ABSTRACT
Recently, a genome-wide association study using plasma HIV RNA from antiretroviral therapy-naive patients reported that 14 naturally occurring nonsynonymous single-nucleotide polymorphisms (SNPs) in HIV derived from antiretrovirus drug-naive patients were associated with virus load (VL). Those SNPs were detected in reverse transcriptase, RNase H, integrase, envelope, and Nef. However, the impact of each mutation on viral fitness was not investigated. Here, we constructed a series of HIV variants encoding each SNP and examined their replicative abilities. An HIV variant containing a Met-to-Ile change at codon 50 in integrase [HIV(IN:M50I)] was found as an impaired virus. Despite the mutation being in integrase, the virus release was significantly suppressed (P < 0.001). Transmission electron microscopy analysis revealed that abnormal bud accumulation on the plasma membrane and the released virus particles retained immature forms. Western blot analysis demonstrated a defect in autoprocessing of GagPol and Gag polyproteins' autoprocessing in the HIV(IN:M50I) particles, although Förster resonance energy transfer (FRET) assay displayed that GagPol containing IN:M50I forms a homodimer with a similar efficiency with GagPol (wild type). The impaired maturation and replication were rescued by two other VL-associated SNPs, Ser-to-Asn change at codon 17 of integrase and Asn-to-Ser change at codon 79 of RNase H. These data demonstrate that Gag and GagPol assembly, virus release, and autoprocessing are regulated by not only integrase but also RNase H.
IMPORTANCE Nascent HIV-1 is a noninfectious viral particle. Cleaving Gag and GagPol polyproteins in the particle by mature HIV protease (PR), the nascent virus becomes an infectious virus. PR is initially translated as an inactive embedded enzyme in a GagPol polyprotein. The embedded PR in homodimerized GagPol polyproteins catalyzes a proteolytic reaction to release the mature PR. This excision step by self-cleavage is called autoprocessing. Here, during the evaluation of the roles of naturally emerging nonsynonymous SNPs in HIV RNA, we found that autoprocessing is inhibited by Met-to-Ile change at codon 50 in integrase GagPol. Other coexisting SNPs, Ser-to-Asn change at codon 17 in integrase or Asn-to-Ser mutation at codon 79 in RNase H, recovered this defect, suggesting that autoprocessing is regulated by not only integrase but also RNase H in GagPol polyprotein.
KEYWORDS: RNase H, SNPs, autoprocessing, human immunodeficiency virus, integrase
INTRODUCTION
Nascent human immunodeficiency virus type 1 (HIV-1) particles are released from infected host cells as immature and noninfectious viruses (1). These immature viral particles contain Gag and GagPol polyproteins, accessory proteins including Nef precursor, and viral genomic RNAs. The Gag and GagPol polyproteins are composed of viral structural proteins and the viral enzymes: protease (PR), reverse transcriptase (RT), RNase H (RH), and integrase (IN). During proteolytic maturation, the polyproteins in the immature particle are cleaved by the viral PR at 11 sites on the Gag and GagPol polyproteins and one site on the Nef precursor (1–3), and then the immature particle is converted into its infectious mature form. While these catalytic activities of PR are well-described, there are other aspects of PR that may play a role in viral replication. During viral Gag and GagPol assembly and budding at the cell membrane, GagPol polyproteins dimerize, followed by dimerization of the PR domains of the GagPol dimer, leading to the release of a functional dimerized PR in the viral particles. Inhibition of dimerization of the GagPol polyproteins has been considered a unique therapeutic target (4). Autoprocessing, which is the initial activity of releasing the embedded PR in the GagPol polyprotein by its processing, results in the formation of mature PR (5–7) that can catalyze the other cleavage reactions. Therefore, the homodimerization of the GagPol polyproteins in the immature virion is required for this excision of PR. IN is a component of the GagPol polyprotein, and a biologic role of the integrase protein of HIV-1 has been thought to be limited to inducing integration of proviral DNA to the host cell genome following infection. However, it is demonstrated that a truncated GagPol lacking IN suppresses viral release (8), and drug resistance mutations to an experimental integrase inhibitor, KF116, interfere with autoprocessing (9, 10). These reports have suggested a role for IN in viral release and autoprocessing.
In a recent substudy of the Strategic Timing of Antiretroviral Therapy (START) study (11), the investigators identified a series of 14 naturally occurring nonsynonymous single-nucleotide polymorphisms (SNPs) in HIV-1 that correlated with different levels of viremia in treatment-naive patients (12). However, the specific impact of each amino acid substitution associated with the SNP on viral fitness was not investigated. In the present study, we created a series of recombinant viruses containing each of the SNPs using site-directed mutagenesis and characterized the role of each mutation in viral fitness. Of all the mutants, only the Met-to-Ile substitution at codon 50 of IN (IN:M50I) resulted in the loss of replication capability, leading to the release of abnormally shaped virions and interfering with autoprocessing. Of note, this “off-target” effect was restored by compensatory mutations in IN(IN:S17N) or RH(RH:N79S) in the 14 SNPs. These results identify critical roles for RH and IN in the autoprocessing of the HIV-1 GagPol polyprotein.
RESULTS
Impact of SNPs on virus replication fitness.
A total of 14 nonsynonymous SNPs were reported to correlate with plasma viral loads in a cohort of 3,592 anti-retroviral drug-naive HIV-infected patients (12). To characterize the impact of each amino acid substitution associated with each SNP (Table 1) on viral fitness, we constructed a series of HIV variants containing each change using site-directed mutagenesis using the cloned HIV laboratory strain HIVNL4.3. Since this strain contains an Asn-to-Ser substitution at codon 79 of RH (RH:N79S), this site was back-mutated from Ser to Asn to yield a clone equivalent to the RH of the clade B consensus sequence (RH:N79). We used this construct, pNL(RH:N79WT), as the backbone wild-type HIV, HIV(WT), as a control in the current study. Each nonsynonymous SNP was introduced onto the WT control backbone using site-directed mutagenesis.
TABLE 1.
Natural polymorphic mutations associated with viral loads in antiviral drug-naive HIV-infected patients
| No. | HIV proteina | Codon no. | Amino acid change | Designationb |
|---|---|---|---|---|
| 1 | RT | 322 | Ser>Thr | RT:S322T |
| 2 | RH | 79 | Asn>Ser | RH:N79S |
| 3 | IN | 17 | Ser>Asn | IN:S17N |
| 4 | IN | 50 | Met>Ile | IN:M50I |
| 5 | Gp120 | 252 | Arg>Lys | G120:R252K |
| 6 | Gp120 | 448 | Asn>Arg | G120:N448R |
| 7 | Gp41 | 24 | Met>Val | G41:M24V |
| 8 | Gp41 | 24 | Met>Leu | G41:M24L |
| 9 | Gp41 | 254 | Leu>Arg | G41:L254R |
| 10 | Gp41 | 340 | Leu>Ala | G41:L340A |
| 11 | Nef | 92 | Lys>Arg | NF:K92R |
| 12 | Nef | 98 | Glu>Asp | NF:E 98D |
| 13 | Nef | 133 | Val>Ile | NF:V133I |
| 14 | Nef | 143 | Tyr>Phe | NF:Y143F |
RT, reverse transcriptase; RH, RNase H; IN, integrase; G120, gp120 envelope; G41, gp41 envelope; NF, nef.
Name of designated mutation in this study.
Plasmids encoding each SNP were transfected into HEK293T cells, and then viral stocks were prepared as described in Materials and Methods. Pelleted viral particles were resuspended in a 1/100 volume of the starting materials and used as viral stocks. To determine the amounts of virus in each variant stock, we measured HIV p24 antigen concentrations in each stock. Almost all variants except HIV(IN:M50I) demonstrated amounts of p24 concentrations comparable with that of HIV(WT) (89.0 ± 12 μg/ml, n = 7) (see Table S1 in the supplemental material). In striking contrast, the p24 concentrations of HIV(IN:M50I) stocks were 0.30 ± 0.087 μg/ml (n = 7) and, thus, around 0.3% of that in HIV(WT) (P < 0.001). To define whether the suppression of HIV(IN:M50I) release is caused by only HEK293T cells, HeLa cells were also transfected with the HIV(WT) or HIV(IN:M50I) construct, and then released HIV amounts were quantified. HIV(IN:M50I) virus was released by 0.3% of HIV(WT) (Table S1) from the cells, indicating that the inhibition is not caused in an HEK293T-specific manner. It is known that HeLa cells produce interferons (IFNs) in response to transfected plasmid DNA as an innate immune response (13), while HEK293T cells lack the inducing activity (14). To avoid any impact of the produced IFNs in downstream experiments for HIV production, we used HEK293T cells in entire studies as an HIV-producing cell.
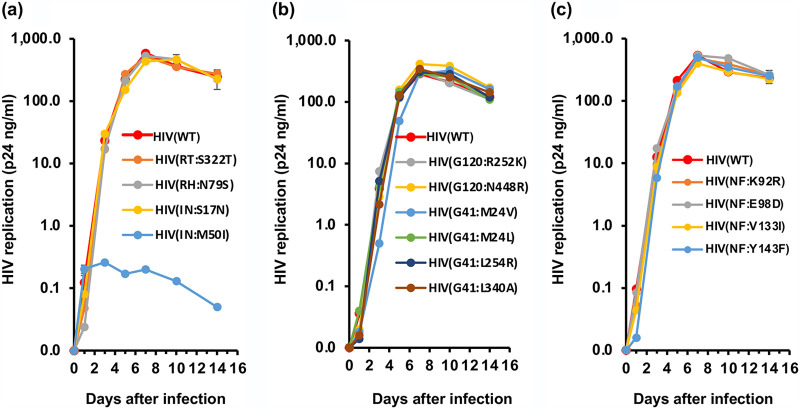
To investigate the impact of each mutation on viral replication, we infected PHA-stimulated primary CD4+ T cells from three independent healthy donors with each HIV variant, and viral replication was monitored for 14 days. All mutants except HIV(IN:M50I) replicated comparably to HIV(WT), and the HIV(IN:M50I) variant showed little to no replication (Fig. 1a to c, Fig. S1a to d). Compared to HIV(WT), in the presence of IN:M50I mutation, HIV replication was suppressed by 99.9% ± 0.030% (n = 7, P < 0.01) on the 7th day after infection, and even when the HIV(IN:M50I)-infected cells were cultured for an additional 7 days, significant viral replication was not detected.
