Abstract
Asbestos-associated diseases remain a social burden worldwide. Our previous studies identified asbestos-induced iron-rich milieu for mesothelial cells with ceaseless macrophage ferroptosis. However, molecular mechanisms how this mutagenic milieu influences mesothelial cells have not been elucidated yet. Here, we propose a novel mechanism that extracellular vesicles (EVs) mediate asbestos-associated mutagenic factors to mesothelial cells. In a mice model of intraperitoneal crocidolite injection, mutagenic milieu highly expressed CD63, an exosomal marker. We then used a GFP-CD63 labeled THP-1 macrophage model exposed to crocidolite/iron, which generated EVs under ferroptotic process. We observed that MeT-5A mesothelial cells can receive and internalize these EVs. Furthermore, we comprehensively analyzed the ferroptosis-dependent EVs (FedEVs) for transported proteins and identified ferritin heavy/light chains as major components. Therefore, we inferred that FedEVs transport iron from ferroptotic macrophages to mesothelial cells. RNA sequencing revealed that the mesothelial cells receiving higher amounts of the FedEVs were mitotic, especially at the S and G2/M phases, by the use of Fucci mesothelial cells. Nuclear 8-hydroxy-2′-deoxyguanosine and γ-H2AX were significantly increased in the recipient mesothelial cells after exposure to FedEVs. Collectively, we here demonstrate a novel mechanism that FedEVs act as a key mutagenic mediator by transporting iron, which contribute to asbestos-induced mesothelial carcinogenesis.
Keywords: Extracellular vesicle, Asbestos, Ferroptosis, CD63
Graphical abstract
Highlights
-
•
Asbestos induces ferroptosis in macrophages, which secrete extracellular vesicles.
-
•
Extracellular vesicles carry asbestos-derived mutagenic factor to mesothelial cells.
-
•
Major mutagenic factor transported by extracellular vesicle (EV) is ferritin.
-
•
Mesothelial cells receiving EVs started mitosis with increased oxidative DNA damage.
Abbreviations
- BFP
Blue-fluorescent protein
- CDK
Cyclin-dependent kinase
- DAVID
The Database for Annotation, Visualization and Integrated Discovery
- DEGs
Differentially expressed genes
- DFO
Desferal (desferrioxamine)
- EMEM
Eagle's minimum essential medium
- EMT
Epithelial mesenchymal transition
- FACS
Fluorescence-activated cell sorting
- FBS
Fetal bovine serum
- FedEVs
Ferroptosis-dependent extracellular vesicles
- FDR
False discovery rate
- GFP
Green fluorescent protein
- GPX4
Glutathione peroxidase 4
- GSEA
Gene set enrichment analysis
- HPLC
High-pressure liquid chromatography
- IF
Immunofluorescence
- ip
Intraperitoneally
- LC-MS
Liquid chromatography-mass spectrometry
- NTA
Nano tracking analysis
- PBS
Phosphate-buffered saline
- PCA
Principal component analysis
- SEM
Standard error of means
- SDS-PAGE
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TEM
Transmission electron microscopy
1. Introduction
Pleural and peritoneal malignant mesotheliomas have been associated with occupational and/or environmental exposures to asbestos, such as crocidolite, amosite, and chrysotile [1]. Crocidolite may be responsible for more deaths than any other type of asbestos due to its rigid linearity and thinness, with the ratio of potency for mesothelioma (pleural and peritoneal combined) being 500:100:1 for crocidolite, amosite and chrysotile, respectively [2]. Asbestos-associated carcinogenesis has been studied from two distinct standpoints, direct and/or indirect effects [3,4]. Direct effects suggest that asbestos fibers phagocytosed by mesothelial cells [5], reach inside the nucleus and generate mutations almost physically [[6], [7], [8]]. On the other hand, indirect effects hypothesize that asbestos localizes inside macrophages, and the oxidative stress based on frustrated phagocytosis induces mesothelial genetic alteration through activation of Nalp3 inflammasome and tissue remodeling processes [9,10]. Our previous study revealed that a month after the injection the asbestos fibers are located within macrophages and not in mesothelial cells whereas DNA damage in mesothelial cells was increased close to the asbestos-induced mutagenic milieu [11]. However, little is known on the molecular mechanisms how this meutagenic milieu influences mesothelial cells toward carcinogenesis.
Extracellular vesicles (EVs) are complex structures, composed of a lipid bilayer that deliver various DNA, RNA and proteins to their recipient cells and mediate multiple physiological phenomena. Exosomes are, among them, endosomal-derived and are 10–150 nm in diameter [12,13]. In contrast, large extracellular vesicles are generated by direct outward budding of the plasma membrane, producing microvesicles, microparticles and large vesicles ranging from ∼100 nm to 1 μm in diameter [14]. Munson and colleagues firstly reported on exosomes from asbestos-exposed lung epithelial cells and macrophages. The exosomes after asbestos exposure contained an abundance of vimentin, thrombospondin, superoxide dismutase and glypican-1, where its exposure to mesothelial cells have resulted in alteration of gene expression, associated with epithelial to mesenchymal transition (EMT) and cancer-related genes, including EGR1 and CCNB2 [15]. Our previous reports showed that the asbestos-related inflammation induced an excess of iron in macrophages, and that this iron excess is the trigger of ferroptosis in macrophages engulfing asbestos [11]. Further, we recently found that ferritin secretion is mediated by CD63-positive EVs under the regulation of IRE-IRP system [16]. Thus, we hypothesized that ferroptosis-dependent EVs (FedEVs) may mediate iron and mutagenic factors, as a novel mechanism of mutagenic niche.
Recently researchers have focussed on exosomes and microvesicles derived from viable cells or apoptotic cells. However, EVs derived from necrotic cells are poorly elucidated. Baxter and colleagues analyzed extracellular vesicles generated from several types of cell death and showed that the secretion of EVs were increased in cell death conditions, especially under membrane permeabilization [17]. We previously observed asbestos-induced lysosome permeabilization in macrophages, which thereafter initiated ferroptosis [11]. Thus, the mutagenic microenvironment derived from asbestos may result in FedEVs production.
Here, we hypothesized that macrophages that engulfed asbestos generate FedEVs, which harbor mutagenic and pathogenic factors associated with asbestos, and that recipient mesothelial cells located a distance away from asbestos would have substantial genomic damage. Our findings suggest that indirect effects are also crucial for mesothelial cell mutagenesis and that asbestos mutagenic factors ride on FedEVs.
2. Experimental procedures
2.1. Reagents
EMEM (Wako, 051–07615), FBS (Biowest, S1810-500; Nuaillé, France), exosome-depleted FBS (Exo-FBS™, Gibco, A27208-03), erastin (Selleck Chemicals, S7242), RSL3 (MedChemExpress, HY-100218A), ferric ammonium citrate (FAC; Sigma Aldrich, F5879), RhoNox-4 (Goryo Chemical; FerroOrange, GC904-01), ExoSparkler Exosome Membrane Labeling kit-Deep Red (Dojindo, EX03), phorbol 12-myristate 13-acetate (PMA; Wako, 162–23591), polyethylene glycol 6000 (Wako, 167–22941), Autofluorescence Quenching Kit (Vector, SP-8400), Aurum™ Total RNA Mini Kit (BioRad, 64337836), NEBNext Ultra II Directional RNA Library prep kit for Illumina (BioLabs, E7760S), NEBNext Multiplex Oligos for Illumina (BioLabs, E6440S), desferal (DFO, Novartis), anti-CD63 (Thermo Fischer Scientific, #10628D), anti-mesothelin (IBL, #28127), anti-αSMA (Abcam, ab5694), anti-CD68 (CST, 97778S), anti-GFP (MBL life science, #598), anti-TfR1 (Thermo Fischer Scientific, #13–6800), anti-FtL (abcam, ab69090), anti-FtH (SantaCruz, sc-H53), anti-IRP1 (CST, #20272), anti-IRP2 (Novus, NB100-1798), Cell Cycle Regulation Antibody Sampler Kit (CST, #9932), anti-mouse IgG Alexa488 (Thermo Fischer Scientific, A11108) and anti-rabbit IgG Alexa 568 (Thermo Fischer Scientific, A-11011) were used.
