ABSTRACT
Rift Valley fever virus (RVFV) is an arbovirus found throughout Africa. It causes disease that is typically mild and self-limiting; however, some infected individuals experience severe manifestations, including hepatitis, encephalitis, or even death. Reports of RVFV encephalitis are notable among immunosuppressed individuals, suggesting a role for adaptive immunity in preventing this severe complication. This phenomenon has been modeled in C57BL/6 mice depleted of CD4 T cells prior to infection with DelNSs RVFV (RVFV containing a deletion of nonstructural protein NSs), resulting in late-onset encephalitis accompanied by high levels of viral RNA in the brain in 30% of animals. In this study, we sought to define the specific type(s) of CD4 T cells that mediate protection from RVFV encephalitis. The viral epitopes targeted by CD4 and CD8 T cells were defined in C57BL/6 mice, and tetramers for both CD4 and CD8 T cells were generated. RVFV-specific CD8 T cells were expanded and of a cytotoxic and proliferating phenotype in the liver following infection. RVFV-specific CD4 T cells were identified in the liver and spleen following infection and phenotyped as largely Th1 or Tfh subtypes. Knockout mice lacking various aspects of pathways important in Th1 and Tfh development and function were used to demonstrate that T-bet, CD40, CD40L, and major histocompatibility complex class II (MHC-II) mediated protection from RVFV encephalitis, while gamma interferon (IFN-γ) and interleukin-12 (IL-12) were dispensable. Virus-specific antibody responses correlated with protection from encephalitis in all mouse strains, suggesting that Tfh/B cell interactions modulate clinical outcome in this model.
IMPORTANCE The prevention of RVFV encephalitis requires intact adaptive immunity. In this study, we developed reagents to detect RVFV-specific T cells and provide evidence for Tfh cells and CD40/CD40L interactions as critical mediators of this protection.
KEYWORDS: CD4 T cells, CD40, CD40L, RVFV, Tfh, Th1, encephalitis, Rift Valley fever virus
INTRODUCTION
Rift Valley fever virus (RVFV) is an arbovirus endemic to Africa that poses a serious threat to public health. RVFV causes disease in both humans and livestock; hence, outbreaks have economic, agricultural, and human health consequences (1). RVFV has an estimated overall mortality rate of 1 to 3%, and severe disease typically manifests as hepatitis, hemorrhagic fever, or encephalitis (2). RVFV encephalitis has been reported in 10 to 20% of cases that present for clinical care (3) and manifests as seizures, altered mental status, paresis, hyperreflexia, and ataxia (4–6). Not only are patients with central nervous system (CNS) disease 7 times more likely to die than those without CNS involvement, but those who survive RVFV encephalitis can be left with devastating neurologic sequalae. Despite these consequences, very little is known about RVFV CNS disease. Risk factors for developing RVFV CNS disease in humans have not been rigorously studied. However, individuals with altered adaptive immunity (e.g., those with chronic myelogenous leukemia or HIV) are notable among the cases of RVFV CNS disease (5, 7, 8), suggesting a role for adaptive immunity in preventing RVFV encephalitis in humans.
Adaptive immunity consists of humoral (antibody-mediated) and cellular arms. CD8 T cells, also known as cytotoxic T cells, control viral infections by identifying and destroying infected cells. CD4 T cells are multifaceted and have different functions, depending on the subtype (9). Typically, specific pathogens or antigens induce the development of different CD4 T cell subtypes. Viral infections generally activate Th1 cells, fungal infections or autoimmunity activates Th17, and allergic responses induce Th2, and providing B cell help in most antigen-specific responses falls to Tfh cells (9).
In studies to date, wild-type (WT) RVFV is almost uniformly lethal in inbred mice, which tend to succumb to fulminant hepatitis within a week of infection (10–16). Conversely, RVFV missing the interferon (IFN)-inhibiting nonstructural protein NSs, known as DelNSs RVFV, does not cause any pathology in mice when administered peripherally (17). However, when given intranasally, the same DelNSs RVFV causes acute, lethal encephalitis (18). This demonstrates that the mutant virus is still pathogenic, but the host immune response can control the infection and prevent disease after peripheral exposure. Prior studies have evaluated the role of various host innate and adaptive immune mechanisms that render DelNSs RVFV infection apathogenic (17, 19, 20). These studies have collectively demonstrated that while type I IFN prevents acute hepatitis, cellular immunity is largely responsible for preventing late-onset encephalitis. When CD4 T cells were depleted prior to infection with DelNSs RVFV, 30% of mice succumbed to late-onset encephalitis. Since CD4 T cells can have many different subtypes and functions, we sought to determine which aspects of CD4 T cell function were most important for providing protection from RVFV encephalitis. In order to define the CD4 T cell subtype(s) involved, the RVFV epitopes targeted by CD4 and CD8 T cells in C57BL/6 mice were defined, and then tetramers were developed to identify virus-specific cells. After phenotyping virus-specific cells, various mouse knockout (KO) models of Th1- and Tfh-focused pathways were used to identify the activities of those CD4 T cell subtypes that were important for preventing encephalitis.
RESULTS
Epitope mapping and tetramer development.
Epitope mapping was performed by intracellular cytokine staining of splenocytes from mice vaccinated with attenuated strains of RVFV using overlapping peptide pools in an ex vivo stimulation assay. Positive pools were deconvoluted to generate a list of individual peptides that were positive. No peptides were identified that stimulated the production of interleukin-4 (IL-4) or IL-17 from CD4 T cells, but many peptides were identified that stimulated the production of gamma interferon (IFN-γ) from CD4 and CD8 T cells. These peptides were compared to a list of predicted binding epitopes from www.IEDB.org (21) for both major histocompatibility complex class I (MHC-I) (H-2-Db or H-2-Kb) and MHC-II (H-2-IAb). Twenty-eight peptides that were both in the top 30% predicted for MHC-II binding using the IEDB consensus algorithm and positive in the empirical screening assay were selected for IAb in vitro binding assays. For class I, 49 peptides of 8 to 11 residues with percentile scores of <1.5% for either or both H-2-Kb and H-2-Db using either the IEDB consensus or NetMHCpan algorithms, and derived from a set of 14 different 15-mers positive in the empirical screening assay, were selected for H-2-Kb and -Db in vitro binding assays. If no H-2-Db or H-2-Kb was identified with the requisite threshold for a cognate 15-mer, the peptide with the highest predicted affinity was selected. The results of the binding assays for both MHC-II and MHC-I are depicted in Tables 1 and 2, respectively. Based upon the binding assay data, 4 peptides with affinities of 1,000 nM or better (boldface in Table 1) were chosen for MHC-II tetramer generation. For MHC-I tetramer generation, 9 peptides with affinity of 500 nM or better (boldface in Table 2) were chosen; in cases where multiple candidates were nested within a contiguous sequence, the optimal binder was selected. The 1,000 nM and 500 nM selection thresholds utilized for class II and class I, respectively, were chosen based on previous analyses that showed them to be associated with the vast majority of CD4 and CD8 epitopes (22, 23).
TABLE 1.
