Abstract
Iron-sulfur (Fe–S) clusters are essential cofactors most commonly known for their role mediating electron transfer within the mitochondrial respiratory chain. The Fe–S cluster pathways that function within the respiratory complexes are highly conserved between bacteria and the mitochondria of eukaryotic cells. Within the electron transport chain, Fe–S clusters play a critical role in transporting electrons through Complexes I, II and III to cytochrome c, before subsequent transfer to molecular oxygen. Fe–S clusters are also among the binding sites of classical mitochondrial inhibitors, such as rotenone, and play an important role in the production of mitochondrial reactive oxygen species (ROS). Mitochondrial Fe–S clusters also play a critical role in the pathogenesis of disease. High levels of ROS produced at these sites can cause cell injury or death, however, when produced at low levels can serve as signaling molecules. For example, Ndufs2, a Complex I subunit containing an Fe–S center, N2, has recently been identified as a redox-sensitive oxygen sensor, mediating homeostatic oxygen-sensing in the pulmonary vasculature and carotid body. Fe–S clusters are emerging as transcriptionally-regulated mediators in disease and play a crucial role in normal physiology, offering potential new therapeutic targets for diseases including malaria, diabetes, and cancer.
Keywords: Fe-S cluster, Mitochondria, Electron transport chain, Drug target, Oxygen-sensing, Epigenetics
Graphical abstract
Highlights
-
•
Fe–S clusters are essential cofactors, evolutionarily conserved across taxa.
-
•
Fe–S clusters mediate electron transfer in the mitochondrial respiratory complexes.
-
•
Fe–S clusters are implicated in ROS production by Complexes I, II and III.
-
•
Cluster N2 required for oxygen-sensing in the pulmonary arteries and carotid body.
-
•
Fe–S clusters emerging as novel therapeutic targets in the treatment of diseases.
1. Introduction
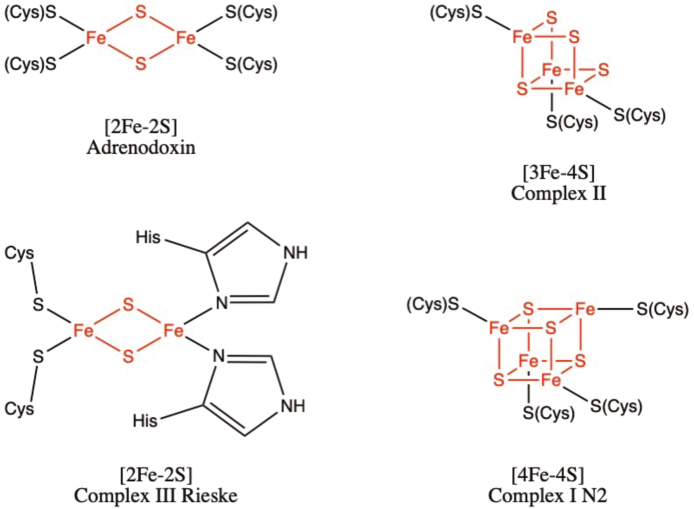
Iron-sulfur (Fe–S) clusters are among the most common cofactors observed across nature and comprise the largest class of metalloproteins. These clusters are structurally diverse, existing in simple forms such as [1Fe–0S], where a single iron atom is coordinated by four cysteine groups, found in rubredoxins of sulfur-metabolizing archaea, and in complex forms such as the [8Fe–7S] found in nitrogenases of nitrogen-fixing bacteria [1,2]. Most commonly, these clusters exist as [2Fe–2S], [3Fe–4S] or [4Fe–4S]. The simplest of these, [2Fe–2S] is a rhombic structure containing two iron atoms bridged by two sulfur atoms, and is the cluster commonly found within ferredoxins, such as adrenodoxin, an Fe–S protein involved in the synthesis of steroids within the adrenal glands (Fig. 1) [3]. Although typically ligated by four cysteine residues within the Fe–S protein, ligation involving both cysteine and histidine residues is also observed, as seen in the Rieske iron-sulfur protein within Complex III of the electron transport chain (ETC) [4]. The [4Fe–4S] cluster exists in a cubic structure, with iron and sulfur atoms found in alternating corner positions. These clusters are found in bacterial ferredoxins, and within the mitochondrial respiratory complexes, such as with cluster N2 located within Complex I of the ETC [5]. The [3Fe–4S] clusters, although less common then the [4Fe–4S] subtype, are also ubiquitous, existing in a variety of ferredoxins and in the quinone binding site of Complex II [6].
Fig. 1.
The most common forms of Fe–S clusters found in biological systems: [2Fe–2S], [3Fe–4S] and [4Fe–4S]. In all structures shown above, the iron and sulfur atoms that are a part of the cluster are shown in red and ligating amino acid residues are shown in black. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fe–S clusters are versatile protein prosthetic groups and serve a variety of functions in biological systems. They function as cofactors in enzyme catalysis, and are often situated in active sites to assist in Lewis acid reactions, as observed with mitochondrial aconitase and radical S-adenosylmethionine (SAM) enzymes [7,8]. These clusters also have regulatory roles, modulating gene expression in response to oxidative stress (via superoxide response (SoxR) proteins), oxygen levels (via fumarate-nitrate reduction (FNR) proteins), as well as iron levels (via Iron Response Proteins including IRP1 and IRP2) [[9], [10], [11]]. Fe–S clusters have also been observed to play important roles in DNA metabolism, coordinating protein conformational changes during DNA replication and repair, as found in primases, helicases, nucleases, and polymerases [[12], [13], [14], [15], [16]]. Within these enzymes, these clusters are involved in maintaining structural stability, and are also known to play roles in DNA binding, unwinding, and exonuclease activity. For example, an Fe–S cluster within the helicase-nuclease AddAB is found adjacent to the enzyme's wedge domain and is essential for DNA unwinding, although its exact function is unknown [17].
Despite these diverse roles, Fe–S clusters are most commonly known for their role in electron transfer, with their arrangements defining the pathways of electron transport within the systems that drive photosynthesis and mitochondrial respiration. Within the mitochondria, Fe–S centers play vital roles in both the tricarboxylic acid cycle (TCA) and the ETC. Electrons donated by both NADH and FADH2 are transferred through numerous Fe–S clusters found in Complexes I, II and III of the ETC, with molecular oxygen serving as the terminal electron acceptor, forming water. In this review, we describe the evolution of Fe–S clusters across taxa, their roles in mitochondrial respiration, with an emphasis on their composition within Complex I of the ETC, their emerging roles as oxygen sensors within the pulmonary arteries, the carotid body, and the ductus arteriosus, as well as emerging targets of therapeutics.
2. Mitochondria and endosymbiosis
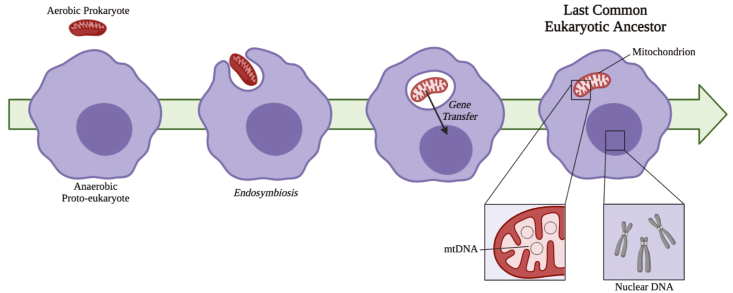
It is important to explore the origins of mitochondria to provide context for the roles of mitochondrial Fe–S clusters. The structure of the mitochondrion was first described in 1888, with Kölliker concluding that all mitochondria, then considered “granules”, have a membrane [18,19]. The origins of mitochondria as bacterial endosymbionts were first proposed by Wallin in 1927 [20]. However, the endosymbiont hypothesis, outlined in Fig. 2, was popularized by Lynn Margulis (then Lynn Sagan) in her 1967 publication [21], and in more detail in her subsequent books [22,23]. Margulis synthesized ideas from a variety of sources to create a clear theory of endosymbiosis [20,[24], [25], [26], [27]]. There was previously a great deal of debate between the opposing hypotheses of endosymbiosis and autogenous origins of mitochondria [[28], [29], [30], [31], [32], [33], [34]], with the autogenous origin theory postulating that mitochondria arose from within a single cell via functional specialization and intracellular compartmentalization. However, endosymbiosis is now universally accepted as the evolutionary origin of mitochondria; that a proto-eukaryote (heterotrophic anaerobe) engulfed a prokaryotic proto-mitochondrion [21]. Genetic analyses have determined that mitochondria are likely descended from α-proteobacteria [35,36], with the closest relative of the proto-mitochondrion belonging to the order Rickettsiales [37,38].
Fig. 2.
Overview of endosymbiont hypothesis. This universally accepted theory of mitochondrial origins posits that the anaerobic proto-eukaryote engulfed the aerobic prokaryote, most closely related to the order Rickettsiales. There was sizeable transfer of genes encoding mitochondrial proteins to the nuclear genome and elimination of redundancy. with current mitochondrial genomes encoding as few as 5 genes. The acquisition of the mitochondria resulted in the last common eukaryotic ancestor, from which all current eukaryotes evolved.
Debate continues surrounding the initial nature of the relationship of the proto-mitochondrion and proto-eukaryote, and about the selective advantage provided by the proto-mitochondrion immediately following endosymbiosis [39,40]. Some suggest the first endosymbiont was an aerobic heterotroph that provided an advantage to the host by supporting its energy need via secretion of ATP [22], removing the fermentation waste of the host [41,42], and/or removing intracellular oxygen that could be damaging or toxic to the host [43]. There is also evidence indicating that the benefit provided by the first mitochondrial endosymbiont was the generation of heat, enabling the host to colonize cooler environments with a higher internal temperature maintaining membrane fluidity, DNA helical tension, enzymatic reaction rates and other temperature-dependent cellular processes [40]. It has been postulated that assembly of Fe–S clusters was the essential contribution of the endosymbiont and the reason for the existence of the mitochondrion [44,45], as Fe–S cluster synthesis is essential for eukaryotic cell survival [[46], [47], [48]]. Others have proposed a non-mutualistic scenario, where the proto-mitochondrion was a predator invading the host [49,50]. These theories are based on current knowledge and understanding of α-proteobacterial diversity [39]. While these theories are not mutually exclusive, no consensus has been reached as to the precise nature of the initial host-symbiont relationship.
While a primary function of the organelle remains the generation of ATP via the respiratory chain, mitochondria exhibit significant diversity between cells and tissues and display tissue heterogeneity in their noncanonical functions (beyond generation of ATP), including energy production, calcium homeostasis, and coordination of programmed cell death (apoptosis). The mammalian respiratory chain is composed of five complexes, each with catalytic subunits highly conserved from bacteria [[51], [52], [53], [54], [55]]. These subunits function to couple electron transport between subunits to proton translocation across the inner mitochondrial membrane (IMM), creating the proton motive force required for ATP synthesis. While mitochondrial subunits are highly conserved, there still exists a great deal of mitochondrial and respiratory chain diversity within the eukaryotic domain. While all eukaryotes are descended from a common ancestor, some now lack subunits of the respiratory chain, and others, such as Giardia muris [56] and Entamoeba histolytica [57], lack mitochondria altogether, instead relying on hydrogenosomes for the anaerobic generation of ATP [58].
The fungal respiratory chain is similar to that of other eukaryotes, with a few notable differences. Complex I has been found in nearly all species of fungi, with Complex I having a similar structure to its counterpart in other forms of life. Notably, Saccharomyces cerevisiae lacks Complex I, as do certain other fungi [59]; these organisms possess alternative NADH:ubiquinone oxidoreductases that enable direct oxidation of external NADH or act as a parallel, rotenone-insensitive route of internal NADH oxidation [60]. Alternative NADH:ubiquinone oxidoreductases are widely distributed in fungi and plants, and lack Fe–S clusters [61]. These alternative NADH dehydrogenases are nuclear-encoded and, unlike traditional Complex I, do not couple electron transport to proton translocation across the IMM [62,63]. These alternative pathways in plants and fungi can function separate from or in conjunction with the canonical pathway [60].
In addition to alternative forms of Complex I, ‘Alternative Oxidase’ is present in most fungal genomes tested [64] and acts in parallel to Complex III to oxidize ubiquinol [65]. Alternative Oxidase also catalyzes the reduction of molecular oxygen to water [65]. Alternative Oxidase does not contain any Fe–S clusters and is not susceptible to the Complex III inhibitors antimycin A and myxothiazol, nor to the Complex IV inhibitor cyanide [[64], [65], [66]]. In certain plant species, alternative oxidase plays a vital role for heat production, whether to volatilize insect attractants, as is the case in the Arum plant, or to permit growth at lower temperatures, as seen in the American and Asian skunk cabbage [67]. However, it is generally considered to be a stress protein induced when factors such as growth inhibition or exposure to wounding, drought, or adverse salinity impair the main respiratory chain [67].
3. Mitochondrial Fe–S clusters
3.1. Aconitase Fe–S clusters
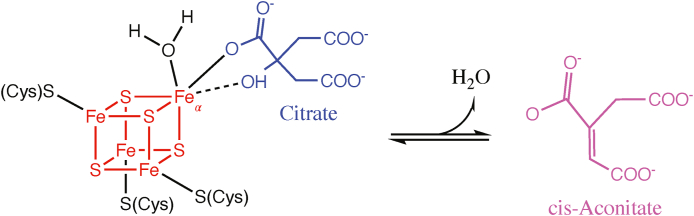
Aconitases are metalloenzymes containing a [4Fe–4S] cluster, acting in both the cytosol and mitochondria in different capacities. In the cytosol, the iron response protein (IRP-1) acts as a sensor of iron levels within the cell. When iron concentrations are high, IRP-1 acts as an aconitase and contains a [4Fe–4S] Fe–S cluster. When iron levels are low, IRP-1 loses its Fe–S cluster and the apoprotein subsequently binds to iron-responsive elements (IREs) within the 5′ or 3′ UTR of target mRNA to promote iron uptake, and reduce iron storage and utilization [68,69]. Within the mitochondria, aconitase acts in the second step of the TCA cycle, converting citrate to isocitrate via the intermediate cis-aconitate. The enzyme exists in two forms: an inactive form containing a [3Fe–4S] cluster, and upon acquiring another iron atom, an active form containing a [4Fe–4S] cluster. This additional labile iron atom (Feα) is coordinated by water molecules and is essential for carrying out the enzyme's catalytic function (Fig. 3) [70]. The Feα atom within active aconitase coordinates with oxygen atoms of citrate and water and acts as a Lewis acid to activate the hydroxyl group of citrate to facilitate the isomerization reaction.
Fig. 3.
Aconitase reaction mechanism. The isomerization reaction of citrate (shown in blue) to cis-Aconitate (magenta) is facilitated by the [4Fe–4S] cluster found within aconitase (shown in red). The addition of another labile iron (Feα) transforms inactive aconitase into its active form, and coordinates with water molecules and oxygen atoms of citrate to facilitate enzyme catalysis. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Aconitase, as well as other Fe–S cluster-containing dehydratases, are sensitive to both reactive oxygen species (ROS), including superoxide (O2·-) and hydrogen peroxide (H2O2), and the production of the reactive nitrogen species (RNS) peroxynitrite (ONOO−) [[71], [72], [73]]. The ETC is a major source of both ROS and RNS within the mitochondria. When levels of ROS/RNS rise, these molecules are capable of oxidizing the Fe–S cluster in active m-aconitase, causing the loss of the labile Feα atom and enzyme inactivation [74]. This susceptibility of m-aconitase to ROS/RNS-induced inactivation makes it a candidate sensor for redox changes within the mitochondria. If levels of oxidative stress increase, aconitase inactivation slows the flow of metabolites through the TCA cycle, thereby decreasing the availability of substrates for the ETC, and possibly promoting fatty acid synthesis through the export of citrate out of the mitochondrion [74]. In this way, the sensitivity of Fe–S cluster inactivation in m-aconitase may allow the enzyme to act as a sensor of ROS/RNS production by controlling metabolic flux through the mitochondria.
