ABSTRACT
Human pegivirus (HPgV) infects peripheral leukocytes but was recently shown to be a neurotropic virus associated with leukoencephalitis in humans. In the present study, we investigated the neural cell tropism of HPgV as well as its effects on host immune responses. HPgV wild type (WT) and a mutant virus with a deletion in the HPgV NS2 gene (ΔNS2) were able to productively infect human astrocytes and microglia but not neurons or an oligodendrocyte-derived cell line. Of note, the ΔNS2 virus replicated better than WT pegivirus in astrocytes, with both viruses being able to subsequently infect and spread in fresh human astrocyte cultures. Infection of human glia by HPgV WT and ΔNS2 viruses resulted in suppression of peroxisome-associated genes, including PEX11B, ABCD1, PEX7, ABCD3, PEX3, and PEX5L, during peak viral production, which was accompanied by reduced expression of IFNB, IRF3, IRF1, and MAVS, particularly in ΔNS2-infected cells. These data were consistent with analyses of brain tissue from patients infected with HPgV in which we observed suppression of peroxisome and type I interferon gene transcripts, including PEX11B, ABCD3, IRF1, and IRF3, with concurrent loss of PMP70 immunoreactivity in glia. Our data indicate that human astrocytes and microglia are permissive to HPgV infection, resulting in peroxisome injury and suppressed antiviral signaling that is influenced by viral diversity.
IMPORTANCE Human pegiviruses are detected in 1 to 5% of the general population, principally infecting leukocytes, although their effects on human health remain uncertain. Here, we show that human pegivirus infects specific neural cell types in culture and human brain and, like other neurotropic flaviviruses, causes suppression of peroxisome and antiviral signaling pathways, which could favor ongoing viral infection and perhaps confer susceptibility to the development of neurological disease.
KEYWORDS: human pegivirus, glia, peroxisome, type I interferon, human pegivirus
INTRODUCTION
Human pegivirus (HPgV) is a positive-sense, single-stranded RNA virus belonging to the family Flaviviridae. Since its discovery in 1967, HPgV has alternatively been known as GB virus C (GBV-C) and hepatitis G virus (HGV) (1). The classification of HGV was predicated on the assumption that the virus was hepatotropic, although later evidence showed that the virus did not replicate in the liver and, therefore, HGV was renamed human pegivirus, in which “pegivirus” represents “persistent G” virus (2). There are at least 7 different HPgV genotypes known to infect humans, with HPgV genotype 2 being most prevalent in North America and Europe (3). HPgV genotypes 3, 4, 6, and 7 are predominantly found in Asian countries, including Japan, China, and the Philippines (3). HPgV can be transmitted vertically (maternal-fetal transmission) or horizontally (sexual exposure or exposure through contaminated blood products) (4). The prevalence of HPgV in the general population ranges from 1 to 4% in high-income countries but can be as high as 20% in low- and middle-income countries (5 to 7). Previous studies suggest that women are twice as likely as men to be infected with HPgV, although conclusive evidence regarding sex differences influencing viral infection has yet to be established (8). Based upon the transfusion frequency worldwide, at least 7,000 people are predicted to receive HPgV-contaminated blood products daily (9). HPgV/HIV-1 coinfection is common, especially among at-risk individuals, including those with other chronic viral infections, which creates chronically dysregulated systemic antiviral immune responses (10). Data from several studies suggest that 20 to 45% of people infected with HIV-1 are coinfected with HPgV (11, 12). HPgV infection may exert beneficial effects in the setting of HIV-1 infection; in contrast, among HCV-infected individuals, HPgV coinfection is occasionally associated with worsened outcomes, including slower HCV clearance (13). Flaviviruses can suppress antiviral signaling pathways involving mitochondrial antiviral signaling protein (MAVS) and type I interferon-associated signaling pathways through their actions on peroxisome functions (14). Thus, it is plausible that HPgV coinfection amplifies HCV’s inhibitory effects on peroxisomal antiviral signaling mechanisms, prompting greater HCV production and associated disease (15, 16).
The HPgV genome (approximately 9.5 kb in size) contains a large open reading frame (ORF) flanked by 5′ and 3′ untranslated regions (UTRs) and is organized similarly to the HCV genome (5, 10, 17), although the capsid-encoding domain remains to be defined. The ORF is translated to form a single polyprotein that is posttranslationally cleaved by both viral and host proteases to generate two envelope proteins, E1 and E2, along with six nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B), including a helicase, a protease, and an RNA-dependent RNA polymerase (17, 18). HPgV infects the spleen and bone marrow, primarily B and T lymphocytes (CD4+ and CD8+) (19). The latter data suggest that HPgV is a lymphotropic virus. Of note, peripheral blood mononuclear cells (PBMCs) isolated from HPgV+ patients display very low levels of viral negative-strand RNA, suggesting restricted infection (20).
While HPgV RNA was not detected in CSF from 17 encephalitis patients in an early study (21), more recently, viral RNA was detected in the brains of patients with multiple sclerosis or encephalitis (6, 22, 23). HPgV RNA was also detected in the CSF from patients with HIV/AIDS and encephalitis or HIV-associated neurocognitive disorder (24, 25) and in CSF from four patients with encephalitis and no other identified infectious agents (24, 26). Given the increasing number of patients with detectable HPgV in brain tissue or CSF with associated viral molecular diversity, we hypothesized that HPgV was neurotropic with pathogenic actions that may be affected by viral molecular diversity. Our findings indicate HPgV preferentially infects glial cells with associated injurious effects on peroxisomes and related antiviral signaling pathways.
RESULTS
HPgV infection of astrocytes.
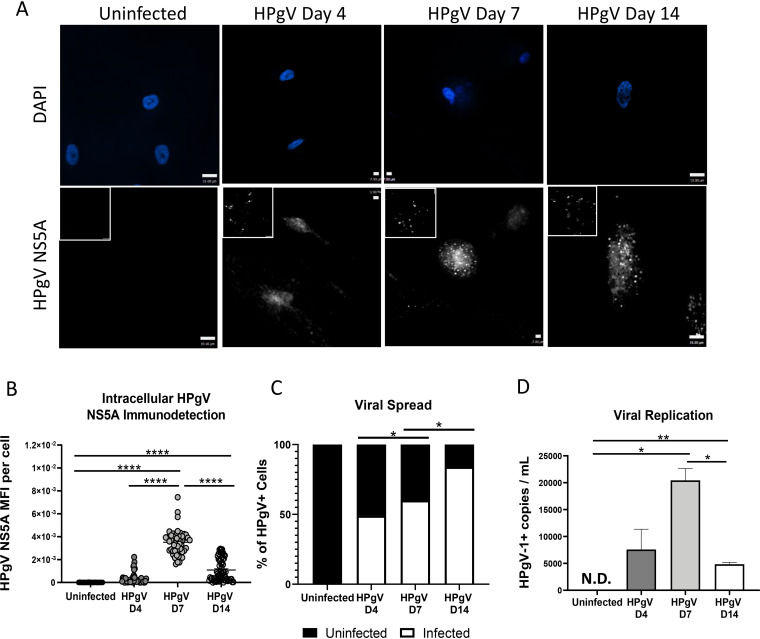
We previously reported that HPgV antigen and genome were detected in the brains of patients with fatal encephalitis, chiefly in cells resembling astrocytes that were GFAP immunopositive (23). These findings prompted us to examine the tropism of HPgV in different neural cell types. Primary human astrocytes were infected (multiplicity of infection [MOI], 0.1) with HPgV derived from U251 cells transfected with RNA encoding an infectious wild-type (WT) HPgV molecular clone. These experiments revealed that viral antigen (HPgV NS5A) was detectable at days 4, 7, and 14 postinfection (Fig. 1A; low-magnification images are presented as insets). Quantitation of intracellular viral antigen showed that immunoreactivity was evident at day 4 and peaked at day 7, with the subsequent decline in relative expression by day 14. (Fig. 1B). Analysis of HPgV NS5A immunoreactive cells indicated there was a steady increase in the number of infected cells from day 4 to day 14 (Fig. 1C). These studies were complemented by the detection of HPgV RNA in culture supernatants from infected cells, which showed positive-strand viral RNA present at day 4 but peaking at day 7 with the decline at day 14 (Fig. 1D). These data suggested that WT HPgV derived from transfected cells can infect and spread to 75% of cells in astrocyte cultures over a 2-week period.
