, respectively. Previously, it was suggested that the Tm of the AAV capsids is charge dependent (72). However, none of the substituted amino acids introduced or removed charged residues (Fig. 5C). The lower Tm values of the variants are comparable to AAV3 and AAV8 (∼71 and ∼72°C) but higher than that for AAV2 (68°C) (72). Furthermore, having different Tm values allows an easy differentiation of the identity of the different AAVrh.10 variant capsids.
FIG 6.
Characterization of the AAVrh.10 variants. (A) Analysis of AVB-purified AAVrh.10-WT and the 7x variant by SDS-PAGE. VP1, VP2, and VP3 are indicated. Cryo-electron micrographs of the purified samples (scale bar, 50 nm) are shown. (B) Determination of the melting temperature (Tm) for the different AAVrh.10 variants and AAV5 by DSF analysis. The thermal profile is shown as normalized relative fluorescence units (RFUs) versus temperature (°C). The experiments were performed in triplicate (n = 3), and each thermal profile is shown. The Tm for each capsid variant is displayed as the mean plus the SD.
Structural characterization of the AAVrh.10-7x variant capsid.
To further characterize the newly developed AAVrh.10-7x variant, its capsid structure was determined by cryo-EM. Utilizing 17,861 individual capsids images, the 7x capsid was reconstructed to 2.71-Å resolution (Table 1). The resulting density map of the 7x variant is very similar to the previously described wild-type AAVrh.10 capsid (54). However, minor differences were observed at the 3-fold protrusions, which were more “pointed” in the 7x variant capsid (Fig. 7A). At a 2.7-Å resolution, amino acid side chains were clearly visible, and a model was built for this variant based on wild-type AAVrh.10 capsid structure (PDB 6O9R). The overall Cα-RMSD for the entire VP comparing AAVrh.10-WT to the 7x variant is only 0.44 Å. This small Cα-RMSD is comparable to other highly similar AAV capsid variants such as AAV1 (3NG9) versus AAV6 (3OAH) that differ by five amino acids in their ordered observed VP structure with a Cα-RMSD of 0.33 Å (73) and AAVrh.10 (6O9R) versus AAVrh.39 (6V1T) that differ by 10 amino acids with a Cα-RMSD of 0.55 Å (54). The only region with significant structural variability is VR-IV (Fig. 7B). This difference is caused by the deletion of serine 453, which makes VR-IV shorter and less slanted to the side compared to wtAAVrh.10, which results in the pointed appearance of the 3-fold protrusions of the 7x variant (Fig. 7A and C). For the remaining amino acid substitutions, the differences in the side chain densities are visible, with the exception of T719V (same side chain size), but they do not lead to major structural differences between the two capsid types (Fig. 7C).
FIG 7.
Determination of the AAVrh.10-7x capsid structure. (A) Reconstructed AAVrh.10-7x capsid map colored according to radial distance from the capsid center (blue) to outermost regions (red) as in Fig. 1. The positions of the 5-fold channel, 3-fold and 2-fold axes, and 2/5-fold wall are indicated. (B) Structural superposition of AAVrh.10-WT (olive green) and AAVrh.10-7x (yellow) shown as ribbon diagrams. The positions of the N and C termini, the core β-sheets, α-helix, and variable regions (VRs) are indicated. This figure was generated using PyMOL (85). (C) The modeled AAVrh.10-7x VRs are shown in stick representation inside the mesh density map. AAVrh.10-WT (PDB 6V1G) is superposed onto the 7x variant. The amino acids are as labeled. The amino acid substitutions of the 7x variant are displayed in red.
Conclusions.
The capsid of AAVrh.10 shares characteristics with both AAV8 and AAV9. Structurally, the AAVrh.10 capsid is very similar to the AAV8 capsid. As a consequence, many antibodies capable of binding AAV8 also cross-react and neutralize AAVrh.10. The AAVrh.10 variants generated to escape the AAV8 MAbs can likely also be adapted for AAV8 escape variants. In addition, AAVrh.10 shares functional similarities to AAV9. Both are able to bind terminal galactoses, using a conserved pocket on the capsid. In addition, both AAV9 and AAVrh.10 have been described to be capable of crossing the blood-brain barrier (BBB) (54, 74, 75). The suggested amino acids responsible for this phenotype overlap the galactose-binding pocket and extend to VR-I. Thus, it is possible that crossing the BBB is dependent on the ability of the capsid to bind terminal galactose glycans. However, this hypothesis needs to be studied in more detail to be confirmed.
The continued characterization and annotation of the receptor and antigenic interactions of the AAVrh.10 capsid provides information for future vector engineering efforts, such as the variant generated in this study that is capable of escaping neutralizing antibodies while maintaining infectivity.
MATERIALS AND METHODS
Cell culture.
