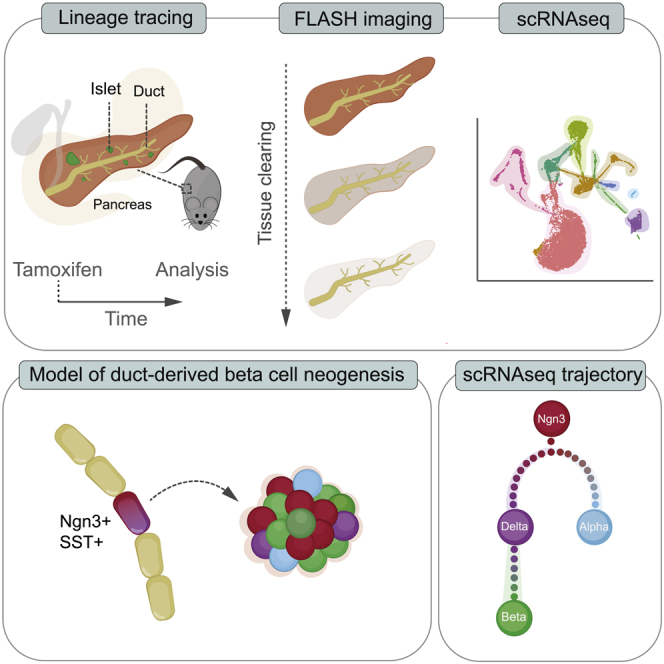
Summary
Ductal cells have been proposed as a source of adult β cell neogenesis, but this has remained controversial. By combining lineage tracing, 3D imaging, and single-cell RNA sequencing (scRNA-seq) approaches, we show that ductal cells contribute to the β cell population over time. Lineage tracing using the Neurogenin3 (Ngn3)-CreERT line identified ductal cells expressing the endocrine master transcription factor Ngn3 that were positive for the δ cell marker somatostatin and occasionally co-expressed insulin. The number of hormone-expressing ductal cells was increased in Akita+/− diabetic mice, and ngn3 heterozygosity accelerated diabetes onset. scRNA-seq of Ngn3 lineage-traced islet cells indicated that duct-derived somatostatin-expressing cells, some of which retained expression of ductal markers, gave rise to β cells. This study identified Ngn3-expressing ductal cells as a source of adult β cell neogenesis in homeostasis and diabetes, suggesting that this mechanism, in addition to β cell proliferation, maintains the adult islet β cell population.
Keywords: β cells, pancreas, diabetes, δ cells, lineage tracing, Ngn3, progenitor, regeneration
Graphical abstract
Highlights
-
•
Ductal Ngn3+ cells contribute to the beta cell population during homeostasis
-
•
Duct-resident endocrine progenitor cells express somatostatin
-
•
Somatostatin-positive ductal cells are increased in Akita+/− diabetic mice
-
•
scRNA-seq analysis suggests ductal somatostatin+ cells give rise to beta cells
A better understanding of adult beta cell neogenesis would open new approaches for diabetes treatment. Gribben et al. used lineage tracing and scRNA-seq to characterize a duct-resident somatostatin-positive endocrine progenitor cell population that is a source of beta cells in homeostasis and during diabetes.
Introduction
The pancreas is central to energy homeostasis, playing essential roles in digestion and in regulation of blood glucose. Glucose homeostasis is controlled by endocrine cells of the islets of Langerhans, which contain mostly insulin-secreting β cells. Type 1 and late-stage type 2 diabetes involve β cell loss (Kharroubi and Darwish, 2015). With 1 in 11 people being diabetic, there is a great need to understand how the adult β cell population is maintained to guide new therapies.
The β cell population is believed to be maintained mainly through proliferation (Dor et al., 2004). However, there is evidence of β cell neogenesis from non-β cell sources in the injured adult murine pancreas (Chera et al., 2014; Thorel et al., 2010; Xu et al., 2008), although the existence and significance of endocrine progenitors in homeostasis of the adult pancreas remain disputed.
Expression of Ngn3, a key transcription factor expressed in endocrine progenitors of the developing pancreas (Gradwohl et al., 2000), has been strongly implicated in adult β cell neogenesis. Ductal cells have been proposed as a source of adult Ngn3+ cells. Deletion of the E3 ubiquitin ligase Fbw7, which controls the stability of Ngn3, in adult ductal cells (Sancho et al., 2014) and overexpression of the transcription factor Pax4 in α (Al-Hasani et al., 2013) and δ cells (Druelle et al., 2017) has uncovered ductal plasticity, where ductal cells can become endocrine cells via Ngn3 expression. However, lineage tracing strategies have produced conflicting evidence for the ability of ductal cells to generate β cells in homeostasis and in injury paradigms. Use of carbonic anhydrase II (CAII)-CreERT with a R26-LacZ tracer suggests that endocrine cells can be derived from ductal cells following the injury model of pancreatic duct ligation (PDL) (Inada et al., 2008). Use of the Sox9-CreERT with a Rosa26-mT/mG tracer labels 90% of ductal cells, and traced β cells can be observed after hyperglycemia and cytokine treatment (Zhang et al., 2016). In contrast, use of promoters such as Krt19, Mucin1, Sox9, Hes1 and Hnf1b has not shown ductal contribution to β cell regeneration (Kopinke et al., 2011; Kopinke and Murtaugh, 2010; Means et al., 2008; Solar et al., 2009). Likewise, dilution of labeled β cells over time in homeostasis and after PDL has been reported (Van de Casteele et al., 2013), but has not been seen in other studies (Dor et al., 2004; Zhao et al., 2021). Thus, the question of whether ductal cells can differentiate into endocrine cells is controversial and unresolved. Differences in expression of promoters used to drive Cre expression in ductal cells and the recombination efficiencies of the tracers used results in great discrepancy of labeling efficiency. Moreover, the duration for which the labeled cells were followed has been highly variable. These technical issues may explain some of the contradictory results observed. Thus, although there is some evidence of adult duct-derived endocrine differentiation, its role and significance are unclear.
Here, by taking advantage of technology improvements in lineage tracing and 3D imaging, we demonstrate that ductal cells contribute to the islet β cell population in homeostasis and during diabetes. Use of the Hnf1b-CreERT driver and R26-CAG-TdTomato lineage tracer enabled efficient, nearly complete labeling of ductal cells. A subpopulation of ductal cells has been found to express Ngn3 (Ngn3+) and also expressed somatostatin (SST+). Additionally, through the use of 3D imaging, Ngn3-traced cells co-expressing insulin (INS) and SST (INS+/SST+) were observed within and between ducts and islets. Single-cell RNA sequencing (scRNA-seq) analysis of Ngn3-traced islets predicted a trajectory where Ngn3+ cells become SST+ cells, which, in turn, become INS+ β cells. Our analyses demonstrate the existence of a rare ductal Ngn3+ population that contributes to the maintenance of islet cell populations.
