Abstract
Immune dysregulation is commonly observed in patients with coronavirus disease 2019 (COVID-19). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces severe lung inflammation and innate immune cell dysregulation. However, the precise interaction between SARS-CoV-2 and the innate immune system is currently unknown. To understand the interaction between SARS-CoV-2 and natural killer (NK) cells, several SARS-CoV-2 S protein peptides capable of binding to the NKG2D receptor were screened by in silico analysis. Among them, two peptides, cov1 and cov2, bound to NK cells and NKG2D receptors. These cov peptides increased NK cytotoxicity toward lung cancer cells, stimulated interferon gamma (IFN-γ) production by NK cells, and likely mediated these responses through the phosphorylation of Vav1, a key downstream-signaling molecule of NKG2D and NK activation genes. The direct interaction between SARS-CoV-2 and NK cells is a novel finding, and modulation of this interaction has potential clinical application as a therapeutic target for COVID-19.
Keywords: SARS-Cov-2, NK, NKG2D, Peptide, Cytotoxicity, IFN-γ
1. Introduction
Coronavirus disease 2019 (COVID-19) is a global pandemic threatening millions of lives worldwide, and the rapid development of effective therapeutics and vaccines is urgently required. Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can lead to immune system dysregulation and macrophage activation syndrome in severe cases [1]. The dysregulation of cytotoxic T cells (CTLs) and natural killer (NK) cells has also been reported [2], [3], [4].
NK cells are one of the major innate immune cells, and are responsible for controlling transformed cells, such as cancer and virus-infected cells. NK cells have receptors that transmit activating signals into the cells in response to stimuli from transformed cells. Moreover, they have inhibitory receptors that deliver signals to inhibit various activation pathways. The balance between the two signals is determined by specific ligands and signals from their corresponding receptors. NKG2A, an inhibitory receptor that is overexpressed in NK cells in COVID-19 patients, induces NK cell exhaustion, and serves as a potential therapeutic target [5]. However, there is limited understanding of the role of NK receptors and their interaction with SARS-CoV-2 in COVID-19 pathogenesis.
NKG2D is one of the major activating receptors that delivers critical activating signals. In humans, there are at least eight ligands, including MICA, MICB, and ULBP1-6, that can bind to NKG2D [6], [7]. NKG2D is associated with DAP10, a signal-delivering adapter in humans, and with DAP10 and DAP12 in mice [8], [9]. Tumors and viruses have devised mechanisms to evade NKG2D recognition by modulating its ligand expression or by secreting soluble forms of its ligands [10].
Several viruses, including the influenza virus, cytomegalovirus, and human immunodeficiency virus, interact with NK cells and exhibit a variety of evasion strategies [11], [12], [13]. Viruses interfere with NK function by direct infection, regulating NK receptor signaling, and modulating cytokine secretion. Some viruses interfere with the recognition between NK cell receptors and ligands. Cowpox virus-encoded OMCP has a similar structure to NKG2D ligands, and it binds directly to NKG2D receptors to inhibit the binding of NKG2D ligands [14]. SARS-CoV-2 likely exhibits similar strategies to overcome antiviral NK cell responses [15]. Potential strategies include the upregulation of NK inhibitory receptors and the HLA-I molecule to induce NK exhaustion, and/or the downregulation of cytotoxic granules such as perforin and granzymes to decrease NK cytotoxicity. Alternatively, it may reduce the NK cell population by increasing cell apoptosis.
The aim of the present study was to use in silico analysis to screen for SARS-CoV-2 S protein peptides that could bind to NKG2D receptors, and evaluate the downstream effects on NK cell activity.
2. Results
2.1. Selection of NKG2D-binding peptides from the SARS-CoV-2 S protein
To identify the NKG2D-binding peptides from the S protein of SARS-CoV-2, the sequence of SARS-CoV-2 S protein was downloaded from UniPort and scanned for NKG2D binding peptides through the PepSite2 program. Initially, five 5-mer peptides, four 6-mer peptides, and three 7-mer peptides were selected (Table 1 ).
Table 1.
