ABSTRACT
Sin Nombre orthohantavirus (SNV), a negative-sense, single-stranded RNA virus that is carried and transmitted by the North American deer mouse Peromyscus maniculatus, can cause infection in humans through inhalation of aerosolized excreta from infected rodents. This infection can lead to hantavirus cardiopulmonary syndrome (HCPS), which has an ∼36% case-fatality rate. We used reverse transcriptase quantitative PCR (RT-qPCR) to confirm SNV infection in a patient and identified SNV in lung tissues in wild-caught rodents from potential sites of exposure. Using viral whole-genome sequencing (WGS), we identified the likely site of transmission and discovered SNV in multiple rodent species not previously known to carry the virus. Here, we report, for the first time, the use of SNV WGS to pinpoint a likely site of human infection and identify SNV simultaneously in multiple rodent species in an area of known host-to-human transmission. These results will impact epidemiology and infection control for hantaviruses by tracing zoonotic transmission and investigating possible novel host reservoirs.
IMPORTANCE Orthohantaviruses cause severe disease in humans and can be lethal in up to 40% of cases. Sin Nombre orthohantavirus (SNV) is the main cause of hantavirus disease in North America. In this study, we sequenced SNV from an infected patient and wild-caught rodents to trace the location of infection. We also discovered SNV in rodent species not previously known to carry SNV. These studies demonstrate for the first time the use of virus sequencing to trace the transmission of SNV and describe infection in novel rodent species.
KEYWORDS: Sin Nombre, hantavirus, reservoir, sequencing, transmission
INTRODUCTION
Orthohantavirus is a genus of negative-sense, single-stranded RNA viruses from the order Bunyavirales and the family Hantaviridae (1). Orthohantavirus genomes are trisegmented and encode four proteins. The small (S) segment encodes the nucleoprotein (N) and the nonstructural protein NSs, the medium (M) segment encodes a polyprotein that is a precursor for glycoproteins Gn and Gc (known previously as G1 and G2), and the large (L) segment encodes the viral RNA-dependent RNA polymerase (2). The New World orthohantavirus Sin Nombre orthohantavirus (SNV) is the primary etiological agent of hantavirus cardiopulmonary syndrome (HCPS) in North America. The primary host reservoir for SNV is thought to be Peromyscus maniculatus (North American deer mouse). Other Peromyscus species have been reported to be seropositive for SNV and could serve as potential secondary reservoirs (3–5). Previous studies have also shown antibody reactivity with multiple small mammals throughout the Southwest, including species of the genera Microtus, Onychomys, Reithrodontomys, and Spermophilus (4, 6–9). However, such findings were largely based on host antibodies, which may cross-react with endemic nonpathogenic hantaviruses (10). Furthermore, sequencing of large portions of SNV genomes in rodents other than P. maniculatus has not been reported.
HCPS was first documented in North America during the 1993 outbreak in the Four Corners region (New Mexico, Arizona, Utah, and Colorado) of the United States, with additional previously suspected cases reported retrospectively (11, 12). Symptoms of HCPS begin 1 to 7 weeks (median of ∼3 weeks) after exposure and consist of fever, headache, and myalgia, with severe cases progressing to cardiopulmonary syndrome. The overall case-fatality rate is approximately 36% but can be higher in individual outbreaks (13). Peripheral blood smear criteria consistent with SNV infection include hemoconcentration, left shift in the white blood cell count (WBC), thrombocytopenia, >10% circulating immunoblasts, and the absence of toxic granules in neutrophils (14).
There are currently no approved therapeutics or vaccines for SNV, making it important to understand the distribution of this virus in associated host reservoirs to enhance risk reduction strategies. Some states have been identified as being disproportionately burdened with HCPS cases, potentially linked to favorable breeding conditions of rodent hosts and increased circulation of the virus (13). New Mexico has reported the highest number of HCPS cases (15), and the question of whether P. maniculatus is the only host that can carry and spread SNV or whether additional reservoirs exist has not been thoroughly explored.
To date, direct tracing of SNV transmission by sequencing viral genomes of infected rodents and then correlating these data with patient infection has not been examined. Studies have suggested that the sizes of the host populations are dependent on a bottom-up, trophic cascade in which the prevalence of hantavirus transmission is primarily based on aspects of the biology of the primary host (P. maniculatus), such as food availability and population dynamics, which are heavily influenced by periodic climatic events (e.g., the El Niño Southern Oscillation) (16). However, if multiple rodent species serve as SNV hosts, then the dynamics of viral circulation and increased zoonotic transmission to humans could be more complex. Therefore, SNV needs to be explored as an emerging/reemerging disease in the context of a greater understanding of the distribution of host reservoirs to help develop more accurate risk models for the prevention of SNV outbreaks (17).
In this study, we investigated the source of human SNV infection from a patient in New Mexico. We used reverse transcriptase quantitative PCR (RT-qPCR) to confirm infection in both the patient and wild-caught rodents near two sites of potential zoonotic transmission. We then sequenced the genomes of SNV from the potential hosts and compared them to the genome sequence of SNV from the patient to investigate the likely site of patient infection. In addition, we conducted genome sequencing of SNV PCR-positive specimens obtained from rodents other than P. maniculatus. Altogether, this is the first study to use genomic epidemiology to trace the origin of an SNV infection and identify SNV in multiple rodent species at the site of a human infection.
RESULTS AND DISCUSSION
SNV confirmation and disease progression.
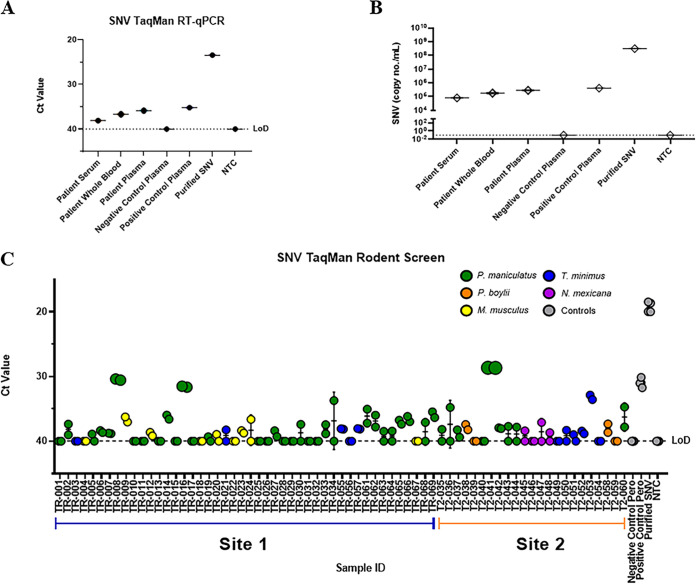
In 2020, a 57-year-old Caucasian male from northern New Mexico without known relevant medical history presented to a local clinic with symptoms of fever, myalgias, and headache for 1 to 2 days. Symptoms progressed to nausea, diarrhea, and hypoxia, with laboratory tests showing thrombocytopenia but normal white blood cell (WBC) count and hematocrit (Hct). Amid the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, the patient had been practicing social distancing, with no travel or encounters with sick individuals. The patient was tested and found to be negative for SARS-CoV-2 RNA, suggesting that he was suffering from another respiratory syndrome. The patient was transferred to the University of New Mexico Hospital and placed on a high-flow nasal cannula, and additional laboratory testing was performed. Interstitial markings were observed on subsequent chest X rays. The peripheral blood smear met 4 of 5 criteria (all except hemoconcentration) for a presumptive diagnosis of HCPS in the cardiopulmonary stage. The patient developed acute kidney injury, and sheaths were placed for potential extracorporeal membrane oxygenation (ECMO), but he did not progress to require that therapy. Serology for SNV infection was positive for IgM, but IgG was equivocal. He was discharged on oxygen after 10 days of hospitalization. RT-qPCR was used to confirm the diagnosis by the detection of SNV RNA. Using SNV-specific TaqMan primers and probe (18), patient serum, whole blood, and plasma were tested for SNV, and all three samples were positive (Fig. 1A). A standard curve with an R2 value of 0.998 of known concentrations of a plasmid containing the N gene for SNV was used to calculate copies per milliliter (Fig. 1B). Copy numbers of SNV genomes in the patient’s samples were comparable to previously published findings from other infected individuals (19).