2.2. Cell experiments and DNA construction
MeT-5A human mesothelial cell line (ATCC), THP1 human macrophage cell line and HEK293T cell line (RIKEN Cell Bank) and HT1080 human sarcoma cell line (JCRB cell bank) were grown under standard sterile cell culture conditions (37 °C, humidified atmosphere, 5% CO2) in EMEM containing 10% FBS and 1% antibiotic-antimycotic (Gibco). Cells were used within two months after thawing. To establish macrophage cells producing GFP-labeled EVs, human CD63, CD9 and CD81 cDNA were cloned via PCR amplification into FUGW-modified lentiviral vector (#14883, Addgene [18]) with EcoRI. THP1 and HT1080 cells were transduced by lentivirus encoding CD63, CD9 and CD81 cDNA, and stable cell lines were obtained by the limiting dilution method. The genes, encoding Orange or Green Fucci as cell cycle indicators [19], were amplified with PCR from the pFucci-G1 orange vector or pFucci-G2/M green vector (MBL Life Sciences) and cloned into FUGW-modified lentiviral vector (#14883, Addgene) with EcoRI and XbaI. MeT-5A cells were transduced by lentivirus, encoding Fucci orange (G1-phase) and green (G2/M-phase), and stable cell lines were cloned by limiting dilution method.
2.3. Animal experiments
Asbestos-induced peritonitis model was produced as previously described [11]. Briefly, male C57BL/6 N mouse (SLC Japan, Shizuoka, Japan; 6-wks old; n = 9) were injected intraperitoneally (ip) with a suspension of 3 mg crocidolite (UICC). After six months, we collected and dissected organs for histological analysis at autopsy after euthanasia. The animal experiment committee of Nagoya University Graduate School of Medicine approved the experiments.
2.4. Isolation and quantification of EVs
To prepare EV samples, the culture medium of THP1 and HT1080 cells were changed to the fresh culture medium containing 1% or 10% Exosome-depleted FBS (Exo-FBS™, Gibco) and incubated for 96 h or 24 h, respectively. Crocidolite exposure under iron-rich condition was 15 μg/cm2 of crocidolite and 100 μg/ml of ferric ammonium citrate up to 96 h. HT1080 fibrosarcoma cells were exposed to RSL3 (10 μM, 24 h) as a representative ferroptosis inducer. The collected conditioned culture media were centrifuged at 500×g for 5 min and 2000×g for 20 min sequentially to remove cell debris and large EVs (mainly microvesicles), finally to obtain supernatants, containing EVs (mainly exosomes). To isolate large EVs, the pellet of large EVs were suspended with PBS and centrifugated at 2000×g for 20 min. To isolate small and/or FedEVs, two methods were performed. Regarding the ultracentrifugation method, the supernatants were further centrifuged at 100,000×g for 90 min. The pellets were washed with phosphate-buffered saline (PBS) and recentrifuged at 100,000×g for 90 min. The pellets were suspended in PBS and stored at −80 °C until use for transmission electron microscopy (TEM) analysis [20] and proteomic analysis [21,22] as described. Regarding the polymer method, the supernatants were mixed to an equal volume of 2 x polyethylene glycol (PEG; PEG6000, Wako) solution at 4 °C, to achieve a desired final PEG concentration (8%) as previously described [23]. Briefly, after overnight incubation, samples were centrifuged at 2900×g for 80 min at 4 °C. Conical tubes were then decanted and allowed to drain for 5 min to remove excess PEG. The resulting pellet was suspended in 100 μl PBS (pH 7.4) and stored at −80 °C until use for immunoblots and EV uptake analysis.
2.5. Nano Tracking analysis (NTA)
Particle size distribution in all EV samples was determined by NTA using a NanoSight NS300 system (Malvern Technologies, Malvern, UK) configured with a 488 nm laser and high sensitivity scientific CMOS camera. Samples were diluted (1:20–1:500) in particle-free PBS to an acceptable concentration, according to the manufacturer's recommendations. Samples were analyzed under constant flow conditions at 25 °C. For bootstrapped samples, 30 s × 60 successive videos were captured with a camera level of 16. Data were analyzed using NTA 3.1.54 software with a detection threshold of 5.
2.6. Mass spectrometry
This was performed as described [21,22]. Briefly, proteins were digested with trypsin for 16 h at 37 °C after reduction and alkylation. The peptides were analyzed by liquid chromatography-mass spectrometry (LC−MS) using an Orbitrap Fusion mass spectrometer (ThermoFisher Scientific Inc., Waltham, MA), coupled with an UltiMate3000 RSLCnano LC system (Dionex Co., Amsterdam, The Netherlands) using a nano HPLC capillary column, 150 mm × 75 μm i.d. (Nikkyo Technos Co., Japan) via a nanoelectrospray ion source. Reversed-phase chromatography was performed with a linear gradient (0 min, 5% B; 100 min, 40% B) of solvent A (2% acetonitrile with 0.1% formic acid) and solvent B (95% acetonitrile with 0.1% formic acid) at a flow rate of 300 L/min. A precursor ion scan was carried out using a 400–1600 mass to charge ratio (m/z) prior to MS/MS analysis. Tandem MS was performed by isolation at 0.8 Th with the quadrupole HCD fragmentation with a normalized collision energy of 30% and rapid scan MS analysis in the ion trap. Only precursors with charge states 2–6 were sampled for MS2. The dynamic exclusion duration was set to 15 s with a 10-ppm tolerance. The instrument was run in top speed mode with 3 s-cycles. The raw data was processed using Proteome Discoverer 1.4 (Thermo Fisher Scientific) in conjunction with the MASCOT search engine, version 2. 6.0 (Matrix Science Inc., Boston, MA) for protein identification. Peptides and proteins were identified against the human or mammal protein database in UniProt (release 2020_04), with a precursor mass tolerance of 10 ppm, and a fragment ion mass tolerance of 0.8 Da. The fixed modification was set to carbamidomethylation of cysteine, and variable modifications were set to oxidation of methionine. Two missed cleavages by trypsin were allowed.
2.7. FedEV uptake assay
1 × 106 cells were cultured in a 6-well plate with media containing 1% EV-depleted FBS prior to FedEVs loading. After overnight incubation, FedEVs from THP1 cells of 10 cm2 dish were loaded in a total volume of 100 μl in PBS. After 24 h of incubation, the supernatant was removed. Then, these cell samples were used for FACS analysis, IF analysis and immunoblot analysis. To detect FedEV uptake in each cell cycle, the FedEVs suspended in PBS were fluorescently labeled using an ExoSparker Exosome Membrane Labeling kit according to the manufacture's protocols. Then, the labeled FedEVs were loaded at the same concentration and volume with other FedEV uptake assay. The effect of iron chelator on FedEV uptake was evaluated by incubating MeT-5A mesothelial cells with DFO (0–100 μM) for 24 h.
2.8. Image acquisition and quantitative analysis
Fluorescent images of granuloma sections and CD63-positive FedEVs were acquired using a SpinSR10 confocal laser scanning microscope with CellSens Dimension software (Olympus). ImageJ Fiji and photoshop (version 22.0, Adobe) were used for image processing.
2.9. RNA sequencing and data analysis
MeT-5A cells that have taken up FedEVs [GFP-CD63(+)] were isolated by FACS Melody (∼7 × 105 cells). Total RNA was isolated, using Aurum™ Total RNA Mini Kit (BioRad) according to the manufacture's protocol. RNA quality was assessed, using a 2100 BioAnalyzer (Agilent). RNA of 100 ng was used for the preparation of sequencing libraries with NEBNext Ultra II Directional RNA Library Prep with Sample Purification Beads (NEB) according to the manufacturer's instructions. Libraries were validated with the BioAnalyzer and quantified by qPCR and Qubit Fluorometric Quantitation (Thermo Fischer). We used the Novaseq (Illumina) which generates 150-bp pair-end reads. Experiments were performed in triplicate.
RNA-seq data analysis was performed as described with some modifications [24]. The quality assessment of raw sequence data was performed using FastQC (Version: FastQC 0.10.0). We performed transcript quantification of paired-end sequence reads using Salmon (v1.3.0) [25] in the mapping-based mode with the transcriptome index created from the Homo sapiens genome assembly (GRCh38.p13 Ensembl release 103). Transcript-level count data was aggregated at the gene-level using tximport (version 1.20.0) [26]. Differentially gene expression analysis between FedEVs-high and -low was performed using the DESeq2 package (version 1.32.0) on protein-coding genes [27]. Differentially expressed genes were selected, using an FDR-adjusted p-value cut-off < 0.05 and an absolute log2 fold change >0.5. Gene symbols were annotated using biomaRt (BioConductor version 3.13). Gene ontology analysis was performed using online bioinformatic tools, Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.8). Heat maps were generated with iDEP.93 or the function pheatmap from the R package gplots. GSEA on specific gene sets was performed with GSEA Desktop v3.0 and version 6.1 of the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb, Broad Institute, MIT) using the following collections: hallmark (H), curated (C2) and GO (C5).