FIG 1.
Characterization of 14 SNPs on HIV replication. (a to c) PHA-stimulated primary CD4+ T cells from healthy donors were infected with 10 ng p24 amounts of HIV(WT) or variants containing mutations in GagPol (a), envelope (b), or Nef (c) as described in Materials and Methods. The infected cells were cultured for 14 days with medium changed every 3 to 4 days. HIV replication was monitored using a p24 antigen capture kit. Representative data from three independent assays are presented as means ± standard deviations (SD) (n = 3).
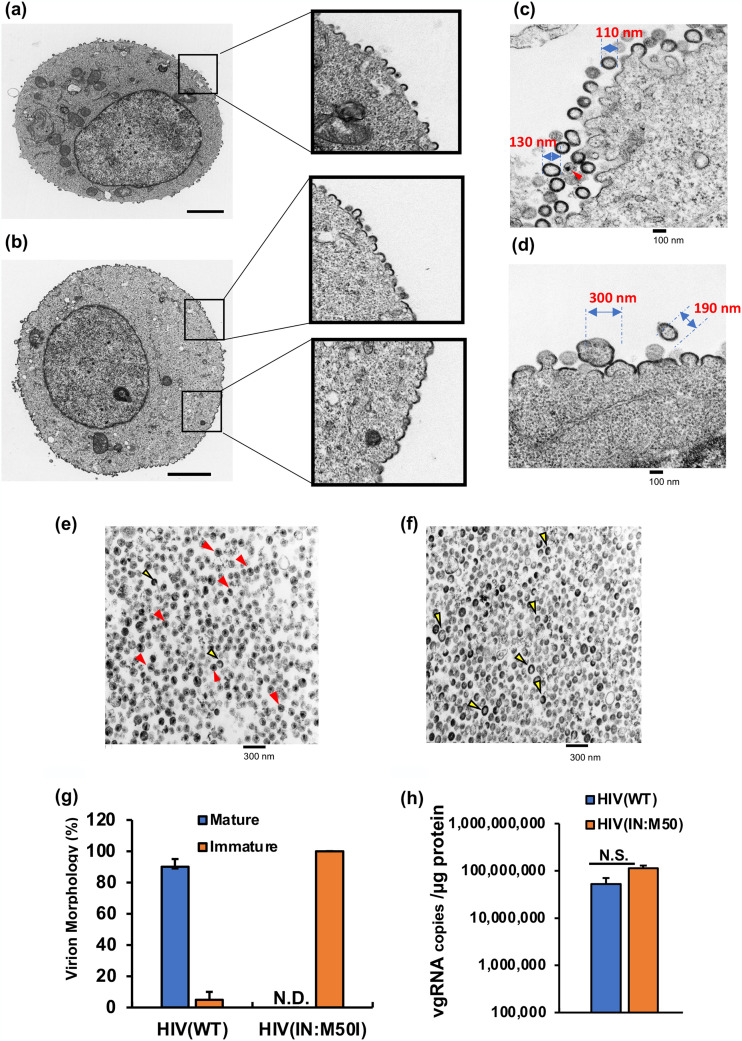
To further investigate the biologic properties of the IN:M50I mutant, we examined the morphology of the transfected HEK293T cells and virus particles 24 h after transfection using transmission electron microscopy (TEM). The cell surface of an HIV(WT)-producing cell demonstrated buds, viral neck formation, and virus release, while HIV(IN:M50I)-producing cells showed accumulation of virus buds on the cell surface (Fig. 2a and b), indicating that HIV(IN:M50I) release is suppressed. While the WT particles were relatively uniform in size (110 to 130 nm in diameter), the M50I particles were highly variable in size (190 to 300 nm in diameter) (Fig. 2c and d). TEM of HIV(WT) particles revealed virions with the expected outer envelope (Fig. 2e and f), with 90 to 95% having an electron-dense core (Fig. 2g). In contrast, TEM of HIV(IN:M50I) particles revealed a similar outer envelope with no evidence of core formation. These findings indicated that HIV(IN:M50I)-producing cells exhibited abnormalities in budding formations on the cell surface and releasing of viral particles from the cells. Unlike HIV(WT) with its dense core, the released mutant viruses had ring, doughnut-shaped, or teardrop structures. These results indicated that HIV(IN:M50I) was a typical defective virus, suggesting a lack of PR activity in the HIV(IN:M50I) virus particles. Gag and GagPol polyproteins play an important role in the intracytoplasmic trafficking of viral genomic RNAs (vgRNAs) and their eventual incorporation into the immature budding virions (1, 15–17). To determine whether or not the mutant phenotype of IN:M50I virions was due to a defect in the delivery of vgRNAs to the budding viral particles, we measured vgRNA copy numbers using purified HIV(WT) and HIV(IN:M50I) particles. No significant differences were observed in vgRNA copy number per microgram of virion protein between HIV(WT) and HIV(IN:M50I) (P > 0.05, n = 3) (Fig. 2h), indicating that the IN:M50I mutation had no impact on the incorporation of vgRNAs into virions.
FIG 2.
Characterization of viral and cell morphologies. HEK293T cells were transfected with plasmid encoding HIV(WT) (a) or HIV(IN:M50I) (b) and cultured for 48 h; each cell was fixed and then subjected to TEM as described in Materials and Methods. Scale bars indicate 2 μm. Boxes indicate the magnified cell surface images. (c and d) HEK293T cells were transfected with plasmids encoding HIV(WT) (c) or HIV(IN:M50I) (d) and then cultured for 24 h. Cells were fixed and then TEM images were taken as described in Materials and Methods. (e and f) TEM image analyses of sucrose-purified HIV particles of HIV(WT) (e) and HIV(IN:M50I) (f). Red and yellow arrowheads indicate typical electron-dense particles with a conical core (red) and electron-dense particles without a core (yellow). Scale bars show 100 nm (c and d) and 200 nm (e and f). (g) The relative population of viral morphology was calculated from 1,000 particle images by three independent assays. Data indicate means ± standard errors (SE) (n = 3). (h) Viral genomic RNA copy numbers in viral stocks were compared between HIV(WT) and HIV(IN:M50I). Copy numbers were determined using the real-time HIV-1 assay kit from three independent virus stocks, and data were normalized to total viral protein. Data show means ± SE (n = 3).
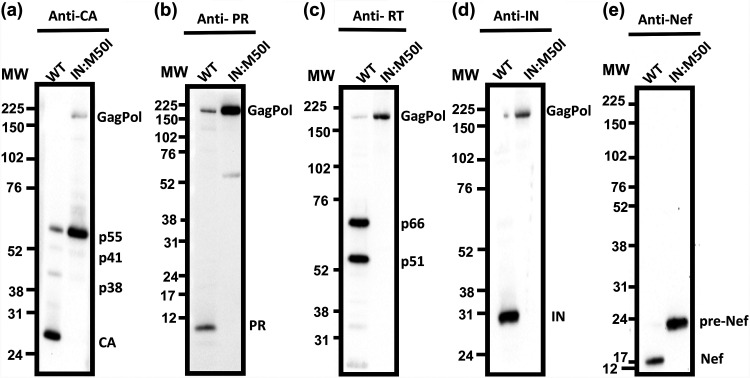
Given the immature appearance of IN:M50I virions, we next sought to determine whether the mutation led to any changes in autoprocessing and proteolytic cleavage of the GagPol polyproteins. Purified particles were analyzed for viral proteins by Western blotting (WB) using a series of antibodies specific to p24 capsid protein (CA), PR, RT, IN, or Nef. In contrast to the wild-type virions, HIV(IN:M50I) virions contained uncleaved Gag and GagPol polyproteins and immature Nef and did not contain the cleaved fragments of CA, PR, RT, and IN as detected in HIV(WT) (Fig. 3a to e). These results indicated that the IN:M50I mutation was somehow interfering with the activity of the HIV-1 PR, likely at the initial GagPol cleavage step, autoprocessing, and suppresses the production of mature CA, PR, RT, IN, and Nef.
FIG 3.
Evaluation of Gag and GagPol processing. A total of 1 μg of viral proteins from HIV(WT) and HIV(IN:M50I) particles was subjected to WB using monoclonal anti-CA or polyclonal anti-PR, RT, IN, or Nef antibodies. Protein bands were detected by using the ECL assay as described in Materials and Methods. The protein gel for detecting PR was run in MES running buffer (Thermo Fisher), and other gels were run in MOPS running buffer (Thermo Fisher). Data are representative of two independent experiments.
Impact of IN:M50I mutation on GagPol dimerization.
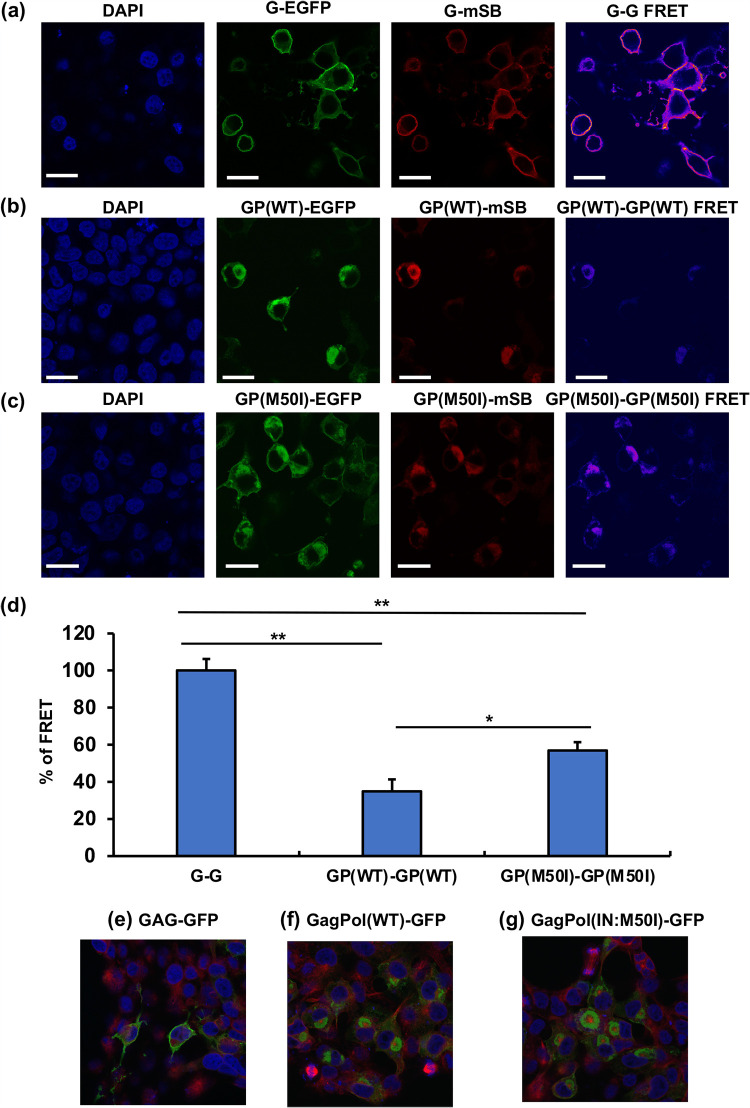
The autoprocessing of GagPol polyprotein depends on the formation of GagPol polyprotein homodimers (5, 18); thus, we hypothesized that the IN:M50I mutation leads to a structural hindrance in GagPol interfering with homodimerization. It is presumed that GagPol homodimerization initiated at the p2 (SP1) region of the Gag domain like the dimerization of Gag polyprotein (19). To determine whether the IN:M50I mutation suppresses homodimer formation, we employed a Förster resonance energy transfer (FRET) assay using the plasmids pGag(MA/EGFP/CA), pGagPol(MA/mSB/CA), pGag(MA/mSB/CA), and pGagPol(MA/EGFP/CA) (20). Since pGag(MA/EGFP/CA) and pGagPol(MA/mSB/CA) contained RH:N79S, they were back-mutated to RH:S79N, and IN:M50I was induced in both plasmids.