FIG 1.
HPgV infects and spreads in human astrocytes in vitro. Astrocytes were infected with HPgV WT (MOI, 0.1) for 4, 7, or 14 days, after which cells were fixed and immunolabeled for HPgV NS5A antigen (white) and labeled with DAPI (blue); both high- and low (inset)-magnification images are shown. To quantify viral infectivity, HPgV NS5A mean fluorescence intensity (MFI) per cell (B) was assessed for a minimum of n = 50 astrocytes per condition using confocal microscopy and compared to uninfected cells (A). (C) Viral spread was determined by quantifying the number of HPgV+ cells per field of view (FoV) under each condition by immunofluorescence. (D) ddPCR of cell supernatants showed an increase in viral replication between D4 and D7, displayed by number of HPgV+ copies. Data shown represent data from two separate donors. (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; one-way ANOVA.)
Construction and expression of HPgV ΔNS2 mutant virus.
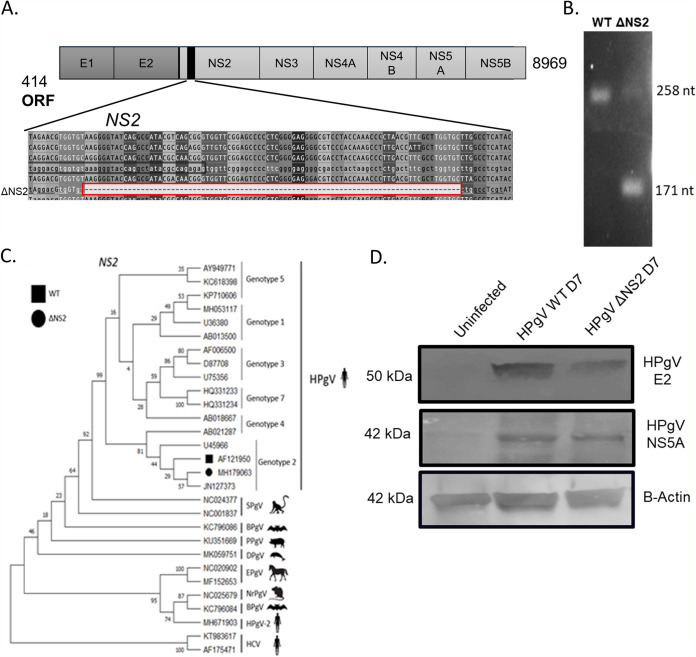
Our previous studies (23) indicated that the HPgV genome detected in brains from patients with leukoencephalitis contained an in-frame 87-nucleotide sequence (ΔNS2) deletion in the NS2 domain of the virus (Fig. 2A). PCR amplification of the NS2 domain of the WT versus the ΔNS2 viral genomes indicated that the resulting amplicon differed in size by 87 nucleotides (Fig. 2B). Construction of a phylogenetic tree using the NS2 domain revealed that the WT and the ΔNS2 mutant viral sequences clustered with HCV NS2 (Fig. 2C). To construct a ΔNS2 virus, an 87-nucleotide deletion was introduced into the plasmid encoding the infectious clone, followed by generation of viral RNA, which was then transfected into U251 cells. Viral protein expression was analyzed by Western blotting, revealing the presence of the envelope2 (E2) protein as well as the NS5A protein for both WT and ΔNS2 viruses in cellular lysates (Fig. 2D).
FIG 2.
HPgV protein detection in human astrocytes following infection. (A) The HPgV ΔNS2 mutant was characterized by an 87-nucleotide deletion located in the NS2 gene. The presence of the NS2 deletion was observed by gel electrophoresis following qPCR compared to cDNA prepared from HPgV WT RNA. (B) To verify the presence of a deletion, viral cDNAs created from WT and ΔNS2 plasmids were analyzed by Sanger sequencing. (C) Using HPgV WT (GenBank accession no. AF121950) and ΔNS2 sequences from LE-1 (GenBank accession no. MH179063), a phylogenetic tree was constructed for the mutation site in NS2. (D) Astrocytes were infected with HPgV WT or ΔNS2 (MOI, 0.1) for 7 days before lysates were harvested and immunoblotted for HPgV NS5A and E2 as indicated. This experiment was repeated in two separate biological donors.
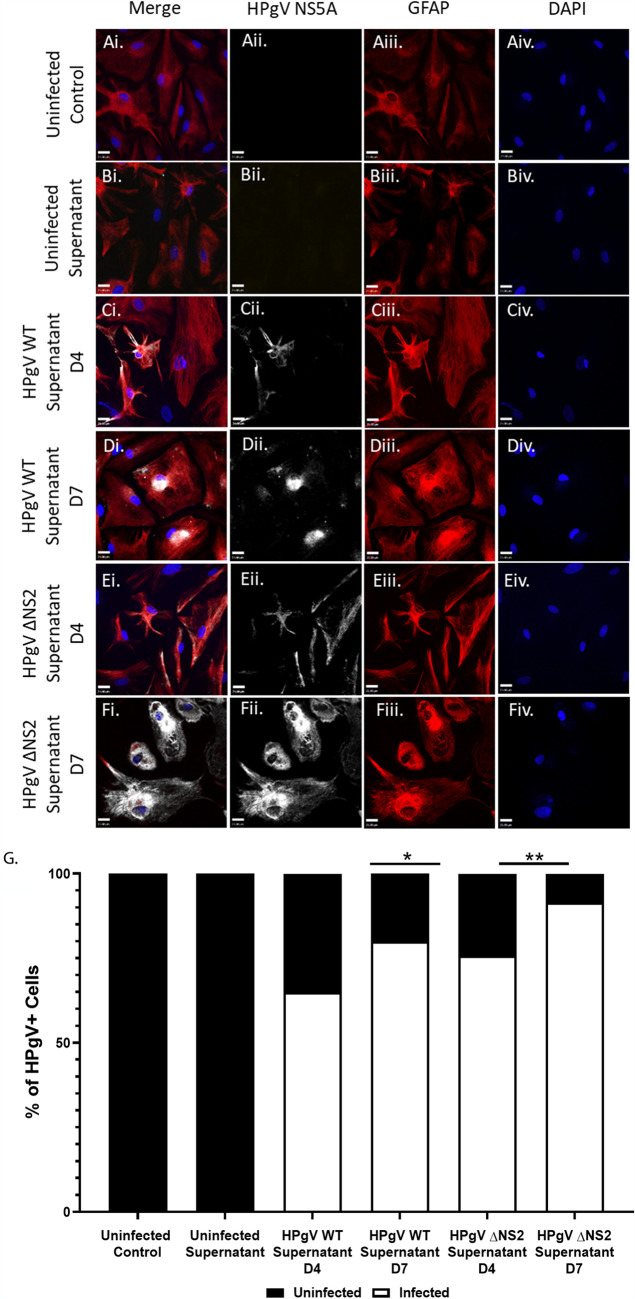
Media containing HPgV virions from previously infected astrocyte cultures were subsequently used to infect primary human astrocytes derived from a different donor (Fig. 3). Infection of human primary astrocytes with the WT or ΔNS2 viruses resulted in detectable NS5A immunoreactivity at days 4 and 7 postinfection for both viruses (Fig. 3C to F). Quantitation of viral antigen-immunopositive cells at days 4 and 7 postinfection showed viral antigen was not detectable in control or mock-infected cells, while the WT and ΔNS2 virus-infected human astrocytes exhibited viral antigen immunoreactivity with higher levels of immunopositive cells at day 7 postinfection. These studies recapitulated the studies described above, suggesting that passaged HPgV could replicate productively in infected astrocytes and spread to uninfected cells over time in primary astrocytes. Further, this study showed that a productive infection of human astrocytes was present, as supernatants from HPgV WT- and ΔNS2-infected cells were capable of infecting and spreading in fresh astrocyte cultures.
FIG 3.