2.10. Immunoblot analysis
Cell line samples were lysed by RIPA buffer containing Pierce™ Protease and Phosphatase inhibitor (Thermo Scientific) and homogenized by super sonication and clarified by centrifugation at 18,000 × g for 30 min at 4 °C. Total protein concentration in lysates was quantified, using Protein Assay Bicinchoninate kit (Nacalai tesque). Protein extraction, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as previously described [28].
2.11. FACS analysis
MeT-5A cells which took up FedEVs were harvested by trypsinization, and resuspended in PBS supplemented with 1% FBS and the cell suspension was passed through a 100 μm mesh filter. For cell sorting, cells were treated with Hoechst33342 to exclude dead cells. Cell sorting was performed with a FACS Melody (BD Biosciences), using a 100 μm nozzle and FACS Chorus software. FACS data were analyzed, using Flowing Software version 2.5.1.
2.12. Statistical analyses
Statistical analyses were performed, using Graphpad Prism software (version 8.4.3) or the R statistical environment (http://r-project.org). All the data are presented as means ± SEM unless indicated otherwise. Data was tested for normal distribution with unpaired two-tailed Student t-test (if two samples have equal variances) or Welch's t-tests (if two samples have unequal variances) to determine statistical significance between the two groups. For the analysis of statistical significance among more than two groups, we performed one-way ANOVA with Tukey's multiple comparison test to assess statistical significance with a 95% confidence interval. P < 0.05 was considered significant unless stated otherwise. All the experiments were performed at least three times independently unless otherwise stated.
2.13. Data availability
All relevant data supporting the results of the present study are included within the article can be obtained from the corresponding author on reasonable request. The original RNA-seq data is available from the Gene Expression Omnibus (GEO) database under accession code GSE184368. A source Data file is included which contains the raw data underlying the reported averages in all figures and supplementary figures.
3. Results
3.1. Ferroptosis-dependent extracellular vesicles (FedEVs) are condensed in the asbestos-induced mutagenic milieu
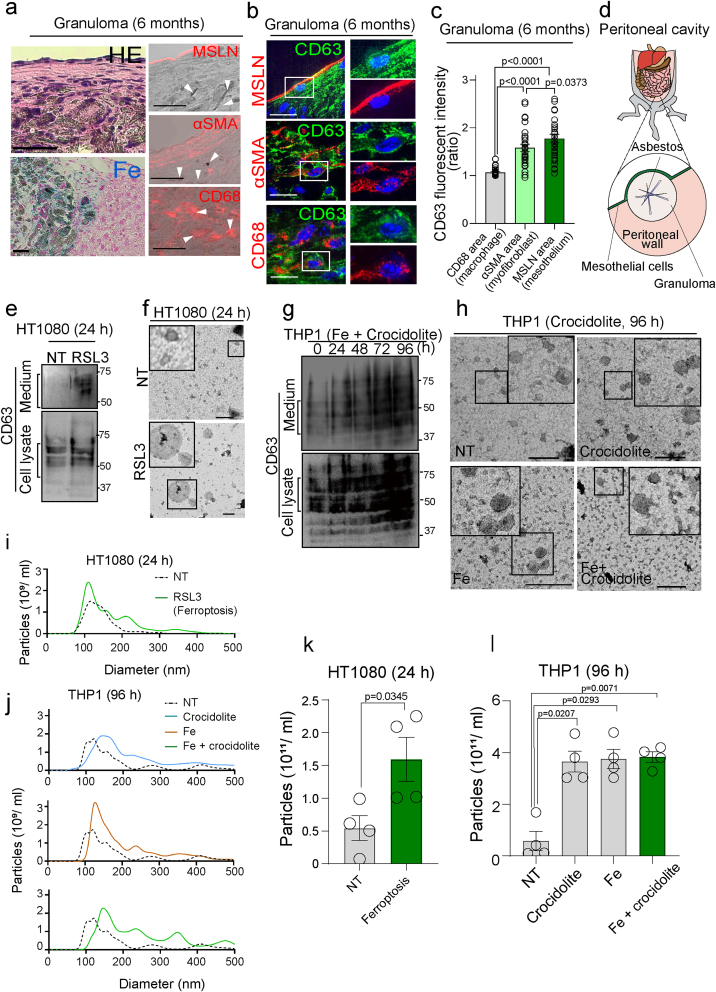
We first performed histological evaluation, using a mouse model of ip crocidolite injection to elucidate the tissue localization of FedEVs in the asbestos-induced granuloma. Six months after the asbestos injection, we observed chronic granulomatous peritonitis as a reaction to foreign material with excess iron, similar to those in our previous report [11] (Fig. 1ad). Immunofluorescence analysis revealed that the asbestos fibers were inside CD68-positive macrophages and neither in the α-SMA-positive myofibroblasts nor mesothelin-positive mesothelial cells (Fig. 1a and Fig. S1ab). Of note, CD63, a marker for EVs, were abundant with a granular pattern in mesothelial cells and myofibroblast cells in proximity to the granuloma (Fig. 1bc). In our previous report, the asbestos-induced macrophage death under iron-rich environment included ferroptosis as a major fraction [11]. Thus, we sought to assess whether typical ferroptosis enhances EVs secretion. Immunoblotting analysis of CD63 revealed that HT1080 cells under ferroptosis with a GPX4 inhibitor, RSL-3, significantly secreted EVs (Fig. 1ef). Similarly, asbestos exposure to THP1 macrophage cells under iron-rich environment increased the production and secretion of EVs (Fig. 1gh and Fig. S1cd and S3bc). Furthermore, we found with Nano Tracking analysis (NTA) assay that the number of EVs increased the most with crocidolite exposure under iron-rich condition (Fig. 1kl), with the diameter of 150 nm and the peak of size shifted at 250 nm and 350 nm (Fig. 1ij). These results indicate that asbestos-dependent macrophage ferroptosis is directly associated with EVs secretion in vivo, suggesting that asbestos-induced granuloma is an outgoing base of ferroptosis-dependent extracellular vesicles (FedEVs).
Fig. 1.
