Abstract
Genetic testing using reverse transcriptase real-time polymerase chain reaction (rRT-PCR) is the mainstay of diagnosis of COVID-19. However, it has not been fully investigated whether infectious viruses are contained in SARS-CoV-2 genome-positive specimens examined using the rRT-PCR test. In this study, we examined the correlation between the threshold Cycle (Ct) value obtained from the rRT-PCR test and virus isolation in cultured cells, using 533 consecutive clinical specimens of COVID-19 patients. The virus was isolated from specimens with a Ct value of less than 30 cycles, and the lower the Ct value, the more efficient the isolation rate. A cytopathic effect due to herpes simplex virus type 1 contamination was observed in one sample with a Ct value of 35 cycles. In a comparison of VeroE6/TMPRSS2 cells and VeroE6 cells used for virus isolation, VeroE6/TMPRSS2 cells isolated the virus 1.7 times more efficiently than VeroE6 cells. There was no significant difference between the two cells in the mean Ct value of the detectable sample.
In conclusion, Lower Ct values in the PCR test were associated with higher virus isolation rates, and VeroE6/TMPRSS2 cells were able to isolate viruses more efficiently than VeroE6 cells.
Keywords: SARS-CoV-2, COVID-19, Virus isolation, Threshold cycle (Ct) value, VeroE6/TMPRSS2, Clinical specimens
Coronavirus disease 2019 (COVID-19), which was caused by a newly identified coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), caused a pandemic in March 2020 [1]. A rapid and widespread infection of COVID-19 has become a national concern in Japan. In Toyama Prefecture, 422 cases of suspected COVID-19 infection were reported during the period from March 30 to October 1, 2020. In our institute, a genetic test using reverse transcription real-time polymerase chain reaction (rRT-PCR) [2] was conducted on 299 infected individuals, and 533 samples (nasopharyngeal swab, 507 samples; sputum, 3 samples; saliva, 23 samples), including the retested ones, were judged to be positive. However, even if the PCR test is positive, it cannot be determined whether or not SARS-CoV-2 is infectious in clinical specimens [3]. Therefore, a virus isolation test was performed on cultured cells using clinical specimens that were positive in the PCR test. Generally, if the virus can be isolated in cultured cells, it can be determined to be infectious. In this study, we addressed the relationships between the results of virus isolation, the number of days for a sample collection from the confirmation of PCR positivity, and the Ct value in the real-time PCR. This study was approved by the Ethics Review Committee of the Toyama Institute of Health (R2-12). For real-time PCR testing, we used SARS-CoV-2 Direct Detection RT-qPCR kit (Takara Bio Inc., Shiga, Japan), which targets the N genes of SARS-CoV-2, and the assays were performed using QuantStudio 5 (Thermo Fisher Scientific Inc., Waltham, MA). The methods and Ct values of the real-time PCR tests were determined according to the manufacturer's instructions.