HEK293 and CHO Pro5/Lec2 cells were maintained adherent in Dulbecco modified Eagle medium or minimum essential medium (Thermo Fisher, Waltham, MA), respectively, supplemented with 10% heat-inactivated fetal calf serum and 100 U of penicillin/ml and 100 μg of streptomycin (Caisson Laboratories, Smithfield, UT) at 37°C in 5% CO2.
Site-directed mutagenesis.
The pAAV2/rh.10 and pXR8 plasmids containing AAV2 rep and either AAVrh.10 or AAV8 cap served as the template for site-directed mutagenesis PCRs. For each mutant, complementary PCR primers were designed that contained the desired mutation, which was flanked on both sides by 10 to 15 homologous base pairs. Primers were ordered from Sigma-Aldrich (Houston, TX) and used in PCR amplification reactions using a C1000 Touch thermal cycler (Bio-Rad, Hercules, CA) and Pfu Ultra high-fidelity DNA polymerase (Agilent, Santa Clara, CA). PCR products were incubated at 37°C for 1 h with DpnI restriction enzyme (NEB, Ipswich, MA) to degrade the methylated template plasmid. The reactions were then transformed into DH5α competent cells (NEB, Ipswich, MA), which were cultured on LB-ampicillin selective media and further amplified to isolate the plasmid. Clones were submitted for Sanger sequencing (Genewiz, South Plainfield, NJ) to verify the introduced mutations.
Recombinant AAV production and purification.
Recombinant AAV vectors with a packaged luciferase gene were produced by triple transfection of HEK293 cells, utilizing pTR-UF3-Luciferase, pHelper (Stratagene), and either pAAV2/rh.10, pXR2, pR2V3, pXR5, pXR8, pXR9, or variants thereof. The transfected cells were harvested 72 h posttransfection and washed with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 100 mM Na2HPO4, 2 mM KH2PO4), and the cells were then pelleted and resuspended in PBS with 1 mM MgCl2 and 2.5 mM KCl. The resuspended cells were subjected to three freeze-thaw cycles (−80°C to 37°C) and subsequently incubated with 125 U/ml benzonase for 1 h at 37°C before centrifugation at 10,000 × g for 15 min to pellet the cell debris. AAV vectors or variants thereof were purified by AVB Sepharose affinity chromatography and concentrated as previously described (71).
IgG production and purification.
IgGs were produced and purified as described previously (50). Briefly, to produce sufficient quantities of MAbs for purification, hybridoma cells of the selected antibody were cultivated in 500 ml of medium in surface roller bottles (Sigma-Aldrich, St. Louis, MO) for 12 days. Cells were centrifuged at 4,000 × g for 10 min, and the supernatant was collected. For preservation, 0.01% sodium azide was added to the supernatant. The MAbs were purified from the hybridoma supernatants by affinity chromatography using protein G-Sepharose columns (GE Healthcare). The hybridoma culture was diluted 1:1 (vol/vol) in PBS and applied to the column at room temperature below the manufacturer-recommended flow rate of 1 ml/min. The column was washed with PBS, and the antibody was eluted with 15 ml of 0.1 M citric acid (pH 2.0). The eluted antibody was neutralized with 1 M Tris-HCl (pH 10.0) to a final pH of 7.4. The elution fractions were concentrated using an Apollo 7-ml centrifugal concentrator (Orbital Biosciences, Topsfield, MA). The total MAb yield was determined at an optical density of 280 nm, along with estimation by SDS-PAGE with bovine serum albumin concentration standards.
Fab generation and purification.
For the generation of Fabs from the purified MAbs, immobilized papain was activated with l-cysteine according to the manufacturer’s instructions (Pierce, Rockford, IL) and mixed with purified sample at a suggested enzyme/substrate ratio of 1:160 (wt/wt). The slurry was incubated with moderate shaking at 37°C overnight. The reaction was stopped with sample buffer (1.5 ml, 10 mM Tris-HCl [pH 7.5]) and then gently centrifuged (200 × g, 5 min) to pellet the immobilized papain-agarose beads. The aqueous mixture was carefully removed and diluted in 20 mM sodium phosphate buffer (pH 8.5) and applied to a Hi-Trap protein A column (GE Healthcare, Uppsala, Sweden) using a peristaltic pump at a rate of 1 ml/min. The Fabs were collected in the flowthrough and concentrated on Amicon-Ultra concentrators (Millipore, Billerica, MA). Their purity was monitored by using SDS-PAGE.
Cryo-electron microscopy data collection.
