Abstract
To catalyze severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) research, including development of novel interventive and preventive strategies, the progression of disease was characterized in a robust coronavirus disease 2019 (COVID-19) animal model. In this model, male and female golden Syrian hamsters were inoculated intranasally with SARS-CoV-2 USA-WA1/2020. Groups of inoculated and mock-inoculated uninfected control animals were euthanized at 2, 4, 7, 14, and 28 days after inoculation to track multiple clinical, pathology, virology, and immunology outcomes. SARS-CoV-2–inoculated animals consistently lost body weight during the first week of infection, had higher lung weights at terminal time points, and developed lung consolidation per histopathology and quantitative image analysis measurements. High levels of infectious virus and viral RNA were reliably present in the respiratory tract at days 2 and 4 after inoculation, corresponding with widespread necrosis and inflammation. At day 7, when the presence of infectious virus was rare, interstitial and alveolar macrophage infiltrates and marked reparative epithelial responses (type II hyperplasia) dominated in the lung. These lesions resolved over time, with only residual epithelial repair evident by day 28 after inoculation. The use of quantitative approaches to measure cellular and morphologic alterations in the lung provides valuable outcome measures for developing therapeutic and preventive interventions for COVID-19 using the hamster COVID-19 model.
In December 2019, a novel β coronavirus was isolated from patients who presented with severe and ultimately fatal pneumonia in Wuhan, China.1 The virus was designated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and rapidly spread through human-to-human transmission, causing the current global pandemic of coronavirus disease 2019 (COVID-19). As of September 2021, there have been >218 million confirmed cases and >4.5 million deaths globally attributed to SARS-CoV-2 infection [World Health Organization: Coronavirus Disease (COVID-19) Pandemic, https://www.who.int/emergencies/diseases/novel-coronavirus-2019, last accessed September 2, 2021).
Although many organ systems can be affected by SARS-CoV-2 infection, pulmonary disease has been most frequently associated with severe and fatal cases of COVID-19.2 The earliest stage of disease is characterized by edema and vascular damage, including endothelial cell degeneration and necrosis, with neutrophilic infiltration of alveolar septa and capillaries (endothelialitis and capillaritis) and microthrombosis.2, 3, 4, 5 This is followed by an exudative phase of diffuse alveolar damage, with fibrinous edema in the alveolar spaces, increased numbers of macrophages and epithelial multinucleated giant cells, hyaline membrane formation, and epithelial necrosis, followed by type 2 pneumocyte hyperplasia. In addition, vascular changes occur, including endothelial necrosis, hemorrhage, thrombosis of capillaries and small arteries, and vasculitis.4,6 In turn, the organizing stage of diffuse alveolar damage and the final fibrotic stage of diffuse alveolar damage ensue, which may include proliferation of myofibroblasts within the lung interstitium and deposition of collagen, leading to fibrosis. Squamous metaplasia has also been observed.2,7
The emergent and widespread nature of this pandemic necessitated the rapid development of multiple animal models and biological systems to study various aspects of pathogenesis, treatment, and prevention of disease. To date, reported animal models of COVID-19 pathology include human angiotensin-converting enzyme 2 transgenic mice,8, 9, 10, 11 golden Syrian hamsters,11, 12, 13, 14, 15, 16, 17 nonhuman primates,18,19 and ferrets.20,21 Recent comprehensive reviews of animal models of COVID-19 were provided by Zeiss et al22 and Veenhuis and Zeiss23 in 2021. Each model species has advantages and limitations with respect to similarity to disease in humans, expense, and practicality. The hamster model offers several advantages over other animal models: it is a relatively small, immunocompetent animal that is susceptible to infection with varied SARS-CoV-2 clinical isolates and readily develops pulmonary disease. Specifically, hamsters consistently develop moderate to severe bronchointerstitial pneumonia characterized by acute inflammation, edema, and necrosis 2 to 4 days after SARS-CoV-2 challenge, progressing to proliferative interstitial pneumonia with type II pneumocyte hyperplasia by 7 days after challenge. Pulmonary lesions have been reported to resolve around 10 to 14 days after inoculation, with little to no evidence of residual damage.12,17,19,24
Although several studies have provided an overview of pulmonary pathology during acute infection, comprehensive longitudinal assessments of pulmonary pathology are lacking, including chronic time points. Likewise, there is a dearth of information integrating clinical, pathology, virology, and immunology findings or reporting systemic pathologic findings associated with SARS-CoV-2 infection in hamsters. Accordingly, the current study provides in-depth, longitudinal, pathologic characterization of multisystemic disease manifestation caused by SARS-CoV-2 infection in male and female golden Syrian hamsters. Furthermore, tissue damage and inflammatory responses were measured by digital image analysis using an open-source platform, QuPath.25,26 The current results show that inoculating hamsters intranasally with SARS-CoV-2 reliably induces acute damage to the respiratory tract with initial viral replication, followed by a macrophage-dominant pulmonary immune response. In turn, a reparative phase follows, with abundant type II pneumocyte hyperplasia restoring the alveolar lining, mirroring SARS-CoV-2 infection in humans.
Materials and Methods
Animals
The 7- to 8-week–old male and female golden Syrian hamsters, Mesocricetus auratus, HsdHan:AURA, were obtained from colonies negative for tested viruses, bacteria, Mycoplasma, fungi, and parasites (Envigo, Indianapolis, IN). Hamsters were singly housed in the Johns Hopkins University Cancer Research Building Animal Biosafety Level-3 facility. After acclimation (7 days minimum), hamsters were sedated intramuscularly with xylazine and ketamine and then inoculated intranasally with 105 50% tissue culture infectious dose (TCID50) of SARS-CoV-2 USA-WA1/2020 (BEI Resources, NR#52281; NIAID, Bethesda, MD) diluted in 100 μL Dulbecco’s modified Eagle’s medium, virus stocks. Uninfected animals were mock inoculated with 100 μL of Dulbecco’s modified Eagle’s medium intranasally to serve as controls. Groups of 12 animals (4 mock and 8 SARS-CoV-2 inoculated; equal numbers of males and females) were euthanized at 2, 4, 7, and 14 days post inoculation (DPI). An additional group of 4 mock (2 male and 2 female) and 19 SARS-CoV-2–inoculated hamsters (10 male and 9 female) were euthanized at 28 DPI. Body weights were measured daily until 10 DPI, then on 14, 21, and 28 DPI. Blood samples were collected at 0, 7, 14, 21, and 28 DPI, depending on group. Terminal blood samples obtained via cardiac puncture were saved for serology and fluorescence-activated cell sorting analysis. At study end points, animals were euthanized with i.p. sodium pentobarbital. A complete post-mortem examination with comprehensive tissue harvest (flash frozen and 10% neutral buffered formalin immersion fixed samples) was performed on all hamsters. The animal procedures in this study were approved by the Institutional Animal Care and Use Committee at Johns Hopkins University and in accordance with the National Research Council's Guide for the Care and Use of Laboratory Animals (eighth edition).27 The Johns Hopkins program of animal care and use is accredited by Association for Assessment and Accreditation of Laboratory Animal Care International.
SARS-CoV-2 Measurements
Infectious virus titers in the respiratory tissue homogenates were determined by the TCID50 assay.28 Briefly, tissue homogenates were serially diluted 10-fold, transferred in sextuplicate into 96-well plates confluent with Vero-E6-TMPRSS2 cells (obtained from the Japan Institute of Infectious Diseases), incubated at 37°C for 4 days, and then stained with naphthol blue-black solution for visualization. Infectious viral titers (TCID50/mL) were determined by Reed and Muench method. For detection of SARS-CoV-2 genome copies, RNA was extracted from lungs using the Qiagen viral RNA extraction kit (Qiagen, Hilden, Germany), and reverse transcriptase PCR was performed as previously described.29 Viral data from a subset of these animals have been included in a report on humoral responses in this model.28
Post-Mortem Examination
Standard necropsy procedures included gross examination of all major organs. After obtaining organ weights, samples were either frozen or immersion fixed in 10% neutral-buffered formalin. The lungs were weighed en bloc, and then the left lobe was removed at the level of the left bronchus. Formalin was gently infused into the left bronchus to inflate the left lobe, and then the entire lobe was submerged in formalin. The right lung lobes were divided into different frozen sample collection tubes for preparing tissue homogenates and RNA extraction.