CD63-positive ferroptosis-dependent EVs (FedEVs) derived from macrophages simultaneously exposed to asbestos and iron in vivo and in vitro. (a) Histological images of peritoneal wall in situ in the murine model 6 months after ip injection of 3 mg crocidolite; hematoxylin and eosin (HE) image (left upper), Berlin blue staining (insoluble iron as bluish green, left lower), phase-contrast with red fluorescent image (right panels); cell-specific marker: MSLN (mesothelin), mesothelial cell; αSMA (smooth muscle actin), myofibroblast; CD68, macrophage; white arrowheads, crocidolite (bar = 100 μm). (b) High-resolution fluorescent image in granuloma; red, cell specific marker; green, CD63; blue, Hoechst33342 nuclear staining (bar = 20 μm). (c) Quantification of fluorescent intensity of CD63 in different cells surrounding granuloma (n = 30; mean ± SEM). (d) Schematic image of peritoneal wall surrounding the asbestos-induced granuloma. (e, g) Immunoblot analysis of CD63 in cells under ferroptosis and their media; HT1080 fibrosarcoma cells were exposed to RSL3 (10 μM, 24 h), and phorbol myristate acetate (PMA)-primed THP1 macrophage cells were exposed to crocidolite (15 μg/cm2) in the presence of absence of ferric ammonium citrate (Fe, 100 μg/ml) up to 96 h. (f, h) Membrane-catch transmission electronmicroscopy (TEM) images of EVs on carbon grids (bar = 200 nm). (i–l) The precision of nanoparticle tracking analysis (NTA) concentration measurements counted size-distribution of EVs derived from HT1080 (24 h) and THP1 (96 h) in different conditions in i and j. NTA concentration measurements counted number of EVs in k and l (mean ± SEM). Refer to text for details. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Mesothelial cells take up FedEVs
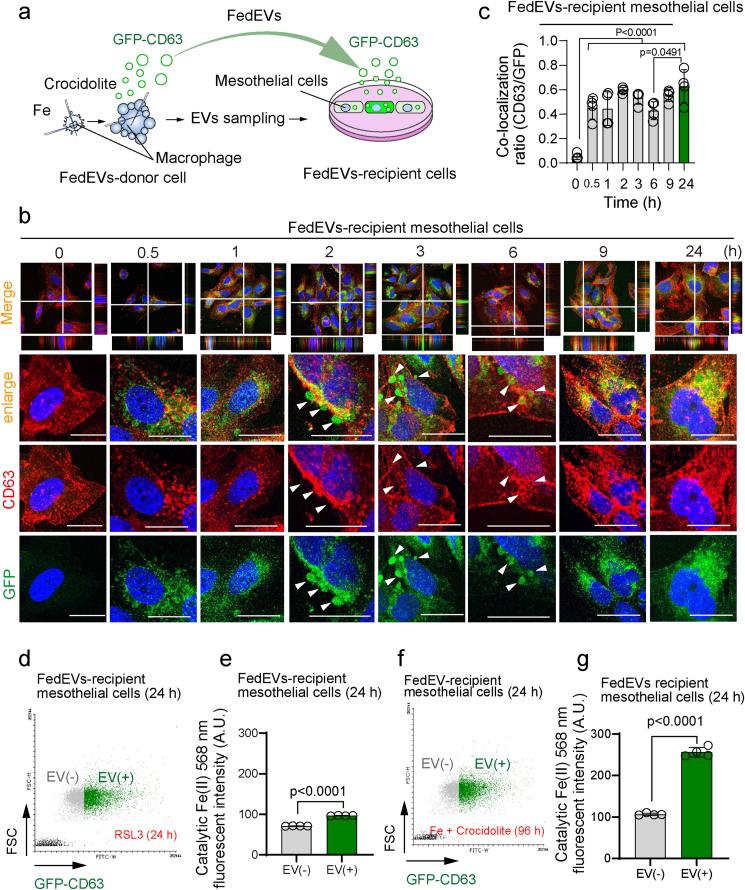
We next studied the possibility of FedEV uptake by the mesothelial cells. We established the THP1 and HT1080 cells, stably expressing GFP-CD63, as FedEV donor cells (Fig. 2a, Fig. S1cd). After addition of GFP-labeled FedEVs to MeT-5A mesothelial cells, GFP-CD63 originating from donor cells were observed on the plasma membrane of recipient MeT-5A cells, starting from 0.5 h, which continued till 3 h with the formation of bleb-like structures. FedEVs were completely internalized thereafter by the recipient mesothelial cells at 6–9 h, as seen by the coexistence of GFP-CD63 and endogenous CD63 in the cytoplasm (endosome ∼ late endosome ∼ lysosome) evaluated by the z-axis (Fig. 2bc). We then evaluated the amounts of catalytic Fe(II) in the mesothelial cells taking up FedEVs at 24 h, which showed significantly increased intracellular catalytic Fe(II) by FACS analysis (Fig. 2d–g). These results suggested that FedEVs may transport iron from ferroptotic macrophages to mesothelial cells.
Fig. 2.
Mesothelial cells phagocytose FedEVs derived from asbestos-exposed macrophages under ferroptosis. (a) Schema of culture system for the uptake by MeT-5A (recipient) cells of FedEVs from THP1 cells stably expressing GFP-CD63. (b) Fluorescent cellular images of recipient mesothelial cells phagocytosing GFP-positive FedEVs derived from THP1 macrophage cells exposed to asbestos; red, endogenous CD63; bar = 10 μm. (c) Time-course of quantitative colocalization ratio between GFP-CD63 and endogenous CD63 (mean ± SEM). (d, e) FACS analysis of MeT-5A mesothelial cells for catalytic Fe(II) (RhoNox-4, red) after exposure to GFP-CD63-labeled FedEVs derived from HT1080 by ferroptosis-stimulation with RSL3; mean ± SEM). (f, g) FACS analysis of MeT-5A mesothelial cells for catalytic Fe(II) (RhoNox-4, red) after exposure to GFP-CD63-labeled FedEVs derived from THP1 cells by ferroptosis-stimulation with [crocidolite + Fe]; mean ± SEM). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. FedEVs include ferritin as a major component
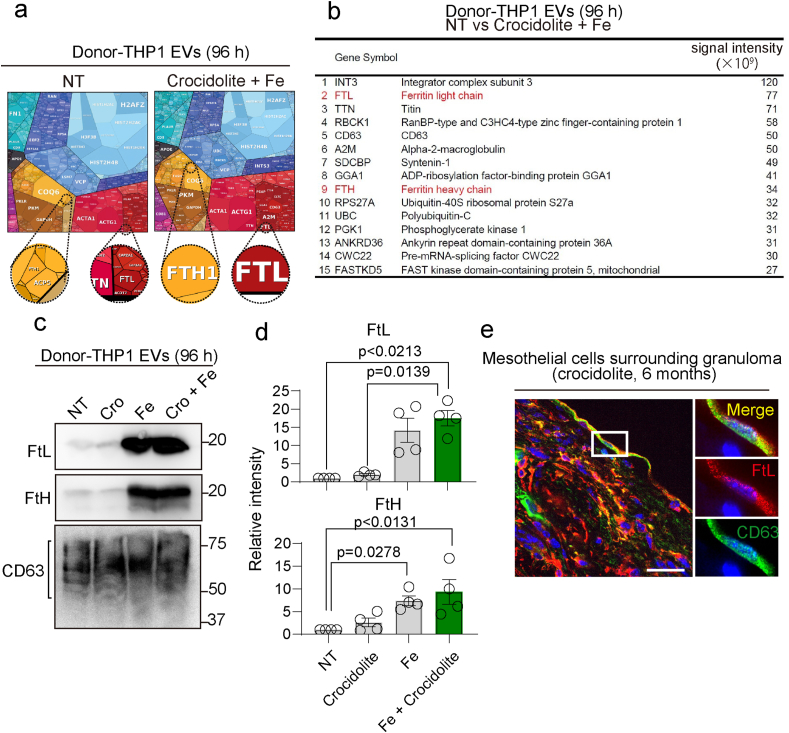
To comprehensively identify the protein components in FedEVs, we performed a proteomic analysis using LC-MS. We extracted proteins from FedEVs secreted from macrophage cells exposed to iron and/or crocidolite for 96 h. We integrated differentially expressed proteins (non-treated vs crocidolite and Fe) as proteomap images, accounting for the entire items and listed the top 15 differentially increased proteins (Fig. 3ab). Ferritin light and heavy chains (FTL and FTH) were the proteins, identified to be associated with iron storage in the FedEVs. Of note, only FedEVs from iron- or iron and crocidolite-treated groups contained a high amount of FtL and FtH (Fig. 3cd), which was associated with iron excess. We also confirmed co-localization in vivo with the FtL and CD63 on the mesothelial cells surrounding granuloma derived from the crocidolite (Fig. 3e). These results indicate that FedEVs can transport iron through ferritin.
Fig. 3.