Results
Non-β cells contribute to the islet β cell population during homeostasis
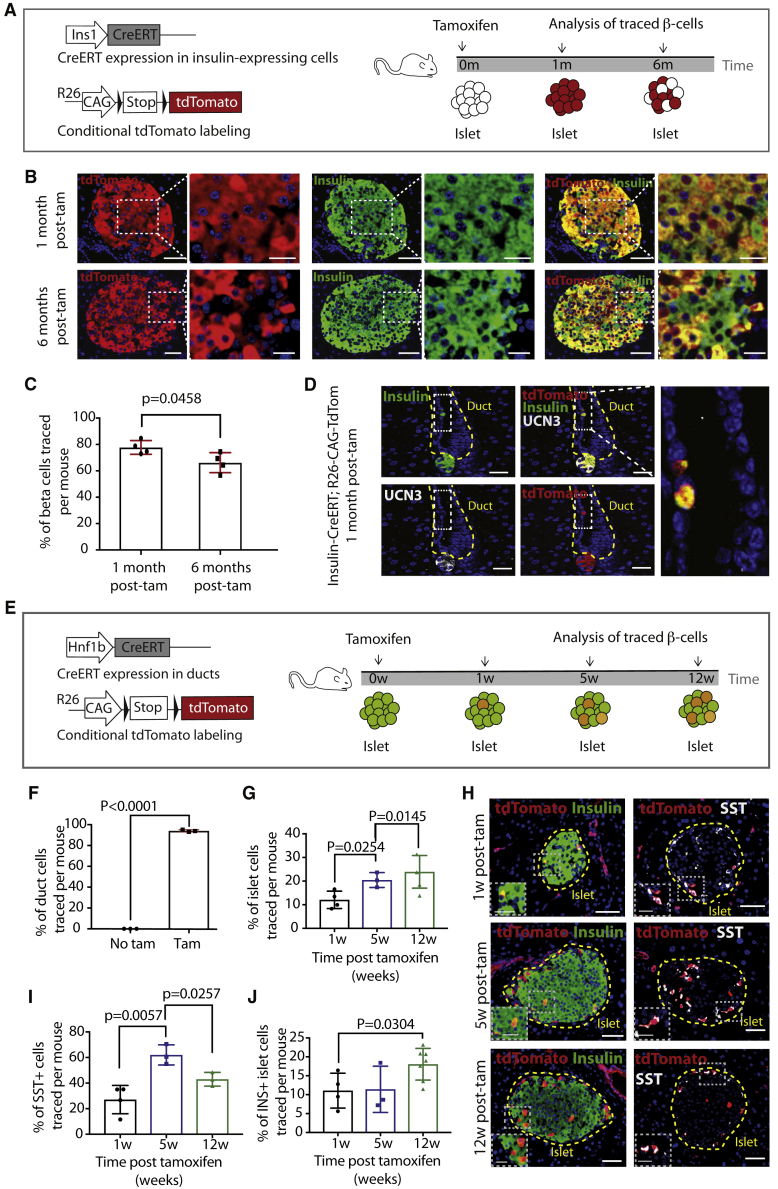
To follow INS+ cells over time, we crossed the Ins1-CreERT line (Wicksteed et al., 2010) to the sensitive lineage tracer R26-CAG-TdTomato (Madisen et al., 2010; Figure 1A). Adult mice were analyzed 1 and 6 months after tamoxifen (Figure 1A), and the pancreas was analyzed by antibody staining, scanning, and automated quantification to determine the percentage of islet β cells traced (Figures S1A and S1B; Tables S1 and S2). We observed a significant decrease in tracing from 78% to 66% between 1 and 6 months after tamoxifen injection (Figures 1B and 1C), suggesting dilution of existing islet β cells of 0.68% per week. Traced INS+ cells were found outside of the islets (Figure 1D), including within ducts, but they were negative for the mature β cell marker Urocortin3 (UCN3), suggesting that these duct-resident INS+ cells may be immature. This indicates that INS+/UCN3− cells exist outside of the islets and may contribute to the β cell population over time.
Figure 1.
The β cell population is not solely maintained by existing β cell proliferation
(A) Lineage tracing strategy and experimental timeline.
(B) Double immunofluorescence (IF) for tdTomato and INS in INS-CreERT; R26-CAG-tdTomato mice 1 and 6 months after tamoxifen, with high magnification shown on the right. Scale bars, 50 μm (left) and 20 μm (right).
(C) Quantification of the percentage of INS+ islet cells traced per mouse (N = 4) at both time points (10–26 weeks of age at injection). Data are represented as mean with SD and p value is indicated.
(D) Triple IF for tdTomato, INS and Urocortin3 (UCN3). A traced ductal INS+/UCN3− (boxed) is shown at high magnification. Scale bars, 25 μm (left) and 10 μm (right).
(E) Lineage tracing strategy and experimental workflow.
(F) Quantification of the percentage of ductal cells labeled 1 week after tamoxifen (N = 3 at 8 weeks of age at injection). Data are represented as mean with SD and p values are indicated.
(G) Quantification of the percentage of islet cells traced per mouse. N = 4 at each time point (8–16 weeks of age at injection). Data are represented as mean with SD and p values are indicated.
(H) Double IF for tdTomato/INS and tdTomato/SST. High magnification is shown in the inset (white dashed line). Scale bars, 50 μm.
(I) Quantification of the percentage of SST+ islet cells traced per mouse. N = 4 at each time point (8–16 weeks of age at injection). Data are represented as mean with SD and p values are indicated.
(J) Quantification of the percentage of INS+ islet cells traced per mouse. N = 4 at each time point (8–16 weeks of age at injection). Data are represented as mean with SD and p values are indicated.
See also Figure S1.
Ductal cells contribute to the β cell population over time
Ductal cells are a candidate population that may account for the dilution of the INS+ islet population over time (Sancho et al., 2014; Figure 1C). To circumvent potential issues caused by the low lineage tracing efficiencies seen in earlier studies, we used Hnf1b-CreERT; R26-CAG-TdTomato mice (Figure 1E). 94% of ductal cells were labeled following tamoxifen admimistration (Figure 1F). Labeling of some δ (SST+) cells was observed (Figures 1H and 1I), as well as labeling of isolated cells found outside of the islets and ducts (Figures S1C and S1D). Islets were analyzed 1, 5, and 12 weeks after tamoxifen (Figures 1G and 1H). We observed a significant increase in the percentage of lineage-traced islet cells between 1 and 5 weeks, which only slightly increased further at the 12-week time point (Figure 1G). More than 90% of traced cells in the islets expressed INS or SST. Traced SST+ cells increased significantly between 1 and 5 weeks and accounted for the majority of tdTomato+ traced cells 5 weeks after tamoxifen, but a sharp decrease was observed 12 weeks after tamoxifen (Figure 1I). The percentage of traced β (INS+) cells was unaltered between 1 week and 5 weeks after tamoxifen but showed a significant increase between 5 and 12 weeks after tamoxifen (Figure 1J), corresponding to the decrease in traced δ cells. Over 12 weeks, this accounted for an increase of 0.66% of traced β cells per week, very similar to the rate of dilution found in the Ins1-CreERT; R26-CAG-tdTomato experiment (Figure 1C). These data suggest that an influx of Hnf1b-traced cells into the islet occurs over time, which contributes to maintaining the β cell population.
