Abstract
Saccharomyces cerevisiae septin mutants have pleiotropic defects, which include the formation of abnormally elongated buds. This bud morphology results at least in part from a cell cycle delay imposed by the Cdc28p-inhibitory kinase Swe1p. Mutations in three other genes (GIN4, encoding a kinase related to the Schizosaccharomyces pombe mitotic inducer Nim1p; CLA4, encoding a p21-activated kinase; and NAP1, encoding a Clb2p-interacting protein) also produce perturbations of septin organization associated with an Swe1p-dependent cell cycle delay. The effects of gin4, cla4, and nap1 mutations are additive, indicating that these proteins promote normal septin organization through pathways that are at least partially independent. In contrast, mutations affecting the other two Nim1p-related kinases in S. cerevisiae, Hsl1p and Kcc4p, produce no detectable effect on septin organization. However, deletion of HSL1, but not of KCC4, did produce a cell cycle delay under some conditions; this delay appears to reflect a direct role of Hsl1p in the regulation of Swe1p. As shown previously, Swe1p plays a central role in the morphogenesis checkpoint that delays the cell cycle in response to defects in bud formation. Swe1p is localized to the nucleus and to the daughter side of the mother bud neck prior to its degradation in G2/M phase. Both the neck localization of Swe1p and its degradation require Hsl1p and its binding partner Hsl7p, both of which colocalize with Swe1p at the daughter side of the neck. This localization is lost in mutants with perturbed septin organization, suggesting that the release of Hsl1p and Hsl7p from the neck may reduce their ability to inactivate Swe1p and thus contribute to the G2 delay observed in such mutants. In contrast, treatments that perturb actin organization have little effect on Hsl1p and Hsl7p localization, suggesting that such treatments must stabilize Swe1p by another mechanism. The apparent dependence of Swe1p degradation on localization of the Hsl1p-Hsl7p-Swe1p module to a site that exists only in budded cells may constitute a mechanism for deactivating the morphogenesis checkpoint when it is no longer needed (i.e., after a bud has formed).
The septins are a family of conserved filament-forming proteins that appear to form scaffolds for the localized assembly of various other proteins at the cell surface (21, 39, 40, 42). In Saccharomyces cerevisiae, the septins encoded by CDC3, CDC10, CDC11, and CDC12 are localized to a band at the cytoplasmic face of the plasma membrane in the mother bud neck (20, 25, 31, 39), to which they recruit proteins involved in bud site selection (14, 56), asymmetric chitin deposition (17), and cytokinesis (9, 18, 38). Temperature-sensitive mutations in any of these genes cause the complete disassembly of the septin-based scaffold at the restrictive temperature, resulting in severe pleiotropic defects in all of the aforementioned processes (20, 25, 26, 31). A striking perturbation of the septin scaffold also results from deletion of GIN4 (40), which encodes one of three S. cerevisiae protein kinases related to Nim1p (a mitotic inducer in Schizosaccharomyces pombe [55]). Gin4p localizes to the neck in a septin-dependent manner (40, 48), and in gin4Δ cells, the septins and associated proteins are frequently found as a set of five to eight bars that traverse the neck. However, these cells display only mild defects in the various septin-dependent processes (40).
Cells containing septin or gin4 mutations also display an elongated-bud morphology associated with a hyperpolarization of the actin cytoskeleton towards the tip of the bud (1, 26, 40). In wild-type cells, bud formation involves an initial period of apical growth, during which new cell wall is inserted primarily at the bud tip, followed by a longer period of isotropic growth during which new cell wall is inserted all over the bud surface (19, 34). The shape of the bud reflects the relative times spent in the apical and isotropic growth stages, and the switch from one to the other is triggered by the cell cycle-regulatory kinase Cdc28p in association with the B-type cyclins (primarily Clb2p) (34). Inactivation of Clb-Cdc28p complexes results in prolonged apical growth and the formation of elongated buds (34). Therefore, the elongated buds of septin mutants and gin4 mutants could arise from a delay in the activation of Clb-Cdc28p complexes, from a defect in the ability to execute the switch to isotropic growth in response to Clb-Cdc28p activation, or from a combination of these mechanisms.
Activation of Clb-Cdc28p complexes is subject to multiple layers of regulation, including the inhibitory phosphorylation of Cdc28p at Tyr 19 by the kinase Swe1p (35, 47). Swe1p is dispensable for normal cell cycle progression in unperturbed cells, but it is essential for the morphogenesis checkpoint response (11, 33, 60). This checkpoint is triggered by perturbations that disrupt the process of bud formation, and it introduces a G2 delay in the nuclear cycle that provides time for further bud growth prior to nuclear division (33, 46). In unperturbed cells, Swe1p is stable during G1 and S phases but becomes quite unstable during G2/M (59). Rapid degradation of Swe1p requires a second Nim1p family kinase, Hsl1p (or Nik1p), the conserved protein Hsl7p, and the ubiquitination complex SCFMet30 (28, 43, 44, 58). Overexpression of Swe1p results in G2 arrest accompanied by the formation of highly elongated buds (11). Thus, one hypothesis to explain the elongated buds in septin mutants and gin4 mutants is that Swe1p accumulates and/or is activated in these mutants (6).
A different hypothesis has been proposed by Kellogg and colleagues (2, 13, 62), who have suggested that Clb-Cdc28p complexes trigger the switch from apical to isotropic bud growth through a signal transduction cascade that involves Gin4p, the septins, and at least two other proteins, Nap1p and Cla4p. Nap1p was identified as a protein that binds to the cyclin Clb2p (30), whereas Cla4p is a member of the p21-activated kinase family that bind to and are activated by Cdc42p-type small GTPases in the GTP-bound form (5, 16). In this hypothesis, the septins contribute to bud morphogenesis by helping Clb-Cdc28p complexes to activate Gin4p (and possibly other components of the signaling pathway), leading ultimately to the switch to isotropic growth.
In this paper, we report studies of the link between septin organization and cell cycle control. Our results suggest that perturbations of septin organization do indeed result in a Swe1p-dependent G2 delay associated with a delayed switch from apical to isotropic bud growth and hence an elongated-bud phenotype. Gin4p, Cla4p, and Nap1p make partially redundant contributions to normal septin organization, which is a prerequisite for the hierarchical assembly of a cell cycle-regulatory module involving Hsl1p, Hsl7p, and Swe1p at the daughter side of the mother-bud neck. Release of this module from the neck is associated with, but cannot entirely account for, the Swe1p-dependent G2 delay in the mutants with perturbed septin organization.
MATERIALS AND METHODS
Strains, plasmids, and PCR manipulations.
Escherichia coli strain DH12S (Life Technologies, Gaithersburg, Md.) and standard media and methods (3) were used for plasmid manipulations. High Fidelity Expand DNA polymerase (Boehringer Mannheim, Indianapolis, Ind.) was used for the PCR synthesis of DNA fragments for cloning and for PCR-mediated epitope tagging; Taq DNA polymerase (Promega, Madison, Wis.) was used in other PCR applications. Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, Iowa). Transformation of yeast was performed as described previously (22), and other yeast genetic manipulations were performed by standard procedures (24).
The S. cerevisiae strains used in this study are listed in Table 1. Construction of deletion and tagged alleles in the YEF473 background was carried out by first modifying the target gene in the diploid strain YEF473. All other strains were obtained by crosses among segregants derived from these original transformants. The successful deletion or tagging of target genes was verified by PCR with genomic DNA from transformants as template and the oligonucleotide primers shown in Table 2, as well as by demonstrating that the marker and any associated phenotype segregated 2:2 after sporulation.
TABLE 1.
