Abstract
The multienzyme complex prolyl 4-hydroxylase catalyzes the hydroxylation of proline residues and acts as a chaperone during collagen synthesis in multicellular organisms. The β subunit of this complex is identical to protein disulfide isomerase (PDI). The free-living nematode Caenorhabditis elegans is encased in a collagenous exoskeleton and represents an excellent model for the study of collagen biosynthesis and extracellular matrix formation. In this study, we examined prolyl 4-hydroxylase α-subunit (PHY; EC 1.14.11.2)- and β-subunit (PDI; EC 5.3.4.1)-encoding genes with respect to their role in collagen modification and formation of the C. elegans exoskeleton. We identified genes encoding two PHYs and a single associated PDI and showed that all three are expressed in collagen-synthesizing ectodermal cells at times of maximal collagen synthesis. Disruption of the pdi gene via RNA interference resulted in embryonic lethality. Similarly, the combined phy genes are required for embryonic development. Interference with phy-1 resulted in a morphologically dumpy phenotype, which we determined to be identical to the uncharacterized dpy-18 locus. Two dpy-18 mutant strains were shown to have null alleles for phy-1 and to have a reduced hydroxyproline content in their exoskeleton collagens. This study demonstrates in vivo that this enzyme complex plays a central role in extracellular matrix formation and is essential for normal metazoan development.
The Caenorhabditis elegans exoskeleton (or cuticle) is a true extracellular matrix (ECM) (21) that is predominantly composed of highly cross-linked collagens (8). This exoskeleton, which is synthesized from the ectoderm (hypodermis) five times during nematode development, is responsible for the maintenance of postembryonic body shape, protection from the environment, and locomotion via opposed muscles (21). Approximately 1% of the C. elegans genome encodes cuticle collagens, representing over 150 small collagen genes (7), which encode short interrupted collagens most like vertebrate FACIT type IX cartilage collagens (30). This complex mixture of collagens constitutes >80% of the proteins in this resilient matrix (17). Mutations in individual cuticle collagen genes can result in dramatic morphological defects in this nematode, as seen for the dumpy, blister, squat, and roller mutations (21).
Prolyl 4-hydroxylase (EC 1.14.11.2) is an endoplasmic reticulum (ER) enzyme responsible for the co- and posttranslational hydroxylation of proline in the Xaa-Pro-Gly repeats of procollagen. In vitro studies have demonstrated that 4-hydroxyproline is required for the thermal stability of the folded triple helix at physiological temperatures (19). An additional function of this enzyme is to act as a chaperone by retaining unfolded procollagen chains in the ER, releasing them for secretion only when they have folded correctly (37). In humans and other vertebrates, two isoforms of prolyl 4-hydroxylase exist (type I and type II) (19); these associate with a single β subunit to form a catalytically active [α(I)]2β2 or [α(II)]2β2 tetramer (1), with catalytic activity residing in the α subunits of the complex. The β subunit is identical to protein disulfide isomerase (PDI; EC 5.3.4.1) (28). The role of PDI in the complex is not related to its disulfide isomerase activity but is hypothesized to retain the α subunits in the ER (36) in a catalytically active, nonaggregated form (14). On its own, PDI plays additional roles in procollagen biosynthesis, including disulfide bond formation and molecular chaperone functions (9).
Two conserved genes for prolyl 4-hydroxylase α subunits, referred to as phy-1 and phy-2, are expressed in the model organism C. elegans. Two potential β-subunit-encoding genes, called pdi-1 and pdi-2 (for protein disulfide isomerase), are also present. Since determination of the genome sequence for this free-living nematode is essentially complete (7), these genes represent the full complement of conserved prolyl 4-hydroxylase complex genes. Coexpression of the recombinant C. elegans PHY-1 subunit with C. elegans PDI-2 in insect cells revealed that the active enzyme forms an αβ dimer (34, 35), rather than the more common α2β2 tetramer of vertebrates (1) and Drosophila (2). The catalytic properties of the C. elegans dimer are similar to those of the vertebrate type II tetramer, both being relatively insensitive to inhibition by poly(l-proline) (34). PHY-1 also forms an active recombinant enzyme when coexpressed with human PDI polypeptide, but again the enzyme is an αβ dimer (35). In addition, an active type I prolyl 4-hydroxylase complex is formed from the coexpression of C. elegans PDI-2 and the human α(I) subunit, the resulting enzyme being an α2β2 tetramer. This finding demonstrates that the formation of a tetramer or dimer is dependent on the properties of the PHY-1 α subunit. The second PDI isoform from C. elegans, PDI-1, homodimerizes and does not form an active enzyme complex with either the PHY-1 or the human α(I) subunit (34). The pdi-1 gene is not considered in this analysis. Temporal and spatial expression patterns for pdi-1 are consistent with a role in collagen biosynthesis (24). However, based on coexpression studies and the genomic organization of this gene in a functionally conserved operon (24, 25), a prolyl 4-hydroxylase-independent role is suggested for the PDI-1 isoform.
In this study, we examined the function and expression of prolyl 4-hydroxylase and associated PDI class of procollagen-modifying enzymes in C. elegans and assessed their role in the formation of the cuticular ECM. Disruption of prolyl 4-hydroxylase activity via RNA interference (RNAi) resulted in embryonic lethality, a direct result of the cuticle being unable to maintain normal worm morphology. Single disruption of phy-1 yielded viable nematodes with altered body morphology, consistent with cuticle collagen defects. This study reveals that these enzymes are essential for development and morphology. phy-1 was shown to correspond to the uncharacterized dpy-18 locus (dumpy, short fat phenotype). This represents the first reported example of a prolyl 4-hydroxylase mutant and firmly establishes an essential role for this class of enzymes in the formation of ECMs.
MATERIALS AND METHODS
C. elegans strains and culture conditions.
C. elegans strains were cultured as described elsewhere (6). Strains used in this study were the wild-type Bristol N2 strain, dpy-10 mutant strain CB128 (e128 II), unc-76 mutant strain DR96 (unc-76 e911 V), and dpy-18 mutant strains CB364 (dpy-18 e364 III) and CB2590 (tra-1 e1099/dpy-18 e1096 III). The Caenorhabditis Genetics Center provided these strains. Cloning of dumpy nematodes from strain CB2590 gave dpy-18 (e1096).
Genomic DNA isolation and generation of cDNA.
Genomic DNA was prepared as described previously (26). Total RNA was isolated from C. elegans N2 mixed-stage cultures with TRIzol reagent (Gibco BRL) by following the manufacturer's recommendations. A Poly(A)quik mRNA isolation kit (Stratagene) was used to prepare mRNA from which cDNA was produced with a 1st strand cDNA synthesis kit (Amersham) by following the manufacturer's instructions.
dsRNA-mediated interference.
The RNAi procedure followed is described by Fire and coworkers (11). cDNA clones for the in vitro transcription of phy-1 (Z81134), phy-2 (Z69637), and pdi-2 (U41542) were generated by PCR on mixed-stage cDNA with Taq polymerase (Applied Biosystems). Full-length clones for each gene (minus the signal peptide-encoding regions) were produced using the following primer combinations (artificial restriction enzyme sites are in lowercase and underlined): phy-1, Phy-1F (EcoRI), sense, 5′ ggcgaattcGATCTGTTCACCTCGATTGC 3′, and Phy-1R (PstI), antisense, 5′ ggcctgcagTTAGAGGGTCTCCCAGACGT 3′; phy-2, Phy-2F (XbaI), sense, 5′ ggctctagaGATTTGTTCACTGCAATTGC 3′, and Phy-2R (PstI), antisense, 5′ ggcctgcagCTATGGATCATTGGCATATG 3′; and pdi-2, Pdi-2F (XmnI), sense, 5′ ggcgaaggatttcGCCGTCATTGAAGAAGAAGAG 3′, and Pdi-2R (PstI), antisense, 5′ ggcctgcagTTAGAGCTCGGTGTGTCCCT 3′. Products were ligated into the pPCR Script cloning vector (Stratagene). Clones were linearized with appropriate restriction enzymes (NotI for T7 reactions and SmaI for T3 reactions), and T7 or T3 Ribomax kits (Promega) were used to generate sense and antisense RNAs from each gene by following the manufacturer's protocols. RNA mixtures (sense plus antisense) were annealed for 30 min at 37°C, and the presence of double-stranded RNA (dsRNA) was confirmed by agarose gel electrophoresis.
dsRNA was microinjected into the syncytial gonad of N2 or dpy-18 (e364) adult hermaphrodites at final concentrations of 0.5 to 1 mg/ml. Following overnight recovery, animals were singly transferred to plates and then sequentially transferred at 24-h periods. Nematode progeny were scored 24 to 72 h postinjection. phy-1 and phy-1–phy-2 RNAi in strain N2 was photographed with Fujichrome T64 film under Zeiss Axioplan Nomarski optics. After overnight recovery of injected hermaphrodites, embryonic progeny were collected from dpy-18 (e364) (phy-2 RNAi) and N2 (pdi-2 RNAi) backgrounds and monitored throughout development. Images were captured at 20- to 30-min intervals with a Hamamatsu digital camera attached to a Zeiss Axioskop 2 microscope by use of Improvision Openlab software.
