Abstract
MicroRNAs (miRNAs) are small non-coding RNA that regulate the expression of messenger RNA and are implicated in almost all cellular processes. Importantly, miRNAs are can be released extracellularly and are stable in these matrices where they may serve as indicators of organ or cell-specific toxicity, disease, and biological status. There has thus been great enthusiasm for developing miRNAs as biomarkers of adverse outcomes for scientific, regulatory, and clinical purposes. Despite advances in measurement capabilities for miRNAs, miRNAs are still not routinely employed as non-invasive biomarkers. This is in part due to the lack of standard approaches for sample preparation and miRNA measurement and uncertainty in their biological interpretation. Members of the microRNA Biomarkers Workgroup within the Health and Environmental Sciences Institute’s (HESI) Committee on Emerging Systems Toxicology for the Assessment of Risk (eSTAR) are a consortium of private- and public-sector scientists dedicated to developing miRNAs as applied biomarkers. Here, we explore major impediments to routine acceptance and use of miRNA biomarkers and case examples of success and deficiencies in development. Finally, we provide insight on miRNA measurement, collection, and analysis tools to provide solid footing for addressing knowledge gaps toward routine biomarker use.
Keywords: microRNAs, biomarkers, biofluids, qualification, safety assessment, regulation, clinical, review
INTRODUCTION
In the almost three decades since the discovery of the small non-coding regulatory RNA by Ambros and Ruvkun (Lee et al. 1993; Wightman et al. 1993), microRNAs (miRNAs) have been under intense investigations to understand their role in disease processes, as putative targets of therapeutics, and as biomarkers of these underlying biological processes. The latter has been of particular interest when it was discovered that miRNAs were stably detected extracellularly in biofluids, including blood, urine, cerebrospinal fluid, saliva, and tears, among others (Valadi et al. 2007; Chen et al. 2008; Wang et al. 2010; Weber et al. 2010; Arroyo et al. 2011; Turchinovich et al. 2011; Vickers et al. 2011). There are a number of reasons why miRNAs are released from the cell into these matrices, including speculation that miRNAs are effectors of cell-to-cell communication (Valadi et al. 2007; Wang et al. 2010; Vickers et al. 2011). In addition, they may also be passively released during cell injury and content release into biofluids, thereby potentially serving as biomarkers of injury and/or toxicity (Harrill et al. 2016). The added value of miRNAs as biomarkers is that some display tissue specificity or enrichment, which has been demonstrated in mouse, rat, dog, and human (Lagos-Quintana et al. 2002; Babak et al. 2004; Liang et al. 2007; Mestdagh et al. 2011; Minami et al. 2014; Yu et al. 2014; Koenig et al. 2016; Smith et al. 2016), as well as high conservation across other model species (Landgraf et al. 2007; Koenig et al. 2016), allowing for human translation of findings in animal and alternative model experimentation (Wang et al. 2009, 2010; Kong et al. 2010; Starkey Lewis et al. 2011; Antoine et al. 2013; Usborne et al. 2014; Liu et al. 2015; Nishimura et al. 2015). Recent studies have indicated strong correlations between environmental pollutants and radiation, thereby altering levels of miRNAs linked to metabolic and vascular disorder in exposed human populations (Ruiz-Vera et al., 2019; Martinez-Ibarra et al., 2019), further indicating direct utility of these biomarkers in environmental hazard evaluations. Not coincidentally, the recent development of methodologies to reproducibly measure miRNAs in small volumes of bodily fluids has also increased the feasibility of further developing biofluid-based miRNA biomarkers (Thompson et al. 2016; Lee et al. 2017; Thorsen et al. 2017; Detassis et al. 2019).
The stage has been seemingly set for robust development of biofluid-based miRNA biomarkers. Although there has been significant investment in prognostic and diagnostic miRNA biomarkers, the adoption of miRNAs beyond discovery and basic scientific applications has not been fully realized. To date, there are no U.S. Food and Drug Administration (FDA)-qualified miRNA biomarkers, which include categories of risk, diagnosis, monitoring, prognosis, predictiveness, response, and safety. This is partially due to the costly and the arduous process of biomarker qualification, although recent initiatives such as the Center for Drug Evaluation and Research’s (CDER) Biomarker Qualification Program (BQP) and published guidance have provided a pathway for early development (Amur et al. 2015). The TGx-28.65 transcriptomic biomarker panel for distinguishing agents that induce DNA damage from those that do not recently received a letter of support by FDA (Li et al. 2019) and is currently under regulatory review for qualification, which holds promise for future molecular-based biomarker qualification such as miRNAs. Evidence of high enthusiasm is also reflected in the scientific literature with citations on biofluid-based miRNA biomarkers nearly doubling annually between 2009 and 2015; however, interest has levelled off in the years since this period (Figure 1). The slow progress toward qualified miRNA biomarkers, and the apparent recent cooling-off period in related scientific publications, is likely attributable to an overall uncertainty associated with biofluid-based miRNA measurements.
Figure 1.
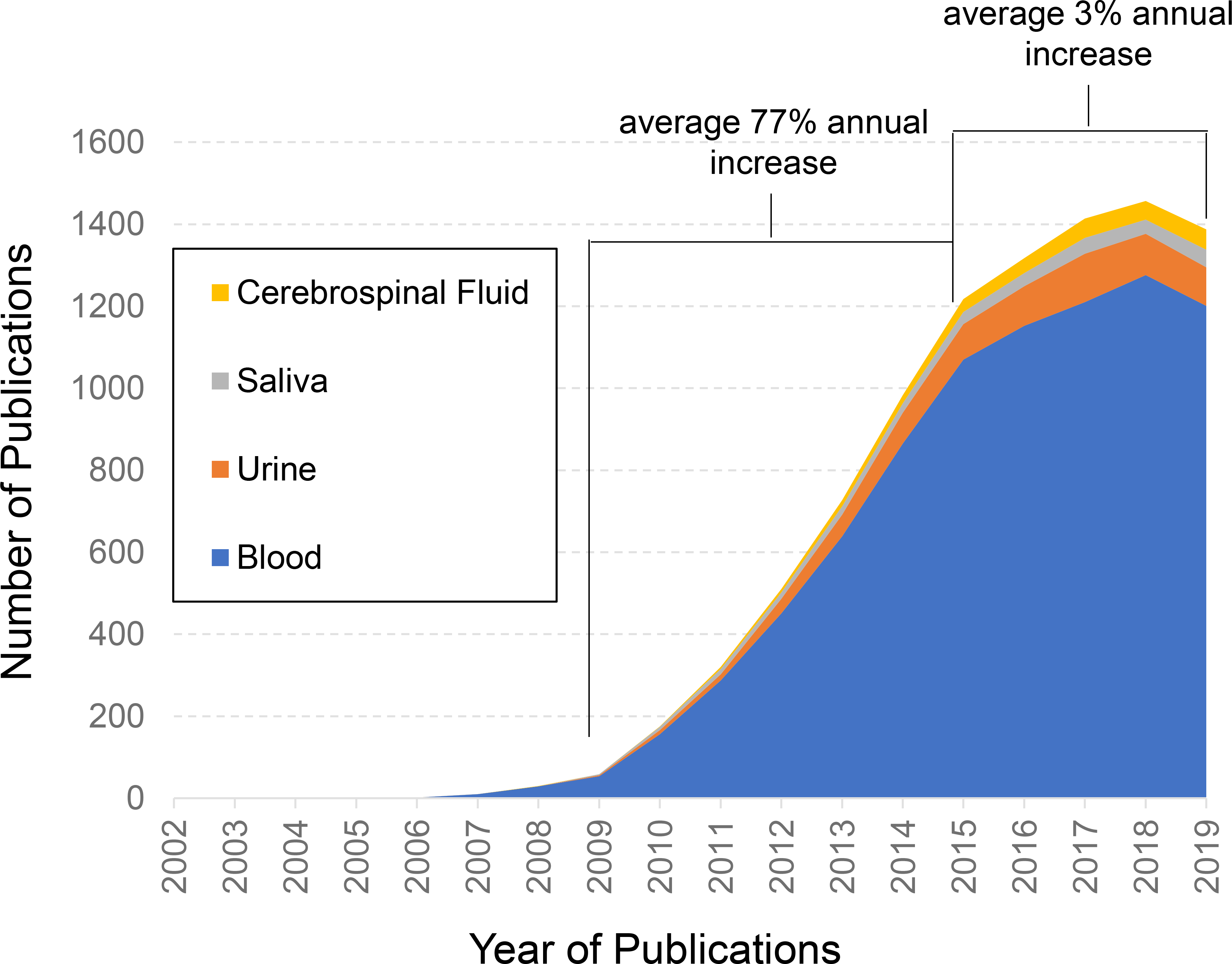
Leveling trend of published microRNA biomarker studies in common biofluids. PubMed was searched for the terms microRNA and biomarker in addition to blood (blue), urine (orange), saliva (grey), or cerebrospinal fluid (yellow). The graph displays the number of total annual publications over the past 17 years. A near annual doubling of publications occurred from the years 2009 until 2015, whereas only a mere 3% increase in annual publications from 2015 until 2019 was noted. While several factors can contribute to these publication trends, this indicates an overall cooling of research interest in biofluid-based microRNA biomarker development.
The Health and Environmental Sciences Institute’s (HESI) Committee on Emerging Systems Toxicology for the Assessment of Risk (eSTAR) encompasses a workgroup of experienced toxicologists and life scientists in the government, academic, non-profit, and commercial sectors who are collectively involved in developing miRNA biomarkers for regulatory, drug development, and clinical applications. For nearly a decade, eSTAR collaborative efforts have focused on key questions surrounding the development of miRNA biomarkers, including tissue specificity (Pavkovic et al. 2015; Koenig et al. 2016; Smith et al. 2016; Bushel et al. 2018) and measurement techniques in biofluids (Harrill et al. 2016; Thompson et al. 2016; Wolfinger et al. 2018). There remain multiple utilization challenges for miRNAs, including but not limited to biases associated with biological sample collection, storage, and isolation, the non-standardization of miRNA quantification, the lack of understanding of normal biological variation of biofluid miRNAs, and understanding the functional and prognostic role of these molecules (Figure 2). The inherent qualities of miRNAs suggest these unique small RNAs are strong biomarker candidates, but a clear understanding of the strengths and weaknesses of the sampling, the analyses performed, and the context of use, are necessary before widespread implementation. The purpose of this manuscript is to review the literature to inform the reader on specific knowledge gaps to enable a more certain path forward in applying biofluid-based miRNA biomarkers in a variety of decision-making contexts.
Figure 2.
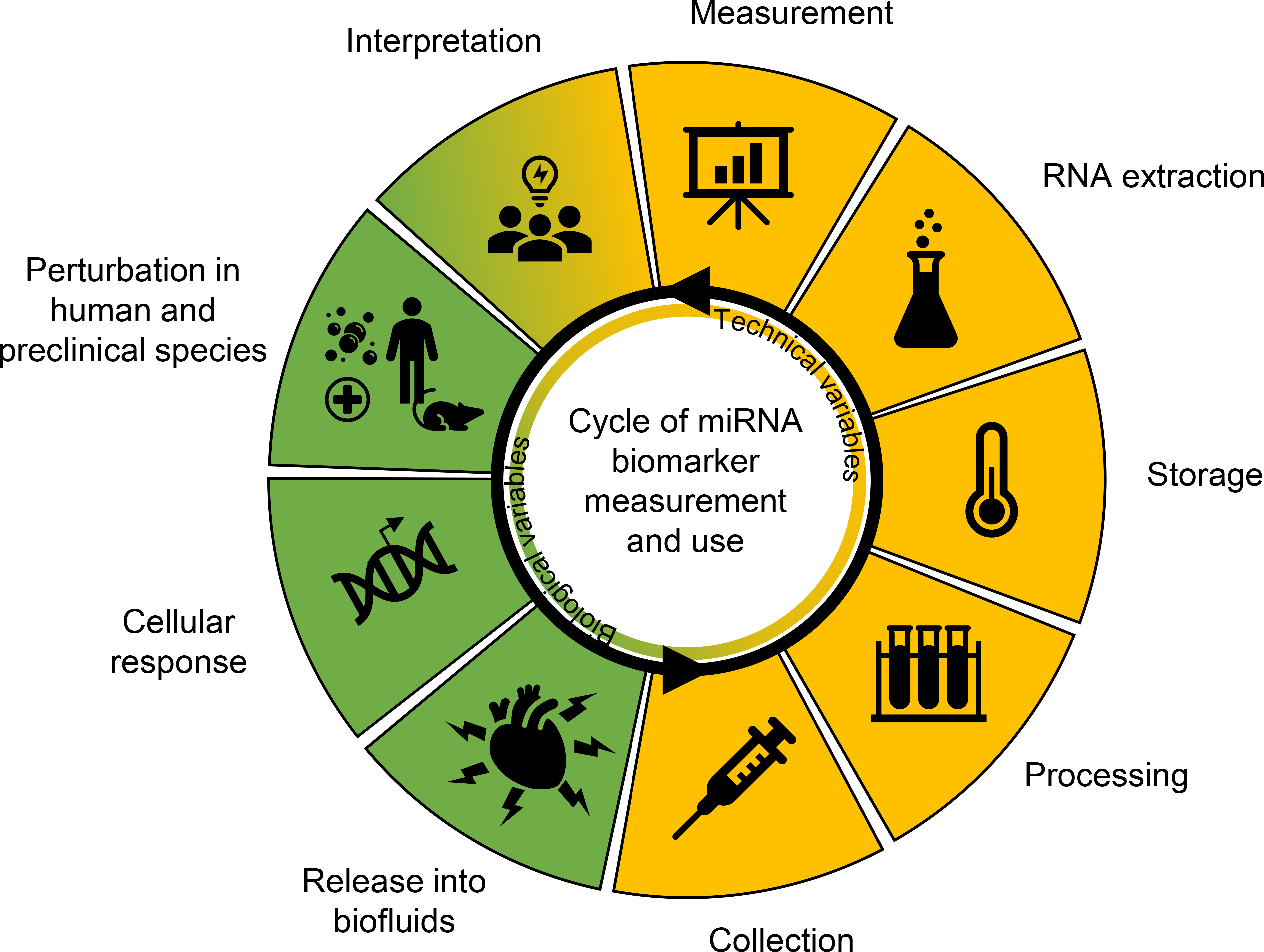
Challenges associated with microRNA (miRNA) biomarker development. A cycle of technical (yellow) and biological (green) features feed into biofluid-based miRNA biomarker development. Each slice indicates stepwise considerations of miRNA collection, processing, storage, extraction, measurement, interpretation, and biological application and understanding. Ultimately, knowledge from these steps feeds back into the primary purpose of these biomarkers, which is to assess human and model species adverse outcomes due to toxicity, disease, and other perturbations. Challenges are associated with each of these steps and need to be addressed before miRNA biomarkers can be established.