S. cerevisiae strains used in this studya
| Strain | Relevant genotypeb | Source |
|---|---|---|
| BF264-15Du | a ade1 his2 leu2-3,112 trp1-1 ura3Δns | 53 |
| YEF473 | a/α his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 lys2-801/lys2-801 trp1-Δ63/trp1-Δ63 ura3-52/ura3-52 | 10 |
| DLY1 | a bar1 | 60 |
| M-905 | a/α cdc11-6/cdc11-6 | This studyc |
| M-1207 | a/α cdc11-6/cdc11-6 swe1::LEU2/swe1::LEU2 | This studyd |
| M-1208 | a/α cdc11-6/cdc11-6 mih1::LEU2/mih1::LEU2 | This studyd |
| M-990 | a/α cdc11Δ::TRP1/CDC11 swe1::LEU2/SWE1 | See text |
| M-991 | a/α cdc11Δ::TRP1/CDC11 mih1::LEU2/MIH1 | See text |
| M-1077 | a/α swe1::LEU2/swe1::LEU2 | See text |
| M-600 | a/α mih1::LEU2/mih1::LEU2 | See text |
| M-272 | a/α gin4-Δ9/gin4-Δ9 | 40 |
| M-825 | a/α gin4-Δ9/gin4-Δ9 swe1::LEU2/swe1::LEU2 | See text |
| M-829 | a/α gin4-Δ9/gin4-Δ9 mih1::LEU2/mih1::LEU2 | See text |
| M-515 | a/α cla4Δ::HIS3/cla4Δ::HIS3 | See text |
| M-1029 | a/α cla4Δ::HIS3/cla4Δ::HIS3 swe1::LEU2/swe1::LEU2 | See text |
| M-522 | a/α cla4Δ::HIS3/cla4Δ::HIS3 gin4-Δ9/gin4-Δ9 | See text |
| M-1025 | a cla4Δ::HIS3 gin4-Δ9 swe1::LEU2 | See text |
| M-546 | a/α nap1Δ::HIS3/nap1Δ::HIS3 | See text |
| M-544 | a/α nap1Δ::HIS3/nap1Δ::HIS3 gin4-Δ9/gin4-Δ9 | See text |
| M-976 | a/α nap1Δ::HIS3/nap1Δ::HIS3 gin4-Δ9/gin4-Δ9 swe1::LEU2/swe1::LEU2 | See text |
| M-601 | a/α hsl1Δ1::URA3/hsl1Δ1::URA3 | See text |
| M-286 | a/α kcc4Δ::HIS3/kcc4Δ::HIS3 | 40e |
| M-652 | a/α GIN4-GFP:kan/GIN4-GFP:kan | See text |
| M-1204 | a/α cla4Δ::HIS3/cla4Δ::HIS3 GIN4-GFP:kan/GIN4-GFP:kan | See text |
| M-1293 | a/α nap1Δ::HIS3/nap1Δ::HIS3 GIN4-GFP:kan/GIN4-GFP:kan | See text |
| M-1203 | a/α hsl1Δ1::URA3/hsl1Δ1::URA3 GIN4-GFP:kan/GIN4-GFP:kan | See text |
| M-1163 | a/α swe1::LEU2/swe1::LEU2 GIN4-GFP:kan/GIN4-GFP:kan | See text |
| M-1224 | a/α mih1::LEU2/mih1::LEU2 GIN4-GFP:kan/GIN4-GFP:kan | See text |
| M-602 | a/α hsl1Δ1::URA3/hsl1Δ1::URA3 kcc4Δ::HIS3/kcc4Δ::HIS3 | See text |
| M-603 | a/α gin4-Δ9/gin4-Δ9 hsl1Δ1::URA3/hsl1Δ1::URA3 | See text |
| M-460 | a/α gin4-Δ9/gin4-Δ9 kcc4Δ::HIS3/kcc4Δ::HIS3 | See text |
| M-604 | a/α gin4-Δ9/gin4-Δ9 hsl1Δ1::URA3/hsl1Δ1::URA3 kcc4Δ::HIS3/kcc4Δ::HIS3 | See text |
| M-1031 | a/α kcc4Δ::HIS3/kcc4Δ::HIS3 mih1::LEU2/mih1::LEU2 | See text |
| JMY1441 | a bar1 SWE1myc:URA3 SWE1myc:TRP1 | See text |
| JMY1477 | a hsl1Δ1::URA3 SWE1myc:TRP1 SWE1myc:HIS2 | 44 |
| JMY1475 | a hsl7Δ1::URA3 SWE1myc:TRP1 SWE1myc:HIS2 | 44 |
| JMY1479 | a hsl1Δ1::URA3 hsl7Δ1::URA3 SWE1myc:TRP1 SWE1myc:HIS2 | 44 |
| M-1427 | a/α HSL1-13myc:kan/HSL1-13myc:kan | See text |
| M-1423 | a/α HSL7-3HA:kan/HSL7-3HA:kan | See text |
| M-1552 | a cdc12-6:LEU2 HSL1-13myc:kan | See text |
| M-1554 | a cdc12-6:LEU2 HSL7-3HA:kan | See text |
| JMY1498 | a bar1 cdc12-6:LEU2 SWE1myc:HIS2 SWE1myc:TRP1(3x) | See text |
| M-1272 | a/α hsl7Δ::kan/hsl7Δ::kan HSL1-GFP:kan/HSL1-GFP:kan | See text |
| M-1426 | a/α swe1::LEU2/swe1::LEU2 HSL1-GFP:kan/HSL1-GFP:kan | See text |
| M-1158 | a/α mih1::LEU2/mih1::LEU2 HSL1-GFP:kan/HSL1-GFP:kan | See text |
| M-1445 | a/α hsl1Δ1::URA3/hsl1Δ1::URA3 HSL7-3HA:kan/HSL7-3HA:kan | See text |
| M-1440 | a/α swe1::LEU2/swe1::LEU2 HSL7-3HA:kan/HSL7-3HA:kan | See text |
| M-1421 | a/α mih1::LEU2/mih1::LEU2 HSL7-3HA:kan/HSL7-3HA:kan | See text |
| M-1443 | a/α gin4-Δ9/gin4-Δ9 HSL1-13myc:kan/HSL1-13myc:kan | See text |
| M-1451 | a/α cla4Δ::HIS3/cla4Δ::HIS3 HSL1-13myc:kan/HSL1-13myc:kan | See text |
| M-1442 | a/α gin4-Δ9/gin4-Δ9 HSL7-3HA:kan/HSL7-3HA:kan | See text |
| M-1454 | a/α cla4Δ::HIS3/cla4Δ::HIS3 HSL7-3HA:kan/HSL7-3HA:kan | See text |
| M-1452 | a/α nap1Δ::HIS3/nap1Δ::HIS3 HSL1-13myc:kan/HSL1-13myc:kan | See text |
| M-1453 | a/α nap1Δ::HIS3/nap1Δ::HIS3 HSL7-3HA:kan/HSL7-3HA:kan | See text |
| M-1439 | a/α hsl7Δ:kan/hsl7Δ:kan | See text |
Strains other than BF264-15Du and YEF473 are listed in the order in which they appear in Results.
Except where noted, strains with DLY or JMY designations are congenic to BF264-15Du, and strains with M- designations are congenic to YEF473.
Derived by HO-induced diploidization (24) of Pringle lab strain M-27 (α cdc11-6 his4 leu2 trp1 ura3); the cdc11-6 allele has been described previously (1).
Derived by mating segregants obtained from strain M-905 after transformation with fragments containing swe1::LEU2 or mih1::LEU2 (see text).
The allele previously referred to as ycl024W-Δ2 (40) has been renamed kcc4Δ::HIS3.
TABLE 2.
Oligonucleotide PCR primers used in this study
| Name | Sequence |
|---|---|
| ML86a | 5′-CGAACAGTGAAACTGAAACATAAAAGAAATAGTGCAAAATGATTGTACTGAGAGTGCACC-3′ |
| ML87ab | 5′-TGTAGTATGTATGATATGCTTATAGAAATAGTTGTGTGCTCTGTGCGTATTTCACACCG-3′ |
| ML96b | 5′-AAAAAGATCTACCAAAAAAGGATAGTTTCC-3′ |
| ML111a | 5′-TTTAGTGAGCCCAGTGGGACCAAGAGCAAAGCGCAAGATCGATTGTACTGAGAGTGCACC-3′ |
| ML112ab | 5′-ATGAATACGGTTATGACCACTCTTGCGAAGTGCGTTGGGACTGTGCGGTATTTCACACCG-3′ |
| ML113b | 5′-CTTTGGAATCCAAAGGAGGCGCTTT-3′ |
| ML130b | 5′-GCTAGGATACAGTTCTCACATCACATCCG-3′ |
| ML133b | 5′-AAATGCCTAATGCATTACTATCACG-3′ |
| ML136cd | 5′-AGTTGAGAACGTCCTGAATAAGGAAGGCGTTCTACAAAAACGGATCCCCGGGTTAATTAA-3′ |
| ML137c | 5′-AAATATTATGGCAGAACAAAACGAAGGAGACAAAACATGAGAATTCGAGCTCGTTTAAAC-3′ |
| ML139b | 5′-TTGGGTTTGTTGCCATCTGCGTCCTTGG-3′ |
| ML140b | 5′-CAAACCGCTAACAATACCTGGGCCCACCACACC-3′ |
| ML142d | 5′-TGTATACACTTATTTTTTTTATAACTTATT-3′ |
| ML165c | 5′-TGATGATGTGGAGAGAGTAATTCGAAATGCCGGACGTTCACGGATCCCCGGGTTAATTAA-3′ |
| ML166c | 5′-GTTAAATTTTTCAAATTATTGTTGTATAATTATATAACATGAATTCGAGCTCGTTTAAAC-3′ |
| ML200c | 5′-GCTACATAATGTCTGTGGCAGAGCCTTTTCCCTGCCTCTGCGGATCCCCGGGTTAATTAA-3′ |
| ML201c | 5′-TTTTACGGTGCGGCGCCGACACCCCGCTCGATCGATGACGGAATTCGAGCTCGTTTAACC-3′ |
| ML202d | 5′-AATCTAGAAACTGGTTGGCAAAGCG-3′ |
| ML203b | 5′-TCGCGATGAGTTAGATGAAAATGAA-3′ |
| ML204b | 5′-ACCTGCATTGCGTGTCATTTCTA-3′ |
| ML205a | 5′-AAGTACGCGAGGTTTGTTGGAATCAATAAGCTAATATTGGCGGATCCCCGGGTTAATTAA-3′ |
| ML206a | 5′-CTCGATCGATGACGCTGACTTGTGGTACTCAGGACGTATGGAATTCGAGCTCGTTTAAAC-3′ |
| ML207b | 5′-GATCCATTTACCATTGAAGAGTTCC-3′ |
| ML276d | 5′-GAATCTTCTCAACTTGTGTTGCCG-3′ |
| ML277e | 5′-AAGCTCAGAGCCCTGTTACTGAGGAC-3′ |
| ML278e | 5′-ATTTTACGCAATAGGTCACAGCCCTG-3′ |
| MLP54-5d | 5′-GGTAAATATAATAGTACCTCGAGTCCAGTACCTCC-3′ |
Primers used to generate deletion cassettes for replacement of CLA4 (ML86 and ML87) and NAP1 (ML111 and ML112) with HIS3 and of HSL7 with kanMX6 (ML205 and ML206). Underlined nucleotides correspond to those flanking the gene to be deleted; the remaining nucleotides correspond to regions upstream and downstream of the selectable marker genes.
Primers used to verify replacement of SWE1 by LEU2 (ML139 and ML204), of MIH1 by LEU2 (ML139 and ML203), of HSL1 by URA3 (ML133 and ML140), of HSL7 by kanMX6 (ML130 and ML207), and of CLA4 (ML87 and ML96) and NAP1 (ML112 and ML113) by HIS3.
Primers used to generate cassettes for tagging the 3′ end of GIN4 with sequences encoding GFP (ML136 and ML137), the 3′ end of HSL1 with sequences encoding GFP or 13 copies of the myc epitope (ML165 and ML166), and the 3′ end of HSL7 with sequences encoding three copies of the HA epitope (ML200 and ML201). Underlined nucleotides correspond to those flanking the site of tagging; the remaining nucleotides correspond to regions upstream and downstream of the selectable marker genes (41).
Primers used to verify tagging of GIN4 (ML136 and MLP54-5), HSL1 (ML142 and ML276), and HSL7 (ML142 and ML202).
Primers used to clone the 3′ end of cdc12-6 into YIplac128 (see text).