In vitro binding affinity of each downselected RVFV peptide for H-2-IAb
| Sequence | Length (aa) | Protein | Position | IC50 (nM) for H-2-IAba |
|---|---|---|---|---|
| MDNYQELAIQFAAQA | 15 | N | 1 | 2,605 |
| VREFAYQGFDARRVI | 15 | N | 25 | 578 |
| AYQGFDARRVIELLK | 15 | N | 29 | 533 |
| YGGADWEKDAKKMIV | 15 | N | 45 | 3,304 |
| DAKKMIVLALTRGNK | 15 | N | 53 | 19,430 |
| MIVLALTRGNKPRRM | 15 | N | 57 | - |
| ALTRGNKPRRMMMKM | 15 | N | 61 | - |
| AALAGWTCQALVVLS | 15 | N | 109 | - |
| QALVVLSEWLPVTGT | 15 | N | 117 | - |
| VLSEWLPVTGTTMDG | 15 | N | 121 | 20,396 |
| WLPVTGTTMDGLSPA | 15 | N | 125 | - |
| AATFTQPMNAAVNSN | 15 | N | 193 | 189 |
| NSNFISHEKRREFLK | 15 | N | 205 | 12,821 |
| KRREFLKAFGLVDSN | 15 | N | 213 | - |
| FLKAFGLVDSNGKPS | 15 | N | 217 | - |
| FGLVDSNGKPSAAVM | 15 | N | 221 | 20,243 |
| DSNGKPSAAVMAAAQ | 15 | N | 225 | 8,048 |
| LPALAVFALAPVVFA | 15 | Gn | 9 | 2,776 |
| EDATCKPVTYAGACS | 15 | Gn | 45 | - |
| YECTAQYANAYCSHA | 15 | Gn | 293 | - |
| STEITLKYPGISQSS | 15 | Gn | 389 | 386 |
| AHGLINYQCHTALSA | 15 | Gn | 441 | 38,705 |
| EGVNTKCRLSGTALI | 15 | Gc | 17 | - |
| GAEACLMLKGVKEDQ | 15 | Gc | 37 | - |
| CLMLKGVKEDQTKFL | 15 | Gc | 41 | - |
| GSLPQTRNDKTFAAS | 15 | Gc | 289 | - |
| SGLMSWFGGPLKTIL | 15 | Gc | 457 | 2,261 |
| TGLSKMWLAATKKAS | 15 | Gc | 493 | 31,329 |
Dashes indicate IC50s of >50,000 nM. Boldface indicates peptides with IC50s of <1,000 nM.
TABLE 2.
In vitro binding affinity of each downselected RVFV peptide for H-2-Db and H-2-Kb
| Sequence | Protein | Position | Length (aa) | IC50 (nM) fora: |
|
|---|---|---|---|---|---|
| H-2-Db | H-2-Kb | ||||
| LAIQFAAQAV | N | 7 | 10 | - | 2,087 |
| IQFAAQAV | N | 9 | 8 | 12,222 | 679 |
| MHPSFAGM | N | 145 | 8 | 29,217 | 119 |
| FAGMVDPSL | N | 149 | 9 | - | 20,949 |
| AGMVDPSL | N | 150 | 8 | - | - |
| PSLPGDYL | N | 155 | 8 | - | - |
| YLRAILDAHSL | N | 161 | 11 | - | - |
| RAILDAHSL | N | 163 | 9 | 278 | 1,227 |
| AILDAHSL | N | 164 | 8 | 18,411 | 196 |
| TQPMNAAV | N | 197 | 8 | 501 | - |
| TQPMNAAVN | N | 197 | 9 | 20,234 | - |
| PMNAAVNSNFI | N | 199 | 11 | 7,781 | - |
| MNAAVNSNFI | N | 200 | 10 | 876 | - |
| MNAAVNSNFIS | N | 200 | 11 | - | - |
| NAAVNSNFI | N | 201 | 9 | 4.2 | 28,721 |
| NAAVNSNFIS | N | 201 | 10 | 1,198 | - |
| AAVNSNFI | N | 202 | 8 | 68 | 10,678 |
| ISHEKRREFL | N | 209 | 10 | 1,750 | 418 |
| VVFAEDPH | Gn | 20 | 8 | - | - |
| VVFAEDPHL | Gn | 20 | 9 | - | 1,895 |
| NRPGKGHNYI | Gn | 160 | 10 | - | - |
| GISGSNSFSFI | Gc | 206 | 11 | 162 | 25,930 |
| ISGSNSFSF | Gc | 207 | 9 | 70 | 8,419 |
| ISGSNSFSFI | Gc | 207 | 10 | 12 | - |
| GSNSFSFI | Gc | 209 | 8 | - | 1,206 |
| NSFSFIES | Gc | 211 | 8 | - | 653 |
| RAPNLISYKPM | Gc | 259 | 11 | 1,409 | 902 |
| PNLISYKPM | Gc | 261 | 9 | - | 7,457 |
| SGSWNFFDWF | Gc | 447 | 10 | 2,459 | - |
| GSWNFFDWF | Gc | 448 | 9 | 3,484 | 12,537 |
| GSWNFFDWFSG | Gc | 448 | 11 | 12,334 | 26,518 |
| SWNFFDWF | Gc | 449 | 8 | 2,200 | - |
| SWNFFDWFSGL | Gc | 449 | 11 | - | 5,217 |
| WNFFDWFSGL | Gc | 450 | 10 | 28,283 | 9,131 |
| WNFFDWFSGLM | Gc | 450 | 11 | - | 230 |
| NFFDWFSGL | Gc | 451 | 9 | - | 488 |
| NFFDWFSGLM | Gc | 451 | 10 | - | 5,190 |
| NFFDWFSGLMS | Gc | 451 | 11 | - | - |
| FFDWFSGL | Gc | 452 | 8 | 23,705 | 197 |
| IGLFFLLI | Gc | 481 | 8 | 19,200 | 63 |
| IGLFFLLIYL | Gc | 481 | 10 | 1,930 | 4,590 |
| LFFLLIYL | Gc | 483 | 8 | - | - |
| LIYLGRTGL | Gc | 487 | 9 | - | 178 |
Dashes indicate IC50s of >30,000 nM. Boldface indicates peptides with IC50s of <500 nM.
