Abstract
Viral-deletion mutants that conditionally replicate and inhibit the wild-type virus (i.e., defective interfering particles, DIPs) have long been proposed as single-administration interventions with high genetic barriers to resistance. However, theories predict that robust, therapeutic DIPs (i.e., therapeutic interfering particles, TIPs) must conditionally spread between cells with R0 >1. Here, we report engineering of TIPs that conditionally replicate with SARS-CoV-2, exhibit R0 >1, and inhibit viral replication 10- to 100-fold. Inhibition occurs via competition for viral replication machinery, and a single administration of TIP RNA inhibits SARS-CoV-2 sustainably in continuous cultures. Strikingly, TIPs maintain efficacy against neutralization-resistant variants (e.g., B.1.351). In hamsters, both prophylactic and therapeutic intranasal administration of lipid-nanoparticle TIPs durably suppressed SARS-CoV-2 by 100-fold in the lungs, reduced pro-inflammatory cytokine expression, and prevented severe pulmonary edema. These data provide proof of concept for a class of single-administration antivirals that may circumvent current requirements to continually update medical countermeasures against new variants.
Keywords: SARS-CoV-2, coronavirus, variants, evolution, defective interfering particles, lipid nanoparticle, therapeutic interfering particles, RNA, intranasal, virus-like particle
Graphical abstract
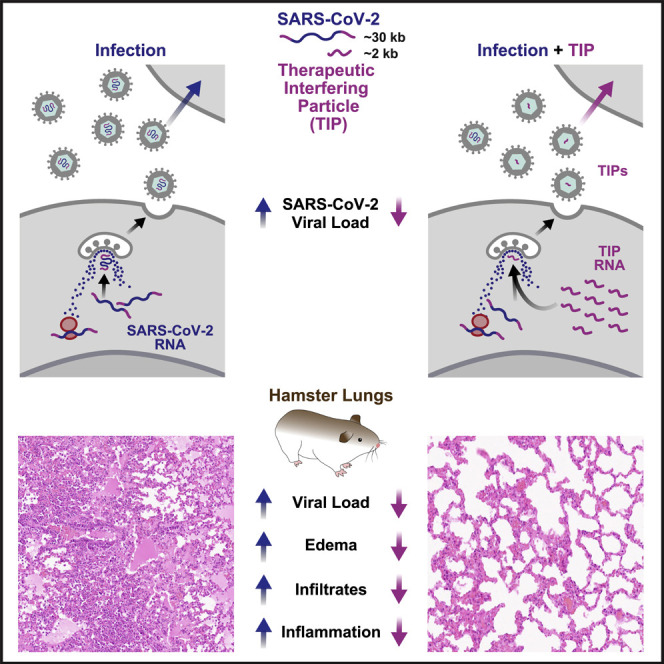
A defective viral particle derived from SARS-CoV-2 competes with the full virus for resources to replicate, showing therapeutic potential by inhibiting viral proliferation in culture and reducing viral load and pathology in animal models for infection.
Introduction
The evolution of resistance to both antimicrobials and vaccines is common across pathogens (Goldberg et al., 2012; Meylan et al., 2018; Petrova and Russell, 2018). Evidence over the past year indicates that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is unlikely to be an exception as viral genetic variability has resulted in variants with increasing resistance to antibody-mediated neutralization (Cele et al., 2021; Wang et al., 2021). In particular, SARS-CoV-2 variants of concern (e.g., B.1.1.7, B.1.351, P.1, B.1.617.2) exhibit increased transmissibility and appear to circumvent natural convalescence and show diminution of vaccine efficacy (Wibmer et al., 2021; Wu et al., 2021). The continued emergence of viral variants suggests parallels to seasonal influenza, where protracted arms races between immunity, waning immunity, and less susceptible viruses occur with a considerable lag between variant emergence and redesigned vaccines.
SARS-CoV-2 is a beta coronavirus with a large ∼30-kb positive-sense, single-stranded RNA genome that replicates with an ∼8-h intracellular replication cycle (Kim et al., 2020). The large genome encodes a suite of nonstructural genes, including the RNA-dependent RNA polymerase (RdRp) within the ORF1ab gene, as well as structural genes—including spike (S), matrix (M), envelope (E), and nucleocapsid (N)—that are required for replication and packaging. In addition to these viral trans elements, the genome is flanked at the 5′ and 3′ termini by untranslated regions (UTRs), which encode regulatory cis elements, including the putative packaging element in the 5′ UTR. Like many RNA viruses, beta coronaviruses also generate sub-genomic deletion mutants that are defective, but, if these RNAs retain obligate cis elements while carrying mutations in trans elements, they can act as defective interfering particles (DIPs) (Makino et al., 1990).
DIPs—originally observed as “autointerference” for influenza virus in the 1940s by Von Magnus (1954) but since reported and studied for many viruses (Akpinar et al., 2015; Holland, 1990; Tapia et al., 2019)—were historically considered cell-culture artifacts with utility for molecular-mechanistic dissection of viruses. However, recent years have seen a renaissance in the study of DIPs following proposals that they could serve as chassis for a class of single-administration antivirals with a high barrier to the evolution of resistance (Metzger et al., 2011; Weinberger et al., 2003). Since DIPs lack self-replication but conditionally replicate with their cognate virus, they have the potential to act as molecular parasites of the wild-type virus within infected cells. Parasitism is mediated by competitive inhibition where viral cis elements encoded by DIPs interact with and “steal” essential trans elements from wild-type virus (e.g., replication or packaging proteins). Consequently, DIPs suppress wild-type viral burst size (the number of viral particles released from an infected cell) and conditionally mobilize their own genomes, spreading their antiviral properties to new cells. Theoretical models predicted that DIPs engineered to have a basic reproductive ratio [R0] >1 could act as durable therapeutics, termed therapeutic interfering particles (TIPs) (Metzger et al., 2011; Weinberger et al., 2003) (Figure 1 A). The predicted high genetic barrier to resistance arises from two considerations: (1) the nature of the cis-trans mechanism of inhibition (Rouzine and Weinberger, 2013) and (2) the R0 TIP >1 enabling TIPs to establish co-evolutionary arms races with wild-type virus (Metzger et al., 2011; Rouzine and Weinberger, 2013; Weinberger et al., 2003).
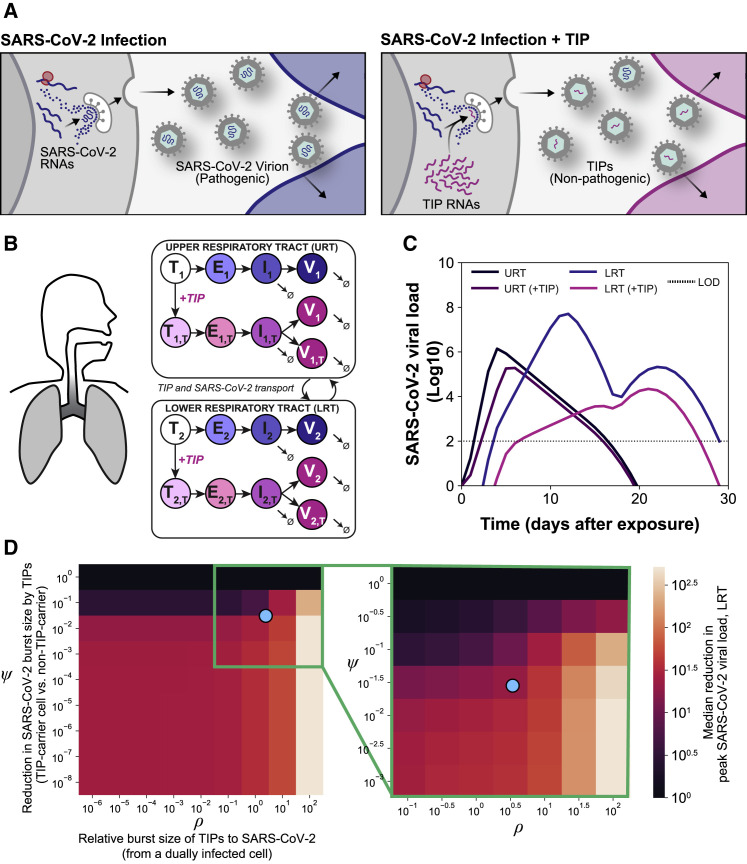
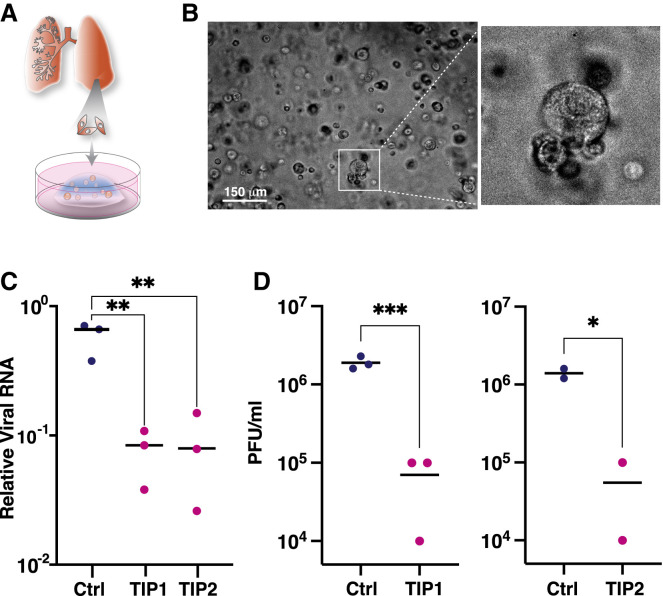
Figure 1.
In silico models predict that a single TIP administration would generate multi-fold reductions in SARS-CoV-2
(A) Schematic of the TIP concept with putative mechanism of action for reducing wild-type virus transmission. Sustained therapeutic efficacy (i.e., molecular parasitism) requires efficient conditional propagation with a basic reproduction ratio (R0) >1.
(B) In silico patient-validated model of SARS-CoV-2 dynamics for upper respiratory tract (URT) and lower respiratory tract (LRT).
(C) Viral-load dynamics in URT and LRT in the presence and absence of a single TIP administration on day 0 showing an R0 >1 TIP is predicted to substantially lower SARS-CoV-2 in both URT and LRT.
(D) Parameter sensitivity analysis for ρ and ψ showing reduction in peak viral load in LRT across eight orders of magnitude (left) and zoomed region (green box, right); the blue dot represents empirically measured values (see Figures 2C and 4E).
See also Figure S1.
Here, we first determined theoretical constraints for a TIP for SARS-CoV-2 and then built TIP candidates, using synthetic SARS-CoV-2 sub-genomic RNAs, that satisfy the constraints and inhibit viral replication in cell culture and donor-derived human lung organoids. The RNAs conditionally propagated with R0 >1—satisfying criteria for a TIP—and mechanistic analyses suggested this occurred via competitive inhibition. We then tested whether TIP antiviral effects were recalcitrant to mutational escape, as predicted. Finally, to test efficacy in vivo, we used Syrian golden hamsters to determine whether TIPs (administered pre- or post-infection) propagate and durably suppress viral replication and associated disease pathology in the lungs. Overall, the data demonstrated that TIPs have robust antiviral efficacy, with a high genetic barrier to the evolution of resistance, indicating their potential as an antiviral countermeasure for respiratory viruses, including SARS-CoV-2.
Results
Theoretical constraints for TIP-mediated inhibition of SARS-CoV-2
We first determined whether TIPs had the theoretical potential to suppress SARS-CoV-2 viral load in vivo. We used in silico modeling approaches that informed therapeutic regimens for other viral pathogens (Perelson, 2002; Perelson et al., 1997) and capitalized on a recent in silico patient-validated model of SARS-CoV-2 within-host viral dynamics (Ke et al., 2020). The model tracks viral loads in two compartments (Figure 1B)—upper respiratory tract (URT) and lower respiratory tract (LRT)—with parameter estimates determined by fitting to longitudinal viral titer data from individual infected patients. We expanded this model to include TIPs using a previously established modeling approach (Weinberger et al., 2003) and numerically solved (Figure 1C) under a range of parameter values for individual patients (Figure S1 A), a range of single-administration dose values (Figure S1B), and a range of viral inoculums (Figure S1C). This showed that TIPs would produce a median predicted knockdown in viral load of ∼1–2 Log in the LRT, and similar viral-load reductions were predicted for the URT.
Figure S1.
Predictive computational modeling of SARS-CoV-2 within-host dynamics in the presence of putative TIPs, related to Figure 1
(a) Representative numerical solutions to within-host model (Ke et al., 2020) in presence of TIPs for nine individual patient parameter sets. (b) Reduction in peak viral load for a range of TIP single-administration dose values (each point represents one patient parameter set) in upper and lower respiratory tracts (URT and LRT). (c) Reduction in peak viral load for 2-Log higher viral inoculum. (d) Relative viral load reduction of a candidate TIP versus a putative non-replicating TIP. (e) Parameter sensitivity analysis for reduction in peak viral load in URT. (f) Reduction in viral loads integrated over time (i.e., area under the viral load curve). (g) Cross-validation with a second patient-parameterized model of SARS-CoV-2 (Kim et al., 2021) showing qualitatively similar effects of TIPs on viral load. (h) Direct and indirect administration of TIPs to a primary index patient can reduce SARS-CoV-2 secondary transmission (direct: R0 = 1.8 to R0 = 0.07; indirect: R0 = 1.8 to R0 = 0.4). Transmission of directly administered TIPs to secondary cases dilutes out (R0 = 0.4 → R00).
To understand whether this reduction in viral load was specific to a particular parameter choice for TIPs, we explored the sensitivity to two molecular parameters previously found to be critical determinants of TIP R0 and efficacy: (1) the TIP competitive efficiency (ρ), which describes the relative difference in production of TIP particles versus wild-type virus particles from a dually infected cell, and (2) the interference efficiency (ψ), which describes the reduction in wild-type virus production by TIPs competing intracellularly for viral proteins. In terms of viral burst size, ρ reflects the relative burst size of TIPs compared to wild-type virus (from a dually infected cell), whereas ψ reflects the reduction in wild-type virus burst size between singly versus dually infected cells.
The parameter sensitivity analysis showed a wide range of ρ and ψ values where TIPs generate a 1-Log reduction in peak viral load in the LRT but that >1-Log reductions in viral load were specific to larger values of ρ and moderate values of ψ (Figure 1D). The phenomenon of larger values of ψ having lesser viral-load reduction has been previously predicted (Weinberger et al., 2003) and results from the TIP “shooting itself in the molecular foot” and limiting efficient mobilization when interference is too strong. Consistent with previous analyses of TIPs for other viruses (Metzger et al., 2011), larger values of ρ were the key factor enabling TIPs to generate larger viral-load reductions (Figure S1D). Similar TIP-mediated reductions in URT viral load were also observed (Figure S1E). To be sure that knockdown of peak viral load reflected a true reduction in viral load, we also analyzed viral loads integrated over time (i.e., area under the curve) and found similar reductions by TIPs (Figure S1F).
The simulations showed that TIPs could yield on average 1-Log reductions in SARS-CoV-2 in a dose-dependent fashion, with substantially larger 6-Log reductions for some patient-parameter estimates (Figure S1A). To be sure this was not model dependent, we also analyzed a second patient-parameterized model of SARS-CoV-2 (Kim et al., 2021) and observed qualitatively similar effects of TIPs on viral load (Figure S1G). Based upon prior work linking SARS-CoV-2 viral load and infectiousness (Goyal et al., 2021; Jones et al., 2021), we also analyzed how TIPs might affect SARS-CoV-2 transmission and found that a TIP-mediated reduction in viral load would generate a substantial reduction in SARS-CoV-2 secondary infections (from R = 1.8 to R = 0.07) (Figure S1H). In summary, these in silico analyses showed that TIPs have the theoretical capacity to suppress SARS-CoV-2 in vivo after a single administration, provided that ρ and ψ surpass specific thresholds.