Plasmids containing the swe1::LEU2, mih1::LEU2, and hsl1Δ1::URA3 null alleles have been described previously (11, 43, 54). To replace the wild-type alleles with these mutant alleles, yeast cells were transformed with appropriate fragments from these plasmids. The chromosomal cla4Δ::HIS3, nap1Δ::HIS3, and hsl7Δ::kan alleles were constructed by the PCR method (7) with plasmid pRS303 (61) or pFA6a-kanMX6 (41) as template and oligonucleotide primers as described in Table 2. The PCR method was also used to generate alleles of GIN4, HSL1, and HSL7 encoding proteins tagged at their C termini with GFP(S65T), 13 tandem myc epitopes, or 3 tandem hemagglutinin (HA) epitopes. The plasmids described previously (41) were used as templates, and the primers used are listed in Table 2. Gin4p-GFP appeared to be fully functional by the criterion that the GIN4-GFP:kan strains had normal cell morphology at 37°C (unpublished results), in contrast to the abnormal morphology of gin4Δ strains (40). Western blotting showed that Hsl1p-13myc and Hsl7p-3HA migrated near the predicted sizes of ∼190 kDa and ∼100 kDa, respectively (unpublished results). Moreover, Hsl1p-13myc, Hsl1p-GFP, and Hsl7p-3HA appeared to be fully functional by the criteria that HSL1-13myc mih1Δ, HSL1-GFP mih1Δ, and HSL7-3HA mih1Δ strains were all viable and had normal cell morphology (unpublished results), in contrast to hsl1Δ mih1Δ and hsl7Δ mih1Δ strains, which are inviable and arrest in G2 with extremely elongated buds (44). Construction of the chromosomal SWE1myc:URA3, SWE1myc:TRP1, SWE1myc:HIS2, and SWE1myc:TRP1(3x) alleles has been described previously (44, 59). The myc-tagged Swe1p appeared to be fully functional by the criterion that it provided a morphogenesis checkpoint-dependent G2 delay in a cdc24 mutant (59).
To introduce the cdc12-6 allele (1) into different strain backgrounds, primers ML277 and ML278 (Table 2) were used to amplify the region encoding the cdc12-6 C terminus with DNA isolated from the cdc12-6 strain M-238 (40) as template. The PCR product was digested with MfeI and PstI (sites in the CDC12 region sequences) and ligated into EcoRI/PstI-digested YIplac128 (23), resulting in plasmid YIplac128/cdc12-6. DNA sequencing verified that the cloned insert contains the cdc12-6 mutation, which is an insertion of an adenine into a stretch of seven adenines spanning nucleotides 1167 to 1173 of the CDC12 open reading frame (B. K. Haarer and J. R. Pringle, unpublished data). Strains were then transformed with YIplac128/cdc12-6 after digestion at the unique HpaI site (upstream of the cdc12-6 mutation), yielding Leu+ transformants. Dissection of tetrads from these transformants yielded 2Ts+:2Ts− segregants, and the Ts− lethality was rescued by introduction of a low-copy-number CDC12 plasmid.
Growth conditions and synchronization.
Yeast media (YM-P and YPD rich media, synthetic complete medium [SC] lacking specific nutrients, and sporulation medium) have been described previously (24, 37). Cells were synchronized by α-factor arrest-release or by centrifugal elutriation as described previously (34, 45). Latrunculin-A (Molecular Probes, Eugene, Oreg.) was added from a 20 mM stock solution in dimethyl sulfoxide that was stored at −20°C.
Protein analysis.
Yeast proteins were isolated by resuspending whole cells in 2× Laemmli sample buffer (32) and boiling for 5 min. Western blot analysis was performed by standard procedures (3) with mouse monoclonal anti-myc antibodies (9E10; Santa Cruz Biotechnology, Santa Cruz, Calif.), mouse monoclonal anti-HA antibodies (HA.11; Berkeley Antibody Co., Richmond, Calif.), and the ECL chemiluminescence detection system (Amersham, Arlington Heights, Ill.).
Immunofluorescence and other microscopic analysis.
Except as noted below, all microscopy was performed using a Nikon Mikrophot SA microscope. Overall cell morphologies were examined by differential interference contrast (DIC) microscopy, and cells were stained with 4′,6′-diamidino-2-phenylindole (DAPI; Sigma Chemical Co., St. Louis, Mo.) to visualize DNA or with Calcofluor (Sigma) to visualize chitin (50). Green fluorescent protein-tagged proteins were visualized using the fluorescein isothiocyanate filter set. To determine whether cells had completed cytokinesis (52), cells were fixed by adding formaldehyde directly to the growth medium to a 3.7% final concentration and incubated for 2 h at the growth temperature. After being washed three times with phosphate-buffered saline (PBS), cells were suspended in PBS with 0.1% β-mercaptoethanol and digested with 0.2 mg of Lyticase (ICN Pharmaceuticals, Costa Mesa, Calif.) per ml for 1 h at room temperature with continuous gentle rolling.
Immunofluorescence localization of Cdc11p, Hsl1p-13myc, and Hsl7p-3HA was performed essentially as described previously (51), using bisBenzamide (Sigma) in the mounting medium to stain DNA. Anti-Cdc11p antibodies were purified as previously described (20) and used at a 1:10 dilution. Mouse anti-myc antibodies, mouse anti-HA antibodies, and rat monoclonal anti-HA antibodies (3F10; Boehringer Mannheim) were all used at a 1:300 dilution. Fluorescein isothiocyanate- or Cy3-labeled secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, Pa.), and Alexa-labeled secondary antibodies were purchased from Molecular Probes; both were used at a 1:200 dilution.
Immunofluorescence detection of Swe1p in a majority of the cells required the use of a four-antibody sandwich protocol. Strains carrying two or four integrated copies of SWE1myc were grown to 5 × 106 cells/ml in YPD medium, fixed by adding formaldehyde to the growth medium to a 4.5% final concentration, and incubated for 45 min at 23°C. Following a 30-min treatment with 12.6 μg of Zymolyase (ICN) per ml at 30°C, the samples were washed with PBS, pH 7.5, and affixed to polylysine-coated slides (51). Cells were then incubated successively with the following antibodies at the indicated dilutions: 9E10 mouse anti-myc, 1:10; rabbit anti-mouse-IgG (Jackson ImmunoResearch), 1:100; mouse anti-rabbit IgG (Jackson ImmunoResearch), 1:100; and Cy3-labeled goat anti-mouse IgG (Jackson ImmunoResearch), 1:25. All antibodies were diluted in PBS containing 1 mg of bovine serum albumin (BSA) per ml (BSA-PBS) and incubated with the cells for 1 h at 23°C in a dark, humid chamber. Between incubations, cells were washed 10 times with 10 μl of BSA-PBS. DNA was visualized by including DAPI in the mounting medium (51). Microscopy was performed with a Zeiss Axioskop with standard fluorescence optics. Images were captured with a cooled charge-coupled device camera (Princeton Instruments, Princeton, N.J.) interfaced with Metamorph software (Universal Imaging Corp., Silver Spring, Md.).
RESULTS
Swe1p-dependent cell cycle delay in septin mutants.
Temperature-sensitive septin mutants arrest as multibudded, multinucleate cells after prolonged incubation at restrictive temperature, indicating that cell cycle progression continues in these mutants (26, 39). However, it seemed possible that there might be a small cell cycle delay that could contribute to the characteristic elongated-bud morphology (1, 26) of septin mutants. Indeed, in agreement with others (6), we found that nuclear division was delayed by 30 to 45 min in septin mutant strains as compared to isogenic wild-type strains (unpublished results). The delay was eliminated by deletion of SWE1 (6; unpublished results), indicating that it depended on Swe1p. Similar results were observed with cdc12-6 and cdc10-1 mutants and by using either α-factor or centrifugal elutriation to obtain synchronized cells (unpublished results).
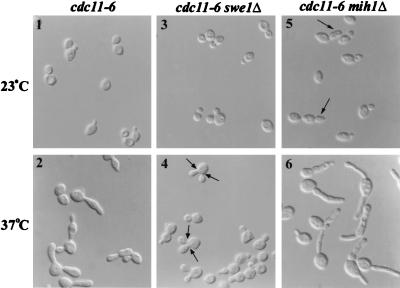
To ask if the Swe1p-dependent cell cycle delay contributes to the elongated-bud phenotype, we examined the morphologies of septin mutants that contained or lacked Swe1p or Mih1p (the phosphatase that reverses the Swe1p-catalyzed phosphorylation of Cdc28p [54]). In agreement with others (6), we found that deletion of SWE1 largely eliminated the elongated-bud phenotype of a septin mutant at restrictive temperature (Fig. 1, panels 2 and 4). In contrast, deletion of MIH1 exacerbated the elongated-bud phenotype (Fig. 1, panel 6), and these cells arrested permanently with a single nucleus (unpublished results). The cdc11-6 mih1Δ double-mutant cells even displayed a slight elongated-bud phenotype at permissive temperature (Fig. 1, panel 5). In contrast to its effect on bud morphology, deletion of SWE1 neither restored septin localization nor rescued the cytokinesis defect in the septin mutant cells, as judged by immunofluorescence (unpublished results) and by the accumulation of multiple buds (Fig. 1, panel 4) that did not detach upon Lyticase treatment of fixed cells (unpublished results) (see Materials and Methods). Introduction of the nonphosphorylatable CDC28Y19F allele (using plasmid pJM1046 [45]) into septin mutant strains also restored nearly normal bud morphology (unpublished results), indicating that the effect of Swe1p was mediated largely, at least, by phosphorylation of Cdc28p at Tyr 19, as expected.
FIG. 1.
Role of Swe1p-dependent inhibition of Cdc28p in the elongated-bud morphology of septin mutants. Homozygous diploid strains M-905 (cdc11-6), M-1207 (cdc11-6 swe1Δ), and M-1208 (cdc11-6 mih1Δ) were grown to exponential phase at 23°C in YM-P medium and examined by DIC microscopy before (panels 1, 3, and 5) and after (panels 2, 4, and 6) a shift to 37°C for 6 h. Arrows indicate multibudded cells (panel 4) or elongated buds (panel 5), as discussed in the text.
Taken together, the data suggest that septin disassembly triggers a Swe1p-dependent inhibition of Cdc28p that leads to a cell cycle delay associated with continued apical bud growth. This response appears to be functionally important: when we dissected tetrads from the doubly heterozygous strains M-990 and M-991, we observed synthetic lethality between the cdc11Δ mutation (21) and either swe1Δ or mih1Δ.