Candidate NKG2D-binding SARS-COV-2 S protein peptides.
| Sequence | P value | |
|---|---|---|
| 5-mers | QNAQA | 0.04885 |
| LPFQQ | 0.0405 | |
| NQNAQ | 0.04885 | |
| FQQFG | 0.0437 | |
| PFQQF | 0.0405 | |
| 6-mers | CNDPFL | 0.04716 |
| FQFCND | 0.03844 (peptide #1, cov1) | |
| EFQFCN | 0.03844 | |
| FCNDPF | 0.04716 | |
| 7-mers | KCVNFNF | 0.03903 (peptide #2, cov2) |
| KVCEFQF | 0.04854 | |
| CVNFNFN | 0.03903 |
Among the candidate peptides, we selected two based on their predicted binding scores: one 6-mer peptide (FQFCND, cov1 peptide #1) and one 7-mer peptide (KCVNFNF, cov2 peptide #2). Peptide #1 (cov1) covers the 133th–138th amino acid sequences of the S protein of SARS-CoV-2, and peptide #2 (cov2) covers the 537th–543th. A control peptide, a 7-mer peptide (1201th–1207th amino acid, QELGKYE) was also designed (Fig. 1 A). The predicted binding of the peptides to the protein surface of NKG2D was visualized using the protein-peptide interaction prediction program of PepSite2 (http://pepsite2.russelllab.org, Fig. 1B, C) [16].
Fig. 1.
Prediction of NKG2D-binding peptides from SARS-CoV-2 S protein. A. Full amino acid sequence of the SARS-CoV-2 S protein. Cov1 peptide #1 (FQFCND, 133th–138th), cov2 peptide #2 (KCVNFNF, 537th–543th), and control peptide (QELGKYE, 1201th–1207th) are in red. Receptor binding domain (331th-524th) for ACE2 is underlined. B. Predicted binding model of cov1 peptide #1 (circle) and NKG2D (ribbon) was analyzed using the protein-peptide interaction prediction program of PepSite. C. Predicted binding model of cov2 peptide #2 (circle) and NKG2D (ribbon) was analyzed using the protein-peptide interaction prediction program of PepSite.
To assess the binding capacity of peptides to NK cells by flow cytometry, we synthesized FITC-labeled cov1, FITC-labeled cov2 and control peptides. FITC-cov1 peptides and FITC-cov2 peptides bound to NK92 cells in a concentration-dependent manner, while the control peptides did not bind at all (Fig. 2 A). In addition, the FITC-cov2 peptides bound to primary NK cells, and the FITC-control peptides did not (Fig. 2B).
Fig. 2.
Binding of S protein NKG2D-binding peptides to NK cells. A. NK92 cells were incubated with FITC-labeled control peptide, cov1, or cov2 and binding was analyzed by flow cytometry. (n = 3), (*p < 0.05, **p < 0.01, student t-test) B. Primary NK cells were incubated with FITC-labeled control peptide or cov2 and binding was analyzed by flow cytometry. C. For competition, soluble NKG2D (500 nM, 2500 nM, 5000 nM) was pre-incubated with FITC-labeled cov peptides (50 nM) or FITC-labeled control peptide (50 nM) for 20 min, then the binding was analyzed by flow cytometry.
To prove this binding is specific for NKG2D, competition between soluble recombinant NKG2D and cov peptides were analyzed. The FITC-cov peptides (50 nM) were pre-incubated with soluble NKG2D protein (500 nM, 2500 nM, 5000 nM) for 20 min before incubating with NK cells. Soluble NKG2D competed with the cov2 peptide for binding to NK cells, but did not with the cov1 peptide (Fig. 2C). It seems that cov1 binds to other site of NKG2D which does not compete with soluble NGK2D (Phe78-Val216) or its binding is non-specific. Next, FITC-labeled cov2 peptides or FITC-labeled control peptides were injected into mice. It was observed that only FITC-labeled cov2 peptides bound to NK cells in vivo (Fig. S1).
2.2. SARS-CoV-2 S protein peptides increase NK activity
Next, the effects of peptides on NK function were analyzed. NK92 cells were incubated with the peptides for 2 h before NK cytotoxicity was analyzed using H460 lung cancer cells. The cov1 and cov2 peptides significantly increased NK cytotoxicity in a dose-dependent manner compared to the control peptide (Fig. 3 A). Furthermore, the cov1 and cov2 peptides increased NK cytotoxicity against other lung cancer cells (Fig. 3B). Incubation of primary NK cells with the peptides increased NK cytotoxicity against H460 cells relative to the control peptide (Fig. 3C).