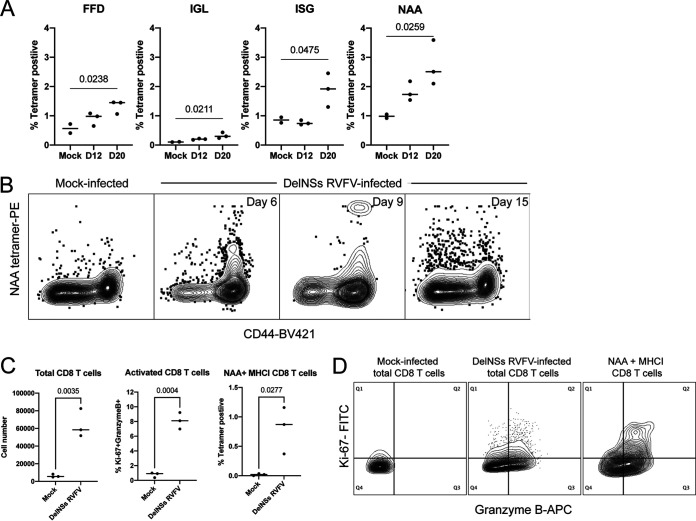
MHC-I tetramers were evaluated using cryopreserved splenocytes from DelNSs RVFV-infected mice isolated at various days postinfection (dpi). Three low-frequency MHC-I tetramers (FFD, IGL, and ISG; all located in Gc) and one high-frequency MHC-I tetramer (NAA, located in N) were identified as epitopes recognized by CD8 T cells in C57BL/6 mice (Fig. 1A and B). A previous study identified a peak of functionally activated (Ki-67+ granzyme B+) CD8 T cells in the liver 7 days after DelNSs RVFV infection (19), so RVFV-specific CD8 T cells (NAA MHC-I tetramer positive) in the liver were examined for Ki-67 and granzyme B expression. There was a marked expansion in total CD8 T cells, activated CD8 T cells, and NAA MHC-I tetramer-positive CD8 T cells in the livers of DelNSs RVFV-infected mice compared to mock-infected mice 7 dpi (Fig. 1C). The NAA MHC-I tetramer-positive cells were highly represented in the Ki-67+ granzyme B+ population, but not all tetramer-positive cells expressed these activation markers (Fig. 1D).
FIG 1.
Use of MHC-I tetramers reveals that RVFV-specific CD8 T cells are activated and proliferating in the livers of DelNSs RVFV-infected mice. Four MHC-I tetramers (named according to the first 3 amino acids present in the peptide sequence) that recognize RVFV-specific CD8 T cells were identified in the spleens of DelNSs RVFV-infected C57BL/6 mice (A). The P values from a two-way ANOVA with post hoc comparisons to mock infected are shown for statistically significant associations. Each symbol represents one mouse. The NAA tetramer exhibited a high CD44+ tetramer-positive frequency in the spleen at various time points during acute infection (B). Expansion of total CD8 T cells and activation of CD8 T cells and virus-specific CD8 T cells were all noted in the livers of DelNSs RVFV-infected mice 7 dpi (C). The P values from Student’s t test are given for statistically significant associations. Each symbol represents one mouse. While many CD8 T cells in the livers of infected mice demonstrated an activated phenotype (Ki-67+ granzyme B+ [middle panel]) compared to mock-infected mice (left panel), NAA MHC-I tetramer-positive cells were highly represented among the activated CD8 T cells (right panel) (D).
Two high-frequency MHC-II tetramers, AYQ and VRE, were identified using cryopreserved splenocytes obtained 36 dpi with DelNSs RVFV (Fig. 2A). Since these tetramers contained the same core epitope, VRE was used in all subsequent studies. Fresh cell preparations from the liver, spleen, and popliteal lymph node of DelNSs-infected mice were obtained 6 and 13 dpi for tissue-specific MHC-II tetramer analysis (Fig. 2B). The highest frequency of RVFV-specific CD4 T cells was found in the liver, consistent with the liver being a main target of RVFV infection (24). The spleen contained the highest numbers of RVFV-specific CD4 T cells: the average mouse spleen contains approximately 1 × 108 total splenocytes, and 2 × 106 were used in the assay.
FIG 2.
MHC-II tetramers reveal that RVFV-specific CD4 T cells in the liver and spleen are Th1 and Tfh focused. Two MHC-II tetramers (AYQ and VRE) with the same core epitope identified RVFV-specific CD4 T cells from the spleen of an infected mouse, while a control tetramer (PVS) was negative (A). The MHC-II VRE tetramer was used to quantitate the frequency and number of virus-specific CD4 T cells in various tissues following DelNSs RVFV infection (B). Each symbol represents one mouse. Liver mononuclear cells and splenocytes isolated 7 days postinfection with DelNSs RVFV were stimulated ex vivo with PMA/ionomycin, followed by staining with IL-13, IFN-γ, CXCR5, and FoxP3. VRE+ CD4 T cells from both the liver and the spleen were identified as largely Th1 and Tfh subtypes. Phenotyping of total CD4 T cells is shown for comparison.
Immunophenotyping of RVFV-specific CD4 T cells.
With the ability to identify RVFV-specific CD4 T cells in hand, the next step was to define the subtype of RVFV-specific CD4 T cells generated following infection. Regulatory T (Treg) (CD44+ FoxP3+), Tfh (CD44+ CXCR5+), Th1 (CD44+ IFN-γ+), and Th2 (CD44+ IL-13+) cells were the focus of the subsequent studies. RVFV-specific cells from the liver and spleen were identified by allophycocyanin (APC) and phycoerythrin (PE) double-labeled VRE MHC-II tetramers as in Fig. 2A Stimulation using phorbol myristate acetate (PMA)/ionomycin identified >50% of tetramer-positive CD4 T cells in the liver as CD44+ IFN-γ+, but <5% were CD44+ IL-13+ (Fig. 2C), consistent with a Th1 phenotype among the majority of virus-specific CD4 T cells. Approximately 16% of tetramer-positive CD4 T cells in the liver were CD44+ CXCR5+, but <1% were CD44+ FoxP3+ (Fig. 2C), demonstrating that some of the virus-specific CD4 T cells in the liver were of a Tfh phenotype. Evaluation of RVFV-specific CD4 T cells in the spleen demonstrated the same predominance of Tfh and Th1 cells, but with a skewing toward the Tfh phenotype.
Evaluation of Th1 pathways in preventing RVFV encephalitis.
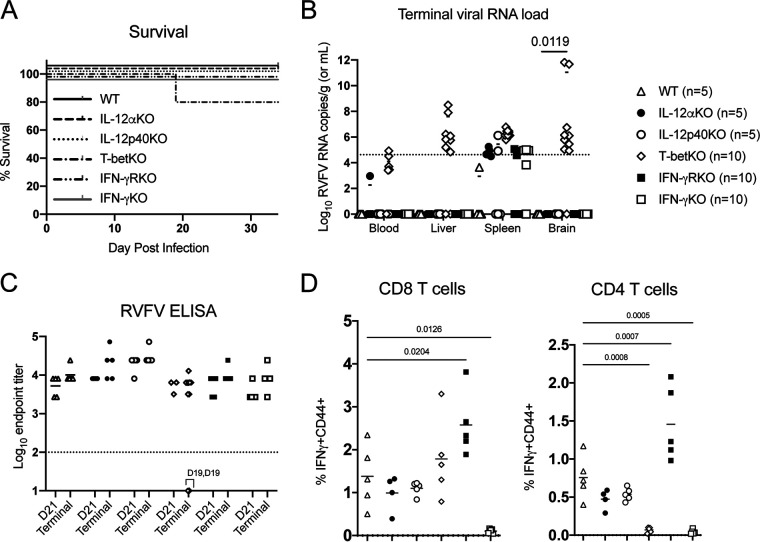
The majority of the RVFV-specific CD4 T cells were identified as Th1, so various genetic mouse models containing gene-specific knockouts of proteins important for mediating the Th1 phenotype were evaluated. IL-12 signaling leads to activation of the transcription factor T-bet, which reprograms naive CD4 T cells into Th1 CD4 T cells (25). Th1 CD4 T cells often exert their effects via cytokine signaling, most prominently IFN-γ. Therefore, initial studies focused on IL-12α KO, IL-12p40 KO, T-bet KO, IFN-γ KO, and IFN-γR KO mice. Animals were infected with DelNSs RVFV and monitored for weight loss and survival. Two (20%) DelNSs RVFV-infected T-bet KO mice required euthanasia at 19 dpi: one for >20% weight loss and one for LT hindlimb paralysis (Fig. 3A). Both mice had high viral RNA loads in the brain consistent with encephalitis (Fig. 3B), and all T-bet KO mice exhibited a trend toward poorer viral load control than other KO strains. All other mice survived infection with DelNSs RVFV without any clinical signs or symptoms. No statistically significant difference in outcome was seen between the different groups of mice, and all mice had RVFV-specific enzyme-linked immunosorbent assay (ELISA) titers similar to those observed in WT mice, with the exception of the 2 mice that required euthanasia on day 19 (Fig. 3C).