We assessed the efficiency of homodimerization among Gag-to-Gag (G-G), GagPol(WT)-to-GagPol(WT) [GP(WT)-GP(WT)], and GagPol(IN:M50I)-to-GagPol(IN:M50I) [GP(M50I)-GP(M50I)] by transfection of equal amounts of each plasmid DNA. Consistent with another report (20), FRET signals from G-G (Fig. 4a) were more robust than the signals from other pairs (Fig. 4b and c). The efficiency of the homodimerization of GP(WT)-to-GP(WT) and GP(IN:M50I)-to-GP(IN:M50I) was 34.9% ± 6.4% (n = 3, P < 0.01) and 57% ± 4.4% (n = 3, P < 0.01) of that of the G-G homodimerization, respectively (Fig. 4d). Interestingly, the efficiency of GagPol(IN:M50I) homodimerization was 1.7- ± 0.45-fold higher than that of GagPol(WT) homodimerization (n = 3, P < 0.05). These data indicated that the emergence of IN:M50I has no interference on the formation of GagPol homodimerization; however, it suppresses the autoprocessing. It was considered that even the IN:M50I mutation might change the distribution of GagPol proteins in the cells. To compare the distribution of each Gag, GagPol(WT), and GagPol(IN:M50) in cells, we transfected each green fluorescent protein (GFP) construct into HEK293T cells and analyzed protein distribution. Gag proteins were distributed throughout cells (the cytosol and the plasma membrane) (Fig. 4e). On the contrary, both GagPol(WT) and GagPol(IN:M50I) were predominantly located in the cytosol with polarization (Fig. 4f and g), indicating that IN:M50I mutation has no impact on GagPol distribution in the cells. Taken together, GagPol(IN:M50I) distributes in a similar location with GagPol(WT) and can form a homodimer at the membrane. Therefore, it was speculated that IN:M50I suppresses the autoprocessing after the homodimerization.
FIG 4.
Evaluation of Gag and GagPol dimerization using FRET assay. HEK293T cells were cotransfected with pGag-EGFP and pGag-mSB (a), pGagPol(WT)-EGFP and pGagPol(WT)-mSB (b), or pGagPol(IN:M50I)-EGFP and pGagPol (IN:M50I)-mSB (c), and then FRET assays were conducted as described in Materials and Methods. Gag-EGFP and Gag-mSB are displayed G-EGFP and G-mSB. GagPol(WT)-EGFP, GagPol(WT)-mSB, GagPol(IN:M50I)-GFP, and GagPol(IN:M50I)-mSB are depicted GP(WT)-EGFP, GP(WT)-mSB, GP(M50I)-EGFP, and GP(M50I)-mSB, respectively. (d) FRET efficiencies were calculated as described in Materials and Methods. Gag and GagPol homodimerization are described as G-G and GP-GP, respectively. The efficiencies of GP-GP of WT and M50I were compared with that of the G-G pair. Data indicate means ± SE from five independent assays. *, P < 0.05; ***, P < 0.001. (e to g) HEK293T cells were transfected with pGag-GFP (a), pGagPol(WT)-GFP (b), or pGagPol(IN:M50I)-GFP (c) construct and the protein distribution was observed. After transfection, cells were fixed with a fixation buffer (Abcam), and tubulins were stained with Alexa Fluor 555 anti-beta tubulin antibody (EPR16774) (ab206627). Scale bars indicate 20 μm.
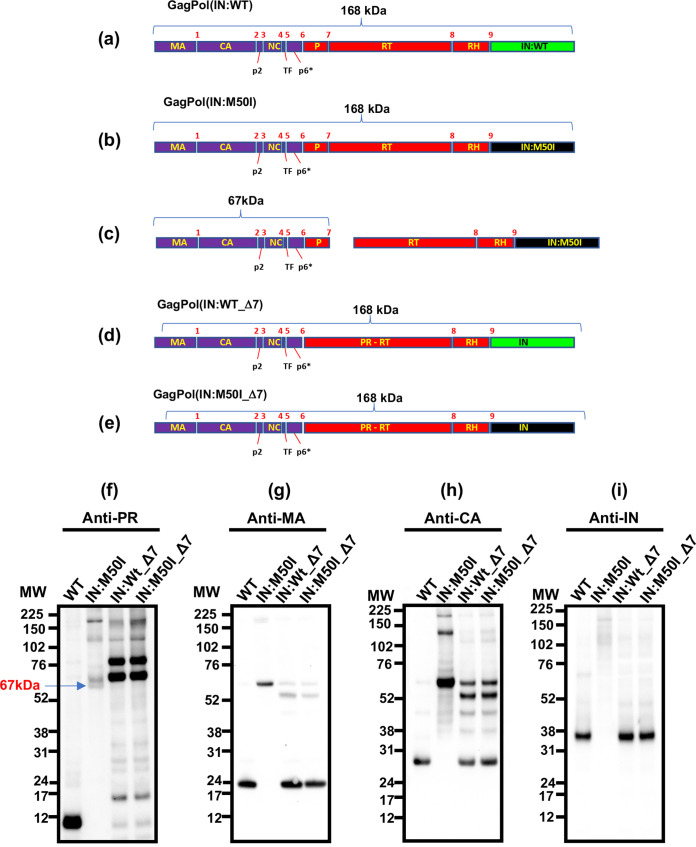
HIV PR cleaves a total of 9 cleavage sites in GagPol polyprotein (Fig. 5a) (1). PR cleaves GagPol in an ordered sequence, leading to virus maturation (21). Generally it is considered that the initial cleavage, as the autoprocessing occurs between the p2 spacer peptide and the NC protein (at cleavage site 3 in Fig. 5a and b) in HIV (22–28), is an intramolecular reaction (cis-reaction) (5, 18, 29, 30). Since, in the presence of IN:M50I mutation, GagPol suppressed the autoprocessing even when it forms homodimers, it was speculated that the mutation changes the conformation of GagPol. Crystal analysis of full-length GagPol, as well as the entire length of IN, was not completed due to disorder at the C-terminal domain of IN and an unclear resulting crystal structure; instead, many partial structure analyses were conducted (31–35). Thus, we attempted to perform in silico structure analysis of full-length IN using the RosettaCM protocol to predict a structure difference. We have not observed any difference in the predicted IN models (Fig. S2), suggesting that IN:M50I suppresses the initiation of autoprocessing via an uncharacterized manner.
FIG 5.
Effect of the deletion of the cleavage site between PR and RT in GagPol on autoprocessing. (a to e) Diagrams indicate the structure of GagPol(WT) and variants. Numbers above the diagrams indicate cleavage sites 1 to 9 by mature PR. Molecular size of each domain: MA, 17 kDa; CA, 24 kDa; p2, 2 kDa; NC, 7 kDa; TF, 1 kDa; P (PR),10 kDa; RT, 51 kDa; RH, 10 kDa; IN, 35 kDa. Cleavage site 7 between PR and RT in HIV(WT) and HIV(IN:M50I) was deleted by point mutagenesis as described in Materials and Methods, and the resulting clones contain a fusion gene of PR and RT (PR-RT). (f to i) Virus particles are isolated using ultracentrifugation as described in Materials and Methods, and 5 μg of viral lysates was subjected to WB. PR, CA, and IN were detected by anti-PR (f), anti-MA (g), anti-CA (h), and anti-IN antibodies (i).
