Abstract
Wolbachia are maternally transmitted intracellular bacteria that can naturally and artificially infect arthropods and nematodes. Recently, they were applied to control the spread of mosquito-borne pathogens by causing cytoplasmic incompatibility (CI) between germ cells of females and males. The ability of Wolbachia to induce CI is based on the prevalence and polymorphism of Wolbachia in natural populations of mosquitoes. In this study, we screened the natural infection level and diversity of Wolbachia in field-collected mosquitoes from 25 provinces of China based on partial sequence of Wolbachia surface protein (wsp) gene and multilocus sequence typing (MLST). Among the samples, 2489 mosquitoes were captured from 24 provinces between July and September, 2014 and the remaining 1025 mosquitoes were collected month-by-month in Yangzhou, Jiangsu province between September 2013 and August 2014. Our results showed that the presence of Wolbachia was observed in mosquitoes of Aedes albopictus (97.1%, 331/341), Armigeres subalbatus (95.8%, 481/502), Culex pipiens (87.0%, 1525/1752), Cx. tritaeniorhynchus (17.1%, 14/82), but not Anopheles sinensis (n = 88). Phylogenetic analysis indicated that high polymorphism of wsp and MLST loci was observed in Ae. albopictus mosquitoes, while no or low polymorphisms were in Ar. subalbatus and Cx. pipiens mosquitoes. A total of 12 unique mutations of deduced amino acid were identified in the wsp sequences obtained in this study, including four mutations in Wolbachia supergroup A and eight mutations in supergroup B. This study revealed the prevalence and polymorphism of Wolbachia in mosquitoes in large-scale regions of China and will provide some useful information when performing Wolbachia-based mosquito biocontrol strategies in China.
Author summary
The mosquitoes Aedes albopictus, Anopheles sinensis, Armigeres subalbatus, Culex pipiens and Cx. tritaeniorhynchus are native to China and the major vectors in the transmission of arboviruses, protozoans and nematodes. Recently, an innovative biocontrol strategy has been developed and evaluated based on the ability of Wolbachia to induce cytoplasmic incompatibility (CI), as well as interfere with the infection and replication of pathogens. Since the ability to induce CI largely depends on the density and diversity of Wolbachia, we investigated and characterized the natural infection of Wolbachia in above-mentioned five species of field-collected mosquitoes in 25 provinces of China. The results showed that the positive rates of Wolbachia infection were high in mosquitoes of Ae. albopictus, Ar. subalbatus and Cx. pipiens in large-scale regions of China and low in Cx. tritaeniorhynchus in Guizhou province. Phylogenetic analysis based on Wolbachia surface protein (wsp) gene and five multilocus sequence typing (MLST) loci indicated the high polymorphism of Wolbachia in Ae. albopictus, and low polymorphisms in Ar. subalbatus and Cx. pipiens. This finding contributes to the understanding of the nationwide distribution of Wolbachia and the potential application of this biocontrol strategy in China.
Introduction
Wolbachia is a genus of Gram-negative intracellular bacteria, belonging to the α-subphylum Proteobacteria, which is the most widely distributed symbiotic bacteria in the world and commonly found in arthropods and some nematodes. It was first reported by Hertig and Wolbach in reproductive tissues of Culex pipiens in 1924 [1]. In mosquitoes and many other arthropods, Wolbachia infection causes sperm-egg incompatibility called cytoplasmic incompatibility (CI) that leads to a reduction in egg-hatch frequencies when a Wolbachia-infected male mates with uninfected females [2]. The consequence of CI provides a reproductive advantage to infected females and contributes to the rapid spread of Wolbachia among the host population [3].
Mosquitoes are the most important vector of numerous arthropod-borne viruses (arboviruses), for example, Zika virus, dengue fever virus, West Nile virus and Japanese encephalitis virus. In order to slow the spread of mosquito-borne diseases, insecticides have been used extensively, but with limited success. At the same time, with the abuse of insecticides, mosquitoes are becoming more and more resistant. In the recent two decades, Wolbachia infection was demonstrated to reduce the potential transmission of mosquito-borne diseases by shortening adult lifespan, affecting mosquito reproduction and interfering with pathogen replication [4–6]. In 2013, Bian et al. established a stable infection of Wolbachia strain wAlbB in Anopheles stephensi, which is one of the most important vectors of malaria. The infection of wAlbB conferred resistance in the mosquitoes to the human malaria parasite Plasmodium falciparum [7].
The major lineages of Wolbachia have different host specificity and type of symbiosis, and are currently clustered into 20 supergroups (A–T) according to the order of their description [8,9], while only supergroups A and B have been found in mosquitoes. Initially, 16S rRNA-based PCR and sequencing were used for the detection and molecular characterization of Wolbachia [10], followed by the employment of the more variable Wolbachia surface protein (wsp) gene [11]. Subsequently, a reproducible and portable method, multilocus sequence typing (MLST), was established targeting five alleles (gatB, coxA, hcpA, ftsZ and fbpA) [12]. Although with limitations, it was gradually used for strain differentiation in the community of Wolbachia researchers. Bleidorn et al. recommended that whole-genome typing methods should be applied to the characterization of Wolbachia [8]. Several studies have instigated the prevalence and/or molecular characterization of Wolbachia in certain species of mosquitoes in China, particularly Aedes albopictus [13,14]. However, up to date, more than 45 species of mosquitoes have been identified in 31 provinces, distributed in the east, south, north, central, northeast, southwest and northwest regions of China. Among them, Ae. albopictus, Armigeres subalbatus, Cx. pipiens, Cx. quinquefasciatus and Cx. tritaeniorhynchus were most widely distributed [15]. Our knowledge of the prevalence and diversity of Wolbachia in mosquitoes of different species and geographic regions remains limited, especially in China. Therefore, in this study, we screened the prevalence and diversity of Wolbachia in approximately 3500 field-collected mosquitoes of Ae. albopictus, An. sinensis, Ar. subalbatus, Cx. pipiens and Cx. tritaeniorhynchus from 25 provinces of China with wsp-based real-time PCR and MLST analyses.
Materials and methods
Ethics statement
All experimental protocols in this study were reviewed and approved by the Institutional Animal Care and Use Committee of Yangzhou University.
Mosquito collection
Convenience whole-body mosquito samples (n = 3514) were trapped in summer (July and September, 2014) in 24 provinces of China, and in the whole year (September 2013 to August 2014) in Yangzhou, Jiangsu province with hand nets, for an epidemiological survey of Rickettsia (Fig 1) [16,17]. Once captured, the mosquitoes were immediately protected separately in sterile tubes containing 400 μL of DNA/RNA stabilization reagent (Roche, Basel, Switzerland). To prevent cross contamination, disposable gloves were changed before the collection of each mosquito. Then the samples were transported at room temperature to the laboratory for species and gender identification and DNA extraction.
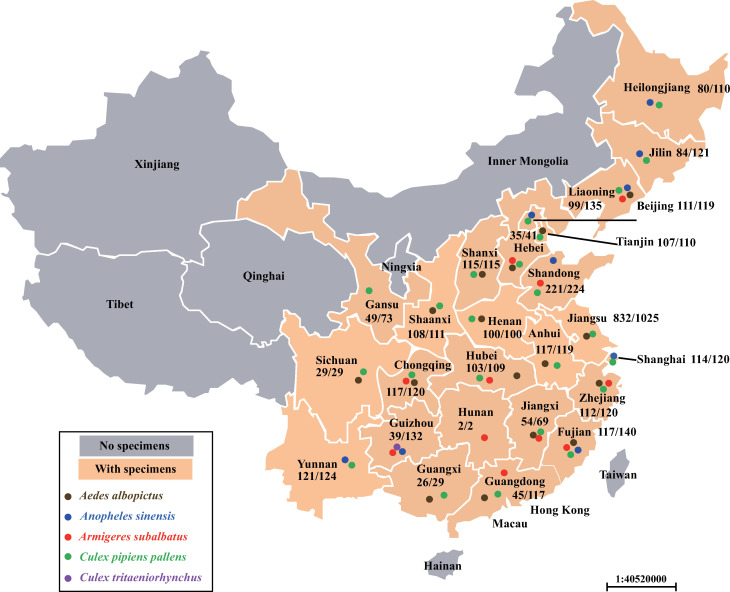
Fig 1. The distributions of field-collected mosquitoes in this study.