RELIABLY DISTINGUISHING SIGNAL FROM NOISE
In established protein biomarkers measured in blood, low baseline values and variability are preferable to detect small changes (Shlipak et al. 2005); however, higher baseline or signal variability can generally only be overcome by a robust responsive signal (Tanno et al. 2007). The latter baseline expression pattern reflects what is commonly observed in circulating miRNA candidate biomarkers, where nearly half of the measured miRNAs in serum have 30- to 1000-fold baseline variation (Cheng et al. 2013). We have determined that variability is likely miRNA-specific (Thompson et al. 2016), and several processes can contribute to this observation, including the mechanism(s) of cellular release and accessibility to biofluids (Valadi et al. 2007; Tominaga et al. 2015), as well as different macromolecule interactions within biofluids which may affect stability (Li et al. 2007; Mitchell et al. 2008). In addition, miRNAs near the lower limit of detection further confound the statistical calls that distinguish measurements from baseline (Wolfinger et al. 2018). While biological variation that is not attributable to perturbation cannot be avoided entirely, some confounding technical factors can be minimized by reducing variables introduced at collection, storage, isolation, measurement, and analysis (Table 1).
Table 1.
Summary of recommendations for sample collection and preparation for common accessible biofluids.
| Sample | Fraction | Preparation | Storage/transport | Timing | Key information |
|---|---|---|---|---|---|
| Blood | miRNA abundances differ in serum, plasma, and whole blood | Use EDTA or citrate collection tubes and avoid heparin | Chill, use within 6 hours otherwise ship on dry ice | Standardize timing of collection across samples | Track freeze/thaw cycles |
| Centrifuge to isolate serum or plasma fraction; store desired fraction within 1–3 hours after collection | If shipping conditions only available at ambient temperature, transfer whole blood | ||||
|
| |||||
| Urine | N/A | 16–18 hours collection (experimental) or spot urine (clinical) | Chill, for immediate use <6 hours, otherwise ship on dry ice | Standardize timing of collection across samples | Track freeze/thaw cycles |
| Centrifuge to eliminate exogenous and/or cellular material | If shipping conditions at ambient temperature, transfer urine supernatant after centrifugation | Track total volume collected | |||
| Saliva | Acellular fraction | Subjects should refrain from eating, drinking, smoking, or oral hygiene procedures for 1 h prior to collection Rinse mouth well with distilled water for 1 min prior to collection on ice. Briefly vortex and centrifuge to separate and collect acellular fraction |
Add 1 μL SUPERase Inhibitor for every mL of saliva collected Freeze at −80°C immediately |
Standardize timing of collection across samples | Track freeze/thaw cycles RNA polymerase III-transcribed U6 snRNA has been used successfully as an endogenous control normalizer in saliva |
Abbreviations: miRNA, microRNA; N/A, not applicable.
Consistent collection, storage, and extraction from sample
Normal biological variation can be dictated by both internal and external biological cues that may not be directly attributable to the disease state or phenotype of interest. For example, sexual dimorphic patterns of miRNAs are present in peripheral blood (Meder et al., 2014; Cui et al., 2018). Also, normal circadian rhythms can influence diurnal patterns of blood- and salivary-based miRNAs, and some miRNAs that display such cyclic patterns have ubiquitous expression throughout the body and release into biofluids (Heegaard et al. 2016; Hicks et al. 2018). Circadian cycles also influence small extracellular vesicle release into urine (Koritzinsky et al. 2019) and presumably packaged miRNAs. Other normal biological cycles such as the menstrual cycle and prandial states may also contribute to specific miRNA variation in biofluids (Vickers et al. 2011; Marzi et al. 2016). The current literature on the biological periodicities of circulating miRNAs is not extensive and there is not a lot known about cyclic expression of most miRNAs. Therefore, timing of sample collection in studies should be consistent to minimize potential variability in not only clinical samples, but also preclinical safety assessments.
As with time of collection, consistent sample types and storage will reduce measurement variability and increase the capability to compare separate studies and should therefore be carefully predetermined for comparability of samples within and across study samples. Consistency in the location of blood sampling (e.g., retro-orbital, sublingual, aortic, jugular, etc) may affect levels of cellular miRNA contamination from surrounding tissue during collection (Mikaelian et al., 2013). Also, different types of blood fractions can exhibit differing patterns of miRNA detection (Mompeon et al., 2020). A contributing factor is plasma which retains of fibrinogen, whereas serum does not contain clotting factors. Importantly, platelets and red and white blood cells also contain a broad array of miRNAs that can be released into the serum during coagulation (Wang et al. 2012) and may contribute to inter-sample variation in serum samples, if pre-processing centrifugation is not performed (McDonald et al. 2011; Wang et al. 2012; Page et al. 2013). Related to this, miRNA measurements should be performed in serum and plasma samples to determine the extent of hemolysis present, in particular for those miRNAs that may have unwanted contribution from blood cells which may be more than the majority of detectible miRNAs in blood fractions (Kirschner et al., 2013). Since this contribution may occur at levels of hemolysis not detectable by eye, UV-Vis absorbance readings of hemoglobin (Kirschner et al., 2013; Kirschner et al., 2011) or measurements of blood cell miRNAs indicative of hemolysis (Blondal et al., 2013) should be considered. In whole blood samples, blood cell miRNAs may also be released during post-collection storage (Kannan et al. 2009), increasing measurable miRNAs after freezing/thaw cycle (Glinge et al. 2017). Hence, rapid processing after collection is preferred to avoid unwanted contribution of extracellular miRNAs as well as haemoglobin and lactoferrin, which may inhibit downstream applications (Al-Soud and Radstrom 2001).
The associations of extracellular miRNAs with proteins such as Argonaute, lipoproteins, or encapsulation in extracellular vesicles (EVs) are hypothesized to confer microRNA stability in biofluids, in addition to potentially mediating their release across cell membranes (Hunter et al. 2008; Arroyo et al. 2011; Turchinovich et al. 2011; Vickers et al. 2011). At room temperature, some miRNAs can be stable for at least 24 hours in whole blood, serum, and plasma (Zhao et al. 2014; Mitchell et al. 2008), but specific miRNAs may show both more and less resilience due to different levels of protection afforded by these interactions (Koberle et al. 2013; Marzi et al. 2016). Long term, miRNAs in these samples can be stable for nearly 2 years when stored at −20°C or −80°C (Grasedieck et al. 2012; Glinge et al. 2017) but may be markedly decreased during longer storage durations. Freeze/thaw cycles can have a significant impact on stability; however, there are disagreements in the literature with some studies showing minimal degredation of serum miRNAs eve after eight freeze/thaw cycles (Mitchell et al. 2008). Similar to sample pre-processing, the inherent stability of miRNA in biofluids may differ depending on protective interactions of proteins and the collection matrices (Chen et al. 2008; Mitchell et al. 2008; Grasedieck et al. 2012; Glinge et al. 2017). The consensus across datasets suggests that, for applications where long incubation and/or storage times are unavoidable, systematic miRNA stability analysis should be performed. Similarly, miRNAs in cell-free urine are relatively stable, which could be attributable to EVs that encapsulate miRNAs and protect from high levels of RNase activity (Lv et al. 2013). Therefore, miRNAs in isolated EVs may be somewhat more stable, considering that only small losses in miRNA levels were observed after 24 hours at 4°C and for at least five freeze-thaw cycles. Additives can improve this stability (Hanke et al. 2010), but immediate chilling or freezing the sample at −80°C is important for ensuring longer-term miRNA sample storage. Standardization of collection and storage practices is imperative for consistency in miRNA measurements; and pilot studies should be leveraged to determine if the miRNA biomarkers of interest may be more susceptible to degradation, thereby reducing chances of measurement bias.
RNA isolation is required for many miRNA measurement protocols and can contribute to technical variability in samples (Marzi et al. 2016; Khan et al. 2017; Max et al. 2018). A significant number of comparison studies have been published to determine differences in efficiency and performance for commercially available miRNA extraction methods. While some studies report that the choice of extraction method is generally comparable (Hantzsch et al. 2014; Brunet-Vega et al. 2015; Aarem et al. 2016), these studies only measured a limited number of miRNAs; therefore, failing to represent sufficient numbers of supportive independent studies and datasets for validation. Other reports noted that phenol-based extraction methods may bias for miRNAs that are GC enriched (Lam et al. 2016). In addition, Brown et al. (2018) cautioned that total RNA extraction methods are optimized for longer RNA yield rather than small RNA recovery and noted 2- to 4-fold differences in abundance for the subset of miRNAs measured by different extraction techniques from homogenized tissue. In biofluids, there is a low concentration of miRNAs in the aqueous phase and high levels of contaminating protein (such as albumin, immunoglobulins, coagulants, and complement components), further complicating extraction. Techniques to remove protein from samples (Kim and Jung 2011) and increase miRNA yield with added carriers or concentrating RNA post-isolation (Ramon-Nunez et al. 2017) can improve recovery; however, inter-sample variability can be introduced as a result of these methods (Bartram et al. 2009; Jarry et al. 2014; Ramon-Nunez et al. 2017). Quality control (e.g., consistent standard operating procedures, documentation and consistent terminology, spike-in miRNAs during isolation, assessment of hemolysis, potential inclusion of reference samples or treatments leading to known miRNA alterations, etc.) and optimized methods for the specific biofluid of interest are imperative for high confidence in measurements (Tan et al. 2015). Notably, there are methods that simply directly measure miRNAs from biofluids (Glineur et al. 2015; Shah et al. 2018; Bailey et al. 2019; López-Longarela et al. 2019), which eliminates bias due to isolation procedures; however, these procedures may be susceptible to inhibitory contaminants within biofluids; thus, consistent quality control steps are warranted.
Selecting methods for measurement
The methodologies currently available to quantitate miRNAs vary by throughput, cost, dynamic range, and sensitivity. These approaches include quantitative polymerase chain reaction (qPCR), digital-based PCR (dPCR), bead or particle detection, microarrays, and next generation sequencing (NGS; Table 2). The choice of which technique to utilize depends on the user’s goals and application, expertise, and capabilities. In general, the choice of technique depends on the number of miRNAs and samples that need to be assessed in tandem. A small body of work exists that describes correlations between platforms that reproducibly demonstrate directionality of differential abundance of miRNA in samples (Mestdagh et al. 2014). Moreover, any primary disagreements in these measurements were due to different calculated absolute fold-changes; however, each platform demonstrated good intra-platform reproducibility. Conversely, consistent best practices for miRNA measurement should not introduce uncertainty in the data generated; the limitations of methods need to be understood before they are applied to biomarker-based application. For example, developing biomarkers to understand disease prognosis, and in particular regulated diagnostics, requires that the methods used meet regulatory standards for consistency of performance, ideally coefficient of variation less than 25% between replicates (https://www.fda.gov/regulatory-information/search-fda-guidance-documents/bioanalytical-method-validation-guidance-industry), and that sufficient technical controls and reference standards are available to measure assay performance. It would be preferable for this application to choose methods that require minimum samples handling, e.g. preparation of RNA from blood, that can be easily automated or performed by a technician, and that can be performed on approved instrumentation (e.g., a 510k cleared qPCR machine). As such, existing next generation sequencing and microarray platforms may not be the best candidates for prognostic biomarker development.
Table 2.
Current methodologies to measure miRNA
| Method | Strengths | Limitations | Applications | Metric | Dynamic range | Sensitivitya |
|---|---|---|---|---|---|---|
| SYBR-based qPCR | Sensitivity Broad target compatibility Detection reproducibility Common standard operating procedure Cheaper than probe-based technologies |
Multi-step workflows Commonly requires purified RNA Input required for many miRNA screenings can be too high for scarce samples Non-specific amplification difficult to measure IsomiR sequences difficult to discriminate |
Discovery development, Validation, Safety assessment/Preclinical studies, Clinical | Cycle threshold (Ct) | 7 logs | ++++ |
| Hydrolysis probe-based (“TaqMan”) qPCR | Sensitivity Broad target compatibility Detection reproducibility Specificity vs. SYBR qPCR Common standard operating procedure |
Multi-step workflows Commonly requires purified RNA Input required for many miRNA screenings can be too high for scarce samples Non-specific amplification difficult to measure IsomiR sequences difficult to discriminate |
Discovery and development, Validation, Safety assessment/Preclinical studies, Clinical | Cycle threshold (Ct) | 7 logs | ++++ |
| Digital PCR | Digital quantification without standard curve Sensitivity due to end-point amplification |
Multi-step workflows Commonly requires purified RNA Non-specific amplification difficult to measure IsomiR sequences difficult to discriminate |
Discovery and development, Validation, Safety assessment/Preclinical studies, Clinical | Copy number | 5 logs | ++++ |
| Bead hybridization | Can be compatible with biofluids and tissues without RNA purification Low input with amplification |
Number of miRNA detected Sensitivity without amplification is poor IsomiR sequences difficult to discriminate |
Discovery, Validation, Safety assessment/Preclinical studies, Clinical | Abundance (with curve) | 5 logs | ++++ |
| Next generation sequencing | Breadth of small RNA species detected Single nucleotide specificity Can distinguish isomiRs |
Laborious multi-step workflow Input for workflows too high for scarce samples |
Discovery and development | Reads per unit | 5–6 logs | ++ |
| Digital counting | Lack of amplification bias Digital quantification without standard curve |
Input requirements higher than qPCR Generally lower specificity within miRNA families IsomiR sequences difficult to discriminate |
Discovery and development, Validation, Safety assessment/Preclinical studies, Clinical | Counts | 5–6 logs | ++ |
| Microarray | Breadth of miRNA detected | Workflows are labor intensive Input for workflows is high, requires purified RNA Generally lower specificity within miRNA families IsomiR sequences difficult to discriminate |
Discovery and development | Relative fluorescence | 3–4 logs | + |
Abbreviations: miRNA, microRNA; qPCR, quantitative PCR.
Sources: Jensen et al. (2011), Sah et al. (2010), Tan et al. (2015), and Godoy et al. (2019).
++++ = <100 copies, +++ = 100–1000 copies, ++ = 1000–5000 copies, and + = ≥5001 copies.