HPgV WT and ΔNS2 viral particles released from infected astrocytes are infectious and spread in subsequent infection of human astrocytes. (A to G) Astrocytes were exposed to supernatants from HPgV WT (MOI, 0.1), ΔNS2 (MOI, 0.1), or uninfected supernatant. Cells were harvested and immunolabeled for HPgV NS5A (white) and GFAP (red) and labeled with DAPI at both 4 and 7 days postinfection and imaged using confocal microscopy. (G) Viral spread was determined by quantifying the number of HPgV+ cells per field of view (FoV) under each condition by immunofluorescence and the relative distribution of infected GFAP+ cells compared to uninfected GFAP+ cells is shown. (*, P < 0.05; **, P < 0.01; Fisher’s exact test.)
Neurotropism of HPgV in vitro.
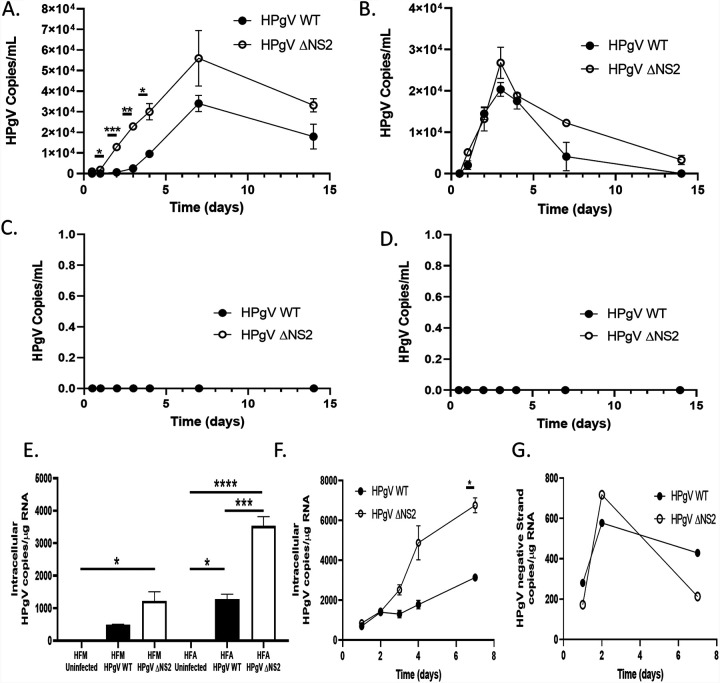
Given that HPgV antigen was detected in glial cells in vivo (23), we explored the cellular tropism of the WT and ΔNS2 viruses in different neural cell types. Both viruses replicated productively in human astrocytes, with peak viral RNA release in supernatants at day 7, although the ΔNS2 virus replicated to significantly higher levels at this time point, while viral production diminished thereafter but was still evident at day 14 postinfection (Fig. 4A). In contrast, viral production peaked at day 4 postinfection in human microglia without a significant difference between the two viruses and rapidly declined thereafter (Fig. 4B). Importantly, human primary neurons (Fig. 4C) and a human oligodendrocyte cell line (MO3.13) (Fig. 4D) were not permissive to HPgV infection based on the absence of detectable viral RNA in corresponding supernatants. Measurement of viral RNA copy numbers by droplet digital PCR (ddPCR) in lysates from infected microglia and astrocytes revealed that the ΔNS2 mutant virus replicated to higher levels than WT virus at day 14 postinfection (Fig. 4E). Similarly, intracellular positive-strand viral RNA continued to increase over time during infection of human astrocytes for both viruses, albeit at higher levels for the ΔNS2 virus (Fig. 4F). Negative-strand viral RNA was detectable at days 1, 2, and 7 postinfection in RNA from infected astrocytes, albeit at lower levels than positive-strand viral levels, for both the WT and ΔNS2 viruses (Fig. 4G). These data implied that astrocytes and microglia were permissive to HPgV infection, whereas human neurons and oligodendrocytes did not support replication of this virus.
FIG 4.
HPgV productively infects human astrocytes and microglia. Human astrocytes (A), microglia (B), neurons (C), and a human oligodendrocyte cell line (MO3.13 cells) (D) were infected with HPgV WT or ΔNS2 (MOI, 0.1), and supernatants were harvested after 12, 24, 48, 72, and 96 h as well as 7 and 14 days postinfection. Cell supernatants were analyzed to determine the number of HPgV copies present in supernatants. (E) Cell lysates from the human astrocytes and microglia were harvested for RNA and the number of intracellular HPgV copies was measured by ddPCR and normalized to the input of 1 μg of cellular RNA. Astrocytes were infected with either HPgV WT or ΔNS2 (MOI, 0.1) for 24 h, 48 h, 72 h, 96 h, or 7 days. At each time point postinfection, cells were harvested and RNA was collected. ddPCR analysis was performed using an input of 1 μg of cellular RNA to determine the number of positive (F)- or negative (G)-strand viral RNA copies intracellularly at each time point. (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001; one-way ANOVA.)
Effects of HPgV infection on host gene expression.
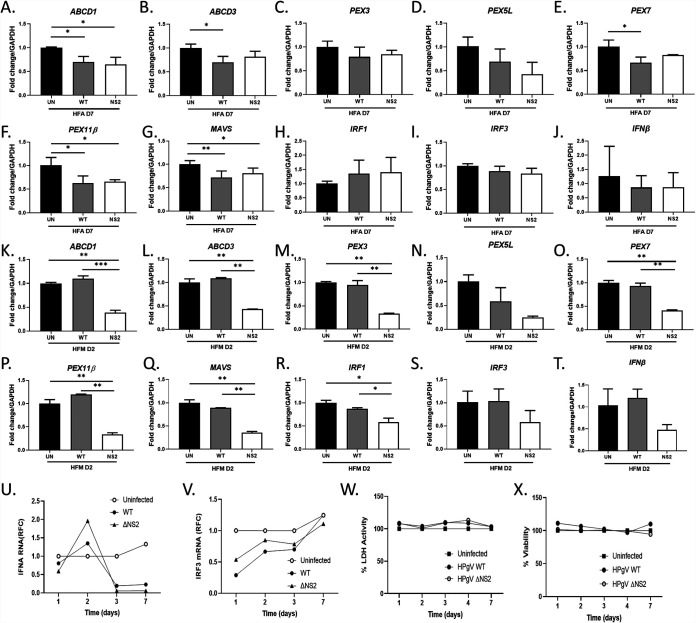
Host immune responses were examined at peak viral production times in glial cells infected with HPgV WT and ΔNS2 (MOI, 0.1) (Fig. 5A to J). Our recent studies showed that other flaviviruses affect peroxisome biogenesis and associated antiviral signaling (27); therefore, we examined the effects of HPgV infection on peroxisome-associated as well as antiviral signaling genes. The results indicated that at 7 days postinfection of astrocytes with HPgV, levels of peroxisome-associated transcripts, including ABCD1 (Fig. 5A), ABCDB (Fig. 5B), PEX7 (Fig. 5C), PEX5L (Fig. 5D), PEX7 (Fig. 5E), and PEX11B (Fig. 5F), were significantly reduced compared to mock-infected cells. Of note, peroxisome gene expression was comparatively unaffected at day 2 postinfection except for a modest induction of ABCD1 transcript levels in WT-infected cells (data not shown). Immunoblotting of cell lysates showed that HPgV infection resulted in modest reduction of PEX11β and PEX3 proteins but not PMP70 (encoded by ABCD3) (data not shown). Since peroxisome function has been linked to antiviral signaling, expression of several antiviral genes was examined, revealing that MAVS transcript levels were also suppressed (Fig. 5G) in HPgV-infected astrocytes while IRF1 (Fig. 5H), IRF3 (Fig. 5I), and IFNB (Fig. 5J) were unchanged.
FIG 5.