3.2. Electron transport chain Fe–S clusters
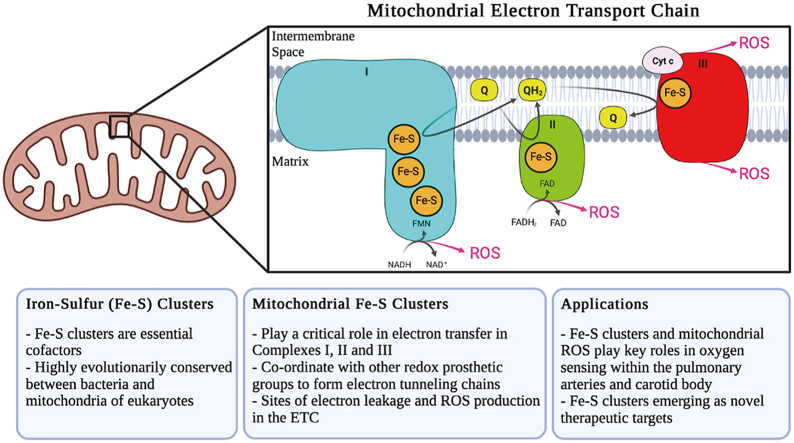
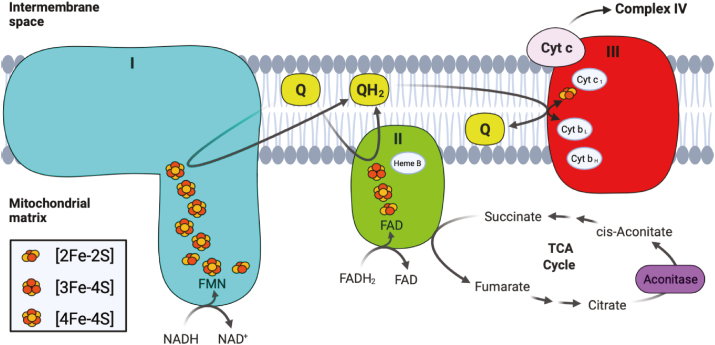
Oxidative phosphorylation describes a series of metabolic reactions that take place within the mitochondria, responsible for the majority of ATP production within aerobic eukaryotes. The electron carriers NADH and FADH2 formed from glycolysis, fatty acid oxidation, and the TCA cycle act as electron donors, passing electrons through four respiratory complexes of the ETC, to eventually reduce molecular oxygen to water. As electrons pass through the ETC complexes, protons are pumped from the mitochondrial matrix into the intermembrane space (IMS), creating a transmembrane electrical potential that is used to drive ATP production by ATP synthase (Complex V). Various redox groups exist within the respiratory complexes to facilitate the movement of electrons from NADH and FADH2 through the ETC, including flavins, quinones, hemes, and Fe–S clusters (Fig. 4). Within the mitochondria, Fe–S clusters play a variety of roles, acting not only in the main electron transfer pathways required for ATP generation, but also acting in mitochondrial DNA (mtDNA) catabolism. Within the pathways which drive mitochondrial respiration, Fe–S clusters are found within three of the four complexes which make up the ETC. In coordination with other redox groups, Fe–S clusters within Complexes I and II form electron tunneling chains, transferring electrons one at a time from the electron carriers FADH2 and NADH to ubiquinone. Although not found in a chain, an additional Fe–S cluster found within Complex III plays an integral role in passing electrons from ubiquinol (the reduced version of ubiquinone) to cytochrome c, to facilitate further transfer of electrons to Complex IV. In the mitochondria, Fe–S biogenesis involves the coordination of several proteins, including iron-sulfur cluster scaffold (ISCU) which provides the cysteine ligands where upon new Fe–S clusters are synthesized, cysteine desulfurase (NFS1) which provides the inorganic sulfur to ISCU, and frataxin (FXN) a protein that regulates desulfurase activity and has been suggested to act as an iron chaperone, delivering iron to the scaffold. After synthesis of [2Fe–2S] clusters in the mitochondria, they are either transferred to apoproteins, exported into the cytosol for incorporation into the cytosolic iron-sulfur cluster assembly (CIA) pathway, or further processed into [4Fe–4S] clusters. Although not the subject of this review, biogenesis of Fe–S clusters has been extensively studied and recently review by Alfadhel et al. and Maio et al. [75,76].
Fig. 4.
Simplified version of the mitochondrial ETC, showing Complexes I (blue), II (green), and III (red). The electron donors NADH and FADH2 are shown reducing Complexes I and respectively, as well as the electron carriers' ubiquinone (Q) and ubiquinol (QH2). Besides the Fe–S clusters, other redox prosthetic groups within each complex are also shown, including FMN and FAD, as well as the heme moieties a part of the cytochrome proteins. Intermediates within the TCA cycle that interact with Complex II and aconitase (purple) are also shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Fe–S clusters in Complex I
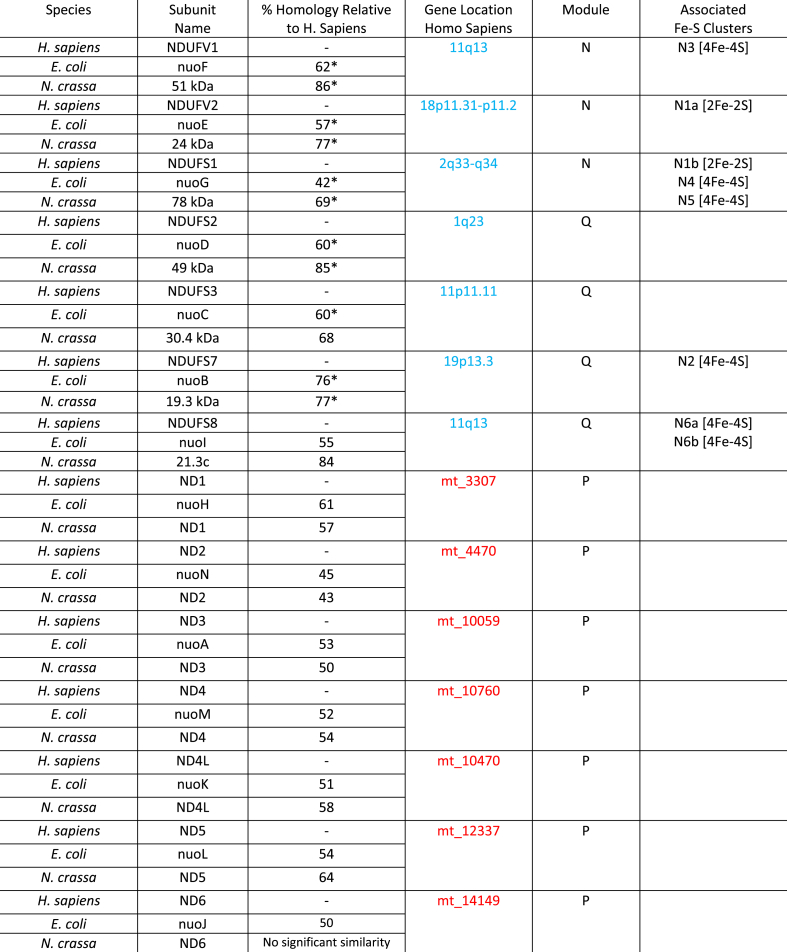
Complex I, also referred to as NADH dehydrogenase, is L-shaped, containing a membrane portion connected to a hydrophilic peripheral portion which protrudes into the mitochondrial matrix (or cytoplasm in bacteria) [[77], [78], [79]]. Although a total of 45 different subunits comprise the mammalian Complex I, a core of 14 subunits is sufficient to drive energy production in the prokaryotic version of the complex [[80], [81], [82]]. These core 14 subunits are highly conserved across both prokaryotes and eukaryotes, and are considered to be the minimal assembly of Complex I [83]. In the mammalian complex, these subunits can be split into two groups, hydrophobic subunits encoded by the mitochondrial genome and hydrophilic subunits encoded by the nuclear genome. These subunits are further subdivided into the three functional modules of Complex I: the electron input/dehydrogenase (N) module that accepts electron via the oxidation of NADH, the electron output/hydrogenase (Q) module that reduces ubiquinone, and the proton translocation (P) module that pumps protons into the IMS [84]. Table 1 summarizes the 14 core subunits found across three species: Homo sapiens, Escherichia coli and Neurospora crassa. The location of the genes encoding each subunit in the human nuclear and mitochondrial genomes, the module where each subunit is found, as well as the presence of any ligated Fe–S clusters are also listed. Note that in E. coli, as with a few other bacteria, two of the core 14 subunits are fused together (nuoC and nuoD), and the complex therefore contains 13 subunits total [85].
Table 1.
Complex I minimal assembly. Subunit names for H. sapiens, E. coli, and N. crassa shown. The location of the genes encoding H. sapiens subunits in the nuclear and mitochondrial genomes are shown in blue and red, respectively. Note that clusters N1a and N5 have not been identified in N. crassa. [[85], [86], [87], [88], [89], [90], [91]] Percent homology values are reported as ‘positives’ (i.e. amino acids that are identical or have similar chemical properties); where multiple (H. sapiens) subunit isoforms exist, percent homology has been averaged across isoforms and indicated with an asterisk.
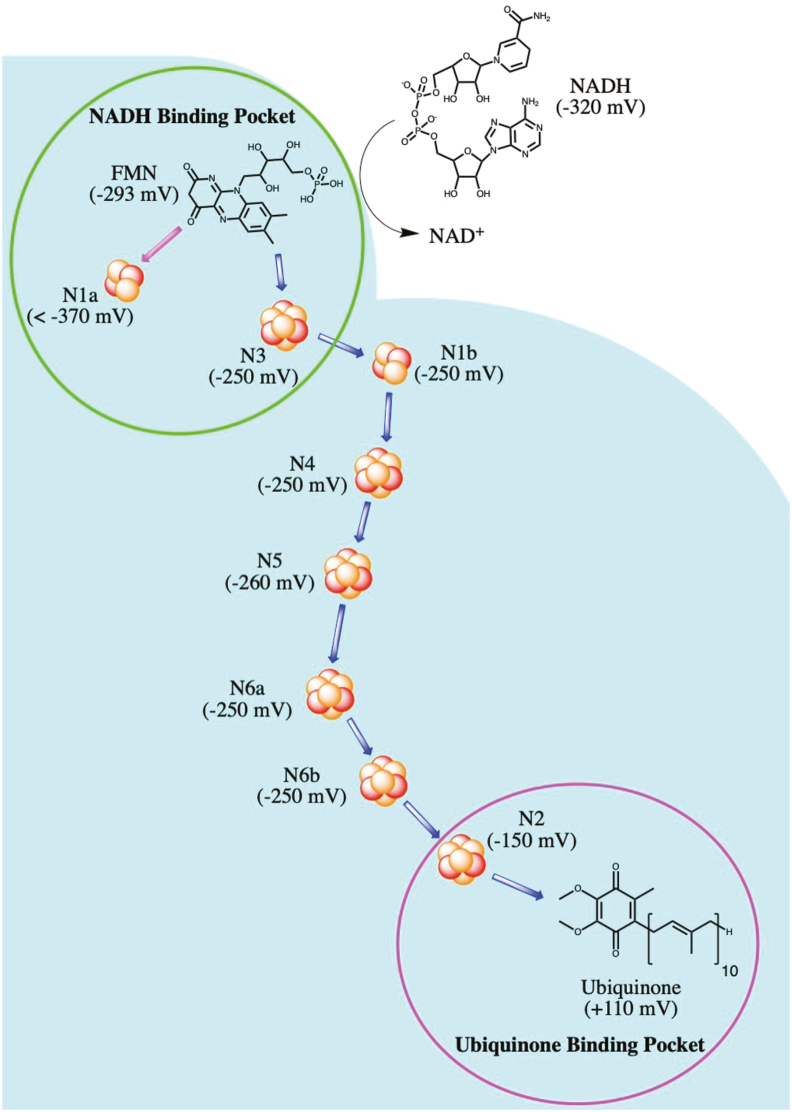
Of the 12 Fe–S clusters found within the ETC complexes, Complex I contains the majority, carrying eight clusters within five of its 14 core subunits. The Fe–S clusters found within Complex I are all located within the peripheral arm, in both the N and Q modules. The cluster chain begins at the NADH binding site near the flavin mononucleotide (FMN), and terminates at the ubiquinone binding site, at the interface of the peripheral and membrane arms. Fig. 5 shows the Fe–S cluster chain within Complex I from B. taurus, with the midpoint redox potentials (a measure of the tendency of a chemical species to gain/lose electrons, with more positive values corresponding to a higher affinity for electrons) at pH 7 (Em7) also shown in parentheses. Of these eight clusters, N1a and N1b are of the [2Fe–2S] form, while the remaining six (N2, N3, N4, N5, N6a and N6b) are of the [4Fe–4S] type. Note that the “N” prefix refers to the Fe–S clusters found within NADH dehydrogenase, whereas the numbering of each cluster corresponds to their spin relaxation rates measured through electron paramagnetic resonance (EPR); higher spin relaxation rates correspond to higher cluster numbers. In some bacteria, including E. coli, the additional N7 Fe–S cluster is found within the nuoG subunit. Based on their positions within the peripheral arm, it is believed that the main pathway of electron movement after transfer from FMN is: N3–N1b-N4-N5-N6a-N6b-N2. Due to the position of cluster N1a (and N7 in bacteria), it is not believed to be directly involved in this pathway. The midpoint redox potential difference between NADH (Em7 = −320 mV) and ubiquinone (Em7 = +110 mV) drives electrons through this Fe–S cluster chain (a distance of more than 95 Å), and provides the energy required to pump protons for ATP production [89].
Fig. 5.
Fe–S chain within Complex I. The midpoint redox potential at pH of 7 (as found in B. taurus) is listed for each cluster, as well as NADH, FMN, and ubiquinone. The NADH and ubiquinone binding sites are depicted in green and magenta circles, respectively, and the main route of electron transfer is depicted by blue arrows. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The N module contains the NADH dehydrogenase [ubiquinone] iron-sulfur protein 1 (NDUFS1), NADH dehydrogenase [ubiquinone] flavoprotein 1 (NDUFV1), and NDUFV2 subunits and is responsible for initiating the transfer of electrons down the chain of Fe–S clusters via NADH and FMN [88]. The first cluster within this chain is N3, found within the N module, ligated to NDUFV1 near its C terminus. The N3 cluster is located 7.6 Å from FMN, which is also non-covalently bound to NDUFV1 [92,93]. N3 receives single electrons from FMN and begins the electron transfer pathway, passing electrons to N1b. This is an intersubunit transfer, as electrons jump from NDUFV1 to NDUFS1, and is sensitive to internal water within the complex: the presence of water increases the rate of transfer by three orders of magnitude [94]. N1b, N4 and N5 are also found within the N module, bound to the NDUFS1 subunit [89,92]. After accepting an electron from N3, electrons from N1b are passed on to N4, then to N5. Unlike the other [4Fe–4S] clusters found within Complex I, N5 is ligated by three cysteines and one histidine, resulting in slightly altered EPR properties [5].
The N1a cluster is the last remaining Fe–S cluster within the N module. This cluster is not believed to be a part of the main electron shuttling pathway due to its position relative to the other clusters, as well as its low redox potential (Em7 < −370 mV) compared to fully reduced FMN, flavohydroquinone (FMNH2, Em7 = −293 mV) [95,96]. However, this cluster is conserved in Complex I across taxa, suggestive of its functional significance. It has been postulated that it may prevent ROS production by reducing the lifetime of reduced FMN species. FMN is situated 7.6 Å from N3 and 11.3 Å from N1a [89]. After accepting two electrons simultaneously from NADH, it is only capable of passing electrons one at a time down the Fe–S cluster chain of Complex I. It has been proposed that N1a may accept an electron from flavosemiquinone (FMNH·, Em7 = −389 mV), shortening the lifetime of this unstable intermediate and possibly preventing electron leakage and ROS production [89]. Recently, N1a has been shown to prevent over-reduction of Complex I by stabilizing bound NAD+, thereby preventing further binding of NADH [97]. The redox state of N1a regulates NADH oxidation in E. coli Complex I by controlling the orientation of a peptide bond close to the NADH binding site. If the rates of N2 cluster oxidation are low due to a predominantly reduced quinone pool, this causes reduction of N1a by FMNH· and a conformational switch within the NADH binding pocket that favours NAD+. This reduced N1a cluster thereby prevents additional binding of NADH and further reduction of the complex, which may provide the complex with an intramolecular feedback mechanism to prevent ROS production [97].
The Q module contains the NDUFS2, NDUFS3, NDUFS7 and NDUFS8 subunits as well as the N6a, N6b and N2 Fe–S clusters [88]. Both the N6a and N6b clusters are ligated within NDUFS8. The N6a cluster is located near the interface of the N and Q modules near the zinc-binding NDUFS6 subunit, and accepts electrons from N5, passing them onwards to N6b and N2 [98]. The longest edge-to-edge distance between clusters within Complex I is the electron transfer from the N to Q modules, between N5 and N6a (14 Å), and is believed to be a rate-limiting step within the complex. Similar to the intersubunit electron transfer between clusters N3 and N1b, the rate of transfer between N6b and N2 is also enhanced by the presence of molecules of water between subunits NDUFS8 and NDUFS7 [94].
The last cluster within the Q module/Complex I is the N2 cluster, ligated by four cysteine residues within the NDUFS7 subunit near its interface with NDUFS2 [92]. This cluster is located ∼12 Å from the ubiquinone binding site, and is responsible for reducing ubiquinone to ubiquinol within the Q module of Complex I, thereby transferring electrons to downstream complexes [99]. In comparison to the other Fe–S clusters in Complex I, N2 exhibits a higher redox potential (Em7 = ∼ −150 mV) in most organisms, including B. taurus, N. crassa, and E. coli, compared to the other clusters within the Complex I cluster chain (Em7 = ∼ −250 mV), making it the electron sink of the complex [89,90,100]. Besides its unusually high redox potential, N2's reduction potential also exhibits pH dependence due to a hydrogen bond formed between a histidine residue in NDUFS2 (His226) and the Fe–S cluster [101]. Remarkably, a similar hydrogen bond with this cluster is also observed in water-soluble [NiFe] hydrogenases in analogous subunits. Mutations at this residue within NDUFS2 to either alanine, glutamine, or cysteine, have shown to reduce the EPR signal of the cluster shifting the Em7 by −80 mV, while only eliciting moderate effects on the catalytic activity of the complex [102]. These findings suggest that the energy conversion mechanism driving proton translocation across the IMM is not associated with one of the electron transfers between the Fe–S clusters upstream but is perhaps linked to the protonation/deprotonations of ubiquinone intermediates. Interestingly, NDUFS2 has been shown to be critical in the assembly and/or stability of the N2 cluster in Complex I, and is also implicated in oxygen-sensing mechanisms within the homeostatic oxygen-sensing system, as discussed in a later section [103].