The low-throughput method of choice for nucleic acid quantitation is qPCR; however, specific challenges are faced with measuring small RNA. These methods require reverse transcription of miRNA to complementary DNA (cDNA) and amplifying the miRNA candidate so that it may be measured typically by fluorometrically labelled hydrolysis probes (e.g., Thermo Fisher Scientific TaqMan) or intercalating dyes (e.g., Qiagen SYBR Green). Given the small size of miRNAs, primers commonly must generate binding specificity from 10 to 15 nucleotides of miRNA sequence. Increased specificity in qPCR can be obtained using primers containing locked nucleic acids (LNA), where the 2′-O is “locked” while the 4′-C atom uses a methylene bridge, which increases thermal stability of the nucleotide and enables high-affinity base pairing to the cDNA strand. Despite this, qPCR may have trouble distinguishing isomeric forms of canonical miRNA, or “isomiRs,” that are present in cells and biofluids. Most isomiRs are 3′ additions to the canonical sequence, most likely reflecting differential nuclease processing from the primary transcript (Wu et al. 2018) and may have functional consequences. Results from the miRNA Quality Control (miRQC) study and others indicated that discrimination varied markedly between measurement platforms and assessments may be problematic when distinguishing isomiRs and closely related miRNA families (Mestdagh et al. 2014; Magee et al. 2017).
Because of low amounts of miRNAs isolated from biofluids, enzymatic pre-amplification of cDNA may be necessary (e.g., Applied Biosystems Megaplex™ Primer pools). Pre-amplification has been reported to enhance the detection of low-expressed species; however, there is evidence that pre-amplification bias of target DNA can occur (Sanders et al. 2011). To potentially measure lower levels of miRNA with greater sensitivity, dPCR is an alternative method that may help alleviate pre-amplification bias and save assessment time. dPCR segregates individual DNA molecules into discrete volumes (such as emulsion droplets) followed by end-point PCR. Hydrolysis probes or intercalating dyes are used to identify count target miRNAs and estimate the original copy number. Despite the advantage of absolute quantitation and higher sensitivity for low copy targets, some limitations exist for miRNA measurements. Given the dynamics of single molecule amplification as well as increased sensitivity to measure off-target and no template amplification, careful optimization of procedures is required for successful and reproducible dPCR measurements (Huggett et al. 2013; Redshaw et al. 2013; Emslie et al. 2019). Importantly, primer design and demonstration of optimal performance (e.g., no off-target amplification, primer efficiency, annealing temperatures) should be documented.
For more medium-throughput applications such as assessing a panel of miRNAs (measuring 10s to 100s of miRNAs in a multiplexed manner), particle or bead-based measurement platforms may be a practical solution. For example, NanoString Technologies employs fluorescent color-coded “molecular barcode” oligonucleotides that hybridize directly to the target molecules. This method does not rely on amplification and therefore offers reduced potential bias for miRNA quantification. As a result, RNA input must be significantly higher than many analogous assays, which may be problematic with limited miRNA amounts derived from biofluids. The Luminex bead-based assays detect miRNAs from purified RNAs and biofluids with a PCR-free workflow. While specific, the assay often lacks sensitivity due to the requirement of a certain number of probe molecules to achieve signal and related binding inefficiencies. Abcam’s FirePlex assay integrates cDNA preparation and amplification into a hydrogel particle-based enrichment workflow and allows higher sensitivity of detection than bead-based platforms (Pregibon et al. 2007). The hydrogel beads can be incubated directly with biofluid and tissue lysates to capture miRNAs present, thereby allowing direct measurement of miRNA without any loss associated with isolation.
For measuring global amounts of miRNAs (1000s at a time), microarray and NGS are commonly used. While currently cost-prohibitive for screening purposes in clinical or regulatory settings, these methods are invaluable assets for biomarker discovery. Microarrays allow comprehensive coverage of known miRNA sequences. However, they cannot be used for absolute quantification and they have a significantly poorer sensitivity and specificity than other methods. For miRNA, the intra-platform reproducibility is typically good; however, the agreement between various microarray technologies is low, especially for the less abundant miRNAs that may be of interest when examining biofluids (Mestdagh et al. 2014). NGS offers several additional advantages compared to microarray profiling, including greater sensitivity and dynamic range. An advantage of NGS relative to other technologies is that 5′ and 3′ sequences are defined, allowing the investigator to distinguish isomeric miRNAs (isomiRs) from miRNAs that can differ by a single nucleotide, including changes related to RNA editing that may influence miRNA stability. Unbiased NGS may also generate a profile of all small RNAs in a sample, including non-coding RNAs, such as short interfering RNA (siRNA), piwi-RNA (piRNA), and repeat-associated siRNA (rasiRNA). Despite the powerful advantages of NGS to profile miRNA, data analysis requires considerable computational and bioinformatics support, and sequencing platform-specific artifacts may occur. As these limitations become minimized over time, sequencing-based quantitation or analogous methods may serve as a complementary follow-up method for more in-depth analysis of complex individuals or those with borderline values.
A number of analyses have been performed to compare different measurement platforms versus common samples to determine relative performance (Giraldez et al., 2018; Mestdagh et al., 2014; Yauk et al., 2010; Git et al., 2010). Common with these studies is the requirement for synthetic miRNA controls and/or abundant samples that can be split and run on each platform. Careful normalization of data between platforms is required before determining whether data generated on each correlate. Firstly, and obviously, it must be determined which miRNAs are efficiently detected on each platform, for instance above a threshold value. Secondly, it is necessary to measure the correlation of fold changes relative to a second sample or an in silico generated benchmark vector from the average expression of common miRNA (Mestdagh et al., 2014; miRQC) in order to measure “fold change” of each miRNA relative to this value. Reports indicate correlations generally between 0.6 and 0.95 depending on the method used (Giraldez et al., 2018; Mestdagh et al., 2014; Yauk et al., 2010; Git et al., 2010), and platforms with similar protocols (e.g., ligation based protocols) anecdotally are more comparable than significantly different methods (e.g., microarray vs RT-qPCR).
From the perspective of biomarker development, one should consider evaluation of markers on multiple platforms a valuable exercise to generate high confidence in the differential abundance detected (Mestdagh et al., 2014). For routine application a single platform should suffice for measurements, providing that sufficient potential signal variables, e.g. isomiRs and cross-reacting sequences, have been well characterized. Reference standards for miRNAs may be purchased from commercial vendors and used for technical controls, albeit consideration must be given limiting degradation from RNases during sample preparation (Shiotsu et al., 2018; Schlosser et al., 2015).
Normalization approaches and recognized limitations
One of the principal limitations for consistently reliable miRNA application is a lack of standardized normalization (Etheridge et al. 2011; Creemers et al. 2012; Meyer et al. 2012). In general, an ideal normalization scheme would result in absolute measures of miRNAs relative to internal reference markers such that data from two different technologies could be compared. Further, measurement of identical samples at different sites would produce identical data, when using sufficiently calibrated equipment. Hence, progress can be made toward high-confidence comparisons between technologies through a combination of technical and computational normalization approaches (see Table 3).
Table 3.
Summary of approaches to normalize miRNA measurement data
| Method (reference) | Compatible platforms | Approach | Strengths | Limitations | Dataset size |
|---|---|---|---|---|---|
| Endogenous RNA | qPCR/dPCR | Design probes to detect endogenous small RNAs within sample | Endogenous probes provide biological normalizer for sample input | Assumes consistent expression of controls | 1–1000 |
| NGS | Normalize to some function of average signal per sample | Some biofluids may not contain control RNAs | |||
| Microarray | Assumes equivalent probe detection to miRNAs | ||||
| Hybridization methods | |||||
|
| |||||
| Exogenous synthetic RNAs | qPCR/dPCR | Design probes to detect endogenous small RNAs within sample | Synthetic RNAs can be added at defined concentrations | Synthetic probe stability may vary in biological samples | 1–1000 |
| NGS | Normalize to some function of average signal per sample | Enables loading controls for multiple assay steps | Chemical modifications can change detection performance | ||
| Microarray | |||||
| Hybridization methods | |||||
|
| |||||
| Target mean normalization (E.g., geNorm, NormFinder, BestKeeper) | qPCR/dPCR | Determine normalization vector by most stable probes between samples | Identifies controls within uncharacterized samples | Technical variation must be removed before stability calculated | 10–1000 |
| NGS | Requires miRNA expression to be stable between samples | ||||
| Microarray | |||||
| Hybridization methods | |||||
|
| |||||
| Sample mean normalization | qPCR/dPCR | Adjust per probe signal by overall sample mean | Effectively removes some sources of technical variation (e.g., sample input) | Requires most detected miRNA to behave similarly. Highly variant small miRNA signatures are harder to normalize | 10–10000 |
| NGS | |||||
| Microarray | |||||
| Hybridization methods | |||||
| Distribution adjustment | qPCR | Adjust per probe signal by overall sample mean (e.g., quantile normalization) | Can adjust for technical variation in amplification efficiency | Requires high number of probes for reliable estimation of distribution | ~≥5000 |
| NGS | |||||
Abbreviations: dPCR, digital-based PCR; miRNA, microRNA; NGS, next generation sequencing; qPCR, quantitative polymerase chain reaction.
Critical evaluation of reference miRNA for normalization is an important task. There are limited idealized “housekeeping” miRNAs currently annotated for specific sample types and the heterogeneity of a sample should be considered. Related miRNA sequences (such as miR family members including miR-17–92 family or let-7 family) should be avoided to reduce the potential for technical crosstalk within platforms from influencing the normalization vector (Mestdagh et al., 2014). In addition, reference miRNAs should be chosen that can be measured to be consistently abundant using multiple platforms, increasing the likely reliability of those measurements.
Quantitative (q)PCR and other targeted approaches are the methods of choice for most application settings (e.g., clinical, preclinical safety assessments) and the recommendation for normalization would be the use of experimentally determined invariant miRNA or other small RNA molecules that are resistant to exposure or physiologic perturbation (Figure 3). Other methods of normalization may increase measurement variability, or worse, lead to misinterpretation of the measurements. Unfortunately, characterizing miRNA expression in biofluids is challenging as no universally invariant calibrator miRNA or any other small RNA molecule has been identified. There are a few invariant miRNAs in solid tissues (Peltier and Reddy 2018) but their consistent expression in various pathological instances has not yet been demonstrated for any biofluid. Endogenous reference RNAs, such as small nuclear (snRNA) and small nucleolar (snoRNAs) or specific miRNAs (Eisenberg and Levanon 2003; Vigneron et al. 2016), have been identified by using mean normalization methods such as geNorm, NormFinder, and Bestkeeper (Vandesompele et al. 2002; Andersen et al. 2004; Pfaffl et al. 2004).The literature indicates that endogenous RNAs are subjected to multiple sources of variation across individuals such as inter-sample heterogeneity and pre-isolations steps (Schwarzenbach et al. 2015). Therefore, putative reference RNAs must be optimized for individual experimental models as well as storage procedures, but these efforts have proven difficult (Vigneron et al. 2016). One commonly referenced solution to the lack of endogenous control is to spike-in exogenous, synthetic miRNA that allow for normalization and estimate the efficiency of miRNA extraction and the reverse transcription step (Mitchell et al. 2008). A concern of the spike-in approach is its reliability for normalization when quantification of extracted RNA is not possible because of RNase activity that is present in the biofluid. To avoid this, addition of known quantities of non-homologous species miRNA spike-ins during the first extraction step or additional spike-ins after extraction. Regardless, spiking-in synthetic miRNAs after collection does not account for biological-based variables (e.g., RNA concentration different due to several biological factors such as diurnal cycles, as noted previously) or urine from variably hydrated subjects (Vickers et al. 2011). As an alternative, established protein and other biomarkers could be used to assess input such as creatinine concentration in urine (Pavkovic et al. 2014; Chen et al. 2017; Ichii et al. 2018), but until this approach has been validated for normalizing urine miRNA values, the rate of urinary miRNA excretion (with timed sample collection) should be used. Population-based studies are further complicating as many factors can influence normalization, including disease state (Levin-Schwartz et al. 2021).
Figure 3.
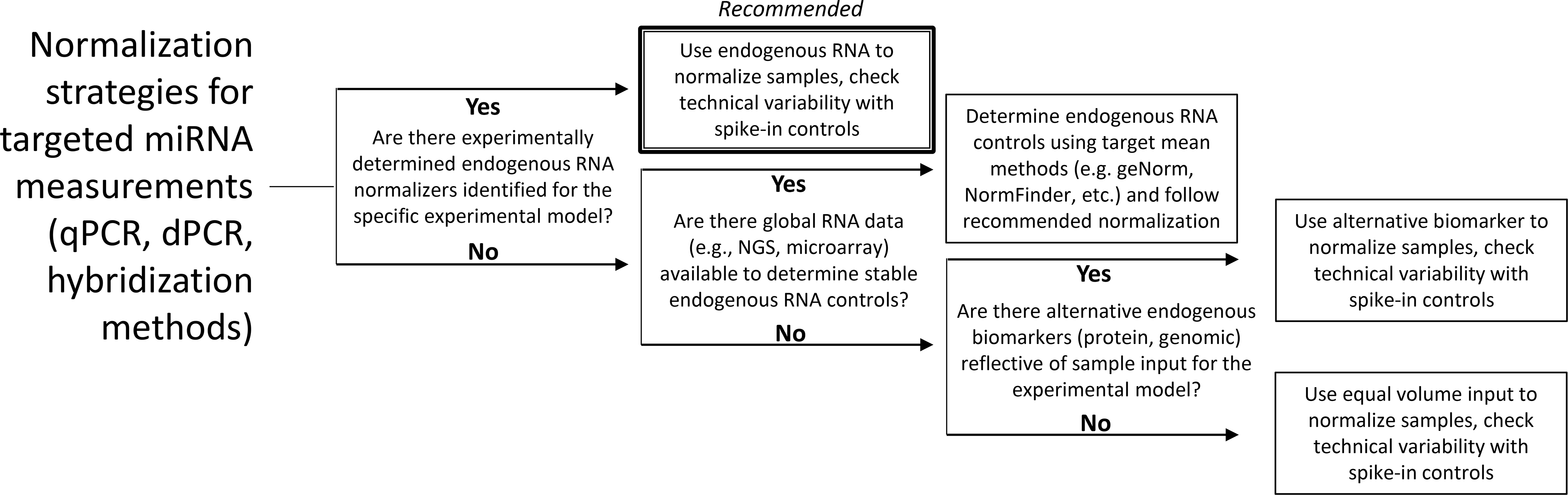
Decision tree for selecting normalization methods based on available information.