Altered transcription of key peroxisome and interferon-related genes are observed following HPgV WT or ΔNS2 infection of human astrocytes and microglia. (A to J) Astrocytes were infected with HPgV WT or ΔNS2 (MOI, 0.1) or left uninfected, and cell lysates were harvested for RNA following 7 days of infection (n = 3 to 5). Select peroxisome- and interferon-related transcripts, including ABCD1 (A), ABCD3 (B), PEX3 (C), PEX5L (D), PEX7 (E), PEX11B (F), MAVS (G), IRF1 (H), IRF3 (I), and IFNB (J), were analyzed. Microglia were infected with HPgV WT or ΔNS2 (MOI, 0.1) or left uninfected, and cell lysates were harvested for RNA following 2 days of infection. Select peroxisome transcripts, including ABCD1 (K), ABCD3 (L), PEX3 (M), PEX5L (N), PEX7 (O), PEX11B (P), MAVS (Q), IRF1 (R), IRF3 (S), and IFNB (T), were analyzed. The time courses were assessed for IFNA (U) and IRF3 (V) transcript levels as well as LDH release (W) and cell viability (V) in astrocytes following infection by either WT and ΔNS2 viruses and compared to mock-infected cells (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ANOVA followed by Tukey’s post hoc test).
As microglia can play pivotal roles in both free radical metabolism and antiviral signaling, we examined peroxisome and antiviral gene expression in HPgV-infected microglia at the time of peak viral production (Fig. 5K to T). These studies showed that infection by the ΔNS2 virus resulted in marked suppression of ABCD1 (Fig. 5K), ABCD3 (Fig. 5L), PEX3 (Fig. 5M), PEX5L (Fig. 5N), PEX7 (Fig. 5O), and PEX11B (Fig. 5P). Similarly, HPgV ΔNS2-infected microglia exhibited suppressed MAVS (Fig. 5Q), IRF1 (Fig. 5R), IRF3 (Fig. 5S), and IFNB (Fig. 5T) compared to both mock-infected controls and HPgV WT-infected cells. To examine the time course of type I interferon-related genes in HPgV-infected astrocytes, IFNA (Fig. 5U) and IRF3 (Fig. 5V) transcript levels were measured at days 1, 2, 3, and 7 postinfection, revealing that both genes displayed nonsignificant changes in expression over time, although IFNA showed a trend toward sustained reduced expression by day 7 for both viruses, in contrast to mock-infected cells. Lactate dehydrogenase release and cell viability assays (Fig. 5W and X) showed that suppression of peroxisome gene expression and relative lack of immune responses in HPgV-infected astrocytes were not a consequence of cell injury or death. These studies suggested that HPgV WT and ΔNS2 differentially suppress peroxisome gene expression in glia, which is associated with diminished antiviral gene expression with more pronounced effects in microglia than astrocytes.
HPgV detection in brain.
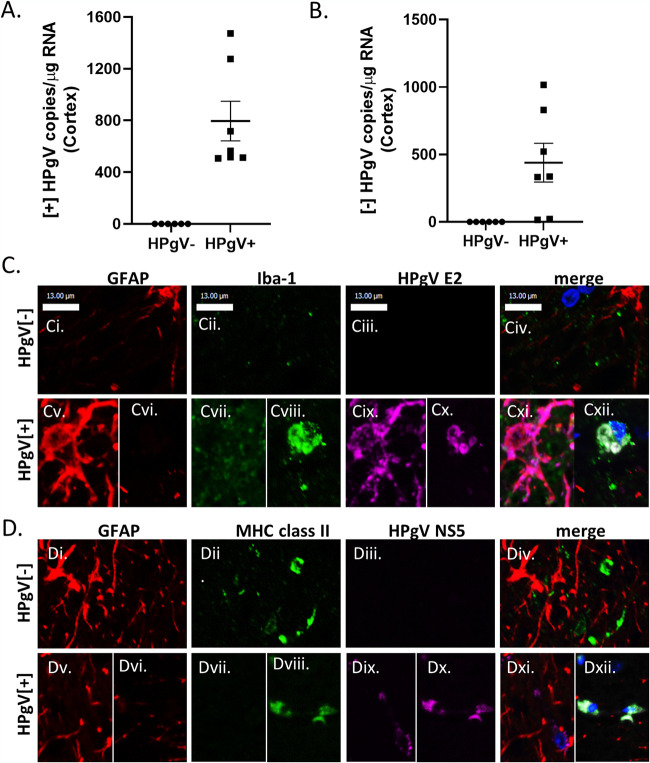
Earlier studies of patients with leukoencephalitis showed HPgV antigen and genome in the brains of patients with marked neuroinflammation. Next, we examined over 70 brains by quantitative reverse transcription-PCR (qRT-PCR) and detected viral genome in seven patients (Table 1). The brains containing HPgV RNA (HPgV+) showed positive (Fig. 6A)- and negative (Fig. 6B)-strand RNA (n = 7) in brain compared to patients without detectable viral RNA (n = 6, HPgV−). Of interest, among all patients with HPgV RNA in brain, none exhibited the ΔNS2 mutation, HIV-1 coinfection, or premortem neurological disorders. We also examined HPgV E2 and NS5A immunoreactivity in brain sections from HPgV+ and HPgV− patients, which disclosed the presence of HPgV E2 immunoreactivity in GFAP (astrocyte) and Iba-1 (microglia/macrophage) immunopositive cells (Fig. 6C). Similarly, HPgV NS5A immunoreactivity was evident in both GFAP and major histocompatibility complex (MHC) class II immunopositive cells (Fig. 6D). These studies showed that both HPgV RNA and protein were detectable in the brains without leukoencephalitis or associated premortem neurological disorders.
TABLE 1.
Patient clinical and demographic features of patients with (HPgV+) and without (HPgV−) brain detection by HPgV
| Characteristic | Value(s) for: |
|
|---|---|---|
| HPgV− (n = 6) | HPgV+ (n = 7) | |
| Mean age [yr (+SD)] | 49.5 (±6.2) | 48 (±10.9) |
| Gender | 5 male, 1 female | 5 male, 2 female |
| Neuropathology | No anomalies (n = 4), white matter microinfarcts (n = 2) | No anomalies (n = 4), acute neuronal necrosis (CA1) (n = 1), ALS (n = 1), atherosclerosis with infarction (n = 1) |
| Substance use history | None reported (n = 4), occasional marijuana use (n = 1), tobacco use (n = 1) | Tobacco use, alcohol, cocaine, methamphetamine, LSD, and crystal meth use (n = 1), alcoholism with tobacco use (n=1), none reported (n = 5) |
| Comorbidities | None reported (n = 5), metastatic cancer (without central nervous system involvement) (n = 1) | None reported (n = 7) |
FIG 6.
HPgV positive- and negative-strand RNA and viral protein detection in human brain. (A) HPgV+ NS3 copies were detected in HPgV+ (n = 7) brain samples but not in HPgV− (n = 6) cortex samples. (B) Similarly, HPgV− E2 copies were detected in the same samples in HPgV+ (n = 7) and was not detected in HPgV− (n = 6) samples. Frontal lobe tissue sections from HPgV+ or HPgV− patients were immunolabeled for GFAP (red), Iba-1 (green), and viral protein HPgV E2 (purple) (C) or HPgV NS5A (D) and imaged using confocal microscopy.
HPgV infection suppresses peroxisome and antiviral gene expression in human brain.
