Abstract
Signal transducer and activator of transcription 3 (STAT3) activation is associated with drug resistance induced by anti-epidermal growth factor receptor (anti-EGFR) therapy in the treatment of colon cancer. Thus, the combined inhibition of EGFR and STAT3 may prove beneficial for this type of cancer. STAT3 has been proven to play a critical role in colon cancer initiation and progression, and is considered the primary downstream effector driven by interleukin-6 (IL-6). A disintegrin and metalloproteinase 17 (ADAM17), documented as an oncogene, catalyzes the cleavage of both EGF and IL-6R, inducing EGFR signaling and enabling IL-6 trans-signaling to activate STAT3 in a wide range of cell types to promote inflammation and cancer development. As a natural product, shikonin (SKN) has been found to function as an antitumor agent; however, its role in the regulation of ADAM17 and IL-6/STAT3 signaling in colon cancer cells remains unknown. In the present study, it was found that SKN inhibited colon cancer cell growth, suppressed both constitutive and IL-6-induced STAT3 phosphorylation, and downregulated the expression of ADAM17. ADAM17 expression was not altered in response to STAT3 knockdown, while IL-6-induced STAT3 activation did not induce ADAM17 transcripts. Furthermore, it was demonstrated that SKN did not affect the expression of key proteins involved in the maturation and degradation of ADAM17. SKN decreased ADAM17 expression possibly through reactive oxygen species (ROS)-mediated translational inhibition, as evidenced by the increased ADAM17 mRNA and phosphorylation levels of eukaryotic initiation factor 2α (eIF2α). The expression of ADAM17 and p-eIF2α was reversed by N-acetylcysteine (NAC, a ROS scavenger). Taken together, these results indicate that the concurrent inhibition of ADAM17 and IL-6/STAT3 signaling by SKN may synergistically contribute to the suppression of colon cancer cell growth.
Keywords: A disintegrin and metalloproteinase 17, IL-6, STAT3, colon cancer, shikonin
Introduction
Cancer has become an increasingly global life-threatening disease with the advancing age of the global population. Based on estimations from the American Cancer Society and the National Cancer Institute, >16.9 million cancer survivors (individuals with a history of all types of cancer) were alive in America on January 1, 2019 (1), among whom ~1.54 million patients suffered from colon cancer (1). The incidence of colon cancer varies up to 10-fold globally, being associated with the human development index and different lifestyles. In developed regions, the incidence rate of colon cancer is >32.2 per 100,000 individuals (2). In 2018 in Europe, colon cancer was the leading cause of new cancer diagnoses, accounting for 44.4 per 100,000 of estimated new cancer cases (3). Despite progress being made in the development of mechanisms and treatment strategies, including colectomy, radiotherapy and chemotherapy, 35% of patients with colorectal cancer do not survive 5 years following the diagnosis (1). Bladder dysfunction or sexual dysfunction ascribed to colostomy (4-6) and tumor chemoresistance induced by current therapies suggest that novel therapeutic avenues and detailed mechanistic explorations are required.
Numerous studies have explored the detailed mechanisms responsible for tumorigenesis, including cell arrest or death, tumor metabolism and enhancing the effect of traditional anti- tumor agents, to combat cancer (7-11). The present perspectives of cancer appear to promote the influence of inflammation, which contributes to tumor initiation, progression and metastasis (12). Interleukin (IL)-6 is an inflammatory cytokine and has been documented to be involved in cancer-related inflammation, cell proliferation, tumor invasion, angiogenesis, metastasis and chemoresistance (13). The binding of IL-6 and its membrane receptor (mIL-6R) allows the recruitment of glycoprotein 130 (gp130), resulting in a trimer comprised of IL-6, mIL-6R and gp130 (14). The protein complex promotes the close contact of Janus kinases and tyrosine kinase with gp130, and facilitates a signaling cascade via phosphorylated tyrosine residues of gp130 (IL-6 classical signaling) (14). In the context of cell signaling without mIL-6R, IL-6 can alternatively bind to the soluble form of IL-6R (sIL-6R) and then recruit gp130, inducing the formation of the aforementioned protein complex to promote the downstream signaling cascade (IL-6 trans-signaling) (15). Signal transducer and activator of transcription 3 (STAT3) is the major downstream transcription factor upon IL-6 induction in both pathways, which drives the transcription of several survival-associated genes (16,17). The constitutive activation of STAT3 plays a pivotal role in colon cancer (18-20). Previously, the authors' research group demonstrated that STAT3 activation was necessary for tumor survival and for the proliferation of cancer-initiating and stem cell-like colon cancer cells (21,22). Agents targeting IL-6/STAT3 signaling exert potential inhibitory effects on colon cancer (23-25). Moreover, in the current treatment for colon cancer, agents targeting epidermal growth factor receptor (EGFR) signaling have significantly contributed to the advancements made in this field. However, a number of patients who receive anti-EGFR therapy experience drug resistance, which has been found to be associated with STAT3 activation. The combined inhibition of EGFR and STAT3 may thus prove beneficial for this type of cancer (26).
A disintegrin and metalloproteinases (ADAMs) are transmembrane metalloproteinases capable of shedding membrane anchoring proteins. ADAM17 was originally identified to contribute to the cleavage of tumor necrosis factor-α (TNF-α) on the cell membrane, allowing the release of soluble TNF-α and promoting the inflammatory cascade by triggering TNF-α receptor signaling (27,28). The expression of ADAM17 is under strict orchestration by a series of factors. Rhomboid family membe 2 (RHBDF2, also known as iRhom2) and furin protein mediate the trafficking and maturation of ADAM17 (29), while RHBDF2 combined with phosphofurin acidic cluster sorting protein 2 (PACS-2) control the degradation or preservation of ADAM17 (ADAM17 recycling) (30,31). When progress in the field of elucidating the role of ADAM17 was made, additional substrates, including ligands of EGFR, were described for ADAM17 (32), highlighting the involvement of ADAM17 in various pathological processes, including cancer (33). It has been reported that ADAM17 promotes tumor development and may thus be a therapeutic target in colon cancer (34). Of note, considered not only a tumor promoter, ADAM17 cleaves mIL-6R and EGF, promoting EGFR signaling and IL-6 trans-signaling, enabling amplified cellular responses to EGF and IL-6 in a wide range of cell types (33). Both ADAM17 and STAT3 contribute to the progression of colon cancer and may exert synergistic effects. Therefore, the discovery of agents targeting these two proteins may benefit the development of colon cancer therapeutics, particularly in overcoming the drug resistance associated with anti-EGFR treatments.
Previously, natural product extracts have received increasing attention for their antitumor properties via diverse mechanisms, including autophagy, immune modulation and apoptosis etc., in various types of cancer (35-37). In a previous study, the authors demonstrated that ursolic acid, a natural triterpenoid compound, inhibited IL-6/STAT3 signaling and suppressed tumor growth in hepatocellular carcinoma (38). Shikonin (SKN) is the major component of extracts from the roots of Lithospermum erythrorhizon Sieb. Et Zucc. that belongs to the Boraginaceae family (39). The applications of this plant include several pathologies, such as burns, carbuncles and measles (40). The antitumor properties of SKN include the induction of apoptosis/necroptosis, promotion of autophagy, induction of oxidative stress, etc. (41,42). The inhibition of phosphorylated (p-)STAT3 by SKN in lung, breast, melanoma and skin cancer types has been reported (43-46); however, the potential effects of SKN on p-STAT3 and ADAM17 expression in colon cancer remain unclear.
The present study used HCT116 and SW480 colon cancer cell lines for in vitro experiments. As STAT3 contributes to tumor initiation and progression, serving as a point of convergence for numerous oncogenic signaling pathways, while ADAM17 controls the cleavage of several tumor-associated ligands (21,47-49), cancer cell growth was assessed in a series of experiments, including cell viability, colony formation, wound healing and apoptosis assays. It was found that SKN suppressed the growth of both cancer cell lines. Importantly, SKN exerted an inhibitory effect on constitutive/IL-6-induced p-STAT3 and ADAM17 expression in these two colon cancer cell lines. Furthermore, it was found that the reactive oxygen species (ROS)-mediated suppression of ADAM17 translation may contribute to the decreased expression of ADAM17 induced by SKN.
Materials and methods
Reagents
SKN was purchased from MedChemExpress (cat. no. HY-N0822) and was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 20 mM as a stock solution. IL-6 (cat. no. 200-06, PeproTech China was dissolved in water at 25 µg/ml for storage. Crystal violet (Thermo Fisher Scientific, Inc.) was dissolved in methyl alcohol (0.5 g/ml) and prepared for staining. N-acetylcysteine (NAC) was purchased from Sigma-Aldrich; Merck KGaA (cat. no. A0737).