Histopathology Analysis
After immersion fixation in 10% neutral-buffered formalin for 72 hours, tissues were trimmed, processed, and embedded in paraffin. Sections (5 μm thick) were mounted onto glass slides and stained with hematoxylin and eosin. Histopathologic analysis was performed independently by two veterinary pathologists (S.B. and K.M.) blinded to animal identification and infection status. Tissues evaluated included nasal cavity, trachea, lung (left lobe), esophagus, stomach, small intestine, cecum, large intestine, brain, heart, kidney, liver, gallbladder, spleen, adrenal gland, reproductive organs, urinary bladder, lymph nodes, salivary glands, bone, haired skin, skeletal muscle, bone marrow, and decalcified cross-sections of the head.
ISH and Immunohistochemistry
ISH to Detect SARS-CoV-2 RNA
In situ hybridization (ISH) was performed on sections (5 μm thick) of formalin-fixed lung mounted onto charged glass slides using the Leica Bond RX automated system (Leica Biosystems, Richmond, IL). Heat-induced epitope retrieval was conducted by heating slides to 95°C for 15 minutes in EDTA-based ER2 buffer (Leica Biosystems). The SARS-CoV-2 probe (catalog number 848568; Advanced Cell Diagnostics, Newark, CA) was used with the Leica RNAScope 2.5 LS Assay-RED kit and a hematoxylin counterstain (Leica Biosystems). Slides were treated in protease (Advanced Cell Diagnostics) for 15 minutes, and probes were hybridized to RNA for 1 minute. An RNApol2 probe served as a hamster gene control to ensure ISH sensitivity; a probe for the bacterial dap2 gene was used as a negative control ISH probe.
Immunohistochemistry
The 10% neutral buffered formalin–fixed lung sections of SARS-CoV-2–infected and control animals were immunostained with anti–ionized calcium binding adaptor molecule-1 (Iba-1) antibody (1:1000; 019-19741; Wako, Richmond, VA), anti-CD3 antibody (1:200; reference A0452; Dako, Santa Clara, CA), or anti–pan-cytokeratin antibody (1:1000; sc-8018; Santa Cruz, Dallas, TX). Heat-induced epitope retrieval was conducted by heating slides to 95°C for 20 minutes in sodium citrate–based ER1 buffer (Leica Biosystems) before immunostaining. Dual immunostaining for the epithelial marker pan-cytokeratin and the macrophage marker Iba-1 was performed on lung sections of SARS-CoV-2–infected animals. For epitope retrieval, slides were heated to 100°C in sodium citrate–based ER1 buffer for 20 minutes (Leica Biosystems). Slides were then stained with anti–Iba-1 antibody (1:2000; 019-19741; Wako) using the Bond Polymer Refine Kit (catalog number DS9800; Leica Biosystems). The slides were stained using a pan-cytokeratin antibody (1:1000; Sc-8018; Santa Cruz) with the Bond Polymer Refine Red Kit (catalog number DS9390; Leica Biosystems). Immunostaining was performed using the Bond RX automated system (Leica Biosystems). Positive immunostaining was visualized using diaminobenzidine and Bond Red detection kit, and slides were counterstained with hematoxylin.
Digital Image Analysis
Whole slides containing sections of the entire left lung lobe cut through the long axis were scanned at ×20 magnification on the Zeiss Axio Scan.Z1 platform (Zeiss, Jena, Germany) using automatic tissue detection with manual verification. Lung sections were analyzed using QuPath version 0.2.2 (Supplemental Script S1). To measure Iba-1 and CD3 immunostaining, the create thresholder function was applied to detect levels of diaminobenzidine above a threshold that was designated as positive within a given annotated area, or region of interest (ROI). The percentage positive ROI was calculated using positive area quantitated by the thresholder divided by total area of the ROI. For SARS-CoV-2 ISH quantitation, the train pixel classifier tool was used. Within an ROI, annotations were generated and designated as either positive or ignore, which allowed QuPath to correctly identify areas of positive staining. Percentage positive ROI was calculated using positive area detected by the classifier divided by total area of the ROI.
To quantitate consolidation of lungs to include infiltrating inflammatory cells (macrophages, neutrophils, and lymphocytes), type II pneumocyte hyperplasia, necrotic cellular debris, and proteinaceous exudate while excluding artefactually collapsed or poorly insufflated regions (atelectasis), the QuPath wand tool was used to outline each scanned section of lung, generating an annotation. Superpixels were generated using the DoG superpixel segmentation function in QuPath. These detections were then selected, and intensity features were added. Next, areas within the ROI were annotated using multiple slides of infected and control animals that were designated as consolidation, nonconsolidated, atelectasis (ie, artefactually collapsed or poorly insufflated regions), or ignore. This allowed the classifier to successfully detect areas of affected tissue, while ignoring areas that were unaffected, densely stained due to normal tissue architecture, or collapsed due to variable formalin infusion of the lungs. The consolidation classifier (artificial intelligence classifier) was assembled using multiple iterations that were qualitatively assessed after each analysis by a veterinary anatomic pathologist. Percentage consolidation was calculated using the number of superpixels identified as consolidated divided by the total number of superpixels detected for a given slide.
Flow Cytometry
Freshly collected whole blood in EDTA was stained for fluorescence-activated cell sorting analysis with the limited antibodies available that cross-react in the hamster model at the initiation of the study. Cell surface markers for CD4 (anti-mouse; clone GK1.5; Biolegend, San Diego, CA), CD8 (anti-rat; clone 341; BD Biosciences, San Jose, CA), major histocompatibility complex (MHC) II (anti-mouse; clone 14-4-4S; BD Biosciences), and a Live/Dead discriminator (Fixable Near-IR Dead Cell Stain Kit; ThermoFisher, Waltham, MA) consistently stained populations of lymphocytes or monocytes, allowing for a limited understanding of the proportion of these cell types. Whole-blood samples were stained with pretitered amounts of the indicated monoclonal antibodies using 50 μL of whole blood at room temperature for 20 minutes. Whole blood samples were then lysed and fixed in 2 mL of FACS Lysing Solution (BD Biosciences) for 10 minutes at room temperature. Samples were collected in a centrifuge at 400 × g for 5 minutes, washed in 2 mL of 1× phosphate-buffered saline, and then resuspended in 0.5 mL of phosphate-buffered saline for analysis. Flow cytometry was performed on a BD LSRFortessa (BD Biosciences). Data were analyzed using FlowJo 10.0.8 software (FlowJo, LLC, Ashland, OR). Whole blood was first gated on size and complexity using forward scatter-A and side scatter-A to remove debris, followed by using forward scatter-H and forward scatter-A and side scatter-H and side scatter-A measurements to remove doublet populations. Single cells were then gated into lymphocyte or monocyte populations based on size and complexity profiles, and then gated to include only live cells using a Live/Dead viability stain. Live lymphocyte-sized cells were gated as MHC II negative/dim (T cells) or MHC II+ (B cells). The MHC II–negative cells were further gated on their expression of CD4 or CD8. Populations of MHC II+ B cells, CD4+ lymphocytes, and CD8+ lymphocytes were calculated as a proportion of total viable lymphocytes. Live monocyte-sized cells were further gated as MHC II+.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism version 7.04 (Graph Pad Software, San Diego, CA). One-way analysis of variance was used to detect significant differences between groups of animals sampled at different time points after inoculation. The two-sample t-test was used to evaluate differences between two groups. Infectious virus titers and viral RNA copies were log transformed and compared using two-way analysis of variance with mixed-effects analysis followed by the Bonferroni multiple comparison test.