An engineered TIP RNA conditionally propagates and inhibits SARS-CoV-2 in cell culture
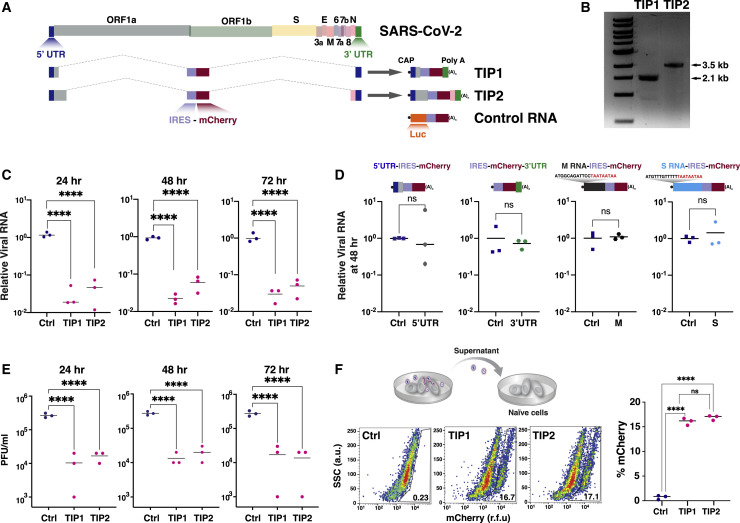
Given the challenges of engineering TIPs for other viruses (Notton et al., 2021), we set out to construct a TIP with appropriate parameters. As a starting point, we used previous high-throughput analyses of the cis genetic elements required for efficient propagation of sub-genomic transcripts in other RNA viruses (Notton et al., 2021) and historical data from murine hepatitis virus (Baric et al., 1988; Makino et al., 1990), both of which argued that, at a minimum, the viral 5′ and 3′ UTRs were essential for conditional replication and construction of a TIP. Consequently, we designed two minimal sub-genomic synthetic constructs, encoding different lengths of the 5′ and 3′ regions of the viral genome and tested whether they met the threshold values to act as TIPs. Both putative TIPs encompass stem loop 5 in the 5′ UTR, which encodes a predicted packaging signal (Chen and Olsthoorn, 2010; Rangan et al., 2020), as well as the entirety of the 3′ UTR and a 1,280 nucleotide (nt) reporter cassette (Figure 2 A) encoding an internal ribosome entry sequence (IRES) driving expression of a fluorescent reporter protein (mCherry). TIP1 (∼2.1 kb) encodes the first 450 nt of the 5′ UTR plus part of polyprotein ORF1ab and the last 328 nt of the 3′ UTR plus the reporter cassette, whereas TIP2 (∼3.5 kb) encodes 1,540 nt encompassing the 5′ UTR and part of ORF1ab and the last 713 nt of the genome containing part of N protein, ORF 10, and the 3′ UTR, along with the reporter cassette. All TIP and control mRNAs were in vitro transcribed (Figure 2B) and a 5′ methyl cap and ∼100-nt 3′ poly(A) tail were added following in vitro transcription.
Figure 2.
Identification of SARS-CoV-2 TIPs that conditionally replicate, inhibit CoV-2, and propagate with R0 >1
(A) Schematic of SARS-CoV-2 genome versus sub-genomic TIP constructs TIP1 (2.1 kb), TIP2 (3.5 kb), and Ctrl (luciferase) RNA (3 kb). All RNAs were 5′ methyl capped and poly(A) tailed and encode an IRES-mCherry reporter cassette.
(B) Gel electrophoresis of in vitro-transcribed TIP1 and TIP2 mRNAs.
(C) Virus yield-reduction assay in Vero cells transfected with either TIP1, TIP2, or Ctrl RNA; cells were infected with SARS-CoV-2 WA-1 isolate (MOI = 0.05) at 24 h after RNA transfection and titered at 24, 48, and 72 h following infection. Viral transcripts were quantified by qRT-PCR for the E gene.
(D) Schematics and viral yield-reduction assay (at 48 h post-infection) for four alternative viral RNA controls: (1) 5′ UTR-IRES-mCherry (1.73 kb), (2) IRES-mCherry-3′ UTR (1.64 kb), (3) M(3xTAA)-IRES-mCherry (1.98 kb), and (4) S(3xTAA)-IRES-mCherry (2.16 kb) compared to Ctrl RNA (luciferase-IRES-mCherry). Viral RNA quantification by qRT-PCR for N gene.
(E) SARS-CoV-2 viral titers quantified by PFU/mL.
(F) Supernatant transfer assay for cells transfected with either TIP1 RNA, TIP2 RNA, or Ctrl RNA and then infected with SARS-CoV-2 WA-1 isolate (MOI = 0.05) at 24 h after RNA transfection. The supernatant was clarified and transferred to naive cells, which were then analyzed by flow cytometry 24 h post-transfer. For all panels: ns, not significant and ∗∗∗∗p < 0.0001, from Student’s t test. See also Figure S2.
To evaluate the interference potential of the putative TIPs, virus yield-reduction assays were performed. Vero cells were transfected with the purified TIP mRNA or a similarly sized control mRNA encoding a luciferase-IRES-mCherry reporter cassette (hereafter referred to as “Ctrl RNA”) and then infected with SARS-CoV-2 virus (WA-1 isolate) at a multiplicity of infection (MOI) of 0.05. Viral gene expression was quantified by qRT-PCR at 24, 48, and 72 h using primers specific to SARS-CoV-2 genes not present in the TIPs—to assay at these time points, the supernatant needed to be transferred to naive target cells. The data show a significant 1.5-Log inhibition of SARS-CoV-2 replication at all time points for both TIP1 and TIP2 (Figure 2C). To verify that the observed interference was not due to off-target or cell-mediated innate immune effects (e.g., cellular RNA interference), we examined an expanded panel of control RNAs encoding the following: (1) 5′ UTR RNA alone, (2) 3′ UTR RNA alone—which lacks the putative 5'UTR packaging signal, (3) an RNA encoding SARS-CoV-2 Matrix (M) with 5' stop codons, and (4) an RNA encoding the SARS-CoV-2 Spike (S) with 5' stop codons. Each of these control RNAs also encoded IRES-mCherry to match the TIP RNAs and were 5'-methyl capped and poly(A) tailed. None of the control RNAs generated a significant reduction in SARS-CoV-2 viral gene expression (Figure 2D).
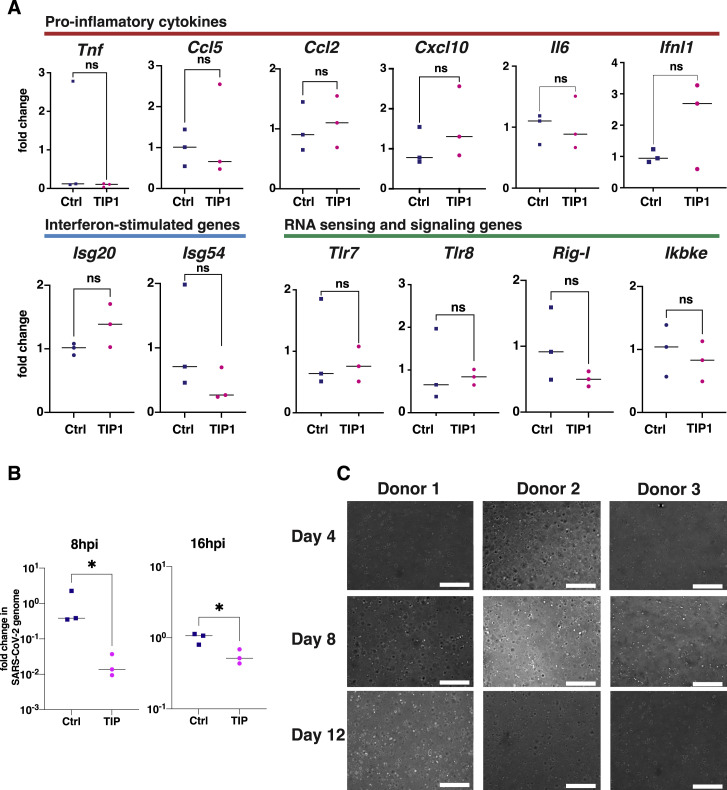
To determine whether innate antiviral mechanisms specific to the TIP RNA sequence were responsible for viral interference, we also analyzed expression of 12 common innate-immune responsive genes including interferon-stimulated genes (ISGs) in cells nucleofected with either TIP RNA or Ctrl RNA in the absence of infection (Figure S2 A). These data show no significant upregulation of pro-inflammatory cytokines, downstream ISGs, or upstream RNA-sensing or signaling genes by TIP RNAs, inconsistent with an RNA-induced innate antiviral mechanism.
Figure S2.
Expression analysis of inflammatory pathways in TIP-treated cells, TIP post-infection activity, and culturing of lung organoids from primary human small-airway epithelial cells, related to Figures 2 and 3
(a) Analysis of relative expression (by qRT-PCR) of a panel of inflammatory genes based on (Nelson et al., 2020) in TIP RNA and Ctrl RNA transfected Vero cells in the absence of SARS-CoV-2 infection. No significant overexpression of inflammatory pathways, interferon-stimulated genes, or RNA sensors. (b) Efficacy of TIP in post-infection setting in cell culture. Cells were infected at MOI = 0.05, transfected with TIP or Ctrl RNA at 8hrs or 16hrs post-infection, and harvested at 48hrs post-infection. Viral RNA copies were quantified by qRT-PCR using N gene primers (normalized to GAPDH). (c) Transmission micrographs of human small airway epithelial cells from three donors cultured in matrigel in the presence of HAO (human airway organoid) medium. Organoids were monitored under transmission microscope at 10x magnification (size bar: 300 μm). Representative images shown for days 4, 8 and 12. [For all panels: ns denotes not significant, ∗ denotes p < 0.05 from Student’s t test].
We next tested whether TIP administration could exert an antiviral effect following infection in cell culture. Vero cells were first infected with SARS-CoV-2 (MOI = 0.05) and TIP RNA was then administered at either 8 h post-infection and 16 h post-infection and, as above, viral gene expression quantified by qRT-PCR at 48 h post-infection using primers specific to SARS-CoV-2 genes not present in the TIPs. These post-infection TIP-administration data show a significant 1-Log inhibition of SARS-CoV-2 replication by TIPs (Figure S2B).
To confirm that TIP RNAs inhibit virus output, we used a plaque-forming unit (PFU) assay, which detects and quantifies infectious virus produced from cells, to analyze at the same time points as in Figure 2C (i.e., 24, 48, and 72 h post-infection). The PFU assay confirmed that TIPs generate a 1.5-Log inhibition of infectious SARS-CoV-2 particles (Figure 2E).
To test whether the putative TIP RNAs conditionally propagate in the context of SARS-CoV-2 infection, we used a supernatant-transfer assay. The cell-free supernatant, collected at 8 h post-infection of SARS-CoV-2 (MOI = 0.05), was transferred undiluted to naive target cells, which were then analyzed for mCherry expression. Flow-cytometry analysis showed significant mCherry expression in target cells after supernatant transfer (Figure 2F). Thus, for SARS-CoV-2, the initial TIP prototypes (i.e., starting points) appear to satisfy the requisite constraints.
TIPs inhibit SARS-CoV-2 in primary human lung organoids
To test whether TIPs interfered with SARS-CoV-2 in a more physiological setting, we employed a human lung organoid model (Figure 3 A) (Sachs et al., 2019; Zhou et al., 2018). Previous analysis of SARS-CoV-2-infected lung organoids (Han et al., 2021) showed they are valuable for screening candidate COVID-19 therapeutics and revealed cytokine/chemokine and interferon signatures consistent with inflammatory changes observed in primary human COVID-19 pulmonary infections (Blanco-Melo et al., 2020). We established and characterized organoids using primary human small-airway epithelial cells (Figure 3B), obtained from three donors (Figure S2C). The organoids were transfected with either TIP1, TIP2, or Ctrl RNA and then infected with SARS-CoV-2 virus at MOI = 0.5 (as these cultures are known to be challenging to infect) 24 h later. Viral titers in lung organoids were assayed by qRT-PCR (Figure 3C) and PFU analysis (Figure 3D) 24 h post-infection, which confirmed that TIPs reduced SARS-CoV-2 by ∼1-Log compared to Ctrl RNA.
Figure 3.
TIPs inhibit SARS-CoV-2 in donor-derived lung organoids
(A) Schematic of primary human small-airway epithelial cell organoids.
(B) Bright-field micrograph of organoids at day 2 following establishment from one representative donor. Scale bar, 150 μm.
(C) Viral transcripts in SARS-CoV-2-infected (MOI = 0.5) lung organoids transfected with Ctrl, TIP1, or TIP2 RNA assayed by qRT-PCR to E gene at 24 h post-infection.
(D) Viral titer quantification by plaque assay (PFU/mL) for samples in (C).
For all panels: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05 from Student’s t test. See also Figure S2.
TIPs generate virus-like particles, compete for viral trans elements, and mobilize with R0 >1
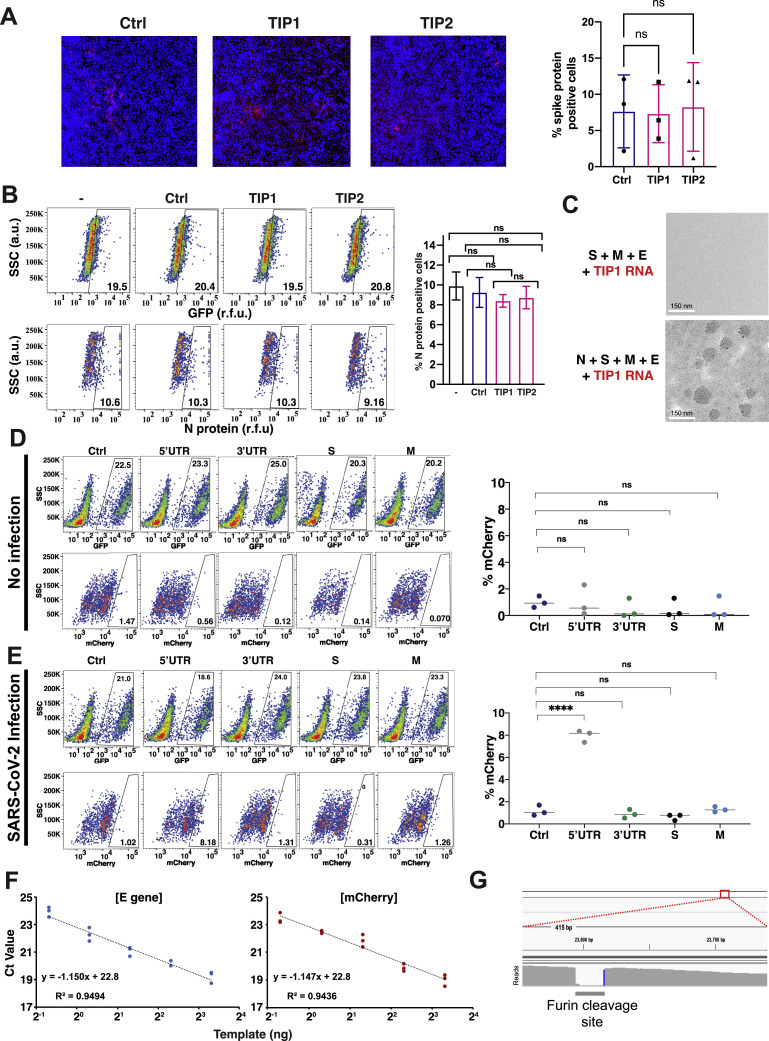
To understand the mechanism of action of TIP interference, we first determined whether TIPs were restricting incoming virus infection, which might indicate interference via induction of innate cellular immune responses (e.g., interferon response) or similar cellular restrictions. Viral entry was analyzed by immunofluorescence staining for S protein at 2 h post-SARS-CoV-2 infection (MOI = 20), and no significant effect of TIP RNA on viral entry was detected (Figure S3 A).
Figure S3.
Controls for TIP mechanism of action and mobilization in cell culture, related to Figure 4
(a) TIPs do not interfere with SARS-CoV-2 virus entry. Entry analysis of Vero cells nucleofected with Ctrl, TIP1 or TIP2 RNAs. 24 hours post nucleofection, cells were infected with SARS-CoV-2 (WA-1 isolate; MOI = 20). Two hours post infection, cells were fixed, stained for S protein and nucleus (DAPI), and subjected to high-throughput confocal microscopy. Left: representative images for overlay of S and nucleus. Right: quantification of % S protein positive cells in all three samples. ns, not significant. (b) TIPs do not interfere with early events in SARS-CoV-2 life cycle. Flow cytometry of Vero cells nucleofected with Ctrl, TIP1 or TIP2 RNAs and infected (MOI = 0.05) at 24 hours post nucleofection. Two hours post-infection cells were thoroughly washed and naive GFP+ cells were added. Cells were harvested at 8 hours post infection (∼one replication cycle), stained for N protein and subjected to flow cytometry. Left: flow cytometry dot plots; Right: bar graph representing % N protein positive cells. ns, not significant. (c) Transmission EM analysis of concentrated supernatant from cells transfected with S, E, and M and TIP RNA showing no VLP (top) and from concentrated supernatant from cells transfected with N, S, E, and M and TIP RNA showing VLPs (bottom), Scale bar = 150nm. (d, e) R0 estimation via 1st round supernatant transfer for Ctrl RNA, 5′UTR, 3′ UTR, S(3xTAA) and M(3xTAA) controls in the absence (d) and presence (e) of infection (MOI = 0.05). TIP-transfected cells were infected with SARS-CoV-2 or mock, thoroughly washed, and at two hours post-infection or mock infection, GFP+ reporter cells were introduced to the culture. At 12 hours post infection, GFP+ cells were analyzed by flow cytometry to quantify % mCherry+ cells (by indirect immunofluorescence) within the GFP+ population. (f) Standard curve of E-gene and mCherry using qPCR at the mentioned concentrations. (g) Alignment of sequencing from long-term culture showing furin cleavage site mutation. (For all panels: Error bars represent standard deviation from three biological replicates; ns denotes not significant, ∗∗∗∗ denotes p < 0.0001, Student’s t test.)