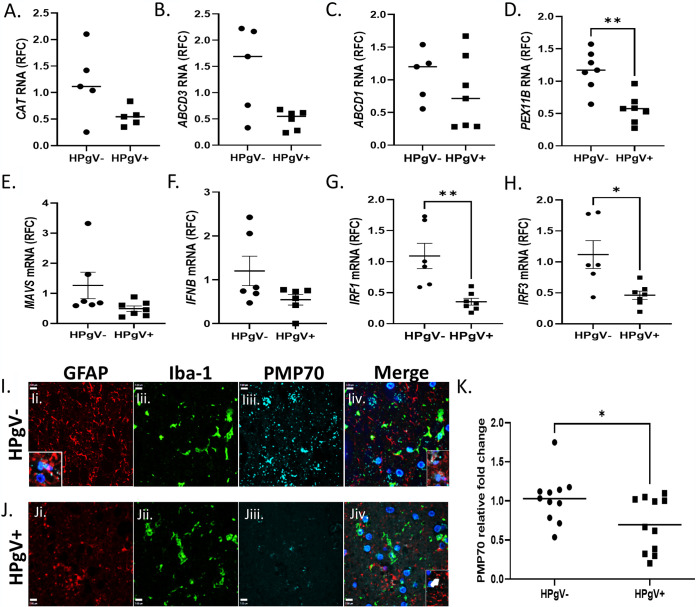
HPgV infection of astrocytes and microglia resulted in suppression of peroxisome gene transcription in vitro in a gene-specific manner (Fig. 5), prompting us to examine the expression of human peroxisome and antiviral genes in brain in conjunction with the presence or absence of HPgV infection. Analyses of peroxisome transcript abundance in patients’ brains indicated that CAT (Fig. 7A), ABCD3 (Fig. 7B), and ABCD1 (Fig. 7C) did not differ between groups, although PEX11B (Fig. 7D) was suppressed in the HPgV-infected patients. Likewise, antiviral transcript levels including, MAVS (Fig. 7E) and IFNB (Fig. 7F), were similar between groups, although IRF1 (Fig. 7G) and IRF3 (Fig. 7H) were significantly reduced in HPgV+ brains. Immunofluorescence studies showed that PMP70 was detected in the HPgV− brains, especially in GFAP and Iba-1 immunopositive cells without HPgV infection (Fig. 7I), while PMP70 immunoreactivity in glia was suppressed in the HPgV+ brains (Fig. 7J). In HPgV+ brains, PMP70 immunodetection was notably reduced in cells where immunoreactivity was colocalized with HPgV NS5A immunoreactivity, compared to nearby glia that lacked HPgV NS5A immunoreactivity (data not shown). This finding describes the appearance of 2 separate populations of PMP70 immunodetection in the HPgV+ group (Fig. 7K). Quantitation of PMP70 immunoreactivity confirmed that its expression was reduced in HPgV+ brains (Fig. 7K). These studies complemented the current in vitro studies suggesting that HPgV infection resulted in diminished peroxisome-associated gene expression.
FIG 7.
Reduced peroxisome and interferon-related gene expression in brain from HPgV-infected patients. (A to H) Analyses of select peroxisome genes, including CAT (A), ABCD3 (B), ABCD1 (C), PEX11B (D), MAVS (E), IFNB (F), IRF1 (G), and IRF3 (H). (I to K) Human patient tissue slides from HPgV+ or HPgV− control patients were immunolabeled for GFAP (red), Iba-1 (green), and PMP70 (cyan) and colabeled with DAPI (blue). (*, P < 0.05, Student's t test.)
DISCUSSION
This is the first study to examine the neural cell tropism of HPgV, revealing that both the wild-type and a mutant virus associated with leukoencephalitis can productively infect and replicate in human astrocytes and microglia. Of note, the ΔNS2 mutant virus was more infectious than WT virus in these cell types, and it more robustly suppressed expression of peroxisome and antiviral genes, particularly in microglia. Given the present findings of different infection levels and associated altered immune responses for the WT versus ΔNS2 viruses, the protease domain encoded by the NS2 domain might exert effects on innate immune responses, as reported for other flaviviruses (28, 29). HPgV RNA and protein were detected in the brains of a human cohort with evidence of viral antigen localized in astrocytes and microglia. These studies were complemented by findings of reduced in vitro peroxisome gene expression in infected astrocytes and microglia. Suppression of antiviral signaling was also apparent during HPgV infection and perhaps related to peroxisomal dysfunction. In summary, these observations suggested that HPgV can infect the central nervous system in a cell type-specific manner and might contribute to brain disease through suppression of peroxisome functions, including antiviral signaling.
Previous studies have reported that other RNA viruses, including flaviviruses such as Zika virus as well as HCV, can infect astrocytes (30) as well as microglia (31), causing suppression of peroxisome-associated gene expression (14). Viral infections can also exert downstream effects on antiviral (type I and III interferon) signaling, because peroxisomes can serve as platforms for innate immune responses (27). Nonetheless, suppression of peroxisome functions might have adverse effects in terms of diminished transport or production of cholesterol and/or the elimination of reactive oxygen species, which also contribute to viral infections. In keeping with our previous study of HPgV-associated leukoencephalitis, the cell tropism for HPgV was restricted to astrocytes and, to a lesser extent, microglia. While immune responses in HPgV-infected cultured astrocytes were limited, innate responses in microglia were greater and more diverse. These circumstances might reflect differences in cell proliferation and differentiation; astrocytes tend to proliferate (favoring viral replication) more than microglia, which differentiate and constitutively express immune genes upon exposure to plastic ware. It is unlikely infection of microglia was merely phagocytosis of free virus, because viral RNA was produced and released into the supernatant by infected microglia that was apparent at several time points.
Several challenges were encountered in the current studies, including the low MOI used here (MOI, 0.1). Despite different approaches to concentrate the virus, this MOI was the highest that could be generated consistently. Another obstacle was the paucity of patients with encephalitis from which we could obtain tissue for HPgV characterization. Several patients in the current cohort of HPgV-infected patients were at risk of HPgV infection because of their histories of substance use, although in our prior study we screened over 65 human brain samples without detecting any HPgV RNA by RT-PCR (23). A more focused search for patients with immunosuppression might increase the yield of patients with HPgV infection of the brain. These studies were also hampered by the lack of an animal model. Perhaps examining pegivirus infections or adapting a mutant virus in other host species might yield an in vivo model for future studies.
While the present studies established that HPgV infects astrocytes and microglia to a lesser extent, with suppressed peroxisome and antiviral gene expression, the full breadth of HPgV’s effects on host neuroimmune responses remain to be investigated. Moreover, its putative interactions with potential chemokine receptors, including CXCR4 and CCR5, which have been proposed as viral receptors, warrants further investigations. Finally, given that HPgV might cause leukoencephalitis, therapeutic approaches are important avenues for future translational research efforts.
MATERIALS AND METHODS
Human brain tissues.
The use of autopsied brain tissues was approved by the University of Alberta Human Research Ethics Board (Biomedical), and written informed consent was received for all samples (Pro0002291). Frontal cortex sections from HPgV+ and HPgV− patients were examined. Brain tissue from patients was obtained from the National Neuro-AIDS Tissue Consortium (NNTC) (32) and the BrainPowerlab tissue archives collection (33). Human fetal tissues were obtained from 15- to 22-week aborted fetuses that were collected with the written informed consent from the donor (Pro00027660), approved by the University of Alberta Human Research Ethics Board (Biomedical).
Cell culture.
Primary fetal human microglia, astrocytes, and neurons were isolated based on differential culture conditions, as previously described (34, 35). Fetal brain tissues from 15- to 22-week fetuses were dissected, meninges were removed, and a single-cell suspension was prepared through enzymatic digestion for 1 h with 2.5% trypsin and 0.2 mg/ml DNase I, followed by trituration through a 70-μm cell strainer. Cells were washed twice with fresh medium and plated in T-75 flasks. Cultures were maintained in minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 1 mM sodium pyruvate, 1× MEM nonessential amino acids, 0.1% dextrose, 100 U/ml penicillin, 100 μg/ml streptomycin, 0.5 μg/ml amphotericin B, and 20 μg/ml gentamicin. For microglial cells, mixed cultures were maintained for 1 to 2 weeks, at which point astrocytes and neurons formed an adherent cell layer with microglia loosely attached or floating in the medium. Cultures were gently rocked for 20 to 30 min to resuspend the weakly adhering microglia in medium, which were then decanted, washed, and plated. Astrocyte cultures were passaged once per week for 4 to 6 weeks until the neurons were eliminated. Human fetal neurons were cultured in medium containing cytosine arabinoside and used within 2 weeks of culture (36). The purity of cultures was verified by immunofluorescence quantitation, as previously reported by our group (34–37). Astrocytoma U251 cell cultures (ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS and 100 U/ml penicillin, 100 μg/ml streptomycin. MO3.13 cell cultures (ATCC) were maintained in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Before experimentation, cells were differentiated for 3 days using 50 ng/ml phorbol-12-myristate 13-acetate (PMA) in serum-free RPMI before medium was removed, cells were washed with PBS, and fresh medium (serum-free) was added for future experimentation.
Viral infection.