FIG 1.
SNV RT-qPCR detection in clinical samples and wild-caught rodents. (A) Singleplex SNV-specific amplification of patient samples using cycle threshold (CT) values plotted along with negative- and positive-control patient plasma and SNV-infected Vero E6 cells (Purified SNV). (B) Copies per milliliter were calculated using an SNV N gene plasmid standard curve with an R2 value of 0.998. (C) Lung tissues from wild-caught rodents were plotted using CT values, with each species labeled a color according to the key, along with controls from infected or uninfected rodents and purified SNV (gray). Limits of detection (LoD) for plots were determined using a no-template control (NTC).
Assessment of potential sites of rodent-to-human SNV transmission.
The patient lived on a ranch in northern New Mexico and had exposure to chickens, dogs, cats, and cattle. Activities prior to infection included cleaning out sheds on two properties 2 to 3 weeks prior to the onset of symptoms, where the patient noted rodent infestation that included mouse droppings. Two weeks after the patient’s diagnosis, we conducted field trapping of rodents from two potential sites of exposure, site 1 (TR) and site 2 (T2), which were in rural areas separated by approximately 15 miles. Trapping locations consisted of several outdoor buildings that were found near woodpiles and vegetation that showed signs of rodent infestation and activity. Rodent nests were found at both sites, inside sheds and on the outskirts of the buildings. We hypothesized site 1 to be the source of transmission as this was located near the primary residence; however, both sites were trapped over a 2-night period. A total of 69 rodents were obtained, and tissues were collected and stored until screening. To identify SNV within potential host reservoirs, we used RT-qPCR to detect the SNV S segment in RNA isolated from the lung tissues of all 69 rodents.
High SNV positivity rate in wild-caught rodents and novel potential rodent reservoirs.
We found that 38% (26/69) of trapped rodents were positive for SNV RNA overall, with 39% of site 1 rodents being positive and 35% of rodents being positive at site 2 (Fig. 1C and Table 1). The identity of all SNV-positive rodents was confirmed by cytochrome b Sanger sequencing of lung tissue and analysis by BLAST (data not shown). In total, we trapped 46 rodents at site 1 and 23 at site 2, with most rodents being P. maniculatus (59%). In addition, we found rodents of another species of Peromyscus, Peromyscus boylii (pinyon mouse), which were trapped only at site 2. Mus musculus (common house mouse) was caught only at site 1. Neotoma mexicana (Mexican woodrat) rodents were primarily found at site 2 and associated with middens, or nests, found on the outside of some of the buildings. We trapped several Tamias minimus (least chipmunk) rodents at both sites; these animals were very active both inside and outside some of the structures suspected for exposure. Surprisingly, we detected SNV not only in P. maniculatus but also in multiple samples from P. boylii and M. musculus (Table 1). Unexpectedly, we also found that of the 11 captured T. minimus rodents, 4 were positive for SNV. We did not find any apparent signs of illness in the SNV-positive rodents, suggesting that, like previous reports for P. maniculatus, other SNV-infected rodents may be largely asymptomatic as well. Furthermore, we found that of captured P. maniculatus rodents, 9 of 20 (45%) males tested positive for SNV, while 8 of 21 (38%) females were positive; previous publications have shown a higher SNV antibody prevalence in male than in female P. maniculatus rodents (20–23).
TABLE 1.
Summary of rodents captured from both potential sites of exposure in northern New Mexico and tested for SNVa
| Species | No. of rodents captured (%) | No. of rodents at location | No. of M/F rodents (% SNV positive) | No. of rodents with SNV-positive lung tissue (%) |
|---|---|---|---|---|
| Peromyscus maniculatus | 41 (59) | 32, site 1 | 16 (44)/16 (38) | 13 (41) |
| 9, site 2 | 4 (50)/5 (40) | 4 (44) | ||
| Peromyscus boylii | 4 (6) | 0, site 1 | ||
| 4, site 2 | 1 (100)/3 (33) | 2 (50) | ||
| Mus musculus | 9 (13) | 9, site 1 | 5 (20)/4 (50) | 3 (33) |
| 0, site 2 | ||||
| Neotoma mexicana | 4 (6) | 0, site 1 | ||
| 4, site 2 | 0 (0)/4 (0) | |||
| Tamias minimus | 11 (16) | 5, site 1 | 3 (33)/2 (50) | 2 (40) |
| 6, site 2 | 2 (0)/4 (50) | 2 (33) | ||
| Total | 69 (100) | 26 (38) | ||
M/F represents the number (percentage) of SNV-positive rodents of each sex (male/female).
Sequencing of SNV from clinical and rodent samples.
To examine the genetic relationships between the clinical isolate and the viruses found in the rodent samples, we sequenced the genomes of SNVs from the patient’s whole blood and plasma, 8 rodent samples (4 from each site), and a cell culture-passaged control (SN77734) reported previously (18). We developed a multiplex PCR-tiling scheme to sequence the three segments of SNV based on protocols utilized for Zika virus (24). To achieve higher genome coverage, some gaps were filled using Sanger sequencing. Viral genome coverage across the samples ranged between 47% and 99% completeness when mapped against the SNV reference genome NMH10 (GenBank accession numbers NC_005215, NC_005216, and NC_005217), which was the first sequence for a human SNV infection (25).