Results
Clinical Outcomes
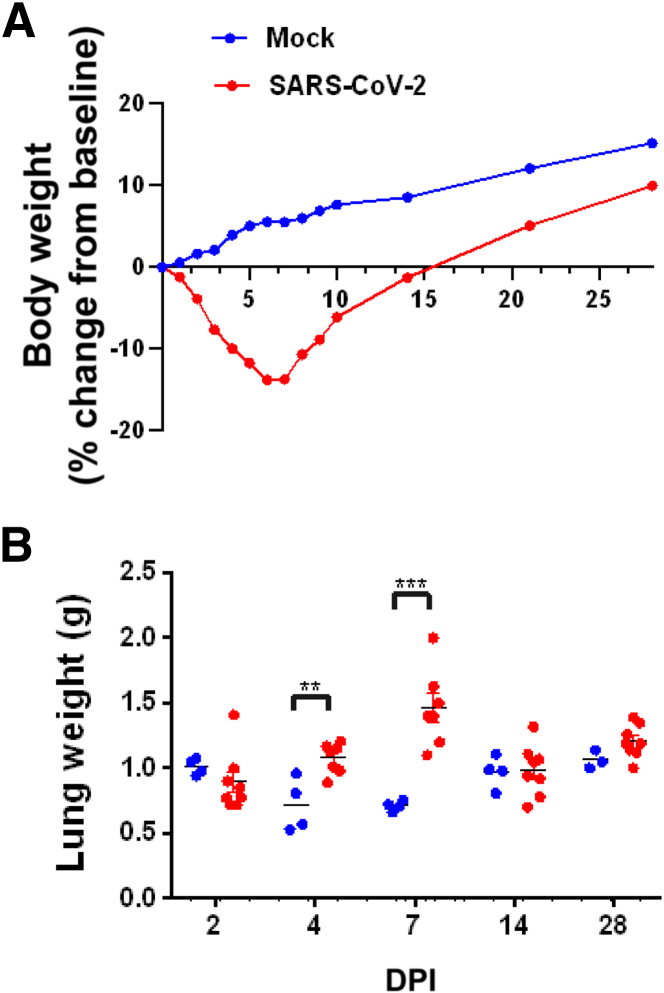
The most robust finding following SARS-CoV-2 inoculation was decreased body weight (Figure 1A). Animals inoculated with SARS-CoV-2 progressively lost body weight in the first week of infection until the 6 DPI nadir (–13.8% decline in group mean body weight from baseline; 19.3% lower than control animal group mean at 6 DPI; P < 0.001) before gradually rebounding over the course of the following 3 weeks. At day 28 DPI, control and infected group body weights were not significantly different (P = 0.095). Sex differences in body mass loss have been documented in hamsters infected with SARS-CoV-2, and those differences have been explored previously.28
Figure 1.
Body weight change over time and post-mortem lung weights. A: Body weight of control mock-inoculated hamsters (blue) compared with SARS-CoV-2–inoculated hamsters (red). Mock-inoculated animals demonstrated a steady increase typical of young growing animals. In contrast, SARS-CoV-2–inoculated animals had a sharp decline in body weight until 6 days post inoculation (DPI) when body weight began to increase. B: At the time of necropsy, lung weights of SARS-CoV-2–inoculated (red) hamsters had significantly higher total lung weights at both 4 and 7 DPI compared with mock-inoculated animals, consistent with lesion severity and consolidation. ∗∗P < 0.01, ∗∗∗P < 0.001 (unpaired t-test).
SARS-CoV-2–inoculated hamsters did not consistently develop clinical signs of respiratory disease; only mild nasal discharge or slight increased respiratory effort was observed intermittently in few animals.
SARS-CoV-2 Infectious Virus and Viral RNA Measurements in the Respiratory Tract
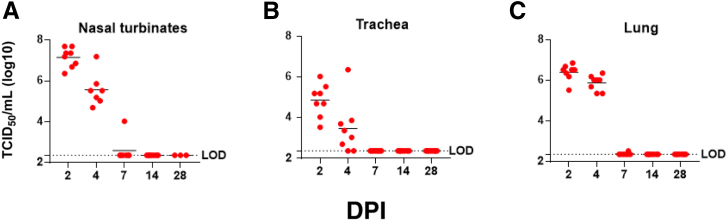
Peak infectious viral titers in the nasal turbinates, trachea, and lungs were present at 2 DPI, which decreased at 4 DPI (Figure 2). The highest levels of infectious SARS-CoV-2 were found in nasal turbinates at 2 DPI (5.0 × 107 TCID50/mL); lung levels were highest at 2 DPI (7.33 × 106 TCID50). Although infectious virus was cleared from the respiratory tract of most of the hamsters by 7 DPI, infectious virus was detected in the lungs of a single animal. No sex differences in viral load were detected in infected hamsters.28
Figure 2.
Viral loads throughout the respiratory system. Tissue culture infectious dose 50 (TCID50) assay revealed the highest levels of infectious virus present in the nasal turbinates (A), trachea (B), and lungs (C) at 2 days post inoculation (DPI). Detectable levels of infectious SARS-CoV-2 were still present in all these tissues at 4 DPI; only a single animal had low-level infectious virus at 7 DPI. LOD, limit of detection.
Gross Pathology
On gross necropsy examination, lungs from both infected and control animals were indistinguishable, with variably mottled red to brown coloration randomly distributed in all lung lobes, suggesting that this change was due to perimortem procedures and not a reliable indicator of pathology related to SARS-CoV-2 infection. Two animals had adhesions between the lung lobes and the diaphragm; both were infected animals euthanized 7 DPI. Compared with uninfected control animals, average post-mortem lung weights were 50% higher in infected animals at 4 DPI and 105% higher at 7 DPI (P = 0.014 and P < 0.0001, respectively; two-way analysis of variance multiple comparisons) (Figure 1B). All other tissues were unremarkable in both control and SARS-CoV-2–infected animals.
Histopathology: Longitudinal Evaluation of Respiratory System Alterations
2 DPI
The nasal cavity contained eosinophilic proteinaceous exudate with abundant degenerate neutrophils in all infected animals (eight of eight) and no uninfected animals (zero of four). In seven of eight infected animals, olfactory epithelium damage ranged from degeneration to necrosis with erosion accompanied by infiltrating neutrophils classified as rhinitis. At this time point, six of eight slides had trachea present to evaluate. Of those six animals, three had mild infiltrates of mononuclear cells within the tracheal submucosa, which were not present in any uninfected animals.
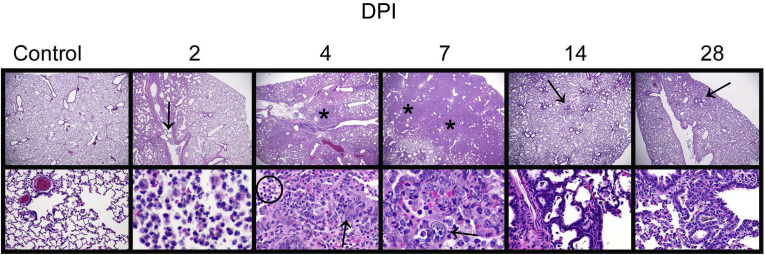
The following changes were observed in infected animal lungs (2 DPI) (Figure 3 and Supplemental Figure S1): minimal to moderate suppurative bronchitis and bronchiolitis (eight of eight), bronchial and bronchiolar intraluminal neutrophilic necrotic cellular debris (seven of eight), bronchial epithelial syncytia (two of eight), alveolar septa expanded by inflammatory cells and eosinophilic proteinaceous material (four of eight), and intra-alveolar macrophages, neutrophils, necrotic cellular debris, and fibrinous exudate (six of eight). Vascular changes in lungs included vascular endothelial hypertrophy within small to medium arteries (four of eight), perivascular edema, and leukocytes infiltrating through the vascular walls (two of eight).
Figure 3.
Progressive morphologic alterations in lung over time. Mock-inoculated (intranasal saline alone) animals that served as uninfected controls had no lesions identified by histologic examination. At 2 days post inoculation (DPI): Lung lesions included intraluminal neutrophilic infiltrates and necrotic cellular debris (arrow). At 4 DPI: Lesions included areas of consolidation (asterisk) consisting of type II pneumocyte hyperplasia (arrow), numerous intra-alveolar macrophages, neutrophils, necrotic cellular debris, and eosinophilic fibrinous exudate (circled). At 7 DPI: Extensive areas of pulmonary consolidation were present (asterisks) with atypical proliferative type II pneumocyte hyperplasia (arrow), abundant intra-alveolar macrophages, degenerate neutrophils, and fewer lymphocytes. At 14 DPI: There were multiple scattered areas of residual type II pneumocyte hyperplasia (arrow). At 28 DPI: Scattered small clusters of type II pneumocyte hyperplasia with cuboidal epithelial cells remained (arrow). Original magnification, ×2 (top panels); ×20 (bottom panel, control); ×40 (bottom panels, hematoxylin and eosin staining).
The lungs of the uninfected control animals had no significant findings. Intra-alveolar hemorrhage was observed intermittently in both uninfected and infected groups at all time points, attributed to tissue collection artifact.
4 DPI
The nasal cavity still contained eosinophilic proteinaceous exudate mixed with abundant degenerate neutrophils in all infected animals and none of the uninfected animals (Supplemental Figure S2). Olfactory epithelium was multifocally eroded with degenerate and necrotic epithelium; inflammatory cells consisted of predominantly neutrophils infiltrating into the mucosa. The tracheal submucosa only contained mild scattered infiltrates of mononuclear cells in one infected animal.