FIG 3.
Most Th1 CD4 T cell pathways are not critical for preventing RVFV encephalitis. Mice with genetic deletions of proteins important in the generation and activity of Th1 CD4 T cells did not exhibit a statistically significant difference in survival following DelNSs RVFV infection (Mantel-Cox test, P = 0.3977) (A). Twenty percent of T-bet KO mice met euthanasia criteria and were found to have viral RNA loads in the brain (B). Each symbol represents one mouse. The number of animals included in each group is noted next to the group name. Geometric means are depicted with a horizontal bar, and the P values from a two-way ANOVA with post hoc comparisons to WT C57BL/6 mice are shown for statistically significant associations. RVFV-specific endpoint ELISA titers were not statistically different between the groups (mixed-effects analysis interaction, P = 0.4926). Geometric mean titers are depicted with a horizontal bar. Notably, the two mice that met euthanasia criteria had undetectable RVFV-specific ELISA titers on the day of euthanasia (day 19 for both animals, as noted on the graph) (C). Ex vivo stimulation of cryopreserved splenocytes obtained at time of euthanasia with an RVFV peptide pool revealed a statistically significant difference in IFN-γ expression between WT C57BL/6 mice and IFN-γR KO mice and IFN-γ KO mice in both CD4 and CD8 T cells, but only CD4 T cells demonstrated decreased RVFV-specific IFN-γ production in T-bet KO mice (D). The mean value is plotted with a horizontal line, and P values from a one-way ANOVA with post hoc comparisons to WT C57BL/6 mice are shown for statistically significant associations.
RVFV-specific T cell function was measured by IFN-γ expression following ex vivo stimulation of splenocytes with an RVFV peptide mix. RVFV-specific IFN-γ expression in both CD4 and CD8 T cells was increased in IFN-γR KO mice, as might have been anticipated given the lack of a negative feedback loop, while, appropriately, no IFN-γ expression was detected in IFN-γ KO mice in either cell type (Fig. 3D). In contrast, only CD4 T cells, and not CD8 T cells, showed a lack of RVFV-specific IFN-γ expression in T-bet KO mice, demonstrating a true loss of CD4 Th1 function in the absence of T-bet. While there was no statistically significant difference in outcome in T-bet KO versus WT mice, the fact that 20% of T-bet KO succumbed to late-onset encephalitis suggests that T-bet-directed activity does play some role in preventing RVFV encephalitis. In contrast, IFN-γ production by Th1 CD4 T cells was not necessary for preventing RVFV encephalitis.
Evaluating Tfh pathways in prevention of RVFV encephalitis.
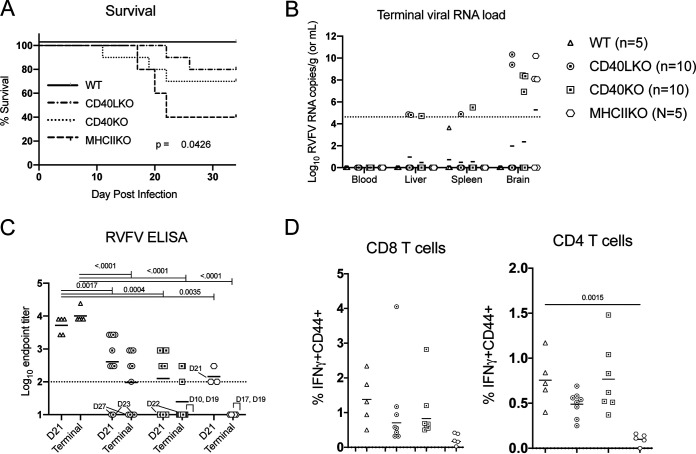
Tfh cells are known to induce class switching and affinity maturation by providing signals to antigen-specific B cells (25). These signals largely consist of cell-cell contacts, primarily MHC-II/T cell receptor (TCR), augmented by costimulation via CD40/CD40L, CD80/CD28, and/or ICOSL/ICOS. Cytokine signaling from T cells to B cells via IL-21 is required for memory B cell formation but is dispensable for acute-phase responses (26). Readily available CD40L KO, CD40 KO, and MHC-II KO mice were infected with DelNSs RVFV and monitored for weight loss and survival; 60% of MHC-II KO, 30% of CD40 KO, and 20% of CD40L KO mice required euthanasia for hindlimb paralysis or weight loss of >20% (Fig. 4A). All mice euthanized for disease had high viral RNA loads in the brain consistent with encephalitis (Fig. 4B). All other mice survived infection with DelNSs RVFV without any clinical signs. Notably, RVFV-specific ELISA titers were significantly lower in CD40L KO, CD40 KO, and MHC-II KO mice (Fig. 4C). In WT C57BL/6 mice, RVFV-specific ELISA titers tended to increase over time, while in CD40L KO, CD40 KO, and MHC-II KO mice, mean RVFV-specific titers declined between blood collection on day 21 and the terminal blood collection. All animals that required euthanasia are noted by the day of euthanasia on the graph, and the corresponding data points are indicated by a connecting line. When an animal was euthanized prior to the day 21 bleed, only the terminal bleed is indicated. For all mice euthanized due to disease, RVFV-specific ELISA titers both on day 21 (when available) or at the time of euthanasia were largely undetectable.
FIG 4.
Disruption of cellular pathways that alter the production of RVFV-specific antibodies leads to increased frequency of RVFV encephalitis. Mice with genetic deletions of proteins important for the interaction of CD4 T cells with B cells exhibited a statistically significant difference in survival following DelNSs RVFV infection (with Mantel-Cox test P value shown on the graph), most notable in MHC-II KO mice (A). Animals that required euthanasia had high viral RNA loads in the brain (B). Each symbol represents one mouse. The number of animals included in each group is noted next to the group name. Geometric mean data are shown for each strain with a horizontal line; there were no statistically significant associations by a two-way ANOVA with post hoc comparison to WT C57BL/6 mice. In blood samples collected at day 21 and terminal time points, CD40L KO, CD40 KO, and MHC-II KO mice all had lower RVFV-specific ELISA titers than WT C57BL/6 mice (C). The geometric mean for each data set is depicted with a horizontal line, and P values from a two-way ANOVA with post hoc comparisons to WT C57BL/6 mice are shown for statistically significant associations. The day of euthanasia for any mouse that required it is noted next to the respective data points. Ex vivo stimulation of cryopreserved splenocytes (obtained at the time of euthanasia) with an RVFV peptide pool demonstrated a statistically significant difference in IFN-γ expression between WT C57BL/6 mice and MHC-II KO mice in CD4 T cells (D). The mean value is plotted with a horizontal line, and the P value from a one-way ANOVA with post hoc comparisons to WT C57BL/6 mice is plotted for statistically significant associations.