To test whether TIPs affected the early events of SARS-CoV-2 infection, cells were infected with SARS-CoV-2 (MOI = 0.05), and GFP positive cells were added (20% of total number of cells infected) at 2 h post-infection. Flow-cytometry analysis of viral N expression was performed at 8 h following infection, within the first round of viral replication. The 8-h time point was chosen to optimize the dynamic range for detection of early restrictions as 8 h is near the end of the first round of the viral life cycle, thereby allowing sufficient time for viral transcripts to accumulate, but is prior to substantial viral egress and 2nd-round infection. We observed no significant impact of TIPs on SARS-CoV-2 viral N protein expression at 8 h post-infection in the GFP positive cells (Figure S3B), indicating that TIP interference could not be explained by early cellular restriction events and that interference occurred during later times in the viral life cycle (e.g., viral packaging).
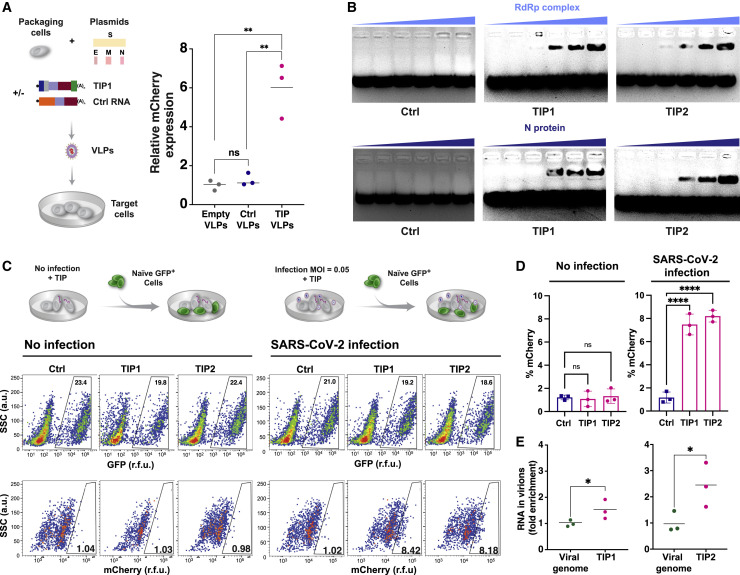
Next, to determine whether TIP RNAs are packaged into virus-like particles (VLPs), we performed reconstitution assays (Figure 4 A). Cells were co-transfected with expression vectors each encoding a cDNA for the matrix (M), envelope (E), spike (S), or nucleocapsid (N) protein of SARS-CoV-2 together with TIP RNA, Ctrl RNA, or no RNA. The supernatant was concentrated (ultracentrifuged) and imaged for the presence of VLPs by transmission electron microscopy (EM) and, in parallel, analyzed for functional VLP transduction of naive cells. EM analysis showed the presence of abundant ∼100 nm-diameter VLPs (Figure S3C). qRT-PCR for mCherry showed substantial TIP transduction of naive cells when VLPs where reconstituted using TIP RNA but not Ctrl RNA (Figure 4A).
Figure 4.
TIP RNAs form functional VLPs, bind SARS-CoV-2 RdRp and N trans elements, and mobilize with R0 >1
(A) Reconstitution assay: schematic and quantification of VLP reconstitution for TIP1 and Ctrl RNA. Quantification in target cells by qRT-PCR for mCherry as compared to empty (RNA-free) VLPs.
(B) EMSA of TIP RNA or Ctrl RNA incubated with increasing concentrations of N protein or RdRp complex from cell extracts.
(C) R0 estimation via 1st-round supernatant transfer. TIP-transfected cells were infected with SARS-CoV-2 (MOI = 0.05) and then thoroughly washed to remove virus, and at 2 h post-infection GFP+ reporter cells were introduced to the culture. At 12 h post-infection, GFP+ cells were analyzed by flow cytometry to quantify the percentage mCherry+ cells (via indirect immunofluorescence staining) within the GFP+ population. Uninfected cells were used as an experimental control to confirm that TIP mobilization only occurred in the presence of SARS-CoV-2.
(D) Flow-cytometry quantification of (C).
(E) Relative packaging of TIP RNA in virions. Cells were nucleofected with TIP1 or TIP2 followed by SARS-CoV-2 infection (MOI = 0.05), and the supernatant was harvested at 24 h post-infection and analyzed by qRT-PCR for TIP RNA (using mCherry qPCR primers) versus viral genomic RNA (using E gene qPCR primers). Standard curves (see Figure S3F) were statistically indistinguishable for both primer sets.
For all panels: ns, not significant, ∗∗∗∗p < 0.0001, ∗∗p < 0.01, ∗p < 0.05 from Student’s t test. See also Figure S3.
To test whether TIP mRNAs directly bind and compete for SARS-CoV-2 viral proteins, we performed electromobility shift assays (EMSAs) on TIP RNA and viral proteins. Since the RdRp complex and the N protein can directly interact with viral RNAs (Baric et al., 1988; Iserman et al., 2020), we hypothesized that these proteins were the most likely to be competition substrates for the TIP. EMSA analysis of cell extracts expressing either RdRp complex or N protein, incubated with purified TIP1 or TIP2 RNA, show that TIP RNAs bind both RdRp complex and N proteins, whereas Ctrl RNA does not bind either of these proteins (Figure 4B).
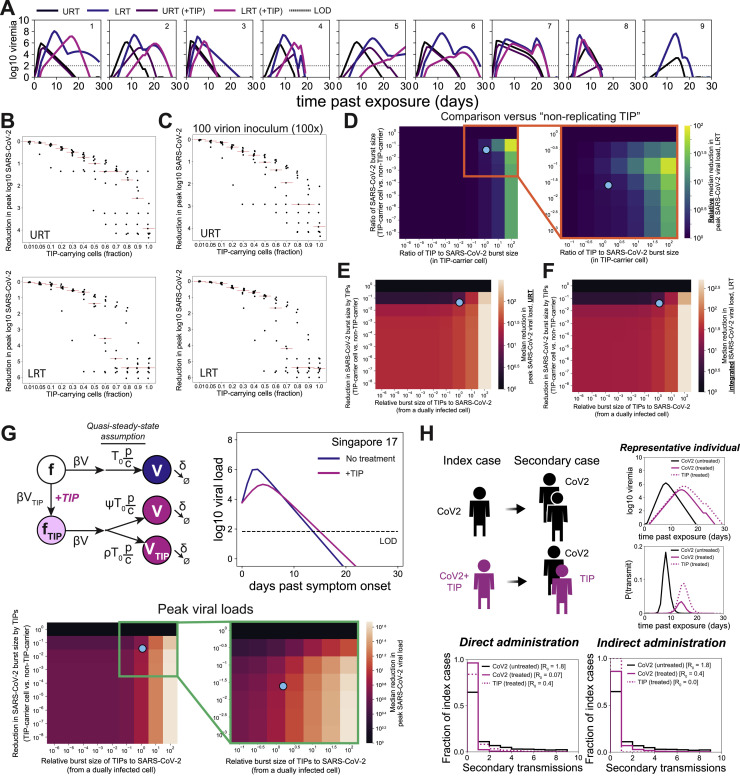
To quantify the R0 of TIPs in the context of SARS-CoV-2 infection, we modified the supernatant-transfer assay into a “1st-round supernatant transfer assay.” TIP-transfected cells were infected at a low MOI (MOI = 0.05) and then thoroughly washed to remove virus, and at 2 h post-infection GFP+ reporter cells were introduced to the culture (at ∼20% of total cells). TIP mobilization into reporter cells was quantified using the percentage of mCherry+ cells within the GFP+ population at 12 h post-infection. Infection-dependent mobilization was confirmed by comparing to uninfected samples for all RNAs (Figures 4C and 4D), and the control RNAs did not mobilize in either the absence or presence of virus (Figures S3D and S3E), with the exception of 5′ UTR, as expected, given that it carries the putative packaging signal (Iserman et al., 2020; Makino et al., 1990). The fraction of TIP+ cells, approximately 8%, was corrected for background autofluorescence, to yield 6.3% TIP+ cells (as compared to approximately 5% infected cells for the original SARS-CoV-2 infection at MOI = 0.05), and this translated to 4% infected cells after accounting for the addition of 20% GFP+ cells in the assay. TIPs propagating into 6.3% of new cells from the initial wild-type infection of 4% of cells represents a roughly 50% increase or roughly an R0 = 1.57; for comparison, R0 = 2 would require a doubling, from 4% to 8%, of cells being mCherry+. This R0 >1 finding for TIPs is further verified below using a continuous serial-passage approach (see Figure 5 ).
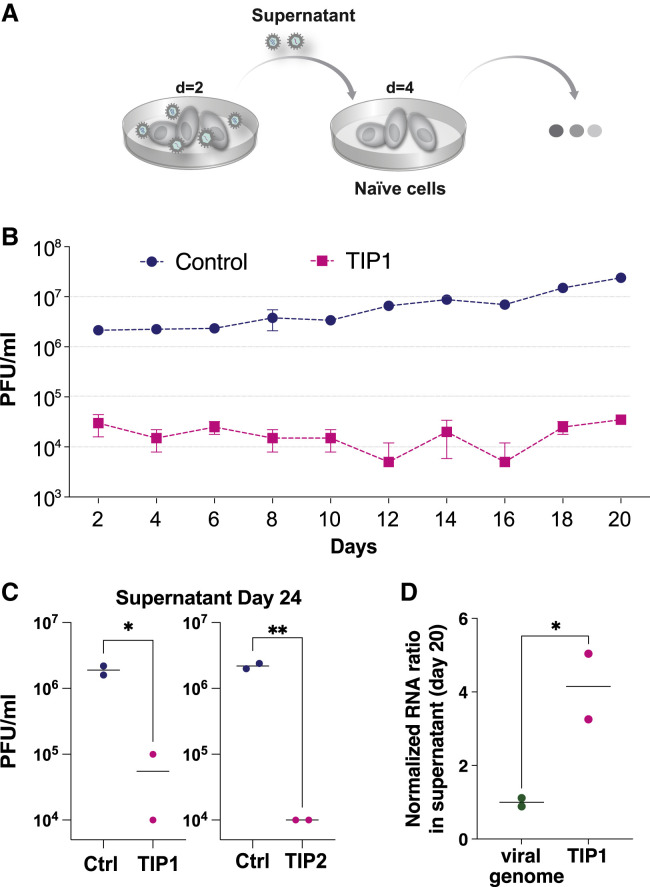
Figure 5.
TIPs have a high barrier to the evolution of resistance in long-term cultures
(A) Schematic of the continual culture serial-passage system for SARS-CoV-2 propagation. Cells were transfected with TIP1 or Ctrl RNA and infected 24 h later with SARS-CoV-2 WA-1 isolate (at MOI = 0.05). The cell-free supernatant was collected every 2 days for titering and transferred to naive cells.
(B) Viral titers of SARS-CoV-2 WA-1 by plaque assay (PFU/mL) from continuous cultures. Error bars represent three biological replicates.
(C) Yield-reduction assay of virus isolated from day 24 of continuous culture tested in naive cells transfected with TIP RNA or Ctrl RNA.
(D) Quantification of TIP and SARS-CoV-2 from day 20 of the continuous culture.
Supernatants from day 20 of the continuous culture were analyzed by qRT-PCR for mCherry and E gene (i.e., SARS-CoV-2 genome) and the mCherry:E ratio calculated (for all panels: ∗∗p < 0.01, ∗p < 0.05 from Student’s t test).
To verify that TIP RNA was packaged into virions at a high level, we quantified the relative fraction of TIP RNA versus SARS-CoV-2 genomic RNA in virions isolated from the supernatant by qRT-PCR (Figure 4E). Analysis showed that the TIP RNA was significantly enriched (1.5- to 2-fold) compared to SARS-CoV-2 viral genomes (for standard curves, see Figure S3F).
Overall, these data indicate the TIPs do not restrict viral entry or early viral expression (i.e., via induction of a cellular response), that TIP RNA generates functional TIP VLPs in the presence of M, N, E, and S, that TIP RNAs bind to and may compete for SARS-CoV-2 proteins in cells, and that competition for packaging and replication resources is sufficient to quantitatively account for the measured TIP-mediated yield reduction.
The SARS-CoV-2 TIP exhibits a high barrier to evolution of resistance
Previous theoretical analyses for other rapidly evolving RNA viruses (e.g., HIV-1) (Metzger et al., 2011; Rast et al., 2016; Rouzine and Weinberger, 2013) predicted that TIPs would have a higher genetic barrier to the evolution of resistance. Based on modeling and comparative sequence analysis indicating that the SARS-CoV-2 UTRs are highly conserved and evolve relatively slowly (Chan et al., 2020; Rangan et al., 2020), we hypothesized that TIP-mediated interference would carry a relatively high barrier to the evolution of viral resistance. To test this hypothesis, we established long-term virus cultures where the viral supernatant was continually serially passaged to new naive cells to sustain high-level viral infection and selected for viral escape mutants (Figure 5A). The continuous viral cultures were initiated using cells transfected with either Ctrl RNA or TIP1 RNA, and cells were then infected and the viral supernatant was serially passaged onto naive, non-transfected cells every 48 h for ∼3 weeks, with virus titered at each passage.
SARS-CoV-2 replicative fitness was enhanced by ∼1-Log over 3 weeks in the Ctrl RNA continuous culture (Figure 5B). This fitness increase was likely due to the furin cleavage mutation in the S gene, which we confirmed via sequencing was overrepresented in the day 20 culture (Figure S3G), and which has previously been reported to arise rapidly in Vero cells (Johnson et al., 2021).
In contrast, the continuous cultures initiated in the presence of TIP RNA exhibited an immediate ∼2-Log decrease in viral titer by PFU (Figure 5B), consistent with single-round yield reduction data (Figures 2 and 3). This reduction in viral titer was sustained over the course of the 20-day culture.
To verify that this viral-load reduction in the continuous culture was due to TIP interference and not a cellular peculiarity, the supernatant from a parallel control culture after day 20 was used to infect cells in the presence of TIP RNA, and the 2-Log decrease in viral titer was recapitulated (Figure 5C). qRT-PCR analysis of the culture supernatants indicated that TIP RNA exhibited a 4-fold increase relative to SARS-CoV-2 RNA on day 20 (Figure 5D). These continuous culture data indicate conditional amplification and sustained transmission of the TIP, i.e., R0 >1, since the TIP RNA was only added to the infected culture once (i.e., a single administration on day 0). The data are not consistent with recombination hypotheses (e.g., of the TIP reporter into the wild-type virus). Indeed, recombination leading to extinction of TIPs would result in a fitness increase in the TIP continuous culture (i.e., rescue), whereas a recombinant virus with reduced fitness would be selectively outcompeted by a wild-type with a 2–3 Log replicative advantage. We further analyze recombination potential below.
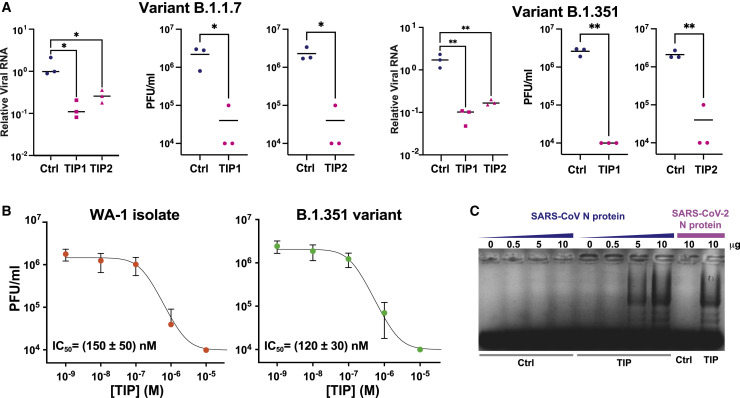
Given the TIP mechanism of action and relatively high barrier to resistance, we hypothesized that TIPs would robustly inhibit SARS-CoV-2 variants of concern, including variants that are resistant to antibody neutralization (Wang et al., 2021). To test this hypothesis, we performed yield-reduction assays for the beta variant of concern (501Y.V2, a.k.a., B.1.351) as well as the alpha variant of concern (B.1.1.7) (Figure 6 A). TIP RNA generated ∼2-Log reduction in viral titers (PFU/mL) for both variants and qRT-PCR showed significant yield reductions. Notably, the TIP-mediated reduction in viral RNA was reduced compared to the effect on PFU, likely due to known complications of excess sub-genomic RNA (Sia et al., 2020). Remarkably, dose-response analysis showed that the half-maximal inhibitory concentrations (IC50) of TIP1 for the WA-1 historical isolate and neutralization-resistant B.1.351 variant were indistinguishable (1.5 ± 0.5 × 10−7 M versus 1.2 ± 0.3 × 10−7 M) (Figure 6B), indicating no loss in TIP interference potential. This IC50 of ∼100 nM is similar to a recently reported broad-spectrum SARS-CoV-2 antiviral advancing to clinical trials (Sheahan et al., 2020).
Figure 6.
TIP inhibition is robust to variants of concern
(A) Viral yield-reduction of SARS-CoV-2 B.1.1.7 and B.1.351 by qRT-PCR for E gene (left) and plaque assay (middle and right) upon transfection of either Ctrl, TIP1, or TIP2 RNA.