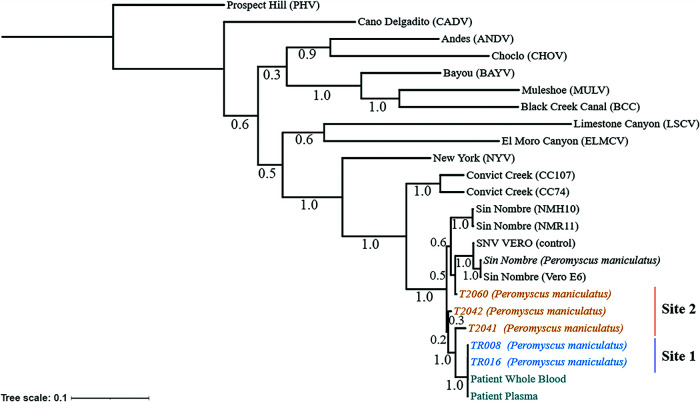
We placed these genomes into a phylogenetic framework via alignment and compared individual rodent sample genomes with the patient sample genomes (Fig. 2). Our analysis shows that all SNV genomes sequenced in this study clustered together in the phylogeny. The patient SNV genomes formed a tight cluster with genomes found in two rodents trapped at site 1 (Fig. 2): P. maniculatus TR008 was trapped near a greenhouse, and P. maniculatus TR016 was captured inside a shed frequently visited by the patient, suggesting that site 1 may be the site of infection rather than site 2, although we did not find that the rodent SNV genomes clustered based on trapping site.
FIG 2.
Phylogenetic analysis of captured P. maniculatus and patient samples suggests transmission at the residential site (site 1). A maximum likelihood tree was generated based on concatenated S, M, and L segments. Bootstrap support is indicated. Both sites were within a 15-mile radius and are indicated as residential site (TR [site 1]) (blue) or second site (T2 [site 2]) (orange). Patient plasma and whole blood were sequenced and are shown (teal). The tree was rooted to the Prospect Hill reference sequence. Additional hantavirus reference sequences were obtained through the NCBI. SNV from infected Vero E6 cells (SNV VERO) was used as a positive sequencing control. The bar represents the number of substitutions per site.
Next, we analyzed specific amino acid variations within the N, Gn/Gc, and L genes in two samples from site 1 (TR008 and TR016), one sample from site 2 (T2041), SNV isolate SN77734 expanded in Vero E6 cells, and the patient samples and compared them to the published SNV reference sequence (NMH10). We found 21 amino acid polymorphisms, with 6 of these being conserved across all the samples and an additional 4 being found only in the cell culture-passaged SN77734 strain (Table 2). The N protein showed two amino acid variants, whereas more variants were found in the glycoprotein Gn/Gc and the viral RNA-dependent RNA polymerase. A total of six single-amino-acid changes were found only in the site 2 virus, whereas four were found only in the site 1 viruses (all of which were also found in the patient virus), compared to the reference sequence. Further investigation on whether these variants are relevant for pathogenesis or rodent-to-human transmission is warranted. Additional experiments are required to assess if there are multiple SNV networks circulating in this population or whether this represents a single introduction into the population.
TABLE 2.
SNV amino acid variants detected between patient and rodent samples, controls, and the SNV reference sequence
| Amino acid position | Amino acid change |
Position in protein structure | Sample(s) that contains amino acid change relative to reference sequence | |
|---|---|---|---|---|
| Reference sequence | Sample | |||
| 89 | H | D | Nucleocapsid | T2041 (site 2) |
| 110 | E | Q | Nucleocapsid | T2041 (site 2) |
| 47 | I | T | Glycoprotein (Gn) | All |
| 85 | K | R | Glycoprotein (Gn) | T2041 (site 2) |
| 217 | V | I | Glycoprotein (Gn) | Patient, TR008, TR016 (site 1) |
| 313 | E | D | Glycoprotein (Gn) | T2041 (site 2) |
| 504 | A | T | Glycoprotein (Gn) | All |
| 1120 | I | V | Glycoprotein (Gc) | SNV Vero |
| 1140 | N | T | Glycoprotein (Gc) | SNV Vero |
| 11 | V | I | RNA polymerase | SNV Vero |
| 51 | K | E | RNA polymerase | SNV Vero |
| 242 | N | S | RNA polymerase | Patient, TR008/TR016 (site 1) |
| 260 | K | R | RNA polymerase | Patient, TR008/TR016 (site 1) |
| 265 | N | S | RNA polymerase | T2041 (site 2) |
| 276 | S | D | RNA polymerase | All |
| 645 | I | V | RNA polymerase | T2041 (site 2) |
| 954 | I | T | RNA polymerase | All except SNV Vero |
| 1791 | R | K | RNA polymerase | All |
| 1806 | K | R | RNA polymerase | Patient, TR008/TR016 (site 1) |
| 1883 | A | T | RNA polymerase | All |
| 1940 | R | K | RNA polymerase | All |
Multiple rodent species as potential reservoirs for SNV.
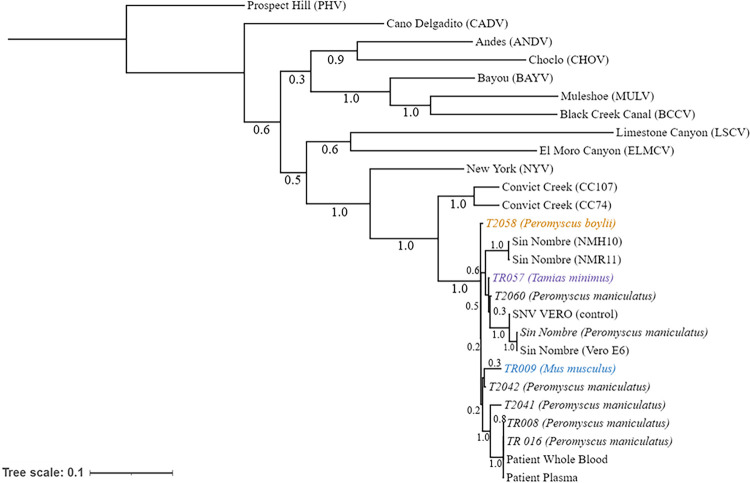
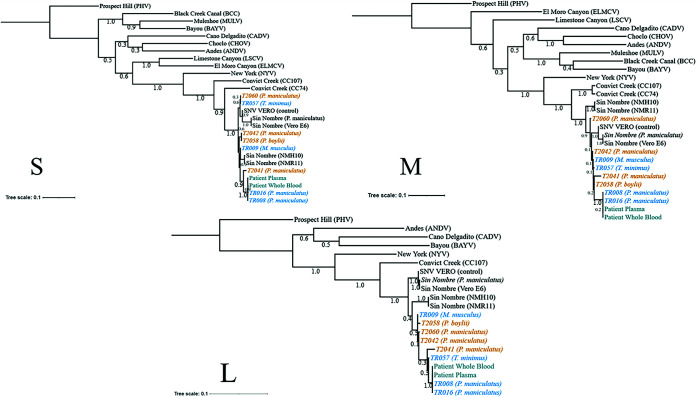
As shown in Table 1, we discovered SNV in rodents other than the known carrier P. maniculatus, including P. boylii, M. musculus, and T. minimus. We sequenced SNV from one animal for each of these rodents and were able to generate a partial genome (50 to 60%) for these samples (TR009, TR057, and T2058). Based on these sequences, we found that the SNV sequences in these rodents grouped more closely with the other SNV sequences from this study than with previously reported SNV samples, and these sequences did not group with previously published non-SNV hantavirus sequences (Fig. 3). Furthermore, we confirmed this finding by analyzing individual segment sequences (Fig. 4). These data provide the first evidence that SNV can be harbored by multiple rodent species at the geographical site of a human SNV infection, suggesting that multiple rodent species may act as hosts for SNV and potentially transmit the virus to humans.