When we performed WB using viral particles of HIV(IN:M50I) and polyclonal anti-PR antibodies, the antibody detected uncleaved GagPol polyproteins, but a mature PR band was not detected (Fig. 3b and 5f); however, in addition to the uncleaved GagPol band, unexpected additional bands were detected (Fig. 5f, Fig. S3). The molecular sizes of the bands were ∼67 and ∼74 kDa. The detection levels of each band varied by samples and antibody lot-dependent manners; however, an ∼67-kDa band was consistently detected among assays, implicating that, in HIV(IN:M50I), GagPol was partially digested at a lower level. GagPol autoprocessing is initiated at site 3 (5, 22, 30, 36), followed by site 5 (30) or site 1 (36). Since the 67-kDa polypeptide band was consistently detected by anti-PR antibody, we speculated that the 67-kDa band contains PR, and the polypeptide might result from the cleaved product between PR and RT at cleavage site 7 in HIV(IN:M50I) (Fig. 5b) and be composed of MA/CA/p2/NC/TF/p6*/PR (Fig. 5c). To define the component, WB using anti-MA or anti-CA antibodies was conducted. The antibodies detected the 67-kDa band (Fig. 5g and h), indicating that the 67-kDa band contains MA, CA, and PR and is most likely a cleaved product at site 7. To address this hypothesis, we created mutant viruses in which cleavage site 7 in HIV(WT) and HIV(IN:M50I) was changed from Phe-Pro to Val-Pro by point mutagenesis, and the resulting viruses were designated HIV(WT_Δ7) and HIV(M50I_Δ7), respectively (Fig. 5d and e). A virus that lacks cleavage site 7, which makes a PR and RT fusion protein (PR-RT), still possesses functional PR activity (37). We therefore expected that if the IN:M50I mutation had no direct impact on PR activity, HIV (M50I_Δ7) would function as well as HIV (WT_Δ7) at GagPol processing. Those constructs were transfected into HEK293T cells, viral particles were collected, and then WB analysis was performed using those virus lysates with a polyclonal anti-PR antibody. As anticipated, the 67-kDa band was no longer present in HIV (M50I_Δ7) (Fig. 5f); instead, dominant bands at 73 kDa and 90 kDa with other minor bands were detected, which were also detected at a comparable level in HIV(WT_Δ7). WB analysis using anti-MA, -CA, and -IN antibodies were also conducted, and comparable levels of mature MA-, CA-, and IN-sized bands were detected in both HIV(WT_Δ7) and HIV(IN:M50I_Δ7) (Fig. 5g, h, and i). These findings indicated that the IN:M50I mutation alters the order of the autoprocessing rather than directly inhibiting PR function and, consequently, maturation of the released virions fails due to inhibition of the initial cleavage at cleavage site 3.
Identification of compensatory mutations.
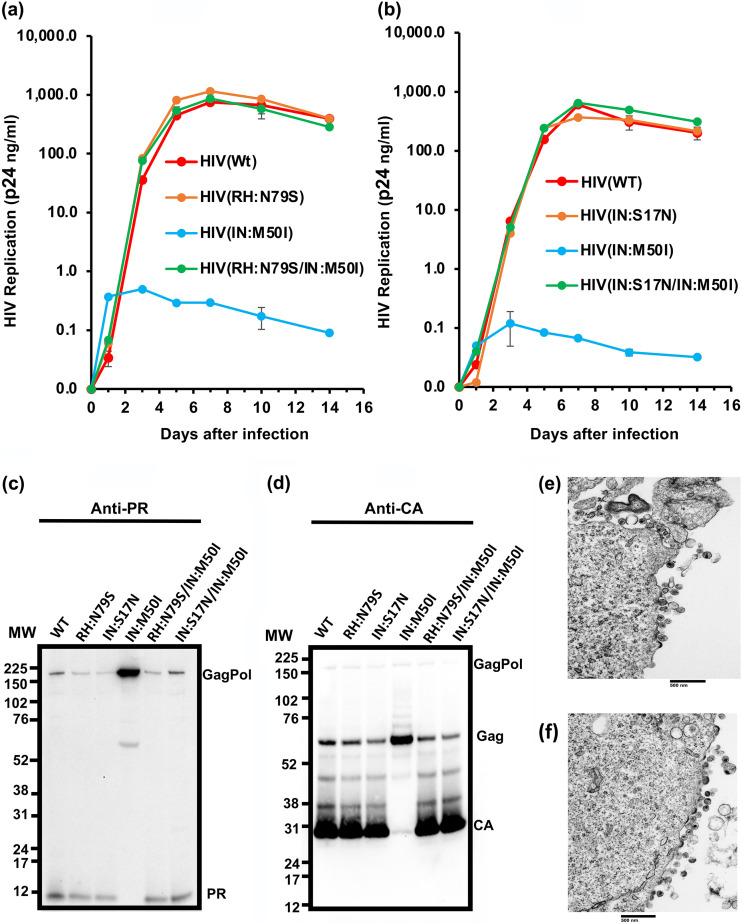
Our viral fitness results demonstrated that the IN:M50I mutation was a lethal mutation when introduced as a single change; however, since it was identified from a study of circulating virions in antiretroviral drug treatment-naive patients, we postulated that the circulating viruses must also contain a compensatory mutation(s) elsewhere in the genome. It has been reported that HIVNL4.3 carrying the IN:M50I mutation is replication competent in vitro (38), and HIVNL4.3 exhibits RH:N79S polymorphism (39). This variant was also found among the quasispecies from the patient pool from which IN:M50I was identified (Table 1). Thus, we presumed that RH:N79S is a compensatory mutation for IN:M50I. Another mutation in IN, IN:S17N, was also present among the swarm of replicating viruses, suggesting that this also is a compensatory mutation. To test these hypotheses, we constructed variants carrying a combination of IN:M50I and RH:N79S [HIV(RH:N79S/IN:M50I)] or IN:M50I and IN:S17N [HIV (IN:S17N/IN:M50I)] and analyzed their ability to replicate. Both double mutant virions restored the ability to replicate (Fig. 6a and b, Fig. S4a and b) and led to the formation of mature virions containing the cleaved PR and CA proteins (Fig. 6c and d) in WB, and amounts of released virus from the producing cells were comparable to that of the wild type (Table S1). As anticipated, TEM demonstrated that the released viral particles of the double mutants were indistinguishable from that of HIV(WT) containing core proteins, and in the presence of the compensatory mutation, we did not detect the accumulated buds on the cell surface (Fig. 6e and f).
FIG 6.
Evaluation of RH:N79S or IN:S17N mutation on HIV(IN:M50I) virus replication and GagPol processing. (a and b) PHA-stimulated primary CD4+ T cells were infected with HIV(WT) or HIV(IN:M50I) with HIV(RH:N79S) or HIV(RH:N79S/IN:M50I) (a) or with HIV(IN:S17N) or HIV(IN:S17N/IN:M50I) (b). The infected cells were cultured for 14 days with medium changed every 3 to 4 days. HIV replication was monitored using a p24 antigen capture kit. Representative data from two independent assays are presented. Data are presented as means ± SD (n = 3). (c and d) Viral lysates of HIV(WT), HIV(RH:N79S), HIV(IN:S17N), HIV(IN:M50I), HIV(RH:N79S/IN:M50I), and HIV(IN:S17N/IN:M50I) underwent WB analyses using anti-PR (c) and anti-CA antibodies (d). (e and f) Comparison of the morphology of viral particles using TEM. HEK293T cells were transfected with plasmid DNA encoding HIV(RH:N79S/IN:M50I) (e) or HIV(IN:S17N/M50I) (f) and then cultured for 24 h. Cells were fixed for TEM analysis.
To determine the potential clinical relevance of these findings, we analyzed the population of viruses carrying these mutations using the Los Alamos HIV Sequence Database (40), since IN:M50I has been observed in clinical studies (41, 42). Of 5,100 HIV sequences, 401 (7.9% of the total sequences) carried the IN:M50I mutation (Tables S2 and S3). Two hundred fifty-two sequences carried either RN:N79S or IN:S17N (the populations of IN:M50I with RH:N79S and IN:S17N were 3.8% and 1.1%, respectively). Thus, additional compensatory mutations for IN:M50I may be present in the remaining 149 variants.
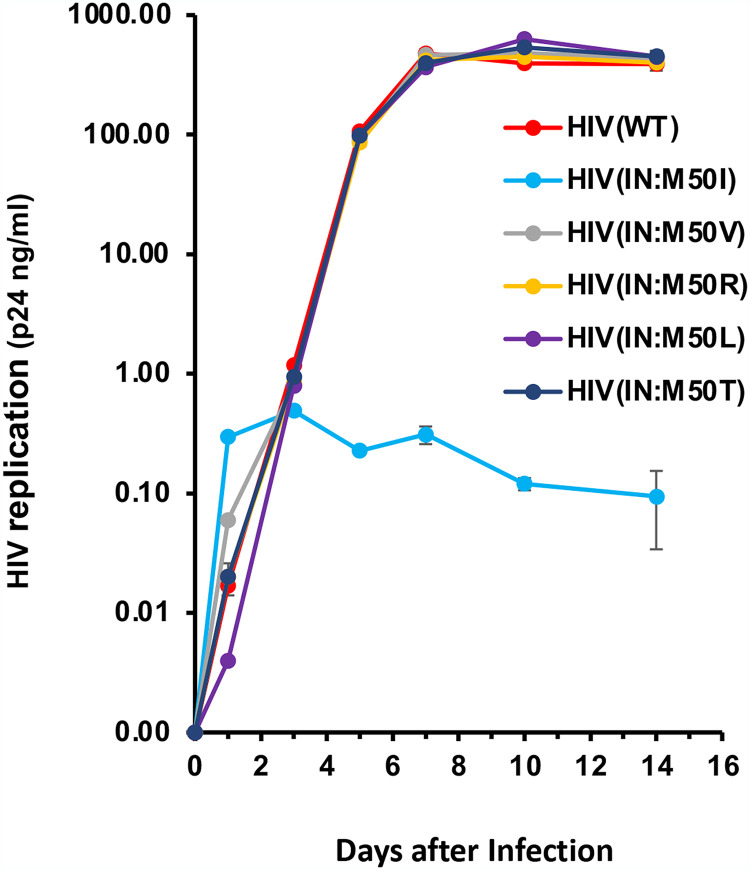
This population analysis also revealed several changes at IN:M50. In addition to IN:M50I, we observed IN:M50L, IN:M50R, IN:M50T, and IN:M50V mutations. To define the role of these mutations in viral fitness, we constructed variants containing each mutation [HIV(IN:M50V), HIV(IN:M50R), HIV(IN:M50L), and HIV(IN:M50T)] and assessed replication fitness. Unlike M50I, those mutants replicated at the same level as HIV(WT) (Fig. 7), highlighting the uniqueness of the M50I variant.
FIG 7.
Impact of mutations at codon 50 in IN on HIV replication. PHA-stimulated primary CD4+ T cells were infected with HIV(WT) or variants containing a different mutation at codon 50 as described in Materials and Methods. The infected cells were cultured for 14 days, changing medium every 3 to 4 days. HIV replication was monitored using a p24 antigen capture kit. Representative data from two independent assays are presented as means ± SD (n = 3).
Population analysis.