Cell culture and treatment
The HCT116 (cat. no. CCL-247) and SW480 (cat. no. CCL-228) colon cancer cell lines were purchased from the American Type Culture Collection (ATCC). All cell lines were authenticated via STR profiling and mycoplasmas were tested monthly. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Nanjing KeyGen Biotech Co., Ltd.) supplemented with high glucose and fetal bovine serum (10X; Gibco; Thermo Fisher Scientific, Inc.) and penicillin/streptomycin (100X, Sigma-Aldrich; Merck KGaA). Prior to treatment, the cells were cultured for 12 h in an incubator (37°C, 5% CO2). To explore STAT3 phosphorylation induced by IL-6, serum-starved cells were pre-treated with SKN (10, 15 and 20 µM, 3 h) and then stimulated with IL-6 (25 ng/ml) for a further 30 min. Various concentrations (10, 15 and 20 µM) SKN (DMSO as a control) were added to the medium, and following 10 h of culture, the cells were collected. For experiments involving NAC, cells were pre-treated with or without NAC (5 mM) for 2 h, and shikonin (20 µM) was then added for a further 10 h.
Western blot analysis
Cell lysate (RIPA lysis buffer containing 1 mmol/l protease inhibitor and 1 mmol/l phosphatase inhibitor, Shanghai Sangon Biotech Co., Ltd.) preparation was performed as previously described (50). The lysates were centrifuged at 13,800 × g for 20 min at 4°C, and the supernatant was collected. The concentration of protein was determined by bicinchoninic acid (BCA) protein assay kit (Shanghai Sangon Biotech Co., Ltd.). Equivalent amounts (15-25 µg) of protein (pre-stained protein ladder; cat. no. 26617, Thermo Fisher Scientific Inc.) were loaded on a 5% Bis-Tris SDS-PAGE gel and separated using 10% gel electrophoresis, transferred to a PVDF membrane, blocked with TBS-T (Tris-buffer saline containing 0.1% Tween-20) containing 5% powdered milk for 60 min (at room temperature), and consequently probed with the following primary antibodies (1:1,000 dilution) overnight at 4°C: Anti-p-STAT3 tyrosine 705 (cat. no. 9145), anti-t-STAT3 (cat. no. 4904), anti-cleaved caspase-3 (cat. no. 9661), anti-cleaved PARP (cat. no. 9544), anti-cyclin D1 (cat. no. 55506), anti-cyclin E1 (cat. no. 20808), anti-p-JAK1 (cat. no. 3331), anti-p-JAK2 (cat. no. 3776), anti-p-eukaryotic initiation factor 2α (eIF2α; cat. no. 3398) (all from Cell Signaling Technology, Inc.), anti-ADAM17 (cat. no. ab2051; Abcam), anti-GAPDH (cat. no. 10494-1-AP), anti-β-tubulin (cat. no. 66240-1-Ig), anti-PACS-2 (cat. no. 19508-1-AP) (all from ProteinTech Group, Inc.), anti-JAK1 (cat. no. 29261), anti-JAK2 (cat. no. 3230) (both from Cell Signaling Technology, Inc.), anti-eIF2α (cat. no. ET7111-34; HuaBio) and anti-RHBDF2 (cat. no. AP13588A; Abcepta, Biotech Ltd., Co.). Horseradish peroxidase-conjugated (HRP-conjugated) antibodies (1:5,000 dilution; cat. no. QSJ-005/QSJ-006, Promoter Co., Ltd.) and Immobilon Western Chemiluminescent HRP Substrate (AntGene Co., Ltd.) were used for protein detection, operated on a ChemiDoc-It 510 Imager with VisionWorks software (version 8.17.16133.9147; Ultra-Violet Products, Ltd.) according to the manufacturer's instructions.
Wound healing assay
The cells were plated in six-well plates and cultured until they reached 100% confluency. The serum-starved cell layers were then scratched using a sterile 10 µl pipette tip and washed with PBS to discard floating cells. Wounded cells were imaged at baseline followed by 3 h of treatment with SKN (10 and 20 µM) or DMSO. The medium was replaced to remove the test compound or DMSO and images were captured using a fluorescence microscope (Guangzhou Micro-shot Technology Co., Ltd.) at the indicated time-points. The wound surface area was assessed using ImageJ software (version 1.53e), and the wound healing activity of the colon cancer cells was determined by the quantification of wound healing progression. Wound healing activity = 1-(wound surface area at the indicated time-point/wound surface area at baseline).
Cell viability assay
After adhering to a 96-well plate for 24 h, cells (6,000 cells per well) were treated with or without various concentrations (5, 10, 15, 20 and 25 µM) of SKN (DMSO as a control) for 10 h, and cell viability was determined using a Cell Counting Kit-8 (cat. no. HY-K0301MedChemExpress) according to the manufacturer's instructions.
Colony formation assay
Following treatment with SKN (10 or 20 µM) or DMSO for 3 h, the cells were collected, counted and seeded (4,000 cells per plate) in 10-cm plates in an incubator for 10 days (37°C, 5% CO2). Crystal violet (0.5 g/ml) was used for staining as previously described (50).
Public database
The differential analysis of transcripts of human IL-6 and ADAM17 in colon cancer ('colon adenocarcinoma', 'colon mucinous adenocarcinoma' and 'colorectal cancer') compared to normal counterparts was performed and downloaded from the public Oncomine database (www.oncomine.org; access date: December 15, 2020, threshold P-value: 0.05; fold change, 2; gene rank, top 10%). For the comparison of genes of interest across different analyses, studies that met the aforementioned criteria were excluded when the total sample size was <40.
RNA interference and transfection
Negative control (NC) and small interfering RNA targeting STAT3 (siSTAT3; sense, CCACUUUGGUGUUUCAUAATT; antisense, UUAUGA AACACCAAAGUGGTT) were purchased from RiboBio (Guangzhou RiboBio Co., Ltd.). Lipofectamine 2000® (cat. no. 11668030, Thermo Fisher Scientific, Inc.) and Opti-MEM® I reduced serum medium (#31985070; Gibco; Thermo Fisher Scientific, Inc.) were used for transfection. Seeded cells were cultured overnight and then cultured in mixed Opti-MEM medium containing 50 nM siSTAT3 and Lipofectamine 2000® according to the manufacturer's instructions. The medium was replaced with complete medium after 6 h of transfection.
Reverse transcription-quantitative PCR (RT-qPCR)
The cells were treated with SKN (20 µM) or IL-6 (25 ng/ml) for 10 h and then prepared for RNA extraction using a HiPure Total RNA Mini kit (Magen Biotechnology Co., Ltd.) according to the manufacturer's instructions. A ReverTra Ace qPCR RT kit (Toyobo Co., Ltd.) was used for cDNA production. qPCR was performed as previously described (50) (initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 sec, annealing at 60°C for 4 sec and extension at 72°C for 45 sec.). The primer sequences were as follows: Human ADAM17 forward, ATCAAACCTTTCCTGCG and reverse, CAAACCCATCCTCGTCCA; and human β-actin forward, CTGGAACGGTGAAGGTGACA and reverse, AAGGGACTT CCTGTAACAATGCA.
Statistical analysis
Data are presented as the mean ± SEM. All data are from three independent experiments. Statistical analysis was performed using SPSS software (version 22.0; SPSS, Inc.). Comparisons of multiple groups were analyzed using one-way analysis of variance (ANOVA) with Bonferroni's post hoc test, and comparisons of two groups were analyzed using paired t-tests. For the comparison of the wound healing area, two-way ANOVA (time and group) was used with Bonferroni's post hoc test. For the quantification of the western blots, ImageJ software (version 1.53e) was used for assessment. A P-value <0.05 was considered to indicate a statistically significant difference.