Fig. 3.
Effects of S protein NKG2D-binding peptides on NK cytotoxicity. A. Effects of peptides on NK92 cytotoxicity against H460 lung cancer cells. (n = 8) B. Effects of peptides on NK92 cytotoxicity against lung cancer cell lines. (n = 3) C. Effects of peptides on primary NK cell cytotoxicity against H460 lung cancer cells. (n = 2), (*p < 0.05, **p < 0.01, student t-test).
Then, the effects of the peptides on NK cell interferon gamma (IFN-γ) production were analyzed. Secreted IFN-γ from NK92 cells after peptide treatment was measured by ELISA (Fig. 4 ). Cov1 and cov2 peptides increased IFN-γ secretion at concentrations between 10 and 100 nM.
Fig. 4.

Effects of S protein NKG2D-binding peptides on NK cell IFN-γ production. NK92 cells were incubated with various concentrations of peptides for 24 h, and secreted IFN-γ production was measured by ELISA. Data represent the average of three separate experiments.
Activation-induced cell death was assessed [17], and peptide-treated NK cells were incubated up to 48 h in the absence (Fig. S2A) or presence (Fig. S2B) of tumor cells. There was no difference in the levels of NK cell apoptosis between control or cov peptide treatment.
2.3. SARS-CoV-2 S protein peptides induces NK activating signals
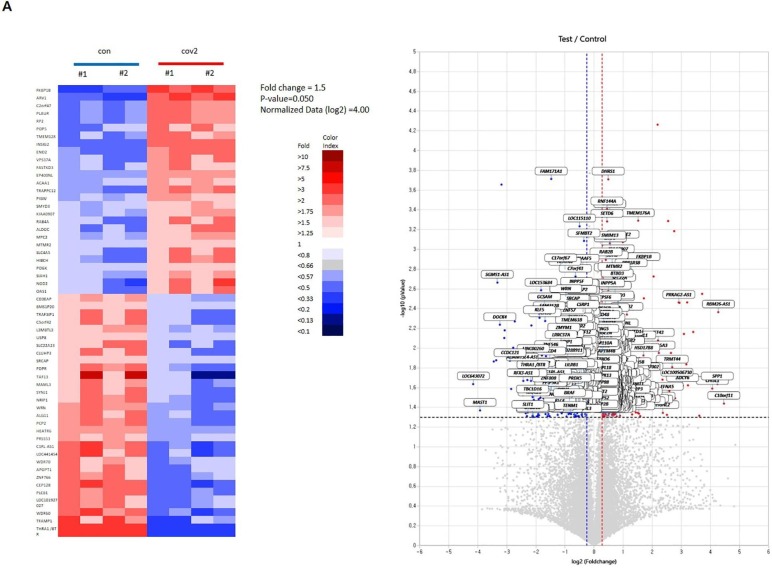
To understand the signaling events induced by the cov peptides, protein phosphorylation of NKG2D downstream molecules was assayed. Vav1, a key molecule for NKG2D signaling [9], [18], [19], was phosphorylated by cov1 and cov2, and not by the control peptide (Fig. 5 ). Furthermore, the change of molecular signature was analyzed by RNA sequencing after peptide treatment. Heat map clustering demonstrated that the 110 genes were quite differentially regulated by the peptide treatment, exhibiting a homogenous expression pattern within the same sample group (Fig. 6 A, Table S1). A volcano plot demonstrating the ratios of cov2 peptide/con peptide showed that 259 activated genes (red spots) and 215 inhibited genes (blue spots) were regulated by cov2 treatment. Based on the Quick GO program, the number of genes including cell activation, defense response, and immune response regulated by cov2 treatment was clustered (Fig. 6B, Table S2). In addition, the number of genes related to NK function such as natural killer cell activation, natural killer cell cytotoxicity, ether lipid metabolism, antigen processing and presentation and natural killer cell differentiation was clustered (Fig. 6C). The correlation analysis revealed that several NK cell activation genes such as IFNG, FADD, CD48, CCL4, MDM2 and IL2RA were identified as cov2 response genes (Fig. 6D).
Fig. 5.