For preparation of infectious viruses, we obtained a clone of HPgV (NIH AIDS Reagent Program 9450), which was subsequently transformed into DH5α (library efficiency) cells (Invitrogen), and plasmids were generated (geneJET plasmid kit; Thermo Scientific) as viral plasmid stocks for downstream applications (18). Several rounds of quantitative PCR and Sanger sequencing were performed on the infectious clones to ensure in-house clones had not undergone mutagenesis during transformation. The mutant HPgV ΔNS2 virus was generated from the HPgV clone described above by targeting the 87 nucleotides in the HPgV NS2 gene for site-directed mutagenesis (New England Biolabs). Primers for site-directed mutagenesis were created using NEBaseChanger online software. Plasmids were subjected to Sanger sequencing to ensure that no additional mutations were introduced during the mutagenesis procedure. For the purposes of viral transfection, plasmids were linearized using the BcuI (SpeI) restriction enzyme (Fisher Scientific) and subjected to T7 RNA synthesis (New England Biolabs) (18). Newly transcribed RNA was quantified (NanoDrop) and transfected (jetPRIME transfection reagent; Polyplus transfection) into U251 astrocytoma-derived cells (ATCC). Virus-containing culture supernatants were harvested at 7 and 10 days postinfection, and RNA was prepared for subsequent viral RNA copy number quantitation (viral RNA minikit; Qiagen). Subsequent cDNA synthesis and droplet digital PCR (ddPCR) were performed to quantify HPgV positive-strand RNA copies/ml (supernatant). To assess infection of primary human neural cells, cells were plated for 24 h and then infected with HPgV for 6 h at MOIs ranging from 0.1 to 0.5, followed by washing with phosphate-buffered saline (PBS) and incubation for the duration of the experiment. Confirmation of infection was assessed by detection of HPgV RNA in cell culture supernatants by ddPCR.
cDNA synthesis.
A volume of 8 μl of prepared viral RNA (QIAamp viral RNA minikit; Qiagen) or 1 μg of total cellular RNA (RNeasy minikit; Qiagen) was prepared and used for first-strand cDNA using Superscript III reverse transcriptase (Invitrogen, Carlsbad CA, USA) and with random hexamer primers (Roche).
qRT-PCR.
Specific genes were quantified by real-time reverse transcriptase PCR (qRT-PCR) using the CFX 96 real-time system (Bio-Rad, Mississauga, ON, Canada). The specific primers used during qRT-PCR (Table 2). Semiquantitative RT-PCR analysis was performed by monitoring, in real time, the increase of fluorescence of the SYBR green dye (iQ SYBR green supermix; Bio-Rad) on the Bio-Rad detection system as previously reported (38) and was expressed as relative fold change (RFC) of the gene of interest relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) compared to uninfected cells and uninfected controls, respectively.
TABLE 2.
PCR primers (forward and reverse sequences)
| Gene name | Species | Forward sequence | Reverse sequence |
|---|---|---|---|
| IRF1 | Human | 5′-TGC CTC CTG GGA AGA TGA-3′ | 5′-CCT GGG ATT GGT GTT ATG C-3′ |
| IFNB | Human | 5′-CAT CTA GCA CTG GCT GGA ATG-3′ | 5′-ACT CCT TGG CCT TCA GGT AAT G-3′ |
| IRF3 | Human | 5′- GCA CAG GAG GAT TTC G-3′ | 5′-AGC CGC TTC AGT GGG TTC-3′ |
| MAVS | Human | 5′-CAG GAG CAG GAC ACA GAA C-3′ | 5′-AGG AGA CAG ATG GAG ACA CAG-3′ |
| DDX58 | Human | 5′-AAA CCA GAA TTA TCC CAA CCG A-3′ | 5′-TGA TCT GAG AAG GCA TTC CAC-3′ |
| ABCD1 | Human | 5′-AGA AGC AGA GAA TCG GCA TG-3′ | 5′-ATG GAG AGC AGG GCA ATG-3′ |
| ABCD3 | Human | 5′-GGC CTG CAC GGT AAG AAA AG-3′ | 5′-CGA GAC ACC AGC ATA ACA GC-3′ |
| PEX3 | Human | 5′-AAG GGA GGC TGC AGA ATA CA-3′ | 5′-AGG CCT CTC TCA GTG TTG GA-3′ |
| PEX5L | Human | 5′-TCT CAA ACC CAA ACC AAA GC-3′ | 5′-AGA TCC ATC GGC CTT TTT CT-3′ |
| PEX7 | Human | 5′-CAT GTC CTC ATC ACC TGT AGT G-3′ | 5′-CCT CCT GAG CGT GTT CTT TAT-3′ |
| PEX11B | Human | 5′-CCC AGT CTG TTC TTA CTC CTT TG-3′ | 5′-GGC ATA GGC CAC AGC TAT TT-3′ |
| GAPDH | Human | 5′-AGC CTT CTC CAT GGT GAA GAC-3′ | 5′-CGG AGT CAA CGG ATT TGG TCG -3′ |
| NS3(+) | HPgV | 5′-CAG ATG GGG CAA CCT CGT T-3′ | 5′-GTC CAC GGC CTA GTG AAC C-3′ |
| E2(-) | HPgV | 5′-GCC ACC GGA AAT ACA CC-3′ | 5′-CTC GGT TGG TCC CGC TTA TC-3′ |
ddPCR.
Viral quantitation by ddPCR was performed using either the ddPCR supermix for probes or QX200 ddPCR EvaGreen supermix on the QX200 droplet digital PCR system (Bio Rad) and analyzed with Quantasoft (1863024, 1864035, 1864001; Bio-Rad). To calculate the number of HPgV copies per microgram of RNA, droplet numbers were first converted to the corresponding amount of cDNA (5 μl used out of 150 μl prepared) and then to the amount of RNA generated from the tissue/cell cultures (8 μl used out of 30 μl prepared). This number was then used in future calculations to arrive at the final concentration of HPgV copies/ml of supernatant or HPgV copies/μg of RNA. The specific primers used during ddPCR reactions are in Table 2.
Cell culture immunofluorescence.
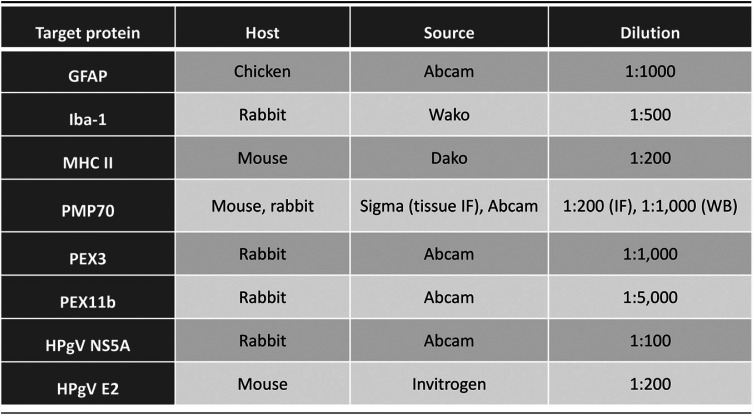
Detection of cellular proteins was performed using immunofluorescence as described previously (36). Cells were cultured on 8-well 180-μm-thick polymer coverslips (μ-Slide ibiTreat, number 80826). After 24 h, cells were fixed using 4% formaldehyde. Cells were permeabilized using 0.1% Triton in PBS, blocked using Odyssey blocking buffer (927-40000; LICOR), and incubated with primary antibody (Table 3) overnight at 4°C. Primary antibody binding was detected using species-matched secondary antibodies (Table 3). Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted using Prolong Gold antifade reagent (P36934; Invitrogen). Slides were imaged using a Wave FX spinning disc confocal microscope (Quorum Technologies) with Volocity 6.3 acquisition and analysis software (Perkin Elmer), and basic contrast enhancement was performed using automatic black-point calculation. Composite z-stack images including 10 xy planes over a total vertical distance of 5 to 10 μm were generated using the Improvision Focus Drive. All cells were imaged using a 20 or 40× oil immersion objective lens unless otherwise indicated.
TABLE 3.
Antibodies (target epitope, source, and dilution)
Immunoblot analysis.