Results
SKN decreases the viability, promotes the apoptosis, disrupts the cell cycle, and suppresses the wound healing and colony formation of colon cancer cells
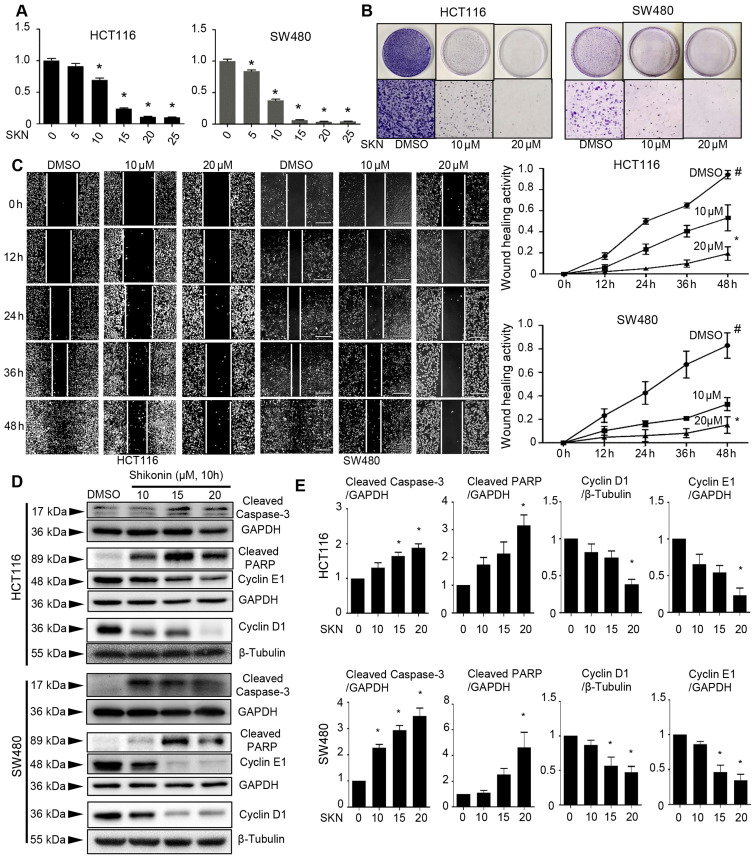
ADAM17 and STAT3 play a key role in inflammation and cancer, and have been found to be associated with colon cancer initiation, progression and metastasis (47-49). Moreover, it has been shown that the inhibition of IL-6/STAT3 signaling by small molecular compounds results in a wide range of impeded cancer cell growth properties (21,22,25). Herein, it was found that SKN exerted inhibitory effects on the viability, colony formation and wound healing ability of colon cancer cells; generally, a suppression of cell growth was observed following treatment with SKN. SKN induced the concentration-dependent suppression of cell viability (Fig. 1A) and colony formation (Fig. 1B) ability of the HCT116 and SW480 cells. Moreover, it was found that treatment with a higher concentration of SKN (20 µM) tended exert a more prominent effect on the wound healing ability of the cells than treatment with 10 µM SKN (Fig. 1C). Treatment with SKN promoted cell apoptosis, as evidenced by increased expression levels of cleaved caspase-3 and cleaved PARP in both cell lines (Fig. 1D and E), as well as by enhanced staining-positive spots in the Annexin V/PI immunofluorescence staining assay (Fig. S1). Moreover, SKN decreased the expression of cyclin D1 and cyclin E1, thus suggesting the disruption of the cell cycle and the suppression of cell growth (Fig. 1D and E).
Figure 1.
SKN inhibits the viability, colony formation and wound healing, and disrupts the cell cycle and promotes the apoptosis of colon cancer cells. (A) SKN (concentration as indicated, 10 h of treatment) suppressed the viability of the HCT116 and SW480 colon cancer cells in a concentration-dependent manner. The cell viability of the DMSO group (0 µM SKN) was arbitrarily set as 1. *P<0.05 vs. DMSO group. (B) SKN (10 µM) inhibited the colony formation of the two cancer cell lines. SKN at 20 µM exerted a more intense antitumor effect. Upper panel, cells cultured in plates and stained with crystal violet; lower panel, high-power field of the plates (magnification, x10). (C) SKN (10 and 20 µM) inhibited the wound healing ability of the HCT116 and SW480 cells. Wound healing activity = 1-(wound surface area at the indicated time-point/wound surface area at baseline). #P<0.05 vs. wound healing activity at the time-point of 0 h. *P<0.05 vs. wound healing activity of DMSO group at 48 h. White scale bars, 400 µm. (D) SKN (concentration as indicated, 10 h of treatment) increased the expression of cleaved caspase-3 and cleaved PARP in the two cancer cell lines. SKN also decreased the expression of cyclin D1 and cyclin E1. (E) Quantitative data analysis of proteins of interest normalized to those of the loading control. All data represent the mean ± SEM. *P<0.05 vs. DMSO group (0 µM SKN). SKN, shikonin.
Shikonin inhibits IL-6-induced expression and constitutive p-STAT3 activation in colon cancer cells
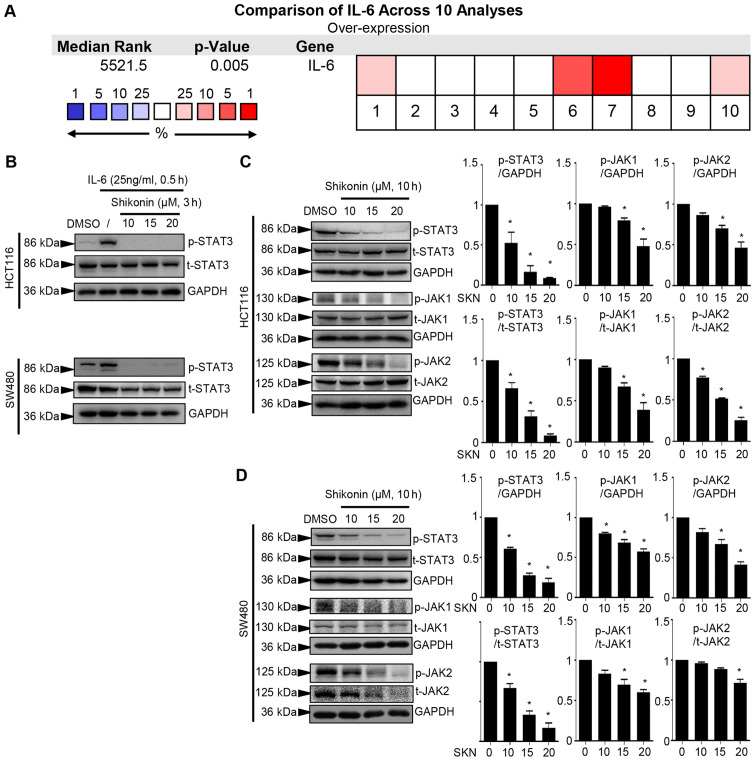
A comparison across 10 analyses performed using Oncomine suggested that IL-6 transcripts were increased in colon cancer (P=0.005, Fig. 2A). It has been reported that IL-6 triggers the formation of the IL-6/IL-6R/EGFR complex. The protein interaction involving EGFR facilitates the phosphorylation of Y1068 of EGFR, which plays a critical role in the biphasic pattern of STAT3 phosphorylation (51). The present study examined whether the concentration of IL-6 (25 ng/ml) used could activate EGFR. As shown in Fig. S2, 25 ng/ml IL-6 did not significantly induce the phosphorylation of Y1068 on EGFR (Fig. S2). Moreover, in a previous study, the authors used this concentration of IL-6 to induce the phosphorylation of STAT3, aiming to determine the inhibitory effect on p-STAT3 of the compound of interest (23,24). Herein, the concentration of 25 ng/ml IL-6 induced the robust phosphorylation of STAT3, and 3 h of pre-treatment with SKN prominently suppressed the cellular response to this trigger in the HCT116 and SW480 cancer cell lines (Fig. 2B). The persistent activation of STAT3 plays a crucial role in colon cancer cells. It was demonstrated that SKN affected the constitutive activation of STAT3 in two types of cancer cell lines. The downregulation of p-STAT3 in response to 10 h of treatment with SKN tended to occur in a concentration-dependent manner (10, 15 and 20 µM) in the HCT116 and SW480 cancer cell lines (Fig. 2C and D). Moreover, JAK1 and JAK2 were activated in response to IL-6; these are potential upstream regulators of STAT3 (52). The inhibitory effects of SKN on the phosphorylation of both JAK1 and JAK2 in the two cell lines were also observed (Fig. 2C and D).
Figure 2.
SKN downregulates p-JAK levels, as well as the constitutive and IL-6-induced activation of p-STAT3 in colon cancer cells. (A) Comparison across 10 analyses based on Oncomine data revealed increased IL-6 transcripts in colon cancer. Blocks in red depict overexpression and blocks in blue represent underexpression. (B) SKN treatment for 3 h prominently inhibited IL-6-induced p-STAT3 expression in both cancer cell lines. (C and D) Western blot analysis revealed that constitutive p-STAT3, p-JAK1 and p-JAK2 levels were decreased following 10 h of treatment with SKN in a concentration-dependent manner. Cells were treated as indicated and quantitative data of proteins of interest were normalized to GAPDH. The quantitative data of the DMSO group (0 µM SKN) were arbitrarily set as 1. All data represent the mean ± SEM. *P<0.05 vs. DMSO group. SKN, shikonin; STAT3, signal transducer and activator of transcription 3; IL, interleukin.