Effects of S protein NKG2D-binding peptides on Vav1 phosphorylation. NK92 cells were treated with 100 nM peptides for 5, 10, and 30 min. Then, cell lysates were analyzed by immunoblotting using vav1 and p-vav1 antibodies.
Fig. 6.
Analysis of cov peptide response genes. A. Heat map clustering of 57 gene sets (fold change = 1.5, p-value = 0.050, normalized data (log2) = 4.00). Red represents a higher level of expression and blue represents a lower level expression. B. Gene ontology categories of cov peptide response genes. Selected genes were classified according to biological processes. C. Pathway categories of cov peptide response. B and C processes significantly enriched in the set of genes identified by RNA-seq as up- or down-regulated and categorized using the Quick Go program. Numbers in the bars indicate the log value assigned to each gene ontology term and pathway term. D. A list of NK-related gene (IFNG, FADD, CD48, CCL4, MDM2 and IL2RA) expression and heat map by cov peptide treatment. con (control peptide treatment); cov2 (cov2 peptide treatment).
3. Discussion
It has been reported that SARS-CoV-2 infection induces a delayed or suppressed type I IFN response with a reduction in T cell and NK cell numbers [20], [21]. Moreover, there is a correlation between the decreased number of NK cells and the severity of COVID-19 [21]. NKG2A expression in NK cells is induced in COVID-19 patients, suggesting that the functional exhaustion of NK cells is associated with the severity of SARS-CoV-2 infection. In addition, NK cells are responsible for the accumulation of neutrophils during SARS-CoV-2 infection [17].
The mechanisms underlying reduced cell number and NK cell dysfunction in COVID-19 patients remain unclear. SARS-CoV-2 may infect and kill NK cells or promote activation-induced cell death. NK cell dysregulation leads to a decrease in IFN-γ production and activation of cytolytic T cells, which are critical in viral defense.
In this study, SARS-CoV-2 S protein peptides, cov1 and cov2, which bind to NK cells and NKG2D receptors were identified by in silico analysis, protein-peptide interaction modeling, and flow cytometric binding assays. Functionally, these peptides increased NK cytotoxicity and IFN-γ production. The NKG2D ligands, MICA and ULPB3, bind to NKG2D through the 180th-200th amino acid of NKG2D [7], [22]. Structural binding prediction showed that the binding sites of the two peptides to NKG2D (120th, 121th, 158th, 188th, 189th, 202th amino acid) partially overlapped with NKG2D ligand binding sites. These two peptides can activate NKG2D signaling, as shown in our study, or potentially compete with NKG2D ligands for NKG2D binding sites. However, further competitive binding assays are necessary to confirm this strategy.
The cov1 and cov2 peptides activated NK cells through the NKG2D receptor and Vav1 phosphorylation. Activation-induced NK cell death is typically dependent on IL-2 priming and granzyme B leakage [23]. However, activation by cov1 and cov2 did not lead to activation-induced NK cell death, suggesting an alternative mode of activation.
Recently, it was reported that SARS-CoV-2 S1 and S2 proteins increased NK cell migration and IFN-γ secretion [24]. This implies that the SARS-CoV-2 S protein may interact with NK cells directly. However, lung epithelial cells transfected with SARS-CoV-2 S1 protein reduced NK cell degranulation and IFN-γ secretion. Transfected SARS-CoV-2 increased HLA-E expression in lung epithelial cells and NKG2A expression in NK cells after co-culture with lung epithelial cells transfected with SP1 protein. This interaction may compromise the anti-viral activity of NK cells through their exhaustion. At the same time, it was reported that a CD3-CD56dimCD16- regulatory NK subset were significantly increased in COVID-19 patients with severe symptoms, while cytotoxic CD3-CD56dimCD16+ NK subsets were decreased. Similarly, PD-1+ NK cells were higher in COVID-19 patients compared to heathy controls, indicating that functional exhaustion and subset alteration of NK cells may contribute to the progression of COVID-19 [25]. Taken together, SARS-CoV-2 S protein can increase NK activity by direct binding or decrease NK activity by modulating HLA expression in transfected cells.