Purified AAVrh.10 capsids were mixed with Fabs at a ratio of ∼2 Fabs per potential VP binding site in the 60-meric capsid, giving a final ratio of ∼1:120 (capsid to Fab). In the case of the sulfated LacNAc, the capsids and the glycans were mixed at a ratio of ∼100 glycan molecules per potential VP binding site. Aliquots (3.5 μl) of the purified samples either with or without glycans or Fabs were applied to glow-discharged Quantifoil copper grids with 2-nm continuous carbon support over holes (Quantifoil R 2/2 400 mesh), blotted, and vitrified using a Vitrobot Mark 4 (FEI Co.) at 95% humidity and 4°C. The particle distribution and ice quality of the grids were screened in-house using a FEI Tecnai G2 F20-TWIN microscope (FEI) operated under low-dose conditions (200 kV, ∼20 e−/Å2). Images were collected on a GatanUltraScan 4000 charge-coupled device camera (Gatan) at a pixel size of 1.82 Å. For the individual AAVrh.10-Fab complexes, data sets comprising ∼50 micrographs were collected under these conditions. Data for the samples intended for high-resolution was obtained by collecting micrograph movie frames using the Leginon application (76) on a Titan Krios electron microscope. This microscope was operated at 300 kV, and data were collected on a DE64 (for AAVrh.10 complexed with LacNAc) or Gatan K2 Summit (for AAVrh.10-7x) direct electron detector (Direct Electron). During data collection, a total dose of ∼60 e−/Å2 was utilized for 43 to 50 movie frames per micrograph. MotionCor2 was used for aligning the movie frames with dose weighting (77). The AAVrh.10-LacNAc data set was collected as part of the NIH “Southeastern Center for Microscopy of MacroMolecular Machines (SECM4)” project. The AAVrh.10-7x data set was collected as part of the National Institutes of Health (NIH) “West/Midwest Consortium for High-Resolution Cryo Electron Microscopy” project.
Icosahedral 3D image reconstruction.
For the 3D image reconstruction, the cisTEM software package was utilized (78), and the data were processed as described previously (13). The sharpened density maps were inspected using Coot and Chimera (79, 80). The −90-Å2/0-Å2 sharpened maps of the high-resolution data were utilized for assignment of the amino acid main and side chains. The resolution of the cryo-reconstructed density maps was estimated based on a Fourier shell correlation of 0.143 (Table 1).
Model building and structure refinement.
The 60-mer capsid model of AAVrh.10 (PDB 6O9R) was docked into the cryo-reconstructed density maps by rigid body rotations and translations using the “fit in map” subroutine within UCSF-Chimera (80) that uses a correlation coefficient (CC) calculation to assess the quality of the fit between the map generated from the model and the reconstructed map. During the model fitting, the voxel (pixel) size of each reconstructed map was adjusted to optimize the CC between the models and maps. The fitted models were exported relative to the respective map for further use. Each map was resized to the voxel size determined in Chimera using the “e2proc3D.py” subroutine in EMAN2 (81) and then converted to the CCP4 format using the program MAPMAN (82). A VP monomer was extracted from each 60-mer, and the side and main chains were adjusted into the maps by manual building and use of the real-space-refinement subroutine in Coot (79). In the case of the AAVrh.10-7x variant the amino acids substituted or deleted relative to the wild-type capsid were changed in Coot. The adjusted capsid model was refined against the map utilizing the rigid body, real space, and B-factor refinement subroutines in Phenix (83). Model refinement was alternated with visualization and adjustment using Coot to maintain model geometry as well as rotamer and Ramachandran constraints (79). The CC and refinement statistics, including the root mean square deviation (RMSD) from ideal bond lengths and angles (Table 1), were analyzed using Phenix (83).
AAV capsid structure comparison.
The Cα positions of all ordered residues within the VP3 atomic coordinates of AAVrh.10, and its variant were superposed using secondary structure matching (SSM) in Coot (84). The SSM subroutine generated RMSD values between the structures and calculated distances (Å) between the aligned amino acid Cα positions.
Native dot immunoblot analysis.
AAV capsids were adsorbed onto nitrocellulose membranes (Bio-Rad) in a dot blot manifold (Schleicher & Schuell, Dassel, Germany). Excess fluid was drawn through the membrane by vacuum filtration. The membrane was removed from the manifold and blocked with 6% milk in PBS (pH 7.4) for 1 h. Primary antibody in the form of hybridoma supernatant was applied to the membrane at a 1:50 dilution in PBS with 6% milk–0.1% Tween 20, followed by incubation for 1 h. The membrane was then washed with PBS, and horseradish peroxidase-linked secondary antibody (GE Healthcare) was applied at a 1:3,000 dilution in PBS, followed by incubation for 1 h. The membrane was next washed with PBS. Immobilon chemiluminescent substrate (Millipore, Darmstadt, Germany) was then applied to the membrane, and the signal was detected on X-ray film.
Determining the transduction efficiency in presence or absence of MAbs or glycans.
Purified AAV vectors were used to infect HEK293 or CHO Pro5/Lec2 cells at a 105 multiplicity of infection. After 48 h, the cells were lysed, and the luciferase activity was assayed using a luciferase assay kit (Promega, Madison, WI) as described in the manufacturer’s protocol. Uninfected cells of the same plate were used as a negative control. In the case of the neutralization assays, the vectors were preincubated for 30 min at 37°C with either purified MAbs, keratan sulfate, or PBS as the negative control at variable ratios relative to the capsids prior to infection. The transduction efficiency was calculated as a percentage to AAV vectors in the absence of antibodies or glycans, respectively.
