Highlights
-
•
Insight into the role of surfactin on B. subtilis cell differentiation.
-
•
Insight into the molecular genetics of surfactin and its production.
-
•
Graphical presentation of surfactin mediated signaling cascades via quorum sensing.
Keywords: Surfactin, srfA operon, Cell differentiation, Bacillus subtilis
Abstract
Surfactin is a biosurfactant produced by Bacillus subtilis. The srfA operon, Sfp gene, and two quorum sensing systems are required for its production. The master regulator spo0A also plays an indispensable role in proper surfactin synthesis. Upon production, surfactin itself acts as a signaling molecule and triggers the activation of Spo0A gene which in turn regulates cell differentiation. Interestingly, surfactin producing cells are immune to the action of surfactin but trigger other cells to differentiate into non-motile cells, matrix producing cells, cannibals, and spores. In case of competent cell differentiation, comS, which resides within the srfA operon, is co-expressed along with surfactin and plays a vital role in competent cell differentiation in response to quorum sensing signal. Surfactin inhibits the motility of certain cell subpopulations, although it helps the non-motile cells to swarm. Thus, surfactin plays significant roles in the differentiation of different subpopulations of specialized cell types of B. subtilis.
1. Introduction
Surfactants are amphiphilic compounds composed of hydrophilic and hydrophobic parts and function by lowering the surface tension between two phase systems. Surfactants can be chemical or biological in origin. Surfactants that are produced by biological systems like bacteria, yeasts, and fungi are known as biosurfactants [1]. The biosurfactants can be classified according to their properties and origin. Some of the major classes of biosurfactants are glycolipids, lipopeptides, lipoproteins, phospholipids, fatty acids, and polymeric surfactants [2]. Biosurfactants have more advantages than synthetic surfactants as biosurfactants possess properties like biodegradability, low toxicity, and less sensitivity to extreme environments [1, 3]. Surfactin, a cyclic lipopeptide is one of the biosurfactants that was identified in 1968 from the culture media of B. subtilis [4]. It is known as one of the most potent biosurfactants because of its strong activity as a surfactant. It can effectively reduce the surface tension of two phase systems at very low concentrations [5]. Structurally surfactin consists of an amino acid chain attached to a fatty acid chain. It is known to possess a range of biological activities, such as it can lyse red blood cells, inhibit blood clot formation, lyse bacterial cells, and inhibit cyclic 3′,5-monophosphate-di-esterase [6].
Knowledge of genetics required for the production of biosurfactants like surfactin is vital for their effective industrial applications. Currently, because of high production cost, biosurfactants are unable to compete with chemically synthesized surfactants. If the genes and their interactions that are associated with biosurfactant production are identified, the production process can be enhanced by applying various advanced techniques, such as placing the genes under the regulation of strong promoters. Concrete understanding of both the genetics and functions can help us to use surfactin more effectively in different industrial fields [7, 8].
Bacteria are capable to differentiate into different subpopulations of cells that together form complex multicellular communities [9]. These subpopulations can be phenotypically distinct, although they are genetically identical [10]. B. subtilis is called the master of differentiation because of its ability to differentiate into a number of cell types. Extensive studies have been done to explore this unique molecular characteristic of the bacteria in cell culture systems. For instance, at the beginning of the stationary phase, B. subtilis can either differentiate into surfactin producers, which may or may not be competent cells that can uptake DNA from the extracellular environment [11, 12], or it can enter into the sporulation phase that is extremely resilient to environmental impacts. The bacteria might also differentiate into matrix-producing cells that form biofilm [13, 14, 15]. These bacterial cells can also differentiate in the exponential growth phase, for example, a portion of cells can develop flagella, as a result they differentiate into motile cells [16]. The ultimate fate of these subpopulations depends on either gene expression that results in morphological differences or on reporter-receptor interaction. The cell differentiation into multiple subpopulations in a stress condition requires a certain level of cell-cell communication that is cell density dependent and known as quorum sensing system [17]. Usually this type of cell-cell communication is accomplished with the help of a particular type of signaling molecules (called autoinducers) that are produced and secreted by the cells of the bacterial community and in turn these signaling molecules can induce cell differentiation [18]. Molecular mechanisms of these types of autoinducer molecules of B. subtilis have already been investigated [19, 20]. Surfactin is one of the signaling molecules that play significant roles in B. subtilis cell differentiation [21].
This review sheds light on the detailed knowledge on molecular genetics of surfactin production and how its production is controlled or regulated by two separate quorum sensing systems in B. subtilis. This review also highlights the gene regulatory pathways that are induced or blocked by surfactin to determine the cell fate of different subpopulations of B. subtilis under stress condition.
1.1. srfA operon–sfp gene cluster system
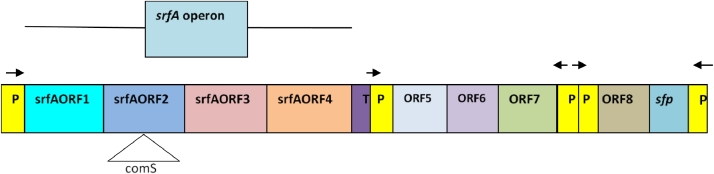
srfA operon-sfp gene cluster plays the most important role in surfactin production. Cosmina et al. sequenced the srfA operon–sfp gene cluster completely. Fig. 1 illustrates the nine major open reading frames (ORFs) of srfA operon along with the sfp gene. The gene cluster is flanked by the srfA promoter at one side and the sfp gene with its promoter at the other side. The distance between the transcriptional starting point of srfA operon and sfp gene was reported to be 30.5 kb. At the end of the forth ORF, a putative transcriptional terminator was detected. ORFs lying between the srfAORF4 and the sfp gene were named as ORF5 to ORF8 [22].
Fig. 1.
A map of the ORFs of srfA operon and the sfp gene
1.2. srfA operon
According to Fabret et al., the length of srfA operon is 27 kb, although Hamoen et al. reported it is to be of 25 kb in length and Cosmina et al. reported that the operon has a length of 30 kb [22, 23, 24]. The srfA gene comprises of four ORFs (srfAORF1 to srfA0RF4) with a promoter sequence at 5′ upstream of srfAORF1 and an adjacent transcriptional terminator sequence at the end of srfAORF4 [22, 25]. Polypeptides of 402, 401, and 144 kDa are encoded by srfAORF1, srfAORF2, and srfAORF3, respectively and Steller et al. reported that the srfORF4 encoded protein has 44 kDa of molecular weight [26]. Surfactin synthetase is encoded by these four ORFs of srfA operon [23]. Later, this surfactin synthetase synthesizes the peptide chain of surfactin by a process known as non-ribosomal peptide synthesis [27].
The surfactin synthetase comprises of four enzymatic modules. Four open reading frames of srfA operon encodes these four modules. These four enzyme subunits are SrfAA (encoded by srfAORF1), SrfAB (encoded by srfAORF2), SrfAC (encoded by srfAORF3), and SrfAD (encoded by srfAORF4) [26]. Each module of the synthetase enzyme consists of numerous domains and each of them incorporate and modify one specific amino acid into the peptide chain of surfactin [6]. Previously it was thought that SrfAD (encoded by srfAORF4) was not required for surfactant production [22, 3]. Later, it was proved that SrfAA alone has the capability to initiate the lipopeptide formation; however, the formation of the products gets efficient support and stimulation from SrfAD [26]. Several research groups reported SrfAD as a repair enzyme because it efficiently regenerates misacylated cofactors attached to peptidyl carrier protein (PCP) domains during non-ribosomal protein synthesis (NRPS) [28, 29, 30]. SrfA is responsible for surfactin synthesis as well as for competence development [24]. Competence development is dependent on the gene comS, located within srfAORF2 [31]
2. ORFs between the srfA operon and sfp gene
According to the study performed by Cosmina et al., the region between the srfA operon and the sfp gene is about 4 kb in length and contains four ORFs (ORF5 to ORF8) (Fig. 1). These ORFs have no effects on surfactin production (neither enhance or repress surfactin production) [22].