Non-selective proteome analysis of asbestos-induced FedEVs reveals ferritins as major components. (a) Proteomaps depicting the fold changes and associated functions for all the identified proteins within FedEVs derived from THP1 macrophage cells (NT or crocidolite + Fe exposure). (b) Relative abundance of proteins in FedEVs (crocidolite + Fe) in comparison to EVs from untreated control THP1 cells. (c) Immunoblot analysis of FedEVs from THP1 macrophage. (d) Quantification of band intensity (n = 4; mean ± SEM). (e) Immunofluorescent analysis of peritoneal wall in situ in the murine model 6 months after ip injection of 3 mg crocidolite; CD63 (FedEVs, green); FtL (ferritin light chain, red) with blue Hoechst33342 nuclear staining (bar = 50 μm). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Coincidence of FedEVs uptake and mitosis in mesothelial cells
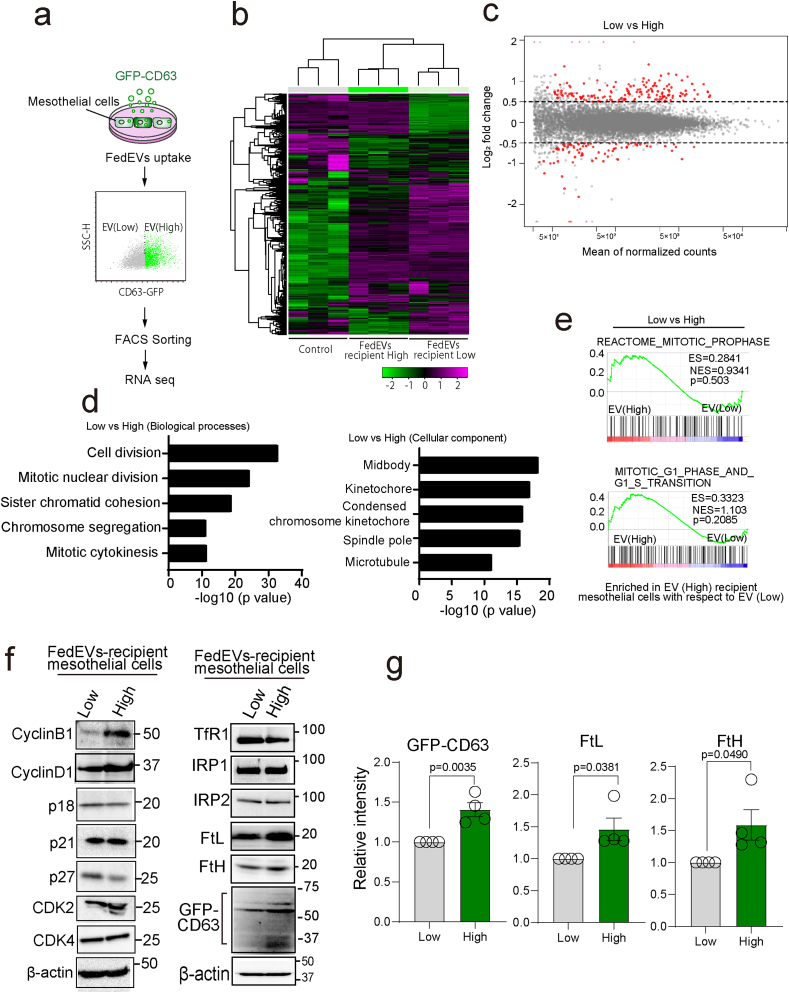
To gain more insight into the biological significance of FedEVs, we performed RNA sequencing and transcript analysis of the recipient mesothelial cells. This analysis was performed in triplicate on FedEVs-high or -low mesothelial cells upon FedEVs exposure, which was differentiated through GFP-CD63 of ferroptotic macrophage origin in comparison to non-treated control mesothelial cells (Fig. 4a). Differential gene expression analysis with an FDR-adjusted p-value < 0.01 and absolute log2 fold change >0.5 identified 245 differentially expressed genes, of which 163 were upregulated and 82 were downregulated (Fig. 4bc). Gene ontology (GO) analysis for biological process linked the gene expression changes in FedEVs recipient cells to cell cycle of mitotic phase, while the midbody and kinetochore were overrepresented cellular component (Fig. 4d). Gene set enrichment analysis (GSEA) of upregulated genes also revealed a significant upregulation of mitosis-associated gene set in FedEVs-high recipient mesothelial cells (Fig. 4e). Particularly, expression of Cyclin B1 and Cyclin D1 was increased in the recipient mesothelial cells with high-FedEVs (Fig. 4fg). These results indicate that FedEVs uptake and mitotic process were coincident in the recipient mesothelial cells.
Fig. 4.
FedEVs-high mesothelial cells are under mitosis. (a) Schema of culture system and FACS sorting for RNA-sequencing analysis of MeT-5A mesothelial cells with differential phagocytosis of FedEVs. (b) Comparison of transcriptional profiles of mesothelial cells with non-treatment (NT), FedEVs low-uptake (Low) and FedEVs high-uptake (High). Magenta and green colors indicate upregulation and downregulation, respectively. (c) MA-plots of differentially expressed genes (DEGs) between low FedEVs-uptake (Low), high FedEVs-uptake (High) mesothelial cells. The X-axis represents the mean of normalized counts and the Y-axis shows the log2 fold change >0.5. Red dots correspond to statistically significant DEGs. (d) Top gene ontology (GO) biological process and cellular components terms related to DEGs in FedEVs-high MeT-5A cells. (e) Gene-set enrichment analysis revealed significant upregulation of mitosis-associated pathways with MSigDB gene set. (f) Immunoblot analysis of FedEVs-recipient mesothelial cells (Low and High). (g) Quantification of band intensity (n = 4; mean ± SEM). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Mitotic mesothelial cells internalize FedEVs leading to iron overload
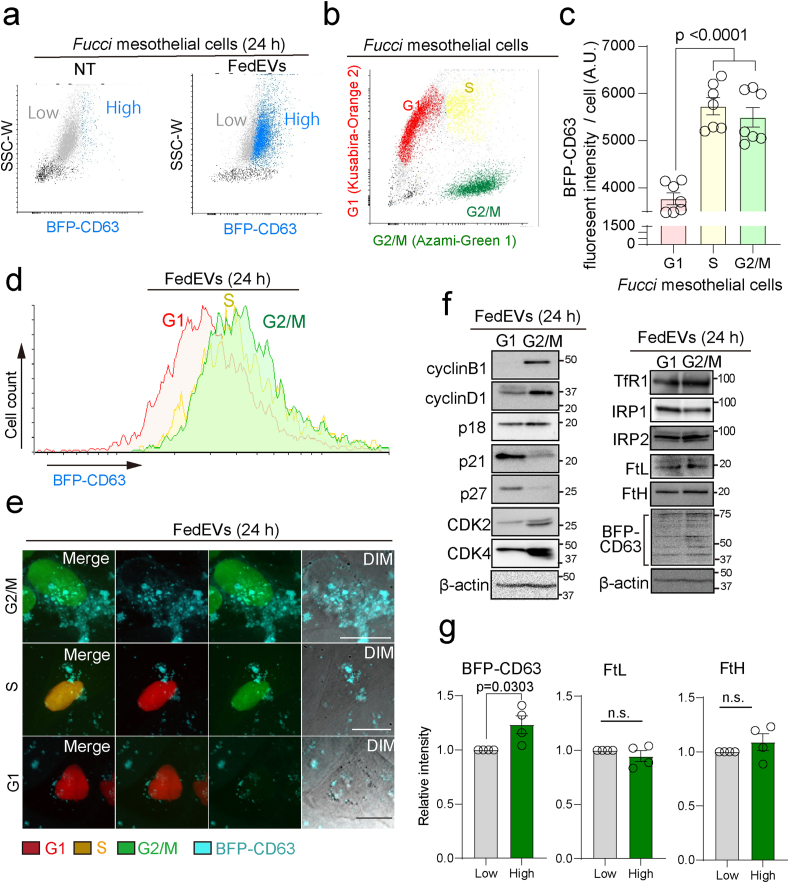
To determine whether mitotic mesothelial cells indeed internalize FedEVs, we established FedEVs donor BFP-CD63 macrophage cells (THP1) and Fucci-expressing recipient mesothelial cells, where G1 (red), S (yellow) and G2/M (green) phases show different fluorescence (fMeT-5A). A combination of FACS and immunoblot analyses could simultaneously detect FedEVs-high recipient cells with BFP-CD63 (Fig. 5a) and the G1, S and G2/M phases of mesothelial cells (Fig. 5be). After FedEVs uptake, BFP-CD63 increased in the fMeT-5A cells of S and G2/M phases (Fig. 5c–e). Furthermore, protein levels of FtL and FtH in FedEVs recipient mesothelial cells increased whereas TfR1 and IRP2 expression were decreased in comparison to the untreated cells, indicating an iron-sufficient status (Fig. 4fg and S4ab). In contrast, the expression of iron-associated proteins revealed no significant change between the G1 and G2/M phases of FedEVs recipient mesothelial cells (Fig. 5fg). Further, the cell cycle-associated proteins, including cyclins and cyclin-dependent kinases (CDKs), showed no expressional alterations as well between the untreated control and FedEVs recipient mesothelial cells (Fig. S4ef). These data indicate that FedEVs uptake directly lead to iron excess without overt modification of cell cycles. As possible EV receptors or ligand-associated proteins, ITGβ1, CD44 and ICAM1 protein expression were evaluated and found increased in the FedEVs uptake group (Fig. S4cd).
Fig. 5.