(B) Half-maximal inhibitory concentration (IC50) of TIP1 for variant B.1.351 compared to WA-1 historical isolate. Dose-response analysis was performed by a plaque assay.
(C) EMSA of cell extracts expressing N protein from SARS-CoV incubated with Ctrl or TIP1 RNA.
For all panels: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05 from Student’s t test.
To further determine limits for TIP robustness to mutational escape, we tested whether TIP RNA could efficiently bind to proteins of the evolutionarily distinct beta coronavirus lineage SARS-CoV. Analyses indicate that the SARS-CoV virus (Guan et al., 2003) and SARS-CoV-2 diverge by about 20% in sequence (Kaur et al., 2021). Given that the mechanism of TIP interference for SARS-CoV-2 involves competition for N protein, we assayed whether TIP RNA could efficiently bind to SARS-CoV N protein. The N protein from both SARS-CoV-2 and SARS-CoV were expressed from cDNAs (Gordon et al., 2020a) in cell culture, and cell extracts were co-incubated with purified TIP1 RNA and analyzed by EMSA (Figure 6C). The EMSAs show that TIP RNA binds SARS-CoV N protein about as efficiently as SARS-CoV-2 N protein, suggesting that the virus would need to evolve farther away in sequence space than SARS-CoV to escape competitive inhibition by the TIP.
In hamsters, intranasal TIP delivery inhibits SARS-CoV-2, reduces pro-inflammatory cytokines, and prevents pulmonary edema
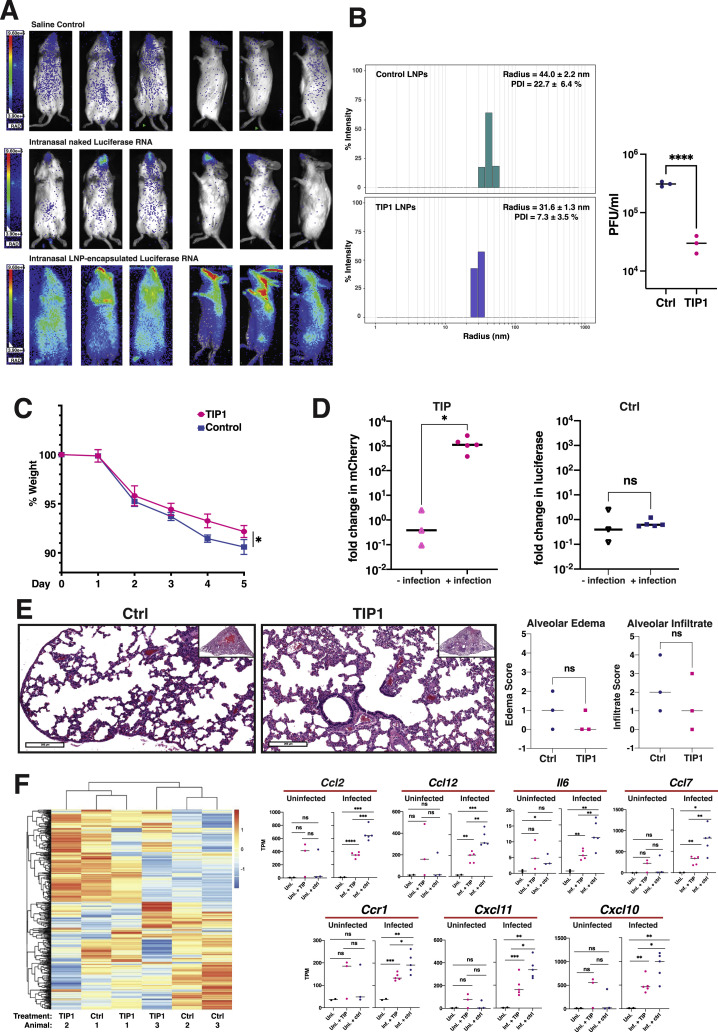
To assay the in vivo efficacy of TIPs, we utilized the Syrian Golden Hamster model of SARS-CoV-2 infection (Sia et al., 2020). First, we tested intranasal administration of various RNA delivery approaches for their ability to efficiently deliver RNA to the respiratory tract of rodents. Using an in vitro-transcribed luciferase-expressing RNA, we tested purified RNA alone (“naked RNA”), RNA encapsulated into cationic polymer nanocarriers (i.e., polyethylenimine), and RNA encapsulated in lipid nanoparticles (LNPs) (Zhang et al., 2020). LNPs exhibited efficient in vivo RNA delivery to the lungs after intranasal administration (Figure S4 A). We generated LNPs containing either TIP1 RNA or Ctrl RNA, characterized them, and confirmed that LNP-encapsulated TIP RNA retained antiviral efficacy using yield-reduction assays in Vero cells (Figure S4B).
Figure S4.
Characterization of lipid nanoparticles (LNPs) for intranasal delivery and analysis of LNP TIPs in infected and uninfected hamsters, related to Figure 7
(a) Bioluminescence imaging of mice six hours after intranasal administration of in vitro transcribed RNA encoding firefly luciferase. Mice were given either saline, purified RNA alone (‘naked RNA’), or LNP-encapsulated RNA. (b) Left: Dynamic light scattering (DLS) characterization of LNPs carrying TIP RNA to measure radius and polydispersity (left). Right: Validation of antiviral activity (yield reduction) of LNP TIPs in infected Vero cells. Confluent cells were incubated for two hours with TIP LNPs or Ctrl LNPs (at an RNA concentration of 0.3ng/μL) and cells were then infected with SARS-CoV-2 (MOI = 0.05). Supernatant was harvested at 48 hours post infection and virus titered by plaque assay (PFU/ml). (c) Weight change of hamsters over time after infection with SARS-CoV-2 in Ctrl- or TIP LNP treated animals. (d) Quantification of TIP and Ctrl RNA in the presence and absence of infection in hamsters. Syrian golden hamsters were treated twice with TIP or Ctrl RNA at 24hrs apart in the presence and absence of SARS-CoV-2 (106 PFU). Lungs were harvested at day 5, RNA was extracted, and qRT-PCR was performed for either mCherry or luciferase. Quantification of TIP and Ctrl RNAs was performed between the infected and uninfected lung samples. (e) H&E staining of lung section of one representative Ctrl-treated and one representative TIP-treated animal in the absence of infection at 5 days post treatment (left). Histopathological scoring of lung sections from the uninfected hamsters (n = 3 for each group of animals) treated with TIP or Ctrl RNA LNPs for alveolar edema and cellular infiltrates to alveolar space (right).(f) Heatmap showing expression level of DEGs in uninfected samples. DEGs were defined by comparing infected samples treated with TIP or Ctrl RNA LNPs. Representative proinflammatory genes are shown on the right in the presence and absence of infection. [For all panels: ns denotes not significant, ∗∗∗∗ denotes p < 0.0001 ∗∗∗ denotes p < 0.001, ∗∗ denotes p < 0.01, ∗ denotes p < 0.05 from Student’s t test].
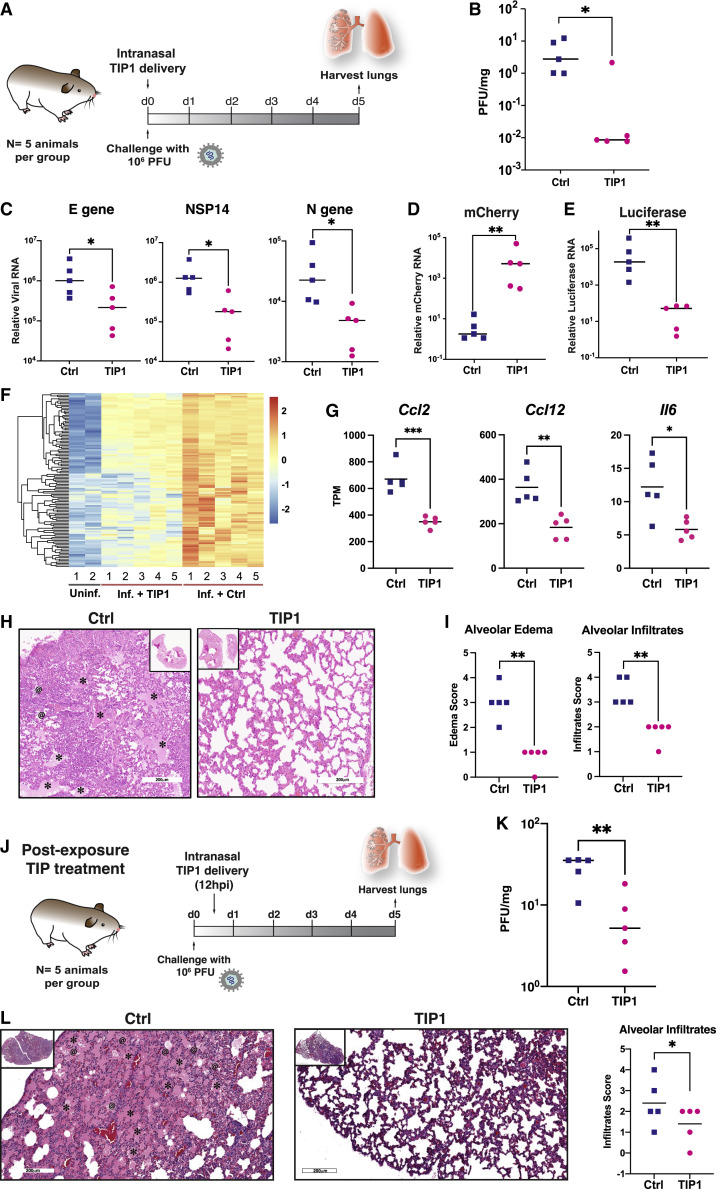
Next, we administered the TIP or Ctrl RNA LNPs intranasally to Syrian Golden hamsters and then challenged them with SARS-CoV-2 (106 PFUs) (Figure 7 A). As expected, control-treated hamsters showed weight loss following infection, but this was significantly ameliorated by TIP treatment (Figure S4C). Previous studies indicated that the observed weight retention would quantitatively correlate with a 2–3 Log reduction in SARS-CoV-2 viral load in the lungs (Rogers et al., 2020). Analysis of infectious virus in lung tissue harvested on day 5 from hamsters confirmed a significant ∼2-Log reduction in SARS-CoV-2 viral load in TIP-treated animals (Figure 7B). One animal did not exhibit a reduction in viral load, which may be consistent with inefficient TIP dosing/delivery. qRT-PCR analysis of viral transcripts in the lung exhibited a correlated, but lesser, 1-Log reduction in viral load for TIP-treated animals (Figure 7C), consistent with previous studies in SARS-CoV-2-infected hamsters (Sia et al., 2020).
Figure 7.
TIPs inhibit SARS-CoV-2 replication in vivo in Syrian golden hamsters and protect against severe pulmonary edema
(A) Timeline of SARS-CoV-2 challenge experiment in Syrian golden hamsters. At 6 h pre-infection, intranasally administration of TIP LNPs (n = 5) or Ctrl RNA LNPs (n = 5) was performed. Animals were then infected with SARS-CoV-2 (106 PFU), and an intranasal LNP booster was administration delivered at 18 h post-infection. Lungs were harvested at 5 days post-infection.
(B) SARS-CoV-2 viral titers from lungs harvested on day 5 by plaque assay.
(C) SARS-CoV-2 viral transcript levels by qRT-PCR for N, NSP14, and E from lungs harvested on day 5 post-infection.
(D) mCherry RNA levels in lungs of TIP and Ctrl RNA-treated animals on day 5 by qRT-PCR.
(E) Luciferase RNA levels from lungs harvested from TIP and Ctrl RNA-treated animals on day 5 by qRT-PCR.
(F) Differential gene expression in lungs on day 5 by RNA-seq analysis. Each column represents one animal clustered by expression profiles and uninfected hamster data obtained from GSE157058 (Sahoo et al., 2021). Cluster III genes are shown in the heatmap (see Figure S5A for all DEGs).
(G) Expression levels for a subset of pro-inflammatory cytokines and IFN-response genes.
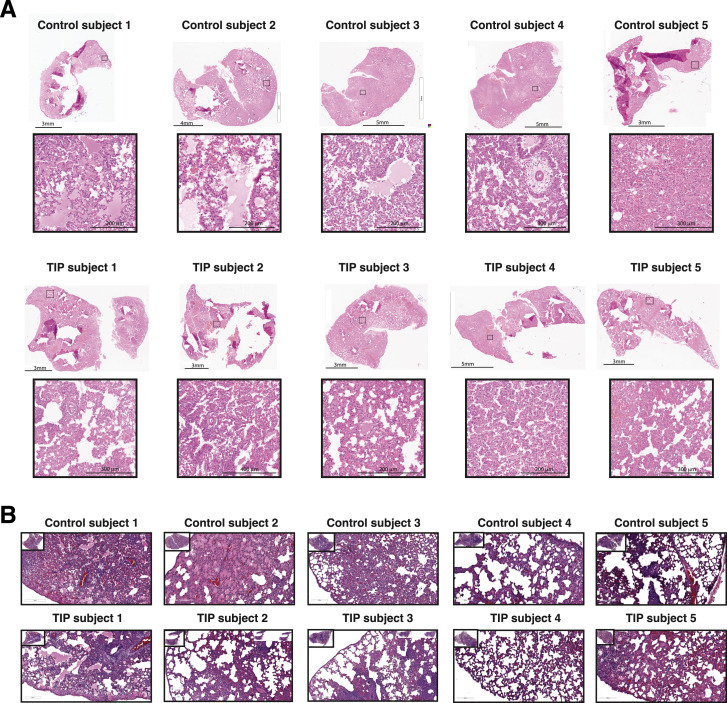
(H) H&E staining of lung section of one representative Ctrl- and TIP-administered animal. Asterisks indicate alveolar edemas, and at signs indicate cellular infiltrates to alveolar space.
(I) Histopathological scoring of lung sections for alveolar edema (left) and cellular infiltrates to alveolar space (right).
(J) Schematic of post-infection treatment experiment. Animals were infected with SARS-CoV-2 (106 PFU) and, at 12 h post-infection, a single-administration of TIP or Ctrl RNA LNPs (n = 5 each) was intranasally administered.
(K) SARS-CoV-2 viral titers in lungs on day 5 of post-infection treatment experiment by plaque assay.
(L) H&E staining of lung section of one representative post-infection Ctrl- and TIP-treated animal. The asterisks indicate alveolar edemas, and the at signs indicate cellular infiltrates to alveolar space.
(M) Histopathological scoring of lung sections for cellular infiltrates to alveolar space.
For (B)–(G), ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05 from Student’s t test; for (I) and (M), ∗∗p < 0.01, obtained from a permutation test (Manly, 2006; Neuhäuser and Manly, 2004). See also Figure S4, Figure S5, Figure S6, Figure S7.
To determine whether conditional propagation of TIPs correlated with SARS-CoV-2 inhibition in vivo, we also analyzed TIP expression in the lungs on day 5 by qRT-PCR and observed high levels of TIP RNA (Figure 7D), whereas Ctrl RNA on day 5 was present at substantially lower levels (Figure 7E). Moreover, to confirm that the presence of SARS-CoV-2 infection is obligatory for conditional propagation of TIPs, we compared the amount of TIP or Ctrl RNA in the presence versus absence of virus on day 5 in hamster lungs. Ctrl RNA levels in the lungs were unaffected by SARS-CoV-2 infection, which starkly contrasted with TIP RNA that was significantly amplified by 4-Log in the presence of SARS-CoV-2 infection (Figure S4D). All qRT-PCR threshold cycle (Ct) values for luciferase in the TIP-treated animals and mCherry in the control animals were >30, indicating negligible non-specific amplification; nevertheless, by convention, normalized RNA values for these samples were reported.