Decreased expression of ADAM17 by SKN
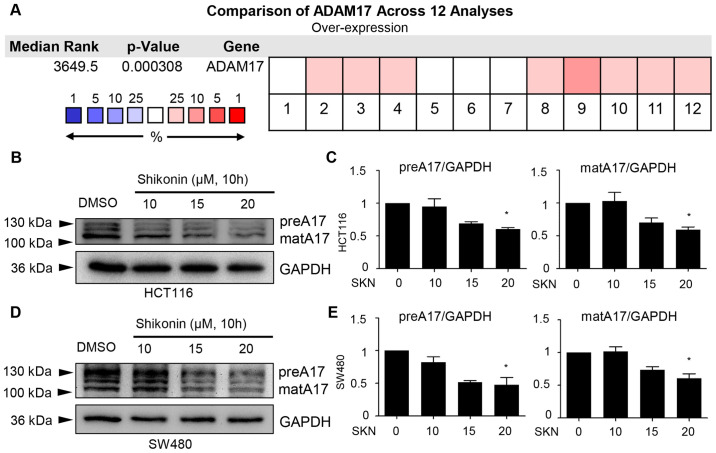
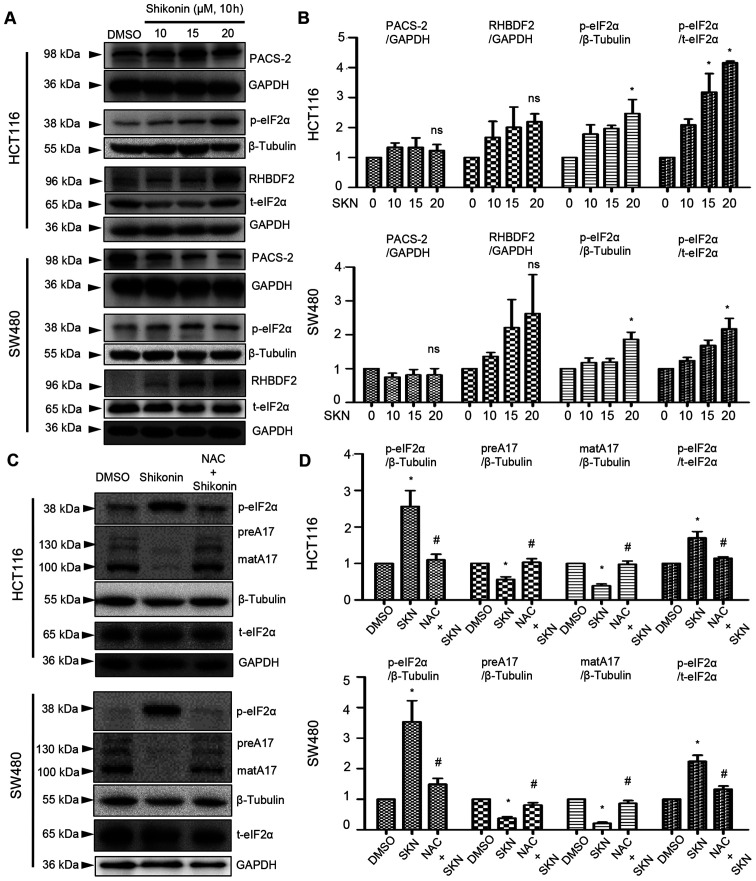
Due to the specific role of ADAM17 in the regulation of IL-6/STAT3 and EGFR signaling, the potential effects of SKN on ADAM17 were further explored. First, a comparison of ADAM17 was performed across 12 analyses, data of which were derived from the Oncomine database. The results mined from this public database revealed the overexpression of ADAM17 mRNA in colon cancer (P=0.000308; Fig. 3A). Of note, it was demonstrated that 10 h of treatment with 20 µM SKN downregulated the expression of both the precursor form and mature form of ADAM17 in the HCT116 and SW480 cancer cell lines (Fig. 3B-E).
Figure 3.
Both the precursor and mature forms of ADAM17 are downregulated by SKN. (A) Comparison across 12 analyses revealed the overexpression of ADAM17 transcripts in colon cancer. Blocks in red depict overexpression and blocks in blue represent underexpression. (B-E) Western blot analysis and quantification revealed that the expression of two forms of ADAM17 was reduced by SKN (concentration as indicated, 10 h of treatment) in HCT116 and SW480 colon cancer cells. Quantitative data of ADAM17 (precursor and mature) were normalized to GAPDH. The quantitative data of the DMSO group (0 µM SKN) were arbitrarily set as 1. All data represent the mean ± SEM. *P<0.05 vs. DMSO group. SKN, shikonin; ADAM17, A disintegrin and metalloproteinase 17; preA17, precursor form of ADAM17; matA17, mature form of ADAM17.
IL-6/STAT3 signaling may not control ADAM17 expression
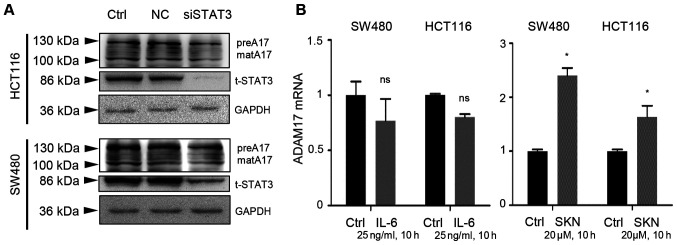
By transfection using a negative control and siSTAT3, it was found that the expression of both the precursor and mature forms of ADAM17 was not markedly altered following STAT3 knockdown, despite a prominent decrease in t-STAT3 expression in the two colon cancer cell lines (Fig. 4A). Additionally, IL-6 did not activate the transcripts of ADAM17 (Fig. 4B, left panel). However, SKN (20 µM, 10 h of treatment) significantly increased the mRNA expression of ADAM17 in the two types of colon cancer cells (Fig. 4B, right panel).
Figure 4.
IL-6/STAT3 signaling does not contribute to the SKN-induced increase in ADAM17 mRNA expression. (A) Transfection with negative control (NC) did not exert an effect on HCT116 or SW480 colon cancer cells. Transfection with siSTAT3 impeded STAT3 expression, but did not reduce the expression of the precursor or mature form ○ ADAM17. (B) IL-6 (25 ng/ml, 10 h of treatment) had no significant effect on ADAM17 mRNA expression in HCT116 or SW480 cells; however, SKN (20 µM, 10 h of treatment) significantly increased the mRNA expression of ADAM17 in the two colon cancer cell lines. The transcripts of ADAM17 in the control group was set as 1. All data represent the mean ± SEM. *P<0.05 vs. control group; ns, no significance. SKN, shikonin; ADAM17, A disintegrin and metalloproteinase 17; preA17, precursor form of ADAM17; matA17, mature form of ADAM17; STAT3, signal transducer and activator of transcription 3; IL, interleukin.
SKN decreases ADAM17 expression via ROS-mediated post-transcriptional regulation
As demonstrated above (Fig. 4B, right panel), 20 µM SKN increased the mRNA expression of ADAM17. However, ADAM17 protein expression was decreased by SKN, as illustrated in Fig. 3B-E. Thus, further analysis was performed involving critical steps in the life cycle of ADAM17. The results of western blot analysis revealed that the expression of PACS-2 was not significantly altered by SKN (Fig. 5A and B), while SKN tended to increase RHBDF2 expression, although not significantly (Fig. 5A and B). The phosphorylation levels of eIF2α were enhanced by SKN (20 µM) in the HCT116 and SW480 colon cancer cells (Fig. 5A and B). ROS are known as inducers of eIF2α phosphorylation, while NAC functions as a ROS scavenger. It was further found that NAC decreased SKN-induced p-eIF2α expression and reversed the SKN-mediated downregulation of ADAM17 protein expression (Fig. 5C and D).
Figure 5.
SKN decreases ADAM17 expression by inducing ROS-mediated p-eIF2α expression. (A and B) Western blot analysis and quantification revealed an increase in p-eIF2α levels by SKN (20 µM). PACS-2 and RHBDF2 expression was not significantly altered by SKN treatment of the HCT116 or SW480 colon cancer cells. (C and D) NAC preserved the SKN-induced downregulation of ADAM17 expression and decreased p-eIF2α expression induced by SKN. Quantitative data of the proteins of interest were normalized to GAPDH or β-tubulin. The quantitative data of the DMSO group (0 µM SKN) were arbitrarily set as 1. All data represent the mean ± SEM. *P<0.05 vs. DMSO group, #P<0.05 vs. SKN group; ns, no significance. SKN, shikonin; ADAM17, A disintegrin and metalloproteinase 17; preA17, precursor form of ADAM17; matA17, mature form of ADAM17; eIF2α, eukaryotic initiation factor 2α; PACS-2, phosphofurin acidic cluster sorting protein 2; RHBDF2, rhomboid family member 2.