Construction and expression of promoter-reporter gene fusions.
Promoter-reporter gene fusions were constructed using the C. elegans nucleus-localized lacZ reporter gene vectors pPD95-03 (with expression-enhancing multi-intron sequences) and pPD21-28 (without multi-intron sequences) (10). PCR was performed on C. elegans N2 genomic DNA with Taq polymerase and primer combinations spanning the putative promoter region for each gene: phy-1, Phy-1PF (PstI), sense, 5′ gcgctgcagGGTCTGCTGGCCGTTTCGTCAG 3′, and Phy-1PR (BamHI), antisense, 5′ gcaggatccCGCATTCTGAAAAATTGAGAG 3′; phy-2, Phy-2PF2 (PstI), sense, 5′ ggcgctgcagAGACTATAGTCTATAGCTGAAAACG 3′; and Phy-2PR2 (BamHI), antisense, 5′ gcgggatccACTGCTCTCATTCTGAAAGACAAATC 3′; and pdi-2, Pdi-2PF (SphI), sense, 5′ GATGGAGAGCATGCATGTTTTG 3′, and Pdi-2PR (BamHI), antisense, 5′ cgcgggatccAACATCACGATGAATAGCGAATGG 3′. PCR products were initially cloned into either pPCR Script or pTAg (R&D Systems), digested with PstI and BamHI for phy-1 and phy-2, and ligated with similarly digested pPD95-03 vector; for pdi-2, they were digested with SphI and BamHI and ligated with similarly digested pPD21-28 and pPD95-03 vectors. For phy-1, sequences from −2755 to +5 relative to ATG were incorporated to generate a translational fusion with lacZ. For phy-2, sequences from −1715 to +11 relative to ATG were analyzed. For pdi-2, sequences from −2620 to +5 relative to ATG were included. For all promoter fragments, the nearest predicted genes are transcribed in the opposite orientation and do not overlap.
Transformations were performed by microinjection into the syncytial gonad of adult nematodes. Reporter gene plasmids (20 μg/ml) were coinjected with either pRF-4 rol-6 (su1006) at 100 μg/ml into wild-type nematodes or p7616B (wild-type unc-76) at 100 μg/ml into DR96 (unc-76 e911) strain. rol-6 conferred a right-hand roller phenotype, thereby allowing visual identification of transgenic animals. The p7616B rescue plasmid repaired the uncoordinated phenotype resulting from the mutated, neuronally expressed protein UNC-76 of strain DR96. Semistable transgenic lines were maintained, fixed, and stained for β-galactosidase activity by following published methods (10). At least three independent lines were maintained and examined for each construct and marker combination. Stained nematodes were viewed and photographed with Fujichrome T64 film under Nomarski optics. The transgenic expression patterns detailed in this study remain to be confirmed by more direct methods.
Semiquantitative reverse transcriptase PCR.
The staged cDNA samples used in this procedure were kindly supplied by Iain Johnstone (Wellcome Centre for Molecular Parasitology [WCMP], Glasgow, United Kingdom). Detailed descriptions of methods used to generate synchronous C. elegans cultures and to synthesize cDNA are given elsewhere (15). For each gene tested (phy-1, phy-2, and pdi-2), PCR was used to amplify cDNA samples from synchronized nematode populations, representing 2-h intervals throughout the postembryonic life cycle at 25°C. Primers corresponding to the test gene and the control gene ama-1 (for the constitutively expressed large subunit of RNA polymerase II) were simultaneously used. ama-1 primer pairs were as follows: Ama-1F, sense, 5′ TTCCAAGCGCCGCTGCGCATTGTCTC 3′, and Ama-1R, antisense, 5′ CAGAATTTCCAGCACTCGAGGAGCG 3′. phy-1 primers were Phy-1F and Phy-1R; phy-2 primers were Phy-2F and Phy-2R; and for pdi-2, primers Pdi-2F and Pdi-2R were used. PCR conditions permitted reactants to remain in excess, and primer combinations were engineered to span introns, thereby distinguishing transcript signals from possible genomic DNA amplification. The products were electrophoresed, Southern blotted, and probed with the appropriate oligonucleotides end labeled with [γ-32P]ATP. Following autoradiography, bands corresponding to the respective genes were excised and counted by scintillation. The relative abundance of test gene transcripts was calculated from the ratio of test gene signal to ama-1 signal and plotted in arbitrary units. This value is not a true measure of real expression levels but does permit the fluctuations of transcript levels to be measured within a single experiment; thus, abundance values cannot be compared directly between individual experiments.
Rescue of the dpy-18 phenotype.
PCR was performed on N2 genomic DNA with a mixture (10:1) of Taq plus Pfu (Stratagene) polymerases and the following primer combination: Phy-1NotIF, sense, 5′ gcggcggccgcTTGGCTCTCCTAAGTTTCAGC 3′, and Phy-1SalIR, antisense, 5′ gcgtcgacGGCTTGCAGCCATCACTTCACAGG 3′. The product was ligated in vector pGEM-T (Promega) to generate a clone containing the phy-1 genomic region extending from position −2006 relative to the ATG translational start site to position 227 after the TAA translational stop signal and including the predicted polyadenylation site. This phy-1 genomic rescue clone (5 μg/ml) was microinjected into the dpy-18 e364 and e1096 strains together with a green fluorescent protein (GFP) transformation marker construct (10 μg/ml) and pBluescript SK (Stratagene) at 100 μg/ml. The GFP marker plasmid is a dpy-7 cuticle collagen promoter in the GFP fusion vector pPD95-67 (a gift from J. Muriel and I. Johnstone, WCMP). This marker was also injected with pBluescript SK in the absence of a rescue plasmid to ensure that the effects seen on body morphology were not conferred by the marker and pBluescript SK plasmids. The F1 progeny from rescue injections were selected for GFP expression and repair of the dumpy phenotype. Lines in which the F2 and subsequent generations continued to display this phenotype were viewed as live specimens using a Zeiss Axioskop 2 microscope with the filter set for GFP fluorescence, and images were captured as described above by use of Openlab software.
Characterization of dpy-18 alleles.
A genomic clone of phy-1 from the dpy-18 (e364) strain was amplified by PCR from genomic DNA with a 10:1 mixture of Taq and Pfu polymerases and the following primer pair: Phy-1HSCF, sense, 5′ gcggatatcATGCGCCTGGCACTCCTTGTAC 3′, and Phy-1HSCR, antisense, 5′ gcggatatcTTAGAGGGTCTCCCAGACGTC 3′. cDNA clones were generated with Taq polymerase and with the same combinations of primers from dpy-18 (e364) mixed-stage cDNA. PCR products were cloned into vector pGEM-T, and two independent cDNA clones were sequenced to identify the mutation and exclude PCR-generated changes. The genomic clone was sequenced on both strands over the area of the mutation. A genomic clone of phy-1 from the dpy-18 (e1096) strain was produced by PCR from genomic DNA with primers Phy-1NotIF and Phy-1SalIR and a cloning strategy like that used for the dpy-18 (e364) strain. The presence of a deletion in strain e1096 was demonstrated by PCR and restriction analysis, and sequencing was applied to confirm the extent of the deletion.
Hydroxyproline analysis.