Lung changes within the medium to large airways in infected animals (4 DPI) (Figure 3 and Supplemental Figure S3) included minimal to moderate suppurative bronchitis and bronchiolitis (eight of eight), bronchial and bronchiolar intraluminal neutrophilic necrotic cellular debris (eight of eight), bronchial epithelial syncytia (eight of eight), and bronchial epithelial hyperplasia (eight of eight). Inflammatory changes extended into the alveolar parenchyma, and included increased numbers of intra-alveolar macrophages, neutrophils, necrotic cellular debris, and eosinophilic fibrinous exudate (eight of eight), fibrin lining alveoli (seven of eight), and alveolar septal necrosis (two of eight). In addition, small foci of type II pneumocyte hyperplasia were present with large, atypical cells (eight of eight). Atypical cells were characterized by large, 15- to 20-μm diameter nuclei, with lacy chromatin and one to three prominent nucleoli, and moderate to large amounts of basophilic vacuolated cytoplasm. Vascular changes included endothelial hypertrophy within small to medium arteries (eight of eight), perivascular lymphocytic aggregates (eight of eight), and mild to severe vasculitis (seven of eight).
7 DPI
The nasal cavity contained mild to moderate amounts of eosinophilic proteinaceous exudate with some degenerate neutrophils in eight of eight infected animals and zero of four uninfected animals.
At this stage, the changes in the lungs reflect a culmination of the inflammatory and reparative processes. All infected animals (eight of eight) developed large regions of pulmonary consolidation encompassing most of the lung (7 DPI) (Figure 3 and Supplemental Figure S4) characterized by type II pneumocyte hyperplasia and bronchial epithelial hyperplasia that was frequently atypical, with large nuclei in variably sized cells, and frequent multinucleated cells and mitotic figures, resulting in massive thickening of the alveolar septa. Remaining alveolar air spaces in these affected areas of lung contained large numbers of macrophages with fewer degenerate neutrophils and multifocal necrotic debris. Aggregates of lymphocytes and plasma cells occasionally surrounded (eight of eight) and infiltrated (two of eight) medium sized pulmonary arteries.
14 DPI
The nasal cavity and trachea morphology were unremarkable in all infected (eight of eight) and control (four of four) animals.
In infected animal lungs (14 DPI) (Figure 3 and Supplemental Figure S5), there were scattered tufts of cuboidal, type II pneumocyte hyperplasia (eight of eight), rare randomly distributed clusters of macrophages that occasionally admixed with lymphocytes and neutrophils (eight of eight), small perivascular aggregates of lymphocytes (five of eight), and multifocal small clusters of pigmented macrophages (four of eight).
28 DPI
The nasal cavity and trachea morphology were unremarkable in all infected (eight of eight) and control (four of four) animals.
In infected animals, the lungs (28 DPI) (Figure 3) contained multifocal residual areas of cuboidal, typical type II pneumocyte hyperplasia (eight of eight) often centered on terminal bronchioles, small clusters of pigmented macrophages (six of eight), and rare aggregates of perivascular lymphocytes with pigmented macrophages (two of eight). SARS-CoV-2–infected hamsters could be distinguished from uninfected animals by the presence of persistent type II pneumocyte hyperplasia in the infected animals. In contrast, lung inflammation had resolved, and fibrosis had not developed.
Tracking SARS-CoV-2 RNA by in Situ Hybridization
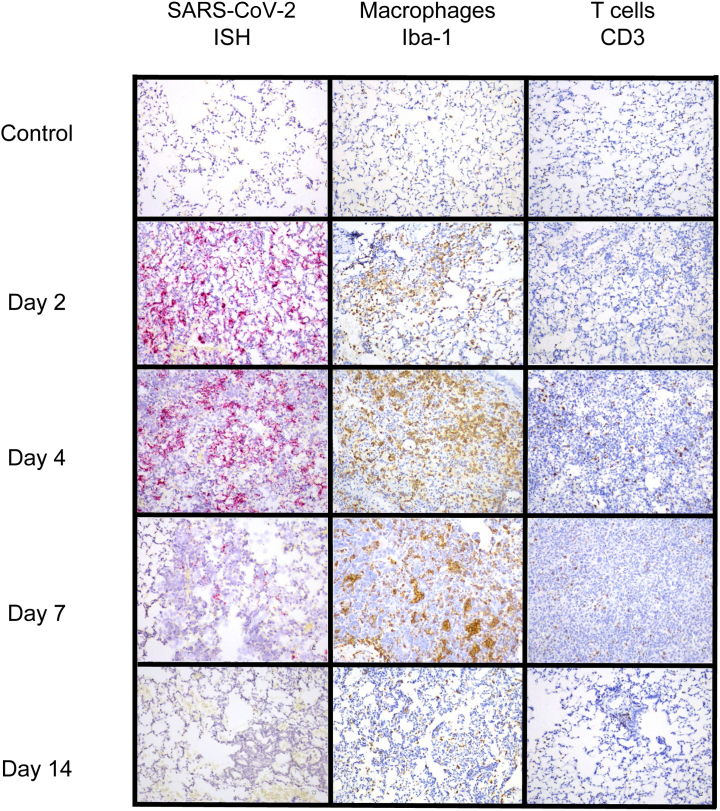
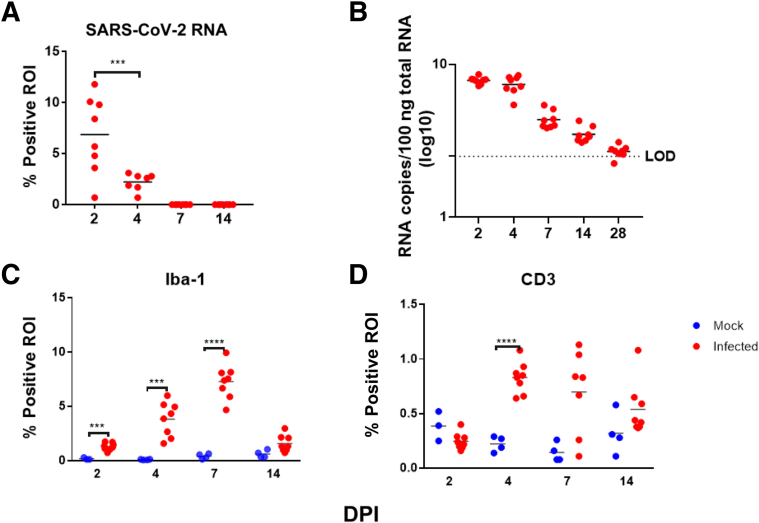
RNAScope in situ hybridization was used to map distribution and amount of viral RNA in the lungs of infected animals throughout infection. Viral RNA expression was quantitated using QuPath pixel classification. Expression of viral RNA was significantly different over time in infected animals (one-way analysis of variance; P < 0.0001): 2 DPI lungs demonstrated the highest expression in alveolar epithelial cells, bronchiolar and bronchial epithelial cells, and alveolar macrophages. At 4 DPI, lung expression was decreased compared with 2 DPI, with positive staining for viral RNA largely restricted to the alveolar epithelial cells and alveolar macrophages in lung (Figure 4 and Supplemental Figure S6). At 7 DPI, lungs were largely negative; only one animal had multifocal low-level staining for viral RNA within scattered alveolar epithelial cells. In addition to lung, scattered positive staining for viral RNA was found within tracheobronchial lymph nodes and tracheal epithelium at 2 DPI (seven of seven animals with tissue present), and less frequently at 4 DPI (two of five animals with tissue present) (Supplemental Figure S6). Viral RNA was detected by reverse transcriptase PCR for up to 28 DPI in SARS-CoV-2–infected hamsters (Figure 5), suggesting that this may be the most sensitive method for viral detection, although not indicative of infectivity.
Figure 4.
In situ hybridization (ISH) and immunohistochemistry findings in lung. RNAScope in situ hybridization showed widespread, punctate staining in the lungs at 2 and 4 days post inoculation (DPI), but was present only in a small focus within one animal at 7 DPI. No staining was present at 14 DPI (RNAScope in situ hybridization with red chromogen and hematoxylin counterstain). Immunostaining to detect Iba-1 revealed that macrophages comprised a major portion of the inflammatory infiltrate beginning at 2 DPI and peaking at 7 DPI. CD3+ lymphocytes represented a much smaller portion of the inflammatory population in the lungs but also increased from 2 to 7 DPI and then decreased at 14 DPI. (Immunostaining was performed with brown chromogen and hematoxylin counterstain.) Original magnification, ×20.