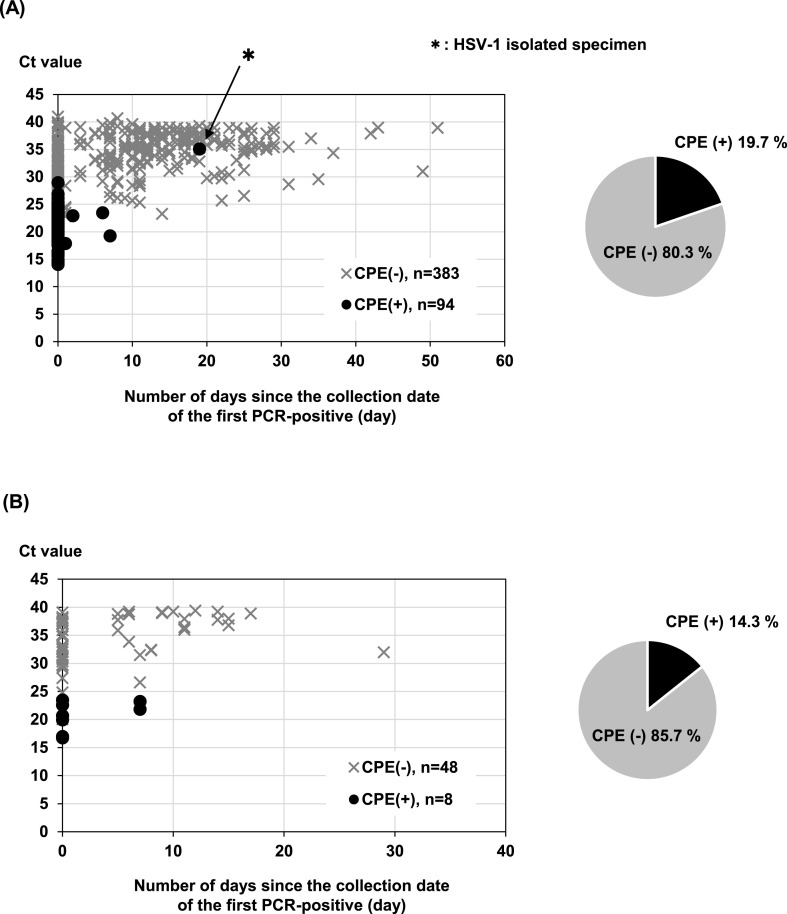
For a virus isolation test, VeroE6 cells overexpressing TMPRSS2 (VeroE6/TMPRSS2) (JCRB1819), which is considered to have a high efficiency of SARS-CoV-2 infection [4], were obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank (Osaka, Japan). VeroE6 cells, which were kindly provided from the National Institute of Infectious Diseases, Japan, were also used to compare the isolation efficiency of SARS-CoV-2. Both cell lines were grown in Dulbecco's Modified Eagle Medium (DMEM; Nacalai Tesque, Inc., Kyoto, Japan) containing 10% heat-inactivated fetal bovine serum (FBS). All specimens are stored at −80 °C until used for a virus isolation test. Approximately 50 μL centrifugal supernatant of nasopharyngeal swab suspension, which exhibited rRT-PCR positivity for SARS-CoV-2, was added to VeroE6/TMPRSS2 cells seeded a day before on a 24-well plate and cultured at 37 °C for 5 days, and the cytopathic effect (CPE) was confirmed by visual observation under a microscope. The treated cells in which CPE was not confirmed were treated with trypsin and then subcultured on a new 24-well plate and cultured for another 5 days, which was used as the final judgment. Of the 533 specimens added to the cultured cells, 102 specimens (19.1%) were confirmed to have a CPE in the first culture. The cells supplemented with 431 specimens in which CPE was not confirmed were passaged and cultured for another 5 days, but no virus was isolated. Generally, if CPE is not confirmed in the virus isolation test, it is necessary to continue culturing by passage. In the SARS-CoV-2 isolation test performed in this study, it was found that if the amount of virus that can be isolated is sufficient, the virus can be isolated in the first culture without repeated passage, which is consistent with previous study [5]. Additionally, infectious viruses can be isolated from asymptomatic individuals at the same rate as symptomatic individuals (19.7% of symptomatic persons, 14.3% of asymptomatic persons) (Fig. 1 ). The tested specimens we used for viral isolation included confirming PCR negativity in the same patient from the second time onward. When limited to first-time study of target patients, the virus could be isolated in 98 of the 299 specimens with a positive rate of 32.8%.
Fig. 1.
The rate of virus isolation and number of days since the collection date of the first PCR positivity. All the specimens of suspected COVID-19 were tested for virus isolation in VeroE6/TMPRSS2 cells. The collection date of the first PCR-positive specimen is indicated on the horizontal axis and the virus isolation rate on the vertical axis. (A) Results of virus isolation test using specimens from symptomatic individuals. (B) Results of virus isolation test using specimens from asymptomatic individuals, ●; Specimens with cytopathic effect (CPE) on virus isolation test, × ; Specimens without CPE on virus isolation test. The pie chart shows the ratio by the number of results.