In contrast to the clear association between clinical RVFV disease and a lack of RVFV-specific ELISA titers, no statistically significant difference was seen in RVFV-specific CD8 T cell activity between WT C57BL/6 mice and CD40L KO, CD40 KO, or MHC-II KO mice, although there was a trend toward lower RVFV-specific IFN-γ production in CD8 T cells in MHC-II KO mice. RVFV-specific IFN-γ production in CD4 T cells was significantly lower in MHC-II KO mice than in WT C57BL/6 mice (Fig. 4D). This of course occurs in the setting of significantly reduced total CD4 T cells due to lack of MHC-II-mediated selection in the thymus (27). In fact, the typical 23% of CD8 T cells and 40% of CD4 T cells observed in the spleens of WT C57BL/6 mice changed to 45% CD8 T cells and 7% CD4 T cells in MHC-II KO mice. Additionally, CD4 T cells identified in DelNSs RVFV-infected MHC-II KO mice that managed to escape thymic selection in the absence of MHC-II exhibited higher levels of expression of CD25, CD44, and CD69, suggestive of autoreactivity (data not shown), similar to original reports of this model (27).
Given the importance of Tfh cells in inducing class switching of antibodies (28), we evaluated RVFV-specific antibody isotype levels in strains of KO mice that exhibited a clinical phenotype in these studies, including a historical sample from CD4-depleted mice (19) (Fig. 5). All mice included in this analysis had been infected with DelNSs RVFV but had survived to the end of the experiment with detectable RVFV-specific antibodies (4 to 5 weeks postinfection). The IgG locus in C57BL/6 mice is ordered in the following way: IgG3, IgG1, IgG2b, then IgG2c. Thus, the most class-switched antibodies would be of the IgG2c class. We saw no differences in RVFV-specific IgG3 levels between WT mice or any of the other strains examined, but WT mice had significantly more IgG2b and IgG2c than CD4-depleted, T-bet KO, CD40 KO, and CD40L KO mice. T-bet KO mice had higher levels of RVFV-specific IgG1 than WT mice and similar levels of total RVFV-specific IgG, despite having very little IgG2c. Consistent with observations in other models of viral infection, this finding suggests a role for T-bet in class switching to IgG2c (29).
FIG 5.

Class switching of RVFV-specific antibodies is altered following DelNSs RVFV infection in CD4-depleted, T-bet KO, CD40 KO, and CD40L KO mice. Terminal serum from mice that survived DelNSs RVFV infection was tested on isotype-specific RVFV ELISAs. Raw optical density at 450 nm (OD450) values are shown at a 1:300 dilution for each sample with mean titers and standard deviations. Each symbol represents one mouse. In the tested mouse strains, a two-way ANOVA with post hoc comparisons to WT demonstrated statistically significant differences (*, P < 0.05) in IgG1, IgG2b, IgG2c, and total IgG titers, but not IgG3 titers.
DISCUSSION
Previously, CD4 T cells were identified as a critical component of adaptive immunity that prevented RVFV-mediated encephalitis (17). In this study, we sought to define the CD4 T cell subtypes that were generated following peripheral DelNSs RVFV infection in mice, with the goal of identifying specific functions of CD4 T cells that mediate protection from RVFV encephalitis. To do so, we identified RVFV-specific epitopes in the C57BL/6 mouse and generated tetramers. Given the magnitude of the work involved in identifying virus-specific epitopes, MHC-I epitopes were included as well as MHC-II epitopes. One high-frequency MHC-I epitope was identified (NAAVNSNFI); this epitope is contained within the viral nucleocapsid protein and had the highest binding affinity for H-2-Db of all of the tested peptides (Table 2). The NAA MHC-I tetramer identified a high frequency of virus-specific CD8 T cells in the livers of mice infected with DelNSs RVFV. These virus-specific CD8 T cells were all CD44+, and many were also Ki-67+ and granzyme B+, suggesting that these activated CD8 T cells might play a role in controlling DelNSs RVFV infection in the liver to render the virus nonpathogenic in the C57BL/6 mouse. Identification of this MHC-I tetramer could be useful to the field for assessing correlates of protection in vaccine studies.
One immunodominant core MHC-II epitope was identified; this epitope is also located within the nucleocapsid protein but, notably, was not the peptide with the highest binding affinity for H-2-IAb (Table 1). Two peptides (VRE and AYQ) with the same core epitope (AYQGFDARRVI) gave similar results in both in vitro binding assays and ex vivo tetramer assays. Using the VRE tetramer, RVFV-specific CD4 T cells were identified. They expressed high levels of IFN-γ or CXCR5 but little IL-13 or FoxP3, indicating that they were largely Th1 or Tfh cells. Further studies using various genetic KO mouse models suggested that Tfh cells, with their role in augmenting antigen-specific humoral immunity, were likely the CD4 T cell type critical for preventing RVFV encephalitis. The increased frequency of encephalitis in MHC-II KO, CD40 KO, and CD40L KO mice supports this conclusion, since all of these molecules modulate the interaction between Tfh and B cells, and these interactions control the generation of high-affinity, class-switched antibodies. CD40/CD40L and MHC-II/TCR interactions also play a role in other functions of CD4 T cells, such as their role in activating macrophages, so this role cannot be excluded. In contrast, while CD4 T cells are known to help CD8 T cells form memory (30), a study in a lymphocytic choriomeningitis virus model demonstrated that CD40L/CD40 interactions between CD4 T and CD8 T cells were completely dispensable for forming memory CD8 T cells (31). This is consistent with our detection of RVFV-specific memory CD8 T cells in CD40L KO and CD40 KO mice. Initially, the finding that a subset of T-bet KO mice succumbed to DelNSs RVFV encephalitis was confusing, since T-bet is the transcription factor associated with Th1 skewing. However, work by Weinstein et al. from 2018 demonstrated that T-bet expression occurs in Tfh cells during viral infection and is required for proper follicular migration and CD40L expression, thus influencing the quality and quantity of virus-specific antibodies (29). We were able to confirm a unique class-switching defect in T-bet KO mice: these mice generated RVFV-specific IgG1 and IgG2b antibodies, but had very few RVFV-specific IgG2c antibodies. These data, combined with the observation that all mice that developed encephalitis had low or undetectable levels of virus-specific antibodies at the time of euthanasia, suggest that Tfh CD4 T cells play a greater role than Th1 cells in preventing RVFV encephalitis.
Early induction of type I IFNs has been associated with Tfh polarization and the production of highly neutralizing antibodies in mice infected with vesicular stomatitis virus (VSV) (32). Certainly, an early and robust type I IFN response occurs during DelNSs RVFV infection (20). Similar to what we report in this study, a subset of CD40L-deficient mice succumbed to VSV-induced meningoencephalitis, presumably due to reduced antibody responses (33), since WT mice normally survive VSV infection just as WT mice survive DelNSs RVFV infection. This suggests that similar antibody-mediated protective mechanisms work to prevent CNS infection by other viruses.