Nematode N2 (wild type), dpy-18 (e364), dpy-18 (e1096), and dpy-10 (e128) strains were cultured and allowed to starve. The resulting dauer-stage larvae were induced to develop in a semisynchronous fashion by feeding; a mixture of late L4 and early adult stages was collected by gentle centrifugation. After extensive washes, the nematode cuticle collagens were purified by previously published methods (8). Briefly, cellular noncollagenous material was solubilized by sonication in S buffer (10 mM Tris [pH 7.4], 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride) and discarded following centrifugation. This step was followed by similar extractions in S buffer supplemented with 1% sodium dodecyl sulfate. After extensive washes, the cuticle collagens were solubilized by boiling in S buffer plus 1% sodium dodecyl sulfate and 5% 2-mercaptoethanol, separated from the insoluble noncollagenous material by centrifugation, and concentrated by acetone precipitation at −20°C. The purified cuticle collagens were dissolved in 50% acetic acid and hydrolyzed with 6 M HCl in the vapor phase under argon at 160°C for 35 min. The amino acid derivatization was carried out by Ian Davidson (Aberdeen University) using an Applied Biosystems 420A amino acid analyzer.
RESULTS
Two PHY α-subunit-encoding genes and an associated PDI β-subunit-encoding gene are expressed by C. elegans.
The free-living nematode C. elegans is the first metazoan for which the genome has been completely sequenced (7). Analysis of the sequence data identified two conserved PHY- and two conserved PDI-encoding genes. The two C. elegans PHY α subunits have a domain structure similar to those of the human α(I) and α(II) isoforms (Fig. 1), the highest conservation being found in the catalytic domains (23). Major differences exist at the extreme C-terminal domains of the proteins. In PHY-1, this region is directly associated with dimer formation, as its deletion prevents complex formation (35). At the protein level, PHY-1 is 44% identical to the human α(I) subunit and 43% identical to the human α(II) subunit over a 484-amino-acid region. Catalytically, PHY-1 most closely resembles α(II), both being insensitive to inhibition by poly(l-proline) (35). PHY-2 remains to be biochemically characterized but is 46% identical to α(I) and 45% identical to α(II) over a 515-amino-acid region. Similar 482-amino-acid regions of the two C. elegans proteins are 57% identical to each other. Although these represent the only highly conserved phy genes from C. elegans, an additional three genes encoding proteins with low homology to the PHY catalytic domains are present in the genome (7). The most similar of these predicted gene products are 25 and 30% identical over a 200-amino-acid C-terminal region to PHY-1 and PHY-2, respectively. Likewise, additional genes expressing thioredoxin-like PDI domains are found in the genome, encoding related ER proteins and thioredoxin proteins.
FIG. 1.
Alignment of the C. elegans (C prefix) prolyl 4-hydroxylase α subunits PHY-1 (GenBank no. Z81134) and PHY-2 (GenBank no. Z69637) to the human (H prefix) α(I) (M24487) and α(II) (U90441) subunits using ClustalW. Gaps (dashes) were introduced for maximal alignment, and signal peptides were removed. Highly conserved cysteine and active-site histidine, aspartic acid, and lysine residues (23) are indicated by an asterisk. The conserved tryptophan76, converted to a stop codon in the phy-1 gene of the dpy-18 (e364) allele, is indicated by a dollar sign.
The combined phy genes and single pdi gene are essential for embryonic development and the maintenance of nematode body shape.
Disruption of activity encoded by the phy and pdi genes was achieved using the specific reverse-genetics method of double-stranded RNAi (11). This is a potent technique that temporarily mimics loss-of-function defects. The disruption of the phy-1 gene was carried out by microinjection of dsRNA corresponding to the phy-1 coding sequence. This resulted in a dumpy (dpy, short and fat) phenotype visible in the L4 and adult stages of the F1 progeny of injected nematodes (Fig. 2A). Dumpy animals were observed in approximately 90% of the F1 progeny and were characteristically 40 to 60% shorter and fatter than wild-type worms. This phenotype is strikingly similar to that of the medium dumpy mutants in C. elegans (6), which predominantly result from mutations in cuticle collagen genes (21). phy-1 was subsequently shown to correspond to the uncloned dpy-18 locus (see below). RNAi of the second α-subunit-encoding gene, phy-2, consistently produced no visible phenotype (Table 1). Our observations imply that nonspecific disruption of the less conserved prolyl 4-hydroxylase α-subunit-related genes would be unlikely, since phy-2 is most similar to phy-1 and failed to phenocopy the phy-1 RNAi effect. The difference in phenotype between phy-1 and phy-2 single disruptions perhaps indicates variations in the substrate and/or cosubstrate specificity of the enzymes.
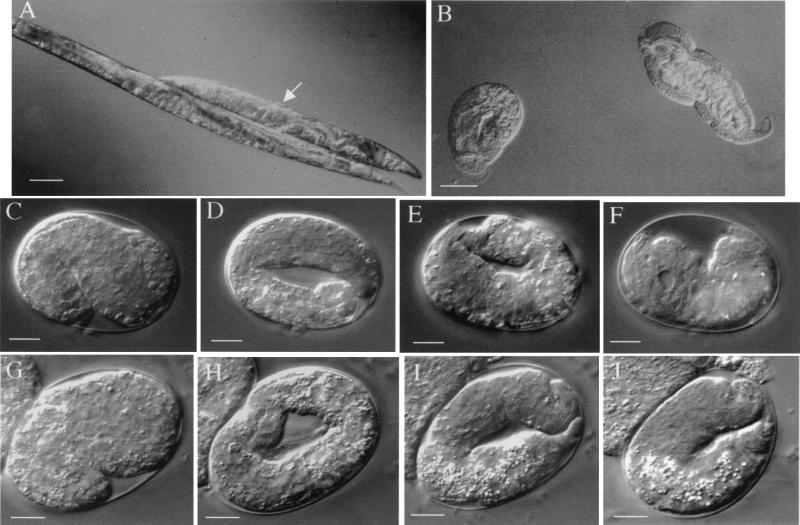
FIG. 2.
Double-stranded RNAi of phy-1, phy-2, and pdi-2. (A) Medium dumpy phy-1 RNAi in wild-type N2 background (arrow) compared to wild-type N2. Adults are depicted. Bar, 100 μm. (B) phy-1–phy-2 combined RNAi in wild-type N2 background. A range of severe dumpy phenotypes is represented by coiled larva (left) and adult nematode (right). Bar, 100 μm. (C to F) phy-2 RNAi in dpy-18 (e364) background. The same individual embryo is depicted in all images. Bars, 10 μm. (C) Beginning of elongation (1.5-fold, 440 min). (D) Elongated embryo (3-fold, 570 min). Head and tail of coiled embryo are out of the focal plane. (E) Retracting embryo (710 min). Head and tail of uncoiled embryo are now visible in the same focal plane. (F) Terminal phenotype (approximately 1,800 min), a fully retracted dying embryo with visible vacuoles. (G to J) pdi-2 RNAi in wild-type N2 background. The same individual embryo is depicted in all images. Bars, 10 μm. (G) Beginning of elongation (1.5-fold, 430 min). (H) Elongated embryo (3-fold, 560 min). Head and tail of coiled embryo are out of the focal plane. (I) Retracting embryo (700 min). Head and tail of uncoiled embryo are now visible in the same focal plane. (J) Terminal phenotype (approximately 960 min), a retracted dying embryo with evident small vacuoles.
TABLE 1.
Double-stranded RNAi of phy-1, phy-2, and pdi-2
| Gene (genetic background) | No. of eggs laid (from n adults) | No. (%) of eggs hatched | Effect |
|---|---|---|---|
| phy-1 (N2)a | Dumpy phenotype in late L4 larvae and adults (short fat worms) | ||
| phy-2 (N2) | No visible morphological effect | ||
| phy-1 and phy-2 (N2)b | 1,232 (20) | 1,208 (98) | Severe dumpy phenotype in 1,189 (97%) |
| phy-2 [dpy-18 (e364)] | 1,250 (16) | 132 (11) | Embryonic lethality in 1,118 (89%) |
| pdi-2 (N2) | 1,432 (11) | 1 (0.1) | Embryonic lethality in 1,431 (99.9%) |
Combined interference of both α subunits was achieved using RNAi with a mixture of both phy-1 and phy-2 dsRNAs in a wild-type genetic background. This resulted in phenotypes ranging from embryonic lethality, through severe dumpy phenotypes (Fig. 2B), to the aforementioned medium dumpy phenotype. The severe dumpy phenotype was the principal effect noted (97% of progeny; Table 1). Many of these mutants remained as coiled, inactive larvae, while some developed into adults (both forms depicted in Fig. 2B). Severe dumpy adults were approximately 50% smaller than single phy-1 (dpy-18) mutants (approximately 75% smaller than the wild type) and possessed the normal complement of organs, which were highly convoluted and folded in the restricted body space. Many bulges and abnormalities, including extra unshed cuticles, were present on the external surfaces of these nematodes. All of these effects are consistent with an effect on the development of the collagen-rich cuticle.