Immunoblot analysis of tissue and cell lysates was performed as described previously (36, 39). Following protein extraction using radioimmunoprecipitation assay (RIPA) buffer, samples were quantified using a DC protein assay kit (5000112; Bio-Rad) and then suspended in Laemmli buffer (161-0747; Bio-Rad) and incubated at 95°C for 8 to 10 min. Samples were loaded onto 4 to 20% Precast SDS-PAGE gels (456-1094; Bio-Rad) and run for 1 h at 100 to 120 V. Following electrophoresis, gels were transferred onto 0.2-μm nitrocellulose (1620112; Bio-Rad) membranes using a Bio-Rad Mini Trans-Blot wet transfer system for 55 min at 0.12 A. Membranes were blocked for 1 to 4 h with Odyssey blocking buffer (927-40000; LICOR), followed by overnight incubation at room temperature with primary antibody (Table 3). Membranes were then washed 3 times for 5 min each time with PBS-T (1× PBS–0.05% Tween 20) and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (Table 2) (Jackson ImmunoResearch) for 1 h, followed by three 5-min washes. Membranes were developed with ECL reagent (32132; Thermo Scientific) and imaged using an ImageQuant LAS4000 Biomolecular Imager (GE Life Sciences). Band intensity was quantified using ImageStudioLite and normalized to β-actin.
Phylogenetic analyses.
HPgV sequences were aligned using Mega-X software, and phylogenetic trees were constructed using reported pegivirus and other related sequences by the Clustal X N‐J bootstrap method and evaluated with MEGA-X software with 10,000 replicates. Viral sequences were obtained from GenBank for the HPgV WT clone (GenBank accession no. AF121950) and from a patient from our original case report, LE-1 (GenBank accession no. MH179063), which represented HPgV ΔNS2.
Tissue immunofluorescence.
For immunofluorescence studies in brain tissues, as described previously (39), brain tissue sections were deparaffinized by incubation for 1 h at 60°C, followed by 10-min (once) and 5-min (twice) incubations in toluene baths through decreasing concentrations of ethanol to distilled water. Antigen retrieval was performed by boiling in 10 mM sodium citrate (pH 6.0). Slides were blocked with 1 mM HEPES buffer, 2% (vol/vol) horse serum, 5% (vol/vol) FBS, 0.1% (wt/vol) sodium azide in Hanks’ balanced salt solution for 4 h at room temperature. Slides were incubated with primary antibodies at 4°C overnight. Primary antibody was removed by PBS washes (5 min thrice), and slides were incubated for 3 min in Trueblack Lipofuscin autofluorescence quencher (23007; Biotium) and washed an additional three times in PBS. Slides were incubated in a mixture of 1:500 fluorescent secondary antibodies as appropriate for 2 h, stained with DAPI for 10 min, washed three times in PBS, and mounted with Prolong Gold. Slides were imaged with an inverted Wave FX spinning disc confocal microscope (Quorum Technologies). To quantify the intracellular mean fluorescence intensity (MFI) of viral antigen in GFAP+ cells, the freehand tool in Volocity 6.3 was used to delineate each GFAP-immunopositive cell as a region of interest (ROI), and the MFI of viral antigen within that ROI was recorded. To quantify the number of immunopositive cells per field of view (FOV), a threshold defined as 4× the background MFI was established separately for each FOV. FOVs were then individually contrast enhanced by setting the black-point to the threshold value (4× background) calculated above. Each GFAP-immunopositive cell was then categorized as either immunopositive or immunonegative for viral antigen based upon whether the intracellular viral signal exceeded the 4× background threshold. Isotype secondary antibody controls were used to gauge tissue autofluorescence. For PMP70 quantification, a threshold similar to that mentioned above was used, and the number of PMP70 immunopositive foci per cell was determined and expressed as fold change relative to the mean number of foci per cell in all uninfected tissues measured.
Statistical analyses.
Comparisons between two or more groups were performed by unpaired Student's t test or by analysis of variance (ANOVA) with Dunnett’s post hoc test, Fisher’s exact test, or Mann-Whitney U test in GraphPad InStat 3.0.
ACKNOWLEDGMENTS
We thank Brienne McKenzie, Jason Fernandes, and Zaikun Xu for helpful discussions.
These studies were supported by the UHF Kaye Fund. C.P. and T.C.H. also received funding from the Canada Foundation for Innovation. NNTC sites were supported by National Institutes of Health (NIMH and NINDS) grants U24MH100931, U24MH100930, U24MH100929, U24MH100928, U24MH100925).
The authors report no conflicts of interest.
Contributor Information
C. Power, Email: chris.power@ualberta.ca.
Bryan R.G. Williams, Hudson Institute of Medical Research
REFERENCES
- 1.Deinhardt F, Holmes AW, Capps RB, Popper H. 1967. Studies on the transmission of human viral hepatitis to marmoset monkeys. I. Transmission of disease, serial passages, and description of liver lesions. J Exp Med 125:673–688. 10.1084/jem.125.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams MJ, King AM, Carstens EB. 2013. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013). Arch Virol 158:2023–2030. 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- 3.Slavov SN, Maraninchi Silveira R, Hespanhol MR, Sauvage V, Rodrigues ES, Fontanari Krause L, Bittencourt HT, Caro V, Laperche S, Covas DT, Kashima S. 2019. Human pegivirus-1 (HPgV-1) RNA prevalence and genotypes in volunteer blood donors from the Brazilian Amazon. Transfus Clin Biol 26:234–239. 10.1016/j.tracli.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 4.de Lima ABF, Sa KSG, Torres M, da Silva Soares L, Resques HR, Azevedo VN, Martins Feitosa RN, Monteiro JC, Rangel da Silva ANM, Ribeiro ALR, Oliveira-Filho AB, Vallinoto ACR, Machado LFA. 2020. Low prevalence of human pegivirus 1 (HPgV-1) in HTLV-1 carriers from Belem, Para, North Region of Brazil. PLoS One 15:e0232783. 10.1371/journal.pone.0232783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. 2011. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol 92:233–246. 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kriesel JD, Hobbs MR, Jones BB, Milash B, Nagra RM, Fischer KF. 2012. Deep sequencing for the detection of virus-like sequences in the brains of patients with multiple sclerosis: detection of GBV-C in human brain. PLoS One 7:e31886. 10.1371/journal.pone.0031886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattarai N, Stapleton JT. 2012. GB virus C: the good boy virus? Trends Microbiol 20:124–130. 10.1016/j.tim.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giulivi A, Slinger R, Tepper M, Sher G, Scalia V, Kessler G, Gill P. 2000. Prevalence of GBV-C/hepatitis G virus viremia and anti-E2 in Canadian blood donors. Vox Sang 79:201–205. 10.1159/000056731. [DOI] [PubMed] [Google Scholar]
- 9.Stapleton JT, Chaloner K. 2010. GB virus C infection and non-Hodgkin lymphoma: important to know but the jury is out. Int J Cancer 126:2759–2761. 10.1002/ijc.25194. [DOI] [PubMed] [Google Scholar]