Swe1p-dependent cell cycle delay in gin4, cla4, and nap1 mutants.
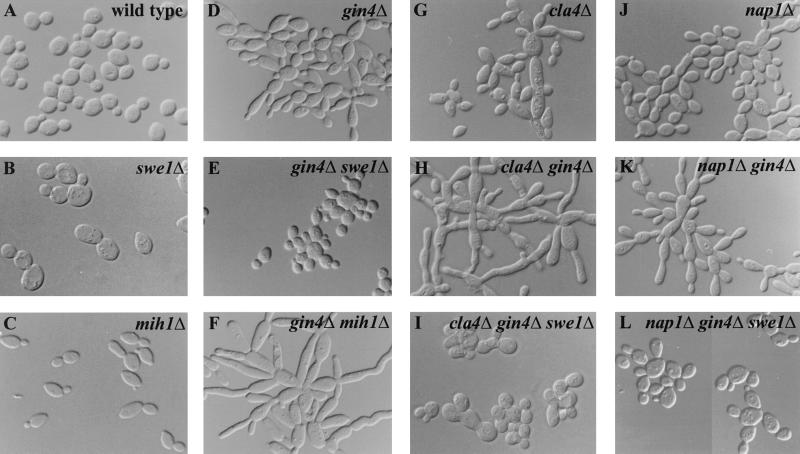
Mutation of GIN4 leads to an alteration of septin organization at the mother-bud neck associated with modest effects on septin function (see Fig. 3A and B) (40). In addition, gin4 mutant cells display an elongated-bud phenotype (Fig. 2D) particularly at elevated temperatures (40). Deletion of SWE1 largely eliminated, and deletion of MIH1 greatly exacerbated, the elongated-bud phenotype of gin4 mutants (Fig. 2E and F; for quantitation of the bud morphology and other phenotypes of these and other mutant strains, see Table 3). Immunofluorescence analysis and staining of chitin showed that septin organization and function were similar in the gin4Δ and gin4Δ swe1Δ strains and only slightly more abnormal in the gin4Δ mih1Δ strain (Table 3), indicating that the effects of deleting SWE1 or MIH1 on bud morphology did not simply reflect effects on septin organization. The nonphosphorylatable CDC28Y19F allele also restored nearly normal bud morphology to gin4Δ cells (unpublished results), indicating that the elongated-bud phenotype of gin4 mutants, like that of septin mutants, is due largely to Swe1p-dependent inhibition of Cdc28p.
FIG. 3.
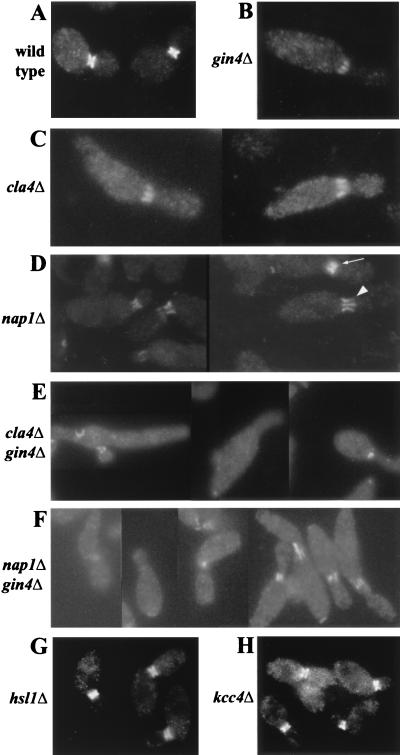
Effects of gin4, cla4, and nap1 mutations, but not of hsl1 or kcc4 mutations, on septin organization. Cells of the indicated strains were grown to exponential phase in YM-P medium at 30°C and examined by immunofluorescence microscopy to localize Cdc11p. (A) YEF473; (B) M-272; (C) M-515; (D) M-546; (E) M-522; (F) M-544; (G) M-601; (H) M-286. The arrow and arrowhead in panel D indicate cells displaying septin misorganization, classified as “fuzzy” and “bars,” respectively, as discussed in Table 3.
FIG. 2.
Role of Swe1p-dependent inhibition of Cdc28p in the elongated-bud morphology of gin4, cla4, and nap1 mutants. The indicated strains were grown to exponential phase in YM-P medium at 30°C and examined by DIC microscopy. (A) YEF473; (B) M-1077; (C) M-600; (D) M-272; (E) M-825; (F) M-829; (G) M-515; (H) M-522; (I) M-1025; (J) M-546; (K) M-544; (L) M-976.
TABLE 3.
Bud morphology, septin organization, and pattern of chitin deposition in wild-type and mutant strainsa
| Strain | Bud morphologyb
|
Septin organizationc
|
Chitin depositiond
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Normal | Elongated | Wild-type | Fuzzy | Bars | Absent | Asymmetric | Symmetric | Delocalized | |
| Wild type | 97 | 3 | 95 | 5 | 0 | 0 | 97 | 3 | 0 |
| swe1Δ | 99 | 1 | 87 | 12 | 1 | 0 | 92 | 3 | 5 |
| mih1Δ | 96 | 4 | 90 | 9 | 0 | 1 | 96 | 4 | 0 |
| gin4Δ | 71 | 29 | 35 | 25 | 28 | 12 | 17 | 66 | 17 |
| gin4Δ swe1Δ | 97 | 3 | 23 | 35 | 32 | 3 | 11 | 76 | 13 |
| gin4Δ mih1Δ | 34 | 66 | 12 | 33 | 37 | 18 | 1 | 53 | 46 |
| cla4Δ | 31 | 69 | 31 | 32 | 19 | 18 | 9 | 50 | 41 |
| cla4Δ swe1Δ | 94 | 6 | 28 | 31 | 23 | 18 | 5 | 61 | 34 |
| nap1Δ | 84 | 16 | 41 | 36 | 13 | 10 | 9 | 78 | 13 |
| cla4Δ gin4Δ | 6 | 94 | 3 | 17 | 47 | 33 | 0 | 22 | 79 |
| cla4Δ gin4Δ swe1Δ | 96 | 4 | 8 | 33 | 33 | 26 | 0 | 28 | 72 |
| nap1Δ gin4Δ | 40 | 60 | 12 | 31 | 38 | 19 | 6 | 76 | 18 |
| nap1Δ gin4Δ swe1Δ | 98 | 2 | 27 | 35 | 27 | 12 | 8 | 82 | 10 |
| hsl1Δ | 94 | 6 | 89 | 11 | 0 | 0 | 95 | 5 | 0 |
| kcc4Δ | 96 | 4 | 95 | 5 | 0 | 0 | 96 | 4 | 0 |
| hsl1Δ kcc4Δ | 96 | 4 | 94 | 6 | 0 | 0 | 96 | 4 | 0 |
| gin4Δ hsl1Δ | 72 | 28 | 28 | 30 | 32 | 10 | 9 | 79 | 12 |
| gin4Δ hsl1Δ kcc4Δ | 73 | 27 | 22 | 35 | 35 | 8 | 10 | 82 | 8 |
| kcc4Δ mih1Δ | 97 | 3 | 90 | 7 | 2 | 1 | 92 | 8 | 0 |
| hsl7Δ | 94 | 6 | 92 | 8 | 0 | 0 | 95 | 5 | 0 |
All strains were grown to exponential phase in YM-P medium at 30°C, and ≥200 cells were counted per sample. The strains used were the wild type, YEF473; swe1Δ, M-1077; mih1Δ, M-600; gin4Δ, M-272; gin4Δ swe1Δ, M-825; gin4Δ mih1Δ, M-829; cla4Δ, M-515; cla4Δ swe1Δ, M-1029; nap1Δ, M-546; cla4Δ gin4Δ, M-522; cla4Δ gin4Δ swe1Δ, M-1025; nap1Δ gin4Δ, M-544; nap1Δ gin4Δ swe1Δ, M-976; hsl1Δ, M-601; kcc4Δ, M-286; hsl1Δ kcc4Δ, M-602; gin4Δ hsl1Δ, M-603; gin4Δ hsl1Δ kcc4Δ, M-604; kcc4Δ mih1Δ, M-1031; and hsl7Δ, M-1439.
Buds were scored as elongated if the length of the bud was more than twice the width of the mother cell.
Septin organization was scored as wild type (localization throughout the neck with well-defined end points; e.g., Fig. 3A), fuzzy (localization throughout the neck but with ill-defined end-points; e.g., Fig. 3D), bars (localization to several bars at the neck, oriented along the mother-bud axis; e.g., Fig. 3B and D), or absent (no localized staining seen), as described previously (40).
Chitin deposition was scored as asymmetric (chitin localized to the mother side of the neck, as in wild-type cells), symmetric (chitin localized with similar staining intensity on both mother and daughter sides of the neck), or delocalized (chitin dispersed more or less uniformly around the cell periphery), as described previously (40).
Cla4p and Nap1p have been suggested to function in a common pathway with Gin4p, based on both genetic data (similar phenotypes were observed for cells lacking these proteins in combination with loss of G1 or mitotic cyclins) and biochemical data (Gin4p binds to Nap1p affinity columns, and both Nap1p and Cla4p are required for maximal phosphorylation of Gin4p in G2/M) (2, 8, 16, 62). We therefore examined whether cla4Δ and nap1Δ mutants also displayed a Swe1p-dependent elongated-bud phenotype. Indeed, both mutants displayed mild elongated-bud phenotypes (Fig. 2G and J) that were eliminated upon deletion of SWE1 or introduction of CDC28Y19F (Table 3) (unpublished results). The elongated-bud phenotype was dramatically exacerbated in cla4Δ gin4Δ and nap1Δ gin4Δ double mutants (Fig. 2H and K), but even these extreme phenotypes were suppressed by deletion of SWE1 (Fig. 2I and L; Table 3) or introduction of CDC28Y19F (unpublished results).
Roles of Cla4p and Nap1p in septin organization and function.