Differential scanning fluorescent stability assay.
Purified AAV capsids diluted in PBS to a total volume of 22.5 μl were utilized for the DSF stability assay and mixed with 2.5 μl of 1% SYPRO-Orange dye (Molecular Probes, Invitrogen), as previously described (72). The assay was conducted in a thermocycler (Bio-Rad CFX Connect) with the temperature ramped from 30 to 99°C, increasing by 0.5° every 30 s. The melting temperature (Tm) for each sample was defined as the vertex of the first derivative (dF/dT) as relative fluorescence unit (RFU) values.
Data availability.
The cryo-EM reconstructed density maps and models built were deposited in the Electron Microscopy Data Bank (EMDB) with under accession numbers EMD-24513, PDB ID 7RL1 (AAVrh.10-7x), EMD-24806, and PDB ID 7S1W (AAVrh.10:LacNAc), respectively. The reconstructed density maps for the capsid:MAb complexes were deposited under accession numbers EMD-24808 (AAVrh.10:ADK8), EMD-24809 (AAVrh.10:ADK8/9), EMD-24810 (AAVrh.10:HL2381), and EMD-24811 (AAVrh.10:HL2383), respectively.
ACKNOWLEDGMENTS
We thank the UF-ICBR Electron Microscopy Core (RRID:SCR_019146) for access to electron microscopes utilized for negative-stain electron microscopy and cryo-electron micrograph screening. The Spirit and TF20 cryo-electron microscopes were provided by the UF College of Medicine (COM) and Division of Sponsored Programs (DSP). Data collection at Florida State University was made possible by NIH grant S10 OD018142-01, purchase of a direct electron camera for the Titan-Krios at FSU (P. I. Taylor), S10 RR025080-01, purchase of a FEI Titan Krios for 3D EM (P. I. Taylor), and U24 GM116788. The Southeastern Consortium for Microscopy of MacroMolecular Machines (P. I. Taylor). The University of Florida COM and NIH GM082946 (to M.A.-M. and R.M.) provided funds for the research efforts at the University of Florida. P.H.S. and F.B. gratefully acknowledge generous financial support from the Max Planck Society, the German Federal Ministry of Education and Research (grant 0315447 to P.H.S.), and the Deutsche Forschungsgemeinschaft (No. SFB-TR84 C8 to P.H.S.). R.H. thanks the Sonnenfeld-Stiftung, Berlin, Germany, for funding of a high-speed rotor and the German Academic Exchange Service (DAAD) for travel grants to the Agbandje-McKenna lab at UF and acknowledges financial support from the Deutsche Forschungsgemeinschaft (He1598/9-1).
The University of Florida Research Foundation has filed and licensed patent applications based on the findings described here.
Contributor Information
Robert McKenna, Email: rmckenna@ufl.edu.
Colin R. Parrish, Cornell University
REFERENCES
- 1.Cotmore SF, Agbandje-McKenna M, Canuti M, Chiorini JA, Eis-Hubinger AM, Hughes J, Mietzsch M, Modha S, Ogliastro M, Penzes JJ, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, ICTV Report Committee . 2019. ICTV virus taxonomy profile: Parvoviridae. J Gen Virol 100:367–368. 10.1099/jgv.0.001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott LJ. 2015. Alipogene tiparvovec: a review of its use in adults with familial lipoprotein lipase deficiency. Drugs 75:175–182. 10.1007/s40265-014-0339-9. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd A, Piglowska N, Ciulla T, Pitluck S, Johnson S, Buessing M, O’Connell T. 2019. Estimation of impact of RPE65-mediated inherited retinal disease on quality of life and the potential benefits of gene therapy. Br J Ophthalmol 103:1610–1614. 10.1136/bjophthalmol-2018-313089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waldrop MA, Kolb SJ. 2019. Current treatment options in neurology-SMA therapeutics. Curr Treat Options Neurol 21:25. 10.1007/s11940-019-0568-z. [DOI] [PubMed] [Google Scholar]
- 5.Agbandje-McKenna M, Kleinschmidt J. 2011. AAV capsid structure and cell interactions. Methods Mol Biol 807:47–92. 10.1007/978-1-61779-370-7_3. [DOI] [PubMed] [Google Scholar]
- 6.Snijder J, van de Waterbeemd M, Damoc E, Denisov E, Grinfeld D, Bennett A, Agbandje-McKenna M, Makarov A, Heck AJ. 2014. Defining the stoichiometry and cargo load of viral and bacterial nanoparticles by Orbitrap mass spectrometry. J Am Chem Soc 136:7295–7299. 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girod A, Wobus CE, Zadori Z, Ried M, Leike K, Tijssen P, Kleinschmidt JA, Hallek M. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J Gen Virol 83:973–978. 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- 8.Popa-Wagner R, Porwal M, Kann M, Reuss M, Weimer M, Florin L, Kleinschmidt JA. 2012. Impact of VP1-specific protein sequence motifs on adeno-associated virus type 2 intracellular trafficking and nuclear entry. J Virol 86:9163–9174. 