A total of 3514 samples of mosquitoes’ whole bodies (convenience samples) were collected in 25 provinces of China, including Aedes albopictus (n = 349), Anopheles sinensis (n = 88), Armigeres subalbatus (n = 502), Culex pipiens (n = 2495) and Cx. tritaeniorhynchus (n = 80). Different colored dots represented different species of mosquitoes, including Ae. albopictus (brown), An. sinensis (blue), Ar. subalbatus (red), Cx. pipiens (green) and Cx. tritaeniorhynchus (purple). The numbers in the region of each province indicated the positive/ total numbers of mosquitoes.
Mosquito species and gender identification
The species and genders of most mosquitoes employed in this study have been identified in our previous study [16]. The remaining samples were identified with standard morphological criteria (https://www.wrbu.si.edu/) with an ordinary microscope (Olympus, Tokyo, Japan). Each mosquito was removed from DNA/RNA stabilization reagent and placed on to a glass slide for morphological identification. In addition, the mosquitoes of each species from each province were subjected to further identification using a genomic approach targeting mitochondrial cytochrome c oxidase subunit I (COI) gene [18] (Table 1). After species and gender identification, the mosquitoes were stored at -80°C until genomic DNA extraction.
Table 1. Primer pairs used in this study.
| Gene | Primer Sequence (5’- 3’) | Annealing Temperature | Amplicon Length (bp) | Reference |
|---|---|---|---|---|
| COI | GGTCAACAAATCATAAAGATATTGG | 51°C | ~650 | [16] |
| TAAACTTCAGGGTGACCAAAAAATCA | ||||
| wsp | TGGTCCAATAAGTGATGAAGAAAC | 53°C | ~610 | [11] |
| AAAAATTAAACGCTACTCCA | ||||
| 16S rRNA | CATACCTATTCGAAGGGATAG | 60°C | ~438 | [19] |
| AGCTTCGAGTGAAACCAATTC | ||||
| gatB | GAKTTAAAYCGYGCAGGBGTT | 54°C | ~471 | [12] |
| TGGYAAYTCRGGYAAAGATGA | ||||
| coxA | TTGGRGCRATYAACTTTATAG | 54°C | ~487 | [12] |
| CTAAAGACTTTKACRCCAGT | ||||
| hcpA | GAAATARCAGTTGCTGCAAA | 54°C | ~515 | [12] |
| GAAAGTYRAGCAAGYTCTG | ||||
| ftsZ | ATYATGGARCATATAAARGATAG | 54°C | ~524 | [12] |
| TCRAGYAATGGATTRGATAT | ||||
| fbpA | GCTGCTCCRCTTGGYWTGAT | 59°C | ~509 | [12] |
| CCRCCAGARAAAAYYACTATTC |
Genomic DNA extraction and molecular detection of Wolbachia
After thawing at room temperature for approximate 15 min, the samples were homogenized individually with Precellys 24 homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) at 5800 x rpm (two short durations of 15 s with a 15-s break in between) and subjected to DNA extraction with High-Pure PCR Template Preparation Kit (Roche, Basel, Switzerland) following the manufacturer’s instructions.
The presence of Wolbachia was screened using SYBR Green real-time PCR targeting a unique and highly conserved fragment of outer surface protein (wsp) gene [11], and the amplicon contained all the four hypervariable regions (HVR1-4) of wsp. In addition, the negative samples for wsp gene were further confirmed with the amplification of 16S rRNA gene [19] (Table 1). The real-time PCR were performed in a Roche LightCycler 480 II real-time PCR platform (Roche, Basel, Switzerland) with 20-μL volumes composed of 10 μL of 2 x ChamQ Universal SYBR qPCR Master (Vazyme, Nanjing, China), 0.4 μL of forward primer, 0.4 μL of reverse primer, 2 μL of DNA template and 7.2 μL of double-distilled water (ddH2O) [20]. Thermal cycling consisted of a 2-min denaturation step at 94°C followed by 37 cycles of 94°C for 30 s, 53°C (wsp) or 60°C (16S rRNA) for 30 s and 72°C for 1 min, final elongation at 72°C for 10 min and final holding at 37°C. The PCR products of wsp (~610 bp) with different melting temperatures were gel purified using a QIAquick Gel Extraction Kit (Qiagen, Valencia, USA) and sequenced by a commercial sequencing laboratory (Tsingke Biotechnology, Tianjin, China) with sanger dideoxy sequencing. Using the gel-purified PCR products as quantitative standards, the concentrations of DNAs were determined with the Quanti-iT PicoGreen dsDNA Assay Kit (Invitrogen Corporation, Carlsbad, USA). The 10-fold dilutions were performed to give the reaction system containing 100,000, 10,000, 1,000, 100 and 10 copies of the wsp gene of Wolbachia supergroup A or B, respectively. The DNA samples of Rickettsia bellii and R. felis were stored in our laboratory and served as negative controls.
Phylogenetic analysis with wsp and MLST
In order to investigate the molecular characterization of the mosquitoes captured from 25 provinces, one to four positive samples of each mosquito species in each province were selected to Wolbachia polymorphism identification with wsp gene sequencing and MLST targeting five alleles (gatB, coxA, hcpA, ftsZ and fbpA). A total of 109 Wolbachia-positive PCR products amplified from female or male mosquitoes of different species and different geographic regions were gel purified with the QIAquick Gel Extraction Kit (QIAGEN, Dusseldorf, Germany) and sequenced and verified by Sanger dideoxy sequencing (Sangon Biotech, Shanghai, China).
The five MLST loci were amplified with specific primers published in a previous study [12] (Table 1). PCR reaction were performed in a final volume of 25 μL containing 1 μL of DNA, 12.5 μL of 2 x Phanta Max Master Mix (Vazyme, Nanjing, China), 1 μL of forward primer, 1 μL of reverse primer and 9.5 μL of ddH2O. Thermal cycling consisted of a 2-min denaturation step at 94°C, followed by 37 cycles of 94°C for 30 s, optimal annealing temperature for 45 s (54°C for gatB, coxA, hcpA, ftsZ and 59°C for fbpA) and 72°C for 1.5 min, final elongation at 72°C for 10 min and final holding at 4°C. The PCR products (gatB: ~471 bp, coxA: ~487 bp, hcpA: ~515 bp, ftsZ: ~524 bp and fbpA: ~509 bp) were bi-directional sequenced by a commercial sequencing laboratory (Sangon Biotech, Shanghai, China) with sanger dideoxy method.
The sequences of partial wsp gene and MLST loci of Wolbachia obtained in this study were assembled with DNASTAR Lasergen 15.2 (DNASTAR Inc., Madison, WI) and aligned using CLUSTAL W in MEGA 7.0 (MEGA, Pennsylvania State University, University Park) along with those of Wolbachia strains represented different supergroups obtained from previous studies or GenBank [11,13]. A neighbor-joining phylogenetic tree was constructed using the Tamura-Nei model and the robustness of clusters was assessed by bootstrapping 1000 replicates. Maximum-likelihood phylogenetic analysis was performed to confirm the results [21]. The sequences of five MLST loci of each strain were submitted to a public database (PubMLST) to obtain allelic profiles and sequence type (ST).
Statistical analysis
Statistical analyses were performed with the Statistica 7.0 software package (StatSoft Inc., USA). Positive rates of Wolbachia in different sampling time or locations were compared with the Chi-squared or Fisher’s exact test. P-value<0.05 indicated a significant difference.
Results
Mosquito species and gender identified
The identification of species and genders were performed on most mosquitoes (77.0%, 2706/3514) employed in this study, revealing the presence of five species of mosquitoes, including Ae. albopictus (n = 341, female = 258, male = 83), An. sinensis (n = 88, female = 62, male = 26), Ar. subalbatus (n = 502, female = 236, male = 266), Cx. pipiens (n = 1752, female = 992, male = 760) and Cx. tritaeniorhynchus (n = 82, female = 72, male = 10) (Table 2) with the accession numbers of OK465203~OK465358. Species identification was not performed on the samples collected in Jiangsu province from September 2013 to March 2014, because the limited number of researchers at that time.