Single-cell analysis of bronchial samples from severe COVID-19 patients showed a strong enrichment in NK cells compared to both healthy controls and patients with moderate COVID-19 [26]. In addition, robust NK cell activation was observed in peripheral blood and bronchioalveolar lavage from COVID-19 patients [27]. A higher expression of Ki-67 was monitored in both NKG2A+CD65L+CD56dim NK cells. Gene expression and signaling profiles of NK cells from bronchioalveolar fluid of severe COVID-19 patients displayed an activated and inflamed pattern, suggesting robust NK cell activation toward SARS-CoV-2 infection and a role for NK cells in the early acute phase of SARS-CoV-2 infection and in COVID-19 pathogenesis. Finally, we searched for differentially expressed genes and signaling cascades following cov peptide treatment. The upregulated genes were related to NK cell cytotoxicity and immunity, and included CCRL2, TNFSF9, EBI3, ACAA1 and IFNG (>1.5-fold upregulation). Pathway-based analysis showed that natural killer cell cytotoxicity, natural killer cell activation pathways were upregulated. Specifically, FADD, CD48, MDM2, IL2RA and IFNG were identified in this pathway analysis.
The appearance of new SARS-CoV-2 variants is increasing. We checked the possible amino acid change in cov1 and cov2 of SARS-CoV-2 S protein. We found one variant (D138Y) in cov1 reported from Brazil (https://bv-brc.org/view/VariantLineage/#view_tab=overview), which may increase transmissibility of SARS-CoV-2.
Based on our observations, cov peptides or these regions of S protein can interact with NKG2D of NK cells to modulate NK function. By blocking or enhancing this interaction, we can regulate viral and immune interaction and this interaction will be a potential target for SARS-CoV-2 treatment.
In conclusion, SARS-CoV-2 S protein NKG2D-binding peptides regulate NK cytotoxicity and IFN-γ production. This interaction induced Vav1 phosphorylation and NK activating genes in NK cells. Modulation of this interaction is a key event linking SARS-CoV-2 and innate immunity, and it serves as a possible therapeutic target for SARS-CoV-2 treatment.
4. Materials and methods
4.1. Reagents and antibodies
Control peptide (Q-E-L-K-Y-E), cov1 peptide (F-Q-F-C-N-D), and cov2 peptide (K-C-V-N-F-N-F) were synthesized by Peptron Co (Daejeon, Korea) and stored at −80 °C. Control peptide FITC (FITC-Ahx-Q-E-L-K-Y-E) and cov2 peptide FITC (FITC-Ahx-K-C-V-N-F-N-F) were also synthesized by Peptron Co (Daejeon, Korea). Vav1 and p-Vav antibodies were obtained from Santa Cruz Biotechnology (Dallas, USA), and the human IFN-γ ELISA kit was purchased from Invitrogen (Carlsbad, USA). Soluble recombinant NKG2D (Ph278-Val216) was obtained from Biolegend (San Diego, USA).
4.2. In silico analysis of SARS-CoV-2 peptides binding to NKG2D
The sequence of SARS-CoV-2 spike protein was downloaded from UniPort KB-P59594(SPIKE_CVHSA)(https://www.uniprot.org/uniprot/P59594). The full protein sequence was scanned for k-mers with k = 5, 6, 7, 8 and unique peptides were identified. The PepSite2 algorithm [28] was run for each k-mer as described in the PepSite API: ‘curl -s \“http://pepsite2.russelllab.org/match? dB = 1mpu&&ligand=” + kmer +“&format = best_pval\” “ + ”> “ + outputfile’. Peptides with a p-value < 0.05 were considered potential candidate targets for NKG2D. With this setting, 5, 4, 3, and 0 peptides were selected for each 5, 6, 7, and 8-mers, respectively.
4.3. Cell culture
The human NK92 cell line was obtained from the American Type Culture Collection (ATCC, Rockville, MD) and cultured in α-minimal essential medium (Welgene) supplemented with 12.5% fetal bovine serum (FBS), 12.5% horse serum, 0.2 mM myo inositol, 0.1 mM 2-mercaptoethanol, 0.02 mM folic acid, 1% antibiotics, and 20 ng/ml IL-2 in a humidified atmosphere with 5% CO2 at 37 °C. The human lung cancer lines (H358, H460, and H3122) were obtained from ATCC and cultured in RPMI medium supplemented with 10% FBS and 1% antibiotics. Primary NK cells were obtained from human cord blood provided by a local hospital (IRB approval number P01-201610-31-002), as described previously [29]. Briefly, CD3 cells and red blood cells were depleted from mononuclear cells with a Rosettesep cocktail (STEMCELL technologies) and CD3-depleted cells were cultured in α-minimal essential medium (Welgene) supplemented with human IL-15 (10 ng/mL), IL-21 (10 ng/mL), and 10−6 M hydrocortisone (StemCell Technologies).