Ductal Ngn3+ cells are present in the adult pancreas and express SST
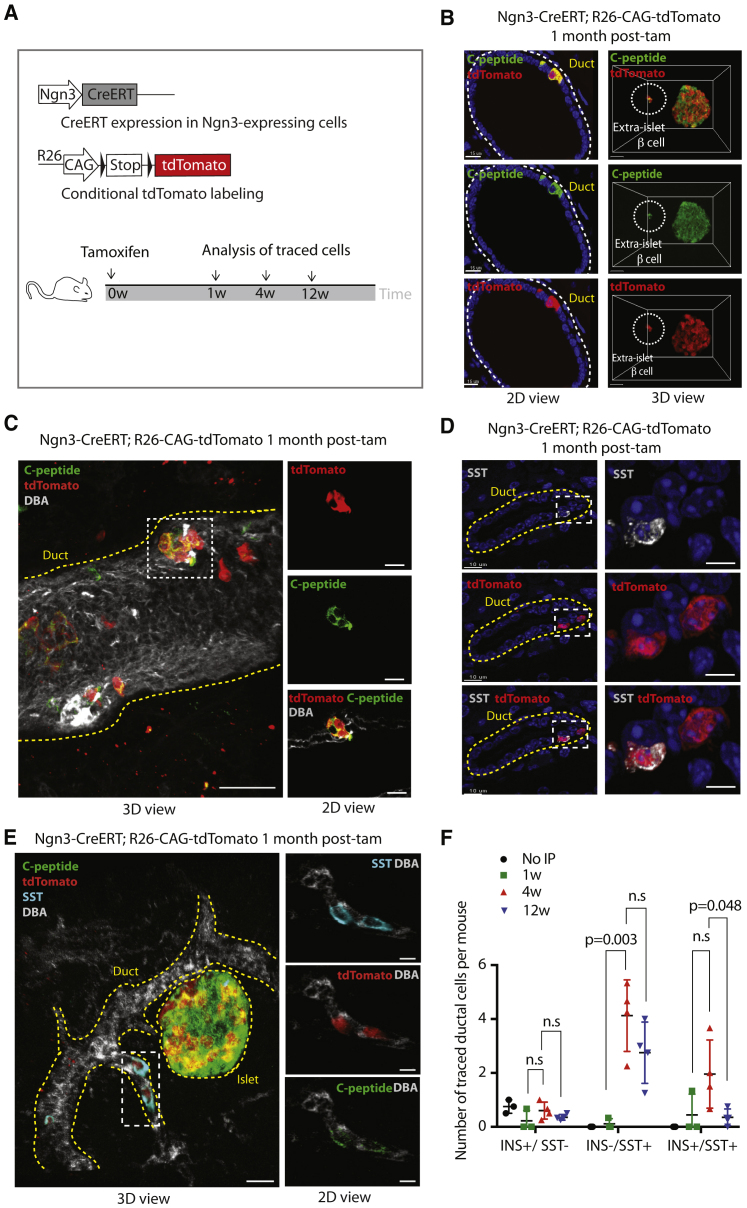
Ductal cells have been shown to be able to transdifferentiate to β cells upon forced expression of transcription factors, including Ngn3 (Al-Hasani et al., 2013; Sancho et al., 2014; Vieira et al., 2018), so we reasoned that, if ductal cells were to give rise to β cells, then Ngn3 expression should be necessary. To analyze the differentiation potential and progeny of Ngn3+ cells, we used Ngn3-CreERT; R26-CAG-tdTomato mice (Figure 2A). Ngn3-traced cells were observed in the ducts and islets and often expressed endocrine markers (Figure 2B, left panel). 3D fast light-microscopic analysis of antibody-stained whole organs(FLASH) imaging of the pancreas (Messal et al., 2019) revealed traced cells located outside of ducts and islets (Figures 2B, right panel, and S2A). In the ducts, mini-islet-like clusters of cells positive for the β cell marker C-peptide were found (Figures 2C and S2B). Moreover, imaging revealed cells in ducts that expressed the δ cell marker SST (Figure 2D) and often co-expressed C-peptide and INS (Figure 2E; Video S1). Quantification of traced ductal cells revealed that the large majority of duct-resident traced cells were positive for SST and only very rarely INS+/SST− (Figure 2F). More hormone-expressing Ngn3-traced ductal cells were observed 4 weeks after tamoxifen compared with 12 weeks after tamoxifen, suggesting that these cells may leave the ducts over time (Figure 2F). 3D imaging revealed SST+ traced cells that lowly expressed C-peptide located between a duct and islet (Figure S2A; Video S2). The location and elongated morphology of these cells supports the possibility that these cells might be migratory. These data suggest that a small population of ductal cells expresses Ngn3 and endocrine markers, predominantly SST, and that these may have the ability to leave the ductal epithelium.
Figure 2.
Ngn3-labeled cells found in ducts in homeostasis express SST
(A) Lineage tracing and experimental timeline.
(B) Left: double IF for tdTomato and C-peptide in Ngn3-CreERT; R26-CAG-tdTomato mice, showing a C-peptide+/tdTomato+ cell in a duct. Scale bar, 15 μm. Right: 3D view following FLASH for tdTomato and C-peptide, showing an extra-islet C-peptide+/tdTomato+ cell. Scale bar, 20 μm.
(C) 3D view following FLASH for tdTomato, C-peptide, and Dolichos biflorus agglutinin (DBA). An islet-like structure associated with the ductal epithelium is boxed. 3D view scale bar, 40 μm; 2D view scale bar, 20 μm.
(D) Double IF for tdTomato and SST, with enlargement shown on the right. Scale bars, 10 μm (left) and 5 μm (right).
(E) 3D view following FLASH for tdTomato, C-peptide, SST, and DBA in an Ngn3-CreERT; R26-CAG-tdTomato mouse pancreas 1 month after tamoxifen. Quadruple-positive cells within the duct (white box) are shown enlarged as a single slice. Scale bars, 40 μm (3D view) and 7 μm (2D views).
(F) Quantification of traced ductal cells per mouse at the listed time points after tamoxifen that are INS+/SST−, INS−/SST+, or INS+/SST+. N = 3 “no intraperitoneal (IP) tamoxifen injection ,” N = 3 at 1 week, N = 4 at 4 weeks, and N = 4 at the 12 week time point. The average of 4 sections per mouse is shown. Data are represented as mean with SD and p values are indicated; n.s (p>0.05).
Ngn3+ ductal cell numbers are increased in diabetes
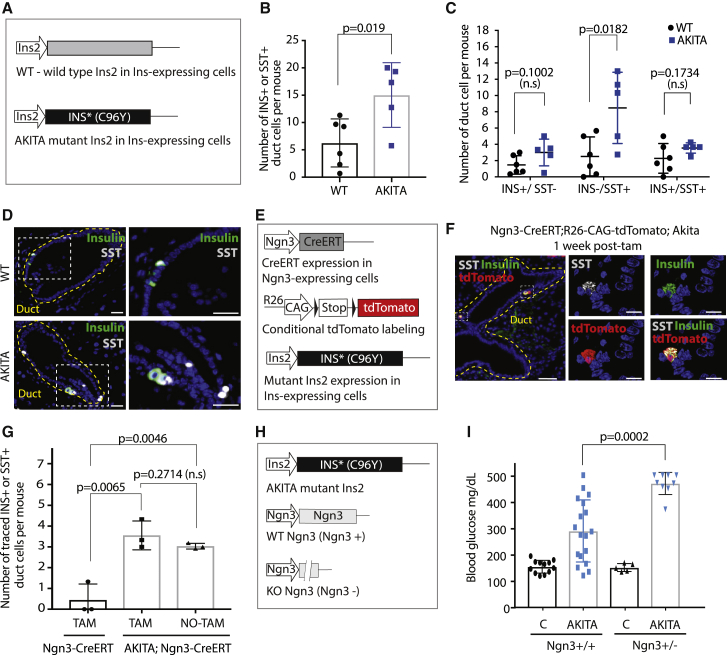
The putative contribution of ductal Ngn3+ cells to maintaining the β cell population in homeostasis is likely minor compared with proliferation because the dilution of traced β cells was small (Figure 1C). To determine whether Ngn3+ cells play a larger role in responding to increased β cell demand, we used the AKITA model of diabetes. AKITA mice harbor a point mutation in the Ins2 gene (Yoshioka et al., 1997; Figure 3A) that causes misfolding of INS and gradual β cell death (Ron, 2002). SST+ or INS+ ductal cells were more common in diabetic mice compared with controls (Figure 3B), with more INS−/SST+ ductal cells found in AKITA mice (Figures 3C and 3D). To further investigate the importance of Ngn3+ cells in the diabetic context, Ngn3-CreERT; R26-CAG-TdTomato; AKITA mice were generated (Figure 3E). Following tamoxifen administration, a substantial increase in traced INS+ or SST+ cells found in ducts was observed in diabetic mice (Figures 3F and 3G). This suggested that the number of duct-resident Ngn3+ cells increases during diabetes. To assess the relevance of Ngn3+ cells in diabetes, one allele of ngn3 was inactivated in an AKITA background (Figure 3H). ngn3 heterozygosity did not affect blood glucose levels or β cell area in wild-type mice (Figures 3I, S2C, and S2D). As expected, 80% of AKITA males were hyperglycemic at 7 weeks of age, with an average blood glucose level of 292 mg/dL (Figure 3I). Heterozygous loss of ngn3 in AKITA mice worsened the phenotype, with all animals being diabetic and exhibiting very high average blood glucose values of 473 mg/dL (Figures 3H and 3I). These data suggest that endocrine neogenesis from Ngn3+ ductal cells is enhanced during diabetes, which may be a regenerative response to β cell loss.