Table 2. The presence and positive rates of Wolbachia in mosquitoes collected from 24 provinces of China.
| Mosquito species | Province | Coordinate | Positive of Wolbachia | ||
|---|---|---|---|---|---|
| Female | Male | Total | |||
| Ae. albopictus | Anhui | 31.5°N, 117.2°E | 100% (2/2) | NAa | 100% (2/2) |
| Chongqing | 29.3°N, 106.3°E | 100% (10/10) | NA | 100% (10/10) | |
| Fujian | 26.1°N, 119.2°E | 100% (1/1) | NA | 100% (1/1) | |
| Guangdong | 23.1°N, 113.1°E | 80.0% (8/10) | 100% (3/3) | 84.6% (11/13) | |
| Guangxi | 22.5°N, 108.2°E | 100% (17/17) | NA | 100% (17/17) | |
| Hebei | 38.0°N, 114.3°E | 100% (30/30) | 100% (1/1) | 100% (31/31) | |
| Henan | 34.5°N, 113.4°E | 100% (15/15) | NA | 100% (15/15) | |
| Hubei | 30.4°N, 114.2°E | 100% (10/10) | 100% (3/3) | 100% (13/13) | |
| Jiangsu | 32.0°N, 118.5°E | 92.6% (25/27) | 95.0% (19/20) | 93.6% (44/47) | |
| Jiangxi | 28.4°N, 115.6°E | 94.7% (18/19) | NA | 94.7% (18/19) | |
| Liaoning | 41.5°N, 123.3°E | 40.0% (2/5) | 100% (3/3) | 62.5% (5/8) | |
| Shaanxi | 34.2°N, 108.6°E | 100% (3/3) | NA | 100% (3/3) | |
| Shandong | 36.4°N, 117.0°E | 100% (55/55) | 100% (42/42) | 100% (97/97) | |
| Shanxi | 37.5°N, 112.3°E | 100% (1/1) | NA | 100% (1/1) | |
| Sichuan | 30.4°N, 104.0°E | 100% (25/25) | NA | 100% (25/25) | |
| Tianjing | 39.0°N, 117.1°E | 95.0% (19/20) | 100% (7/7) | 96.3% (26/27) | |
| Zhejiang | 30.2°N, 120.1°E | 100% (8/8) | 100% (4/4) | 100% (12/12) | |
| Total | 96.5% (249/258) | 98.8% (82/83) | 97.1% (331/341) | ||
| An. sinensis | Beijing | 39.6°N, 116.2°E | 0% (0/2) | NA | 0% (0/2) |
| Fujian | 26.1°N, 119.2°E | 0% (0/14) | 0% (0/8) | 0% (0/22) | |
| Guizhou | 26.4°N, 106.4°E | 0% (0/15) | 0% (0/10) | 0% (0/25) | |
| Heilongjiang | 45.4°N, 126.4°E | 0% (0/3) | NA | 0% (0/3) | |
| Jilin | 43.5°N, 125.2°E | 0% (0/5) | NA | 0% (0/5) | |
| Liaoning | 41.5°N, 123.3°E | 0% (0/15) | 0% (0/6) | 0% (0/21) | |
| Shandong | 36.4°N, 117.0°E | 0% (0/2) | NA | 0% (0/2) | |
| Shanghai | 31.1°N, 121.3°E | 0% (0/3) | NA | 0% (0/3) | |
| Yunnan | 25.0°N, 102.4°E | 0% (0/3) | NA | 0% (0/3) | |
| Zhejiang | 30.2°N, 120.1°E | NA | 0% (0/2) | 0% (0/2) | |
| Total | 0% (0/62) | 0% (0/26) | 0% (0/88) | ||
| Ar. subalbatus | Chongqing | 29.4°N, 106.3°E | 97.6% (40/41) | 79.0% (64/66) | 97.2% (104/107) |
| Fujian | 26.1°N, 119.2°E | 100% (77/77) | 100% (38/38) | 100% (115/115) | |
| Guangdong | 23.1°N, 113.1°E | 100% (5/5) | 100% (28/28) | 100% (33/33) | |
| Guizhou | 26.4°N, 106.4°E | 100% (16/16) | 100% (9/9) | 100% (25/25) | |
| Hebei | 38.0°N, 114.3°E | 100% (2/2) | NA | 100% (2/2) | |
| Hubei | 30.4°N, 114.2°E | 97.1% (33/34) | 95.0% (19/20) | 96.3% (52/54) | |
| Hunan | 28.1°N, 112.6°E | 100% (2/2) | NA | 100% (2/2) | |
| Jiangxi | 28.4°N, 115.6°E | NA | 100% (1/1) | 100% (1/1) | |
| Liaoning | 41.5°N, 123.3°E | 63.6% (21/33) | 100% (35/35) | 82.4% (56/68) | |
| Shandong | 36.4°N, 117.0°E | 100% (4/4) | 100% (1/1) | 100% (5/5) | |
| Zhejiang | 30.2°N, 120.1°E | 90.9% (20/22) | 97.1% (66/68) | 95.6% (86/90) | |
| Total | 93.2% (220/236) | 98.1% (261/266) | 95.8% (481/502) | ||
| Cx. pipiens | Anhui | 31.5°N, 117.2°E | 98.1% (53/54) | 98.4% (62/63) | 98.3% (115/117) |
| Beijing | 39.6°N, 116.2°E | 95.2% (60/63) | 94.4% (51/54) | 94.9% (111/117) | |
| Chongqing | 29.4°N, 106.3°E | NA | 100% (3/3) | 100% (3/3) | |
| Fujian | 26.1°N, 119.2°E | 0% (0/1) | 100% (1/1) | 50.0% (1/2) | |
| Gansu | 36.0°N, 103.5°E | 59.3% (35/59) | 100% (14/14) | 67.1% (49/73) | |
| Guangdong | 23.1°N, 113.1°E | 0% (0/68) | 33.3% (1/3) | 1.4% (1/71) | |
| Guangxi | 22.5°N, 108.2°E | 70.0% (7/10) | 100% (2/2) | 75.0% (9/12) | |
| Hebei | 38.0°N, 114.3°E | 16.7% (1/6) | 50.0% (1/2) | 25.0% (2/8) | |
| Heilongjiang | 45.4°N, 126.4°E | 42.2% (19/45) | 98.4% (61/62) | 74.8% (80/107) | |
| Henan | 34.5°N, 113.4°E | 100% (35/35) | 100% (50/50) | 100% (85/85) | |
| Hubei | 30.4°N, 114.2°E | 81.0% (17/21) | 100% (21/21) | 90.5% (38/42) | |
| Jiangsu | 32.0°N, 118.5°E | 92.6% (113/122) | 82.4 (89/107) | 88.2% (202/229) | |
| Jiangxi | 28.4°N, 115.6°E | 50.0% (10/20) | 86.2% (25/29) | 71.4% (35/49) | |
| Jilin | 43.5°N, 125.2°E | 72.2% (83/115) | 100% (1/1) | 72.4% (84/116) | |
| Liaoning | 41.5°N, 123.3°E | 100% (7/7) | 100% (31/31) | 100% (38/38) | |
| Shaanxi | 34.2°N, 108.6°E | 98.0% (97/99) | 88.9% (8/9) | 97.2% (105/108) | |
| Shandong | 36.4°N, 117.0°E | 98.2% (54/55) | 100% (65/65) | 99.2% (119/120) | |
| Shanghai | 31.1°N, 121.3°E | 96.3% (77/80) | 100% (37/37) | 97.4% (114/117) | |
| Shanxi | 37.5°N, 112.3°E | 100% (17/17) | 100% (97/97) | 100% (114/114) | |
| Sichuan | 30.4°N, 104.0°E | 100% (4/4) | NA | 100% (4/4) | |
| Tianjing | 39.0°N, 117.1°E | 98.7% (76/77) | 83.3% (5/6) | 97.6% (81/83) | |
| Yunnan | 25.0°N, 102.4°E | 100% (27/27) | 100% (94/94) | 100% (121/121) | |
| Zhejiang | 30.2°N, 120.1°E | 85.7% (6/7) | 88.9% (8/9) | 87.5% (14/16) | |
| Total | 80.4% (798/992) | 95.7% (727/760) | 87.0% (1525/1752) | ||
| Cx. tritaeniorhynchus | Guizhou | 26.4°N, 106.4°E | 5.6% (4/72) | 100% (10/10) | 17.1% (14/82) |
| Unidentified | Jiangsu | 32.0°N, 118.5°E | 78.2% (586/749) | ||
| Total | 83.6% (2937/3514) | ||||
a NA: not available
Presence of Wolbachia in mosquitoes from 25 provinces of China
All the mosquitoes captured in 25 provinces were screened for the presence of Wolbachia with wsp-based PCR. The detection limit of this PCR was 10 copies of the wsp gene per reaction for both Wolbachia supergroups A and B, and nonspecific amplifications were not observed (S1 Fig). Overall, 83.6% of the mosquitoes (2937/3514) were positive for Wolbachia, including 97.1% (331/341) of Ae. albopictus, 95.8% (481/502) of Ar. subalbatus, 87.0% (1525/1752) of Cx. pipiens, 17.1% (14/82) of Cx. tritaeniorhynchus and 78.2% (586/749) of unidentified mosquitoes, while both wsp and 16S rRNA genes of Wolbachia was unavailable to be detected in An. sinensis (n = 88). In detail, the positive rates of Wolbachia were 62.5~100% in Ae. albopictus, 82.4~100% in Ar. subalbatus, 1.4~100% in Cx. pipiens and 17.1% in Cx. tritaeniorhynchus sampled in different provinces of China (Table 2). The positive rate of Wolbachia in Cx. pipiens was significantly lower than that in Ae. albopictus and Ar. subalbatus (P<0.01), and significantly higher than that in Cx. tritaeniorhynchus (P<0.01).