Given the phenotypic similarities and genetic and biochemical interactions among Gin4p, Cla4p, and Nap1p, it seemed possible that Cla4p and Nap1p might also play roles in septin organization. Indeed, both cla4Δ and nap1Δ mutants exhibited aberrant septin organization similar to that seen in gin4 mutants (Fig. 3A to D). As in gin4Δ mutants, the abnormal septin organization in cla4Δ and nap1Δ mutants was typically reflected in a loss of the normal asymmetry of chitin deposition at the mother-bud neck (Table 3).
As both Cla4p and Nap1p are required for maximal phosphorylation of Gin4p (2, 62), it seemed possible that the roles of Cla4p and Nap1p in septin organization are simply to localize and/or activate Gin4p. Consistent with this hypothesis, deletion of CLA4 or NAP1 resulted in the loss of detectable localization of Gin4p to the neck in a large fraction of the cells (Table 4). However, an alternative possibility is that Cla4p and Nap1p promote normal septin organization through a pathway(s) separate from that of Gin4p and that the septin misorganization in cla4Δ and nap1Δ cells precludes the efficient recruitment of Gin4p to the neck. Consistent with this hypothesis, both cla4Δ gin4Δ and nap1Δ gin4Δ double mutants displayed more severe defects in septin organization (and in the associated localization of chitin deposition) than did the single mutants (Fig. 3E and F; Table 3).
TABLE 4.
Gin4p localization in wild-type and mutant strainsa
| Strain | % Neck-localized Gin4p-GFP
|
|
|---|---|---|
| Detected | Not detected | |
| Wild type (M-652) | 80 | 20 |
| cla4Δ (M-1204) | 18 | 82 |
| nap1Δ (M-1293) | 4 | 96 |
| hsl1Δ (M-1203) | 72 | 28 |
| swe1Δ (M-1163) | 73 | 27 |
| mih1Δ (M-1224) | 72 | 28 |
Strains with an integrated GIN4-GFP allele were grown to mid-exponential phase in YM-P medium at 23°C, and the percentages of budded cells in which Gin4p could be detected at the neck were determined by fluorescence microscopy. More than 200 cells were counted for each sample.
Although deletion of SWE1 restored normal bud morphology to the gin4Δ, cla4Δ, nap1Δ, cla4Δ gin4Δ, and nap1Δ gin4Δ strains (see above), it did not restore normal septin organization or localization of chitin deposition (Table 3). Thus, taken together, the data suggest that Cla4p and Nap1p, like Gin4p, have roles in proper septin organization and that one consequence of the defective septin organization observed in their absence is the Swe1p-dependent inhibition of Cdc28p.
Distinct roles of Nim1p family kinases in septin organization and cell cycle control.
In addition to Gin4p, S. cerevisiae contains two other kinases, Hsl1p (or Nik1p) and Kcc4p (or Ycl024Wp), in the family defined by S. pombe Nim1p (6, 27, 40, 48). These proteins have closely related kinase domains at their N termini and long, generally dissimilar C-terminal regions. We asked whether Hsl1p and Kcc4p, like Gin4p, play roles in septin organization. However, hsl1Δ and kcc4Δ single mutants and hsl1Δ kcc4Δ double mutants all displayed essentially normal septin organization and localization of chitin deposition (Fig. 3G and H; Table 3). Moreover, the gin4Δ hsl1Δ and gin4Δ kcc4Δ double-mutant strains and even the triple-mutant gin4Δ hsl1Δ kcc4Δ strain did not display a significant exacerbation of the defects seen in the gin4Δ single mutant (Table 3) (unpublished results). Thus, of the three Nim1p family kinases in S. cerevisiae, only Gin4p appears to play a major role in septin organization or function.
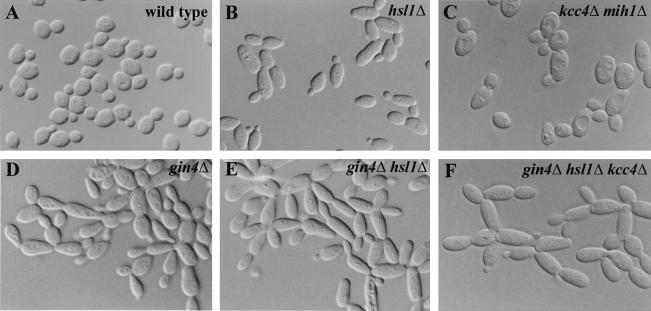
In S. pombe, Nim1p directly phosphorylates the Swe1p-related kinase Wee1p, inhibiting its catalytic activity (15, 49, 64). Given the similarities between the kinase domains of Nim1p, Gin4p, Hsl1p, and Kcc4p, it was recently suggested that the three S. cerevisiae kinases play redundant roles in the down-regulation of Swe1p (6). Consistent with other evidence for a direct role of Hsl1p in the down-regulation of Swe1p (43, 44), we observed a mild elongated-bud phenotype in hsl1Δ mutants grown to high cell density (Fig. 4B), although not in exponential-phase cells (Table 3). Moreover, the hsl1Δ mutation was lethal when combined with deletion of MIH1 (to enhance any effect of a partial loss of Swe1p regulation [44]). In contrast, no such effects were seen in the kcc4Δ mutant, which grew well and formed buds of normal shape even when MIH1 was also deleted (Fig. 4C; Table 3). In addition, the role of Gin4p in Swe1p regulation seems likely to be an indirect consequence of its role in septin organization, as similar degrees of bud elongation were observed upon deletion of GIN4, CLA4, or NAP1, all of which affect septin organization (see above). Moreover, bud elongation was neither more frequent nor more pronounced in the gin4Δ hsl1Δ double mutant or the gin4Δ hsl1Δ kcc4Δ triple mutant than in the gin4Δ single mutant (Fig. 4D to F; Table 3), suggesting that these proteins do not independently inhibit Swe1p. Thus, of the three Nim1p family kinases in S. cerevisiae, only Hsl1p appears to play a direct role in Swe1p regulation.
FIG. 4.
Effects of gin4, hsl1, and kcc4 mutations on bud morphology. The indicated strains were grown to exponential phase or (for the hsl1 strain) to a high cell density in YM-P medium at 30°C and examined by DIC microscopy. (A) YEF473; (B) M-601; (C) M-1031; (D) M-272; (E) M-603; (F) M-604.
Interestingly, the mutations that we have examined that affect septin organization caused a considerably greater Swe1p-dependent elongated-bud phenotype than did deletion of HSL1 in the same genetic background (see above). Thus, perturbing septin organization appears to promote both the loss of Hsl1p-mediated Swe1p down-regulation (so that once septins have been perturbed, deletion of HSL1 has little or no further effect) and an additional Hsl1p-independent pathway(s) that impinges on Swe1p-mediated Cdc28p phosphorylation.
Septin-dependent, hierarchical localization of Swe1p, Hsl1p, and Hsl7p to the neck.
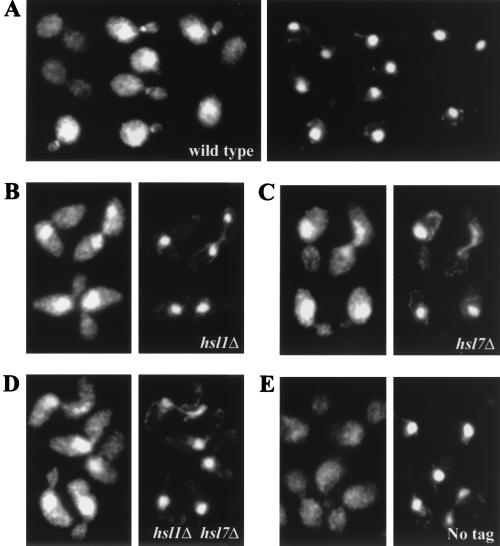
To investigate the basis for the Swe1p-dependent cell cycle delay observed upon perturbation of septin organization, we examined the localization of Swe1p, Hsl1p, and Hsl7p (another negative regulator of Swe1p; see Introduction) using epitope-tagged versions of these proteins that appeared to be fully functional (44, 59) (see Materials and Methods). In unbudded wild-type cells, Swe1p either was not detected or was detected only in the nucleus (32% of the cells examined) (Fig. 5A). In budded wild-type cells, Swe1p was detected only in the nucleus (12% of the cells examined), only at the neck (23% of the cells), or at both locations (39% of the cells) (Fig. 5A). The Swe1p nuclear staining varied greatly in intensity, being most intense in unbudded and small-budded cells and essentially undetectable in postanaphase budded cells. The Swe1p neck staining was observed predominantly in cells with medium-size or large buds, where Swe1p appeared to form a ring on the daughter side of the neck (Fig. 5A). As recently reported by others (6, 58), Hsl1p and Hsl7p were also localized primarily to a ring on the daughter side of the neck (unpublished results). Double-label immunofluorescence experiments showed that Hsl1p and Hsl7p precisely colocalized at all stages and that both Hsl1p and Hsl7p were always localized entirely within the septin-containing region of the neck (unpublished results).
FIG. 5.
Localization of Swe1p in wild-type, hsl1 mutant, and hsl7 mutant cells. The indicated strains were grown to exponential phase in YPD medium at 30°C and stained for Swe1p (left-hand panels) or DNA (right-hand panels) as described in Materials and Methods. (A) JMY1441; (B) JMY1477; (C) JMY1475; (D) JMY1479; (E) DLY1. Strains JMY1441, JMY1477, JMY1475, and JMY1479 all contain two integrated copies of SWE1myc, whereas strain DLY1 lacks SWE1myc as a negative control for the antibody sandwich protocol.
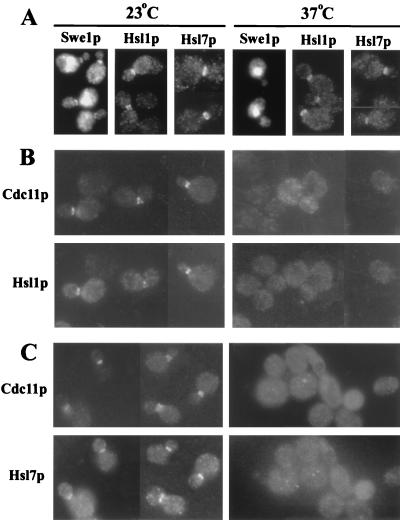
To determine if the neck localization of Swe1p, Hsl1p, and Hsl7p depends on the septins, we examined the localization of these proteins in a temperature-sensitive septin mutant. After a 30-min shift to restrictive temperature, Hsl1p and Hsl7p were both delocalized from the neck in the mutant cells (Fig. 6B and C) but not in control wild-type cells (Fig. 6A). Swe1p was delocalized from the neck even in wild-type cells under these conditions (Fig. 6A). However, by 60 min after the temperature shift, Swe1p was again localized to the necks of wild-type cells but not of septin mutant cells (strain JMY1498) (unpublished results). Thus, Swe1p, Hsl1p, and Hsl7p are all localized to the neck in a septin-dependent manner.