For microarray or sequencing data, adjusting measurements to the mean or the distribution of miRNAs in samples underlies many of the methods of normalization. A common normalization method used is Trimmed Mean of M values (TMM) (Anders and Huber 2010). The underlying assumption of this method is that the condition or the treatment changes only a few of the miRNA targets, so most targets should be constitutively expressed. As such, miRNA levels are adjusted from sample to sample according to calculated proportions of mean expression. The method is effective when there is a large set of unperturbed miRNA targets in samples for a study, so that the presence of a few differentially expressed targets between case and control do not appreciably change the average. Most distribution adjustment methods work under a similar assumption. The distribution of target expression levels in a single sample will often have a log-normal, Poisson, or a negative binomial distribution in controls that is then used to fit the identified distribution to case examples. A particularly elegant implementation of the method is quantile normalization (Amaratunga and Cabrera 2001), which makes no assumptions about the actual distribution in the reference but instead directly forces a match between the quantiles of each sample to the quantiles of the reference sample. Unlike the mean-adjusting methods, quantile normalization can adjust different targets in the same sample by different amounts. Unfortunately, the assumption underlying these methods likely does not apply for miRNAs present in biofluids, where the absolute quantity and variety of measurable miRNAs is more limited and overall more likely influenced by biological conditions than in tissues (e.g., the assumption that most miRNAs are invariant from sample to sample is likely not true). Alternative strategies must be investigated to standardize normalization efforts, which depends on the method of measurement. Indeed, a combination of absolute quantitation with standard curves of synthetic miRNAs and internal biological controls is necessary for direct comparison of samples between platforms and between analysis sites (Thompson et al. 2016).
INTERPRETING THE BIOLOGICAL MEANING OF MIRNA ALTERATIONS
A major challenge in biofluid-based miRNA studies is interpreting the biology behind the changes that are measured. If there is high confidence in the measurements of miRNAs in a biofluid, there still exists “variability” in understanding of the underlying pathology or toxicity of interest that influences these measurements. Without this information, the mechanistic-based information afforded by miRNA biomarkers is not realized. It is key to understand why these values may change in a matrix where contributions are derived from many sources. Some miRNAs are tissue or cellular specific and may serve as an injury or disease biomarker for that target tissue. Other miRNAs may be specific to a process or cellular pathway and could serve as a biomarker of a biological response. Both aspects that influence the presence of miRNAs in biofluids must both be understood to garner full utility as biomarkers.
Tissue specificity of miRNA expression
Early evidence established that some miRNAs expressed in adults are tissue-specific, even more as compared to mRNA expression (Babak et al. 2004). Other miRNAs are not tissue-specific but display restricted expression for specific organs (Ludwig et al. 2016). Families of miRNAs can also display a more restricted expression pattern. For example, the miR-378 family is preferentially expressed in muscle tissues and the myocardium, with very low or no expression in other tissues. Similarly, members of the miR-506 family are preferentially expressed in testis (Ludwig et al. 2016). This expression pattern is highly associated with the involvement of miRNAs in normal physiological functions, during disease development, and in response to injuries (Akamatsu et al. 2015). Thus, the identification of the tissue of origin of miRNA is critical in order to identify the source of perturbation when these miRNAs are identified in blood or any other body fluids (Xu et al. 2011; Krauskopf et al. 2015; Pirola et al. 2015).
An example of a well-characterized, blood-based, single miRNA biomarker candidate to detect liver injury because of its organ-specific expression is miR-122. Landgraf et al. published a survey of small RNA sequencing data from mammalian tissues that indicated miR-122 to be expressed almost exclusively in the liver of mammals (Landgraf et al. 2007; Koenig et al. 2016). It was shown to be present at approximately 66,000 copies per adult mouse liver cell, which comprises nearly 70% of the entire mature miRNA pool in the liver (Lagos-Quintana et al. 2002; Chang et al. 2004). Qualitatively, liver injury leads to an increase in detectable miR-122 in circulating fluids (i.e., plasma and serum), most often through apoptosis/necrosis of liver cells, the majority of which are hepatocytes. Changes in circulating miR-122 levels have been shown in different physiological processes in hepatic function as well as a variety of liver pathologies (Trebicka et al. 2013; Clarke et al. 2014; Park et al. 2016; Howell et al. 2018). Up to a 6000-fold increase in plasma miR-122 levels has been observed in rats treated with the hepatotoxicant, chlorobromomethane (Laterza et al. 2009). In the clinic, multiple studies demonstrated that circulating miR-122 becomes elevated prior to the increase of the traditional clinical chemistry biomarker for liver injury, alanine transaminase (ALT), and has a greater sensitivity in predicting acetaminophen liver toxicity than serum aminotransferases (Wang et al. 2009; Antoine et al. 2013; Dear et al. 2014; Thulin et al. 2014; Ward et al. 2014). It was also shown to be more accurate in detecting drug-induced liver injury (DILI) upon first presentation at the hospital in patients with normal ALT levels (Antoine et al. 2013; Dear et al. 2014). Combining high levels of expression, tissue specificity, homology between species, and release into biofluids, miR-122 has been proposed as a novel, non-invasive highly specific translatable biomarker of liver homeostasis including liver injury.
Given this scientific enthusiasm, one may question why miR-122 has not been adopted as a replacement biomarker for liver toxicity and disease. The utility of a new biomarker can also be assessed through estimation of the additional value it brings when quantified concomitantly with existing biomarkers. It has been demonstrated in pre-clinical studies that the addition of miR-122 to the traditional clinical chemistry panel (ALT, aspartate aminotransferase [AST], and glutamate dehydrogenase [GLDH]) improves the diagnostic accuracy by 4%, although miR-122 alone outperformed ALT as a minimally-invasive biomarker and was equivalent in performance to AST and GLDH relative to microscopic evidence of the liver histopathology findings (Sharapova et al. 2016). A systematic review of the miR-122 as a biomarker of DILI showed that despite a moderate heterogeneity between datasets, there was high sensitivity and specificity overall for miR-122 as a biomarker (0.85 and 0.93, respectively) but only a modest ability to detect DILI (Liu et al. 2018). Although the addition of miR-122 to the biomarker panel was shown to be beneficial in both nonclinical and clinical settings, the added cost to monitor for changes in miR-122 in may not provide an overall benefit given the modest improvements in predictivity and assessment of DILI. Therefore, economics is a major factor in the development of miR-122 as a biofluid-based liver safety biomarker.
Broader insight with miRNA panels
While many studies have focused on single, tissue-specific miRNA to infer physiologic changes, a panel of miRNA biomarkers that includes both tissue-specific and tissue-enriched miRNAs can overcome some limitations of single miRNA biomarkers. Indeed, NGS has expanded the scope of discovery of candidate miRNA biomarkers in biofluids. Further refinement of these panels may be gleaned from histopathology. For example, techniques such as in situ hybridization of miRNAs, laser capture microdissection-guided PCR, and single-cell transcriptomics can help prioritize biofluid-based miRNAs for biomarkers of tissue-specific damage and potentially severity of injury and disease processes.
One example of where a panel of miRNA biomarkers may inform underlying tissue pathology is urinary miRNA biomarkers of kidney injury. Select miRNAs are characteristically expressed in the kidney and involved in many renal physiological and pathological processes (Bhatt et al. 2011). Urinary miRNAs are relatively stable (Mall et al. 2013) and have been shown to be differentially expressed in nephron segments in kidney (Kriegel et al. 2013). Additionally, given the highly conserved nature of miRNA sequences across species (Friedman et al. 2009) and the overlap in kidney miRNA expression between laboratory animal models and humans (Pavkovic and Vaidya 2016), there is potential translational context of use for miRNAs as kidney-specific injury markers when monitoring for acute, progressive and/or reversible kidney injury.
A cross-laboratory program was conducted by the Health and Environmental Sciences Institute (HESI) Biomarkers of Nephrotoxicity Committee to identify and characterize miRNA patterns (i.e., changes in individual miRNAs and/or a panel of differentially expressed miRNAs) indictive of kidney injury in rats (Chorley et al., 2020). The overall strategy of this program is depicted in Figure 4. The program combined miRNA profiling in urine from rats treated with nephron segment-selective toxicants and experiments on nephron segments isolated by laser capture microdissection to further characterize the localization of miRNA candidates directly in the kidney. Results from the individual rat studies showed that several urinary miRNAs were increased with doses of compounds resulting in minimal to moderate degeneration/necrosis in the expected nephron segments; although the traditional clinical pathology parameters serum creatinine and blood urea nitrogen (BUN) were either not or only slightly changed. Interestingly, in the rat study with the glomerular toxicant doxorubicin, urinary miR-34c-3p was found to be increased earlier than albuminuria (Church et al. 2014). It was demonstrated that several miRNAs changed early in glomerular and urine samples in two different rat models of glomerular injury (Nassirpour et al. 2015). In a rat study with the proximal tubular toxicant cisplatin, other urinary miRNAs were increased earlier than BUN and serum creatinine and were comparable to the more specific and sensitive kidney injury biomarker, KIM-1, with respect to timing (Pavkovic et al. 2014). Similarly, miR-210–3p was reported in another study as displaying the most robust changes in urine from rats treated with collecting duct toxicant, N-phenylanthranylic acid (NPAA) (Glineur et al. 2018). In contrast, another set miRNAs were found to be changed in abundance in urine from rats treated with nephrotoxicants affecting different nephron segments, thus suggesting that these miRNAs are expressed generally in the kidney across all locations and lack diagnostic capacity for site-specificity of injury. This was especially the case for mechanistic miRNAs like miR-34c-5p and miR-155–5p, which were increased in urine as a consequence of their biological function, regardless of the localization of the kidney lesions (Glineur et al. 2018). Taken together, these results suggest that a panel of urinary miRNAs with preferential localization in the glomerulus and the proximal and distal tubules may be useful to discriminate the localization of drug-induced lesions in the kidney.
Figure 4.
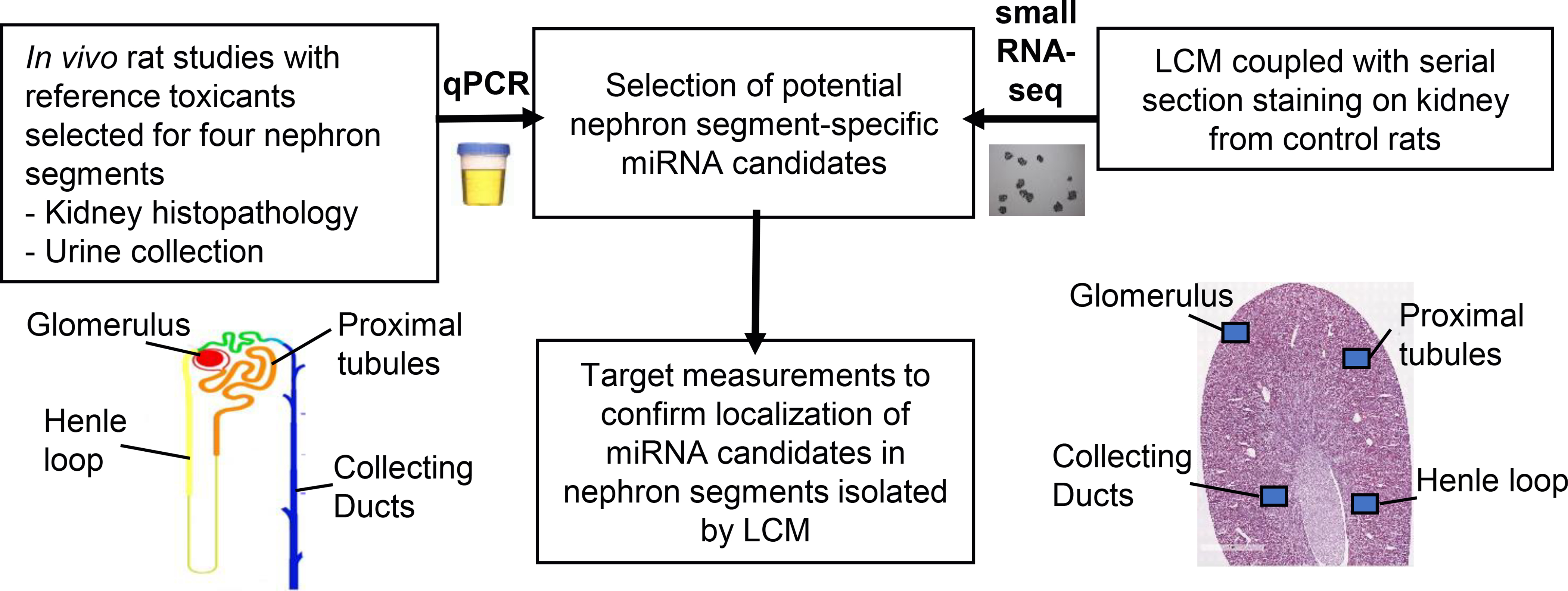
Strategy to identify nephron segment-specific urinary microRNAs (miRNAs) within a cross-laboratory research program conducted by the Health and Environmental Sciences Institute Biomarkers of Nephrotoxicity Committee. Rats were treated with nephron segment selective toxicants causing injury to glomeruli, proximal tubules, Henle’s loops, and collecting ducts. Urinary miRNAs were profiled by quantitative polymerase chain reaction (qPCR). A meta-analysis was then performed to identify significantly modulated urinary miRNAs that were common and/or specific across studies. Given that some compounds produced a mixture of lesions in several nephron segments, small RNA sequencing and other confirmatory experiments were conducted in nephron segments isolated by laser capture microdissection (LCM) to further characterize the localization of miRNA candidates.
Interestingly, liver and cardiac injury outside of the kidney may also be assessed in urine (Cheng et al. 2012; Yang et al. 2012; Zhou et al. 2013; Wolenski et al. 2017). In the kidney, it is plausible that glomerular damage may facilitate the passage of plasma miRNAs through the glomerulus into urine and thus may complicate the interpretation of results. However, it remains to be confirmed that plasma miRNAs can be excreted into the urine and by which mechanism(s) if any in the absence of kidney damage. Conversely, it was reported that certain miRNA levels were altered in plasma in correlation with kidney injury (Lorenzen et al. 2011; Gutierrez-Escolano et al. 2015; Zhang et al. 2017). The origin of these plasma miRNAs (extra-renal versus renal) is not known. Additional studies are needed to identify pathophysiological mechanisms associated with these plasma miRNA changes.