FIG 3.
Phylogenetic analysis of captured rodents indicates that multiple rodents can be infected with SNV. A maximum likelihood tree was generated based on S, M, and L segments combined. The numbers below the branches indicate bootstrap values from 500 replicates. Chipmunk (T. minimus) (purple), common house mouse (M. musculus) (blue), and pinyon mouse (P. boylii) (orange) genomes from both sites are displayed. The tree was rooted to the Prospect Hill reference sequence. Additional reference sequences were obtained through the NCBI and combined. SNV-infected Vero E6 cells (SNV VERO) were used as positive sequencing controls. The bar represents the number of substitutions per site.
FIG 4.
Phylogenetic analysis of individual segments. A maximum likelihood tree was generated based on S, M, and L segments individually. The numbers below the branches indicate bootstrap values from 500 replicates. Rodents from site 1 (orange) and site 2 (blue) are labeled accordingly, along with patient samples (teal). The tree was rooted to the Prospect Hill reference sequence. Additional reference sequences were obtained through the NCBI. SNV-infected Vero E6 cells (SNV VERO) were used as positive sequencing controls. The bars represent the number of substitutions per site.
Identifying host reservoirs that contribute to zoonosis for emerging and reemerging pathogens has implications for human health (26–30). Molecular tools such as RT-qPCR and viral sequencing can help diagnose and understand these spillover events in humans by allowing the rapid screening of infected host and human samples while comparing viral genome sequences. With advances in technology, sequencing has become essential in tracking transmission and detecting novel variants in several zoonotic viruses (31–35). Spontaneous outbreaks of SNV and its high case-fatality rate raise concerns that additional host reservoirs may exist that could contribute to increased infection rates in humans.
Here, we sequenced large portions of SNV genomes in both infected wild-caught rodents and patient samples. The SNV sequences from the samples that we report here did not group in the same clade as SNV CC107 (GenBank accession numbers KT885044, KT885045, and KT885046), which was isolated from P. maniculatus in northern California, suggesting that regional differences exist in SNVs circulating in P. maniculatus populations, as previously shown (36, 37). It has also been shown that partial sequencing of the S and M segments is sufficient to identify genotypes between SNVs in P. maniculatus, which generated individual clades for SNVs from mice caught in New Mexico compared to CC74 (GenBank accession numbers L33816.1, L33684.1, and L35009.1), another isolate from a P. maniculatus rodent captured in northern California (37). Previous studies have conducted serum antibody tests to measure SNV prevalence in rodents, with a range from as low as 2% up to 33% depending on the rodent and study (3, 4, 6, 38). Our results indicate a high rate of SNV prevalence (39%) based on the detection of SNV genomic RNA in lung tissue from rodents trapped near the patient’s residence. The use of RT-qPCR rather than serum antibody tests may be more sensitive and less prone to false-positive results, which can occur from cross-reactive antibodies induced by infection with other hantaviruses known to circulate in rodents.
One limitation of our study is the incomplete genome coverage (47 to 99%) from the SNV tissue samples. Sequencing of SNV expanded in tissue culture results in much-improved genome coverage; however, in vitro culture can introduce mutations to SNV that can affect pathogenesis (39). We found that a higher cycle threshold (CT) value (greater than ∼35) resulted in lower genome coverage (24, 40). Future experiments will focus on the optimization of the SNV sequencing protocol from wild-caught rodent tissue samples.
Our findings are the first to report the sequencing of large portions of SNV in rodents other than P. maniculatus. This is notable since the primary host of SNV is thought to be P. maniculatus, but an understanding of the barriers that restrict orthohantavirus spillover into additional host populations remains elusive. Antibody-based tests can help monitor and survey SNV prevalence in populations of small mammals, including P. maniculatus (3, 5, 37, 41–43). However, to date, only a few studies have reported serum antibodies against SNV in the genus Tamias (chipmunk) (3, 4, 41), and few reports of detection of SNV genomes by specific RT-qPCR have been demonstrated (6). Furthermore, no genome sequencing of SNV in non-P. maniculatus species has been reported. Given the cross-reactivity of serum antibodies against multiple hantaviruses and the widespread distribution of avirulent hantaviruses in rodent populations, sequencing is vital to confirm SNV infection in rodents (3, 4, 6). The grouping of SNV samples from multiple rodents such as P. boylii, M. musculus, and T. minimus with SNV samples from P. maniculatus raises concerns that additional rodent reservoirs can potentially cause zoonoses. It is unknown whether these represent spillover events from P. maniculatus to other rodents and are dead-end hosts or if these rodents are reservoirs for SNV. Interestingly, the overlap of rodent populations due to their distribution in this study may contribute to this phenomenon of spillover. It has been shown that multiple Peromyscus species reside in New Mexico along with T. minimus in the northern parts of the state, which may contribute to higher competition for resources and interactions between different small mammals allowing increased spillover of SNV (44, 45). There are reports of Peromyscus species that can travel over 8 miles from original trap sites (46). Factors such as geographical barriers, resources, and population density can affect rodent dispersal and migration. Possibly, the SNV spillover is due to multiple conditions that affect the overlap of rodent habitats. A critical next step is to assess whether any of these rodents can transmit SNV and whether the SNVs from these rodents are virulent in humans. Multiple studies have analyzed SNV RNA in feces, urine, or saliva/oropharyngeal fluid in laboratory-infected or naturally infected P. maniculatus. In general, SNV-infected animals had undetectable viral RNA in feces and a low prevalence in urine but a potentially higher frequency in saliva or oropharyngeal fluid (47–50). Future work should address whether SNV is found at a higher level in the excreta of P. maniculatus than in other infected rodents. Additional surveying and sequencing will help to further understand the role of these hosts in hantavirus ecology and transmission.
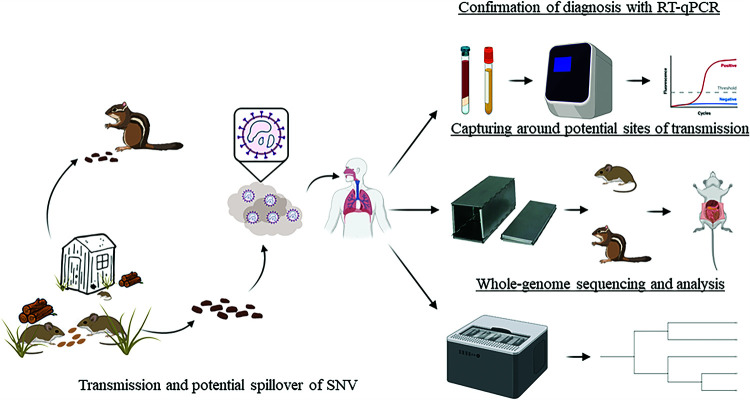
In conclusion, we report a case study for an SNV-infected patient, followed by rodent capture and the use of molecular tools and viral genome sequencing technology to trace transmission to a potential site and activity (cleaning of the shed at site 1). While doing so, we also discovered that multiple rodents, including T. minimus, P. boylii, and M. musculus, can be infected with SNV (Fig. 5).