Figure 5.
Quantitative digital image analysis of in situ hybridizaton and immunohistochemical staining in the lungs using QuPath analysis. A: Quantitation of staining for SARS-CoV-2 RNA within the lungs showed highest levels at 2 days post inoculation (DPI), with virus still detectable at 4 DPI. No measurable virus was detected at 7 or 14 DPI. B: Copy numbers of viral RNA within the lungs, detected by reverse transcriptase PCR, reached highest levels at 2 and 4 DPI, with progressively lower amounts detected at 7 DPI, extending through 28 DPI. C: Quantitation of Iba-1 immunolabeling within the lungs revealed an increase in macrophages in infected animals until 7 DPI, with a decrease at 14 DPI, although still above levels found in mock-inoculated animals. D: Quantitation of CD3+ immunolabeling within the lungs revealed an increase in lymphocytes until 4 to 7 DPI. Numbers of lymphocytes were 10-fold lower than macrophages. Region of interest (ROI) was entire left lung lobe. ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. LOD, limit of detection.
Immunophenotypic Characterization of Lung Inflammation
To establish the immunophenotype and progression of inflammatory cell infiltrates in the lung, lung sections from mock-inoculated and SARS-CoV-2–infected animals were evaluated for expression of the macrophage marker Iba-1 and the T-cell marker CD3 at 2, 4, 7, and 14 DPI. Following immunostaining, QuPath was used to quantitate percentage positive ROI in whole slide scanned images of the left lung lobe, the largest hamster lung lobe. Iba-1 expression was significantly different in infected animals versus uninfected animals over time (one-way analysis of variance; P < 0.0001) (Figure 5). Specifically, Iba-1 immunostaining was higher in infected animals compared with uninfected control animals at 2, 4, 7, and 14 DPI (t-tests; P < 0.001, P < 0.001, P < 0.0001, and P = 0.04, respectively).
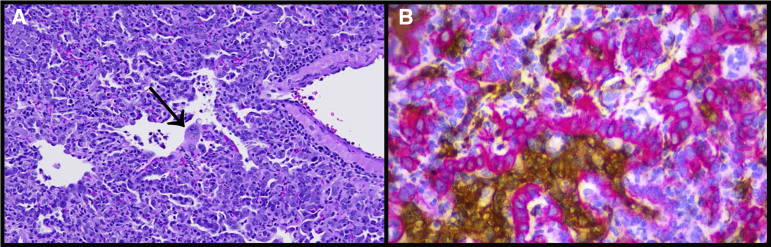
CD3 expression, reflecting T-cell responses, was more variable than Iba-1 expression. CD3 expression in infected animals was different over time (one-way analysis of variance; P < 0.0001) (Figure 5). CD3 expression was significantly greater in infected animals compared with uninfected control animals at 4 and 7 DPI (t-tests; P < 0.0001 and P = 0.023, respectively). Expression of both Iba-1 and CD3 increased until 7 DPI, and then decreased at 14 DPI (Figure 4); however, IBA-1 expression was much more abundant than CD3, indicating that macrophages comprised a greater portion of the inflammatory response in this model, consistent with previous reports.16 Many of the atypical and multinucleated cells in the day 4 and day 7 animals were immunonegative for Iba-1 expression. Immunostaining for pan-cytokeratin revealed that numerous atypical and multinucleated cells were immunopositive, indicating that these cells were epithelial, consistent with proliferative type II pneumocytes contributing to alveolar repair (Figure 6). Double immunostaining demonstrated that both macrophages and proliferative epithelial cells were present within the densely cellular areas of the lungs of 7 DPI animals, confirming that inflammatory and reparative responses were contributing to the vast areas of consolidation at this time point.
Figure 6.
Consolidation in the lungs composed of both large numbers of macrophages and extensive type II pneumocyte hyperplasia. A: Representative image of lung at 7 days post inoculation (DPI), showing severe consolidation, including type II pneumocyte hyperplasia with large atypical epithelial cells and multinucleated syncytial cells (arrow). Hematoxylin and eosin staining was used. B: Similar area of consolidation within the lungs at 7 DPI with immunostaining to detect pan-cytokeratin (red chromogen) and Iba-1 (brown chromogen) revealed that the large, atypical cells and multinucleated cells were epithelial, whereas the macrophages were within the alveolar and interstitial spaces. Hematoxylin counterstain was used. Original magnification, ×40 (A and B).
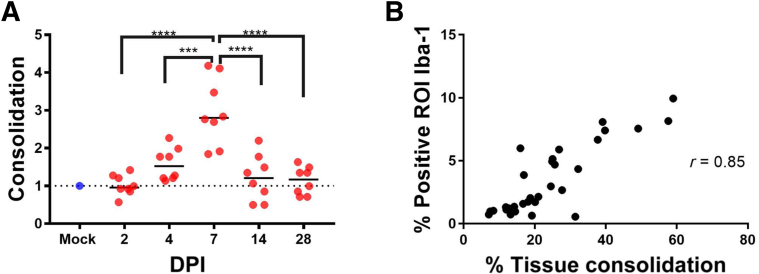
Quantitation of Lung Consolidation
QuPath was used to perform digital image analysis to quantitate the percentage of lung affected with inflammation and reparative processes in SARS-CoV-2–infected animals (Supplemental Figure S7). The goal of this consolidation scoring approach was to encompass inflammatory cells (macrophages, neutrophils, and scattered lymphocytes), type II pneumocyte hyperplasia, necrotic cellular debris, and proteinaceous exudate while excluding artifactual collapsed or poorly insufflated regions (atelectasis). Because atelectatic areas in which alveolar septa are collapsed are common in post-mortem lung examples, there was a baseline level of detection above zero in control animals. When comparing the percentage of consolidation in infected animals with the baseline level in control animals, day 7 animals had a significant increase in fold change (Figure 7). Infected animals at 7 DPI had a mean value of 3.0-fold change above baseline (P = 0.02; Tukey multiple comparisons test). The highest percentage of affected lung was 59% and occurred in a 7 DPI animal. Consolidation scores were highly correlated with IBA-1 quantitation by digital image analysis (Pearson r = 0.84; P < 0.0001) (Figure 7).
Figure 7.
Quantitation of consolidation within lungs using QuPath. A: Infected animals at 7 days post inoculation (DPI) had significantly higher percentage of tissue consolidation compared with mock animals, as well as animals at all other time points (fold change versus control). B: Percentage of tissue consolidation was highly correlated with percentage positive Iba-1 labeling within the lungs. Dashed line represents average consolidation value of uninfected animals. ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001. ROI, region of interest.
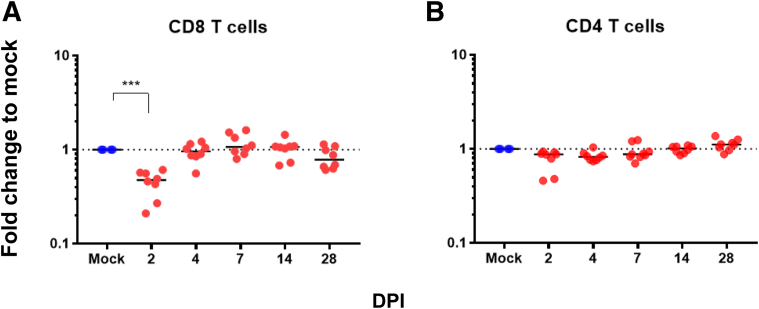
Flow Cytometry to Detect Alterations in Circulating Cells
Fold change, relative to mock-infected hamsters at each time point, was used to assess changes in the proportions of T-lymphocyte populations circulating in blood. A significant early decrease in the proportion of CD8+ lymphocytes at 2 DPI was observed (P < 0.001) (Figure 8). No significant changes were observed in CD4+ lymphocytes (P = 0.06) (Figure 8); however, the two animals with the largest decrease in CD8+ lymphocytes also had a detectable decrease in CD4+ lymphocytes. No changes were observed in either B cells or monocyte-sized MHC II+ cells in any animals (data not shown).
Figure 8.
Flow cytometry of whole blood. Whole blood fluorescence-activated cell sorting revealed an early decrease in the proportion of circulating CD8+ lymphocytes at 2 days post inoculation (DPI; A), whereas CD4 lymphocytes do not differ from baseline (B). Dashed lines represent average values (CD8 and CD4 T cells) of uninfected animals. ∗∗∗P < 0.001.