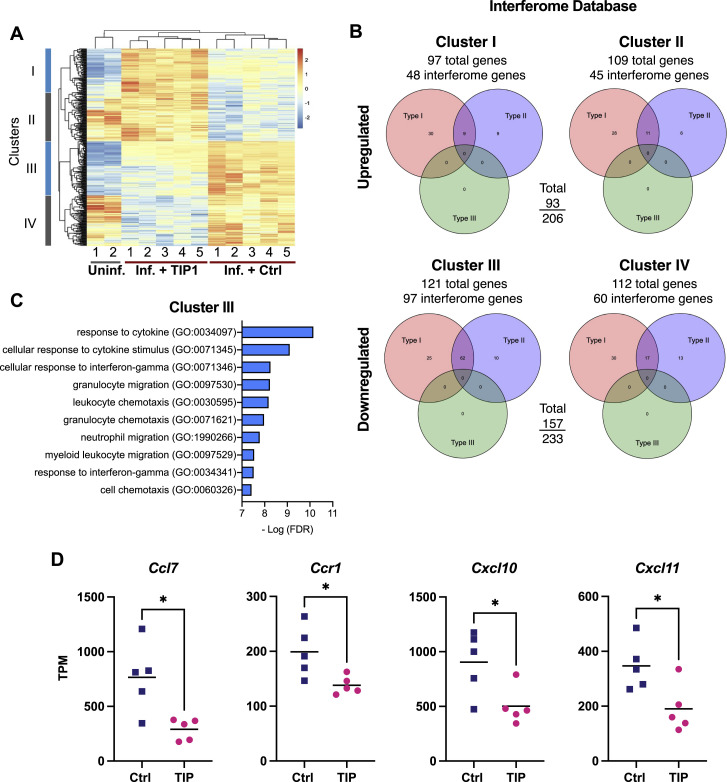
Since inflammation has been implicated in SARS-CoV-2 pathogenesis (Lucas et al., 2020), we assessed cytokine and interferon responses in the lungs of infected animals by performing RNA sequencing (RNA-seq). Analysis of hamster lung samples showed that TIP-treated animals could be clearly differentiated from control-treated animals, with 206 upregulated genes and 233 downregulated genes (Figure S5 A). These differentially expressed genes (DEGs) form four clusters when analyzed together with uninfected hamster lung samples (Figure S5A). The majority of downregulated genes in TIP-treated animals were ISGs (157 out of 233; Figure S5B), especially for genes in cluster III (97 out of 121; Figure S5B). Gene ontology (GO) analysis showed that TIP treatment significantly downregulated pro-inflammatory immune response pathways, which are significantly enriched in cluster III (Figure S5C). The reduced expression of cluster III genes in TIP-treated samples (Figure 7F) suggested alleviated immune responses. Specifically, expression levels of proinflammatory cytokines and receptors previously reported to be upregulated in COVID-19 patients—including Il6, Ccl2, Ccl7, Cxcl10, and Ccr1 (Blanco-Melo et al., 2020; Zhou et al., 2020)—were significantly reduced in TIP-treated animals (Figure 7G; Figure S5D). Importantly, DEGs that can distinguish TIP-treated from Ctrl-treated in infected animals cannot separate TIP from control in uninfected animals (Figure S5A versus Figure S4F), indicating the alleviated proinflammatory immune response is infection dependent and not solely due to TIP RNAs.
Figure S5.
TIP-treatment mitigates overexpression of pro-inflammatory genes in SARS-CoV-2-infected hamster lungs, related to Figure 7
(a) Differential gene expression in hamster lungs on day 5 post infection by RNaseq analysis. Each column represents one animal clustered by expression profiles. Uninfected hamster data was obtained from GSE157058 (Sahoo et al., 2021). DEGs were defined by comparing infected samples treated with TIP RNA or Ctrl RNA LNPs, and are grouped in four clusters. (b) Venn diagram of RNA sequencing of hamster lungs summarizing differentially expressed genes (DEGs) in TIP versus Ctrl-treated animals using the Interferome database http://www.interferome.org with parameters Mus musculus to approximate Syrian golden hamster. According to the Interferome database, the majority of the DEGs in cluster III are interferon-stimulated genes (ISGs), regulated by either Type I or Type II interferons (IFNs). (c) Gene ontology (GO) analysis showing the top ten biological processes enriched in cluster III. (d) Expression levels in terms of transcripts per million (TPM) for additional representative genes belonging to cytokine/chemokine pathways (individual animals are shown as individual data points). These proinflammatory cytokines (Ccl7, Ccr1, Cxcl10, Cxcl11) were previously reported to be upregulated in COVID-19 patients but are significantly reduced in TIP-treated animals. [For all panels: ∗ denotes p < 0.05 from Student’s t test].
Given the reduced inflammatory profile in TIP-treated animals, we performed histological analysis of day 5 hamster lung tissue samples. This revealed dramatic differences in the lungs of Ctrl versus TIP-treated animals (Figure 7H), with control animals exhibiting signs of severe pulmonary edema not present in TIP-treated animals. Specifically, despite all animals exhibiting some signs of inflammation consistent with infection, control animals evidenced pronounced alveolar edema and conspicuous cell infiltrates in alveolar spaces (Figure 7I), indicating severe vascular leakage. In stark contrast, lungs of TIP-treated animals showed substantially less edema and cell infiltration, which is linked to heart failure (Cotter et al., 2001). Histopathological scoring of the images (Figure S6 A) indicated significant reductions in alveolar edema and cell infiltrates in the TIP-treated hamsters (Figure 7I). Uninfected hamsters treated with either TIP or Ctrl RNA LNPs were used as control and showed non-significant difference in the alveolar edema and infiltrates, confirming the severe vascular leakage is due to viral infection (Figure S4E).
Figure S6.
Histopathology imaging of Syrian hamster lungs following pre- and post-infection treatment, related to Figure 7
(a) Animals were treated intranasally with TIP or Ctrl RNA LNPs 6 hr prior to and 18hrs following SARS-CoV-2 infection (106 PFU). Lungs were harvested at 5 days post-infection and histopathology was performed. Micrographs of brightfield imaging of H&E-stained lung sections from all animals (top: Ctrl RNA treated hamsters; bottom: TIP-treated hamsters). Stitched images were analyzed using Leica Aperio ImageScope software. For each animal, whole lung shown on above and a representative zoomed-in section to visualize histopathology shown below. (b) Animals were infected with SARS-CoV-2 (106 PFU) and, 12hrs later, were intranasally treated with either TIP or Ctrl RNA LNPs. Lungs were harvested at 5 days post-infection and histopathology performed. Micrographs of brightfield imaging of H&E-stained lung sections from all animals (top: Ctrl RNA treated hamsters; bottom: TIP-treated hamsters). Stitched images were analyzed using Leica Aperio ImageScope software. For each animal, whole lung shown on above and a representative zoomed-in section to visualize histopathology shown below. Scale bars are as indicated, n = 5 for each group. Labels that were added to the raw images during sectioning were covered during figure preparation, and size bars were added to the image.
To test the efficacy of TIPs in a post-exposure therapeutic setting, hamsters were inoculated with SARS-CoV-2 (106 PFUs) and then given a single intranasal administration of LNP TIP or LNP Ctrl RNA at 12 h post-infection (Figure 7J)—based on previous post-exposure timing used for other therapeutic antivirals now in clinical development (Sheahan et al., 2020), and in vitro analysis that a 12 h post-infection administration could yield viral knockdown (Figure S2B). In agreement with the above results, we observed a significant reduction in SARS-CoV-2 viral load (Figure 7K) as well as reduced pathogenesis in the lungs of animals at day 5 (Figures 7L–7M and S6B).
Discussion
Together, these data demonstrate that a synthetic sub-genomic viral-deletion mutant can conditionally replicate to durably suppress a virus infection (i.e., SARS-CoV-2) in vivo, thereby constituting a therapeutic interfering particle (TIP). If successfully translated to the clinic, TIPs could represent a class of single-administration antiviral with a high genetic barrier to the evolution of resistance. Below, we discuss potential clinical translational paths for this class of antiviral intervention in comparison to other interventions, the longer-term evolutionary considerations, and the limitations of the present study.
Unmet medical need and comparison to other interventions
The therapeutic and vaccine landscape has migrated significantly throughout the course of the COVID-19 pandemic with the emergence of variants that appear to have increased ability to evade immunity and vaccines. Whereas early after the introduction of vaccines the perceived unmet need was largely for post-infection hospital administration therapeutics for unvaccinated individuals, the recent emergence of vaccine-resistant escape variants coupled with the increased availability of at-home rapid tests has highlighted the unmet need for pre- and post-exposure prophylactics. Previously, this unmet clinical need focused primarily on certain individuals (i.e., immunosuppressed from chemotherapy, biologics, transplants, etc.) who could not mount an immune response to the vaccines and thus required a prophylactic. However, the general consensus now is that this need has expanded to the general population at large, and active development of such prophylactic treatments for people once they have been exposed remains a goal as there is no agent to prevent infection after someone has been exposed to SARS-CoV-2.
Monoclonal antibodies (mAbs) (e.g., Bamlanivimab and the REGN COV cocktail) are being tested as prophylactic agents in nursing home patients (Cohen et al., 2021) and in at-risk household settings (O’Brien et al., 2021), but the requirement for infusion has created difficulty in clinical deployment. Several other mAb cocktails specifically indicated for prophylaxis (administered via intramuscular injection) were abandoned after they did not meet clinical endpoints. Other agents that are oral (e.g., MK-4482/Molnupiravir [Merck]) are in development for post-exposure therapy and exhibit efficacy when administered 12 h post-infection in animal models (Sheahan et al., 2020).
Relative to these interventions, TIPs exhibit substantial reduction in SARS-CoV-2 viral load per dose delivered. Specifically, for a 0.44 mg/kg dose in hamsters, TIPs exhibit a ∼2-Log reduction in infectious SARS-CoV-2 viral load (Figure 7B). In comparison, mAb studies in mice show that a 2 mg/kg dose can produce a 4-Log viral-load decrease (Chen et al., 2021), or that a 10 mg/kg dose can produce a 3-Log reduction (Martinez et al., 2021). In hamsters, mAb studies have reported that a 18 mg/kg dose can produce a 1-Log reduction (Kreye et al., 2020), a 16.5 mg/kg dose can produce a 2.5-Log reduction (Rogers et al., 2020), or that a 0.5–50 mg/kg dose produces no significant effect on viral load (Baum et al., 2020). Similarly, hamster studies of small-molecule inhibitors of SARS-CoV-2 (i.e., Molnupiravir) have reported that a 250 mg/kg 12-h repeated dose generates a 2-Log viral-load reduction (Rosenke et al., 2021), or a 200 mg/kg dose generates a 2-Log reduction (Abdelnabi et al., 2021a)—with similar results for Favipiravir (Abdelnabi et al., 2021b). Notably, these studies in SARS-CoV-2 infected rodents consistently show that qRT-PCR analysis generates a lesser (by 1-Log) reduction in viral load, likely due to residual non-infectious RNA fragments—hence the focus on infectious viral-load reduction as measured by PFU. Overall, relative to mAbs and small-molecule antivirals, TIPs appear to have a comparable antiviral effect at a substantially reduced dose (i.e., 0.44 mg/kg for TIPs versus 4–250 mg/kg for mAbs and small molecules).
Like these other antivirals under development, TIPs could serve as similar pre/post-exposure prophylactic therapies based on the current precedent of Oseltamivir (Tamiflu) pre/post-exposure prophylaxis for influenza in at-risk household-exposure settings (Hayden et al., 2004), and the data above show that intranasal TIP delivery could act as a single-administration intervention. Notably, TIP-mediated reduction in SARS-CoV-2 viral load, like mAbs and small-molecule inhibitors, could generate long-term protection to re-infection akin to protection from natural infection, which may offer more durable protection against SARS-CoV-2 reinfection than current vaccines (Gazit et al., 2021). Historically, natural immunity and live-attenuated vaccines provide more durable and effective protection than subunit vaccines for diverse viruses including influenza, rubella, and others (Christenson and Böttiger, 1994; Cox et al., 2004; Horstmann et al., 1985; Johnson et al., 1986).
Evolutionary considerations
It remains unclear whether TIP-like deletions of SARS-CoV-2 have spontaneously arisen and spread. From an evolutionary perspective, it is also not immediately obvious why endogenous TIPs have not naturally evolved to limit virus infections and the considerations are complex, particularly in a virus’s natural hosts (Daugherty and Malik, 2012). For lentiviruses such as HIV, the barrier appears more straightforward, as R0 >1 variants appear to require recombination “acrobatics” since a cis element necessary for efficient transmission is within the TIP deletion and must be recovered and repositioned outside the deleted region (Notton et al., 2021). However, for flaviviruses there is evidence that natural DIPs arise and transmit through host populations (Aaskov et al., 2006), and historical hypotheses have asserted that such DIPs have biological fitness roles aiding the parent wild-type virus. For example, it has been postulated that such DIPs may enhance virus persistence, serve as immunological decoys, or reduce pathogenicity of wild-type virus to enable transmission (Vignuzzi and López, 2019), although our analysis (Figure S1G) indicates that TIPs would reduce transmission of SARS-CoV-2 and bring the R value below 1, leading to contraction of epidemic spread.
Similar to gene drives (Burt, 2003), there are understandable fears that TIPs may drive evolution of increased wild-type virus virulence. This theoretical possibility has been previously addressed at both the host and population scales (Rast et al., 2016), and the hypothesis of TIPs driving increased virulence does not appear consistent with selection theory, but additional in vivo empirical testing will be the best way to address this concern. There is also the possibility that the virus will evolve to escape from TIP. For example, one proposed escape mechanism is upregulation of viral packaging proteins in order to outcompete the TIP and diminish interference; however, this particular compensation mechanism was previously found to be evolutionarily non-robust as it also provides an equivalent excess of trans elements for TIP packaging (Rouzine and Weinberger, 2013).
Optimization and further development of TIPs
Whereas the current TIPs generate a significant protection from disease via a 2-Log reduction in SARS-CoV-2 viral load (Figure 7B), computational models (Figure 1) argue that this viral-load reduction could be enhanced by engineering to optimize TIP transmission (ρ) and interference (ψ) parameters. Specifically, the parameter ρ helps reduce the viral load by spreading the TIP to more cells in the tissue. This is the reason for the seemingly counterintuitive effect that moderate values of ψ result in the largest reduction in patient viral load (Figure 1E). Since the TIP requires wild-type virus to mobilize, if ψ is too large (generating too much inhibition) the TIP essentially shoots itself in the molecular foot and less virus is available to mobilize the TIP. This phenomenon was previously predicted for HIV (Metzger et al., 2011; Rouzine and Weinberger, 2013; Weinberger et al., 2003). In essence, ρ and ψ generate a type of synergistic effect at the whole-tissue scale. As such, optimization of TIPs by enhancing ρ via addition of specific packaging signals (Iserman et al., 2020) could generate more effective TIPs.
Limitations of the study
As with all models, the computational analysis herein is a relatively simple representation of a complex system and necessarily makes certain assumptions. Nevertheless, despite these limitations, this model of SARS-CoV-2 replication in the human airway generates predictions of TIP efficacy (Figure 1) that appear roughly in line with the in vivo hamster qRT-PCR data from challenge experiments (Figure 7C)—we note that the human model is calibrated on patient qRT-PCR data, not PFU data, so the comparison to PFU data is arguably less relevant.
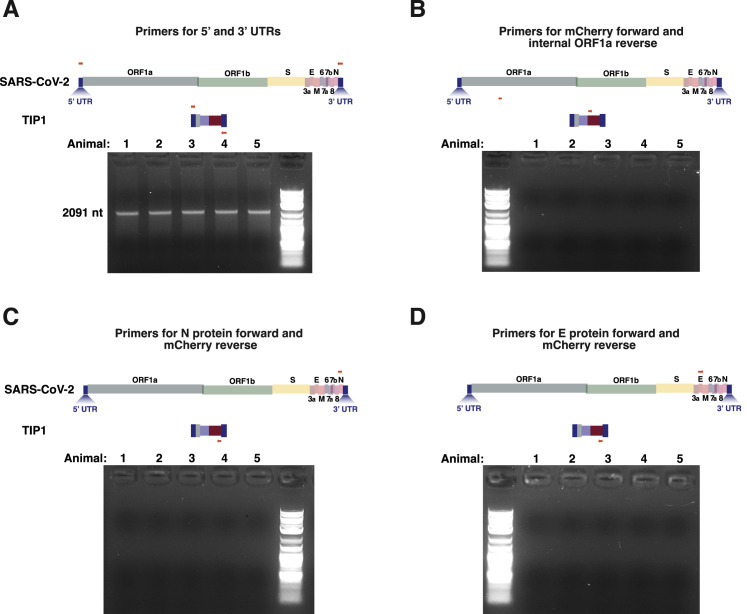
While our cell-culture measurements show that mobilization of TIPs in the presence of SARS-CoV-2 infection exhibit an R0 >1 (Figure 4C), different cell types and contexts outside of tissue culture may alter this result, though the hamster data appear to show substantial mobilization of TIPs in animal lungs in vivo (Figure 7D). The use of reporter expression (mCherry) to quantify transmission does raise concerns regarding recombination of the reporter into the wild-type virus, thereby causing the reporter to be an indicator of virus rather than TIP transmission, but we have not seen evidence of recombination even in vivo (Figures S7 A–S7D).
Figure S7.
No evidence of recombination between SARS-CoV-2 genome and TIP, related to Figure 7
RNA from lungs of SARS-CoV-2-infected TIP-treated animals on day 5 analyzed by RT-PCR and analyzed by gel electrophoresis. Primer sets were: (a) 5′ (forward) and 3′UTRs (reverse) showing expected band for TIP at ∼2kb, (b) mCherry (forward) and ORF1a (reverse), (c) N (forward) and mCherry (reverse) and (d) E (forward) and mCherry (reverse).
Moreover, persistent TIP-mediated knockdown of viral load in continuous cultures (Figures 5A–5C) is not parsimonious with recombination of mCherry, which could only occur if a less fit recombinant was carrying mCherry. Such a less-fit recombinant would be selected against, and a higher fitness non-mCherry virus would dominate, but this was not observed in our system.
One concern is that the continuous culture may not have been run for sufficient time to enable selection of escape mutants, although for many viruses (Coffin, 1995; Schnipper and Crumpacker, 1980; Turner and Chao, 1999), the time frames we used are sufficient for evolution of resistance. Further, the 1-Log increase in fitness in the control (and the emergence of the furin cleavage mutation; Figure S3G) argues that 20 days is sufficient for selection. Moreover, the binding of TIP RNA to the N protein from SARS-CoV (Figure 6C) argues that there may be an extremely high barrier to the evolution of viral escape and a correspondingly long time frame needed to select for such putative escape variants.