Future work will focus on the role of antibodies in preventing RVFV encephalitis. Do antibodies function to prevent CNS access, or can they work after the virus has entered the nervous system? This has implications for when antibody therapy might be useful clinically. Postexposure antibody prophylaxis seems likely to be successful to treat RVFV, as it has for other viral infections, such as varicella-zoster and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, treatment with antibodies once symptoms have developed is a much higher bar. Antibodies as therapy have shown efficacy in treating Ebola virus disease, so they are certainly worth evaluating further in the case of RVFV.
MATERIALS AND METHODS
Biosafety and ethics.
Research with live virus was conducted at the Center for Vaccine Research’s Regional Biocontainment Laboratory or at the Centers for Disease Control and Prevention (CDC) under biosafety level 3 (BSL3) conditions. Institutional Animal Care and Use Committee approval was provided by the University of Pittsburgh (protocol no. 17080998 and 19044158) or CDC (protocol no. 2577MCEMOUC or 2629MCEMOUC). All recombinant virus work was approved by the respective institutional biosafety committees.
Animals, sample collection, and qRT-PCR.
Mice were purchased from the Jackson Laboratory (Bar Harbor, ME). All mice were female (no sex-based differences in outcome following RVFV infection were noted in a prior study [10]) and between 4 and 12 weeks of age at time of infection. The following strains were used: C57BL/6J (000664), B6.129S6-Tbx21tm1Glm/J (004648), B6.129S1-Il12atm1Jm/J (002692), B6.129S1-Il12btm1Jm/J (002693), B6.129S7-Ifngtm1Ts/J (002287), B6.129S7-Ifngr1tm1Agt/J (003288), B6.129S2-Cd40lgtm1lmx/J (002770), B6.129P2-Cd40tm1Kik/J (002928), and B6.129S2-H2dlAb1-Ea/J (003584). Mice were group-housed in HEPA-filtered containment caging with ad lib access to food and water. After infection, mice were weighed and assessed for clinical signs of disease daily. Any mouse with a clinical score greater than 10 (scoring system previously described [10]) was euthanized, and liver, spleen, brain, and blood were collected for various assays. Twenty-one days post-infection (dpi), blood was drawn via a lateral saphenous bleed. Surviving mice were euthanized 32 to 36 dpi, and the same samples were collected. Quantitative reverse transcription-PCR (qRT-PCR) using a primer/probe set specific to the RVFV L segment (34) was run, and results were analyzed as previously described (10). Splenocytes, liver mononuclear cell preparations, and lymph node preparations were performed as previously described (19).
Virus.
Using a reverse-genetics system based upon RVFV ZH501 (35), viruses missing either the NSs gene (DelNSs RVFV) or both NSs and NSm genes (DelNSs/DelNSm RVFV) were generated. Viruses were propagated to passage 2, quantitated using the 50% tissue culture infective dose (TCID50) method (36), and fully sequence confirmed. For peptide mapping studies, mice were infected with 2 × 105 TCID50 DelNSs/DelNSm RVFV either subcutaneously (s.c.) or via footpad (FP) injection. For all other experiments, mice were infected with 2 × 105 TCID50 DelNSs RVFV via FP injection.
Epitope mapping.
Overlapping peptides (15-mers with 11-amino-acid [aa] overlap) were generated based on the ZH501 strain sequence of the viral structural proteins, nucleocapsid (N) and glycoproteins (Gn and Gc), purified to >70%, and suspended in dimethyl sulfoxide (DMSO) (GenScript). Two-dimensional maps were made using the peptides for N (59 peptides), Gn (140 peptides), and Gc (124 peptides), and peptide pools were generated as described previously (37). The pools were evaluated in an ex vivo stimulation assay using splenocytes from C57BL/6 mice that had been infected with either DelNSs RVFV or DelNSs/DelNSm RVFV 5 to 6 weeks prior. Approximately 7 × 106 splenocytes were incubated for 6 h in the presence of 5 μg/ml of brefeldin A (BioLegend), with each pool containing each peptide at a final concentration of at least 2 μg/ml. Following ex vivo stimulation, staining was performed as described in the “Flow cytometry” section. A peptide pool was considered positive if any of the cytokine readouts (IL-4, IL-17, or IFN-γ) was 3 standard deviations above the average of 4 negative-control (DMSO alone) samples. Staphylococcus enterotoxin B (SEB) stimulation was included as a positive control.
Flow cytometry.
Cells were washed in phosphate-buffered saline (PBS) and then stained with LIVE/DEAD e780 (eBioscience) or LIVE/DEAD Near IR (Thermo Fisher) for 10 min, washed in PBS with 2% fetal bovine serum (FBS), and incubated in Fc block (BioLegend). Surface stain was added for 20 min, and cells were washed, fixed in BD Fix/Perm, washed again, and then incubated for 45 min in intracellular stain. Events were acquired on either a S1000EXi (Stratedigm) or an LSRII (BD) flow cytometer, and data were analyzed using Flow Jo. Antibodies were obtained from BioLegend or BD and include the following: CD3, Alexa Fluor 488 (AF488)-, APC-, or BV510-conjugated 17A2; CD4, peridinin chlorophyll protein (PerCP)-Cy5.5- or fluorescein isothiocyanate (FITC)-conjugated RM4-4; CD8, BV510- or PE-Cy7-conjugated 53-6.7; IFN-γ, PE- or BV421-conjugated XMG1.2; IL-4, APC-conjugated 11B11; IL-17, BV605-conjugated TC11-18H10; CD19, APC-Cy7-conjugated 6D5; CD14, APC-Cy7-conjugated Sa14-2; CD44, BV421- or AF700-conjugated IM7; CD4, BUV395-conjugated RM4-5; Ki-67, FITC-conjugated 16A.8; granzyme B, APC-conjugated QA18.A28; streptavidin conjugated with PE-Cy7; CXCR5, biotin-conjugated 2G8; IL-13, AF488-conjugated eBio13A; and FoxP3, CF594-conjugated MF23.
Binding assays.
Classical competition assays to quantitatively measure peptide binding to mouse class I H-2-Kb and -Db and class II H-2-IAb MHC molecules were based on the inhibition of binding of high-affinity radiolabeled peptides to purified MHC molecules and performed as detailed elsewhere (38). Briefly, 0.1 to 1 nM radiolabeled peptide was coincubated at room temperature with purified MHC in the presence of a cocktail of protease inhibitors and, for class I assays, B2-microglobulin. Following a 2-day incubation, MHC-bound radioactivity was determined by capturing MHC/peptide complexes on Lumitrac 600 plates (Greiner Bio-One, Frickenhausen, Germany) coated with either Y3 (anti-H-2-Kb), 28-14-8s (anti-H-2-Db, Ld, and Dq), or Y3JP (anti-H-2-IAb,s,u) monoclonal antibodies; bound counts per minute were measured using the TopCount (Packard Instrument Co., Meriden, CT) microscintillation counter. The concentration of peptide yielding 50% inhibition (IC50) of binding of the radiolabeled peptide was calculated: under the conditions utilized, where [label] < [MHC] and IC50 ≥ [MHC], measured IC50 values are reasonable approximations of the true dissociation constant (Kd) (39, 40). Each competitor peptide was tested at 6 different concentrations covering a 100,000-fold range and in 3 or more independent experiments. As a positive control, the unlabeled version of the radiolabeled probe was also tested in each experiment.