The effect of combined phy disruption was examined further by RNAi of phy-2 in a dpy-18 strain. This strain was found to be a null mutant of phy-1 (see below). Disruption resulted in a postelongation embryonic lethal phenotype, the time course of which is depicted in Fig. 2C to F. From the 1,250 embryos laid by 16 injected hermaphrodites, 89% failed to hatch (Table 1). Affected embryos were observed to develop normally through gastrulation and elongation (Fig. 2C and D) and were motile throughout elongation. Distinct internal structures, including a discernible pharynx, were also noted. The abnormal phenotype became evident after cuticle synthesis (710 min into development; see also reference 31). The fully elongated embryos (threefold; Fig. 2D) began to retract to a shorter, fatter form (twofold; Fig. 2E), which ultimately failed to hatch. Animals then died slowly, with a corresponding breakdown in structural organization and the appearance of internal vacuoles (Fig. 2F). This phenotype is consistent with the generation of a dysfunctional or poorly formed exoskeleton that fails to maintain worm body shape. This result demonstrates that the prolyl 4-hydroylase class of enzymes is essential for normal embryonic development.
The effects of disrupting the associated β-subunit gene pdi-2 were also examined via RNAi. From 11 injected hermaphrodites, 1,432 embryos were examined, 99.9% of which failed to hatch (Table 1). A developmental time course of affected embryos is depicted in Fig. 2G to J. These embryos exhibited a phenotype identical to that of the lethal phy-2 disruption in the phy-1 null strain (dpy-18 strain e364). Embryos elongated normally (Fig. 2G and H) with associated movement and the formation of well-defined structures. The mutant phenotype became evident after cuticle synthesis occurred (700 min into development) and was characterized by the retraction of the fully elongated nematode (Fig. 2I). As described for the phy-2 disruption in the dpy-18 strain, embryos slowly become disorganized (Fig. 2J), failed to hatch, and ultimately died. These results support the proposal that PDI-2 represents the single β subunit for conserved forms of prolyl 4-hydroxylase complexes in C. elegans and confirm the previous biochemical association noted between PHY-1 and PDI-2 (34).
The severe dumpy phenotype and embryonic death described in this study are comparable to the effects of the prolyl 4-hydroxylase-inhibitory compounds pyridine 2,4-dicarboxylic acid and pyridine 2,5-dicarboxylic acid on dpy-18 (e364) nematodes (unpublished observations). Both compounds are competitive-analogue inhibitors of the essential cosubstrate 2-oxaloglutarate (18) and result in a severe dumpy phenotype and embryonic death.
Both phy genes and the single pdi gene are expressed in oscillating waves of abundance in the cuticle-synthesizing hypodermis.
To examine further the function of the prolyl 4-hydroxylase complex, the detailed spatial expression patterns of the phy and pdi-2 genes were studied via reporter transgene analysis (Fig. 3). The promoter regions of the three genes were fused in frame to a lacZ reporter plasmid construct containing a nuclear localization signal. Constructs were transformed into the C. elegans germ line via microinjection, and transformants were selected via the expression of phenotypic markers, fixed, and stained for β-galactosidase activity. The injection of plasmid DNA into the hermaphrodite gonad results in the formation of extrachromosomal arrays. These semistable arrays are then transmitted at 10 to 90% to subsequent generations. Individual nematodes do, however, display mosaicism; therefore, at least three independent lines were selected for each construct and marker combination. Many individual nematodes encompassing all the different life cycle stages were examined to establish which individual nuclei were reproducibly expressing the reporter gene constructs. Both hypodermally and nonhypodermally expressed transformation markers were used, thereby excluding the possibility of reporter gene expression being driven by the transcriptional regulatory units of the marker plasmid. No differences were noted in expression patterns between transformants generated with either marker, and no significant differences were observed between the independent lines generated in this study.
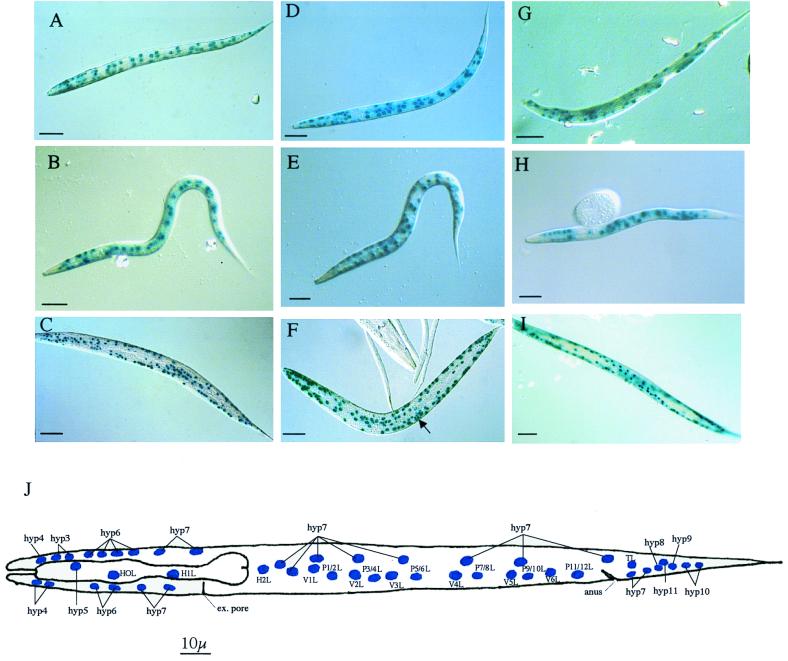
FIG. 3.