There remain limitations, such as in being able to induce comparable severity of injury in only one specific location of the kidney at a time. Thus, more systematic meta-analyses across studies reporting development of a panel of miRNA changes in the urine in both animal models and human patients with kidney injury, differentiating between acute or chronic settings, are still needed. These evaluations may be followed by specifically designed measurements and exploration of several analysis approaches. Such investigations may require collaborations between many stakeholders yet may then be expected to yield miRNA panels in reasonable time for focused kidney injury settings.
Interpreting non-canonical sequence
Another advantage of sequencing is the ability to detect modifications of the canonical sequence of miRNAs. As discussed previously, some of these detected sequence changes are due to technical sequencing errors; however, some have been consistently identified and validated by other methods (Morin et al. 2008; Neilsen et al. 2012; Gomes et al. 2013). Biologically, the existence of directed modification, such as adenylation or uridylation at the 3′ or some microRNAs, suggests that this region may be a focal point of regulation (Rissland et al. 2007; Menezes et al. 2018). In addition, isomiRs may be a result of differential cleavage of precursor miRNAs (Seitz et al. 2008; Hu et al. 2009). The endogenous biological functions of most isomiRs has proven elusive to date despite some evidence of regulatory roles (Cloonan et al. 2011). Most isomiR variants occur at the 3′ of the miRNA sequences, with templated and non-templated additions common (Wang et al. 2016; Wu et al. 2018). 5′ variants are predicted to change the miRNA seed and therefore may have significant influence on miRNA specificity (e.g., mammalian miR-142–3p, miR-101–5p, or mmu-miR-223–3p), whereas some 3′ variants may modulate miRNA stability or subcellular location (Telakivi and Flink 1985). Not surprisingly, 5′ isomiR variants are significantly less common than 3′ variants in most sequencing datasets (Wang et al. 2016) if it is speculated that these seed alterations may have greater biological impact.
The association of isomiRs with certain biological or disease states is intriguing biologically and has implications for biofluid-based biomarker development. IsomiR species appear to be more abundant in tumor cells, indicating a potential role for dysregulation of miRNA processing (Wang et al. 2016). These examples indicate that isomiR function and presence in biofluids must be considered and may prove to be a better indicator of adverse pathology than canonical sequence alone. Like advantages of assessing miRNA panels, the distinction of isomeric miRNAs may be critical.
One example of this phenomenon is that the source of variability in the baseline levels of circulating miR-122 in controls may result from the presence of various isomiRs. Multiple isoforms of miR-122 have been detected in the liver, and their distribution between different conditions, types of injury, as well as animal strains may vary. Using NGS technology, it has been shown that in the plasma of subjects with acetaminophen toxicity, only 20% of all detected mature miR-122–5p represent a canonical sequence referenced in the miRBase; the majority of miR-122–5p was present as trimmed isomiRs (Krauskopf et al. 2017). Primer design for qPCR based on sequence data in miRBase may not detect all isoforms present, thus affecting the accuracy of miRNA results in given sample. Applying NGS methods for miRNA analysis may provide more accurate quantification of miR-122 as well as aid in getting useful information on isomiR distribution, thereby increasing the utility of this putative liver biomarker.
miRNAs as biomarkers of active cell signalling versus passive leakage
Like ALT, the release of miR-122 into the circulation was initially attributed to passive leakage of miRNA-protein complexes from injured hepatocytes (Arroyo et al. 2011; Hornby et al. 2014). However, multiple studies have confirmed that miR-122 is also actively released by hepatocytes as cargo of EVs (specifically exosomes) (Cho et al. 2018). Moreover, its enrichment and half-life in each fraction varies greatly based on liver condition or mechanism of injury. In vitro, miR-122 was detected in hepatocyte-derived exosomes at doses of acetaminophen in the absence of the overt hepatotoxicity, possibly serving as an early damage associated signalling molecule (Holman et al. 2016; Mosedale et al. 2018). In animal studies, circulating miR-122 was localized to protein-rich and exosomal fractions in a time- and injury-specific manner (Bala et al. 2012; Motawi et al. 2018). In animals with acetaminophen-induced toxicity, the early (3–12 hours after dosing) release of miR-122 was associated with exosomes. During late release, which was accompanied by massive hepatocyte necrosis, miR-122 was primarily detected in the protein-rich fraction of serum and not exosomes (Bala et al. 2012; Holman et al. 2016). In rodents with alcohol-induced liver injury, miR-122 was highly enriched in the exosomal fraction while just a minimal increase was observed in protein-bound fraction. This same study showed that in an inflammatory liver disease model, miR-122 was detected almost exclusively in exosomes and decreased in protein fraction compared to the controls (Bala et al. 2012; Yang et al. 2014). Another study reported that exosomal miR-122 derived from ethanol-treated hepatocytes, when up-taken by monocytes and Kupffer cells, can modulate liver inflammatory response, positioning miR-122 as an important signalling molecule in cell-cell interaction (Momen-Heravi et al. 2015). The signalling role of miR-122 is supported by the evidence that exosomal miR-122 can also be taken up by tissues outside of the liver, including kidneys and muscle (Rivkin et al. 2016; Chai et al. 2017). Finally, decreased exosomal levels of miR-122 were associated with hepatocellular carcinoma, which agrees with reported downregulation of miR-122 in liver cancer (Gramantieri et al. 2007; Sohn et al. 2015). Therefore, serum-based miRNAs do not always reflect the presence or extent of organ damage and may be regarded as an extracellular stress signal. The reported findings on longitudinal and spatial miR-122 distribution in the circulation may account for the miR-122 variability in healthy individuals observed in studies of liver injury and make the interpretation of miR-122 results more complex but also enticing as a putative biomarker beyond overt organ injury.
MOVING FORWARD AND MEETING THE CHALLENGES
Shortly after it was discovered that miRNAs were present in biofluids, qualities of ideal biomarkers were noted (Etheridge et al. 2011) such as specificity for tissue of origin, sensitivity to distinguish adverse from transient cellular responses, patterns of expression predictive for an apical pathological state, robust response and stability in biofluids, and orthology across human and non-clinical species. These promising molecular-based biomarkers may not only enhance the utility but also fill gaps that are present with the use of existing biomarkers. Despite this promise, both technical and biological challenges add variability to the measurements and uncertainty to the interpretation of miRNA biomarkers in biofluids, and the full application for safety, clinical, and clinical use has yet to be realized. Several challenges have been listed within this summary which may seem overwhelming, but a holistic view of existing research can serve as a guide for what areas need most attention (Table 4). In general, issues associated with technical variables (such as sampling, storage, extraction, and quantitation) can be minimized with careful planning, standard operating procedures, and experience, whereas the interpretation of these measurements is less well defined and will require focused research to better connect to the biological phenotype or outcome that the miRNAs serve as biomarkers.
Table 4.
Areas of highest research need for biofluid-based miRNA biomarker development
| Type of variable | Area of focus | Degree of impacta | Current efforts to address need | References |
|---|---|---|---|---|
| Technical | Determine pre-analytical stability of miRNA biomarker | + | Individual laboratory research | Glinge et al. (2017), Enelund et al. (2017), Mitchell et al. (2008), Grasedieck et al. (2012) |
| Technical | Establish Standard Operating Procedures for sample collection and measurements | +++ | Individual laboratory research, commercial, EDRN | Exiqon (http://www.exiqon.com/ls/Documents/Scientific/microRNA-serum-plasma-guidelines.pdf), Farina et al. (2014), National Cancer Institute EDRN (https://prevention.cancer.gov/major-programs/early-detection-research) |
| Technical | Establish a framework for reporting of data | ++ | Individual laboratory research, commercial | Gant et al. (2017) |
| Technical/ biological | Determine mechanism of action of EV-derived miRNAs | ++ | Individual laboratory research, commercial, ERCC | Das et al. (2019), Liu et al. (2019) |
| Technical/ biological | isomiR identification and linkage to outcomes | ++ | Individual laboratory research, IsomiR Bank, miR-isomiRExp | Zhang et al. (2016), Guo et al. (2016) |
| Biological | Identify tissue/cellular source of miRNA release | ++ | Individual laboratories research, atlas efforts | Landgraf et al. (2007), Smith et al. (2016), de Rie et al. (2017), Koenig et al. (2016) |
Abbreviations: EDRN, Early Detection Research Network; ERCC, Extracellular RNA Communication Consortium; EV, extracellular vesicle; miRNA, microRNA.
+ = isolated, ++ = widespread, +++ = foundational.
As highlighted in the previous HESI effort, biofluid-based evaluation of a robust miRNA response to a toxicant using a defined method can be replicated across different laboratories and sites (Thompson et al. 2016). After removing measurement data from sites that had clear outlying results, variability in the measurements was primarily due to the biofluid source (in the study, urine versus plasma) and inherent biological range in the miRNA biomarker response not due to technical bias. Standardized methods for sample processing and qPCR-based detection significantly minimized technical variability and were selected for the specific purpose of robust, consistent measurements and uniform available equipment. This work and many others tested the consistency and robustness of miRNA measurements (McDonald et al. 2011; Mestdagh et al. 2014; Bailey et al. 2019; López-Longarela et al. 2019) and underscores the importance of controlling uncertainty by minimizing technical factors through consistent collection, processing, storage/transport, isolation, and measurement protocols with experienced staff and technicians. It is also important to keep in mind that these procedures are fit-for-purpose, and certain caveats exist for specific miRNAs measured. For example, some miRNAs may not be as stable during collection and storage conditions, which may influence results if measurements cannot be immediately performed after collection (Glinge et al. 2017). This is particularly important in clinical and safety settings if an adverse value is determined by a defined normal range. Therefore, a general standard operating procedure is not defined as part of this review, but rather is encouraged to be developed for specific contexts of use with respect to the caveats of costs and availability of technology, stability of the miRNA biomarkers measured, experimentally determined normalizers, and other factors appropriate for the model and perturbation being assessed. As one example for a specific application, namely targeted measurement of certain miRNAs, we do propose a workflow for normalization methods (Figure 3). Future research investment should be made for promising candidate miRNA biomarkers to optimize technical procedures to reduce any potential bias before qualification and/or routine use in diagnostic, prognostic, and predictive situations. Additionally, standard and minimal reporting procedures for measurements made for a published study should be developed, similar to current efforts for general gene expression studies such as the Transcriptomics Reporting Framework (TRF) (Gant et al., 2017). Setting such reporting standards would increase replicability of published data, reduce technical variability associated with different measurement approaches, and ultimately increase the confidence in biomarker results and use.
Importantly in the HESI study, result variability was introduced by the biofluid that was being assessed due to the tissue origin of the miRNAs. There have been multiple efforts to determine tissue and cellular specificity of miRNAs (Landgraf et al. 2007; Koenig et al. 2016; Smith et al. 2016; de Rie et al. 2017) that could serve as toxicity or “release” biomarkers in human and other non-clinical species which present obvious roads for development. However, sub-toxic, predictive/prognostic markers of disease or therapeutic response require more in-depth knowledge of the source of cellular response and release but also hold the most promising use of mechanism-based miRNA biomarkers. One strategy to improve the signal to noise ratio uses fractionation to isolate a subset of miRNAs, such as from EVs (apoptotic bodies, microparticles, microvesicles, and/or exosomes) (Duttagupta et al. 2011; Turchinovich et al. 2011). Currently, the best examples of the predictive power of EV-derived miRNA scans assessed the presence of cancer and response to therapy (Hannafon et al. 2016; Qin et al. 2019; Rodriguez-Martinez et al. 2019); however, one can envision the use of these paracrine signals as a way to measure cellular response prior to adverse health outcomes. Even with this enrichment, signal may still be an issue. Several reports indicate that a small fraction of circulating EVs contain miRNAs, whereas other reports imply that selection of RNA containing EVs may improve yield (Chen et al. 2014; Chevillet et al. 2014). Specific EV enrichment may be achieved using markers correlated with tumor cells or specific miRNA signatures, such as CD63, epithelial cell adhesion molecule (EpCAM), and prostate-specific membrane antigen (PSMA) in the case of prostate tumor-derived exosomes (Meng et al. 2016; Fang et al. 2017) or TSG101 in urine exosomes (Koritzinsky et al. 2019). Investment in large-scale efforts, such as the Extracellular RNA Communication Consortium (ERCC), which collate existing research, provide protocols for isolation, house databases for analysis, and tools for discovery and validation is key to our better understanding of the role of RNA, including miRNA, in cell-to-cell communication (Das et al. 2019). Such information will guide the development of specific panels of biofluid-based miRNA biomarkers for specific prognostic and diagnostic uses.
There are additional challenges for developing clinical biomarkers. As post-transcriptional regulators, miRNAs can affect multiple genes that represent an orchestrated response, but they are not “master controllers.” With multiple potential sources of any given miRNA, traditional downstream bioinformatics approaches to infer biologically meaningful information (e.g., Ingenuity Pathway Analysis, miRPathDB, etc.) may be obscured, diluted and/or misinterpreted due to mixed signals. Furthermore, any given miRNA is working in concert with other miRNAs in a given response. This distributed control makes it unlikely that single miRNAs would be adequate biomarkers and complicates the path toward biomarker qualification for these purposes. Normal human subjects tend to be homogeneous, both in biomarker levels and demographics/characteristics, whereas patients with injury or disease exhibit wide ranges of biomarker values with a large array of confounding factors that make translation to the clinic nearly impossible without large cohorts. Several clinical studies demonstrate that multiple miRNAs change during disease progression such as obesity, diabetes (Mori et al. 2019), sepsis (Rahmel et al. 2018), or cancer (Zorofchian et al. 2019), and it remains to be seen whether these can be leveraged to provide a template for tissue injury by toxicants or can be used to reduce the variability of disease-specific miRNA signals in clinical cohorts that contain subjects with subclinical disease. Fortunately, blueprints for success exist, such as the Predictive Safety Testing Consortium (PSTC) (Stephenson and Sauer 2014), where several protein biomarkers have been FDA qualified to serve as biomarkers for nephrotoxicity. Key features that can be applied to miRNAs are as follows: a) a large consortium of investigators who each have access to limited technical/analytical resources and samples, but when pooled become adequate to approach a common problem; b) multiple platforms to perform unbiased searches of potential biomarkers; and c) a critical mass of stakeholders whose consensus can influence and guide the field toward common standards, from sample preparation to measurement to analysis and interpretation of data. When combined with a biorepository of longitudinal or cross-sectional natural history studies such as the African American Study of Kidney Disease and Hypertension (AASK) (Almaani et al. 2017) or the Chronic Renal Insufficiency Cohort (CRIC) (Denker et al. 2015), the larger cohorts can provide the needed statistical power to develop panels of biomarkers. Additionally, miRNA biomarker panels can be assembled by comparing their performance, then combining the top performers, but these panels should not be limited to miRNAs. Other classes of biomarkers, such as proteins, other nucleic acids, or lipids, could provide complementary information that would synergize with the top miRNAs.