FIG 5.
Tracing SNV infection in both the patient and rodents. Patient samples were confirmed for SNV infection, and wild-caught rodents caught at two potential sites of infection were tested for SNV. A whole viral genome sequencing approach was used on patient and rodent samples to determine site 1 as a possible source of SNV patient exposure. This study reports for the first time the use of whole viral genome sequencing to study SNV epidemiology.
MATERIALS AND METHODS
Plasmid construction, cultured virus, and clinical samples.
A plasmid carrying Sin Nombre virus N was generated by GenScript using the N gene from the S segment sequence in a pFastBac1 backbone and was used for the RT-qPCR standard curve. For positive virus controls, Vero E6 cells were infected with Sin Nombre virus (SN77734) at a multiplicity of infection (MOI) of 1 in a biosafety level 3 (BSL-3) facility, and RNA was extracted after lysis. Patient serum, whole blood, and plasma were obtained after patient consent in accordance with institutional review board (IRB) protocol 17-382. SNV infection was confirmed by RT-qPCR using primers and a probe that were adopted from previously published sets based on the detection of the S segment of the Sin Nombre virus genome (18). Human ACTB (beta-actin) primers with a VIC/TAMRA (6-carboxytetramethylrhodamine) dye probe (catalog number 4310881E; Applied Biosystems) were used as endogenous positive controls.
Rodent sample collection.
A total of 69 rodents were collected over 2 consecutive nights (160 trap nights), which included two sites in northern New Mexico, with permission of the property owner. Sherman live traps (3 by 3.5 by 9 in.; H. B. Sherman Co., Tallahassee, FL) baited with peanut butter and oats were used. All field procedures were performed according to the animal care and use guidelines of the American Society of Mammalogists (54, 55) and approved by the University of New Mexico Institutional Animal Care and Use Committee, and animals were collected under a New Mexico Department of Game and Fish permit (authorization number 3300). Holistic museum specimens were prepared according to best practices for emerging pathogen research and databased in a relational collection management system (https://arctosdb.org) to facilitate linkage of host specimen data and derived pathogen data (51, 52). Measurements (total length, tail length, hind foot [with claw], ear [from notch], and weight), reproductive data (sex, reproductive status, and testes and embryo crown-rump measurements), and age were recorded. Species identifications were determined through a combination of measurement data and morphological characteristics and confirmed by cytochrome b sequence analysis. Collected tissues were snap-frozen in liquid nitrogen and included brown fat, spleen, heart, lung, kidney, liver, colon (with feces), urinary bladder (with urine if present), and serum from blood centrifuged in the field. Animal specimens were deposited in the University of New Mexico, Museum of Southwestern Biology (MSB) with catalog numbers MSB:Mamm:332811 to -332874, -332702, -332703, -332706, -332711, and -332767.
RNA extraction.
RNA extraction of rodent lung tissue was performed using the QIAamp viral RNA minikit (Qiagen) according to the manufacturer’s instructions, with slight modifications. Briefly, an average of 40 mg of frozen lung tissue was added to a bead beater tube preloaded with 1.0 g of 1.0-mm-diameter zirconia beads (catalog number 1107911zx; BioSpec), 1.0 g of 2.0-mm-diameter zirconia beads (catalog number 11079124zx; BioSpec), and 800 μl of AVL buffer. The tissue was bead beaten using a Benchmark Bead Bug-6 instrument at a speed of 4,350 rpm for 30 s for 1 cycle. Homogenates were centrifuged, pipetted into a microcentrifuge vial, and centrifuged to remove debris, and the clear lysate was pipetted into a fresh microcentrifuge vial. The RNA carrier was then added to the clear lysate, and RNA extraction proceeded according to the manufacturer’s instructions.
Reverse transcriptase quantitative PCR.
Two-step reverse transcription (RT) using SuperScript II (Invitrogen, Thermo Fisher Scientific) was established for the QuantStudio5 series system (Applied Biosystems). RT was performed using 5 μl of RNA (∼500 ng) with 1 μl of SuperScript II containing 4 μl 5× first strand buffer, 2 μl 0.1 M dithiothreitol (DTT), 1 μl RNaseOUT, 1 μl random primers, 1 μl deoxynucleoside triphosphate (dNTP) mix (10 mM), and 5 μl of reverse transcriptase quantitative PCR (RT-qPCR)-grade water. This reaction mixture was incubated at 65°C for 5 min and placed on ice briefly, followed by 10 min at room temperature for binding with a 50-min reaction at 42°C, which was then terminated by 15 min at 70°C. RT-qPCRs were carried out by using 10 μl TaqMan Fast advanced master mix (catalog number 4444965; Applied Biosystems, Thermo Fisher Scientific) containing 2 μl cDNA (∼200 ng), 2 μl primer (2.5 μM), 1 μl of either probe (10 μM), and 4 to 5 μl diethyl pyrocarbonate (DEPC)-treated water (Nalgene) in a 20-μl final reaction volume. For each sample, duplicate or triplicate wells were tested using the following cycling conditions: 20 s at 95°C for the hold stage, while the PCR stage used 1 s at 95°C followed by 52°C for 20 s for a total of 40 cycles. Controls included a no-template control (NTC), a positive SNV sample, a plasmid used for the standard curve, and positive and negative patient samples or wild-caught rodent samples. Additionally, β-actin primers (forward [F] primer ATG TAC GTA GCC ATC CAG GC and reverse [R] primer TCT TGC TCG AAG TCT AGG GC) specific for P. maniculatus or human ACTB were used as internal controls (53). QuantStudio Design and Analysis software v1.5.1 and GraphPad Prism v9.0.2 were used for analysis and graphs.
Whole-genome sequencing of Sin Nombre virus.
Genome sequencing of SNV was conducted on cDNA of RNA isolated from clinical samples and tissues obtained from wild-caught rodents that tested positive by RT-qPCR. SNV genomes were amplified using a PCR-tiling approach with the generation of primer pools designed using PrimalScheme (https://primalscheme.com/) (24) against SNV genomes available through the National Center for Biotechnology Information (NCBI). The primers are listed in Table S1 in the supplemental material. Primers were tested using GoTaq green master mix (catalog number M7123; Promega) and SNV-infected Vero E6 cells with an adjusted annealing temperature of 62°C for 40 cycles. DNA gel electrophoresis was then performed using 2% agarose gels run at 80 V (120 V depending on the size of the gel) with a 100-bp DNA ladder and imaged using an Analytik Jena gel imager. Using a slightly modified version of a SARS-CoV-2 genome sequencing protocol (https://github.com/CDCgov/SARS-CoV-2_Sequencing/blob/master/protocols/ONT-COVID-19_Tiling/PCR%20tiling%20of%20COVID-19%20virus-minion.pdf), samples were sequenced using an Oxford Nanopore GridION X5 system, with gaps partially filled by Sanger sequencing (Sequetech).
Bioinformatics, phylogenetic analysis, and additional software used.