MHC-I tetramer assays.
Peptides were generated by Genscript at >70% purity and sent to the NIH Tetramer Core for tetramer generation. MHC-I tetramers were labeled with PE. Splenocytes or liver mononuclear cell preparations were obtained at various times after C57BL/6 mice were infected with DelNSs RVFV as previously described (19). Following LIVE/DEAD stain, cells were washed and incubated for 30 min with PE-labeled MHC-I tetramers diluted to 26 to 28 μg/ml in PBS. Cells were washed, and staining proceeded as described in the “Flow cytometry” section.
MHC-II tetramer assays.
Peptides were generated by Genscript at >70% purity and sent to the NIH Tetramer Core for tetramer generation. MHC-II tetramers were generated with either a PE or an APC label. Splenocytes (2 × 106), entire liver mononuclear cell preparations, or entire draining popliteal lymph node preparations were obtained at various time points after infection of C57BL/5 mice with DelNSs RVFV as previously described (19). Cells were incubated for 2 h at 37°C with both PE- and APC-labeled MHC-II tetramers at concentrations of 13 to 15 μg/ml in RPMI 10 medium. Cells were washed, and staining proceeded as described in the “Flow cytometry” section.
Phenotyping of MHC-II tetramer-positive cells.
Seven days after mock infection or infection with DelNSs RVFV, livers, and spleens were processed from 5 mice. The combined liver mononuclear cell preparations or 2 × 106 splenocytes were incubated with Cell Activation Cocktail (PMA plus ionomycin; BioLegend) containing brefeldin A or a negative DMSO control for 4 h at 37°C. MHC-II VRE or control PVS tetramer was then added, and cells were incubated for another 2 h at 37°C. Following LIVE/DEAD stain, cells were incubated with CXCR5 biotin for 45 min before proceeding with the rest of staining as described in the “Flow cytometry” section with the following modifications: surface stains included streptavidin with PE-Cy7, and FoxP3 Fix/Perm was used.
ELISA.
Enzyme-linked immunosorbent assays (ELISAs) were performed and analyzed as previously described (10) using the following secondary antibodies (all from Jackson Immunoresearch, West Grove, PA) at a concentration of 1:5,000: donkey anti-mouse total IgG (715-035-150), goat anti-mouse IgG Fcγ subclass 1 (115-035-205), goat anti-mouse IgG Fcγ subclass 2b (115-035-207), goat anti-mouse IgG Fcγ subclass 2c (115-035-208), and goat anti-mouse IgG Fcγ subclass 3 (115-035-209).
Ex vivo stimulation assays.
Cryopreserved splenocytes were thawed, and 2 × 106 cells were incubated with DMSO (negative control), Cell Activation Cocktail without brefeldin A (positive control), or RVFV stim mix (a panel of 13 RVFV peptides that encompass the MHC-I and MHC-II immunodominant epitopes targeted in the C57BL/6 mouse [in boldface in Tables 1 and 2]) for 6 h at 37°C in the presence of 5 μg/ml of brefeldin A. Staining was performed as described in “Flow cytometry” section.
Data presentation and statistical analyses.
All graphs were generated using GraphPad Prism. The specific statistical test applied to each data set is noted in the respective figure legend.
ACKNOWLEDGMENTS
We thank DLAR for animal husbandry support and Tanya Klimova for editorial review.
Funding was provided through NIH K08 AI119448 (A.K.M.), a Burroughs Wellcome Career Award for Medical Scientists 1013362.02 (A.K.M.), and NIH-NIAID contract 75N9301900065 (A.S.). The NIH Tetramer Facility is supported by contract 75N93020D00005 from NIAID.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the U.S. Centers for Disease Control and Prevention.
Footnotes
Supplemental material is available online only.
Contributor Information
Anita K. McElroy, Email: mcelroya@pitt.edu.
Stacey Schultz-Cherry, St. Jude Children's Research Hospital.
REFERENCE
- 1.McMillen CM, Hartman AL. 2018. Rift Valley fever in animals and humans: current perspectives. Antiviral Res 156:29–37. doi: 10.1016/j.antiviral.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meegan JM, Watten RH, Laughlin LW. 1981. Clinical experience with Rift Valley fever in humans during the 1977 Egyptian epizootic. Contr Epidem Biostatist 3:114–123. [Google Scholar]
- 3.Madani TA, Al-Mazrou YY, Al-Jeffri MH, Mishkhas AA, Al-Rabeah AM, Turkistani AM, Al-Sayed MO, Abodahish AA, Khan AS, Ksiazek TG, Shobokshi O. 2003. Rift Valley fever epidemic in Saudi Arabia: epidemiological, clinical, and laboratory characteristics. Clin Infect Dis 37:1084–1092. doi: 10.1086/378747. [DOI] [PubMed] [Google Scholar]
- 4.Laughlin LW, Girgis NI, Meegan JM, Strausbaugh LJ, Yassin MW, Watten RH. 1978. Clinical studies on Rift Valley fever. Part 2. Ophthalmologic and central nervous system complications. J Egypt Public Health Assoc 53:183–184. [PubMed] [Google Scholar]
- 5.Alrajhi AA, Al-Semari A, Al-Watban J. 2004. Rift Valley fever encephalitis. Emerg Infect Dis 10:554–555. doi: 10.3201/eid1003.020817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Velden DJ, Meyer JD, Olivier J, Gear JH, McIntosh B. 1977. Rift Valley fever affecting humans in South Africa: a clinicopathological study. S Afr Med J 51:867–871. [PubMed] [Google Scholar]
- 7.Mohamed M, Mosha F, Mghamba J, Zaki SR, Shieh WJ, Paweska J, Omulo S, Gikundi S, Mmbuji P, Bloland P, Zeidner N, Kalinga R, Breiman RF, Njenga MK. 2010. Epidemiologic and clinical aspects of a Rift Valley fever outbreak in humans in Tanzania, 2007. Am J Trop Med Hyg 83:22–27. doi: 10.4269/ajtmh.2010.09-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jansen van Vuren P, Shalekoff S, Grobbelaar AA, Archer BN, Thomas J, Tiemessen CT, Paweska JT. 2015. Serum levels of inflammatory cytokines in Rift Valley fever patients are indicative of severe disease. Virol J 12:159. doi: 10.1186/s12985-015-0392-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, Paul WE. 2008. CD4 T cells: fates, functions, and faults. Blood 112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cartwright HN, Barbeau DJ, McElroy AK. 2020. Rift Valley fever virus is lethal in different inbred mouse strains independent of sex. Front Microbiol 11:1962. doi: 10.3389/fmicb.2020.01962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.do Valle TZ, Billecocq A, Guillemot L, Alberts R, Gommet C, Geffers R, Calabrese K, Schughart K, Bouloy M, Montagutelli X, Panthier JJ. 2010. A new mouse model reveals a critical role for host innate immunity in resistance to Rift Valley fever. J Immunol 185:6146–6156. doi: 10.4049/jimmunol.1000949. [DOI] [PubMed] [Google Scholar]