Figure 3.
Ngn3+ ductal cell contribution to the endocrine cell population is increased during diabetes
(A) Schematic of the AKITA model of diabetes.
(B) Quantification of SST+ or INS+ duct cells per mouse in age-matched (8–12 weeks of age) control (wild type [WT]) or diabetic (AKITA) mice. N = 6 WT and N = 5 AKITA. Diabetic mice exhibited non-fasting blood glucose of 350–500 mg/dL. Data are represented as mean with SD and p values are indicated.
(C) Quantification of ductal cells per mouse in WT or AKITA (diabetic) mice that are INS+/SST−, INS−/SST+, or INS+/SST+. N = 6 WT and N = 5 AKITA. The average of 4 sections per mouse is shown. Data are represented as mean with SD and p values are indicated
(D) Double IF for INS and SST in WT and AKITA. Hormone-expressing ductal cells are shown enlarged (boxed). Scale bars, 25 μm.
(E) Schematic of strategy to label Ngn3-expressing cells in AKITA mice.
(F) Triple IF for INS, SST, and tdTomato. Hormone-expressing cells near the ductal epithelium are shown enlarged (boxed). Scale bars, 50 μm (left) and 10 μm (right).
(G) Quantification of traced SST+ or INS+ duct cells per mouse in age-matched Ngn3-CreERT tamoxifen-injected (N = 3), AKITA; Ngn3-CreERT tamoxifen-injected (N = 3), and Ngn3CreERT non-injected (N = 4) mice. Mice were injected at 8–12 weeks of age and analyzed 1 week after tamoxifen. Data are represented as mean with SD and p values are indicated.
(H) Schematic of the breeding strategy. Akita+/− mice were crossed to Ngn3+/− mice.
(I) Non-fasted blood glucose levels in mice of listed genotypes measured at 7 weeks of age. Control Ngn3+/+, N = 12, AKITA Ngn3+/+, N = 18; control Ngn3+/−, N = 5; AKITA Ngn3+/−, N = 9. Data are represented as mean with SD and p values are indicated.
See also Figure S2.
β Cell mass is known to increase during pregnancy (Kim et al., 2010). Lineage tracing was initiated in Hnf1b-CreERT; R26-CAG-TdTomato mice, followed by mating to induce pregnancy (Figures S2E and S2F). However, the percentages of lineage-traced INS+ β cells were actually decreased by pregnancy, suggesting that the β cell mass increase in pregnancy is not mediated by ductal endocrine progenitors (Figures S2E–S2H).
Single-cell transcriptomics identifies a Ngn3+/SST+ population able to differentiate to β cells
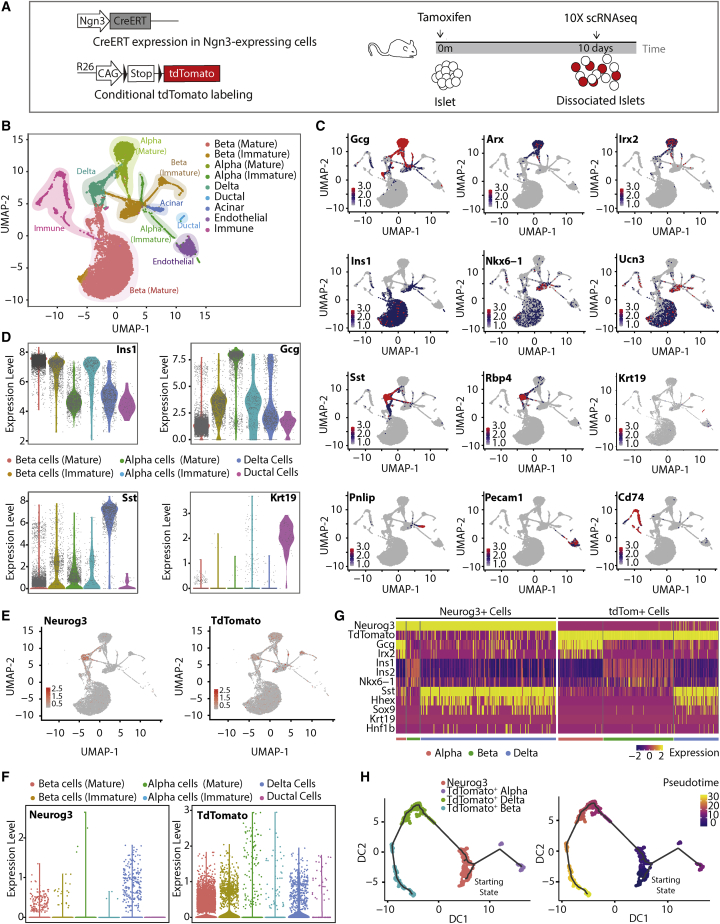
Because our data suggest that Ngn3+ ductal cells contribute to the islet endocrine cell population over time (Figures 1 and 2), we decided to analyze Ngn3-traced islet cells to determine their origin. To this end, we performed scRNA-seq of islets isolated from Ngn3-CreERT; R26-CAG-tdTomato mice 10 days after tamoxifen (Figure 4A), a time point when traced cells are observed in the islets and neogenesis may have occurred.
Figure 4.
scRNA-seq analysis reveals an adult Ngn3+ population able to differentiate into endocrine cells
(A) scRNA-seq experimental design.
(B) UMAP projection of 21,813 pancreatic islet cells. Cells are colored by assigned cluster.
(C) Expression of α (Gcg, Arx, and Irx2), β (Ins1, Nkx6-1, and Ucn3), δ (Sst and Rbp4), ductal (Krt19), acinar (Pnlip), endothelial (Pecam1), and immune (Cd74) cell markers across the scRNA-seq dataset.
(D) Violin plots for expression of Ins1, Gcg, Sst, and Krt19 across identified clusters.
(E) UMAP projection of Ngn3 (Neurog3) and tdTomato expression across a single-cell dataset.
(F) Violin plots for expression of Ngn3 in Ngn3+ cells (left) and tdTomato in tdTomato+ cells (right) across the identified clusters.
(G) Expression heatmap of α, β, δ, and ductal cells markers across Ngn3+ and tdTomato+ cells.
(H) Monocle 3.12 pseudotime analysis of Ngn3+ and tdTomato+ cells (left, predicted lineage trajectories; right, pseudotime projection in the predicted trajectories).
See also Figure S3.