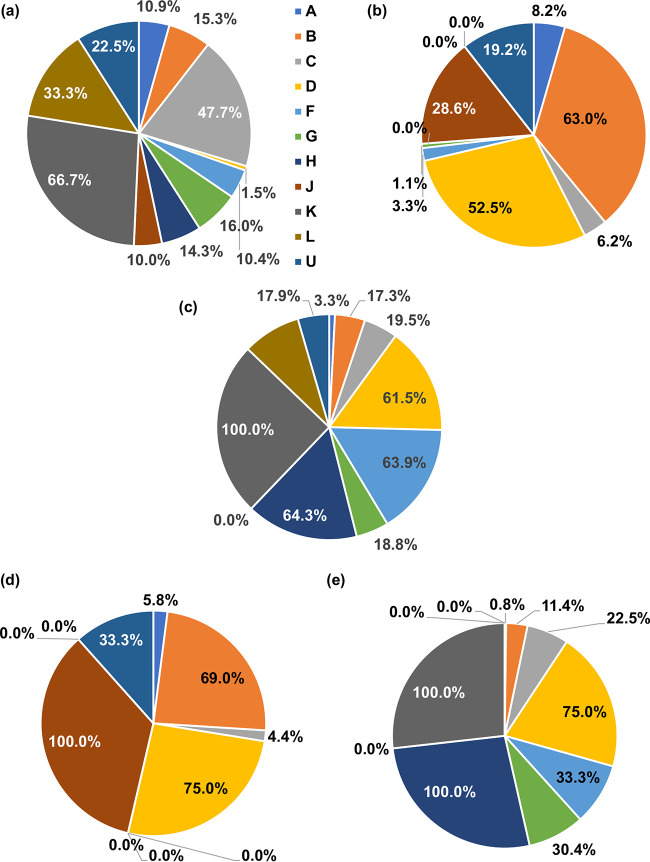
To further define whether the variants RH:N79S, IN:S17N, and IN:M50I emerge in specific HIV-1 subtypes or unique subtypes, we performed a population analysis using the Los Alamos Database. Although the IN:M50I population in subtype D (1.5%) was relatively smaller than those of other subtypes, overall, the mutation was detected in all analyzed subtypes, including CRFs (Fig. 8a, Table S4); thus, IN:M50I emerges without any subtype specificity. Of interest, the compensatory mutations RH:N79S and IN:S17N were not detected in subtypes H, K, and L (Fig. 8b, Table S5) and subtype J (Fig. 8c, Table S6), respectively, suggesting that those subtypes contain other compensatory mutations. To define the frequency of RH:N79S/IN:M50I or IN:S17N/IN:M50I in all subtypes, further population analysis was conducted. Even though the combination of RH:N79S/IN:M50I or IN:S17N/IN:M50 was detected in most subtypes, the frequency was not 100% (Fig. 8d and e, Tables S7 and S8), suggesting the presence of other compensatory mutations against IN:M50I in all subtypes.
FIG 8.
Population analysis in HIV subtypes. A total of 15,453 HIV sequences were obtained from the Los Alamos HIV database, and amino acid (aa) sequences were compared to that in the corresponding HXB2 codon sequence. Population (%) of IN:M50I (a), RH:N79S (b), IN:S17N(c), and the frequency of the combination of RH:N79S with IN:M50I (d) and IN:S17N with IN:M50I (e) in each subtype is depicted using pie charts.
DISCUSSION
Recently, a total of 14 nonsynonymous SNPs correlating with different levels of viremia in treatment-naive HIV-infected patients were reported (12). However, the specific impact of each amino acid substitution on viral fitness was not investigated. In the present study, we generated HIV-1 variants carrying each of the 14 SNPs to examine the impact of these mutations on viral fitness. We found that the IN:M50I mutation is associated with the loss of replicative capacity. This defect appears to be due to the inhibition of PR autoprocessing by the embedded PR in GagPol polyprotein. Furthermore, we demonstrated that compensatory mutations in IN and RH rescue the loss of replicative capacity and maturation.
It has been reported that GagPol processing is sensitive to the dynamics of viral assembly and that PR activation is required for correct polyprotein processing (43).
Interestingly, our TEM images of HIV(IN:M50I) demonstrated the abnormal assembly of virions at the cell membrane. WB studies illustrated that HIV(IN:M50I) mutant lacked mature PR, implicating inhibition of autoprocessing. A series of FRET analyses illustrated that GagPol(IN:M50I) could form homodimers, and the dimerization efficiency was higher than that of GagPol(WT). Therefore, IN:M50I suppresses autoprocessing after homodimerization, most likely at the initial cleavage step, which is regulated by the mutation. We presumed this suppression was caused via a structural hindrance; however, in silico predicted modeling did not indicate any significant difference in the structure between WT and IN:M50I; therefore, IN:M50I mutation may alter the structure in GagPol polyprotein or increase the stiffness in the protein dynamics, followed by decreasing the flexibility of the polyprotein or enhancing the efficiency of oligomerization of GagPol polymerization. This change may subsequently suppress the initial autoprocessing. Further study defined the mechanism of the inhibition. Crystal analysis of the entire length of GagPol polyprotein, even full-length IN, was not completed due to unclear structure at the C-terminal domain (31, 33). Amino acid substitution of IN in the crystal structure analysis had greatly improved solubility (44); thus, the M50I mutation may contribute to the structure analysis of IN.
The molecular events leading to viral assembly and release have been extensively investigated using Gag polyproteins (45–49). Using sequentially deleted IN mutated virus, it is reported that IN plays a role in assembly (8, 31, 50–53). A role for IN in proteolytic processing of GagPol and Gag polyproteins has been reported using several mutants, i.e., amino acid-substituted or domain-truncated IN (8, 50, 51). Selected mutations associated with resistance to integrase inhibitor KF116 regulated proteolytic processing of Gag and GagPol (9, 54), and quinoline-based allosteric integrase inhibitors (ALLINIs) not only suppress integration but also block the formation of mature virions by impairing their maturation (55, 56). These results indicate that the GagPol structure and the activity of autoprocessing by the embedded PR are intimately linked with the structure of IN; however, the mechanism of regulation is poorly investigated. In the present study, we demonstrated that in addition to the IN domain, the RH domain in the GagPol polyprotein is also involved in the regulation of autoprocessing. To define the molecular mechanism of the role of RH:N79S mutation in GagPol, further studies are needed to elucidate the mechanism of the RH-mediated regulation of autoprocessing. This study may reveal a novel target to regulate HIV assembly and budding.
When we performed Western blotting using a different lot of HIV(IN:M50I) lysates with different lot numbers of polyclonal antibodies, we consistently observed the ∼67-kDa band but never detected a mature PR band (the cleaved product at sites 6 and 7 in Fig. 5b). Therefore, we assumed that PR is still functional in the presence of IN:M50I, but the mutations may interrupt the conventional initial autoprocessing by PR and induce a disorder in the processing. The order of autoprocessing by PR is well-investigated. It is generally agreed that initial autocleavage occurred at site 3 (Fig. 5a) (5, 23, 36), followed by cleavage at site 5 (5) or site 1 (36) as an intramolecular process (5). The enzymatic digestion at site 7 in HIV(WT) is considered a late step in the processing (36). Using HIV(IN:M50I_Δ7), we demonstrated that the profile of GagPol-cleaved products in the mutant was comparable to that in HIV(IN:WT_Δ7), in essence restoring the defect created by the mutation in IN and identifying this as a critical site for the interaction of IN with the GagPol polyproteins and autoprocessing. Since the suppression was restored in HIV(IN:M50I_Δ7), it may be caused by an indirect effect at site 3, but it is regulated by site 7. We need further study to delineate the mechanism of the inhibition of the initiation and regulation by site 7. This study may further define the regulation mechanism in the initiation of autoprocessing.
In summary, the current study has characterized naturally occurring SNPs in IN- and RH-regulated assembly, leading to abnormal virion formations and suppressing autoprocessing in the released virions. These findings illustrate the multifunctional aspects of several HIV-1 proteins and indicate that the overall structure-function relationships of the intact virion proteins involve more than Gag. Further study of functional and structural analyses of RH and IN in GagPol polyproteins and the GagPol interaction mechanism may provide new insights into the regulation of viral assembly, budding, and viral maturation in HIV. The study may disclose new targets to develop novel anti-HIV drugs that may be effective against HIV variants, especially multiclass drug-resistant viruses.
MATERIALS AND METHODS
Approval for these studies, including all sample materials, was granted by the National Institute of Allergy and Infectious Diseases Institutional Review Board, and participants provided informed written consent prior to blood being drawn. All experimental procedures in these studies were approved by the National Cancer Institute at Frederick and Frederick National Laboratory for Cancer Research and performed in accordance with the relevant guidelines and regulations.
Cells.
Peripheral blood mononuclear cells (PBMCs) were isolated from healthy donors’ apheresis packs using lymphocyte separation medium (ICN Biomedical, Aurora, OH, USA) (57). CD4+ T cells were purified from PBMCs using CD4 MicroBeads (Miltenyi Biotec, Auburn, CA, USA) according to the manufacturer’s instructions. The purity of the cell types was at least 90% based on flow-cytometric analysis. Cell viability was determined using a trypan blue (Thermo Fisher, Waltham, MA, USA) exclusion method. HEK293T cells and HeLa cells were obtained from the ATCC (Manassas, VA, USA) and maintained as previously described (57).
Site-directed mutagenesis.
Mutations of interest were induced on pNL4.3, a plasmid encoding full-length HIVNL4.3 (39) (the plasmid was obtained from M. Martin through the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health) by the QuikChange lightning kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s protocol. Briefly, ApaI (New England Biolab [NEB], Ipswich, MA, USA) and EcoRI (NEB) fragments of pNL4.3 were cloned into the pCR2.1 vector (Thermo Fisher). This clone was used as a shuttle vector and served as the backbone for mutagenesis studies. Primers used in the mutagenesis assays are listed in Table S9 in the supplemental material. All mutagenesis results were confirmed by Sanger DNA sequencing using the BigDye Terminator v3 (Thermo Fisher) and SeqStudio genetic analyzer (Thermo Fisher). After confirming the sequence, the intended clones were digested with ApaI and EcoRI, and then the fragments from the ApaI and EcoRI digestion were used to replace with the corresponding fragment of pNL4.3. DNA sequencing was used to ascertain that each clone possessed the intended mutations.
Recombinant HIV-1 viruses.