Presence of Wolbachia in mosquitoes in Yangzhou, Jiangsu province
From September 2013 to August 2014, a total of 1025 mosquito samples were collected in a campus in Yangzhou, Jiangsu province. The monthly positive rates of Wolbachia ranged from 50.8% (66/130) to 98.4% (182/185). Compared with March 2014, the mosquito samples collected from September to December (2013), April (2014) and July (2014) harbored significant lower positive rates (P<0.01) of Wolbachia. Among the samples collected from April to August (2014), of which the species and gender information is available, significant higher infection rates were found with female samples of Cx. pipiens in April (P<0.05) (Table 3).
Table 3. The presence and positive rates of Wolbachia in mosquitoes collected in Yangzhou, Jiangsu province between September 2013 and August 2014.
| Sampling time | Daily mean Temperature (°C) | Species | Positive rate | ||
|---|---|---|---|---|---|
| Female | Male | Total | |||
| Sep 2013 | 20~27 | NA | NA | NA | 50.8% (66/130) |
| Oct 2013 | 13~23 | NA | NA | NA | 76.0% (225/296) |
| Nov 2013 | 7~16 | NA | NA | NA | 82.1% (78/95) |
| Dec 2013 | 0~9 | NA | NA | NA | 81.4% (35/43) |
| Mar 2014 | 6~15 | NA | NA | NA | 98.4% (182/185) |
| Apr 2014 | 11~20 | Cx. pipiens | 97.9% (47/48) | 79.5% (35/44) | 89.1% (82/92) |
| May 2014 | 17~27 | Cx. pipiens | 95.7% (22/23) | 79.5% (23/23) | 97.8% (45/46) |
| Jun 2014 | 20~28 | Cx. pipiens | 96.2% (25/26) | 95.0% (19/20) | 95.7% (44/46) |
| Jul 2014 | 23~30 | Ae. albopictus | 0% (0/1) | NA | 0% (0/1) |
| Cx. pipiens | 76.0% (19/25) | 60.0% (12/20) | 68.9% (31/45) | ||
| Total | 73.1% (19/26) | 60.0% (12/20) | 67.4% (31/46) | ||
| Aug 2014 | 22~28 | Ae. albopictus | 96.2% (25/26) | 95.0% (19/20) | 95.7% (44/46) |
| Total | 0~30 | 81.2% (832/1025) | |||
a NA: not available
Molecular characterization of Wolbachia with wsp and MLST
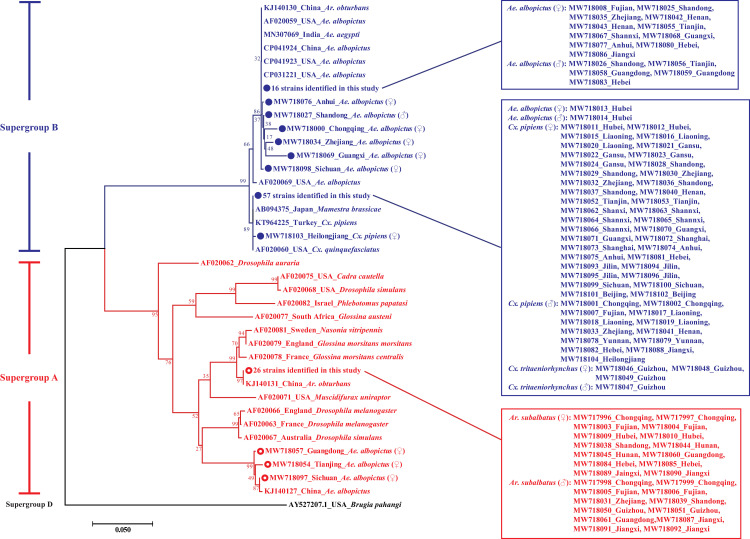
The sequences of partial Wolbachia wsp gene identified in this study were submitted to GenBank with the accession numbers of MW717996~MW718104. The neighbor-joining and maximum-likelihood phylogenetic trees both showed that 26.6% (29/109) and 73.4% (80/109) of the sequenced samples were clustered in supergroup A and B, respectively (Figs 2 and S2). The Wolbachia was available to be detected in four species of mosquitoes, including Ae. albopictus, Ar. subalbatus, Cx pipiens and Cx. tritaeniorhynchus. Only the positive samples of Ae. albopictus shared both supergroups, while the Wolbachia in Ar. subalbatus (supergroup A), Cx. pipiens (supergroup B) and Cx. tritaeniorhynchus (supergroup B) belonged to a single supergroup, respectively. In supergroup A, the partial sequences of wsp gene amplified from 26 Ar. subalbatus (14 females and 12 males) were identical to KJ140131 identified in Ae. albopictus in China. The remaining three strains (MW718054, MW718057 and MW718097) in supergroup A were closest to KJ140127 (Ae. albopictus, China). In supergroup B, the Wolbachia wsp sequences identified in this study were clustered into two sub-clades. The first sub-clade consisted the 81.5% (22/27) of Wolbachia sequences amplified from Ae. albopictus in this study. The second sub-clade consisted of all the Wolbachia wsp sequences obtained from Cx. pipiens (n = 52) and Cx. tritaeniorhynchus (n = 4), and two sequences obtained from Ae. albopictus (Fig 2).
Fig 2. Neighbor-joining phylogenetic tree based on Wolbachia wsp gene partial sequences (~610 bp) obtained in this study and GenBank database.
Strains identified in this study are identified with open circles (○) for supergroup A (in red) and filled circles (●) for supergroup B (in blue). The numbers at the branches show bootstrap support (1000 replicates). The bar at the bottom of the figure denotes distance.