Tissue-specific localization of phy-1, phy-2, and pdi-2. L1 staining patterns correspond to the positions of labeled hypodermal nuclei in panel J, which are particularly apparent at the anterior and posterior ends of the nematode. Staining patterns may be complicated by the corresponding nuclei on the opposite lateral focal plane of the depicted nematode. (A) 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining of L1 larvae showing phy-1 (pPD95-03, with a nuclear localization signal [NLS])-driven lacZ expression using the rol-6 marker plasmid. The anterior is to the left, dorsal is at the top, and focus is in the lateral plane. All anterior, posterior, and midbody hyp cells are evident. The lateral seam (H, T, and V) and P cells are stained. Bar, 10 μm. (B) X-Gal staining of L1 larvae showing phy-1 (pPD95-03, with an NLS)-driven lacZ expression using the unc-76 marker plasmid. The anterior is to the left, dorsal is at the top, and focus is in the lateral plane. Most anterior hyp cells (hyp5 to hyp7) are evident; only hyp7 nuclei are visible in the posterior. Some of the lateral seam (H, T, and V) and P cells are visible. Bar, 10 μm. (C) X-Gal staining of adult nematode showing phy-1 (pPD95-03, with an NLS)-driven lacZ expression using the rol-6 marker plasmid. The anterior is to the left, and the body is twisted due to expression of the rol-6 phenotype. Many anterior and posterior hyp cells and lateral hypodermal cells are visible. Bar, 100 μm. (D) X-Gal staining of L1 larvae showing phy-2 (pPD95-03, with an NLS)-driven lacZ expression using the rol-6 marker plasmid. The anterior is to the left, ventral is at the top, and focus is in the lateral plane. All anterior hyp cells are evident; only hyp7 nuclei are present in the tail. The midbody lateral seam cells, P cells, and hyp cells are visible. Additional midbody nonhypodermal cells are also evident. Bar, 10 μm. (E) X-Gal staining of L1 larvae showing phy-2 (pPD95-03, with an NLS)-driven lacZ expression using the unc-76 marker plasmid. The anterior is to the left, dorsal is at the top, and focus is in the lateral plane. Most anterior hyp cells are evident, with weak, incomplete staining of posterior hyp cells. Midbody hyp7 cells, lateral seam cells, and P cells are conspicuous. Additional midbody nonhypodermal cells are present. Bar, 10 μm. (F) X-Gal staining of adult nematode revealing phy-2 (pPD95-03, with an NLS)-driven lacZ expression using the rol-6 marker plasmid. The anterior is to the left, and the body is helically twisted due to expression of the rol-6 phenotype. Anterior, posterior, and lateral hypodermal cells are discernible. Vulval cell nuclear staining is indicated by an arrow. Additional nonhypodermal cell nuclei are also conspicuous. Bar, 100 μm. (G) X-Gal staining of L1 larvae showing pdi-2 (pPD21-28, with an NLS)-driven lacZ expression using the rol-6 marker plasmid. The anterior is to the left, dorsal is at the top, and focus is in the lateral plane. Some of the anterior hyp cells are evident; hyp7 nuclei are present in the tail. Some of the midbody lateral seam cells, P cells, and hyp cells are visible. Bar, 10 μm. (H) X-Gal staining of L1 larvae showing pdi-2 (pPD95-03, with an NLS)-driven lacZ expression using the unc-76 marker plasmid. The anterior is to the left, dorsal is at the top, and focus is in the lateral plane. Most anterior hyp cells are evident (hyp3, hyp6, and hyp7); only hyp7 nuclei are evident in the posterior (partially out of focus). Some of the lateral seam (H, T, and V) and P cells are stained. Bar, 10 μm. (I) X-Gal staining of immature adult nematode showing pdi-2 (pPD21-28, with an NLS)-driven lacZ expression using the rol-6 marker plasmid. The anterior is to the left, and the body is twisted due to expression of the rol-6 phenotype. Anterior and posterior hyp cells and lateral hypodermal cells are visible. Bar, 100 μm. (J) Diagrammatic representation of the L1 left lateral aspect depicting the hypodermal cell nuclei. An identical pattern is present on the right lateral view, with additional hyp7 nuclei dorsal to H2R nuclei. ex, excretory. Panel J is based on the original L1 hypodermal cell nucleus designation (31).
For all three genes, all stages from embryo to adult consistently expressed the reporter gene constructs in at least hypodermal cell nuclei. phy-1 and phy-2 were expressed from the midelongation stage of embryonic development (data not shown). pdi-2 was expressed earlier during embryonic elongation, at approximately 1.5-fold stage (data not shown). Expression of all three genes was examined in detail in the first larval (L1) stage, since the position of all hypodermal cell nuclei can be most accurately determined in this stage (Fig. 3J) (31). For phy-1, phy-2, and pdi-2, expression was detected in L1 hypodermal cells, including the anterior H0L, H1L, and hyp3 to hyp7, the posterior TL and hyp7 to hyp11, the midbody hyp7, and the lateral P, V, H2R, and H2L cells (Fig. 3A, B, D, E, G, and H). Mosaicism in expression was evident, especially in the posterior hyp and anterior hyp3 and hyp4 cells. In accordance with the increase in the numbers of hypodermal cells in the late larval and adult stages (31), the expression pattern became increasingly more complex (Fig. 3C, F, and I). For phy-1, phy-2, and pdi-2, most of the identifiable stained nuclei were of hypodermal origin; additional nuclei were, however, apparent, particularly for phy-2. The hypodermal pattern for phy-1 and phy-2 included the vulval cell nuclei (Fig. 3F); vulval cell staining was absent for pdi-2. In addition, phy-2 expression was occasionally detected in the body wall muscle cells, a pattern which became increasingly evident when sensitive staining methods were applied (data not shown). Expression in the cuticle collagen-synthesizing hypodermal cells is consistent with the cuticle-related defects generated by RNAi for these three genes.
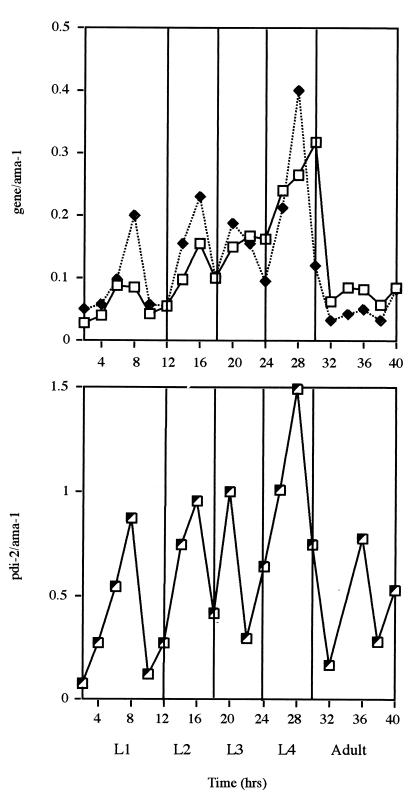
The temporal expression patterns for the three transcripts were examined by applying a semiquantitative reverse transcriptase PCR approach (15). This method permitted the abundance of the individual genes to be quantified via mRNA isolated from synchronized populations of C. elegans sampled at 2-h intervals throughout postembryonic development. These values were then expressed as ratios normalized against the constitutively expressed gene ama-1 (for the RNA polymerase II large subunit) (4). The temporal expression patterns of the three transcripts were consistent with their expected roles in cuticle collagen modification and their association in the enzyme complex. All three enzymes had very similar transcript profiles (Fig. 4), displaying an overall increase throughout larval development, with distinct peaks of abundance corresponding to the midlarval stages. Expression was highest in the L4 larvae and was followed by a dramatic decrease in the adult stage (Fig. 4). Comparable oscillating expression patterns have been described for a number of individual cuticle collagen genes (15) and two potential collagen-folding enzymes, including PDI-1 (24). The four larval stages are characterized by the shedding and resynthesis of the cuticle, a structure in which more than 80% of the proteins are collagenous (8). As the exoskeleton progressively increases in size, greater pulses of collagen-folding enzymes will be required to assemble correctly this complex ECM. The transcript abundance profiles for the two hypodermally expressed phy genes are virtually identical (Fig. 4), indicating that they may have shared or common roles, a point supported by the genetic and spatial expression pattern data (Fig. 2 and 3). The single β-subunit-encoding gene pdi-2 had an oscillating pattern similar to that of the two α-subunit-encoding genes. pdi-2 displayed a further peak of expression in the adult stage which was absent for the phy genes, perhaps indicating an additional adult-specific role for this multifunctional PDI enzyme.
FIG. 4.
Temporal expression pattern of the phy-1 (□), phy-2 (⧫), and pdi-2 (┘) transcripts during postembryonic development. A semiquantitative reverse transcriptase PCR approach was applied to examine the ratio of expression (y axes) of the individual enzyme-encoding genes to that of the constitutively expressed gene ama-1. The arbitrary values for phy-1 and phy-2 were plotted together for comparison. All values were obtained from mRNA of synchronously maintained larval and early adult stages at 25°C. L1 to L4, first to fourth larval stages.
The dpy-18 mutant phenotype is rescued by the α-subunit-encoding gene phy-1.