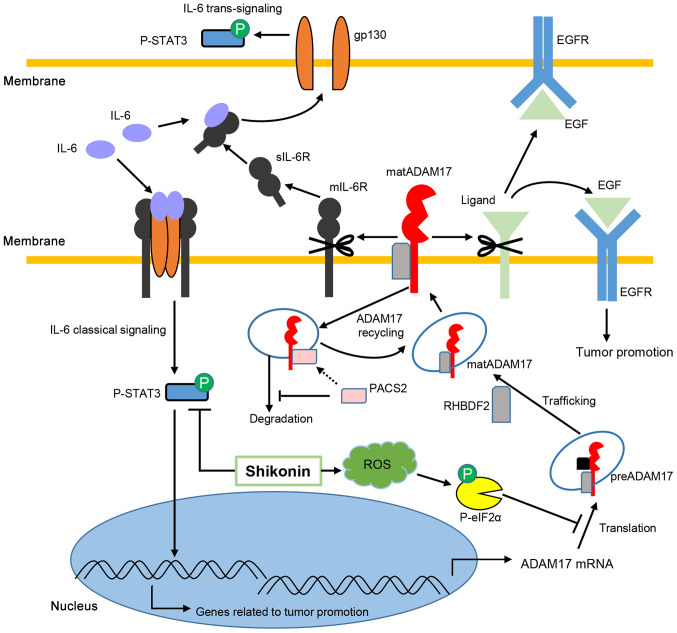
On the whole, IL-6 classical and trans-signaling contribute to cancer development, while ADAM17 cleaves its substrates, including mIL-6R, which controls the balance of two pathways of IL-6 and EGF, allowing EGFR signaling to promote cancer. In the present study, SKN decreased the phosphorylation of STAT3 and suppressed the expression of ADAM17 mediated by ROS-associated p-eIF2α expression in the HCT116 and SW480 colon cancer cells (Fig. 6).
Figure 6.
Potential mechanisms through which shikonin regulates ADAM17 and IL-6/STAT3 signaling. IL-6 binds to membrane IL-6R to activate classical signaling and binds to sIL-6R to transduce trans-signaling, both of which result in STAT3 phosphorylation and contribute to cancer development. ADAM17 mRNA is translated to the precursor form of ADAM17, which is modified to mature ADAM17 mediated by RHBDF2. RHBDF2 also stabilizes mature ADAM17, enabling substrate cleavage. PACS-2 plays an important role in ADAM17 recycling, and PACS-2 deficiency leads to the degradation of mature ADAM17 by endosomes. ADAM17 cleaves mIL-6R to enable IL-6 trans-signaling by releasing more sIL-6R and cleaves EGF, allowing EGFR signaling, to promote cancer. Shikonin decreases p-STAT3 and suppresses the expression of ADAM17 by inducing ROS-mediated p-eIF2α. Translational inhibition by increased p-eIF2α results in ADAM17 defects in HCT116 and SW480 colon cancer cells. Concurrent inhibition of ADAM17 and IL-6/STAT3 signaling may synergistically contribute to the suppression of colon cancer cells in response to shikonin. STAT3, signal transducer and activator of transcription 3; IL, interleukin; A disintegrin and metalloproteinase 17; preA17, precursor form of ADAM17; matA17, mature form of ADAM17; eIF2α, eukaryotic initiation factor 2α; PACS-2, phosphofurin acidic cluster sorting protein 2; RHBDF2, rhomboid family member 2.
Discussion
Although significant advancements in the treatment of colon cancer have been made, further efforts are still required. IL-6/STAT3 signaling and ADAM17 play important roles in colon cancer. Herein, it was demonstrated that the antitumor effects of SKN on colon cancer cells were associated with its inhibition of the IL-6/STAT3 signaling pathway. Moreover, the concomitant suppression of ADAM17 expression may also contribute to the inhibition of cancer cell growth.
As shown using the Oncomine database, transcripts of IL-6 and ADAM17 are increased in colon cancer. IL-6 activates downstream signaling and promotes the transcription of STAT3-related genes, which aids tumor growth. Of note, IL-6R is expressed on specific cell types, such as hepatocytes, epithelial cells and leukocytes, revealing that IL-6 classical signaling is found only in cells expressing IL-6R (53). However, IL-6 trans-signaling may occur in almost all cells where gp130 exists. Moreover, the expression level of gp130 is higher than that of IL-6R (54), allowing the synergistic stimulation of both IL-6 signaling and amplifying the inflammatory cascade in chronic inflammation, which critically contributes to perturbed tissue homeostasis, including tumor development and therapy (55). Notably, ADAM17 not only cleaves proliferative ligands, such as EGF, to trigger downstream signaling and controlling a wide range of cell biological functions, but also modulates the balance between IL-6 classical and trans-signaling pathways. The enhanced activity of ADAM17 produces higher levels of sIL-6R, enabling trans-signaling, which is considered to be associated with the malignant proliferation of epithelial and colon cancer cells (33,56). The benefit conferred by anti-human IL-6 receptor monoclonal antibody has been documented with downregulated colon stem markers and increased chemosensitivity (57). Therefore, simultaneously elevated levels of IL-6 signaling and ADAM17 may result in increased p-STAT3 levels in a wide set of cell types.
On the one hand, STAT3 activation has been documented to be associated with a poor prognosis of patients with in stage II colon cancer (58). On the other hand, IL-6/STAT3 signaling, regulated by secretory RAB GTPase 3C (RAB3C), leads to a poor prognosis of patients with an advanced pathological stage and distant metastasis (59). Moreover, the IL-6 rs2069837 genotype has been documented as a clinically relevant prognostic factor in colon cancer patients treated with bevacizumab-based chemotherapy (60), all of which indicate that IL-6/STAT3 signaling activity may have clinical significance. However, to date, studies on the prognostic role of ADAM17 in cohorts appear to lack abundant evidence. Although ADAM17 functions as an oncogene and its expression is increased in patients with colon cancer, as previously reported (61) and as shown in Oncomine, the activity of ADAM17 is difficult to detect in patients. Further data are still required to disclose the role of ADAM17 in cancer in detail.
In previous studies, the authors found that raloxifene targeted the interface of IL-6 and gp130 (62), inhibiting p-STAT3 and colon cancer cell growth (23). SKN prominently suppressed p-STAT3 expression in colon cancer cells. The underlying mechanisms may include the predicted binding of SKN with STAT3 in the pY-X and pY+0 sub-pockets located in the SH2 domains of STAT3, through which STAT3 dimers are formed to facilitate subsequent phosphorylation and translocation into the cell nucleus (63). As aforementioned, ADAM17 is considered an oncogene, and targeting ADAM17 represents another strategy for the treatment of cancer. However, to date, different STAT3 inhibitors are being developed (e.g., FLLL11, FLLL12 and LY5) (64,65), however, few agents to inhibit ADAM17 expression are available. The majority of compounds targeting ADAM17 involve catalytic sites and decrease the cleavage activity. Li et al (66) found that ZLDI-8 (ADAM17 inhibitor) downregulated the expression of ADAM17 and synergistically promoted the antitumor effects of 5-fluorouracil on colon cancer. Regardless, the comprehensive inhibitory effect on both constitutive p-STAT3, IL-6-induced p-STAT3 and ADAM17 expression may indicate a potential broader spectrum of the antitumor effects of SKN on colon cancer cells. Moreover, SKN did not tend to alter the expression of β-catenin, indicating that β-catenin-dependent tumorigenesis may not significantly contribute to the inhibitory effects of SKN on STAT3 and ADAM17 (Fig. S3).
As illustrated in Fig. 4, the expression of ADAM17 was not significantly altered by STAT3 knockdown in either colon cancer cell line examined. Moreover, transcripts of ADAM17 were not associated with IL-6 administration in these two cell lines, suggesting that the basal expression of ADAM17 may be independent of IL-6/STAT3 signaling in the HCT116 and SW480 cells. As previously reported (33), ADAM17 enables IL-6 trans-signaling and EGF signaling by mIL-6R and EGF cleavage, possibly promoting STAT3 activation in a wide range of cell types, which may not increase the transcripts of ADAM17 in turn; however, whether activated IL-6/STAT3 signaling enhances the activity of this cleavage enzyme is unknown. In addition to the other obscure potential mechanisms, the association between STAT3 and ADAM17 warrants further investigation in order to be fully elucidated. Of note, in contrast to the downregulated protein expression of ADAM17, SKN increased the mRNA expression of ADAM17. Therefore, post-transcriptional regulation has attracted increasing attention.