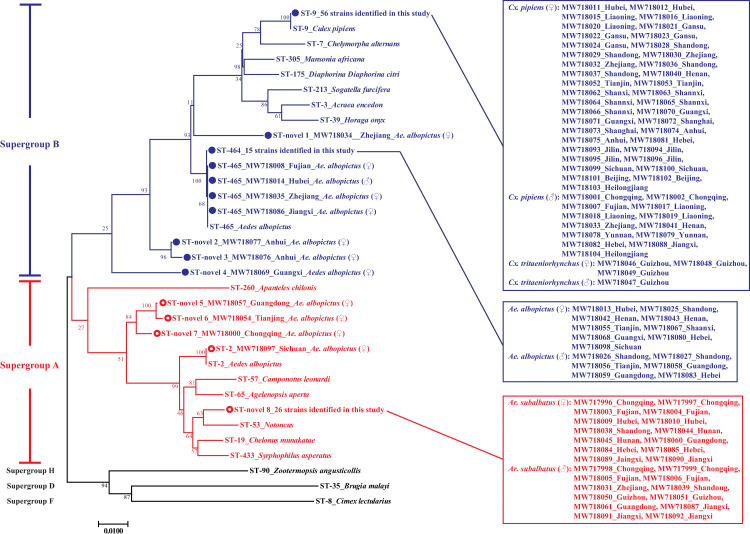
Submissions to the MLST database of Wolbachia are currently suspended until a new curator can be appointed (https://pubmlst.org/organisms/wolbachia-spp). So for the strains with novel sequence types, we are unavailable to receive the ST codes immediately, and “ST-novel #” was marked in front of these strains instead of the ST codes (Fig 3). There were 12 different sequence types of the Wolbachia strains in this study, including four archived sequence types, ST-2 (n = 1), ST-9 (n = 56), ST-464 (n = 15) and ST-465 (n = 4), and eight novel ones (n = 33). The performance of MLST-based phylogenetic tree showed a high consistency with that constructed with wsp. Based on MLST, the Wolbachia strains identified in this study belonged to supergroups A (n = 30) and B (n = 79). Interestingly, a Wolbachia strain identified in an Ae. albopictus mosquito from Chongqing was clustered into supergroup A and B in the phylogenetic trees of MLST and wsp, respectively (Figs 2 and 3). There were three main clusters in both wsp and MLST-based phylogenetic trees, and it worth noting that most strains in these clusters corresponded to each other in the two phylogenetic trees (Figs 2 and 3).
Fig 3. Neighbor-joining phylogenetic tree based on five MLST loci (gatB, coxA, hcpA, ftsZ and fbpA) of the strains obtained in this study and GenBank database.
Strains identified in this study are identified with open circles (○) for supergroup A (in red) and filled circles (●) for supergroup B (in blue). The GenBank accession number of wsp sequences of each strain identified in this study was marked on the right side of ST code. The numbers at the branches show bootstrap support (1000 replicates). The bar at the bottom of the figure denotes distance.
Alignment of nucleotide and deduced amino acid sequences of wsp gene
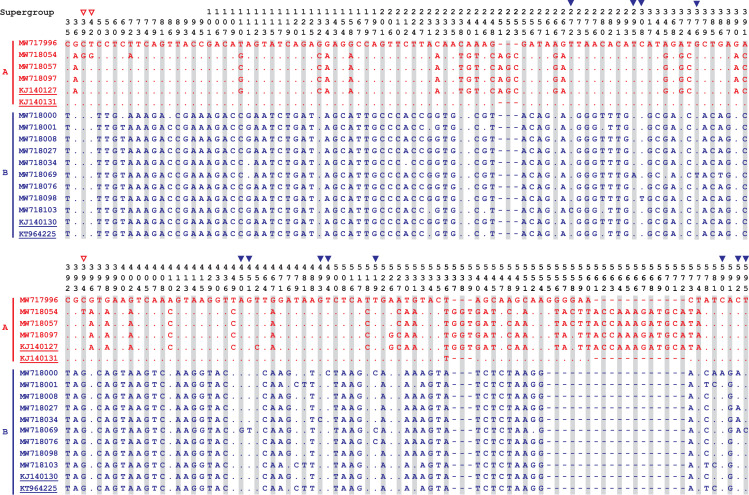
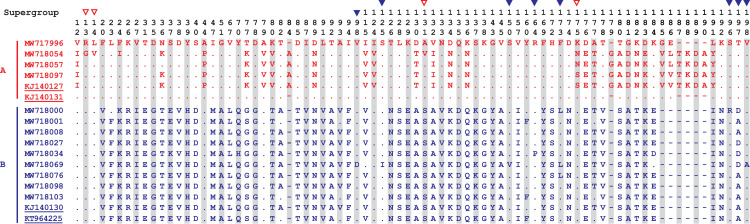
When compared with the closest reference sequences (KJ140127, KJ140131, KJ140130 and KT964225) deposited in GenBank, we found that the sequences obtained in this study had a total of 15 unique mutations in partial wsp gene. Among them, three mutations (C39G, T42G and C/G394T) were identified from supergroup A and the remaining 12 mutations (T272C, T295A, C308T, G376T, A450G, G451T, G493C, T494C, T519C, C590A, C592A and T595C) were included in supergroup B. Alignment of deduced amino acid sequences demonstrated that there were four (R13G, L14V, A/S131V and K175N/S) and eight (V98D, S125I, S150V, R164P, F173L, S196R, T197D/A and V198A) mutations in supergroups A and B, respectively (Figs 4 and 5).
Fig 4. Alignment of partial-length of Wolbachia wsp gene nucleotide (~610 bp) sequences identified in this study (MW717996, MW718054, MW718057, MW718097, MW718000, MW718001, MW718008, MW718027, MW718034, MW718069, MW718076, MW718098, MW718103) together with 4 mostly identical reference sequences (KJ140127, KJ140131, KJ140130 and KT964225) obtained in GenBank database.
Strains belonging to supergroup A and B are shown in red and blue, respectively, while the reference sequences are indicated with underlines. Numbers, read from top to bottom, above the sequences are nucleotide positions on the wsp gene of MW717996. Dots indicate nucleotides identical to the reference sequences. The triangles above nucleotide position numbers indicate sites of mutations in our isolates (supergroup A: ▽ in red, and supergroup B: ▼ in blue) relative to the one or both of the reference sequences.
Fig 5. Alignment of partial-length of Wolbachia wsp gene deduced amino acid (~200 AA) sequences identified in this study (MW717996, MW718054, MW718057, MW718097, MW718000, MW718001, MW718008, MW718027, MW718034, MW718069, MW718076, MW718098, MW718103) together with 4 mostly identical reference sequences (KJ140127, KJ140131, KJ140130 and KT964225) obtained in GenBank database.
Strains belonging to supergroup A and B are shown in red and blue, respectively, while the reference sequences are indicated with underlines. Numbers, read from top to bottom, above the sequences are deduced amino acid positions on the wsp gene of MW717996. Dots indicate nucleotides identical to the reference sequences. The triangles above nucleotide position numbers indicate sites of mutations in our isolates (supergroup A: ▽ in red, and supergroup B: ▼ in blue) relative to the one or both of the reference sequences.