Next, the correlation between the Ct values of PCR-positive specimens and virus isolation results was analyzed (Fig. 1). The virus was isolated from the specimens collected within 7 days after the first PCR positivity. The virus was not isolated from the specimens except for one specimen regardless of the Ct value in the specimens after 8 days of the first PCR positivity. No significant difference was found between those who were symptomatic at the first PCR positivity [Fig. 1 (A)] and those who were asymptomatic at the first PCR positivity [Fig. 1 (B)]. It was also found that the virus was isolated from specimens with a Ct value of less than 30 (Fig. 1 and Table 1 ). No correlation was found between the patient's clinical characteristics, such as age, sex, the severity of disease, and the rate of virus isolation (data not shown). Recently, a similar report was published from South Korean gruoup [6]. They described that viral culture was positive only in samples with a Ct value of 28.4 or less, which are supporting in our results. Analysis of the Ct value and the rate of virus isolation showed that the rate of virus isolation tended to decrease as the Ct value increased (Table 1; p < 0.001). It was found that the virus can be isolated with a high frequency of 91% or more (94.7% less than 20 and 65.1% more than 20 and less than 25) when the Ct value is less than 25. Alternatively, the rate of virus isolation decreased significantly (p < 0.001) when the Ct value was 25 or more and less than 30 (12.0%). Furthermore, virus isolation was not observed at Ct values of 30 or more (25 or more and less than 30 vs. 30 or more and less than 35; p < 0.001). The positive rate of virus isolation is difficult to judge because it is affected by the quality and quantity of the specimens used and the culture conditions. According to the results of virus isolation using the Ct value as an indicator, the Ct value of the specimens from which the virus was isolated was approximately less than 30. It has been reported that the virus is isolated even if the Ct value is 30 or higher, but it is considered as a rare case [5,7,8]. In the analysis using the collection date of the specimens as an indicator, all the specimens after 8 days from the first PCR positivity date were negative for virus isolation. Based on these results, it was considered that there was no infectious virus in the specimens collected from patients 8 days after the confirmation of PCR positivity regardless of whether individuals were symptomatic or asymptomatic. In this study, cells were inoculated with 50 μL of the specimens, and the genome copy number of Ct value 30 calculated from positive control RNA is considered to be about 1 × 104 copies/50 μL. Assuming that the infectious virus particles are 1/100 to 1/1000 of the genome amount, it is predicted that about 10–100 infectious viruses are contained in the inoculated specimens to the cells. In this virus isolation test, it is considered that hundreds of viruses (Ct value less than 25) are sufficient to isolate the viruses.
Table 1.
Ct value of rRT-PCR and the rate of virus isolation.
| Ct value | PCR-positive specimens | Positive for virus isolation | Ratio of virus isolation (%)a) | 95% CI | p valueb) |
|---|---|---|---|---|---|
| <20 | 38 | 36 | 94.7 | 82.3–99.4 | 0.002 |
| 20–25 | 86 | 56 | 65.1 | 54.1–75.1 | <0.001 |
| 25–30 | 75 | 9 | 12.0 | 5.6–21.6 | <0.001 |
| 30–35 | 141 | 0 | 0.0 | 0.0–2.6 | NA |
| ≧35 | 192 | 0 | 0.0 | 0.0–1.9 | |
| Total | 532 | 101 | 19.0 | 15.7–22.6 |
The relationship between Ct value in rRT-PCR and the rate of virus isolation is divided into five groups (less than 20, 20, or more, and less than 25, 25, or more, and less than 30, 30, or more, and less than 35, 35, or more; n = 532, except for HSV-1 isolated specimen) based on the Ct value. The rate of virus isolation and 95% confidence interval were determined. The Mantel–Haenszel test for trend was conducted for the tendency of the rate of virus isolation. The comparison between the two groups was conducted by the χ2 test or Fisher's exact test, and p values were corrected using the Bonferroni method. All statistical tests were performed using IBM SPSS Statistics 24.0, and p < 0.05 was considered significant.
p < 0.001.
Comparison with the group with next highest Ct value.
It is known that the efficiency of cell entry in VeroE6/TMPRSS2 cells with SARS-CoV-2 is higher than that in VeroE6 cells [4]. Therefore, to investigate the virus isolation efficiency in VeroE6/TMPRSS2 cells, 102 specimens that could isolate the virus in VeroE6/TMPRSS2 cells were performed with the same viral isolation test to see if they could also isolate viruses in VeroE6 cells. Consequently, it was found that the virus can be isolated for 60 specimens (approximately 58.8%) in VeroE6 cells [Fig. 2 (A) and (B)]. The efficiency of virus isolation in VeroE6/TMPRSS2 cells was higher than that in VeroE6 cells in the virus isolation test for clinical specimens. No significant difference in the Ct value of the specimens that could be isolated viruses were found between VeroE6/TMPRSS2 cells and VeroE6 cells [Fig. 2 (C)], suggesting that the quantity of virus did not simply affect the success rate of the virus isolation.
Fig. 2.
Comparison of the Ct value and virus isolation between VeroE6/TMPRSS2 and VeroE6 cells. (A) Graphs blotted with Ct values of each specimen isolated in VeroE6/TMPRSS2 (●) or VeroE6 (▲) cells. (B) Rate of total specimens isolated in VeroE6/TMPRSS2 or VeroE6 cells. (C) Ct values of each specimen isolated in VeroE6/TMPRSS2 or VeroE6 cells. n.s.; not significant.