2.1. sfp gene
The sfp gene is positioned about 4 kb downstream of the srfA operon. A study performed by Quadri et al. showed that the sfp gene has significant roles in surfactin production. It creates docking sites in surfactin synthetase protein for loading specific amino acids to form peptide chain of surfactin [32]. Hamoen et al. reported that sfp gene activates the PCP domains by converting their inactive forms into their active forms [24].
2.2. comQXPA gene cluster
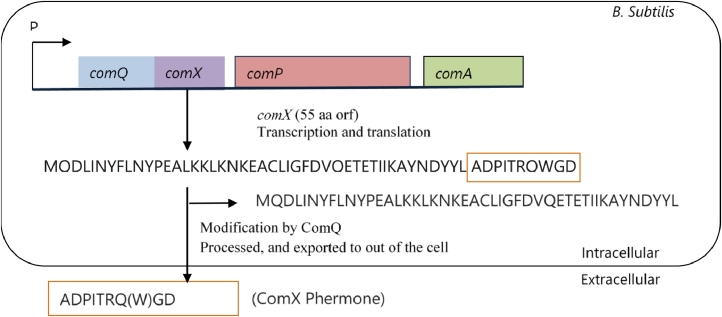
comQXPA gene cluster of B. subtilis is composed of comQ, comX, comP, and comA genes and has already been sequenced in the year 2000 [33]. comQ is found to be positioned adjacent to comX where comP and comA is located at the downstream of comX in the bacterial chromosome (Fig. 2). comQ and comX genes are required for ComX pheromone production. ComX pheromone regulates one of the two quorum sensing systems that controls surfactin production [19]. The comX encodes a precursor of ComX pheromone which consists of a 55-amino acid long peptide. Production of mature ComX pheromone (10 amino acid long peptide) requires the processing, modification, and secretion of this peptide outside the cell. comQ helps in the processing and modification of precursor ComX pheromone. Studies showed that, comQ null mutant strains are unable to produce comX pheromone and have decreased expression of srfA. Transcriptional regulatory protein ComA and sensor histidine kinase ComP (response regulators) is produced by the two-component regulatory system ComP/ComA. After secretion from cell, ComX interact with the N-terminal sensory domain of ComP and activates it [34, 35]. In turn, this activated ComP phosphorylates and activates ComA which then (ComA) directly attaches to the promoter region of srfA operon and initiates the transcription of the adjacent gene [36, 25].
Fig. 2.
The production process of ComX pheromone. At first a precursor ComX (encoded by comX) is produced that is 55 amino acids long. It is then modified at position 53, processed, and exported outside from the cell by ComQ. The mature ComX pheromone is of 10 amino acids long. ComP and ComA are the two proteins encoded by comP and comA, respectively and they respond to the ComX pheromone
2.3. rapC-phrC operon
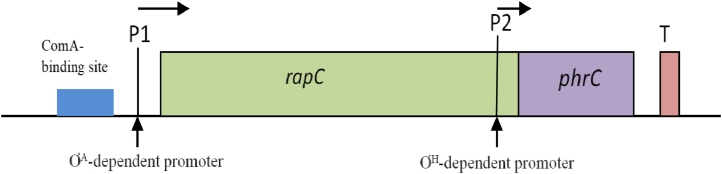
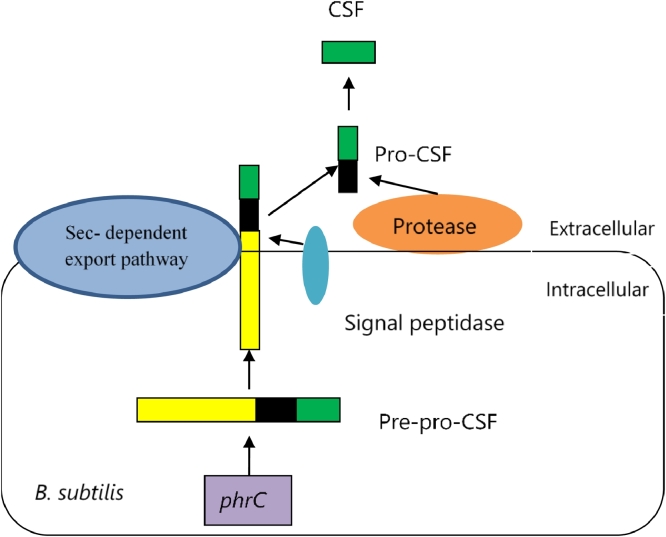
The rapC-phrC operon consists of rapC and phrC genes (illustrated in Fig. 3). This operon has two promoters, P1 and P2. The first one, promoter P1, is situated immediately upstream of the rapC gene, with the recognition sequence for the vegetative sigma factor, ƠA. The second one, promoter P2, is positioned inside rapC and upstream of phrC, and is ƠH-dependent [37], (Fig. 2A). Lazazzera proposed that, both P1 and P2 might control the expression of phrC gene [38, 39]. The phrC gene is responsible for the expression of an extracellular signaling peptide called competence and sporulation factor (CSF). The phrC gene encodes a pre-pro-peptide which is excreted outside of the cell by the Sec-dependent exporting pathway and a peptidase enzyme removes the signaling peptide. The secreted pro-peptide is then cleaved by an enzyme to release a mature five amino acid signaling peptide [39]. Fig. 4 shows a model of the mechanism of production of CSF from phrC gene. The rapC encodes a putative phosphatase [40] that constrains the activation of ComA and thus negatively regulates the expression of srfA [41]. On the other hand, CSF enters into cell with the help of oligopeptide permease, which is also known as Spo0K. Upon entering, it inhibits the phosphatase RapC and stimulates expression of srfA [42, 41].
Fig. 3.
A map of the rapC-phrC operon. It possess two promoters, both of them are indicated by the arrows. There are a terminator (T) positioned downstream of phrC which is Rho-independent, and a binding site for ComA (ComA Box) positioned upstream of rapC.
Fig. 4.
The mechanism of competence and sporulation factor (CSF) production from phrC gene. The phrC encodes a pre-pro-peptide, the yellow portion of which represents signal sequence, the black portion of which represents secreted non-signaling peptide sequence, and the Green portion of which represent the mature signaling peptide sequence. The signaling sequence is cut off and the pro-CSF is exported outside of the cell where a protease cuts the pro-CSF and transforms it into active CSF.