Discussion
Approximately 3500 species of mosquitoes belonging to 41 genera are widely distributing in the world, of which at least 350 species have been found in China [22–24], and Ae. albopictus, An. sinensis, Ar. subalbatus, Cx. pipiens and Cx. tritaeniorhynchus were dominant populations [25,26]. In this study, we investigated the prevalence of Wolbachia among these four genera of mosquitoes captured in 25 provinces of China. Ae. albopictus, the major vector of arboviruses, is the natural host with a prevalence of almost 100% of Wolbachia [13,27,28]. In this study, the total infection rate of Wolbachia was 97.1% (331/341) among Ae. albopictus collected from 17 provinces. The prevalence rates were above 94.7% in most of the sampled provinces, except Guangdong and Liaoning provinces, where the infection rate was significantly lower in the females of Ae. albopictus in Liaoning province (P<0.05). To the best of our knowledge, this study revealed the high prevalence of natural Wolbachia infection in Ar. subalbatus (95.8%, 481/502), Cx. pipiens (87.0%, 1525/1752) and Cx. tritaeniorhynchus (17.1%, 14/82) in vast areas of China for the first time (Fig 1, Tables 2 and 3). A few studies have demonstrated the presence of natural Wolbachia infection in Ar. subalbatus [29–31], however, the size and geographical diversity of samples were very limited. In this study, a total of 502 mosquitoes of Ar. subalbatus were collected in ten provinces, locating in eastern (Fujian, Jiangxi, Shandong and Zhejiang provinces), northern (Hebei province), northeastern (Liaoning province), central (Hubei and Hunan provinces), southwestern (Chongqing and Guizhou provinces) and southern (Guangdong province) of China. The overall infected rate of Wolbachia in Ar. subalbatus in this study (95.8%) is similar to the previous epidemiological survey conducted in Sri Lanka (100%) [30], indicating that Ar. subalbatus is the naturally harbor of Wolbachia (Fig 1 and Table 2). Cx. pipiens complex mosquitoes are widely distributed throughout China, including at high altitudes, such as Tibet [16,32,33]. Although considered as an important disease vector, there is little information on the distribution and diversity of Wolbachia in this kind of mosquitoes in China. In this study, samples of Cx. pipiens mosquitoes were identified in 23 out of 25 sampled provinces (except for Guizhou and Hunan provinces). The prevalence of Wolbachia varied greatly among these mosquitoes from different provinces. For example, only one mosquito of Cx. pipiens (1.4%, 1/71) was infected in Guangdong province, while there were 100% positive rates in Chongqing, Henan, Liaoning, Shanxi, Sichuan and Yunnan provinces (Fig 1, Tables 2 and 3). We noticed that Cx. pipiens is not a native mosquito species in Guangdong [15]. Was there a research group that artificially released mosquitoes, resulting in a low rate of Wolbachia infection? Cx. tritaeniorhynchus was only identified in the samples from Guizhou province. Our results showed a prevalence of 17.1% for Wolbachia in natural populations of Cx. tritaeniorhynchus in Guizhou province, China, considerably lower than the positive rates of Ae. albopictus, Ar. subalbatus and Cx. pipiens (Fig 1 and Table 2). For decades, Anopheles mosquitoes were considered resistant to Wolbachia. Although have been experimental infected in the laboratory, natural infections of Wolbachia in wild-collected Anopheles mosquitoes have barely been reported [34,35]. Anopheles mosquitoes were always considered to be exempt of Wolbachia until some recent studies showed evidence of natural Wolbachia infections in field populations of several Anopheles mosquitoes (An. arabiensis, An. coluzzii, An. demeilloni, An. funestus, An. gambiae and An. moucheti) targeting 16S rRNA [36–40], but not wsp gene. In addition, Walker et al. determined a heavy infection of Wolbachia in the ovaries of An. moucheti that could induce maternal transmission [41]. However, in this study, the mosquitoes of An. sinensis sampled from ten provinces in this study were all negative for Wolbachia by the screening of wsp and 16S rRNA genes (Table 2).
The temporal distribution of Wolbachia in arthropods is largely unknown. In this study, mosquitoes were captured at the first day of every month from September 2013 to August 2014, except for January and February, when the temperature was below zero degrees Celsius in average. Although the information on the species and genders of the captured mosquitoes was only identified and noted from April to August, 2014, it was available for us to find a significant reduction of the infected rates of Wolbachia in Cx. pipiens in July (P<0.01), in both females and males. Previous studies have indicated that the rearing temperature have significant effects on the fertility, frequency and density of mosquitoes and other arthropods infected with Wolbachia, especially wMel and wAlbB [42–47]. Further studies are needed to determine the effect on the infection of Wolbachia at more extreme temperature conditions (Table 3).
Although do not perform particularly well in the differentiation of closely related Wolbachia genomes [8], compared with 16S rRNA and cell division protein gene (ftsZ), phylogenetic tree of improved resolution can be constructed with wsp gene, which is evolving at a much faster rate [11]. Molecular phylogeny based on wsp sequences revealed that most Wolbachia infection in Ae. albopictus clustered with wAlbB (supergroup B) (88.9%, 24/27), while 11.1% (3/27) belonged to wAlbA (supergroup A). The genetic variation was observed within both wAlbA and wAlbB strains, including some strains sampled from the same location (Figs 2 and 4). Compared with previous studies, our results suggested that Wolbachia could have invaded and spread throughout populations of these mosquitoes for a long time [48–51]. The information of the diversity of Wolbachia infection in Ar. subalbatus is quite limited. Nugapola et al. reported the presence of Wolbachia supergroup A in two mosquitoes in Sri Lanka in 2017 [30]. To the best of our knowledge, this was the only published study regarding the diversity of Wolbachia infection in Ar. subalbatus, until we demonstrated the presence of Wolbachia supergroup A in 11 provinces of China. Interestingly, no polymorphism was observed among these strains based on both wsp and MLST loci (Figs 2 and 3). In Cx. pipiens, 98.1% (51/52) of samples were infected with wPip with non-polymorphism based on wsp sequence, except for an individual collected from Heilongjiang province (Fig 2). Further studies are needed to investigate the diversity of wPip with higher-resolution molecular biological assays, for example, whole-genome typing methods, more accurate markers and PCR-restriction fragment length polymorphism (RFLP) [8,52]. Bleidorn and Gerth have provided a characterization of 252 single copy loci for the application of few loci approaches, and some of these loci have the potential to perform better than wsp and MLST. However, some of these loci may not be harbored by mosquitoes or have no polymorphism in mosquitoes. Further studies are still needed to screen accurate markers from these loci. Although Rickettsia were screened in Cx. tritaeniorhynchus with a low prevalence [53], this mosquito specie was not considered to harbor natural Wolbachia infections [54,55]. Interestingly, in this study, Wolbachia-positive samples were detected in Cx. tritaeniorhynchus mosquitoes in Guizhou province and identical with wPip strains (Figs 1–3, S2 and Table 2). Although there were a few outliers, molecular phylogeny based on five MLST loci was generally consistent with that of wsp (Figs 2 and 3).
The present study identified a total of 12 unique mutations of the deduced amino acid (AA) with wsp gene sequencing and aligning, including four mutations in Wolbachia supergroup A and eight mutations in supergroup B, respectively. Two insertions (G179 and TKDA187-190) were identified among the three strains (MW718054, MW718057 and MW718097) of Wolbachia supergroup A. Both the phylogenetic tree and nucleotide/ AA alignment showed that these three strains were mostly closed to the reference sequence (KJ140127) with low polymorphism, although the mosquito vectors were sampled from three far separate provinces (Tianjin, Guangdong and Sichuan provinces) (Figs 1, 2, 4 and 5).
Supporting information
The 10-fold dilutions were performed to give solutions containing 100,000, 10,000, 1,000, 100 and 10 gene copies per PCR reaction system. The DNA samples of Rickettsia bellii and R. felis and double-distilled water were served as negative controls.
(TIF)
Strains identified in this study are identified with open circles (○) for supergroup A and filled circles (●) for supergroup B. The numbers at the branches show bootstrap support (1000 replicates). The bar at the bottom of the figure denotes distance.