Histopathology of Organ Systems
Minimal to mild multifocal foci of necrosis, accompanied by neutrophils within the liver, were occasionally observed in all groups and were considered a background lesion. The bone marrow in all animals was densely cellular, with abundant myeloid cells, including numerous band cells, as well as erythroid and megakaryocyte lineages. The spleen in all animals was composed of predominantly red pulp, with no obvious signs of lymphoid depletion or lymphoid hyperplasia. Abdominal lymph nodes occasionally contained germinal centers in control and infected animals. No significant changes were noted in the gastrointestinal tract, including the oral cavity, esophagus, small intestine, cecum, and colon. Kidneys in animals across all groups (29/45) had minimal protein and mineral within the tubules. Adrenal glands within hamsters of all groups had variable numbers of pigmented cells within the cortex, which has been reported to be a change in aged hamsters.30 Other organs that were examined without significant changes on histopathology included brain, heart, gallbladder, male and female reproductive organs, urinary bladder, salivary glands, bone, haired skin, skeletal muscle, and decalcified cross-sections of the head.
Discussion
This study conducted a longitudinal, in-depth comprehensive histopathologic analysis of all tissues and organ systems within male and female golden Syrian hamsters inoculated intranasally with SARS-CoV-2, spanning 2 to 28 DPI. In addition, it characterized the immunophenotype of the inflammatory infiltrates within the lungs across all stages of disease and assessed viral load within the lungs via in situ hybridization, virus titration, and reverse transcriptase PCR for viral RNA. Morphologic findings were quantified using QuPath, an open-source digital image analysis platform applied to analyze an entire lung lobe imaged via whole slide scanning. This objective approach provided robust outcome measures for immunohistochemistry, ISH, and pulmonary consolidation, enhancing rigor of this COVID-19 model.
The main lesion type within the lungs was proliferative bronchointerstitial pneumonia, which started as acute necrosis of airway epithelium (eight of eight animals) and alveolar septa (six of eight animals) coincident with SARS-CoV-2 replication. Cellular immune responses were first dominated by recruited neutrophils but switched to a macrophage-dominant phenotype accompanied by a robust atypical type II pneumocyte reparative response beginning by day 4 and peaking at day 7 after inoculation, resembling other reports of disease timing in this animal model.12, 13, 14, 15, 16, 17 Elements of pathology-associated SARS-CoV-2 infection in hamsters have been reviewed and summarized previously: at 2 to 3 DPI, previous reports describe necrosuppurative bronchitis,31 and variable percentage of affected area from 5% to 10%12 to more widespread diffuse alveolar damage.13 Reports range from nonspecific inflammation,12,14 to suppurative and/or mononuclear infiltration13,16,31 within the lungs. At 4 to 5 DPI, increasing lung consolidation is reported, with proliferation of epithelial cells, and syncytia. Changes within vasculature, including endothelialitis, are reported at 5 DPI.15,31 Various studies consistently report increase in severity of inflammation up to 7 DPI, with variable specificity on the cell types involved, and include lymphocytes, neutrophils, and macrophages.12,15,31 Inflammatory changes in the lungs are reported to resolve by 10 to 14 DPI, with the exception of some residual nonspecific inflammatory changes in aged hamsters.12,16,31 In this study, residual areas of type II pneumocyte hyperplasia were present in all infected animals at 14 and 28 DPI.
In this model, pulmonary pathology emulated findings reported in humans, and included features of diffuse alveolar damage, intra-alveolar macrophage infiltration, and type II pneumocyte hyperplasia.32 Diffuse alveolar damage was not a dominant aspect in this study, but foci of septal necrosis and organized fibrinous exudate developed in some animals at early time points. Atypical proliferation of type II pneumocytes in the hamster model during the reparative phase of disease has been inconsistently reported but was a consistent and prominent finding in this study.13 The striking proliferation was more robust than what has been reported in humans, with frequent anisocytosis and anisokaryosis and numerous mitotic figures. Type II pneumocyte hyperplasia in hamsters has been reported in other experimentally induced respiratory viral infections,33,34 but is not usually this florid. Hamsters were evaluated from initial stages of infection at progressive time points, whereas reports of human COVID-19 lung pathology are cross-sectional, typically at autopsy representing the most severe disease; thus, the timing of lung sampling may explain this disparity.
Vascular changes are consistently described in human SARS-CoV-2 infection and are thought to be a key factor in the pathogenesis of severely affected individuals.3,32 Within the lungs, vascular changes in humans include endothelial damage, thrombosis, microangiopathy, congestion, and angiogenesis.3 In this study, within the pulmonary vasculature, there was prominent endothelial hypertrophy in days 2 to 7, as well as overt vasculitis in small- and medium-sized arteries predominantly at day 4. Further exploration and characterization of these changes in future studies is warranted given the important role that these factors may play in human disease. Interstitial fibrosis is a chronic sequela to SARS-CoV-2 infection in some individuals.2,32 Interestingly, although interstitial fibrosis was not observed in this study, there was residual type II pneumocyte hyperplasia at day 14 and 28, frequently present adjacent to terminal bronchioles, which has not been reported in COVID-19 animal models or in human COVID-19 cases, likely because these changes develop during the reparative stage time points when post-mortem samples are not typically available.
In this model, a significant decrease in the proportion of CD8+ lymphocytes in peripheral blood of infected hamsters was detected at day 2 DPI. Further investigation is warranted to characterize this change to determine if this is representative of a CD8-dominant lymphopenia, as this could have important implications for pathogenesis and may be representative of human disease.23,35, 36, 37, 38 In human COVID-19 patients, CD8-dominant lymphopenia has been consistently documented, and critical patients have more severe lymphopenia than individuals with mild cases.36,38,39 The mechanisms underlying this change are unclear. In this study, we did not observe morphologic changes in the thymus, spleen, lymph nodes, or gut-associated lymphoid tissue, suggestive of lymphoid depletion.
Overall, hamsters developed consistent pulmonary disease associated with SARS-CoV-2 infection progressing though distinct stages: acute viral replication and cell necrosis accompanied by infiltrating neutrophils, a transition to macrophage-dominant inflammation with control of viral replication and robust reparative epithelial responses when lung consolidation was most severe 7 DPI, and resolution of inflammation with residual evidence of epithelial repair still evident 28 DPI with SARS-CoV-2. Hamsters are susceptible to infection with an intact immune system, in contrast to mouse model systems in which the mouse or virus needs to be altered to study this pathogenesis. Hamsters also have the advantage of being a small animal model, which are less expensive and easier to maintain than larger animals, such as nonhuman primates. The consistency of disease in this model further emphasizes the need to develop additional antibodies and reagents to characterize immunopathologic changes.
This study provides detailed and objectively quantified longitudinal histopathologic analysis of the respiratory system, and describes associated changes observed in all tissues and organ systems. In addition, the methods for objective quantification of immunohistochemically labeled cell types as well as percentage of lung consolidation are outlined using QuPath, an open-source platform for digital image analysis. Residual type II pneumocyte hyperplasia was observed at 14 and 28 DPI, which is a novel finding in this model. This study shows that lung weights are greater in infected animals at peak disease time points, which provides another simple and objective measurable outcome that has not been previously reported. These comprehensive and integrated findings on disease progression and resolution serve as a baseline of SARS-CoV-2 infection outcome measures in this model and will be valuable for determining the efficacy of therapeutic and preventive interventions for COVID-19.
Acknowledgments
Additional Johns Hopkins COVID-19 Hamster Study Group members include Michael J. Betenbaugh, Bess Carlson, Natalie Castell, Jennie Ruelas Castillo, Kelly Flavahan, Eric K. Hutchinson, Kirsten Littlefield, Monika M. Looney, Maggie Lowman, Natalia Majewski, Amanda Maxwell, Filipa Mota, Alice L. Mueller, Alvaro A. Ordonez, Lisa Pieterse, Darla Quijada, Camilo A. Ruiz-Bedoya, Mitchel Stover, Rachel Vistein, and Melissa Wood.
A.S.P. would like to dedicate this article to the memory of R. Mark Buller, whose collaborations on the golden Syrian hamster model for SARS-CoV infection formed the basis for this study.