- 10.Chivero ET, Stapleton JT. 2015. Tropism of human pegivirus (formerly known as GB virus C/hepatitis G virus) and host immunomodulation: insights into a highly successful viral infection. J Gen Virol 96:1521–1532. 10.1099/vir.0.000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horemheb-Rubio G, Ramos-Cervantes P, Arroyo-Figueroa H, Avila-Rios S, Garcia-Morales C, Reyes-Teran G, Escobedo G, Estrada G, Garcia-Iglesias T, Munoz-Saucedo N, Kershenobich D, Ostrosky-Wegman P, Ruiz-Palacios GM. 2017. High HPgV replication is associated with improved surrogate markers of HIV progression. PLoS One 12:e0184494. 10.1371/journal.pone.0184494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackard JT, Ma G, Welge JA, Taylor LE, Mayer KH, Klein RS, Celentano DD, Sobel JD, Jamieson DJ, King CC. 2017. Cytokine/chemokine expression associated with human Pegivirus (HPgV) infection in women with HIV. J Med Virol 89:1904–1911. 10.1002/jmv.24836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng KT, Takebe Y, Chook JB, Chow WZ, Chan KG, Abed Al-Darraji HA, Kamarulzaman A, Tee KK. 2015. Co-infections and transmission networks of HCV, HIV-1 and HPgV among people who inject drugs. Sci Rep 5:15198. 10.1038/srep15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You J, Hou S, Malik-Soni N, Xu Z, Kumar A, Rachubinski RA, Frappier L, Hobman TC. 2015. Flavivirus infection impairs peroxisome biogenesis and early antiviral signaling. J Virol 89:12349–12361. 10.1128/JVI.01365-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira AR, Magalhaes AC, Camoes F, Gouveia A, Vieira M, Kagan JC, Ribeiro D. 2016. Hepatitis C virus NS3-4A inhibits the peroxisomal MAVS-dependent antiviral signalling response. J Cell Mol Med 20:750–757. 10.1111/jcmm.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bender S, Reuter A, Eberle F, Einhorn E, Binder M, Bartenschlager R. 2015. Activation of type I and III interferon response by mitochondrial and peroxisomal MAVS and inhibition by hepatitis C virus. PLoS Pathog 11:e1005264. 10.1371/journal.ppat.1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papageorgiou L, Loukatou S, Sofia K, Maroulis D, Vlachakis D. 2016. An updated evolutionary study of Flaviviridae NS3 helicase and NS5 RNA-dependent RNA polymerase reveals novel invariable motifs as potential pharmacological targets. Mol Biosyst 12:2080–2093. 10.1039/c5mb00706b. [DOI] [PubMed] [Google Scholar]
- 18.Xiang J, Wunschmann S, Schmidt W, Shao J, Stapleton JT. 2000. Full-length GB virus C (hepatitis G virus) RNA transcripts are infectious in primary CD4-positive T cells. J Virol 74:9125–9133. 10.1128/jvi.74.19.9125-9133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fama A, Larson MC, Link BK, Habermann TM, Feldman AL, Call TG, Ansell SM, Liebow M, Xiang J, Maurer MJ, Slager SL, Nowakowski GS, Stapleton JT, Cerhan JR. 2020. Human pegivirus infection and lymphoma risk: a systematic review and meta-analysis. Clin Infect Dis 71:1221–1228. 10.1093/cid/ciz940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellor J, Haydon G, Blair C, Livingstone W, Simmonds P. 1998. Low level or absent in vivo replication of hepatitis C virus and hepatitis G virus/GB virus C in peripheral blood mononuclear cells. J Gen Virol 79(Pt 4):705–714. 10.1099/0022-1317-79-4-705. [DOI] [PubMed] [Google Scholar]
- 21.Radkowski M, Przyjalkowski W, Lipowski D, Wang LF, Laskus T. 1998. Lack of GB virus C/hepatitis G virus sequences in cerebrospinal fluid in patients with central nervous system infections. Scand J Infect Dis 30:539. 10.1080/00365549850161647. [DOI] [PubMed] [Google Scholar]
- 22.Tuddenham R, Eden JS, Gilbey T, Dwyer DE, Jennings Z, Holmes EC, Branley JM. 2020. Human pegivirus in brain tissue of a patient with encephalitis. Diagn Microbiol Infect Dis 96:114898. 10.1016/j.diagmicrobio.2019.114898. [DOI] [PubMed] [Google Scholar]
- 23.Balcom EF, Doan MAL, Branton WG, Jovel J, Blevins G, Edguer B, Hobman TC, Yacyshyn E, Emery D, Box A, van Landeghem FKH, Power C. 2018. Human pegivirus-1 associated leukoencephalitis: clinical and molecular features. Ann Neurol 84:781–787. 10.1002/ana.25343. [DOI] [PubMed] [Google Scholar]
- 24.Hardie D, Smuts H. 2017. Human pegivirus-1 in the CSF of patients with HIV-associated neurocognitive disorder (HAND) may be derived from blood in highly viraemic patients. J Clin Virol 91:58–61. 10.1016/j.jcv.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Zhang Y, Wei F, Xu M, Mou D, Zhang T, Li W, Chen D, Wu H. 2016. Detection of GB virus C genomic sequence in the cerebrospinal fluid of a HIV-infected patient in China: a case report and literature review. Epidemiol Infect 144:106–112. 10.1017/S0950268815001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bukowska-Ośko I, Perlejewski K, Pawełczyk A, Rydzanicz M, Pollak A, Popiel M, Cortés KC, Paciorek M, Horban A, Dzieciątkowski T, Radkowski M, Laskus T. 2018. Human pegivirus in patients with encephalitis of unclear etiology, Poland. Emerg Infect Dis 24:1785–1794. 10.3201/eid2410.180161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong CP, Xu Z, Hou S, Limonta D, Kumar A, Power C, Hobman TC. 2019. Interplay between Zika virus and peroxisomes during infection. Cells 8:725. 10.3390/cells8070725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green AM, Beatty PR, Hadjilaou A, Harris E. 2014. Innate immunity to dengue virus infection and subversion of antiviral responses. J Mol Biol 426:1148–1160. 10.1016/j.jmb.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Wu Z, Wang M, Cheng A. 2017. Innate immune evasion mediated by Flaviviridae non-structural proteins. Viruses 9:291. 10.3390/v9100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vivithanaporn P, Maingat F, Lin LT, Na H, Richardson CD, Agrawal B, Cohen EA, Jhamandas JH, Power C. 2010. Hepatitis C virus core protein induces neuroimmune activation and potentiates human immunodeficiency virus-1 neurotoxicity. PLoS One 5:e12856. 10.1371/journal.pone.0012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potokar M, Jorgacevski J, Zorec R. 2019. Astrocytes in flavivirus infections. Int J Mol Sci 20:691. 10.3390/ijms20030691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgello S, Gelman BB, Kozlowski PB, Vinters HV, Masliah E, Cornford M, Cavert W, Marra C, Grant I, Singer EJ. 2001. The National NeuroAIDS Tissue Consortium: a new paradigm in brain banking with an emphasis on infectious disease. Neuropathol Appl Neurobiol 27:326–335. 10.1046/j.0305-1846.2001.00334.x. [DOI] [PubMed] [Google Scholar]
- 33.Xu Z, Asahchop EL, Branton WG, Gelman BB, Power C, Hobman TC. 2017. MicroRNAs upregulated during HIV infection target peroxisome biogenesis factors: implications for virus biology, disease mechanisms and neuropathology. PLoS Pathog 13:e1006360. 10.1371/journal.ppat.1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mamik MK, Hui E, Branton WG, McKenzie BA, Chisholm J, Cohen EA, Power C. 2017. HIV-1 viral protein R activates NLRP3 inflammasome in microglia: implications for HIV-1 associated neuroinflammation. J Neuroimmune Pharmacol 12:233–248. 10.1007/s11481-016-9708-3. [DOI] [PubMed] [Google Scholar]
- 35.Mamik MK, Asahchop EL, Chan WF, Zhu Y, Branton WG, McKenzie BA, Cohen EA, Power C. 2016. Insulin treatment prevents neuroinflammation and neuronal injury with restored neurobehavioral function in models of HIV/AIDS neurodegeneration. J Neurosci 36:10683–10695. 10.1523/JNEUROSCI.1287-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh JG, Reinke SN, Mamik MK, McKenzie BA, Maingat F, Branton WG, Broadhurst DI, Power C. 2014. Rapid inflammasome activation in microglia contributes to brain disease in HIV/AIDS. Retrovirology 11:35. 10.1186/1742-4690-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenzie BA, Mamik MK, Saito LB, Boghozian R, Monaco MC, Major EO, Lu JQ, Branton WG, Power C. 2018. Caspase-1 inhibition prevents glial inflammasome activation and pyroptosis in models of multiple sclerosis. Proc Natl Acad Sci USA 115:E6065–E6074. 10.1073/pnas.1722041115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maingat FG, Polyak MJ, Paul AM, Vivithanaporn P, Noorbakhsh F, Ahboucha S, Baker GB, Pearson K, Power C. 2013. Neurosteroid-mediated regulation of brain innate immunity in HIV/AIDS: DHEA-S suppresses neurovirulence. FASEB J 27:725–737. 10.1096/fj.12-215079. [DOI] [PubMed] [Google Scholar]
- 39.Boghozian R, McKenzie BA, Saito LB, Mehta N, Branton WG, Lu J, Baker GB, Noorbakhsh F, Power C. 2017. Suppressed oligodendrocyte steroidogenesis in multiple sclerosis: implications for regulation of neuroinflammation. Glia 65:1590–1606. 10.1002/glia.23179. [DOI] [PubMed] [Google Scholar]