(TIF)
Acknowledgments
We thank Jianqiang Ye and Jianzhong Zhu (College of Veterinary Medicine, Yangzhou University) for sharing the laboratory space with us. We also thank Chan Ding (Shanghai Veterinary Research Institute, Chinese Academy of Agriculture Sciences) and Shuo Su (College of Veterinary Medicine, Nanjing Agriculture University) for valuable discussions.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) (to YY and SS) and Shenzhen strategic emerging industry development special funds (JCYJ20170816143646446 to JS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hertig M, Wolbach SB. Studies on Rickettsia-Like Micro-Organisms in Insects. J Med Res. 1924;44(3):329–74 7. ; PubMed Central PMCID: PMC2041761. [PMC free article] [PubMed] [Google Scholar]
- 2.Stouthamer R, Breeuwer JA, Hurst GD. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71 . [DOI] [PubMed] [Google Scholar]
- 3.Turelli M, Hoffmann AA. Microbe-induced cytoplasmic incompatibility as a mechanism for introducing transgenes into arthropod populations. Insect Mol Biol. 1999;8(2):243–55. doi: 10.1046/j.1365-2583.1999.820243.x . [DOI] [PubMed] [Google Scholar]
- 4.McMeniman CJ, Lane RV, Cass BN, Fong AW, Sidhu M, Wang YF, et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323(5910):141–4. doi: 10.1126/science.1165326 . [DOI] [PubMed] [Google Scholar]
- 5.Jiggins FM. The spread of Wolbachia through mosquito populations. PLoS Biol. 2017;15(6):e2002780. doi: 10.1371/journal.pbio.2002780 ; PubMed Central PMCID: PMC5453404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Hurk AF, Hall-Mendelin S, Pyke AT, Frentiu FD, McElroy K, Day A, et al. Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PLoS Negl Trop Dis. 2012;6(11):e1892. doi: 10.1371/journal.pntd.0001892 ; PubMed Central PMCID: PMC3486898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bian G, Joshi D, Dong Y, Lu P, Zhou G, Pan X, et al. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340(6133):748–51. doi: 10.1126/science.1236192 . [DOI] [PubMed] [Google Scholar]
- 8.Bleidorn C, Gerth M. A critical re-evaluation of multilocus sequence typing (MLST) efforts in Wolbachia. FEMS Microbiol Ecol. 2018;94(1). doi: 10.1093/femsec/fix163 . [DOI] [PubMed] [Google Scholar]
- 9.Laidoudi Y, Levasseur A, Medkour H, Maaloum M, Ben Khedher M, Sambou M, et al. An Earliest Endosymbiont, Wolbachia massiliensis sp. nov., Strain PL13 from the Bed Bug (Cimex hemipterus), Type Strain of a New Supergroup T. International journal of molecular sciences. 2020;21(21). doi: 10.3390/ijms21218064 ; PubMed Central PMCID: PMC7662661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Neill SL, Giordano R, Colbert AM, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci U S A. 1992;89(7):2699–702. doi: 10.1073/pnas.89.7.2699 ; PubMed Central PMCID: PMC48729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W, Rousset F, O’Neil S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc Biol Sci. 1998;265(1395):509–15. doi: 10.1098/rspb.1998.0324 ; PubMed Central PMCID: PMC1688917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72(11):7098–110. doi: 10.1128/AEM.00731-06 ; PubMed Central PMCID: PMC1636189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Xi Z, Liu X, Wang J, Guo Y, Ren D, et al. Identification and molecular characterization of Wolbachia strains in natural populations of Aedes albopictus in China. Parasites & vectors. 2020;13(1):28. doi: 10.1186/s13071-020-3899-4 ; PubMed Central PMCID: PMC6961339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang EYY, Wong AYP, Lee IHT, Qu Z, Yip HY, Leung CW, et al. Infection patterns of dengue, Zika and endosymbiont Wolbachia in the mosquito Aedes albopictus in Hong Kong. Parasites & vectors. 2020;13(1):361. doi: 10.1186/s13071-020-04231-x ; PubMed Central PMCID: PMC7372788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atoni E, Zhao L, Hu C, Ren N, Wang X, Liang M, et al. A dataset of distribution and diversity of mosquito-associated viruses and their mosquito vectors in China. Sci Data. 2020;7(1):342. doi: 10.1038/s41597-020-00687-9 ; PubMed Central PMCID: PMC7555486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Lu G, Li J, Kelly P, Li M, Wang J, et al. Molecular Detection of Rickettsia felis and Rickettsia bellii in Mosquitoes. Vector Borne Zoonotic Dis. 2019;19(11):802–9. doi: 10.1089/vbz.2019.2456 . [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Lu G, Kelly PJ, Wang C. Seasonal and Gender Differences in Presence of Rickettsia felis and Blood meals Provide Additional Evidence of a Vector Role for Mosquitoes. Can J Infect Dis Med Microbiol. 2019;2019:8543460. doi: 10.1155/2019/8543460 ; PubMed Central PMCID: PMC6481102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3(5):294–9. . [PubMed] [Google Scholar]
- 19.Werren JH, Windsor DM. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc Biol Sci. 2000;267(1450):1277–85. doi: 10.1098/rspb.2000.1139 ; PubMed Central PMCID: PMC1690679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braig HR, Zhou W, Dobson SL, O’Neill SL. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia pipientis. J Bacteriol. 1998;180(9):2373–8. doi: 10.1128/JB.180.9.2373-2378.1998 ; PubMed Central PMCID: PMC107178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Chu S, Shang S, Yang Z, Wang C. Short communication: Genotyping and single nucleotide polymorphism analysis of bovine leukemia virus in Chinese dairy cattle. J Dairy Sci. 2019;102(4):3469–73. doi: 10.3168/jds.2018-15481 . [DOI] [PubMed] [Google Scholar]
- 22.Xia H, Wang Y, Atoni E, Zhang B, Yuan Z. Mosquito-Associated Viruses in China. Virol Sin. 2018;33(1):5–20. doi: 10.1007/s12250-018-0002-9 ; PubMed Central PMCID: PMC5866263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G, Li C, Guo X, Xing D, Dong Y, Wang Z, et al. Identifying the main mosquito species in China based on DNA barcoding. PLoS One. 2012;7(10):e47051. doi: 10.1371/journal.pone.0047051 ; PubMed Central PMCID: PMC3468562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Y, Song Z, Luo L, Wang Q, Zhou G, Yang D, et al. Molecular evidence for new sympatric cryptic species of Aedes albopictus (Diptera: Culicidae) in China: A new threat from Aedes albopictus subgroup? Parasites & vectors. 2018;11(1):228. doi: 10.1186/s13071-018-2814-8 ; PubMed Central PMCID: PMC5885320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng X, Zhong D, He Y, Zhou G. Seasonality modeling of the distribution of Aedes albopictus in China based on climatic and environmental suitability. Infect Dis Poverty. 2019;8(1):98. doi: 10.1186/s40249-019-0612-y ; PubMed Central PMCID: PMC6889612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu G, Xia H, Zhou H, Li J, Lu F, Liu Y, et al. Susceptibility of Anopheles sinensis to Plasmodium vivax in malarial outbreak areas of central China. Parasites & vectors. 2013;6:176. doi: 10.1186/1756-3305-6-176 ; PubMed Central PMCID: PMC3695883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das B, Satapathy T, Kar SK, Hazra RK. Genetic structure and Wolbachia genotyping in naturally occurring populations of Aedes albopictus across contiguous landscapes of Orissa, India. PLoS One. 2014;9(4):e94094. doi: 10.1371/journal.pone.0094094 ; PubMed Central PMCID: PMC3979767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitrayapong P, Baimai V, O’Neill SL. Field prevalence of Wolbachia in the mosquito vector Aedes albopictus. Am J Trop Med Hyg. 2002;66(1):108–11. doi: 10.4269/ajtmh.2002.66.108 . [DOI] [PubMed] [Google Scholar]
- 29.Jamnongluk W, Kittayapong P, Baisley KJ, O’Neill SL. Wolbachia infection and expression of cytoplasmic incompatibility in Armigeres subalbatus (Diptera: Culicidae). J Med Entomol. 2000;37(1):53–7. doi: 10.1603/0022-2585-37.1.53 . [DOI] [PubMed] [Google Scholar]
- 30.Nugapola N, De Silva W, Karunaratne S. Distribution and phylogeny of Wolbachia strains in wild mosquito populations in Sri Lanka. Parasites & vectors. 2017;10(1):230. doi: 10.1186/s13071-017-2174-9 ; PubMed Central PMCID: PMC5424329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai KH, Lien JC, Huang CG, Wu WJ, Chen WJ. Molecular (sub) grouping of endosymbiont Wolbachia infection among mosquitoes of Taiwan. J Med Entomol. 2004;41(4):677–83. doi: 10.1603/0022-2585-41.4.677 . [DOI] [PubMed] [Google Scholar]