Following isolation, islets were dissociated to single cells and processed for transcriptomic analysis using 10X Chromium technology. After quality control steps, our dataset contained 21,813 cells (Figure S3). Following principal-component analysis (PCA), uniform manifold approximation and projection (UMAP) was performed, which revealed distinct clusters (Figure 4B). The identity of these clusters was allocated based on the expression of bona fide markers for β (Ins1), α (Gcg), δ (Sst), ductal (Krt19), immune (Cd74), and endothelial cells (Pecam1) (Figures 4B–4D and S3). Further analysis of the expression of markers for each cell type confirmed accurate cluster identities (Figure 4C). Interestingly, the β cell cluster (Ins1+ cells) subdivided into Ucn3low/Ins1low (immature β cells) and Ucn3high/Ins1high (mature β cell) populations, and the α cell cluster contained Gcghigh (mature) and Gcglow (immature) cells, suggesting heterogeneity in these endocrine populations (Figures 4D and S3). Ngn3+ cells (n = 227) and tdTomato+ cells (n = 365) were identified within the dataset (Figure 4E). Strikingly, although Ngn3+ cells were mostly found within the δ cell cluster, tdTomato+ cells were mainly clustered in the mature β cell group (Figure 4F). The tdTomato+ β cell transcriptome was indistinguishable from the transcriptome of mature tdTomato− β cells from our dataset and from previous published datasets (Figures S4A and S4B) and they expressed the mature β cell markers C-peptide and UCN3 (Figures S4C and S4D). Expression of Ngn3 in tdTomato cells was minimal 10 days after tamoxifen, indicating that expression of Ngn3 is transient. Further analysis of gene expression associated with the different islet cell types in Ngn3+ or tdTomato+ cells demonstrated that both groups exhibit distinct signatures, with the ductal markers Hnf1b and Sox9 found to be expressed in some Ngn3+ and tdTomato+ cells, suggesting a ductal origin (Figure 4G). To address whether we could infer the lineage path of the traced β cells based on individual single-cell transcriptomic signatures, we performed a pseudotime trajectory analysis (Trapnell et al., 2014) on Ngn3+/tdTomato+ cells. Interestingly, a lineage trajectory was predicted where Ngn3+ cells (starting state) give rise to tdTomato+ α (Gcg+) cells and tdTomato+ δ (Sst+) cells and where the latter are precursors of tdTomato+ β (Ins+) cells (Figure 4H). Thus, this analysis is in agreement with our lineage-tracing studies (Figures 1I–1L), which suggested that duct-derived SST+ δ cells give rise to β cells in the adult pancreas.
Discussion
Here we show that β cell neogenesis occurs in the adult pancreas from Ngn3-traced ductal cells. Immunofluorescence analysis of paraffin sections and analysis of the pancreas by 3D FLASH imaging, complemented by scRNA-seq of islets containing cells traced from Ngn3+ cells, indicates that Ngn3+ cells can become INS+ cells via an intermediate SST+ state. Higher numbers of hormone-positive ductal cells traced from Ngn3+ cells were observed in diabetic mice, and heterozygous loss of Ngn3 accelerated diabetes onset. The adult β cell population is believed to be maintained mainly by proliferation, but our study identified a second physiologically important mechanism of β cell neogenesis.
Our lineage tracing of β cells suggested that some β cells are not derived from β cell replication over time (0.68% of β cells per week) (Figure 1C). Use of Hnf1b-CreERT; R26-CAG-tdTomato mice resulted in labeling of 94% of ductal cells (Figure 1F), and an increased contribution of Hnf1b-expressing cells to the β cell population over time was observed (Figures 1H and 1J). This accounted for 0.66% of β cells per week, strikingly similar to the rate of dilution of β cells traced by Ins1-CreERT (Figure 1C).
Because δ cell labeling was also observed using the Hnf1b-CreERT lineage tracer (Figure 1I), we cannot rule out a contribution of islet δ cells to the traced β cell population over time. However, the percentage of tdTomato-labeled islet cells increased significantly over time (Figure 1G), suggesting that an influx of traced cells from outside of the islet occurred. The percentage of SST+-labeled cells dropped 12 weeks after tamoxifen injection, which coincided with a significant increase of labeled β cells observed at 12 weeks (Figures 1I and J). This is consistent with differentiation of duct-derived SST+ cells into β cells, corroborated by the lineage trajectory inferred from scRNA-seq (Figure 4H). It has been reported that ectopic expression of Pax4 in pancreatic δ cells results in β-like cell neogenesis (Druelle et al., 2017); our results suggest that this transdifferentiation process also occurs physiologically to maintain the β cell population.
We observed Ngn3-traced bi-hormonal (INS+/SST+) and mono-hormonal (INS+ or SST+) cells within ducts that were sometimes aggregated in small islet-like clusters (Figures 2C and S2B). Furthermore, scRNA-seq suggested a ductal origin of some of the traced islet cells and predicted INS+/SST+ cells as intermediates to the newly formed β cells (Figure 4H). This suggests that the pancreatic ducts harbor a progenitor cell population capable of differentiation into different endocrine cell types.
Contribution of ductal cells to the islet population requires cell migration. 3D imaging revealed traced Ngn3+ cells and INS+/SST+ cells located between ducts and islets (Figure S2A; Video S2), as would be predicted for duct-derived endocrine progenitor cells migrating toward the islets. Thus, our study suggests that β cell neogenesis can occur from the ductal compartment in homeostasis (Figure S4E).
The acceleration of diabetic onset in Akita+/− Ngn3+/− mice, combined with increased numbers of Ngn3-traced INS+/SST+ ductal cells in diabetic mice (Figure 3), suggests that Ngn3-mediated adult β cell neogenesis is enhanced in diabetes. Ductal cells have been considered a potential source of β cells in humans because of observations of endocrine cells within the ductal epithelium, with INS+ ductal cells observed, especially in obese individuals (Butler et al., 2003). Further characterization of adult endocrine neogenesis may reveal mechanisms and enable harnessing of this physiological β cell source, which could be employed for the benefit of individuals with diabetes.
Limitations of study
In this study, we identified Ngn3+ ductal progenitors as a source of adult β cell neogenesis. We only analyzed these cells up to 8 months in age in mice, and it is conceivable that the contribution of duct-derived β cell neogenesis changes with age. Longer-term lineage tracing is needed to address this.