Complex I is considered to be the main producer of reactive oxygen species within mitochondria [104,105]. The crystal structure of Complex I shows that the majority of redox cofactors within the enzyme are shielded from the solvent and are unlikely to react with molecular oxygen to form ROS. In Complex I, ROS production occurs at either end of the Fe–S chain, at the FMN site (IF) or the quinone binding site (IQ) and can occur when electrons travel in either the forward or reverse directions. Due to the positions of these sites within the peripheral arm, it is believed that Complex I generates ROS exclusively into the mitochondrial matrix [106].
When electrons move in the forward direction, i.e. from FMN to N2, blocking the IQ site via rotenone causes a backlog of electrons in the Fe–S cluster chain, and an over-reduction of the IF site [107]. This over-reduction of Complex I increases the lifetime of FMNH·, causing electrons to leak and produce O2·-. The effects of rotenone on ROS production appears to be tissue-specific, showing different responses in various cell types. Although rotenone increases ROS in renal artery smooth muscle cells (SMC), within the pulmonary artery SMC, rotenone decreases ROS production [108]. This effect is attributed to decreased overall electron flux through the complex and is explored in further detail below.
Complex I can also produce ROS via reverse electron transport (RET) [106]. RET occurs when there is an overabundance of ubiquinol in the Q/QH2 pool due to reduction by electrons from Complex II. When this occurs, electrons can be transferred from ubiquinol to N2. In isolated mitochondria supplemented with succinate, RET can produce NADH from NAD+ via RET through Complex I, however this process is also associated with a significant production of ROS, which is believed to come from the IQ site [109,110]. Unlike the forward direction, RET is dependent on both the redox state of the Q/QH2 pool as well as the proton motive force, with inhibition of Complex I by rotenone (an IQ site inhibitor) or mitochondrial uncoupling by FCCP reducing ROS production via RET.
ROS production from RET is known to be an important pathophysiological mechanism and a major contributor to oxidative stress during heart attack and stroke, via ischaemia-reperfusion (I/R) injury in the heart and the brain, respectively. Ischaemia is characterized by an overabundance of fumarate which causes reversal of succinate dehydrogenase activity and accumulation of succinate. After reperfusion, the accumulated succinate is rapidly oxidized by Complex II causing increased ROS production via RET and initiating oxidative damage [109]. A new generation of antioxidants that are capable of suppressing electron leak specifically from the IQ site have been shown to protect against ischaemia-reperfusion injury in in vitro modeling of cardiac I/R, providing further evidence of RET-induced ROS production during reperfusion and providing new therapeutic leads for protecting against reperfusion induced oxidative damage [111].
3.4. Fe–S clusters in Complex II
Of the remaining four Fe–S clusters outside of Complex I, three are within ETC Complex II, also called succinate dehydrogenase (SDH) or succinate-coenzyme Q reductase. SDH is an IMM bound protein with a hydrophilic head that protrudes into the mitochondrial matrix. SDH serves dual functions in mitochondrial respiration: oxidizing succinate to fumarate in the TCA cycle, and reducing ubiquinone (Q) to ubiquinol (QH2) as a result of electron transfer from FADH2 in the ETC (Fig. 4) [112]. Complex II is the only ETC complex without any mtDNA-encoded subunits, and is comprised of four nuclear-encoded core proteins: the flavoprotein (SDHA) containing a covalently attached flavin adenine dinucleotide (FAD) cofactor, an Fe–S protein (SDHB) housing the clusters required for electron transfer from FADH2 to ubiquinone, and two hydrophobic membrane-spanning subunits (SDHC and SDHD) which anchor the catalytic SDHA-SDHB dimer to the IMM and contain the ubiquinone binding site [113]. Within the SDHB subunit, three Fe–S clusters exist, all in different forms: [2Fe–2S], [4Fe–4S], and [3Fe–4S]. The rhombic cluster is found in the N-terminal domain of SDHB, adjacent to the FAD bound to SDHA, whereas the other two clusters are found within the C-terminal domain [114]. All three clusters are aligned nearly linearly, each spaced less than 14 Å from the other (the distance limit for productive electron transfer) [114,115]. The first cluster in this chain [2Fe–2S] accepts electrons from FAD, passing them through the [4Fe–4S] cluster and finally to [3Fe–4S], located 7.1 Å from the ubiquinone binding site [114].
Mitochondrial SDH also contains a fifth redox prosthetic group, heme b, located between subunits SDHC and SDHD. Although this heme was originally believed to transfer electrons from the [3Fe–4S] group to ubiquinone, the edge-to-edge distance between the [3Fe–4S] cluster and bound ubiquinone is smaller (7.1 Å) than the distance between the cluster and the heme (13.3 Å). Additionally, the redox potential of heme b (Em7 = −185 mV) is much lower than both the [3Fe–4S] cluster (Em7 = +60 mV) and the ubiquinone reduction (Em7 = +110 mV), suggesting transfer from the cluster directly to heme b would be unfavourable [114]. In E. coli Complex II, the heme group is closer to both the [3Fe–4S] cluster and bound quinone, and also has a more comparable redox potential (Em7 = +36 mV) [116]. It is believed that electrons transferred to ubiquinone are in equilibrium with heme b, an effect which may stabilize ubisemiquinone radicals and reduce production of ROS from the enzyme.
Under normal conditions, production of ROS by Complex II is considered to be negligible, although mutated Complex II is known to cause oxidative stress due to ROS production at the flavin site (IIF) [[117], [118], [119]]. In isolated mitochondria, Complex II is capable of producing ROS at IIF, as the FAD within the flavoprotein is known to be a potent site of electron leak [110]. Similar to Complex I, ROS production by Complex II occurs exclusively in the matrix, and this electron leakage can result from electrons traveling in the forward direction after being donated by succinate/FADH2, or in the reverse direction after transfer from ubiquinol [120]. ROS production at site IIF is dependent on the occupancy of the substrate oxidation site, with significant ROS production only occurring in the presence of an open flavin site. Occupiers of this site, including the dicarboxylic acids oxaloacetate and malate, as well as succinate itself, are postulated to bind at site IIF, preventing oxygen from entering and being reduced to produce ROS in vivo [120,121].
3.5. Fe–S clusters in Complex III
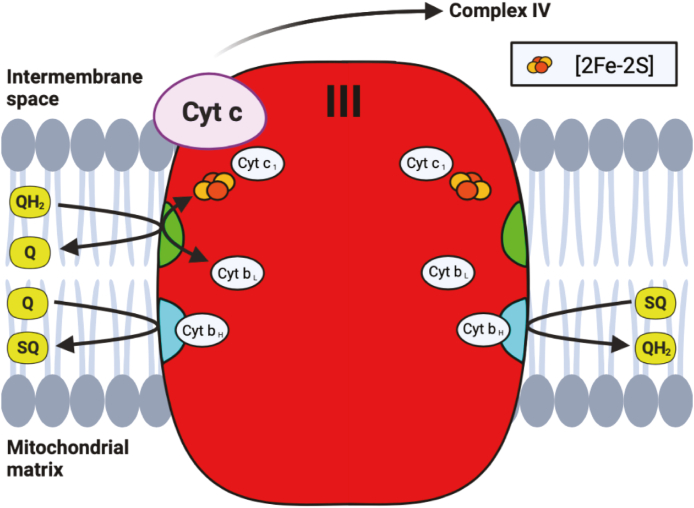
Electrons are eventually shuttled from Complexes I and II via QH2, to Complex III, where the final Fe–S cluster within the ETC is found. Complex III, also called cytochrome bc-1 complex or coenzyme Q: cytochrome c-oxidoreductase, is a homodimer containing three essential redox subunits: cytochrome b, cytochrome c1, and the iron sulfur protein (ISP) containing the sole Fe–S cluster [122]. Although in its simplest form this complex exists with only these three essential subunits, in bovine and humans, this complex includes an additional 8 supernumerary subunits in each monomer which are believed to contributed to the structure's stability [[123], [124], [125]]. At this complex, ubiquinol is oxidized in a bifurcated fashion (commonly known as the Q-cycle) at the Qo site, where two electrons from QH2 are passed through two separate pathways (Fig. 6) [125]. The “low potential pathway” involves electrons passing through two heme units within cytochrome b: a low potential heme cyt bL (Em7 = −30 mV), and a high potential heme cyt bH (Em7 = +100 mV) [125]. This electron will either reduce ubiquinone to ubisemiquinone (SQ), or after a subsequent round of this bifurcated reaction, reduce SQ to QH2 at the quinone reductase site (Qi). This low potential pathway therefore regenerates a molecule of ubiquinol every two rounds of ubiquinone oxidation at the Qo site, with uptake of two protons from the mitochondrial matrix [126].
Fig. 6.
Q-cycle within Complex III. Ubiquinol (QH2) is oxidized at the Qo site (shown in green), passing electrons to either cytochrome c (cyt c) through the [2Fe–2S] cluster within the ISP and cytochrome cl (cyt c1), or to the Qisite (shown in blue) through cyt bL and bH. As shown on the left side of the complex, ubiquinone (Q) is reduced to ubisemiquinone (SQ) at the Qi site after a single round of the Q-cycle, while a second round of the cycle reduces SQ to QH2, as shown on the right side of the complex. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The “high potential pathway” involves electron transfer from ubiquinone at the Qo site to cytochrome c, via the Fe–S cluster within the iron sulfur protein (Em7 = +250 mV) and cytochrome c1 (Em7 = +230 mV) [125]. In contrast to other clusters found within the respiration complexes, the cluster within ISP has a Rieske-type [2Fe–2S] structure, where the iron atoms are coordinated by two cysteine and 2 histidine residues (Fig. 1) [127]. Both ISP and cytochrome c1 are anchored to the IMM via transmembrane helices, and both contain soluble extramembrane domains within the IMS [123,128]. This extramembrane domain of ISP (ISP-ED) contains the Rieske Fe–S cluster within the IMS, and moves between the Qo site and cytochrome c1 to facilitate electron transfer through Complex III [129]. Although substantial conformational change within ISP-ED is required for the movement between these redox centers, the exact mechanism is still unknown, although different models have been proposed to explain its movement [130,131].
Aside from Complex I, Complex III is the other primary site of mitochondrial ROS production. The Qo site is known to be the major source of ROS production by Complex III [110]. The production of ubisemiquinone radicals associated with the bifurcated nature of electron movement at this complex increases its susceptibility to generate ROS. The Qi site inhibitor antimycin A prevents electron transfer from the Qo site, increasing the lifetime of the ubisemiquinone species which readily reacts with oxygen to form O2·- [132]. Inhibition of Complex III by antimycin A also increases the QH2/Q ratio, which can lead to electron backlog in Complexes I and II. Similar to the effects of rotenone on Complex I, the effects of antimycin A on Complex III also appear to be tissue-specific, causing opposing changes in ROS production in renal and pulmonary artery SMC [133]. In the rat lungs and pulmonary artery SMC, antimycin A decreases ROS production, an effect attributed to decrease electron flux through the complex. The tissue-specific effects of proximal ETC inhibitors rotenone and antimycin on ROS production are discussed in more detail in the next section [134]. Unlike both Complexes I and II, Complex III releases O2·- into both the IMS as well as the mitochondrial matrix [135].
4. Fe–S clusters and the homeostatic oxygen-sensing system
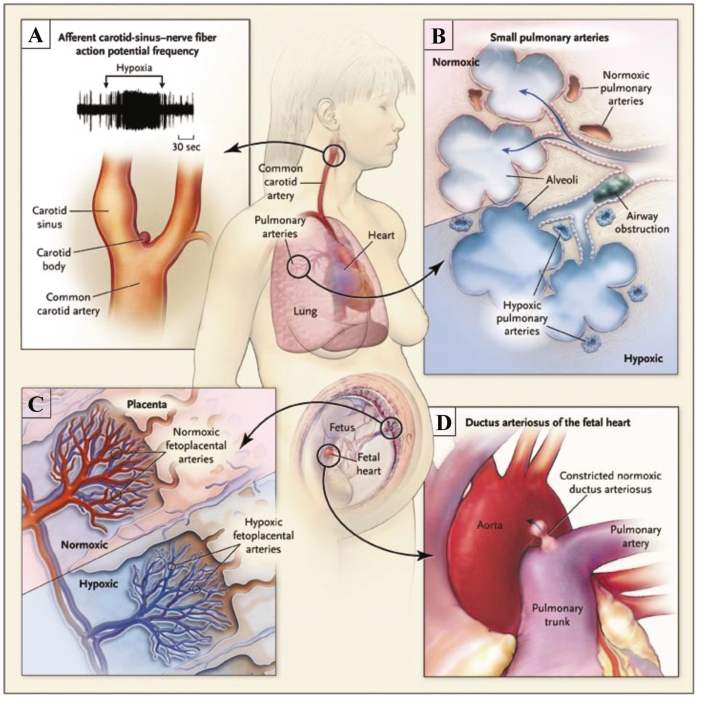
Electron tunneling through the ETC complexes and the associated ROS production that occurs at several distinct sites has been implicated in both pathophysiological diseases and normal physiological processes. Although the sites of electron leak are typically attributed to the flavin sites or quinone binding sites within the respiratory complexes, the role of Fe–S clusters in ROS production and signaling is a novel and emerging area of investigation, and in recent years has been shown to be important within oxygen-sensing systems. Changes in oxygen tension are accompanied by redox changes, with Fe–S clusters being highly susceptible to redox changes and known to function as redox switches within cells [136,137]. This is of particular interest in the highly specialized tissues designed to sense and respond to changes in oxygen. The homeostatic oxygen-sensing system (HOSS), outlined in Fig. 7, is comprised of specialized tissues throughout the body, specifically adapted to sense small changes in both arterial oxygen content and airway oxygen levels [138]. In response to changes in environmental oxygen levels or localized changes in pO2, these tissues can elicit changes in vascular tone, respiration, and neurosecretion [139]. The chromaffin cells of the fetal adrenal medulla, the type 1 cells within the carotid body, as well as the neuroepithelial bodies within the lungs, are all members of the HOSS which respond to changes in oxygen via transmitter release. Also included within HOSS are the smooth muscle cells (SMC) of the fetoplacental arteries, the ductus arteriosus, and the systemic and resistance pulmonary arteries (PAs), all of which respond to changes in oxygen content by modulating vascular tone [138,140]. In all of these tissues, the cellular response to changes in oxygen tension are mediated by changes in ROS production, largely by the mitochondria. The modulation of redox sensitive K+ channels causes changes in membrane polarization and influences Ca2+ entry into the cell [[140], [141], [142]]. This is followed either by changes in vascular tone or secretion of transmitters, depending on the tissue type. (See Fig. 8).
Fig. 7.
The specialized tissues of the Homoeostatic Oxygen Sensing System (HOSS). A. The carotid body, (located at the bifurcation of the carotid artery) increases the frequency of action potentials upon exposure to hypoxia. B. Pulmonary arteries constrict in hypoxia to divert blood to regions of the lung with better oxygenation (ventilation-perfusion matching). C. Hypoxic Fetoplacental vasoconstriction optimizes maternal and fetal perfusion matching. D. The ductus arteriosus rapidly constricts in response to nomioxia postnatally, properly separating the pulmonary and systemic circulations. Adapted with permission from [138] Weir et al. (2005) NEJM.
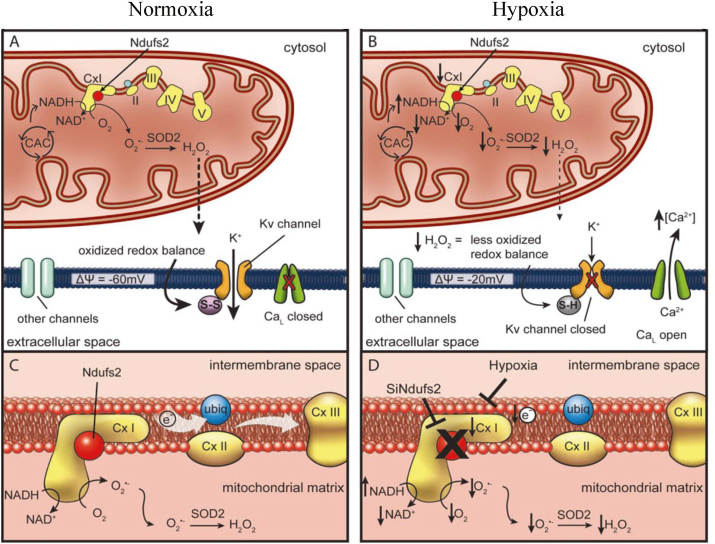
Fig. 8.
PASMC mitochondria during normoxia and hypoxia. A. Under normoxic conditions, oxygen radicals produced at ETC Complex I are converted to H2O2 by SOD2, with this ROS production leading to oxidation of sulfhydryl groups in the Kv channel, forming disulfide bridges (S–S) and increasing the channel's open-state probability. B. With a switch to hypoxia, production of superoxide is decreased due to decreased uncoupled electron transport. The resultant decrease in H2O2 and the accumulation of NADH depolarize the cell, closing the Kv channels, with subsequent opening of calcium channels causing vasoconstriction. C. Optimal function of Complex I requires intact Ndufs2. D. Inhibition of Complex I, via hypoxia or siRNA targeting Ndufs2, results in a more reduced redox state. Adapted with permission from [173] Dunham-Snary et al. (2019) Circ. Res.