- 32.Wang ZM, Li CX, Xing D, Yu YH, Liu N, Xue RD, et al. Detection and widespread distribution of sodium channel alleles characteristic of insecticide resistance in Culex pipiens complex mosquitoes in China. Med Vet Entomol. 2012;26(2):228–32. doi: 10.1111/j.1365-2915.2011.00985.x . [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Baimaciwang, Wu H, Pengcuociren, Guo Y, Cirenwangla, et al. Breeding Site Characteristics and Associated Factors of Culex pipiens Complex in Lhasa, Tibet, P. R. China. International journal of environmental research and public health. 2019;16(8). doi: 10.3390/ijerph16081407 ; PubMed Central PMCID: PMC6517927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joshi D, Pan X, McFadden MJ, Bevins D, Liang X, Lu P, et al. The Maternally Inheritable Wolbachia wAlbB Induces Refractoriness to Plasmodium berghei in Anopheles stephensi. Front Microbiol. 2017;8:366. doi: 10.3389/fmicb.2017.00366 ; PubMed Central PMCID: PMC5340780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomes FM, Hixson BL, Tyner MDW, Ramirez JL, Canepa GE, Alves ESTL, et al. Effect of naturally occurring Wolbachia in Anopheles gambiae s.l. mosquitoes from Mali on Plasmodium falciparum malaria transmission. Proc Natl Acad Sci U S A. 2017;114(47):12566–71. doi: 10.1073/pnas.1716181114 ; PubMed Central PMCID: PMC5703331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baldini F, Segata N, Pompon J, Marcenac P, Shaw WR, Dabire RK, et al. Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nat Commun. 2014;5:3985. doi: 10.1038/ncomms4985 ; PubMed Central PMCID: PMC4059924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niang EHA, Bassene H, Makoundou P, Fenollar F, Weill M, Mediannikov O. First report of natural Wolbachia infection in wild Anopheles funestus population in Senegal. Malar J. 2018;17(1):408. doi: 10.1186/s12936-018-2559-z ; PubMed Central PMCID: PMC6219158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong ML, Liew JWK, Wong WK, Pramasivan S, Mohamed Hassan N, Wan Sulaiman WY, et al. Natural Wolbachia infection in field-collected Anopheles and other mosquito species from Malaysia. Parasites & vectors. 2020;13(1):414. doi: 10.1186/s13071-020-04277-x ; PubMed Central PMCID: PMC7425011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw WR, Marcenac P, Childs LM, Buckee CO, Baldini F, Sawadogo SP, et al. Wolbachia infections in natural Anopheles populations affect egg laying and negatively correlate with Plasmodium development. Nat Commun. 2016;7:11772. doi: 10.1038/ncomms11772 ; PubMed Central PMCID: PMC4895022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeffries CL, Lawrence GG, Golovko G, Kristan M, Orsborne J, Spence K, et al. Novel Wolbachia strains in Anopheles malaria vectors from Sub-Saharan Africa. Wellcome Open Res. 2018;3:113. doi: 10.12688/wellcomeopenres.14765.2 ; PubMed Central PMCID: PMC6234743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker T, Quek S, Jeffries CL, Bandibabone J, Dhokiya V, Bamou R, et al. Stable high-density and maternally inherited Wolbachia infections in Anopheles moucheti and Anopheles demeilloni mosquitoes. Curr Biol. 2021;31(11):2310–20 e5. doi: 10.1016/j.cub.2021.03.056 ; PubMed Central PMCID: PMC8210651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ant TH, Herd CS, Geoghegan V, Hoffmann AA, Sinkins SP. The Wolbachia strain wAu provides highly efficient virus transmission blocking in Aedes aegypti. PLoS Pathog. 2018;14(1):e1006815. doi: 10.1371/journal.ppat.1006815 ; PubMed Central PMCID: PMC5784998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lau MJ, Ross PA, Hoffmann AA. Infertility and fecundity loss of Wolbachia-infected Aedes aegypti hatched from quiescent eggs is expected to alter invasion dynamics. PLoS Negl Trop Dis. 2021;15(2):e0009179. doi: 10.1371/journal.pntd.0009179 ; PubMed Central PMCID: PMC7909672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau MJ, Ross PA, Endersby-Harshman NM, Hoffmann AA. Impacts of Low Temperatures on Wolbachia (Rickettsiales: Rickettsiaceae)-Infected Aedes aegypti (Diptera: Culicidae). J Med Entomol. 2020;57(5):1567–74. doi: 10.1093/jme/tjaa074 ; PubMed Central PMCID: PMC7566743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nazni WA, Hoffmann AA, NoorAfizah A, Cheong YL, Mancini MV, Golding N, et al. Establishment of Wolbachia Strain wAlbB in Malaysian Populations of Aedes aegypti for Dengue Control. Curr Biol. 2019;29(24):4241–8 e5. doi: 10.1016/j.cub.2019.11.007 ; PubMed Central PMCID: PMC6926472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross PA, Axford JK, Yang Q, Staunton KM, Ritchie SA, Richardson KM, et al. Heatwaves cause fluctuations in wMel Wolbachia densities and frequencies in Aedes aegypti. PLoS Negl Trop Dis. 2020;14(1):e0007958. doi: 10.1371/journal.pntd.0007958 ; PubMed Central PMCID: PMC6977724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu YX, Song ZR, Zhang YY, Hoffmann AA, Hong XY. Spider Mites Singly Infected With Either Wolbachia or Spiroplasma Have Reduced Thermal Tolerance. Front Microbiol. 2021;12:706321. doi: 10.3389/fmicb.2021.706321 ; PubMed Central PMCID: PMC8292952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Armbruster P, Damsky WE Jr., Giordano R, Birungi J, Munstermann LE, Conn JE. Infection of New- and Old-World Aedes albopictus (Diptera: Culicidae) by the intracellular parasite Wolbachia: implications for host mitochondrial DNA evolution. J Med Entomol. 2003;40(3):356–60. doi: 10.1603/0022-2585-40.3.356 . [DOI] [PubMed] [Google Scholar]
- 49.Kambhampati S, Rai KS, Burgun SJ. Unidirectional Cytoplasmic Incompatibility in the Mosquito, Aedes Albopictus. Evolution. 1993;47(2):673–7. doi: 10.1111/j.1558-5646.1993.tb02121.x . [DOI] [PubMed] [Google Scholar]
- 50.Sinkins SP, Braig HR, O’Neill SL. Wolbachia superinfections and the expression of cytoplasmic incompatibility. Proc Biol Sci. 1995;261(1362):325–30. doi: 10.1098/rspb.1995.0154 . [DOI] [PubMed] [Google Scholar]
- 51.de Albuquerque AL, Magalhaes T, Ayres CF. High prevalence and lack of diversity of Wolbachia pipientis in Aedes albopictus populations from Northeast Brazil. Mem Inst Oswaldo Cruz. 2011;106(6):773–6. doi: 10.1590/s0074-02762011000600021 . [DOI] [PubMed] [Google Scholar]
- 52.Dumas E, Atyame CM, Milesi P, Fonseca DM, Shaikevich EV, Unal S, et al. Population structure of Wolbachia and cytoplasmic introgression in a complex of mosquito species. BMC Evol Biol. 2013;13:181. doi: 10.1186/1471-2148-13-181 ; PubMed Central PMCID: PMC3846486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo WP, Tian JH, Lin XD, Ni XB, Chen XP, Liao Y, et al. Extensive genetic diversity of Rickettsiales bacteria in multiple mosquito species. Sci Rep. 2016;6:38770. doi: 10.1038/srep38770 ; PubMed Central PMCID: PMC5146937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeffries CL, Walker T. The Potential Use of Wolbachia-Based Mosquito Biocontrol Strategies for Japanese Encephalitis. PLoS Negl Trop Dis. 2015;9(6):e0003576. doi: 10.1371/journal.pntd.0003576 ; PubMed Central PMCID: PMC4472807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tiawsirisup S, Sripatranusorn S, Oraveerakul K, Nuchprayoon S. Distribution of mosquito (Diptera: Culicidae) species and Wolbachia (Rickettsiales: Rickettsiaceae) infections during the bird immigration season in Pathumthani province, central Thailand. Parasitol Res. 2008;102(4):731–5. doi: 10.1007/s00436-007-0825-z . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 10-fold dilutions were performed to give solutions containing 100,000, 10,000, 1,000, 100 and 10 gene copies per PCR reaction system. The DNA samples of Rickettsia bellii and R. felis and double-distilled water were served as negative controls.
(TIF)
Strains identified in this study are identified with open circles (○) for supergroup A and filled circles (●) for supergroup B. The numbers at the branches show bootstrap support (1000 replicates). The bar at the bottom of the figure denotes distance.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.