Although the concept of exploiting this progenitor population for diabetes treatment is attractive, these SST+ ductal endocrine progenitors remain uncharacterized. Approaches like scRNA-seq of the ductal compartment, focusing on SST+ ductal cells, is necessary to further characterize these cells before our findings can be exploited therapeutically.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-C-peptide | Cell Signaling | 4593 (RRID: AB_10691857) |
| Goat anti-GFP | Abcam | ab6673 (RRID: AB_305643) |
| Mouse anti-Insulin | Sigma-Aldrich | I2018 (RRID: AB_260137) |
| Rabbit anti-RFP | TebuBio | 600-401-379 (RRID: AB_2209751) |
| Rat-anti-Somatostatin | Abcam | ab30788 (RRID: AB_778010) |
| Goat anti-TdTomato | Biorbyt | Orb182397 (RRID: AB_2687917) |
| Mouse anti-Urocortin3 | Phoenix Pharmaceuticals | Cat# H-019-29 |
| Alexa Fluor 488 donkey anti-mouse | Thermo | A-21202 (RRID: AB_141607) |
| Alexa Fluor 546 donkey anti-rabbit | Thermo | A10040 (RRID: AB_2534016) |
| Alexa Fluor 647 goat anti-rat | Thermo | A-21247 (RRID: AB_141778) |
| Alexa Fluor 488 goat anti-goat | Thermo | A-11055 (RRID: AB_2534102) |
| Chemicals, peptides, and recombinant proteins | ||
| Fluorescein labeled Dolichos Biflorus Agglutinin (DBA) | Vector labs | FL-1031 |
| Tamoxifen | Sigma | T5648 |
| Peanut oil | Sigma | P2144 |
| Formalin solution, neutral buffered, 10% | Sigma | HT501128 |
| Bovine Serum Albumin | Sigma | A7906 |
| Trisodium citrate dihydrate | Sigma | S1804 |
| Methyl salicylate | Sigma | M6752 |
| Triton X-100 | Sigma | T9284 |
| Sudan Black B | Sigma | 199664 |
| Methanol | Sigma | 32213 |
| Histo-Clear | Fisher | 12358637 |
| Deposited data | ||
| scRNA-seq of islet cells from Ngn3-CreERT; R26-CAG-tdTomato mice | This paper | GEO: GSE138038 |
| Experimental models: Organisms/strains | ||
| Mouse: Hnf1bTg(Hnf1b-cre/ERT2)1Jfer | Solar et al., 2009 | (RRID: IMSR_JAX:027681) |
| Mouse: (Tg)(Ins1-cre/ERT)1Lphi | The Jackson Laboratory | Jax: 024709 (RRID: IMSR_JAX:024709) |
| Mouse: Tg(Neurog3-cre/Esr1∗)1Dam | The Jackson Laboratory | Jax: 008119 (RRID: IMSR_JAX:008119) |
| Mouse: C57BL/6-Ins2Akita/J | The Jackson Laboratory | Jax: 003548 (RRID: IMSR_JAX:003548) |
| Mouse: tm9.1(CAG-tdTomato)Hze | The Jackson Laboratory | Jax: 007909 (RRID: IMSR_JAX:007909) |
| Mouse: Neurog3tm1(EGFP)Khk | MMRRC | 000344-UCD (RRID: MMRRC_000344-UCD) |
| Software and algorithms | ||
| StrataQuest Analysis Software | Tissue Gnostics | https://tissuegnostics.com/en/products/analysing-software/strataquest |
| Zen 2.3 | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen-lite.html |
| Imaris 8.3.1 | Bitplane | https://imaris.oxinst.com/packages |
| Cell Ranger 3.2.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| RStudio 3.6.1 | RStudio | https://www.rstudio.com/ |
| Prism 7 for MacOS | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| FastQC | Andrews, 2010 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc |
| Cutadapt | Martin, 2011 | https://github.com/marcelm/cutadapt |
| Scater | McCarthy et al., 2017 | https://bioconductor.org/packages/release/bioc/html/scater.html |
| Seurat | Satija et al., 2015 | https://satijalab.org/seurat/ |
| TSCAN | Ji and Ji, 2016(Ji and Ji, 2016) | https://bioconductor.org/packages/release/bioc/html/TSCAN.html |
| SCE (Single Cell Experiment) | Amezquita et al., 2020(Amezquita et al., 2020) | https://bioconductor.org/packages/release/bioc/html/SingleCellExperiment.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Axel Behrens (axel.behrens@icr.ac.uk).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animal models
All animal experiments were approved by the Francis Crick Institute’s Animal Welfare and Ethical Review Body and conformed to UK Home Office regulations under the Animals (Scientific Procedures) Act 1986 including Amendment Regulations 2012. Mouse lines used were described previously: Hnf1b-CreERT (MGI: 4420452 Tg(Hnf1b-cre/ERT2)1Jfer) (Solar et al., 2009), Ngn3-CreERT (MGI: 3796205 Neurog3tm1(Neurog3,cre/ERT)Ggu) (Gu et al., 2002), Ins1-CreERT (MGI: 4410453 Tg(Ins1-cre/ERT)1Lphi (Wicksteed et al., 2010), Ngn3+/− (MGI: 2651394 Neurog3tm1(EGFP)Khk) (Lee et al., 2002), AKITA (MGI: 1857572 Ins2Akita) (Yoshioka et al., 1997) and Rosa26-CAG-tdTdtomato (MGI: 3809523 Gt(ROSA)26Sortm9(CAG-tdTomato)Hze) (Madisen et al., 2010). For lineage tracing, adult mice (over 8 weeks of age, male and female) were injected intraperitoneally (I.P) with 20 mg/ml tamoxifen at a volume of 5 μL per gram of body weight once daily for 2 days. For blood glucose sampling, a small incision was made at the saphenous vein. Only 1 drop of blood was required. Measurements were recorded using a FreeStyle glucose meter (Abbott), using a FreeStyle disposable strip (Abbott). Non-fasting glucose measurements were performed. Glucose levels were measured at 7 weeks of age in AKITA +/− males and WT male litter mates were used as controls. Details of age and sex of the mice used can be found in Table S1.
Method details
Processing tissue for immunofluorescent (IF) staining
The pancreas was dissected and fixed in 10% NBF (Neutral buffered formalin) overnight. Samples were transferred to 70% ethanol, dehydrated and embedding in paraffin. 4 μm tissue sections were cut using a microtome (RM2235 Leica) and placed in a water bath at 37°C. Sections were then mounted on Superfrost Ultra Plus charged slides and incubated in an oven at 37°C for 15 minutes to encourage adherence.
Immunofluorescent (IF) staining of sections
Slides were dewaxed in HistoClear twice for 5 minutes before being washed in 100% ethanol for 5 minutes. Slides were then passed through a re-hydration series for 5 minutes in each of 95%, 90%, 80% and 50% ethanol, then dH2O. Heat-mediated antigen retrieval was performed using 10mM citrate buffer pH 6.2. The buffer was pre-warmed in a microwave until gently bubbling, before slides were submerged and heated for 15 minutes in the microwave on 50% power to maintain the temperature of the buffer. Slides were then cooled and washed twice briefly in PBS. Blocking solution containing 1% (w/v) BSA, 5% (v/v) donkey serum and 0.1% (v/v) Triton X-100 was applied to the sections for at least 30 minutes at room temperature in a humidified chamber. Primary antibodies were then diluted in blocking solution and incubated overnight at 4°C in a humidified chamber. Slides were washed for 5 minutes in PBS three times before fluorophore-conjugated secondary antibodies plus DAPI were applied for 45 minutes at room temperature in the humidified chamber. Following this, slides were washed for 5 minutes in PBS three times before being submerged in 0.1% (w/v) Sudan Black B in 70% ethanol (filtered) for 15 minutes at room temperature to reduce autofluorescence. Slides were washed for 5 minutes in PBS three times and mounted in 1 drop of DAKO fluorescent mounting media.
FLASH (3D-IF staining)
FLASH (fast light-microscopic analysis of antibody-stained whole organs) was performed as described (Messal et al., 2019). Cardiac perfusion was performed with 20ml of PBS before tissue was dissected and fixed overnight in 10% NBF at room temperature. The sample was washed in PBT (0.4% Triton X-100 in PBS) 3 times for 1 hour per wash and incubated in antigen retrieval buffer (200mM boric acid plus 4% SDS pH7.0) overnight at 54°C. The sample was washed in PBT 3 times for 1 hour per wash, before being moved to blocking buffer (1% BSA, 5% DMSO, 10% FCS, 0.02% sodium azide and 0.2% Triton X-100) in PBS and incubated overnight at room temperature. Primary antibodies were then incubated in blocking buffer for at least 2 nights at room temperature on a nutator. Samples were washed in PBT 3 times for 1 hour per wash before fluorophore-conjugated secondary antibodies were applied for 2 nights at room temperature on a nutator. Samples were washed in PBS 3 times for 30 minutes per wash and passed through a dehydration series of 30%, 50%, 75% and then 2x 100% methanol for 1h in each solution, protected from light. Dehydrated samples were then gradually cleared by submerging in methyl salicylate diluted in methanol at 25%, 50%, 75% and 2x 100% methyl salicylate for 30 minutes each in a glass dish protected from light. Cleared samples were then mounted on a glass slide in 100% methyl salicylate.
Imaging
For pancreas sections, entire tissue sections were imaged using a Zeiss Axio Scan Z1 slide scanner. A 20x objective was used, detecting signal from Alexa Fluor 488, Alexa Fluor 546 and Alexa Fluor 647-conjugated secondary antibodies, along with DAPI. Multiple images were stitched to form 1 large image file of the complete section that can be navigated using Zen software. Regions of interest were often further imaged manually using a Zeiss upright 710 confocal microscope and a 63x objective. For imaging following FLASH, a Zeiss LSM 780 confocal microscope with a 10x and 25x objective was used. Three-dimensional image analysis was performed using Imaris software. In FLASH images ducts were identified as DBA positive structures. In tissue sections ducts were identified by Hnf1b-tracing and by DAPI staining, which enables identification by ductal morphology. For display purposes images were cropped to display relevant areas.