10.1128/JVI.00282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, Wilson JM. 2004. Clades of Adeno-associated viruses are widely disseminated in human tissues. J Virol 78:6381–6388. 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt M, Katano H, Bossis I, Chiorini JA. 2004. Cloning and characterization of a bovine adeno-associated virus. J Virol 78:6509–6516. 10.1128/JVI.78.12.6509-6516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bossis I, Chiorini JA. 2003. Cloning of an avian adeno-associated virus (AAAV) and generation of recombinant AAAV particles. J Virol 77:6799–6810. 10.1128/jvi.77.12.6799-6810.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mietzsch M, Jose A, Chipman P, Bhattacharya N, Daneshparvar N, McKenna R, Agbandje-McKenna M. 2021. Completion of the AAV structural atlas: serotype capsid structures reveals clade-specific features. Viruses 13. 10.3390/v13010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mietzsch M, Li Y, Kurian J, Smith JK, Chipman P, McKenna R, Yang L, Agbandje-McKenna M. 2020. Structural characterization of a bat adeno-associated virus capsid. J Struct Biol 211:107547. 10.1016/j.jsb.2020.107547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mietzsch M, Penzes JJ, Agbandje-McKenna M. 2019. Twenty-five years of structural parvovirology. Viruses 11. 10.3390/v11040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett A, Keravala A, Makal V, Kurian J, Belbellaa B, Aeran R, Tseng YS, Sousa D, Spear J, Gasmi M, Agbandje-McKenna M. 2020. Structure comparison of the chimeric AAV2.7m8 vector with parental AAV2. J Struct Biol 209:107433. 10.1016/j.jsb.2019.107433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Q, Yoshioka CK, Chapman MS. 2020. Adeno-associated virus (AAV-DJ)-cryo-EM structure at 1.56-Å resolution. Viruses 12. 10.3390/v12101194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaelber JT, Yost SA, Webber KA, Firlar E, Liu Y, Danos O, Mercer AC. 2020. Structure of the AAVhu.37 capsid by cryoelectron microscopy. Acta Crystallogr F Struct Biol Commun 76:58–64. 10.1107/S2053230X20000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu H-L, Brown A, Loveland AB, Lotun A, Xu M, Luo L, Xu G, Li J, Ren L, Su Q, Gessler DJ, Wei Y, Tai PWL, Korostelev AA, Gao G. 2020. Structural characterization of a novel human adeno-associated virus capsid with neurotropic properties. Nat Commun 11:3279. 10.1038/s41467-020-17047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govindasamy L, Padron E, McKenna R, Muzyczka N, Kaludov N, Chiorini JA, Agbandje-McKenna M. 2006. Structurally mapping the diverse phenotype of adeno-associated virus serotype 4. J Virol 80:11556–11570. 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett A, Mietzsch M, Agbandje-McKenna M. 2017. Understanding capsid assembly and genome packaging for adeno-associated viruses. Future Virol 12:283–297. 10.2217/fvl-2017-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Summerford C, Samulski RJ. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol 72:1438–1445. 10.1128/JVI.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Handa A, Muramatsu S, Qiu J, Mizukami H, Brown KE. 2000. Adeno-associated virus (AAV)-3-based vectors transduce haematopoietic cells not susceptible to transduction with AAV-2-based vectors. J Gen Virol 81:2077–2084. 10.1099/0022-1317-81-8-2077. [DOI] [PubMed] [Google Scholar]
- 23.Halbert CL, Allen JM, Miller AD. 2001. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J Virol 75:6615–6624. 10.1128/JVI.75.14.6615-6624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt M, Govindasamy L, Afione S, Kaludov N, Agbandje-McKenna M, Chiorini JA. 2008. Molecular characterization of the heparin-dependent transduction domain on the capsid of a novel adeno-associated virus isolate, AAV(VR-942). J Virol 82:8911–8916. 10.1128/JVI.00672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mietzsch M, Broecker F, Reinhardt A, Seeberger PH, Heilbronn R. 2014. Differential adeno-associated virus serotype-specific interaction patterns with synthetic heparins and other glycans. J Virol 88:2991–3003. 10.1128/JVI.03371-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J Virol 75:6884–6893. 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z, Miller E, Agbandje-McKenna M, Samulski RJ. 2006. Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J Virol 80:9093–9103. 10.1128/JVI.00895-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walters RW, Yi SM, Keshavjee S, Brown KE, Welsh MJ, Chiorini JA, Zabner J. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J Biol Chem 276:20610–20616. 10.1074/jbc.M101559200. [DOI] [PubMed] [Google Scholar]