To confirm whether the CPE of the virus-isolated cells was due to the SARS-CoV-2 infection, the cell culture supernatants were collected. The genomic RNA was extracted, and then rRT-PCR was performed. As a result, it was found that 101 of the 102 specimens in which CPE was confirmed were positive for the SARS-CoV-2 gene, with the remaining one specimen being negative for the same gene. In this specimen, CPE was observed in the cultured cells, even though the Ct value of the rRT-PCR test 19 days after the confirmation of positivity was about 35. Therefore, since it was predicted that the CPE was caused by a virus other than SARS-CoV-2, rRT-PCR was performed on this specimen with various virus species (enterovirus, rhinovirus, respiratory syncytial virus A, respiratory syncytial virus B, parainfluenza virus 1–4, human bocavirus, human coronavirus OC43, human coronavirus NL63, human metapneumovirus, influenza A virus, influenza B virus). Consequently, positivity for herpes simplex virus was obtained from rRT-PCR. Genomic sequence analysis revealed that the specimen contained the herpes simplex virus type 1 (HSV-1) gene. Additionally, since this culture supernatant was isolated from the specimen of nasopharyngeal swab of the patient collected at the second period, the presence of HSV-1 in that collected at the first period was confirmed by rRT-PCR. HSV-1 was not detected in the specimen collected at the first period. These results suggested that the SARS-CoV-2 gene was detected by rRT-PCR test from the patient-derived specimen, but HSV-1 was isolated and showed CPE in the virus isolation test using VeroE6/TMPRSS2 cells. In this case, it was suggested that HSV-1, initially latently infected, was reactivated and amplified after SARS-CoV-2 infection. If virus isolation is exceptionally detected in the virus isolation test, it is considered necessary to confirm whether or not the target virus has been isolated. Analysis by a virus isolation test from COVID-19-infected patients is vital for preventing infection spread and establishing therapeutic agents, and it is necessary to continue investigation and research in the future.
In conclusion, we verified the relation between the rRT-PCR test and virus isolation in cultured cells of 533 consecutive clinical specimens of COVID-19 patients. The virus was isolated at a higher rate in samples with low Ct values. The virus isolation rate was low in asymptomatic specimens. In virus isolation, VeroE6/TMPRSS2 cells were able to isolate the virus in 1.7 times as many samples as VeroE6 cells, but there was no significant difference in the mean Ct value of the rRT-PCR test. From the standpoint of virus infection prevention, more research is needed on PCR positivity and virus infectivity.
Author contribution
Conceptualization and methodology, E.I., H.T., K.T., and K.O.; sample collection, H.K., S.N., H.I., S.T., Y.Y., S.Y., T.I., Y.H., K.H., N.H., and Toyama COVID-19 Epidemiological Research Group; investigation. E.I., H.T., M.I., T.S., Y.S., N.I., S.H., and S.Y.; writing–original draft presentation, H.T.; writing–review and editing, E.I., H.T., K.T., T.I., and K.O.; supervision, H.T., H.S., C.K., and K.O.; funding acquisition, H.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by a grant-in-aid from the Japan Agency for Medical Research and Development (AMED) (Grant No. JP20he0622035) and in part by a grant-in-aid from the Tamura Science and Technology Foundation.
Declaration of competing interest
None.
Acknowledgments
We sincerely thank Emi Maenishi, Kaoru Uchida, Jun-ichi Kanatani, Junko Isobe, Masanori Watahiki, and Keiko Kimata, Department of Bacteriology, Toyama Institute of Health, for the conduction of PCR analysis, and Toyama City Public Health Center, Takaoka Welfare Center, Tonami Welfare Center, Niikawa Welfare Center, Chubu Welfare Center, and Health Division of Toyama Prefectural Government, for the collection, transportation, and arrangement of clinical specimens. We also thank Yoko Kanamori and Izumi Kawaguchi for technical and secretarial assistance.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shirato K., Nao N., Katano H., Takayama I., Saito S., Kato F., et al. Development of genetic diagnostic methods for detection for novel coronavirus 2019(ncov-2019) in Japan. Jpn J Infect Dis. 2020;73:304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 3.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., et al. Duration of infectiousness and correlation with rt-pcr cycle threshold values in cases of covid-19, england, january to may 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., et al. Enhanced isolation of sars-cov-2 by tmprss2-expressing cells. Proc Natl Acad Sci U S A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada S., Fukushi S., Kinoshita H., Ohnishi M., Suzuki T., Fujimoto T., et al. Assessment of sars-cov-2 infectivity of upper respiratory specimens from covid-19 patients by virus isolation using veroe6/tmprss2 cells. BMJ Open Respir Res. 2021;8 doi: 10.1136/bmjresp-2020-000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim M.C., Cui C., Shin K.R., Bae J.Y., Kweon O.J., Lee M.K., et al. Duration of culturable sars-cov-2 in hospitalized patients with covid-19. N Engl J Med. 2021;384:671–673. doi: 10.1056/NEJMc2027040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., et al. Viral rna load as determined by cell culture as a management tool for discharge of sars-cov-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]