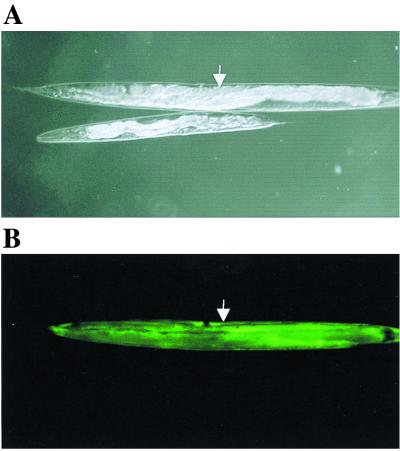
Single gene RNAi of phy-1 resulted in a medium dumpy phenotype (Fig. 2A), and the approximate physical map position of the phy-1-encoding cosmid T28D6 (chromosome III, map position 7.8) was in the vicinity of the genetic locus dpy-18 (chromosome III, map position 8.62). Mutant dpy-18 alleles were originally derived from ethyl methanesulfonate-mutagenized nematodes (6) and exhibited medium dumpy phenotypes and associated temperature-sensitive male tail abnormalities (3). To date, these alleles remain to be molecularly defined. To examine the possible correlation between phy-1 and dpy-18, two strains of dpy-18 (e1096 and e364) were obtained from the Caenorhabditis Genetics Center, and attempts were made to rescue the phenotypes of the alleles. A wild-type copy of the phy-1 gene which included the 5′ promoter, the genomic coding sequence, and the 3′ untranslated region (incorporating the polyadenylation signal) was cloned by PCR from wild-type (N2) genomic template DNA. The cloned phy-1 fragment was then coinjected into dpy-18 strains with a visible marker, namely, the dpy-7 cuticle collagen (16) promoter fused to GFP. This step allowed transformants to be selected on the basis of fluorescence using a UV dissecting microscope. These experiments revealed that the dpy-18 phenotype was rescued by phy-1, since transformed fluorescent progeny were restored to the wild-type phenotype (Fig. 5). The repair to wild-type body shape and the corresponding fluorescence of the dpy-18 e364 strain are shown in Fig. 5. Identical rescue results were obtained with the second mutant allele, dpy-18 (e1096) (data not shown). A control injection of the dpy-7 promoter–GFP construct eliminated the involvement of this marker in phenotype repair (data not shown).
FIG. 5.
Rescue of medium dumpy phenotype of a dpy-18 strain by coinjection of a wild-type copy of phy-1 and the selectable marker dpy-7–GFP. (A) Rescued progeny (arrow) and nonrescued progeny of a dpy-18 strain (e364) viewed by Nomarski optics. (B) Corresponding fluorescence in the rescued dpy-18 strain (e364) (arrow) viewed under a UV filter. Adult stages are depicted.
Mutations in phy-1 result in the dpy-18 phenotype.
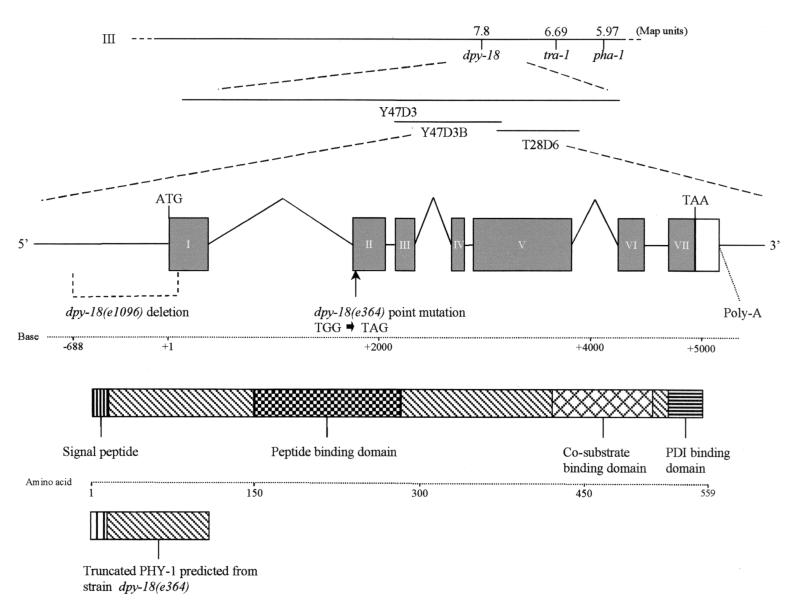
The correlation between dpy-18 alleles and the α-subunit-encoding gene phy-1 was confirmed by sequencing the phy-1 gene from the dpy-18 mutant alleles e364 and e1096. Cloning was achieved by PCR of both mRNA and genomic DNA for e364 and of genomic DNA for e1096. These experiments were repeated on two independent occasions to confirm that changes from the wild-type copy of the gene were not PCR-induced artifacts. Sequencing of e364 allele revealed the presence of a single point mutation in the 5′ coding sequence of exon 2 in phy-1, cloned from both mRNA and genomic DNA. This mutation was a TGG-TAG (tryptophan76-amber stop codon) (Fig. 1 and 6) and probably represented a null mutant, as translation is predicted to terminate before the functional peptide-, cosubstrate-, and PDI-binding domains of the encoded enzyme (Fig. 6). Cloning of phy-1 from strain e1096 was achieved only from genomic DNA, with the resulting product being notably smaller than the wild-type copy. Sequencing revealed a 776-bp 5′ deletion from positions −688 to +88 relative to the initiation methionine (Fig. 6). This deletion removed part of the promoter region, signal peptide, and N-terminal coding region of PHY-1 and therefore represented a true null mutation.
FIG. 6.
Gene structure of phy-1 showing physical and genetic map locations. Exons are shown as filled boxes. Introns and promoter regions are represented by lines, and 3′ untranslated regions are indicated by an open box. ATG and TAA indicate the positions of the translational start and stop signals, respectively. The 776-bp deletion in phy-1 from dpy-18 (e1096), extending from positions −688 to +88 (relative to the ATG), is depicted. The tryptophan76 (TGG)-to-amber stop codon (TAG) point mutation found in phy-1 from dpy-18 (e364) is also indicated. The domain structure for PHY-1 is shown along with the truncated protein predicted for dpy-18 (e364), revealing missing functional domains.
The cuticle collagens of two dpy-18 mutant strains have reduced hydroxyproline contents.
In order to confirm biochemically the role that PHY-1 plays in proline hydroxylation of the cuticle collagens of C. elegans, the hydroxyproline contents of these proteins from the two dpy-18 strains were examined. For comparison, identical cuticle collagen extracts were prepared from wild-type strain N2 and a collagen dumpy mutant, namely, dpy-10 (e128). This analysis revealed a significant reduction in the hydroxyproline contents of cuticle collagens from the dpy-18 strains compared with wild-type cuticle extracts. dpy-18 (e364) (phy-1 amber) and dpy-18 (e1096) (phy-1 deletion) strains expressed mercaptoethanol-soluble cuticle extracts with 18 and 31% reductions, respectively—hydroxyproline contents of 73 residues (standard error [SE], 3.5) and 61 residues (SE, 2.88) per 1,000 total residues, respectively. The unrelated dpy-10 dumpy mutant did not exhibit a similar reduction in hydroxyproline content compared with the wild-type strain (N2)—hydroxyproline contents of 93 and 89 residues per 1,000 total residues, respectively. This biochemical analysis validates the findings from the genetic analysis of phy-1 in dpy-18 mutants.
DISCUSSION
The multienzyme complex prolyl 4-hydroxylase has been proposed to be an important collagen-specific catalyst (19) and chaperone (37) of multicellular organisms. In this study, we demonstrate conclusively in vivo that the conserved forms of the prolyl 4-hydroxylase complex from C. elegans are essential during development and play a critical role in the biosynthesis of the collagenous cuticular ECM in this nematode. Analysis of the expression pattern of each of the subunits supports this role for the complex. Expression was demonstrated in the cuticular collagen-synthesizing hypodermal tissue in a developmental pattern mirroring that of the cuticle collagen substrate. We have also characterized the dpy-18 locus, which corresponds to the α-subunit-encoding gene phy-1.
ECM-modifying enzymes affect nematode development.
In C. elegans, the cuticle collagens are arranged into an exoskeleton, which imparts structural integrity and maintains the worm shape after embryonic elongation (29). Abnormalities in this structure have dramatic effects on embryogenesis and nematode body morphology (22). Elongation of the spherical embryo to worm shape is initiated by the cytoskeletal organization of the outermost surrounding layer of embryonic cells, the hypodermis (29). While these cells determine the shape of the embryo during elongation, the cuticle performs this function after elongation is completed (29).