FIG. 6.
Septin dependence of Hsl1p and Hsl7p localization. Cells of the indicated strains were grown to exponential phase in YM-P medium at 23°C and fixed either before (left-hand panels) or 30 min after (right-hand panels) a shift to 37°C. (A) Cells of wild-type strains JMY1441 (SWE1-myc), M-1427 (HSL1-13myc), and M-1423 (HSL7-3HA) were stained with antibodies to the tagged proteins. (B and C) Cells of cdc12-6 strains M-1552 (HSL1-13myc) (B) and M-1554 (HSL7-3HA) (C) were double stained with antibodies to Cdc11p and to the tagged protein.
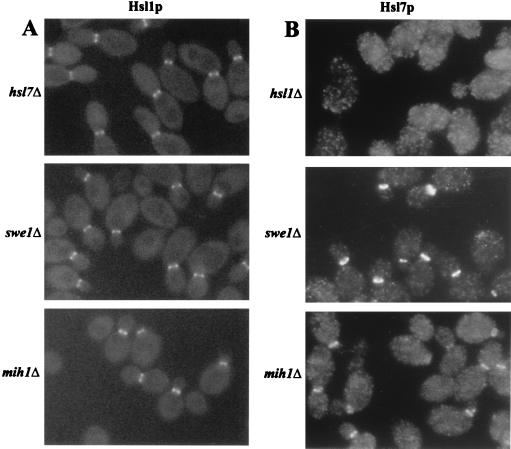
In addition to colocalizing at the neck, Hsl1p, Hsl7p, and Swe1p appear to interact physically with one another (44, 58). Thus, we determined if the neck localization of each of these proteins depends on the others. Deletion of either HSL1 or HSL7 eliminated any detectable neck localization of Swe1p (Fig. 5B, C), whereas deletion of SWE1 (or of MIH1) did not affect neck localization of either Hsl1p or Hsl7p (Fig. 7A and B). In addition, deletion of HSL1 eliminated the neck localization of Hsl7p (Fig. 7B) (58), whereas deletion of HSL7 did not affect the neck localization of Hsl1p (Fig. 7A) (6). These data suggest the existence of a localization hierarchy in which the septins link Hsl1p to the neck, Hsl1p links Hsl7p to the neck, and Hsl7p links Swe1p to the neck.
FIG. 7.
Localization of Hsl1p and Hsl7p in hsl7, hsl1, swe1, and mih1 mutant cells. Cells of the indicated strains were grown to exponential phase in YM-P medium at 30°C and examined by fluorescence microscopy to localize Hsl1p-GFP (A) or fixed and examined by immunofluorescence microscopy to detect Hsl7p-HA (B). (A) HSL1-GFP strains M-1272 (hsl7Δ), M-1426 (swe1Δ), and M-1158 (mih1Δ). (B) HSL7-3HA strains M-1445 (hsl1Δ), M-1440 (swe1Δ), and M-1421 (mih1Δ).
In the hsl1Δ and hsl7Δ single mutants, as well as in an hsl1Δ hsl7Δ double mutant, Swe1p was still localized to the nucleus (Fig. 5B to D). However, in these strains, Swe1p remained detectable in the nuclei of the majority (∼90%) of anaphase and postanaphase cells, where it was rarely (<3%) detected in wild-type cells (Fig. 5A).
Effects of abnormal septin organization and of actin depolymerization on the localization of Hsl1p and Hsl7p.
The abnormally organized septins present in gin4, cla4, and nap1 mutants appear to retain much of their function, as judged by the ability of the mutant cells to localize bud site-selection markers and components of the chitin synthase III complex (40) (Table 3). Nonetheless, it seemed possible that changes in the localization of Hsl1p and/or Hsl7p might contribute to the apparent activation of Swe1p in these mutant strains. Indeed, both Hsl1p and Hsl7p were undetectable at the neck in ≥73% of gin4Δ, cla4Δ, or nap1Δ cells (Fig. 8; Table 5). Double labeling showed that Hsl1p and Hsl7p were delocalized from the neck even in a majority of the cells with relatively normal septin-staining patterns (unpublished results). In contrast to the effect of GIN4 deletion on Hsl1p localization, deletion of HSL1 (or SWE1 or MIH1) did not affect Gin4p localization to the neck (Table 4). Thus, not only a loss of septin localization (such as seen in septin mutants; see above) but also more subtle perturbations of septin organization are associated with delocalization of Hsl1p and Hsl7p (and thus presumably of Swe1p) from the neck. In contrast, deletion of HSL1 (Fig. 3G), HSL7 (Table 3), or SWE1 (unpublished results) produced no obvious perturbation of septin organization.
FIG. 8.
Loss of Hsl1p and Hsl7p localization in gin4 and cla4 mutants. Strains M-1443 (gin4Δ HSL1-13myc) (A), M-1451 (cla4Δ HSL1-13myc) (B), M-1442 (gin4Δ HSL7-3HA) (C), and M-1454 (cla4Δ HSL7-3HA) (D) were grown to exponential phase in YM-P medium at 30°C and examined by immunofluorescence microscopy to localize Hsl1p-myc or Hsl7p-HA. Arrows indicate the minority of cells that show localization of the tagged proteins to the neck.
TABLE 5.
Effects of perturbing septin organization on the neck localization of Hsl1p and Hsl7pa
| Strain | % of cells with neck localization of:
|
||
|---|---|---|---|
| Septinsb | Hsl1p | Hsl7p | |
| Wild type (M-1427) | 96 | 92 | NA |
| gin4Δ (M-1443) | 94 | 25 | NA |
| cla4Δ (M-1451) | 91 | 27 | NA |
| nap1Δ (M-1452) | 90 | 23 | NA |
| Wild type (M-1423) | 100 | NA | 90 |
| gin4Δ (M-1442) | 86 | NA | 27 |
| cla4Δ (M-1454) | 86 | NA | 23 |
| nap1Δ (M-1453) | 89 | NA | 20 |
Wild-type and homozygous mutant diploid strains were grown to exponential phase in YM-P medium at 23°C, and the localizations of septins, Hsl1p, and Hsl7p were determined by double-label immunofluorescence microscopy. More than 200 budded cells were counted per sample. NA, not applicable.
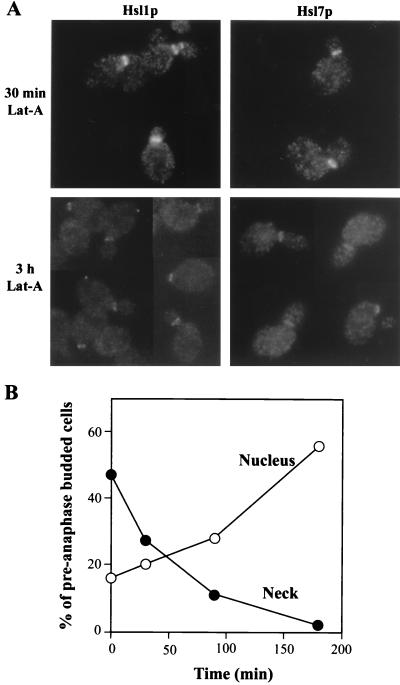
In contrast to the dramatic effects of septin perturbations, complete depolymerization of F actin using latrunculin-A (4) had little effect on the localization of Hsl1p or Hsl7p to the neck (Fig. 9A, 30 min), although prolonged exposure to the drug caused an eventual reduction in the staining intensity (Fig. 9A, 3 h). In contrast, Swe1p disappeared progressively and relatively rapidly from the necks of cells exposed to latrunculin-A, with a concomitant increase in the proportion of cells displaying nuclear staining (Fig. 9B). Thus, although perturbations of both actin and the septins promote Swe1p-dependent delays of the cell cycle, such perturbations have very different effects on the localization of Hsl1p, Hsl7p, and Swe1p. In this regard, it is also of interest that temperature shock, which produces a transient depolarization of the actin cytoskeleton (36), also affects the localization of Swe1p while having no detectable effect on the localizations of Hsl1p and Hsl7p (Fig. 6A).
FIG. 9.
Localization of Hsl1p, Hsl7p, and Swe1p following actin depolymerization. (A) HSL1-13myc (M-1427) and HSL7-3HA (M-1423) cells growing exponentially in YM-P medium at 30°C were treated with 100 μM latrunculin-A for 30 min or 3 h, as indicated, and Hsl1p-myc and Hsl7p-HA were localized by immunofluorescence. Equal exposure times were used for all four panels. (B) SWE1myc cells (JMY1441) were grown to exponential phase in YPD medium at 30°C and treated with 100 μM latrunculin-A. The proportions of preanaphase budded cells displaying Swe1p staining at the neck (●) or in the nucleus (○) were determined at the indicated times. (Cells with Swe1p at both locations were counted in both categories.) More than 200 cells were counted per sample.
DISCUSSION
Roles for Gin4p, Cla4p, and Nap1p in septin organization.