ADAM17 mRNA is driven to produce the precursor form of ADAM17. RHBDF2 binds to the immature form of ADAM17 and promotes its trafficking from the endoplasmic reticulum to the Golgi apparatus. During this process, the precursor form of ADAM17 is cleaved by furin to generate mature ADAM17. RHBDF2 contributes to the post-translational regulation of ADAM17 in almost every step, including trafficking, maturation, membrane stabilization and degradation (31). Of note, the amino terminus of RHBDF2 is critical in this reported biological process (31); thus, herein, an antibody targeting the N-terminal region of human RHBDF2 was used to detect its expression level. On the other hand, PACS-2 has been reported to divert endocytosed mature ADAM17 away from degradation pathways, resulting in an increased ADAM17 membrane availability. In the absence of PACS-2, the disrupted ADAM17 recycling leads to decreased expression levels of membrane mature ADAM17 (30). In the present study, SKN treatment did not significantly alter the expression of PACS-2 and appeared to increase detectable RHBDF2 levels in the HCT116 and SW480 cells. In fact, the expression levels of precursor and mature ADAM17 were both downregulated by SKN, which revealed that SKN may not exert a prominent inhibitory effect on the maturation and degradation process in these two cancer cell lines.
Notably, SKN significantly increased the levels of p-eIF2α. In brief, eIF2 delivers the initiator methionyl tRNA (Met-tRNAiMet) to the 40S subunit of the ribosome to form the 43S complex, permitting subsequent translation (67). The phosphorylation of the α-subunit of eIF2 blocks translational initiation and attenuates general translation, which is considered to help cells overcome extracellular stress by easing the burden on protein folding and amino acid consumption. Oxidative stress, viral infection, amino acid deprivation and endoplasmic reticulum stress (ERS)-mediated PERK activation are considered inducers of α-subunit phosphorylation (68). Previous studies have found that SKN is capable of inducing oxidative stress in colon cancer cells (69) and activates ERS, leading to PERK activation in SNU-407 cells (another colon cancer cell line) (70). In the present study, NAC, a ROS scavenger, reversed the expression of ADAM17 and p-eIF2α in both cell lines, indicating that ROS-induced p-eIF2α expression contributes to the decreased expression of ADAM17 induced by SKN. However, eIF2α-associated general translation inhibition is only the first step of translation, and of note, the suppressive effect of SKN was observed at a concentration of 10 µM (Fig. 1), while 10 µM SKN appeared to exert no overt effect on p-eIF2α (Fig. 5A and B) or ADAM17 (Fig. 3B-E) expression. This suggests that the inhibition of cancer cells may involve other mechanisms where a low concentration of SKN was used. Overall, the antitumor effect of shikonin is likely more intricate than what is currently known.
Therefore, on the whole, IL-6 classical and trans-signaling contribute to cancer development, while ADAM17 cleaves its substrates, including mIL-6R, which controls the balance of two pathways of IL-6 and EGF, allowing EGFR signaling to promote cancer. In the present study, SKN decreased the phosphorylation of STAT3 and suppressed the expression of ADAM17 mediated by ROS-associated p-eIF2α expression in the HCT116 and SW480 colon cancer cells (Fig. 6). Shikonin shares the same backbone with LLL-12, which has been previously described as an effective STAT3 inhibitor (21,63), indicating the promise of developing SKN and its derivatives.
However, similar to other natural products in Chinese herbal medicine, the potential toxicity and unknown side-effects of SKN merit accelerated investigations. Indeed, SKN can affect multiple biological processes, including protein translation, folding, modification and translocation, as well as exosome secretion and energy production (71). To date, the antitumor properties of SKN have been demonstrated in several in vivo studies on colon cancer (71-73). Moreover, it has been proven that SKN functions as a complement to enhance the antitumor effect of cisplatin in colon cancer in vivo (74). The present study also examined the effects of SKN on non-cancer cells (NCM460 cells), as described in Data S1 and as shown in Fig. S4. Although 15 and 20 µM SKN decreased cell viability, SKN appeared to exhibit a weaker toxicity in human normal colonic epithelial cells (NCM460) than in colon cancer cells (please see Fig. 1A). Importantly, SKN has been reported to decrease tumor volume and exert antitumor effects with minimal toxicity to non-cancer cells without liver or kidney injury in xenograft tumor models (69,71). Although the present study did not perform animal experiments, the authors aimed to determine the potential effects of SKN on STAT3 and ADAM17 expression in vitro, which may provide evidence of the use of SKN under certain circumstances in which STAT3 and/or ADAM17 overactivation occurs. In the era of modern medicine, combined therapy is the current trend. Due to its pleiotropic effects on cancer, SKN may be studied in further investigations where it is administered in combination with other known chemo- therapies. Moreover, although the present study demonstrated that SKN-induced p-eIF2α expression was decreased by the ROS scavenger, NAC, which may disrupt general translation, resulting in the decreased expression of ADAM17, the alteration in transcripts of other oncogenes remains unknown in response to SKN. Furthermore, RHBDF2 knockdown warrants further investigations in order to elucidate the regulatory mechanisms involved in protein processing. Further studies are also required to determine the potential effects of SKN on the components constituting transcription factor complexes, the stability of mRNA, and the regulation of other types of RNAs.
Supplementary Data
Acknowledgments
Not applicable.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant nos. 82070396 and 81974032), the Hubei Province Health and Family Planning Scientific Research Project (grant no. WJ2019M120), the Science and Technology Project Foundation of Wuhan (grant no. 2019020701011439) and the Natural Science Foundation of Hubei Province (grant no. 2019CFB668).
Availability of data and materials
All the datasets generated and analyzed in the present study are included in this article.
Authors' contributions
WS and LM designed the study and wrote the manuscript. WS, LM, XP, DP, SH, PL and MW performed the biological molecular experiments. TJ, LM and JG assisted with data analysis. WS, YJ and LP interpreted the data and revised the manuscript. LL, SL and JL were involved in the conception and design of the study, and provided extensive support and reviewed the manuscript. SL and JL confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363–385. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, Gavin A, Visser O, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356–387. doi: 10.1016/j.ejca.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Den Oudsten BL, Traa MJ, Thong MS, Martijn H, De Hingh IH, Bosscha K, van de Poll-Franse LV. Higher prevalence of sexual dysfunction in colon and rectal cancer survivors compared with the normative population: A population-based study. Eur J Cancer. 2012;48:3161–3170. doi: 10.1016/j.ejca.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Liu L, Herrinton LJ, Hornbrook MC, Wendel CS, Grant M, Krouse RS. Early and late complications among long-term colorectal cancer survivors with ostomy or anastomosis. Dis Colon Rectum. 2010;53:200–212. doi: 10.1007/DCR.0b013e3181bdc408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schover LR, van der Kaaij M, van Dorst E, Creutzberg C, Huyghe E, Kiserud CE. Sexual dysfunction and infertility as late effects of cancer treatment. EJC Suppl. 2014;12:41–53. doi: 10.1016/j.ejcsup.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammad N, Malvi P, Meena AS, Singh SV, Chaube B, Vannuruswamy G, Kulkarni MJ, Bhat MK. Cholesterol depletion by methyl-β-cyclodextrin augments tamoxifen induced cell death by enhancing its uptake in melanoma. Mol Cancer. 2014;13:204. doi: 10.1186/1476-4598-13-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohammad N, Singh SV, Malvi P, Chaube B, Athavale D, Vanuopadath M, Nair SS, Nair B, Bhat MK. Strategy to enhance efficacy of doxorubicin in solid tumor cells by methyl-β-cyclodextrin: Involvement of p53 and Fas receptor ligand complex. Sci Rep. 2015;5:11853. doi: 10.1038/srep11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rashmi R, Jayachandran K, Zhang J, Menon V, Muhammad N, Zahner M, Ruiz F, Zhang S, Cho K, Wang Y, et al. Glutaminase inhibitors induce Thiol-mediated oxidative stress and radiosensitization in treatment-resistant cervical cancers. Mol Cancer Ther. 2020;19:2465–2475. doi: 10.1158/1535-7163.MCT-20-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh S, Chouhan S, Mohammad N, Bhat MK. Resistin causes G1 arrest in colon cancer cells through upregulation of SOCS3. FEBS Lett. 2017;591:1371–1382. doi: 10.1002/1873-3468.12655. [DOI] [PubMed] [Google Scholar]