In response to hypoxia, the pulmonary arteries constrict, whereas the renal arteries (RAs) dilate. In a study by Michelakis et al., diversity in mitochondrial function was proposed as a possible explanation for the observed differences in PAs and RAs responding to hypoxia. Several differences are observed in mitochondria between these tissues, with lung mitochondria being more depolarized, having lower oxygen consumption rates, and producing more ROS at baseline [108]. Proximal ETC inhibitors, like rotenone (an inhibitor of Complex I) and antimycin A (an inhibitor of Complex III), were found to mimic hypoxia in both tissues, causing inhibition of K+ channels in PAs and activation of K+ channels in RAs. Interestingly, other modulators of vascular tone in these vessels, such as endothelin-1 and 4-AP (a Kv blocker), constrict both vessel types, suggesting the mechanism behind the opposing effects of hypoxia on both tissue types may reside in the mitochondrial ETC, rather than in downstream ion channels [108]. These effects of hypoxia on these vessels were also observed in the absence of endothelium, suggesting the sensor of oxygen may reside within the SMC themselves.
As electrons flow through the IMM, electron leakage occurs at several sites and upon exiting the ETC, electrons can combine with molecular oxygen to produce O2·- radicals [143]. These anions are quickly converted into H2O2, a species with a larger diffusion radius, via the actions of superoxide dismutase [144]. H2O2 is a well-established signaling molecule, and has the ability to cause numerous structural/functional changes within proteins, including oxidizing methionine residues, and forming disulfide bridges via oxidation of cysteine residues [145]. Among these redox-sensitive proteins are ion channels, such as the potassium channel Kv1.5; changes in redox state of the cell changes the open state probability of the K+ channel, thus controlling membrane potential and in-turn modulating certain voltage-sensitive channels (e.g. L-type Ca2+ channels) [146,147]. Redox changes also modulate activation of Rho-kinase which, when active, maintains SMC constriction independent of intracellular calcium [138,148,149]. In this way, it is believed that alterations in electron leak and ROS production may provide a sensing mechanism whereby changes in rates of mitochondrial respiration (dependent on oxygen availability), alter the redox status of the cell, and modulate downstream ion channels. Recent findings suggest that ROS production from Fe–S clusters plays an important role in redox signaling within oxygen sensitive tissues. These changes in ROS production may also alter the Fe–S clusters themselves, with reversible redox changes to Fe–S clusters potentially playing a vital role in sensing changes in oxygen tension. The role of Fe–S clusters and the effects of ROS signaling in the fetoplacental arteries, carotid body, pulmonary arteries, and ductus arteriosus will be explored in the following sections.
4.1. Oxygen-sensing in fetoplacental arteries
The fetoplacental arteries are highly sensitive to changes in oxygen tension [150]. The placental cotyledons contain the fetal capillaries and are surrounded by maternal blood, thus serving as the site of exchange for nutrients and oxygen from the maternal blood to fetal circulation. Fetoplacental arteries constrict under hypoxic conditions as a result of hypoxic inhibition of Kv channels, similarly to the PAs [151]. Distribution of maternal blood flow is variable amongst placental cotyledons, with hypoxic fetoplacental artery vasoconstriction acting to divert fetal blood flow to better perfused cotyledons to increase efficiency of placental oxygen exchange [151,152]. The role of mitochondria and mitochondrial Fe–S clusters in fetoplacental artery oxygen-sensing requires further investigation. However, while the acute placental response to oxygen has yet to be studied, chronic hypoxia of the placenta from under-perfusion is a key pathologic feature of pre-eclampsia [153,154]. Pre-eclampsia occurs in 3–8% of all pregnancies [155]; it can present as a maternal syndrome (hypertension and proteinuria), or as a fetal syndrome (fetal growth restriction, abnormal oxygenation, reduced amniotic fluid), and is associated with increased maternal and perinatal morbidity and mortality [153,155,156]. Pre-eclampsia occurs more frequently at high altitudes [157,158], with high altitude pregnancies having lower levels of Complex I and ISCU in placental tissue [159]. Additionally mitochondrial dysfunction has been reported in pre-eclampsia, with decreased expression of ISCU [153]: the transcription factor HIF-1α (hypoxia inducible factor 1α) acts to decrease ISCU via the HIF-1a-responsive miR210 [153,159]. Though the role of mitochondrial Fe–S clusters in the placental oxygen response has yet to be studied, the modulation of an essential Fe–S cluster assembly pathway component by hypoxia suggests that the role of Fe–S clusters in placental oxygen-sensing is a valuable area of further investigation.
4.2. Oxygen-sensing in the carotid body
Acute oxygen-sensing by chemoreceptors within the carotid body is essential for mammalian adaptations to changing pO2 levels, as occurs in response to disease and to changes in atmospheric oxygen supply with altitude. In response to decreased blood oxygen, activation of chemoreceptors within the carotid body glomus cells mediates cardiorespiratory reflexes, causing both sympathetic activation and hyperventilation. Within the glomus cells, O2-sensitive K+ channels close in response to hypoxia, causing subsequent cell depolarization and release of neurotransmitters which activate sensory fibers that terminate at the respiratory center within the brain stem [146,160,161]. Fernández-Agüera et al. demonstrated that ablation of the Ndufs2 gene within the carotid body glomus cells of mice results in the loss of the hypoxic ventilatory response, while maintaining responses to other stimuli such as hypercapnia and hypoglycemia [162]. Using this same model of knockout mice, deletion of Ndufs4 was observed to reduce Complex I activity, while maintaining normal cellular responses to hypoxia, suggesting an important role for Ndufs2 and ubiquinone oxidoreductase activity in oxygen-sensing within glomus cells. In this model of Ndufs2 deficient mice, glomus cells exhibited increased levels of NADH and ROS, suggesting the loss of the hypoxia response within these cells may be a result of alterations in mitochondrial homeostasis, rather than a consequence of Complex I dysfunction specifically. To address this, Ndufs2-null cells were supplemented with pyruvate and α-ketoglutarate to regenerate NAD+, as well as exogenous succinate to bolster the Complex II through Complex IV pathway. Despite these supplements, these cells were still found to exhibit an almost complete absence of hypoxic response, suggesting an essential role of Complex I derived NADH and ROS in acute oxygen-sensing [163]. This same group also found that in response to hypoxia, glomus cells showed an increase in ROS production in the IMS and a decrease in ROS production within the mitochondrial matrix. One could speculate that this may be the result of O2·- being electrostatically channeled into the IMS by its positive potential, causing the subcellular compartmentalization observed [163]. The authors suggest that during hypoxia, an increase in ratio of reduced to oxidized ubiquinone may cause decrease rates of electron flux through Complex I, possibly increasing the lifetime of the reduced form of the N2 cluster, causing increased electron leak and O2·- production. Although not speculated by the authors, these findings may also suggest that alterations in the redox midpoint potential of the N2 cluster due to the absence of the hydrogen bond formed between the cluster and Ndufs2 may cause less controlled movement of electrons through the Fe–S chain, possibly increasing the chances of leakage and ROS production.
4.3. Oxygen-sensing in the pulmonary artery
Oxygen-sensing within the pulmonary arteries tightly matches ventilation to perfusion, optimizing oxygen uptake and systemic delivery [138]. In response to hypoxia, small resistance pulmonary arteries constrict, diverting blood away from poorly ventilated portions of the lung and towards better oxygenated portions, optimizing overall oxygen uptake [164]. This hypoxic pulmonary vasoconstriction (HPV) mechanism is intrinsic to the SMC of the resistance pulmonary arteries, and similar to other tissues included in the HOSS, involves the coordination of redox and voltage sensitive K+ and Ca2+ ion channels. In response to hypoxia, inhibition of K+ channels leads to cell depolarization, Ca2+ uptake through large conductance voltage sensitive channels, and subsequent vasoconstriction in response to increased intracellular Ca2+ [165]. Although like other members of the HOSS the sensor responsible for this cascade is believed to reside in the mitochondria, the exact mechanism of oxygen-sensing, as well as the influence of hypoxia on ROS production within pulmonary artery SMC (PASMC), is still a matter of debate [133,[166], [167], [168], [169], [170], [171], [172]]. Similar to the findings in the carotid body glomus cells, a recent study by our group has shown that the HPV response within PASMC is also dependent on Ndufs2 [173]. Ndufs2 expression was observed to be greater in rat PASMC in comparison to renal artery SMC, providing a possible reasoning for the discrepancy in oxygen-sensing abilities between these cell types as described by Michelakis et al. [108]. In isolated PASMC treated with siRNA to knockdown Ndufs2, the hypoxia induced rise in intracellular Ca2+ was not observed, whereas responses to other stimuli such as hyperkalemia remained intact. siNdufs2 was also observed to mimic aspects of chronic normoxia, including decreased Complex I activity, elevated NADH/NAD+ ratios, and decreased expression of the O2-sensitive Kv1.5 channel. Nebulization of siNdufs2 resulted in partial knockdown of subunit expression (∼35% knockdown in whole lung homogenate), but completely eliminated HPV in a rat model. Similarly, an attenuated HPV response was observed in mice treated with siNdufs2 as measured using intravital microscopy and confocal imaging of excised lungs. Collectively, these in vitro and in vivo findings suggest a key role for this subunit in the oxygen-sensing pathway within the resistance pulmonary arteries. As in the carotid body, these findings further suggest an importance of the interaction between Ndufs2 and the N2 Fe–S cluster, and suggest that the redox midpoint potential of this cluster is vital for maintaining normal oxygen signaling pathways through the control of ROS production from Complex I.
4.4. Oxygen-sensing in the ductus arteriosus
Oxygen-sensing in the ductus arteriosus (DA) is crucial for circulatory transition into neonatal life. The ductus arteriosus is a fetal vessel connecting the pulmonary artery and aorta, diverting blood in the pulmonary circulation, oxygenated by the placenta, away from the developing lungs to the systemic circulation. The DA rapidly constricts in response to the changes in the arterial partial pressure of oxygen (PaO2) that occur with the transition from the hypoxic fetal environment (PaO2 < 40 mmHg) to the normoxic neonatal environment (PaO2 ∼100 mmHg) [174]. The immediate constriction of the DA, occurring within minutes of birth, and permanent closure, which typically occurs within 72 h, is crucial for proper separation of the pulmonary and systemic circulation [175,176]. Failure of DA closure is a common complication of extreme preterm birth and can have serious adverse outcomes if not treated [174,177]. DA response to oxygen occurs via an oxygen-induced increase in mitochondrial ROS in DA smooth muscle cells, with ROS inhibiting voltage-gated potassium channels (Kv) [[178], [179], [180]]. Kv channel inhibition leads to cell depolarization, opening of L-type Ca2+ channels, and smooth muscle cell constriction [181]. Mitochondrial ROS are mainly produced by Complexes I and III of the ETC, with inhibition of these complexes inhibiting DA response to oxygen without affecting constriction induced by vasoconstrictors [182]. A recent transcriptomic study by Bentley et al. [183] explored the gene expression profiles of human DA smooth muscle cells under hypoxia and following 96 h oxygen exposure. This study found that, among genes significantly differentially expressed between the oxygen condition, mitochondrial pathways were highly enriched [183]. Furthermore, 20 genes from Complex I were significantly upregulated by oxygen exposure [183]. While transcriptomic data cannot shed light on the mechanistic pathways involved, these findings point to a role of Complex I in the oxygen response of DA smooth muscle cells. The specific role of Complex I subunits and Fe–S clusters is a promising area that requires further investigation as the DA is a historically under researched model of oxygen-sensing.
5. Fe–S clusters as emerging targets of therapeutics
Fe–S clusters play important roles in a variety of cellular functions, and as such have increasingly been of interest in drug research, either as potential therapeutic targets or as the source of drug toxicity that can be mitigated. Fe–S clusters can be the subject of therapeutic targeting in three primary domains: targeting of Fe–S clusters within a protein, targeting Fe–S cluster biogenesis, and drug-induced ROS production modulating Fe–S clusters (reviewed in detail by Vernis et al. [184]). Fe–S clusters frequently play a critical role in the catalytic subunits of proteins and are highly sensitive to redox changes, making them useful potential targets that could have large effects [136]. Fe–S clusters are particularly sensitive to redox changes within the cell, and play important roles serving as redox switches, sensing changes in ROS [136,137]. Redox changes can cause proteins to lose their cluster, interconvert between [4Fe–4S] and [2Fe–2S], or change the redox state of the cluster by giving or receiving an electron [137]. Proteins using Fe–S redox switches are found in bacteria, yeast, and mammals, and are involved in a variety of functions, including: enzyme protection, Fe–S cluster transfer/repair (mitoNEET), DNA repair, and regulation of gene expression [137]. MitoNEET is an Fe–S protein anchored to the outer mitochondrial membrane [185]. MitoNEET, a homodimer with a [2Fe–2S] cluster in each monomer, plays a role in Fe–S cluster shuttling and in redox reactions [[185], [186], [187]]. As a general rule, when the Fe–S cluster is reduced, the protein is in a dormant state, with a switch to active state occurring when a signal induces oxidation of the cluster [9,137,188].
Drugs acting on Fe–S targets either directly or indirectly have been shown to be effective in a wide variety of treatments (outlined in Table 2), from anticancer drugs to antibacterial/antiparasitic drugs. One such drug is β-phenethyl isothiocyanate (PEITC), a natural anticancer product highly effective against human leukemia [189]. PEITC increases production of ROS with depletion of mitochondrial antioxidant glutathione [189]. At least part of the anticancer activity of PEITC is due to this increase in ROS degrading the Fe–S center of NDUFS3 of ETC Complex I [184,189].
Table 2.
Summary of drugs targeting Fe–S clusters and their mechanisms of action.
| Drug | Target | Effect |
|---|---|---|
| PEITC | Glutathione | Glutathione depletion increases ROS, degrading Fe–S cluster of NADH Dehydrogenase 3 [189] |
| Cluvenone | MitoNEET & NAF-1 | Cluvenone-derivative MAD28 destabilizes Fe–S clusters of mitoNEET and NAF-1, inhibiting cell proliferation, targeting cancer cells [[190], [191], [192], [193]] |
| Pioglitazone | MitoNEET | Stabilizes Fe–S clusters and protects from damaging effects of NADP(H) binding [[194], [195], [196]] |
| Primaquine | Aconitase & Rli1 | Targeting Fe–S clusters decreases activity of aconitase and RNase L inhibitor (Rli1), with inhibitory effect on respiration and growth of malaria parasites [197] |
| l-serine + fluoroquinolones | Bacterial Fe–S clusters | Increases ROS production, destabilizing Fe–S clusters. Combination of l-serine with ofloxacin or moxifloxacin increases bactericidal efficiency [[198], [199], [200]] |
| ‘882 | SUF biosynthesis complex | Inhibition of bacterial Fe–S biosynthesis (SUF), with subsequent decrease in aconitase activity [201] |
| – | SUF | Malaria parasites necessitate Fe–S clusters as cofactors: targeting SUF Fe–S cluster biosynthesis for potential anti-malarial drugs [[202], [203], [204], [205]] |
| Doxorubicin | Increased ROS | Cardiac toxicity of the anticancer drug results from iron-mediated mitochondrial ROS production [[206], [207], [208], [209]]. Adjuvant therapies can modulate adverse effects [[210], [211], [212]] |
Cluvenone is a mitochondria-targeted molecule, which has demonstrated anticancer properties and decent tumor selectivity [213]. MAD-28, a cluvenone derivative, acts by binding and destabilizing two [2Fe–2S] proteins overexpressed in several cancer lines: mitoNEET (mitochondria), and nutrient deprivation autophagy factor-1 (NAF-1, endoplasmic reticulum) [[190], [191], [192]]. MAD-28 destabilizes the Fe–S clusters by breaking the coordinative bond between the histidine ligand and the iron of the mitoNEET and NAF-1 Fe–S clusters [190,193]. Due to this cluster destabilization, MAD-28 strongly inhibits cell proliferation and has high specificity in targeting cancer cells [190,193,214,215].
In addition to its overexpression in cancer lines, mitoNEET has been implicated in diabetes and is the target of the thiazolidinedione class of insulin-sensitizing drugs such as pioglitazone [196,216,217]. MitoNEET primarily interacts with NADP(H), with NADP(H) binding destabilizing the Fe–S cluster and facilitating cluster release [195]. Pioglitazone acts to enhance the stability of mitoNEET Fe–S clusters [194], binding with a much higher affinity than NADP(H) and protecting the cells from damaging effects [195].
There are a number of anti-bacterial and antiparasitic drugs that function through Fe–S cluster targeting. Fe–S clusters are targeted by the antimalarial drug Primaquine, as shown in yeast [197]. Primaquine decreases the activity of aconitase and RNase L inhibitor (Rli1), both of which are sensitive to oxidative damage due to their reliance on Fe–S clusters for activity [197]. Rli1 is an essential protein involved in a variety of cellular processes, including ribosome biogenesis [218] and translation initiation and termination [219,220]. The Fe–S clusters of aconitase and Rli1 are proposed to be the primary target of primaquine in vivo, with the growth inhibitory effect reliant on respiration and the subsequent ROS production [197].
Another antibacterial treatment of interest is the combination of l-serine with the two fluoroquinolones ofloxacin or moxifloxacin, with combined treatment having higher bactericidal efficiency regardless of growth phase [184,198,199]. These effects are due to an increase in ROS production and rapid disruption of Fe–S clusters, though whether the clusters are directly targeted in addition to their redox-mediated disruption was not explored [200].
The examination of novel antimicrobial strategies has become increasingly vital as antibiotic resistance emerges. This was the focus of a study by Choby et al. [201] where the attempt to circumvent antimicrobial resistance in Staphylococcus aureus strains identified a new molecule named ‘882. The toxicity of ‘882 was due to its inhibition of the sulfur mobilization (SUF) biosynthesis complex. The SUF operon encodes six major proteins that function in Fe–S cluster assembly (SufB, SufC, SufD), with sulfur mobilization from cysteine mediated by SufE and SufS, and with the use of carrier protein SufA [44,[221], [222], [223], [224]]. ‘882 physically interacts with the SUF Fe–S cluster synthesis machinery and subsequently decreases the activity of aconitase, an Fe–S cluster-dependent enzyme [201].