FASTQ files were concatenated and adapters were trimmed using TrimGalore v0.6.1 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Using the artic-ncov2019 pipeline (https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html), FASTQ files were processed for individual segments and then combined to generate a full consensus genome. Consensus bases were called at a minimum threshold of a 10× depth of coverage, and the consensus genomes were viewed through SnapGene v5.2. SNV genomes were aligned with CLUSTAL Omega and Jalview. Molecular Evolutionary Genetics Analysis (MEGA-X) was used to construct a maximum likelihood tree with 500 bootstraps. Phylogenetic tree figures were generated using Interactive Tree of Life (iTOL). CLUSTAL Omega was also used to produce percent identity matrices. Reference sequences (GenBank accession numbers) used in the analysis are as follows: Sin Nombre NMR11 (L37902 to L37904.1), Sin Nombre NMH10 (NC_005215 to NC_005217.1), Sin Nombre (P. maniculatus) (KF537001 to KF537003.1), Sin Nombre Vero E6 (KF537004 to KF537006.1), Convict Creek 74 (L33816.1, L33684.1, and L35009.1), Convict Creek 107 (KT885044 to KT885046.1), Prospect Hill (NC_038938 to NC_038940.1), Cano Delgadito (NC_034528.1, NC_034525.1, and NC_034515.1), Andes (NC_003466 to NC_003468.1), Choclo (NC_038373 to NC_038374.1), Bayou (NC_038298 to NC_038300.1), Muleshoe (KX066124 to KX066125.1), Black Creek Canal (NC_043073 to NC_043075.1), New York (MG717391 to MG717393.1), El Moro Canyon (NC_038423 to NC_038424.1), and Limestone Canyon (AF307322 to AF307323.1). ExPASy was used to convert nucleotides into amino acids to view variants. Figure 5 was created with BioRender.
Data availability.
Sequences have been deposited in GenBank (accession numbers MZ787939 to MZ787941, MZ787943 to MZ787945, and MZ851449 to MZ851475).
ACKNOWLEDGMENTS
This work was supported by a University of New Mexico School of Medicine Research Allocation Committee (UNM SOM RAC) grant (S.B.B.). S.M.G. is supported by UNM HSC Infectious Disease and Inflammation Program National Institutes of Health (NIH) grant T32AI007. Research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103451 (S.M.G.). The sequencing of the patient isolates was supported by an NIH/National Institute of Allergy and Infectious Diseases (NIAID) Prometheus grant (AI-I7-042 U19). We thank the UNM Center for Advanced Research Computing, supported in part by the National Science Foundation, for providing the research computing resources used in this work.
We thank Mariel Campbell for assistance in tissue loans through the Museum of Southwestern Biology and the Division of Genomic Resources.
S.M.G., R.A.N., D.B.D., D.L.D., Y.G., and K.C.S. were involved in viral sequencing and analyses. S.M.G., R.A.N., C.Y., J.A.C., and J.L.D. performed the animal collection, tissue processing, and permitting processes. M.H. and G.J.M. participated in patient care and/or clinical course description. S.M.G., R.A.N., K.C., and S.B.B. designed the study and wrote the paper. All authors contributed to and approved the final version of the manuscript.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Daryl B. Domman, Email: DDomman@salud.unm.edu.
Darrell L. Dinwiddie, Email: DLDinwiddie@salud.unm.edu.
Steven B. Bradfute, Email: sbradfute@salud.unm.edu.
Stacey Schultz-Cherry, St. Jude Children’s Research Hospital.
REFERENCES
- 1.Laenen L, Vergote V, Calisher CH, Klempa B, Klingstrom J, Kuhn JH, Maes P. 2019. Hantaviridae: current classification and future perspectives. Viruses 11:788. 10.3390/v11090788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldsmith CS, Elliott LH, Peters CJ, Zaki SR. 1995. Ultrastructural characteristics of Sin Nombre virus, causative agent of hantavirus pulmonary syndrome. Arch Virol 140:2107–2122. 10.1007/BF01323234. [DOI] [PubMed] [Google Scholar]
- 3.Mills JN, Johnson JM, Ksiazek TG, Ellis BA, Rollin PE, Yates TL, Mann MO, Johnson MR, Campbell ML, Miyashiro J, Patrick M, Zyzak M, Lavender D, Novak MG, Schmidt K, Peters CJ, Childs JE. 1998. A survey of hantavirus antibody in small-mammal populations in selected United States National Parks. Am J Trop Med Hyg 58:525–532. 10.4269/ajtmh.1998.58.525. [DOI] [PubMed] [Google Scholar]
- 4.Mills JN, Ksiazek TG, Ellis BA, Rollin PE, Nichol ST, Yates TL, Gannon WL, Levy CE, Engelthaler DM, Davis T, Tanda DT, Frampton JW, Nichols CR, Peters CJ, Childs JE. 1997. Patterns of association with host and habitat: antibody reactive with Sin Nombre virus in small mammals in the major biotic communities of the Southwestern United States. Am J Trop Med Hyg 56:273–284. 10.4269/ajtmh.1997.56.273. [DOI] [PubMed] [Google Scholar]
- 5.Burns JE, Metzger ME, Messenger S, Fritz CL, Vilcins IE, Enge B, Bronson LR, Kramer VL, Hu R. 2018. Novel focus of Sin Nombre virus in Peromyscus eremicus mice, Death Valley National Park, California, USA. Emerg Infect Dis 24:1112–1115. 10.3201/eid2406.180089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Childs JE, Ksiazek TG, Spiropoulou CF, Krebs JW, Morzunov S, Maupin GO, Gage KL, Rollin PE, Sarisky J, Enscore RE, Frey JK, Peters CJ, Nichol ST. 1994. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the Southwestern United States. J Infect Dis 169:1271–1280. 10.1093/infdis/169.6.1271. [DOI] [PubMed] [Google Scholar]
- 7.Otteson EW, Riolo J, Rowe JE, Nichol ST, Ksiazek TG, Rollin PE, St Jeor SC. 1996. Occurrence of hantavirus within the rodent population of northeastern California and Nevada. Am J Trop Med Hyg 54:127–133. 10.4269/ajtmh.1996.54.127. [DOI] [PubMed] [Google Scholar]
- 8.Dearing MD, Mangione AM, Karasov WH, Morzunov S, Otteson E, St Jeor S. 1998. Prevalence of hantavirus in four species of Neotoma from Arizona and Utah. J Mammal 79:1254–1259. 10.2307/1383016. [DOI] [Google Scholar]