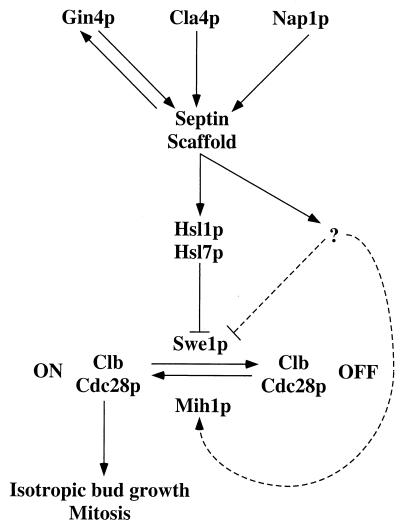
Previous observations had suggested that both Gin4p and Cla4p are important for normal organization of the mother-bud neck and that Gin4p, Cla4p, and Nap1p all function in related processes (2, 8, 16, 40, 62). Consistent with these observations, we found that both cla4 and nap1 single mutants exhibited striking defects in septin organization similar to those observed previously in gin4 mutants. Moreover, the cla4 gin4 and nap1 gin4 double-deletion mutants displayed significantly more severe defects in septin organization than did any of the single mutants. It was reported previously that cla4 nap1 double-deletion mutants display a synthetic growth defect compared to cla4 or nap1 single mutants (62). Taken together, these synthetic effects appear to rule out a linear pathway such as that proposed by Tjandra et al. (62). Instead, the results suggest that Gin4p, Cla4p, and Nap1p function at least partially in parallel to promote normal septin organization (Fig. 10).
FIG. 10.
Model summarizing the results presented in this paper. See text for details.
Gin4p becomes hyperphosphorylated as cells progress through the cell cycle, with maximal phosphorylation in G2/M; this hyperphosphorylation requires (and appears to stimulate) Gin4p kinase activity and depends also on the septins, Cla4p, and Nap1p (2, 13, 62). Because Gin4p localization occurs early in the cell cycle and does not depend on Gin4p kinase activity (40), the hyperphosphorylation presumably does not affect Gin4p localization. In contrast, as efficient localization of Gin4p to the neck requires Cla4p and Nap1p (Table 4) as well as the septins (40), it seems possible that Gin4p localization to the neck might be important for its subsequent hyperphosphorylation. In this case, the role of the septins, Gin4p, Cla4p, and Nap1p in activating Gin4p may be indirect by means of their promotion of a septin organization that allows Gin4p localization (Fig. 10).
Swe1p-dependent cell cycle delay upon perturbation of septin organization.
Septin mutants display a modest G2 delay accompanied by the formation of highly elongated buds. Similarly, the disorganization of the septins in gin4, cla4, nap1, cla4 gin4, and nap1 gin4 mutants is also accompanied by the formation of elongated buds. In all cases, the elongated-bud phenotypes, but not the defects in septin organization and function, were suppressed by deleting SWE1 or by introducing the nonphosphorylatable CDC28Y19F allele. The simplest interpretation of these results is that defects in septin organization promote the accumulation and/or activation of Swe1p, resulting in lowered Cdc28p activity and thus in a cell cycle delay associated with continued apical growth of the bud (Fig. 10).
Barral and coworkers recently reported similar observations on septin mutants and reached similar conclusions (6). In contrast, Kellogg and coworkers have suggested that the cell cycle delays in septin and gin4, cla4, and nap1 mutants occur after Cdc28p activation, and therefore that the septins, Gin4p, Cla4p, and Nap1p must function downstream of Cdc28p in a pathway for executing the switch from apical to isotropic bud growth (2, 13, 30, 62). However, this conclusion was based largely on measurements of Clb2p-associated histone H1 kinase activity in vitro, which might not faithfully reflect the levels of relevant activity in vivo; a further caveat is that the specific activities of Cdc28p were not measured in the various mutants. Moreover, without additional assumptions, the model of Kellogg and coworkers seems very difficult to reconcile with the observations that either deletion of SWE1 or introduction of CDC28Y19F suppresses the elongated-bud phenotypes of the various mutants. However, it should also be noted that none of the models proposed to date explains why the apical bud growth of septin mutants apparently continues even after the cells have undergone nuclear division (presumably marking the end of the Swe1p-dependent G2 delay) (1, 26).
Distinct roles for Nim1p family kinases in septin organization and cell cycle control.
Gin4p, Hsl1p, and Kcc4p all contain closely related kinase domains and are localized to the neck, and at least for Gin4p and Hsl1p, neck localization appears to be important for function. Based on these similarities, it has been suggested that these three kinases play redundant roles in both septin organization and cell cycle control (6). However, we found no evidence for a role of either Hsl1p or Kcc4p in septin organization, even in the absence of Gin4p, and other tests for redundancy in function between Gin4p and Kcc4p (which contain large related nonkinase domains that have little sequence similarity to the corresponding domain in Hsl1p) were also negative (40). In this regard, it is also worth noting that the localization patterns of the three kinases at the neck are distinct: Gin4p typically colocalizes with the septins on both mother and bud sides of the neck (40), whereas Hsl1p and Kcc4p localize specifically to the bud side of the neck (reference 6 and this study). In addition, we found no evidence for a direct role of either Gin4p or Kcc4p in cell cycle control. Swe1p is fully stabilized (i.e., is as stable in G2/M as it is in G1) in cells deleted only for HSL1, suggesting that neither Gin4p nor Kcc4p targets Swe1p for degradation (44), and kcc4 deletion cells showed no evidence of a G2 delay even when MIH1 was also deleted (Fig. 4C). Moreover, although gin4 mutant cells did display a G2 delay that was actually more pronounced than that seen in hsl1 mutant cells, this delay seems likely to be an indirect effect of the septin perturbation in the gin4 mutant cells (Fig. 10). Thus, it appears that the three Nim1p-related kinases of S. cerevisiae play distinct roles in the cell: Gin4p promotes normal septin organization (40), Hsl1p down-regulates Swe1p (6, 43, 44), and Kcc4p plays a role that remains to be discovered.
In this context, recent observations on a second S. pombe Nim1p family kinase, Cdr2p, are also of interest. Deletion of cdr2 caused a pronounced Wee1p-dependent G2 delay, but simultaneous deletion of nim1 and cdr2 did not cause a greater G2 delay in proliferating cells than deletion of cdr2 alone (12, 29). This result seems inconsistent with the hypothesis that Nim1p and Cdr2p play redundant roles in cell cycle control. Instead, it suggests that cdr2 mutations (like gin4 mutations) may cause a cell cycle delay indirectly, consistent with the hypothesis that Nim1p family kinases may generally play distinct roles as specified by their large nonkinase domains.
Septin-dependent recruitment of Hsl1p, Hsl7p, and Swe1p to the neck.
A clue as to how septin perturbations might affect Swe1p came from the observations that Hsl1p (6; this study), Hsl7p (58; this study), and a fraction of cellular Swe1p (Fig. 5) are all colocalized to the neck. The neck localization of Hsl1p requires the septins but not Hsl7p or Swe1p; the neck localization of Hsl7p requires the septins and Hsl1p but not Swe1p; and the neck localization of Swe1p requires both Hsl1p and Hsl7p, as well as the septins. Consistent with these findings, recent biochemical evidence indicates that the septin Cdc3p interacts physically with Hsl1p (6), that Hsl1p interacts physically with Hsl7p (58), and that Hsl7p interacts physically with Swe1p (44, 58). Thus, Hsl1p and Hsl7p appear to function as part of a septin-dependent hierarchy for localization of Swe1p to the neck, providing another illustration of the general role of the septins in organizing other proteins at the cell surface (40, 42).
The colocalization of Swe1p and its negative regulators at the neck raised the possibility that mislocalization of these proteins might contribute to the apparent increase in Swe1p activity in mutants with septin defects. Indeed, we found that even the relatively mild perturbations of septin organization observed in gin4, cla4, and nap1 mutant cells were associated with greatly reduced localization of Hsl1p and Hsl7p (and hence, presumably, of Swe1p) to the neck. In contrast, two functionally unrelated proteins, Bni4p and Bud4p, were still localized efficiently to the neck in gin4 mutant cells (40), suggesting that Hsl1p and Hsl7p localization may be particularly sensitive to perturbations of septin organization. The involvement of both Hsl1p and Hsl7p in targeting Swe1p for degradation during G2/M (44, 58) might simply reflect a role for Hsl1p and Hsl7p in delivering Swe1p to a location at which the ubiquitination complex SCFMet30 (28) can target it for degradation. Alternatively (or in addition), Hsl1p and Hsl7p may play a direct role in targeting Swe1p for degradation (e.g., through phosphorylation of Swe1p by Hsl1p [44, 58]). In this case, the localization of the proteins to the neck might be important for the activity of Hsl1p and/or Hsl7p. Consistent with this possibility, the autophosphorylation activity of Hsl1p was reduced in extracts from a septin mutant strain relative to that in extracts from a wild-type strain (6). However, it should also be noted that cells with delocalized Hsl1p appear to retain at least partial Hsl1p activity, because gin4, cla4, and nap1 mutations are not lethal in combination with deletion of MIH1 (Fig. 2) (unpublished results), whereas hsl1 mih1 double mutants undergo a lethal G2 arrest (44).
The stabilized Swe1p in hsl1 and hsl7 mutants accumulated in the nucleus, which presumably facilitates its inhibition of nuclear Clb-Cdc28p complexes. It seems likely that septin perturbations, by delocalizing Hsl1p and Hsl7p, also cause Swe1p stabilization and nuclear accumulation, contributing to the observed G2 delay. It is also possible that changes in Swe1p specific activity contribute to the G2 delay. Indeed, the Swe1p-dependent cell cycle delays observed in mutants with perturbed septin organization were more severe than those observed upon deletion of HSL1 or HSL7, suggesting that septin organization affects Swe1p (and/or Mih1p) function through at least one additional Hsl1p/Hsl7p-independent pathway (Fig. 10).
Why are Hsl1p, Hsl7p, and Swe1p localized to the daughter side of the neck?
There appear to be at least four possible models to explain the surprising localization of this regulatory module to the septin ring at the mother-bud neck. These models are not mutually exclusive.
First, as suggested previously (6, 58), Hsl1p and Hsl7p might serve as sensors that monitor septin organization as part of a checkpoint pathway that delays nuclear division if the neck is not well organized for subsequent cytokinesis. However, it is not known whether yeast cells in their natural environment ever experience perturbations of septin organization (for instance, neither temperature shock nor osmotic shock produces obvious alterations of septin organization [unpublished results]), and it is unclear how introduction of a short G2 delay might help cells to cope with such perturbations. Nevertheless, the finding that deletion of SWE1 exacerbates the growth defects of certain septin mutants (6; this study) does suggest that under some circumstances the delay is beneficial, supporting the hypothesis of a septin-monitoring checkpoint.