2.4. spo0A gene
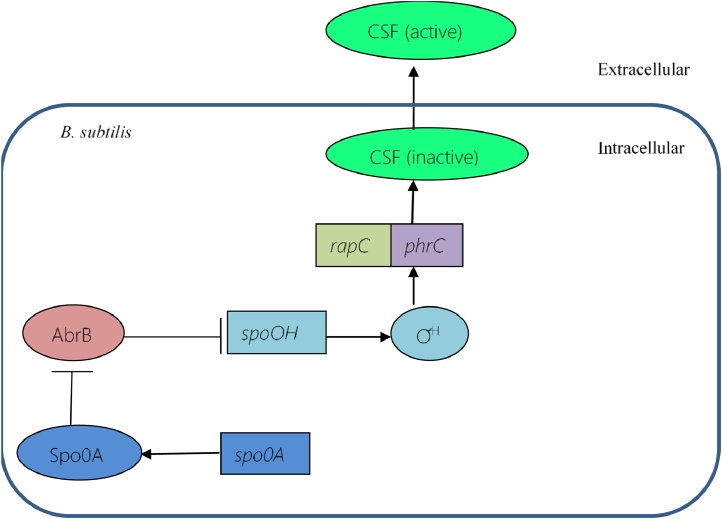
Spo0A regulates the expression of genes that are required for the onset of sporulation in B. subtilis [43]. A study performed by Nakano et al. indicates that spo0A gene has significant role in surfactin production too (Illustrated in Fig. 5) [44]. Different studies reported that deletion of spo0A gene results in absolutely no surfactin production [45, 46] Spo0A protein negatively regulates the gene abrB and thus the synthesis of AbrB protein. This Spo0A-mediated inhibition of abrB results in the reduction of the AbrB protein in the cell and in turn, the genes that are usually negatively controlled by AbrB protein, get activated [47]. The spo0H gene is one of those genes that are negatively controlled by AbrB protein. So, any reduction in the cellular level of AbrB protein leads to activation of spo0H gene. This gene encodes a sigma factor, ƠH, which is involved in expression of many different genes [48, 20]. For instance, ƠH is required in the transcription of CSF from phrC [49, 50]. ƠH binds to the second promoter of racC-phrC operon, which is a ƠH-dependent promoter and partly regulates CSF transcription [51]. So, the repression of AbrB protein by spo0A leads to the expression of CSF and thus indirectly regulates the expression of srfA.
Fig. 5.
Regulation of the expression of competence and sporulation factor (CSF) by spo0A. CSF is encoded by the phrC gene. spo0H encodes the ƠH, which is required by phrC to produce CSF. This spo0H is negatively regulated by the AbrB protein, which is antagonised by Spo0A protein. Deactivation of AbrB by Spo0A protein leads to optimal production of CSF.
3. Surfactin production network
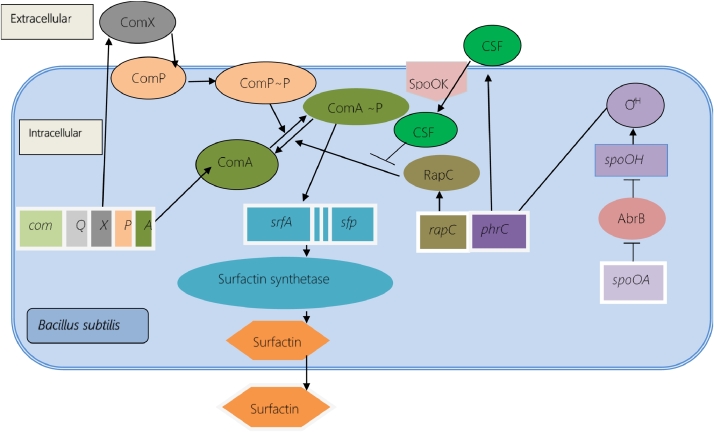
Two quorum sensing systems regulates surfactin production in B. subtilis. Regulation initiates with the production of inactive ComX pheromone, modification is done by the ComQ, and finally get activated to become the signaling peptide ComX. ComX pheromone is secreted outside of the cell which interacts with membrane-bound histidine kinase ComP. This interaction causes ComP to autophosphorylate and get activated. Phosphorylated ComP donates a phosphate group to ComA that in turn gets phosphorylated and activated. Activated ComA binds directly to the promoter region of the operon srfA and initiates gene expression to produce surfactin synthetase enzyme. The gene sfp, situated downstream of the srfA operon is also needed for the surfactin production process. The phrC encodes CSF, which regulates the second quorum sensing system. Optimal CSF production requires ƠH that is encoded by spo0H. The AbrB protein represses the spo0H that is antagonizes the spo0A gene by repressing the abrB gene. This is how spo0A, which is also called the master regulator, regulates the production of surfactin by regulating the expression of phrC. However, mature CSF enters into the cell with the help of Spo0K and induces srfA expression by inhibiting the activity of RapC protein. Surfactin synthetase later synthesize surfactin using its various domains and finally, surfactin is excreted outside the cell. The overall pathway of surfactin production and regulation is illustrated in Fig. 6.
Fig. 6.
A model illustrating the genes, proteins, and their interactions required for surfactin production and regulation. ComA, which is mainly responsible for srfA operon expression, gets activated and remains in activated form by the help of ComX and CSF.
4. Effect of surfactin on cell differentiation
Bacterial communities can perceive environmental changes and adjust gene expression accordingly via a system, known as quorum sensing. It is a bacterial cell-density-dependent process which is performed with the help of the signaling molecules called autoinducers e.g. surfactin in B. subtilis. Concentration of signaling molecules increases in the extracellular environment as the bacterial cell density increases and when it reaches a threshold level, this group of bacterial cells acts in synchrony in response to the signaling molecule. These signaling molecules lead to an alteration in gene expression, coordination of bacterial behavioural changes, and differentiation into different cell types to adapt environmental changes [18]. In B. subtilis, surfactin helps to regulate genes that exerts cellular differentiation into different cell types via quorum sensing according to need to adapt adverse condition[49, 17].According to some studies, along with cell differentiation surfactin may also have substantial role as a quorum sensing molecule in carbon metabolism [52, 53].
5. Surfactin producing cells are immune to surfactin
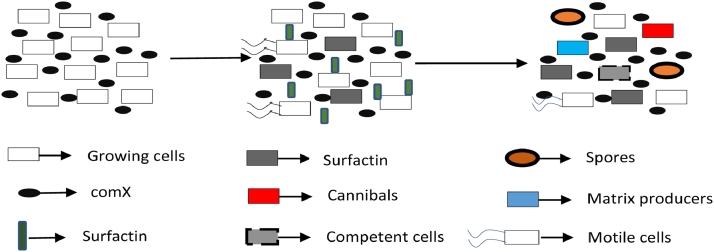
Production and secretion of specific signaling molecules that can trigger diverse signaling pathways, regulate the quorum sensing in bacteria (illustrated in Fig. 7) [18]. There are several secreted signaling molecules that activate specific signaling pathways in B. subtilis. Surfactin is one of them. Surfactin is produced by surfactin producing cells, a subpopulation of B. subtilis and other subpopulations respond to it. As the signal-producing and receptive cells are different, this is an example of paracrine signaling. Surfactin has significant roles on matrix producing cells, cannibals, competent cells, spores, and motile cells. However, surfactin producing cells are immune to its product, surfactin. It is not known for sure how they gain this immunity. But according to Lopez et al., ComS might be responsible for this immunity [54]. ComS protein is encoded by the gene comS, which is positioned within the srfA operon and this is why the gene is co-expressed with the surfactin production [31]. ComS might inactivate Spo0A, which is needed to respond to surfactin by interacting with MecA. It allows MecA to bind to and inhibit Spo0A by freeing MecA from ComK [17]. Inhibition of Spo0A might be the reason that allows the surfactin producing cells to remain immune to surfactin.
Fig. 7.
The pheromone, ComX is produced and secreted by B. subtilis. After that, a subpopulation of cells responds to this pheromone and produces surfactin. Other subpopulations except the surfactin producing ones, responds to this surfactin differently and become different types of specialized cells.