Mitotic mesothelial cells specifically phagocytosed FedEVs. (a) FedEVs-uptake analysis using MeT-5A mesothelial (recipient) and THP1 macrophage (donor, BFP-CD63 expressed) cells. (b–e) FACS/imaging analysis of mesothelial cells transduced with the Fucci cell cycle sensor; G1 phase, red; S phase, yellow; G2/Mitosis phase, green. Fucci mesothelial cells taken up BFP-CD63-labeled FedEVs after 24 h (mean ± SEM). (e) Live cell imaging of fucci mesothelial cells after 24 h. (f) Immunoblot analysis of Fucci mesothelial cells taken up BFP-CD63-labeled FedEVs after 24 h incubations and sorting; G1, red (Kusabira-orange2) in (b); G2/M: green (Azami-green1) in (b) for cell cycle-associated proteins and iron metabolism-associated proteins. (g) Quantification of band intensity (n = 4; mean ± SEM). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We also evaluated the effects of an redox-inactive iron chelator, desferal (DFO), on this FedEVs uptake by mesothelial cells (Fig. S5ab). DFO dose-dependently decreased the catalytic Fe(II) in the mesothelial cells though catalytic Fe(II) was notably higher than the control level even in the presence of 100 μM DFO (Fig. S5c), suggesting that exogenous iron transfer through FedEVs contribute to a high level of intracellular catalytic fe(II) in mesothelial cells. Further, catalytic Fe(II) was present inside FedEVs, which was decreased in the presence of DFO (Fig. S5d). Finally, ferritin in FedEVs indeed contained insoluble Fe(III), which was not affected by DFO (Fig. S5e).
3.6. Uptake of FedEVs enhances DNA damage in mesothelial cells
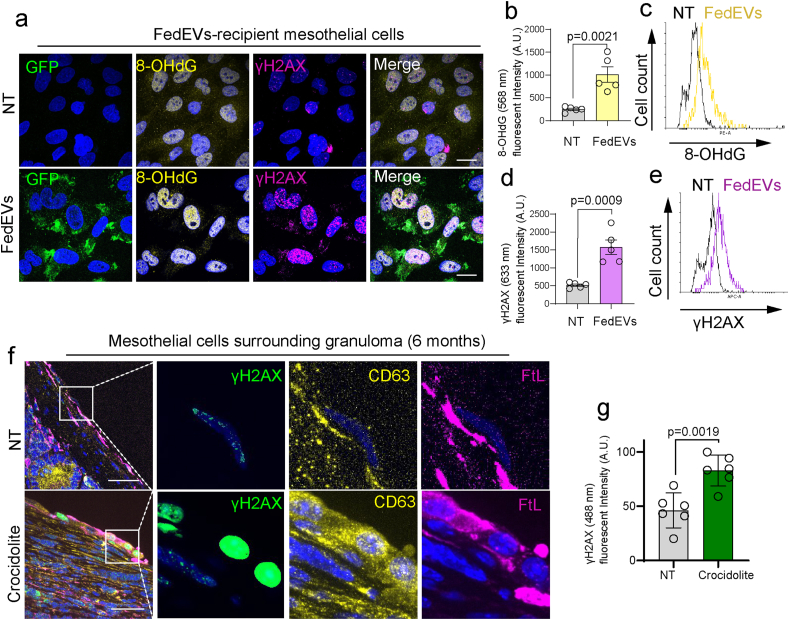
To evaluate the DNA damage in the FedEVs recipient mesothelial cells, we performed the FACS and imaging analysis of oxidative DNA base modification and DNA double-strand breaks. 8-Hydroxy-2′-deoxyguanosine (8-OHdG) is one of the most abundant DNA modifications caused either by hydroxyl radical, single oxygen or peroxynitrite [29,30]. γH2AX is rapidly recruited to the DNA double-strand breaks for repair, thus working as a DNA damage marker [31]. Mesothelial cells were exposed to crocidolite-derived FedEVs from THP1 cells, which were evaluated 24 h afterwards. We observed the increase both in nuclear 8-OHdG and γH2AX in the recipient cells of FedEVs (Fig. 6a–e), which was confirmed in the peritoneum of mice model with CD63 and FtL increasing in the mesothelial cells surrounding granuloma (Fig. 6fg).
Fig. 6.
Phagocytosis of FedEVs causes oxidative DNA base modification (8-hydroxy-2′-deoxyguanosine, 8-OHdG) and DNA double-strand breaks detected by γH2AX in the recipient mesothelial cells. (a) Immunofluorescent images of mesothelial cells after exposure of THP1-derived FedEVs for 24 h; green, CD63-positive FedEVs from THP1 cells; yellow, 8-OHdG; magenda, γH2AX with blue Hoechst33342 nuclear staining (bar = 10 μm). (b–e) FACS analysis, detecting 8-OHdG and γH2AX in mesothelial cells exposed to FedEVs. (f) Immunofluorescent images of mesothelial cells surrounding granuloma in vivo 6 months after ip injection of crocidolite to mice; green, γH2AX; yellow, CD63; magenta, FtL (bar = 20 μm at the left panels; 20 μm at the right 3 panels). (g) Quantification of the γH2AX in mesothelial cells. Refer to text for details (mean ± SEM). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
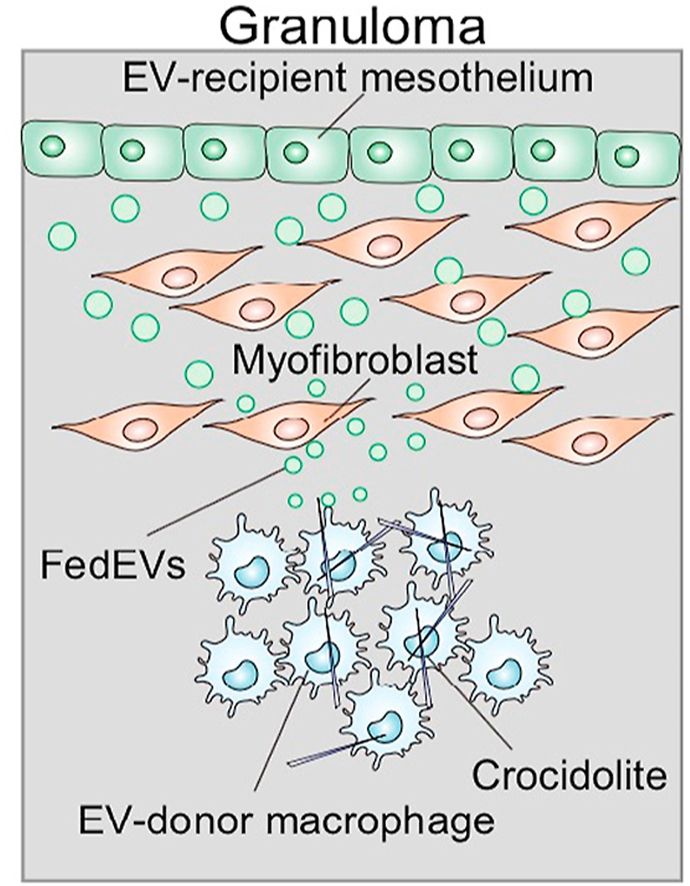
We for the first time demonstrated a completely novel mechanism involved in asbestos-induced carcinogenesis from the viewpoint of iron metabolism. Namely, extracellular vesicles derived from ferroptotic macrophages (FedEVs) are loaded with a high amount of ferritin and are indeed received by mesothelial cells, leading to significant oxidative DNA damage, such as 8-OHdG and double-strand breaks.
We started from a simple in vivo model of injecting crocidolite to the peritoneal cavity of mice, which indeed causes malignant mesothelioma [32]. In the model we found massive expression of CD63, a marker of EVs [12,13], inside and also outside of various cells, including macrophages, myofibroblasts and mesothelial cells. This lesion is full of iron in addition to the injected asbestos fibers, which we call mutagenic stromal milieu and recognized ferroptosis of macrophages as a major death mode in the previous study [11]. Then, we used two ferroptosis cell models of GPX4 inhibition and asbestos/iron loading, both of which showed increased secretion of EVs. Thus, we named the generated EVs as ferroptosis-dependent EVs (FedEVs). We also found the diameter of FedEV was increased, which may a significant feature of EVs derived from cell death (Fig. 1ij and Fig. S2a). The EVs derived from macrophage exposed only to crocidolite contained little ferritin that we assume present no genotoxicity (Fig. 3cd and Fig. S3ab). We believe that EVs including ferritin is essentially critical for mutagenic milieu.