4.4. Flow cytometry
NK cells were washed with ice-cold PBS and stained with the indicated antibodies or FITC-labeled peptides in a staining buffer (PBS containing 1% FBS and 0.01% NaN3) for 20 min at 4 °C. After washing, flow cytometry was performed on a FACS Canto II (BD Biosciences) and data were analyzed using FlowJo software (Tree Star).
4.5. NK cytotoxicity
NK92 or primary NK cells were incubated for 12 h in NK92 culture media supplemented with 0.1% BSA instead of 12.5% FBS and 12.5% horse serum. After washing, cells were incubated with various concentrations of peptides (control, cov1, cov2) for 2 h, and then NK cytotoxicity was evaluated by a calcein-AM release assay [30]. Briefly, target cells were labeled with calcein-AM (Invitrogen, Carlsbad, CA, USA) for 1 h. Then, calcein-labeled target cells (1 × 104 cells per well) and serially diluted NK cells were co-cultured in 96-well round-bottom plates for 4 h. “Maximum release” was simulated by adding 2% Triton X-100 to the target cells, and “spontaneous release” was simulated by adding culture medium to the target cells. The calcein released into the supernatant was measured using a multi-mode microplate reader (Molecular Devices, San Jose, CA, USA). The percent specific lysis was calculated according to the formula ((test release-spontaneous release)/(maximum release-spontaneous release)) × 100.
4.6. IFN-γ assay
NK cells, preincubated in 0.1% BSA for 12 h, were treated with peptides (control, cov1, cov2) for 2 h. Then, peptide-treated NK cells were co-cultured with H460 lung cancer cell for 24 h. The culture supernatants were collected and analyzed for IFN-γ by using a human IFN-γ ELISA kit (Invitrogen).
4.7. Vav1 phosphorylation
NK cells, preincubated in 0.1% BSA for 12 h, were treated with peptides (control, cov1, cov2) for the indicated times. Then, cells were washed and lysed in RIPA II lysis buffer (genDEPOT) containing protease and phosphatase inhibitors. Ten micrograms of sample was separated by SDS-PAGE, and transferred to a membrane. The membrane was immunoblotted with Vav1 and p-Vav1 antibodies.
4.8. RNA sequencing
For control and test RNAs, library construction was performed using QuantSeq 3′mRNA-Seq Library Prep Kit (Lexogen, Inc., Austria) according to the manufacturer’s instructions. In brief, 500 ng of total RNA was prepared and an oligo-dT primer containing an Illumina-compatible sequence at its 5′end was hybridized to the RNA, and reverse transcription was performed. After degradation of the RNA template, second strand synthesis was initiated by a random primer containing an Illumina-compatible linker sequence at its 5′end. The double-stranded library was purified using magnetic beads to remove all reaction components. The library was amplified to add the complete adapter sequences required for cluster generation. The finished library was purified from PCR components. High-throughput sequencing was performed as single-end 75 sequencing using NextSeq 500 (Illumina, Inc., USA).
4.9. Data analysis
QuantSeq 3′mRNA-Seq reads were aligned using Bowtie2. Differentially expressed genes were determined based on counts from unique and multiple alignments using coverage in Bedtools. The read count data were processed based on the quantile normalization method using EdgeR within R development Core Team using Bioconductor. Gene classification was based on searches done by Quick GO (https://www.ebi.ac.uk/QuickGO/) and Medline databases (http://www.ncbi.nlm.nih.gov/).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by grants from the National Research Foundation of Korea (NRF) (2019R1A2C3002034, 2019R1A2C1007906, and 2020R1A2C2012467) and the KRIBB Research Initiative Program (KGM1211231), Republic of Korea.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cellimm.2021.104454.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Supplementary figure 2.