6. Competence
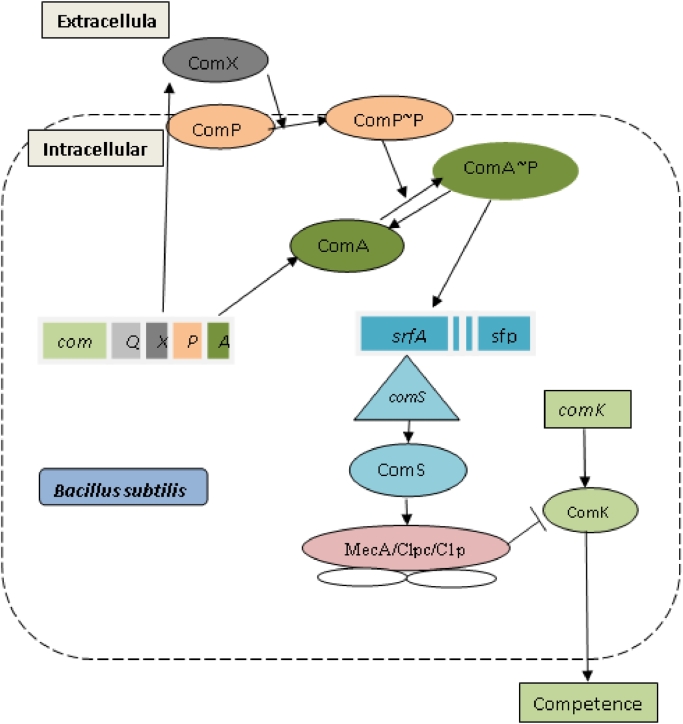
Developing natural competence is a very well-known phenomenon of B. subtilis. Competence is a state of cells when they can uptake and assimilate extracellular DNA into the cells. In environmental stress conditions, natural competency grow in a subpopulation via quorum sensing system to better adapt to the stress conditions. However, surfactin does not directly regulate competence development in B. subtilis. But the induction of competence and surfactin production follow the same pathway (Fig. 8) [25, 55]. The comS, a small ORF positioned within the srfA operon and is regulated by the same promoter (the promoter of the srfA operon). Therefore, the ComS protein is expressed along with surfactin. ComK, a vital factor for developing competence in B. subtilis, gets activated by ComS protein [31, 19, 56]. Both the ComS and ComK proteins are degraded by the protease, Clpc/C1pP [57]. MecA, which is an adapter protein, binds to either ComS or ComK and helps Clpc/C1pP to degrade them [58]. The ComS has a higher affinity than ComK to bind with MecA adapter. So, if there is a higher concentration of ComS, MecA binds to ComS instead of ComK and concentration of ComK increases by autoregulation. In this way, increased ComS promotes ComK to increase its concentration and thus the cell develops competence [59].
Fig. 8.
The process of competence development in a subpopulation of B. subtilis. Expression of comS and srfA promoter starts together after the binding of the activated ComA to the promoter of srfA operon. The MecA/Clpc/C1pP protease complex binds and degrades ComK that prevents cells from entering into comptence state. However, ComS competes with ComK for binding with MecA which allows ComK to increase in concentration and thus leads to the onset of competence state development.
All surfactin-producing cells produce ComS, so competence might develop in all of them. But generally this does not occur. A bimodal regulation system of ComK activation [60], allows only a small part of surfactin-producing cells to develop competence. Competence and surfactin production are induced by the same signaling molecules, pheromones ComX and CSF. Among the strains of B. subtilis, a polymorphism is found in the amino acid chains of ComX and CSF. ComP and Spo0K can precisely identify these signaling molecules that belong to a specific strain and rejects the other variants with the help of their extracellular recognition domains. [34, 33]. This is how B. subtilis controls the competence development only in the presence of some specific strains.
7. Matrix production
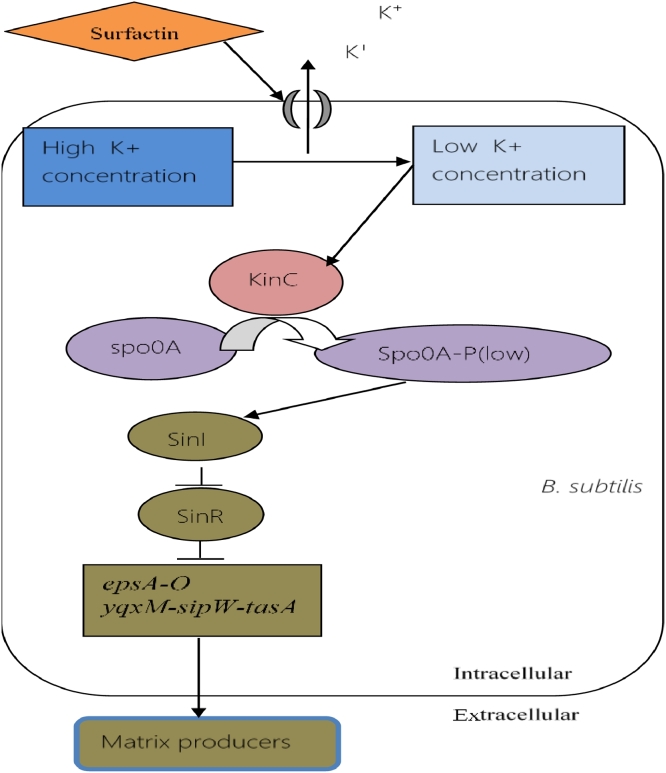
Matrix-producing cells are one of the well-known and well-studied specialised cell types of B. subtilis [54]. These cell types produce and secret extracellular proteins that are required to hold the cells together to form biofilm [9, 61]. According to a study conducted by Lopez et al. surfactin produced by B. subtilis act as an quorum sensing signaling molecule and induce genes required to form biofilm [21]. Once produced, surfactin forms pores in the membrane and causes potassium leakage [62]. This potassium leakage leads to the decrease of the intracellular concentration of potassium. Low concentration of intracellular potassium is detected by KinC (a sensor histidine kinase) through its PAS–PAC sensory domain [21]. In response to this, KinC phosphorylates Spo0A and phosphorylated Spo0A induced the activation of genes that are involved in matrix production [21, 17, 54]. Two operons are required to be activated for the production of the extracellular matrix. The first one, epsA-O operon, composed of 15 genes, usually produces the exo-polysaccharide component. The second one, yqxM-sipW-tasA operon, produces and secrets TasA, a major protein component of the biofilm matrix [63, 16]. During exponential growth, SinR acts as an inhibitor of the eps and yqxM operons, and represses them in throughout the cell population. A subpopulation of cells with activated Spo0A expresses the SinI gene, which can antagonize SinR [64]. The SinI protein binds to the SinR protein and represses it. Thus SinR, the repressor, become disabled to inhibit the operons [65, 66]. In this manner, surfactin triggers the phosphorylation of Spo0A, concentration of Spo0A gets increased in the cell, which in turn ends the repression of operons and allows them to produce the proteins needed for biofilm formation (illustrated in Fig. 9).
Fig. 9.
Regulation of matrix production. Low concentration of phosphorylated spo0A helps to express the two operons that produce proteins needed for the development of biofilm.