- 9.Abbott KD, Ksiazek TG, Mills JN. 1999. Long-term hantavirus persistence in rodent populations in central Arizona. Emerg Infect Dis 5:102–112. 10.3201/eid0501.990112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindkvist M, Naslund J, Ahlm C, Bucht G. 2008. Cross-reactive and serospecific epitopes of nucleocapsid proteins of three hantaviruses: prospects for new diagnostic tools. Virus Res 137:97–105. 10.1016/j.virusres.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, Sanchez A, Childs J, Zaki S, Peters CJ. 1993. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science 262:914–917. 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 12.Hjelle B, Jenison S, Torrez-Martinez N, Yamada T, Nolte K, Zumwalt R, MacInnes K, Myers G. 1994. A novel hantavirus associated with an outbreak of fatal respiratory disease in the Southwestern United States: evolutionary relationships to known hantaviruses. J Virol 68:592–596. 10.1128/JVI.68.2.592-596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonsson CB, Figueiredo LT, Vapalahti O. 2010. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev 23:412–441. 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koster F, Foucar K, Hjelle B, Scott A, Chong YY, Larson R, McCabe M. 2001. Rapid presumptive diagnosis of hantavirus cardiopulmonary syndrome by peripheral blood smear review. Am J Clin Pathol 116:665–672. 10.1309/CNWF-DC72-QYMR-M8DA. [DOI] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. 2019. Reported cases of hantavirus disease. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/hantavirus/surveillance/index.html. Accessed 2 September 2021. [Google Scholar]
- 16.Yates TL, Mills JN, Parmenter CA, Ksiazek TG, Parmenter RR, Vande Castle JR, Calisher CH, Nichol ST, Abbott KD, Young JC, Morrison ML, Beaty BJ, Dunnum JL, Baker RJ, Salazar-Bravo J, Peters CJ. 2002. The ecology and evolutionary history of an emergent disease: hantavirus pulmonary syndrome. Evidence from two El Niño episodes in the American Southwest suggests that El Niño-driven precipitation, the initial catalyst of a trophic cascade that results in a delayed density-dependent rodent response is sufficient to predict heightened risk for human contraction of hantavirus pulmonary syndrome. Bioscience 52:989–998. 10.1641/0006-3568(2002)052[0989:TEAEHO]2.0.CO;2. [DOI] [Google Scholar]
- 17.Konda M, Dodda B, Konala VM, Naramala S, Adapa S. 2020. Potential zoonotic origins of SARS-CoV-2 and insights for preventing future pandemics through One Health approach. Cureus 12:e8932. 10.7759/cureus.8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Botten J, Mirowsky K, Kusewitt D, Bharadwaj M, Yee J, Ricci R, Feddersen RM, Hjelle B. 2000. Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus). Proc Natl Acad Sci USA 97:10578–10583. 10.1073/pnas.180197197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terajima M, Hendershot JD, III, Kariwa H, Koster FT, Hjelle B, Goade D, DeFronzo MC, Ennis FA. 1999. High levels of viremia in patients with the hantavirus pulmonary syndrome. J Infect Dis 180:2030–2034. 10.1086/315153. [DOI] [PubMed] [Google Scholar]
- 20.Bagamian KH, Towner JS, Mills JN, Kuenzi AJ. 2013. Increased detection of Sin Nombre hantavirus RNA in antibody-positive deer mice from Montana, USA: evidence of male bias in RNA viremia. Viruses 5:2320–2328. 10.3390/v5092320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calisher CH, Wagoner KD, Amman BR, Root JJ, Douglass RJ, Kuenzi AJ, Abbott KD, Parmenter C, Yates TL, Ksiazek TG, Beaty BJ, Mills JN. 2007. Demographic factors associated with prevalence of antibody to Sin Nombre virus in deer mice in the Western United States. J Wildl Dis 43:1–11. 10.7589/0090-3558-43.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Lehmer EM, Jones JD, Bego MG, Varner JM, St Jeor S, Clay CA, Dearing MD. 2010. Long-term patterns of immune investment by wild deer mice infected with Sin Nombre virus. Physiol Biochem Zool 83:847–857. 10.1086/656215. [DOI] [PubMed] [Google Scholar]
- 23.Lonner BN, Douglass RJ, Kuenzi AJ, Hughes K. 2008. Seroprevalence against Sin Nombre virus in resident and dispersing deer mice. Vector Borne Zoonotic Dis 8:433–441. 10.1089/vbz.2007.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, Oliveira G, Robles-Sikisaka R, Rogers TF, Beutler NA, Burton DR, Lewis-Ximenez LL, de Jesus JG, Giovanetti M, Hill SC, Black A, Bedford T, Carroll MW, Nunes M, Alcantara LC, Jr, Sabino EC, Baylis SA, Faria NR, Loose M, Simpson JT, Pybus OG, Andersen KG, Loman NJ. 2017. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc 12:1261–1276. 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spiropoulou CF, Morzunov S, Feldmann H, Sanchez A, Peters CJ, Nichol ST. 1994. Genome structure and variability of a virus causing hantavirus pulmonary syndrome. Virology 200:715–723. 10.1006/viro.1994.1235. [DOI] [PubMed] [Google Scholar]
- 26.Fauci AS. 2005. Emerging and reemerging infectious diseases: the perpetual challenge. Acad Med 80:1079–1085. 10.1097/00001888-200512000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Hjelle B, Torres-Perez F. 2010. Hantaviruses in the Americas and their role as emerging pathogens. Viruses 2:2559–2586. 10.3390/v2122559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morens DM, Fauci AS. 2013. Emerging infectious diseases: threats to human health and global stability. PLoS Pathog 9:e1003467. 10.1371/journal.ppat.1003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cook JA, Arai S, Armien B, Bates J, Bonilla CAC, Cortez MBS, Dunnum JL, Ferguson AW, Johnson KM, Khan FAA, Paul DL, Reeder DM, Revelez MA, Simmons NB, Thiers BM, Thompson CW, Upham NS, Vanhove MPM, Webala PW, Weksler M, Yanagihara R, Soltis PS. 2020. Integrating biodiversity infrastructure into pathogen discovery and mitigation of emerging infectious diseases. Bioscience 70:531–534. 10.1093/biosci/biaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson CW, Phelps KL, Allard MW, Cook JA, Dunnum JL, Ferguson AW, Gelang M, Khan FAA, Paul DL, Reeder DM, Simmons NB, Vanhove MPM, Webala PW, Weksler M, Kilpatrick CW. 2021. Preserve a voucher specimen! The critical need for integrating natural history collections in infectious disease studies. mBio 12:e02698-20. 10.1128/mBio.02698-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardy J, Loman NJ, Rambaut A. 2015. Real-time digital pathogen surveillance—the time is now. Genome Biol 16:155. 10.1186/s13059-015-0726-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houldcroft CJ, Beale MA, Breuer J. 2017. Clinical and biological insights from viral genome sequencing. Nat Rev Microbiol 15:183–192. 10.1038/nrmicro.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shrivastava S, Puri V, Dilley KA, Ngouajio E, Shifflett J, Oldfield LM, Fedorova NB, Hu L, Williams T, Durbin A, Amedeo P, Rashid S, Shabman RS, Pickett BE. 2018. Whole genome sequencing, variant analysis, phylogenetics, and deep sequencing of Zika virus strains. Sci Rep 8:15843. 10.1038/s41598-018-34147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim WK, No JS, Lee D, Jung J, Park H, Yi Y, Kim JA, Lee SH, Kim Y, Park S, Cho S, Lee GY, Song DH, Gu SH, Park K, Kim HC, Wiley MR, Chain PSG, Jeong ST, Klein TA, Palacios G, Song JW. 2020. Active targeted surveillance to identify sites of emergence of hantavirus. Clin Infect Dis 70:464–473. 10.1093/cid/ciz234. [DOI] [PubMed] [Google Scholar]