- 29.Bell CL, Vandenberghe LH, Bell P, Limberis MP, Gao GP, Van Vliet K, Agbandje-McKenna M, Wilson JM. 2011. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J Clin Invest 121:2427–2435. 10.1172/JCI57367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen S, Bryant KD, Brown SM, Randell SH, Asokan A. 2011. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J Biol Chem 286:13532–13540. 10.1074/jbc.M110.210922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahm HS, Broecker F, Kawasaki F, Mietzsch M, Heilbronn R, Fukuda M, Seeberger PH. 2017. Automated glycan assembly of oligo-N-acetyllactosamine and keratan sulfate probes to study virus-glycan interactions. Chem 2:114–124. 10.1016/j.chempr.2016.12.004. [DOI] [Google Scholar]
- 32.Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, Jae LT, Wosen JE, Nagamine CM, Chapman MS, Carette JE. 2016. An essential receptor for adeno-associated virus infection. Nature 530:108–112. 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asokan A, Hamra JB, Govindasamy L, Agbandje-McKenna M, Samulski RJ. 2006. Adeno-associated virus type 2 contains an integrin α5β1 binding domain essential for viral cell entry. J Virol 80:8961–8969. 10.1128/JVI.00843-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Summerford C, Bartlett JS, Samulski RJ. 1999. αVβ5 integrin: a coreceptor for adeno-associated virus type 2 infection. Nat Med 5:78–82. 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- 35.Akache B, Grimm D, Pandey K, Yant SR, Xu H, Kay MA. 2006. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J Virol 80:9831–9836. 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashiwakura Y, Tamayose K, Iwabuchi K, Hirai Y, Shimada T, Matsumoto K, Nakamura T, Watanabe M, Oshimi K, Daida H. 2005. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J Virol 79:609–614. 10.1128/JVI.79.1.609-614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blackburn SD, Steadman RA, Johnson FB. 2006. Attachment of adeno-associated virus type 3H to fibroblast growth factor receptor 1. Arch Virol 151:617–623. 10.1007/s00705-005-0650-6. [DOI] [PubMed] [Google Scholar]
- 38.Di Pasquale G, Davidson BL, Stein CS, Martins I, Scudiero D, Monks A, Chiorini JA. 2003. Identification of PDGFR as a receptor for AAV-5 transduction. Nat Med 9:1306–1312. 10.1038/nm929. [DOI] [PubMed] [Google Scholar]
- 39.Opie SR, Warrington KH, Jr, Agbandje-McKenna M, Zolotukhin S, Muzyczka N. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J Virol 77:6995–7006. 10.1128/jvi.77.12.6995-7006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell CL, Gurda BL, Van Vliet K, Agbandje-McKenna M, Wilson JM. 2012. Identification of the galactose binding domain of the adeno-associated virus serotype 9 capsid. J Virol 86:7326–7333. 10.1128/JVI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lerch TF, Chapman MS. 2012. Identification of the heparin binding site on adeno-associated virus serotype 3B (AAV-3B). Virology 423:6–13. 10.1016/j.virol.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R, Cao L, Cui M, Sun Z, Hu M, Zhang R, Stuart W, Zhao X, Yang Z, Li X, Sun Y, Li S, Ding W, Lou Z, Rao Z. 2019. Adeno-associated virus 2 bound to its cellular receptor AAVR. Nat Microbiol 4:675–682. 10.1038/s41564-018-0356-7. [DOI] [PubMed] [Google Scholar]
- 43.Zhang R, Xu G, Cao L, Sun Z, He Y, Cui M, Sun Y, Li S, Li H, Qin L, Hu M, Yuan Z, Rao Z, Ding W, Rao Z, Lou Z. 2019. Divergent engagements between adeno-associated viruses with their cellular receptor AAVR. Nat Commun 10:3760. 10.1038/s41467-019-11668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang LY, Patel A, Ng R, Miller EB, Halder S, McKenna R, Asokan A, Agbandje-McKenna M. 2016. Characterization of the adeno-associated virus 1 and 6 sialic acid binding site. J Virol 90:5219–5230. 10.1128/JVI.00161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Afione S, DiMattia MA, Halder S, Di Pasquale G, Agbandje-McKenna M, Chiorini JA. 2015. Identification and mutagenesis of the adeno-associated virus 5 sialic acid binding region. J Virol 89:1660–1672. 10.1128/JVI.02503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emmanuel SN, Mietzsch M, Tseng YS, Smith JK, Agbandje-McKenna M. 2021. Parvovirus capsid-antibody complex structures reveal conservation of antigenic epitopes across the family. Viral Immunol 34:3–17. 10.1089/vim.2020.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boutin S, Monteilhet V, Veron P, Leborgne C, Benveniste O, Montus MF, Masurier C. 2010. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum Gene Ther 21:704–712. 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 48.Calcedo R, Vandenberghe LH, Gao G, Lin J, Wilson JM. 2009. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis 199:381–390. 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Havlik LP, Simon KE, Smith JK, Klinc KA, Tse LV, Oh DK, Fanous MM, Meganck RM, Mietzsch M, Kleinschmidt J, Agbandje-McKenna M, Asokan A. 2020. Coevolution of adeno-associated virus capsid antigenicity and tropism through a structure-guided approach. J Virol 94. 10.1128/JVI.00976-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jose A, Mietzsch M, Smith JK, Kurian J, Chipman P, McKenna R, Chiorini J, Agbandje-McKenna M. 2019. High-resolution structural characterization of a new adeno-associated virus serotype 5 antibody epitope toward engineering antibody-resistant recombinant gene delivery vectors. J Virol 93. 10.1128/JVI.01394-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennett AD, Wong K, Lewis J, Tseng YS, Smith JK, Chipman P, McKenna R, Samulski RJ, Kleinschmidt J, Agbandje-McKenna M. 2018. AAV6 K531 serves a dual function in selective receptor and antibody ADK6 recognition. Virology 518:369–376. 10.1016/j.virol.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zerah M, Piguet F, Colle MA, Raoul S, Deschamps JY, Deniaud J, Gautier B, Toulgoat F, Bieche I, Laurendeau I, Sondhi D, Souweidane MM, Cartier-Lacave N, Moullier P, Crystal RG, Roujeau T, Sevin C, Aubourg P. 2015. Intracerebral gene therapy using AAVrh.10-hARSA recombinant vector to treat patients with early-onset forms of metachromatic leukodystrophy: preclinical feasibility and safety assessments in nonhuman primates. Hum Gene Ther Clin Dev 26:113–124. 10.1089/humc.2014.139. [DOI] [PubMed] [Google Scholar]
- 53.Tardieu M, Zerah M, Husson B, de Bournonville S, Deiva K, Adamsbaum C, Vincent F, Hocquemiller M, Broissand C, Furlan V, Ballabio A, Fraldi A, Crystal RG, Baugnon T, Roujeau T, Heard JM, Danos O. 2014. Intracerebral administration of adeno-associated viral vector serotype rh.10 carrying human SGSH and SUMF1 cDNAs in children with mucopolysaccharidosis type IIIA disease: results of a phase I/II trial. Hum Gene Ther 25:506–516. 10.1089/hum.2013.238. [DOI] [PubMed] [Google Scholar]
- 54.Mietzsch M, Barnes C, Hull JA, Chipman P, Xie J, Bhattacharya N, Sousa D, McKenna R, Gao G, Agbandje-McKenna M. 2020. Comparative analysis of the capsid structures of AAVrh.10, AAVrh.39, and AAV8. J Virol 94. 10.1128/JVI.01769-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lochrie MA, Tatsuno GP, Christie B, McDonnell JW, Zhou S, Surosky R, Pierce GF, Colosi P. 2006. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J Virol 80:821–834. 10.1128/JVI.80.2.821-834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dudek AM, Pillay S, Puschnik AS, Nagamine CM, Cheng F, Qiu J, Carette JE, Vandenberghe LH. 2018. An alternate route for adeno-associated virus (AAV) entry independent of AAV receptor. J Virol 92. 10.1128/JVI.02213-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eckhardt M, Gotza B, Gerardy-Schahn R. 1998. Mutants of the CMP-sialic acid transporter causing the Lec2 phenotype. J Biol Chem 273:20189–20195. 10.1074/jbc.273.32.20189. [DOI] [PubMed] [Google Scholar]
- 58.Lu Y, Ai J, Gessler D, Su Q, Tran K, Zheng Q, Xu X, Gao G. 2016. Efficient transduction of corneal stroma by adeno-associated viral serotype vectors for implications in gene therapy of corneal diseases. Hum Gene Ther 27:598–608. 10.1089/hum.2015.167. [DOI] [PubMed] [Google Scholar]
- 59.Fu L, Sun X, He W, Cai C, Onishi A, Zhang F, Linhardt RJ, Liu Z. 2016. Keratan sulfate glycosaminoglycan from chicken egg white. Glycobiology 26:693–700. 10.1093/glycob/cww017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caterson B, Melrose J. 2018. Keratan sulfate, a complex glycosaminoglycan with unique functional capability. Glycobiology 28:182–206. 10.1093/glycob/cwy003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Melrose J. 2019. Keratan sulfate (KS)-proteoglycans and neuronal regulation in health and disease: the importance of KS-glycodynamics and interactive capability with neuroregulatory ligands. J Neurochem 149:170–194. 10.1111/jnc.14652. [DOI] [PubMed] [Google Scholar]
- 62.Conrad AH, Zhang Y, Tasheva ES, Conrad GW. 2010. Proteomic analysis of potential keratan sulfate, chondroitin sulfate A, and hyaluronic acid molecular interactions. Invest Ophthalmol Vis Sci 51:4500–4515. 10.1167/iovs.09-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thwaite R, Pages G, Chillon M, Bosch A. 2015. AAVrh.10 immunogenicity in mice and humans: relevance of antibody cross-reactivity in human gene therapy. Gene Ther 22:196–201. 10.1038/gt.2014.103. [DOI] [PubMed] [Google Scholar]