- 12.Gray KK, Worthy MN, Juelich TL, Agar SL, Poussard A, Ragland D, Freiberg AN, Holbrook MR. 2012. Chemotactic and inflammatory responses in the liver and brain are associated with pathogenesis of Rift Valley fever virus infection in the mouse. PLoS Negl Trop Dis 6:e1529. doi: 10.1371/journal.pntd.0001529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikegami T, Balogh A, Nishiyama S, Lokugamage N, Saito TB, Morrill JC, Shivanna V, Indran SV, Zhang L, Smith JK, Perez D, Juelich TL, Morozov I, Wilson WC, Freiberg AN, Richt JA. 2017. Distinct virulence of Rift Valley fever phlebovirus strains from different genetic lineages in a mouse model. PLoS One 12:e0189250. doi: 10.1371/journal.pone.0189250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lathan R, Simon-Chazottes D, Jouvion G, Godon O, Malissen M, Flamand M, Bruhns P, Panthier JJ. 2017. Innate immune basis for Rift Valley fever susceptibility in mouse models. Sci Rep 7:7096. doi: 10.1038/s41598-017-07543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reed C, Steele KE, Honko A, Shamblin J, Hensley LE, Smith DR. 2012. Ultrastructural study of Rift Valley fever virus in the mouse model. Virology 431:58–70. doi: 10.1016/j.virol.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 16.Smith DR, Steele KE, Shamblin J, Honko A, Johnson J, Reed C, Kennedy M, Chapman JL, Hensley LE. 2010. The pathogenesis of Rift Valley fever virus in the mouse model. Virology 407:256–267. doi: 10.1016/j.virol.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Dodd KA, McElroy AK, Jones ME, Nichol ST, Spiropoulou CF. 2013. Rift Valley fever virus clearance and protection from neurologic disease are dependent on CD4+ T cell and virus-specific antibody responses. J Virol 87:6161–6171. doi: 10.1128/JVI.00337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodd KA, McElroy AK, Jones TL, Zaki SR, Nichol ST, Spiropoulou CF. 2014. Rift Valley fever virus encephalitis is associated with an ineffective systemic immune response and activated T cell infiltration into the CNS in an immunocompetent mouse model. PLoS Negl Trop Dis 8:e2874. doi: 10.1371/journal.pntd.0002874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmon JR, Spengler JR, Coleman-McCray JD, Nichol ST, Spiropoulou CF, McElroy AK. 2018. CD4 T cells, CD8 T cells, and monocytes coordinate to prevent Rift Valley fever virus encephalitis. J Virol 92:e01270-18. doi: 10.1128/JVI.01270-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouloy M, Janzen C, Vialat P, Khun H, Pavlovic J, Huerre M, Haller O. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J Virol 75:1371–1377. doi: 10.1128/JVI.75.3.1371-1377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, Peters B. 2019. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. 2011. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul S, Weiskopf D, Angelo MA, Sidney J, Peters B, Sette A. 2013. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J Immunol 191:5831–5839. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gommet C, Billecocq A, Jouvion G, Hasan M, Zaverucha do Valle T, Guillemot L, Blanchet C, van Rooijen N, Montagutelli X, Bouloy M, Panthier JJ. 2011. Tissue tropism and target cells of NSs-deleted Rift Valley fever virus in live immunodeficient mice. PLoS Negl Trop Dis 5:e1421. doi: 10.1371/journal.pntd.0001421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duckworth BC, Groom JR. 2021. Conversations that count: cellular interactions that drive T cell fate. Immunol Rev 300:203–219. doi: 10.1111/imr.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rankin AL, MacLeod H, Keegan S, Andreyeva T, Lowe L, Bloom L, Collins M, Nickerson-Nutter C, Young D, Guay H. 2011. IL-21 receptor is critical for the development of memory B cell responses. J Immunol 186:667–674. doi: 10.4049/jimmunol.0903207. [DOI] [PubMed] [Google Scholar]
- 27.Madsen L, Labrecque N, Engberg J, Dierich A, Svejgaard A, Benoist C, Mathis D, Fugger L. 1999. Mice lacking all conventional MHC class II genes. Proc Natl Acad Sci USA 96:10338–10343. doi: 10.1073/pnas.96.18.10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crotty S. 2019. T follicular helper cell biology: a decade of discovery and diseases. Immunity 50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinstein JS, Laidlaw BJ, Lu Y, Wang JK, Schulz VP, Li N, Herman EI, Kaech SM, Gallagher PG, Craft J. 2018. STAT4 and T-bet control follicular helper T cell development in viral infections. J Exp Med 215:337–355. doi: 10.1084/jem.20170457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun JC, Bevan MJ. 2003. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science 300:339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun JC, Bevan MJ. 2004. Cutting edge: long-lived CD8 memory and protective immunity in the absence of CD40 expression on CD8 T cells. J Immunol 172:3385–3389. doi: 10.4049/jimmunol.172.6.3385. [DOI] [PubMed] [Google Scholar]
- 32.De Giovanni M, Cutillo V, Giladi A, Sala E, Maganuco CG, Medaglia C, Di Lucia P, Bono E, Cristofani C, Consolo E, Giustini L, Fiore A, Eickhoff S, Kastenmuller W, Amit I, Kuka M, Iannacone M. 2020. Spatiotemporal regulation of type I interferon expression determines the antiviral polarization of CD4(+) T cells. Nat Immunol 21:321–330. doi: 10.1038/s41590-020-0596-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borrow P, Tishon A, Lee S, Xu J, Grewal IS, Oldstone MB, Flavell RA. 1996. CD40L-deficient mice show deficits in antiviral immunity and have an impaired memory CD8+ CTL response. J Exp Med 183:2129–2142. doi: 10.1084/jem.183.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bird BH, Bawiec DA, Ksiazek TG, Shoemaker TR, Nichol ST. 2007. Highly sensitive and broadly reactive quantitative reverse transcription-PCR assay for high-throughput detection of Rift Valley fever virus. J Clin Microbiol 45:3506–3513. doi: 10.1128/JCM.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bird BH, Albarino CG, Nichol ST. 2007. Rift Valley fever virus lacking NSm proteins retains high virulence in vivo and may provide a model of human delayed onset neurologic disease. Virology 362:10–15. doi: 10.1016/j.virol.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 36.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 37.Hoffmeister B, Kiecker F, Tesfa L, Volk HD, Picker LJ, Kern F. 2003. Mapping T cell epitopes by flow cytometry. Methods 29:270–281. doi: 10.1016/s1046-2023(02)00349-3. [DOI] [PubMed] [Google Scholar]
- 38.Sidney J, Southwood S, Moore C, Oseroff C, Pinilla C, Grey HM, Sette A. 2013. Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. Curr Protoc Immunol Chapter 18:Unit 18 3. doi: 10.1002/0471142735.im1803s100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng Y, Prusoff WH. 1973. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol 22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 40.Gulukota K, Sidney J, Sette A, DeLisi C. 1997. Two complementary methods for predicting peptides binding major histocompatibility complex molecules. J Mol Biol 267:1258–1267. doi: 10.1006/jmbi.1997.0937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1<br>. Download jvi.01506-21-s0001.pdf, PDF file, 1.3 MB (1.3MB, pdf)