- 35.Hodcroft EB, Zuber M, Nadeau S, Vaughan TG, Crawford KHD, Althaus CL, Reichmuth ML, Bowen JE, Walls AC, Corti D, Bloom JD, Veesler D, Mateo D, Hernando A, Comas I, Gonzalez-Candelas F, SeqCOVID-SPAIN Consortium, Stadler T, Neher RA. 2021. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature 595:707–712. 10.1038/s41586-021-03677-y. [DOI] [PubMed] [Google Scholar]
- 36.Li D, Schmaljohn AL, Anderson K, Schmaljohn CS. 1995. Complete nucleotide sequences of the M and S segments of two hantavirus isolates from California: evidence for reassortment in nature among viruses related to hantavirus pulmonary syndrome. Virology 206:973–983. 10.1006/viro.1995.1020. [DOI] [PubMed] [Google Scholar]
- 37.Black WC, IV, Doty JB, Hughes MT, Beaty BJ, Calisher CH. 2009. Temporal and geographic evidence for evolution of Sin Nombre virus using molecular analyses of viral RNA from Colorado, New Mexico and Montana. Virol J 6:102. 10.1186/1743-422X-6-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feldmann H, Sanchez A, Morzunov S, Spiropoulou CF, Rollin PE, Ksiazek TG, Peters CJ, Nichol ST. 1993. Utilization of autopsy RNA for the synthesis of the nucleocapsid antigen of a newly recognized virus associated with hantavirus pulmonary syndrome. Virus Res 30:351–367. 10.1016/0168-1702(93)90101-r. [DOI] [PubMed] [Google Scholar]
- 39.Safronetz D, Prescott J, Feldmann F, Haddock E, Rosenke R, Okumura A, Brining D, Dahlstrom E, Porcella SF, Ebihara H, Scott DP, Hjelle B, Feldmann H. 2014. Pathophysiology of hantavirus pulmonary syndrome in rhesus macaques. Proc Natl Acad Sci USA 111:7114–7119. 10.1073/pnas.1401998111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.No JS, Kim WK, Cho S, Lee SH, Kim JA, Lee D, Song DH, Gu SH, Jeong ST, Wiley MR, Palacios G, Song JW. 2019. Comparison of targeted next-generation sequencing for whole-genome sequencing of Hantaan orthohantavirus in Apodemus agrarius lung tissues. Sci Rep 9:16631. 10.1038/s41598-019-53043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turell MJ, Korch GW, Rossi CA, Sesline D, Enge BA, Dondero DV, Jay M, Ludwig GV, Li D, Schmaljohn CS, Jackson RJ, Ascher MS. 1995. Short report: prevalence of hantavirus infection in rodents associated with two fatal human infections in California. Am J Trop Med Hyg 52:180–182. 10.4269/ajtmh.1995.52.180. [DOI] [PubMed] [Google Scholar]
- 42.Yang DS, Kenagy G. 2011. Population delimitation across contrasting evolutionary clines in deer mice (Peromyscus maniculatus). Ecol Evol 1:26–36. 10.1002/ece3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guralnick R, Hantak MM, Li D, McLean BS. 2020. Body size trends in response to climate and urbanization in the widespread North American deer mouse, Peromyscus maniculatus. Sci Rep 10:8882. 10.1038/s41598-020-65755-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bedford NL, Hoekstra HE. 2015. Peromyscus mice as a model for studying natural variation. Elife 4:e06813. 10.7554/eLife.06813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montana Field Guide. 3 September 2021. Least chipmunk—Tamias minimus. Montana Natural Heritage Program and Montana Fish, Wildlife and Parks, Helena, MT. https://fieldguide.mt.gov/speciesDetail.aspx?elcode=AMAFB02020. [Google Scholar]
- 46.Maier TJ. 2002. Long-distance movements by female white-footed mice, Peromyscus leucopus, in extensive mixed-wood forest. Can Field Nat 116:108–111. [Google Scholar]
- 47.Botten J, Mirowsky K, Ye C, Gottlieb K, Saavedra M, Ponce L, Hjelle B. 2002. Shedding and intracage transmission of Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) model. J Virol 76:7587–7594. 10.1128/jvi.76.15.7587-7594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warner BM, Stein DR, Griffin BD, Tierney K, Leung A, Sloan A, Kobasa D, Poliquin G, Kobinger GP, Safronetz D. 2019. Development and characterization of a Sin Nombre virus transmission model in Peromyscus maniculatus. Viruses 11:183. 10.3390/v11020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Safronetz D, Lindsay R, Dibernardo A, Hjelle B, Xiao R, Artsob H, Drebot MA. 2005. A preliminary study of the patterns of Sin Nombre viral infection and shedding in naturally infected deer mice (Peromyscus maniculatus). Vector Borne Zoonotic Dis 5:127–132. 10.1089/vbz.2005.5.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Safronetz D, Drebot MA, Artsob H, Cote T, Makowski K, Lindsay LR. 2008. Sin Nombre virus shedding patterns in naturally infected deer mice (Peromyscus maniculatus) in relation to duration of infection. Vector Borne Zoonotic Dis 8:97–100. 10.1089/vbz.2007.0113. [DOI] [PubMed] [Google Scholar]
- 51.Dunnum JL, Yanagihara R, Johnson KM, Armien B, Batsaikhan N, Morgan L, Cook JA. 2017. Biospecimen repositories and integrated databases as critical infrastructure for pathogen discovery and pathobiology research. PLoS Negl Trop Dis 11:e0005133. 10.1371/journal.pntd.0005133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galbreath KE, Hoberg EP, Cook JA, Armien B, Bell KC, Campbell ML, Dunnum JL, Dursahinhan AT, Eckerlin RP, Gardner SL, Greiman SE, Henttonen H, Jimenez FA, Koehler AVA, Nyamsuren B, Tkach VV, Torres-Perez F, Tsvetkova A, Hope AG. 2019. Building an integrated infrastructure for exploring biodiversity: field collections and archives of mammals and parasites. J Mammal 100:382–393. 10.1093/jmammal/gyz048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davenport BJ, Willis DG, Prescott J, Farrell RM, Coons TA, Schountz T. 2004. Generation of competent bone marrow-derived antigen presenting cells from the deer mouse (Peromyscus maniculatus). BMC Immunol 5:23. 10.1186/1471-2172-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mills JN, Childs JE, Ksiazek TG, Peters CJ. 1995. Methods for trapping and sampling small mammals for virologic testing. US Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA. https://stacks.cdc.gov/view/cdc/11507. [Google Scholar]
- 55.Sikes RS, Animal Care and Use Committee of the American Society of Mammalogists . 2016. Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J Mammal 97:663–688. 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 (Primers used for SNV sequencing.)<br>. Download jvi.01534-21-s0001.xlsx, XLSX file, 0.02 MB (18KB, xlsx)
Data Availability Statement
Sequences have been deposited in GenBank (accession numbers MZ787939 to MZ787941, MZ787943 to MZ787945, and MZ851449 to MZ851475).