Recombinant HIV-1s were prepared by transfection of pNL4.3 by following a method previously reported (58). Briefly, 4 × 106 HEK293T cells or 2 × 106 HeLa cells seeded in a 100-mm dish were transfected with 10 μg of each plasmid purified using the endotoxin-free plasmid isolation kit (Qiagen, Germantown, MD, USA) with TransIT-293 (Mirus, Houston, TX, USA) for HEK293T cells or TransIT-HeLa MONSTER (Mirus) for HeLa cells. Culture supernatants were collected at 48 h after transfection. After centrifugation at 500 × g for 5 min, the supernatants were filtered through a 0.45-μm-pore-size membrane filter (Millipore-Sigma, Louis, MO, USA). Viral particles in the filtrate (8 ml) were ultracentrifuged on 20% sucrose on an HBS (10 mM HEPES [Quality Biochemical Inc, QBI, Gaithersburg, MD, USA]–150 mM NaCl) cushion for 2 h at 4°C and then resuspended in 80 μl of RP10 or phosphate-buffered saline (PBS) (QBI) and stored at −80°C until use. Viruses were only used after a single thaw. The concentration of HIV in each stock was determined by a p24 antigen capture kit (PerkinElmer, Waltham, MA, USA). As the HIV(IN:M50I) variant demonstrated a lower level of virus production from HEK293T cells (Table S1), for Western blot and TEM analyses, the variant was produced using three to five 100-mm dishes at a time and combined the produced particles from the multiple dishes for downstream assays.
HIV replication assay.
The levels of HIV-1 replication were determined using primary CD4+ T cells as follows. CD4+ T cells were stimulated with 5 μg/ml phytohemagglutinin (PHA; Millipore-Sigma) in complete RPMI 1640 (Thermo Fisher) supplemented with 10 mM HEPES, 10% (vol/vol) fetal bovine serum (FBS; Thermo Fisher), and 50 μg/ml gentamicin (Thermo Fisher) (RP10). The PHA-stimulated CD4+ T cells (10 × 106 cells) were infected with 10 ng of p24 of HIV in 1 ml for 2 h at 37°C and then cultured at 1 × 106 cells/ml in complete RPMI 1640 supplemented with 20 U/ml of recombinant IL-2 (Millipore-Sigma) (RP10) for 14 days at 37°C in T25 flasks. Half of the culture supernatants were exchanged with fresh RP10 every 3 or 4 days of incubation. HIV-1 replication activity was determined by measuring p24 antigen levels in the culture supernatants using the p24 antigen capture assay as described above.
Western blotting.
Virus lysates for Western blot analysis were prepared using radioimmunoprecipitation assay (RIPA) lysis buffer (Boston Biology, Boston, MA, USA) with a proteinase inhibitor cocktail (Millipore-Sigma). Total protein concentrations in the samples were quantified using a bicinchoninic acid (BCA) protein assay kit (Thermo Fisher), and 1 or 5 μg of total viral protein was subjected to experimentation. SDS-PAGE was run using 4–12% NuPAGE Bis-Tris gels (Thermo Fisher) in morpholinepropanesulfonic acid (MOPS) buffer (Thermo Fisher) for most cases (58); however, to detect PR, SDS-PAGE was run in morpholineethanesulfonic (MES) acid buffer (Thermo Fisher), and Western blot analyses were conducted using the ECL Prime Western blot detection system (Thermo Fisher) as previously described (57). Mouse monoclonal anti-HIV-1 CA (p24) antibody (ab9071), rabbit polyclonal anti-HIV PR antibody (ab211627), rabbit polyclonal anti-HIV1 RT antibody (ab63911), and mouse monoclonal anti-HIV IN antibody (ab66645) were obtained from Abcam (Cambridge, MA, USA). Goat polyclonal anti-MA(p17) antibody and rabbit polyclonal anti-Nef antibody were kindly provided by R. Gorelick at the Frederick National Laboratory for Cancer Research and R. Swanstrom (through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH [no. 2949]) (59), respectively. Horseradish peroxidase-conjugated anti-rabbit IgG and anti-mouse IgG antibodies (NA931V and NA934V) were obtained from Thermo Fisher.
TEM analysis.
Plasmid-transfected HEK293T cells were cultured for 24 h. Cell-free transection supernatants containing recombinant viruses or the transfected cells were fixed with 2.5% glutaraldehyde (E.M. Sciences, Warrington, PA) in Millonig’s sodium phosphate buffer (Tousimis Research, Rockville, MD) (G-MPB) for 1 min at room temperature. Viruses were pelleted as described above, and the fixed cells were harvested using a scraper and centrifuged at 500 × g for 15 min. The virus and the cell pellets were stored in fresh G-MPB at 4°C overnight. Virus particles were pelleted as described above, and then pelleted particles were fixed at 4°C using G-MPB without disturbing the pellets. All fixed samples were washed repeatedly in Millonig’s buffer and then incubated for 2 h in 1.0% osmium tetroxide (E.M. Sciences) in Millonig’s buffer. Following rinsing steps in ultrapure water and en bloc staining with 2.0% uranyl acetate (E.M. Sciences), the samples were dehydrated in a series of graded ethanol, infiltrated, and embedded in Spurr’s plastic resin (E.M. Sciences). Embedded blocks were sectioned using a Leica UC7 Ultramicrotome. Seventy- to 80-nm sections were collected on 150-mesh copper grids and poststained with Reynold’s lead citrate. Samples were examined in an FEI Tecnai Spirit twin transmission electron microscope, operating at 80 kV.
RNA copy assay.
To quantify copy numbers of HIV genomic RNA in virus stocks, the RealTime HIV-1 assay kit (Abbot Laboratories, Abbott Park, IL, USA) was used. Briefly, 100-μl aliquots of HIV stocks were 10-fold serially diluted and then combined with an internal control RNA from the kit. All assays were conducted on the automated m2000 System (Abbot) with Abbott mSample preparation system reagents (Abbot). The detection range of the assay system was 40 × 106 to 10 × 106 copies/ml. A BCA protein assay was used to determine total protein concentrations in each sample, and the results from the copy assay were normalized by the amounts of protein.
FRET assay.
Interactions of Gag-Gag, Gag-GagPol, and GagPol-GagPol were measured by FRET assays using a series of the modified expression plasmids encoding HIVNL4.3 Gag or GagPol (lacking frame-shifting signal and containing inactive PR with D25N mutation) fused with fluorescent protein genes (EGFP or mSB), generating pGag(MA/EGFP/CA), pGag(MA/mSB/CA), pGagPol(MA/EGFP/CA), and pGagPol(MA/mSB/CA) (20). pGagPol constructs lacking the frameshifting signal and gag and pol were placed in-frame, and Gag-Pol polyproteins were translated without the frameshift (20). To induce an inactive PR in pNL(WT) and pNL(IN:M50I), a D25N mutation was induced in the PR of each plasmid using point mutagenesis with D25N primers (Table S9). pGagPol(MA/EGFP/CA) and pGagPol(MA/mSB/CA) were digested with ApaI and EcoRI, and the ApaI-EcoRI region was replaced with a corresponding ApaI and EcoRI fragment of PR-inactive pNL(WT) or pNL(IN:M50I). The subcloned plasmids were termed pGP(WT)GFP, pGP(WT)mSB, pGP(IN:M50I)GFP, and pGP(IN:M50I)mSB. HEK293T cells (50 × 103 cells) were seeded for 24 h on μ-Slide 8-well glass-bottom chambers (ibidi, GmbH, Planegg, Germany) and then transfected with a total of 0.26 μg of DNA (GFP vector:mSB vector, 1:1) using the TransiT-293. Twenty-four hours after transfection, cell images were taken. All images were acquired on a Zeiss AxioObserver.Z1 equipped with the LSM800 confocal module, using a Plan-Apochromat 63×/1.40 objective (Carl Zeiss Microscopy, White Plains, NY, USA); cells were maintained at 37°C and 5% CO2 during the experiments. For FRET analysis, a combination of three images was taken for each field of view, with respective excitation/emission wavelengths of 488/491 to 509 nm (donor [EGFP] channel), 561/587 to 603 nm (acceptor [mSB] channel) and 488/587 to 603 nm (FRET channel). The pinhole was set at 35 μm throughout all three FRET channels, i.e., 0.82 arbitrary units (AU) for donor channel and 0.70 AU for acceptor and FRET channels. Scan zoom was kept at 1.0×, resulting in a 0.099-μm pixel size. At least 24 images from three independent experiments per experimental condition were subjected to FRET analysis using the FRET and Colocalization Analyzer plugin of the FiJi app as follows. For each field of view, a bleed-through-corrected FRET index image was generated using the previously described formula (60):
where αD and αA are signal bleed-through coefficients from donor-only and acceptor-only transfection conditions, generated for each independent experiment, and IFRET, IDonor, and IAcceptor are signal intensities from FRET, donor, and acceptor channels, respectively; a 1-pixel-radius median filter was subsequently applied to resulting FRET index images to smooth signal dispersion. Resulting background levels on filtered images were determined for each independent experiment and filtered out, thereby creating masks mapping every above-background FRET-positive pixel. These masks were then applied to unfiltered FRET index, donor, and acceptor images, allowing for unbiased analysis of FRET efficiency for each field of view as a whole and not singled-out cells. Apparent FRET efficiencies were calculated as previously described (20, 61) as a function of acceptor (EA):
Tubulin immunostaining.
HEK293T cells (3 × 105 cells) were plated on sterilized round 15-mm no. 1 glass coverslips (VWR) in 12-well plates and seeded for 24 h prior to transfection. Transfection was conducted using 1.5 μg of pG-GFP, pGP(WT)GFP, or pGP(IN:M50I)GFP as described above, and 24 h after transfection, culture medium was removed. The cells were fixed with methanol-free 4% formaldehyde (E.M. Sciences) for 15 min at room temperature; fixative was subsequently removed, and cells were washed four times in PBS (Quality Biological). Cells were then permeabilized using 0.1% Triton X-100 (Calbiochem, Millipore-Sigma) in PBS for 5 min at room temperature and then washed four times in PBS. After cells were blocked using BlockAid blocking solution (Thermo Fisher Scientific) for 1 h at room temperature, coverslips were recovered from plates and placed upside-down on a 100-ml drop of antibody-containing BlockAid (ab204686, recombinant Alexa Fluor 647 anti-beta tubulin antibody at 1:100; ab206627, recombinant Alexa Fluor 555 anti-beta tubulin antibody at 1:100) and incubated in a humidity chamber overnight at 4°C in the dark. Coverslips were then washed 4 times in PBS and mounted on glass slides in a drop of ProLong diamond antifade mountant with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific). Imaging was carried out on a Zeiss AxioObserver.Z1 motorized microscope using a Plan-Apochromat 63×/1.40 objective and an LSM800 confocal module.