To obtain solid data, we constructed several distinct fluorescence-labeled vector systems for precise morphological analysis, such as GFP/BFP-CD63 and Fucci. These systems unequivocally differentiated not only between donor and recipient cell components but also cell cycle phases. FedEVs, regardless of classical (RSL3) or non-classical (asbestos/iron) derivation of ferroptosis, were efficiently accepted by MeT-5A mesothelial cells, leading to significant increase in catalytic Fe(II) (Fig. 2d–g). The uptake took 6–9 h with the z-axis observation by confocal microscopy and we observed bleb-like structure at 3 h. CD63 is well known as a ubiquitously expressed protein and the major pool of CD63 resides in late endosomes and lysosomes [33,34]. We observed that the endogenous CD63 surrounded FedEVs at 6 h after exposure (Fig. 2b), suggesting that FedEVs may be taken up by endocytosis. Mechanisms for internalization of various EVs remain unelucidated whereas exosome internalization occurs either by vesicle-cell fusion, receptor-mediated endocytosis, macropinocytosis or phagocytosis [12]. Reportedly, vesicle-cell fusion and receptor-mediated endocytosis are associated with cellular response and the other two pathways are for the clearance of EVs [14]. Our RNA-sequencing analysis revealed that some of the candidate EV receptor genes, such as ITGB1, CD44, CD11a and ICAM1, are overexpressed in the high FedEVs-uptake group, which was confirmed with immunoblot (Fig. S4b) but needs further investigation. This indicates that FedEVs may internalize to cytoplasm of mesothelial cells via the receptor-mediated endocytosis.
The most particular finding in this study was that ferritins were the major transported proteins to mesothelial cells via FedEVs, revealed by proteomic analysis and RNA sequencing (Fig. 3ab and Fig. S4ab). We demonstrated that ferritin inside FedEVs indeed contained insoluble Fe(III) (Fig. S5e). Inability of DFO to completely block the intracellular increase in catalytic Fe(II) confirms the importance of ferritin uptake through FedEVs. This may be partially from the presence of catalytic Fe(II) inside FedEVs (Fig. S5d). Inclusion of ferritin in exosomes have been previously reported in normal human urine [35] and in murine RAW264.7 macrophage cells [36]. Here we think that iron-rich stromal environment elicited by asbestos exposure is critical for the presence of this abundant FedEVs rich in ferritins, based on in vitro experiments. In the case of crocidolite alone in in vitro experiments, the amounts of ferritins inside EVs were rather low. Thus, biopersistent foreign fibrous materials of certain physical dimension are important to mature carcinogenic stroma [37]. In this sense, iron removal either by iron chelating drug, such as deferasirox [38] or repeated phlebotomy [39] is expected to work as a malignant mesothelioma prevention strategy in people already exposed to asbestos.
We further recognized that macrophage cell death produces larger EVs as minor components, such as microvesicles (MVs) and apoptotic vesicles (Fig. S2). We identified FtL and FtH in larger- and smaller-size EVs (Fig. 3c, S2b-d and S3ab). Membrane-catch TEM analysis and NTA assay revealed that the large vesicles contain various small vesicular structures, such as lysosome, mitochondrial fragment, exosome and crocidolite. The membrane of MVs was often damaged and leaky in the iron and crocidolite-treated group, suggesting that these EVs remain nearby granuloma and would work to establish iron-rich mutagenic milieu.
In the previous study, we reported that β-catenin is overexpressed in the mesothelial cells in rats after ip administration of crocidolite and that β-catenin increases intracellular catalytic Fe(II) highest at the G2/M phase [11]. Here we sought for the significance of receiving FedEVs in mesothelial cells by RNA sequencing, which showed the involvement of mitosis via multiple pathways. Major portion of these were confirmed with immunoblot analysis (Fig. 4fg) as well as Fucci system (Fig. 5). We believe that G2/M-phase mesothelial cells prefer to uptaking FedEVs rather than that mesothelial cells which received FedEVs would be stuck in G2/M-phase because exposure to iron and crocidolite did not alter the fraction of each phase for the whole mesothelial cells (Fig. S4c).
No life on earth can live without iron [30,40]. However, iron presents a double-edged sword and excess iron has been associated with human carcinogenesis, as in asbestos exposure, genetic hemochromatosis, viral hepatitis and ovarian endometriosis [41]. In conclusion, we demonstrated a novel remote mutagenic mechanism of iron loading to mesothelial cells via FedEVs containing ferritin. Iron-rich stroma is thus critical in asbestos-induced mesothelial carcinogenesis as an indirect effect, which is conversely a basis for prevention target of malignant mesothelioma with iron reduction. Finally, scientists are responsible for paying persistent attention to novel materials and chemicals. As an example, multi-walled carbon nanotube of ∼50-nm diameter is also a risk for malignant mesothelioma [42,43]. Further studies are warranted to confirm the current results. Nanomaterials, including asbestos, can be a carcinogen but some of them also can be drugs [44]. FedEVs may be used as a biomarker in the extracellular fluid in the near future.
Author contributions
F.I. and S.T. contributed to the conception and design of the work and drafted the work. F.I. and Y.I. contributed to the acquisition and analysis of the EV western blotting and sampling. F.I. and K.K. and T.M. contributed to the acquisition and analysis of the RNA-sequence sample preparation and data analysis; S.T. substantially revised the manuscript.
Declaration of competing interest
All the authors declare no conflict of interest.
Acknowledgments
This work was supported, in part, by JST CREST (Grant Number JPMJCR19H4), JSPS Kakenhi (Grant Number JP19H05462 and JP20H05502) and Research Grant of the Princess Takamatsu Cancer Research Fund (19–251). Electron microscopic analysis and proteomic analysis were supported by Koji Itakura and Kentaro Taki, Division of Medical Research Engineering, Nagoya University Graduate School of Medicine. We thank Nobuaki Misawa (Department of Pathology and Biological Responses, Nagoya University Graduate School of Medicine) for excellent technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102174.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Asciak R., George V., Rahman N.M. Update on biology and management of mesothelioma. Eur. Respir. Rev. 2021;30(159) doi: 10.1183/16000617.0226-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO, IARC . IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. A Review of Human Carcinogens; Part C: Arsenic, Metals, Fibres, and Dusts. 2012. Asbestos (chrysotile, amosite, crocidolite, tremolite, actinolite, and anthophyllite) pp. 219–309. Lyon, France. [PMC free article] [PubMed] [Google Scholar]
- 3.Donaldson K., Murphy F.A., Duffin R., Poland C.A. Asbestos, carbon nanotubes and the pleural mesothelium: a review of the hypothesis regarding the role of long fibre retention in the parietal pleura, inflammation and mesothelioma. Part. Fibre Toxicol. 2010;7:5. doi: 10.1186/1743-8977-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toyokuni S. Iron addiction with ferroptosis-resistance in asbestos-induced mesothelial carcinogenesis: toward the era of mesothelioma prevention. Free Radic. Biol. Med. 2019;133:206–215. doi: 10.1016/j.freeradbiomed.2018.10.401. [DOI] [PubMed] [Google Scholar]
- 5.Yamashita K., Nagai H., Toyokuni S. Receptor role of the annexin A2 in the mesothelial endocytosis of crocidolite fibers. Lab. Invest. 2015;95(7):749–764. doi: 10.1038/labinvest.2015.28. [DOI] [PubMed] [Google Scholar]
- 6.Sincock A., Seabright M. Induction of chromosome changes in Chinese hamster cells by exposure to asbestos fibres. Nature. 1975;257(5521):56–58. doi: 10.1038/257056a0. [DOI] [PubMed] [Google Scholar]
- 7.Pricejones M.J., Gubbings G., Chamberlain M. The genetic-effects of crocidolite asbestos - comparison of chromosome-abnormalities and sister-chromatid exchanges. Mutat. Res. 1980;79(4):331–336. [Google Scholar]
- 8.Jiang L., Nagai H., Ohara H., Hara S., Tachibana M., Hirano S., Shinohara Y., Kohyama N., Akatsuka S., Toyokuni S. Characteristics and modifying factors of asbestos-induced oxidative DNA damage. Cancer Sci. 2008;99(11):2142–2151. doi: 10.1111/j.1349-7006.2008.00934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donaldson K., Slight J., Hannant D., Bolton R.E. Increased release of hydrogen peroxide and superoxide anion from asbestos-primed macrophages. Effect of hydrogen peroxide on the functional activity of alpha 1-protease inhibitor. Inflammation. 1985;9(2):139–147. doi: 10.1007/BF00917586. [DOI] [PubMed] [Google Scholar]