The SUF Fe–S synthesis pathway has also been suggested as a promising target in malaria parasites. Malaria parasites contain a plastid organelle called the apicoplast which “harbors biochemical pathways of prokaryotic origin” [202], thought to have arisen from a separate endosymbiotic event [225]. The apicoplast is necessary for survival of malaria parasites during the liver and blood stages of development [203]. Apicoplast enzymes are predicted to necessitate Fe–S cluster cofactors [204,[226], [227], [228], [229]] and the SUF pathways play a crucial role in maintaining the apicoplast [203], thus the SUF pathway is a promising target for potential antimalarial treatments. This idea has been extended to suggestions that the SUF pathway and apicoplast proteins are promising drug targets for anti-parasitic drugs against apicomplexan parasites in general, as these proteins are “among the most divergent relative to the mammalian proteins” [205] and thus could likely be tolerated by humans. This target/pathway is included in Table 2, though a named drug has yet to be developed.
The targeting of Fe–S clusters has also been found as the cause for adverse outcomes from certain drugs. For example, doxorubicin is an anticancer drug of the anthracycline family [230] that can also lead to development of cardiac toxicity and cardiomyopathy [231,232]. The accumulation of doxorubicin in mitochondria can initiate mitochondrial production of ROS and RNS [233], activating apoptotic pathways [209,234]. Additionally, it has been proposed that an increase in ROS is induced by doxorubicin in an iron-mediated pathway [[206], [207], [208],235]. Doxorubicin functions as an iron chelator, with the resultant complex between iron and the drug catalyzing conversion of hydrogen peroxide to hydroxyl radicals [208]. With a cardioprotective adjuvant treatment such as dexrazoxane, which chelates free iron [210], the doxorubicin-induced depletion of mitochondrial DNA due to oxidative stress can be avoided [211]. Other mitochondrial targeted antioxidants, mitoTEMPOL and MitoQ, were effective in rodents at preventing cardiac injury while maintaining the anti-tumor efficacy of doxorubicin [212]. By understanding the mechanism of the adverse drug outcome, a targeted adjuvant therapy can prevent cardiotoxicity while not interfering with the anti-cancer efficacy of doxorubicin.
6. Conclusions
Fe–S clusters are multi-functional protein prosthetic groups highly conserved between bacteria and eukaryotes. In the mitochondria, Fe–S clusters play essential roles in the enzyme aconitase, while also being integral to electron transfer within the respiratory complexes. During mitochondrial respiration, electron transfer through these clusters is associated with ROS production at several distinct sites with Complexes I, II and III. These ROS act as cell signaling molecules, acting within oxygen-sensitive tissues to mediate changes in vascular tone and respiration. Cluster N2 of Complex I has been identified as a potential redox-sensitive oxygen sensor, regulating the production of ROS to initiate cell-signaling cascades within the pulmonary artery and carotid body. Fe–S containing proteins are also suggested to be transcriptionally-regulated mediators in fetal vascular development and are emerging as therapeutic targets in the treatment of diabetes, malaria, and cancer. Numerous questions regarding the role of Fe–S centers in health and disease still remain. For example, it is not known if there is a biochemical threshold for Complex I substrates (i.e., NADH) in oxygen-sensing tissues. Dunham-Snary et al. demonstrated that PASMC exposed to chronic hypoxia (to mimic group 3 pulmonary hypertension) lowered expression of NDUFS2, reduced Complex I activity, and increased the ratio of NADH/NAD+ [173]. Further investigation is required to determine if this relationship is bidirectional, and if NADH deprivation would inhibit oxygen-sensing in PASMC. Further investigation is also warranted to determine the effect of mitochondrial dysfunction due to mitochondrial disease on oxygen-sensing. Jain et al. reported that chronic hypoxia exposure activates the HIF signaling cascade, resulting in improved survival and reduced disease severity in a mouse model of Leigh syndrome, one of the most common mitochondrial diseases [236]. However, it remains unknown whether impaired acute oxygen-sensing is linked to mitochondrial diseases, and what HOSS tissues (if any) are affected. With the central role of Fe–S clusters in mitochondria, future investigations into potential links between mitochondrial diseases, acute oxygen-sensing, and Fe–S assembly/function/dysfunction could reveal new therapeutic avenues.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by a Queen’s University Research Initiation Grant (389308; KDS), the Garfield Kelly Cardiovascular Research & Development Fund (374166; KDS), and the Queen’s University Department of Medicine and Translational Institute of Medicine (374180; KDS). Figures were created using BioRender and ChemDraw.
References
- 1.Burén S., Jiménez-Vicente E., Echavarri-Erasun C., Rubio L.M. Biosynthesis of nitrogenase cofactors. Chem. Rev. 2020;120(12):4921–4968. doi: 10.1021/acs.chemrev.9b00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo Y., Ergenekan C.E., Fischer J.T., Tan M.L., Ichiye T. The molecular determinants of the increased reduction potential of the rubredoxin domain of rubrerythrin relative to rubredoxin. Biophys. J. 2010;98(4):560–568. doi: 10.1016/j.bpj.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ewen K.M., Ringle M., Bernhardt R. Adrenodoxin–a versatile ferredoxin. IUBMB Life. 2012;64(6):506–512. doi: 10.1002/iub.1029. [DOI] [PubMed] [Google Scholar]
- 4.Ali M.E., Nair N.N., Retegan M., Neese F., Staemmler V., Marx D. The iron–sulfur core in Rieske proteins is not symmetric. JBIC Journal of Biological Inorganic Chemistry. 2014;19(8):1287–1293. doi: 10.1007/s00775-014-1185-7. [DOI] [PubMed] [Google Scholar]
- 5.Ohnishi T. Iron–sulfur clusters/semiquinones in Complex I, Biochimica et Biophysica Acta (BBA) Bioenergetics. 1998;1364(2):186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 6.Yankovskaya V., Horsefield R., Törnroth S., Luna-Chavez C., Miyoshi H., Léger C., Byrne B., Cecchini G., Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299(5607):700. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 7.Robbins A.H., Stout C.D. The structure of aconitase. Proteins. 1989;5(4):289–312. doi: 10.1002/prot.340050406. [DOI] [PubMed] [Google Scholar]
- 8.Lanz N.D., Booker S.J. Auxiliary iron-sulfur cofactors in radical SAM enzymes. Biochim. Biophys. Acta. 2015;1853(6):1316–1334. doi: 10.1016/j.bbamcr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi K., Fujikawa M., Kozawa T. Oxidative stress sensing by the iron-sulfur cluster in the transcription factor, SoxR. J. Inorg. Biochem. 2014;133:87–91. doi: 10.1016/j.jinorgbio.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Kiley P.J., Beinert H. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 1998;22(5):341–352. doi: 10.1111/j.1574-6976.1998.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 11.Hentze M.W., Kühn L.C. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 1996;93(16):8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White M.F. Structure, function and evolution of the XPD family of iron-sulfur-containing 5'-->3' DNA helicases. Biochem. Soc. Trans. 2009;37(Pt 3):547–551. doi: 10.1042/BST0370547. [DOI] [PubMed] [Google Scholar]
- 13.Stiban J., Farnum G.A., Hovde S.L., Kaguni L.S. The N-terminal domain of the Drosophila mitochondrial replicative DNA helicase contains an iron-sulfur cluster and binds DNA. J. Biol. Chem. 2014;289(35):24032–24042. doi: 10.1074/jbc.M114.587774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariotti L., Wild S., Brunoldi G., Piceni A., Ceppi I., Kummer S., Lutz R.E., Cejka P., Gari K. The iron-sulphur cluster in human DNA2 is required for all biochemical activities of DNA2. Commun Biol. 2020;3(1):322. doi: 10.1038/s42003-020-1048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White M.F., Dillingham M.S. Iron–sulphur clusters in nucleic acid processing enzymes. Curr. Opin. Struct. Biol. 2012;22(1):94–100. doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Baranovskiy A.G., Siebler H.M., Pavlov Y.I., Tahirov T.H. Iron-sulfur clusters in DNA polymerases and primases of eukaryotes. Methods Enzymol. 2018;599:1–20. doi: 10.1016/bs.mie.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saikrishnan K., Yeeles J.T., Gilhooly N.S., Krajewski W.W., Dillingham M.S., Wigley D.B. Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex. EMBO J. 2012;31(6):1568–1578. doi: 10.1038/emboj.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altmann R. Die Elementarorganismen und ihre Beziehungen zu den Zellen. Veit. 1894 [Google Scholar]
- 19.Kölliker A. Zur Kenntnis der quergestreiften Muskelfasern. Wilh. Engelmann. 1888 [Google Scholar]
- 20.Wallin I.E. Williams & Wilkins Company; Baltimore: 1927. Symbionticism and the Origin of Species. [Google Scholar]
- 21.Sagan L. On the origin of mitosing cells. J. Theor. Biol. 1967;14(3):255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 22.Margulis L. Yale University Press; 1970. Origin of Eukaryotic Cells: Evidence and Research Implications for a Theory of the Origin and Evolution of Microbial, Plant and Animal Cells on the Precambrian Earth. [Google Scholar]
- 23.Margulis L. 1981. Symbiosis in cell evolution: life and its environment on the early earth. [Google Scholar]
- 24.Lederberg J. Cell genetics and hereditary symbiosis. Physiol. Rev. 1952;32(4):403–430. doi: 10.1152/physrev.1952.32.4.403. [DOI] [PubMed] [Google Scholar]
- 25.Ris H., Plaut W. Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas. J. Cell Biol. 1962;13:383–391. doi: 10.1083/jcb.13.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson E.B. The cell in development and heredity. Macmillan. 1925 [Google Scholar]
- 27.Haldane J.B.S. In: New Biology, No. 16. Johnson M.L., Abercrombie M., Fogg G.E., editors. Penguin Books; London: 1954. The Origins of Life. [Google Scholar]
- 28.Gray M.W., Doolittle W.F. Has the endosymbiont hypothesis been proven? Microbiol. Rev. 1982;46(1):1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavalier-Smith T. The origin of nuclei and of eukaryotic cells. Nature. 1975;256(5517):463–468. doi: 10.1038/256463a0. [DOI] [PubMed] [Google Scholar]
- 30.Dodson E.O. Crossing the prokaryote–eucaryote border: endosymbiosis or continuous development? Can. J. Microbiol. 1979;25(6):651–674. doi: 10.1139/m79-096. [DOI] [PubMed] [Google Scholar]
- 31.Doolittle W.F. Evolutionary molecular biology: where is it going? Can. J. Biochem. 1982;60(2):83–90. [PubMed] [Google Scholar]
- 32.Taylor F. Autogenous theories for the origin of eukaryotes. Taxon. 1976:377–390. [Google Scholar]
- 33.Baum D.A. A comparison of autogenous theories for the origin of eukaryotic cells. Am. J. Bot. 2015;102(12):1954–1965. doi: 10.3732/ajb.1500196. [DOI] [PubMed] [Google Scholar]
- 34.Jékely G. Origin of eukaryotic endomembranes: a critical evaluation of different model scenarios. Eukaryotic Membranes and Cytoskeleton. 2007:38–51. doi: 10.1007/978-0-387-74021-8_3. [DOI] [PubMed] [Google Scholar]
- 35.Yang D., Oyaizu Y., Oyaizu H., Olsen G.J., Woese C.R. Mitochondrial origins. Proc. Natl. Acad. Sci. U. S. A. 1985;82(13):4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z., Wu M. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci. Rep. 2015;5:7949. doi: 10.1038/srep07949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray M.W., Spencer D.F. Cambridge University Press; 1996. Evolution of Microbial Life. [Google Scholar]
- 38.Fitzpatrick D.A., Creevey C.J., McInerney J.O. Genome phylogenies indicate a meaningful alpha-proteobacterial phylogeny and support a grouping of the mitochondria with the Rickettsiales. Mol. Biol. Evol. 2006;23(1):74–85. doi: 10.1093/molbev/msj009. [DOI] [PubMed] [Google Scholar]
- 39.Roger A.J., Muñoz-Gómez S.A., Kamikawa R. The origin and diversification of mitochondria, Curr. Biol. 2017;27(21):R1177–R1192. doi: 10.1016/j.cub.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 40.Dunn C.D. Some liked it hot: a hypothesis regarding establishment of the proto-mitochondrial endosymbiont during eukaryogenesis. J. Mol. Evol. 2017;85(3–4):99–106. doi: 10.1007/s00239-017-9809-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whatley J.M., John P., Whatley F.R. From extracellular to intracellular: the establishment of mitochondria and chloroplasts. Proc. R. Soc. Lond. B Biol. Sci. 1979;204(1155):165–187. doi: 10.1098/rspb.1979.0020. [DOI] [PubMed] [Google Scholar]
- 42.Speijer D. Alternating terminal electron-acceptors at the basis of symbiogenesis: how oxygen ignited eukaryotic evolution. Bioessays 39(2) 2017 doi: 10.1002/bies.201600174. [DOI] [PubMed] [Google Scholar]
- 43.Andersson S.G., Kurland C.G. Origins of mitochondria and hydrogenosomes. Curr. Opin. Microbiol. 1999;2(5):535–541. doi: 10.1016/s1369-5274(99)00013-2. [DOI] [PubMed] [Google Scholar]
- 44.Tsaousis A.D. On the origin of iron/sulfur cluster biosynthesis in eukaryotes. Front. Microbiol. 2019;10:2478. doi: 10.3389/fmicb.2019.02478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lill R., Diekert K., Kaut A., Lange H., Pelzer W., Prohl C., Kispal G. 1999. The essential role of mitochondria in the biogenesis of cellular iron-sulfur proteins. [DOI] [PubMed] [Google Scholar]
- 46.Braymer J.J., Lill R. Iron–sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 2017;292(31):12754–12763. doi: 10.1074/jbc.R117.787101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouault T.A., Maio N. Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem. 2017;292(31):12744–12753. doi: 10.1074/jbc.R117.789537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peña-Diaz P., Lukeš J. Fe–S cluster assembly in the supergroup Excavata. JBIC Journal of Biological Inorganic Chemistry. 2018;23(4):521–541. doi: 10.1007/s00775-018-1556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davidov Y., Huchon D., Koval S.F., Jurkevitch E. A new alpha-proteobacterial clade of Bdellovibrio-like predators: implications for the mitochondrial endosymbiotic theory. Environ Microbiol. 2006;8(12):2179–2188. doi: 10.1111/j.1462-2920.2006.01101.x. [DOI] [PubMed] [Google Scholar]
- 50.Davidov Y., Jurkevitch E. Predation between prokaryotes and the origin of eukaryotes. Bioessays. 2009;31(7):748–757. doi: 10.1002/bies.200900018. [DOI] [PubMed] [Google Scholar]
- 51.Walker J.E. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 1992;25(3):253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 52.Yagi T., Matsuno-Yagi A. The proton-translocating NADH-quinone oxidoreductase in the respiratory chain: the secret unlocked. Biochemistry. 2003;42(8):2266–2274. doi: 10.1021/bi027158b. [DOI] [PubMed] [Google Scholar]
- 53.Burger G., Lang B.F., Reith M., Gray M.W. Genes encoding the same three subunits of respiratory complex II are present in the mitochondrial DNA of two phylogenetically distant eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 1996;93(6):2328–2332. doi: 10.1073/pnas.93.6.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hunte C., Koepke J., Lange C., Rossmanith T., Michel H. Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;8(6):669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Z., Huang L., Shulmeister V.M., Chi Y.I., Kim K.K., Hung L.W., Crofts A.R., Berry E.A., Kim S.H. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392(6677):677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 56.Friend D.S. The fine structure of Giardia muris. J. Cell Biol. 1966;29(2):317–332. doi: 10.1083/jcb.29.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rosenbaum R.M., Wittner M. Ultrastructure of bacterized and axenic trophozoites of Entamoeba histolytica with particular reference to helical bodies. J. Cell Biol. 1970;45(2):367–382. doi: 10.1083/jcb.45.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Giezen M. Hydrogenosomes and mitosomes: conservation and evolution of functions. J. Eukaryot. Microbiol. 2009;56(3):221–231. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- 59.Friedrich T., Steinmuller K., Weiss H. The proton-pumping respiratory complex I of bacteria and mitochondria and its homologue in chloroplasts. FEBS Lett. 1995;367(2):107–111. doi: 10.1016/0014-5793(95)00548-n. [DOI] [PubMed] [Google Scholar]