Reducing the uncertainty associated with these challenges in biomarker development for miRNAs will require systematic focus and large-scale investment. The challenges are well defined and the added benefits of these mechanistic-based biomarkers for pre-clinical, clinical, regulatory, and scientific uses are a promise that can still be fulfilled.
ACKNOWLEDGMENTS
The authors sincerely thank Ms. Carolina Morell-Perez of HESI for her kind assistance with preparation and external peer review of the manuscript prior to submission. The authors acknowledge Drs. Sheau-Fung Thai, Weichun Huang, Katie O’Shaughnessy, and Maureen Gwinn of the US EPA and Liwen Liu and Kevin Gerrish of the NIEHS and Alison Sanders of the Icahn School of Medicine at Mount Sinai for constructive comments on this manuscript.
FUNDING
This work was supported, in part, by internal funding from the Intramural Research Program of the National Institutes of Health; the National Institute of Diabetes and Digestive and Kidney Diseases [grant Z01 DK043400] and the National Institute of Environmental Health Sciences, the U.S. Environmental Protection Agency Office of Research and Development, and the US Army Medical Research and Development Command, Military Operational Medicine Research Program.
Footnotes
DISCLAIMER
The research described in this article has been reviewed by the U.S. Environmental Protection Agency and approved for publication. Approval does not signify that the contents necessarily reflect the views and the policies of the Agency. This material has been reviewed by the National Institute of Environmental Health Sciences and the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense, the National Institutes of Health, or the US Government. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
REFERENCES
- Aarem J, Brunborg G, Aas KK, Harbak K, Taipale MM, Magnus P, Knudsen GP, Duale N. 2016. Comparison of blood RNA isolation methods from samples stabilized in Tempus tubes and stored at a large human biobank. BMC Res Notes. 9:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu S, Hayes CN, Tsuge M, Miki D, Akiyama R, Abe H, Ochi H, Hiraga N, Imamura M, Takahashi S, et al. 2015. Differences in serum microRNA profiles in hepatitis B and C virus infection. J Infect. 70:273–287. [DOI] [PubMed] [Google Scholar]
- Al-Soud WA, Radstrom P. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 39:485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaani S, Hebert LA, Rovin BH, Birmingham DJ. 2017. The urine preservative acetic acid degrades urine protein: implications for urine biorepositories and the AASK cohort study. J Am Soc Nephrol. 28:1394–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaratunga D, Cabrera J. 2001. Analysis of data from viral DNA microchips. J Am Stat Assoc. 96:1161–1170. [Google Scholar]
- Amur SG, Sanyal S, Chakravarty AG, Noone MH, Kaiser J, McCune S, Buckman-Garner SY. 2015. Building a roadmap to biomarker qualification: challenges and opportunities. Biomark Med. 9:1095–1105. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Orntoft TF. 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64:5245–5250. [DOI] [PubMed] [Google Scholar]
- Antoine DJ, Dear JW, Lewis PS, Platt V, Coyle J, Masson M, Thanacoody RH, Gray AJ, Webb DJ, Moggs JG, et al. 2013. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 58:777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. 2011. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 108:5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. 2004. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 10:1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey WJ, Barnum JE, Erdos Z, LaFranco-Scheuch L, Lane P, Vlasakova K, Sistare FD, Glaab WE. 2019. A performance evaluation of liver and skeletal muscle-specific miRNAs in rat plasma to detect drug-induced injury. Toxicol Sci. 168:110–125. [DOI] [PubMed] [Google Scholar]
- Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. 2012. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced and inflammatory liver diseases. Hepatology. 56:1946–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartram A, Poon C, Neufeld J. 2009. Nucleic acid contamination of glycogen used in nucleic acid precipitation and assessment of linear polyacrylamide as an alternative co-precipitant. Biotechniques. 47:1019–1022. [DOI] [PubMed] [Google Scholar]
- Bhatt K, Mi QS, Dong Z. 2011. microRNAs in kidneys: biogenesis, regulation, and pathophysiological roles. Am J Physiol Renal Physiol. 300:F602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, Dahlsveen IK. (2013).Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods, 59, S1–6 [DOI] [PubMed] [Google Scholar]
- Brown RAM, Epis MR, Horsham JL, Kabir TD, Richardson KL, Leedman PJ. 2018. Total RNA extraction from tissues for microRNA and target gene expression analysis: not all kits are created equal. BMC Biotechnol. 18:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet-Vega A, Pericay C, Quilez ME, Ramirez-Lazaro MJ, Calvet X, Lario S. 2015. Variability in microRNA recovery from plasma: comparison of five commercial kits. Anal Biochem. 488:28–35. [DOI] [PubMed] [Google Scholar]
- Bushel PR, Caiment F, Wu H, O’Lone R, Day F, Calley J, Smith A, Li J. 2018. RATEmiRs: the rat atlas of tissue-specific and enriched miRNAs database. BMC Genomics. 19:825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai C, Rivkin M, Berkovits L, Simerzin A, Zorde-Khvalevsky E, Rosenberg N, Klein S, Yaish D, Durst R, Shpitzen S, et al. 2017. Metabolic circuit involving free fatty acids, microRNA 122, and triglyceride synthesis in liver and muscle tissues. Gastroenterology. 153:1404–1415. [DOI] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 1:106–113. [DOI] [PubMed] [Google Scholar]
- Chen C, Lu C, Qian Y, Li H, Tan Y, Cai L, Weng H. 2017. Urinary miR-21 as a potential biomarker of hypertensive kidney injury and fibrosis. Sci Rep. 7:17737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WX, Zhong SL, Ji MH, Pan M, Hu Q, Lv MM, Luo Z, Zhao JH, Tang JH. 2014. MicroRNAs delivered by extracellular vesicles: an emerging resistance mechanism for breast cancer. Tumour Biol. 35:2883–2892. [DOI] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. 2008. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18:997–1006. [DOI] [PubMed] [Google Scholar]
- Cheng HH, Yi HS, Kim Y, Kroh EM, Chien JW, Eaton KD, Goodman MT, Tait JF, Tewari M, Pritchard CC. 2013. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS One. 8:e64795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Wang X, Yang J, Duan X, Yao Y, Shi X, Chen Z, Fan Z, Liu X, Qin S, et al. 2012. A translational study of urine miRNAs in acute myocardial infarction. J Mol Cell Cardiol. 53:668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, et al. 2014. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 111:14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y-E, Song B-J, Akbar M, Baek M-C. 2018. Extracellular vesicles as potential biomarkers for alcohol- and drug-induced liver injury and their therapeutic applications. Pharmacol Ther. 187:180–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorley B, Ellinger-Ziegelbauer H, Tackett M, Simutis FJ, Harrill AH, McDuffie J, Atabakhsh E, Nassirpour R, Whiteley LO, Leonard JF, Carswell GK, Harpur E, Chen CL, Gautier JC. (2020).Urinary miRNA biomarkers of drug-induced kidney injury and their site specificity within the nephron. Toxicological sciences : an official journal of the Society of Toxicology [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church RJ, McDuffie JE, Sonee M, Otieno M, Ma JY, Liu X, Watkins PB, Harrill AH. 2014. MicroRNA-34c-3p is an early predictive biomarker for doxorubicin-induced glomerular injury progression in male Sprague-Dawley rats. Toxicol Res. 3:384–394. [Google Scholar]
- Clarke JD, Sharapova T, Lake AD, Blomme E, Maher J, Cherrington NJ. 2014. Circulating microRNA 122 in the methionine and choline deficient mouse model of nonalcoholic steatohepatitis. J Appl Toxicol. 34:726–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Wani S, Xu Q, Gu J, Lea K, Heater S, Barbacioru C, Steptoe AL, Martin HC, Nourbakhsh E, et al. 2011. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 12:R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers EE, Tijsen AJ, Pinto YM. 2012. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 110:483–495. [DOI] [PubMed] [Google Scholar]
- Cui C, Yang W, Shi J, Zhou Y, Yang J, Cui Q, Zhou Y. (2018).Identification and Analysis of Human Sex-biased MicroRNAs. Genomics Proteomics Bioinformatics, 16, 200–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Ansel KM, Bitzer M, Breakefield XO, Charest A, Galas DJ, Gerstein MB, Gupta M, Milosavljevic A, McManus MT, et al. 2019. The Extracellular RNA Communication Consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell. 177:231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rie D, Abugessaisa I, Alam T, Arner E, Arner P, Ashoor H, Astrom G, Babina M, Bertin N, Burroughs AM, et al. 2017. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat Biotechnol. 35:872–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear JW, Antoine DJ, Starkey-Lewis P, Goldring CE, Park BK. 2014. Early detection of paracetamol toxicity using circulating liver microRNA and markers of cell necrosis. Br J Clin Pharmacol. 77:904–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker M, Boyle S, Anderson AH, Appel LJ, Chen J, Fink JC, Flack J, Go AS, Horwitz E, Hsu CY, et al. 2015. Chronic Renal Insufficiency Cohort Study (CRIC): overview and summary of selected findings. Clin J Am Soc Nephrol. 10:2073–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detassis S, Grasso M, Tabraue-Chavez M, Marin-Romero A, Lopez-Longarela B, Ilyine H, Ress C, Ceriani S, Erspan M, Maglione A, et al. 2019. New platform for the direct profiling of microRNAs in biofluids. Anal Chem. 91:5874–5880. [DOI] [PubMed] [Google Scholar]
- Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. 2011. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS One. 6:e20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. 2003. Human housekeeping genes are compact. Trends Genet. 19:362–365. [DOI] [PubMed] [Google Scholar]
- Emslie KR, JL HM, Griffiths K, Forbes-Smith M, Pinheiro LB, Burke DG. 2019. Droplet volume variability and impact on digital PCR copy number concentration measurements. Anal Chem. 91:4124–4131. [DOI] [PubMed] [Google Scholar]
- Enelund L, Nielsen LN, Cirera S. 2017. Evaluation of microRNA stability in plasma and serum from healthy dogs. Microrna. 6:42–52. [DOI] [PubMed] [Google Scholar]
- Etheridge A, Lee I, Hood L, Galas D, Wang K. 2011. Extracellular microRNA: a new source of biomarkers. Mutat Res. 717:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Tian H, Li X, Jin D, Li X, Kong J, Yang C, Yang X, Lu Y, Luo Y, et al. 2017. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS One. 12:e0175050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina NH, Wood ME, Perrapato SD, Francklyn CS, Stein GS, Stein JL, Lian JB. 2014. Standardizing analysis of circulating microRNA: clinical and biological relevance. J Cell Biochem. 115:805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gant TW, Sauer UG, Zhang SD, Chorley BN, Hackermuller J, Perdichizzi S, Tollefsen KE, van Ravenzwaay B, Yauk C, Tong W, Poole A. (2017).A generic Transcriptomics Reporting Framework (TRF) for ‘omics data processing and analysis. Regul Toxicol Pharmacol, 91 Suppl 1, S36–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez MD, Spengler RM, Etheridge A, Godoy PM, Barczak AJ, Srinivasan S, De Hoff PL, Tanriverdi K, Courtright A, Lu S, Khoory J, Rubio R, Baxter D, Driedonks TAP, Buermans HPJ, Nolte-’t Hoen ENM, Jiang H, Wang K, Ghiran I, Wang YE, Van Keuren-Jensen K, Freedman JE, Woodruff PG, Laurent LC, Erle DJ, Galas DJ, Tewari M. (2018).Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat Biotechnol, 36, 746–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, Bertone P, Caldas C. (2010).Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA, 16, 991–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glineur SF, De Ron P, Hanon E, Valentin J-P, Dremier S, da Costa AN. 2015. Paving the route to plasma miR-208a-3p as an acute cardiac injury biomarker: preclinical rat data supports its use in drug safety assessment. Toxicol Sci. 149:89–97. [DOI] [PubMed] [Google Scholar]
- Glineur SF, Hanon E, Dremier S, Snelling S, Berteau C, De Ron P, Nogueira da Costa A. 2018. Assessment of a urinary kidney microRNA panel as potential nephron segment-specific biomarkers of subacute renal toxicity in preclinical rat models. Toxicol Sci. 166:409–419. [DOI] [PubMed] [Google Scholar]
- Glinge C, Clauss S, Boddum K, Jabbari R, Jabbari J, Risgaard B, Tomsits P, Hildebrand B, Kaab S, Wakili R, et al. 2017. Stability of circulating blood-based microRNAs - pre-analytic methodological considerations. PLoS One. 12:e0167969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy PM, Barczak AJ, DeHoff P, Srinivasan S, Das S, Erle DJ, Laurent LC. 2019. Comparison of miRNA profiling methods using synthetic miRNA pools and standardized exRNA samples reveals substantial performance differences. bioRxiv:645762. [Google Scholar]
- Gomes CP, Cho JH, Hood L, Franco OL, Pereira RW, Wang K. 2013. A review of computational tools in microRNA discovery. Front Genet. 4:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu C-G, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, et al. 2007. Cyclin G1 Is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 67:6092–6099. [DOI] [PubMed] [Google Scholar]
- Grasedieck S, Scholer N, Bommer M, Niess JH, Tumani H, Rouhi A, Bloehdorn J, Liebisch P, Mertens D, Dohner H, et al. 2012. Impact of serum storage conditions on microRNA stability. Leukemia. 26:2414–2416. [DOI] [PubMed] [Google Scholar]
- Guo L, Yu J, Liang T, Zou Q. 2016. miR-isomiRExp: a web-server for the analysis of expression of miRNA at the miRNA/isomiR levels. Sci Rep. 6:23700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Escolano A, Santacruz-Vazquez E, Gomez-Perez F. 2015. Dysregulated microRNAs involved in contrast-induced acute kidney injury in rat and human. Ren Fail. 37:1498–1506. [DOI] [PubMed] [Google Scholar]
- Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, Warnecke JM, Sczakiel G. 2010. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 28:655–661. [DOI] [PubMed] [Google Scholar]
- Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC, Ding WQ. 2016. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 18:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantzsch M, Tolios A, Beutner F, Nagel D, Thiery J, Teupser D, Holdt LM. 2014. Comparison of whole blood RNA preservation tubes and novel generation RNA extraction kits for analysis of mRNA and MiRNA profiles. PLoS One. 9:e113298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill AH, McCullough SD, Wood CE, Kahle JJ, Chorley BN. 2016. MicroRNA biomarkers of toxicity in biological matrices. Toxicol Sci. 152:264–272. [DOI] [PubMed] [Google Scholar]
- Heegaard NH, Carlsen AL, Lilje B, Ng KL, Ronne ME, Jorgensen HL, Sennels H, Fahrenkrug J. 2016. Diurnal variations of human circulating cell-free micro-RNA. PLoS One. 11:e0160577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SD, Khurana N, Williams J, Dowd Greene C, Uhlig R, Middleton FA. 2018. Diurnal oscillations in human salivary microRNA and microbial transcription: implications for human health and disease. PLoS One. 13:e0198288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman NS, Mosedale M, Wolf KK, LeCluyse EL, Watkins PB. 2016. Subtoxic alterations in hepatocyte-derived exosomes: an early step in drug-induced liver injury? Toxicol Sci. 151:365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby RJ, Starkey Lewis P, Dear J, Goldring C, Park BK. 2014. MicroRNAs as potential circulating biomarkers of drug-induced liver injury: key current and future issues for translation to humans. Expert Rev Clin Pharmacol. 7:349–362. [DOI] [PubMed] [Google Scholar]
- Howell LS, Ireland L, Park BK, Goldring CE. 2018. MiR-122 and other microRNAs as potential circulating biomarkers of drug-induced liver injury. Expert Rev Mol Diagn. 18:47–54. [DOI] [PubMed] [Google Scholar]
- Hu HY, Yan Z, Xu Y, Hu H, Menzel C, Zhou YH, Chen W, Khaitovich P. 2009. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics. 10:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, Hellemans J, Kubista M, Mueller RD, Nolan T, et al. 2013. The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin Chem. 59:892–902. [DOI] [PubMed] [Google Scholar]
- Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, et al. 2008. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 3:e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichii O, Ohta H, Horino T, Nakamura T, Hosotani M, Mizoguchi T, Morishita K, Nakamura K, Sasaki N, Takiguchi M, et al. 2018. Urinary exosome-derived microRNAs reflecting the changes in renal function in cats. Front Vet Sci. 5:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarry J, Schadendorf D, Greenwood C, Spatz A, van Kempen LC. 2014. The validity of circulating microRNAs in oncology: five years of challenges and contradictions. Mol Oncol. 8:819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen SG, Lamy P, Rasmussen MH, Ostenfeld MS, Dyrskjot L, Orntoft TF, Andersen CL. 2011. Evaluation of two commercial global miRNA expression profiling platforms for detection of less abundant miRNAs. BMC Genomics. 12:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan M, Mohan KV, Kulkarni S, Atreya C. 2009. Membrane array-based differential profiling of platelets during storage for 52 miRNAs associated with apoptosis. Transfusion. 49:1443–1450. [DOI] [PubMed] [Google Scholar]
- Khan J, Lieberman JA, Lockwood CM. 2017. Variability in, variability out: best practice recommendations to standardize pre-analytical variables in the detection of circulating and tissue microRNAs. Clin Chem Lab Med. 55:608–621. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Jung JS. 2011. Methods for analyzing microRNA expression and function during osteogenic differentiation of human adipose tissue-derived mesenchymal stem cells. Methods Mol Biol. 702:401–418. [DOI] [PubMed] [Google Scholar]
- Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, Reid G. (2011).Haemolysis during sample preparation alters microRNA content of plasma. PLoS One, 6, e24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MB, Edelman JJ, Kao SC, Vallely MP, van Zandwijk N, Reid G. (2013).The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front Genet, 4, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberle V, Pleli T, Schmithals C, Augusto Alonso E, Haupenthal J, Bonig H, Peveling-Oberhag J, Biondi RM, Zeuzem S, Kronenberger B, et al. 2013. Differential stability of cell-free circulating microRNAs: implications for their utilization as biomarkers. PLoS One. 8:e75184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig EM, Fisher C, Bernard H, Wolenski FS, Gerrein J, Carsillo M, Gallacher M, Tse A, Peters R, Smith A, et al. 2016. The beagle dog microRNA tissue atlas: identifying translatable biomarkers of organ toxicity. BMC Genomics. 17:649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong XY, Du YQ, Li L, Liu JQ, Wang GK, Zhu JQ, Man XH, Gong YF, Xiao LN, Zheng YZ, et al. 2010. Plasma miR-216a as a potential marker of pancreatic injury in a rat model of acute pancreatitis. World J Gastroenterol. 16:4599–4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koritzinsky EH, Street JM, Chari RR, Glispie DM, Bellomo TR, Aponte AM, Star RA, Yuen PST. 2019. Circadian variation in the release of small extracellular vesicles can be normalized by vesicle number or TSG101. Am J Physiol Renal Physiol. 317:F1098–F1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I, Parameswaran J, Shapira I, Lovecchio J, Menzin A, Whyte J, Dos Santos L, Liang S, Bhuiya T, Keogh M, Khalili H, Pond C, Liew A, Shih A, Gregersen PK, Lee AT. (2017).Two microRNA signatures for malignancy and immune infiltration predict overall survival in advanced epithelial ovarian cancer. J Investig Med, 65, 1068–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauskopf J, Caiment F, Claessen SM, Johnson KJ, Warner RL, Schomaker SJ, Burt DA, Aubrecht J, Kleinjans JC. 2015. Application of high-throughput sequencing to circulating microRNAs reveals novel biomarkers for drug-induced liver injury. Toxicol Sci. 143:268–276. [DOI] [PubMed] [Google Scholar]
- Krauskopf J, de Kok TM, Schomaker SJ, Gosink M, Burt DA, Chandler P, Warner RL, Johnson KJ, Caiment F, Kleinjans JC, et al. 2017. Serum microRNA signatures as “liquid biopsies” for interrogating hepatotoxic mechanisms and liver pathogenesis in human. PLoS ONE. 12:e0177928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegel AJ, Liu Y, Liu P, Baker MA, Hodges MR, Hua X, Liang M. 2013. Characteristics of microRNAs enriched in specific cell types and primary tissue types in solid organs. Physiol Genomics. 45:1144–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. 2002. Identification of tissue-specific microRNAs from mouse. Curr Biol. 12:735–739. [DOI] [PubMed] [Google Scholar]
- Lam B, Simkin M, Rghei N, Haj-Ahmad Y. (2016).Revised Guidelines for RNA Quality Assessment for Diverse Biological Sample Input (Application Note 47 RNA Sample Preparation). Thorold, Ontario, Canada, Norgen Biotek Corp. [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 129:1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laterza OF, Lim L, Garrett-Engele PW, Vlasakova K, Muniappa N, Tanaka WK, Johnson JM, Sina JF, Fare TL, Sistare FD, et al. 2009. Plasma microRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 55:1977–1983. [DOI] [PubMed] [Google Scholar]
- Lee I, Baxter D, Lee MY, Scherler K, Wang K. 2017. The importance of standardization on analyzing circulating RNA. Mol Diagn Ther. 21:259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75:843–854. [DOI] [PubMed] [Google Scholar]
- Levin-Schwartz Y, Curtin P, Flores D, Aushev VN, Tamayo-Ortiz M, Svensson K, Pantic I, Estrada-Gutierrez G, Pizano-Zarate ML, Gennings C, Satlin LM, Baccarelli AA, Tellez-Rojo MM, Wright RO, Sanders AP. (2021).Exosomal miRNAs in urine associated with children’s cardiorenal parameters: a cross-sectional study. Epigenomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-H, Yauk CL, Chen R, Hyduke DR, Williams A, Frötschl R, Ellinger-Ziegelbauer H, Pettit S, Aubrecht J, Fornace AJ. 2019. TGx-DDI, a transcriptomic biomarker for genotoxicity hazard assessment of pharmaceuticals and environmental chemicals. Front Big Data. 2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, Guenther SM, O’Leary JJ, Sheils O. 2007. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ridzon D, Wong L, Chen C. 2007. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 8:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhang Q, Zhang J, Li C, Miao YR, Lei Q, Li Q, Guo AY. 2019. EVmiRNA: a database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 47:D89–D93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fan Z, Zhao T, Cao W, Zhang L, Li H, Xie Q, Tian Y, Wang B. 2015. Plasma miR-1, miR-208, miR-499 as potential predictive biomarkers for acute myocardial infarction: an independent study of Han population. Exp Gerontol. 72:230–238. [DOI] [PubMed] [Google Scholar]
- Liu Y, Li P, Liu L, Zhang Y. 2018. The diagnostic role of miR-122 in drug-induced liver injury: a systematic review and meta-analysis. Medicine (Baltimore). 97:e13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Longarela B, Morrison EE, Tranter JD, Chahman-Vos L, Léonard J-F, Gautier J-C, Laurent S, Lartigau A, Boitier E, Sautier L, et al. 2019. Direct detection of circulating microRNA-122 using dynamic chemical labelling with single molecule detection overcomes stability and isomiR challenges for biomarker qualification. bioRxiv:777458. [Google Scholar]
- Lorenzen JM, Kielstein JT, Hafer C, Gupta SK, Kumpers P, Faulhaber-Walter R, Haller H, Fliser D, Thum T. 2011. Circulating miR-210 predicts survival in critically ill patients with acute kidney injury. Clin J Am Soc Nephrol. 6:1540–1546. [DOI] [PubMed] [Google Scholar]
- Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stahler C, Meese E, et al. 2016. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 44:3865–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv LL, Cao Y, Liu D, Xu M, Liu H, Tang RN, Ma KL, Liu BC. 2013. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int J Biol Sci. 9:1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee R, Telonis AG, Cherlin T, Rigoutsos I, Londin E. 2017. Assessment of isomiR discrimination using commercial qPCR methods. Noncoding RNA. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall C, Rocke DM, Durbin-Johnson B, Weiss RH. 2013. Stability of miRNA in human urine supports its biomarker potential. Biomark Med. 7:623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Ibarra A, Martinez-Razo LD, Vazquez-Martinez ER, Martinez-Cruz N, Flores-Ramirez R, Garcia-Gomez E, Lopez-Lopez M, Ortega-Gonzalez C, Camacho-Arroyo I, Cerbon M. (2019).Unhealthy Levels of Phthalates and Bisphenol A in Mexican Pregnant Women with Gestational Diabetes and Its Association to Altered Expression of miRNAs Involved with Metabolic Disease. Int J Mol Sci, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi MJ, Montani F, Carletti RM, Dezi F, Dama E, Bonizzi G, Sandri MT, Rampinelli C, Bellomi M, Maisonneuve P, et al. 2016. Optimization and standardization of circulating microRNA detection for clinical application: the miR-test case. Clin Chem. 62:743–754. [DOI] [PubMed] [Google Scholar]
- Max KEA, Bertram K, Akat KM, Bogardus KA, Li J, Morozov P, Ben-Dov IZ, Li X, Weiss ZR, Azizian A, et al. 2018. Human plasma and serum extracellular small RNA reference profiles and their clinical utility. Proc Natl Acad Sci U S A. 115:E5334–E5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. 2011. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 57:833–840. [DOI] [PubMed] [Google Scholar]
- Menezes MR, Balzeau J, Hagan JP. 2018. 3’ RNA uridylation in epitranscriptomics, gene regulation, and disease. Front Mol Biosci. 5:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng FD, Wang S, Jiang YH, Sui CG. 2016. Antitumor effect of dendritic cells transfected with prostate-specific membrane antigen recombinant adenovirus on prostate cancer: an in vitro study. Mol Med Rep. 13:2124–2134. [DOI] [PubMed] [Google Scholar]
- Meder B, Backes C, Haas J, Leidinger P, Stahler C, Grossmann T, Vogel B, Frese K, Giannitsis E, Katus HA, Meese E, Keller A. (2014).Influence of the confounding factors age and sex on microRNA profiles from peripheral blood. Clin Chem, 60, 1200–8 [DOI] [PubMed] [Google Scholar]
- Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, Cheo D, D’Andrade P, DeMayo M, Dennis L, et al. 2014. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 11:809–815. [DOI] [PubMed] [Google Scholar]
- Mestdagh P, Lefever S, Pattyn F, Ridzon D, Fredlund E, Fieuw A, Ongenaert M, Vermeulen J, De Paepe A, Wong L, et al. 2011. The microRNA body map: dissecting microRNA function through integrative genomics. Nucleic Acids Res. 39:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SU, Kaiser S, Wagner C, Thirion C, Pfaffl MW. 2012. Profound effect of profiling platform and normalization strategy on detection of differentially expressed microRNAs--a comparative study. PLoS One. 7:e38946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikaelian I, Scicchitano M, Mendes O, Thomas RA, Leroy BE. (2013).Frontiers in preclinical safety biomarkers: microRNAs and messenger RNAs. Toxicol Pathol, 41, 18–31 [DOI] [PubMed] [Google Scholar]
- Minami K, Uehara T, Morikawa Y, Omura K, Kanki M, Horinouchi A, Ono A, Yamada H, Ohno Y, Urushidani T. 2014. miRNA expression atlas in male rat. Sci Data. 1:140005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momen-Heravi F, Bala S, Kodys K, Szabo G. 2015. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Scientific Reports. 5:9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mompeon A, Ortega-Paz L, Vidal-Gomez X, Costa TJ, Perez-Cremades D, Garcia-Blas S, Brugaletta S, Sanchis J, Sabate M, Novella S, Dantas AP, Hermenegildo C. (2020).Disparate miRNA expression in serum and plasma of patients with acute myocardial infarction: a systematic and paired comparative analysis. Sci Rep, 10, 5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori MA, Ludwig RG, Garcia-Martin R, Brandao BB, Kahn CR. (2019).Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab, 30, 656–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale M, Eaddy JS, Trask JOJ, Holman NS, Wolf KK, LeCluyse E, Ware BR, Khetani SR, Lu J, Brock WJ, et al. 2018. miR-122 release in exosomes precedes overt tolvaptan-induced necrosis in a primary human hepatocyte micropatterned coculture model. Toxicol Sci. 161:149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motawi TK, Mohamed MR, Shahin NN, Ali MAM, Azzam MA. 2018. Time-course expression profile and diagnostic potential of a miRNA panel in exosomes and total serum in acute liver injury. Int J Biochem Cell Biol. 100:11–21. [DOI] [PubMed] [Google Scholar]