While clinical translation of this technology would still need to overcome significant regulatory and other challenges, the data above demonstrate the potential of intranasal lipid-nanoparticle TIPs, and mRNA technology faced similar challenges and skepticism until it was successfully adopted as the basis for SARS-CoV-2 vaccines.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| N protein antibody | Abcam | cat# ab273434; RRID:AB_2893371 |
| goat anti-mouse IgG H&L-Alexa fluor 488 | Abcam | cat# ab150113; RRID:AB_2576208 |
| mouse anti-Spike antibody | Abcam | cat# ab273433; RRID: AB 2891068) |
| Rabbit anti-mCherry antibody | Novus Biologicals | cat# nbp2-43720; RRID:AB_2893372 |
| Goat anti-rabbit IgG (H+L) cross-adsorbed secondary antibody, Alexa fluor 405 | Invitrogen | cat# a-31556, RRID:AB_221605 |
| Bacterial and virus strains | ||
| SARS-CoV-2 isolate (USA-WA1/2020) | SARS-CoV-2 isolate (USA-WA1/2020) was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020. | NR-52281 |
| SARS-CoV-2 variant 501Y.V2 | SARS-CoV-2 variant 501Y.V2 was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/USA/MD-HP01542/2021 (Lineage B.1.351), in Homo sapiens Lung Adenocarcinoma (Calu-3) Cells, NR-55282, contributed by Andrew S. Pekosz. | NR- 54008 |
| SARS-CoV-2 variant B1.1.7 | SARS-CoV-2 variant B1.1.7 was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/England/204820464/2020, contributed by Bassam Hallis. | NR- 54000 |
| SARS-CoV-2 variant 501YV2 HV Delta | SARS-CoV-2 variant 501YV2 HV Delta was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/USA/MD-HP01542/2021 (Lineage B.1.351), in Homo sapiens Lung Adenocarcinoma (Calu-3) Cells, NR-55282, contributed by Andrew S. Pekosz. | NR- 54009 |
| Chemicals, peptides, and recombinant proteins | ||
| Advanced DMEM/F12 | Invitrogen | 12634010 |
| HEPES | Invitrogen | 15630-056 |
| GlutaMAX | Invitrogen | 35050061 |
| Penicillin-Streptomycin | Invitrogen | 15140-122 |
| B27 supplement | Invitrogen | 17504-044 |
| N-acetylcysteine | Sigma | A9165 |
| Nicotinamide | Sigma | N0636 |
| Y-27632 | Tocris | 1254 |
| A8301 | Tocris | 2939 |
| FGF-7 | Peprotech | 100-19 |
| FGF-10 | Peprotech | 100-26 |
| Primocin | InvivoGen | ant-pm-1 |
| Heregulin beta-1 | Peprotech | 100-03 |
| SB202190 | Sigma | S7067 |
| Uranyl Formate | VWR International | 16984-59-1 |
| Hoechst stain | Thermofisher Scientific | H1399 |
| Experimental models: Cell lines | ||
| Vero cells | ATCC | CRL-1586 |
| Human small airway epithelial cells (HSAEpC) | PromoCell | C12642 |
| Experimental models: Organisms/strains | ||
| Hamster / Golden Syrian Hamster / Male / 6-8 weeks old | Charles River | Strain Code 049 |
| NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice/ female/ 7 weeks old | Jackson Laboratory | 005557 |
| Oligonucleotides | ||
| Gene block for TIP1 | Integrated DNA Technology | N/A |
| Gene blocks for TIP2 | Integrated DNA Technology | N/A |
| Primers: See Table S1 | Integrated DNA Technology | N/A |
| Critical commercial assays | ||
| SE cell line 4D nucleofector kit | Lonza | V4XC-1012 |
| Direct-zol RNA extraction kit | Zymo Research | R2070T |
| Illumina Stranded total RNA library prep kit | Illumina | 20040525 |
| HiScribe T7 high yield RNA synthesis kit | New England Biolabs | E2040S |
| Vaccinia Capping System | New England Biolabs | M2080S |
| E.coli Poly(A) polymerase | New England Biolabs Inc | M0276S |
| RNase free DNase-I | Thermofisher Scientific | EN0521 |
| SuperScript II Reverse Transcriptase | Thermofisher Scientific | 12574026 |
| SYBR green PCR master mix | Thermofisher Scientific | 4309155 |
| TRIzol LS Reagent | Invitrogen | 10296010 |
| Lipofectamine 3000 transfection reagent | Thermofisher Scientific | L3000001 |
| Recombinant DNA | ||
| pLVX-EF1alpha-SARS-CoV-2-N-2xStrep-IRES-Puro | (Gordon et al., 2020b) | N/A |
| pLVX-EF1alpha-SARS-CoV-2-nsp7-2xStrep-IRES-Puro | (Gordon et al., 2020b) | N/A |
| pLVX-EF1alpha-SARS-CoV-2-nsp8-2xStrep-IRES-Puro | (Gordon et al., 2020b) | N/A |
| pLVX-EF1alpha-SARS-CoV-2-nsp12-2xStrep-IRES-Puro | (Gordon et al., 2020b) | N/A |
| pLVX-EF1alpha-SARS-CoV-2-S-2xStrep-IRES-Puro | (Gordon et al., 2020b) | N/A |
| pLVX-EF1alpha-SARS-CoV-2-M-2xStrep-IRES-Puro | (Gordon et al., 2020b) | N/A |
| pLVX-EF1alpha-SARS-CoV-2-E-2xStrep-IRES-Puro | (Gordon et al., 2020b) | N/A |
| pMAX GFP vector | Lonza | V4XC-1012 |
| Software and algorithms | ||
| FlowJo | FlowJo | https://www.flowjo.com/ |
| Prism | GraphPad | https://www.graphpad.com/ |
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| MetaXpress | Molecular Devices | https://www.moleculardevices.com/products/cellular-imaging-systems/acquisition-and-analysis-software/metaxpress |
| Integrated Genome Browser | Broad Institute | https://software.broadinstitute.org/software/igv/ |
| Lago in-vivo Imaging System | Spectral Instruments Imaging | https://spectralinvivo.com/software/ |
| Code and Raw Data | This paper | https://zenodo.org/record/5579847 Zenodo: 5579847 |
| Deposited data | ||
| RNAseq Data | This paper | GEO: GSE184447https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE184447 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Leor Weinberger (leor.weinberger@gladstone.ucsf.edu).
Materials availability
All unique reagents generated in this study are available from the Lead Contact with a completed Materials Transfer agreement.
Experimental model and subject details
Animals
All Syrian golden hamster (Hamster / Golden Syrian Hamster / Male / 6-8 weeks old/ Charles River / Strain Code 049) were approved by the Scripps Research Institute Institutional Animal Care and Use Committee (Protocol 20-0003) and experiments were carried out in accordance with recommendations. NOD.Cg-Prkdc scid Il2rg tm1Wjl/SzJ (NSG) mice (Female/ 7 weeks old) (JAX stock #005557) were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were maintained in accordance with the Guide for the Care and Use of Laboratory Animals and were housed in an SPF conditions. All experiments were performed according to the guidelines of the Institutional Animal Committee of the Beckman Research Institute of the City of Hope, IACUC protocol #16095. The experiments were also approved by ACURO (MTEC). Hamsters and mice were maintained in pathogen-free facilities at Scripps Research Institute and the City of Hope, respectively.
Cell lines
Vero cells (Vero C1008) were obtained from ATCC and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin (P/S). Human small airway epithelial cells (HSAEpC) (cat# C12642, PromoCell Inc.) from three different donors were cultured in Advanced DMEM/F12 containing 1 × GlutaMAX, 10 mM HEPES and, antibiotics (AdV+++, Invitrogen, 12634010). All the cells were cultured under 5% CO2 in a humidified incubator at 37°C.
Method details
Within-host computational model of SARS-CoV-2 TIPs
A previous ordinary differential equation model of within-host SARS-CoV-2 infection (Ke et al., 2020) was extended to include TIPs, by introducing two new parameters (burst size of TIP particles relative to SARS-CoV-2 particles from dually infected cells, i.e., infected with both TIP and wild-type virus) and (burst size of SARS-CoV-2 from dually infected cells, compared to the burst size of SARS-CoV-2 from cells infected with only wild-type virus). Similar to previous approaches (Rouzine and Weinberger, 2013; Weinberger et al., 2003), the model was also expanded to include state variables for TIP infected cells, dually infected cells, and TIP particles. To simulate treatment with TIPs, a fraction of target cells and in the upper and lower respiratory tracts are converted into TIP-carrier cells and , as in previous TIP models. TIP entry into cells, clearance of TIPs and TIP-containing cells, and transport between the upper and lower respiratory tract are assumed to be equivalent for TIP and wild-type virus. The model equations are provided below; parameter descriptions and values are described in Table S2.
The model also includes both a “target cell extension” step and an adaptive immune response model. “Target cell extension” is an approximation of the spatial spread of infection in the lungs: is increased by new cells at the time . The adaptive immune response is assumed to have an increase in viral clearance at 2 weeks post infection according to:
For analytical expediency, a minor modification is made to the initial conditions from (Ke et al., 2020), setting the initial virus in the URT () rather than the initial infected cells in the URT (). Since viral entry is fast (minutes) relative to the timescales of patient infection (days-weeks), the initial exposure is well-captured by specifying initial virions instead of infected cells. This expediency simplified our sensitivity analysis for dose regimens. That is, we do not have to vary both and . Importantly, this modification does not change our overall results: the no-TIP predictions match well to the original model, and the efficacy of TIPs are robust to the initial viral inoculum.
Cross-validation of the within-host model
We assessed the robustness of the within-host model predictions by incorporating TIPs into a second simpler within-host model, also validated on clinical samples (Kim et al., 2021). This simpler model only has four state variables:
where denotes the fraction of target cells remaining, denotes the fraction of TIP-carrier target cells remaining, is SARS-CoV-2 per ml, and is TIPs per ml. First, and set TIP efficacy as described above. This model assumes SARS-CoV-2 (and TIPs) are in quasi-steady-state with respect to the number of infected cells. As a result, we do not explicitly model the infected cell state, and instead have the aggregated parameter where is the per cell viral production rate, is the number of initial target cells, and is the rate of viral clearance. The other parameters and represent infectivity and infected cell clearance. Sensitivity analysis allowed and to vary over several orders of magnitude, though we focus on and . Half the target cells were converted to TIP carriers prior to virion exposure (). These simulations were performed for thirty individual parameter sets calibrated to patient samples from Singapore, China, Germany, and Korea by (Kim et al., 2021), reproduced in Table S3 for completeness.
Computational model of secondary transmissions by a primary infectious individual
A probabilistic model of SARS-CoV-2 transmissions using viral dynamics (Goyal et al., 2021) to calculate transmission risk was extended and recalibrated to predict TIP efficacy and TIP transmission. Transmission model calibration was done as follows: SARS-CoV-2 infections were simulated using a patient-validated model, producing viral load dynamics of 10,000 untreated infections based on per-patient parameter uncertainty (Ke et al., 2020). Viral load dynamics were converted to transmission risk dynamics using , with time-varying log10 viral load and parameters and set based on (Jones et al., 2021). Each individual had simulated daily contacts drawn from the Gamma distribution , whose parameters were fit by minimizing the error between measured and predicted secondary transmission distributions (resulting in = 10, = 10). New secondary transmission events were estimated using a one-day timestep.
After calibration to untreated transmissions, we simulated 10,000 TIP-treated index patients and predicted changes in both SARS-CoV-2 secondary transmissions and TIP secondary transmissions (TIP parameters = 1.5 and = 0.02 as above). We assumed identical transmission risk parameters for TIP and SARS-CoV-2, so transmission differences were purely driven by changes in viral load.
To model direct administration of TIP to patients, we assumed that half the target cells in the upper and lower respiratory tract received TIP. To model indirect administration of TIP (i.e., due to person-to-person transmission) we assume an equivalent dose of TIP as to SARS-CoV-2.
All simulations were performed in Python. NumPy (v1.19.4) and SciPy (v1.5.4) were used with additional optimization for solving ODEs using Numba (v0.51.2). Data tabulation and visualizations were done with Pandas (v1.1.4), Seaborn (v0.11.0) and Matplotlib (v3.3.3).
Virus and cell culture conditions
SARS-CoV-2 isolate (USA-WA1/2020), SARS-CoV-2 variants 501Y.V2, 501YV2 HV Delta, and B1.1.7 were obtained from Biodefense and Emerging Infections (BEI) Resources. Viral stocks were prepared by propagating in Vero cells (Vero C1008, obtained from ATCC) in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin (P/S). All the cells were cultured under 5% CO2 in a humidified incubator at 37°C. For the R0 calculation in the 1st round transfer experiment, GFP+ cells were prepared by transfecting naive Vero cells with pMAX GFP vector from Lonza (1 μg plasmid for 1 million cells; see below for the transfection protocol). All live virus experiments were performed at the Gladstone Institutes in a Biosafety Level 3 (BSL3) containment facility under an approved Biosafety Use Authorization from UCSF and in compliance with UCSF guidelines and procedures.
Molecular cloning, in vitro transcription, and transfection of RNA
All gene fragments and PCR primers used (see Table S1) were obtained from Integrated DNA Technology and assembled using standard molecular cloning techniques. RNA was in vitro transcribed from 1 μg of agarose gel-purified band corresponding to the intended size using the HiScribe™ T7 high yield RNA synthesis kit (cat#E2040S, New England Biolabs Inc.). 5′ capping was carried out using the Vaccinia Capping System (cat#M2080S, New England Biolabs Inc.) and a poly-A tail added using E.coli Poly(A) polymerase (cat#M0276S, New England Biolabs Inc.). Transcribed RNA was purified using phenol-chloroform extraction. Briefly, an equal amount of phenol:chloroform was added to the RNA, followed by vortexing for 10 s and centrifugation at 10,000 rpm for 10 minutes at room temperature. The aqueous phase was harvested and one volume of isopropanol was added to the RNA, incubated for 5 minutes at room temperature, centrifuged for 10 minutes at 10,000 r.p.m, washed twice with ice-cold 70% ethanol and the pellet was resuspended in nuclease free water. Naive Vero cells were transfected with either Ctrl or TIP RNAs at a concentration of 1 μg RNA per 1 million cells using 4D-Nucleofector (cat# AAF-1002B, Lonza Inc.) and the SE cell-line 4D nucleofector kit (cat#V4XC-1012, Lonza Inc). For post-infection therapy experiment, we transfected SARS-CoV-2 infected cells using Lipofectamine 3000 transfection reagent (cat# L3000001, Thermofisher Scientific) at 8 or 16 hr post infection at the concentration of (1 μg RNA per 1 million cells).
RNA extraction and qRT-PCR titering of virus
At indicated time points, infected cells (in the BSL-3 lab) were lysed in TRIzol LS (cat# 10296010, Invitrogen) solution (0.75 mL TRIzol LS for 0.25ml of sample volume). RNA was extracted using the Direct-zol RNA extraction kit (cat# R2070T, Zymo Research Inc.). RNA was DNase treated using RNase free DNase-I (cat# EN0521, Thermofisher Scientific). 1 μg of RNA was used for each SuperScript II Reverse Transcriptase reaction with oligo d(T) primers (cat#12574026, Thermofisher Scientific), and cDNA was analyzed by quantitative real-time polymerase chain reaction (qRT-PCR) analysis using SYBR green PCR master mix (cat#4309155, Thermofisher Scientific) with sequence specific primers. All the qRT-PCR measurements were normalized to GAPDH or beta-actin (Table S1). For quantification of relative packaging of RNA in virions, Vero cells were nucleofected with TIP1 or TIP2 RNA (1 μg RNA for 1 million cells), and infected with SARS-CoV-2 (MOI = 0.05) at 24 hours post nucleofection. Supernatants were harvested at 24 hours post infection, followed by qRT-PCR using mCherry and E gene primers. TIP fold enrichment was quantified relative to viral genome.
Quantification of viral titers by plaque assay (PFU/ml)
Infectious virus titers were quantified by plaque forming unit (PFU/ml) assay on Vero cells. Briefly, Vero cells were prepared by plating as a confluent monolayer in 12- or 24-well plates 24 hours before performing the plaque assay. On the day of the plaque assay, media was aspirated, cells were washed with 2ml of PBS, 250 μL of diluted virus in modified DMEM media (DMEM, 2%FBS, L-glut, P/S) was added to confluent monolayer followed by incubation at 37°C for 1 hour with gentle rocking every 15 minutes. After one hour of incubation, 2 mL of overlay media (1.2% Avicel in 1X MEM) was added to each well. At 72h post infection, overlay media was aspirated, monolayer was washed with PBS and fixed with 10% formalin for 1 hour. Plaques were stained with 0.1% crystal violet for 10 minutes and washed with cell culture grade water three times, followed by enumeration of plaques using ImageJ (Schneider et al., 2012) and viral titer calculation to pfu/ml.