8. Cannibalism
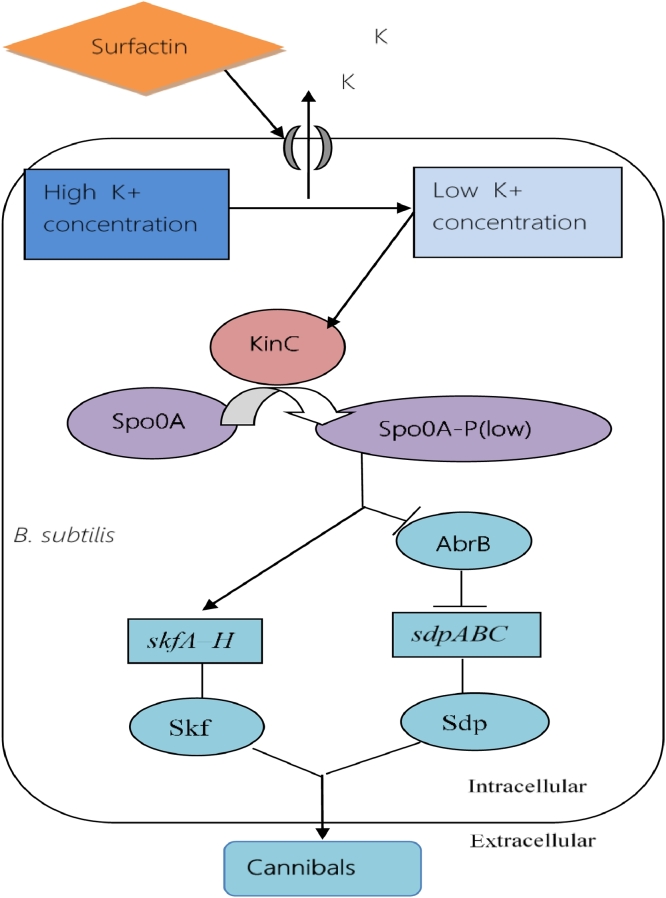
Under environmental stress condition, surfactin triggers the development of cannibalistic cells. A subpopulation of B. subtilis that kill and lyse some other cells, is known as cannibals. The expression of cannibalism is triggered by low levels of phosphorylated Spo0A (illustrated in Fig. 10). This subpopulation secretes specialized toxins that lyse or kill neighboring other cells but the cells that produce cannibalistic toxins, are immune to the effects of toxins [67, 17]
Fig. 10.
The regulation of cannibalism. Low concentration of phosphorylated spo0A helps to produce Skf and Sdp proteins that work as cannibalistic proteins.
Cannibals produce and secret two toxins, sporulation killing factor (Skf) and sporulation-delaying protein (Sdp). Neighboring susceptible cells are killed by these two toxins in a process, termed as cannibalism. These dead cells are used by the living cells as food at the time of nutritional deficiency and to delay the onset of spore formation [68, 69, 70]. Expression of the Skf and Sdp toxins is positively regulated by low intracellular concentration of phosphorylated Spo0A (Spo0A∼P) [71]. Expression of the skfA–H operon, producer of the Skf toxin, is directly promoted by Spo0A∼P. On the other hand, Spo0A∼P indirectly promotes the expression of the sdpABC operon, the producer of the Sdp toxin, by repressing AbrB (repressor of sdpABC operon) [[17], [21], [54], [67]]. The living cells use the dead cells as their food and competent cells take up the released DNA of the dead cells. However, competent cells are immune to these harmful toxins because they can undergo in K-state (a semi-dormant state) that is toxin and antibiotic resistant [72, 73, 67, 74].
9. Motility
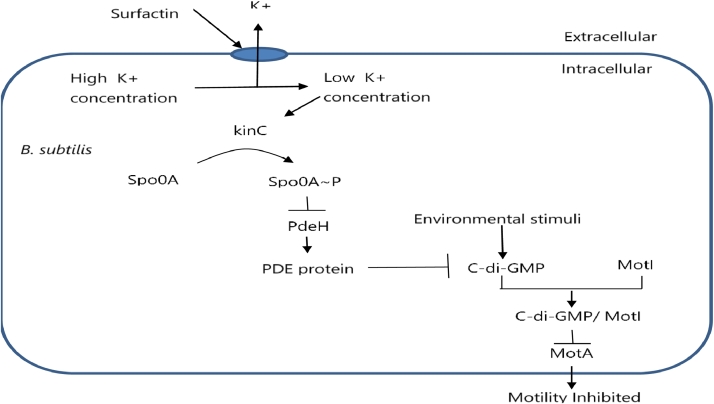
In non-motile subpopulation of B. subtilis, surfactin inhibits motility via spo0A∼P (illustrated in Fig. 11). Spo0A promotes the transcription of the genes required for motility. Concentration of c-di-GMP increases in bacterial cells in response to environmental stress condition. Concentration of c-di-GMP is very important in regulating motility (inhibit motility) [75]. Phosphodiesterase (PDE) proteins enhance motility by degrading c-di-GMP. In response to surfactin, Spo0A gets phosphorylated and this phosphorylated Spo0A represses the PDE proteins, therefore concentration of c-di-GMP remains high in environmental stress condition [76, 77, 78]. High concentration of c-di-GMP leads to attachment with MotI. Bound c-di-GMP and MotI repress MotA, the flagellar motor component. Thus motor-rotor interactions of the flagellar machinery gets disturbed and finally motility gets inhibited. [79].
Fig. 11.
The regulation of motility. Phosphorylated spo0A increases C-di-GMP concentration by inhibiting PdeH. High concentration of c-di-GMP inhibits MotA protein and thus inhibits motility.
In motile subpopulation of B. subtilis, surfactin is responsible for increasing swarming motility. Swarming motility refers to a prompt movement of bacteria over a surface with the help of its flagellar rotation [80]. High cellular concentrations and presence of a surfactant promote swarming motility [81]. Many swarming bacteria like B. subtilis secrete surfactants like surfactin, that reduce the surface tension between the substrate and bacterial cell and thus, this phenomenon permits bacterial movement across surfaces [82, 83, 84, 85]. Mutations in surfactin-producing machinery abolish swarming, which can be regained by adding purified surfactin in the system [83, 86]. Surfactin contains fatty-acyl tails of various lengths and they become distributed across the swarming tendrils. A study conducted by Debois et al. suggests that surfactin with the longest fatty-acyl chain are the most hydrophobic (14–16 carbon atoms) and can spread the fastest [87, 88].
10. Sporulation
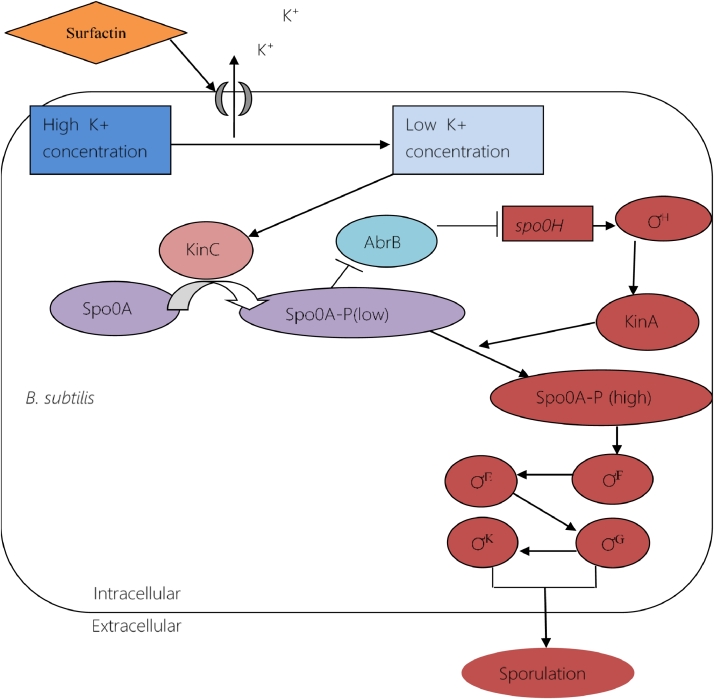
Spores are usually dormant cells that are not active metabolically. Cell sporulation occurs when environmental conditions become tough and nutritional scarcity arises [13]. B. subtilis can measure the concentration of important metabolites inside the cell. They stimulate the activation of the master regulator Spo0A as soon as it senses scarcity of these intracellular metabolites [71, 89]. Phosphorylation of the master regulator, Spo0A triggers sporulation [64, 90]. Many types of genes get activated in accordance with different Spo0A∼P concentrations. For instance, high concentration of Spo0A∼P triggers sporulation (illustrated in Fig. 12), whereas low concentration induces expression of genes are required for matrix production [71]. Surfatin plays a fundamental role in sporulation. Chen et al. reported that surfactin mutant strain showed serious defects in developing biofilm and spore formation in B. amyloliquefaciens, a close relative to B. subtilis [52].