- 11.Kumar B, Chand V, Ram A, Usmani D, Muhammad N. Oncogenic mutations in tumorigenesis and targeted therapy in breast cancer. Curr Mol Biol Rep. 2020;6:116–125. doi: 10.1007/s40610-020-00136-x. [DOI] [Google Scholar]
- 12.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 13.Unver N, McAllister F. IL-6 family cytokines: Key inflammatory mediators as biomarkers and potential therapeutic targets. Cytokine Growth Factor Rev. 2018;41:10–17. doi: 10.1016/j.cytogfr.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cron L, Allen T, Febbraio MA. The role of gp130 receptor cytokines in the regulation of metabolic homeostasis. J Exp Biol. 2016;219:259–265. doi: 10.1242/jeb.129213. [DOI] [PubMed] [Google Scholar]
- 15.Rose-John S, Heinrich PC. Soluble receptors for cytokines and growth factors: Generation and biological function. Biochem J. 1994;300:281–290. doi: 10.1042/bj3000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 17.Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, Nam S, Eweis I, Diaz N, Sullivan D, et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12:11–19. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]
- 18.Ma XT, Wang S, Ye YJ, Du RY, Cui ZR, Somsouk M. Constitutive activation of Stat3 signaling pathway in human colorectal carcinoma. World J Gastroenterol. 2004;10:1569–1573. doi: 10.3748/wjg.v10.i11.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corvinus FM, Orth C, Moriggl R, Tsareva SA, Wagner S, Pfitzner EB, Baus D, Kaufmann R, Huber LA, Zatloukal K, et al. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia. 2005;7:545–555. doi: 10.1593/neo.04571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusaba T, Nakayama T, Yamazumi K, Yakata Y, Yoshizaki A, Nagayasu T, Sekine I. Expression of p-STAT3 in human colorectal adenocarcinoma and adenoma; correlation with clinicopathological factors. J Clin Pathol. 2005;58:833–838. doi: 10.1136/jcp.2004.023416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin L, Liu A, Peng Z, Lin HJ, Li PK, Li C, Lin J. STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res. 2011;71:7226–7237. doi: 10.1158/0008-5472.CAN-10-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin L, Fuchs J, Li C, Olson V, Bekaii-Saab T, Lin J. STAT3 signaling pathway is necessary for cell survival and tumorsphere forming capacity in ALDH+/CD133+ stem cell-like human colon cancer cells. Biochem Biophys Res Commun. 2011;416:246–251. doi: 10.1016/j.bbrc.2011.10.112. [DOI] [PubMed] [Google Scholar]
- 23.Shi W, Yan D, Zhao C, Xiao M, Wang Y, Ma H, Liu T, Qin H, Zhang C, Li C, et al. Inhibition of IL-6/STAT3 signaling in human cancer cells using Evista. Biochem Biophys Res Commun. 2017;491:159–165. doi: 10.1016/j.bbrc.2017.07.067. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Zhao C, Jou D, Lü J, Zhang C, Lin L, Lin J. Ursolic acid inhibits the growth of colon cancer-initiating cells by targeting STAT3. Anticancer Res. 2013;33:4279–4284. [PubMed] [Google Scholar]
- 25.Zhao C, Wang W, Yu W, Jou D, Wang Y, Ma H, Xiao H, Qin H, Zhang C, Lü J, et al. A novel small molecule STAT3 inhibitor, LY5, inhibits cell viability, colony formation, and migration of colon and liver cancer cells. Oncotarget. 2016;7:12917–12926. doi: 10.18632/oncotarget.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ung N, Putoczki TL, Stylli SS, Ng I, Mariadason JM, Chan TA, Zhu HJ, Luwor RB. Anti-EGFR therapeutic efficacy correlates directly with inhibition of STAT3 activity. Cancer Biol Ther. 2014;15:623–632. doi: 10.4161/cbt.28179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 28.Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, Clay WC, Didsbury JR, Hassler D, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature. 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 29.Khokha R, Murthy A, Weiss A. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat Rev Immunol. 2013;13:649–665. doi: 10.1038/nri3499. [DOI] [PubMed] [Google Scholar]
- 30.Dombernowsky SL, Samsøe-Petersen J, Petersen CH, Instrell R, Hedegaard AM, Thomas L, Atkins KM, Auclair S, Albrechtsen R, Mygind KJ, et al. The sorting protein PACS-2 promotes ErbB signalling by regulating recycling of the metalloproteinase ADAM17. Nat Commun. 2015;6:7518. doi: 10.1038/ncomms8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grieve AG, Xu H, Künzel U, Bambrough P, Sieber B, Freeman M. Phosphorylation of iRhom2 at the plasma membrane controls mammalian TACE-dependent inflammatory and growth factor signalling. eLife. 2017;6:6. doi: 10.7554/eLife.23968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavlenko E, Cabron AS, Arnold P, Dobert JP, Rose-John S, Zunke F. Functional characterization of colon cancer-associated mutations in ADAM17: Modifications in the pro-domain interfere with trafficking and maturation. Int J Mol Sci. 2019;20:20. doi: 10.3390/ijms20092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schumacher N, Rose-John S. ADAM17 activity and IL-6 trans-signaling in inflammation and cancer. Cancers (Basel) 2019;11:11. doi: 10.3390/cancers11111736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mustafi R, Dougherty U, Mustafi D, Ayaloglu-Butun F, Fletcher M, Adhikari S, Sadiq F, Meckel K, Haider HI, Khalil A, et al. ADAM17 is a tumor promoter and therapeutic target in western diet-associated colon cancer. Clin Cancer Res. 2017;23:549–561. doi: 10.1158/1078-0432.CCR-15-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muhammad N, Steele R, Isbell TS, Philips N, Ray RB. Bitter melon extract inhibits breast cancer growth in preclinical model by inducing autophagic cell death. Oncotarget. 2017;8:66226–66236. doi: 10.18632/oncotarget.19887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharya S, Muhammad N, Steele R, Kornbluth J, Ray RB. Bitter melon enhances natural killer-mediated toxicity against head and neck cancer cells. Cancer Prev Res (Phila) 2017;10:337–344. doi: 10.1158/1940-6207.CAPR-17-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhattacharya S, Muhammad N, Steele R, Peng G, Ray RB. Immunomodulatory role of bitter melon extract in inhibition of head and neck squamous cell carcinoma growth. Oncotarget. 2016;7:33202–33209. doi: 10.18632/oncotarget.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu T, Ma H, Shi W, Duan J, Wang Y, Zhang C, Li C, Lin J, Li S, Lv J, et al. Inhibition of STAT3 signaling pathway by ursolic acid suppresses growth of hepatocellular carcinoma. Int J Oncol. 2017;51:555–562. doi: 10.3892/ijo.2017.4035. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Cui JH, Meng QQ, Li SS, Zhou W, Xiao S. Advance in anti-tumor mechanisms of shikonin, alkannin and their derivatives. Mini Rev Med Chem. 2018;18:164–172. doi: 10.2174/1389557517666170228114809. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Yang L, Oppenheim JJ, Howard MZ. Cellular pharmacology studies of shikonin derivatives. Phytother Res. 2002;16:199–209. doi: 10.1002/ptr.1100. [DOI] [PubMed] [Google Scholar]
- 41.Guo C, He J, Song X, Tan L, Wang M, Jiang P, Li Y, Cao Z, Peng C. Pharmacological properties and derivatives of shikonin-A review in recent years. Pharmacol Res. 2019;149:104463. doi: 10.1016/j.phrs.2019.104463. [DOI] [PubMed] [Google Scholar]
- 42.Boulos JC, Rahama M, Hegazy MF, Efferth T. Shikonin derivatives for cancer prevention and therapy. Cancer Lett. 2019;459:248–267. doi: 10.1016/j.canlet.2019.04.033. [DOI] [PubMed] [Google Scholar]