Disruption of the conserved forms of the prolyl 4-hydroxylase complex results in embryonic lethality, which is proposed to be a direct result of cuticular defects due to the loss of hydroxylation and perhaps associated chaperone activities. Embryos develop normally, elongate to their full length (threefold), and then retracted back to the gross morphology of the twofold stage. Retraction occurs after synthesis of the first cuticle, at a time when this collagenous structure normally maintains the elongated form (31). We propose that the cuticle is malformed and/or thermally unstable and is therefore unable to fulfill its central role in morphogenesis. These observations are similar to those noted for severe mutant alleles in the gene sqt-3, which encodes the cuticle collagen COL-1 (33). Embryos of the temperature-sensitive allele of sqt-3 (e2117) elongate normally and then retract to approximately their original length at the restrictive temperature (29). The range of cuticular defects, including the severe dumpy phenotype, observed with phy-1–phy-2 simultaneous RNAi is analogous to that seen with the less severe mutant alleles of sqt-3 (sc63 and e24). These sqt-3 mutations are caused by glycine substitutions in the Gly-Xaa-Yaa repeat regions which are hypothesized to result in longer noncollagenous domains with a consequent decrease in thermal stability (33). A reduction in the hydroxyproline content of collagen and the secretion of misfolded or mutant trimers due to reduced chaperone function are likewise proposed to reduce the thermal stability of collagens. It is significant that the deformed cuticular ray phenotype of the male tail in dpy-18 (e364) is also temperature sensitive (3). From the RNAi experiments, we were unable to distinguish whether our results were due to loss of enzymatic activity or loss of chaperone activity. Prolyl 4-hydroxylase may, however, represent a major collagen chaperone in C. elegans, as the genome does not contain a single homologue of the gene encoding the collagen-specific chaperone Hsp47 (unpublished results). Recent in vitro studies also support a central chaperone role for prolyl 4-hydroxylase, not Hsp47, in the retention of improperly folded type III collagen (37).
The disruption results were obtained using the specific technique of RNAi (11), which accurately phenocopies loss-of-function defects. For phy-1, the RNAi-induced phenotype was directly confirmed using classical genetics, and phy-1 was found to be identical to the previously identified mutation dpy-18 (6). The less severe phenotypes produced by RNAi with the combined phy genes (compared to RNAi of phy-2 in a dpy-18 background) reveal an interesting aspect of this interference method. The range of phenotypes observed in this experiment suggests a dose-dependent effect that occurs within an injected population. Repeating simultaneous RNAi at reduced dsRNA concentrations (0.5 mg/ml rather than 1 mg/ml) substantiated this observation, as only the least severe medium dumpy phenotype was noted, similar to the results for phy-1 single knockouts.
Two forms of collagenous ECM exist in nematodes: the cuticle that forms the exoskeleton and the basement membranes that surround the tissues. Our results provide direct evidence for a central role of collagen-modifying enzymes in the formation of the cuticular ECM. There are several additional examples of C. elegans collagen-modifying enzymes which affect development and morphology; the enzymes BLI-4, GON-1, and LET-268 are also proposed to modify the ECM. Mutations in the bli-4 locus result in a viable cuticular defect and an embryonic lethal phenotype (27). The bli-4 gene encodes a serine endoprotease belonging to the kex2–subtilisin-like family (32), and C. elegans cuticle collagens have a potential N-terminal kex2–subtilisin-like protease processing site (20, 38). Both bli-4 mutant phenotypes are consistent with a role in collagen modification, as large fluid-filled blisters are present on the cuticle of nematodes expressing the viable allele, while embryonic death is thought to result from weakening of the cuticle due to improperly processed collagens. GON-1 is an ECM-modifying enzyme affecting organ morphogenesis (5); gon-1 mutants display severe malformation of the gonad. GON-1 contains a metalloprotease domain and multiple thrombospondin-like repeats, a domain structure observed in a family of enzymes that includes the collagen-processing enzyme bovine procollagen I N-protease. Thus, GON-1 is proposed to be crucial for gonadal morphogenesis and is thought to achieve this process through remodeling of basement membrane collagens.
An additional cotranslational modification that is important in animal collagens is the hydroxylation of lysine residues. This reaction is catalyzed by the lysyl hydroxylases, a second class of 2-oxaloglutarate dioxygenases which shares many features with the prolyl 4-hydroxylases (19). The hydroxylysine residues serve as attachment sites for carbohydrate and are necessary for the stability of intermolecular collagen cross-link formation. The C. elegans locus let-268 encodes a lysyl hydroxylase enzyme (K. Norman and D. Moerman, personal communication) and is the only predicted member of this class found in the genome (7). let-268 mutants arrest during embryogenesis, showing disorganization of muscle and basement membranes. Type IV basement membrane collagens connect body wall muscle to the hypodermis and are essential in the process of embryo elongation (12). These observations indicate an essential role for lysyl hydroxylase in ECM modification and development.
A role for 4-hydroxyproline in basement membrane collagens in C. elegans remains to be established. The expression patterns of the conserved prolyl 4-hydroxylase subunit genes support their direct role in cuticle collagen biosynthesis. Time course examination of RNAi-disrupted embryos revealed normal elongation with associated embryo motility, indicating that basement membranes are unaffected. Additionally, the potential basement membrane-related expression shown by phy-2 (in muscle cells; data not shown) was determined not to be essential by RNAi. However, we cannot rule out the possibility that these observations may be due to RNAi not efficiently disrupting potential basement membrane prolyl 4-hydroxylase activity. Alternatively, these findings may imply a significant difference in the modification of these two types of collagen in C. elegans. Hydroxylysine is found in the cuticle only in the environmentally resistant dauer-stage larvae (8) but is essential for basement membrane formation. Different mechanisms, possibly involving hydroxylysine or 3-hydroxyproline, may therefore stabilize the basement membrane type IV collagens from this nematode. Alternatively, the as-yet-uncharacterized divergent prolyl 4-hydroxylase isoforms may play a role in the modification of these collagens.
Inhibition of prolyl 4-hydroxylase.
Multicellular organisms produce an array of ECMs. The model nematode C. elegans is an excellent experimental system for the study of the ECM due to the wide range of molecular genetic and biochemical techniques applicable to this organism. From this study we conclude that prolyl 4-hydroxylase is a critical modulator of ECM formation and morphogenesis and may have a similar essential function throughout the animal kingdom. A greater understanding of prolyl 4-hydroxylase function and design of specific inhibitors would have an impact on human health issues in two major ways. First, excessive accumulation of collagens in the ECM plays a critical role in fibrotic disease. Prolyl 4-hydroxylase activity represents the most suitable target for the inhibition of collagen biosynthesis and as such provides a direct means to control disease due to fibrotic alterations in the ECM and excessive collagen deposition (13), such as liver cirrhosis, lung fibrosis, and scleroderma. Second, the essential role of this enzyme complex in the formation of the nematode cuticular ECM, as evidenced by this study, may provide a selective drug target for the control of parasitic nematode species.
ACKNOWLEDGMENTS
We thank Iain Johnstone (Glasgow) for stimulating discussions and advice regarding this research and for the provision of staged C. elegans mRNA and the dpy-7–GFP marker. We thank Ian Davidson (Aberdeen) for carrying out the amino acid analysis. We thank Don Moerman (Vancouver) for communicating unpublished results. The Caenorhabditis Genetics Center provided some nematode strains used in this work. We thank Dave Barry and Iain Johnstone for critical comments on the manuscript.
This work was funded by the MRC through a senior fellowship award to A.P.P.