Footnotes
Supported through the generosity of the collective community of donors to the Johns Hopkins University School of Medicine for coronavirus disease research (J.L.M.), NIH OD T32OD011089 (J.L.M.), The Johns Hopkins Center of Excellence in Influenza Research and SurveillanceHHSN272201400007C AP (S.L.K.), the NIH/National Cancer Institute COVID-19 Serology Center of Excellence U54CA260492 (S.L.K.), the NIH/Office of Research on Women’s Health/National Institue on Aging Specialized Center of Research Excellence in Sex Differences U54AG062333 (S.L.K.) and R01AI153349 (S.K.J.), and the Center for Infection and Inflammation Imaging Research (Johns Hopkins University; S.K.J.).
Disclosures: None declared.
Current address of K.A.M.P., Division of Comparative Medicine and Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2021.10.009.
Contributor Information
Joseph L. Mankowski, Email: jmankows@jhmi.edu.
Johns Hopkins COVID-19 Hamster Study Group:
Michael J. Betenbaugh, Bess Carlson, Natalie Castell, Jennie Ruelas Castillo, Kelly Flavahan, Eric K. Hutchinson, Kirsten Littlefield, Monika M. Looney, Maggie Lowman, Natalia Majewski, Amanda Maxwell, Filipa Mota, Alice L. Mueller, Alvaro A. Ordonez, Lisa Pieterse, Darla Quijada, Camilo A. Ruiz-Bedoya, Mitchel Stover, Rachel Vistein, and Melissa Wood
Supplemental Data
Additional images of lung pathology at 2 days post inoculation. A: Bronchiolar epithelial degeneration, necrosis, and loss. B: An artery with vascular endothelial hypertrophy (arrow) and leukocyte exocytosis (inset, neutrophils migrating through hypertrophied vascular endothelial cells). C: Eosinophilic fibrin lining alveoli and expanding alveolar septa, and organized fibrinous exudate within alveoli (arrow). D: Inflammatory cells, including macrophages and degenerate neutrophils, were within the alveolar lumen (asterisk). Scale bars: 50 μm (A, C, and D); 100 μm (B).
Pathology of the nasal cavity in SARS-CoV-2–infected hamsters. Representative images at 4 days post inoculation. A: Abundant suppurative exudate (asterisk) admixed with proteinaceous material within the nasal cavity. B: Inflammatory cells infiltrated into the nasal mucosal epithelium (asterisk), with epithelial necrosis and ulceration (arrow). Scale bar = 50 μm (A and B).
Additional images of lung pathology at 4 days post inoculation. A: Atypical type II pneumocyte hyperplasia (arrow). B: Bronchial epithelial syncytial cell (arrow). C: Numerous intra-alveolar macrophages, degenerate neutrophils along with fibrinous exudate and thickened alveolar septa. D: Vasculitis, demonstrated by inflammatory cells effacing the vessel wall and necrosis of the vascular wall. Scale bars: 50 μm (A and B); 100 μm (C and D).
Additional images of lung pathology at 7 days post inoculation. A: Degenerate neutrophils, eosinophilic exudate, and macrophages were within alveolar spaces. B: Many large, atypical cells were present lining the alveolar septa that contained large nuclei with prominent nucleoli and lacy chromatin. C: Frequent multinucleated epithelial cells were present throughout affected areas of lung. D: Numerous mitotic figures were present within areas of type II pneumocyte hyperplasia, indicating a robust reparative response. Scale bars: 50 μm (A); 25 μm (B); 30 μm (C); 10 μm (D).
Additional images of lung pathology at 14 days post inoculation. A: Representative tertiary bronchiole from an uninfected animal. B: Residual type II pneumocyte hyperplasia in an arborizing pattern. Scale bar = 100 μm (A and B).
In situ hybridization of SARS-CoV-2 at 2 days post inoculation. SARS-CoV-2 RNA (red chromogen) is detectable within the tracheal epithelium (A), bronchial lymph node (B), bronchiolar epithelium (C), and within alveolar epithelial cells and macrophages (arrow; D). Scale bars: 50 μm (A and B); 100 μm (C); 30 μm (D).
Consolidation classifier of the lungs at 7 days post inoculation (DPI) using QuPath. A: Representative image of SARS-CoV-2–infected animal at 7 DPI, showing multiple coalescing areas of consolidation. B: Example of color-coded classifier of affected area using QuPath-generated superpixel classified as consolidation (green), unaffected (blue), or atelectasis (yellow). Areas where tissue is visible (bronchiolar epithelium and blood vessels) represent superpixels classified as ignore. The red line in each image indicates the annotation marker to generate the region of interest in QuPath. Original magnification, ×2 (A and B).
References
- 1.Ren L.-L., Wang Y.-M., Wu Z.-Q., Xiang Z.-C., Guo L., Xu T., et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. 2020;133:1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bösmüller H., Matter M., Fend F., Tzankov A. The pulmonary pathology of COVID-19. Virchows Archiv. 2021;478:137–150. doi: 10.1007/s00428-021-03053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., Li W.W., Li V.W., Mentzer S.J., Jonigk D. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menter T., Haslbauer J.D., Nienhold R., Savic S., Hopfer H., Deigendesch N., Frank S., Turek D., Willi N., Pargger H., Bassetti S., Leuppi J.D., Cathomas G., Tolnay M., Mertz K.D., Tzankov A. Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology. 2020;77:198–209. doi: 10.1111/his.14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bösmüller H., Traxler S., Bitzer M., Häberle H., Raiser W., Nann D., Frauenfeld L., Vogelsberg A., Klingel K., Fend F. The evolution of pulmonary pathology in fatal COVID-19 disease: an autopsy study with clinical correlation. Virchows Arch. 2020;477:349–357. doi: 10.1007/s00428-020-02881-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lax S.F., Skok K., Zechner P., Kessler H.H., Kaufmann N., Koelblinger C., Vander K., Bargfrieder U., Trauner M. Pulmonary arterial thrombosis in COVID-19 with fatal outcome: results from a prospective, single-center, clinicopathologic case series. Ann Intern Med. 2020;173:350–361. doi: 10.7326/M20-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carsana L., Sonzogni A., Nasr A., Rossi R.S., Pellegrinelli A., Zerbi P., Rech R., Colombo R., Antinori S., Corbellino M., Galli M., Catena E., Tosoni A., Gianatti A., Nebuloni M. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect Dis. 2020;20:1135–1140. doi: 10.1016/S1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., Ritter J.H., Kang L.-I., Dort S., Robichaud A., Head R., Holtzman M.J., Diamond M.S. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., Ritter J.H., Kang L.-I., Dort S., Robichaud A., Head R., Holtzman M.J., Diamond M.S. SARS-CoV-2 infection in the lungs of human ACE2 transgenic mice causes severe inflammation, immune cell infiltration, and compromised respiratory function. bioRxiv. 2020 10.1101/2020.07.09.196188 [Preprint] doi: [Google Scholar]
- 10.Sun S.-H., Chen Q., Gu H.-J., Yang G., Wang Y.-X., Huang X.-Y., Liu S.-S., Zhang N.-N., Li X.-F., Xiong R., Guo Y., Deng Y.-Q., Huang W.-J., Liu Q., Liu Q.-M., Shen Y.-L., Zhou Y., Yang X., Zhao T.-Y., Fan C.-F., Zhou Y.-S., Qin C.-F., Wang Y.-C. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28:124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdel-Moneim A.S., Abdelwhab E.M. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens. 2020;9:529. doi: 10.3390/pathogens9070529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sia S.F., Yan L.-M., Chin A.W.H., Fung K., Choy K.-T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., Peiris M., Yen H.-L. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan J.F.-W., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Chan W.-M., Fan Z., Tsoi H.-W., Wen L., Liang R., Cao J., Chen Y., Tang K., Luo C., Cai J.-P., Kok K.-H., Chu H., Chan K.-H., Sridhar S., Chen Z., Chen H., To K.K.-W., Yuen K.-Y. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., Yamada S., Fan S., Chiba S., Kuroda M., Guan L., Takada K., Armbrust T., Balogh A., Furusawa Y., Okuda M., Ueki H., Yasuhara A., Sakai-Tagawa Y., Lopes T.J.S., Kiso M., Yamayoshi S., Kinoshita N., Ohmagari N., Hattori S.-I., Takeda M., Mitsuya H., Krammer F., Suzuki T., Kawaoka Y. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osterrieder N., Bertzbach L.D., Dietert K., Abdelgawad A., Vladimirova D., Kunec D., Hoffmann D., Beer M., Gruber A.D., Trimpert J. Age-dependent progression of SARS-CoV-2 infection in Syrian hamsters. Viruses. 2020;12:779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat Med. 2020;26:1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreye J., Reincke S.M., Kornau H.