References
- 1.Wang F., Nie J., Wang H., Zhao Q., Xiong Y., Deng L., Song S., Ma Z., Mo P., Zhang Y. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J. Infect. Dis. 2020;221:1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song J.W., Zhang C., Fan X., Meng F.P., Xu Z., Xia P., Cao W.J., Yang T., Dai X.P., Wang S.Y., Xu R.N., Jiang T.J., Li W.G., Zhang D.W., Zhao P., Shi M., Agrati C., Ippolito G., Maeurer M., Zumla A., Wang F.S., Zhang J.Y. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020;11:3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., Koufargyris P., Karageorgos A., Katrini K., Lekakis V., Lupse M., Kotsaki A., Renieris G., Theodoulou D., Panou V., Koukaki E., Koulouris N., Gogos C., Koutsoukou A. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1003. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Market M., Angka L., Martel A.B., Bastin D., Olanubi O., Tennakoon G., Boucher D.M., Ng J., Ardolino M., Auer R.C. Flattening the COVID-19 curve with natural killer cell based immunotherapies. Front. Immunol. 2020;11:1512. doi: 10.3389/fimmu.2020.01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yaqinuddin A., Kashir J. Innate immunity in COVID-19 patients mediated by NKG2A receptors, and potential treatment using Monalizumab, Cholroquine, and antiviral agents. Med. Hypotheses. 2020;140 doi: 10.1016/j.mehy.2020.109777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Mariuzza R.A. Structural basis for recognition of cellular and viral ligands by NK cell receptors. Front. Immunol. 2014;5:123. doi: 10.3389/fimmu.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radaev S., Rostro B., Brooks A.G., Colonna M., Sun P.D. Conformational plasticity revealed by the cocrystal structure of NKG2D and its class I MHC-like ligand ULBP3. Immunity. 2001;15:1039–1049. doi: 10.1016/s1074-7613(01)00241-2. [DOI] [PubMed] [Google Scholar]
- 8.Lanier L.L. NKG2D receptor and its ligands in host defense. Cancer Immunol. Res. 2015;3:575–582. doi: 10.1158/2326-6066.CIR-15-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Upshaw J.L., Arneson L.N., Schoon R.A., Dick C.J., Billadeau D.D., Leibson P.J. NKG2D-mediated signaling requires a DAP10-bound Grb2-Vav1 intermediate and phosphatidylinositol-3-kinase in human natural killer cells. Nat. Immunol. 2006;7:524–532. doi: 10.1038/ni1325. [DOI] [PubMed] [Google Scholar]
- 10.Chávez-Blanco A., Chacón-Salinas R., Dominguez-Gomez G., Gonzalez-Fierro A., Perez-Cardenas E., Taja-Chayeb L., Trejo-Becerril C., Duenas-Gonzalez A. Viral inhibitors of NKG2D ligands for tumor surveillance. Expert Opin. Ther. Targets. 2016;20:1375–1387. doi: 10.1080/14728222.2016.1202928. [DOI] [PubMed] [Google Scholar]
- 11.Schafer J.L., Ries M., Guha N., Connole M., Colantonio A.D., Wiertz E.J., Wilson N.A., Kaur A., Evans D.T., Silvestri G. Suppression of a natural killer cell response by simian immunodeficiency virus peptides. PLoS Pathog. 2015;11:e1005145. doi: 10.1371/journal.ppat.1005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mancini M., Vidal S.M. Mechanisms of natural killer cell evasion through viral adaptation. Annu. Rev. Immunol. 2020;38:511–539. doi: 10.1146/annurev-immunol-082619-124440. [DOI] [PubMed] [Google Scholar]
- 13.Mandelboim O., Lieberman N., Lev M., Paul L., Arnon T.I., Bushkin Y., Davis D.M., Strominger J.L., Yewdell J.W., Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 14.Lazear E., Peterson L.W., Nelson C.A., Fremont D.H. Crystal structure of the cowpox virus-encoded NKG2D ligand OMCP. J. Virol. 2013;87:840–850. doi: 10.1128/JVI.01948-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouayad A. Innate immune evasion by SARS-CoV-2: comparison with SARS-CoV. Rev. Med. Virol. 2020 doi: 10.1002/rmv.2135. [DOI] [PubMed] [Google Scholar]