Preparing islet cells for single cell RNA-sequencing
For the single cell RNA-seq, islets were pooled from six Ngn3-CreERT; R26-CAG-TdTomato mice at 12 weeks of age, at 10 days post-tamoxifen injection. Data was pooled from 2 separate experiments (batch 1 and 2), which included 6 mice in total. Mice were culled by cervical dislocation and the pancreas was perfused with 1mg/ml Collagenase type XI in MEM media. The inflated pancreas was then collected and digested in 1mg/ml Collagenase type XI at 37°C for 10 minutes. Digestion was terminated by adding ice cold complete MEM (MEM plus 10% FCS and 1% P/S) and the sample was centrifuged at 1,400 rpm for 90 s. The pellet was resuspended in complete MEM and spun again to wash the sample. This was repeated 3 times. The suspension was then sieved, centrifuged and resuspended in 15ml of histopaque-1077. 10ml of complete MEM was gently added and the sample was centrifuged at 3,500 rpm for 24 minutes at 4°C with no break. Islets were collected at the histopaque/media interface and washed 3 times in 50ml of complete MEM. Approximately 300 islets were then manually picked using a dissection microscope to increase purity. Islets were pelleted by spinning at 1000 rpm for 5 minutes, resuspended in 1ml of TrypLE express and incubated at 37°C for 20 minutes with occasional pipetting to mix. Reaction was stopped with ice cold PBS and the sample was pelleted. Cells were resuspended in 0.04% BSA and filtered through a 35 μm nylon mesh and processed immediately for single cell RNA-sequencing.
scRNaseq Library Preparation and Sequencing
Cell viability and number were assessed by Trypan blue staining and the Eve automatic cell counter respectively. Approximately 10,000-15,000 cells were loaded into a channel of the 10x Chromium Chip B - according to the 10x Genomics Single Cell 3′ Reagent Kit v3 GEM protocol. RNA was reverse transcribed via 11 PCR cycles, followed by a further 12 PCR cycles for cDNA amplification. Library size was quantified via Agilent TapeStation. scRNaseq library sequencing was performed on the HiSeq 4000 (Illumina) platform, using sequencing parameters: 8-98-28.
Generation of single cell expression matrix
The 10X Genomics Single Cell Software Suite, Cell Ranger 5.0.0 was used to process the scRNaseq data (https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome). Demultiplexing was performed based on the 8 bp sample index read to generate FASTQ files for the Read1 and Read2 paired-end reads. Raw reads were pre-processed with the quality control tools FastQC (Andrews, 2010) and Cutadapt (Martin, 2011). Cell Ranger Count aligned reads to the mm10 reference genome via STAR (Dobin et al., 2013). To enable mapping of tdTomato-derived transcripts, the sequence for tdTomato was added to the reference genome and GTF file. Aligned sequence reads with valid cell barcodes mapping to exons (Ensembl GTF GRCm38.93) were used to generate the gene expression matrix.
scRNaseq Quality Control and Cell Filtering
Using the Bioconductor package Scater 3.9 (McCarthy et al., 2017) low quality cells were removed based upon library size and the number of expressed gene transcripts (Figure S3A). We also removed cells with high mitochondrial read proportion (Figure S3B), indicative of apoptosis. Finally, total cell features were plotted against log10 transformed number of total reads and visible outliers removed (Figure S3C). The scRNaseq dataset (post QC 21813 cells) was a combination of post-QC cells from batch 1 (post QC 1183 cells) and batch 2 (post QC 20630 cells).
Dimensionality Reduction, UMAP Visualization and Clustering Analysis
Cells which passed QC metrics were passed to Seurat 4.0.0 (Satija et al., 2015) for downstream analysis. Normalization was performed using ‘regularized negative binomial regression’ and the data scaled based upon the top 5000 variable genes. Dimensionality reduction was performed via principal-component analysis (PCA) to enable cell clustering. The top 15 principal components (PCs) were selected – indicated via the inbuilt permutation-based test and passed to Uniform Manifold Approximation and Projection (UMAP) for clustering visualization (van der Maaten and Hinton, 2008). The clustering analysis was performed with a resolution value of 0.5 and cluster number validated by scree plot (Figure S3D).
Differential Expression Analysis and Cluster Identification
Differentially expressed genes between clusters were identified by comparing average intra-cluster expression values against expression in cells of all other clusters using the Seurat DeSeq2 workflow (McDavid et al., 2013) with standard parameters used. The top 10 differentially expressed genes per cluster were used for identification and compared to known marker genes (Muraro et al., 2016) for validation.
Pseudotime Trajectory Analysis
Pseudotime trajectory analysis of Ngn3+ and tdTomato+ cells was performed using the Bioconductor package Monocle 3.12 (Ji and Ji, 2016). In brief, raw counts for Ngn3+ and tdTomato+ cells were subsetted and pre-processed prior to dimensionality reduction via PCA. Cells were clustered to form a minimal spanning tree enabling cell ordering along their predicted trajectory as previously described.
Quantification and statistical analysis
All statistics were calculated using GraphPad Prism 7, where unpaired t tests were performed to compare time points. P values are specified in figures.
Percentage of islet beta cells traced
The percentage of INS+ islet cells traced was determined by using a custom-made analysis profile using Strataquest software. Analysis was performed on scanned sections. The software defined multicellular INS+ structures as islets, TdTomato+ cells as traced cells and used DAPI to define single cells. By performing an overlay, the percentage INS+/tdTomato+ islet cells was determined. Data from 4 sections per mouse were averaged and presented per mouse.
Ductal labeling efficiency
To determine the ductal recombination efficiency in Hnf1b-CreERT; R26-CAG-tdTomato mice, manual quantification was performed. Whole pancreas sections were analyzed following IF staining for tdTomato. Ducts were identified based on DAPI staining and morphology. For each duct, each cell was counted and the percentage of cells within a duct that were tdTomato positive was calculated.
INS+ and SST+ cells in ducts
To quantify the number of INS+ and SST+ cells in ducts, whole pancreas sections were scanned following IF for TdTomato, Insulin and SST. The number of ductal cells expressing the markers were then manually counted.
Acknowledgments
We thank Dr. J. Ferrer for the Hnf1b-CreERT mouse line. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001039), the UK Medical Research Council (FC001039), and the Wellcome Trust (FC001039); by King’s College London; by a UK Medical Research Council grant (MR/S000011/1 to R.S.); and by a Wellcome Seed Award in Science (207529/Z/17/Z to R.S.). C.L. is supported by the MRC-KCL Doctoral Training Partnership in Biomedical Sciences (MR/N013700/1).
Author contributions
C.G., C.L., R.S., and A.B. conceived the study, analyzed data, and wrote the paper. C.G. performed lineage tracing, C.G. and C.L. performed immunofluorescence, C.L. performed the single-cell analysis, I.E. assisted with the in vivo experiments, H.A.M. developed and performed the FLASH imaging, C.R. and E.-L.H. performed islet isolation for RNA-seq, and H.H. and P.J. assisted with study design and interpretation.
Declaration of interests
The authors declare no competing interests.
Published: September 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stem.2021.08.003.
Contributor Information
Rocio Sancho, Email: rocio.sancho@kcl.ac.uk.
Axel Behrens, Email: axel.behrens@icr.ac.uk.
Supplemental information
Data and code availability
All high-throughput sequencing data, both raw and processed files, have been deposited in NCBI’s Gene Expression Omnibus and are accessible under accession number GEO: GSE138038.