Second, as also suggested previously (6), septin misorganization might serve as a sensitive indicator for more general morphogenetic problems, so that the septin sensor postulated above could in fact cause a checkpoint delay in response to perturbations of actin organization or of bud formation. However, there is no evidence suggesting that actin perturbations affect septin organization; for example, septin rings form normally in cells lacking F actin because of treatment with latrunculin-A (4), and we found that the actin perturbations induced by heat shock or by latrunculin-A did not significantly alter the neck localizations of Hsl1p and Hsl7p (Fig. 6A and 9). These observations suggest that the mechanisms underlying Swe1p regulation in response to actin perturbations and to septin perturbations are distinct. Consistent with this hypothesis, actin perturbations can promote much longer Swe1p-dependent cell cycle delays (>15 h [46]) than those observed upon septin perturbation (30 to 45 min).
Third, consistent with other evidence for the septins serving as a scaffold for the assembly of functional complexes (40, 42), the neck may simply be a convenient place to concentrate Swe1p together with its regulators. In this regard, it is interesting that another cell cycle regulator, Cdc14p, is localized to the nucleolus, where it is prevented from acting for much of the cell cycle (57, 63). It may be that the neck and the nucleolus provide convenient sites to store (or inactivate or degrade) cell cycle regulators at times when they are not needed.
A final model is particularly attractive because it also rationalizes the strikingly asymmetric localization of the regulatory module to the daughter side of the neck. In particular, this model proposes that the ability of the module to assemble in this location serves as an indicator that a bud has been formed. Whereas unbudded and small-budded cells undergo a Swe1p-dependent G2 arrest upon depolymerization of F actin, larger-budded cells do not (46). However, constitutive but modest Swe1p overexpression, which is insufficient to delay the normal cell cycle, allows larger-budded cells to arrest in G2 in response to actin depolymerization (46). This observation suggests that large-budded cells retain the ability to sense actin perturbations but do not normally contain sufficient Swe1p to enforce a G2 arrest. Perhaps, once a bud has been formed, the cells no longer require the morphogenesis checkpoint, and Swe1p is then degraded so that subsequent actin perturbations do not affect the cell cycle. Assembly of the module on the daughter side of the neck presumably relies on a particular septin organization that is unique to budded cells. Activation of Hsl1p and Hsl7p upon such assembly could therefore couple Swe1p degradation to bud formation.
ACKNOWLEDGMENTS
We thank the other members of the Pringle and Lew laboratories for providing great working environments and many valuable discussions. We also thank Yves Barral, Mike Snyder, Doug Kellogg, Mark Shulewitz, and Jeremy Thorner for valuable discussions and communication of unpublished results. We also thank Susan Whitfield for her usual outstanding assistance with the illustrations.
This work was supported by NIH grant GM31006 to J.R.P. and by NIH grant GM53050 and funds from the Searle Scholars Program/The Chicago Community Trust to D.J.L.
REFERENCES
- 1.Adams A E M, Pringle J R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman R, Kellogg D. Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol. 1997;138:119–130. doi: 10.1083/jcb.138.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1995. [Google Scholar]
- 4.Ayscough K R, Stryker J, Pokala N, Sanders M, Crews P, Drubin D G. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagrodia S, Cerione R A. Pak to the future. Trends Cell Biol. 1999;9:350–355. doi: 10.1016/s0962-8924(99)01618-9. [DOI] [PubMed] [Google Scholar]
- 6.Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benton B K, Tinkelenberg A, Gonzalez I, Cross F R. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol. 1997;17:5067–5076. doi: 10.1128/mcb.17.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi E, Maddox P, Lew D J, Salmon E D, McMillan J N, Yeh E, Pringle J R. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi E, Pringle J R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booher R N, Deshaies R J, Kirschner M W. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breeding C S, Hudson J, Balasubramanian M K, Hemmingsen S M, Young P G, Gould K L. The cdr2+ gene encodes a regulator of G2/M progression and cytokinesis in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9:3399–3415. doi: 10.1091/mbc.9.12.3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll C W, Altman R, Schieltz D, Yates III J R, Kellogg D. The septins are required for the mitosis-specific activation of the Gin4 kinase. J Cell Biol. 1998;143:709–717. doi: 10.1083/jcb.143.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle J R. Role of Bud3p in producing the axial budding pattern of yeast. J Cell Biol. 1995;129:767–778. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coleman T R, Tang Z, Dunphy W G. Negative regulation of the Wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell. 1993;72:919–929. doi: 10.1016/0092-8674(93)90580-j. [DOI] [PubMed] [Google Scholar]
- 16.Cvrčková F, De Virgilio C, Manser E, Pringle J R, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 17.DeMarini D J, Adams A E M, Fares H, De Virgilio C, Valle G, Chuang J S, Pringle J R. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Epp J A, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7:921–929. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- 19.Farkaš V, Kovařik J, Košinová A, Bauer Š. Autoradiographic study of mannan incorporation into the growing cell walls of Saccharomyces cerevisiae. J Bacteriol. 1974;117:265–269. doi: 10.1128/jb.117.1.265-269.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ford S K, Pringle J R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev Genet. 1991;12:281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- 21.Frazier J A, Wong M L, Longtine M S, Pringle J R, Mann M, Mitchison T J, Field C. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J Cell Biol. 1998;143:737–749. doi: 10.1083/jcb.143.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 24.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [PubMed] [Google Scholar]
- 25.Haarer B K, Pringle J R. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol Cell Biol. 1987;7:3678–3687. doi: 10.1128/mcb.7.10.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartwell L H. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- 27.Hunter T, Plowman G D. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser P, Sia R A L, Bardes E G S, Lew D J, Reed S I. Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 1998;12:2587–2597. doi: 10.1101/gad.12.16.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanoh J, Russell P. The protein kinase Cdr2, related to Nim1/Cdr1 mitotic inducer, regulates the onset of mitosis in fission yeast. Mol Biol Cell. 1998;9:3321–3334. doi: 10.1091/mbc.9.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kellogg D R, Murray A W. NAP1 acts with Clb2 to perform mitotic functions and to suppress polar bud growth in budding yeast. J Cell Biol. 1995;130:675–685. doi: 10.1083/jcb.130.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H B, Haarer B K, Pringle J R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Lew D J, Reed S I. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lew D J, Weinert T, Pringle J R. Cell cycle control in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 607–695. [Google Scholar]
- 36.Lillie S H, Brown S S. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesin-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lillie S H, Pringle J R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980;143:1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longtine M S, DeMarini D J, Valencik M L, Al-Awar O S, Fares H, De Virgilio C, Pringle J R. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- 40.Longtine M S, Fares H, Pringle J R. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longtine M S, McKenzie III A, DeMarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 42.Longtine M S, Pringle J R. Septins. In: Kreis T, Vale R, editors. Guidebook to the cytoskeletal and motor proteins. Oxford, England: Oxford University Press; 1999. pp. 359–363. [Google Scholar]
- 43.Ma X-J, Lu Q, Grunstein M. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- 44.McMillan J N, Longtine M S, Sia R A, Theesfeld C L, Bardes E S, Pringle J R, Lew D J. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol Cell Biol. 1999;19:6929–6939. doi: 10.1128/mcb.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMillan J N, Sia R A L, Bardes E S G, Lew D J. Phosphorylation-independent inhibition of Cdc28p by the tyrosine kinase Swe1p in the morphogenesis checkpoint. Mol Cell Biol. 1999;19:5981–5990. doi: 10.1128/mcb.19.9.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McMillan J N, Sia R A L, Lew D J. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J Cell Biol. 1998;142:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 48.Okuzaki D, Tanaka S, Kanazawa H, Nojima H. Gin4 of S. cerevisiae is a bud neck protein that interacts with the Cdc28 complex. Genes Cells. 1997;2:753–770. doi: 10.1046/j.1365-2443.1997.1590358.x. [DOI] [PubMed] [Google Scholar]
- 49.Parker L L, Walter S A, Young P G, Piwnica-Worms H. Phosphorylation and inactivation of the mitotic inhibitor Wee1 by the nim1/cdr1 kinase. Nature. 1993;363:736–738. doi: 10.1038/363736a0. [DOI] [PubMed] [Google Scholar]
- 50.Pringle J R. Staining of bud scars and other cell wall chitin with Calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- 51.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 52.Pringle J R, Mor J-R. Methods for monitoring the growth of yeast cultures and for dealing with the clumping problem. Methods Cell Biol. 1975;11:131–168. doi: 10.1016/s0091-679x(08)60320-9. [DOI] [PubMed] [Google Scholar]
- 53.Richardson H E, Wittenberg C, Cross F, Reed S I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 54.Russell P, Moreno S, Reed S I. Conservation of mitotic controls in fission and budding yeast. Cell. 1989;57:295–303. doi: 10.1016/0092-8674(89)90967-7. [DOI] [PubMed] [Google Scholar]
- 55.Russell P, Nurse P. The mitotic inducer nim1+ functions in a regulatory network of protein kinase homologs controlling the initiation of mitosis. Cell. 1987;49:569–576. doi: 10.1016/0092-8674(87)90459-4. [DOI] [PubMed] [Google Scholar]
- 56.Sanders S L, Herskowitz I. The Bud4 protein of yeast, required for axial budding, is localized to the mother/bud neck in a cell cycle-dependent manner. J Cell Biol. 1996;134:413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shou W, Seol J H, Shevchenko A, Baskerville C, Moazed D, Chen Z W, Jang J, Charbonneau H, Deshaies R J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 58.Shulewitz M J, Inouye C J, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sia R A L, Bardes E S G, Lew D J. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 1998;17:6678–6688. doi: 10.1093/emboj/17.22.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sia R A L, Herald H A, Lew D J. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol Biol Cell. 1996;7:1657–1666. doi: 10.1091/mbc.7.11.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tjandra H, Compton J, Kellogg D. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr Biol. 1998;8:991–1000. doi: 10.1016/s0960-9822(07)00419-8. [DOI] [PubMed] [Google Scholar]
- 63.Visintin R, Hwang E S, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 64.Wu L, Russell P. Nim1 kinase promotes mitosis by inactivating Wee1 tyrosine kinase. Nature. 1993;363:738–741. doi: 10.1038/363738a0. [DOI] [PubMed] [Google Scholar]