-C., Sánchez-Sendin E., Corman V.M., Liu H., et al. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., Avanzato V.A., Rosenke R., Hanley P.W., Saturday G., Scott D., Fischer E.R., de Wit E. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y.-I., Kim S.-G., Kim S.-M., Kim E.-H., Park S.-J., Yu K.-M., Chang J.-H., Kim E.J., Lee S., Casel M.A.B., Um J., Song M.-S., Jeong H.W., Lai V.D., Kim Y., Chin B.S., Park J.-S., Chung K.-H., Foo S.-S., Poo H., Mo I.-P., Lee O.-J., Webby R.J., Jung J.U., Choi Y.K. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.-S., Riveros-Balta A.X., Albrecht R.A., et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeiss C.J., Compton S., Veenhuis R.T. Animal models of COVID-19, I: comparative virology and disease pathogenesis. ILAR J. 2021 doi: 10.1093/ilar/ilab007. 10.1093/ilar/ilab007 [Epub ahead of print] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veenhuis R.T., Zeiss C.J. Animal models of COVID-19, II: comparative immunology. ILAR J. 2021 doi: 10.1093/ilar/ilab010. 10.1093/ilar/ilab010 [Epub ahead of print] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.-T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R.K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M.J., Rawlings S.A., Wu N.C., Yuan M., Smith D.M., Nemazee D., Teijaro J.R., Voss J.E., Wilson I.A., Andrabi R., Briney B., Landais E., Sok D., Jardine J.G., Burton D.R. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G., James J.A., Salto-Tellez M., Hamilton P.W. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morriss N.J., Conley G.M., Ospina S.M., Meehan W.P., III, Qiu J., Mannix R. Automated quantification of immunohistochemical staining of large animal brain tissue using QuPath software. Neuroscience. 2020;429:235–244. doi: 10.1016/j.neuroscience.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Committee for the Update of the Guide for the Care and Use of Laboratory Animals; National Research Council . Eighth Edition. National Academies Press; Washington, DC: 2011. Guide for the Care and Use of Laboratory Animals: [Google Scholar]
- 28.Dhakal S., Ruiz-Bedoya C.A., Zhou R., Creisher P., Villano J., Littlefield K., Castillo J., Marinho P., Jedlicka A., Ordonez A., Majewska N., Betenbaugh M., Flavahan K., Mueller A., Looney M., Quijada D., Mota F., Beck S.E., Brockhurst J.K., Braxton A., Castell N., D'Alessio F., Metcalf Pate K.A., Karakousis P.C., Mankowski J.L., Pekosz A., Jain S.K., Klein S.L. Sex differences in lung imaging and SARS-CoV-2 antibody responses in a COVID-19 golden Syrian hamster model. bioRxiv. 2021 doi: 10.1128/mBio.00974-21. 10.1101/2021.04.02.438292 [Preprint] doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gniazdowski V., Morris C.P., Wohl S., Mehoke T., Ramakrishnan S., Thielen P., Powell H., Smith B., Armstrong D.T., Herrera M., Reifsnyder C., Sevdali M., Carroll K.C., Pekosz A., Mostafa H.H. Repeat COVID-19 molecular testing: correlation of SARS-CoV-2 culture with molecular assays and cycle thresholds. Clin Infect Dis. 2021;73:860–869. doi: 10.1093/cid/ciaa1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyers M.W., Charipper H.A. A histological and cytological study of the adrenal gland of the golden hamster (Cricetus auratus) in relation to age. Anat Rec. 1956;124:1–25. doi: 10.1002/ar.1091240102. [DOI] [PubMed] [Google Scholar]
- 31.Gruber A.D., Osterrieder N., Bertzbach L.D., Vladimirova D., Greuel S., Ihlow J., Horst D., Trimpert J., Dietert K. Standardization of reporting criteria for lung pathology in SARS-CoV-2-infected hamsters: what matters? Am J Respir Cell Mol Biol. 2020;63:856–859. doi: 10.1165/rcmb.2020-0280LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polak S.B., Van Gool I.C., Cohen D., von der Thüsen J.H., van Paassen J. A systematic review of pathological findings in COVID-19: a pathophysiological timeline and possible mechanisms of disease progression. Mod Pathol. 2020;33:2128–2138. doi: 10.1038/s41379-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baseler L., de Wit E., Scott D.P., Munster V.J., Feldmann H. Syrian hamsters (Mesocricetus auratus) oronasally inoculated with a Nipah virus isolate from Bangladesh or Malaysia develop similar respiratory tract lesions. Vet Pathol. 2015;52:38–45. doi: 10.1177/0300985814556189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prescott J., Safronetz D., Haddock E., Robertson S., Scott D., Feldmann H. The adaptive immune response does not influence hantavirus disease or persistence in the Syrian hamster. Immunology. 2013;140:168–178. doi: 10.1111/imm.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J., Li S., Liu J., Liang B., Wang X., Wang H., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F., Nie J., Wang H., Zhao Q., Xiong Y., Deng L., Song S., Ma Z., Mo P., Zhang Y. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020;221:1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J., Liu Y., Xiang P., Pu L., Xiong H., Li C., Zhang M., Tan J., Xu Y., Song R., Song M., Wang L., Zhang W., Han B., Yang L., Wang X., Zhou G., Zhang T., Li B., Wang Y., Chen Z., Wang X. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J Transl Med. 2020;18:206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.-Q., Wang Q., Miao H. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional images of lung pathology at 2 days post inoculation. A: Bronchiolar epithelial degeneration, necrosis, and loss. B: An artery with vascular endothelial hypertrophy (arrow) and leukocyte exocytosis (inset, neutrophils migrating through hypertrophied vascular endothelial cells). C: Eosinophilic fibrin lining alveoli and expanding alveolar septa, and organized fibrinous exudate within alveoli (arrow). D: Inflammatory cells, including macrophages and degenerate neutrophils, were within the alveolar lumen (asterisk). Scale bars: 50 μm (A, C, and D); 100 μm (B).
Pathology of the nasal cavity in SARS-CoV-2–infected hamsters. Representative images at 4 days post inoculation. A: Abundant suppurative exudate (asterisk) admixed with proteinaceous material within the nasal cavity. B: Inflammatory cells infiltrated into the nasal mucosal epithelium (asterisk), with epithelial necrosis and ulceration (arrow). Scale bar = 50 μm (A and B).
Additional images of lung pathology at 4 days post inoculation. A: Atypical type II pneumocyte hyperplasia (arrow). B: Bronchial epithelial syncytial cell (arrow). C: Numerous intra-alveolar macrophages, degenerate neutrophils along with fibrinous exudate and thickened alveolar septa. D: Vasculitis, demonstrated by inflammatory cells effacing the vessel wall and necrosis of the vascular wall. Scale bars: 50 μm (A and B); 100 μm (C and D).
Additional images of lung pathology at 7 days post inoculation. A: Degenerate neutrophils, eosinophilic exudate, and macrophages were within alveolar spaces. B: Many large, atypical cells were present lining the alveolar septa that contained large nuclei with prominent nucleoli and lacy chromatin. C: Frequent multinucleated epithelial cells were present throughout affected areas of lung. D: Numerous mitotic figures were present within areas of type II pneumocyte hyperplasia, indicating a robust reparative response. Scale bars: 50 μm (A); 25 μm (B); 30 μm (C); 10 μm (D).
Additional images of lung pathology at 14 days post inoculation. A: Representative tertiary bronchiole from an uninfected animal. B: Residual type II pneumocyte hyperplasia in an arborizing pattern. Scale bar = 100 μm (A and B).
In situ hybridization of SARS-CoV-2 at 2 days post inoculation. SARS-CoV-2 RNA (red chromogen) is detectable within the tracheal epithelium (A), bronchial lymph node (B), bronchiolar epithelium (C), and within alveolar epithelial cells and macrophages (arrow; D). Scale bars: 50 μm (A and B); 100 μm (C); 30 μm (D).
Consolidation classifier of the lungs at 7 days post inoculation (DPI) using QuPath. A: Representative image of SARS-CoV-2–infected animal at 7 DPI, showing multiple coalescing areas of consolidation. B: Example of color-coded classifier of affected area using QuPath-generated superpixel classified as consolidation (green), unaffected (blue), or atelectasis (yellow). Areas where tissue is visible (bronchiolar epithelium and blood vessels) represent superpixels classified as ignore. The red line in each image indicates the annotation marker to generate the region of interest in QuPath. Original magnification, ×2 (A and B).