Fig. 12.
The development of sporulation. High concentration of phosphorylated spo0A activates a cascade of sigma factors. Later, these sigma factors promote the activation of a set of genes required for sporulation.
Regulation of the master regulator Spo0A is highly complicated and involves many feedback loops. Regulation begins with the production of surfactin by surfactin producing subpopulation of B. subtilis. Surfactin later indirectly induces KinC to phosphorylate Spo0A. After the phosphorylation of Spo0A, it promotes the expression of the sigma factor ƠH [91]. Later, ƠH stimulates the expression of both the genes needed for phosphorylation and expression of Spo0A (e.g. KinA and Spo0A itself). Kinase KinA possesses three PAS–PAC sensory domains, PAS-A, PAS-B, and PAS-C. KinA can observe the changes in concentration of critical factors and metabolites inside the cell by using these domains and phosphorylates Spo0A when required [92]. That's how a cycle of both synthesis and phosphorylation gets turned on by the phosphorylation of Spo0A [93]. Finally, the high level of phosphorylated Spo0A initiates the expression of genes required for sporulation [94].
The process of sporulation starts by dividing the cells unequally and producing two distinct sections, one is a mother cell and another one is a forespore. This unequal division activates a cascade of sigma factors. Different groups of genes are activated by the sigma factors in different sections. In forespore, the sigma factor ƠF is activated and later in mother cell, it activates ƠE. The products of genes under the control of these two sigma factors regulate the genes that are required for the forespore to be engulfed. Sigma factor ƠG becomes activated inside the forespore after the engulfment and later in the mother cell it activates ƠK. Finally, the cortex and the spore coat are produced that are essential for the maturation of the spore [13].
11. Conclusion
Surfactin is one of the most potential and well-known biosurfactants till now. However, currently, due to its high production costs, surfactin is not able to challenge the chemically synthesized surfactants economically. Therefore, novel strategies have to be discovered to improve its yields to lower the production cost. Better understanding of its genetic regulations may lead us toward the creation of high-yielding recombinant strains (for industrial production) that would make the usage of surfactin cost efficient compared to its other synthetic counterparts. The discussion summarized in this study gives a detailed review on genetic regulation of surfactin production and also demonstrates how surfactin works as a signaling compound and triggers B. subtilis population heterogeneity. We now know that, surfactin works as a quorum sensing molecule and affects cell differentiation to different subpopulation. Better understanding of B. subtilis cell heterogeneity will help us to manipulate its growth kinetics more efficiently and to use these different types of subpopulations according to our needs.
Compliance with ethical standards
Funding Information: This study did not receive any funding.
Ethical Approval: There are no studies with human participants or animals in this work, performed by any of the authors.
CRediT authorship contribution statement
Faisal Bin Rahman: Conceptualization, Data curation, Writing – original draft. Bishajit Sarkar: Data curation, Formal analysis, Writing – review & editing. Ripa Moni: Data curation, Formal analysis, Writing – review & editing. Mohammad Shahedur Rahman: Conceptualization, Formal analysis.
Declaration of Competing Interest
The authors declare that they have no conflict of interest among themselves. All of the authors have read and accepted the final version of the manuscript.
References
- 1.Mulligan C.N. Environmental applications for biosurfactants. Environ. Pollut. 2005;133:183–198. doi: 10.1016/j.envpol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Desai J.D., Banat I.M. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 1997;61:47–64. doi: 10.1128/mmbr.61.1.47-64.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan E.R. Molecular genetics of biosurfactant production. Curr. Opin. Biotechnol. 1998;9:263–269. doi: 10.1016/s0958-1669(98)80057-8. [DOI] [PubMed] [Google Scholar]
- 4.Arima K. Surfactin, acrystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 1968;31:488–494. doi: 10.1016/0006-291x(68)90503-2. [DOI] [PubMed] [Google Scholar]
- 5.Peypoux F., Bonmatin J.M., Wallach J. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 1999;51:553–563. doi: 10.1007/s002530051432. [DOI] [PubMed] [Google Scholar]
- 6.Shaligram N.S., Singhal R.S. Surfactin–a review on biosynthesis, fermentation, purification and applications. Food Technol Biotechnol. 2010;48:119–134. [Google Scholar]
- 7.Ku J., Mirmira R.G., Liu L., Santi D.V. Expression of a functional non-ribosomal peptide synthetase module in Escherichia coli by coexpression with a phosphopantetheinyl transferase. Chem. Biol. 1997;4:203–207. doi: 10.1016/s1074-5521(97)90289-1. [DOI] [PubMed] [Google Scholar]
- 8.Stachelhaus T., Schneider A., Marahiel M.A. Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science (80-) 1995;269:69–72. doi: 10.1126/science.7604280. [DOI] [PubMed] [Google Scholar]
- 9.Aguilar C., Vlamakis H., Losick R., Kolter R. Thinking about Bacillus subtilis as a multicellular organism. Curr. Opin. Microbiol. 2007;10:638–643. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veening J., Igoshin O.A., Eijlander R.T., Nijland R., Hamoen L.W., Kuipers O.P. Transient heterogeneity in extracellular protease production by Bacillus subtilis. Mol. Syst. Biol. 2008;4:184. doi: 10.1038/msb.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubnau D. Genetic competence in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 1991;55:395–424. doi: 10.1128/mr.55.3.395-424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubnau D., Provvedi R. Internalizing DNA. Res. Microbiol. 2000;151:475–480. doi: 10.1016/s0923-2508(00)00166-2. [DOI] [PubMed] [Google Scholar]
- 13.Piggot P.J., Hilbert D.W. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Rudner D.Z., Losick R. Morphological coupling in development: lessons from prokaryotes. Dev. Cell. 2001;1:733–742. doi: 10.1016/s1534-5807(01)00094-6. [DOI] [PubMed] [Google Scholar]
- 15.Vlamakis H., Aguilar C., Losick R., Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kearns D.B., Chu F., Branda S.S., Kolter R., Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 17.López D., Vlamakis H., Losick R., Kolter R. Paracrine signaling in a bacterium. Genes Dev. 2009;23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camilli A., Bassler B.L. Bacterial small-molecule signaling pathways. Science (80-) 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnuson R., Solomon J., Grossman A.D. Biochemical and genetic characterization of a competence pheromone from B. subtilis. CellCell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 20.Weir J., Predich M., Dubnau E., Nair G., Smith I. Regulation of spo0H, a gene coding for the Bacillus subtilis sigma H factor. J. Bacteriol. 1991;173:521–529. doi: 10.1128/jb.173.2.521-529.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López D., Fischbach M.A., Chu F., Losick R., Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl Acad. Sci. 2009;106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cosmina P., Rodriguez F., de Ferra F., Grandi G., Perego M., Venema G., van Sinderen D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol. 1993;8:821–831. doi: 10.1111/j.1365-2958.1993.tb01629.x. [DOI] [PubMed] [Google Scholar]