- 60.Joseph-Horne T., Hollomon D.W., Wood P.M. Fungal respiration: a fusion of standard and alternative components. Biochim. Biophys. Acta. 2001;1504(2–3):179–195. doi: 10.1016/s0005-2728(00)00251-6. [DOI] [PubMed] [Google Scholar]
- 61.Yagi T. Bacterial NADH-quinone oxidoreductases. J. Bioenerg. Biomembr. 1991;23(2):211–225. doi: 10.1007/BF00762218. [DOI] [PubMed] [Google Scholar]
- 62.De Vries S., Van Witzenburg R., Grivell L.A., Marres C.A. Primary structure and import pathway of the rotenone-insensitive NADH-ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Eur. J. Biochem. 1992;203(3):587–592. doi: 10.1111/j.1432-1033.1992.tb16587.x. [DOI] [PubMed] [Google Scholar]
- 63.Luttik M.A., Overkamp K.M., Kotter P., de Vries S., van Dijken J.P., Pronk J.T. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH, J. Biol. Chem. 1998;273(38):24529–24534. doi: 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- 64.Minagawa N., Yoshimoto A. The induction of cyanide-resistant respiration in Hansenula anomala. J. Biochem. 1987;101(5):1141–1146. doi: 10.1093/oxfordjournals.jbchem.a121978. [DOI] [PubMed] [Google Scholar]
- 65.Moore A.L., Siedow J.N. The regulation and nature of the cyanide-resistant alternative oxidase of plant mitochondria. Biochim. Biophys. Acta. 1991;1059(2):121–140. doi: 10.1016/s0005-2728(05)80197-5. [DOI] [PubMed] [Google Scholar]
- 66.Albury M.S., Dudley P., Watts F.Z., Moore A.L. Targeting the plant alternative oxidase protein to Schizosaccharomyces pombe mitochondria confers cyanide-insensitive respiration. J. Biol. Chem. 1996;271(29):17062–17066. doi: 10.1074/jbc.271.29.17062. [DOI] [PubMed] [Google Scholar]
- 67.Nicholls D.G., Fergusion S.J. 4 ed. Academic Press; 2013. Bioenergetics. [Google Scholar]
- 68.Anderson C.P., Shen M., Eisenstein R.S., Leibold E.A. Mammalian iron metabolism and its control by iron regulatory proteins, Biochimica et Biophysica Acta (BBA) Molecular Cell Research. 2012;1823(9):1468–1483. doi: 10.1016/j.bbamcr.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rouault T.A. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006;2(8):406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 70.Beinert H., Kennedy M.C. Aconitase, a two-faced protein: enzyme and iron regulatory factor. Faseb. J. 1993;7(15):1442–1449. doi: 10.1096/fasebj.7.15.8262329. [DOI] [PubMed] [Google Scholar]
- 71.Hausladen A., Fridovich I. Superoxide and peroxynitrite inactivate aconitases, but nitric oxide does not. J. Biol. Chem. 1994;269(47):29405–29408. [PubMed] [Google Scholar]
- 72.Gardner P.R., Fridovich I. Superoxide sensitivity of the Escherichia coli aconitase. J. Biol. Chem. 1991;266(29):19328–19333. [PubMed] [Google Scholar]
- 73.Bulteau A.-L., Ikeda-Saito M., Szweda L.I. Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry. 2003;42(50):14846–14855. doi: 10.1021/bi0353979. [DOI] [PubMed] [Google Scholar]
- 74.Castro L., Tórtora V., Mansilla S., Radi R. Aconitases: non-redox iron–sulfur proteins sensitive to reactive species. Acc. Chem. Res. 2019;52(9):2609–2619. doi: 10.1021/acs.accounts.9b00150. [DOI] [PubMed] [Google Scholar]
- 75.Maio N., Rouault T.A. Outlining the complex pathway of mammalian Fe-S cluster biogenesis. Trends Biochem. Sci. 2020;45(5):411–426. doi: 10.1016/j.tibs.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alfadhel M., Nashabat M., Ali Q.A., Hundallah K. Mitochondrial iron-sulfur cluster biogenesis from molecular understanding to clinical disease. Neurosciences Journal. 2017;22(1):4. doi: 10.17712/nsj.2017.1.20160542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leonard K., Haiker H., Weiss H. Three-dimensional structure of NADH: ubiquinone reductase (complex I) from Neurospora mitochondria determined by electron microscopy of membrane crystals. J. Mol. Biol. 1987;194(2):277–286. doi: 10.1016/0022-2836(87)90375-5. [DOI] [PubMed] [Google Scholar]
- 78.Hofhaus G., Weiss H., Leonard K. Electron microscopic analysis of the peripheral and membrane parts of mitochondrial NADH dehydrogenase (Complex I) J. Mol. Biol. 1991;221(3):1027–1043. doi: 10.1016/0022-2836(91)80190-6. [DOI] [PubMed] [Google Scholar]
- 79.Guénebaut V., Vincentelli R., Mills D., Weiss H., Leonard K.R. Three-dimensional structure of NADH-dehydrogenase from Neurospora crassa by electron microscopy and conical tilt reconstruction11Edited by K. Nagai. J. Mol. Biol. 1997;265(4):409–418. doi: 10.1006/jmbi.1996.0753. [DOI] [PubMed] [Google Scholar]
- 80.Janssen R.J.R.J., Nijtmans L.G., Heuvel L.P.v.d., Smeitink J.A.M. Mitochondrial complex I: structure, function and pathology. J. Inherit. Metab. Dis. 2006;29(4):499–515. doi: 10.1007/s10545-006-0362-4. [DOI] [PubMed] [Google Scholar]
- 81.Carroll J., Shannon R.J., Fearnley I.M., Walker J.E., Hirst J. Definition of the nuclear encoded protein composition of bovine heart mitochondrial complex I: identification OF two new subunits*. J. Biol. Chem. 2002;277(52):50311–50317. doi: 10.1074/jbc.M209166200. [DOI] [PubMed] [Google Scholar]
- 82.Ugalde C., Vogel R., Huijbens R., van den Heuvel B., Smeitink J., Nijtmans L. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum. Mol. Genet. 2004;13(20):2461–2472. doi: 10.1093/hmg/ddh262. [DOI] [PubMed] [Google Scholar]
- 83.Vogel R.O., Smeitink J.A.M., Nijtmans L.G.J. Human mitochondrial complex I assembly: a dynamic and versatile process, Biochimica et Biophysica Acta (BBA) Bioenergetics. 2007;1767(10):1215–1227. doi: 10.1016/j.bbabio.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 84.Mimaki M., Wang X., McKenzie M., Thorburn D.R., Ryan M.T. Understanding mitochondrial complex I assembly in health and disease, Biochimica et Biophysica Acta (BBA) Bioenergetics. 2012;1817(6):851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 85.Friedrich T., Dekovic D.K., Burschel S. Assembly of the Escherichia coli NADH:ubiquinone oxidoreductase (respiratory complex I), Biochimica et Biophysica Acta (BBA) Bioenergetics. 2016;1857(3):214–223. doi: 10.1016/j.bbabio.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 86.Videira A. Complex I from the fungus Neurospora crassa, Biochimica et Biophysica Acta (BBA) Bioenergetics. 1998;1364(2):89–100. doi: 10.1016/s0005-2728(98)00020-6. [DOI] [PubMed] [Google Scholar]
- 87.Marques I., Duarte M., Assunção J., Ushakova A.V., Videira A. Composition of complex I from Neurospora crassa and disruption of two “accessory” subunits, Biochimica et Biophysica Acta (BBA) Bioenergetics. 2005;1707(2):211–220. doi: 10.1016/j.bbabio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 88.Brandt U. Energy converting NADH: quinone oxidoreductase (complex I) Annu. Rev. Biochem. 2006;75(1):69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 89.Nakamaru-Ogiso E. In: A Structural Perspective on Respiratory Complex I: Structure and Function of NADH:ubiquinone Oxidoreductase. Sazanov L., editor. Springer Netherlands; Dordrecht: 2012. Iron–sulfur clusters in complex I; pp. 61–79. [Google Scholar]
- 90.Wang D.-C., Meinhardt S.W., Sackmann U., Weiss H., Ohnishi T. The iron-sulfur clusters in the two related forms of mitochondrial NADH: ubiquinone oxidoreductase made by Neurospora crassa. Eur. J. Biochem. 1991;197(1):257–264. doi: 10.1111/j.1432-1033.1991.tb15906.x. [DOI] [PubMed] [Google Scholar]
- 91.Rasmussen T., Scheide D., Brors B., Kintscher L., Weiss H., Friedrich T. Identification of two tetranuclear FeS clusters on the ferredoxin-type subunit of NADH:ubiquinone oxidoreductase (complex I) Biochemistry. 2001;40(20):6124–6131. doi: 10.1021/bi0026977. [DOI] [PubMed] [Google Scholar]
- 92.Sazanov L.A., Hinchliffe P. Structure of the hydrophilic domain of respiratory complex I from <em>Thermus thermophilus</em>. Science. 2006;311(5766):1430. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 93.Holt P.J., Efremov R.G., Nakamaru-Ogiso E., Sazanov L.A. Reversible FMN dissociation from Escherichia coli respiratory complex I, Biochimica et Biophysica Acta (BBA) Bioenergetics. 2016;1857(11):1777–1785. doi: 10.1016/j.bbabio.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 94.Hayashi T., Stuchebrukhov A. vol. 660. J Electroanal Chem; Lausanne: 2011. pp. 356–359. (Electron Tunneling Pathways in Respiratory Complex I. The Role of the Internal Water between the Enzyme Subunits). 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yagi T., Matsuno-Yagi A. The proton-translocating NADH−Quinone oxidoreductase in the respiratory Chain: the secret unlocked. Biochemistry. 2003;42(8):2266–2274. doi: 10.1021/bi027158b. [DOI] [PubMed] [Google Scholar]
- 96.Sled V.D., Rudnitzky N.I., Hatefi Y., Ohnishi T. Thermodynamic analysis of flavin in mitochondrial NADH:ubiquinone oxidoreductase (complex I) Biochemistry. 1994;33(33):10069–10075. doi: 10.1021/bi00199a034. [DOI] [PubMed] [Google Scholar]
- 97.Schulte M., Frick K., Gnandt E., Jurkovic S., Burschel S., Labatzke R., Aierstock K., Fiegen D., Wohlwend D., Gerhardt S., Einsle O., Friedrich T. A mechanism to prevent production of reactive oxygen species by Escherichia coli respiratory complex I. Nat. Commun. 2019;10(1) doi: 10.1038/s41467-019-10429-0. 2551–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kmita K., Wirth C., Warnau J., Guerrero-Castillo S., Hunte C., Hummer G., Kaila V.R.I., Zwicker K., Brandt U., Zickermann V. Accessory NUMM (NDUFS6) subunit harbors a Zn-binding site and is essential for biogenesis of mitochondrial complex I. Proc. Natl. Acad. Sci. Unit. States Am. 2015;112(18):5685. doi: 10.1073/pnas.1424353112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yano T., Dunham W.R., Ohnishi T. Characterization of the delta muH+-sensitive ubisemiquinone species (SQ(Nf)) and the interaction with cluster N2: new insight into the energy-coupled electron transfer in complex I. Biochemistry. 2005;44(5):1744–1754. doi: 10.1021/bi048132i. [DOI] [PubMed] [Google Scholar]
- 100.Sled V.D., Friedrich T., Leif H., Weiss H., Meinhardt S.W., Fukumori Y., Calhoun M.W., Gennis R.B., Ohnishi T. Bacterial NADH-quinone oxidoreductases: iron-sulfur clusters and related problems. J. Bioenerg. Biomembr. 1993;25(4):347–356. doi: 10.1007/BF00762460. [DOI] [PubMed] [Google Scholar]
- 101.Le Breton N., Wright J.J., Jones A.J.Y., Salvadori E., Bridges H.R., Hirst J., Roessler M.M. Using hyperfine electron paramagnetic resonance spectroscopy to define the proton-coupled electron transfer reaction at Fe-S cluster N2 in respiratory complex I. J. Am. Chem. Soc. 2017;139(45):16319–16326. doi: 10.1021/jacs.7b09261. [DOI] [PubMed] [Google Scholar]
- 102.Zwicker K., Galkin A., Dröse S., Grgic L., Kerscher S., Brandt U. The redox-bohr group Associated with iron-sulfur cluster N2 of complex I*. J. Biol. Chem. 2006;281(32):23013–23017. doi: 10.1074/jbc.M603442200. [DOI] [PubMed] [Google Scholar]
- 103.Hameedi M.A., Grba D.N., Richardson K.H., Jones A.J.Y., Song W., Roessler M.M., Wright J.J., Hirst J. A conserved arginine residue is critical for stabilizing the N2 FeS cluster in mitochondrial complex I, J. Biol. Chem. 2021;296:100474. doi: 10.1016/j.jbc.2021.100474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kowaltowski A.J., de Souza-Pinto N.C., Castilho R.F., Vercesi A.E. Mitochondria and reactive oxygen species. Free Radic. Biol. Med. 2009;47(4):333–343. doi: 10.1016/j.freeradbiomed.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 105.Brand M.D. The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 2010;45(7–8):466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scialò F., Fernández-Ayala D.J., Sanz A. Role of mitochondrial reverse electron transport in ROS signaling: potential roles in health and disease. Front. Physiol. 2017;8(428) doi: 10.3389/fphys.2017.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Murphy Michael P. How mitochondria produce reactive oxygen species. Biochem. J. 2008;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Michelakis E.D., Hampl V., Nsair A., Wu X., Harry G., Haromy A., Gurtu R., Archer S.L. Diversity in mitochondrial function explains differences in vascular oxygen sensing. Circ. Res. 2002;90(12):1307–1315. doi: 10.1161/01.res.0000024689.07590.c2. [DOI] [PubMed] [Google Scholar]
- 109.Chouchani E.T., Pell V.R., Gaude E., Aksentijević D., Sundier S.Y., Robb E.L., Logan A., Nadtochiy S.M., Ord E.N.J., Smith A.C., Eyassu F., Shirley R., Hu C.-H., Dare A.J., James A.M., Rogatti S., Hartley R.C., Eaton S., Costa A.S.H., Brookes P.S., Davidson S.M., Duchen M.R., Saeb-Parsy K., Shattock M.J., Robinson A.J., Work L.M., Frezza C., Krieg T., Murphy M.P. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515(7527):431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brand M.D. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 2016;100:14–31. doi: 10.1016/j.freeradbiomed.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 111.Brand M.D., Goncalves R.L.S., Orr A.L., Vargas L., Gerencser A.A., Borch Jensen M., Wang Y.T., Melov S., Turk C.N., Matzen J.T., Dardov V.J., Petrassi H.M., Meeusen S.L., Perevoshchikova I.V., Jasper H., Brookes P.S., Ainscow E.K. Suppressors of superoxide-H(2)O(2) production at site I(Q) of mitochondrial complex I protect against stem cell hyperplasia and ischemia-reperfusion injury. Cell Metabol. 2016;24(4):582–592. doi: 10.1016/j.cmet.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang S., Millar A.H. Succinate dehydrogenase: the complex roles of a simple enzyme. Curr. Opin. Plant Biol. 2013;16(3):344–349. doi: 10.1016/j.pbi.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 113.Rasheed M.R.H.A., Tarjan G. Succinate dehydrogenase complex: an updated review. Arch. Pathol. Lab Med. 2018;142(12):1564–1570. doi: 10.5858/arpa.2017-0285-RS. [DOI] [PubMed] [Google Scholar]
- 114.Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121(7):1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 115.Page C.C., Moser C.C., Chen X., Dutton P.L. Natural engineering principles of electron tunnelling in biological oxidation–reduction. Nature. 1999;402(6757):47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 116.Anderson R.F., Shinde S.S., Hille R., Rothery R.A., Weiner J.H., Rajagukguk S., Maklashina E., Cecchini G. Electron-transfer pathways in the heme and quinone-binding domain of complex II (succinate dehydrogenase) Biochemistry. 2014;53(10):1637–1646. doi: 10.1021/bi401630m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.St-Pierre J., Buckingham J.A., Roebuck S.J., Brand M.D. Topology of superoxide production from different sites in the mitochondrial electron transport chain *. J. Biol. Chem. 2002;277(47):44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 118.Ackrell B.A.C. Cytopathies involving mitochondrial complex II. Mol. Aspect. Med. 2002;23(5):369–384. doi: 10.1016/s0098-2997(02)00012-2. [DOI] [PubMed] [Google Scholar]
- 119.Hoekstra A.S., Bayley J.-P. The role of complex II in disease, Biochimica et Biophysica Acta (BBA) Bioenergetics. 2013;1827(5):543–551. doi: 10.1016/j.bbabio.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 120.Quinlan C.L., Orr A.L., Perevoshchikova I.V., Treberg J.R., Ackrell B.A., Brand M.D. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J. Biol. Chem. 2012;287(32):27255–27264. doi: 10.1074/jbc.M112.374629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ackrell B.A., Kearney E.B., Singer T.P. Mammalian succinate dehydrogenase. Methods Enzymol. 1978;53:466–483. doi: 10.1016/s0076-6879(78)53050-4. [DOI] [PubMed] [Google Scholar]
- 122.Crofts A.R. The cytochrome bc1 complex: function in the context of structure. Annu. Rev. Physiol. 2004;66(1):689–733. doi: 10.1146/annurev.physiol.66.032102.150251. [DOI] [PubMed] [Google Scholar]