Virus yield-reduction assay
Virus yield-reduction assays were performed by transfecting Vero cells with TIP or Ctrl RNAs (1 μg/1 millioncells) 24 hours prior to infection with SARS-CoV-2 at the indicated MOI, and harvesting culture supernatants for titration at various time-points (24, 48, or 72 hours post infection). Post-exposure yield-reduction assays were analogous except transfections were performed at specified times (8 or 16 hours post infection). Quantification of virus titer in supernatant was then performed by plaque assay on Vero cells or by qRT-PCR (as above) using E, NSP14 or N protein primers and normalized with beta-actin or GAPDH primers as mentioned in figure legends.
Flow cytometry
Cells were harvested at indicated time points, fixed with 4% formaldehyde for 15 minutes, washed three times with PBS and analyzed by flow cytometry on a BD LSRFortessa (BD Bioscience Inc.) in the Gladstone-UCSF Flow Cytometry Core facility for respective fluorophores. Data was analyzed in FlowJo. For N protein antibody staining, cells were permeabilized with ethanol after 4% formaldehyde treatment for 15 minutes, treated with N protein antibody (cat#ab273434, Abcam inc; RRID:AB_2893371) at 1:500 dilution or rabbit anti-mcherry antibody (cat# nbp2_43720, Novus Biologicals; RRID:AB_2893372) at 1:200 dilution for 2 hours at room temperature followed by three washes in PBS buffer. Cells were then treated with 1:2000 dilution of secondary antibody (goat anti-mouse IgG H&L-Alexa Fluor 488 (cat# ab150113, Abcam Inc; RRID:AB_2576208) or goat anti-rabbit (H+L) cross adsorbed secondary antibody, alexa fluor 405 (cat# a-31556, Invitrogen; RRID:AB_221605) for three hours at room temperature, washed three times with PBS and subjected to flow cytometry.
‘1st round supernatant transfer assay’ and R0 calculation
To quantify the R0 of TIPs in the context of SARS-CoV-2 infection, a supernatant-transfer assay was modified to a ‘1st round supernatant transfer assay’. 80K TIP-transfected cells were infected (MOI = 0.05), then thoroughly washed to remove virus, and at two hours post infection 20K GFP+ reporter cells (prepared by nucleofecting 1 million 1Vero cells with 1 μg of pMAX-GFP vector obtained in the SE cell line 4D-nucleofector kit [cat# V4XC-1012 Lonza]) were introduced to the culture (at ∼20% of total cells). 12 hours post infection, GFP+ cells were analyzed by flow-cytometry to quantify % mCherry+ cells within the GFP+ population. To estimate R0 of TIP, the fraction of TIP+ cells, after correcting for background autofluorescence, was compared to % infected cells for the original SARS-CoV-2 infection after accounting for the addition of 20% GFP+ cells in the assay.
Virus entry analysis
Cells were nucleofected with Ctrl, TIP1 or TIP2 RNA and plated with 80% confluency in a 96-well black/clear bottom plate. At 24 hours post nucleofection, cells were infected with SARS-CoV-2 (WA-1 strain) at MOI = 20, cells were washed three times with 100 μL sterile PBS buffer after 1 hour of infection, at 2 hours post infection, cells were fixed with 4% paraformaldehyde for 15 minutes, followed by washing wells twice with 100 μL of PBS buffer. Cells were permeabilized by adding 0.5% Triton X-100 in PBS for 5 minutes, followed by 1x washing with PBS, blocking with 3% BSA in PBS buffer for 30 minutes at room temperature. Wells were washed with PBS, followed by addition of 30 μL of primary antibody (mouse anti-Spike antibody, cat# ab273433, Abcam, RRID:AB 2891068) for one hour, followed by 100 μL of secondary antibody (anti-mouse Alexa flour 488) in 3% BSA in PBS buffer for 30 minutes, wells were washed twice with PBS and 100 μL of 1:2000 Hoechst stain was added for 10 minutes, followed by two washes in PBS. High-throughput microscopy was performed on an ImageXpress-Confocal Microscope, and images were analyzed using MetaXpress software.
Human primary lung organoid cultures and infections
Organoids were generated as described previously (Sachs et al., 2019; Zhou et al., 2018) with slight modifications. Human small airway epithelial cells (HSAEpC) (cat# C12642, PromoCell Inc.) from three different donors were cultured in Advanced DMEM/F12 containing 1 × GlutaMAX, 10 mM HEPES and, antibiotics (AdV+++, Invitrogen, 12634010). 2% Fetal Bovine Serum was added and cells were centrifuged at 400 × g. If present, red blood cells were lysed by adding 2 mL of red blood cell lysis buffer (Roche, 11814389001) for 5 min at room temperature. Then, 10 mL AdV+++ was added and cells centrifuged again at 400 × g. The pellet was resuspended in ice-cold Cultrex BME type 2 (Trevigen, 3533-010-02) diluted with cold AdV+++ at a 4:1 ratio. Cells were diluted to 1.2 million/ml and 50 μL drops of BME-cell suspension were allowed to solidify on pre-warmed 24-well suspension culture plates (Greiner, M9312) at 37°C for 10–60 min. Upon gelation, 500 μL of human airway organoid (HAO) medium was added to each well and plates transferred to humidified 37°C/5% CO2 incubators at ambient O2.
Human airway organoid (AO) medium is composed of Advanced DMEM/F12 (cat#12634010, Invitrogen), 1% HEPES (cat#15630-056, Invitrogen), 1% GlutaMAX (cat#35050061, Invitrogen), 1% Penicillin-Streptomycin (cat#15140-122, Invitrogen), 10% Rspondin1 conditioned medium, 10% Noggin conditioned medium, 2% B27 supplement (cat#17504-044, Invitrogen), 1.25mM N-acetylcysteine (cat#A9165, Sigma Aldrich), 10mM nicotinamide (cat#N0636, Sigma Aldrich), 5 μM Y-27632 (cat#1254, Tocris inc.), 500nM A8301 (cat#2939, Tocris inc.), 1 μM SB202190 (cat# S7067, Sigma Aldrich), 5ng/ml FGF-7 (cat#100-19, Peprotech), 20ng/ml FGF-10 (cat#100-26, Peprotech), 100 μg/ml primocin (ant-pm-1) and 5nM heregulin beta-1 (cat#100-03, Peprotech).
Medium was changed every 4 days and organoids were passaged when drops became too dense. Media was carefully aspirated and BME drops were collected in with ice-cold AdV+++ media, and transferred to 15 mL tubes. Drops were manually dissociated using sterile 1ml pipette tip and organoids were pelleted at 500 g, 4°C for 5 min. Cells were washed with ice-cold AdV+++ to remove any remaining BME and centrifuged again. Then, the pellet was dissociated with 10x TrypLE (Invitrogen, A1217701) and incubated in a 37°C incubator for up to 10 min. AdV+++ was added, cells were centrifuged and resuspended in ice cold 3:1 BME in AdV+++. Finally, 50 μL drops were formed and covered in AO medium as described. Organoids were imaged every other day to observe their morphology using transmission microscopy. For transfecting organoids, organoids were trypsinized and cells spun at 800 g for 5 minutes, supernatant was removed and cells were plated at high density in organoid media, followed by preparation of RNA-lipofectamine 3000 complexes using standard lipofectamine protocol. 4ml of Lipofectamine 3000 reagent (in Opti-MEM media) and 1 μg of RNA were mixed together, incubated for 5 minutes, and added to cells (50 μL per well). Plates were centrifuged at 600 g at 32°C for 1 hour, and incubated for 4 hours in 37°C incubator with 5% CO2, followed by resuspending in AO medium with ice-cold BME diluted 1:500 (2%) to a final concentration of 0.5 million cells/ml. Then, 200 μL of cell suspension/well was added to 8-well chamber slides pre-coated with BME 1:100. Cells were allowed to attach for at least 1 day. In BSL3, media was removed and fresh media with SARS-CoV-2 virus was added at MOI = 0.5. Cells were incubated at 37°C for 2 hours and then media was replaced with warm AO medium without virus. Slides were incubated for one day until harvested for measuring viral load by qRT-PCR and plaque assay.
Reconstitution of VLPs and functional analysis
Vero cells were co-transfected with 0.5 μg of expression plasmids for nucleocapsid (N), matrix (M), and envelop (E) proteins plus 25 ng of expression plasmid for spike (S) protein (all kindly provided by Nevan Krogan and as described previously [Gordon et al., 2020b]) and 1 μg of TIP1 RNA (per 1 million cells). 24 hours later, supernatant was harvested, filtered using a 0.2-μm filter, subjected to sucrose density gradient ultracentrifugation (gradient: 10%–40% sucrose in PBS), stored at 4°C and analyzed by electron microscopy (below). For functional analysis, 10 μL of supernatant containing VLPs was added to Vero cells in individual confluent wells of a 24-well plate, and flow cytometry analysis performed as above to detect mCherry.
Negative staining and electron microscopy
VLPs were visualized under electron microscopy by performing negative staining. Briefly, 37.5 μg of uranyl formate (Sigma Aldrich) was added to 5ml of sterile water, and stirred until uranyl formate was dissolved, followed by dropwise addition of 5M NaOH until the stain solution becomes slightly darker yellow, the stain was filtered with 0.2 μm filter. Carbon grids were glow discharged for 30 s, 10 μL of VLPs were pipetted onto the grid, washed twice with water, uranyl formate stained and dried. EM imaging was performed on a FEI Tecnai T12 electron microscope.
Electrophoretic mobility shift assay
Vero cells were transiently transfected with plasmids (1 μg plasmid used per 1 million cells) expressing N protein or co-transfected with plasmids expressing RdRP complex proteins NSP7, NSP8 and NSP12 in equimolar ratios (kindly provided by Nevan Krogan and described previously (Gordon et al., 2020b) using a 4D-Nucleofector (cat# AAF-1002B, Lonza Inc.) with the SE cell line 4D nucleofector kit (cat#V4XC-1012, Lonza Inc). One day post transfection, cells were harvested, washed three times in ice-cold phosphate-buffered saline, harvested in 300 μL of lysis buffer (10mM Tris, pH 7.05; 50mM NaCl; 30mM sodium pyrophosphate; 50mM NaF; 5mM ZnCl2; 0.1mM Na3VO4; 1% Triton X-100; 1mM phenylmethylsulfonyl fluoride; pierce protease inhibitor cocktail (cat#A32963, Thermofisher Scientific). Cells were incubated on ice for 10 minutes while vortexing intermittently every 2 minutes, supernatant was harvested after centrifugation at 14,000 rpm for 10 minutes at 4°C. Supernatant was passed through amicon filters (Sigma-Aldrich), followed by binding experiment (at concentration range of 0-20 μM) with 1 μg of respective RNAs (i.e., Ctrl, TIP1 or TIP2) in binding buffer (20mM HEPES, pH 7.3; 50mM KCl, 3mM MgCl2, 1mM EDTA, 4% glycerol, 1mM β-mercaptoethanol, 10mM dithiothreitol) in the presence of 1 μg of yeast RNA for 30 minutes at room temperature. Samples were run on an agarose gel in 1x tris acetate EDTA buffer at 4°C at 20V and imaged.
Continuous serial-passage cultures
Vero cells were nucleofected with either Ctrl RNA or TIP RNA (1 μg per 1 million cells) as described above, and 24 hours following transfection, cells were infected with SARS-CoV-2 (WA-1 strain) at an MOI = 0.05. Supernatant was removed after 48hrs with half added to naive Vero cells while the other half was stored at −80°C for titering by plaque assay. 48 hours later, the supernatant from the new culture was removed and half added to a new naive culture and half frozen for titering. This supernatant passage process repeated every 48 hours for 20 days.
LNP formulation and characterization
RNA was packaged into LNPs using a NanoAssemblr microfluidic system (Precision Nanosystems, Vancouver, Canada) according to manufacturer instructions. Briefly, LNP formulations were formed by injecting 12.5 mM of the lipid solution and 0.173 μg/μl of RNA in formulation buffer at a flow rate of 12 ml/min. After mixing, LNP suspension was immediately diluted in PBS (Corning, Manassas, USA). Then, the formulation was reconcentrated by centrifugation at 2000 g in Amicon filters (30,000 MWCO, Amicon Ultra-15 Centrifugal Filter Unit, Millipore Sigma, Burlington, USA). Finally, the LNP suspension was filtered using a 0.22 micron syringe filter and kept at 4°C until use. Free and total RNA concentration were determined by Ribogreen assay (Quant-iT RiboGreen RNA, Invitrogen, Carlsbad, USA). For obtaining total RNA concentration, LNPs were lysed for 30 min at 37°C in Triton X-100 1%. Encapsulation was calculated as (total RNA-Free RNA)/(total RNA x 100). Particle sizes were measured by Dynamic Light Scattering in a DynaPro NanoStar instrument (Wyatt Technology, Santa Barbara, USA) and analyzed with Dynamics 8.0 software (Wyatt Technology, Santa Barbara, USA). Samples were diluted in PBS (Corning, Manassas, USA) until full laser power could be used to record the signal. For each sample, 3 measurements were conducted, each consisting of 10 recordings of 10 s.
In vivo bioluminescence imaging of mice
NOD.Cg-Prkdc scid Il2rg tm1Wjl/SzJ (NSG) mice (JAX stock #005557) were purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were maintained in accordance with the Guide for the Care and Use of Laboratory Animals and were housed in an SPF condition. All experiments were performed according to the guidelines of the Institutional Animal Committee of the Beckman Research Institute of the City of Hope, IACUC protocol #16095. Mice were 4-6 weeks old at time of experiments. A total of 9 mice were injected either with lipid nano particle (LNP), naked RNA, or saline control. n = 3 mice were intranasally instilled with 20 μL (10ul each nostril) LNP, n = 3 mice were intranasally instilled with 20 μL (10 μL each nostril) naked RNA, and n = 3 mice were intranasally instilled with 20 μL (10 μL each nostril) saline control. All mice were injected or instilled within 30 mins and bioluminescence was analyzed at 6 hours post injection. The in vivo imaging system (Lago in vivo Imaging System, Spectral Instruments Imaging) was used for bioluminescence. D-Luciferin potassium salt (PerkinElmer), the substrate for firefly luciferase, was dissolved in phosphate-buffered saline at a concentration of 30 μg/ml and filtered through a 0.22-μm-pore-size filter before use. Mice were injected with 100 μL of luciferin (3 μg) and immediately anesthetized in an oxygen-rich induction chamber with 3%–5% isoflurane. The mice were transferred to imaging box and positioned in ventrodorsal and lateral positions for imaging. Mice were maintained for at least 5 min so that there was adequate dissemination of the injected substrate and so that the animals were fully anesthetized. Images were taken using Aura imaging software (Spectral Instruments Imaging) using the following settings: 120-180 s acquisition time and heavy binding. Imaging analysis was done using the Aura imaging software; quantitation of signal was done using regions of interest (ROI) over mice and recorded as photons per second (flux).
Syrian golden hamster infection studies
8-week old Syrian golden hamsters were infected through intranasal installation of 106 PFU per animal of SARS-CoV-2 (USA-WA1/2020) in 100 μL of DMEM, as described (Rogers et al., 2020). At 6 hours pre-infection and 18 hours post-infection animals were given an intranasal dose of 100 μL of LNP solution containing 60 μg of either TIP RNA or Ctrl RNA. Hamsters were then weighed daily for the duration of the study. At day-5 post-infection, animals were sacrificed and lungs were harvested. For the post-infection therapy experiment, 8-week old Syrian golden hamsters (n = 10) were intranasally infected with 106 PFU, and at 12 hours post infection, 100 μL of LNP solution containing either TIP RNA (n = 5) or Ctrl RNA(n = 5) was administered intranasally. Lungs were harvested at five days post infection. For the uninfected control experiment, 8-week old Syrian golden hamsters were intranasally treated with 100 μL of LNP solution containing either TIP RNA (n = 3) or Ctrl RNA (n = 3), and lungs were harvested at five days post treatment. The research protocol was approved and performed in accordance with Scripps Research IACUC Protocol #20-0003. Lung tissue was analyzed by viral titering (by PFU/ml and qRT-PCR), RNA sequencing, and histology imaging. Briefly, formalin-fixed lung from each animal group were processed and paraffin embedded, and tissue sections were stained with hematoxylin and eosin (H&E) as described (Fischer et al., 2008), imaged, and images were analyzed using Leica Aperio ImageScope software. To assess the statistical significance of the observed variation between the scoring of control and therapeutic histopathology microscopy, we performed a permutation test (Manly, 2006; Neuhäuser and Manly, 2004), an extension of the Fisher exact test for multinomial variables via random sampling. We randomly partitioned concatenated control and variable observations 100,000 times, then tabulated the null hypothesis probability that the mean of control and variable were drawn from the same population.
RNA-seq analysis
RNA was extracted from the lung lysates using Direct-zol RNA extraction kit (cat# R2070T Zymo Research inc.). RNA libraries were prepared by the Gladstone Institutes Genomics Core using Illumina Stranded total RNA library prep kit (cat# 20040525, Illumina inc.) and sequencing performed on a NextSeq 500 sequencer (cat# SY-415-1001, Illumina Inc). Read quality was checked via FASTQC, followed by mapping to MesAur1.0 (GCA_000349665.1) reference genome using STAR (STAR-2.7.9a) with default parameters. Transcript level quantification was performed using featureCounts (subread-2.0.2) with default parameters. The quantification matrix was then imported into R and analyzed via DESeq2. Differentially expressed genes (DEGs) were identified with adjusted p < 0.05. For IFN-stimulated gene identification, http://www.interferome.org was used with parameters -Mus musculus, -fold change up 2 and down 2.