- 23.Fabret C., Quentin Y., Guiseppi A., Busuttil J., Haiech J., Denizot F. Analysis of errors in finished DNA sequences: the surfactin operon ofBacillus subtilisas an example. Microbiology. 1995;141:345–350. doi: 10.1099/13500872-141-2-345. [DOI] [PubMed] [Google Scholar]
- 24.Hamoen L.W., Venema G., Kuipers O.P. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003;149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- 25.Nakano M.M., Magnuson R., Myers A., Curry J., Grossman A.D., Zuber P. srfA is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis. J. Bacteriol. 1991;173:1770–1778. doi: 10.1128/jb.173.5.1770-1778.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steller S., Sokoll A., Wilde C., Bernhard F., Franke P., Vater J. Initiation of surfactin biosynthesis and the role of the SrfD-thioesterase protein. Biochemistry. 2004;43:11331–11343. doi: 10.1021/bi0493416. [DOI] [PubMed] [Google Scholar]
- 27.Vater J., Stein T., Vollenbroich D., Kruft V., Wittmann-Liebold B., Franke P., Liu L., Zuber P. The modular organization of multifunctional peptide synthetases. J. Protein Chem. 1997;16:557–564. doi: 10.1023/a:1026386100259. [DOI] [PubMed] [Google Scholar]
- 28.Kraas F.I., Helmetag V., Wittmann M., Strieker M., Marahiel M.A. Functional dissection of surfactin synthetase initiation module reveals insights into the mechanism of lipoinitiation. Chem. Biol. 2010;17:872–880. doi: 10.1016/j.chembiol.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 29.Schwarzer D., Mootz H.D., Linne U., Marahiel M.A. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl Acad. Sci. 2002;99:14083–14088. doi: 10.1073/pnas.212382199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeh E., Kohli R.M., Bruner S.D., Walsh C.T. Type II thioesterase restores activity of a NRPS module stalled with an aminoacyl-S-enzyme that cannot be elongated. Chem.BioChem. 2004;5:1290–1293. doi: 10.1002/cbic.200400077. [DOI] [PubMed] [Google Scholar]
- 31.D'Souza C., Nakano M.M., Zuber P. Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc. Natl Acad. Sci. 1994;91:9397–9401. doi: 10.1073/pnas.91.20.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quadri L.E.N., Weinreb P.H., Lei M., Nakano M.M., Zuber P., Walsh C.T. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 33.Tran L.P., Nagai T., Itoh Y. Divergent structure of the ComQXPA quorum-sensing components: molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol. Microbiol. 2000;37:1159–1171. doi: 10.1046/j.1365-2958.2000.02069.x. [DOI] [PubMed] [Google Scholar]
- 34.Piazza F., Tortosa P., Dubnau D. Mutational analysis and membrane topology of ComP, a quorum-sensing histidine kinase of Bacillus subtilis controlling competence development. J. Bacteriol. 1999;181:4540–4548. doi: 10.1128/jb.181.15.4540-4548.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinrauch Y., Penchev R., Dubnau E., Smith I., Dubnau D. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 1990;4:860–872. doi: 10.1101/gad.4.5.860. [DOI] [PubMed] [Google Scholar]
- 36.Comella N., Grossman A.D. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 2005;57:1159–1174. doi: 10.1111/j.1365-2958.2005.04749.x. [DOI] [PubMed] [Google Scholar]
- 37.Carter III H.L., Tatti K.M., Moran Jr C.P. Cloning of a promoter used by sigma H RNA polymerase in Bacillus subtilis. Gene. 1990;96:101–105. doi: 10.1016/0378-1119(90)90347-t. [DOI] [PubMed] [Google Scholar]
- 38.Lazazzera B.A. (1999) Cell-density control of gene expression and development in Bacillus subtilis. Cell-Cell Signal Bact 27–47.
- 39.Pottathil M., Lazazzera B.A. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front. Biosci. 2003;8:d32–d45. doi: 10.2741/913. [DOI] [PubMed] [Google Scholar]
- 40.Perego M., Glaser P., Hoch J.A. Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis. Mol. Microbiol. 1996;19:1151–1157. doi: 10.1111/j.1365-2958.1996.tb02460.x. [DOI] [PubMed] [Google Scholar]
- 41.Solomon J.M., Lazazzera B.A., Grossman A.D. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 42.Lazazzera B.A., Solomon J.M., Grossman A.D. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. CellCell. 1997;89:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 43.Burbulys D., Trach K.A., Hoch J.A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. CellCell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 44.Nakano M.M., Marahiel M.A., Zuber P. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 1988;170:5662–5668. doi: 10.1128/jb.170.12.5662-5668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klausmann P., Hennemann K., Hoffmann M., Treinen C., Aschern M., Lilge L., Heravi K.M., Henkel M., Hausmann R. Bacillus subtilis high cell density fermentation using a sporulation-deficient Strain for the production of surfactin. Appl. Microbiol. Biotechnol. 2021;105:4141–4151. doi: 10.1007/s00253-021-11330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M., Yu H., Li X., Shen Z. Single-gene regulated non-spore-forming Bacillus subtilis: construction, transcriptome responses, and applications for producing enzymes and surfactin. Metab. Eng. 2020;62:235–248. doi: 10.1016/j.ymben.2020.08.008. [DOI] [PubMed] [Google Scholar]
- 47.Strauch M., Webb V., Spiegelman G., Hoch J.A. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl Acad. Sci. 1990;87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaacks K.J., Healy J., Losick R., Grossman A.D. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krätzschmar J., Krause M., Marahiel M.A. Gramicidin S biosynthesis operon containing the structural genes grsA and grsB has an open reading frame encoding a protein homologous to fatty acid thioesterases. J. Bacteriol. 1989;171:5422–5429. doi: 10.1128/jb.171.10.5422-5429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cosby W.M., Vollenbroich D., Lee O.H., Zuber P. Altered srf expression in bacillus subtilisResulting from changes in culture pH is dependent on the Spo0K oligopeptide permease and the ComQX System of extracellular control. J. Bacteriol. 1998;180:1438–1445. doi: 10.1128/jb.180.6.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lazazzera B.A., Kurtser I.G., McQuade R.S., Grossman A.D. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J. Bacteriol. 1999;181:5193–5200. doi: 10.1128/jb.181.17.5193-5200.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen B., Wen J., Zhao X., Ding J., Qi G. Surfactin: a quorum-sensing signal molecule to relieve CCR in Bacillus amyloliquefaciens. Front. Microbiol. 2020;11:631. doi: 10.3389/fmicb.2020.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen J., Zhao X., Si F., Qi G. Surfactin, a quorum sensing signal molecule, globally affects the carbon metabolism in Bacillus amyloliquefaciens. Metab. Eng. Commun. 2021;12:e00174. doi: 10.1016/j.mec.2021.e00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.López D., Vlamakis H., Losick R., Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol. Microbiol. 2009;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakano M.M., Xia L.A., Zuber P. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J. Bacteriol. 1991;173:5487–5493. doi: 10.1128/jb.173.17.5487-5493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Sinderen D., Galli G., Cosmina P., de Ferra F., Withoff S., Venema G., Grandi G. Characterization of the srfA locus of Bacillus subtilis: only the valine-activating domain of srfA is involved in the establishment of genetic competence. Mol. Microbiol. 1993;8:833–841. doi: 10.1111/j.1365-2958.1993.tb01630.x. [DOI] [PubMed] [Google Scholar]