- 123.Iwata S., Lee J.W., Okada K., Lee J.K., Iwata M., Rasmussen B., Link T.A., Ramaswamy S., Jap B.K. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281(5373):64. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 124.Yu C.-A., Yu L., Xia D. In: Encyclopedia of Biophysics. Roberts G.C.K., editor. Springer Berlin Heidelberg; Berlin, Heidelberg: 2013. Ubiquinol-cytochrome c oxidoreductase (complex III) pp. 2679–2684. [Google Scholar]
- 125.Xia D., Esser L., Tang W.-K., Zhou F., Zhou Y., Yu L., Yu C.-A. Structural analysis of cytochrome bc1 complexes: implications to the mechanism of function. Biochim. Biophys. Acta. 2013;1827(11–12):1278–1294. doi: 10.1016/j.bbabio.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cape J.L., Bowman M.K., Kramer D.M. A semiquinone intermediate generated at the Qo site of the cytochrome <em>bc</em>1 complex: importance for the Q-cycle and superoxide production. Proc. Natl. Acad. Sci. Unit. States Am. 2007;104(19):7887. doi: 10.1073/pnas.0702621104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Iwata S., Saynovits M., Link T.A., Michel H. Structure of a water soluble fragment of the ‘Rieske’ iron–sulfur protein of the bovine heart mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5 å resolution. Structure. 1996;4(5):567–579. doi: 10.1016/s0969-2126(96)00062-7. [DOI] [PubMed] [Google Scholar]
- 128.Smith J.L., Zhang H., Yan J., Kurisu G., Cramer W.A. Cytochrome bc complexes: a common core of structure and function surrounded by diversity in the outlying provinces. Curr. Opin. Struct. Biol. 2004;14(4):432–439. doi: 10.1016/j.sbi.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 129.Zhang Z., Huang L., Shulmeister V.M., Chi Y.-I., Kim K.K., Hung L.-W., Crofts A.R., Berry E.A., Kim S.-H. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392(6677):677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 130.Esser L., Gong X., Yang S., Yu L., Yu C.-A., Xia D. Surface-modulated motion switch: capture and release of iron–sulfur protein in the cytochrome <em>bc</em>1 complex. Proc. Natl. Acad. Sci. Unit. States Am. 2006;103(35):13045. doi: 10.1073/pnas.0601149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Berry E.A., De Bari H., Huang L.S. Unanswered questions about the structure of cytochrome bc1 complexes. Biochim. Biophys. Acta. 2013;1827(11–12):1258–1277. doi: 10.1016/j.bbabio.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 132.Quinlan C.L., Gerencser A.A., Treberg J.R., Brand M.D. The mechanism of superoxide production by the antimycin-inhibited mitochondrial Q-cycle *. J. Biol. Chem. 2011;286(36):31361–31372. doi: 10.1074/jbc.M111.267898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Archer S.L., Nelson D.P., Weir E.K. Simultaneous measurement of O2 radicals and pulmonary vascular reactivity in rat lung. J. Appl. Physiol. 1985;67(5):1903–1911. doi: 10.1152/jappl.1989.67.5.1903. 1989. [DOI] [PubMed] [Google Scholar]
- 134.Archer S.L., Huang J., Henry T., Peterson D., Weir E.K. A redox-based O2 sensor in rat pulmonary vasculature. Circ. Res. 1993;73(6):1100–1112. doi: 10.1161/01.res.73.6.1100. [DOI] [PubMed] [Google Scholar]
- 135.Muller F.L., Liu Y., Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J. Biol. Chem. 2004;279(47):49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 136.Beinert H., Kiley P.J. Fe-S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol. 1999;3(2):152–157. doi: 10.1016/S1367-5931(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 137.Golinelli-Cohen M.-P., Bouton C. Fe-S proteins acting as redox switch: new key actors of cellular adaptive responses. Curr. Chem. Biol. 2017;11(2):70–88. [Google Scholar]
- 138.Weir E.K., López-Barneo J., Buckler K.J., Archer S.L. Acute oxygen-sensing mechanisms. N. Engl. J. Med. 2005;353(19):2042–2055. doi: 10.1056/NEJMra050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Weir E.K., Archer S.L. The role of redox changes in oxygen sensing. Respir. Physiol. Neurobiol. 2010;174(3):182–191. doi: 10.1016/j.resp.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dunham-Snary K.J., Hong Z.G., Xiong P.Y., Del Paggio J.C., Herr J.E., Johri A.M., Archer S.L. A mitochondrial redox oxygen sensor in the pulmonary vasculature and ductus arteriosus. Pflügers Archiv. 2016;468(1):43–58. doi: 10.1007/s00424-015-1736-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nurse C.A., Salman S., Scott A.L. Hypoxia-regulated catecholamine secretion in chromaffin cells. Cell Tissue Res. 2018;372(2):433–441. doi: 10.1007/s00441-017-2703-z. [DOI] [PubMed] [Google Scholar]
- 142.Peers C., Kemp P.J. Acute oxygen sensing: diverse but convergent mechanisms in airway and arterial chemoreceptors. Respir. Res. 2001;2(3):145–149. doi: 10.1186/rr51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jastroch M., Divakaruni A.S., Mookerjee S., Treberg J.R., Brand M.D. Mitochondrial proton and electron leaks. Essays Biochem. 2010;47:53–67. doi: 10.1042/bse0470053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Palma F.R., He C., Danes J.M., Paviani V., Coelho D.R., Gantner B.N., Bonini M.G. Mitochondrial superoxide dismutase: what the established, the intriguing, and the novel reveal about a key cellular redox switch. Antioxidants Redox Signal. 2020;32(10):701–714. doi: 10.1089/ars.2019.7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Di Marzo N., Chisci E., Giovannoni R. The role of hydrogen peroxide in redox-dependent signaling: homeostatic and pathological responses in mammalian cells. Cells. 2018;7(10):156. doi: 10.3390/cells7100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lopez-Barneo J., Lopez-Lopez J.R., Urena J., Gonzalez C. Chemotransduction in the carotid body: K+ current modulated by PO2 in type I chemoreceptor cells. Science. 1988;241(4865):580–582. doi: 10.1126/science.2456613. [DOI] [PubMed] [Google Scholar]
- 147.Archer S.L., Wu X.-C., Thébaud B., Nsair A., Bonnet S., Tyrrell B., McMurtry M.S., Hashimoto K., Harry G., Michelakis E.D. Preferential expression and function of voltage-gated, O2-sensitive K+ channels in resistance pulmonary arteries explains regional heterogeneity in hypoxic pulmonary vasoconstriction: ionic diversity in smooth muscle cells. Circ. Res. 2004;95(3):308–318. doi: 10.1161/01.RES.0000137173.42723.fb. [DOI] [PubMed] [Google Scholar]
- 148.Kajimoto H., Hashimoto K., Bonnet S.N., Haromy A., Harry G., Moudgil R., Nakanishi T., Rebeyka I., Thébaud B., Michelakis E.D. Oxygen activates the Rho/Rho-kinase pathway and induces RhoB and ROCK-1 expression in human and rabbit ductus arteriosus by increasing mitochondria-derived reactive oxygen species: a newly recognized mechanism for sustaining ductal constriction. Circulation. 2007;115(13):1777–1788. doi: 10.1161/CIRCULATIONAHA.106.649566. [DOI] [PubMed] [Google Scholar]
- 149.Uehata M., Ishizaki T., Satoh H., Ono T., Kawahara T., Morishita T., Tamakawa H., Yamagami K., Inui J., Maekawa M. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389(6654):990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 150.Howard R.B., Hosokawa T., Maguire M.H. Hypoxia-induced fetoplacental vasoconstriction in perfused human placental cotyledons. Am. J. Obstet. Gynecol. 1987;157(5):1261–1266. doi: 10.1016/s0002-9378(87)80307-1. [DOI] [PubMed] [Google Scholar]
- 151.Hampl V., J. Bíbová. Z. Straňák. Wu X., Michelakis E.D., K. Hashimoto. Archer S.L. Hypoxic fetoplacental vasoconstriction in humans is mediated by potassium channel inhibition. Am. J. Physiol. Heart Circ. Physiol. 2002;283(6):H2440–H2449. doi: 10.1152/ajpheart.01033.2001. [DOI] [PubMed] [Google Scholar]
- 152.Power G.G., Longo L.D., Wagner H.N., Kuhl D.E., Forster R.E. Uneven distribution of maternal and fetal placental blood flow, as demonstrated using macroaggregates, and its response to hypoxia. J. Clin. Invest. 1967;46(12):2053–2063. doi: 10.1172/JCI105693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lee D.-C., Romero R., Kim J.-S., Tarca A.L., Montenegro D., Pineles B.L., Kim E., Lee J., Kim S.Y., Draghici S. miR-210 targets iron-sulfur cluster scaffold homologue in human trophoblast cell lines: siderosis of interstitial trophoblasts as a novel pathology of preterm preeclampsia and small-for-gestational-age pregnancies. Am. J. Pathol. 2011;179(2):590–602. doi: 10.1016/j.ajpath.2011.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Khong T., De Wolf F., Robertson W., Brosens I. Inadequate maternal vascular response to placentation in pregnancies complicated by pre-eclampsia and by small-for-gestational age infants. BJOG: An International Journal of Obstetrics & Gynaecology. 1986;93(10):1049–1059. doi: 10.1111/j.1471-0528.1986.tb07830.x. [DOI] [PubMed] [Google Scholar]
- 155.Bauer S.T., Cleary K.L. Seminars in Perinatology. Elsevier; 2009. Cardiopulmonary complications of pre-eclampsia; pp. 158–165. [DOI] [PubMed] [Google Scholar]
- 156.Sibai B., Dekker G., Kupferminc M. Pre-eclampsia. Lancet. 2005;365(9461):785–799. doi: 10.1016/S0140-6736(05)17987-2. [DOI] [PubMed] [Google Scholar]
- 157.Keyes L.E., Armaza F.J., Niermeyer S., Vargas E., Young D.A., Moore L.G. Intrauterine growth restriction, preeclampsia, and intrauterine mortality at high altitude in Bolivia. Pediatr. Res. 2003;54(1):20–25. doi: 10.1203/01.PDR.0000069846.64389.DC. [DOI] [PubMed] [Google Scholar]
- 158.Palmer S.K., Moore L.G., Young D.A., Cregger B., Berman J.C., Zamudio S. Altered blood pressure course during normal pregnancy and increased preeclampsia at high altitude (3100 meters) in Colorado. Am. J. Obstet. Gynecol. 1999;180(5):1161–1168. doi: 10.1016/s0002-9378(99)70611-3. [DOI] [PubMed] [Google Scholar]
- 159.Colleoni F., Padmanabhan N. H.-w. Yung, E.D. Watson, I. Cetin, M.C. Tissot van Patot, G.J. Burton, A.J. Murray, Suppression of mitochondrial electron transport chain function in the hypoxic human placenta: a role for miRNA-210 and protein synthesis inhibition. PLoS One. 2013;8(1):e55194. doi: 10.1371/journal.pone.0055194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.López-Barneo J., González-Rodríguez P., Gao L., Fernández-Agüera M.C., Pardal R., Ortega-Sáenz P. Oxygen sensing by the carotid body: mechanisms and role in adaptation to hypoxia. Am. J. Physiol. Cell Physiol. 2016;310(8):C629–C642. doi: 10.1152/ajpcell.00265.2015. [DOI] [PubMed] [Google Scholar]
- 161.Peers C. Hypoxic suppression of K+ currents in type I carotid body cells: selective effect on the Ca2+-activated K+ current. Neurosci. Lett. 1990;119(2):253–256. doi: 10.1016/0304-3940(90)90846-2. [DOI] [PubMed] [Google Scholar]
- 162.Fernández-Agüera M.C., Gao L., González-Rodríguez P., Pintado C.O., Arias-Mayenco I., García-Flores P., García-Pergañeda A., Pascual A., Ortega-Sáenz P., López-Barneo J. Oxygen sensing by arterial chemoreceptors depends on mitochondrial complex I signaling. Cell Metabol. 2015;22(5):825–837. doi: 10.1016/j.cmet.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 163.Arias-Mayenco I., González-Rodríguez P., Torres-Torrelo H., Gao L., Fernández-Agüera M.C., Bonilla-Henao V., Ortega-Sáenz P., López-Barneo J. Acute O2 sensing: role of coenzyme QH2/Q ratio and mitochondrial ROS compartmentalization. Cell Metabol. 2018;28(1):145–158. doi: 10.1016/j.cmet.2018.05.009. e4. [DOI] [PubMed] [Google Scholar]
- 164.Michelakis E.D., Thébaud B., Weir E.K., Archer S.L. Hypoxic pulmonary vasoconstriction: redox regulation of O2-sensitive K+ channels by a mitochondrial O2-sensor in resistance artery smooth muscle cells. J. Mol. Cell. Cardiol. 2004;37(6):1119–1136. doi: 10.1016/j.yjmcc.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 165.López-Barneo J., Pardal R., Montoro R.J., Smani T., García-Hirschfeld J., Ureña J. K+ and Ca2+ channel activity and cytosolic [Ca2+] in oxygen-sensing tissues. Respir. Physiol. 1999;115(2):215–227. doi: 10.1016/s0034-5687(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 166.Freeman B.A., Crapo J.D. Hyperoxia increases oxygen radical production in rat lungs and lung mitochondria. J. Biol. Chem. 1981;256(21):10986–10992. [PubMed] [Google Scholar]
- 167.Berkelhamer S.K., Kim G.A., Radder J.E., Wedgwood S., Czech L., Steinhorn R.H., Schumacker P.T. Developmental differences in hyperoxia-induced oxidative stress and cellular responses in the murine lung. Free Radic. Biol. Med. 2013;61:51–60. doi: 10.1016/j.freeradbiomed.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Michelakis E.D., Archer S.L., Weir E.K. Acute hypoxic pulmonary vasoconstriction: a model of oxygen sensing. Physiol. Res. 1995;44(6):361–367. [PubMed] [Google Scholar]
- 169.Waypa G.B., Marks J.D., Mack M.M., Boriboun C., Mungai P.T., Schumacker P.T. Mitochondrial reactive oxygen species trigger calcium increases during hypoxia in pulmonary arterial myocytes. Circ. Res. 2002;91(8):719–726. doi: 10.1161/01.res.0000036751.04896.f1. [DOI] [PubMed] [Google Scholar]
- 170.Guzy R.D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K.D., Simon M.C., Hammerling U., Schumacker P.T. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metabol. 2005;1(6):401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 171.Gao L., González-Rodríguez P., Ortega-Sáenz P., López-Barneo J. Redox signaling in acute oxygen sensing. Redox Biol. 2017;12:908–915. doi: 10.1016/j.redox.2017.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sommer N., Hüttemann M., Pak O., Scheibe S., Knoepp F., Sinkler C., Malczyk M., Gierhardt M., Esfandiary A., Kraut S., Jonas F., Veith C., Aras S., Sydykov A., Alebrahimdehkordi N., Giehl K., Hecker M., Brandes R.P., Seeger W., Grimminger F., Ghofrani H.A., Schermuly R.T., Grossman L.I., Weissmann N. Mitochondrial complex IV subunit 4 isoform 2 is essential for acute pulmonary oxygen sensing. Circ. Res. 2017;121(4):424–438. doi: 10.1161/CIRCRESAHA.116.310482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dunham-Snary K.J., Wu D., Potus F., Sykes E.A., Mewburn J.D., Charles R.L., Eaton P., Sultanian R.A., Archer S.L. Ndufs2, a core subunit of mitochondrial complex I, is essential for acute oxygen-sensing and hypoxic pulmonary vasoconstriction. Circ. Res. 2019;124(12):1727–1746. doi: 10.1161/CIRCRESAHA.118.314284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Koch J., Hensley G., Roy L., Brown S., Ramaciotti C., Rosenfeld C.R. Prevalence of spontaneous closure of the ductus arteriosus in neonates at a birth weight of 1000 grams or less. Pediatrics. 2006;117(4):1113–1121. doi: 10.1542/peds.2005-1528. [DOI] [PubMed] [Google Scholar]
- 175.Seidner S.R., Chen Y.Q., Oprysko P.R., Mauray F., Mary M.T., Lin E., Koch C., Clyman R.I. Combined prostaglandin and nitric oxide inhibition produces anatomic remodeling and closure of the ductus arteriosus in the premature newborn baboon. Pediatr. Res. 2001;50(3):365–373. doi: 10.1203/00006450-200109000-00012. [DOI] [PubMed] [Google Scholar]
- 176.Kajino H., Chen Y.-Q., Seidner S.R., Waleh N., Mauray F., Roman C., Chemtob S., Koch C.J., Clyman R.I. Factors that increase the contractile tone of the ductus arteriosus also regulate its anatomic remodeling, American Journal of Physiology-Regulatory. Integrative and Comparative Physiology. 2001;281(1):R291–R301. doi: 10.1152/ajpregu.2001.281.1.R291. [DOI] [PubMed] [Google Scholar]
- 177.Hermes-DeSantis E., Clyman R. Patent ductus arteriosus: pathophysiology and management. J. Perinatol. 2006;26(1):S14–S18. doi: 10.1038/sj.jp.7211465. [DOI] [PubMed] [Google Scholar]
- 178.Hong Z., Kutty S., Toth P.T., Marsboom G., Hammel J.M., Chamberlain C., Ryan J.J., Zhang H.J., Sharp W.W., Morrow E. Role of dynamin-related protein 1 (Drp1)-mediated mitochondrial fission in oxygen sensing and constriction of the ductus arteriosus. Circ. Res. 2013;112(5):802–815. doi: 10.1161/CIRCRESAHA.111.300285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Thébaud B., Michelakis E.D., Wu X.-C., Moudgil R., Kuzyk M., Dyck J.R., Harry G., Hashimoto K., Haromy A., Rebeyka I. Oxygen-sensitive Kv channel gene transfer confers oxygen responsiveness to preterm rabbit and remodeled human ductus arteriosus: implications for infants with patent ductus arteriosus. Circulation. 2004;110(11):1372–1379. doi: 10.1161/01.CIR.0000141292.28616.65. [DOI] [PubMed] [Google Scholar]