REFERENCES
- 1.Annunen P, Helaakoski T, Myllyharju J, Veijola J, Pihlajaniemi T, Kivirikko K I. Cloning of the human prolyl 4-hydroxylase alpha subunit isoform alpha(II) and characterization of the type II enzyme tetramer—the alpha(I) and alpha(II) subunits do not form a mixed alpha(I)alpha(II)beta(2) tetramer. J Biol Chem. 1997;272:17342–17348. doi: 10.1074/jbc.272.28.17342. [DOI] [PubMed] [Google Scholar]
- 2.Annunen P, Koivunen P, Kivirikko K I. Cloning of the alpha subunit of prolyl 4-hydroxylase from Drosophila and expression and characterization of the corresponding enzyme tetramer with some unique properties. J Biol Chem. 1999;274:6790–6796. doi: 10.1074/jbc.274.10.6790. [DOI] [PubMed] [Google Scholar]
- 3.Baird S E, Emmons S W. Properties of a class of genes required for ray morphogenesis in Caenorhabditis elegans. Genetics. 1990;126:335–344. doi: 10.1093/genetics/126.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird D M, Riddle D L. Molecular cloning and sequencing of ama-1, the gene encoding the largest subunit of Caenorhabditis elegans RNA polymerase II. Mol Cell Biol. 1989;9:4119–4130. doi: 10.1128/mcb.9.10.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blelloch R, Kimble J. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature. 1999;399:586–590. doi: 10.1038/21196. [DOI] [PubMed] [Google Scholar]
- 6.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.C. elegans Genome Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 8.Cox G N, Kusch M, Edgar R S. Cuticle of Caenorhabditis elegans: its isolation and partial characterisation. J Cell Biol. 1981;90:7–17. doi: 10.1083/jcb.90.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darby N J, Penka E, Vincentelli R. The multi-domain structure of protein disulfide isomerase is essential for high catalytic efficiency. J Mol Biol. 1998;276:239–247. doi: 10.1006/jmbi.1997.1504. [DOI] [PubMed] [Google Scholar]
- 10.Fire A, White Harrison S, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 11.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 12.Gupta M, Graham P, Kramer J M. Characterization of alpha 1(IV) collagen mutations in Caenorhabditis elegans and the effects of alpha 1 and alpha 2(IV) mutations on type IV collagen distribution. J Cell Biol. 1997;137:1185–1196. doi: 10.1083/jcb.137.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanauske-Abel H. Prolyl 4-hydroxylase, a target enzyme for drug development—design of suppressive agents and the in vitro effects of inhibitors and proinhibitors. J Hepatol. 1991;13:S8–S16. doi: 10.1016/0168-8278(91)90003-t. [DOI] [PubMed] [Google Scholar]
- 14.John D C A, Grant M E, Bulleid N J. Cell-free synthesis and assembly of prolyl 4-hydroxylase—the role of the beta-subunit (PDI) in preventing misfolding and aggregation of the alpha-subunit. EMBO J. 1993;12:1587–1595. doi: 10.1002/j.1460-2075.1993.tb05803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnstone I L, Barry J D. Temporal reiteration of a precise gene expression pattern during nematode development. EMBO J. 1996;15:3633–3639. [PMC free article] [PubMed] [Google Scholar]
- 16.Johnstone I L, Shafi Y, Barry J D. Molecular analysis of mutations in the Caenorhabditis elegans collagen gene dpy-7. EMBO J. 1992;11:3857–3863. doi: 10.1002/j.1460-2075.1992.tb05478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kingston B I. Nematode collagen genes. Parasitol Today. 1991;7:11–15. doi: 10.1016/0169-4758(91)90077-2. [DOI] [PubMed] [Google Scholar]
- 18.Kivirikko K I, Myllyharju J. Prolyl 4-hydroxylases and their protein disulfide isomerase subunit. Matrix Biol. 1998;16:357–368. doi: 10.1016/s0945-053x(98)90009-9. [DOI] [PubMed] [Google Scholar]
- 19.Kivirikko K I, Pihlajaniemi T. Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv Enzymol Related Areas Mol Biol. 1998;72:325–400. doi: 10.1002/9780470123188.ch9. [DOI] [PubMed] [Google Scholar]
- 20.Kramer J M. Structures and functions of collagens in Caenorhabditis elegans. FASEB J. 1994;8:329–336. doi: 10.1096/fasebj.8.3.8143939. [DOI] [PubMed] [Google Scholar]
- 21.Kramer J M. Extracellular matrix. In: Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. C. elegans II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 471–500. [PubMed] [Google Scholar]
- 22.Kusch M, Edgar R S. Genetic studies of unusual loci that affect body shape of the nematode Caenorhabditis elegans and may code for cuticle structural proteins. Genetics. 1986;113:621–639. doi: 10.1093/genetics/113.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myllyharju J, Kivirikko K I. Characterization of the iron- and 2-oxoglutarate-binding sites of human prolyl 4-hydroxylase. EMBO J. 1997;16:1173–1180. doi: 10.1093/emboj/16.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Page A P. Cyclophilin and protein disulphide isomerase genes are co-transcribed in a functionally related manner in Caenorhabditis elegans. DNA Cell Biol. 1997;16:1335–1343. doi: 10.1089/dna.1997.16.1335. [DOI] [PubMed] [Google Scholar]
- 25.Page A P. A highly conserved nematode protein folding operon in Caenorhabditis elegans and Caenorhabditis briggsae. Gene. 1999;230:267–275. doi: 10.1016/s0378-1119(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 26.Page A P, MacNiven K, Hengartner M O. Cloning and biochemical characterisation of the cyclophilin homologues from the free-living nematode Caenorhabditis elegans. Biochem J. 1996;317:179–185. doi: 10.1042/bj3170179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters K, McDowall J, Rose A M. Mutations in the bli-4 (I) locus of Caenorhabditis elegans disrupt both adult cuticle and early larval development. Genetics. 1991;129:95–102. doi: 10.1093/genetics/129.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pihlajaniemi T, Helaakoski T, Tasanen K, Myllyla R, Huhtal M-L, Koivu J K, Kivirikko K I. Molecular cloning of the β-subunit of human prolyl 4-hydroxylase. This subunit and protein disulphide isomerase are products of the same gene. EMBO J. 1987;6:643–649. doi: 10.1002/j.1460-2075.1987.tb04803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Priess J, Hirsh D. Caenorhabditis elegans morphogenesis—the role of the cytoskeleton in elongation of the embryo. Dev Biol. 1986;117:156–173. doi: 10.1016/0012-1606(86)90358-1. [DOI] [PubMed] [Google Scholar]
- 30.Prockop D J, Kivirikko K I. Collagens: molecular biology, diseases, and potential for therapy. Annu Rev Biochem. 1995;64:403–434. doi: 10.1146/annurev.bi.64.070195.002155. [DOI] [PubMed] [Google Scholar]
- 31.Sulston J, Schierenberg E, White J, Thomason J. The embryonic-cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 32.Thacker C, Peters K, Srayko M, Rose A M. The bli-4 locus of Caenorhabditis elegans encodes structurally distinct kex2/subtilisin-like endoproteases essential for early development and adult morphology. Genes Dev. 1995;9:956–971. doi: 10.1101/gad.9.8.956. [DOI] [PubMed] [Google Scholar]
- 33.Vanderkeyl H, Kim H, Espey R, Oke C V, Edwards M K. Caenorhabditis elegans sqt-3 mutants have mutations in the COL-1 collagen gene. Dev Dynamics. 1994;201:86–94. doi: 10.1002/aja.1002010109. [DOI] [PubMed] [Google Scholar]
- 34.Veijola J, Annunen P, Koivunen P, Page A P, Pihlajaniemi T, Kivirikko K I. Baculovirus expression of two protein disulphide isomerase isoforms from Caenorhabditis elegans and characterisation of prolyl 4-hydroxylases containing one of these polypeptides as their β-subunit. Biochem J. 1996;317:721–729. doi: 10.1042/bj3170721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veijola J, Koivunen P, Annunen P, Pihlajaniemi T, Kivirikko K I. Cloning, baculovirus expression, and characterization of the α-subunit of prolyl 4-hydroxylase from the nematode Caenorhabditis elegans—this α-subunit forms an active αβ dimer with the human protein disulfide-isomerase β-subunit. J Biol Chem. 1994;269:26746–26753. [PubMed] [Google Scholar]
- 36.Vuori K, Pihlajaniemi T, Myllyla R, Kivirikko K I. Site-directed mutagenesis of human protein disulfide isomerase—effect on the assembly, activity and endoplasmic-reticulum retention of human prolyl 4-hydroxylase in Spodoptera frugiperda insect cells. EMBO J. 1992;11:4213–4217. doi: 10.1002/j.1460-2075.1992.tb05515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walmsley A, Batten M, Lad U, Bulleid N. Intracellular retention of procollagen within the endoplasmic reticulum is mediated by prolyl 4-hydroxylase. J Biol Chem. 1999;274:14884–14892. doi: 10.1074/jbc.274.21.14884. [DOI] [PubMed] [Google Scholar]
- 38.Yang J, Kramer J M. In vitro mutagenesis of Caenorhabditis elegans cuticle collagens identifies a potential subtilisin-like protease cleavage site and demonstrates that carboxyl domain disulfide bonding is required for normal function but not assembly. Mol Cell Biol. 1994;14:2722–2730. doi: 10.1128/mcb.14.4.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]