- 57.Maamar H., Raj A., Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science (80-) 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turgay K., Hahn J., Burghoorn J., Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stiegelmeyer S.M., Giddings M.C. Agent-based modeling of competence phenotype switching in Bacillus subtilis. Theor. Biol. Med. Model. 2013;10:1–21. doi: 10.1186/1742-4682-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smits W.K., Eschevins C.C., Susanna K.A., Bron S., Kuipers O.P., Hamoen L.W. Stripping Bacillus: comK auto-stimulation is responsible for the bistable response in competence development. Mol. Microbiol. 2005;56:604–614. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- 61.Branda S.S., Vik Å., Friedman L., Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 62.Sheppard J.D., Jumarie C., Cooper D.G., Laprade R. Ionic channels induced by surfactin in planar lipid bilayer membranes. Biochim. Biophys. Acta (BBA)-Biomembranes. 1991;1064:13–23. doi: 10.1016/0005-2736(91)90406-x. [DOI] [PubMed] [Google Scholar]
- 63.Branda S.S., González-Pastor J.E., Dervyn E., Ehrlich S.D., Losick R., Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J. Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chai Y., Chu F., Kolter R., Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewis R.J., Brannigan J.A., Offen W.A., Smith I., Wilkinson A.J. An evolutionary link between sporulation and prophage induction in the structure of a repressor: anti-repressor complex. J. Mol. Biol. 1998;283:907–912. doi: 10.1006/jmbi.1998.2163. [DOI] [PubMed] [Google Scholar]
- 66.Lewis R.J., Brannigana J.A., Smith I., Wilkinson A.J. Crystallisation of the Bacillus subtilis sporulation inhibitor SinR, complexed with its antagonist. Sinl. FEBS Lett. 1996;378:98–100. doi: 10.1016/0014-5793(95)01432-2. [DOI] [PubMed] [Google Scholar]
- 67.Lopez D., Vlamakis H., Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 2008;33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 68.Claverys J.-.P., Håvarstein L.S. Cannibalism and fratricide: mechanisms and raisons d'etre. Nat. Rev. Microbiol. 2007;5:219–229. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- 69.Ellermeier C.D., Hobbs E.C., Gonzalez-Pastor J.E., Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 70.González-Pastor J.E., Hobbs E.C., Losick R. Cannibalism by sporulating bacteria. Science (80-) 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- 71.Fujita M., González-Pastor J.E., Losick R. High-and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berka R.M., Hahn J., Albano M., Draskovic I., Persuh M., Cui X., Sloma A., Widner W., Dubnau D. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 2002;43:1331–1345. doi: 10.1046/j.1365-2958.2002.02833.x. [DOI] [PubMed] [Google Scholar]
- 73.Leisner M., Stingl K., Rädler J.O., Maier B. Basal expression rate of comK sets a ‘switching-window'into the K-state of Bacillus subtilis. Mol. Microbiol. 2007;63:1806–1816. doi: 10.1111/j.1365-2958.2007.05628.x. [DOI] [PubMed] [Google Scholar]
- 74.Maamar H., Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bange G., Bedrunka P. Physiology of guanosine-based second messenger signaling in Bacillus subtilis. Biol. Chem. 2020;401:1307–1322. doi: 10.1515/hsz-2020-0241. [DOI] [PubMed] [Google Scholar]
- 76.Gao X., Mukherjee S., Matthews P.M., Hammad L.A., Kearns D.B., Dann C.E. Functional characterization of core components of the Bacillus subtilis cyclic-di-GMP signaling pathway. J. Bacteriol. 2013;195:4782–4792. doi: 10.1128/JB.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Molle V., Fujita M., Jensen S.T., Eichenberger P., González-Pastor J.E., Liu J.S., Losick R. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 78.Weiss C.A., Hoberg J.A., Liu K., Tu B.P., Winkler W.C. Single-cell microscopy reveals that levels of cyclic di-GMP vary among Bacillus subtilis subpopulations. J. Bacteriol. 2019;201 doi: 10.1128/JB.00247-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Y., Chai Y., Guo J., Losick R. Evidence for cyclic di-GMP-mediated signaling in Bacillus subtilis. J. Bacteriol. 2012;194:5080–5090. doi: 10.1128/JB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Henrichsen J. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 1972;36:478. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kearns D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alberti L., Harshey R.M. Differentiation of Serratia marcescens 274 into swimmer and swarmer cells. J. Bacteriol. 1990;172:4322–4328. doi: 10.1128/jb.172.8.4322-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Julkowska D., Obuchowski M., Holland I.B., Séror S.J. Branched swarming patterns on a synthetic medium formed by wild-type Bacillus subtilis strain 3610: detection of different cellular morphologies and constellations of cells as the complex architecture develops. Microbiology. 2004;150:1839–1849. doi: 10.1099/mic.0.27061-0. [DOI] [PubMed] [Google Scholar]
- 84.Rashid M.H., Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Senesi S., Celandroni F., Salvetti S., Beecher D.J., Wong A.C.L., Ghelardi E. Swarming motility in Bacillus cereus and characterization of a fliY mutant impaired in swarm cell differentiationThe EMBL accession number for the sequence reported in this paper is Y08031. Microbiology. 2002;148:1785–1794. doi: 10.1099/00221287-148-6-1785. [DOI] [PubMed] [Google Scholar]
- 86.Kearns D.B., Losick R. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 87.Debois D., Hamze K., Guérineau V., Le Caër J., Holland I.B., Lopes P., Ouazzani J., Séror S.J., Brunelle A., Laprévote O. In situ localisation and quantification of surfactins in a Bacillus subtilis swarming community by imaging mass spectrometry. Proteomics. 2008;8:3682–3691. doi: 10.1002/pmic.200701025. [DOI] [PubMed] [Google Scholar]
- 88.Raaijmakers J.M., De Bruijn I., Nybroe O., Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol. Rev. 2010;34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 89.Kudoh J., Ikeuchi T., Kurahashi K. Identification of the sporulation gene spoOA product of Bacillus subtilis. Biochem. Biophys. Res. Commun. 1984;122:1104–1109. doi: 10.1016/0006-291x(84)91205-1. [DOI] [PubMed] [Google Scholar]
- 90.Dubnau D., Losick R. Bistability in bacteria. Mol. Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 91.Hamoen L.W., Eshuis H., Jongbloed J., Venema G., van Sinderen D. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol. Microbiol. 1995;15:55–63. doi: 10.1111/j.1365-2958.1995.tb02220.x. [DOI] [PubMed] [Google Scholar]
- 92.Stephenson K., Hoch J.A. PAS-A domain of phosphorelay sensor kinase A: a catalytic ATP-binding domain involved in the initiation of development in Bacillus subtilis. Proc. Natl Acad. Sci. 2001;98:15251–15256. doi: 10.1073/pnas.251408398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chung J.D., Stephanopoulos G., Ireton K., Grossman A.D. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J. Bacteriol. 1994;176:1977–1984. doi: 10.1128/jb.176.7.1977-1984.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang M., Grau R., Perego M. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J. Bacteriol. 2000;182:303–310. doi: 10.1128/jb.182.2.303-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]