Homology modeling of HIV-1 integrase wild-type and M50I mutation structures.
The wild-type HIV-1 integrase protein sequence was based on the NL4.3 genome sequence. The full-length structure was predicted using the Baker lab-hosted Robetta server, which follows the RosettaCM protocol, which recombines fragments from the aligned template structures and de novo fragments, building the unaligned regions, followed by structural optimization and refinement (62). To construct the homology model of IN based on the DNA-free templates, the sequences of three domains of IN, i.e., N-terminal domain (NTD), catalytic core domain (CCD), and C-terminal domain (CTD), were separately used as query sequences. To identify the corresponding templates we used BLAST searches of the PDB database (63), where three structures (PDB entries 1WJB [64], 1K6Y [34], and 1WJA [64]) were identified as the templates of CCD with an identity of 99.4%, final structures (PDB entries 2ITG [65], 1B9D [66], and 1ITG [44]) were identified as the templates of CCD with identity of 99.4%, and, finally, two structures (PDB entries 5HOT [67] and 1EX4 [32]) were chosen as the CTD templates, with identity of 100% and 94.7%, respectively. These eight templates were then used as the input for Robetta to perform the multiple-template modeling protocol. Specifically, the WT sequence with a length of 288 amino acids was submitted to the Robetta server. The default options were used for the model-building procedure, and the eight templates were uploaded. The comparative model protocol was selected with 1,000 models sampled. Finally, the best WT model was determined by carefully comparing the structures of templates. After that, the mutated sequences of M50I and the WT sequence were submitted to Robetta with the same protocol as that described above, using the predicted WT structure as the single template for modeling corresponding structures. The two final predicted models (IN:WT and IN:M50I) and the alignment between them are shown in Fig. 1a to c. It appears that the structures are aligned well, and no significant conformational change can be detected by Robetta between the wild-type and M50I mutated sequences.
Population analysis.
All subtypes of HIV-1 Pol sequences of group M (subtype A, B, C, D, F, G, H, J, K, L, U, and circular recombinant form [CRF]) were obtained by downloading from Los Alamos HIV sequence database (https://www.hiv.lanl.gov/components/sequence/HIV/search/search.html) for the genomic region covering RH and IN. One sequence per patient was downloaded. A total of 15,453 sequences were downloaded. In summary, there were 1,194 sequences of subtype A, 6,357 for subtype B, 2,880 for C, 260 for D, 144 for F, 144 for G, 14 for H, 10 for J, 3 for K, 3 for L, 40 for U, and 4,404 for CRF. Sequences that do not provide RH codon 79 (RH:79), IN codon 17 (IN:17), or IN codon 50 (IN:50) were excluded. The final number of sequences used for the analysis was RH:N79 (748 for subtype A, 5,239 for B, 2,609 for C, 202 for D, 90 for F, 89 for G, 10 for H, 7 for J, 3 for K, 3 for L and 26 for U, 3,483 for CRF), IN:S17 (1,194 for subtype A, 6,352 for B, 2,877 for C, 260 for D, 144 for F, 144 for G, 14 for H, 10 for J, 3 for K, 3 for L, 39 for U, and 4,404 for CRF), and IN:M50 (1,194 for subtype A, 6,351 for B, 2,875 for C, 260 for D, 144 for F, 144 for G, 14 for H, 10 for J, 3 for K, 3 for L, 40 for U, and 4,403 for CRF).
Statistical analysis.
Intergroup comparisons were performed using two-tailed unpaired t tests using Prism 8 software (GraphPad, San Diego, CA, USA). P values of <0.05 were considered statistically significant.
Data availability.
All data are available in the main text or the supplemental material.
ACKNOWLEDGMENTS
We thank M. Bennedbæk and J. D. Lundgren for providing information on the 14 SNPs to initiate this study, Y. Morikawa for providing plasmid constructs for FRET assay, R. Gorelick for providing anti-MA antibody, J. Kovacs for discussions, and L. Huzella, X. Jiao, and F. Scrimieri for critical readings of the manuscript.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. This research was supported in part by the National Institute of Allergy and Infectious Diseases. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
T.I. designed all studies, performed assays, analyzed data, oversaw assays, supervised, and wrote the manuscript. Q.C. performed assays. J.G.B., S.L., J.Y., H.H., M.H., and H.S. contributed assays and wrote a draft of the manuscript. R.D. and W.C. supervised and reviewed the draft manuscript, and H.C.L. conceptualized this project and wrote the manuscript. All authors reviewed the final manuscript.
We declare that we have no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Tomozumi Imamichi, Email: timamichi@mail.nih.gov.
Guido Silvestri, Emory University.
REFERENCES
- 1.Swanstrom R, Wills JW. 1997. Synthesis, assembly, and processing of viral proteins. In Coffin JM, Hughes SH, Varmus HE (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 2.Freund J, Kellner R, Konvalinka J, Wolber V, Kräusslich HG, Kalbitzer HR. 1994. A possible regulation of negative factor (Nef) activity of human immunodeficiency virus type 1 by the viral protease. Eur J Biochem 223:589–593. 10.1111/j.1432-1033.1994.tb19029.x. [DOI] [PubMed] [Google Scholar]
- 3.Gaedigk-Nitschko K, Schön A, Wachinger G, Erfle V, Kohleisen B. 1995. Cleavage of recombinant and cell derived human immunodeficiency virus 1 (HIV-1) Nef protein by HIV-1 protease. FEBS Lett 357:275–278. 10.1016/0014-5793(94)01370-g. [DOI] [PubMed] [Google Scholar]
- 4.Davis DA, Soule EE, Davidoff KS, Daniels SI, Naiman NE, Yarchoan R. 2012. Activity of human immunodeficiency virus type 1 protease inhibitors against the initial autocleavage in Gag-Pol polyprotein processing. Antimicrob Agents Chemother 56:3620–3628. 10.1128/AAC.00055-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pettit SC, Everitt LE, Choudhury S, Dunn BM, Kaplan AH. 2004. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J Virol 78:8477–8485. 10.1128/JVI.78.16.8477-8485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis JM, Clore GM, Gronenborn AM. 1999. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat Struct Biol 6:868–875. 10.1038/12327. [DOI] [PubMed] [Google Scholar]
- 7.Huang L, Li Y, Chen C. 2011. Flexible catalytic site conformations implicated in modulation of HIV-1 protease autoprocessing reactions. Retrovirology 8:79. 10.1186/1742-4690-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukovsky A, Göttlinger H. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J Virol 70:6820–6825. 10.1128/JVI.70.10.6820-6825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoyte AC, Jamin AV, Koneru PC, Kobe MJ, Larue RC, Fuchs JR, Engelman AN, Kvaratskhelia M. 2017. Resistance to pyridine-based inhibitor KF116 reveals an unexpected role of integrase in HIV-1 Gag-Pol polyprotein proteolytic processing. J Biol Chem 292:19814–19825. 10.1074/jbc.M117.816645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balakrishnan M, Yant SR, Tsai L, O'Sullivan C, Bam RA, Tsai A, Niedziela-Majka A, Stray KM, Sakowicz R, Cihlar T. 2013. Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PLoS One 8:e74163. 10.1371/journal.pone.0074163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lundgren JD, Babiker AG, Gordin F, Emery S, Grund B, Sharma S, Avihingsanon A, Cooper DA, Fätkenheuer G, Llibre JM, Molina JM, Munderi P, Schechter M, Wood R, Klingman KL, Collins S, Lane HC, Phillips AN, Neaton JD. 2015. Initiation of antiretroviral therapy in early asymptomatic HIV infection. N Engl J Med 373:795–807. 10.1056/NEJMoa1506816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabrielaite M, Bennedbaek M, Ekenberg C, Kan VL, Touloumi G, Vandekerckhove L, Turner D, Neaton J, Lane HC, Lundgren JD, Marvig RL. 2020. Association of HIV and host genetic variants in antiretroviral therapy-naive persons. Abstr Conf Retroviruses Opportun Infect, Boston, MA. 8 to 11 March, 2020. [Google Scholar]
- 13.Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang DW, Lempicki RA, Baseler MW, Veenstra TD, Young HA, Lane HC, Imamichi T. 2011. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol 186:4541–4545. 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sui H, Zhou M, Imamichi H, Jiao X, Sherman BT, Lane HC, Imamichi T. 2017. STING is an essential mediator of the Ku70-mediated production of IFN-λ1 in response to exogenous DNA. Sci Signal 10:aah5054. 10.1126/scisignal.aah5054. [DOI] [PubMed] [Google Scholar]
- 15.Rein A. 2019. RNA packaging in HIV. Trends Microbiol 27:715–723. 10.1016/j.tim.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dilley KA, Nikolaitchik OA, Galli A, Burdick RC, Levine L, Li K, Rein A, Pathak VK, Hu WS. 2017. Interactions between HIV-1 Gag and viral RNA genome enhance virion assembly. J Virol 91:e02319-16. 10.1128/JVI.02319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundquist WI, Kräusslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettit SC, Lindquist JN, Kaplan AH, Swanstrom R. 2005. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology 2:66. 10.1186/1742-4690-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datta SA, Clark PK, Fan L, Ma B, Harvin DP, Sowder RC, Jr, Nussinov R, Wang YX, Rein A. 2016. Dimerization of the SP1 region of HIV-1 Gag induces a helical conformation and association into helical bundles: implications for particle assembly. J Virol 90:1773–1787. 10.1128/JVI.02061-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takagi S, Momose F, Morikawa Y. 2017. FRET analysis of HIV-1 Gag and GagPol interactions. FEBS Open Biol 7:1815–1825. 10.1002/2211-5463.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettit SC, Sheng N, Tritch R, Erickson-Viitanen S, Swanstrom R. 1998. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv Exp Med Biol 436:15–25. 10.1007/978-1-4615-5373-1_2. [DOI] [PubMed] [Google Scholar]
- 22.Pettit SC, Moody MD, Wehbie RS, Kaplan AH, Nantermet PV, Klein CA, Swanstrom R. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol 68:8017–8027. 10.1128/JVI.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pettit SC, Henderson GJ, Schiffer CA, Swanstrom R. 2002. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J Virol 76:10226–10233. 10.1128/jvi.76.20.10226-10233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erickson-Viitanen S, Manfredi J, Viitanen P, Tribe DE, Tritch R, Hutchison CA, III, Loeb DD, Swanstrom R. 1989. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res Hum Retroviruses 5:577–591. 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- 25.Gowda SD, Stein BS, Engleman EG. 1989. Identification of protein intermediates in the processing of the p55 HIV-1 gag precursor in cells infected with recombinant vaccinia virus. J Biol Chem 264:8459–8462. 10.1016/S0021-9258(18)81809-9. [DOI] [PubMed] [Google Scholar]