Quantification and statistical analysis
Statistical differences were determined by using the two-tailed unpaired Student’s t test and 1-way ANOVA followed by Tukey’s multiple comparisons test (GraphPad Prism). A p value less than 0.05 was considered statistically significant: ∗ < 0.05, ∗∗ < 0.01, ∗∗∗ < 0.001, ∗∗∗∗ < 0.0001, ns: not significant. To assess the statistical significance of the observed variation between the scoring of histopathology, a permutation test (Manly, 2006; Neuhäuser and Manly, 2004), an extension of the Fisher exact test for multinomial variables via random sampling, was used. Statistical details of experiments can be found in the Figure Legends and Results section.
Acknowledgments
We thank the late Dr. John Holland for many wonderful insightful discussions, J. Gimlett and S. Kumar for initiating the DARPA INTERCEPT program, which drove significant expansion of the TIP initiative, and the US Army Medical Infectious Diseases Research Program (MIDRP) for invaluable support without which this work would not have been possible. We thank D. Finzi (NIAID) for bridge funding enabling this work. We thank K. Claiborn for editing; S. Larkin for administrative support; L. Minall, F.N.N. Pitchai, T. Tabata, L. Buie, S. Abdullah, and J. Doudna for technical guidance; and the Gladstone-UCSF CFAR flow-cytometry core, funded by NIH grants P30 AI027763 and S10 RR028962 and the James B. Pendleton Trust, as well as the Gladstone Genomics and Histology Cores, and D. Bulkley (UCSF EM Core). This work was supported by Pamela and Edward Taft (to L.S.W.), US Army Medical Infectious Disease Research Program (MTEC 2020-492) (to L.S.W.), and NIH NIDA DP1DA051144-01S1 (to L.S.W.).
Author contributions
S.C. and L.S.W. conceived and designed the study. G.V., D.R., S.I., R.R., J.B., L.H., D.M.S., T.R., N.B., B.N., and L.S.W. conceived, designed, performed, and analyzed the lipid-nanoparticle hamster and mouse studies. S.C., E.J.T., J.B., L.H., X.C., G.V., D.R., A.K., T.R., D.M.S., M.P., R.R., S.I., and P.-Y.C. designed and performed the experiments and curated the data. R.R., M.O., and L.S.W. provided reagents and resources. S.C., G.V., R.R., S.I., and L.S.W. wrote the paper.
Declaration of interests
L.S.W., S.C., and R.R. are co-inventors on a patent application filed for therapeutic interfering particles for SARS-CoV-2. L.S.W. is a scientific co-founder of VxBiosciences.
Published: November 10, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.11.004.
Supplemental information
Data and code availability
-
•
RNAseq data have been deposited to GEO with accession number GSE184447 and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources Table.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the Key Resources Table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aaskov J., Buzacott K., Thu H.M., Lowry K., Holmes E.C. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science. 2006;311:236–238. doi: 10.1126/science.1115030. [DOI] [PubMed] [Google Scholar]
- Abdelnabi R., Foo C.S., De Jonghe S., Maes P., Weynand B., Neyts J. Molnupiravir Inhibits Replication of the Emerging SARS-CoV-2 Variants of Concern in a Hamster Infection Model. J. Infect. Dis. 2021;224:749–753. doi: 10.1093/infdis/jiab361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelnabi R., Foo C.S., Kaptein S.J.F., Zhang X., Do T.N.D., Langendries L., Vangeel L., Breuer J., Pang J., Williams R., et al. The combined treatment of Molnupiravir and Favipiravir results in a potentiation of antiviral efficacy in a SARS-CoV-2 hamster infection model. EBioMedicine. 2021;72:103595. doi: 10.1016/j.ebiom.2021.103595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akpinar F., Timm A., Yin J. High-Throughput Single-Cell Kinetics of Virus Infections in the Presence of Defective Interfering Particles. J. Virol. 2015;90:1599–1612. doi: 10.1128/JVI.02190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric R.S., Nelson G.W., Fleming J.O., Deans R.J., Keck J.G., Casteel N., Stohlman S.A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 2003;270:921–928. doi: 10.1098/rspb.2002.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Gazy I., Jackson L., Hwa S.-H., Tegally H., Lustig G., Giandhari J., Pillay S., Wilkinson E., Naidoo Y., et al. Network for Genomic Surveillance in South Africa. COMMIT-KZN Team Escape of SARS-CoV-2 501Y.V2 from neutralization by convalescent plasma. Nature. 2021;593:142–146. doi: 10.1038/s41586-021-03471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.P., Choi Y., Schork N.J. Conserved Genomic Terminals of SARS-CoV-2 as Co-evolving Functional Elements and Potential Therapeutic Targets. bioRxiv. 2020 doi: 10.1128/mSphere.00754-20. 2020.2007.2006.190207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.C., Olsthoorn R.C. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology. 2010;401:29–41. doi: 10.1016/j.virol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Winkler E.S., Case J.B., Aziati I.D., Bricker T.L., Joshi A., Darling T.L., Ying B., Errico J.M., Shrihari S., et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature. 2021;596:103–108. doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenson B., Böttiger M. Measles antibody: comparison of long-term vaccination titres, early vaccination titres and naturally acquired immunity to and booster effects on the measles virus. Vaccine. 1994;12:129–133. doi: 10.1016/0264-410x(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Coffin J.M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Cohen M., Nirula A., Mulligan M., Novak R., Marovich M., Stemer A., Adams A., Schade A., Knorr J., Tuttle J. Bamlanivimab prevents COVID-19 morbidity and mortality in nursing-home setting. Top. Antivir. Med. 2021;29:32–33. [Google Scholar]
- Cotter G., Kaluski E., Moshkovitz Y., Milovanov O., Krakover R., Vered Z. Pulmonary edema: new insight on pathogenesis and treatment. Curr. Opin. Cardiol. 2001;16:159–163. doi: 10.1097/00001573-200105000-00001. [DOI] [PubMed] [Google Scholar]
- Cox R.J., Brokstad K.A., Ogra P. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand. J. Immunol. 2004;59:1–15. doi: 10.1111/j.0300-9475.2004.01382.x. [DOI] [PubMed] [Google Scholar]
- Daugherty M.D., Malik H.S. Rules of engagement: molecular insights from host-virus arms races. Annu. Rev. Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- Fischer A.H., Jacobson K.A., Rose J., Zeller R. Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc. 2008;2008:prot4986. doi: 10.1101/pdb.prot4986. [DOI] [PubMed] [Google Scholar]
- Gazit S., Shlezinger R., Perez G., Lotan R., Peretz A., Ben-Tov A., Cohen D., Muhsen K., Chodick G., Patalon T. Comparing SARS-CoV-2 natural immunity to vaccine-induced immunity: reinfections versus breakthrough infections. medRxiv. 2021 doi: 10.1101/2021.08.24.21262415. Published online August 25, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg D.E., Siliciano R.F., Jacobs W.R., Jr. Outwitting evolution: fighting drug-resistant TB, malaria, and HIV. Cell. 2012;148:1271–1283. doi: 10.1016/j.cell.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., et al. QCRG Structural Biology Consortium. Zoonomia Consortium Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370:eabe9403. doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A., Reeves D.B., Cardozo-Ojeda E.F., Schiffer J.T., Mayer B.T. Viral load and contact heterogeneity predict SARS-CoV-2 transmission and super-spreading events. eLife. 2021;10:e63537. doi: 10.7554/eLife.63537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J., et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden F.G., Belshe R., Villanueva C., Lanno R., Hughes C., Small I., Dutkowski R., Ward P., Carr J. Management of influenza in households: a prospective, randomized comparison of oseltamivir treatment with or without postexposure prophylaxis. J. Infect. Dis. 2004;189:440–449. doi: 10.1086/381128. [DOI] [PubMed] [Google Scholar]
- Holland J. In: Virology. Bn F., Dm K., editors. Raven Press; 1990. Generation and replication of defective viral genomes; pp. 77–99. [Google Scholar]
- Horstmann D.M., Schluederberg A., Emmons J.E., Evans B.K., Randolph M.F., Andiman W.A. Persistence of vaccine-induced immune responses to rubella: comparison with natural infection. Rev. Infect. Dis. 1985;7(Suppl 1):S80–S85. doi: 10.1093/clinids/7.supplement_1.s80. [DOI] [PubMed] [Google Scholar]
- Iserman C., Roden C.A., Boerneke M.A., Sealfon R.S.G., McLaughlin G.A., Jungreis I., Fritch E.J., Hou Y.J., Ekena J., Weidmann C.A., et al. Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2 Nucleocapsid. Mol. Cell. 2020;80:1078–1091. doi: 10.1016/j.molcel.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P.R., Feldman S., Thompson J.M., Mahoney J.D., Wright P.F. Immunity to influenza A virus infection in young children: a comparison of natural infection, live cold-adapted vaccine, and inactivated vaccine. J. Infect. Dis. 1986;154:121–127. doi: 10.1093/infdis/154.1.121. [DOI] [PubMed] [Google Scholar]
- Johnson B.A., Xie X., Bailey A.L., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.C., Biele G., Mühlemann B., Veith T., Schneider J., Beheim-Schwarzbach J., Bleicker T., Tesch J., Schmidt M.L., Sander L.E., et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021;373:373. doi: 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur N., Singh R., Dar Z., Bijarnia R.K., Dhingra N., Kaur T. Genetic comparison among various coronavirus strains for the identification of potential vaccine targets of SARS-CoV2. Infect. Genet. Evol. 2021;89:104490. doi: 10.1016/j.meegid.2020.104490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke R., Zitzmann C., Ribeiro R.M., Perelson A.S. Kinetics of SARS-CoV-2 infection in the human upper and lower respiratory tracts and their relationship with infectiousness. medRxiv. 2020 2020.2009.2025.20201772. [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.S., Ejima K., Iwanami S., Fujita Y., Ohashi H., Koizumi Y., Asai Y., Nakaoka S., Watashi K., Aihara K., et al. A quantitative model used to compare within-host SARS-CoV-2, MERS-CoV, and SARS-CoV dynamics provides insights into the pathogenesis and treatment of SARS-CoV-2. PLoS Biol. 2021;19:e3001128. doi: 10.1371/journal.pbio.3001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.D., et al. A Therapeutic Non-self-reactive SARS-CoV-2 Antibody Protects from Lung Pathology in a COVID-19 Hamster Model. Cell. 2020;183:1058–1069. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Yale IMPACT Team Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Yokomori K., Lai M.M. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J. Virol. 1990;64:6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly B.F. Volume 70. CRC press; 2006. Randomization, Bootstrap and Monte Carlo Methods in Biology. [Google Scholar]
- Martinez D.R., Schäfer A., Leist S.R., Li D., Gully K., Yount B., Feng J.Y., Bunyan E., Porter D.P., Cihlar T., et al. Prevention and therapy of SARS-CoV-2 and the B.1.351 variant in mice. Cell Rep. 2021;36:109450. doi: 10.1016/j.celrep.2021.109450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger V.T., Lloyd-Smith J.O., Weinberger L.S. Autonomous targeting of infectious superspreaders using engineered transmissible therapies. PLoS Comput. Biol. 2011;7:e1002015. doi: 10.1371/journal.pcbi.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan S., Andrews I.W., Collins J.J. Targeting Antibiotic Tolerance, Pathogen by Pathogen. Cell. 2018;172:1228–1238. doi: 10.1016/j.cell.2018.01.037. [DOI] [PubMed] [Google Scholar]
- Nelson J., Sorensen E.W., Mintri S., Rabideau A.E., Zheng W., Besin G., Khatwani N., Su S.V., Miracco E.J., Issa W.J., et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020;6:eaaz6893. doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhäuser M., Manly B.F. The Fisher-Pitman permutation test when testing for differences in mean and variance. Psychol. Rep. 2004;94:189–194. doi: 10.2466/pr0.94.1.189-194. [DOI] [PubMed] [Google Scholar]
- Notton T., Glazier J.J., Saykally V.R., Thompson C.E., Weinberger L.S. RanDeL-Seq: a High-Throughput Method to Map Viral cis- and trans-Acting Elements. MBio. 2021;12:e01724-20. doi: 10.1128/mBio.01724-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M., Neto E., Chen K., Isa F., Heirman I., Sarkar N., Ramesh D., Cohen M., Barnabas R., Hurt C. Casirivimab with imdevimab antibody cocktail for COVID-19 prevention: Interim results. Top. Antivir. Med. 2021;29:33–34. [Google Scholar]
- Perelson A.S. Modelling viral and immune system dynamics. Nat. Rev. Immunol. 2002;2:28–36. doi: 10.1038/nri700. [DOI] [PubMed] [Google Scholar]
- Perelson A.S., Essunger P., Ho D.D. Dynamics of HIV-1 and CD4+ lymphocytes in vivo. AIDS. 1997;11(Suppl A):S17–S24. [PubMed] [Google Scholar]
- Petrova V.N., Russell C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018;16:60. doi: 10.1038/nrmicro.2017.146. [DOI] [PubMed] [Google Scholar]
- Rangan R., Zheludev I.N., Hagey R.J., Pham E.A., Wayment-Steele H.K., Glenn J.S., Das R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA. 2020;26:937–959. doi: 10.1261/rna.076141.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast L.I., Rouzine I.M., Rozhnova G., Bishop L., Weinberger A.D., Weinberger L.S. Conflicting Selection Pressures Will Constrain Viral Escape from Interfering Particles: Principles for Designing Resistance-Proof Antivirals. PLoS Comput. Biol. 2016;12:e1004799. doi: 10.1371/journal.pcbi.1004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenke K., Hansen F., Schwarz B., Feldmann F., Haddock E., Rosenke R., Barbian K., Meade-White K., Okumura A., Leventhal S., et al. Orally delivered MK-4482 inhibits SARS-CoV-2 replication in the Syrian hamster model. Nat. Commun. 2021;12:2295. doi: 10.1038/s41467-021-22580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine I.M., Weinberger L.S. Design requirements for interfering particles to maintain coadaptive stability with HIV-1. J. Virol. 2013;87:2081–2093. doi: 10.1128/JVI.02741-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N., Papaspyropoulos A., Zomer-van Ommen D.D., Heo I., Böttinger L., Klay D., Weeber F., Huelsz-Prince G., Iakobachvili N., Amatngalim G.D., et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38:e100300. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo D., Katkar G.D., Khandelwal S., Behroozikhah M., Claire A., Castillo V., Tindle C., Fuller M., Taheri S., Rogers T.F., et al. AI-guided discovery of the invariant host response to viral pandemics. EBioMedicine. 2021;68:103390. doi: 10.1016/j.ebiom.2021.103390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnipper L.E., Crumpacker C.S. Resistance of herpes simplex virus to acycloguanosine: role of viral thymidine kinase and DNA polymerase loci. Proc. Natl. Acad. Sci. USA. 1980;77:2270–2273. doi: 10.1073/pnas.77.4.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., 3rd, Stevens L.J., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12:12. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia F., Laske T., Wasik M.A., Rammhold M., Genzel Y., Reichl U. Production of Defective Interfering Particles of Influenza A Virus in Parallel Continuous Cultures at Two Residence Times-Insights From qPCR Measurements and Viral Dynamics Modeling. Front. Bioeng. Biotechnol. 2019;7:275. doi: 10.3389/fbioe.2019.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P.E., Chao L. Prisoner’s dilemma in an RNA virus. Nature. 1999;398:441–443. doi: 10.1038/18913. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M., López C.B. Defective viral genomes are key drivers of the virus-host interaction. Nat. Microbiol. 2019;4:1075–1087. doi: 10.1038/s41564-019-0465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Magnus P. Incomplete forms of influenza virus. Adv. Virus Res. 1954;2:59–79. doi: 10.1016/s0065-3527(08)60529-1. [DOI] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Weinberger L.S., Schaffer D.V., Arkin A.P. Theoretical design of a gene therapy to prevent AIDS but not human immunodeficiency virus type 1 infection. J. Virol. 2003;77:10028–10036. doi: 10.1128/JVI.77.18.10028-10036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., Bennett H., Boyoglu-Barnum S., Shi W., Graham B.S., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021 2021.2001.2025.427948. [Google Scholar]
- Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., Huang W.J., Gao P., Zhou C., Zhang R.R., et al. A Thermostable mRNA Vaccine against COVID-19. Cell. 2020;182:1271–1283. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Sachs N., Chiu M.C., Wong B.H., Chu H., Poon V.K., Wang D., Zhao X., Wen L., et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. USA. 2018;115:6822–6827. doi: 10.1073/pnas.1806308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020;27:883–890. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNAseq data have been deposited to GEO with accession number GSE184447 and are publicly available as of the date of publication. Accession numbers are listed in the Key Resources Table.
-
•
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the Key Resources Table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.