- 43.Tang JC, Ren YG, Zhao J, Long F, Chen JY, Jiang Z. Shikonin enhances sensitization of gefitinib against wild-type EGFR non-small cell lung cancer via inhibition PKM2/stat3/cyclinD1 signal pathway. Life Sci. 2018;204:71–77. doi: 10.1016/j.lfs.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Thakur R, Trivedi R, Rastogi N, Singh M, Mishra DP. Inhibition of STAT3, FAK and Src mediated signaling reduces cancer stem cell load, tumorigenic potential and metastasis in breast cancer. Sci Rep. 2015;5:10194. doi: 10.1038/srep10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao HH, Liu DY, Lai YC, Chen YY, Yu LZ, Shao M, Liu JS. Inhibition of the STAT3 signaling pathway contributes to the anti-melanoma activities of shikonin. Front Pharmacol. 2020;11:748. doi: 10.3389/fphar.2020.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian R, Li Y, Gao M. Shikonin causes cell-cycle arrest and induces apoptosis by regulating the EGFR-NF-κB signalling pathway in human epidermoid carcinoma A431 cells. Biosci Rep. 2015;35:35. doi: 10.1042/BSR20150002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Zou S, Tong Q, Liu B, Huang W, Tian Y, Fu X. Targeting STAT3 in cancer immunotherapy. Mol Cancer. 2020;19:145. doi: 10.1186/s12943-020-01258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chalikonda G, Lee H, Sheik A, Huh YS. Targeting key transcriptional factor STAT3 in colorectal cancer. Mol Cell Biochem. 2021;476:3219–3228. doi: 10.1007/s11010-021-04156-8. [DOI] [PubMed] [Google Scholar]
- 49.Scheller J, Chalaris A, Garbers C, Rose-John S. ADAM17: A molecular switch to control inflammation and tissue regeneration. Trends Immunol. 2011;32:380–387. doi: 10.1016/j.it.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Shi W, Ma H, Liu T, Yan D, Luo P, Zhai M, Tao J, Huo S, Guo J, Li C, et al. Inhibition of Interleukin-6/glycoprotein 130 signalling by Bazedoxifene ameliorates cardiac remodelling in pressure overload mice. J Cell Mol Med. 2020;24:4748–4761. doi: 10.1111/jcmm.15147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, van Boxel-Dezaire AH, Cheon H, Yang J, Stark GR. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc Natl Acad Sci USA. 2013;110:16975–16980. doi: 10.1073/pnas.1315862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 53.Rose-John S. IL-6 trans-signaling via the soluble IL-6 receptor: Importance for the pro-inflammatory activities of IL-6. Int J Biol Sci. 2012;8:1237–1247. doi: 10.7150/ijbs.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peters M, Blinn G, Solem F, Fischer M, Meyer zum Büschenfelde KH, Rose-John S. In vivo and in vitro activities of the gp130-stimulating designer cytokine Hyper-IL-6. J Immunol. 1998;161:3575–3581. [PubMed] [Google Scholar]
- 55.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: Crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 56.Chalaris A, Garbers C, Rabe B, Rose-John S, Scheller J. The soluble Interleukin 6 receptor: Generation and role in inflammation and cancer. Eur J Cell Biol. 2011;90:484–494. doi: 10.1016/j.ejcb.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 57.Ying J, Tsujii M, Kondo J, Hayashi Y, Kato M, Akasaka T, Inoue T, Shiraishi E, Inoue T, Hiyama S, et al. The effectiveness of an anti-human IL-6 receptor monoclonal antibody combined with chemotherapy to target colon cancer stem-like cells. Int J Oncol. 2015;46:1551–1559. doi: 10.3892/ijo.2015.2851. [DOI] [PubMed] [Google Scholar]
- 58.Cross-Knorr S, Lu S, Perez K, Guevara S, Brilliant K, Pisano C, Quesenberry PJ, Resnick MB, Chatterjee D. RKIP phosphorylation and STAT3 activation is inhibited by oxaliplatin and camptothecin and are associated with poor prognosis in stage II colon cancer patients. BMC Cancer. 2013;13:463. doi: 10.1186/1471-2407-13-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang YC, Su CY, Chen MH, Chen WS, Chen CL, Hsiao M. Secretory RAB GTPase 3C modulates IL6-STAT3 pathway to promote colon cancer metastasis and is associated with poor prognosis. Mol Cancer. 2017;16:135. doi: 10.1186/s12943-017-0687-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsusaka S, Hanna DL, Cao S, Zhang W, Yang D, Ning Y, Sunakawa Y, Okazaki S, Berger MD, Miyamato Y, et al. Prognostic impact of IL6 genetic variants in patients with metastatic colorectal cancer treated with bevacizumab-based chemotherapy. Clin Cancer Res. 2016;22:3218–3226. doi: 10.1158/1078-0432.CCR-15-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walkiewicz K, Kozieł P, Bednarczyk M, Błażelonis A, Mazurek U, Muc-Wierzgoń M. Expression of migration-related genes in human colorectal cancer and activity of a disintegrin and metalloproteinase 17. BioMed Res Int. 2016;2016:8208904. doi: 10.1155/2016/8208904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Xiao H, Lin L, Jou D, Kumari V, Lin J, Li C. Drug design targeting protein-protein interactions (PPIs) using multiple ligand simultaneous docking (MLSD) and drug repositioning: Discovery of raloxifene and bazedoxifene as novel inhibitors of IL-6/GP130 interface. J Med Chem. 2014;57:632–641. doi: 10.1021/jm401144z. [DOI] [PubMed] [Google Scholar]
- 63.Qiu HY, Zhu X, Luo YL, Lin HY, Tang CY, Qi JL, Pang YJ, Yang RW, Lu GH, Wang XM, et al. Identification of new shikonin derivatives as antitumor agents targeting STAT3 SH2 domain. Sci Rep. 2017;7:2863. doi: 10.1038/s41598-017-02671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin L, Hutzen B, Ball S, Foust E, Sobo M, Deangelis S, Pandit B, Friedman L, Li C, Li PK, et al. New curcumin analogues exhibit enhanced growth-suppressive activity and inhibit AKT and signal transducer and activator of transcription 3 phosphorylation in breast and prostate cancer cells. Cancer Sci. 2009;100:1719–1727. doi: 10.1111/j.1349-7006.2009.01220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao H, Bid HK, Jou D, Wu X, Yu W, Li C, Houghton PJ, Lin J. A novel small molecular STAT3 inhibitor, LY5, inhibits cell viability, cell migration, and angiogenesis in medulloblastoma cells. J Biol Chem. 2015;290:3418–3429. doi: 10.1074/jbc.M114.616748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li DD, Zhao CH, Ding HW, Wu Q, Ren TS, Wang J, Chen CQ, Zhao QC. A novel inhibitor of ADAM17 sensitizes colorectal cancer cells to 5-fluorouracil by reversing Notch and epithelial-mesenchymal transition in vitro and in vivo. Cell Prolif. 2018;51:e12480. doi: 10.1111/cpr.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ryoo HD, Vasudevan D. Two distinct nodes of translational inhibition in the Integrated Stress Response. BMB Rep. 2017;50:539–545. doi: 10.5483/BMBRep.2017.50.11.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang W, Cui J, Zhang K, Xi H, Cai A, Li J, Gao Y, Hu C, Liu Y, Lu Y, et al. Shikonin induces ROS-based mitochondria-mediated apoptosis in colon cancer. Oncotarget. 2017;8:109094–109106. doi: 10.18632/oncotarget.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han X, Kang KA, Piao MJ, Zhen AX, Hyun YJ, Kim HM, Ryu YS, Hyun JW. Shikonin exerts cytotoxic effects in human colon cancers by inducing apoptotic cell death via the endoplasmic reticulum and mitochondria-mediated pathways. Biomol Ther (Seoul) 2019;27:41–47. doi: 10.4062/biomolther.2018.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Y, Ni J, Gao Y, Zhang J, Liu X, Chen Y, Chen Z, Wu Y. Integrated proteomics and metabolomics reveals the comprehensive characterization of antitumor mechanism underlying Shikonin on colon cancer patient-derived xenograft model. Sci Rep. 2020;10:14092. doi: 10.1038/s41598-020-71116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Y, Si L, Zhang J, Yu H, Liu X, Chen Y, Wu Y. Uncovering the antitumor effects and mechanisms of Shikonin against colon cancer on comprehensive analysis. Phytomedicine. 2021;82:153460. doi: 10.1016/j.phymed.2021.153460. [DOI] [PubMed] [Google Scholar]
- 73.Li MY, Mi C, Wang KS, Wang Z, Zuo HX, Piao LX, Xu GH, Li X, Ma J, Jin X. Shikonin suppresses proliferation and induces cell cycle arrest through the inhibition of hypoxia-inducible factor-1α signaling. Chem Biol Interact. 2017;274:58–67. doi: 10.1016/j.cbi.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 74.He G, He G, Zhou R, Pi Z, Zhu T, Jiang L, Xie Y. Enhancement of cisplatin-induced colon cancer cells apoptosis by shikonin, a natural inducer of ROS in vitro and in vivo. Biochem Biophys Res Commun. 2016;469:1075–1082. doi: 10.1016/j.bbrc.2015.12.100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the datasets generated and analyzed in the present study are included in this article.