- 16.Trabuco L.G., Lise S., Petsalaki E., Russell R.B. PepSite: prediction of peptide-binding sites from protein surfaces. Nucleic Acids Res. 2012;40:W423–W427. doi: 10.1093/nar/gks398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jewett A. The potential effect of novel coronavirus SARS-CoV-2 on NK cells; a perspective on potential therapeutic interventions. Front Immunol. 2020;11:1692. doi: 10.3389/fimmu.2020.01692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.L.G. Meza Guzman, N. Keating, S.E. Nicholson, Natural Killer Cells: Tumor Surveillance and Signaling, Cancers (Basel), 12 (2020). [DOI] [PMC free article] [PubMed]
- 19.Kwon H.-J., Kim N., Kim H.S. Molecular checkpoints controlling natural killer cell activation and their modulation for cancer immunotherapy. Exp. Mol. Med. 2017;49:e311. doi: 10.1038/emm.2017.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 21.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P., Morris D.L., Willcox B.E., Steinle A., Spies T., Strong R.K. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat. Immunol. 2001;2:443–451. doi: 10.1038/87757. [DOI] [PubMed] [Google Scholar]
- 23.Ida H., Nakashima T., Kedersha N., Yamasaki S., Huang M., Izumi Y., Miyashita T., Origuchi T., Kawakami A., Migita K., Bird P., Anderson P., Eguchi K. Granzyme B leakage-induced cell death: a new type of activation-induced natural killer cell death. Eur. J. Immunol. 2003;33:3284–3292. doi: 10.1002/eji.200324376. [DOI] [PubMed] [Google Scholar]
- 24.Bortolotti D., Gentili V., Rizzo S., Rotola A., Rizzo R. SARS-CoV-2 spike 1 protein controls natural killer cell activation via the HLA-E/NKG2A pathway. Cells. 2020;9 doi: 10.3390/cells9091975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M., Guo W., Dong Y., Wang X., Dai D., Liu X., Wu Y., Li M., Zhang W., Zhou H., Zhang Z., Lin L., Kang Z., Yu T., Tian C., Qin R., Gui Y., Jiang F., Fan H., Heissmeyer V., Sarapultsev A., Wang L., Luo S., Hu D. Elevated exhaustion levels of NK and CD8(+) T cells as indicators for progression and prognosis of COVID-19 disease. Front. Immunol. 2020;11:580237. doi: 10.3389/fimmu.2020.580237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thurmann L., Kurth F., Volker M.T., Kazmierski J., Timmermann B., Twardziok S., Schneider S., Machleidt F., Muller-Redetzky H., Maier M., Krannich A., Schmidt S., Balzer F., Liebig J., Loske J., Suttorp N., Eils J., Ishaque N., Liebert U.G., von Kalle C., Hocke A., Witzenrath M., Goffinet C., Drosten C., Laudi S., Lehmann I., Conrad C., Sander L.E., Eils R. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 27.Maucourant C., Filipovic I., Ponzetta A., Aleman S., Cornillet M., Hertwig L., Strunz B., Lentini A., Reinius B., Brownlie D., Cuapio A., Ask E.H., Hull R.M., Haroun-Izquierdo A., Schaffer M., Klingstrom J., Folkesson E., Buggert M., Sandberg J.K., Eriksson L.I., Rooyackers O., Ljunggren H.G., Malmberg K.J., Michaelsson J., Marquardt N., Hammer Q., Stralin K., Bjorkstrom N.K., Karolinska C.-S.-G. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petsalaki E., Stark A., García-Urdiales E., Russell R.B., Rost B. Accurate prediction of peptide binding sites on protein surfaces. PLoS Comput. Biol. 2009;5:e1000335. doi: 10.1371/journal.pcbi.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi I., Yoon S.R., Park S.Y., Kim H., Jung S.J., Jang Y.J., Kang M., Yeom Y.I., Lee J.L., Kim D.Y., Lee Y.S., Kang Y.A., Jeon M., Seol M., Lee J.H., Lee J.H., Kim H.J., Yun S.C., Lee K.H. Donor-derived natural killer cells infused after human leukocyte antigen-haploidentical hematopoietic cell transplantation: a dose-escalation study. Biol. Blood Marrow Transplant. 2014;20:696–704. doi: 10.1016/j.bbmt.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 30.Somanchi S.S., Senyukov V.V., Denman C.J., Lee D.A. Expansion, purification, and functional assessment of human peripheral blood NK cells. J. Vis. Exp. 2011 doi: 10.3791/2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.