- 10.Dostert C., Petrilli V. Asbestos triggers inflammation by activating the Nalp3 inflammasome. Med. Sci. 2008;24(11):916–918. doi: 10.1051/medsci/20082411916. [DOI] [PubMed] [Google Scholar]
- 11.Ito F., Yanatori I., Maeda Y., Nimura K., Ito S., Hirayama T., Nagasawa H., Kohyama N., Okazaki Y., Akatsuka S., Toyokuni S. Asbestos conceives Fe(II)-dependent mutagenic stromal milieu through ceaseless macrophage ferroptosis and beta-catenin induction in mesothelium. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pegtel D.M., Gould S.J. Exosomes. Annu. Rev. Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 13.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478) doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thery C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7(1) doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munson P., Lam Y.W., Dragon J., MacPherson M., Shukla A. Exosomes from asbestos-exposed cells modulate gene expression in mesothelial cells. FASEB J. 2018;32(8):4328–4342. doi: 10.1096/fj.201701291RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanatori I., Richardson D.R., Dhekne H.S., Toyokuni S., Kishi F. CD63 is regulated by iron via the IRE-IRP system and is important for ferritin secretion by extracellular vesicles. Blood. 2021;138(16):1490–1503. doi: 10.1182/blood.2021010995. 2021010995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baxter A.A., Phan T.K., Hanssen E., Liem M., Hulett M.D., Mathivanan S., Poon I.K.H. Analysis of extracellular vesicles generated from monocytes under conditions of lytic cell death. Sci. Rep. 2019;9(1):7538. doi: 10.1038/s41598-019-44021-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lois C., Hong E.J., Pease S., Brown E.J., Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295(5556):868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 19.Sakaue-Sawano A., Yo M., Komatsu N., Hiratsuka T., Kogure T., Hoshida T., Goshima N., Matsuda M., Miyoshi H., Miyawaki A. Genetically encoded tools for optical dissection of the mammalian cell cycle. Mol. Cell. 2017;68(3):626–640 e5. doi: 10.1016/j.molcel.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Jiang L., Zheng H., Lyu Q., Hayashi S., Sato K., Sekido Y., Nakamura K., Tanaka H., Ishikawa K., Kajiyama H., Mizuno M., Hori M., Toyokuni S. Lysosomal nitric oxide determines transition from autophagy to ferroptosis after exposure to plasma-activated Ringer's lactate. Redox Biol. 2021;43 doi: 10.1016/j.redox.2021.101989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagai H., Ishihara T., Lee W.H., Ohara H., Okazaki Y., Okawa K., Toyokuni S. Asbestos surface provides a niche for oxidative modification. Cancer Sci. 2011;102:2118–2125. doi: 10.1111/j.1349-7006.2011.02087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Okazaki Y., Shi L., Kohda H., Tanaka M., Taki K., Nishioka T., Hirayama T., Nagasawa H., Yamashita Y., Toyokuni S. Role of hemoglobin and transferrin in multi-wall carbon nanotube-induced mesothelial injury and carcinogenesis. Cancer Sci. 2016;107:250–257. doi: 10.1111/cas.12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rider M.A., Hurwitz S.N., Meckes D.G., Jr. ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 2016;6 doi: 10.1038/srep23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato K., Dieguez-Hurtado R., Park D.Y., Hong S.P., Kato-Azuma S., Adams S., Stehling M., Trappmann B., Wrana J.L., Koh G.Y., Adams R.H. Pulmonary pericytes regulate lung morphogenesis. Nat. Commun. 2018;9(1) doi: 10.1038/s41467-018-04913-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4 doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito F., Nishiyama T., Shi L., Mori M., Hirayama T., Nagasawa H., Yasui H., Toyokuni S. Contrasting intra- and extracellular distribution of catalytic ferrous iron in ovalbumin-induced peritonitis. Biochem. Biophys. Res. Commun. 2016;476(4):600–606. doi: 10.1016/j.bbrc.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Toyokuni S., Tanaka T., Hattori Y., Nishiyama Y., Ochi H., Hiai H., Uchida K., Osawa T. Quantitative immunohistochemical determination of 8-hydroxy-2'-deoxyguanosine by a monoclonal antibody N45.1: its application to ferric nitrilotriacetate-induced renal carcinogenesis model. Lab. Invest. 1997;76:365–374. [PubMed] [Google Scholar]
- 30.Toyokuni S. The origin and future of oxidative stress pathology: from the recognition of carcinogenesis as an iron addiction with ferroptosisresistance to non-thermal plasma therapy. Pathol. Int. 2016;66:245–259. doi: 10.1111/pin.12396. [DOI] [PubMed] [Google Scholar]
- 31.Dickey J.S., Redon C.E., Nakamura A.J., Baird B.J., Sedelnikova O.A., Bonner W.M. H2AX: functional roles and potential applications. Chromosoma. 2009;118(6):683–692. doi: 10.1007/s00412-009-0234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Funahashi S., Okazaki Y., Akatsuka S., Takahashi T., Sakumi K., Nakabeppu Y., Toyokuni S. Mth1 deficiency provides longer survival upon intraperitoneal crocidolite injection in female mice. Free Radic. Res. 2020;54:195–205. doi: 10.1080/10715762.2020.1743285. [DOI] [PubMed] [Google Scholar]
- 33.Bonifacino J.S., Traub L.M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 34.Pols M.S., Klumperman J. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 2009;315(9):1584–1592. doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 35.Gonzales P.A., Pisitkun T., Hoffert J.D., Tchapyjnikov D., Star R.A., Kleta R., Wang N.S., Knepper M.A. Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol. 2009;20(2):363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Truman-Rosentsvit M., Berenbaum D., Spektor L., Cohen L.A., Belizowsky-Moshe S., Lifshitz L., Ma J., Li W., Kesselman E., Abutbul-Ionita I., Danino D., Gutierrez L., Li H.H., Li K.Y., Lou H.F., Regoni M., Poli M., Glaser F., Rouault T.A., Meyron-Holtz E.G. Ferritin is secreted via 2 distinct nonclassical vesicular pathways. Blood. 2018;131(3):342–352. doi: 10.1182/blood-2017-02-768580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagai H., Toyokuni S. Differences and similarities between carbon nanotubes and asbestos fibers during mesothelial carcinogenesis: shedding light on fiber entry mechanism. Cancer Sci. 2012;103(8):1378–1390. doi: 10.1111/j.1349-7006.2012.02326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagai H., Okazaki Y., Chew S.H., Misawa N., Yasui H., Toyokuni S. Deferasirox induces mesenchymal-epithelial transition in crocidolite-induced mesothelial carcinogenesis in rats. Cancer Prev. Res. 2013;6:1222–1230. doi: 10.1158/1940-6207.CAPR-13-0244. [DOI] [PubMed] [Google Scholar]
- 39.Ohara Y., Chew S.H., Shibata T., Okazaki Y., Yamashita K., Toyokuni S. Phlebotomy as a preventive measure for crocidolite-induced mesothelioma in male rats. Cancer Sci. 2018;109(2):330–339. doi: 10.1111/cas.13460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toyokuni S. Role of iron in carcinogenesis: cancer as a ferrotoxic disease. Cancer Sci. 2009;100(1):9–16. doi: 10.1111/j.1349-7006.2008.01001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toyokuni S., Yanatori I., Kong Y., Zheng H., Motooka Y., Jiang L. Ferroptosis at the crossroads of infection, aging and cancer. Cancer Sci. 2020;111:2665–2671. doi: 10.1111/cas.14496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagai H., Okazaki Y., Chew S., Misawa N., Yamashita Y., Akatsuka S., Yamashita K., Ishihara T., Yoshikawa Y., Jiang L., Ohara H., Takahashi T., Ichihara G., Kostarelos K., Miyata Y., Shinohara H., Toyokuni S. Diameter of multi-walled carbon nanotubes is a critical factor in mesothelial injury and subsequent carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 2011;108(49):E1330–E1338. doi: 10.1073/pnas.1110013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toyokuni S. Genotoxicity and carcinogenicity risk of carbon nanotubes. Adv. Drug Deliv. Rev. 2013;65:2098–2110. doi: 10.1016/j.addr.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 44.Toyokuni S., Kong Y., Cheng Z., Sato K., Hayashi S., Ito F., Jiang L., Yanatori I., Okazaki Y., Akatsuka S. Carcinogenesis as side effects of iron and oxygen utilization: from the unveiled truth toward ultimate bioengineering. Cancers. 2020;12(11) doi: 10.3390/cancers12113320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data supporting the results of the present study are included within the article can be obtained from the corresponding author on reasonable request. The original RNA-seq data is available from the Gene Expression Omnibus (GEO) database under accession code GSE184368. A source Data file is included which contains the raw data underlying the reported averages in all figures and supplementary figures.