- 26.Kräusslich HG, Ingraham RH, Skoog MT, Wimmer E, Pallai PV, Carter CA. 1989. Activity of purified biosynthetic proteinase of human immunodeficiency virus on natural substrates and synthetic peptides. Proc Natl Acad Sci USA 86:807–811. 10.1073/pnas.86.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mervis RJ, Ahmad N, Lillehoj EP, Raum MG, Salazar FH, Chan HW, Venkatesan S. 1988. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol 62:3993–4002. 10.1128/JVI.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich HG. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol 72:2846–2854. 10.1128/JVI.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettit SC, Gulnik S, Everitt L, Kaplan AH. 2003. The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage. J Virol 77:366–374. 10.1128/jvi.77.1.366-374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettit SC, Clemente JC, Jeung JA, Dunn BM, Kaplan AH. 2005. Ordered processing of the human immunodeficiency virus type 1 GagPol precursor is influenced by the context of the embedded viral protease. J Virol 79:10601–10607. 10.1128/JVI.79.16.10601-10607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dar MJ, Monel B, Krishnan L, Shun MC, Di Nunzio F, Helland DE, Engelman A. 2009. Biochemical and virological analysis of the 18-residue C-terminal tail of HIV-1 integrase. Retrovirology 6:94. 10.1186/1742-4690-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen JC-H, Krucinski J, Miercke LJW, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Natl Acad Sci USA 97:8233–8238. 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook NJ, Li W, Berta D, Badaoui M, Ballandras-Colas A, Nans A, Kotecha A, Rosta E, Engelman AN, Cherepanov P. 2020. Structural basis of second-generation HIV integrase inhibitor action and viral resistance. Science 367:806–810. 10.1126/science.aay4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang JY, Ling H, Yang W, Craigie R. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J 20:7333–7343. 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanja M, Cappy P, Levy N, Oladosu O, Schmidt S, Rossolillo P, Winter F, Gasser R, Moog C, Ruff M, Negroni M, Lener D. 2020. NKNK: a new essential motif in the C-terminal domain of HIV-1 group M integrases. J Virol 94:e01035-20. 10.1128/JVI.01035-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Könnyű B, Sadiq SK, Turányi T, Hírmondó R, Müller B, Kräusslich HG, Coveney PV, Müller V. 2013. Gag-Pol processing during HIV-1 virion maturation: a systems biology approach. PLoS Comput Biol 9:e1003103. 10.1371/journal.pcbi.1003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cherry E, Liang C, Rong L, Quan Y, Inouye P, Li X, Morin N, Kotler M, Wainberg MA. 1998. Characterization of human immunodeficiency virus type-1 (HIV-1) particles that express protease-reverse transcriptase fusion proteins. J Mol Biol 284:43–56. 10.1006/jmbi.1998.1968. [DOI] [PubMed] [Google Scholar]
- 38.Wares M, Mesplède T, Quashie PK, Osman N, Han Y, Wainberg MA. 2014. The M50I polymorphic substitution in association with the R263K mutation in HIV-1 subtype B integrase increases drug resistance but does not restore viral replicative fitness. Retrovirology 11:7. 10.1186/1742-4690-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. 10.1128/JVI.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foley B, Apetrei C, Hahn B, Mizrachi I, Mullins J, Rambaut A, Wolinsky S, Korber B. 2018. HIV sequence compendium 2018. Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]
- 41.Ceccherini-Silberstein F, Malet I, Fabeni L, Dimonte S, Svicher V, D'Arrigo R, Artese A, Costa G, Bono S, Alcaro S, Monforte A, Katlama C, Calvez V, Antinori A, Marcelin AG, Perno CF. 2010. Specific HIV-1 integrase polymorphisms change their prevalence in untreated versus antiretroviral-treated HIV-1-infected patients, all naive to integrase inhibitors. J Antimicrob Chemother 65:2305–2318. 10.1093/jac/dkq326. [DOI] [PubMed] [Google Scholar]
- 42.Low A, Prada N, Topper M, Vaida F, Castor D, Mohri H, Hazuda D, Muesing M, Markowitz M. 2009. Natural polymorphisms of human immunodeficiency virus type 1 integrase and inherent susceptibilities to a panel of integrase inhibitors. Antimicrob Agents Chemother 53:4275–4282. 10.1128/AAC.00397-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattei S, Anders M, Konvalinka J, Kräusslich H-G, Briggs JAG, Müller B. 2014. Induced maturation of human immunodeficiency virus. J Virol 88:13722–13731. 10.1128/JVI.02271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981–1986. 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 45.Cashikar AG, Shim S, Roth R, Maldazys MR, Heuser JE, Hanson PI. 2014. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. Elife 3:e02184. 10.7554/eLife.02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raiborg C, Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458:445–452. 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 47.Freed EO. 2015. HIV-1 assembly, release and maturation. Nat Rev Microbiol 13:484–496. 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL, Myszka DG, Sundquist WI. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65. 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 49.Rose KM, Hirsch VM, Bouamr F. 2020. Budding of a retrovirus: some assemblies required. Viruses 12:1188. 10.3390/v12101188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol 69:2729–2736. 10.1128/JVI.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin CG, Taddeo B, Haseltine WA, Farnet CM. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J Virol 68:1633–1642. 10.1128/JVI.68.3.1633-1642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohammed KD, Topper MB, Muesing MA. 2011. Sequential deletion of the integrase (Gag-Pol) carboxyl terminus reveals distinct phenotypic classes of defective HIV-1. J Virol 85:4654–4666. 10.1128/JVI.02374-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Houwer S, Demeulemeester J, Thys W, Taltynov O, Zmajkovicova K, Christ F, Debyser Z. 2012. Identification of residues in the C-terminal domain of HIV-1 integrase that mediate binding to the transportin-SR2 protein. J Biol Chem 287:34059–34068. 10.1074/jbc.M112.387944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma A, Slaughter A, Jena N, Feng L, Kessl JJ, Fadel HJ, Malani N, Male F, Wu L, Poeschla E, Bushman FD, Fuchs JR, Kvaratskhelia M. 2014. A new class of multimerization selective inhibitors of HIV-1 integrase. PLoS Pathog 10:e1004171. 10.1371/journal.ppat.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fontana J, Jurado KA, Cheng N, Ly NL, Fuchs JR, Gorelick RJ, Engelman AN, Steven AC. 2015. Distribution and redistribution of HIV-1 nucleocapsid protein in immature, mature, and integrase-inhibited virions: a role for integrase in maturation. J Virol 89:9765–9780. 10.1128/JVI.01522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jurado KA, Wang H, Slaughter A, Feng L, Kessl JJ, Koh Y, Wang W, Ballandras-Colas A, Patel PA, Fuchs JR, Kvaratskhelia M, Engelman A. 2013. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc Natl Acad Sci USA 110:8690–8695. 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fakruddin JM, Lempicki RA, Gorelick RJ, Yang J, Adelsberger JW, Garcia-Pineres AJ, Pinto LA, Lane HC, Imamichi T. 2007. Noninfectious papilloma virus-like particles inhibit HIV-1 replication: implications for immune control of HIV-1 infection by IL-27. Blood 109:1841–1849. 10.1182/blood-2006-02-001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brann TW, Dewar RL, Jiang MK, Shah A, Nagashima K, Metcalf JA, Falloon J, Lane HC, Imamichi T. 2006. Functional correlation between a novel amino acid insertion at codon 19 in the protease of human immunodeficiency virus type 1 and polymorphism in the p1/p6 Gag cleavage site in drug resistance and replication fitness. J Virol 80:6136–6145. 10.1128/JVI.02212-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shugars DC, Smith MS, Glueck DH, Nantermet PV, Seillier-Moiseiwitsch F, Swanstrom R. 1993. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J Virol 67:4639–4650. 10.1128/JVI.67.8.4639-4650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hachet-Haas M, Converset N, Marchal O, Matthes H, Gioria S, Galzi JL, Lecat S. 2006. FRET and colocalization analyzer–a method to validate measurements of sensitized emission FRET acquired by confocal microscopy and available as an ImageJ Plug-in. Microsc Res Tech 69:941–956. 10.1002/jemt.20376. [DOI] [PubMed] [Google Scholar]
- 61.van Rheenen J, Langeslag M, Jalink K. 2004. Correcting confocal acquisition to optimize imaging of fluorescence resonance energy transfer by sensitized emission. Biophys J 86:2517–2529. 10.1016/S0006-3495(04)74307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, Baker D. 2013. High-resolution comparative modeling with RosettaCM. Structure 21:1735–1742. 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res 28:235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM. 1997. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol 4:567–577. 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 65.Bujacz G, Alexandratos J, Qing ZL, Clément-Mella C, Wlodawer A. 1996. The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant. FEBS Lett 398:175–178. 10.1016/s0014-5793(96)01236-7. [DOI] [PubMed] [Google Scholar]
- 66.Greenwald J, Le V, Butler SL, Bushman FD, Choe S. 1999. The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity. Biochemistry 38:8892–8898. 10.1021/bi9907173. [DOI] [PubMed] [Google Scholar]
- 67.Gupta K, Turkki V, Sherrill-Mix S, Hwang Y, Eilers G, Taylor L, McDanal C, Wang P, Temelkoff D, Nolte RT, Velthuisen E, Jeffrey J, Van Duyne GD, Bushman FD. 2016. Structural basis for inhibitor-induced aggregation of HIV integrase. PLoS Biol 14:e1002584. 10.1371/journal.pbio.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4; Tables S1 to S9<br>. Download jvi.01323-21-s0001.pdf, PDF file, 1.7 MB (1.7MB, pdf)
Data Availability Statement
All data are available in the main text or the supplemental material.