- 180.Tristani-Firouzi M., Reeve H.L., Tolarova S., Weir E.K., Archer S.L. Oxygen-induced constriction of rabbit ductus arteriosus occurs via inhibition of a 4-aminopyridine-, voltage-sensitive potassium channel. J. Clin. Invest. 1996;98(9):1959–1965. doi: 10.1172/JCI118999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Thebaud B., Wu X.-C., Kajimoto H., Bonnet S., Hashimoto K., Michelakis E.D., Archer S.L. Developmental absence of the O 2 sensitivity of L-type calcium channels in preterm ductus arteriosus smooth muscle cells impairs O 2 constriction contributing to patent ductus arteriosus. Pediatr. Res. 2008;63(2):176–181. doi: 10.1203/PDR.0b013e31815ed059. [DOI] [PubMed] [Google Scholar]
- 182.Michelakis E.D., Rebeyka I., Wu X., Nsair A., Thebaud B., Hashimoto K., Dyck J.R., Haromy A., Harry G., Barr A., Archer S.L. O2 sensing in the human ductus arteriosus: regulation of voltage-gated K+ channels in smooth muscle cells by a mitochondrial redox sensor. Circ. Res. 2002;91(6):478–486. doi: 10.1161/01.res.0000035057.63303.d1. [DOI] [PubMed] [Google Scholar]
- 183.Bentley R.E., Hindmarch C.C., Dunham-Snary K.J., Snetsinger B., Mewburn J.D., Thébaud A., Lima P.D., Thébaud B., Archer S.L. The molecular mechanisms of oxygen-sensing in human ductus arteriosus smooth muscle cells: a comprehensive transcriptome profile reveals a central role for mitochondria. Genomics. 2021 doi: 10.1016/j.ygeno.2021.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Vernis L., El Banna N., Baille D., Hatem E., Heneman A., Huang M.E. Fe-S clusters emerging as targets of therapeutic drugs. Oxid Med Cell Longev. 2017;2017:3647657. doi: 10.1155/2017/3647657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wiley S.E., Murphy A.N., Ross S.A., van der Geer P., Dixon J.E. MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc. Natl. Acad. Sci. U. S. A. 2007;104(13):5318–5323. doi: 10.1073/pnas.0701078104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wiley S.E., Paddock M.L., Abresch E.C., Gross L., van der Geer P., Nechushtai R., Murphy A.N., Jennings P.A., Dixon J.E. The outer mitochondrial membrane protein mitoNEET contains a novel redox-active 2Fe-2S cluster. J. Biol. Chem. 2007;282(33):23745–23749. doi: 10.1074/jbc.C700107200. [DOI] [PubMed] [Google Scholar]
- 187.Lin J., Zhou T., Ye K., Wang J. Crystal structure of human mitoNEET reveals distinct groups of iron sulfur proteins. Proc. Natl. Acad. Sci. U. S. A. 2007;104(37):14640–14645. doi: 10.1073/pnas.0702426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Schlesier J., Rohde M., Gerhardt S., Einsle O. A conformational switch triggers nitrogenase protection from oxygen damage by shethna protein II (FeSII) J. Am. Chem. Soc. 2016;138(1):239–247. doi: 10.1021/jacs.5b10341. [DOI] [PubMed] [Google Scholar]
- 189.Chen G., Chen Z., Hu Y., Huang P. Inhibition of mitochondrial respiration and rapid depletion of mitochondrial glutathione by beta-phenethyl isothiocyanate: mechanisms for anti-leukemia activity. Antioxidants Redox Signal. 2011;15(12):2911–2921. doi: 10.1089/ars.2011.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bai F., Morcos F., Sohn Y.S., Darash-Yahana M., Rezende C.O., Lipper C.H., Paddock M.L., Song L., Luo Y., Holt S.H., Tamir S., Theodorakis E.A., Jennings P.A., Onuchic J.N., Mittler R., Nechushtai R. The Fe-S cluster-containing NEET proteins mitoNEET and NAF-1 as chemotherapeutic targets in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2015;112(12):3698–3703. doi: 10.1073/pnas.1502960112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Salem A.F., Whitaker-Menezes D., Howell A., Sotgia F., Lisanti M.P. Mitochondrial biogenesis in epithelial cancer cells promotes breast cancer tumor growth and confers autophagy resistance. Cell Cycle. 2012;11(22):4174–4180. doi: 10.4161/cc.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sohn Y.S., Tamir S., Song L., Michaeli D., Matouk I., Conlan A.R., Harir Y., Holt S.H., Shulaev V., Paddock M.L., Hochberg A., Cabanchick I.Z., Onuchic J.N., Jennings P.A., Nechushtai R., Mittler R. NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc. Natl. Acad. Sci. U. S. A. 2013;110(36):14676–14681. doi: 10.1073/pnas.1313198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Darash-Yahana M., Pozniak Y., Lu M., Sohn Y.S., Karmi O., Tamir S., Bai F., Song L., Jennings P.A., Pikarsky E., Geiger T., Onuchic J.N., Mittler R., Nechushtai R. Breast cancer tumorigenicity is dependent on high expression levels of NAF-1 and the lability of its Fe-S clusters. Proc. Natl. Acad. Sci. U. S. A. 2016;113(39):10890–10895. doi: 10.1073/pnas.1612736113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Paddock M.L., Wiley S.E., Axelrod H.L., Cohen A.E., Roy M., Abresch E.C., Capraro D., Murphy A.N., Nechushtai R., Dixon J.E., Jennings P.A. MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc. Natl. Acad. Sci. U. S. A. 2007;104(36):14342–14347. doi: 10.1073/pnas.0707189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhou T., Lin J., Feng Y., Wang J. Binding of reduced nicotinamide adenine dinucleotide phosphate destabilizes the iron-sulfur clusters of human mitoNEET. Biochemistry. 2010;49(44):9604–9612. doi: 10.1021/bi101168c. [DOI] [PubMed] [Google Scholar]
- 196.Colca J.R., McDonald W.G., Waldon D.J., Leone J.W., Lull J.M., Bannow C.A., Lund E.T., Mathews W.R. Identification of a novel mitochondrial protein ("mitoNEET") cross-linked specifically by a thiazolidinedione photoprobe. Am. J. Physiol. Endocrinol. Metab. 2004;286(2):E252–E260. doi: 10.1152/ajpendo.00424.2003. [DOI] [PubMed] [Google Scholar]
- 197.Laleve A., Vallieres C., Golinelli-Cohen M.P., Bouton C., Song Z., Pawlik G., Tindall S.M., Avery S.V., Clain J., Meunier B. The antimalarial drug primaquine targets Fe-S cluster proteins and yeast respiratory growth, Redox Biol. 2016;7:21–29. doi: 10.1016/j.redox.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kohanski M.A., Dwyer D.J., Hayete B., Lawrence C.A., Collins J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130(5):797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 199.Dwyer D.J., Kohanski M.A., Hayete B., Collins J.J. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Duan X., Huang X., Wang X., Yan S., Guo S., Abdalla A.E., Huang C., Xie J. l-Serine potentiates fluoroquinolone activity against Escherichia coli by enhancing endogenous reactive oxygen species production. J. Antimicrob. Chemother. 2016;71(8):2192–2199. doi: 10.1093/jac/dkw114. [DOI] [PubMed] [Google Scholar]
- 201.Choby J.E., Mike L.A., Mashruwala A.A., Dutter B.F., Dunman P.M., Sulikowski G.A., Boyd J.M., Skaar E.P. A small-molecule inhibitor of iron-sulfur cluster Assembly uncovers a link between virulence regulation and metabolism in Staphylococcus aureus. Cell Chem Biol. 2016;23(11):1351–1361. doi: 10.1016/j.chembiol.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ralph S.A., Van Dooren G.G., Waller R.F., Crawford M.J., Fraunholz M.J., Foth B.J., Tonkin C.J., Roos D.S., McFadden G.I. Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat. Rev. Microbiol. 2004;2(3):203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 203.Gisselberg J.E., Dellibovi-Ragheb T.A., Matthews K.A., Bosch G., Prigge S.T. The suf iron-sulfur cluster synthesis pathway is required for apicoplast maintenance in malaria parasites. PLoS Pathog. 2013;9(9):e1003655. doi: 10.1371/journal.ppat.1003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cicchillo R.M., Lee K.-H., Baleanu-Gogonea C., Nesbitt N.M., Krebs C., Booker S.J. Escherichia coli lipoyl synthase binds two distinct [4Fe− 4S] clusters per polypeptide. Biochemistry. 2004;43(37):11770–11781. doi: 10.1021/bi0488505. [DOI] [PubMed] [Google Scholar]
- 205.Mather M.W., Henry K.W., Vaidya A.B. Mitochondrial drug targets in apicomplexan parasites. Curr. Drug Targets. 2007;8(1):49–60. doi: 10.2174/138945007779315632. [DOI] [PubMed] [Google Scholar]
- 206.Berthiaume J., Wallace K.B. Adriamycin-induced oxidative mitochondrial cardiotoxicity. Cell Biol. Toxicol. 2007;23(1):15–25. doi: 10.1007/s10565-006-0140-y. [DOI] [PubMed] [Google Scholar]
- 207.Ichikawa Y., Ghanefar M., Bayeva M., Wu R., Khechaduri A., Prasad S.V.N., Mutharasan R.K., Naik T.J., Ardehali H. Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation. J. Clin. Invest. 2014;124(2):617–630. doi: 10.1172/JCI72931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Myers C. The role of iron in doxorubicin-induced cardiomyopathy. Semin. Oncol. 1998:10–14. [PubMed] [Google Scholar]
- 209.Pacher P., Liaudet L., Bai P., Virag L., Mabley J., Hasko G., Szabo C. Activation of poly (ADP-ribose) polymerase contributes to development of doxorubicin-induced heart failure. J. Pharmacol. Exp. Therapeut. 2002;300(3):862–867. doi: 10.1124/jpet.300.3.862. [DOI] [PubMed] [Google Scholar]
- 210.Schroeder P.E., Hasinoff B.B. Metabolism of the one-ring open metabolites of the cardioprotective drug dexrazoxane to its active metal-chelating form in the rat. Drug Metab. Dispos. 2005;33(9):1367–1372. doi: 10.1124/dmd.105.005546. [DOI] [PubMed] [Google Scholar]
- 211.Lebrecht D., Geist A., Ketelsen U.P., Haberstroh J., Setzer B., Walker U. Dexrazoxane prevents doxorubicin-induced long-term cardiotoxicity and protects myocardial mitochondria from genetic and functional lesions in rats. Br. J. Pharmacol. 2007;151(6):771–778. doi: 10.1038/sj.bjp.0707294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Dickey J.S., Gonzalez Y., Aryal B., Mog S., Nakamura A.J., Redon C.E., Baxa U., Rosen E., Cheng G., Zielonka J. Mito-tempol and dexrazoxane exhibit cardioprotective and chemotherapeutic effects through specific protein oxidation and autophagy in a syngeneic breast tumor preclinical model. PLoS One. 2013;8(8):e70575. doi: 10.1371/journal.pone.0070575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Batova A., Altomare D., Chantarasriwong O., Ohlsen K.L., Creek K.E., Lin Y.C., Messersmith A., Yu A.L., Yu J., Theodorakis E.A. The synthetic caged garcinia xanthone cluvenone induces cell stress and apoptosis and has immune modulatory activity. Mol. Cancer Therapeut. 2010;9(11):2869–2878. doi: 10.1158/1535-7163.MCT-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Geldenhuys W.J., Funk M.O., Barnes K.F., Carroll R.T. Structure-based design of a thiazolidinedione which targets the mitochondrial protein mitoNEET. Bioorg. Med. Chem. Lett. 2010;20(3):819–823. doi: 10.1016/j.bmcl.2009.12.088. [DOI] [PubMed] [Google Scholar]
- 215.Logan S.J., Yin L., Geldenhuys W.J., Enrick M.K., Stevanov K.M., Carroll R.T., Ohanyan V.A., Kolz C.L., Chilian W.M. Novel thiazolidinedione mitoNEET ligand-1 acutely improves cardiac stem cell survival under oxidative stress. Basic Res. Cardiol. 2015;110(2):19. doi: 10.1007/s00395-015-0471-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Colca J.R. Insulin sensitizers may prevent metabolic inflammation. Biochem. Pharmacol. 2006;72(2):125–131. doi: 10.1016/j.bcp.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 217.Yki-Jarvinen H. Thiazolidinediones. N. Engl. J. Med. 2004;351(11):1106–1118. doi: 10.1056/NEJMra041001. [DOI] [PubMed] [Google Scholar]
- 218.Kispal G., Sipos K., Lange H., Fekete Z., Bedekovics T., Janaky T., Bassler J., Aguilar Netz D.J., Balk J., Rotte C., Lill R. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 2005;24(3):589–598. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Khoshnevis S., Gross T., Rotte C., Baierlein C., Ficner R., Krebber H. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010;11(3):214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Dong J., Lai R., Nielsen K., Fekete C.A., Qiu H., Hinnebusch A.G. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem. 2004;279(40):42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- 221.Hirabayashi K., Yuda E., Tanaka N., Katayama S., Iwasaki K., Matsumoto T., Kurisu G., Outten F.W., Fukuyama K., Takahashi Y. Functional dynamics revealed by the structure of the SufBCD complex, a novel ATP-binding cassette (ABC) protein that serves as a scaffold for iron-sulfur cluster biogenesis. J. Biol. Chem. 2015;290(50):29717–29731. doi: 10.1074/jbc.M115.680934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Loiseau L., Ollagnier-de-Choudens S., Nachin L., Fontecave M., Barras F. Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J. Biol. Chem. 2003;278(40):38352–38359. doi: 10.1074/jbc.M305953200. [DOI] [PubMed] [Google Scholar]
- 223.Fontecave M., De Choudens S.O., Py B., Barras F. Mechanisms of iron–sulfur cluster assembly: the SUF machinery. JBIC Journal of Biological Inorganic Chemistry. 2005;10(7):713–721. doi: 10.1007/s00775-005-0025-1. [DOI] [PubMed] [Google Scholar]
- 224.Wollers S., Layer G., Garcia-Serres R., Signor L., Clemancey M., Latour J.-M., Fontecave M., de Choudens S.O. Iron-sulfur (Fe-S) cluster assembly: the SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor. J. Biol. Chem. 2010;285(30):23331–23341. doi: 10.1074/jbc.M110.127449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Fast N.M., Kissinger J.C., Roos D.S., Keeling P.J. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol. 2001;18(3):418–426. doi: 10.1093/oxfordjournals.molbev.a003818. [DOI] [PubMed] [Google Scholar]
- 226.Lee M., Gräwert T., Quitterer F., Rohdich F., Eppinger J., Eisenreich W., Bacher A., Groll M. Biosynthesis of isoprenoids: crystal structure of the [4Fe–4S] cluster protein IspG. J. Mol. Biol. 2010;404(4):600–610. doi: 10.1016/j.jmb.2010.09.050. [DOI] [PubMed] [Google Scholar]
- 227.Rekittke I., Wiesner J., Röhrich R., Demmer U., Warkentin E., Xu W., K. Troschke. Hintz M., No J.H., Duin E.C. Structure of (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase, the terminal enzyme of the non-mevalonate pathway. J. Am. Chem. Soc. 2008;130(51):17206–17207. doi: 10.1021/ja806668q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zepeck F., Gräwert T., Kaiser J., Schramek N., Eisenreich W., Bacher A., Rohdich F. Biosynthesis of isoprenoids. Purification and properties of IspG protein from Escherichia coli. J. Org. Chem. 2005;70(23):9168–9174. doi: 10.1021/jo0510787. [DOI] [PubMed] [Google Scholar]
- 229.Pierrel F., Björk G.R., Fontecave M., Atta M. Enzymatic modification of tRNAs: MiaB is an iron-sulfur protein. J. Biol. Chem. 2002;277(16):13367–13370. doi: 10.1074/jbc.C100609200. [DOI] [PubMed] [Google Scholar]
- 230.Arcamone F., Cassinelli G., Fantini G., Grein A., Orezzi P., Pol C., Spalla C. Adriamycin, 14-hydroxydaimomycin, a new antitumor antibiotic from S. Peucetius var. caesius, Biotechnology and bioengineering. 1969;11(6):1101–1110. doi: 10.1002/bit.260110607. [DOI] [PubMed] [Google Scholar]
- 231.Lefrak E.A., Piťha J., Rosenheim S., Gottlieb J.A. A clinicopathologic analysis of adriamycin cardiotoxicity. Cancer. 1973;32(2):302–314. doi: 10.1002/1097-0142(197308)32:2<302::aid-cncr2820320205>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 232.Steinherz L.J., Steinherz P.G., Tan C.T., Heller G., Murphy M.L. Cardiac toxicity 4 to 20 years after completing anthracycline therapy. Jama. 1991;266(12):1672–1677. [PubMed] [Google Scholar]
- 233.Mukhopadhyay P., Rajesh M., Yoshihiro K., Haskó G., Pacher P. Simple quantitative detection of mitochondrial superoxide production in live cells. Biochem. Biophys. Res. Commun. 2007;358(1):203–208. doi: 10.1016/j.bbrc.2007.04.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Pacher P., Liaudet L., Mabley J.G., Cziráki A., Haskó G., Szabó C. Beneficial effects of a novel ultrapotent poly (ADP-ribose) polymerase inhibitor in murine models of heart failure. Int. J. Mol. Med. 2006;17(2):369–375. [PMC free article] [PubMed] [Google Scholar]
- 235.Xu X., Persson H., Richardson D. Molecular pharmacology of the interaction of anthracyclines with iron. Mol. Pharmacol. 2005;68(2):261–271. doi: 10.1124/mol.105.013383. [DOI] [PubMed] [Google Scholar]