- 64.Tseng YS, Vliet KV, Rao L, McKenna R, Byrne BJ, Asokan A, Agbandje-McKenna M. 2016. Generation and characterization of anti-adeno-associated virus serotype 8 (AAV8) and anti-AAV9 monoclonal antibodies. J Virol Methods 236:105–110. 10.1016/j.jviromet.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sonntag F, Kother K, Schmidt K, Weghofer M, Raupp C, Nieto K, Kuck A, Gerlach B, Bottcher B, Muller OJ, Lux K, Horer M, Kleinschmidt JA. 2011. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J Virol 85:12686–12697. 10.1128/JVI.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gurda BL, Raupp C, Popa-Wagner R, Naumer M, Olson NH, Ng R, McKenna R, Baker TS, Kleinschmidt JA, Agbandje-McKenna M. 2012. Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J Virol 86:7739–7751. 10.1128/JVI.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Havlik LP, Das A, Mietzsch M, Oh DK, Ark J, McKenna R, Agbandje-McKenna M, Asokan A. 2021. Receptor switching in newly evolved adeno-associated viruses. J Virol 95:e0058721. 10.1128/JVI.00587-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wobus CE, Hugle-Dorr B, Girod A, Petersen G, Hallek M, Kleinschmidt JA. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J Virol 74:9281–9293. 10.1128/jvi.74.19.9281-9293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McCraw DM, O’Donnell JK, Taylor KA, Stagg SM, Chapman MS. 2012. Structure of adeno-associated virus-2 in complex with neutralizing monoclonal antibody A20. Virology 431:40–49. 10.1016/j.virol.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mietzsch M, Grasse S, Zurawski C, Weger S, Bennett A, Agbandje-McKenna M, Muzyczka N, Zolotukhin S, Heilbronn R. 2014. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1–12 vectors for gene therapy. Hum Gene Ther 25:212–222. 10.1089/hum.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mietzsch M, Smith JK, Yu JC, Banala V, Emmanuel SN, Jose A, Chipman P, Bhattacharya N, McKenna R, Agbandje-McKenna M. 2020. Characterization of AAV-specific affinity ligands: consequences for vector purification and development strategies. Mol Ther Methods Clin Dev 19:362–373. 10.1016/j.omtm.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bennett A, Patel S, Mietzsch M, Jose A, Lins-Austin B, Yu JC, Bothner B, McKenna R, Agbandje-McKenna M. 2017. Thermal stability as a determinant of AAV serotype identity. Mol Ther Methods Clin Dev 6:171–182. 10.1016/j.omtm.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ng R, Govindasamy L, Gurda BL, McKenna R, Kozyreva OG, Samulski RJ, Parent KN, Baker TS, Agbandje-McKenna M. 2010. Structural characterization of the dual glycan binding adeno-associated virus serotype 6. J Virol 84:12945–12957. 10.1128/JVI.01235-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Albright BH, Storey CM, Murlidharan G, Castellanos Rivera RM, Berry GE, Madigan VJ, Asokan A. 2018. Mapping the structural determinants required for AAVrh.10 transport across the blood-brain barrier. Mol Ther 26:510–523. 10.1016/j.ymthe.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H, Yang B, Mu X, Ahmed SS, Su Q, He R, Wang H, Mueller C, Sena-Esteves M, Brown R, Xu Z, Gao G. 2011. Several rAAV vectors efficiently cross the blood-brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Mol Ther 19:1440–1448. 10.1038/mt.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. 2005. Automated molecular microscopy: the new Leginon system. J Struct Biol 151:41–60. 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 77.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grant T, Rohou A, Grigorieff N. 2018. cisTEM, user-friendly software for single-particle image processing. Elife 7. 10.7554/eLife.35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 80.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 81.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 82.Kleywegt GJ, Jones TA. 1996. xdlMAPMAN and xdlDATAMAN: programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr D Biol Crystallogr 52:826–828. 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 83.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60:2256–2268. 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 85.DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The cryo-EM reconstructed density maps and models built were deposited in the Electron Microscopy Data Bank (EMDB) with under accession numbers EMD-24513, PDB ID 7RL1 (AAVrh.10-7x), EMD-24806, and PDB ID 7S1W (AAVrh.10:LacNAc), respectively. The reconstructed density maps for the capsid:MAb complexes were deposited under accession numbers EMD-24808 (AAVrh.10:ADK8), EMD-24809 (AAVrh.10:ADK8/9), EMD-24810 (AAVrh.10:HL2381), and EMD-24811 (AAVrh.10:HL2383), respectively.