References
- Al-Hasani K., Pfeifer A., Courtney M., Ben-Othman N., Gjernes E., Vieira A., Druelle N., Avolio F., Ravassard P., Leuckx G., et al. Adult duct-lining cells can reprogram into β-like cells able to counter repeated cycles of toxin-induced diabetes. Dev. Cell. 2013;26:86–100. doi: 10.1016/j.devcel.2013.05.018. [DOI] [PubMed] [Google Scholar]
- Amezquita R.A., Lun A.T.L., Becht E., et al. Orchestrating single-cell analysis with Bioconductor. Nat Methods. 2020;17:137–145. doi: 10.1038/s41592-019-0654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. Babraham Bioinformatics; 2010. FastQC: a quality control tool for high throughput sequence data. [Google Scholar]
- Butler A.E., Janson J., Bonner-Weir S., Ritzel R., Rizza R.A., Butler P.C. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- Chera S., Baronnier D., Ghila L., Cigliola V., Jensen J.N., Gu G., Furuyama K., Thorel F., Gribble F.M., Reimann F., Herrera P.L. Diabetes recovery by age-dependent conversion of pancreatic δ-cells into insulin producers. Nature. 2014;514:503–507. doi: 10.1038/nature13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y., Brown J., Martinez O.I., Melton D.A. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- Druelle N., Vieira A., Shabro A., Courtney M., Mondin M., Rekima S., Napolitano T., Silvano S., Navarro-Sanz S., Hadzic B., et al. Ectopic expression of Pax4 in pancreatic δ cells results in β-like cell neogenesis. J. Cell Biol. 2017;216:4299–4311. doi: 10.1083/jcb.201704044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradwohl G., Dierich A., LeMeur M., Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. USA. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G., Dubauskaite J., Melton D.A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Inada A., Nienaber C., Katsuta H., Fujitani Y., Levine J., Morita R., Sharma A., Bonner-Weir S. Carbonic anhydrase II-positive pancreatic cells are progenitors for both endocrine and exocrine pancreas after birth. Proc. Natl. Acad. Sci. USA. 2008;105:19915–19919. doi: 10.1073/pnas.0805803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z., Ji H. TSCAN: Pseudo-time reconstruction and evaluation in single-cell RNA-seq analysis. Nucleic Acids Res. 2016;44:e117. doi: 10.1093/nar/gkw430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharroubi A.T., Darwish H.M. Diabetes mellitus: The epidemic of the century. World J. Diabetes. 2015;6:850–867. doi: 10.4239/wjd.v6.i6.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Toyofuku Y., Lynn F.C., Chak E., Uchida T., Mizukami H., Fujitani Y., Kawamori R., Miyatsuka T., Kosaka Y., et al. Serotonin regulates pancreatic beta cell mass during pregnancy. 2010. Nat. Med. 2010;16:804–808. doi: 10.1038/nm.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopinke D., Murtaugh L.C. Exocrine-to-endocrine differentiation is detectable only prior to birth in the uninjured mouse pancreas. BMC Dev. Biol. 2010;10:38. doi: 10.1186/1471-213X-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopinke D., Brailsford M., Shea J.E., Leavitt R., Scaife C.L., Murtaugh L.C. Lineage tracing reveals the dynamic contribution of Hes1+ cells to the developing and adult pancreas. Development. 2011;138:431–441. doi: 10.1242/dev.053843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.S., Perreault N., Brestelli J.E., Kaestner K.H. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488–1497. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- McCarthy D.J., Campbell K.R., Lun A.T., Wills Q.F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDavid A., Finak G., Chattopadyay P.K., Dominguez M., Lamoreaux L., Ma S.S., Roederer M., Gottardo R. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013;29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means A.L., Xu Y., Zhao A., Ray K.C., Gu G. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis. 2008;46:318–323. doi: 10.1002/dvg.20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messal H.A., Alt S., Ferreira R.M.M., Gribben C., Wang V.M., Cotoi C.G., Salbreux G., Behrens A. Tissue curvature and apicobasal mechanical tension imbalance instruct cancer morphogenesis. Nature. 2019;566:126–130. doi: 10.1038/s41586-019-0891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro M.J., Dharmadhikari G., Grün D., Groen N., Dielen T., Jansen E., van Gurp L., Engelse M.A., Carlotti F., de Koning E.J.P., van Oudenaarden A. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016;3:385–394.e3. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D. Proteotoxicity in the endoplasmic reticulum: lessons from the Akita diabetic mouse. J. Clin. Invest. 2002;109:443–445. doi: 10.1172/JCI15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho R., Gruber R., Gu G., Behrens A. Loss of Fbw7 reprograms adult pancreatic ductal cells into α, δ, and β cells. Cell Stem Cell. 2014;15:139–153. doi: 10.1016/j.stem.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solar M., Cardalda C., Houbracken I., Martín M., Maestro M.A., De Medts N., Xu X., Grau V., Heimberg H., Bouwens L., Ferrer J. Pancreatic exocrine duct cells give rise to insulin-producing beta cells during embryogenesis but not after birth. Dev. Cell. 2009;17:849–860. doi: 10.1016/j.devcel.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Thorel F., Népote V., Avril I., Kohno K., Desgraz R., Chera S., Herrera P.L. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Casteele M., Leuckx G., Baeyens L., Cai Y., Yuchi Y., Coppens V., De Groef S., Eriksson M., Svensson C., Ahlgren U., et al. Neurogenin 3+ cells contribute to β-cell neogenesis and proliferation in injured adult mouse pancreas. Cell Death Dis. 2013;4:e523. doi: 10.1038/cddis.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L., Hinton G. Visualizing data using t-SNE. Machine Learning Research. Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- Vieira A., Vergoni B., Courtney M., Druelle N., Gjernes E., Hadzic B., Avolio F., Napolitano T., Navarro Sanz S., Mansouri A., Collombat P. Neurog3 misexpression unravels mouse pancreatic ductal cell plasticity. PLoS ONE. 2018;13:e0201536. doi: 10.1371/journal.pone.0201536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicksteed B., Brissova M., Yan W., Opland D.M., Plank J.L., Reinert R.B., Dickson L.M., Tamarina N.A., Philipson L.H., Shostak A., et al. Conditional gene targeting in mouse pancreatic ß-Cells: analysis of ectopic Cre transgene expression in the brain. Diabetes. 2010;59:3090–3098. doi: 10.2337/db10-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., D’Hoker J., Stangé G., Bonné S., De Leu N., Xiao X., Van de Casteele M., Mellitzer G., Ling Z., Pipeleers D., et al. Beta cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell. 2008;132:197–207. doi: 10.1016/j.cell.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Yoshioka M., Kayo T., Ikeda T., Koizumi A. A novel locus, Mody4, distal to D7Mit189 on chromosome 7 determines early-onset NIDDM in nonobese C57BL/6 (Akita) mutant mice. Diabetes. 1997;46:887–894. doi: 10.2337/diab.46.5.887. [DOI] [PubMed] [Google Scholar]
- Zhang M., Lin Q., Qi T., Wang T., Chen C.C., Riggs A.D., Zeng D. Growth factors and medium hyperglycemia induce Sox9+ ductal cell differentiation into β cells in mice with reversal of diabetes. Proc. Natl. Acad. Sci. USA. 2016;113:650–655. doi: 10.1073/pnas.1524200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Huang X., Liu Z., Pu W., Lv Z., He L., Li Y., Zhou Q., Lui K.O., Zhou B. Pre-existing beta cells but not progenitors contribute to new beta cells in the adult pancreas. Nat. Metab. 2021;3:352–365. doi: 10.1038/s42255-021-00364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All high-throughput sequencing data, both raw and processed files, have been deposited in NCBI’s Gene Expression Omnibus and are accessible under accession number GEO: GSE138038.