- Nassirpour R, Homer BL, Mathur S, Li Y, Li Z, Brown T, Carraher D, Warneke J, Bailey S, Percival K, et al. 2015. Identification of promising urinary microRNA biomarkers in two rat models of glomerular injury. Toxicol Sci. 148:35–47. [DOI] [PubMed] [Google Scholar]
- Neilsen CT, Goodall GJ, Bracken CP. 2012. IsomiRs--the overlooked repertoire in the dynamic microRNAome. Trends Genet. 28:544–549. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Kondo C, Morikawa Y, Tonomura Y, Torii M, Yamate J, Uehara T. 2015. Plasma miR-208 as a useful biomarker for drug-induced cardiotoxicity in rats. J Appl Toxicol. 35:173–180. [DOI] [PubMed] [Google Scholar]
- Page K, Guttery DS, Zahra N, Primrose L, Elshaw SR, Pringle JH, Blighe K, Marchese SD, Hills A, Woodley L, et al. 2013. Influence of plasma processing on recovery and analysis of circulating nucleic acids. PLoS One. 8:e77963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-K, Jo W, Choi H-J, Jang S, Ryu J-E, Lee H-J, Lee H, Kim H, Yu E-S, Son W-C. 2016. Time-course changes in the expression levels of miR-122, −155, and −21 as markers of liver cell damage, inflammation, and regeneration in acetaminophen-induced liver injury in rats. J Vet Sci. 17:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavkovic M, Riefke B, Ellinger-Ziegelbauer H. 2014. Urinary microRNA profiling for identification of biomarkers after cisplatin-induced kidney injury. Toxicology. 324:147–157. [DOI] [PubMed] [Google Scholar]
- Pavkovic M, Riefke B, Frisk AL, Groticke I, Ellinger-Ziegelbauer H. 2015. Glomerulonephritis-induced changes in urinary and kidney microRNA profiles in rats. Toxicol Sci. 145:348–359. [DOI] [PubMed] [Google Scholar]
- Pavkovic M, Vaidya VS. 2016. MicroRNAs and drug-induced kidney injury. Pharmacol Ther. 163:48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier D, Reddy P. 2018. Non-coding RNA mediated regulation of allogeneic T cell responses after hematopoietic transplantation. Front Immunol. 9:1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 26:509–515. [DOI] [PubMed] [Google Scholar]
- Pirola CJ, Fernandez Gianotti T, Castano GO, Mallardi P, San Martino J, Mora Gonzalez Lopez Ledesma M, Flichman D, Mirshahi F, Sanyal AJ, Sookoian S. 2015. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 64:800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregibon DC, Toner M, Doyle PS. 2007. Multifunctional encoded particles for high-throughput biomolecule analysis. Science. 315:1393–1396. [DOI] [PubMed] [Google Scholar]
- Qin X, Guo H, Wang X, Zhu X, Yan M, Wang X, Xu Q, Shi J, Lu E, Chen W, et al. 2019. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 20:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmel T, Schafer ST, Frey UH, Adamzik M, Peters J. (2018).Increased circulating microRNA-122 is a biomarker for discrimination and risk stratification in patients defined by sepsis-3 criteria. PLoS One, 13, e0197637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon-Nunez LA, Martos L, Fernandez-Pardo A, Oto J, Medina P, Espana F, Navarro S. 2017. Comparison of protocols and RNA carriers for plasma miRNA isolation. Unraveling RNA carrier influence on miRNA isolation. PLoS One. 12:e0187005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redshaw N, Wilkes T, Whale A, Cowen S, Huggett J, Foy CA. 2013. A comparison of miRNA isolation and RT-qPCR technologies and their effects on quantification accuracy and repeatability. Biotechniques. 54:155–164. [DOI] [PubMed] [Google Scholar]
- Rissland OS, Mikulasova A, Norbury CJ. 2007. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol. 27:3612–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkin M, Simerzin A, Zorde-Khvalevsky E, Chai C, Yuval JB, Rosenberg N, Harari-Steinfeld R, Schneider R, Amir G, Condiotti R, et al. 2016. Inflammation-induced expression and secretion of microRNA 122 leads to reduced blood levels of kidney-derived erythropoietin and anemia. Gastroenterology. 151:999–1010. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Martinez A, de Miguel-Perez D, Ortega FG, Garcia-Puche JL, Robles-Fernandez I, Exposito J, Martorell-Marugan J, Carmona-Saez P, Garrido-Navas MDC, Rolfo C, et al. 2019. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res. 21:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Vera T, Ochoa-Martinez AC, Pruneda-Alvarez LG, Dominguez-Cortinas G, Perez-Maldonado IN. (2019).Expression levels of circulating microRNAs-126, −155, and −145 in Mexican women exposed to polycyclic aromatic hydrocarbons through biomass fuel use. Environ Mol Mutagen, 60, 546–58 [DOI] [PubMed] [Google Scholar]
- Sah S, McCall MN, Eveleigh D, Wilson M, Irizarry RA. 2010. Performance evaluation of commercial miRNA expression array platforms. BMC Res Notes. 3:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders R, Huggett JF, Bushell CA, Cowen S, Scott DJ, Foy CA. 2011. Evaluation of digital PCR for absolute DNA quantification. Anal Chem. 83:6474–6484. [DOI] [PubMed] [Google Scholar]
- Schlosser K, McIntyre LA, White RJ, Stewart DJ. (2015).Customized Internal Reference Controls for Improved Assessment of Circulating MicroRNAs in Disease. PLoS One, 10, e0127443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach H, da Silva AM, Calin G, Pantel K. 2015. Data normalization strategies for microRNA quantification. Clin Chem. 61:1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD. 2008. Argonaute loading improves the 5’ precision of both microRNAs and their miRNA* strands in flies. Curr Biol. 18:147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R, Ziegler O, Yeri A, Liu X, Murthy V, Rabideau D, Xiao Chun Y, Hanspers K, Belcher A, Tackett M, et al. 2018. MicroRNAs associated with reverse left ventricular remodeling in humans identify pathways of heart failure progression. Circulation: Heart Failure. 11:e004278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharapova T, Devanarayan V, LeRoy B, Liguori MJ, Blomme E, Buck W, Maher J. 2016. Evaluation of miR-122 as a serum biomarker for hepatotoxicity in investigative rat toxicology studies. Vet Pathol. 53:211–221. [DOI] [PubMed] [Google Scholar]
- Shiotsu H, Okada K, Shibuta T, Kobayashi Y, Shirahama S, Kuroki C, Ueda S, Ohkuma M, Ikeda K, Ando Y, Matsui H, Kayamori Y, Umemura T. (2018).The Influence of Pre-Analytical Factors on the Analysis of Circulating MicroRNA. Microrna, 7, 195–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlipak MG, Sarnak MJ, Katz R, Fried LF, Seliger SL, Newman AB, Siscovick DS, Stehman-Breen C. 2005. Cystatin C and the risk of death and cardiovascular events among elderly persons. N Engl J Med. 352:2049–2060. [DOI] [PubMed] [Google Scholar]
- Smith A, Calley J, Mathur S, Qian HR, Wu H, Farmen M, Caiment F, Bushel PR, Li J, Fisher C, et al. 2016. The rat microRNA body atlas; Evaluation of the microRNA content of rat organs through deep sequencing and characterization of pancreas enriched miRNAs as biomarkers of pancreatic toxicity in the rat and dog. BMC Genomics. 17:694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn W, Kim J, Kang SH, Yang SR, Cho J-Y, Cho HC, Shim SG, Paik Y-H. 2015. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 47:e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DG, Antoine DJ, French NS, Dhaun N, Webb DJ, Costello EM, et al. 2011. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 54:1767–1776. [DOI] [PubMed] [Google Scholar]
- Stephenson D, Sauer JM. 2014. The predictive safety testing consortium and the coalition against major diseases. Nat Rev Drug Discov. 13:793–794. [DOI] [PubMed] [Google Scholar]
- Tan GW, Khoo AS, Tan LP. 2015. Evaluation of extraction kits and RT-qPCR systems adapted to high-throughput platform for circulating miRNAs. Sci Rep. 5:9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno T, Bhanu NV, Oneal PA, Goh SH, Staker P, Lee YT, Moroney JW, Reed CH, Luban NL, Wang RH, et al. 2007. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat Med. 13:1096–1101. [DOI] [PubMed] [Google Scholar]
- Telakivi T, Flink T. 1985. [Refsum’s disease]. Duodecim. 101:1958–1965. [PubMed] [Google Scholar]
- Thompson KL, Boitier E, Chen T, Couttet P, Ellinger-Ziegelbauer H, Goetschy M, Guillemain G, Kanki M, Kelsall J, Mariet C, et al. 2016. Absolute measurement of cardiac injury-induced microRNAs in biofluids across multiple test sites. Toxicol Sci. 154:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen M, Blondal T, Mouritzen P. 2017. Quantitative RT-PCR for microRNAs in biofluids. Methods Mol Biol. 1641:379–398. [DOI] [PubMed] [Google Scholar]
- Thulin P, Nordahl G, Gry M, Yimer G, Aklillu E, Makonnen E, Aderaye G, Lindquist L, Mattsson CM, Ekblom B, et al. 2014. Keratin-18 and microRNA-122 complement alanine aminotransferase as novel safety biomarkers for drug-induced liver injury in two human cohorts. Liver Int. 34:367–378. [DOI] [PubMed] [Google Scholar]
- Tominaga N, Kosaka N, Ono M, Katsuda T, Yoshioka Y, Tamura K, Lotvall J, Nakagama H, Ochiya T. 2015. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat Commun. 6:6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebicka J, Anadol E, Elfimova N, Strack I, Roggendorf M, Viazov S, Wedemeyer I, Drebber U, Rockstroh J, Sauerbruch T, et al. 2013. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J Hepatol. 58:234–239. [DOI] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Langheinz A, Burwinkel B. 2011. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39:7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usborne AL, Smith AT, Engle SK, Watson DE, Sullivan JM, Walgren JL. 2014. Biomarkers of exocrine pancreatic injury in 2 rat acute pancreatitis models. Toxicol Pathol. 42:195–203. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 9:654–659. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:Research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. 2011. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 13:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron N, Meryet-Figuiere M, Guttin A, Issartel JP, Lambert B, Briand M, Louis MH, Vernon M, Lebailly P, Lecluse Y, et al. 2016. Towards a new standardized method for circulating miRNAs profiling in clinical studies: interest of the exogenous normalization to improve miRNA signature accuracy. Mol Oncol. 10:981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Qin YW, Jing Q. 2010. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 31:659–666. [DOI] [PubMed] [Google Scholar]
- Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. 2012. Comparing the microRNA spectrum between serum and plasma. PLoS One. 7:e41561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. 2009. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 106:4402–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang S, Weber J, Baxter D, Galas DJ. 2010. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 38:7248–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xu Y, Li M, Tu J, Lu Z. 2016. Dysregulation of miRNA isoform level at 5’ end in Alzheimer’s disease. Gene. 584:167–172. [DOI] [PubMed] [Google Scholar]
- Ward J, Kanchagar C, Veksler-Lublinsky I, Lee RC, McGill MR, Jaeschke H, Curry SC, Ambros VR. 2014. Circulating microRNA profiles in human patients with acetaminophen hepatotoxicity or ischemic hepatitis. Proc Natl Acad Sci U S A. 111:12169–12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. 2010. The microRNA spectrum in 12 body fluids. Clin Chem. 56:1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 75:855–862. [DOI] [PubMed] [Google Scholar]
- Wolenski FS, Shah P, Sano T, Shinozawa T, Bernard H, Gallacher MJ, Wyllie SD, Varrone G, Cicia LA, Carsillo ME, et al. 2017. Identification of microRNA biomarker candidates in urine and plasma from rats with kidney or liver damage. J Appl Toxicol. 37:278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfinger RD, Beedanagari S, Boitier E, Chen T, Couttet P, Ellinger-Ziegelbauer H, Guillemain G, Mariet C, Mouritzen P, O’Lone R, et al. 2018. Two approaches for estimating the lower limit of quantitation (LLOQ) of microRNA levels assayed as exploratory biomarkers by RT-qPCR. BMC Biotechnol. 18:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CW, Evans JM, Huang S, Mahoney DW, Dukek BA, Taylor WR, Yab TC, Smyrk TC, Jen J, Kisiel JB, et al. 2018. A comprehensive approach to sequence-oriented isomiR annotation (CASMIR): demonstration with IsomiR profiling in colorectal neoplasia. BMC Genomics. 19:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C, et al. 2011. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 50:136–142. [DOI] [PubMed] [Google Scholar]
- Yang X, Greenhaw J, Shi Q, Su Z, Qian F, Davis K, Mendrick DL, Salminen WF. 2012. Identification of urinary microRNA profiles in rats that may diagnose hepatotoxicity. Toxicol Sci. 125:335–344. [DOI] [PubMed] [Google Scholar]
- Yang X, Weng Z, Mendrick DL, Shi Q. 2014. Circulating extracellular vesicles as a potential source of new biomarkers of drug-induced liver injury. Toxicology Lett. 225:401–406. [DOI] [PubMed] [Google Scholar]
- Yauk CL, Rowan-Carroll A, Stead JD, Williams A. (2010).Cross-platform analysis of global microRNA expression technologies. BMC Genomics, 11, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, et al. 2014. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun. 5:3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xu Y, Xue S, Wang X, Dai H, Qian J, Ni Z, Yan Y. 2017. Implications of dynamic changes in miR-192 expression in ischemic acute kidney injury. Int Urol Nephrol. 49:541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zang Q, Xu B, Zheng W, Ban R, Zhang H, Yang Y, Hao Q, Iqbal F, Li A, et al. 2016. IsomiR Bank: a research resource for tracking IsomiRs. Bioinformatics. 32:2069–2071. [DOI] [PubMed] [Google Scholar]
- Zhao H, Shen J, Hu Q, Davis W, Medico L, Wang D, Yan L, Guo Y, Liu B, Qin M, et al. 2014. Effects of preanalytic variables on circulating microRNAs in whole blood. Cancer Epidemiol Biomarkers Prev. 23:2643–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Mao A, Wang X, Duan X, Yao Y, Zhang C. 2013. Urine and serum microRNA-1 as novel biomarkers for myocardial injury in open-heart surgeries with cardiopulmonary bypass. PLoS One. 8:e62245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorofchian S, Iqbal F, Rao M, Aung PP, Esquenazi Y, Ballester LY. (2019).Circulating tumour DNA, microRNA and metabolites in cerebrospinal fluid as biomarkers for central nervous system malignancies. J Clin Pathol, 72, 271–80 [DOI] [PubMed] [Google Scholar]


